SUPPLEMENTARY INFORMATION
|
|
|
- May Mathews
- 6 years ago
- Views:
Transcription
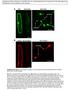
1 doi: /nature09606 Chen Li-Qing et al. A novel class of sugar transporters. Supplementary Figure 1. Confocal imaging of FRET sensor localization in HEK293T cells. The subcellular localization of the FRET sensors was not affected by coexpression of SWEET1 in HEK293T cells. (a) Cytosolic FRET sensor FLIPglu600µΔ13V in cells that do not coexpress SWEET1. (b) Cytosolic FRET sensor FLIPglu600µΔ13V in cells that coexpress SWEET1. (c) ER-targeted FRET sensor FLIPglu600µΔ13V ER in cells that do not coexpress SWEET1. (d) ER-targeted FRET sensor FLIPglu600µΔ13V ER in cells that coexpress SWEET1. WWW NATURE.COM/NATURE 1
2 Supplementary Figure 2. Glucose uptake in HEK293T cells mediated by SWEET1- GFP fusions. (a) Plasma membrane localization of the SWEET1-GFP fusion protein in HEK293T cells by confocal microscopy. (b) SWEET1-GFP mediates glucose uptake in HEK293T cells. (c) SWEET1-GFP mediates glucose efflux into the ER of HEK293T cells. (d) Intracellular localization of the GFP-SWEET1 fusion in HEK293T cells. (e) GFP-SWEET1 did not mediate measurable glucose uptake in HEK293T cells. (f) HEK293T cells expressing only the cytosolic FRET sensor did not show significant glucose uptake. (g) HEK293T cells expressing only the ER-targeted FRET sensor did not show significant glucose efflux into the ER. WWW NATURE.COM/NATURE 2
3 Supplementary Figure 3. Mutant forms of SWEET1 do not induce or mediate glucose-dependent changes in FRET sensor responses and do not complement the yeast glucose uptake mutant EBY4000. (a) Negative control: HEK293T cell expressing only the FRET sensor FLIPglu600µ 13V. (b) Positive control: HEK293T cell coexpressing FLIPglu600µ 13V and SWEET1. (c) HEK293T cell coexpressing FLIPglu600µ 13V and the truncated SWEET1 with a premature stop codon at leucine 198 (L198*). (d) HEK293T cell coexpressing FLIPglu600µ 13V and the truncated SWEET1 (deleted after leucine 197) fused to GFP (SWEET1Δ198-GFP). (e) Localization of SWEET1-GFP in yeast cells. (f) Localization of the truncated SWEET1Δ198-GFP in yeast cells. (g, h) Functional complementation of the glucose uptake deficiency of EBY4000 by SWEET1 and SWEET1-GFP, but not by SWEET1Δ198-GFP. WWW NATURE.COM/NATURE 3
4 Supplementary Figure 4. SWEET1-mediated glucose transport is independent of GLUT activity in HEK293T cells. (a) Inhibition of GLUT1 activity by 20 µm cytochalasin B (Cyt) analyzed using the FLIPglu600µΔ13V sensor co-transfected with GLUT1. (b) Insensitivity of SWEET1 activity to 20 µm cytochalasin B analyzed using the FLIPglu600µΔ13V sensor co-transfected with SWEET1. (c) Expression level of SLC2 (GLUT) and SLC5 (SGLT) glucose transporter genes in HepG2 cells, HEK293T cells, and HEK293T cells coexpressing FLIPglu600µ 13V with or without SWEET1. WWW NATURE.COM/NATURE 4
5 Supplementary Figure 5. Biochemical properties of SWEET1. (a) Complementation of glucose uptake-deficiency of EBY4000 is at acidic and neutral ph. (b) ph optimum for SWEET1 glucose transport activity. Radiotracer uptake was measured at different ph. The ph optimum for uptake is about ph 8.5. Data are mean± S.D, n=3. WWW NATURE.COM/NATURE 5
6 Supplementary Figure 6. Substrate specificity of SWEET1 from Arabidopsis. SWEET1 partially complements the mannose, fructose and galactose uptake in the yeast mutant EBY4000. OsSWEET11 and HsSWEET1/RAG1AP1 do not show detectable complementation of the growth deficiency. WWW NATURE.COM/NATURE 6
7 Supplementary Figure 7. Tissue specific expression of SWEET1 and SWEET8 genes in Arabidopsis. The analysis is based on microarray studies from WWW NATURE.COM/NATURE 7
8 Supplementary Figure 8. Phylogenetic tree for SWEETs from different species. (a) Phylogenetic tree of the SWEET superfamily (PFAM PFO3083). Distances were calculated from a multiple sequence alignment (ClustalW) using the neighbor-joining method. The tree displays bootstrap values (percentage of 1000). SWEET genes fall into four clades. All sequences were obtained from NCBI ( or the Aramemnon database ( The family members were color-coded indicating the species: At, Arabidopsis thaliana (light green); Os, Oryza sativa (blue); Mt, Medicago trunculata (cyan); Chlamydomonas reinhardtii (dark green), Physcomitrella patens (orange). (b) Pairwise comparison of amino acid identity of 17 Arabidopsis SWEETs, OsSWEET11 and OsSWEET14. a WWW NATURE.COM/NATURE 8
9 Supplementary Figure 9. Prediction of transmembrane helices in SWEETs from different species. The TMHMM2.0 program ( was used to predict transmembrane helices in (a) SWEET1, (b) SWEET12, (c) SWEET13 from Arabidopsis, (d) CeSWEET1 from C. elegans, (e) HsSWEET1/ RAG1AP1 from Human, and (f) CiSWEET1/CiRGA from Ciona intestinalis. OsSWEET11 and OsSWEET14 also contain seven predicted TMHs (data not shown). WWW NATURE.COM/NATURE 9
10 Supplementary Figure 10. Functional analysis of SWEET8 in heterologous systems. (a) Cells coexpressing the cytosolic FRET glucose sensor FLIPglu600µΔ13V and SWEET8 accumulated glucose in the cytosol as evidenced by a negative cytosolic FRET ratio change in HEK293T cells 1. (b) Cells efflux glucose from the cytosol into the ER when coexpressing the ER FRET glucose sensor FLIPglu-600µ 13V ER and SWEET8 in HEK293T cells Data points are mean ± SD (n>10). (c) Relative uptake rate of SWEET1, SWEET8 and vector control in the yeast glucose transport-deficient mutant EBY4000 (2 min; 10 mm D-glucose; 0.1µCi [ 14 C]-D-glucose). Values are normalized to SWEET1 (100%). Data are mean ± S.D, n=3. (d) Confocal imaging of SWEET8 localization in leaves using stably transformed Arabidopsis plants (T1 generation). WWW NATURE.COM/NATURE 10
11 Supplementary Figure 11. Arabidopsis SWEET transporters function in glucose uptake in yeast. The five Arabidopsis SWEETs (SWEET1, 4, 5, 7, and 8) function as glucose transporters when expressed in the yeast mutant EBY4000. Note that activity of all Arabidopsis SWEETs that function in EBY4000 can be detected in HEK293T cells, except SWEET13. The other 11 Arabidopsis SWEETs did not complement the yeast mutant to a significant extent (data not shown). WWW NATURE.COM/NATURE 11
12 Supplementary Figure 12. Arabidopsis SWEET transporters function in glucose uptake in HEK293T cells. Five Arabidopsis SWEETs (SWEET1, 4, 5, 7, and 13) function as glucose transporters when expressed in HEK293T cells. Note that SWEET13 did not show significant complementation of the EBY4000 yeast sugar transport mutant. HEK293T cells coexpressed the glucose FRET sensor FLIPglu600µΔ13V with or without a respective SWEET gene. For details on method see Figure 1. In several experiments, SWEET12 showed low glucose uptake activity and the other 10 Arabidopsis SWEETs did not show significant FRET responses relative to the negative control (data not shown). WWW NATURE.COM/NATURE 12
13 Supplementary Figure 13. PthXo1 drives TAL effector-specific host gene induction. (a) Promoter fragments used for transient assays extended eighteen bases upstream of the predicted PthXo1 promoter binding site (underlined) to the start codon of OsSWEET11/Os8N3. Triangle indicates site of insertion/deletion (sequence of insertion is not shown) in the OsSWEET11/Os8N3 promoter of the IRBB13 allele of the xa13 locus. Altered bases in the mutant version of the promoter Os8N3pM1 (OsWEET11pM1) are in lower case letters. Each fragment was fused to the uida gene for β-glucuronidase (GUS) at the ATG codon. (b) Agrobacterium-mediated transient GUS assays of the promoter constructs (from Supplementary Fig. 13a) in N. benthamiana. Each construct was codelivered with 35S pthxo1 (left half of the leaf shown) or empty binary vector (right half). Sites 1 and 7 denote the sites where Os8N3pBB13 (OsSWEET11) was tested; sites 2 and 8, Os8N3pδ1; sites 3 and 9, Os8N3pδ2; sites 4 and 10, Os8N3pM1; and sites 5 and 11, Os8N3pWT. Site 6 was inoculated with 35S pthxo1 alone. (c) GUS activity in leaves co-expressing 35S pthxo1 (Supplementary Fig. 13b). Each combination represents the average of three inoculations and is expressed as µm 4-methylumbelliferyl β-dglucuronide (MUG) min -1 µg protein -1. I-bars indicate one standard deviation. The label Os8N3pWt/AvrXa7 indicates the use of a combination of the wild type OsSWEET11/Os8N3 promoter and 35S-avrXa7 in place of 35S-pthXo1. Vector indicates combination of Os8N3pWt promoter and the CaMV 35S expression. Vector indicates combination of Os8N3pWt promoter and the CaMV 35S expression vector with a TAL effector gene. Assays were performed 48 h after inoculation. Each disk was excised from independent plants and assayed as described 2. WWW NATURE.COM/NATURE 13
14 Supplementary Figure 14. OsSWEET14-mediated [ 14 C]-glucose uptake into Xenopus oocytes. Time-dependent glucose uptake mediated by OsSWEET14 in oocytes (control OsSWEET-F203* mutant; mean ± SD, n 7). WWW NATURE.COM/NATURE 14
15 Supplementary Figure 15. Functionality of GFP fusions of the C. elegans CeSWEET1 transporter. Both N- and C-terminal GFP fusions of CeSWEET1 were functional in cellular glucose uptake in HEK293T cells. WWW NATURE.COM/NATURE 15
16 Supplementary Figure 16. Control experiment testing for HsSWEET1- and CeSWEET1-induced leakiness of oocytes. Efflux measurements were performed with oocytes injected with [ 14 C]-sucrose. Sucrose efflux was unaffected by expression of HsSWEET1 or CeSWEET1. WWW NATURE.COM/NATURE 16
17 doi: /nature09606 Supplementary Figure 17. Immunolocalization of HsSWEET1/RAG1AP1 in the Golgi of HEK293T cells. Immunolocalization in fixed HEK293T cells was performed with a primary HsSWEET1 antiserum (Abcam AB5271; red channel). The Golgi was localized with an anti-human Golgin97 antiserum (Invitrogen A21270, green channel). The nuclei were stained with Hoechst (blue channel). The overlay is derived from all three channels with the transmission channel. Fluorescence imaging was performed on a Leica SP5 confocal microscope. Shown are z-sections. Secondary antiserum alone served as control. Localization was performed either on un-transfected HEK293T cells or HEK293T cells transfected with HsSWEET1 in pcdna3/v5-dest. WWW NATURE.COM/NATURE 17
18 doi: /nature09606 Supplementary Figure 18. Immunolocalization of HsSWEET1/RAG1AP1 variants/mutants in the Golgi of HEK293T cells. Endogenous HsSWEET1 or overexpressed HsSWEET1 were immunolocalized as in Supplementary Fig. 17. The dileucine motif mutant HsSWEET1m (Y216A, L218A, L219A) and the splice variants HsSWEET1-2 (Open Biosystems Clone Id ) were localized using the same approach. Mutation of the potential di-leucine motif at the C-terminus did not lead to increased plasma membrane localization or increased transporter activity. WWW NATURE.COM/NATURE 18
19 Supplementary Figure 19. Localization of HsSWEET1/RAG1AP1-GFP fusions in HEK293T cells. Top panel: GFP-HsSWEET1 fusion (green channel); bottom panel: HsSWEET-GFP fusion (green channel). The Golgi was localized with an anti-human Golgin97 antiserum (Invitrogen A21270, red channel). The nuclei were stained with Hoechst (blue channel). The overlay is derived from all three channels. Fluorescence imaging was performed on a Leica SP5 confocal microscope. Shown are z- sections. HsSWEET1-GFP fusions localize to the ER and Golgi in HEK293T cells. WWW NATURE.COM/NATURE 19
20 Supplementary Figure 20. Expression of HsSWEET1 in human cell lines and tissues. Microarray data suggest ubiquitous expression throughout human tissues and cell lines, with highest expression in the oviduct, the epididymis, and the intestine. Metaprofile analysis was performed using the Anatomy function in Geneinvestigator 3 WWW NATURE.COM/NATURE 20
21 Supplementary Figure 21. Expression of the mouse homolog MmSWEET1 in mammary glands. Expression was induced in the mammary gland during lactation as shown by microarray studies 4. WWW NATURE.COM/NATURE 21
22 Supplementary Figure 22. Cellular glucose efflux model. Glucose is taken up into cell either via the Na + -coupled glucose cotransporters of the SGLT family or via GLUT glucose uniporters. Cytosolic glucose is phosphorylated by hexokinase or directly exported from the cell, either via a GLUT uniporter of via a vesicular pathway as suggested by Hosokawa et al (2002) 1. Golgi-localized SWEETs may function in uptake of glucose into the Golgi as part of a vesicular efflux route. Similarly, glycogenolysisderived glucose can enter the ER, where is dephosphorylated by a membrane-bound phosphatase. Subsequently glucose is exported back into the cytosol, potentially by GLUT uniporters that transit through the ER on the way to the plasma membrane (Takanaga & Frommer 2010) 5. Subsequently, cytosolic glucose can be exported by either of the two potential efflux routes. The model may be relevant for glucose efflux from intestinal, liver or epididymis cells. WWW NATURE.COM/NATURE 22
23 Supplemental Tables Supplementary Table 1. qpcr primers used for amplification of Arabidopsis thaliana SWEET genes (SWEET names and corresponding gene ID). name gene ID primers length SWEET1 AT1G21460 SWEET1F CTTCTCCACTCTCCATCATGAGATT 232 SWEET1R CATCTGCAGATTTCTCTCCTTTGT SWEET2 AT3G14770 SWEET2F AACAGAGAGTTTAAGACAGAGAGAAG 145 SWEET2R ATCCTCCTAAACGTTGGCATTGGT SWEET3 AT5G53190 SWEET3F CCAACTTTTCCCTAATCTTTGTTCTTC 133 SWEET3R AACACCCTTGAAAATGTTACTATTGGA SWEET4 AT3G28007 SWEET4F CCATCATGAGTAAGGTGATCAAGA 128 SWEET4R CAAAATGAAAAGGTCGAACTTAATAAGT G SWEET5 AT5G62850 SWEET5F TGACCCTTATATTTTGATTCCAAATGGT 151 SWEET5R GCCAAGTTCGATTCCAGCATTC SWEET6 AT1G66770 SWEET6F GACTCGGTTACGTTGGTGAAGT 96 SWEET6R CAAACGCCGCTAACTCTTTTGTTTAA SWEET7 AT4G10850 SWEET7F GACCCATTCATGGCTATACCAAAT 240 SWEET7R ATCCCATAATCCGAAGTTTAATAACACT SWEET8 AT5G40260 SWEET8F TTGCTCTCTTCTTCATCAATCTCTCT 150 SWEET8R AGATCCTCCAGAAAGTCTTCGCT SWEET9 AT2G39060 SWEET9F GCAAGAGAAAGAGAGAAAAGTGAAGA 124 SWEET9R CCCATAAAACGTTGGCACTGGT SWEET10 AT5G50790 SWEET10F TAGAGGAAGAGAGAGGGAGAGAGT 201 SWEET10R ATATACGAACGAACGTCGGTATTG SWEET11 AT3G48740 SWEET11F TCCTTCTCCTAACAACTTATATACCATG 131 SWEET11R SWEET12 AT5G23660 SWEET12F SWEET12R TCCTATAGAACGTTGGCACAGGA AAAGCTGATATCTTTCTTACTACTTCGA A 204 CTTACAAATCCTATAGAACGTTGGCAC SWEET13 AT5G50800 SWEET13F CTTCTACGTTGCCCTTCCAAATG 309 SWEET13R CTTTGTTTCTGGACATCCTTGTTGA SWEET14 AT4G25010 SWEET14F ACTTCTACGTTGCGCTTCCAAATA 334 SWEET14R CAGTTCAACATTAAAGTCAATCACTAAT TC WWW NATURE.COM/NATURE 23
24 SWEET15 AT5G13170 SWEET15F CAATGACATATGCATAGCGATTCCAA 153 SWEET15R GGACTCATCACGACAATACTCTTAAG SWEET16 AT3G16690 SWEET16F GAGATGCAAACTCGCGTTCTAGT 114 SWEET16R GCACACTTCTCGTCGTCACA SWEET17 AT4G15920 SWEET17F AGTGACAACAAAGAGCGTGAAATAC 211 SWEET17R ACTTAAACCGTTGCTTAAACCAACC ACTIN8F CACTTTCCAGCAGATGTGGATC 58 ACTIN8R AATGCCTGGACCTGCTTCAT Supplementary Table 2. Naming of SWEET genes from Oryza sativa corresponding to gene ID OsSWEET1a OsSWEET1b OsSWEET2a OsSWEET2b OsSWEET3a OsSWEET3b OsSWEET4 OsSWEET5 OsSWEET6a OsSWEET6b OsSWEET7a OsSWEET7b OsSWEET7c OsSWEET7d OsSWEET7e OsSWEET11 / Os8N3 OsSWEET12 OsSWEET13 OsSWEET14/Os11N3 OsSWEET15 OsSWEET16 Os01g65880 Os05g35140 Os01g36070 Os01g50460 Os05g12320 Os01g12130 Os02g19820 Os05g51090 Os01g42110 Os01g42090 Os09g08030 Os09g08440 Os12g07860 Os09g08490 Os09g08270 Os08g42350 Os03g22590 Os12g29220 Os11g31190 Os02g30910 Os03g22200 WWW NATURE.COM/NATURE 24
25 Supplementary Table 3. qpcr primers used for analysis immune-precipitation of Os8N3/TAL effector complexes. Primer name Os8N3-5 -F Os8N3-5 -R Os8N3-3 -F Os8N3-3 -R Os11N3-5 -F Os11N35 -R Primer sequence CAGAGTGAAAAAGAAATATCAAGC GGTGTTAATCAGTGAGAAGG GGAGAGTGCTGTATGGCGA GTCTGCCCTGTTCAAACACA GTCAGCAGCTGGTCATGTGT GTTTGGTGGGAGGAGATCA WWW NATURE.COM/NATURE 25
26 Supplementary References References 1. Takanaga, H. & Frommer, W. B. Facilitative plasma membrane transporters function during ER transit. FASEB J 24, , (2010). 2. Gallagher, S. R. in The GUS reporter system as a tool to study plant gene expression (eds T. Martin et al.) (San Diego, 1992). 3. Hruz, T. et al. Genevestigator v3: a reference expression database for the metaanalysis of transcriptomes. Adv. Bioinformatics 2008, , (2008). 4. Anderson, S. M., Rudolph, M. C., McManaman, J. L. & Neville, M. C. Key stages in mammary gland development. Secretory activation in the mammary gland: it's not just about milk protein synthesis! Breast Cancer Res 9, 204, (2007). 5. Hosokawa, M. & Thorens, B. Glucose release from GLUT2-null hepatocytes: characterization of a major and a minor pathway. Am. J. Physiol. Enocriol. Metab., E794-E801, (2002). WWW NATURE.COM/NATURE 26
Supplemental Data. Gao et al. (2012). Plant Cell /tpc
 Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011) /tpc
 Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
ACTIVE TRANSPORT AND GLUCOSE TRANSPORT. (Chapter 14 and 15, pp and pp )
 ACTIVE TRANSPORT AND GLUCOSE TRANSPORT (Chapter 14 and 15, pp 140-143 and pp 146-151) Overview Active transport is the movement of molecules across a cell membrane in the direction against their concentration
ACTIVE TRANSPORT AND GLUCOSE TRANSPORT (Chapter 14 and 15, pp 140-143 and pp 146-151) Overview Active transport is the movement of molecules across a cell membrane in the direction against their concentration
Supplemental Data. Chen and Thelen (2010). Plant Cell /tpc
 Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type
 A B 2 3 3 2 1 1 Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type (A) and d27 (B) seedlings at the four
A B 2 3 3 2 1 1 Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type (A) and d27 (B) seedlings at the four
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
Name: TF: Section Time: LS1a ICE 5. Practice ICE Version B
 Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Nature Genetics: doi: /ng Supplementary Figure 1. ssp mutant phenotypes in a functional SP background.
 Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Importance of Protein sorting. A clue from plastid development
 Importance of Protein sorting Cell organization depend on sorting proteins to their right destination. Cell functions depend on sorting proteins to their right destination. Examples: A. Energy production
Importance of Protein sorting Cell organization depend on sorting proteins to their right destination. Cell functions depend on sorting proteins to their right destination. Examples: A. Energy production
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Supplementary Figure 1. Biochemical and sequence alignment analyses the
 Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
SUPPLEMENTARY INFORMATION
 doi:1.138/nature111 cytosol Model: PILS function in cellular auxin homeostasis ER nucleus IAA degradation? sequestration? conjugation? storage? signalling? PILS IAA ER cytosol Supplemental Figure 1 Model
doi:1.138/nature111 cytosol Model: PILS function in cellular auxin homeostasis ER nucleus IAA degradation? sequestration? conjugation? storage? signalling? PILS IAA ER cytosol Supplemental Figure 1 Model
SUPPLEMENTARY INFORMATION
 GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
Supplementary Figure 1.
 Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Transport of glucose across epithelial cells: a. Gluc/Na cotransport; b. Gluc transporter Alberts
 Figure 7 a. Secondary transporters make up the largest subfamily of transport proteins. TAGI 2000. Nature 408, 796 1. Na+- or H+-coupled cotransporters - Secondary active transport 2/7-02 Energy released
Figure 7 a. Secondary transporters make up the largest subfamily of transport proteins. TAGI 2000. Nature 408, 796 1. Na+- or H+-coupled cotransporters - Secondary active transport 2/7-02 Energy released
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
Supplemental Table 1. Primers used for cloning and PCR amplification in this study
 Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
Supplemental Data. Perea-Resa et al. Plant Cell. (2012) /tpc
 Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Heather Currinn, Benjamin Guscott, Zita Balklava, Alice Rothnie and Thomas Wassmer*
 Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Cellular Neuroanatomy I The Prototypical Neuron: Soma. Reading: BCP Chapter 2
 Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang Chen, Qiang Dong, Jiangqi Wen, Kirankumar S. Mysore and Jian Zhao
 New Phytologist Supporting Information Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bhlh transcription factor MtTT8 Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang
New Phytologist Supporting Information Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bhlh transcription factor MtTT8 Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises
 Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Cytokinin. Fig Cytokinin needed for growth of shoot apical meristem. F Cytokinin stimulates chloroplast development in the dark
 Cytokinin Abundant in young, dividing cells Shoot apical meristem Root apical meristem Synthesized in root tip, developing embryos, young leaves, fruits Transported passively via xylem into shoots from
Cytokinin Abundant in young, dividing cells Shoot apical meristem Root apical meristem Synthesized in root tip, developing embryos, young leaves, fruits Transported passively via xylem into shoots from
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing
 Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
7.06 Cell Biology EXAM #3 KEY
 7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
Supporting Information
 Supporting Information Mullins et al. 10.1073/pnas.0906781106 SI Text Detection of Calcium Binding by 45 Ca 2 Overlay. The 45 CaCl 2 (1 mci, 37 MBq) was obtained from NEN. The general method of 45 Ca 2
Supporting Information Mullins et al. 10.1073/pnas.0906781106 SI Text Detection of Calcium Binding by 45 Ca 2 Overlay. The 45 CaCl 2 (1 mci, 37 MBq) was obtained from NEN. The general method of 45 Ca 2
. Supplementary Information
 . Supplementary Information Supplementary Figure S1. Mature embryo sac observations. Supplementary Figure S2. STT observations. Supplementary Figure S3. Comparison of the PTB1 cdna with that of the mutant.
. Supplementary Information Supplementary Figure S1. Mature embryo sac observations. Supplementary Figure S2. STT observations. Supplementary Figure S3. Comparison of the PTB1 cdna with that of the mutant.
Membranes 2: Transportation
 Membranes 2: Transportation Steven E. Massey, Ph.D. Associate Professor Bioinformatics Department of Biology University of Puerto Rico Río Piedras Office & Lab: NCN#343B Tel: 787-764-0000 ext. 7798 E-mail:
Membranes 2: Transportation Steven E. Massey, Ph.D. Associate Professor Bioinformatics Department of Biology University of Puerto Rico Río Piedras Office & Lab: NCN#343B Tel: 787-764-0000 ext. 7798 E-mail:
Waithe et al Supplementary Figures
 Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Reconstructing Mitochondrial Evolution?? Morphological Diversity. Mitochondrial Diversity??? What is your definition of a mitochondrion??
 Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial
Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial
Supplementary Material. An analysis of the role of the ShSUT1 sucrose transporter in sugarcane using RNAi suppression
 10.1071/FP17073_AC CSIRO 2017 Supplementary Material: Functional Plant Biology, 2017, 44(8), 795 808. Supplementary Material An analysis of the role of the ShSUT1 sucrose transporter in sugarcane using
10.1071/FP17073_AC CSIRO 2017 Supplementary Material: Functional Plant Biology, 2017, 44(8), 795 808. Supplementary Material An analysis of the role of the ShSUT1 sucrose transporter in sugarcane using
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells
 Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a
 Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
Nature Genetics: doi: /ng Supplementary Figure 1. Icm/Dot secretion system region I in 41 Legionella species.
 Supplementary Figure 1 Icm/Dot secretion system region I in 41 Legionella species. Homologs of the effector-coding gene lega15 (orange) were found within Icm/Dot region I in 13 Legionella species. In four
Supplementary Figure 1 Icm/Dot secretion system region I in 41 Legionella species. Homologs of the effector-coding gene lega15 (orange) were found within Icm/Dot region I in 13 Legionella species. In four
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Supplemental Data. Yang et al. (2012). Plant Cell /tpc
 Supplemental Figure 1. Mature flowers of P. heterotricha. (A) An inflorescence of P. heterotricha showing the front view of a zygomorphic flower characterized by two small dorsal petals and only two fertile
Supplemental Figure 1. Mature flowers of P. heterotricha. (A) An inflorescence of P. heterotricha showing the front view of a zygomorphic flower characterized by two small dorsal petals and only two fertile
7.06 Problem Set
 7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
targets. clustering show that different complex pathway
 Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Biochemistry. Biochemistry 9/20/ Bio-Energetics. 4.2) Transport of ions and small molecules across cell membranes
 9/20/15 Biochemistry Biochemistry 4. Bio-Energetics 4.2) Transport of ions and small molecules across cell membranes Aquaporin, the water channel, consists of four identical transmembrane polypeptides
9/20/15 Biochemistry Biochemistry 4. Bio-Energetics 4.2) Transport of ions and small molecules across cell membranes Aquaporin, the water channel, consists of four identical transmembrane polypeptides
Membrane transport 1. Summary
 Membrane transport 1. Summary A. Simple diffusion 1) Diffusion by electrochemical gradient no energy required 2) No channel or carrier (or transporter protein) is needed B. Passive transport (= Facilitated
Membrane transport 1. Summary A. Simple diffusion 1) Diffusion by electrochemical gradient no energy required 2) No channel or carrier (or transporter protein) is needed B. Passive transport (= Facilitated
Biochemistry. Biochemistry 7/11/ Bio-Energetics. 4.2) Transport of ions and small molecules across cell membranes
 Biochemistry Biochemistry 4. Bio-Energetics 4.2) Transport of ions and small molecules across cell membranes Aquaporin, the water channel, consists of four identical transmembrane polypeptides Key Energy
Biochemistry Biochemistry 4. Bio-Energetics 4.2) Transport of ions and small molecules across cell membranes Aquaporin, the water channel, consists of four identical transmembrane polypeptides Key Energy
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/452/ra106/dc1 Supplementary Materials for Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots Hyo-Jun Lee, Jun-Ho
www.sciencesignaling.org/cgi/content/full/9/452/ra106/dc1 Supplementary Materials for Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots Hyo-Jun Lee, Jun-Ho
Last time: Obtaining information from a cloned gene
 Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Supplemental Materials Molecular Biology of the Cell
 Supplemental Materials Molecular iology of the Cell Figure S1 Krüger et al. Arabidopsis Plasmodium H. sapiens* 1) Xenopus* 1) Drosophila C.elegans S.cerevisae S.pombe L.major T.cruzi T.brucei DCP5 CITH
Supplemental Materials Molecular iology of the Cell Figure S1 Krüger et al. Arabidopsis Plasmodium H. sapiens* 1) Xenopus* 1) Drosophila C.elegans S.cerevisae S.pombe L.major T.cruzi T.brucei DCP5 CITH
Protein Sorting, Intracellular Trafficking, and Vesicular Transport
 Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Supplemental Data. Perrella et al. (2013). Plant Cell /tpc
 Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Reception The target cell s detection of a signal coming from outside the cell May Occur by: Direct connect Through signal molecules
 Why Do Cells Communicate? Regulation Cells need to control cellular processes In multicellular organism, cells signaling pathways coordinate the activities within individual cells that support the function
Why Do Cells Communicate? Regulation Cells need to control cellular processes In multicellular organism, cells signaling pathways coordinate the activities within individual cells that support the function
Actions of auxin. Hormones: communicating with chemicals History: Discovery of a growth substance (hormone- auxin)
 Hormones: communicating with chemicals History- discovery of plant hormone. Auxin Concepts of hormones Auxin levels are regulated by synthesis/degradation, transport, compartmentation, conjugation. Polar
Hormones: communicating with chemicals History- discovery of plant hormone. Auxin Concepts of hormones Auxin levels are regulated by synthesis/degradation, transport, compartmentation, conjugation. Polar
Genomics and bioinformatics summary. Finding genes -- computer searches
 Genomics and bioinformatics summary 1. Gene finding: computer searches, cdnas, ESTs, 2. Microarrays 3. Use BLAST to find homologous sequences 4. Multiple sequence alignments (MSAs) 5. Trees quantify sequence
Genomics and bioinformatics summary 1. Gene finding: computer searches, cdnas, ESTs, 2. Microarrays 3. Use BLAST to find homologous sequences 4. Multiple sequence alignments (MSAs) 5. Trees quantify sequence
Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA
 Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Molecular Cell Biology 5068 In Class Exam 2 November 8, 2016
 Molecular Cell Biology 5068 In Class Exam 2 November 8, 2016 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Molecular Cell Biology 5068 In Class Exam 2 November 8, 2016 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL
 GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
JMJ14-HA. Col. Col. jmj14-1. jmj14-1 JMJ14ΔFYR-HA. Methylene Blue. Methylene Blue
 Fig. S1 JMJ14 JMJ14 JMJ14ΔFYR Methylene Blue Col jmj14-1 JMJ14-HA Methylene Blue Col jmj14-1 JMJ14ΔFYR-HA Fig. S1. The expression level of JMJ14 and truncated JMJ14 with FYR (FYRN + FYRC) domain deletion
Fig. S1 JMJ14 JMJ14 JMJ14ΔFYR Methylene Blue Col jmj14-1 JMJ14-HA Methylene Blue Col jmj14-1 JMJ14ΔFYR-HA Fig. S1. The expression level of JMJ14 and truncated JMJ14 with FYR (FYRN + FYRC) domain deletion
Figure 18.1 Blue-light stimulated phototropism Blue light Inhibits seedling hypocotyl elongation
 Blue Light and Photomorphogenesis Q: Figure 18.3 Blue light responses - phototropsim of growing Corn Coleoptile 1. How do we know plants respond to blue light? 2. What are the functions of multiple BL
Blue Light and Photomorphogenesis Q: Figure 18.3 Blue light responses - phototropsim of growing Corn Coleoptile 1. How do we know plants respond to blue light? 2. What are the functions of multiple BL
Cellular Transport. 1. Transport to and across the membrane 1a. Transport of small molecules and ions 1b. Transport of proteins
 Transport Processes Cellular Transport 1. Transport to and across the membrane 1a. Transport of small molecules and ions 1b. Transport of proteins 2. Vesicular transport 3. Transport through the nuclear
Transport Processes Cellular Transport 1. Transport to and across the membrane 1a. Transport of small molecules and ions 1b. Transport of proteins 2. Vesicular transport 3. Transport through the nuclear
cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work
 SUPPLEMENTARY INFORMATION Table S1. List of strains Strains Genotype Source B. subtilis PY79 Prototrophic derivative of B. subtilis 168 (60) RL1070 spovid::kan (12) HS176 cotz phs2 (cotz-gfp spc) (22)
SUPPLEMENTARY INFORMATION Table S1. List of strains Strains Genotype Source B. subtilis PY79 Prototrophic derivative of B. subtilis 168 (60) RL1070 spovid::kan (12) HS176 cotz phs2 (cotz-gfp spc) (22)
BIOINFORMATICS LAB AP BIOLOGY
 BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
Figure S1: Extracellular nicotinic acid, but not tryptophan, is sufficient to maintain
 SUPPLEMENTAL INFORMATION Supplemental Figure Legends Figure S1: Extracellular nicotinic acid, but not tryptophan, is sufficient to maintain mitochondrial NAD +. A) Extracellular tryptophan, even at 5 µm,
SUPPLEMENTAL INFORMATION Supplemental Figure Legends Figure S1: Extracellular nicotinic acid, but not tryptophan, is sufficient to maintain mitochondrial NAD +. A) Extracellular tryptophan, even at 5 µm,
Signal Transduction. Dr. Chaidir, Apt
 Signal Transduction Dr. Chaidir, Apt Background Complex unicellular organisms existed on Earth for approximately 2.5 billion years before the first multicellular organisms appeared.this long period for
Signal Transduction Dr. Chaidir, Apt Background Complex unicellular organisms existed on Earth for approximately 2.5 billion years before the first multicellular organisms appeared.this long period for
The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively as positive regulators of salt stress tolerance in rice
 Lou et al. BMC Plant Biology (2018) 18:203 https://doi.org/10.1186/s12870-018-1408-0 RESEARCH ARTICLE Open Access The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively
Lou et al. BMC Plant Biology (2018) 18:203 https://doi.org/10.1186/s12870-018-1408-0 RESEARCH ARTICLE Open Access The sucrose non-fermenting-1-related protein kinases SAPK1 and SAPK2 function collaboratively
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
** * * * Col-0 cau1 CAU1. Actin2 CAS. Actin2. Supplemental Figure 1. CAU1 affects calcium accumulation.
 Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
CHAPTER 3. Cell Structure and Genetic Control. Chapter 3 Outline
 CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
Supplemental Data. Yamamoto et al. (2016). Plant Cell /tpc WT + HCO 3. slac1-4/δnc #5 + HCO 3 WT + ABA. slac1-4/δnc #5 + ABA
 I (pa) I (pa) A WT + HCO 3 B 2 V (mv) 15 1 5 5 2 slac14/δnc #5 + HCO 3 4 6 WT (n = 6) 8 WT + HCO 3 (n = 7) slac14/δnc #5 (n = 6) slac14/δnc #5 + HCO 3 (n = 1) C WT + ABA D 2 V (mv) 15 1 5 5 2 slac14/δnc
I (pa) I (pa) A WT + HCO 3 B 2 V (mv) 15 1 5 5 2 slac14/δnc #5 + HCO 3 4 6 WT (n = 6) 8 WT + HCO 3 (n = 7) slac14/δnc #5 (n = 6) slac14/δnc #5 + HCO 3 (n = 1) C WT + ABA D 2 V (mv) 15 1 5 5 2 slac14/δnc
Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar Compartment tothelyticvacuoleinvivo W
 The Plant Cell, Vol. 18, 2275 2293, September 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar
The Plant Cell, Vol. 18, 2275 2293, September 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%%
 !"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
Supplemental Data. Hou et al. (2016). Plant Cell /tpc
 Supplemental Data. Hou et al. (216). Plant Cell 1.115/tpc.16.295 A Distance to 1 st nt of start codon Distance to 1 st nt of stop codon B Normalized PARE abundance 8 14 nt 17 nt Frame1 Arabidopsis inflorescence
Supplemental Data. Hou et al. (216). Plant Cell 1.115/tpc.16.295 A Distance to 1 st nt of start codon Distance to 1 st nt of stop codon B Normalized PARE abundance 8 14 nt 17 nt Frame1 Arabidopsis inflorescence
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Reading Assignments. A. Genes and the Synthesis of Polypeptides. Lecture Series 7 From DNA to Protein: Genotype to Phenotype
 Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Functional role of oligomerization for bacterial and plant SWEET sugar transporter family
 Functional role of oligomerization for bacterial and plant SWEET sugar transporter family Yuan Hu Xuan a,1,yibinghu a,b,1, Li-Qing Chen a, Davide Sosso a, Daniel C. Ducat c, Bi-Huei Hou a, and Wolf B.
Functional role of oligomerization for bacterial and plant SWEET sugar transporter family Yuan Hu Xuan a,1,yibinghu a,b,1, Li-Qing Chen a, Davide Sosso a, Daniel C. Ducat c, Bi-Huei Hou a, and Wolf B.
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed
 Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport
 Ph.D. thesis Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport Zsigmond Laura Supervisor: Dr. Szabados László Arabidopsis Molecular Genetic Group Institute of Plant
Ph.D. thesis Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport Zsigmond Laura Supervisor: Dr. Szabados László Arabidopsis Molecular Genetic Group Institute of Plant
GFP GAL bp 3964 bp
 Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
CELL BIOLOGY. Which of the following cell structures does not have membranes? A. Ribosomes B. Mitochondria C. Chloroplasts D.
 1 CELL BIOLOGY PROKARYOTIC and EUKARYOTIC SP/1. SP/2. SP/4. Plant and animal cells both have A. ribosomes, cell walls and mitochondria. B. Golgi apparatus, chromosomes and mitochondria. C. Golgi apparatus,
1 CELL BIOLOGY PROKARYOTIC and EUKARYOTIC SP/1. SP/2. SP/4. Plant and animal cells both have A. ribosomes, cell walls and mitochondria. B. Golgi apparatus, chromosomes and mitochondria. C. Golgi apparatus,
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
Genome-wide multilevel spatial interactome model of rice
 Sino-German Workshop on Multiscale Spatial Computational Systems Biology, Beijing, Oct 8-12, 2015 Genome-wide multilevel spatial interactome model of rice Ming CHEN ( 陈铭 ) mchen@zju.edu.cn College of Life
Sino-German Workshop on Multiscale Spatial Computational Systems Biology, Beijing, Oct 8-12, 2015 Genome-wide multilevel spatial interactome model of rice Ming CHEN ( 陈铭 ) mchen@zju.edu.cn College of Life
5- Semaphorin-Plexin-Neuropilin
 5- Semaphorin-Plexin-Neuropilin 1 SEMAPHORINS-PLEXINS-NEUROPILINS ligands receptors co-receptors semaphorins and their receptors are known signals for: -axon guidance -cell migration -morphogenesis -immune
5- Semaphorin-Plexin-Neuropilin 1 SEMAPHORINS-PLEXINS-NEUROPILINS ligands receptors co-receptors semaphorins and their receptors are known signals for: -axon guidance -cell migration -morphogenesis -immune
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family
 Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development
 Supplementary Information: Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development Krishanpal Karmodiya 1, Krishanpal Anamika 1,2, Vijaykumar
Supplementary Information: Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development Krishanpal Karmodiya 1, Krishanpal Anamika 1,2, Vijaykumar
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
Supplemental material
 Supplemental material Table 1- Segregation analysis of sgt1a sgt1b double mutant plants Parental genotype No. of lines Normal seeds Phenotype Abnormal seeds Ratio of abnormal seeds (%) WT 3 171 2 1.16
Supplemental material Table 1- Segregation analysis of sgt1a sgt1b double mutant plants Parental genotype No. of lines Normal seeds Phenotype Abnormal seeds Ratio of abnormal seeds (%) WT 3 171 2 1.16
GFP WxTP-GFP WxTP-GFP. stromules. DsRed WxTP-DsRed WxTP-DsRed. stromules
 Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Cryo-EM data collection, refinement and validation statistics
 1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
Regulation and signaling. Overview. Control of gene expression. Cells need to regulate the amounts of different proteins they express, depending on
 Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
Proportion of Arabidopsis genes in different functions. 5% of genome consists of transporters (>1000 genes)
 Membranes are crucial to the life of the cell 1. PM define boundary of cell, and maintains difference between cytosol and external environment. Inside, organelles are distinct from one another. 2. Membrane
Membranes are crucial to the life of the cell 1. PM define boundary of cell, and maintains difference between cytosol and external environment. Inside, organelles are distinct from one another. 2. Membrane
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
Predominant Golgi Residency of the Plant K/HDEL Receptor Is Essential for its Function in Mediating ER Retention
 Plant Cell Advance Publication. Published on August 2, 2018, doi:10.1105/tpc.18.00426 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41
Plant Cell Advance Publication. Published on August 2, 2018, doi:10.1105/tpc.18.00426 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature17991 Supplementary Discussion Structural comparison with E. coli EmrE The DMT superfamily includes a wide variety of transporters with 4-10 TM segments 1. Since the subfamilies of the
doi:10.1038/nature17991 Supplementary Discussion Structural comparison with E. coli EmrE The DMT superfamily includes a wide variety of transporters with 4-10 TM segments 1. Since the subfamilies of the
