Supporting Information
|
|
|
- Milton Johns
- 5 years ago
- Views:
Transcription
1 Supporting Information Mullins et al /pnas SI Text Detection of Calcium Binding by 45 Ca 2 Overlay. The 45 CaCl 2 (1 mci, 37 MBq) was obtained from NEN. The general method of 45 Ca 2 overlay has been described in detail elsewhere (1). Our specific conditions were as follows: Purified GST-fusion peptides (2 and 10 g per lane) and CaM (0.5 g per lane) were transferred to a PVDF membrane after 13% SDS PAGE. The blot was extensively washed for 3 10 min with binding buffer (60 mm KCl, 5 mm MgCl 2, and 10 mm imidazole-hcl, ph 6.8) to remove electrode buffer. The overlay assay was performed by incubating the blot with 8.8 M 45 CaCl 2 (10 mci/mg; NEN) in 20 ml of binding buffer at 25 C for 1 h, followed by 3 2-min washes in dh 2 O and a 2 min wash in 50% ethanol. After air drying, the blot was exposed to Hyperfilm (GE Healthcare). After autoradiography the blot was stained in 40% methanol, 10% acetic acid, and 0.1% Coomassie brillant blue R250, and destained in 20% methanol and 7% acetic acid. Primers Used in Generating Constructs. Cherry-STIM Forward, CTGGCATCCACTAACTGGTGGCTGC; reverse, gcagc- CACCAGTTAGTGGATGCCAG. GFP-STIM Forward, CGCCCCAACCCTGCTTAGT- TCATCATGACTGAC; reverse, GTCAGTCATGATGAAC- TAAGCAGGGTTGGGGCG. GFP-STIM Forward, CCCTTGTCCATGCAGTGAC- CTAGCCTGCAGAGC; reverse GCTCTGCAGGCTAGGT- CACTGCATGGACAAGGG. GFP-STIM Forward, CATGGCCTGGGATCTCAGTAG- GATTTGACCCATTCC; reverse, GGAATGGGTCAAATC- CTACTGAGATCCCAGGCCATG. GFP-STIM Forward, GGCAGCCACCGGCTGATCTA- GGGGGTCCACCCAGGG; reverse, CCCTGGGTGGAC- CCCCTAGATCAGCCGGTGGCTGCC. GFP-STIM Forward, GACACACCATCTCCAGTTTAG- GACAGCCGAGCCCTG; reverse, CAGGGCTCGGCTGTC- CTAAACTGGAGATGGTGTGTC. Cherry-STIM Forward, GACTCCAGCCCAGGCTGAAA- GAAGTTTCCTCTC; reverse, GAGAGGAAACTTCTT- TCAGCCTGGGCTGGAGTC. YFP-STIM Forward, AACCGTTACTCCAAGGAGCAC; reverse, GAATTCAGTGGATGCCAGGGTTGTTG. YFP-STIM Forward, GGATCCATGTATGCTCCAGAG- GCCCTTC; reverse, TCAAGCAGGGTTGGGGCGTG- TACTGCC. YFP-STIM Forward, GGATCCATGTATGCTCCAGAG- GCCCTTC; reverse, TCAGATCAGCCGGTGGCTGCCATT- GGAAGT. YFP-STIM Forward, GGATCCATGTATGCTCCAGAG- GCCCTTC; reverse, TCAAACTGGAGATGGTGTGTCTGG. myc-stim1 478,481 DD 3 GG. Forward, ATGACTGACGACGT- GGGTGACATGGGTGAGGAGATTGTGTCTCCC; reverse, GGGAGACACAATCTCCTCACCCATGT- CACCCACGTCGTCAGTCAT. myc-stim1 4A2G (from myc-stim1 478,481 DD 3 GG). Forward, CACT- TCATCATGACTGCCGCCGTGGGTGACAT- GGGTGCGGCGATTGTGTCTCCC; reverse, GGGAGACA- CAATCGCCGCACCCATGTCACCCACGGCGGCAGTCA- TGATGAAGTG. GFP-STIM1 475/476 DD 3 AA. Forward, CACTTCATCATGACT- GCAGcCGTGGATGACATGGAT; reverse, ATCCATGT- CATCCACGGCTGCAGTCATGATGAAGTG. GFP-STIM1 482,483 EE 3 AA. Forward, GACGTGGATGACATG- GATGCGGCGATTGTGTCTCCCTTG; reverse, CAAGG- GAGACACAATCGCCGCATCCATGTCATCCACGTC (in myc- and GFP-myc-). Forward, CTTAAAGC- CTCCAGCCGGTGATCGGCTCTGCTCTCC; reverse, GGAGAGCAGAGCCGATCACCGGCTGGAGGCTTTAAG. A73E (in myc- and GFP-myc ). Forward, GAG- CACTCCATGCAGGAGCTGTCCTGGCGCAAG; reverse, CTTGCGCCAGGACAGCTCCTGCATGGAGTGCTC. W76E (in myc- and GFP-myc ). Forward, ATG- CAGGCGCTGTCCGAGCGCAAGCTCTACTTG; reverse, CAAGTAGAGCTTGCGCTCGGACAGCGCCTGCAT. Y80E (in myc- and GFP-myc ). Forward, TCCTG- GCGCAAGCTCGAGTTGAGCCGCGCCAAG; reverse, CT- TGGCGCGGCTCAACTCGAGCTTGCGCCAGGA. W76A (in myc- and GFP-myc ). Forward, ATG- CAGGCGCTGTCCGCGCGCAAGCTCTACTTG; reverse CAAGTAGAGCTTGCGCGCGGACAGCGCCTGCAT. W76S (in myc- and GFP-myc ). Forward, ATG- CAGGCGCTGTCCAGTCGCAAGCTCTACTTG; reverse, CAAGTAGAGCTTGCGACTGGACAGCGCCTGCAT. Y80A (in myc- and GFP-myc ). Forward, TCCTG- GCGCAAGCTCGCGTTGAGCCGCGCCAAG; reverse, CT- TGGCGCGGCTCAACGCGAGCTTGCGCCAGGA. Y80S (in myc- and GFP-myc ). Forward, TCCTG- GCGCAAGCTCAGTTTGAGCCGCGCCAAG; reverse, CT- TGGCGCGGCTCAAACTGAGCTTGCGCCAGGA. Calmodulin (CaM). Forward, GCCACCATGGCTGATCAGCT- GACTGAAGAACAG; reverse, GGATCCTCATTTTG- CAGTCATCATCTGTAC. CaM-T27C. Forward, AAGGATGGAGATGGCTGTATCAC- CACCAAGGAG; reverse, CTCCTTGGTGGTGATACAGC- CATCTCCATCCTT. STIM Forward, GGATCCATGCACTTCATCATGACT- GACGACGTG; reverse, TCACTGCATGGACAAGGGAGA- CAC. 1. Maruyama K, Mikawa T, Ebashi S (1984) Detection of calcium binding proteins by 45 Ca autoradiography on nitrocellulose membrane after sodium dodecyl sulfate gel electrophoresis. J Biochem 95: of10
2 Fig. S1. Inactivation kinetics of heterologously expressed Ca 2 release-activated Ca 2 (CRAC) channels. Currents were elicited in 20 mm Ca 2 o in HEK293 cells cotransfected with myc- GFP-STIM1 as described in Fig. 1A, and fitted by the relation I I o A 1 e t/ 1 A 2 e t/ 2 (see Materials and Methods). (A) Representative fit (red dashed line) to current elicited by a step to 120 mv (black). (B) Fast time constants and amplitudes of inactivation plotted against test potential. Each point shows the mean SEM of five cells. (C) Slow time constants and amplitudes of inactivation plotted against test potential. Each point shows the mean SEM of five cells. 2of10
3 WT (1-685) CAD ( ) A2G 475, 476 DD->AA 478, 481 DD->GG 482, 483 EE->AA A73E W76A Fig. S2. Comparison of current densities for STIM1 and constructs. HEK293 cells were cotransfected with myc- and the indicated STIM1-derived constructs, or GFP-STIM1 and the indicated -derived constructs, and I CRAC was allowed to reach a maximal level of induction. For each cell, peak current was measured 3 ms after stepping to 100 mv in 20 mm Ca 2 o and normalized to the cell capacitance. Each bar shows the mean SEM of 5 10 cells for each construct. Although significant cell cell variability was seen (as expected for transient transfection experiments), average current densities were similar for most constructs tested. W76E W76S Y80A Y80E Y80S 3of10
4 Fig. S3. Effects of extending the CRAC activation domain (CAD) peptide on Ca 2 -dependent inactivation (CDI). Currents were elicited in 20 mm Ca 2 o in HEK293 cells cotransfected with myc- and the indicated CAD-derived construct. All currents were constitutively active, with large currents present at the first pulse after break-in. (A D) Representative currents during pulses to the indicated voltages for YFP-STIM (A), YFP-STIM (B), YFP-STIM (C), and YFP-STIM (D). (E) Extent of inactivation, quantified as the residual current remaining at the end of the test pulse (R 195 ms I 195 ms /I 3ms ), plotted against test potential. Each point represents the mean SEM of five to six cells. Data from Fig. 1E are included in the summary graph for comparison purposes. Extending CAD to amino acids 560 or 630 only partially rescues inactivation relative to STIM or STIM of10
5 ID STIM A B CBD Orai Fig. S4. Alignment of ID STIM and CBD Orai sequences. (A) Full-length STIM1 amino acid sequences from vertebrate and invertebrate species were aligned with CLC Sequence Viewer version 5 using default settings. The region corresponding to amino acids of human STIM1 is shown, and the ID STIM (amino acids ) is annotated (bar). The degree of conservation for each position is shown at the bottom. Although a high degree of sequence conservation is seen in this region among vertebrates, greater variability is seen among invertebrates, leading to the introduction of several gaps. (B) Full-length amino acid sequences from vertebrate and invertebrate species were aligned with CLC Sequence Viewer version 5 using default settings. The region corresponding to amino acids of human is shown. The degree of conservation for each position is shown at the bottom. The residues A73, W76, and Y80, all key for inactivation (Fig. 4), are highly conserved in vertebrates, but variable among invertebrates. 5of10
6 Fig. S5. The 482/483 EE 3 AA mutation in STIM1 increases the calcium sensitivity of I CRAC inactivation. Currents were recorded in HEK293 cells cotransfected with myc- and the indicated STIM1-derived construct, after induction of I CRAC reached a maximum. (A) Representative currents evoked by WT GFP-STIM1 recorded in 2 (Left)or20(Right)mMCa 2 o during pulses to the indicated voltages. Traces in 20 mm Ca 2 o are reproduced from Fig. 1A.(B) Representative currents evoked by STIM1 482/483 EE 3 AA recorded in 2 (Left)or20(Right)mMCa 2 o. Note the presence of strong inactivation even in low [Ca 2 ] o, unlike the WT traces in A. Traces in 20 mm Ca 2 o are reproduced from Fig. 2E. (C) Extent of inactivation for WT and 482/483 EE 3 AA plotted against test potential. Each point represents the mean SEM of five cells (WT, 20 mm Ca 2 o) or 7 cells (482/483 EE 3 AA, 20 mm Ca 2 o).for2mmca 2 o, n 1 cell for both constructs. 6of10
7 GST-ID STIM peptide ( ) CaM GST WT ID STIM DD -> AA DD -> GG EE -> AA 4A2G kda ug Ca 2+ overlay Coomassie blue Fig. S6. Ca 2 binding to the ID STIM domain detected by 45 Ca 2 overlay. Purified CaM (0.5 g) and GST (2 and 10 g) were loaded for controls. WT and mutant GST-STIM1 peptides corresponding to amino acids were tested. (Upper) CaM, WT GST-ID STIM , and GST-fused peptides in which 2 aa were mutated were able to bind to Ca 2 to variable degrees. No Ca 2 binding was detected for GST or the 4A2G mutant. (Lower) Coomassie staining shows loaded proteins. 7of10
8 YFP- NT II-III CT Ca 2+ EGTA Ca 2+ EGTA Ca 2+ EGTA Input ΙΒ:α-GFP Pull-down ΙΒ: α-gfp 34 Fig. S7. CaM Sepharose pull-down confirming Ca 2 -dependent binding of CaM to the N terminus. Lysates from HEK 293T cells expressing YFP-tagged N-terminal, II-III loop, and C-terminal fragments of were applied to CaM-Sepharose in the presence of 2 mm Ca 2 or 4 mm EGTA. 8of10
9 Fig. S8. Acceleration of inactivation kinetics by mutant STIM1 and constructs. (A D) The biexponential function I I o A t/ 1 1e A 2 e t/ 2 was fitted to I CRAC evoked by hyperpolarizing pulses in 20 mm Ca 2 o (representative records are shown in Figs. 1A, 2E, and 4 H and I). Each point represents the mean SEM. (A) Fast time constants of inactivation plotted against test potential. (B) Slow time constants of inactivation plotted against test potential. (C) Fast amplitudes of inactivation plotted against test potential. (D) Slow amplitudes of inactivation plotted against test potential. 9of10
10 Fig. S9. Absence of inactivation for mutant STIM1 constructs transfected at 4:1 STIM1/ mass ratio. Currents were elicited in 20 mm Ca 2 o in HEK293 cells cotransfected with the indicated STIM1 construct in 4-fold excess over myc-. (A and B) Representative currents during pulses to the indicated voltages for GFP-STIM (A) and myc-stim1 4A2G (B). (C) Extent of inactivation, quantified as the residual current remaining at the end of the test pulse (R 195 ms I 195 ms /I 3ms ), plotted against test potential. Each point represents the mean SEM of five cells. 10 of 10
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises
 Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
A single lysine in the N-terminal region of store-operated channels is critical for STIM1-mediated gating
 Published Online: 29 November, 2010 Supp Info: http://doi.org/10.1085/jgp.201010484 Downloaded from jgp.rupress.org on December 25, 2018 A r t i c l e A single lysine in the N-terminal region of store-operated
Published Online: 29 November, 2010 Supp Info: http://doi.org/10.1085/jgp.201010484 Downloaded from jgp.rupress.org on December 25, 2018 A r t i c l e A single lysine in the N-terminal region of store-operated
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a
 Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplemental Data. Gao et al. (2012). Plant Cell /tpc
 Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Supporting information for
 Supporting information for Rewiring multi-domain protein switches: transforming a fluorescent Zn 2+ -sensor into a light-responsive Zn 2+ binding protein Stijn J.A. Aper and Maarten Merkx Laboratory of
Supporting information for Rewiring multi-domain protein switches: transforming a fluorescent Zn 2+ -sensor into a light-responsive Zn 2+ binding protein Stijn J.A. Aper and Maarten Merkx Laboratory of
Waithe et al Supplementary Figures
 Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing
 Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
Differential polyamine sensitivity in inwardly rectifying Kir2 potassium channels
 J Physiol 571.2 (2006) pp 287 302 287 Differential polyamine sensitivity in inwardly rectifying Kir2 potassium channels Brian K. Panama and Anatoli N. Lopatin University of Michigan, Department of Molecular
J Physiol 571.2 (2006) pp 287 302 287 Differential polyamine sensitivity in inwardly rectifying Kir2 potassium channels Brian K. Panama and Anatoli N. Lopatin University of Michigan, Department of Molecular
In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE
 In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE (Lamond Lab / April 2008)! Perform all the pipetting steps in a laminar flow hood. We routinely do our digestions in our TC room hoods.
In-gel digestion of immunoprecipitated proteins separated by SDS-PAGE (Lamond Lab / April 2008)! Perform all the pipetting steps in a laminar flow hood. We routinely do our digestions in our TC room hoods.
Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants. GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
 SUPPLEMENTAL DATA Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants Sense primer (5 to 3 ) Anti-sense primer (5 to 3 ) GAL1 mutants GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
SUPPLEMENTAL DATA Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants Sense primer (5 to 3 ) Anti-sense primer (5 to 3 ) GAL1 mutants GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
Supplementary Information
 Supplementary Information An engineered protein antagonist of K-Ras/B-Raf interaction Monique J. Kauke, 1,2 Michael W. Traxlmayr 1,2, Jillian A. Parker 3, Jonathan D. Kiefer 4, Ryan Knihtila 3, John McGee
Supplementary Information An engineered protein antagonist of K-Ras/B-Raf interaction Monique J. Kauke, 1,2 Michael W. Traxlmayr 1,2, Jillian A. Parker 3, Jonathan D. Kiefer 4, Ryan Knihtila 3, John McGee
Exam I Answer Key: Summer 2006, Semester C
 1. Which of the following tripeptides would migrate most rapidly towards the negative electrode if electrophoresis is carried out at ph 3.0? a. gly-gly-gly b. glu-glu-asp c. lys-glu-lys d. val-asn-lys
1. Which of the following tripeptides would migrate most rapidly towards the negative electrode if electrophoresis is carried out at ph 3.0? a. gly-gly-gly b. glu-glu-asp c. lys-glu-lys d. val-asn-lys
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
A Novel Carbonic Anhydrase II Binding Site Regulates NHE1 Activity
 2414 Biochemistry 2006, 45, 2414-2424 A Novel Carbonic Anhydrase II Binding Site Regulates NHE1 Activity Xiuju Li, Yongsheng Liu, Bernardo V. Alvarez, Joseph R. Casey, and Larry Fliegel* Department of
2414 Biochemistry 2006, 45, 2414-2424 A Novel Carbonic Anhydrase II Binding Site Regulates NHE1 Activity Xiuju Li, Yongsheng Liu, Bernardo V. Alvarez, Joseph R. Casey, and Larry Fliegel* Department of
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplementary Figure 1. Biochemical and sequence alignment analyses the
 Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
HHS Public Access Author manuscript Nat Cell Biol. Author manuscript; available in PMC 2009 September 01.
 SOAR and the polybasic STIM1 domains gate and regulate the Orai channels Joseph P. Yuan 1,3, Weizhong Zeng 1,3, Michael R. Dorwart 1, Young-Jin Choi 1, Paul F. Worley 2,4, and Shmuel Muallem 1,4 1 Department
SOAR and the polybasic STIM1 domains gate and regulate the Orai channels Joseph P. Yuan 1,3, Weizhong Zeng 1,3, Michael R. Dorwart 1, Young-Jin Choi 1, Paul F. Worley 2,4, and Shmuel Muallem 1,4 1 Department
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Quantitation of the binding of pro53 peptide to sorla Vps10p measured by the AP reporter assay. The graph shows tracings of the typical chromogenic AP reaction observed with AP-pro53
Supplementary Figure 1 Quantitation of the binding of pro53 peptide to sorla Vps10p measured by the AP reporter assay. The graph shows tracings of the typical chromogenic AP reaction observed with AP-pro53
TNFα 18hr. Control. CHX 18hr. TNFα+ CHX 18hr. TNFα: 18 18hr (KDa) PARP. Cleaved. Cleaved. Cleaved. Caspase3. Pellino3 shrna. Control shrna.
 Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Supplementary Information to
 Supplementary Information to Wiesner et al.: A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases Supplementary Material and Methods Cloning and Mutagenesis Site-directed
Supplementary Information to Wiesner et al.: A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases Supplementary Material and Methods Cloning and Mutagenesis Site-directed
Voltage-clamp and Hodgkin-Huxley models
 Voltage-clamp and Hodgkin-Huxley models Read: Hille, Chapters 2-5 (best Koch, Chapters 6, 8, 9 See also Hodgkin and Huxley, J. Physiol. 117:500-544 (1952. (the source Clay, J. Neurophysiol. 80:903-913
Voltage-clamp and Hodgkin-Huxley models Read: Hille, Chapters 2-5 (best Koch, Chapters 6, 8, 9 See also Hodgkin and Huxley, J. Physiol. 117:500-544 (1952. (the source Clay, J. Neurophysiol. 80:903-913
Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition
 Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
SUPPLEMENTARY INFORMATION
 5 N 4 8 20 22 24 2 28 4 8 20 22 24 2 28 a b 0 9 8 7 H c (kda) 95 0 57 4 28 2 5.5 Precipitate before NMR expt. Supernatant before NMR expt. Precipitate after hrs NMR expt. Supernatant after hrs NMR expt.
5 N 4 8 20 22 24 2 28 4 8 20 22 24 2 28 a b 0 9 8 7 H c (kda) 95 0 57 4 28 2 5.5 Precipitate before NMR expt. Supernatant before NMR expt. Precipitate after hrs NMR expt. Supernatant after hrs NMR expt.
Supplementary Figure S1. MscS orientation in spheroplasts and liposomes (a) Current-voltage relationship for wild-type MscS expressed in E.
 a b c Supplementary Figure S1. MscS orientation in spheroplasts and liposomes (a) Current-voltage relationship for wild-type MscS expressed in E. coli giant spheroplasts (MJF465) and reconstituted into
a b c Supplementary Figure S1. MscS orientation in spheroplasts and liposomes (a) Current-voltage relationship for wild-type MscS expressed in E. coli giant spheroplasts (MJF465) and reconstituted into
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis):
 SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis): Aim: SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is one of the common methods used in the molecular biology
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis): Aim: SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is one of the common methods used in the molecular biology
Rho1 binding site PtdIns(4,5)P2 binding site Both sites
 localization Mutation site DMSO LatB WT F77A I115A I131A K134A Rho1 binding site PtdIns(4,5)P2 binding site Both sites E186A E199A N201A R84A-E186A-E199A L131A-K136A-E186A L131A-E186A-E199A K136A-E186A-E199A
localization Mutation site DMSO LatB WT F77A I115A I131A K134A Rho1 binding site PtdIns(4,5)P2 binding site Both sites E186A E199A N201A R84A-E186A-E199A L131A-K136A-E186A L131A-E186A-E199A K136A-E186A-E199A
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL Figure S1. Mitochondrial morphology in Fis1-null, Mff-null and Fis1/Mff-null MEF cells. (A) Western blotting of lysates from Fis1-null, Mff-null and Fis1/Mff-null cells. Lysates were
SUPPLEMENTAL MATERIAL Figure S1. Mitochondrial morphology in Fis1-null, Mff-null and Fis1/Mff-null MEF cells. (A) Western blotting of lysates from Fis1-null, Mff-null and Fis1/Mff-null cells. Lysates were
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Voltage-clamp and Hodgkin-Huxley models
 Voltage-clamp and Hodgkin-Huxley models Read: Hille, Chapters 2-5 (best) Koch, Chapters 6, 8, 9 See also Clay, J. Neurophysiol. 80:903-913 (1998) (for a recent version of the HH squid axon model) Rothman
Voltage-clamp and Hodgkin-Huxley models Read: Hille, Chapters 2-5 (best) Koch, Chapters 6, 8, 9 See also Clay, J. Neurophysiol. 80:903-913 (1998) (for a recent version of the HH squid axon model) Rothman
Acta Crystallographica Section D
 Supporting information Acta Crystallographica Section D Volume 70 (2014) Supporting information for article: Structural characterization of the virulence factor Nuclease A from Streptococcus agalactiae
Supporting information Acta Crystallographica Section D Volume 70 (2014) Supporting information for article: Structural characterization of the virulence factor Nuclease A from Streptococcus agalactiae
downstream (0.8 kb) homologous sequences to the genomic locus of DIC. A DIC mutant strain (ro- 6
 A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
Transfer of ion binding site from ether-à-go-go to Shaker: Mg 2+ binds to resting state to modulate channel opening
 A r t i c l e Transfer of ion binding site from ether-à-go-go to Shaker: Mg 2+ binds to resting state to modulate channel opening Meng-chin A. Lin, 1 Jeff Abramson, 1 and Diane M. Papazian 1,2,3 1 Department
A r t i c l e Transfer of ion binding site from ether-à-go-go to Shaker: Mg 2+ binds to resting state to modulate channel opening Meng-chin A. Lin, 1 Jeff Abramson, 1 and Diane M. Papazian 1,2,3 1 Department
Protein separation and characterization
 Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: tech@aladdin-e.com Email EU: eutech@aladdin-e.com Email Asia Pacific: cntech@aladdin-e.com Protein separation
Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: tech@aladdin-e.com Email EU: eutech@aladdin-e.com Email Asia Pacific: cntech@aladdin-e.com Protein separation
Protocol for 2D-E. Protein Extraction
 Protocol for 2D-E Protein Extraction Reagent 1 inside the ReadyPrep TM Sequential Extraction kit (in powder form) 50ml of deionized water is used to dissolve all the Reagent 1. The solution is known as
Protocol for 2D-E Protein Extraction Reagent 1 inside the ReadyPrep TM Sequential Extraction kit (in powder form) 50ml of deionized water is used to dissolve all the Reagent 1. The solution is known as
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A)
 Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Tex 25mer ssrna Binding Stoichiometry
 Figure S. Determination of Tex:2nt ssrna binding stoichiometry using fluorescence polarization. Fluorescein labeled RNA was held at a constant concentration 2-fold above the K d. Tex protein was titrated
Figure S. Determination of Tex:2nt ssrna binding stoichiometry using fluorescence polarization. Fluorescein labeled RNA was held at a constant concentration 2-fold above the K d. Tex protein was titrated
Ca 2+ readdition protocol
 11 th Calcium signaling course May 2-13, 2011 Md. Shahidul Islam, M.D., Ph.D. Associate Professor Department t of fclinical i l Sciences and deducation, Södersjukhuset Karolinska Institutet Forskningscentrum,
11 th Calcium signaling course May 2-13, 2011 Md. Shahidul Islam, M.D., Ph.D. Associate Professor Department t of fclinical i l Sciences and deducation, Södersjukhuset Karolinska Institutet Forskningscentrum,
Supplementary Figure 1.
 Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
SDS-polyacrylamide gel electrophoresis
 SDS-polyacrylamide gel electrophoresis Protein Isolation and Purification Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells,
SDS-polyacrylamide gel electrophoresis Protein Isolation and Purification Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells,
National de la Recherche Scientifique and Université Paris Descartes, Paris, France.
 FAST-RESPONSE CALMODULIN-BASED FLUORESCENT INDICATORS REVEAL RAPID INTRACELLULAR CALCIUM DYNAMICS Nordine Helassa a, Xiao-hua Zhang b, Ianina Conte a,c, John Scaringi b, Elric Esposito d, Jonathan Bradley
FAST-RESPONSE CALMODULIN-BASED FLUORESCENT INDICATORS REVEAL RAPID INTRACELLULAR CALCIUM DYNAMICS Nordine Helassa a, Xiao-hua Zhang b, Ianina Conte a,c, John Scaringi b, Elric Esposito d, Jonathan Bradley
7.06 Cell Biology EXAM #3 KEY
 7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1.
 Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Supplementary Figure 1
 Supplementary Figure 1 Activation of P2X2 receptor channels in symmetric Na + solutions only modestly alters the intracellular ion concentration. a,b) ATP (30 µm) activated P2X2 receptor channel currents
Supplementary Figure 1 Activation of P2X2 receptor channels in symmetric Na + solutions only modestly alters the intracellular ion concentration. a,b) ATP (30 µm) activated P2X2 receptor channel currents
Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc
 OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
SUPPLEMENTARY INFORMATION
 Supplementary Table 1: Amplitudes of three current levels. Level 0 (pa) Level 1 (pa) Level 2 (pa) TrkA- TrkH WT 200 K 0.01 ± 0.01 9.5 ± 0.01 18.7 ± 0.03 200 Na * 0.001 ± 0.01 3.9 ± 0.01 12.5 ± 0.03 200
Supplementary Table 1: Amplitudes of three current levels. Level 0 (pa) Level 1 (pa) Level 2 (pa) TrkA- TrkH WT 200 K 0.01 ± 0.01 9.5 ± 0.01 18.7 ± 0.03 200 Na * 0.001 ± 0.01 3.9 ± 0.01 12.5 ± 0.03 200
(A) Effect of I-EPI-002, EPI-002 or enzalutamide on dexamethasone (DEX, 10 nm)
 Supplemental Figure Legends Supplemental Figure 1. (A) Effect of I-EPI-002, EPI-002 or enzalutamide on dexamethasone (DEX, 10 nm) induced GR transcriptional activity in LNCaP cells that were transiently
Supplemental Figure Legends Supplemental Figure 1. (A) Effect of I-EPI-002, EPI-002 or enzalutamide on dexamethasone (DEX, 10 nm) induced GR transcriptional activity in LNCaP cells that were transiently
Visual pigments. Neuroscience, Biochemistry Dr. Mamoun Ahram Third year, 2019
 Visual pigments Neuroscience, Biochemistry Dr. Mamoun Ahram Third year, 2019 References Webvision: The Organization of the Retina and Visual System (http://www.ncbi.nlm.nih.gov/books/nbk11522/#a 127) The
Visual pigments Neuroscience, Biochemistry Dr. Mamoun Ahram Third year, 2019 References Webvision: The Organization of the Retina and Visual System (http://www.ncbi.nlm.nih.gov/books/nbk11522/#a 127) The
Activated MAP Kinase INSTRUCTION MANUAL. Catalog # Revision A.02. For In Vitro Use Only
 Activated MAP Kinase INSTRUCTION MANUAL Catalog #206110 Revision A.02 For In Vitro Use Only 206110-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this product. No other
Activated MAP Kinase INSTRUCTION MANUAL Catalog #206110 Revision A.02 For In Vitro Use Only 206110-12 LIMITED PRODUCT WARRANTY This warranty limits our liability to replacement of this product. No other
Supplementary Figure 1. Stability constants of metal monohydroxides. The log K values are summarized according to the atomic number of each element
 Supplementary Figure 1. Stability constants of metal monohydroxides. The log K values are summarized according to the atomic number of each element as determined in a previous study 1. The log K value
Supplementary Figure 1. Stability constants of metal monohydroxides. The log K values are summarized according to the atomic number of each element as determined in a previous study 1. The log K value
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
7. N-Terminal and Internal Amino Acid Sequence Analysis and Homology Search of Amino Acid Sequence
 Protocol 1. Sample Preparation 2. Two Dimensional Polyacrylamide Gel Electrophoresis 3. Image acquisition and Data Analysis 4. Cleveland Peptide Mapping 5. Western Blotting 6. Deblocking of Blotted Proteins
Protocol 1. Sample Preparation 2. Two Dimensional Polyacrylamide Gel Electrophoresis 3. Image acquisition and Data Analysis 4. Cleveland Peptide Mapping 5. Western Blotting 6. Deblocking of Blotted Proteins
The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression
 Traffic 2010; 11: 827 842 2010 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2010.01055.x The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression Danming Tang, Hebao Yuan and Yanzhuang
Traffic 2010; 11: 827 842 2010 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2010.01055.x The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression Danming Tang, Hebao Yuan and Yanzhuang
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Ainsliadimer A (1) inhibited I B phosphorylation in spleen extracts. The mice were received intravenous injection of DMSO, 1 (25 mg/kg) or BMS-345541 (25 mg/kg)
Supplementary Figures Supplementary Figure 1. Ainsliadimer A (1) inhibited I B phosphorylation in spleen extracts. The mice were received intravenous injection of DMSO, 1 (25 mg/kg) or BMS-345541 (25 mg/kg)
Supporting Information
 Supporting Information Tuning Supramolecular Structure and Functions of Peptide bola-amphiphile by Solvent Evaporation-Dissolution Anhe Wang,, Lingyun Cui,, Sisir Debnath, Qianqian Dong, Xuehai Yan, Xi
Supporting Information Tuning Supramolecular Structure and Functions of Peptide bola-amphiphile by Solvent Evaporation-Dissolution Anhe Wang,, Lingyun Cui,, Sisir Debnath, Qianqian Dong, Xuehai Yan, Xi
SUPPORTING INFORMATION FOR. SEquence-Enabled Reassembly of β-lactamase (SEER-LAC): a Sensitive Method for the Detection of Double-Stranded DNA
 SUPPORTING INFORMATION FOR SEquence-Enabled Reassembly of β-lactamase (SEER-LAC): a Sensitive Method for the Detection of Double-Stranded DNA Aik T. Ooi, Cliff I. Stains, Indraneel Ghosh *, David J. Segal
SUPPORTING INFORMATION FOR SEquence-Enabled Reassembly of β-lactamase (SEER-LAC): a Sensitive Method for the Detection of Double-Stranded DNA Aik T. Ooi, Cliff I. Stains, Indraneel Ghosh *, David J. Segal
Tellurite resistance protein/ethidium efflux transporter/ proflavin transporter. Putative inner membrane protein: function unknown
 Additional file 1. Table S1 and Figures S1-4 of Zhang et al. High-level production of membrane proteins in E. coli BL21(DE3) by omitting the inducer IPTG Table S1. Properties of the membrane proteins used
Additional file 1. Table S1 and Figures S1-4 of Zhang et al. High-level production of membrane proteins in E. coli BL21(DE3) by omitting the inducer IPTG Table S1. Properties of the membrane proteins used
STIM1 Clusters and Activates CRAC Channels via Direct Binding of a Cytosolic Domain to Orai1
 STIM1 Clusters and Activates CRAC Channels via Direct Binding of a Cytosolic Domain to Orai1 Chan Young Park, 1,7 Paul J. Hoover, 2,7 Franklin M. Mullins, 2,3 Priti Bachhawat, 2,4 Elizabeth D. Covington,
STIM1 Clusters and Activates CRAC Channels via Direct Binding of a Cytosolic Domain to Orai1 Chan Young Park, 1,7 Paul J. Hoover, 2,7 Franklin M. Mullins, 2,3 Priti Bachhawat, 2,4 Elizabeth D. Covington,
EST1 Homology Domain. 100 aa. hest1a / SMG6 PIN TPR TPR. Est1-like DBD? hest1b / SMG5. TPR-like TPR. a helical. hest1c / SMG7.
 hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
Copyright WILEY-VCH Verlag GmbH, D Weinheim, Supporting Information for Angew. Chem. Int. Ed. Z 18050
 Copyright WILEY-VCH Verlag GmbH, D-69451 Weinheim, 2001. Supporting Information for Angew. Chem. Int. Ed. Z 18050 Protein Affinity Labeling Mediated by Genetically Encoded Peptide Tags Frank Amini, Thomas
Copyright WILEY-VCH Verlag GmbH, D-69451 Weinheim, 2001. Supporting Information for Angew. Chem. Int. Ed. Z 18050 Protein Affinity Labeling Mediated by Genetically Encoded Peptide Tags Frank Amini, Thomas
The C Terminus of the L-Type Voltage-Gated Calcium Channel Ca V 1.2 Encodes a Transcription Factor
 The C Terminus of the L-Type Voltage-Gated Calcium Channel Ca V 1.2 Encodes a Transcription Factor Natalia Gomez-Ospina, 1 Fuminori Tsuruta, 1,3 Odmara Barreto-Chang, 1,3 Linda Hu, 2 and Ricardo Dolmetsch
The C Terminus of the L-Type Voltage-Gated Calcium Channel Ca V 1.2 Encodes a Transcription Factor Natalia Gomez-Ospina, 1 Fuminori Tsuruta, 1,3 Odmara Barreto-Chang, 1,3 Linda Hu, 2 and Ricardo Dolmetsch
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Molecular determinants of Ca 2 /calmodulindependent regulation of Ca v 2.1 channels
 Molecular determinants of Ca 2 /calmodulindependent regulation of Ca v 2.1 channels Amy Lee*, Hong Zhou, Todd Scheuer*, and William A. Catterall* *Department of Pharmacology, University of Washington School
Molecular determinants of Ca 2 /calmodulindependent regulation of Ca v 2.1 channels Amy Lee*, Hong Zhou, Todd Scheuer*, and William A. Catterall* *Department of Pharmacology, University of Washington School
Biochemistry 3100 Sample Problems Chemical and Physical Methods
 (1) Describe the function of the following compounds in chemical sequencing, synthesis and modification: (a) Dithiothreitol (c) t-butyloxycarbonyl chloride (b) Dicyclohexyl carbodiimide (d) Cyanogen Bromide
(1) Describe the function of the following compounds in chemical sequencing, synthesis and modification: (a) Dithiothreitol (c) t-butyloxycarbonyl chloride (b) Dicyclohexyl carbodiimide (d) Cyanogen Bromide
BA, BSc, and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes
Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes
T H E J O U R N A L O F C E L L B I O L O G Y
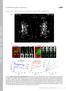 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Eisner et al., http://www.jcb.org/cgi/content/full/jcb.201312066/dc1 Figure S1. Mitochondrial continuity in adult skeletal muscle. (A)
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Eisner et al., http://www.jcb.org/cgi/content/full/jcb.201312066/dc1 Figure S1. Mitochondrial continuity in adult skeletal muscle. (A)
Signaling to the Nucleus by an L-type Calcium Channel- Calmodulin Complex Through the MAP Kinase Pathway
 Signaling to the Nucleus by an L-type Calcium Channel- Calmodulin Complex Through the MAP Kinase Pathway Ricardo E. Dolmetsch, Urvi Pajvani, Katherine Fife, James M. Spotts, Michael E. Greenberg Science
Signaling to the Nucleus by an L-type Calcium Channel- Calmodulin Complex Through the MAP Kinase Pathway Ricardo E. Dolmetsch, Urvi Pajvani, Katherine Fife, James M. Spotts, Michael E. Greenberg Science
Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea nd Semester
 Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea 2018. 2 nd Semester Absorbance Assay (280 nm) Considerations for use Absorbance assays are fast and
Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea 2018. 2 nd Semester Absorbance Assay (280 nm) Considerations for use Absorbance assays are fast and
depends on the translocation assembly module nanomachine
 ARTICLE NUMBER: 16064 DOI: 10.1038/NMICROBIOL.16.64 Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine Christopher Stubenrauch 1, Matthew J. Belousoff
ARTICLE NUMBER: 16064 DOI: 10.1038/NMICROBIOL.16.64 Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine Christopher Stubenrauch 1, Matthew J. Belousoff
Chemical Exchange and Ligand Binding
 Chemical Exchange and Ligand Binding NMR time scale Fast exchange for binding constants Slow exchange for tight binding Single vs. multiple binding mode Calcium binding process of calcium binding proteins
Chemical Exchange and Ligand Binding NMR time scale Fast exchange for binding constants Slow exchange for tight binding Single vs. multiple binding mode Calcium binding process of calcium binding proteins
Structure and RNA-binding properties. of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex
 Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Membrane Protein Channels
 Membrane Protein Channels Potassium ions queuing up in the potassium channel Pumps: 1000 s -1 Channels: 1000000 s -1 Pumps & Channels The lipid bilayer of biological membranes is intrinsically impermeable
Membrane Protein Channels Potassium ions queuing up in the potassium channel Pumps: 1000 s -1 Channels: 1000000 s -1 Pumps & Channels The lipid bilayer of biological membranes is intrinsically impermeable
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
Nature Medicine: doi: /nm.3776
 C terminal Hsp90 inhibitors restore glucocorticoid sensitivity and relieve a mouse allograft model of Cushing s disease Mathias Riebold, Christian Kozany, Lee Freiburger, Michael Sattler, Michael Buchfelder,
C terminal Hsp90 inhibitors restore glucocorticoid sensitivity and relieve a mouse allograft model of Cushing s disease Mathias Riebold, Christian Kozany, Lee Freiburger, Michael Sattler, Michael Buchfelder,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
Supplementary Information. Recognition of the pre-mirna structure by Drosophila Dicer-1
 Supplementary Information Recognition of the pre-mirna structure by Drosophila Dicer-1 Akihisa Tsutsumi 1,2, Tomoko Kawamata 1, Natsuko Izumi 1 Hervé Seitz 3,4 and Yukihide Tomari 1,2 Nature Structural
Supplementary Information Recognition of the pre-mirna structure by Drosophila Dicer-1 Akihisa Tsutsumi 1,2, Tomoko Kawamata 1, Natsuko Izumi 1 Hervé Seitz 3,4 and Yukihide Tomari 1,2 Nature Structural
Supporting Information
 Supporting Information Fera et al. 10.1073/pnas.1409954111 SI Methods Compliance. All work related to human subjects complied with protocols approved by the Duke University Health System Institutional
Supporting Information Fera et al. 10.1073/pnas.1409954111 SI Methods Compliance. All work related to human subjects complied with protocols approved by the Duke University Health System Institutional
Graded activation of CRAC channel by binding of different numbers of STIM1 to Orai1 subunits
 ORIGINAL ARTICLE Cell Research (2011) 21:305-315. 2011 IBCB, SIBS, CAS All rights reserved 1001-0602/11 $ 32.00 www.nature.com/cr npg Graded activation of CRAC channel by binding of different numbers of
ORIGINAL ARTICLE Cell Research (2011) 21:305-315. 2011 IBCB, SIBS, CAS All rights reserved 1001-0602/11 $ 32.00 www.nature.com/cr npg Graded activation of CRAC channel by binding of different numbers of
(Supplementary Information)
 (Supplementary Information) Peptidomimetic-based Multi-Domain Targeting Offers Critical Evaluation of Aβ Structure and Toxic Function Sunil Kumar 1*, Anja Henning-Knechtel 2, Mazin Magzoub 2, and Andrew
(Supplementary Information) Peptidomimetic-based Multi-Domain Targeting Offers Critical Evaluation of Aβ Structure and Toxic Function Sunil Kumar 1*, Anja Henning-Knechtel 2, Mazin Magzoub 2, and Andrew
Supporting Information for: Kinetic Mechanisms Governing Stable Ribonucleotide Incorporation in Individual DNA Polymerase Complexes
 Supporting Information for: Kinetic Mechanisms Governing Stable Ribonucleotide Incorporation in Individual DNA Polymerase Complexes Joseph M. Dahl, Hongyun Wang, José M. Lázaro, Margarita Salas, and Kate
Supporting Information for: Kinetic Mechanisms Governing Stable Ribonucleotide Incorporation in Individual DNA Polymerase Complexes Joseph M. Dahl, Hongyun Wang, José M. Lázaro, Margarita Salas, and Kate
Supplemental Table 1. Primers used for cloning and PCR amplification in this study
 Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
The Journal of Physiology
 J Physiol 593.3 (2015) pp 559 572 559 Anchoring protein AKAP79-mediated PKA phosphorylation of STIM1 determines selective activation of the ARC channel, a store-independent Orai channel Jill L. Thompson
J Physiol 593.3 (2015) pp 559 572 559 Anchoring protein AKAP79-mediated PKA phosphorylation of STIM1 determines selective activation of the ARC channel, a store-independent Orai channel Jill L. Thompson
Contribution of N- and C-terminal channel domains to Kv channel interacting proteins in a mammalian cell line
 J Physiol 568.2 (2005) pp 397 412 397 Contribution of N- and C-terminal channel domains to Kv channel interacting proteins in a mammalian cell line Britta Callsen, Dirk Isbrandt, Kathrin Sauter, L. Sven
J Physiol 568.2 (2005) pp 397 412 397 Contribution of N- and C-terminal channel domains to Kv channel interacting proteins in a mammalian cell line Britta Callsen, Dirk Isbrandt, Kathrin Sauter, L. Sven
Supplementary Information. The protease GtgE from Salmonella exclusively targets. inactive Rab GTPases
 Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Quantification of Protein Half-Lives in the Budding Yeast Proteome
 Supporting Methods Quantification of Protein Half-Lives in the Budding Yeast Proteome 1 Cell Growth and Cycloheximide Treatment Three parallel cultures (17 ml) of each TAP-tagged strain were grown in separate
Supporting Methods Quantification of Protein Half-Lives in the Budding Yeast Proteome 1 Cell Growth and Cycloheximide Treatment Three parallel cultures (17 ml) of each TAP-tagged strain were grown in separate
Supporting information
 Supporting information The L-rhamnose Antigen: a Promising Alternative to α-gal for Cancer Immunotherapies Wenlan Chen,, Li Gu,#, Wenpeng Zhang, Edwin Motari, Li Cai, Thomas J. Styslinger, and Peng George
Supporting information The L-rhamnose Antigen: a Promising Alternative to α-gal for Cancer Immunotherapies Wenlan Chen,, Li Gu,#, Wenpeng Zhang, Edwin Motari, Li Cai, Thomas J. Styslinger, and Peng George
targets. clustering show that different complex pathway
 Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
ydci GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC
 Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R)
 Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R) Shown in cyan and green are two adjacent tetramers from the crystallographic lattice of COP, forming the only unique inter-tetramer
Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R) Shown in cyan and green are two adjacent tetramers from the crystallographic lattice of COP, forming the only unique inter-tetramer
Full-length GlpG sequence was generated by PCR from E. coli genomic DNA. (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
 Supplementary Methods Protein expression and purification Full-length GlpG sequence was generated by PCR from E. coli genomic DNA (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
Supplementary Methods Protein expression and purification Full-length GlpG sequence was generated by PCR from E. coli genomic DNA (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
JMJ14-HA. Col. Col. jmj14-1. jmj14-1 JMJ14ΔFYR-HA. Methylene Blue. Methylene Blue
 Fig. S1 JMJ14 JMJ14 JMJ14ΔFYR Methylene Blue Col jmj14-1 JMJ14-HA Methylene Blue Col jmj14-1 JMJ14ΔFYR-HA Fig. S1. The expression level of JMJ14 and truncated JMJ14 with FYR (FYRN + FYRC) domain deletion
Fig. S1 JMJ14 JMJ14 JMJ14ΔFYR Methylene Blue Col jmj14-1 JMJ14-HA Methylene Blue Col jmj14-1 JMJ14ΔFYR-HA Fig. S1. The expression level of JMJ14 and truncated JMJ14 with FYR (FYRN + FYRC) domain deletion
Exquisite Sequence Selectivity with Small Conditional RNAs
 Supplementary Information Exquisite Sequence Selectivity with Small Conditional RNAs Jonathan B. Sternberg and Niles A. Pierce,, Division of Biology & Biological Engineering, Division of Engineering &
Supplementary Information Exquisite Sequence Selectivity with Small Conditional RNAs Jonathan B. Sternberg and Niles A. Pierce,, Division of Biology & Biological Engineering, Division of Engineering &
Transport of glucose across epithelial cells: a. Gluc/Na cotransport; b. Gluc transporter Alberts
 Figure 7 a. Secondary transporters make up the largest subfamily of transport proteins. TAGI 2000. Nature 408, 796 1. Na+- or H+-coupled cotransporters - Secondary active transport 2/7-02 Energy released
Figure 7 a. Secondary transporters make up the largest subfamily of transport proteins. TAGI 2000. Nature 408, 796 1. Na+- or H+-coupled cotransporters - Secondary active transport 2/7-02 Energy released
Basic Principles of Kinetics and Thermodynamics Arthur L. Haas, Ph.D. Department of Biochemistry and Molecular Biology
 Basic Principles of Kinetics and Thermodynamics Arthur L. Haas, Ph.D. Department of Biochemistry and Molecular Biology Review: Garrett and Grisham, Enzyme Kinetics (Chapt. 13), in Biochemistry, Third Edition,
Basic Principles of Kinetics and Thermodynamics Arthur L. Haas, Ph.D. Department of Biochemistry and Molecular Biology Review: Garrett and Grisham, Enzyme Kinetics (Chapt. 13), in Biochemistry, Third Edition,
Preparing Extension Products for Electrophoresis
 Preparing Extension Products for Electrophoresis Overview Preparation of extension products for electrophoresis will vary depending on the cycle sequencing chemistry used. Dye Terminator Chemistries Unincorporated
Preparing Extension Products for Electrophoresis Overview Preparation of extension products for electrophoresis will vary depending on the cycle sequencing chemistry used. Dye Terminator Chemistries Unincorporated
