Supplementary Figure 1
|
|
|
- Megan Carson
- 5 years ago
- Views:
Transcription
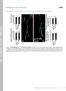
1 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections, we employed an artificial microrna (amirna)-mediated knockdown strategy to generate plants with reduced HSP21 gene activity (see Methods). Schematic presentation of the HSP21 coding sequence showing the positions of different sites targeted by the designed amirnas. Of the three amirna constructs transformed, only amirna1 caused a strong reduction in HSP21 transcript levels (~4- to 16-fold; determined by qrt-pcr in different lines). (b) Reduced HSP21 transcript abundance in all three lines of amirna-1 seedlings (#10, 11 and 12) confirmed by qrt-pcr (n = 2) and end-point PCR. For end-point PCR, primers annealing to the start and stop regions of HSP21 were used. cdna was prepared from RNA of leaves of five-day-old heat-treated (37 C for 90 min, then 22 C for 90 min, then 44 C for 45 min) seedlings. (c) Elevated expression of HSP21 in 35S:HSP21 plants (lines #5 and #9) compared to emptyvector (EV) control plants. Expression was analysed by qrt-pcr (n = 2). Values were expressed as the difference between an arbitrary value of 40 and dct, so that high 40 - dct value indicates high gene expression level. (d) Immunodetection of HSP21 protein in HSP21 transgenics and EV plants exposed to the HS regime shown schematically, using anti-hsp21 antibody (top panel). Reduction and increase in HSP21 protein was confirmed in amirna and overexpression lines, respectively. RbcL, ribulose-1,5-bisphosphate carboxylase/ oxygenase large subunit (loading control; bottom panel). kda, kilo Dalton. 1
2 Supplementary Figure 2. Supplementary Figure 2. Phenotype of HSP21 transgenic and wild-type () plants before and after priming. The phenotype of HSP21-amiRNA, 35S:HSP21 and plants before heat stress priming (upper row of panels), and after 3 d (middle panels) and 4 d (lower panels) of recovery. Growth condition and heat stress regimes are schematically shown on the right of each row of panels. 2
3 Supplementary Figure 3. Supplementary Figure 3. Assessment of basal heat tolerance in and. (a) Seven-day-old and seedlings were subjected to 44 C for 45 min. The seedlings were then transferred to normal growth condition and photographed seven days later. (b) Seedling survival was scored seven days after the recovery period; data are represented as percentage of the initial viable seedlings. Means ± SD are given (n = 7 plates with ~ 50 seedlings each). 3
4 Supplementary Figure 4. Supplementary Figure 4. Protein level and sequence comparison of HSP21 in Arabidopsis accessions, and Tsu-0. (a) Immunoblot analyses of HSP21 during the memory phase (one and three days after priming) shown on the right of each section. Note the much higher abundance of HSP21 protein level in compared to and Tsu-0 at day 4 into the memory phase. kda, kilo Dalton. Bands shown for seedlings have been rearranged for presentation purpose. The original blot image is shown in Supplementary Fig. 10. (b) Comparison of the HSP21 protein sequences (deduced from cdna sequences) in accessions, and Tsu-0. The threonine 77 in is changed to alanine in and Tsu-0 due to an ACC to GCC transition. (c) HSP21 expression in, and Tsu-0 seedlings during the memory phase (24 h and 48 h) compared to unprimed controls. FC, fold change. Error bars indicate means ± SD of three independent biological replicates each containing a pool of ~100 seedlings. 4
5 Supplementary Figure 5. Nucleotide Alignment of FtsH6 Coding Sequences from Arabidopsis Accessions. (a) PCR-amplified FtsH6 coding sequences of accessions and were sequenced and compared. Sequence polymorphisms are shown in yellow. A nucleotide deletion (G) at position 379 of the FtsH6 open reading frame leads to a premature translation stop codon (red). (b) Alignment of FtsH6 amino acid sequences from Arabidopsis accessions. a) FtsH6 sequences from accessions and. 1 1 ATGGCATCATCATCATCAGCTTTGTCTTTCCCTCTCTCCAACATCCCAAC ATGGCATCATCATCATCAGCTTTGTCTTTCCCTCTCTCCAACATCCCAAC CTGTAGTAAGAAGTCTCAACAATTTCAAAAACCGGCTTCTTTATCCAAAT CTGTAGTAAGAAGTCTCAACAATTTCAAAAACCGGCTTCTTTATCCAAAT CCAGTCACACACATAAACCTAGTCTCAAAACCCAAATTTTACACCACAAA CCAGTCACACCCATAAACCTAGTCTCAAAACCCAAACTTTACACCACAAA TTCACTAAGAGAAATTTACTGAGTTTGACGACTGCCTTGGGATTTACGTC CTCACTAAGAGAAATTTACTGAGTTTGACGACTGCCTTGGGATTTACGTC AGCGCTAGGAACTGTTCTTGCTCACCCCGCAAAAGCTGAACCAGAAGCTC AGCGCTAGGAACTGTTCTTGCTCACCCCGCAAAAGCTGAACCAGAAGCTC CCATCGAAGCCACTTCCAATAGAATGTCGTATTCGAGATTCCTGCAGCAT CCATCGAAGCCACTTCCAATAGAATGTCGTATTCGAGATTCCTGCAGCAT CTGAAAGAGAATGAAGTGAAGAAAGTTGACTTGATCGAGAACGGAACGGT CTGAAAGAGAATGAAGTGAAGAAAGTTGACTTGATCGAGAACGGAACGGT TGCGATCGTAGAGATCTCTAATCCAGTAGTAGGAAAGATCCAGAGGGTTA TGCGATCGTAGAGATCTCTAATCCAGTA- TAGGAAAGATCCAGAGGGTTA GGGTTAACCTTCCCGGTTTACCAGTCGATCTGGTGAGGGAGATGAAGGAG GGGTTAACCTTCCCGGTTTACCAGTCGATCTGGTGAGGGAGATGAAGGAG AAGAACGTCGATTTCGCTGCTCATCCCATGAATGTGAACTGGGGAGCTTT AAGAACGTCGATTTCGCTGCTCATCCCATGAATGTGAACTGGGGAGCTTT CTTGCTCAACTTCTTGGGGAATTTAGGGTTTCCTTTGATCTTGCTTGTCT CTTGCTCAACTTCTTGGGGAATTTAGGGTTTCCTTTGATCTTGCTTGTCT CTCTGCTTTTAACATCTTCTTCAAGAAGAAACCCTGCTGGACCTAACTTG CTCTGCTTTTAACATCTTCTTCAAGAAGAAACCCTGCTGGACCTAACTTG CCTTTTGGTCTTGGAAGAAGCAAAGCTAAGTTTCAGATGGAGCCTAACAC CCTTTTGGTCTTGGAAGAAGCAAAGCTAAGTTTCAGATGGAGCCTAACAC AGGGATAACGTTCGAGGATGTAGCAGGAGTAGACGAAGCCAAGCAAGACT AGGGATAACGTTCGAGGATGTAGCAGGAGTAGACGAAGCCAAGCAAGACT TTGAAGAGATCGTTGAATTTTTGAAAACCCCAGAGAAATTCTCAGCTTTG TTGAAGAGATCGTTGAATTTTTGAAAACCCCAGAGAAATTCTCAGCTTTG GGGGCTAAAATCCCAAAAGGCGTCTTGTTGACCGGACCGCCAGGAACCGG GGGGCTAAAATCCCAAAAGGCGTCTTGTTGACCGGACCGCCAGGAACCGG AAAGACACTCTTGGCCAAGGCTATAGCCGGAGAAGCCGGTGTTCCTTTTT AAAGACACTCTTGGCCAAGGCTATAGCCGGAGAAGCCGGTGTTCCTTTTT TTTCATTGTCTGGTTCGGAGTTCATAGAGATGTTTGTTGGTGTAGGAGCA 900 5
6 6 850 TTTCATTGTCTGGTTCGGAGTTCATAGAGATGTTTGTTGGTGTAGGAGCA TCTAGAGCTAGAGACTTGTTTAACAAGGCAAAGGCTAATTCACCTTGTAT TCTAGGGCTAGAGACTTGTTTAACAAGGCAAAGGCTAATTCACCTTGTAT AGTGTTCATTGATGAGATTGATGCTGTTGGGAGAATGAGAGGAACCGGTA AGTGTTCATTGATGAGATTGATGCTGTTGGGAGAATGAGAGGAACCGGTA TAGGAGGAGGAAACGACGAACGTGAGCAGACGCTAAACCAGATATTAACC TAGGAGGAGGAAACGACGAACGTGAGCAGACGCTAAACCAGATATTAACC GAAATGGATGGGTTTGCGGGGAATACCGGAGTAATTGTGATCGCTGCAAC GAAATGGATGGGTTTGCGGGGAATACCGGAGTAATTGTGATCGCTGCAAC GAACCGGCCAGAGATTCTAGACTCTGCTTTGCTCCGACCAGGAAGATTCG GAACCGGCCAGAGATTCTAGACTCTGCTTTGCTCCGACCAGGAAGATTCG ATAGGCAGGTATGTTGGTTAATCCTTAAACCGAATAAATCGAACAGGTTT ATAGGCAGGTATGTTGGTTAATCCTTAAACCGAATAAATCGAACAGGTTT GGTATAATGTCAACATGTTTTAAACAGGTTTCTGTTGGTTTACCGGATAT GGTATAATGTCAACATGTTTTAAACAGGTTTCTGTTGGTTTACCGGATAT AAGAGGAAGAGAGGAGATATTAAAAGTTCACAGCAGAAGCAAGAAACTCG AAGAGGAAGAGAGGAGATATTAAAAGTTCACAGCAGAAGCAAGAAACTCG ACAAAGATGTATCTTTGAGCGTAATTGCCATGAGAACTCCAGGTTTTAGT ACAAAGATGTATCTTCGAGCGTAATTGCCATGAGAACTCCAGGTTTTAGT GGAGCTGACTTGGCCAATCTTATGAACGAAGCAGCGATTCTCGCCGGAAG GGAGCTGACTAGGCCAATCTTATGAACGAAGCAGCGATTCTCGCCGGAAG AAGAGGAAAAGACAAGATTACCCTTACAGAGATCGACGACTCTATCGATC AAGAGGAAAAGACAAGATTACCCTTACAGAGATCGACGACTCTATCGATC GGATAGTCGCCGGAATGGAAGGGACAAAGATGATCGACGGTAAAAGTAAA GGATAGTCGCCGGAATGGAAGGGACAAAGATGATCGACGGTAAAAGTAAA GCGATTGTAGCGTACCATGAAGTAGGACATGCAATTTGTGCGACTTTGAC GCGATTGTAGCGTACCATGAAGTAGGACATGCAATTTGTGCGACTTTGAC GGAGGGTCATGATCCGGTTCAGAAAGTTACGTTGGTTCCTAGAGGTCAAG GGAGGGTCATGATCCGGTTCAGAAAGTTACGTTGGTTCCTAGAGGTCAAG CACGTGGTCTCACGTGGTTCTTACCGGGAGAAGATCCGACATTGGTGTCT CACGTGGTCTCACGTGGTTCTTACCGGGAGAAGATCCGACATTGGTGTCT AAACAGCAATTGTTCGCTAGAATCGTCGGAGGACTCGGAGGCAGAGCCGC AAACAGCAATTGTTCGCTAGAATCGTCGGAGGACTCGGAGGCAGAGCCGC CGAAGATGTAATTTTTGGAGAACCGGAGATAACCACCGGCGCTGCCGGTG CGAAGATGTAATTTTTGGAGAACCGGAGATAACCACCGGCGCTGCCGGTG ATCTCCAGCAAGTCACTGAGATAGCAAGACAGATGGTGACGATGTTTGGT ATCTCCAGCAAGTCACTGAGATAGCAAGACAGATGGTGACGATGTTTGGT ATGTCGGAAATCGGTCCATGGGCACTAACCGACCCAGCGGTTAAGCAAAA ATGTCGGAAATCGGTCCATGGGCACTAACCGACCCAGCGGTTAAGCAAAA CGATGTGGTTCTACGGATGCTAGCGAGGAACTCAATGTCCGAGAAACTTG CGATGTGGTTCTACGGATGCTAGCGAGGAACTCAATGTCCGAGAAACTTG CGGAGGATATTGATTCTTGCGTGAAGAAAATAATCGGTGACGCTTACGAG CGGAGGATATTGATTCTTGCGTGAAGAAAATAATCGGTGACGCTTACGAG
7 GTTGCGAAAAAGCACGTGAGGAATAATAGAGAAGCTATAGACAAGCTTGT GTTGCGAAAAAGCACGTGAGGAATAATAGAGAAGCTATAGACAAGCTTGT TGATGTTTTGTTGGAGAAAGAAACCTTAACGGGAGACGAGTTTCGAGCAA TGATGTTTTGTTGGAGAAAGAAACCTTAACGGGAGACGAGTTTCGAGCAA TTCTGTCTGAGTATACTGATCAACCGTTAAATACCGATGGCGATGTAAGA TTCTGTCTGAGTATACTGATCAACCGTTAAATACCGATGGCGATGTTAGA ATTCGAATCAATGACTTGATTAGTGTCTAA ATTCGAATCAATGACTTGATTAGTGTCTAA FtsH6 protein from Met A S S S S A L S F P L S N I P T C S K K S Q Q F Q K P A S L S K S S H T H K P S L K T Q I L H H K F T K R N L L S L T T A L G F T S A L G T V L A H P A K A E P E A P I E A T S N R Met S Y S R F L Q H L K E N E V K K V D L I E N G T V A I V E I S N P V V G K I Q R V R V N L P G L P V D L V R E Met K E K N V D F A A H P Met N V N W G A F L L N F L G N L G F P L I L L V S L L L T S S S R R N P A G P N L P F G L G R S K A K F Q Met E P N T G I T F E D V A G V D E A K Q D F E E I V E F L K T P E K F S A L G A K I P K G V L L T G P P G T G K T L L A K A I A G E A G V P F F S L S G S E F I E Met F V G V G A S R A R D L F N K A K A N S P C I V F I D E I D A V G R Met R G T G I G G G N D E R E Q T L N Q I L T E Met D G F A G N T G V I V I A A T N R P E I L D S A L L R P G R F D R Q V C W L I L K P N K S N R F G I Met S T C F K Q V S V G L P D I R G R E E I L K V H S R S K K L D K D V S L S V I A Met R T P G F S G A D L A N L Met N E A A I L A G R R G K D K I T L T E I D D S I D R I V A G Met E G T K Met I D G K S K A I V A Y H E V G H A I C A T L T E G H D P V Q K V T L V P R G Q A R G L T W F L P G E D P T L V S K Q Q L F A R I V G G L G G R A A E D V I F G E P E I T T G A A G D L Q Q V T E I A R Q Met V T Met F G Met S E I G P W A L T D P A V K Q N D V V L R Met L A R N S Met S E K L A E D I D S C V K K I I G D A Y E V A K K H V R N N R E A I D K L V D V L L E K E T L T G D E F R A I L S E Y T D Q P L N T D G D V R I R I N D L I S V Stop FtsH6 protein from Met A S S S S A L S F P L S N I P T C S K K S Q Q F Q K P A S L S K S S H T H K P S L K T Q T L H H K L T K R N L L S L T T A L G F T S A L G T V L A H P A K A E P E A P I E A T S N R Met S Y S R F L Q H L K E N E V K K V D L I E N G T V A I V E I S N P V Stop b) Alignment of FtsH6 amino acid sequences from Arabidopsis accessions. The following Arabidopsis accessions were included in our preliminary screen for differences in thermomemory:,, Cvi-0, C24, Ct, Can-0, Lip-0, Mdn-1, Mh-0, Mt-0, Aitba-1, Abd-0, Bl-1, Bur-0, Dog-4, Tsu-0, St-0, Kas-1, Yo-0, Xan-1, Kondara, Ler-0, Ko-2, Ri-0, Fei-0, Leo-1, Yeg-1, Ste-0, ICE1, Kin-0, Le-0, Oy-0, N14, Sah-0, Berk, Hiro, Ita, HEK2, Stepn, Shign, and Borsk2. The FtsH6 amino acid sequence alignment shown below includes sequences from all above accessions with the exception of sequences that were identical to those of (namely Lip-0, Mh-0, Bl-1, Tsu-0, 7
8 Ler-0, Ko-2, Fei-0, Ste-0, and Oy-0), or those for which no sequence information was available (N14, Sah- 0, Berk, Hiro, Ita, HEK2, Stepn, Shign, and Borsk2). Also not included in the alignment is the sequence of, which has a premature stop codon (see above). FtsH6 sequences were extracted from the Arabidopsis 1001 Genomes project database ( For the multi-sequence alignment the MUSCLE tool ( was used. Amino acids divergent from are highlighted in red. Conservative substitutions are indicated as : in the sequence, while non-conservative exchanges are indicated by a free space. Asterisks (*) denote amino acids identical to. Can-0 Cvi-0 Bur-0 Leo-1 Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 Can-0 Cvi-0 Bur-0 Leo-1 Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 MASSSSALSFPLSNIPTCSKKSQQFQNPASLSKSSHTHKPSLKTQILHHKFTKRNLLSLT MASSSSALSFPLSNIPTCSKKSQQFQNPASLSKSSHTHKPSLKTQILHHKFTKRNLLSLT MASSSSALSFPLSNIPTCSKKSQQFQNPASLSKSSHTHKPSLKTQILHHKFTKRNLLSLT MASSSSALSFPLSNIPTCSKKSQQFQNPASLSKSSHTHKPSLKTQILHHKFTKRNLLSLT **************************:********************************* TALGFTSALGTVLAHPAKAEPEAPIEATSNRMSYSRFLQHLKENKVKKVDLIENGTVAIV TALGFTSALGTVLAHPAKAEPEAPIEATSNRMSYSRFLQHLKENKVKKVDLIENGTVAIV 8
9 9 Kas-1 ********************************************:*************** Can-0 Cvi-0 Bur-0 Leo-1 Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 ************************************************************ Can-0 LLVSLLLTSSSRRNPAEPNLPFGLGRSKAKFQMEPNTGITFEDVAGVDEAKQDFEEIVEF Cvi-0 LLVSLLLTSSSRRNPAEPNLPFGLGRSKAKFQMEPNTGITFEDVAGVDEAKQDFEEIVEF Bur-0 Leo-1 LLVSLLLTSSSRRNPAGPNLPFGLGRSKAKFQMEPNTGITFEDLAGVDEAKQDFEEIVEF Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 **************** **************************:**************** Can-0 LKTPEKFSALGAKIPKGVLLTGPPGTGKTLLAKAIAGEAGVPLFSLSGSEFIEMFVGVGA Cvi-0 LKTPEKFSALGAKIPKGVLLTGPPGTGKTLLAKAIAGEAGVPLFSLSGSEFIEMFVGVGA Bur-0 Leo-1 Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1
10 10 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 ******************************************:***************** Can-0 Cvi-0 Bur-0 Leo-1 Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 ************************************************************ Can-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR Cvi-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR Bur-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR Leo-1 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR Le-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRLGIMSTCFKQVSVGLPDIRGR ICE1 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRLGIMSTCFKQVSVGLPDIRGR Yeg-1 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRLGIMSTCFKQVSVGLPDIRGR Kondara VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRLGIMSTCFKQVSVGLPDIRGR Yo-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRLGIMSTCFKQVSVGLPDIRGR Dog-4 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRLGIMSTCFKQVSVGLPDIRGR Aitba-1 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR Kin-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFCIMSTCFKQVSVGLPDIRGR Ri-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFCIMSTCFKQVSVGLPDIRGR St-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFCIMSTCFKQVSVGLPDIRGR Abd-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFCIMSTCFKQVSVGLPDIRGR Mdn-1 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFCIMSTCFKQVSVGLPDIRGR Xan-1 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR Mt-0 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR Ct VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR C24 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR Kas-1 VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR VIVIAATNRPEILDSALLRPGRFDRQVCWLILKPNKSNRFGIMSTCFKQVSVGLPDIRGR ***************************************: ******************* Can-0 Cvi-0
11 11 Bur-0 Leo-1 EEILKVHSRSKKLDKDVSSSVIAMRTPGFSGADLANLMNEAAILAGRRGKDKITLTEIDD Le-0 EEILKVHSRSKKLDKDVSSSVIAMRTPGFSGADLANLMNEAAILAGRRGKDKITLTEIDD ICE1 EEILKVHSRSKKLDKDVSSSVIAMRTPGFSGADLANLMNEAAILAGRRGKDKITLTEIDD Yeg-1 EEILKVHSRSKKLDKDVSSSVIAMRTPGFSGADLANLMNEAAILAGRRGKDKITLTEIDD Kondara EEILKVHSRSKKLDKDVSSSVIAMRTPGFSGADLANLMNEAAILAGRRGKDKITLTEIDD Yo-0 EEILKVHSRSKKLDKDVSSSVIAMRTPGFSGADLANLMNEAAILAGRRGKDKITLTEIDD Dog-4 EEILKVHSRSKKLDKDVSSSVIAMRTPGFSGADLANLMNEAAILAGRRGKDKITLTEIDD Aitba-1 EEILKVHSRSKKLDKDVSSSVIAMRTPGFSGADLANLMNEAAILAGRRGKDKITLTEIDD Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 ****************** ***************************************** Can-0 Cvi-0 Bur-0 Leo-1 SIDRIVAGMEGTKMIDGKSKAIVAYHEVGHAICATLTEGHDPDQKVTLVPRGQARGLTWF Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 SIDRIVAGMEGTKMIDGKSKAIVAYHEVGHAICATLTEGYDPVQKVTLVPRGQARGLTWF ***************************************:** ***************** Can-0 Cvi-0 Bur-0 Leo-1 Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1
12 12 Mt-0 Ct C24 Kas-1 ************************************************************ Can-0 Cvi-0 Bur-0 MSEIGPWALIDPAVKQNDVVLRMLARNSVSEKLAEDIDSCVKKIIGDAYEVAKKHVRNNR Leo-1 Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 ********* ******************:******************************* Can-0 Cvi-0 Bur-0 Leo-1 Le-0 ICE1 Yeg-1 Kondara Yo-0 Dog-4 Aitba-1 Kin-0 Ri-0 St-0 Abd-0 Mdn-1 Xan-1 Mt-0 Ct C24 Kas-1 *************************************************
13 Supplementary Figure 6. 13
14 Supplementary Figure 6. Basal and acquired HS tolerance, and thermomemory of ftsh6 mutants. Seedlings of ftsh6 mutant and wild-type () plants were exposed to different HS regimes (schematically shown on the right of each section). (a) Upper middle and top panels: HS triggering stimulus after extended recovery, i.e., 4 and 3 days, respectively, following the priming treatment. Lower middle panel: acquired HS tolerance. Bottom panel: basal HS tolerance; HS applied to 7-day-old seedlings. Following HS, the seedlings were transferred to normal growth condition and photographed after 10 days (upper middle and top panels), 8 days (lower middle panel), or 5 days (bottom panel). (b) and (c) Quantification of results shown in a. (b) The percentage of seedlings in different phenotype classes. At 3 d recovery, data for ftsh6 mutants are not significantly different from. At 4 d recovery all data for ftsh6 mutants are different from (p < 0.05; Student s t-test). (c) Seedling fresh weight compared to control plants. Means ± SD are given (n = 5 plates with ~25 seedlings each). (d) Hypocotyl elongation assay for ftsh6 mutants and under HS. Six-day-old seedlings were subjected to the HS triggering stimulus without (unprimed) or with (primed) pre-treatment with moderate temperature stress as shown schematically. Photos were taken 5 days after triggering. (e) Hypocotyl lengths measured 5 days after recovery from the HS triggering stimulus and compared between primed and unprimed seedlings. Means ± SD are given (n = 7 plates with ~20 seedlings each). Asterisks indicate statistically significant difference (p < 0.05; Student s t-test) from the control. 14
15 Supplementary Figure 7. Supplementary Figure 7. The effect of cycloheximide on the accumulation of FtsH6 protein in Arabidopsis accession. (a) Immunoblot analyses of FtsH6 protein in at days 2, 3 and 4 of the memory phase upon cycloheximide (CHX) and mock (0.1% DMSO) treatment. CHX and mock treatment was applied to seedlings at 6 h into the memory phase, as shown schematically. The seedlings were harvested for immunoblotting at days 2, 3 or 4. kda, kilo Dalton. (b) Signals of the immunoblot analysis were quantified using ImageJ and normalized to the amount of RbcL in the same samples. Mean ± SD are given (n = 3; independent biological replicates each representing a pool of ~120 seedlings grown on 6 plates; gel blots used for the quantification are shown in Supplementary Fig. 10). Asterisks indicate significant difference in the level of FtsH6 between CHX- and mock-treated samples at each indicated time point (p < 0.05; Student s t-test). 15
16 Supplementary Figure 8. Supplementary Figure 8. FtsH6 expression in 35S:FtsH6 / plants. (a) Elevated expression of FtsH6 in 35S:FtsH6 / plants (lines #1, #2 and #3) compared to empty-vector (EV) control plants. Expression was analysed by qrt-pcr (n = 2). Values are expressed as the difference between an arbitrary value of 40 and dct, so that high 40-dCt value indicates high gene expression level. (b) Immunodetection of FtsH6 protein in 35S:FtsH6 / and EV seedlings exposed to the HS regime shown schematically, using anti-ftsh6 antibody. Note the presence of FtsH6 protein in 35S:FtsH6 Col- 0/ plants. RbcL, ribulose-1,5-bis-phosphate carboxylase/oxygenase large subunit (loading control; bottom panel). kda, kilo Dalton. (c) The phenotype of 35S:FtsH6 / and empty-vector (EV) seedlings exposed to different HS regimes (schematically shown on the right of each section in panels). 16
17 Supplementary Figure 9. Supplementary Figure 9. Immunodetection of HSP101 protein in and ftsh6. Immunoblot analysis of HSP101 protein was performed in and ftsh6 seedlings at days 2, 3 and 4 of the memory phase, using anti-hsp101 antibody (Abcam). Note the decline of HSP101 protein level at day 4 of the memory phase with no difference between and ftsh6. RbcL, ribulose-1,5-bis-phosphate carboxylase/ oxygenase large subunit (loading control). kda, kilo Dalton. 17
18 Supplementary Figure 10. Supplementary Figure 10. Uncropped gel and western blot images presented in this manuscript. Labelling above each image indicates the corresponding figure in the main manuscript or the supplement. M, molecular weight marker. Numbers left and right of images indicate protein molecular weights in kilo Dalton. 18
19 Supplementary Figure 10 (continued). Supplementary Figure 10. Uncropped gel and western blot images presented in this manuscript. Labelling above each image indicates the corresponding figure in the main manuscript or the supplement. M, molecular weight marker. Numbers left and right of images indicate protein molecular weights in kilo Dalton. 19
20 Supplementary Figure 10 (continued). Supplementary Figure 10. Uncropped gel and western blot images presented in this manuscript. Labelling above each image indicates the corresponding figure in the main manuscript or the supplement. M, molecular weight marker. Numbers left and right of images indicate protein molecular weights in kilo Dalton. 20
21 Supplementary Figure 10 (continued). Supplementary Figure 10. Uncropped gel and western blot images presented in this manuscript. Labelling above each image indicates the corresponding figure in the main manuscript or the supplement. M, molecular weight marker. Numbers left and right of images indicate protein molecular weights in kilo Dalton. 21
22 Supplementary Figure 10 (continued). Supplementary Figure 10. Uncropped gel and western blot images presented in this manuscript. Labelling above each image indicates the corresponding figure in the main manuscript or the supplement. M, molecular weight marker. Numbers left and right of images indicate protein molecular weights in kilo Dalton. 22
23 Supplementary Figure 10 (continued). Supplementary Figure 10. Uncropped gel and western blot images presented in this manuscript. Labelling above each image indicates the corresponding figure in the main manuscript or the supplement. M, molecular weight marker. Numbers left and right of images indicate protein molecular weights in kilo Dalton. 23
24 Supplementary Figure 10 (continued). Supplementary Figure 10. Uncropped gel and western blot images presented in this manuscript. Labelling above each image indicates the corresponding figure in the main manuscript or the supplement. M, molecular weight marker. Numbers left and right of images indicate protein molecular weights in kilo Dalton. 24
25 Supplementary Table 1. Primer sequences. Primers for cloning Sequence Sites added 35S:HSP21 HSP21-Forward GTTTAAACATGGCTTCTACACTCTCATTTGC added PmeI cloning site underlined HSP21-Reverse TTAATTAACTACTGAATCTGGACATCGATGA added PacI cloning site underlined HSP21-amiRNA amirna-1 I mir-s-1 II mir-a-1 III mir*s-1 IV mir*a-1 amirna-ii I mir-s-2 II mir-a-2 III mir*s-2 IV mir*a-2 amirna-iii I mir-s-3 II mir-a-3 III mir*s-3 IV mir*a-3 GATTAGTATCTAACATTTGTCGCCTCTCTTTTGTATTCCA AGGCGACAAATGTTAGATACTAATCAAAGAGAATCAATGA AGGCAACAAATGTTACATACTATTCACAGGTCGTGATATG GAATAGTATGTAACATTTGTTGCCTACATATATATTCCTA GATGTATCTAACATTTGTCGCATCTCTCTTTTGTATTCCA AGATGCGACAAATGTTAGATACATCAAAGAGAATCAATGA AGATACGACAAATGTAAGATACTTCACAGGTCGTGATATG GAAGTATCTTACATTTGTCGTATCTACATATATATTCCTA GATATAATGTTGATCGAGTCCTATCTCTCTTTTGTATTCC GATAGGACTCGATCAACATTATATCAAAGAGAATCAATGA GATAAGACTCGATCATCATTATTTCACAGGTCGTGATATG GAAATAATGATGATCGAGTCTTATCTACATATATATTCCT A Primer GTTTAAACCTGCAAGGCGATTAAGTTGGGTAAC added PmeI cloning site underlined (universal for making amirna) B primer TTAATTAAGCGGATAACAATTTCACACAGGAAACAG added PacI cloning site underlined (universal for making amirna) qrt-pcr primers Gene Forward Reverse Actin2 TCCCTCAGCACATTCCAGCAGAT AACGATTCCTGGACCTGCCTCATC HSP21 TGGACGTCTCTCCTTTCGGATTG TTTGTCGCATCGTCCTCATTGG FtsH6 GCCGGAATGGAAGGGACAAAGATG ATCATGACCCTCCGTCAAAGTCG 25
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated
 Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
Supplemental Data. Perrella et al. (2013). Plant Cell /tpc
 Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Supplemental Data. Chen and Thelen (2010). Plant Cell /tpc
 Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field.
 Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field. A, Phenotypes of 30-day old wild-type (WT) and abci8 mutant plants grown in a paddy field under normal sunny
Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field. A, Phenotypes of 30-day old wild-type (WT) and abci8 mutant plants grown in a paddy field under normal sunny
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed
 Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
SUPPLEMENTARY INFORMATION
 reverse 3175 3175 F L C 318 318 3185 3185 319 319 3195 3195 315 8 1 315 3155 315 317 Supplementary Figure 3. Stability of expression of the GFP sensor constructs return to warm conditions. Semi-quantitative
reverse 3175 3175 F L C 318 318 3185 3185 319 319 3195 3195 315 8 1 315 3155 315 317 Supplementary Figure 3. Stability of expression of the GFP sensor constructs return to warm conditions. Semi-quantitative
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering
 Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Ethylene is critical to the maintenance of primary root growth and Fe. homeostasis under Fe stress in Arabidopsis
 Ethylene is critical to the maintenance of primary root growth and Fe homeostasis under Fe stress in Arabidopsis Guangjie Li, Weifeng Xu, Herbert J. Kronzucker, Weiming Shi * Supplementary Data Supplementary
Ethylene is critical to the maintenance of primary root growth and Fe homeostasis under Fe stress in Arabidopsis Guangjie Li, Weifeng Xu, Herbert J. Kronzucker, Weiming Shi * Supplementary Data Supplementary
Supplemental Data. Hou et al. (2016). Plant Cell /tpc
 Supplemental Data. Hou et al. (216). Plant Cell 1.115/tpc.16.295 A Distance to 1 st nt of start codon Distance to 1 st nt of stop codon B Normalized PARE abundance 8 14 nt 17 nt Frame1 Arabidopsis inflorescence
Supplemental Data. Hou et al. (216). Plant Cell 1.115/tpc.16.295 A Distance to 1 st nt of start codon Distance to 1 st nt of stop codon B Normalized PARE abundance 8 14 nt 17 nt Frame1 Arabidopsis inflorescence
** * * * Col-0 cau1 CAU1. Actin2 CAS. Actin2. Supplemental Figure 1. CAU1 affects calcium accumulation.
 Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/452/ra106/dc1 Supplementary Materials for Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots Hyo-Jun Lee, Jun-Ho
www.sciencesignaling.org/cgi/content/full/9/452/ra106/dc1 Supplementary Materials for Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots Hyo-Jun Lee, Jun-Ho
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011) /tpc
 Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Supplemental Data. Wang et al. (2014). Plant Cell /tpc
 Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
GFP GAL bp 3964 bp
 Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Supplemental material
 Supplemental material Table 1- Segregation analysis of sgt1a sgt1b double mutant plants Parental genotype No. of lines Normal seeds Phenotype Abnormal seeds Ratio of abnormal seeds (%) WT 3 171 2 1.16
Supplemental material Table 1- Segregation analysis of sgt1a sgt1b double mutant plants Parental genotype No. of lines Normal seeds Phenotype Abnormal seeds Ratio of abnormal seeds (%) WT 3 171 2 1.16
Supplemental Data. Perea-Resa et al. Plant Cell. (2012) /tpc
 Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1
 Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL Figure S1. Mitochondrial morphology in Fis1-null, Mff-null and Fis1/Mff-null MEF cells. (A) Western blotting of lysates from Fis1-null, Mff-null and Fis1/Mff-null cells. Lysates were
SUPPLEMENTAL MATERIAL Figure S1. Mitochondrial morphology in Fis1-null, Mff-null and Fis1/Mff-null MEF cells. (A) Western blotting of lysates from Fis1-null, Mff-null and Fis1/Mff-null cells. Lysates were
Supplementary Table 1. Primers used in this study.
 Supplementary Tale 1. Primers used in this study. Name Primer sequence (5'-3') Primers of PCR-ased molecular markers developed in this study M1 F M1 R M2 F M2 R M3 F M3 R M4 F M4 R M5 F M5 R M6 F M6 R
Supplementary Tale 1. Primers used in this study. Name Primer sequence (5'-3') Primers of PCR-ased molecular markers developed in this study M1 F M1 R M2 F M2 R M3 F M3 R M4 F M4 R M5 F M5 R M6 F M6 R
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplemental Figure 1. Phenotype of ProRGA:RGAd17 plants under long day
 Supplemental Figure 1. Phenotype of ProRGA:RGAd17 plants under long day conditions. Photo was taken when the wild type plant started to bolt. Scale bar represents 1 cm. Supplemental Figure 2. Flowering
Supplemental Figure 1. Phenotype of ProRGA:RGAd17 plants under long day conditions. Photo was taken when the wild type plant started to bolt. Scale bar represents 1 cm. Supplemental Figure 2. Flowering
Supplementary Figure 1.
 Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner
 ABA represses the transcription of chloroplast genes Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner Supplementary data Supplementary tables Table
ABA represses the transcription of chloroplast genes Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner Supplementary data Supplementary tables Table
Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids
 Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids Plant growth conditions The soil was a 1:1 v/v mixture of loamy soil and organic compost. Initial soil water content was determined
Heterosis and inbreeding depression of epigenetic Arabidopsis hybrids Plant growth conditions The soil was a 1:1 v/v mixture of loamy soil and organic compost. Initial soil water content was determined
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and
 Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
(Lys), resulting in translation of a polypeptide without the Lys amino acid. resulting in translation of a polypeptide without the Lys amino acid.
 1. A change that makes a polypeptide defective has been discovered in its amino acid sequence. The normal and defective amino acid sequences are shown below. Researchers are attempting to reproduce the
1. A change that makes a polypeptide defective has been discovered in its amino acid sequence. The normal and defective amino acid sequences are shown below. Researchers are attempting to reproduce the
BLAST. Varieties of BLAST
 BLAST Basic Local Alignment Search Tool (1990) Altschul, Gish, Miller, Myers, & Lipman Uses short-cuts or heuristics to improve search speed Like speed-reading, does not examine every nucleotide of database
BLAST Basic Local Alignment Search Tool (1990) Altschul, Gish, Miller, Myers, & Lipman Uses short-cuts or heuristics to improve search speed Like speed-reading, does not examine every nucleotide of database
FSC-W FSC-H CD4 CD62-L
 Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
UNIT 5. Protein Synthesis 11/22/16
 UNIT 5 Protein Synthesis IV. Transcription (8.4) A. RNA carries DNA s instruction 1. Francis Crick defined the central dogma of molecular biology a. Replication copies DNA b. Transcription converts DNA
UNIT 5 Protein Synthesis IV. Transcription (8.4) A. RNA carries DNA s instruction 1. Francis Crick defined the central dogma of molecular biology a. Replication copies DNA b. Transcription converts DNA
Supplemental Figures. Supplemental Data. Sugliani et al. Plant Cell (2016) /tpc Clades RSH1. Rsh1[HS] RSH2 RSH3. Rsh4[HS] HYD TGS ACT
![Supplemental Figures. Supplemental Data. Sugliani et al. Plant Cell (2016) /tpc Clades RSH1. Rsh1[HS] RSH2 RSH3. Rsh4[HS] HYD TGS ACT Supplemental Figures. Supplemental Data. Sugliani et al. Plant Cell (2016) /tpc Clades RSH1. Rsh1[HS] RSH2 RSH3. Rsh4[HS] HYD TGS ACT](/thumbs/81/84002827.jpg) Supplemental Figures Clades RSH1 TP - HYD TGS ACT Rsh1[HS] RSH2 TP HYD SYN TGS Rsh2[HS] RSH3 TP HYD SYN TGS CRSH TP - SYN EFh Rsh4[HS] Supplemental Figure 1. Arabidopsis RSH domain structure. Schematic
Supplemental Figures Clades RSH1 TP - HYD TGS ACT Rsh1[HS] RSH2 TP HYD SYN TGS Rsh2[HS] RSH3 TP HYD SYN TGS CRSH TP - SYN EFh Rsh4[HS] Supplemental Figure 1. Arabidopsis RSH domain structure. Schematic
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice Supplementary Figure 2.
 Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Supplementary Information. Drought response transcriptomics are altered in poplar with reduced tonoplast sucrose transporter expression
 Supplementary Information Drought response transcriptomics are altered in poplar with reduced tonoplast sucrose transporter expression Liang Jiao Xue, Christopher J. Frost, Chung Jui Tsai, Scott A. Harding
Supplementary Information Drought response transcriptomics are altered in poplar with reduced tonoplast sucrose transporter expression Liang Jiao Xue, Christopher J. Frost, Chung Jui Tsai, Scott A. Harding
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
P. syringae and E. coli
 CHAPTER 6 A comparison of the recd mutant phenotypes of P. syringae and E. coli 6.1 INTRODUCTION The RecBCD complex is essential for recombination mediated repair of double strand breaks (DSBs) of DNA
CHAPTER 6 A comparison of the recd mutant phenotypes of P. syringae and E. coli 6.1 INTRODUCTION The RecBCD complex is essential for recombination mediated repair of double strand breaks (DSBs) of DNA
Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type
 A B 2 3 3 2 1 1 Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type (A) and d27 (B) seedlings at the four
A B 2 3 3 2 1 1 Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type (A) and d27 (B) seedlings at the four
Development 143: doi: /dev : Supplementary information
 Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
Supplemental Table 1. Primers used for cloning and PCR amplification in this study
 Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Table S1 List of primers used for genotyping and qrt-pcr.
 Table S1 List of primers used for genotyping and qrt-pcr. genotyping! allele! ligomer*! 5'-sequence-3'! rice! d10-2! F! TTGGCTTTGCCTCGTTTC!!! R! AGCCTCCACTTGTACTGTG! Arabidopsis! max2-3, max2-4! F! ACTCTCTCCGACCTCCCTGACG!!!
Table S1 List of primers used for genotyping and qrt-pcr. genotyping! allele! ligomer*! 5'-sequence-3'! rice! d10-2! F! TTGGCTTTGCCTCGTTTC!!! R! AGCCTCCACTTGTACTGTG! Arabidopsis! max2-3, max2-4! F! ACTCTCTCCGACCTCCCTGACG!!!
TNFα 18hr. Control. CHX 18hr. TNFα+ CHX 18hr. TNFα: 18 18hr (KDa) PARP. Cleaved. Cleaved. Cleaved. Caspase3. Pellino3 shrna. Control shrna.
 Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
microrna pseudo-targets
 microrna pseudo-targets Natalia Pinzón Restrepo Institut de Génétique Humaine (CNRS), Montpellier, France August, 2012 microrna target identification mir: target: 2 7 5 N NNNNNNNNNNNNNN NNNNNN 3 the seed
microrna pseudo-targets Natalia Pinzón Restrepo Institut de Génétique Humaine (CNRS), Montpellier, France August, 2012 microrna target identification mir: target: 2 7 5 N NNNNNNNNNNNNNN NNNNNN 3 the seed
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
Sequence analysis and comparison
 The aim with sequence identification: Sequence analysis and comparison Marjolein Thunnissen Lund September 2012 Is there any known protein sequence that is homologous to mine? Are there any other species
The aim with sequence identification: Sequence analysis and comparison Marjolein Thunnissen Lund September 2012 Is there any known protein sequence that is homologous to mine? Are there any other species
Genomics and bioinformatics summary. Finding genes -- computer searches
 Genomics and bioinformatics summary 1. Gene finding: computer searches, cdnas, ESTs, 2. Microarrays 3. Use BLAST to find homologous sequences 4. Multiple sequence alignments (MSAs) 5. Trees quantify sequence
Genomics and bioinformatics summary 1. Gene finding: computer searches, cdnas, ESTs, 2. Microarrays 3. Use BLAST to find homologous sequences 4. Multiple sequence alignments (MSAs) 5. Trees quantify sequence
downstream (0.8 kb) homologous sequences to the genomic locus of DIC. A DIC mutant strain (ro- 6
 A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
SUPPLEMENTARY INFORMATION
 Figure S1. Haploid plant produced by centromere-mediated genome elimination Chromosomes containing altered CENH3 in their centromeres (green dots) are eliminated after fertilization in a cross to wild
Figure S1. Haploid plant produced by centromere-mediated genome elimination Chromosomes containing altered CENH3 in their centromeres (green dots) are eliminated after fertilization in a cross to wild
Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus:
 m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
robustness: revisting the significance of mirna-mediated regulation
 : revisting the significance of mirna-mediated regulation Hervé Seitz IGH (CNRS), Montpellier, France October 13, 2012 microrna target identification .. microrna target identification mir: target: 2 7
: revisting the significance of mirna-mediated regulation Hervé Seitz IGH (CNRS), Montpellier, France October 13, 2012 microrna target identification .. microrna target identification mir: target: 2 7
BLAST Database Searching. BME 110: CompBio Tools Todd Lowe April 8, 2010
 BLAST Database Searching BME 110: CompBio Tools Todd Lowe April 8, 2010 Admin Reading: Read chapter 7, and the NCBI Blast Guide and tutorial http://www.ncbi.nlm.nih.gov/blast/why.shtml Read Chapter 8 for
BLAST Database Searching BME 110: CompBio Tools Todd Lowe April 8, 2010 Admin Reading: Read chapter 7, and the NCBI Blast Guide and tutorial http://www.ncbi.nlm.nih.gov/blast/why.shtml Read Chapter 8 for
Supp- Figure 2 Confocal micrograph of N. benthamiana tissues transiently expressing 35S:YFP-PDCB1. PDCB1 was targeted to plasmodesmata (twin punctate
 Supplemental Data. Simpson et al. (009). n rabidopsis GPI-anchor plasmodesmal neck protein with callosebinding activity and potential to regulate cell-to-cell trafficking. 5 0 stack Supp- Figure Confocal
Supplemental Data. Simpson et al. (009). n rabidopsis GPI-anchor plasmodesmal neck protein with callosebinding activity and potential to regulate cell-to-cell trafficking. 5 0 stack Supp- Figure Confocal
Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr.
 Molecular Cell, Volume 30 Supplemental Data Coupling of Ribosomal L1 Stalk and trna Dynamics during Translation Elongation Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr. Supplemental
Molecular Cell, Volume 30 Supplemental Data Coupling of Ribosomal L1 Stalk and trna Dynamics during Translation Elongation Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr. Supplemental
JMJ14-HA. Col. Col. jmj14-1. jmj14-1 JMJ14ΔFYR-HA. Methylene Blue. Methylene Blue
 Fig. S1 JMJ14 JMJ14 JMJ14ΔFYR Methylene Blue Col jmj14-1 JMJ14-HA Methylene Blue Col jmj14-1 JMJ14ΔFYR-HA Fig. S1. The expression level of JMJ14 and truncated JMJ14 with FYR (FYRN + FYRC) domain deletion
Fig. S1 JMJ14 JMJ14 JMJ14ΔFYR Methylene Blue Col jmj14-1 JMJ14-HA Methylene Blue Col jmj14-1 JMJ14ΔFYR-HA Fig. S1. The expression level of JMJ14 and truncated JMJ14 with FYR (FYRN + FYRC) domain deletion
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Principles of Genetics
 Principles of Genetics Snustad, D ISBN-13: 9780470903599 Table of Contents C H A P T E R 1 The Science of Genetics 1 An Invitation 2 Three Great Milestones in Genetics 2 DNA as the Genetic Material 6 Genetics
Principles of Genetics Snustad, D ISBN-13: 9780470903599 Table of Contents C H A P T E R 1 The Science of Genetics 1 An Invitation 2 Three Great Milestones in Genetics 2 DNA as the Genetic Material 6 Genetics
Role of Mitochondrial Remodeling in Programmed Cell Death in
 Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Supplementary Information for. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria
 Supplementary Information for Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria Isabella Santi 1 *, Neeraj Dhar 1, Djenet Bousbaine 1, Yuichi Wakamoto, John D.
Supplementary Information for Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria Isabella Santi 1 *, Neeraj Dhar 1, Djenet Bousbaine 1, Yuichi Wakamoto, John D.
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A)
 Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Supplementary Figure 1. Nature Genetics: doi: /ng.3848
 Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Waithe et al Supplementary Figures
 Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
** LCA LCN PCA
 % of wild type value % of wild type value a 12 1 8 2 b 12 1 8 2 LCA LCN PCA Col- sod3-1 Supplementary Figure 1 sod3-1 influences cell proliferation. (a) Fifth leaf cell area (LCA) and leaf cell number
% of wild type value % of wild type value a 12 1 8 2 b 12 1 8 2 LCA LCN PCA Col- sod3-1 Supplementary Figure 1 sod3-1 influences cell proliferation. (a) Fifth leaf cell area (LCA) and leaf cell number
Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport
 Ph.D. thesis Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport Zsigmond Laura Supervisor: Dr. Szabados László Arabidopsis Molecular Genetic Group Institute of Plant
Ph.D. thesis Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport Zsigmond Laura Supervisor: Dr. Szabados László Arabidopsis Molecular Genetic Group Institute of Plant
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Mourier et al., http://www.jcb.org/cgi/content/full/jcb.201411100/dc1 Figure S1. Size and mitochondrial content in Mfn1 and Mfn2 knockout hearts. (A) Body
Supplemental material THE JOURNAL OF CELL BIOLOGY Mourier et al., http://www.jcb.org/cgi/content/full/jcb.201411100/dc1 Figure S1. Size and mitochondrial content in Mfn1 and Mfn2 knockout hearts. (A) Body
Growth and development of Arabidopsis thaliana under single-wavelength red
 1 Supplementary Information 2 3 4 Growth and development of Arabidopsis thaliana under single-wavelength red and blue laser light 5 6 7 8 Authors Amanda Ooi 1 *, Aloysius Wong 1 *, Tien Khee Ng 2, Claudius
1 Supplementary Information 2 3 4 Growth and development of Arabidopsis thaliana under single-wavelength red and blue laser light 5 6 7 8 Authors Amanda Ooi 1 *, Aloysius Wong 1 *, Tien Khee Ng 2, Claudius
ydci GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC
 Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
. Supplementary Information
 . Supplementary Information Supplementary Figure S1. Mature embryo sac observations. Supplementary Figure S2. STT observations. Supplementary Figure S3. Comparison of the PTB1 cdna with that of the mutant.
. Supplementary Information Supplementary Figure S1. Mature embryo sac observations. Supplementary Figure S2. STT observations. Supplementary Figure S3. Comparison of the PTB1 cdna with that of the mutant.
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
EST1 Homology Domain. 100 aa. hest1a / SMG6 PIN TPR TPR. Est1-like DBD? hest1b / SMG5. TPR-like TPR. a helical. hest1c / SMG7.
 hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
1. In most cases, genes code for and it is that
 Name Chapter 10 Reading Guide From DNA to Protein: Gene Expression Concept 10.1 Genetics Shows That Genes Code for Proteins 1. In most cases, genes code for and it is that determine. 2. Describe what Garrod
Name Chapter 10 Reading Guide From DNA to Protein: Gene Expression Concept 10.1 Genetics Shows That Genes Code for Proteins 1. In most cases, genes code for and it is that determine. 2. Describe what Garrod
targets. clustering show that different complex pathway
 Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Nature Medicine: doi: /nm.3776
 C terminal Hsp90 inhibitors restore glucocorticoid sensitivity and relieve a mouse allograft model of Cushing s disease Mathias Riebold, Christian Kozany, Lee Freiburger, Michael Sattler, Michael Buchfelder,
C terminal Hsp90 inhibitors restore glucocorticoid sensitivity and relieve a mouse allograft model of Cushing s disease Mathias Riebold, Christian Kozany, Lee Freiburger, Michael Sattler, Michael Buchfelder,
Midterm Review Guide. Unit 1 : Biochemistry: 1. Give the ph values for an acid and a base. 2. What do buffers do? 3. Define monomer and polymer.
 Midterm Review Guide Name: Unit 1 : Biochemistry: 1. Give the ph values for an acid and a base. 2. What do buffers do? 3. Define monomer and polymer. 4. Fill in the Organic Compounds chart : Elements Monomer
Midterm Review Guide Name: Unit 1 : Biochemistry: 1. Give the ph values for an acid and a base. 2. What do buffers do? 3. Define monomer and polymer. 4. Fill in the Organic Compounds chart : Elements Monomer
A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development
 A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development Xuemei Chen Waksman Institute, Rutgers University, Piscataway, NJ 08854, USA. E-mail: xuemei@waksman.rutgers.edu Plant
A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development Xuemei Chen Waksman Institute, Rutgers University, Piscataway, NJ 08854, USA. E-mail: xuemei@waksman.rutgers.edu Plant
Fitness constraints on horizontal gene transfer
 Fitness constraints on horizontal gene transfer Dan I Andersson University of Uppsala, Department of Medical Biochemistry and Microbiology, Uppsala, Sweden GMM 3, 30 Aug--2 Sep, Oslo, Norway Acknowledgements:
Fitness constraints on horizontal gene transfer Dan I Andersson University of Uppsala, Department of Medical Biochemistry and Microbiology, Uppsala, Sweden GMM 3, 30 Aug--2 Sep, Oslo, Norway Acknowledgements:
Supplementary Methods
 Supplementary Methods Microarray analysis Grains of 7 DAP of the wild-type and gif1 were harvested for RNA preparation. Microarray analysis was performed with the Affymetrix (Santa Clara, CA) GeneChip
Supplementary Methods Microarray analysis Grains of 7 DAP of the wild-type and gif1 were harvested for RNA preparation. Microarray analysis was performed with the Affymetrix (Santa Clara, CA) GeneChip
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
Regulation of Phosphate Homeostasis by microrna in Plants
 Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Exploring Evolution & Bioinformatics
 Chapter 6 Exploring Evolution & Bioinformatics Jane Goodall The human sequence (red) differs from the chimpanzee sequence (blue) in only one amino acid in a protein chain of 153 residues for myoglobin
Chapter 6 Exploring Evolution & Bioinformatics Jane Goodall The human sequence (red) differs from the chimpanzee sequence (blue) in only one amino acid in a protein chain of 153 residues for myoglobin
Structure and RNA-binding properties. of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex
 Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/7/345/ra91/dc1 Supplementary Materials for TGF-β induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops Jingyu
www.sciencesignaling.org/cgi/content/full/7/345/ra91/dc1 Supplementary Materials for TGF-β induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops Jingyu
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
Computational Biology: Basics & Interesting Problems
 Computational Biology: Basics & Interesting Problems Summary Sources of information Biological concepts: structure & terminology Sequencing Gene finding Protein structure prediction Sources of information
Computational Biology: Basics & Interesting Problems Summary Sources of information Biological concepts: structure & terminology Sequencing Gene finding Protein structure prediction Sources of information
Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA
 Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
ACCGGTTTCGAATTGACAATTAATCATCGGCTCGTATAATGGTACC TGAAATGAGCTGTTGACAATTAATCATCCGGCTCGTATAATGTGTGG AATTGTGAGCGGATAACAATTTCACAGGTACC
 SUPPLEMENTAL TABLE S1. Promoter and riboswitch sequences used in this study. Predicted transcriptional start sites are bolded and underlined. Riboswitch sequences were obtained from Topp et al., Appl Environ
SUPPLEMENTAL TABLE S1. Promoter and riboswitch sequences used in this study. Predicted transcriptional start sites are bolded and underlined. Riboswitch sequences were obtained from Topp et al., Appl Environ
THE ROLE OF THE PHYTOCHROME B PHOTORECEPTOR IN THE REGULATION OF PHOTOPERIODIC FLOWERING. AnitaHajdu. Thesis of the Ph.D.
 THE ROLE OF THE PHYTOCHROME B PHOTORECEPTOR IN THE REGULATION OF PHOTOPERIODIC FLOWERING AnitaHajdu Thesis of the Ph.D. dissertation Supervisor: Dr. LászlóKozma-Bognár - senior research associate Doctoral
THE ROLE OF THE PHYTOCHROME B PHOTORECEPTOR IN THE REGULATION OF PHOTOPERIODIC FLOWERING AnitaHajdu Thesis of the Ph.D. dissertation Supervisor: Dr. LászlóKozma-Bognár - senior research associate Doctoral
Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures
 Nature Methods Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures Yannick Doyon, Thuy D Vo, Matthew C Mendel, Shon G Greenberg, Jianbin Wang, Danny F Xia, Jeffrey
Nature Methods Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures Yannick Doyon, Thuy D Vo, Matthew C Mendel, Shon G Greenberg, Jianbin Wang, Danny F Xia, Jeffrey
Biology I Level - 2nd Semester Final Review
 Biology I Level - 2nd Semester Final Review The 2 nd Semester Final encompasses all material that was discussed during second semester. It s important that you review ALL notes and worksheets from the
Biology I Level - 2nd Semester Final Review The 2 nd Semester Final encompasses all material that was discussed during second semester. It s important that you review ALL notes and worksheets from the
Table S1. Sequence of primers used in RT-qPCR
 Table S1. Sequence of primers used in RT-qPCR Primer Name P16Ink4a-F P16Ink4a-R P15Ink4b-F P15Ink4b-R P19Arf-F P19Arf-R P53-F P53-R P21cip1-F P21cip1-R P27kip1-F P27kip1-R P18Ink4c-F P18Ink4c-R P19Ink4d-F
Table S1. Sequence of primers used in RT-qPCR Primer Name P16Ink4a-F P16Ink4a-R P15Ink4b-F P15Ink4b-R P19Arf-F P19Arf-R P53-F P53-R P21cip1-F P21cip1-R P27kip1-F P27kip1-R P18Ink4c-F P18Ink4c-R P19Ink4d-F
Videos. Bozeman, transcription and translation: https://youtu.be/h3b9arupxzg Crashcourse: Transcription and Translation - https://youtu.
 Translation Translation Videos Bozeman, transcription and translation: https://youtu.be/h3b9arupxzg Crashcourse: Transcription and Translation - https://youtu.be/itsb2sqr-r0 Translation Translation The
Translation Translation Videos Bozeman, transcription and translation: https://youtu.be/h3b9arupxzg Crashcourse: Transcription and Translation - https://youtu.be/itsb2sqr-r0 Translation Translation The
