Supplementary material
|
|
|
- Bennett Fleming
- 6 years ago
- Views:
Transcription
1 Table 1: The number of protein sequences listed for the BacelLo training data set according to each origin and localization. Localization Plants Animal Fungi chloroplast (ch) cytoplasm (cy) mitochondria (mi) nucleus (nu) secretory pathway (SP) Total Supplementary material Training datasets The number of protein sequences for each localization in the BacelLo and Höglund training data sets are shown in Tab. 1 and Tab. 2. Phylogenetic profiles We downloaded 453 fully sequenced genomes from the National Center for Biotechnology Information (NCBI) ftp site (ftp.ncbi.nih.gov/genomes) consisting of 20 eukaryotes, 33 archaea and 400 bacteria. However, only 78 genomes were used for the calculation of phylogenetic profiles. We used all eukaryotic and archaea genomes and selected only the 25 genetically most distant bacteria genomes in order to get an approximately equal distribution of genomes from the three domains of life. We used the same genome subselection procedure as described in Sun et al., The method uses the NCBI taxonomy information to reconstruct a taxonomy tree and exploits hierarchical information in a top down approach to select a preferably nonredundant set of genomes. The complete set of the genomes used is listed in Tab. 3, Tab. 4 and Tab. 5. 1
2 Table 2: The number of protein sequences listed for the Höglund training data set according to each localization. Localization Nr chloroplast (ch) 449 cytoplasm (cy) 1411 Endoplasmatic Reticulum (er) 198 extracellular space (ex) 843 Golgi apparatus (go) 150 lysosome (ly) 103 mitochondria (mi) 510 nucleus (nu) 837 peroxisome (pe) 157 plasma membrane (pm) 1238 vacuole (va) 63 Total
3 Table 3: The selected fully sequenced eukaryotic genomes used for the calculation of phylogenetic profiles. Kingdom Taxonomy ID Organism name Fungi 4932 Saccharomyces cerevisiae Fungi Eremothecium gossypii Fungi Cryptococcus neoformans var. neoformans JEC21 Fungi Kluyveromyces lactis NRRL Y-1140 Fungi Yarrowia lipolytica CLIB122 Fungi Debaryomyces hansenii CBS767 Fungi Candida glabrata CBS 138 Fungi Schizosaccharomyces pombe 972h- Fungi Encephalitozoon cuniculi GB-M1 Metazoa 6239 Caenorhabditis elegans Metazoa 7227 Drosophila melanogaster Metazoa 9606 Homo sapiens Metazoa Mus musculus Protista 5693 Trypanosoma cruzi Protista Thalassiosira pseudonana Protista Plasmodium falciparum 3D7 Protista Cyanidioschyzon merolae strain 10D Protista Entamoeba histolytica HM-1:IMSS Viridiplantae 3702 Arabidopsis thaliana Viridiplantae Oryza sativa Japonica Group 3
4 Table 4: The selected fully sequenced archaea genomes used for the calculation of phylogenetic profiles. Phylum Taxonomy ID Organism name Crenarchaeota Pyrobaculum aerophilum str. IM2 Crenarchaeota Aeropyrum pernix K1 Crenarchaeota Sulfolobus solfataricus P2 Crenarchaeota Sulfolobus tokodaii str. 7 Crenarchaeota Sulfolobus acidocaldarius DSM 639 Crenarchaeota Thermofilum pendens Hrk 5 Crenarchaeota Pyrobaculum islandicum DSM 4184 Crenarchaeota Hyperthermus butylicus DSM 5456 Euryarchaeota Halobacterium sp. NRC-1 Euryarchaeota Thermococcus kodakarensis KOD1 Euryarchaeota Pyrococcus horikoshii OT3 Euryarchaeota Pyrococcus furiosus DSM 3638 Euryarchaeota Methanothermobacter thermautotrophicus str. Delta H Euryarchaeota Methanosarcina acetivorans C2A Euryarchaeota Methanopyrus kandleri AV19 Euryarchaeota Methanosarcina mazei Go1 Euryarchaeota Archaeoglobus fluorides DSM 4304 Euryarchaeota Methanocaldococcus jannaschii DSM 2661 Euryarchaeota Methanococcoides burtonii DSM 6242 Euryarchaeota Picrophilus torridus DSM 9790 Euryarchaeota Methanococcus maripaludis S2 Euryarchaeota Methanosarcina barkeri str. Fusaro Euryarchaeota Haloarcula marismortui ATCC Euryarchaeota Pyrococcus abyssi GE5 Euryarchaeota Thermoplasma acidophilum DSM 1728 Euryarchaeota Thermoplasma volcanium GSS1 Euryarchaeota Methanospirillum hungatei JF-1 Euryarchaeota Methanosphaera stadtmanae DSM 3091 Euryarchaeota Natronomonas pharaonis DSM 2160 Euryarchaeota Methanosaeta thermophila PT Euryarchaeota Haloquadratum walsbyi DSM Euryarchaeota Methanocorpusculum labreanum Z Nanoarchaeota Nanoarchaeum equitans Kin4-M 4
5 Table 5: The selected fully sequenced bacteria genomes used for the calculation of phylogenetic profiles. Phylum Taxonomy ID Organism name Chloroflexi Dehalococcoides ethenogenes 195 Chloroflexi Dehalococcoides sp. CBDB1 Cyanobacteria 1140 Synechococcus elongatus PCC 7942 Cyanobacteria 1148 Synechocystis sp. PCC 6803 Cyanobacteria Prochlorococcus marinus str. NATL2A Cyanobacteria Synechococcus sp. CC9311 Cyanobacteria Nostoc sp. PCC 7120 Cyanobacteria Thermosynechococcus elongatus BP-1 Cyanobacteria Trichodesmium erythraeum IMS101 Cyanobacteria Anabaena variabilis ATCC Cyanobacteria Gloeobacter violaceus PCC 7421 Firmicutes Symbiobacterium thermophilum IAM Proteobacteria Shewanella sp. MR-4 Proteobacteria Azoarcus sp. BH72 Proteobacteria Acinetobacter sp. ADP1 Proteobacteria Magnetococcus sp. MC-1 Proteobacteria Acidovorax sp. JS42 Proteobacteria Mesorhizobium sp. BNC1 Proteobacteria Jannaschia sp. CCS1 Proteobacteria Silicibacter sp. TM1040 Proteobacteria Polaromonas sp. JS666 Proteobacteria Pseudoalteromonas haloplanktis TAC125 Proteobacteria Baumannia cicadellinicola str. Hc (Homalodisca coagulata) Proteobacteria Candidatus Carsonella ruddii PV Proteobacteria Candidatus Ruthia magnifica str. Cm (Calyptogena magnifica) 5
6 Gene Ontology terms GOLoc uses only the GO terms that can be observed in the training data since for other GO terms we can not make any statements. In addition, it has the advantage that only a small portion of the over available GO terms is used. Table 6 shows the number of selected GO terms for every individual predictor. A list of selected GO terms can be obtained from the MultiLoc2 webpage. Table 6: The selected GO terms for the individual MultiLoc2 predictors. MultiLoc2 version Training dataset Number of selected GO terms MultiLoc2-LowRes Animals BaCelLo animals 712 MultiLoc2-LowRes Fungi BaCelLo fungi 508 MultiLoc2-LowRes Plants BaCelLo plants 408 MultiLoc2-HighRes Animals MultiLoc animals 1097 MultiLoc2-HighRes Fungi MultiLoc fungi 1075 MultiLoc2-HighRes Plants MultiLoc plants
7 MultiLoc2-LowRes architecture The architecture of the animal version of MultiLoc2-LowRes is shown in Fig. 1. Compared with MultiLoc2-HighRes the SVMSA subpredictor is not used because MultiLoc2-LowRes is specialized on globular proteins. Figure 1: The architecture of MultiLoc2-LowRes (animal version). A query sequence is processed by a first layer of five subprediction methods (SVMTarget, SVMaac, PhyloLoc, GOLoc and MotifSearch). The individual output of the layer one methods are collected in the PPV which enters a second layer of SVMs producing probability estimates for each localization. Independent test without GO terms The results of the simulation that no GO terms are available for all proteins of the independent data set are presented in this section. Tab. 7 shows the localization-specific performance results using sensitivity and MCC and Tab. 8 summarizes the overall performances using AVG and ACC. MultiLoc2-LowRes The animal prediction performance of MultiLoc2-LowRes is reduced by only one per cent regarding to AVG and ACC when predicting three classes and by two and four percent when predicting four classes. The reason is that more nuclear proteins are wrongly predicted if we discard the GO terms. The fungal prediction performance is almost unchanged which is mainly caused by the fact that on average only 34% of the fungal proteins are annotated 7
8 with GO terms by InterProScan. The plant ACCs are decreased from 83% to 80% and from 76% to 71% for the prediction of three classes and four classes respectively. This is caused by the dropping sensitivity of the nuclear proteins (from 91% to 77%). The AVGs are reduced by nine per cent which seems to be a very significant performance lost at the first view. The reason is that the SP sensitivity is reduced from 83% to 50%. Only two SP proteins are additionally wrong predicted if we neglect the GO annotation. However, these two proteins have a large impact on the AVGs because the SP cluster contains only six proteins overall. MultiLoc2-HighRes Similar to the MultiLoc2-LowRes, the fungal prediction performance of MultiLoc2-HighRes is almost unchanged. This is the same for the plants in case of the prediction of four classes. The performance reduction by three per cent for the prediction of five plant classes is also moderate. However, very different to MultiLoc2-LowRes, the animal ACCs are reduced by nine percent and 11% respectively. We analyzed the additionally wrong predicted proteins and found out that this was caused by a failure in the clustering procedure performed by the curators of the data set [Casadio et al., 2008]. The nuclear data set contains 56 proteins of the protamine-p1 family. Each protein represents one cluster which biases the prediction towards this overrepresented protein class. The reason for the failed clustering could be the relatively short sequences of the proteins between 50 and 60 amino acids. Therefore, we reclustered the nuclear proteins using BLASTClust and 30% sequence identity. Now, the 56 proteins of the protamine-p1 family are clustered and the new number of clusters is 186 for the nuclear proteins and 277 for the nu/cy class. The comparison of the animal results based on the reclustered nuclear proteins delivers only a slightly performance reduction. We also applied BLASTClust on all other localizations and always received either the same number of clusters or a few more which indicates that the described clustering problem did not appear for the remaining classes. References Sun,J., Xu,J., Liu,Z., Liu,Q., Zhao,A., Shi,T., Li,Y. (2005) Refined phylogenetic profiles method for predicting protein-protein interactions., Bioinformatics, 16, Casadio,R., Martelli,P.,L., Pierleoni,A. (2008) The prediction of protein subcellular localization from sequence: a shortcut to functional genome annotation, Brief Funct Genomic Proteomic., 7(1),
9 Table 7: Comparison of the localization-specific prediction results of the MultiLoc2 predictors using BacelLo independent dataset (BaCelLo IDS) Version Loc Nr ML2-LowRes ML2-LowRes* ML2-HighRes ML2-HighRes* SE MCC SE MCC SE MCC SE MCC Animals SP mi nu cy nu/cy Animals + SP mi nu cy nu/cy Fungi SP mi nu cy nu/cy Plants SP mi ch nu cy nu/cy The sensitivity (SE) and Matthews correlation coefficient (MCC) of MultiLoc2 (ML2) are listed for each localization (Loc). The number of clusters (Nr) per localization is also shown. The results for MultiLoc2-LowRes* and MultiLoc2-HighRes* are obtained by simulating that for all test proteins no GO term is available. The Animals + dataset was obtained by reclustering the nuclear proteins from the original animals dataset. Changes in performance are highlighted in italic. Table 8: Comparison of the overall performance results of the MultiLoc2 predictors using BacelLo independent dataset (BaCelLo IDS) Version Classes ML2-LowRes ML2-LowRes* ML2-HighRes ML2-HighRes* Animals 3 93 (93) 92 (92) 87 (89) 83 (80) 4 80 (73) 78 (69) 75 (68) 70 (57) Animals (93) 91 (92) 87 (89) 87 (88) 4 78 (70) 75 (66) 73 (67) 72 (64) Fungi 3 79 (87) 79 (88) 71 (78) 72 (77) 4 66 (60) 65 (60) 59 (52) 58 (51) Plants 4 80 (83) 71 (80) 74 (70) 74 (70) 5 72 (76) 63 (71) 65 (62) 62 (59) The average sensitivity and the overall accuracy (in parenthesis) of MultiLoc2 (ML2) for the prediction of three and four classes for animals and fungi and four and five classes for plants are shown. The results for MultiLoc2-LowRes* and MultiLoc2-HighRes* are obtained by simulating that for all test proteins no GO term is available. The Animals + dataset was obtained by reclustering the nuclear proteins from the original animals dataset. Changes in performance are highlighted in italic. 9
Whole-genome based Archaea phylogeny and taxonomy: A composition vector approach
 Article SPECIAL TOPIC Bioinformatics August 2010 Vol.55 No.22: 2323 2328 doi: 10.1007/s11434-010-3008-8 Whole-genome based Archaea phylogeny and taxonomy: A composition vector approach SUN JianDong 1*,
Article SPECIAL TOPIC Bioinformatics August 2010 Vol.55 No.22: 2323 2328 doi: 10.1007/s11434-010-3008-8 Whole-genome based Archaea phylogeny and taxonomy: A composition vector approach SUN JianDong 1*,
LIFE AT THE LIMITS - EXTREME ENVIRONMENTS "HOW CAN THEY SURVIVE?"
 COMPARATIVE MICROBIAL GENOMICS ANALYSIS LIFE AT THE LIMITS - EXTREME ENVIRONMENTS "HOW CAN THEY SURVIVE?" Group 5 Kanokwan Inban Lakkhana Khanhayuwa Parwin Tantayapirak Introduction 2 Source: http://pathmicro.med.sc.edu/fox/evol.gif
COMPARATIVE MICROBIAL GENOMICS ANALYSIS LIFE AT THE LIMITS - EXTREME ENVIRONMENTS "HOW CAN THEY SURVIVE?" Group 5 Kanokwan Inban Lakkhana Khanhayuwa Parwin Tantayapirak Introduction 2 Source: http://pathmicro.med.sc.edu/fox/evol.gif
A Method for Identification of Selenoprotein Genes in Archaeal Genomes
 Method A Method for Identification of Selenoprotein Genes in Archaeal Genomes Mingfeng Li, Yanzhao Huang, and Yi Xiao* Biomolecular Physics and Modeling Group, Department of Physics, Huazhong University
Method A Method for Identification of Selenoprotein Genes in Archaeal Genomes Mingfeng Li, Yanzhao Huang, and Yi Xiao* Biomolecular Physics and Modeling Group, Department of Physics, Huazhong University
DNA sequence analysis using Markov chain models
 DNA sequence analysis using Markov chain models Boris Ryabko Siberian University of Telecommunication an Informatics, Institute of Computational Technologies, Siberian Branch of RAS Novosibirsk E-mail:
DNA sequence analysis using Markov chain models Boris Ryabko Siberian University of Telecommunication an Informatics, Institute of Computational Technologies, Siberian Branch of RAS Novosibirsk E-mail:
Procedure to Create NCBI KOGS
 Procedure to Create NCBI KOGS full details in: Tatusov et al (2003) BMC Bioinformatics 4:41. 1. Detect and mask typical repetitive domains Reason: masking prevents spurious lumping of non-orthologs based
Procedure to Create NCBI KOGS full details in: Tatusov et al (2003) BMC Bioinformatics 4:41. 1. Detect and mask typical repetitive domains Reason: masking prevents spurious lumping of non-orthologs based
Building Phylogenetic Trees Based on Biochemical Pathways
 Building Phylogenetic Trees Based on Biochemical Pathways Research Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Science in Computer Science Adi Mano Submitted
Building Phylogenetic Trees Based on Biochemical Pathways Research Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Science in Computer Science Adi Mano Submitted
Using N-terminal targeting sequences, amino acid composition, and sequence motifs for predicting protein subcellular localization
 Using N-terminal targeting sequences, amino acid composition, and sequence motifs for predicting protein subcellular localization Annette Höglund, Pierre Dönnes, Torsten Blum, Hans-Werner Adolph, and Oliver
Using N-terminal targeting sequences, amino acid composition, and sequence motifs for predicting protein subcellular localization Annette Höglund, Pierre Dönnes, Torsten Blum, Hans-Werner Adolph, and Oliver
Supplementary Material and Figures
 Supplementary Material and Figures Nutrient transport suggests an evolutionary basis for charged archaeal surface layer proteins Po-Nan Li et al. Supplementary Figure S1. Numerical convergence of the simulation.
Supplementary Material and Figures Nutrient transport suggests an evolutionary basis for charged archaeal surface layer proteins Po-Nan Li et al. Supplementary Figure S1. Numerical convergence of the simulation.
Review Article Diversity of the DNA Replication System in the Archaea Domain
 Archaea, Article ID 675946, 15 pages http://dx.doi.org/10.1155/2014/675946 Review Article Diversity of the DNA Replication System in the Archaea Domain Felipe Sarmiento, 1 Feng Long, 1 Isaac Cann, 2 and
Archaea, Article ID 675946, 15 pages http://dx.doi.org/10.1155/2014/675946 Review Article Diversity of the DNA Replication System in the Archaea Domain Felipe Sarmiento, 1 Feng Long, 1 Isaac Cann, 2 and
Metagenomics of Kamchatkan hot spring filaments reveal two new major (hyper)thermophilic lineages related to Thaumarchaeota
 Research in Microbiology 164 (2013) 425e438 www.elsevier.com/locate/resmic Metagenomics of Kamchatkan hot spring filaments reveal two new major (hyper)thermophilic lineages related to Thaumarchaeota Laura
Research in Microbiology 164 (2013) 425e438 www.elsevier.com/locate/resmic Metagenomics of Kamchatkan hot spring filaments reveal two new major (hyper)thermophilic lineages related to Thaumarchaeota Laura
Unicellular Marine Organisms. Chapter 4
 Unicellular Marine Organisms Chapter 4 The Cellular Structure of Life: Review Cell wall: firm, fairly rigid structure located outside the plasma membrane of plants, fungi, most bacteria, and some protists;
Unicellular Marine Organisms Chapter 4 The Cellular Structure of Life: Review Cell wall: firm, fairly rigid structure located outside the plasma membrane of plants, fungi, most bacteria, and some protists;
Supplementary Materials for R3P-Loc Web-server
 Supplementary Materials for R3P-Loc Web-server Shibiao Wan and Man-Wai Mak email: shibiao.wan@connect.polyu.hk, enmwmak@polyu.edu.hk June 2014 Back to R3P-Loc Server Contents 1 Introduction to R3P-Loc
Supplementary Materials for R3P-Loc Web-server Shibiao Wan and Man-Wai Mak email: shibiao.wan@connect.polyu.hk, enmwmak@polyu.edu.hk June 2014 Back to R3P-Loc Server Contents 1 Introduction to R3P-Loc
Midterm Exam #1 : In-class questions! MB 451 Microbial Diversity : Spring 2015!
 Midterm Exam #1 : In-class questions MB 451 Microbial Diversity : Spring 2015 Honor pledge: I have neither given nor received unauthorized aid on this test. Signed : Name : Date : TOTAL = 45 points 1.
Midterm Exam #1 : In-class questions MB 451 Microbial Diversity : Spring 2015 Honor pledge: I have neither given nor received unauthorized aid on this test. Signed : Name : Date : TOTAL = 45 points 1.
Supplementary Materials for mplr-loc Web-server
 Supplementary Materials for mplr-loc Web-server Shibiao Wan and Man-Wai Mak email: shibiao.wan@connect.polyu.hk, enmwmak@polyu.edu.hk June 2014 Back to mplr-loc Server Contents 1 Introduction to mplr-loc
Supplementary Materials for mplr-loc Web-server Shibiao Wan and Man-Wai Mak email: shibiao.wan@connect.polyu.hk, enmwmak@polyu.edu.hk June 2014 Back to mplr-loc Server Contents 1 Introduction to mplr-loc
Quantitative Measurement of Genome-wide Protein Domain Co-occurrence of Transcription Factors
 Quantitative Measurement of Genome-wide Protein Domain Co-occurrence of Transcription Factors Arli Parikesit, Peter F. Stadler, Sonja J. Prohaska Bioinformatics Group Institute of Computer Science University
Quantitative Measurement of Genome-wide Protein Domain Co-occurrence of Transcription Factors Arli Parikesit, Peter F. Stadler, Sonja J. Prohaska Bioinformatics Group Institute of Computer Science University
Horizontal gene transfer and archaeal origin of deoxyhypusine synthase homologous genes in bacteria
 Gene 330 (2004) 169 176 www.elsevier.com/locate/gene Horizontal gene transfer and archaeal origin of deoxyhypusine synthase homologous genes in bacteria Céline Brochier a, Purificación López-García b,
Gene 330 (2004) 169 176 www.elsevier.com/locate/gene Horizontal gene transfer and archaeal origin of deoxyhypusine synthase homologous genes in bacteria Céline Brochier a, Purificación López-García b,
Introductory Microbiology Dr. Hala Al Daghistani
 Introductory Microbiology Dr. Hala Al Daghistani Why Study Microbes? Microbiology is the branch of biological sciences concerned with the study of the microbes. 1. Microbes and Man in Sickness and Health
Introductory Microbiology Dr. Hala Al Daghistani Why Study Microbes? Microbiology is the branch of biological sciences concerned with the study of the microbes. 1. Microbes and Man in Sickness and Health
UNIT 3 CP BIOLOGY: Cell Structure
 UNIT 3 CP BIOLOGY: Cell Structure Page CP: CHAPTER 3, Sections 1-3; HN: CHAPTER 7, Sections 1-2 Standard B-2: The student will demonstrate an understanding of the structure and function of cells and their
UNIT 3 CP BIOLOGY: Cell Structure Page CP: CHAPTER 3, Sections 1-3; HN: CHAPTER 7, Sections 1-2 Standard B-2: The student will demonstrate an understanding of the structure and function of cells and their
Archaeal signal peptidases
 Microbiology (2007), 153, 305 314 DOI 10.1099/mic.0.2006/003087-0 Mini-Review Correspondence Ken F. Jarrell jarrellk@post.queensu.ca Archaeal signal peptidases Sandy Y. M. Ng, Bonnie Chaban, David J. VanDyke
Microbiology (2007), 153, 305 314 DOI 10.1099/mic.0.2006/003087-0 Mini-Review Correspondence Ken F. Jarrell jarrellk@post.queensu.ca Archaeal signal peptidases Sandy Y. M. Ng, Bonnie Chaban, David J. VanDyke
Cell Organelles Tutorial
 1 Name: Cell Organelles Tutorial TEK 7.12D: Differentiate between structure and function in plant and animal cell organelles, including cell membrane, cell wall, nucleus, cytoplasm, mitochondrion, chloroplast,
1 Name: Cell Organelles Tutorial TEK 7.12D: Differentiate between structure and function in plant and animal cell organelles, including cell membrane, cell wall, nucleus, cytoplasm, mitochondrion, chloroplast,
Topic 3: Cells Ch. 6. Microscopes pp Microscopes. Microscopes. Microscopes. Microscopes
 Topic 3: Cells Ch. 6 -All life is composed of cells and all cells have a plasma membrane, cytoplasm, and DNA. pp.105-107 - The development of the microscope was the key to understanding that all living
Topic 3: Cells Ch. 6 -All life is composed of cells and all cells have a plasma membrane, cytoplasm, and DNA. pp.105-107 - The development of the microscope was the key to understanding that all living
Identification and Genomic Analysis of Transcription Factors in Archaeal Genomes Exemplifies Their Functional Architecture and Evolutionary Origin
 Identification and Genomic Analysis of Transcription Factors in Archaeal Genomes Exemplifies Their Functional Architecture and Evolutionary Origin Ernesto Pérez-Rueda*,1 and Sarath Chandra Janga*,2 1 Departamento
Identification and Genomic Analysis of Transcription Factors in Archaeal Genomes Exemplifies Their Functional Architecture and Evolutionary Origin Ernesto Pérez-Rueda*,1 and Sarath Chandra Janga*,2 1 Departamento
Function and Illustration. Nucleus. Nucleolus. Cell membrane. Cell wall. Capsule. Mitochondrion
 Intro to Organelles Name: Block: Organelles are small structures inside cells. They are often covered in membranes. Each organelle has a job to do in the cell. Their name means little organ. Just like
Intro to Organelles Name: Block: Organelles are small structures inside cells. They are often covered in membranes. Each organelle has a job to do in the cell. Their name means little organ. Just like
Introduction to Cells- Stations Lab
 Introduction to Cells- Stations Lab Station 1: Microscopes allow scientists to study cells. Microscopes: How do light microscopes differ from electron microscopes? (How does each work? How much can each
Introduction to Cells- Stations Lab Station 1: Microscopes allow scientists to study cells. Microscopes: How do light microscopes differ from electron microscopes? (How does each work? How much can each
Advanced Cell Biology. Lecture 2
 Advanced Cell Biology. Lecture 2 Alexey Shipunov Minot State University January 13, 2012 Outline Questions and answers Microscopy Prokaryotic and eukaryotic cells Outline Questions and answers Microscopy
Advanced Cell Biology. Lecture 2 Alexey Shipunov Minot State University January 13, 2012 Outline Questions and answers Microscopy Prokaryotic and eukaryotic cells Outline Questions and answers Microscopy
Genome feature exploration using hyperbolic Self-Organising Maps
 Genome feature exploration using hyperbolic Self-Organising Maps Christian Martin, Naryttza N. Diaz, Jörg Ontrup and Tim W. Nattkemper Center for Biotechnology Technical Faculty, AG Applied Neuroinformatics
Genome feature exploration using hyperbolic Self-Organising Maps Christian Martin, Naryttza N. Diaz, Jörg Ontrup and Tim W. Nattkemper Center for Biotechnology Technical Faculty, AG Applied Neuroinformatics
Origins of Life. Fundamental Properties of Life. Conditions on Early Earth. Evolution of Cells. The Tree of Life
 The Tree of Life Chapter 26 Origins of Life The Earth formed as a hot mass of molten rock about 4.5 billion years ago (BYA) -As it cooled, chemically-rich oceans were formed from water condensation Life
The Tree of Life Chapter 26 Origins of Life The Earth formed as a hot mass of molten rock about 4.5 billion years ago (BYA) -As it cooled, chemically-rich oceans were formed from water condensation Life
The origins and early evolution of DNA mismatch repair genes multiple horizontal gene transfers and co-evolution
 Published online 26 October 2007 Nucleic Acids Research, 2007, Vol. 35, No. 22 7591 7603 doi:10.1093/nar/gkm921 The origins and early evolution of DNA mismatch repair genes multiple horizontal gene transfers
Published online 26 October 2007 Nucleic Acids Research, 2007, Vol. 35, No. 22 7591 7603 doi:10.1093/nar/gkm921 The origins and early evolution of DNA mismatch repair genes multiple horizontal gene transfers
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11541 Supplementary Figure 1. Genome-wide analysis of the phylogenetic relationships Phylogenetic tree of NDH-2s from Prokaryota, Protista, Plantae, Fungi and Metazoa. The WWW.NATURE.COM/NATURE
doi:10.1038/nature11541 Supplementary Figure 1. Genome-wide analysis of the phylogenetic relationships Phylogenetic tree of NDH-2s from Prokaryota, Protista, Plantae, Fungi and Metazoa. The WWW.NATURE.COM/NATURE
CELL STRUCTURE & FUNCTION
 7-1 Life Is Cellular CELL STRUCTURE & FUNCTION Copyright Pearson Prentice Hall The Discovery of the Cell 1665: Robert Hooke used an early compound microscope to look at a thin slice of cork. Cork looked
7-1 Life Is Cellular CELL STRUCTURE & FUNCTION Copyright Pearson Prentice Hall The Discovery of the Cell 1665: Robert Hooke used an early compound microscope to look at a thin slice of cork. Cork looked
MARLIA SINGGIH WIBOWO SCHOOL OF PHARMACY INSTITUT TEKNOLOGI BANDUNG
 MARLIA SINGGIH WIBOWO SCHOOL OF PHARMACY INSTITUT TEKNOLOGI BANDUNG Farmakope Indonesia edisi IV, 1995. USP 32, 2009. Michael T Madigan, John M Martinko, Brock Biology of Microorganisms, 11 th edition,
MARLIA SINGGIH WIBOWO SCHOOL OF PHARMACY INSTITUT TEKNOLOGI BANDUNG Farmakope Indonesia edisi IV, 1995. USP 32, 2009. Michael T Madigan, John M Martinko, Brock Biology of Microorganisms, 11 th edition,
A Structural Equation Model Study of Shannon Entropy Effect on CG content of Thermophilic 16S rrna and Bacterial Radiation Repair Rec-A Gene Sequences
 A Structural Equation Model Study of Shannon Entropy Effect on CG content of Thermophilic 16S rrna and Bacterial Radiation Repair Rec-A Gene Sequences T. Holden, P. Schneider, E. Cheung, J. Prayor, R.
A Structural Equation Model Study of Shannon Entropy Effect on CG content of Thermophilic 16S rrna and Bacterial Radiation Repair Rec-A Gene Sequences T. Holden, P. Schneider, E. Cheung, J. Prayor, R.
Cell Organelles. Wednesday, October 22, 14
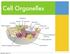 Cell Organelles Cell/Plasma Membrane ALL cells have a cell membrane It is the layer that surrounds the cell and controls what goes in and out Bacteria (Prokaryotic Cell) Cell/Plasma Membrane ALL cells
Cell Organelles Cell/Plasma Membrane ALL cells have a cell membrane It is the layer that surrounds the cell and controls what goes in and out Bacteria (Prokaryotic Cell) Cell/Plasma Membrane ALL cells
Introduction to cells
 Almen Cellebiologi Introduction to cells 1. Unity and diversity of cells 2. Microscopes and visualization of cells 3. Prokaryotic cells, eubacteria and archaea 4. Eucaryotic cells, nucleus, mitochondria
Almen Cellebiologi Introduction to cells 1. Unity and diversity of cells 2. Microscopes and visualization of cells 3. Prokaryotic cells, eubacteria and archaea 4. Eucaryotic cells, nucleus, mitochondria
Visualization of multiple alignments, phylogenies and gene family evolution
 nature methods Visualization of multiple alignments, phylogenies and gene family evolution James B Procter, Julie Thompson, Ivica Letunic, Chris Creevey, Fabrice Jossinet & Geoffrey J Barton Supplementary
nature methods Visualization of multiple alignments, phylogenies and gene family evolution James B Procter, Julie Thompson, Ivica Letunic, Chris Creevey, Fabrice Jossinet & Geoffrey J Barton Supplementary
Cell Structure Vocab. Plasma membrane. Vacuole. Cell wall. Nuclear envelope. Chloroplast. Nucleus. Cytoskeleton. Nucleolus. Cytoplasm.
 The Cell Cell Structure Vocab Plasma membrane Nuclear envelope Nucleus Nucleolus Cytoplasm Mitochondria Endoplasmic reticulum Golgi apparatus Lysosome Vacuole Cell wall Chloroplast Cytoskeleton Centriole
The Cell Cell Structure Vocab Plasma membrane Nuclear envelope Nucleus Nucleolus Cytoplasm Mitochondria Endoplasmic reticulum Golgi apparatus Lysosome Vacuole Cell wall Chloroplast Cytoskeleton Centriole
JEPSLD: A JUDGMENTAL EUKARYOTIC PROTEIN SUBCELLULAR LOCATION DATABASE. Sanjeev Patra
 JEPSLD: A JUDGMENTAL EUKARYOTIC PROTEIN SUBCELLULAR LOCATION DATABASE by Sanjeev Patra A thesis submitted to the Faculty of the University of Delaware in partial fulfillment of the requirements for the
JEPSLD: A JUDGMENTAL EUKARYOTIC PROTEIN SUBCELLULAR LOCATION DATABASE by Sanjeev Patra A thesis submitted to the Faculty of the University of Delaware in partial fulfillment of the requirements for the
Molecular biology and biochemistry of archaeal DNA replication
 Molecular biology and biochemistry of archaeal DNA replication Isaac Cann Institute for Universal Biology, NASA Astrobiology Institute Department of Animal Science & Department of Microbiology University
Molecular biology and biochemistry of archaeal DNA replication Isaac Cann Institute for Universal Biology, NASA Astrobiology Institute Department of Animal Science & Department of Microbiology University
Uncultured Archaea in a hydrothermal microbial assemblage: phylogenetic diversity and characterization of a genome fragment from a euryarchaeote
 Uncultured Archaea in a hydrothermal microbial assemblage: phylogenetic diversity and characterization of a genome fragment from a euryarchaeote Hélène Moussard, David Moreira 2, Marie-Anne Cambon-Bonavita,
Uncultured Archaea in a hydrothermal microbial assemblage: phylogenetic diversity and characterization of a genome fragment from a euryarchaeote Hélène Moussard, David Moreira 2, Marie-Anne Cambon-Bonavita,
Genome-wide multilevel spatial interactome model of rice
 Sino-German Workshop on Multiscale Spatial Computational Systems Biology, Beijing, Oct 8-12, 2015 Genome-wide multilevel spatial interactome model of rice Ming CHEN ( 陈铭 ) mchen@zju.edu.cn College of Life
Sino-German Workshop on Multiscale Spatial Computational Systems Biology, Beijing, Oct 8-12, 2015 Genome-wide multilevel spatial interactome model of rice Ming CHEN ( 陈铭 ) mchen@zju.edu.cn College of Life
Species Level Deconvolution of Metagenome Assemblies with Hi C Based Contact Probability Maps
 Species Level Deconvolution of Metagenome Assemblies with Hi C Based Contact Probability Maps Joshua N. Burton*, Ivan Liachko*, Maitreya J. Dunham, Jay Shendure Department of Genome Sciences, University
Species Level Deconvolution of Metagenome Assemblies with Hi C Based Contact Probability Maps Joshua N. Burton*, Ivan Liachko*, Maitreya J. Dunham, Jay Shendure Department of Genome Sciences, University
Evolutionary Use of Domain Recombination: A Distinction. Between Membrane and Soluble Proteins
 1 Evolutionary Use of Domain Recombination: A Distinction Between Membrane and Soluble Proteins Yang Liu, Mark Gerstein, Donald M. Engelman Department of Molecular Biophysics and Biochemistry, Yale University,
1 Evolutionary Use of Domain Recombination: A Distinction Between Membrane and Soluble Proteins Yang Liu, Mark Gerstein, Donald M. Engelman Department of Molecular Biophysics and Biochemistry, Yale University,
Cell Alive Homeostasis Plants Animals Fungi Bacteria. Loose DNA DNA Nucleus Membrane-Bound Organelles Humans
 UNIT 3: The Cell DAYSHEET 45: Introduction to Cellular Organelles Name: Biology I Date: Bellringer: Place the words below into the correct space on the Venn Diagram: Cell Alive Homeostasis Plants Animals
UNIT 3: The Cell DAYSHEET 45: Introduction to Cellular Organelles Name: Biology I Date: Bellringer: Place the words below into the correct space on the Venn Diagram: Cell Alive Homeostasis Plants Animals
The early evolution of eukaryotes with special reference to ribosome export factors. Hajime Ohyanagi. Doctor of Philosophy
 The early evolution of eukaryotes with special reference to ribosome export factors Hajime Ohyanagi Doctor of Philosophy Department of Genetics School of Life Science The Graduate University for Advanced
The early evolution of eukaryotes with special reference to ribosome export factors Hajime Ohyanagi Doctor of Philosophy Department of Genetics School of Life Science The Graduate University for Advanced
Unit 2: Cells. Students will understand that the organs in an organism are made of cells that have structures & perform specific life functions
 Unit 2: Cells Students will understand that the organs in an organism are made of cells that have structures & perform specific life functions Vocabulary Cell Chloroplast Tissue Cell wall Organ Lysosome
Unit 2: Cells Students will understand that the organs in an organism are made of cells that have structures & perform specific life functions Vocabulary Cell Chloroplast Tissue Cell wall Organ Lysosome
A D A E J (L) J(s) K L
 I A E A D G C B C F G E F H B J (L) H K L J(s) Animal Cells Less Support Needed Large, multicellular organisms made of animal cells often have support systems such as bones or exoskeletons Plant and Animal
I A E A D G C B C F G E F H B J (L) H K L J(s) Animal Cells Less Support Needed Large, multicellular organisms made of animal cells often have support systems such as bones or exoskeletons Plant and Animal
A putative viral defence mechanism in archaeal cells
 Archaea 2, 59 72 2006 Heron Publishing Victoria, Canada A putative viral defence mechanism in archaeal cells REIDUN K. LILLESTØL, 1 PETER REDDER, 1 ROGER A. GARRETT 1 and KIM BRÜGGER 1,2 1 Institute of
Archaea 2, 59 72 2006 Heron Publishing Victoria, Canada A putative viral defence mechanism in archaeal cells REIDUN K. LILLESTØL, 1 PETER REDDER, 1 ROGER A. GARRETT 1 and KIM BRÜGGER 1,2 1 Institute of
SUPPLEMENTARY INFORMATION
 doi:.38/nature464 Table S1: Primers used to amplify a 3 bp part of the Acidianus A1-3 CS 2 gene (CS 2-1bF, 525bR, 40bF and 333bR), as well as the C- and N-terminal ends by inverse PCR (F4inv and R3inv);
doi:.38/nature464 Table S1: Primers used to amplify a 3 bp part of the Acidianus A1-3 CS 2 gene (CS 2-1bF, 525bR, 40bF and 333bR), as well as the C- and N-terminal ends by inverse PCR (F4inv and R3inv);
the cell history cell theory basic properties of cells chapter 1
 history the cell chapter 1 invention of the microscope hans and zacharias janssen - 1590 galileo galilei - 1609 giovanni faber robert hooke -1665 - Micrographia anton van leewenhoek - 1676 schleiden &
history the cell chapter 1 invention of the microscope hans and zacharias janssen - 1590 galileo galilei - 1609 giovanni faber robert hooke -1665 - Micrographia anton van leewenhoek - 1676 schleiden &
Living Things. Chapter 2
 Living Things Chapter 2 Section 1: What is Life? 6 Characteristics of Living Things: 1. cellular vs. cellular 2. Composed of 5 essential chemicals 1. 2. - main energy source 3. 4. (Fats) 5. - genetic material
Living Things Chapter 2 Section 1: What is Life? 6 Characteristics of Living Things: 1. cellular vs. cellular 2. Composed of 5 essential chemicals 1. 2. - main energy source 3. 4. (Fats) 5. - genetic material
Introduction to Bioinformatics Integrated Science, 11/9/05
 1 Introduction to Bioinformatics Integrated Science, 11/9/05 Morris Levy Biological Sciences Research: Evolutionary Ecology, Plant- Fungal Pathogen Interactions Coordinator: BIOL 495S/CS490B/STAT490B Introduction
1 Introduction to Bioinformatics Integrated Science, 11/9/05 Morris Levy Biological Sciences Research: Evolutionary Ecology, Plant- Fungal Pathogen Interactions Coordinator: BIOL 495S/CS490B/STAT490B Introduction
Bio 111 Study Guide Chapter 6 Tour of the Cell
 Bio 111 Study Guide Chapter 6 Tour of the Cell BEFORE CLASS: Reading: Read the whole chapter from p. 93-121, mostly skimming Concept 6.1 on microscopy. Figure 6.8 on pp. 100-101 is really helpful in showing
Bio 111 Study Guide Chapter 6 Tour of the Cell BEFORE CLASS: Reading: Read the whole chapter from p. 93-121, mostly skimming Concept 6.1 on microscopy. Figure 6.8 on pp. 100-101 is really helpful in showing
= Monera. Taxonomy. Domains (3) BIO162 Page Baluch. Taxonomy: classifying and organizing life
 Taxonomy BIO162 Page Baluch Taxonomy: classifying and organizing life species Genus Family Order Class Phylum Kingdom Spaghetti Good For Over Came Phillip King Domains (3) DOMAINS 1. Bacteria 2. Archea
Taxonomy BIO162 Page Baluch Taxonomy: classifying and organizing life species Genus Family Order Class Phylum Kingdom Spaghetti Good For Over Came Phillip King Domains (3) DOMAINS 1. Bacteria 2. Archea
9/11/18. Molecular and Cellular Biology. 3. The Cell From Genes to Proteins. key processes
 Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Biology 160 Cell Lab. Name Lab Section: 1:00pm 3:00 pm. Student Learning Outcomes:
 Biology 160 Cell Lab Name Lab Section: 1:00pm 3:00 pm Student Learning Outcomes: Upon completion of today s lab you will be able to do the following: Properly use a compound light microscope Discuss the
Biology 160 Cell Lab Name Lab Section: 1:00pm 3:00 pm Student Learning Outcomes: Upon completion of today s lab you will be able to do the following: Properly use a compound light microscope Discuss the
CELL PART Expanded Definition Cell Structure Illustration Function Summary Location ALL CELLS DNA Common in Animals Uncommon in Plants Lysosome
 CELL PART Expanded Definition Cell Structure Illustration Function Summary Location is the material that contains the Carry genetic ALL CELLS information that determines material inherited characteristics.
CELL PART Expanded Definition Cell Structure Illustration Function Summary Location is the material that contains the Carry genetic ALL CELLS information that determines material inherited characteristics.
Variation, Evolution, and Correlation Analysis of C+G Content and Genome or Chromosome Size in Different Kingdoms and Phyla
 Variation, Evolution, and Correlation Analysis of C+G Content and Genome or Chromosome Size in Different Kingdoms and Phyla Xiu-Qing Li 1 *, Donglei Du 2 1 Molecular Genetics Laboratory, Potato Research
Variation, Evolution, and Correlation Analysis of C+G Content and Genome or Chromosome Size in Different Kingdoms and Phyla Xiu-Qing Li 1 *, Donglei Du 2 1 Molecular Genetics Laboratory, Potato Research
Parts of the Cell book pgs
 Parts of the Cell book pgs. 12-18 Animal Cell Cytoplasm Cell Membrane Go to Section: Eukaryotic Cell: Organelles & Functions 1. Cell Membrane (Nickname: skin ) Function: A protective layer that covers
Parts of the Cell book pgs. 12-18 Animal Cell Cytoplasm Cell Membrane Go to Section: Eukaryotic Cell: Organelles & Functions 1. Cell Membrane (Nickname: skin ) Function: A protective layer that covers
Unit 3: Cells. Objective: To be able to compare and contrast the differences between Prokaryotic and Eukaryotic Cells.
 Unit 3: Cells Objective: To be able to compare and contrast the differences between Prokaryotic and Eukaryotic Cells. The Cell Theory All living things are composed of cells (unicellular or multicellular).
Unit 3: Cells Objective: To be able to compare and contrast the differences between Prokaryotic and Eukaryotic Cells. The Cell Theory All living things are composed of cells (unicellular or multicellular).
Chapter 7.2. Cell Structure
 Chapter 7.2 Cell Structure Daily Objectives Describe the structure and function of the cell nucleus. Describe the function and structure of membrane bound organelles found within the cell. Describe the
Chapter 7.2 Cell Structure Daily Objectives Describe the structure and function of the cell nucleus. Describe the function and structure of membrane bound organelles found within the cell. Describe the
2. Cellular and Molecular Biology
 2. Cellular and Molecular Biology 2.1 Cell Structure 2.2 Transport Across Cell Membranes 2.3 Cellular Metabolism 2.4 DNA Replication 2.5 Cell Division 2.6 Biosynthesis 2.1 Cell Structure What is a cell?
2. Cellular and Molecular Biology 2.1 Cell Structure 2.2 Transport Across Cell Membranes 2.3 Cellular Metabolism 2.4 DNA Replication 2.5 Cell Division 2.6 Biosynthesis 2.1 Cell Structure What is a cell?
prokaryotic eukaryotic
 Cell Basics Two Basic Cell Types All cells are either prokaryotic or eukaryotic Prokaryotic Cells a.k.a. Bacteria Prokaryotes, which includes all bacteria. They are the simplest cellular organisms. They
Cell Basics Two Basic Cell Types All cells are either prokaryotic or eukaryotic Prokaryotic Cells a.k.a. Bacteria Prokaryotes, which includes all bacteria. They are the simplest cellular organisms. They
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools. Evaluation. Course Homepage.
 CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools Giri Narasimhan ECS 389; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs06.html 1/12/06 CAP5510/CGS5166 1 Evaluation
CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools Giri Narasimhan ECS 389; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs06.html 1/12/06 CAP5510/CGS5166 1 Evaluation
The Basic Unit of Life Copyright Amy Brown Science Stuff
 Cell Structure and Function The Basic Unit of Life Copyright Amy Brown Science Stuff The Discovery of the Cell Robert Hooke looked at thin slices of cork (plant cells) under the microscope. Named it a
Cell Structure and Function The Basic Unit of Life Copyright Amy Brown Science Stuff The Discovery of the Cell Robert Hooke looked at thin slices of cork (plant cells) under the microscope. Named it a
The Cell. The basic unit of all living things
 The Cell The basic unit of all living things 1 Robert Hooke was the first to name the cell (1665) 2 The Cell Theory The cell is the unit of Structure of all living things. The cell is the unit of Function
The Cell The basic unit of all living things 1 Robert Hooke was the first to name the cell (1665) 2 The Cell Theory The cell is the unit of Structure of all living things. The cell is the unit of Function
Evolution of complete proteomes: guanine-cytosine pressure, phylogeny and environmental influences blend the proteomic architecture
 Chen et al. BMC Evolutionary Biology 213, 13:219 RESEARCH ARTICLE Open Access Evolution of complete proteomes: guanine-cytosine pressure, phylogeny and environmental influences blend the proteomic architecture
Chen et al. BMC Evolutionary Biology 213, 13:219 RESEARCH ARTICLE Open Access Evolution of complete proteomes: guanine-cytosine pressure, phylogeny and environmental influences blend the proteomic architecture
Shibiao Wan and Man-Wai Mak December 2013 Back to HybridGO-Loc Server
 Shibiao Wan and Man-Wai Mak December 2013 Back to HybridGO-Loc Server Contents 1 Functions of HybridGO-Loc Server 2 1.1 Webserver Interface....................................... 2 1.2 Inputing Protein
Shibiao Wan and Man-Wai Mak December 2013 Back to HybridGO-Loc Server Contents 1 Functions of HybridGO-Loc Server 2 1.1 Webserver Interface....................................... 2 1.2 Inputing Protein
The Cell Theory. Prokaryotic (Pre) ( Nucleus) Cells 10/28/2013. Types of Cells. All living things have cells. Always single- celled lacks organelles
 Types of Cells Cells 1. I can explain the cell theory The Cell Theory 1. All living things are composed of one or more cells. 2. Cells are the basic units of structure and function. 3. Cells are produced
Types of Cells Cells 1. I can explain the cell theory The Cell Theory 1. All living things are composed of one or more cells. 2. Cells are the basic units of structure and function. 3. Cells are produced
Chapter 26 Phylogeny and the Tree of Life
 Chapter 26 Phylogeny and the Tree of Life Chapter focus Shifting from the process of how evolution works to the pattern evolution produces over time. Phylogeny Phylon = tribe, geny = genesis or origin
Chapter 26 Phylogeny and the Tree of Life Chapter focus Shifting from the process of how evolution works to the pattern evolution produces over time. Phylogeny Phylon = tribe, geny = genesis or origin
microrna Studies Chen-Hanson Ting SVFIG June 23, 2018
 microrna Studies Chen-Hanson Ting SVFIG June 23, 2018 Summary MicroRNA (mirna) Species and organisms studied mirna in mitocondria Huge genome files mirna in human Chromosome 1 mirna in bacteria Tools used
microrna Studies Chen-Hanson Ting SVFIG June 23, 2018 Summary MicroRNA (mirna) Species and organisms studied mirna in mitocondria Huge genome files mirna in human Chromosome 1 mirna in bacteria Tools used
and their organelles
 and their organelles Discovery Video: Cells REVIEW!!!! The Cell Theory 1. Every living organism is made of one or more cells. 2. The cell is the basic unit of structure and function. It is the smallest
and their organelles Discovery Video: Cells REVIEW!!!! The Cell Theory 1. Every living organism is made of one or more cells. 2. The cell is the basic unit of structure and function. It is the smallest
Elements of Bioinformatics 14F01 TP5 -Phylogenetic analysis
 Elements of Bioinformatics 14F01 TP5 -Phylogenetic analysis 10 December 2012 - Corrections - Exercise 1 Non-vertebrate chordates generally possess 2 homologs, vertebrates 3 or more gene copies; a Drosophila
Elements of Bioinformatics 14F01 TP5 -Phylogenetic analysis 10 December 2012 - Corrections - Exercise 1 Non-vertebrate chordates generally possess 2 homologs, vertebrates 3 or more gene copies; a Drosophila
Chapter 4: Cells: The Working Units of Life
 Name Period Chapter 4: Cells: The Working Units of Life 1. What are the three critical components of the cell theory? 2. What are the two important conceptual implications of the cell theory? 3. Which
Name Period Chapter 4: Cells: The Working Units of Life 1. What are the three critical components of the cell theory? 2. What are the two important conceptual implications of the cell theory? 3. Which
Cell Structure and Function
 Cell Structure and Function Cell size comparison Animal cell Bacterial cell What jobs do cells have to do for an organism to live Gas exchange CO 2 & O 2 Eat (take in & digest food) Make energy ATP Build
Cell Structure and Function Cell size comparison Animal cell Bacterial cell What jobs do cells have to do for an organism to live Gas exchange CO 2 & O 2 Eat (take in & digest food) Make energy ATP Build
Cell Is the basic structural, functional, and biological unit of all known living organisms. Cells are the smallest unit of life and are often called
 The Cell Cell Is the basic structural, functional, and biological unit of all known living organisms. Cells are the smallest unit of life and are often called the "building blocks of life". The study of
The Cell Cell Is the basic structural, functional, and biological unit of all known living organisms. Cells are the smallest unit of life and are often called the "building blocks of life". The study of
Cell Structure. Chapter 4. Cell Theory. Cells were discovered in 1665 by Robert Hooke.
 Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
3.2 Cell Organelles. KEY CONCEPT Eukaryotic cells share many similarities.
 KEY CONCEPT Eukaryotic cells share many similarities. ! Cells have an internal structure. ! Cells have an internal structure. The cytoskeleton has many functions. ! Cells have an internal structure. The
KEY CONCEPT Eukaryotic cells share many similarities. ! Cells have an internal structure. ! Cells have an internal structure. The cytoskeleton has many functions. ! Cells have an internal structure. The
Unit 4: Cells. Biology 309/310. Name: Review Guide
 Unit 4: Cells Review Guide LEARNING TARGETS Place a checkmark next to the learning targets you feel confident on. Then go back and focus on the learning targets that are not checked. Identify the parts
Unit 4: Cells Review Guide LEARNING TARGETS Place a checkmark next to the learning targets you feel confident on. Then go back and focus on the learning targets that are not checked. Identify the parts
Double-stranded DNA is the molecule that
 Archaeology of Eukaryotic DNA Replication Kira S. Makarova and Eugene V. Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland
Archaeology of Eukaryotic DNA Replication Kira S. Makarova and Eugene V. Koonin National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Maryland
Chapter 7 Learning Targets Cell Structure & Function
 Name: Chapter 7 Learning Targets Cell Structure & Function a. Define the word cell: 1. I know the history of the cell: b. Who discovered the cell? What did he observe? 2. I can list the three parts of
Name: Chapter 7 Learning Targets Cell Structure & Function a. Define the word cell: 1. I know the history of the cell: b. Who discovered the cell? What did he observe? 2. I can list the three parts of
Modern ionotropic glutamate receptor with a K + selectivity signature sequence
 SUPPLEMENTARY INFORMATION Modern ionotropic glutamate receptor with a K + selectivity signature sequence H. Janovjak, G. Sandoz and E.Y. Isacoff * * Correspondence should be addressed to E.Y.I. (ehud@berkeley.edu).
SUPPLEMENTARY INFORMATION Modern ionotropic glutamate receptor with a K + selectivity signature sequence H. Janovjak, G. Sandoz and E.Y. Isacoff * * Correspondence should be addressed to E.Y.I. (ehud@berkeley.edu).
Cell Structure. Chapter 4
 Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Supporting Information
 Supporting Information Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi Zhen-Na Zhang 1,2,, Qin-Yi Wu 1,, Gui-Zhi Zhang 2, Yue-Yan Zhu 1, Robert W. Murphy 3, Zhen Liu 3, * and
Supporting Information Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi Zhen-Na Zhang 1,2,, Qin-Yi Wu 1,, Gui-Zhi Zhang 2, Yue-Yan Zhu 1, Robert W. Murphy 3, Zhen Liu 3, * and
Classification Cladistics & The Three Domains of Life. Biology Mrs. Flannery
 Classification Cladistics & The Three Domains of Life Biology Mrs. Flannery Finding Order in Diversity Earth is over 4.5 billion years old. Life on Earth appeared approximately 3.5 billion years ago and
Classification Cladistics & The Three Domains of Life Biology Mrs. Flannery Finding Order in Diversity Earth is over 4.5 billion years old. Life on Earth appeared approximately 3.5 billion years ago and
Cell Types. Prokaryotes
 Cell Types Prokaryotes before nucleus no membrane-bound nucleus only organelle present is the ribosome all other reactions occur in the cytoplasm not very efficient Ex.: bacteria 1 Cell Types Eukaryotes
Cell Types Prokaryotes before nucleus no membrane-bound nucleus only organelle present is the ribosome all other reactions occur in the cytoplasm not very efficient Ex.: bacteria 1 Cell Types Eukaryotes
MICROBIOLOGIA GENERALE. The Archaea
 MICROBIOLOGIA GENERALE The Archaea The Archaea s traits 1. The cell wall of Archaea: pseudopeptidoglycan, polysaccharide, glycoprotein 2. The cytoplasmic membrane of Archaea: ether linkage, glycerol
MICROBIOLOGIA GENERALE The Archaea The Archaea s traits 1. The cell wall of Archaea: pseudopeptidoglycan, polysaccharide, glycoprotein 2. The cytoplasmic membrane of Archaea: ether linkage, glycerol
CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools
 CAP 5510: to : Tools ECS 254A / EC 2474; Phone x3748; Email: giri@cis.fiu.edu My Homepage: http://www.cs.fiu.edu/~giri http://www.cs.fiu.edu/~giri/teach/bioinfs15.html Office ECS 254 (and EC 2474); Phone:
CAP 5510: to : Tools ECS 254A / EC 2474; Phone x3748; Email: giri@cis.fiu.edu My Homepage: http://www.cs.fiu.edu/~giri http://www.cs.fiu.edu/~giri/teach/bioinfs15.html Office ECS 254 (and EC 2474); Phone:
UNIVERSITY OF CALIFORNIA SANTA CRUZ CHARACTERIZATION OF ARCHAEAL SPECIES THROUGH RNASE P AND TRANSFER RNAS
 UNIVERSITY OF CALIFORNIA SANTA CRUZ CHARACTERIZATION OF ARCHAEAL SPECIES THROUGH RNASE P AND TRANSFER RNAS A dissertation submitted in partial satisfaction of the requirements for the degree of DOCTOR
UNIVERSITY OF CALIFORNIA SANTA CRUZ CHARACTERIZATION OF ARCHAEAL SPECIES THROUGH RNASE P AND TRANSFER RNAS A dissertation submitted in partial satisfaction of the requirements for the degree of DOCTOR
Name Date Class. This section describes cell structure and function in plant cells, animal cells, and bacteria.
 Name Date Class Cell Structure and Function Guided Reading and Study Looking Inside Cells This section describes cell structure and function in plant cells, animal cells, and bacteria. Use Target Reading
Name Date Class Cell Structure and Function Guided Reading and Study Looking Inside Cells This section describes cell structure and function in plant cells, animal cells, and bacteria. Use Target Reading
Dr. Dina A. A. Hassan Associate Professor, Pharmacology
 Cytology Dr. Dina A. A. Hassan Associate Professor, Pharmacology Email: da.hassan@psau.edu.sa Cells All living things are made up of cells Basic building blocks of life It is the smallest functional and
Cytology Dr. Dina A. A. Hassan Associate Professor, Pharmacology Email: da.hassan@psau.edu.sa Cells All living things are made up of cells Basic building blocks of life It is the smallest functional and
arxiv: v3 [q-bio.gn] 17 Jun 2016
![arxiv: v3 [q-bio.gn] 17 Jun 2016 arxiv: v3 [q-bio.gn] 17 Jun 2016](/thumbs/84/89710698.jpg) SICLE: A high-throughput tool for extracting evolutionary relationships from phylogenetic trees Dan F. DeBlasio 1,* and Jennifer H. Wisecaver 2,3 arxiv:1303.5785v3 [q-bio.gn] 17 Jun 2016 1 Department of
SICLE: A high-throughput tool for extracting evolutionary relationships from phylogenetic trees Dan F. DeBlasio 1,* and Jennifer H. Wisecaver 2,3 arxiv:1303.5785v3 [q-bio.gn] 17 Jun 2016 1 Department of
Eukaryotic cells are more complex than prokaryotic cells. They are identified by the presence of certain membrane-bound organelles.
 Eukaryotic Cells Eukaryotic cells are more complex than prokaryotic cells. They are identified by the presence of certain membrane-bound organelles. Prokaryotic cells have organelles too, but much fewer
Eukaryotic Cells Eukaryotic cells are more complex than prokaryotic cells. They are identified by the presence of certain membrane-bound organelles. Prokaryotic cells have organelles too, but much fewer
9/2/17. Molecular and Cellular Biology. 3. The Cell From Genes to Proteins. key processes
 Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Universal Rules Governing Genome Evolution Expressed by Linear Formulas
 The Open Genomics Journal, 8, 1, 33-3 33 Open Access Universal Rules Governing Genome Evolution Expressed by Linear Formulas Kenji Sorimachi *,1 and Teiji Okayasu 1 Educational Support Center and Center
The Open Genomics Journal, 8, 1, 33-3 33 Open Access Universal Rules Governing Genome Evolution Expressed by Linear Formulas Kenji Sorimachi *,1 and Teiji Okayasu 1 Educational Support Center and Center
Introduction to Botany. Lecture 12
 Introduction to Botany. Lecture 12 Alexey Shipunov Minot State University September 25, 2017 Shipunov (MSU) Introduction to Botany. Lecture 12 September 25, 2017 1 / 26 Outline 1 Questions and answers
Introduction to Botany. Lecture 12 Alexey Shipunov Minot State University September 25, 2017 Shipunov (MSU) Introduction to Botany. Lecture 12 September 25, 2017 1 / 26 Outline 1 Questions and answers
Organelles in Eukaryotic Cells
 Why? Organelles in Eukaryotic Cells What are the functions of different organelles in a cell? The cell is the basic unit and building block of all living things. Organisms rely on their cells to perform
Why? Organelles in Eukaryotic Cells What are the functions of different organelles in a cell? The cell is the basic unit and building block of all living things. Organisms rely on their cells to perform
Cells & Cell Organelles. Doing Life s Work
 Cells & Cell Organelles Doing Life s Work Types of cells bacteria cells Prokaryote Eukaryotes animal cells plant cells Cell size comparison Animal cell Bacterial cell most bacteria 1-10 microns eukaryotic
Cells & Cell Organelles Doing Life s Work Types of cells bacteria cells Prokaryote Eukaryotes animal cells plant cells Cell size comparison Animal cell Bacterial cell most bacteria 1-10 microns eukaryotic
Biology 2.1 Taxonomy: Domain, Kingdom, Phylum. ICan2Ed.com
 Biology 2.1 Taxonomy: Domain, Kingdom, Phylum ICan2Ed.com Taxonomy is the scientific field that catalogs, describes, and names living organisms. The way to divide living organisms into groups based on
Biology 2.1 Taxonomy: Domain, Kingdom, Phylum ICan2Ed.com Taxonomy is the scientific field that catalogs, describes, and names living organisms. The way to divide living organisms into groups based on
11. What are the four most abundant elements in a human body? A) C, N, O, H, P B) C, N, O, P C) C, S, O, H D) C, Na, O, H E) C, H, O, Fe
 48017 omework#1 on VVP Chapter 1: and in the provided answer template on Monday 4/10/17 @ 1:00pm; Answers on this document will not be graded! Matching A) Phylogenetic B) negative C) 2 D) Δ E) TS F) halobacteria
48017 omework#1 on VVP Chapter 1: and in the provided answer template on Monday 4/10/17 @ 1:00pm; Answers on this document will not be graded! Matching A) Phylogenetic B) negative C) 2 D) Δ E) TS F) halobacteria
CELL THEORY, STRUCTURE & FUNCTION
 CELL THEORY, STRUCTURE & FUNCTION History of Cells Robert Hooke (1665) observed cork under a microscope Thought they looked like the rooms monks lived in called cells. History of Cells Antony Van Leeuwenhoek
CELL THEORY, STRUCTURE & FUNCTION History of Cells Robert Hooke (1665) observed cork under a microscope Thought they looked like the rooms monks lived in called cells. History of Cells Antony Van Leeuwenhoek
