Ab1 (57-68) Ab2 ( ) Ab3 ( ) Ab4 ( ) GBP-N domain GBP-C domain CARD domain
|
|
|
- Cathleen Malone
- 5 years ago
- Views:
Transcription
1 MDKPVCLIDTGSDGKLCVQQAALQVLQQIQQPVVVVAVVGLYRTGKSFLMNRLAG 55 KRTGFALSSNIKPKTEGIWMWCVPHPTKAGTSLVLLDTKGLGDVEKGDSKRDTYI 110 FSLTVLLSSTLVYNSRGVIDNKAMEELQYVTELIEHIKVTPDEDADDCTAFAKFF 165 PHFIWCLRDFTLELKLDGKDLTEDEYLEFALKLRPGTLKKVMMYNLPRECIQKFF 220 PCRTCFTFPSPTTPEKRSILESLSPAELDPEFLEVTKRFCKFVFDRSEVKQLKGG 275 HTVTGRVLGNLTKMYVDTISSGAVFCLENAVIAMAQIENEAATQEGLEVYQRGME 330 KLKSSFPLELEQVSSEHQRLSRMATQAFMARSFKDTDGKHLKALEGEMGKLFDAY 385 RSQNKQVGLETHCDLLLYMRCKDPLILHVYFFPVNDARSKEKVEQNERSSLPISH 440 PRPDRPFQVKTPHVLEVPGASVYPEEGISFRTDVEPNFFKVRKLQVDDVQMNLVR 495 QKDKMSVWTTTIWKEEFVHLQQVRDERKLNSEIEKNDFFNAHRVAFIERVTNVKS 550 IADKLHGQRIIHKELYSEITQTNVTRQQIMRKICDSVDSSGRIAKCKFIDILQEE 605 ERCLLEDLKLSES 618 Ab1 (57-68) Ab2 ( ) Ab3 ( ) Ab4 ( ) GBP-N domain GBP-C domain CARD domain Supplementary Figure 1, related to Figure 1. Epitopes recognized by the four monoclonal antibodies against zebrafish Gbp4. Ab1 and Ab2 bind to the GBP, N-terminal domain (yellow), Ab3 to the GBP, C- terminal (grey), and Ab4 to the CARD domain (black). Binding sites are shown in different colours. The essential residues of the nucleotide-binding pocket that have been mutated to obtain the GTPase-deficient mutant are in bold and red font.
2 A B Supplementary Figure 2, related to Figure 2. The T3SS of S. Typhimurium is required for its virulence in zebrafish. Survival (A) and caspase-1 activity (B) were determined as described in Figures 2A and 2B, respectively. Four different strains of ST were used: the wild type (WT) and its isogenic derivative mutants SPI-1, SPI-2 and SPI-1-2. The sample size for each treatment is 280 in A, 30 in B. S.I., ST infection. ns, not significant; **p<0.01; ***p<0.001 according to log rank test (A) or ANOVA followed Tukey multiple range test (B).
3 Supplementary Figure 3, related to Figures 2 and 4: Gbp4 regulates S. Typhimurium clearance. Zebrafish one-cell stage embryos were microinjected with antisense (As), Gbp4 or Gbp4 KS/AA mrnas and infected at 2 dpf with ST:DsRedT3 into the otic vesicle. Fluorescence intensity quantification of ST:DsRedT3 bacteria at 1 and 24 hpi. Each dot represents the fluorescence from a single larva (N=36, 35, 41, 36, 35, 41). The mean ± SEM of the fluorescence for each group is also shown. Scale bars = 100μm. auf, arbitrary units of fluorescence. **p<0.01; ***p<0.001 according to ANOVA followed by Tukey multiple range test.
4 A B C D E F Supplementary Figure 4, related to Figure 3. The Gbp4-mediated resistance to S. Typhimurium is Caspa dependent. (A, B) Zebrafish one-cell embryos were injected with antisense (As) or Gbp4 mrnas and treated by immersion with vehicle alone (DMSO) or 50 µm of a general inhibitor of caspases (Q-VD- OPh, PINH). (C, D) Zebrafish one-cell embryos were injected with standard control (Std) or Asc MOs in combination with antisense (As) or Caspa mrnas. (E, F) Zebrafish one-cell embryos were injected with standard control (Std) or Gbp4 MOs in combination with antisense (As) or Caspb mrnas. At 2 dpf, larvae were infected and survival (A, C, E) and caspase-1 activity (B, D, F) determined as described in Figures 2A and 2B, respectively. The sample size for each treatment is 340 in A, C and E, 30 in B, D and F. S.I., ST infection. ns, not significant; *p<0.05; **p<0.01; ***p<0.001 according to log rank test (A, C and E) or ANOVA followed by Tukey multiple range test (B, D and F).
5 A B C D Supplementary Figure 5, related to Figure 3. Asc is crucial for S. Typhimurium resistance in zebrafish. (A) Schematic representations of wild type and predicted altered spliced transcripts and truncated Asc protein, and RT-PCR analysis of Asc MOinduced altered splicing of asc transcript at the indicated times. Samples from embryos injected with a standard control MO (Std) are shown for comparison. The primer pairs (F and R) used for amplification and the annealing of the MO are indicated with arrows and a dashed line, respectively. The altered splicing of the asc transcripts resulted in three smaller amplification products than the one observed in samples injected with a standard control MO. The highest product contained a complete deletion of exon 2 and the first base pair of exon 3, which resulted in a predicted truncated Asc protein containing the PYD alone and, therefore, may act as a dominant negative (DN) form 1. The smaller transcript contained a deletion of exons 2, 3 and 4 that resulted in a predicted Asc protein lacking the linker between the PYD and CARD, which may be functional, since a spliced variant of mouse ASC lacking this linker was found to activates caspase-1 to a similar degree that the full length ASC 2. Finally, we failed to sequence the medium size asc transcript but, judging from its size, it should contain a deletion of exons 2 and 3 or exons 3 and 4 that would result in Asc proteins with a shortened linker between PYD and CARD. (B, C) Zebrafish one-cell embryos were injected with standard control (Std) or Asc-ATG MOs in combination with antisense (As), GBP4 (B, C) or DN-Asc-GFP (D) mrnas, infected at 2 dpf, and survival (B) and caspase-1 activity (C) determined as described in Figures 2A and 2B, respectively. The sample size for each treatment is 35 in A, 320 in B and D, 30 in C. S.I., ST infection. ns, not significant; *p<0.05; **p<0.01; ***p<0.001 according to log rank test (B and D) or ANOVA followed by Tukey multiple range test in C.
6 A B C D Supplementary Figure 6, related to Figure 5. Gbp4 levels fine-tune inflammasome activation and caspase-1 activity. Zebrafish one-cell embryos were injected with antisense (As) or increasing concentrations of Gbp4 (A), Gbp4 CARD (B), Gbp4 KS/AA (C) or Gbp4 DM (D) mrnas. Caspase-1 activity was determined as described in Figure 2B. The sample size for each treatment is 30 in A-D. S.I., ST infection. ns, not significant; *p<0.05; **p<0.01; ***p<0.001 according to ANOVA followed by Tukey multiple range test (A-D).
7 A hunlrp1_card hunlrp1_card zfasc_card zfgbp3_card zfgbp4_card zfgbp4_card hunlrp1_card hunlrp1_card zfasc_card zfgbp3_card zfgbp4_card zfgbp4_card B C Supplementary Figure 7, related to Figure 6. Gbp4 and Asc CARD domains. (A) ESPript output 3 obtained with representative CARD domains retrieved from Uniprot database: Human NLRP1 (Q9C000, residues ), zebrafish Asc (A8E7Q7, residues ), zebrafish Gbp3 (B0V1H4, residues ) and zebrafish Gbp4 (A4QNT4, residues ). Residues strictly conserved are in red. Springs above or below blocks of sequences represent helices. (B, C) Electrostatic charge surface of the Gbp4- CARD and zfasc-card structures. The electrostatic charge surface patches of the CARD structures obtained from homology models using Chimera 4 are displayed on a scale of -5 kt/e (red) to 5 kt/e (blue).
8 A B C D E Supplementary Figure 8, related to Figure 7. Gbp4 regulates the number of neutrophils. Zebrafish mpx:egfp one-cell embryos were injected with standard control (Std) or Gbp4 MOs in combination with antisense (As), Gbp4 (A-E) or Gcsfa (C-E) mrnas (A, D). Each dot represents the number of neutrophils from a single larva, while the mean ± SEM for each group is also shown. The sample size (n) is indicated for each treatment. (C) At 2 dpf, larvae were infected and survival determined as described in Figures 2A. (B, E) Representative images of green channels of whole larvae for the different treatments. Scale bars, 100 µm. The sample size for each treatment is shown in the graph in A and D, and is 290 in C. ns, not significant; ***p<0.001 according to log rank test (C) or ANOVA followed by Tukey multiple range test (A and D).
9 A F B C D E Supplementary Figure 9, related to Figures 8, 9. The Gbp4-mediated resistance to S. Typhimurium is independent of IL-1β processing and pyroptotic cell death. (A-E) Zebrafish one-cell embryos were injected with standard control (Std), Asc (A), Gbp4 (C), or Il1b (D, E) MOs in combination with antisense (As) or Gbp4 mrnas (B) and zfigluc mrna (A-C), treated by immersion with vehicle alone (DMSO), 100 µm of a general inhibitor of caspases (PINH) or 100 µm of a specific inhibitor of caspase-1 (C1INH) (A), and infected at 2 dpi with ST (MOI of 10). The luciferase activity determined at 24 hpi as described in the Method section (A-C), while survival (D) and caspase-1 activity (E) were determined as described in Figures 2A and 2B, respectively. The Gluc was used as a positive control (C+). (F) Zebrafish lys:dsred one-cell embryos were injected with standard control (Std) or Gbp4 MOs and infected at 2 dpf in the otic vesicle with ST at a MOI of 100. YO-PRO compound was injected at 3 hpi in the otic vesicle and pictures of each larva were taken at 4.5 hpi at the fluorescent microscope to visualize dead cells. Representative pictures are shown for each treatment. As a positive control, a group of cells dying during larvae development in the front part of the head are shown. The sample size for each treatment is 25 in A-C, 300 in D, 30 in E, 40 in F. S.I., ST infection. ns, not significant. **p<0.01; ***p<0.001 according to log rank test (D) or ANOVA followed by Tukey multiple range test (A, B, C and E).
10 Supplementary Figure 10, related to Figures 8, Supplementary (S) Biosynthesis of eicosanoids. The different pharmacological inhibitors and the lipid mediators used in this study are shown.
11 A B C D E F G H I J K Supplementary Figure 11, related to Figures 8, 9. Disparate regulation of the expression of genes encoding the enzymes involved in eicosanoid biosynthesis by S. Typhimurium infection. (A, I) mrna levels of the indicated genes were measured by RT-qPCR in FACS-sorted neutrophils from 3 dpf mpx:egfp larvae (A-G) or whole wild type larvae (H, I), which were previously infected with ST or not at 2 dpf. Data were normalized with uninfected neutrophils. (J, K) Larvae were infected at 2 dpf with ST and the Pla2 activity in whole cell extracts determined using the EnzChek Phospholipase A2 Assay Kit. The sample size for each treatment is 950 in A-G, 25 in H and I, 30 in J and K. ND: non detected. *p<0.05; **p<0.01; ***p<0.001 according to Student t- test (A-G) or ANOVA followed by Tukey multiple range test (H-K).
12 A B C D E F Supplementary Figure 12, related to Figures 8, 9. Impact of pharmacological inhibition of key enzymes involved in eicosanoid biosynthesis on larval resistance to S. Typhimurium infection. (A-F) Zebrafish onecell embryos were injected with antisense (As) or Gbp4 mrnas. Larvae were then treated by immersion with pharmacological inhibitors of cpla2 (CAY10502, 0.5 M) (A, D), Ptgs2 (meloxicam, 10 µm) (B, E), Alox5 (MK-886, 1 µm) (C, F) or vehicle alone (DMSO) at 2 dpf (1 h before infection) (A-C) or 3 dpf (24 hpi) (D-F), infected at 2 dpf and survival determined as described in Figure 2A. The sample size for each treatment is 300 in A-F. S.I., ST infection. *p<0.05; **p<0.01; ***p<0.001 according to log rank test (A-F).
13 A B C Supplementary Figure 13, related to Figures 8, 9. Exogenous addition of AA increases larval resistance to S. Typhimurium infection, while LTB4 does not affect the bacterial clearance phase. (A-C) Zebrafish onecell embryos were injected with control (Std) or Gbp4 MOs. Larvae were then treated by immersion with 20 µm AA (A, B), 1 µm LTB4 (C) or vehicle alone (DMSO) at 2 dpf (1 h before infection) (A) or 3 dpf (24 hpi) (B, C), infected at 2 dpf and survival determined as described in Figure 2A. The sample size for each treatment is 300 in A-C. S.I., ST infection. *p<0.05; **p<0.01; ***p<0.001 according to log rank test (A-C).
14 Supplementary Figure 14, related to Figs. 1 and 6. Full size images of western blots cropped for presentation in Figs. 1E (A) and 6E (B). The molecular weight ladders are also shown.
15 Supplementary Table 1. Lipidomic analysis in 3 dpf whole zebrafish larvae at 8 and 24 h post-injection (hpi) with PBS or S. Typhimurium (ST). Zebrafish one-cell embryos were injected with antisense (As), Gbp4 and Asc mrnas, infected at 2 dpf and the levels of the indicated lipid mediators determined by LC-MS-MS. Lipid concentrations are shown in pg/1000 larvae. LT, leukotriene; LX, lipoxin; TX, thromboxane; PG, prostaglandin. LTB4 LXA4* LXB4 PGD2 PGE2 PGF2α TXB2 As / PBS As / ST Gbp4 / ST As / PBS As / ST Gbp4 / ST Asc / ST Neutrophil recruitment phase (8 hpi) Bacterial clearance phase (24 hpi) *13,14-dihydro-15-oxo LXA4: LXA4 metabolite found in zebrafish 5.
16 Supplementary Table 2. Lipidomic analysis in 3 dpf whole zebrafish larvae at 24 h post-injection (hpi) with PBS or S. Typhimurium (ST). Zebrafish larvae were treated by immersion with vehicle alone (DMSO) or 100 µm of a specific inhibitor of caspase-1 (Ac-YVAD-CMK,C1INH) at 2 dpf (1 h before infection), infected and the levels of the indicated lipid mediators determined by LC-MS-MS at 24 hpi. Lipid concentrations are shown in pg/1000 larvae. LT, leukotriene; LX, lipoxin; TX, thromboxane; PG, prostaglandin. LTB4 LXA4* LXB4 PGD2 PGE2 PGF2α TXB2 DMSO / PBS DMSO / ST C1INH / ST Bacterial clearance phase (24 hpi) *13,14-dihydro-15-oxo LXA4: LXA4 metabolite found in zebrafish 5.
17 Supplementary Table 3. Morpholinos used in this study. The gene symbols followed the Zebrafish Nomenclature Guidelines ( Gene Ensembl ID Target Sequence (5 3 ) Concentration (mm) Reference gbp4 ENSDARG e1/i1 GCTGTTTGTGTGTCTCTAACCTGTT 0.1 This work asc (pycard) ENSDARG e2/i2 AGTGATTCGCTTACTCACCATCAGA 1.68 This work 5 UTR/ATG GCTGCTCCTTGAAAGATTCCGCCAT 0.6 This work cxcr2 ENSDARG UTR/ATG ACTCTGTAGTAGCAGTTTCCATGTT 0.3 il1b ENSDARG i2/e3 CCCACAAACTGCAAAATATCAGCTT 0.5 lta4h ENSDART UTR/ATG AGCTAGGGTCTGAAACTGGAGTCAT 0.2 Deng et al., 2013 López-Muñoz et al., 2011 Tobin et al., 2010
18 Supplementary Table 4. Primers used in this study. The gene symbols followed the Zebrafish Nomenclature Guidelines ( ENA, European Nucleotide Archive ( Gene ENA ID Name Sequence (5 3 ) Use asc (pycard) NM_ F R ATTTTGAGGGCGATCAAGTG GCATCCTCAAGGTCATCCAT RT-PCR rps11 NM_ F1 R1 GGCGTCAACGTGTCAGAGTA GCCTCTTCTCAAAACGGTTG gbp4 NM_ F R ACTGGGAGATGTGGAAAAGGGCG CCATAGCCTTGTTGTCGATCACCCC il1b NM_ F5 R5 GGCTGTGTGTTTGGGAATCT TGATAAACCAACCGGGACA gfp EF F1 R1 ACGTAAACGGCCACAAGTTC AAGTCGTGCTGCTTCATGTG ptgs2b ptgs2a NM_ NM_ F2 R2 F1 R1 CCCCAGAGTACTGGAAACCA ACATGGCCCGTTGACATTAT TGGATCTTTCCTGGTGAAGG GAAGCTCAGGGGTAGTGCAG RT-qPCR ptgs1 AY F R TTTTGCTGCTGAGTGTGTCC CGAACACAGATCCCTTGGTT cpla2 NM_ F1 R1 CTGTTCATGCAGACACGCAG GGTGGGAACCTCTCTTGGTG alox5b.2 NM_ F1 R1 GGACAACGAGCTGTTTTTAGGC CATTTCGGCTCCTGATGGTCT alox5b.3 NM_ F1 R1 TCAGAGGCGTCATCAAGAGC CGTCAATGGAAATCACCGTTCC
19 References Supplementary of Supplementary References Material 1. Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proil-beta. Mol Cell 10, (2002). 2. Matsushita, K. et al. A splice variant of ASC regulates IL-1beta release and aggregates differently from intact ASC. Mediators Inflamm 2009, (2009). 3. Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42, W (2014). 4. Pettersen, E.F. et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, (2004). 5. Tobin, D.M. et al. Host genotype-specific therapies can optimize the inflammatory response to mycobacterial infections. Cell 148, (2012).
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS. Transient infection of the zebrafish notochord triggers chronic inflammation
 SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS Transient infection of the zebrafish notochord triggers chronic inflammation Mai Nguyen-Chi 1,2,5, Quang Tien Phan 1,2,5, Catherine Gonzalez 1,2, Jean-François
SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS Transient infection of the zebrafish notochord triggers chronic inflammation Mai Nguyen-Chi 1,2,5, Quang Tien Phan 1,2,5, Catherine Gonzalez 1,2, Jean-François
FSC-W FSC-H CD4 CD62-L
 Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Supplementary Fig. 1 a SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H CD4 CD62-L b SSC-A FSC-A FSC-W FSC-A FSC-A 7-AAD FSC-A CD4 IL-9 CD4 c SSC-A FSC-A FSC-W FSC-H SSC-W SSC-H 7-AAD KI67 Annexin-V 7-AAD d I L -5
Supporting Information
 Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
SUPPLEMENTARY INFORMATION
 reverse 3175 3175 F L C 318 318 3185 3185 319 319 3195 3195 315 8 1 315 3155 315 317 Supplementary Figure 3. Stability of expression of the GFP sensor constructs return to warm conditions. Semi-quantitative
reverse 3175 3175 F L C 318 318 3185 3185 319 319 3195 3195 315 8 1 315 3155 315 317 Supplementary Figure 3. Stability of expression of the GFP sensor constructs return to warm conditions. Semi-quantitative
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and
 Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A)
 Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Different crystal forms obtained for Sky
 Supplementary Figure 1 Different crystal forms obtained for Sky 1 353. (a) Crystal form 1 obtained in the presence of 20% PEG 3350 and 0.2 M ammonium citrate tribasic ph 7.0. (b) Crystal form 1 of the
Supplementary Figure 1 Different crystal forms obtained for Sky 1 353. (a) Crystal form 1 obtained in the presence of 20% PEG 3350 and 0.2 M ammonium citrate tribasic ph 7.0. (b) Crystal form 1 of the
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Waithe et al Supplementary Figures
 Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Lipidomic Analysis of Dynamic Eicosanoid Responses During the Induction and Resolution of Lyme Arthritis
 Lipidomic Analysis of Dynamic Eicosanoid Responses During the Induction and Resolution of Lyme Arthritis Victoria A. Blaho 1, Matthew W. Buczynski 2, Charles R. Brown 1, and Edward A. Dennis 2 1 Department
Lipidomic Analysis of Dynamic Eicosanoid Responses During the Induction and Resolution of Lyme Arthritis Victoria A. Blaho 1, Matthew W. Buczynski 2, Charles R. Brown 1, and Edward A. Dennis 2 1 Department
m1 m2 m3 m4 m5 m6 m7 m8 wt m m m m m m m m8 - + wt +
 otherwise, you couldn't grow them!) You perform pairwise infections with each of your mutant bacteriophage strains and get the following results: (+) = pair of phages lysed host cells, (-) = pair of phages
otherwise, you couldn't grow them!) You perform pairwise infections with each of your mutant bacteriophage strains and get the following results: (+) = pair of phages lysed host cells, (-) = pair of phages
TNFα 18hr. Control. CHX 18hr. TNFα+ CHX 18hr. TNFα: 18 18hr (KDa) PARP. Cleaved. Cleaved. Cleaved. Caspase3. Pellino3 shrna. Control shrna.
 Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Survival ( %) a. TNFα 18hr b. Control sirna Pellino3 sirna TNFα: 18 18hr c. Control shrna Pellino3 shrna Caspase3 Actin Control d. Control shrna Pellino3 shrna *** 100 80 60 CHX 18hr 40 TNFα+ CHX 18hr
Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system
 Disease Models & Mechanisms 6, 841-854 (2013) doi:10.1242/dmm.010843 RESOURCE ARTICLE Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system
Disease Models & Mechanisms 6, 841-854 (2013) doi:10.1242/dmm.010843 RESOURCE ARTICLE Functional analysis of a zebrafish myd88 mutant identifies key transcriptional components of the innate immune system
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1
 Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its
 Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its transcriptional activity in wild-type embryo. A gradient of canonical
Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its transcriptional activity in wild-type embryo. A gradient of canonical
Supplementary Figure 1. Biochemical and sequence alignment analyses the
 Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Supplementary Figure 1. Nature Genetics: doi: /ng.3848
 Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
downstream (0.8 kb) homologous sequences to the genomic locus of DIC. A DIC mutant strain (ro- 6
 A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
P. syringae and E. coli
 CHAPTER 6 A comparison of the recd mutant phenotypes of P. syringae and E. coli 6.1 INTRODUCTION The RecBCD complex is essential for recombination mediated repair of double strand breaks (DSBs) of DNA
CHAPTER 6 A comparison of the recd mutant phenotypes of P. syringae and E. coli 6.1 INTRODUCTION The RecBCD complex is essential for recombination mediated repair of double strand breaks (DSBs) of DNA
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX.
 Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX. Phase contrast and widefield fluorescence microscopy (20x magnification).
Supplementary Figure 1. AnnexinV FITC and Sytox orange staining in wild type, Nlrp3 /, ASC / and casp1/11 / TEC treated with TNF /CHX. Phase contrast and widefield fluorescence microscopy (20x magnification).
Multiple Choice Review- Eukaryotic Gene Expression
 Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
 DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Chemical structure of LPS and LPS biogenesis in Gram-negative bacteria. a. Chemical structure of LPS. LPS molecule consists of Lipid A, core oligosaccharide and O-antigen. The polar
Supplementary Figure 1 Chemical structure of LPS and LPS biogenesis in Gram-negative bacteria. a. Chemical structure of LPS. LPS molecule consists of Lipid A, core oligosaccharide and O-antigen. The polar
Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct
 SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
3.B.1 Gene Regulation. Gene regulation results in differential gene expression, leading to cell specialization.
 3.B.1 Gene Regulation Gene regulation results in differential gene expression, leading to cell specialization. We will focus on gene regulation in prokaryotes first. Gene regulation accounts for some of
3.B.1 Gene Regulation Gene regulation results in differential gene expression, leading to cell specialization. We will focus on gene regulation in prokaryotes first. Gene regulation accounts for some of
AP Biology. Free-Response Questions
 2018 AP Biology Free-Response Questions College Board, Advanced Placement Program, AP, AP Central, and the acorn logo are registered trademarks of the College Board. AP Central is the official online home
2018 AP Biology Free-Response Questions College Board, Advanced Placement Program, AP, AP Central, and the acorn logo are registered trademarks of the College Board. AP Central is the official online home
Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA
 Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
Supplemental Data. Chen and Thelen (2010). Plant Cell /tpc
 Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON
 PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
targets. clustering show that different complex pathway
 Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
SUPPLEMENTARY INFORMATION
 GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
Nature Genetics: doi: /ng Supplementary Figure 1. ssp mutant phenotypes in a functional SP background.
 Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature1237 a b retinol retinal RA OH RDH (retinol dehydrogenase) O H Raldh2 O R/R.6.4.2 (retinaldehyde dehydrogenase 2) RA retinal retinol..1.1 1 Concentration (nm)
SUPPLEMENTARY INFORMATION doi:1.138/nature1237 a b retinol retinal RA OH RDH (retinol dehydrogenase) O H Raldh2 O R/R.6.4.2 (retinaldehyde dehydrogenase 2) RA retinal retinol..1.1 1 Concentration (nm)
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Hes5. Hes Relative mrna levels
 A a Absolute mrna levels 6 5 4 3 2 1 hjag1 EP cdl1 EP Jag1 E2+1 b d m Hey1 c e EP Hey1 Dl1 E2+1 f h m Hey1 g i EP Hey1 1 a Hes5 Hes5 a Hes5 Hes5 B a b EP e 1.4 1.2 EP GFP GFP E2+1 c m Hey1 d Hey1 Relative
A a Absolute mrna levels 6 5 4 3 2 1 hjag1 EP cdl1 EP Jag1 E2+1 b d m Hey1 c e EP Hey1 Dl1 E2+1 f h m Hey1 g i EP Hey1 1 a Hes5 Hes5 a Hes5 Hes5 B a b EP e 1.4 1.2 EP GFP GFP E2+1 c m Hey1 d Hey1 Relative
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice Supplementary Figure 2.
 Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Rho1 binding site PtdIns(4,5)P2 binding site Both sites
 localization Mutation site DMSO LatB WT F77A I115A I131A K134A Rho1 binding site PtdIns(4,5)P2 binding site Both sites E186A E199A N201A R84A-E186A-E199A L131A-K136A-E186A L131A-E186A-E199A K136A-E186A-E199A
localization Mutation site DMSO LatB WT F77A I115A I131A K134A Rho1 binding site PtdIns(4,5)P2 binding site Both sites E186A E199A N201A R84A-E186A-E199A L131A-K136A-E186A L131A-E186A-E199A K136A-E186A-E199A
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises
 Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
Heather Currinn, Benjamin Guscott, Zita Balklava, Alice Rothnie and Thomas Wassmer*
 Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering
 Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Lecture 18 June 2 nd, Gene Expression Regulation Mutations
 Lecture 18 June 2 nd, 2016 Gene Expression Regulation Mutations From Gene to Protein Central Dogma Replication DNA RNA PROTEIN Transcription Translation RNA Viruses: genome is RNA Reverse Transcriptase
Lecture 18 June 2 nd, 2016 Gene Expression Regulation Mutations From Gene to Protein Central Dogma Replication DNA RNA PROTEIN Transcription Translation RNA Viruses: genome is RNA Reverse Transcriptase
Supplementary Material. Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with
 Supplementary Material Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata Emine Kaplanoglu
Supplementary Material Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata Emine Kaplanoglu
Supplemental Data. Perrella et al. (2013). Plant Cell /tpc
 Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Structure and RNA-binding properties. of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex
 Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
Nature Neuroscience: doi: /nn.2717
 Supplementary Fig. 1. Dendrite length is not secondary to body length. Dendrite growth proceeds independently of the rate of body growth and decreases in rate in adults. n 20 on dendrite measurement, n
Supplementary Fig. 1. Dendrite length is not secondary to body length. Dendrite growth proceeds independently of the rate of body growth and decreases in rate in adults. n 20 on dendrite measurement, n
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Mourier et al., http://www.jcb.org/cgi/content/full/jcb.201411100/dc1 Figure S1. Size and mitochondrial content in Mfn1 and Mfn2 knockout hearts. (A) Body
Supplemental material THE JOURNAL OF CELL BIOLOGY Mourier et al., http://www.jcb.org/cgi/content/full/jcb.201411100/dc1 Figure S1. Size and mitochondrial content in Mfn1 and Mfn2 knockout hearts. (A) Body
Introduction to Molecular and Cell Biology
 Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the molecular basis of disease? What
Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the molecular basis of disease? What
Ethylene is critical to the maintenance of primary root growth and Fe. homeostasis under Fe stress in Arabidopsis
 Ethylene is critical to the maintenance of primary root growth and Fe homeostasis under Fe stress in Arabidopsis Guangjie Li, Weifeng Xu, Herbert J. Kronzucker, Weiming Shi * Supplementary Data Supplementary
Ethylene is critical to the maintenance of primary root growth and Fe homeostasis under Fe stress in Arabidopsis Guangjie Li, Weifeng Xu, Herbert J. Kronzucker, Weiming Shi * Supplementary Data Supplementary
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
2012 Univ Aguilera Lecture. Introduction to Molecular and Cell Biology
 2012 Univ. 1301 Aguilera Lecture Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the
2012 Univ. 1301 Aguilera Lecture Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the
Nature Methods: doi: /nmeth Supplementary Figure 1. In vitro screening of recombinant R-CaMP2 variants.
 Supplementary Figure 1 In vitro screening of recombinant R-CaMP2 variants. Baseline fluorescence compared to R-CaMP1.07 at nominally zero calcium plotted versus dynamic range ( F/F) for 150 recombinant
Supplementary Figure 1 In vitro screening of recombinant R-CaMP2 variants. Baseline fluorescence compared to R-CaMP1.07 at nominally zero calcium plotted versus dynamic range ( F/F) for 150 recombinant
RNA Synthesis and Processing
 RNA Synthesis and Processing Introduction Regulation of gene expression allows cells to adapt to environmental changes and is responsible for the distinct activities of the differentiated cell types that
RNA Synthesis and Processing Introduction Regulation of gene expression allows cells to adapt to environmental changes and is responsible for the distinct activities of the differentiated cell types that
FINAL REPORT For Japan-Korea Joint Research Project AREA
 (Form4-2) FINAL REPORT For Japan-Korea Joint Research Project AREA 1. Mathematics & Physics 2. Chemistry & Material Science 3. Biology 4. Informatics & Mechatronics 5. Geo-Science & Space Science 6. Medical
(Form4-2) FINAL REPORT For Japan-Korea Joint Research Project AREA 1. Mathematics & Physics 2. Chemistry & Material Science 3. Biology 4. Informatics & Mechatronics 5. Geo-Science & Space Science 6. Medical
Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1
 Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1 (a) Fold-change in UCP-1 mrna abundance in white adipocytes upon β-adrenergic stimulation
Supplementary Figure 1 Analysis of beige fat and cells and characteristics of exosome release, related to Figure 1 (a) Fold-change in UCP-1 mrna abundance in white adipocytes upon β-adrenergic stimulation
Regulation and signaling. Overview. Control of gene expression. Cells need to regulate the amounts of different proteins they express, depending on
 Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis Sergio de Cima, Luis M. Polo, Carmen Díez-Fernández, Ana I. Martínez, Javier
SUPPLEMENTARY INFORMATION Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis Sergio de Cima, Luis M. Polo, Carmen Díez-Fernández, Ana I. Martínez, Javier
Nature Structural & Molecular Biology: doi: /nsmb.3194
 Supplementary Figure 1 Mass spectrometry and solution NMR data for -syn samples used in this study. (a) Matrix-assisted laser-desorption and ionization time-of-flight (MALDI-TOF) mass spectrum of uniformly-
Supplementary Figure 1 Mass spectrometry and solution NMR data for -syn samples used in this study. (a) Matrix-assisted laser-desorption and ionization time-of-flight (MALDI-TOF) mass spectrum of uniformly-
A New Method For Molecular Diagnostics In Veterinary Medicine. Dr. Mikael Leijon
 New Method For Molecular Diagnostics In Veterinary Medicine Dr. Mikael Leijon The challenge in genetic detection the third position is often undetermined but variation also exist in the first (L, S, R)
New Method For Molecular Diagnostics In Veterinary Medicine Dr. Mikael Leijon The challenge in genetic detection the third position is often undetermined but variation also exist in the first (L, S, R)
Translation Part 2 of Protein Synthesis
 Translation Part 2 of Protein Synthesis IN: How is transcription like making a jello mold? (be specific) What process does this diagram represent? A. Mutation B. Replication C.Transcription D.Translation
Translation Part 2 of Protein Synthesis IN: How is transcription like making a jello mold? (be specific) What process does this diagram represent? A. Mutation B. Replication C.Transcription D.Translation
GCD3033:Cell Biology. Transcription
 Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Introduction. Gene expression is the combined process of :
 1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
. Supplementary Information
 . Supplementary Information Supplementary Figure S1. Mature embryo sac observations. Supplementary Figure S2. STT observations. Supplementary Figure S3. Comparison of the PTB1 cdna with that of the mutant.
. Supplementary Information Supplementary Figure S1. Mature embryo sac observations. Supplementary Figure S2. STT observations. Supplementary Figure S3. Comparison of the PTB1 cdna with that of the mutant.
Name: SBI 4U. Gene Expression Quiz. Overall Expectation:
 Gene Expression Quiz Overall Expectation: - Demonstrate an understanding of concepts related to molecular genetics, and how genetic modification is applied in industry and agriculture Specific Expectation(s):
Gene Expression Quiz Overall Expectation: - Demonstrate an understanding of concepts related to molecular genetics, and how genetic modification is applied in industry and agriculture Specific Expectation(s):
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells
 Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis
 Title Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis Author list Yu Han 1, Huihua Wan 1, Tangren Cheng 1, Jia Wang 1, Weiru Yang 1, Huitang Pan 1* & Qixiang
Title Comparative RNA-seq analysis of transcriptome dynamics during petal development in Rosa chinensis Author list Yu Han 1, Huihua Wan 1, Tangren Cheng 1, Jia Wang 1, Weiru Yang 1, Huitang Pan 1* & Qixiang
Biological Roles of Cytokinins
 Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
From gene to protein. Premedical biology
 From gene to protein Premedical biology Central dogma of Biology, Molecular Biology, Genetics transcription replication reverse transcription translation DNA RNA Protein RNA chemically similar to DNA,
From gene to protein Premedical biology Central dogma of Biology, Molecular Biology, Genetics transcription replication reverse transcription translation DNA RNA Protein RNA chemically similar to DNA,
MRC-Holland MLPA. Description version 14; 21 January 2015
 SALSA MLPA probemix P229-B2 OPA1 Lot B2-0412. As compared to version B1-0809, two reference probes and the 88 and 96 nt control fragments have been replaced (QDX2). The OPA1 gene product is a nuclear-encoded
SALSA MLPA probemix P229-B2 OPA1 Lot B2-0412. As compared to version B1-0809, two reference probes and the 88 and 96 nt control fragments have been replaced (QDX2). The OPA1 gene product is a nuclear-encoded
1. Supplemental figures and legends
 1. Supplemental figures and legends Figure S1. Phenotypes of the inactivation of the BMP2/4 and BMP5/8 ligands. AD, phenotypes of the inactivation of the BMP2/4 and BMP5/8 ligands at 72h. A, sea urchin
1. Supplemental figures and legends Figure S1. Phenotypes of the inactivation of the BMP2/4 and BMP5/8 ligands. AD, phenotypes of the inactivation of the BMP2/4 and BMP5/8 ligands at 72h. A, sea urchin
Supporting Information
 Supporting Information López et al. 10.1073/pnas.0810940106 1. Ivey DM, et al. (1993) Cloning and characterization of a putative Ca2 /H antiporter gene from Escherichia coli upon functional complementation
Supporting Information López et al. 10.1073/pnas.0810940106 1. Ivey DM, et al. (1993) Cloning and characterization of a putative Ca2 /H antiporter gene from Escherichia coli upon functional complementation
Supporting Information
 Supporting Information Sana et al. 10.1073/pnas.1608858113 Fig. S1. Representation of the SPI-6 type VI secretion system. (A) Representation of the SPI-6 genetic locus starting at STM0266 and ending at
Supporting Information Sana et al. 10.1073/pnas.1608858113 Fig. S1. Representation of the SPI-6 type VI secretion system. (A) Representation of the SPI-6 genetic locus starting at STM0266 and ending at
Supplemental Data. Wang et al. (2014). Plant Cell /tpc
 Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Actinobacteria Relative abundance (%) Co-housed CD300f WT. CD300f KO. Colon length (cm) Day 9. Microscopic inflammation score
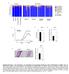 y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Bi 1x Spring 2014: LacI Titration
 Bi 1x Spring 2014: LacI Titration 1 Overview In this experiment, you will measure the effect of various mutated LacI repressor ribosome binding sites in an E. coli cell by measuring the expression of a
Bi 1x Spring 2014: LacI Titration 1 Overview In this experiment, you will measure the effect of various mutated LacI repressor ribosome binding sites in an E. coli cell by measuring the expression of a
Table S1. Sequence of primers used in RT-qPCR
 Table S1. Sequence of primers used in RT-qPCR Primer Name P16Ink4a-F P16Ink4a-R P15Ink4b-F P15Ink4b-R P19Arf-F P19Arf-R P53-F P53-R P21cip1-F P21cip1-R P27kip1-F P27kip1-R P18Ink4c-F P18Ink4c-R P19Ink4d-F
Table S1. Sequence of primers used in RT-qPCR Primer Name P16Ink4a-F P16Ink4a-R P15Ink4b-F P15Ink4b-R P19Arf-F P19Arf-R P53-F P53-R P21cip1-F P21cip1-R P27kip1-F P27kip1-R P18Ink4c-F P18Ink4c-R P19Ink4d-F
Induction of cap-independent BiP (hsp-3) and Bcl-2 (ced-9) translation in response to eif4g (IFG-1) depletion in C. elegans
 Research Paper Translation 2, e28935; April; 2014 Landes Bioscience Research Paper Induction of cap-independent BiP (hsp-3) and Bcl-2 (ced-9) translation in response to eif4g (IFG-1) depletion in C. elegans
Research Paper Translation 2, e28935; April; 2014 Landes Bioscience Research Paper Induction of cap-independent BiP (hsp-3) and Bcl-2 (ced-9) translation in response to eif4g (IFG-1) depletion in C. elegans
ydci GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC
 Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Biology of Salmonella David Holden
 Biology of Salmonella David Holden Lecture 2 life on the inside trafficking and phagolysosomal avoidance PhoP/Q and the SPI-2 T3SS control of SPI-2 effector translocation effector function analysis at
Biology of Salmonella David Holden Lecture 2 life on the inside trafficking and phagolysosomal avoidance PhoP/Q and the SPI-2 T3SS control of SPI-2 effector translocation effector function analysis at
DNA Technology, Bacteria, Virus and Meiosis Test REVIEW
 Be prepared to turn in a completed test review before your test. In addition to the questions below you should be able to make and analyze a plasmid map. Prokaryotic Gene Regulation 1. What is meant by
Be prepared to turn in a completed test review before your test. In addition to the questions below you should be able to make and analyze a plasmid map. Prokaryotic Gene Regulation 1. What is meant by
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
