SUPPLEMENTARY INFORMATION
|
|
|
- Flora Chambers
- 5 years ago
- Views:
Transcription
1 DOI: /ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5, top panel), GFP-tagged human Rab7 (GFP-Rab7, middle panel), or mch-tagged human Rab11 (mch-rab11, bottom panel). These Rabs all localized to distinct vesicular compartments and did not co-localize with the luminal ER marker (Rab, red; KDEL, green). The third and fourth image in each panel shows merged images (ER in green, Rab in red) and zoomed images of boxed regions in merge, respectively. Scale bars = 10 µm. 1
2 Figure S2 Rab10 GTPase localizes to the ER and Golgi. a, Sequence alignment of Xenopus laevis Rab8/10 with Homo sapiens paralogs Rab8 and Rab10. b, Cos-7 cells were co-transfected with an ER luminal marker (KDEL-venus) and low amounts of mch-rab10. c, Cos-7 cells were cotransfected with mch-rab10 and a Golgi marker (GPP130-eGFP), mch- Rab10 and a mitochondrial marker (GFP-mito), BFP-tagged human Rab10 (BFP-Rab10) and an early endosome marker (mch-rab5), or BFP-Rab10 and a recycling endosome marker (mch-rab11), as indicated. Right panels in b and c, show merged images and zoomed images of boxed region (Rab10 in green, organelle markers in red). For b and c, scale bars = 10 µm. 2
3 Figure S3 Rab10 has a similar localization and function in HeLa cells. a, HeLa cells co-expressing mch-rab10 WT or mch-rab10 T23N and a luminal ER protein (KDEL-venus) were localized by confocal FM (top and bottom panels, respectively). Third and fourth panels show merged image and zoom of boxed region (Rab in green, ER in red). Scale bars = 10 µm. b, Immuno-blot analysis with antibodies against Rab10 reveals expression levels of endogenous and expressed Rab10 constructs under various conditions: no transfection, mch-rab10 WT or mch-rab10 T23N. GAPDH protein levels were measured as a loading control. When measured by densitometry, fusion proteins were found to be expressed 2.5 and 2.8- fold over endogenous for WT and T23N, respectively. Arrowhead indicates expressed mch-rab10 constructs. Asterisk indicates endogenous Rab
4 Figure S4 Rab10 sirna does not affect Golgi morphology. a, Cos-7 cells transfected with control sirna (top panel) or Rab10 sirna (bottom panel) and expressing Golgi marker GPP130-eGFP and mch-kdel (GPP130 in green, KDEL in red). b, Cos-7 cell transfected with GDP-locked mcherry tagged human Rab5 (mch-rab5 S34N) and KDEL-venus; note that the ER morphology remains highly tubular, (Rab5 S34N in red, KDEL in green). c, Cos-7 cell transfected with KDEL-venus and mcherry ER integral membrane protein, Climp63 (mch-climp63) and KDEL-venus. Note that expression of mch-climp63 results in cisternal ER, (Climp63 in red, KDEL in green). For a, b and c: the third image in each panel shows merged images. The fourth image in each panel shows zoomed, merged images. Boxes highlight the zoomed region. Scale bars = 10 µm. d, Percentage of the ER comprised of cisternae for cells transfected with KDEL-venus and mch-rab5 WT, mch-rab5 S34N or mch- Climp63. As additional controls for our cisternal ER analysis, we expressed both WT and GDP-locked mch-rab5 (S34N); expression of these constructs did not alter the percentage of ER that was cisternal when compared to other controls, 20% for both mch-rab5 WT and mch-rab5 S34N. Means ± standard errors were calculated from each condition, n = 20 cells, 3 boxes per cell. 4
5 Figure S5 Fusion events per area of ER (µm 2 ) a, Quantification of successful ER fusion events/area of ER (µm 2 ) over a time course of 5 min for KDEL-venus alone or KDEL-venus with mch-rab10 WT, mch-rab10 T23N. b, Quantification of successful ER fusion events/area of ER (µm 2 ) over a time course of 5 min for KDEL-venus with control or Rab10 sirna. Means ± standard errors were calculated for a and b, n = 30 cells for each condition. *** P <
6 Figure S6 Rab10-mediated dynamics occur on microtubules. a, Cos-7 cells expressing BFP-Rab10 WT, mch-tagged α-tubulin (to visualize MTs) and KDELvenus (not shown) were imaged live by confocal fluorescence microscopy. Arrowheads mark dynamic ER domains mediated by BFP-Rab10 punctum (Rab10, green; α-tubulin, red). Times indicated are in seconds. b, Cells as in a were imaged live before and after 5 µm nocodazole (NZ) to depolymerize MTs. Two images, taken 1 minute apart, were collected at 0, 20 and 60 minutes post nocodazole treatment. These images were superimposed in order to visualize the change in ER/Rab10 over a 1 minute time period before and after NZ treatment at each time point. Top panel shows BFP-Rab10 at t = 0 (green) and t = 60 seconds (red), white arrowheads indicate Rab10 rearrangement, blue arrowhead indicates ER fusion event. Bottom panel shows the sensitivity of the MT network to NZ treatment over time. Note that the overlay from samples treated for 60 minutes with NZ, shows very little change in ER/Rab10 positioning, indicating that Rab10 dynamic domains have stopped moving once MTs have depolymerized. Images in a and b, scale bars = 2 µm. 6
7 Figure S7 CEPT1 localizes to Rab10/PIS dynamic domain. a, A Cos-7 cell coexpressing KDEL-venus and mch-tagged human CEPT1 (mch-cept1) (CEPT1 in red, KDEL in green). Scale bar = 10 µm. b, As in a for cells expressing KDEL-venus, mch-cept1 (CEPT1 in red, KDEL in green). Note that CEPT1 localizes at dynamic domains that do not initially label with KDEL. Arrowheads mark position of ER extension that follows the mch-cept1 dynamic domain structure. Times indicated are in seconds. c, A Cos-7 cell co-expressing KDELvenus, mch-cept1 and BFP-Rab10 WT (CEPT1 in red, Rab10 in blue, KDEL in green); top panel shows overlay, bottom panel shows KDEL-venus alone to illustrate the absence of the ER from the Rab10/CEPT1 domain. Note the positions where Rab10 and CEPT1 co-localize at dynamic domains that do not initially label with KDEL. Arrowheads mark positions of successful fusion events that follow the BFP-Rab10/mCh-CEPT1 dynamic domain structures. Times indicated are in seconds. In b and c, scale bars = 2 µm. d, Percentage of co-localization between Rab10, CEPT1 and PIS; note that the majority of puncta have Rab10, CEPT1 and PIS, n = 10 cells. 7
8 Figure S8 Rab10-mediated ER assembly model. a, Model depicting known ER domains (in green) and domain regulating proteins. Rab10 dynamic domains (in pale green) travel along MTs (in gray) to facilitate homotypic ER fusion reactions. Rab10 (in blue) and the lipid synthesizing proteins PIS (in red) and CEPT1 (in purple) localize to dynamic domains at leading edge of nearly half of all ER tubule dynamics events. Atlastin-mediated ER fusion machinery (in orange) localizes to 3-way junctions. The organization of the peripheral ER cisternae is regulated by Climp63 (in yellow) and large protein complexes such as polyribosomes (not shown). Reticulon proteins (in turquois) oligomerize to maintain the tubular ER and curved edges of the cisternal ER. b, Rab10 and lipid synthesizing enzymes (PIS and CEPT1) localize to punctate structures at the dynamic tips of the ER (in red). GTPbound Rab10 facilitates ER assembly on the MT cytoskeleton (in blue), with the lipid synthesizing enzymes PIS and CEPT1 presumably providing the necessary lipids required for ER growth and dynamics. Depletion of endogenous Rab10 or expression of GDP-locked Rab10 results in the inhibition of ER assembly and an increase in ER cisternae at the expense of ER tubules (ER in green). 8
9 Figure S9 Uncropped scans of immunoblots. a, Uncropped immunoblot from Figure 1e. Reactions in Figure 1b were alternatively spun down and separated into soluble (S) and membrane (M) fractions. Soluble and membrane proteins were analyzed by immuno-blotting (IB) with antibodies recognizing Xenopus Rab8/10. Note that Rab 8/10 is displaced from the ER vesicles by concentrations of Rab GDI that inhibit tubule formation (as assayed in Figure 1a and 1b). b, Uncropped immuno-blot from Figure 3b. Immuno-blot analysis with antibodies against Rab10 reveals efficient depletion of Rab10 by sirna. GAPDH protein levels were measured as a loading control. c, Uncropped immuno-blot from Supplementary Figure 3b. Immuno-blot analysis with antibodies against Rab10 reveals expression levels of endogenous and expressed Rab10 constructs under various conditions: no transfection, mch-rab10 WT or mch-rab10 T23N. GAPDH protein levels were measured as a loading control. When measured by densitometry, fusion proteins were found to be expressed 2.5 and 2.8-fold over endogenous for WT and T23N, respectively. Arrowhead indicates expressed mch-rab10 constructs. Asterisk indicates endogenous Rab
10 Supplementary Videos Supplementary Video 1 Relative localization over time of Rab10 WT and an ER luminal protein. Cos-7 cells expressing mch-rab10 WT and KDEL-venus (Rab10 WT, red; KDEL, green) were imaged live by confocal fluorescence microscopy. Scale bar = 10 µm. Supplementary Video 2 Relative localization of PIS, Rab10 WT and an ER luminal protein. Cos-7 cells co-expressing mch-pis, BFP-Rab10 WT and KDELvenus (PIS, red; Rab10 WT, blue; KDEL, green) were imaged live by confocal fluorescence microscopy. Scale bar = 10 µm. Supplementary Video 3 Relative localization of Rab10 WT, CEPT1 and PIS. Cos-7 cells co-expressing BFP-Rab10 WT, mch-cept1 and GFP-PIS (Rab10 WT, blue; CEPT1, red, GFP, green) were imaged live by confocal fluorescence microscopy. Scale bar = 10 µm. Supplementary Video 4 Relative localization and structure of the ER in cells expressing Rab10 T23N, CEPT1 and PIS. Cos-7 cells co-expressing BFP-Rab10 T23N, mch-cept1 and GFP-PIS (Rab10 T23N, blue; CEPT1, red, GFP, green) were imaged live by confocal fluorescence microscopy. Scale bar = 10 µm. Supplementary Video 5 Relative localization of PIS, Rab10 WT and Atl3 puncta. Cos-7 cells expressing mch-pis, BFP-Rab10 WT and GFP-Atl3 (mch-pis, red; BFP-Rab10 WT, blue; GFP-Atl3, green) were imaged live by confocal fluorescence microscopy. Scale bar = 10 µm. Table S1 Rab10 Isoforms PER Counts. Raw data presented in Figure 2d. Percentage of the ER comprised of cisternae for cells expressing KDEL-venus alone or together with mch-rab10 WT, mch-rab10 Q68L, or mch-rab10 T23N. To determine ER morphology composition, three 28 pixel wide line segments were drawn on the images beginning at the NE away from the MT organizing center (yellow boxes), the Reni Entropy Threshold setting was used to select only ER cisternae or the total ER. Dividing the number of ER cisternae pixels (green) by the number of total ER pixels (green + blue) gives the percentage of ER cisternae in each line segment. The remaining ER was defined as ER tubules. Means ± standard errors were calculated from each condition, n = 50 cells for each condition, 3 regions per cell. *** P < Table S2 Controls for PER Counts. Raw data presented in Supplementary Figure 4d. Percentage of the ER comprised of cisternae for cells transfected with KDEL-venus and mch-rab5 WT, mch-rab5 S34N or mch-climp63 (analysis performed as described in Table S1). Means ± standard errors were calculated from each condition, n = 20 cells, 3 boxes per cell. Table S3 Control vs Rab10 sirna PER Counts. Raw data presented in Figure 3c. Percentage of the ER comprised of cisternae for cells transfected with control or Rab10 sirna (analysis performed as described in Table S1). Means ± standard errors were calculated from each condition, n = 50 cells, 3 regions per cell. *** P < Table S4 Rab10 Isoforms Fusion Counts. Raw data presented in Figure 4b, 4c and Supplementary Figure 5a. Cos-7 cells expressing KDEL-venus and mch-rab10 WT or mch-rab10 T23N were imaged live to visualize ER tubule extension and fusion events that occurred in a 10 µm 2 box over a 5 min time course. These tubular extensions were scored as either resulting in a successful or unsuccessful fusion event. The successful and unsuccessful fusion events measured were alternatively assayed as a percentage of events leading to fusion and ER fusion events/area of ER (µm 2 ). Means ± standard errors were calculated from each condition, n = 30 cells, 2 regions per cell. *** P < Table S5 Control vs Rab10 sirna Fusion Counts. Raw data presented in Figure 4e, 4f and Supplementary Figure 5b. Cos-7 cells expressing KDEL-venus to visualize ER extension and fusion were co-transfected with control sirna or Rab10 sirna and imaged live to visualize ER tubule extension and fusion events that occurred in a 10 µm 2 box over a 5 min time course. These tubular extensions were scored as either resulting in a successful or unsuccessful fusion event. The successful and unsuccessful fusion events measured were alternatively assayed as a percentage of events leading to fusion and ER fusion events/ area of ER (µm 2 ). Means ± standard errors were calculated from each condition, n = 30 cells, 2 regions per cell. *** P < Table S6 Puncta Counts WT vs T23N BFP-Rab10 + mch-pis. Raw data presented in Figure 5c. Cos7 cells expressing KDEL-venus, mch-pis and either BFP- Rab10 WT or BFP-Rab10 T23N. Percentage of ER attached puncta that contain both BFP-Rab10 and mch-pis, BFP-Rab10 alone, or mch-pis alone (n = 444 puncta from 10 cells for BFP-Rab10 WT and 492 puncta from 10 cells for BFP-Rab10 T23N). *** P < Table S7 ER Dynamics WT vs T23N BFP-Rab10 + mch-pis. Raw data presented in Figure 5d, 5e. Cos7 cells expressing KDEL-venus, mch-pis and either BFP-Rab10 WT or BFP-Rab10 T23N were imaged live by confocal FM. All ER extension events occurring in a 10 µm 2 box over a 5 min time course were observed. We calculated the percentage of total events clearly led by either a PIS/Rab10 WT or PIS/Rab10 T23N dynamic domain (621 and 437 total events, respectively). For each of the events led by a PIS/Rab10 dynamic domain, we scored if the event led to a successful fusion reaction with adjacent ER domains. The efficiency of fusion was calculated for puncta containing WT or T23N Rab10. Means ± standard errors were calculated for each graph; n = 30 cells, 2 boxes per cell. *** P < Table S8. Rab10 Co-Localization. Raw data presented in Figure 6b. Cos-7 cell expressing BFP-Rab10 with mch-cept1, mch-pis, mch-sec61β or KDELvenus. Percentage of co-localization for cells expressing BFP-Rab10 with mch-cept1, mch-pis, mch-sec61β or KDEL-venus, as measured by Spearman- Pearson s co-localization coefficient in a 10 µm 2 box. Means ± standard errors were calculated from each condition, n = 10 cells for each condition. ** P < Table S9 Puncta Profile. Raw data presented in Supplementary Figure 7d. We determined the presence of BFP-Rab10, mch-cept1 and GFP-PIS at all of the puncta present in the imaged cells; n = 10 cells/condition. Table S10 WT vs T23N BFP-Rab10 T0-T60. Raw data presented in Figure 7d. Percentage of co-localization for cells expressing GFP-PIS withbfp-rab10 WT or BFP-Rab10 T23N, as measured by Spearman-Pearson s co-localization coefficient. Note that the higher the percentage co-localization, the less overall dynamics. Means ± standard errors were calculated from each condition, n = 10 cells for each condition. *** P <
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
Waithe et al Supplementary Figures
 Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL Figure S1. Mitochondrial morphology in Fis1-null, Mff-null and Fis1/Mff-null MEF cells. (A) Western blotting of lysates from Fis1-null, Mff-null and Fis1/Mff-null cells. Lysates were
SUPPLEMENTAL MATERIAL Figure S1. Mitochondrial morphology in Fis1-null, Mff-null and Fis1/Mff-null MEF cells. (A) Western blotting of lysates from Fis1-null, Mff-null and Fis1/Mff-null cells. Lysates were
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
Supplementary Figure 1.
 Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
Supplementary Information
 Supplementary Information MAP2/Hoechst Hyp.-AP ph 6.5 Hyp.-SD ph 7.2 Norm.-SD ph 7.2 Supplementary Figure 1. Mitochondrial elongation in cortical neurons by acidosis. Representative images of neuronal
Supplementary Information MAP2/Hoechst Hyp.-AP ph 6.5 Hyp.-SD ph 7.2 Norm.-SD ph 7.2 Supplementary Figure 1. Mitochondrial elongation in cortical neurons by acidosis. Representative images of neuronal
Supplementary Figure 1. CoMIC in 293T, HeLa, and HepG2 cells. (a) Mitochondrial morphology in 293T, HeLa and HepG2 cells. Cells were transfected with
 Supplementary Figure 1. CoMIC in 293T, HeLa, and HepG2 cells. (a) Mitochondrial morphology in 293T, HeLa and HepG2 cells. Cells were transfected with DsRed-mito. Right panels are time-course enlarged images
Supplementary Figure 1. CoMIC in 293T, HeLa, and HepG2 cells. (a) Mitochondrial morphology in 293T, HeLa and HepG2 cells. Cells were transfected with DsRed-mito. Right panels are time-course enlarged images
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Mourier et al., http://www.jcb.org/cgi/content/full/jcb.201411100/dc1 Figure S1. Size and mitochondrial content in Mfn1 and Mfn2 knockout hearts. (A) Body
Supplemental material THE JOURNAL OF CELL BIOLOGY Mourier et al., http://www.jcb.org/cgi/content/full/jcb.201411100/dc1 Figure S1. Size and mitochondrial content in Mfn1 and Mfn2 knockout hearts. (A) Body
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
Supplementary information
 Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
Heather Currinn, Benjamin Guscott, Zita Balklava, Alice Rothnie and Thomas Wassmer*
 Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
 DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%%
 !"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
targets. clustering show that different complex pathway
 Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
T H E J O U R N A L O F C E L L B I O L O G Y
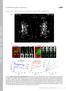 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Eisner et al., http://www.jcb.org/cgi/content/full/jcb.201312066/dc1 Figure S1. Mitochondrial continuity in adult skeletal muscle. (A)
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Eisner et al., http://www.jcb.org/cgi/content/full/jcb.201312066/dc1 Figure S1. Mitochondrial continuity in adult skeletal muscle. (A)
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Selective Targeting of ER Exit Sites Supports Axon Development
 Traffic 2009; 10: 1669 1684 2009 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2009.00974.x Selective Targeting of ER Exit Sites Supports Axon Development Meir Aridor 1 and Kenneth N. Fish 2, 1 Department
Traffic 2009; 10: 1669 1684 2009 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2009.00974.x Selective Targeting of ER Exit Sites Supports Axon Development Meir Aridor 1 and Kenneth N. Fish 2, 1 Department
Protein Sorting, Intracellular Trafficking, and Vesicular Transport
 Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Role of Mitochondrial Remodeling in Programmed Cell Death in
 Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Supplementary information
 Supplementary information General Strategy for Direct Cytosolic Protein Delivery via Protein- Nanoparticle Co-Engineering Rubul Mout, Moumita Ray, Tristan Tay, Kanae Sasaki, Gulen Yesilbag Tonga, Vincent
Supplementary information General Strategy for Direct Cytosolic Protein Delivery via Protein- Nanoparticle Co-Engineering Rubul Mout, Moumita Ray, Tristan Tay, Kanae Sasaki, Gulen Yesilbag Tonga, Vincent
FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell
 FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell All media is on the Instructors Resource CD/DVD JPEG Resources Figures, Photos, and Tables PowerPoint Resources Chapter Outline with Figures Lecture
FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell All media is on the Instructors Resource CD/DVD JPEG Resources Figures, Photos, and Tables PowerPoint Resources Chapter Outline with Figures Lecture
Way to impose membrane curvature
 Way to impose membrane curvature Sar1 reticulons? clathrin and other vesicle coats dynamin BAR domains McMahon, 2005 The ER is continuous with the nuclear envelope and contains tubules and sheets Gia Voeltz
Way to impose membrane curvature Sar1 reticulons? clathrin and other vesicle coats dynamin BAR domains McMahon, 2005 The ER is continuous with the nuclear envelope and contains tubules and sheets Gia Voeltz
Supporting Information
 Supporting Information Fleissner et al. 10.1073/pnas.0907039106 Fig. S1. (A) MAK-2-GFP localized to CATs tips is not bound by membrane. his-3::pccg1 mak-2-gfp; mak-2 strain labeled with membrane dye FM4
Supporting Information Fleissner et al. 10.1073/pnas.0907039106 Fig. S1. (A) MAK-2-GFP localized to CATs tips is not bound by membrane. his-3::pccg1 mak-2-gfp; mak-2 strain labeled with membrane dye FM4
The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression
 Traffic 2010; 11: 827 842 2010 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2010.01055.x The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression Danming Tang, Hebao Yuan and Yanzhuang
Traffic 2010; 11: 827 842 2010 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2010.01055.x The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression Danming Tang, Hebao Yuan and Yanzhuang
Supplemental Data. Gao et al. (2012). Plant Cell /tpc
 Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Cryo-EM data collection, refinement and validation statistics
 1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
CELB40060 Membrane Trafficking in Animal Cells. Prof. Jeremy C. Simpson. Lecture 2 COPII and export from the ER
 CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises
 Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
T H E J O U R N A L O F C E L L B I O L O G Y
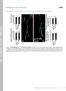 Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Supplementary Figure 1. Biochemical and sequence alignment analyses the
 Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Supplementary material
 Supplementary material Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins Vladimir V. Rogov 1,*, Hironori Suzuki 2,3,*, Mija Marinković 4, Verena Lang
Supplementary material Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins Vladimir V. Rogov 1,*, Hironori Suzuki 2,3,*, Mija Marinković 4, Verena Lang
downstream (0.8 kb) homologous sequences to the genomic locus of DIC. A DIC mutant strain (ro- 6
 A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
Molecular Cell Biology 5068 In Class Exam 1 September 30, Please print your name:
 Molecular Cell Biology 5068 In Class Exam 1 September 30, 2014 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Molecular Cell Biology 5068 In Class Exam 1 September 30, 2014 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Division Ave. High School AP Biology
 Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
SUPPLEMENTARY INFORMATION
 Supplementary Notes Downregulation of atlastin does not affect secretory traffic We investigated whether Atlastin might play a role in secretory traffic. Traffic impairment results in disruption of Golgi
Supplementary Notes Downregulation of atlastin does not affect secretory traffic We investigated whether Atlastin might play a role in secretory traffic. Traffic impairment results in disruption of Golgi
Supplementary Information for. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria
 Supplementary Information for Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria Isabella Santi 1 *, Neeraj Dhar 1, Djenet Bousbaine 1, Yuichi Wakamoto, John D.
Supplementary Information for Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria Isabella Santi 1 *, Neeraj Dhar 1, Djenet Bousbaine 1, Yuichi Wakamoto, John D.
7.06 Cell Biology EXAM #3 KEY
 7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
Reconstructing Mitochondrial Evolution?? Morphological Diversity. Mitochondrial Diversity??? What is your definition of a mitochondrion??
 Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial
Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial
Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX.
 Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX. Phase contrast and widefield fluorescence microscopy (20x magnification).
Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX. Phase contrast and widefield fluorescence microscopy (20x magnification).
Supporting Online Material for
 Originally posted 1 September 2011; corrected 5 October 2011 www.sciencemag.org/cgi/content/full/science.1207385/dc1 Supporting Online Material for ER Tubules Mark Sites of Mitochondrial Division Jonathan
Originally posted 1 September 2011; corrected 5 October 2011 www.sciencemag.org/cgi/content/full/science.1207385/dc1 Supporting Online Material for ER Tubules Mark Sites of Mitochondrial Division Jonathan
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and
 Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
The Ultrastructure of Cells (1.2) IB Diploma Biology
 The Ultrastructure of Cells (1.2) IB Diploma Biology Explain why cells with different functions have different structures. Cells have different organelles depending on the primary function of the cell
The Ultrastructure of Cells (1.2) IB Diploma Biology Explain why cells with different functions have different structures. Cells have different organelles depending on the primary function of the cell
Correct Targeting of Plant ARF GTPases Relies on Distinct Protein Domains
 Traffic 2008; 9: 103 120 Blackwell Munksgaard # 2007 The Authors Journal compilation # 2007 Blackwell Publishing Ltd doi: 10.1111/j.1600-0854.2007.00671.x Correct Targeting of Plant ARF GTPases Relies
Traffic 2008; 9: 103 120 Blackwell Munksgaard # 2007 The Authors Journal compilation # 2007 Blackwell Publishing Ltd doi: 10.1111/j.1600-0854.2007.00671.x Correct Targeting of Plant ARF GTPases Relies
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists. International authors and editors
 We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists 3,900 116,000 120M Open access books available International authors and editors Downloads Our
We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists 3,900 116,000 120M Open access books available International authors and editors Downloads Our
Cytoplasmic control of Rab family small GTPases through BAG6
 Article Cytoplasmic control of Rab family small GTPases through Toshiki Takahashi 1, Setsuya Minami 1, Yugo Tsuchiya 1, Kazu Tajima 1, Natsumi Sakai 1, Kei Suga 2,3, Shin-ichi Hisanaga 4, Norihiko Ohbayashi
Article Cytoplasmic control of Rab family small GTPases through Toshiki Takahashi 1, Setsuya Minami 1, Yugo Tsuchiya 1, Kazu Tajima 1, Natsumi Sakai 1, Kei Suga 2,3, Shin-ichi Hisanaga 4, Norihiko Ohbayashi
UNIT 3 CP BIOLOGY: Cell Structure
 UNIT 3 CP BIOLOGY: Cell Structure Page CP: CHAPTER 3, Sections 1-3; HN: CHAPTER 7, Sections 1-2 Standard B-2: The student will demonstrate an understanding of the structure and function of cells and their
UNIT 3 CP BIOLOGY: Cell Structure Page CP: CHAPTER 3, Sections 1-3; HN: CHAPTER 7, Sections 1-2 Standard B-2: The student will demonstrate an understanding of the structure and function of cells and their
GFP WxTP-GFP WxTP-GFP. stromules. DsRed WxTP-DsRed WxTP-DsRed. stromules
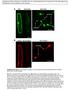 Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
Chapter 6: A Tour of the Cell
 AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10923 Supplementary Figure 1 Ten-a and Ten-m antibody and cell type specificities. a c, Representative single confocal sections of a Drosophila NMJ stained with antibodies to Ten-a (red),
doi:10.1038/nature10923 Supplementary Figure 1 Ten-a and Ten-m antibody and cell type specificities. a c, Representative single confocal sections of a Drosophila NMJ stained with antibodies to Ten-a (red),
Lecture 6 - Intracellular compartments and transport I
 01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
supplementary information
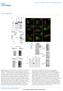 DOI: 10.1038/ncb2119 Figure S1 Analysis of IRGM localization, GST-IRGMd, GST-IRGMdS47N and GST-IRGMb protein purification, and analysis of their binding to cardiolipin. a. Mitochondria were purified by
DOI: 10.1038/ncb2119 Figure S1 Analysis of IRGM localization, GST-IRGMd, GST-IRGMdS47N and GST-IRGMb protein purification, and analysis of their binding to cardiolipin. a. Mitochondria were purified by
Chapter 4 A Tour of the Cell*
 Chapter 4 A Tour of the Cell* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. The Fundamental Units of Life Cells
Chapter 4 A Tour of the Cell* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. The Fundamental Units of Life Cells
Components of a functional cell. Boundary-membrane Cytoplasm: Cytosol (soluble components) & particulates DNA-information Ribosomes-protein synthesis
 Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
Running title: Golgi enzymes in mitosis Key words: Golgi apparatus, endoplasmic reticulum, mitosis, Arf1, Sar1
 Golgi inheritance in mammalian cells is mediated through ER export activities Nihal Altan-Bonnet, Rachid Sougrat, Wei Liu, Erik L. Snapp, Theresa Ward* and Jennifer Lippincott-Schwartz # Cell biology and
Golgi inheritance in mammalian cells is mediated through ER export activities Nihal Altan-Bonnet, Rachid Sougrat, Wei Liu, Erik L. Snapp, Theresa Ward* and Jennifer Lippincott-Schwartz # Cell biology and
SUPPLEMENTARY MATERIAL
 Activity [RLU] Activity [RLU] RAB:TALA TALA:RAB TALB:RAB RAB:TALB RAB:TALB TALB:RAB TALA:RAB RAB:TALA Activity [nrlu] Activity [nrlu] SUPPLEMENTARY MATERIAL a b 100 100 50 50 0 0 5 25 50 5 25 50 0 50 50
Activity [RLU] Activity [RLU] RAB:TALA TALA:RAB TALB:RAB RAB:TALB RAB:TALB TALB:RAB TALA:RAB RAB:TALA Activity [nrlu] Activity [nrlu] SUPPLEMENTARY MATERIAL a b 100 100 50 50 0 0 5 25 50 5 25 50 0 50 50
The Cell. C h a p t e r. PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas
 C h a p t e r 2 The Cell PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas Copyright 2009 Pearson Education, Inc., publishing as Pearson Benjamin Cummings Introduction
C h a p t e r 2 The Cell PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas Copyright 2009 Pearson Education, Inc., publishing as Pearson Benjamin Cummings Introduction
SUPPLEMENTARY INFORMATION
 GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
Biology of Fungi. Fungal Structure and Function. Lecture: Structure/Function, Part A BIOL 4848/ Fall Overview of the Hypha
 Biology of Fungi Fungal Structure and Function Overview of the Hypha The hypha is a rigid tube containing cytoplasm Growth occurs at the tips of hyphae Behind the tip, the cell is aging Diagram of hyphal
Biology of Fungi Fungal Structure and Function Overview of the Hypha The hypha is a rigid tube containing cytoplasm Growth occurs at the tips of hyphae Behind the tip, the cell is aging Diagram of hyphal
Name: Date: Hour:
 Name: Date: Hour: 1 2 3 4 5 6 Comprehension Questions 1. At what level of organization does life begin? 2. What surrounds all cells? 3. What is meant by semipermeable? 4. What 2 things make up the cell
Name: Date: Hour: 1 2 3 4 5 6 Comprehension Questions 1. At what level of organization does life begin? 2. What surrounds all cells? 3. What is meant by semipermeable? 4. What 2 things make up the cell
Cell (Learning Objectives)
 Cell (Learning Objectives) 1. Understand & describe the basic components necessary for a functional cell. 2. Review the order of appearance of cells on earth and explain the endosymbiotic theory. 3. Compare
Cell (Learning Objectives) 1. Understand & describe the basic components necessary for a functional cell. 2. Review the order of appearance of cells on earth and explain the endosymbiotic theory. 3. Compare
Supplementary Information. The protease GtgE from Salmonella exclusively targets. inactive Rab GTPases
 Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization
 Cell Metabolism Supplemental Information Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization Prashant Mishra, Grigor Varuzhanyan,
Cell Metabolism Supplemental Information Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization Prashant Mishra, Grigor Varuzhanyan,
Cells. Steven McLoon Department of Neuroscience University of Minnesota
 Cells Steven McLoon Department of Neuroscience University of Minnesota 1 Microscopy Methods of histology: Treat the tissue with a preservative (e.g. formaldehyde). Dissect the region of interest. Embed
Cells Steven McLoon Department of Neuroscience University of Minnesota 1 Microscopy Methods of histology: Treat the tissue with a preservative (e.g. formaldehyde). Dissect the region of interest. Embed
TNFα 18hr. Control. CHX 18hr. TNFα+ CHX 18hr. TNFα: 18 18hr (KDa) PARP. Cleaved. Cleaved. Cleaved. Caspase3. Pellino3 shrna. Control shrna.
 Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Eukaryotic microorganisms and viruses
 Eukaryotic microorganisms and viruses 2 1 Heribert Cypionka What is the main difference between the prokaryotic and the eukaryotic cell? Compartimentation >> Separation of reaction rooms >> More complex
Eukaryotic microorganisms and viruses 2 1 Heribert Cypionka What is the main difference between the prokaryotic and the eukaryotic cell? Compartimentation >> Separation of reaction rooms >> More complex
Cell Biology Review. The key components of cells that concern us are as follows: 1. Nucleus
 Cell Biology Review Development involves the collective behavior and activities of cells, working together in a coordinated manner to construct an organism. As such, the regulation of development is intimately
Cell Biology Review Development involves the collective behavior and activities of cells, working together in a coordinated manner to construct an organism. As such, the regulation of development is intimately
Movement and Remodeling of the Endoplasmic Reticulum in Nondividing Cells of Tobacco Leaves W
 The Plant Cell, Vol. 21: 3937 3949, December 2009, www.plantcell.org ã 2009 American Society of Plant Biologists Movement and Remodeling of the Endoplasmic Reticulum in Nondividing Cells of Tobacco Leaves
The Plant Cell, Vol. 21: 3937 3949, December 2009, www.plantcell.org ã 2009 American Society of Plant Biologists Movement and Remodeling of the Endoplasmic Reticulum in Nondividing Cells of Tobacco Leaves
Cell Structure and Function. The Development of Cell Theory
 Cell Structure and Function The Development of Cell Theory Hooke's original microscope, that he used at Oxford University, in 1665. Robert Hooke first examined a thin piece of cork (see below), and coined
Cell Structure and Function The Development of Cell Theory Hooke's original microscope, that he used at Oxford University, in 1665. Robert Hooke first examined a thin piece of cork (see below), and coined
Structure and RNA-binding properties. of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex
 Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Chapter 12: Intracellular sorting
 Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
4.1 Cells are the Fundamental Units of Life. Cell Structure. Cells. Fundamental units of life Cell theory. Except possibly viruses.
 Cells 4.1 Cells are the Fundamental Units of Life Fundamental units of life Cell theory All living things are composed of one or more cells. The cell is the most basic unit of life. All cells come from
Cells 4.1 Cells are the Fundamental Units of Life Fundamental units of life Cell theory All living things are composed of one or more cells. The cell is the most basic unit of life. All cells come from
Downloaded from at University of Washington Health Sciences Libraries, on November 12, 2010
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 285, NO. 41, pp. 31590 31602, October 8, 2010 2010 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Rab32 Modulates Apoptosis
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 285, NO. 41, pp. 31590 31602, October 8, 2010 2010 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Rab32 Modulates Apoptosis
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo
 Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Biology 580 Cellular Physiology Spring 2007 Course Syllabus
 Class # 17416 MW 0800-0850 Biology 580 Cellular Physiology Spring 2007 Course Syllabus Course Text: Course Website: Molecular Biology of the Cell by Alberts et al.; 4th edition, Garland Science, 2002 (ISBN
Class # 17416 MW 0800-0850 Biology 580 Cellular Physiology Spring 2007 Course Syllabus Course Text: Course Website: Molecular Biology of the Cell by Alberts et al.; 4th edition, Garland Science, 2002 (ISBN
Heteromultimeric TRPML channel assemblies play a crucial role in the regulation of cell viability models and starvation-induced autophagy
 3112 Research Article Heteromultimeric TRPML channel assemblies play a crucial role in the regulation of cell viability models and starvation-induced autophagy David A. Zeevi 1, *, Shaya Lev 2,3, *, Ayala
3112 Research Article Heteromultimeric TRPML channel assemblies play a crucial role in the regulation of cell viability models and starvation-induced autophagy David A. Zeevi 1, *, Shaya Lev 2,3, *, Ayala
Role of Unc51.1 and its binding partners in CNS axon outgrowth
 Role of Unc51.1 and its binding partners in CNS axon outgrowth Toshifumi Tomoda, Jee Hae Kim, Caixin Zhan, and Mary E. Hatten 1 Laboratory of Developmental Neurobiology, The Rockefeller University, New
Role of Unc51.1 and its binding partners in CNS axon outgrowth Toshifumi Tomoda, Jee Hae Kim, Caixin Zhan, and Mary E. Hatten 1 Laboratory of Developmental Neurobiology, The Rockefeller University, New
Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis
 Article Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis Liang Ge 1,,, Min Zhang 1, Samuel J Kenny 2, Dawei Liu 1, Miharu Maeda 3, Kota Saito 3, Anandita Mathur
Article Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis Liang Ge 1,,, Min Zhang 1, Samuel J Kenny 2, Dawei Liu 1, Miharu Maeda 3, Kota Saito 3, Anandita Mathur
FSC-W FSC-H CD4 CD62-L
 Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Protein Sorting. By: Jarod, Tyler, and Tu
 Protein Sorting By: Jarod, Tyler, and Tu Definition Organizing of proteins Organelles Nucleus Ribosomes Endoplasmic Reticulum Golgi Apparatus/Vesicles How do they know where to go? Amino Acid Sequence
Protein Sorting By: Jarod, Tyler, and Tu Definition Organizing of proteins Organelles Nucleus Ribosomes Endoplasmic Reticulum Golgi Apparatus/Vesicles How do they know where to go? Amino Acid Sequence
White Paper. Linear Range Determination in Empiria Studio Software
 White Paper Linear Range Determination in Empiria Studio Software Published September 2018. The most recent version of this document is posted at licor.com/bio/support. Page 2 - Linear Range Determination
White Paper Linear Range Determination in Empiria Studio Software Published September 2018. The most recent version of this document is posted at licor.com/bio/support. Page 2 - Linear Range Determination
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Breker et al., http://www.jcb.org/cgi/content/full/jcb.201301120/dc1 Figure S1. Single-cell proteomics of stress responses. (a) Using
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Breker et al., http://www.jcb.org/cgi/content/full/jcb.201301120/dc1 Figure S1. Single-cell proteomics of stress responses. (a) Using
Predominant Golgi Residency of the Plant K/HDEL Receptor Is Essential for its Function in Mediating ER Retention
 Plant Cell Advance Publication. Published on August 2, 2018, doi:10.1105/tpc.18.00426 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41
Plant Cell Advance Publication. Published on August 2, 2018, doi:10.1105/tpc.18.00426 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41
Chapter 6: A Tour of the Cell
 Chapter 6: A Tour of the Cell 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when Robert Hooke first looked at dead cells from an oak tree.
Chapter 6: A Tour of the Cell 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when Robert Hooke first looked at dead cells from an oak tree.
Identification and characterization of multiple novel Rab myosin Va interactions
 MBoC ARTICLE Identification and characterization of multiple novel Rab myosin Va interactions Andrew J. Lindsay a,b, Florence Jollivet b, Conor P. Horgan a, Amir R. Khan c, Graça Raposo d,e, Mary W. McCaffrey
MBoC ARTICLE Identification and characterization of multiple novel Rab myosin Va interactions Andrew J. Lindsay a,b, Florence Jollivet b, Conor P. Horgan a, Amir R. Khan c, Graça Raposo d,e, Mary W. McCaffrey
Biological Process Term Enrichment
 Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
Imaging the secretory pathway: The past and future impact of live cell optical techniques
 Biochimica et Biophysica Acta 1744 (2005) 259 272 Review Imaging the secretory pathway: The past and future impact of live cell optical techniques John F. Presley* McGill University, Department of Anatomy
Biochimica et Biophysica Acta 1744 (2005) 259 272 Review Imaging the secretory pathway: The past and future impact of live cell optical techniques John F. Presley* McGill University, Department of Anatomy
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
Syntabulin-mediated anterograde transport of mitochondria along neuronal processes
 JCB: ARTICLE Syntabulin-mediated anterograde transport of mitochondria along neuronal processes Qian Cai, Claudia Gerwin, and Zu-Hang Sheng Synaptic Function Unit, The Porter Neuroscience Research Center,
JCB: ARTICLE Syntabulin-mediated anterograde transport of mitochondria along neuronal processes Qian Cai, Claudia Gerwin, and Zu-Hang Sheng Synaptic Function Unit, The Porter Neuroscience Research Center,
