SUPPLEMENTARY INFORMATION
|
|
|
- Morgan Randall
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTARY INFORMATION doi: /nature13082 Supplementary Table 1. Examination of nectar production in wild-type and atsweet9 flowers. No. of flowers with detectable nectar out of the total observed flowers Wild-type atsweet9-1 atsweet /100 0/100 0/100 Supplementary Table 2. Total nectar glucose content ratio of AtSWEET9 overexpression lines driving by AtSWEET9 promoter to wild type Arabidopsis and the estimation nectar volume. Nectar glucose contents were evaluated in each line and shown relative to wild-type nectar glucose in 8 independent repeats. A single biological replicate for nectar glucose assays consisted of nectar collected from the lateral nectaries of ten stage flowers using wicks cut from Whatman No. 1 filter paper. In each independent repeats, extra AtSWEET9 copies increased nectar glucose contents (n=10, t significant at P < for each overexpression line versus wild-type). Relative nectar glucose concentration was analyzed using Amplex Red Glucose/Glucose Oxidase Assay Kit (life technology). Nectar droplets were typically ovally-shaped, and the nectar volumes in different lines were compared by measuring the longest axis of nectar droplets on the sepals (t significant at P<0.01 for each overexpression line versus wild-type). Arabidopsis Lines Nectar Glucose content Nectar droplet length Ratio to WT (±SEM, n=8) (mm ±SEM, n 12) Col ±0.02 patsweet9:atsweet9-egfp-5/col ± ±0.02 patsweet9:atsweet9-egfp-8/col ± ±
2 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Table 3. Degenerate primers used for amplification of SWEETs orthologs from Nicotiana attenuata SUT1-23 SUT2-23 SUT3-23 SUT4-23 SUT5-25 SUT6-23 SUT7-25 SUT8-25 SUT9-27 SUT GTNTTCYTNKCNCCARYKCCAAC-3 5 -GTSTTCYTRKCWCCAGTKCCRAC-3 5 -GTGTTCTTAGCACCAGTKCCAAC-3 5 -GGCATGWACTCNACRCTYTTYGT-3 5 -AANGGCATGWACTCNACRCTYTTYG-3 5 -GGCATGAACTCYACRCTCTTYGT-3 5 -AAGGGCATGAACTCYACRCTCTTYG-3 5 -AAGGGCATGAACTCTACGCTCTTCG-3 5 -ATGACACTATTAAGTGTTCAAGAATTG-3 5 -TTACTTCAAAGTCAAAACGCTGG-3 Primer pairs for amplification of the same SUT region used for leaves/nectaries screening for sucrose transporter transcripts: SUT SUT4-23 SUT SUT5-25 SUT SUT6-23 SUT SUT7-25 SUT SUT8-25 Primer pairs for amplification of the same SUT region used then for flower part screening for sucrose transporter transcripts: SUT SUT4-23 SUT SUT8-25 Primer pair for the amplification of full length ORF of SWEET9 similar to a SWEET gene from ornamental tobacco (assembled consensus sequence from a number of ESTs from "NecGex_355B09 Ornamental tobacco (LxS8) post-fertilization Floral nectary cdna library Nicotiana langsdorffii x Nicotiana sanderae cdna", which showed homology to NEC1 from Petunia hybrida, and which all obviously originated from the same transcript): SUT SUT
3 SUPPLEMENTARY INFORMATION RESEARCH Supplementary Table 4. Primer list PCR purpose Primer name Primer sequence Truncated version of AtSWEET9-F201* construction in poo2-gw Truncated version of BrSWEET9-L201* construction in poo2-gw AtSWEET9- F201*_F AtSWEET9- F201*_R BrSWEET9- L201*_F BrSWEET9- L201*_R GCCAAACATTCTCGGTTTTCTATGAGGTG TAGCTCAGATGATACT AGTATCATCTGAGCTACACCTCATAGAAA ACCGAGAATGTTTGGC GCTATGCCAAACATTCTTGGATTTTAGTT CGGTATAGCTCAGATG CATCTGAGCTATACCGAACTAAAATCCA AGAATGTTTGGCATAGC size (bp) Truncated version of NaSWEET9-L201* construction in poo2-gw NaSWEET9- L201*_F NaSWEET9- L201*_R CCAAACGTGCTGGGATTTCTGTAGGGAAT TGTTCAAATGATACTA TAGTATCATTTGAACAATTCCCTACAGAA ATCCCAGCACGTTTGG atsweet9-1 mutant sk225 psktaill3 AtSWT9-1LP AtSWT9-1RP ATACGACGGATCGTAATTTGT CTTTGTCGGATTTAGAAGGCC ATTTGCAATGTCGTCTCCAAG 966 NEC1-34 GCGGCGCTGCAGCAAAAAAGGATCATCA GAAGGG NaSWEET9 silencing NEC2-34 NEC3-33 GCGGCGAAGCTTCAATAGCACAAATCCAT CCAAC GCGGCGGAGCTCAAAAAAGGATCATCAG AAGGG NEC4-34 GCGGCGCTCGAGCAATAGCACAAATCCA TCCAAC atsweet9-2 mutant SALK_ AtSWT9-2LP AtSWT9-2RP CGCTGAAGAATCCAATAATGTG ATTTTTGACAATTTGCGATGG 1088 atsweet9-3 mutant SALK_202913C AtSWT9-3LP AtSWT9-3RP TGCCGACCAAAATAAAAAGTG ACTTGGCAGATATCCACGTTG 1192 Arabidopsis SALK LBb1.3 ATTTTGCCGATTTCGGAAC 3
4 RESEARCH SUPPLEMENTARY INFORMATION T-DNA mutants patsweet9:atswee T9-GUS fusion SWEET9cmF SWEET9cm R GGGGACAAGTTTGTACAAAAAAGCAGGC TTAACTCTAAGACACGAGCGGAGATA GGGGACCACTTTGTACAAGAAAGCTGGG TACTTCATTGGCCTCACCGATCC 3237 patsweet9:atswee T9-eGFP fusion constructs in pgtkan3 AtSWT9_ pgtkan3f AtSWT9_ pgtkan3r AAGGATCCACTCTAAGACACGAGCGGA AACTGCAGACTTCATTGGCCTCACCGA 3327 patsweet9:atswee T9 complementation constructs in ptkan2 patsweet9:atswee T11/12 complementation constructs in ptkan2 RT-PCR for AtSWEET9 in atsweet9-1 and atsweet9-2 RT-PCR for AtSWEET9 in atsweet9-3 RT-PCR for AtACTIN2 AtSWT9_ ptkan2f AtSWT9_ ptkan2r KpnI-SWT9p ro F SWT9pro-Ba mhi R BamHI-SWT 11 F SWT11-PstI R BamHI-SWT 12 F SWT12-PstI R AtSWT9_214 5F AtSWT9_qP CR R AT2G39060 RT Fwd AT2G39060 RT Rev AtACT2F AtACT2R AAGGATCCACTCTAAGACACGAGCGGA AACTGCAGCTTCATTGGCCTCACCGAT TAGGTACCACTCTAAGACACGAGCGG ATGGATCCCTTCACTTTTCTCTCTTT TTGGATCCATGAGTCTCTTCAACACT CGCTGCAGTTAATTCTCAAGTCAAAT TAGGATCCATGGCTCTCTTCGACACT TACTGCAGGTTGCTTGAGAATTTTCA GGGTGTTCTTGTCACCAGTGT TGTCGTCTCCCATACATCTTCCC TCTCGCAGTCTTTGCTTCTCCCTT GTTTGCCCTTCACTTCATTGGCCT TCCAAGCTGTTCTCTCCTTG GAGGGCTGGAACAAGACTTC RT-PCR for GAPDH RT AGGGTGGTGCCAAGAAGGTTG
5 SUPPLEMENTARY INFORMATION RESEARCH AtGAPDH brsweet9-1, -2 & -3 AtSPS1F primers for RT PCR AtSPS2F primers for RT PCR AtACTIN2 primers for RT PCR Oligos for generating SPS amirna "TATATGTTTTGCA GTGGTCTG" Fwd GAPDH RT Rev BrSWEET9 TILLING SEQ F BrSWEET9 TILLING SEQ R SPS1F RT-F SPS1F RT-R SPS1F RT-F SPS1F RT-R ACTIN2 RT-F ACTIN2 RT-R AtSPS1/2f-T 1-I AtSPS1/2f-T 1-II AtSPS1/2f-T 1-III AtSPS1/2f-T 1-IV oligo A oligo B GTAGCCCCACTCGTTGTCGTA TGCCGACCAAAATAAAAAGTG ACTTGGCAGATATCCACGTTG AGGAAAAAGAAGCACAGAGGC ACCAGCATCAGCATAGTGTCC TGGCCAGAGCACTGCAAAACA TGAGGTTGCGCTACAGTCCACA GTTGGGATGAACCAGAAGGA GAACCACCGATCCAGACACT gatatatgttttgcagtggtctgtctctcttttgtatt cc gacagaccactgcaaaacatatatcaaagagaa tcaatga gacaaaccactgcaatacatatttcacaggtcgt gatatg gaaatatgtattgcagtggtttgtctacatatatat tcct CTGCAAGGCGATTAAGTTGGGTAAC GCGGATAACAATTTCACACAGGAAACAG N/A
6 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Table 5. Species Full Names. Abbreviation Full Name (Common name) P. hybrida Petunia hybrid (Petunia) N. attenuata Nicotiana attenuate (Coyote Tobacco) S. lycopersicum Solanum lycopersicum (Tomato) V. vinifera Vitis vinifera (Grape Vine) C. sativus Cucumis sativus (Cucumber) C. melo Cucumis melo (Muskmelon) P. persica Prunus persica (Peach) F. vesca Fragaria vesca (Strawberry) M. domestica Malus domestica (Apple) M. truncatula Medicago truncatula (Barrel Medic) L. japonicus Lotus japonicus (Lotus japonicus) P. vulgaris Phaseolus vulgaris (Bean) G. max Glycine max (Soybean) R. communis Ricinus communis (Castor Bean) M. esculenta Manihot esculenta (Cassava) P. trichocarpa Populus trichocarpa (Poplar) T. cacao Theobroma cacao (Cacao) C. papaya Carica papaya (Papaya) A. lyrata Arabidopsis lyrata (Arabidopsis l.) A. thaliana Arabidopsis thaliana (Arabidopsis) B. rapa Brassica rapa (Turnip) C.sinensis Citrus sinensis (Orange) A. caerulea Aquilegia caerulea (Columbine) P. dactylifera Phoenix dactylifera (Palm Date) M. acuminata Musa acuminate (Banana) Z. mays Zea mays (Maize) S. bicolor Sorghum bicolor (Sorghum) B. distachyon Brachypodium distachyon (Brachypodium) H. vulgare Hordeum vulgare (Barley) T. aestivum Triticum aestivum (Wheat) O. sativa Oryzae sativa (Rice) A. trichopoda Amborella trichopoda (Amborella) S. moellendorffii Selaginella moellendorffii (Selaginella) P. patens Physcomitrella patens (Physcomitrella) C. reinhardtii Chlamydomonas reinhardtii (Chlamydomonas) 6
7 SUPPLEMENTARY INFORMATION RESEARCH Supplementary Table 6. SWEET Accessions. Arabidopsis thaliana Oryza sativa Aquilegia caerulea Medigaco truncatula AtSWEET1-At1G21460 OsSWEET1a-Os01g65880 AcSWEET1-Aquca_014_00360 MtSWEET2b - AC235677_9 AtSWEET2-At3G14770 OsSWEET1b-Os05g35140 AcSWEET17-Aquca_017_00148 MtSWEET3c - Medtr1g AtSWEET3-At5G53190 OsSWEET2a-Os01g36070 AcSWEET3b-Aquca_012_00215 MtSWEET1a - Medtr1g AtSWEET4-At3G28007 OsSWEET2b-Os01g50460 AcSWEET13-Aquca_011_00056 MtSWEET15a - Medtr2g AtSWEET5-At5G62850 OsSWEET3a-Os05g12320 AcSWEET12-Aquca_021_00060 MtSWEET6 - Medtr3g AtSWEET6-At1G66770 OsSWEET3b-Os01g12130 AcSWEET4-Aquca_037_00238 MtSWEET1b - Medtr3g AtSWEET7-At4G10850 OsSWEET4-Os02g19820 AcSWEET6-Aquca_001_00818 MtSWEET3a - Medtr3g AtSWEET8-At5G40260 OsSWEET5-Os05g51090 AcSWEET2b-Aquca_468_00002 MtSWEET3b - Medtr3g AtSWEET9-At2G39060 OsSWEET6a-Os01g42110 AcSWEET16-Aquca_003_00698 MtSWEET13 - Medtr3g AtSWEET10-At5G50790 OsSWEET6b-Os01g42090 AcSWEET11-Aquca_003_00877 MtSWEET11 - Medtr3g AtSWEET11-At3G48740 OsSWEET7a-Os09g08030 AcSWEET5-Aquca_002_00199 MtSWEET4 - Medtr4g AtSWEET12-At5G23660 OsSWEET7b-Os09g08440 AcSWEET2c-Aquca_055_00129 MtSWEET15b - Medtr5g AtSWEET13-At5G50800 OsSWEET7c-Os12g07860 AcSWEET2a-Aquca_022_00050 MtSWEET9a - Medtr5g AtSWEET14-At4G25010 OsSWEET7d-Os09g08490 AcSWEET3a-Aquca_013_00133 MtSWEET5a - Medtr6g AtSWEET15-At5G13170 OsSWEET7e-Os09g08270 AcSWEET7-Aquca_025_00283 MtSWEET5c - Medtr6g AtSWEET16-At3G16690 OsSWEET11-Os08g42350 MtSWEET5d - Medtr6g AtSWEET17-At4G15920 OsSWEET12-Os03g22590 Chlamydomonas reinhardtii MtSWEET5b - Medtr6g OsSWEET13-Os12g29220 CrSWEET4-Cre07.g MtSWEET2c - Medtr6g OsSWEET14-Os11g31190 CrSWEET2-Cre06.g27180 MtSWEET9b - Medtr7g OsSWEET15-Os02g30910 CrSWEET3-Cre06.g MtSWEET15d - Medtr7g OsSWEET16-Os03g22200 CrSWEET5-Cre10.g MtSWEET15c - Medtr7g Carica papaya Manioth esculenta Ricinus communis MtSWEET2a - Medtr8g CpSWEET9 - CP00051G00690 MeSweet9 - ME07238G00800 RcSweet9 - RC29647G00280 MtSWEET14 - Medtr8g Malus domestica Fragaria vesca Theobroma cacao MtSWEET12 - Medtr8g MdSweet9 - MD04G FvSweet9 FV0G27360 TcSweet9 - TC01G MtSWEET7 - Medtr8g Solanum lycopersicum Manioth Esculenta Populus trichocarpa MtSWEET16 - Mtr S1_at SlSWEET1a-Solyc04g MeSWEET1a-cassava4.1_014638m PtSWEET1a-Potri.005G Glycine max SlSWEET1b-Solyc04g MeSWEET1b-cassava4.1_014650m PtSWEET17a-Potri.013G GmSWEET1-XP SlSWEET1f-Solyc06g MeSWEEET2a-cassava4.1_015227m PtSWEET16c-Potri.013G GmSWEET2-XP SlSWEET1c-Solyc04g MeSWEET2b-cassava4.1_030719m PtSWEET17b-Potri.013G GmSWEET3a-XP SlSWEET1e-Solyc06g MeSWEET3a-cassava4.1_026477m PtSWEET16b-Potri.013G GmSWEET3b SlSWEET1d-Solyc04g MeSWEET3b-cassava4.1_022559m PtSWEET15a-Potri.003G GmSWEET9-GM18G53250 SlSWEET2c-Solyc03g MeSWEET4-cassava4.1_016815m PtSWEET6-Potri.003G GmSWEET11a-XP SlSWEET2a-Solyc07g MeSWEET5-cassava4.1_026390m PtSWEET2c-Potri.011G GmSWEET11b-XP SlSWEET6-Solyc08g MeSWEET6-cassava4.1_014231m PtSWEET3a-Potri.015G GmSWEET11c-XP SlSWEET2b-Solyc02g MeSWEET7-cassava4.1_028141m PtSWEET5-Potri.015G GmSWEET12a- XP SlSWEET7-Solyc12g MeSWEET9a-cassava4.1_032222m PtSWEET10c-Potri.015G GmSWEET12b-XP SlSWEET17-Solyc01g MeSWEET9b-cassava4.1_031208m PtSWEET11-Potri.015G GmSWEET15a-XP SlSWEET3-Solyc03g MeSWEET10a-cassava4.1_013474m PtSWEET1b-Potri.002G GmSWEET15b-XP SlSWEET4-Solyc03g MeSWEET10b-cassava4.1_015602m PtSWEET9-Potri.019G SlSWEET5-Solyc06g MeSWEET10c-cassava4.1_021350m PtSWEET10a-Potri.015G Amborella trichopoda 7
8 RESEARCH SUPPLEMENTARY INFORMATION SlSWEET9-Solyc09g MeSWEET10d-cassava4.1_013519m PtSWEET16a-Potri.005G AmboSWEET1-scaffold SlSWEET10d-Solyc06g MeSWEET10e-cassava4.1_032927m PtSWEET16d-Potri.008G AmboSWEET2-scaffold SlSWEET10e-Solyc03g MeSWEET11-cassava4.1_028116m PtSWEET2d-Potri.001G AmboSWEET3a-scaffold SlSWEET10f-Solyc03g MeSWEET12a-cassava4.1_017557m PtSWEET2b-Potri.001G AmboSWEET3b-scaffold SlSWEET10g-Solyc03g MeSWEET12b-cassava4.1_018003m PtSWEET2a-Potri.001G AmboSWEET6-scaffold SlSWEET10h-Solyc03g MeSWEET13-cassava4.1_026944m PtSWEET4-Potri.001G AmboSWEET7-scaffold SlSWEET10a-Solyc03g MeSWEET15a-cassava4.1_026251m PtSWEET15b-Potri.001G AmboSWEET11-scaffold SlSWEET10i-Solyc03g MeSWEET15b-cassava4.1_014124m PtSWEET10d-Potri.012G AmboSWEET16-scaffold SlSWEET8b-Solyc02g MeSWEET16a-cassava4.1_014996m PtSWEET3b-Potri.012G Physcomitrella patens SlSWEET10-Solyc03g MeSWEET16b-cassava4.1_015143m Brassica rapa PpSWEET1a-Pp1s127_127V6.1 SlSWEET15-Solyc05g MeSWEET17-cassava4.1_014640m BrSWEET9-AGO61984 PpSWEET1b-Pp1s54_64V6.1 SlSWEET10c-Solyc06g MeSWEET17a-cassava4.1_032999m Nicotiana attenuata PpSWEET2a-Pp1s240_24V6.1 SlSWEET10b-Solyc06g MeSWEET17b-cassava4.1_012690m NaSWEET9/NEC1-Q9FPN0 PpSWEET2b-Pp1s307_21V6.1 SlSWEET16-Solyc01g MeSWEET17c-cassava4.1_014587m PpSWEET4-Pp1s39_291V6.1 SlSWEET8a-Solyc11g PpSWEET8-XP
Supplementary Figure 3
 Supplementary Figure 3 7.0 Col Kas-1 Line FTH1A 8.4 F3PII3 8.9 F26H11 ATQ1 T9I22 PLS8 F26B6-B 9.6 F27L4 9.81 F27D4 9.92 9.96 10.12 10.14 10.2 11.1 0.5 Mb T1D16 Col % RGR 83.3 101 227 93.5 75.9 132 90 375
Supplementary Figure 3 7.0 Col Kas-1 Line FTH1A 8.4 F3PII3 8.9 F26H11 ATQ1 T9I22 PLS8 F26B6-B 9.6 F27L4 9.81 F27D4 9.92 9.96 10.12 10.14 10.2 11.1 0.5 Mb T1D16 Col % RGR 83.3 101 227 93.5 75.9 132 90 375
Supplementary Figure 1. Number of CC- and TIR- type NBS- LRR genes and presence of mir482/2118 on sequenced plant genomes.
 Number of CC- NBS and CC- NBS- LRR R- genes Number of TIR- NBS and TIR- NBS- LRR R- genes 0 50 100 150 200 250 0 50 100 150 200 250 300 350 400 450 mir482 and mir2118 Cajanus cajan Glycine max Hevea brasiliensis
Number of CC- NBS and CC- NBS- LRR R- genes Number of TIR- NBS and TIR- NBS- LRR R- genes 0 50 100 150 200 250 0 50 100 150 200 250 300 350 400 450 mir482 and mir2118 Cajanus cajan Glycine max Hevea brasiliensis
Bioinformatics tools to analyze complex genomes. Yves Van de Peer Ghent University/VIB
 Bioinformatics tools to analyze complex genomes Yves Van de Peer Ghent University/VIB Detecting colinearity and large-scale gene duplications A 1 2 3 4 5 6 7 8 9 10 11 Speciation/Duplicatio n S1 S2 1
Bioinformatics tools to analyze complex genomes Yves Van de Peer Ghent University/VIB Detecting colinearity and large-scale gene duplications A 1 2 3 4 5 6 7 8 9 10 11 Speciation/Duplicatio n S1 S2 1
Cao, J, K Schneeberger, S Ossowski, et al Whole genome sequencing of multiple Arabidopsis thaliana populations. Nat Genet 43:
 Figure S1. Syntenic map of SAE1B duplication. We have used the nucleotide sequences of Arabidopsis thaliana Col-0 gene tandem duplicates AT5G50580 and AT5G506800 as queries in independent BLASTN searches
Figure S1. Syntenic map of SAE1B duplication. We have used the nucleotide sequences of Arabidopsis thaliana Col-0 gene tandem duplicates AT5G50580 and AT5G506800 as queries in independent BLASTN searches
Supplementary Material
 Supplementary Material Supplementary Table S1. Genomes available in build 47 Supplementary Table S2. Counts of putative contiguous gene split models in 39 plant reference genomes in build 47 Supplementary
Supplementary Material Supplementary Table S1. Genomes available in build 47 Supplementary Table S2. Counts of putative contiguous gene split models in 39 plant reference genomes in build 47 Supplementary
Evaluation of Genome Sequencing Quality in Selected Plant Species Using Expressed Sequence Tags
 Evaluation of Genome Sequencing Quality in Selected Plant Species Using Expressed Sequence Tags Lingfei Shangguan 1, Jian Han 1, Emrul Kayesh 1, Xin Sun 1, Changqing Zhang 2, Tariq Pervaiz 1, Xicheng Wen
Evaluation of Genome Sequencing Quality in Selected Plant Species Using Expressed Sequence Tags Lingfei Shangguan 1, Jian Han 1, Emrul Kayesh 1, Xin Sun 1, Changqing Zhang 2, Tariq Pervaiz 1, Xicheng Wen
** LCA LCN PCA
 % of wild type value % of wild type value a 12 1 8 2 b 12 1 8 2 LCA LCN PCA Col- sod3-1 Supplementary Figure 1 sod3-1 influences cell proliferation. (a) Fifth leaf cell area (LCA) and leaf cell number
% of wild type value % of wild type value a 12 1 8 2 b 12 1 8 2 LCA LCN PCA Col- sod3-1 Supplementary Figure 1 sod3-1 influences cell proliferation. (a) Fifth leaf cell area (LCA) and leaf cell number
Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type
 A B 2 3 3 2 1 1 Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type (A) and d27 (B) seedlings at the four
A B 2 3 3 2 1 1 Supplemental Figure 1. Comparison of Tiller Bud Formation between the Wild Type and d27. (A) and (B) Longitudinal sections of shoot apex in wild-type (A) and d27 (B) seedlings at the four
SUPPLEMENTARY INFORMATION
 doi:1.138/nature111 cytosol Model: PILS function in cellular auxin homeostasis ER nucleus IAA degradation? sequestration? conjugation? storage? signalling? PILS IAA ER cytosol Supplemental Figure 1 Model
doi:1.138/nature111 cytosol Model: PILS function in cellular auxin homeostasis ER nucleus IAA degradation? sequestration? conjugation? storage? signalling? PILS IAA ER cytosol Supplemental Figure 1 Model
SUPPLEMENTARY INFORMATION
 doi:1.138/nature11394 Supplementary Note Our study is based on two different tracriptome indices. Both combine tracriptional information with an important evolutionary parameter. For the tracriptome age
doi:1.138/nature11394 Supplementary Note Our study is based on two different tracriptome indices. Both combine tracriptional information with an important evolutionary parameter. For the tracriptome age
AtTIL-P91V. AtTIL-P92V. AtTIL-P95V. AtTIL-P98V YFP-HPR
 Online Resource 1. Primers used to generate constructs AtTIL-P91V, AtTIL-P92V, AtTIL-P95V and AtTIL-P98V and YFP(HPR) using overlapping PCR. pentr/d- TOPO-AtTIL was used as template to generate the constructs
Online Resource 1. Primers used to generate constructs AtTIL-P91V, AtTIL-P92V, AtTIL-P95V and AtTIL-P98V and YFP(HPR) using overlapping PCR. pentr/d- TOPO-AtTIL was used as template to generate the constructs
Supplemental Table 1. Primers used for cloning and PCR amplification in this study
 Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
Supplemental Table 1. Primers used for cloning and PCR amplification in this study Target Gene Primer sequence NATA1 (At2g393) forward GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT CAT GGC GCC TCC AAC CGC AGC
Genome-wide discovery of G-quadruplex forming sequences and their functional
 *Correspondence and requests for materials should be addressed to R.G. (rohini@nipgr.ac.in) Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants Rohini Garg*,
*Correspondence and requests for materials should be addressed to R.G. (rohini@nipgr.ac.in) Genome-wide discovery of G-quadruplex forming sequences and their functional relevance in plants Rohini Garg*,
Stage 1: Karyotype Stage 2: Gene content & order Step 3
 Supplementary Figure Method used for ancestral genome reconstruction. MRCA (Most Recent Common Ancestor), AMK (Ancestral Monocot Karyotype), AEK (Ancestral Eudicot Karyotype), AGK (Ancestral Grass Karyotype)
Supplementary Figure Method used for ancestral genome reconstruction. MRCA (Most Recent Common Ancestor), AMK (Ancestral Monocot Karyotype), AEK (Ancestral Eudicot Karyotype), AGK (Ancestral Grass Karyotype)
Supplemental Data. Chen and Thelen (2010). Plant Cell /tpc
 Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Data. Yang et al. (2012). Plant Cell /tpc
 Supplemental Figure 1. Mature flowers of P. heterotricha. (A) An inflorescence of P. heterotricha showing the front view of a zygomorphic flower characterized by two small dorsal petals and only two fertile
Supplemental Figure 1. Mature flowers of P. heterotricha. (A) An inflorescence of P. heterotricha showing the front view of a zygomorphic flower characterized by two small dorsal petals and only two fertile
Regulatory Change in YABBY-like Transcription Factor Led to Evolution of Extreme Fruit Size during Tomato Domestication
 SUPPORTING ONLINE MATERIALS Regulatory Change in YABBY-like Transcription Factor Led to Evolution of Extreme Fruit Size during Tomato Domestication Bin Cong, Luz Barrero, & Steven Tanksley 1 SUPPORTING
SUPPORTING ONLINE MATERIALS Regulatory Change in YABBY-like Transcription Factor Led to Evolution of Extreme Fruit Size during Tomato Domestication Bin Cong, Luz Barrero, & Steven Tanksley 1 SUPPORTING
Potato Genome Analysis
 Potato Genome Analysis Xin Liu Deputy director BGI research 2016.1.21 WCRTC 2016 @ Nanning Reference genome construction???????????????????????????????????????? Sequencing HELL RIEND WELCOME BGI ZHEN LLOFRI
Potato Genome Analysis Xin Liu Deputy director BGI research 2016.1.21 WCRTC 2016 @ Nanning Reference genome construction???????????????????????????????????????? Sequencing HELL RIEND WELCOME BGI ZHEN LLOFRI
Miloš Duchoslav and Lukáš Fischer *
 Duchoslav and Fischer BMC Plant Biology (2015) 15:133 DOI 10.1186/s12870-015-0523-4 RESEARCH ARTICLE Open Access Parallel subfunctionalisation of PsbO protein isoforms in angiosperms revealed by phylogenetic
Duchoslav and Fischer BMC Plant Biology (2015) 15:133 DOI 10.1186/s12870-015-0523-4 RESEARCH ARTICLE Open Access Parallel subfunctionalisation of PsbO protein isoforms in angiosperms revealed by phylogenetic
Browsing Genomic Information with Ensembl Plants
 Browsing Genomic Information with Ensembl Plants Etienne de Villiers, PhD (Adapted from slides by Bert Overduin EMBL-EBI) Outline of workshop Brief introduction to Ensembl Plants History Content Tutorial
Browsing Genomic Information with Ensembl Plants Etienne de Villiers, PhD (Adapted from slides by Bert Overduin EMBL-EBI) Outline of workshop Brief introduction to Ensembl Plants History Content Tutorial
Supplementary Information for: The genome of the extremophile crucifer Thellungiella parvula
 Supplementary Information for: The genome of the extremophile crucifer Thellungiella parvula Maheshi Dassanayake 1,9, Dong-Ha Oh 1,9, Jeffrey S. Haas 1,2, Alvaro Hernandez 3, Hyewon Hong 1,4, Shahjahan
Supplementary Information for: The genome of the extremophile crucifer Thellungiella parvula Maheshi Dassanayake 1,9, Dong-Ha Oh 1,9, Jeffrey S. Haas 1,2, Alvaro Hernandez 3, Hyewon Hong 1,4, Shahjahan
South Green Bioinformatics activities at CIRAD
 South Green Bioinformatics activities at CIRAD Data Integration Team of the research unit DAP Manuel Ruiz, CIP, Lima, 23rd january The Joint Research Unit DAP (Développement et Amélioration des Plantes
South Green Bioinformatics activities at CIRAD Data Integration Team of the research unit DAP Manuel Ruiz, CIP, Lima, 23rd january The Joint Research Unit DAP (Développement et Amélioration des Plantes
SUPPLEMENTARY MATERIAL SUPPLEMENTARY TABLES
 SUPPLEMENTARY MATERIAL SUPPLEMENTARY TABLES Supplementary Table 1. Genomes available in Gramene build 38 Supplementary Table 2. Ontology associations in Gramene build 38 Supplementary Table 3. Synteny
SUPPLEMENTARY MATERIAL SUPPLEMENTARY TABLES Supplementary Table 1. Genomes available in Gramene build 38 Supplementary Table 2. Ontology associations in Gramene build 38 Supplementary Table 3. Synteny
Supporting Information
 Supporting Information Rossig et al. 10.1073/pnas.1319648110 SI Materials and Methods Athp20 and Athp30 Mutant Characterization and Analysis of the Athp30/30-2 RNAi Lines. Athp20;1 and Athp20;2 (SALK_020671
Supporting Information Rossig et al. 10.1073/pnas.1319648110 SI Materials and Methods Athp20 and Athp30 Mutant Characterization and Analysis of the Athp30/30-2 RNAi Lines. Athp20;1 and Athp20;2 (SALK_020671
gi (NCBI databse: ES frame +2 (NCBI: databse
 Figure legend for supplemental tables (S1-S6). The predicted and annotated amino acid sequences for the different LEA proteins found in the available databases (378) were analyzed using MEME software (Bailey
Figure legend for supplemental tables (S1-S6). The predicted and annotated amino acid sequences for the different LEA proteins found in the available databases (378) were analyzed using MEME software (Bailey
Plant Genome Sequencing
 Plant Genome Sequencing Traditional Sanger Sequencing Genome Sequencing Approach 1. Create sequencing libraries of different insert sizes 2kb o Bulk of sequencing is performed on these libraries 10kb o
Plant Genome Sequencing Traditional Sanger Sequencing Genome Sequencing Approach 1. Create sequencing libraries of different insert sizes 2kb o Bulk of sequencing is performed on these libraries 10kb o
NECTAR COLLECTION AND ANALYSES. One significant consideration when performing nectar collection is timing, both
 NECTAR COLLECTION AND ANALYSES COLLECTION PROCEDURES Important Considerations One significant consideration when performing nectar collection is timing, both developmental and circadian. This is because
NECTAR COLLECTION AND ANALYSES COLLECTION PROCEDURES Important Considerations One significant consideration when performing nectar collection is timing, both developmental and circadian. This is because
Evolution by duplication: paleopolyploidy events in plants reconstructed by deciphering the evolutionary history of VOZ transcription factors
 Gao et al. BMC Plant Biology (2018) 18:256 https://doi.org/10.1186/s12870-018-1437-8 RESEARCH ARTICLE Evolution by duplication: paleopolyploidy events in plants reconstructed by deciphering the evolutionary
Gao et al. BMC Plant Biology (2018) 18:256 https://doi.org/10.1186/s12870-018-1437-8 RESEARCH ARTICLE Evolution by duplication: paleopolyploidy events in plants reconstructed by deciphering the evolutionary
USDA-DOE Plant Feedstock Genomics for Bioenergy
 USDA-DOE Plant Feedstock Genomics for Bioenergy BERAC Thursday, June 7, 2012 Cathy Ronning, DOE-BER Ed Kaleikau, USDA-NIFA Plant Feedstock Genomics for Bioenergy Joint competitive grants program initiated
USDA-DOE Plant Feedstock Genomics for Bioenergy BERAC Thursday, June 7, 2012 Cathy Ronning, DOE-BER Ed Kaleikau, USDA-NIFA Plant Feedstock Genomics for Bioenergy Joint competitive grants program initiated
Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants
 Mohanta et al. BMC Genomics (2015) 16:58 DOI 10.1186/s12864-015-1244-7 RESEARCH ARTICLE Open Access Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel
Mohanta et al. BMC Genomics (2015) 16:58 DOI 10.1186/s12864-015-1244-7 RESEARCH ARTICLE Open Access Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel
Eud! Mag! Chl! Cer! Mon! outgroup! g 88. Eud! Cer! <50. Chl! Mag! Mon! outgroup! Eud! Mon! Cer! Mag! Chl! outgroup!
 a
a
Genomics and Bioinformatics Resources for Crop Improvement
 Genomics and Bioinformatics Resources for Crop Improvement Keiichi Mochida and Kazuo Shinozaki RIKEN Plant Science Center, Yokohama, 230-0045 Japan Corresponding author: E-mail, sinozaki@rtc.riken.jp ;
Genomics and Bioinformatics Resources for Crop Improvement Keiichi Mochida and Kazuo Shinozaki RIKEN Plant Science Center, Yokohama, 230-0045 Japan Corresponding author: E-mail, sinozaki@rtc.riken.jp ;
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed
 Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
SUPPLEMENTARY INFORMATION
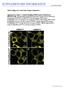 doi:10.1038/nature09606 Chen Li-Qing et al. A novel class of sugar transporters. Supplementary Figure 1. Confocal imaging of FRET sensor localization in HEK293T cells. The subcellular localization of the
doi:10.1038/nature09606 Chen Li-Qing et al. A novel class of sugar transporters. Supplementary Figure 1. Confocal imaging of FRET sensor localization in HEK293T cells. The subcellular localization of the
QENDRA E RESURSEVE GJENETIKE
 QENDRA E RESURSEVE GJENETIKE BANKA GJENETIKE ANALIZA VJETORE Dokumentimi i Resurseve Gjenetike për 6 mujorin e I rw 2011 10/02/2011 Dokumentimi i RGB 1 10/02/2011 Dokumentimi i RGB (B. Gixhari) 2 Organizimi
QENDRA E RESURSEVE GJENETIKE BANKA GJENETIKE ANALIZA VJETORE Dokumentimi i Resurseve Gjenetike për 6 mujorin e I rw 2011 10/02/2011 Dokumentimi i RGB 1 10/02/2011 Dokumentimi i RGB (B. Gixhari) 2 Organizimi
PaleoGenomics & Evolution (group PaleoEVO). UMR1095 INRA UBP Génétique, Diversité & Ecophysiologie des Céréales
 TranscriptomePartitioning Following Polyploidization Events in Grasses Journée Transcriptome URGV 1 Mai 13 Plant and Animal Genomes SPECIES COMMON NAME CHROMOSOMES GENOME (MB) ANNOTATED GENES SYNTENY DUPLICATION
TranscriptomePartitioning Following Polyploidization Events in Grasses Journée Transcriptome URGV 1 Mai 13 Plant and Animal Genomes SPECIES COMMON NAME CHROMOSOMES GENOME (MB) ANNOTATED GENES SYNTENY DUPLICATION
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011) /tpc
 Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Agave Genomics in Support
 Agave Genomics in Support of CAM Engineering cambiodesign.org Xiaohan Yang Biosciences Division Oak Ridge National Laboratory C4-CAM Conference August 09, 2013 Acknowledgements Oak Ridge National Laboratory
Agave Genomics in Support of CAM Engineering cambiodesign.org Xiaohan Yang Biosciences Division Oak Ridge National Laboratory C4-CAM Conference August 09, 2013 Acknowledgements Oak Ridge National Laboratory
Research Article mir156- and mir171-binding Sites in the Protein-Coding Sequences of Several Plant Genes
 BioMed Research International Volume, Article ID, pages http://dx.doi.org/.// Research Article mir- and mir-binding Sites in the Protein-oding Sequences of Several Plant Genes Assyl Bari, Saltanat Orazova,
BioMed Research International Volume, Article ID, pages http://dx.doi.org/.// Research Article mir- and mir-binding Sites in the Protein-oding Sequences of Several Plant Genes Assyl Bari, Saltanat Orazova,
CHAPTER 35:02 - PLANT PROTECTION: SUBSIDIARY LEGISLATION INDEX TO SUBSIDIARY LEGISLATION
 CHAPTER 35:02 - PLANT PROTECTION: SUBSIDIARY LEGISLATION INDEX TO SUBSIDIARY LEGISLATION Plant Protection Regulations PLANT PROTECTION REGULATIONS (section 29) (17th July, 2009) ARRANGEMENT OF REGULATIONS
CHAPTER 35:02 - PLANT PROTECTION: SUBSIDIARY LEGISLATION INDEX TO SUBSIDIARY LEGISLATION Plant Protection Regulations PLANT PROTECTION REGULATIONS (section 29) (17th July, 2009) ARRANGEMENT OF REGULATIONS
Supplemental Figure 1. Comparisons of GC3 distribution computed with raw EST data, bi-beta fits and complete genome sequences for 6 species.
 Supplemental Figure 1. Comparisons of GC3 distribution computed with raw EST data, bi-beta fits and complete genome sequences for 6 species. Filled distributions: GC3 computed with raw EST data. Dashed
Supplemental Figure 1. Comparisons of GC3 distribution computed with raw EST data, bi-beta fits and complete genome sequences for 6 species. Filled distributions: GC3 computed with raw EST data. Dashed
Jill M Duarte 1, P Kerr Wall 1,5, Patrick P Edger 2, Lena L Landherr 1, Hong Ma 1,4, J Chris Pires 2, Jim Leebens-Mack 3, Claude W depamphilis 1*
 RESEARCH ARTICLE Open Access Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels Jill M Duarte 1,
RESEARCH ARTICLE Open Access Identification of shared single copy nuclear genes in Arabidopsis, Populus, Vitis and Oryza and their phylogenetic utility across various taxonomic levels Jill M Duarte 1,
Scientific Names of Common Plants/Trees Scientific Names - Fruits and Vegetables
 OUR OWN HIGH SCHOOL, Al WARQA A, DUBAI BIOLOGY Grade 11 Unit 1 Diversity in the living world Name : Chapter The Living World Class and Section: Topic Binomial Nomenclature Date: s of Common Plants/Trees
OUR OWN HIGH SCHOOL, Al WARQA A, DUBAI BIOLOGY Grade 11 Unit 1 Diversity in the living world Name : Chapter The Living World Class and Section: Topic Binomial Nomenclature Date: s of Common Plants/Trees
Supporting Online Material for
 Supporting Online Material for Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Cora A. MacAlister 1, Soon Ju Park 1, Ke Jiang 1, Fabien Marcel 2, Abdelhafid Bendahmane
Supporting Online Material for Synchronization of the flowering transition by the tomato TERMINATING FLOWER gene. Cora A. MacAlister 1, Soon Ju Park 1, Ke Jiang 1, Fabien Marcel 2, Abdelhafid Bendahmane
AUXIN CONJUGATE HYDROLYSIS DURING PLANT-MICROBE INTERACTION AND EVOLUTION
 AUXIN CONJUGATE HYDROLYSIS DURING PLANT-MICROBE INTERACTION AND EVOLUTION J. Ludwig-Müller 1, A. Schuller 1, A.F. Olajide 2, V. Bakllamaja 2, J.J. Campanella 2 ABSTRACT Plants regulate auxin balance through
AUXIN CONJUGATE HYDROLYSIS DURING PLANT-MICROBE INTERACTION AND EVOLUTION J. Ludwig-Müller 1, A. Schuller 1, A.F. Olajide 2, V. Bakllamaja 2, J.J. Campanella 2 ABSTRACT Plants regulate auxin balance through
Supplementary Information
 Supplementary Information Rice APC/C TE controls tillering through mediating the degradation of MONOCULM 1 Qibing Lin 1*, Dan Wang 1*, Hui Dong 2*, Suhai Gu 1, Zhijun Cheng 1, Jie Gong 2, Ruizhen Qin 1,
Supplementary Information Rice APC/C TE controls tillering through mediating the degradation of MONOCULM 1 Qibing Lin 1*, Dan Wang 1*, Hui Dong 2*, Suhai Gu 1, Zhijun Cheng 1, Jie Gong 2, Ruizhen Qin 1,
Phylogenetic Comparison of F-Box (FBX) Gene Superfamily within the Plant Kingdom Reveals Divergent Evolutionary Histories Indicative of Genomic Drift
 Phylogenetic Comparison of F-Box (FBX) Gene Superfamily within the Plant Kingdom Reveals Divergent Evolutionary Histories Indicative of Genomic Drift Zhihua Hua 1, Cheng Zou 2, Shin-Han Shiu 2, Richard
Phylogenetic Comparison of F-Box (FBX) Gene Superfamily within the Plant Kingdom Reveals Divergent Evolutionary Histories Indicative of Genomic Drift Zhihua Hua 1, Cheng Zou 2, Shin-Han Shiu 2, Richard
PLNT2530 (2018) Unit 5 Genomes: Organization and Comparisons
 PLNT2530 (2018) Unit 5 Genomes: Organization and Comparisons Unless otherwise cited or referenced, all content of this presenataion is licensed under the Creative Commons License Attribution Share-Alike
PLNT2530 (2018) Unit 5 Genomes: Organization and Comparisons Unless otherwise cited or referenced, all content of this presenataion is licensed under the Creative Commons License Attribution Share-Alike
Supplemental Data. Perea-Resa et al. Plant Cell. (2012) /tpc
 Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Host_microbe_PPI - R package to analyse intra-species and interspecies protein-protein interactions in the model plant Arabidopsis thaliana
 Host_microbe_PPI - R package to analyse intra-species and interspecies protein-protein interactions in the model plant Arabidopsis thaliana Thomas Nussbaumer 1,2 1 Institute of Network Biology (INET),
Host_microbe_PPI - R package to analyse intra-species and interspecies protein-protein interactions in the model plant Arabidopsis thaliana Thomas Nussbaumer 1,2 1 Institute of Network Biology (INET),
Phylogenetic Analysis of the Plant-specific Zinc Finger-Homeobox and Mini Zinc Finger Gene Families
 Journal of Integrative Plant Biology 2008, 50 (8): 1031 1045 Phylogenetic Analysis of the Plant-specific Zinc Finger-Homeobox and Mini Zinc Finger Gene Families Wei Hu, Claude W. depamphilis and Hong Ma
Journal of Integrative Plant Biology 2008, 50 (8): 1031 1045 Phylogenetic Analysis of the Plant-specific Zinc Finger-Homeobox and Mini Zinc Finger Gene Families Wei Hu, Claude W. depamphilis and Hong Ma
The evolution of WRKY transcription factors
 Rinerson et al. BMC Plant Biology (2015) 15:66 DOI 10.1186/s12870-015-0456-y RESEARCH ARTICLE Open Access The evolution of WRKY transcription factors Charles I Rinerson 1, Roel C Rabara 1, Prateek Tripathi
Rinerson et al. BMC Plant Biology (2015) 15:66 DOI 10.1186/s12870-015-0456-y RESEARCH ARTICLE Open Access The evolution of WRKY transcription factors Charles I Rinerson 1, Roel C Rabara 1, Prateek Tripathi
Paleo-evolutionary plasticity of plant disease resistance genes
 Zhang et al. BMC Genomics 2014, 15:187 RESEARCH ARTICLE Paleo-evolutionary plasticity of plant disease resistance genes Rongzhi Zhang 1,2, Florent Murat 1, Caroline Pont 1, Thierry Langin 1 and Jerome
Zhang et al. BMC Genomics 2014, 15:187 RESEARCH ARTICLE Paleo-evolutionary plasticity of plant disease resistance genes Rongzhi Zhang 1,2, Florent Murat 1, Caroline Pont 1, Thierry Langin 1 and Jerome
training workshop 2015
 TransPLANT user training workshop 2015 Slides: http://tinyurl.com/transplant2015 Workshop on variation data EMBL-EBI Hinxton-UK 2nd July 2015 Ensembl Genomes Team Notes: This workshop is based on Ensembl
TransPLANT user training workshop 2015 Slides: http://tinyurl.com/transplant2015 Workshop on variation data EMBL-EBI Hinxton-UK 2nd July 2015 Ensembl Genomes Team Notes: This workshop is based on Ensembl
Identification and biochemical characterization of the fructokinase gene family in Arabidopsis thaliana
 Riggs et al. BMC Plant Biology (2017) 17:83 DOI 10.1186/s12870-017-1031-5 RESEARCH ARTICLE Open Access Identification and biochemical characterization of the fructokinase gene family in John W. Riggs 1,
Riggs et al. BMC Plant Biology (2017) 17:83 DOI 10.1186/s12870-017-1031-5 RESEARCH ARTICLE Open Access Identification and biochemical characterization of the fructokinase gene family in John W. Riggs 1,
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Zhenning Liu 1,2, Lijun Kong 1,2, Mei Zhang 1,2, Yanxia Lv 1,2, Yapei Liu 1,2, Minghau Zou 1,2, Gang Lu 1,2, Jiashu Cao 1,2, Xiaolin Yu 1,2 * Abstract
 Genome-Wide Identification, Phylogeny, Evolution and Expression Patterns of AP2/ERF Genes and Cytokinin Response Factors in Brassica rapa ssp. pekinensis Zhenning Liu 1,2, Lijun Kong 1,2, Mei Zhang 1,2,
Genome-Wide Identification, Phylogeny, Evolution and Expression Patterns of AP2/ERF Genes and Cytokinin Response Factors in Brassica rapa ssp. pekinensis Zhenning Liu 1,2, Lijun Kong 1,2, Mei Zhang 1,2,
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Nature Genetics: doi: /ng Supplementary Figure 1. ssp mutant phenotypes in a functional SP background.
 Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Genome-Wide Computational Prediction and Analysis of Core Promoter Elements across Plant Monocots and Dicots
 Genome-Wide Computational Prediction and Analysis of Core Promoter Elements across Plant Monocots and Dicots Sunita Kumari 1, Doreen Ware 1,2 * 1 Cold Spring Harbor Laboratory, Cold Spring Harbor, New
Genome-Wide Computational Prediction and Analysis of Core Promoter Elements across Plant Monocots and Dicots Sunita Kumari 1, Doreen Ware 1,2 * 1 Cold Spring Harbor Laboratory, Cold Spring Harbor, New
Accepted Article. Received Date : 19-Oct Accepted Date : 25-Jan-2017
 Received Date : 19-Oct-2016 Accepted Date : 25-Jan-2017 Article type : Original Article Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules Colin Ruprecht 1,
Received Date : 19-Oct-2016 Accepted Date : 25-Jan-2017 Article type : Original Article Phylogenomic analysis of gene co-expression networks reveals the evolution of functional modules Colin Ruprecht 1,
Lineage specific conserved noncoding sequences in plants
 Lineage specific conserved noncoding sequences in plants Nilmini Hettiarachchi Department of Genetics, SOKENDAI National Institute of Genetics, Mishima, Japan 20 th June 2014 Conserved Noncoding Sequences
Lineage specific conserved noncoding sequences in plants Nilmini Hettiarachchi Department of Genetics, SOKENDAI National Institute of Genetics, Mishima, Japan 20 th June 2014 Conserved Noncoding Sequences
Isohydric and anisohydric characterisation of vegetable crops
 Isohydric and anisohydric characterisation of vegetable crops The classification of vegetables by their physiological responses to water stress Compiled by: Acknowledgements: Sarah Limpus Horticulturist
Isohydric and anisohydric characterisation of vegetable crops The classification of vegetables by their physiological responses to water stress Compiled by: Acknowledgements: Sarah Limpus Horticulturist
Supplemental data. Vos et al. (2008). The plant TPX2 protein regulates pro-spindle assembly before nuclear envelope breakdown.
 Supplemental data. Vos et al. (2008). The plant TPX2 protein regulates pro-spindle assembly before nuclear envelope breakdown. SUPPLEMENTAL FIGURE 1 ONLINE Xenopus laevis! Xenopus tropicalis! Danio rerio!
Supplemental data. Vos et al. (2008). The plant TPX2 protein regulates pro-spindle assembly before nuclear envelope breakdown. SUPPLEMENTAL FIGURE 1 ONLINE Xenopus laevis! Xenopus tropicalis! Danio rerio!
Genome-wide Identification of Lineage Specific Genes in Arabidopsis, Oryza and Populus
 Genome-wide Identification of Lineage Specific Genes in Arabidopsis, Oryza and Populus Xiaohan Yang Sara Jawdy Timothy Tschaplinski Gerald Tuskan Environmental Sciences Division Oak Ridge National Laboratory
Genome-wide Identification of Lineage Specific Genes in Arabidopsis, Oryza and Populus Xiaohan Yang Sara Jawdy Timothy Tschaplinski Gerald Tuskan Environmental Sciences Division Oak Ridge National Laboratory
Genome-wide analysis of TCP family in tobacco
 Genome-wide analysis of TCP family in tobacco L. Chen 1,2, Y.Q. Chen 3, A.M. Ding 1, H. Chen 1,2, F. Xia 1,2, W.F. Wang 1,2 and Y.H. Sun 1 1 Key Laboratory for Tobacco Gene Resources, Tobacco Research
Genome-wide analysis of TCP family in tobacco L. Chen 1,2, Y.Q. Chen 3, A.M. Ding 1, H. Chen 1,2, F. Xia 1,2, W.F. Wang 1,2 and Y.H. Sun 1 1 Key Laboratory for Tobacco Gene Resources, Tobacco Research
The Solute Accumulation: The Mechanism for Drought Tolerance in RD23 Rice (Oryza sativa L) Lines
 R ESEARCH ARTICLE ScienceAsia 27 (2001) : 93-97 The Solute Accumulation: The Mechanism for Drought Tolerance in RD23 Rice (Oryza sativa L) Lines Montakan Vajrabhaya, Warunya Kumpun and Supachitra Chadchawan*
R ESEARCH ARTICLE ScienceAsia 27 (2001) : 93-97 The Solute Accumulation: The Mechanism for Drought Tolerance in RD23 Rice (Oryza sativa L) Lines Montakan Vajrabhaya, Warunya Kumpun and Supachitra Chadchawan*
Evolution of tonoplast P-ATPase transporters involved in vacuolar acidification
 Research Evolution of tonoplast P-ATPase transporters involved in vacuolar acidification Yanbang Li 1,2 *, Sofia Provenzano 1,3 *, Mattijs Bliek 1,2 *, Cornelis Spelt 1,2, Ingo Appelhagen 4, Laura Machado
Research Evolution of tonoplast P-ATPase transporters involved in vacuolar acidification Yanbang Li 1,2 *, Sofia Provenzano 1,3 *, Mattijs Bliek 1,2 *, Cornelis Spelt 1,2, Ingo Appelhagen 4, Laura Machado
Supplementary Information. Drought response transcriptomics are altered in poplar with reduced tonoplast sucrose transporter expression
 Supplementary Information Drought response transcriptomics are altered in poplar with reduced tonoplast sucrose transporter expression Liang Jiao Xue, Christopher J. Frost, Chung Jui Tsai, Scott A. Harding
Supplementary Information Drought response transcriptomics are altered in poplar with reduced tonoplast sucrose transporter expression Liang Jiao Xue, Christopher J. Frost, Chung Jui Tsai, Scott A. Harding
SUPPLEMENTARY INFORMATION
 reverse 3175 3175 F L C 318 318 3185 3185 319 319 3195 3195 315 8 1 315 3155 315 317 Supplementary Figure 3. Stability of expression of the GFP sensor constructs return to warm conditions. Semi-quantitative
reverse 3175 3175 F L C 318 318 3185 3185 319 319 3195 3195 315 8 1 315 3155 315 317 Supplementary Figure 3. Stability of expression of the GFP sensor constructs return to warm conditions. Semi-quantitative
New insights into the evolution and functional divergence of the SWEET family in Saccharum based on comparative genomics
 Hu et al. BMC Plant Biology (2018) 18:270 https://doi.org/10.1186/s12870-018-1495-y RESEARCH ARTICLE Open Access New insights into the evolution and functional divergence of the SWEET family in Saccharum
Hu et al. BMC Plant Biology (2018) 18:270 https://doi.org/10.1186/s12870-018-1495-y RESEARCH ARTICLE Open Access New insights into the evolution and functional divergence of the SWEET family in Saccharum
The Plant Cell, November. 2017, American Society of Plant Biologists. All rights reserved
 The Genetics of Floral Development Teaching Guide Overview The development of flowers in angiosperm plants provided a critical evolutionary advantage, allowing more options for pollen dispersal and seed
The Genetics of Floral Development Teaching Guide Overview The development of flowers in angiosperm plants provided a critical evolutionary advantage, allowing more options for pollen dispersal and seed
** * * * Col-0 cau1 CAU1. Actin2 CAS. Actin2. Supplemental Figure 1. CAU1 affects calcium accumulation.
 Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
The Populus Class III HD ZIP Transcription Factor POPCORONA Affects Cell Differentiation during Secondary Growth of Woody Stems
 The Populus Class III HD ZIP Transcription Factor POPCORONA Affects Cell Differentiation during Secondary Growth of Woody Stems Juan Du 1 a, Eriko Miura 1, Marcel Robischon 1 b, Ciera Martinez 2, Andrew
The Populus Class III HD ZIP Transcription Factor POPCORONA Affects Cell Differentiation during Secondary Growth of Woody Stems Juan Du 1 a, Eriko Miura 1, Marcel Robischon 1 b, Ciera Martinez 2, Andrew
Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants
 Liu et al. BMC Evolutionary Biology (2017) 17:47 DOI 10.1186/s12862-017-0891-5 RESEARCH ARTICLE Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants
Liu et al. BMC Evolutionary Biology (2017) 17:47 DOI 10.1186/s12862-017-0891-5 RESEARCH ARTICLE Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants
An Overview on Stress Signals in Relation to Flowering in Litchi chinensis and Dimocarpus longan
 An Overview on Stress Signals in Relation to Flowering in Litchi chinensis and Dimocarpus longan Biyan Zhou and Houbin Chen College of Horticulture South China Agricultural University Guangzhou, China
An Overview on Stress Signals in Relation to Flowering in Litchi chinensis and Dimocarpus longan Biyan Zhou and Houbin Chen College of Horticulture South China Agricultural University Guangzhou, China
Boyce Thompson Institute
 Joyce Van Eck (BTI) Plant Biotechnology and Translational Research Boyce Thompson Institute Mission: To advance and communicate scientific knowledge in plant biology to improve agriculture, protect the
Joyce Van Eck (BTI) Plant Biotechnology and Translational Research Boyce Thompson Institute Mission: To advance and communicate scientific knowledge in plant biology to improve agriculture, protect the
Journal of Integrative Agriculture 2017, 16(5): Available online at ScienceDirect
 Journal of Integrative Agriculture 2017, 16(5): 60345-7 Available online at www.sciencedirect.com ScienceDirect RESEARCH ARTICLE Characterization and expression analysis of a novel RING-HC gene, ZmRHCP1,
Journal of Integrative Agriculture 2017, 16(5): 60345-7 Available online at www.sciencedirect.com ScienceDirect RESEARCH ARTICLE Characterization and expression analysis of a novel RING-HC gene, ZmRHCP1,
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
Cryopreservation of syn seeds
 COST Action FA1104 Sustainable production of high-quality cherries for the European market Long Term Preservation of Woody Species by Cryo Techniques, March 2015, Firenze Cryopreservation of syn seeds
COST Action FA1104 Sustainable production of high-quality cherries for the European market Long Term Preservation of Woody Species by Cryo Techniques, March 2015, Firenze Cryopreservation of syn seeds
Development 143: doi: /dev : Supplementary information
 Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
European Phenology. Dutch phenological Network Natuurkalender. Network
 European Phenology Network Dutch phenological Network Natuurkalender Arnold J.H. van Vliet Environmental Systems Analysis Group Wageningen University Content European Phenology Network Issues: Observation
European Phenology Network Dutch phenological Network Natuurkalender Arnold J.H. van Vliet Environmental Systems Analysis Group Wageningen University Content European Phenology Network Issues: Observation
Powdery scab management in South Africa
 Powdery scab management in South Africa Prof Jacquie van der Waals, University of Pretoria Spongospora subterranea f. sp. subterranea (Sss) is an increasingly important pathogen of potatoes globally. Infection
Powdery scab management in South Africa Prof Jacquie van der Waals, University of Pretoria Spongospora subterranea f. sp. subterranea (Sss) is an increasingly important pathogen of potatoes globally. Infection
Fractionation resistance of duplicate genes following whole genome duplication in plants as a function of gene ontology category and expression level
 Fractionation resistance of duplicate genes following whole genome duplication in plants as a function of gene ontology category and expression level Eric Chun-Hung Chen Thesis submitted to the Faculty
Fractionation resistance of duplicate genes following whole genome duplication in plants as a function of gene ontology category and expression level Eric Chun-Hung Chen Thesis submitted to the Faculty
State Key Laboratory of Crop Stress Biology in Arid Areas, Northwest A&F University, Yangling, Shaanxi, China
 Molecular cloning and expression of the male sterility-related CtYABBY1 gene in flowering Chinese cabbage (Brassica campestris L. ssp chinensis var. parachinensis) X.L. Zhang 1,2,3 and L.G. Zhang 1,2,3
Molecular cloning and expression of the male sterility-related CtYABBY1 gene in flowering Chinese cabbage (Brassica campestris L. ssp chinensis var. parachinensis) X.L. Zhang 1,2,3 and L.G. Zhang 1,2,3
Available online at International journal of Advanced Biological and Biomedical Research. Volume 2, Issue 2, 2014:
 Available online at http://www.ijabbr.com International journal of Advanced Biological and Biomedical Research Volume 2, Issue 2, 2014: 504-509 Bioinformatics comparison of codon usage of genes encoding
Available online at http://www.ijabbr.com International journal of Advanced Biological and Biomedical Research Volume 2, Issue 2, 2014: 504-509 Bioinformatics comparison of codon usage of genes encoding
Effect of Acetosyringone on Agrobacterium-mediated Transformation of Eustoma grandiflorum Leaf Disks
 JARQ 51 (4), 351-355 (2017) https://www.jircas.go.jp Improvement in Agrobacterium-mediated Transformation of Eustoma grandiflorum by Acetosyringone Effect of Acetosyringone on Agrobacterium-mediated Transformation
JARQ 51 (4), 351-355 (2017) https://www.jircas.go.jp Improvement in Agrobacterium-mediated Transformation of Eustoma grandiflorum by Acetosyringone Effect of Acetosyringone on Agrobacterium-mediated Transformation
Identification and characterization of two wheat Glycogen Synthase Kinase 3/ SHAGGY-like kinases
 Bittner et al. BMC Plant Biology 2013, 13:64 RESEARCH ARTICLE Open Access Identification and characterization of two wheat Glycogen Synthase Kinase 3/ SHAGGY-like kinases Thomas Bittner 1, Sarah Campagne
Bittner et al. BMC Plant Biology 2013, 13:64 RESEARCH ARTICLE Open Access Identification and characterization of two wheat Glycogen Synthase Kinase 3/ SHAGGY-like kinases Thomas Bittner 1, Sarah Campagne
... -: Crop physiology. - : Crop technology. -: Crop improvement. -: Crop production.
 .:.:.: /.: - Thomas Malthus...: : : : : :...: : : : : : (Malthus ) beta."a" "carotene Bt Cotton No Tillage ( ) Minimum Tillage ) ( - : :) : :) : :) : :.( 2012 2.1 :) : : : Field crops science Agro Agronomy
.:.:.: /.: - Thomas Malthus...: : : : : :...: : : : : : (Malthus ) beta."a" "carotene Bt Cotton No Tillage ( ) Minimum Tillage ) ( - : :) : :) : :) : :.( 2012 2.1 :) : : : Field crops science Agro Agronomy
Supplementary Methods
 Supplementary Methods Microarray analysis Grains of 7 DAP of the wild-type and gif1 were harvested for RNA preparation. Microarray analysis was performed with the Affymetrix (Santa Clara, CA) GeneChip
Supplementary Methods Microarray analysis Grains of 7 DAP of the wild-type and gif1 were harvested for RNA preparation. Microarray analysis was performed with the Affymetrix (Santa Clara, CA) GeneChip
Supplement of Ice nucleators, bacterial cells and Pseudomonas syringae in precipitation at Jungfraujoch
 Supplement of Biogeosciences, 14, 1189 1196, 2017 http://www.biogeosciences.net/14/1189/2017/ doi:10.5194/bg-14-1189-2017-supplement Author(s) 2017. CC Attribution 3.0 License. Supplement of Ice nucleators,
Supplement of Biogeosciences, 14, 1189 1196, 2017 http://www.biogeosciences.net/14/1189/2017/ doi:10.5194/bg-14-1189-2017-supplement Author(s) 2017. CC Attribution 3.0 License. Supplement of Ice nucleators,
Supporting Information
 Supporting Information Fawcett et al. 10.1073/pnas.0900906106 SI Text Estimating the Age of Gene Duplication Events: The Use of K S Values. One of the most common methods used to study and visualize gene
Supporting Information Fawcett et al. 10.1073/pnas.0900906106 SI Text Estimating the Age of Gene Duplication Events: The Use of K S Values. One of the most common methods used to study and visualize gene
Curriculum Vitae. development / Plant growth regulators / Tissue culture / Design and analysis of agricultural experiments
 Curriculum Vitae Prof. Dr. Majeed Kadhim Abbas Al-Hamzawi E-mail: Majeed_edu@yahoo.com Mobile: 0780-2344057 Academic Certificate Academic Certificate University Faculty Department Graduation Year Bachelor
Curriculum Vitae Prof. Dr. Majeed Kadhim Abbas Al-Hamzawi E-mail: Majeed_edu@yahoo.com Mobile: 0780-2344057 Academic Certificate Academic Certificate University Faculty Department Graduation Year Bachelor
A dual sgrna approach for functional genomics in Arabidopsis thaliana
 A dual sgrna approach for functional genomics in Arabidopsis thaliana CRISPR AgBio Europe 2017 Laurens Pauwels #CRISPRAgBio @laurenspauwels Outline - Workflow for CRISPR/Cas9 in Arabidopsis thaliana -
A dual sgrna approach for functional genomics in Arabidopsis thaliana CRISPR AgBio Europe 2017 Laurens Pauwels #CRISPRAgBio @laurenspauwels Outline - Workflow for CRISPR/Cas9 in Arabidopsis thaliana -
Supplemental Data. Wang et al. (2014). Plant Cell /tpc
 Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Cloning and expression analysis of an E-class MADS-box gene from Populus deltoides
 African Journal of Biotechnology Vol. 8 (19), pp. 4789-4796, 5 October, 2009 Available online at http://www.academicjournals.org/ajb ISSN 1684 5315 2009 Academic Journals Full Length Research Paper Cloning
African Journal of Biotechnology Vol. 8 (19), pp. 4789-4796, 5 October, 2009 Available online at http://www.academicjournals.org/ajb ISSN 1684 5315 2009 Academic Journals Full Length Research Paper Cloning
Plant Nucleotide Binding Site Leucine-Rich Repeat (NBS-LRR) Genes: Active Guardians in Host Defense Responses
 Int. J. Mol. Sci. 2013, 14, 7302-7326; doi:10.3390/ijms14047302 Review OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Plant Nucleotide Binding Site Leucine-Rich
Int. J. Mol. Sci. 2013, 14, 7302-7326; doi:10.3390/ijms14047302 Review OPEN ACCESS International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Plant Nucleotide Binding Site Leucine-Rich
Ascorbate peroxidase-related (APx-R) is a new hemecontaining protein functionally associated with ascorbate peroxidase but evolutionarily divergent
 Research Ascorbate peroxidase-related (APx-R) is a new hemecontaining protein functionally associated with ascorbate peroxidase but evolutionarily divergent Fernanda Lazzarotto 1 *, Felipe Karam Teixeira
Research Ascorbate peroxidase-related (APx-R) is a new hemecontaining protein functionally associated with ascorbate peroxidase but evolutionarily divergent Fernanda Lazzarotto 1 *, Felipe Karam Teixeira
Integrating Phylogenetic and Network Approaches to Study Gene Family Evolution: The Case of the AGAMOUS Family of Floral Genes
 University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications from the Center for Plant Science Innovation Plant Science Innovation, Center for 2018 Integrating
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications from the Center for Plant Science Innovation Plant Science Innovation, Center for 2018 Integrating
Slovene Plant Gene Bank (SPGB) and Genetic Resources Programme
 Slovene Plant Gene Bank (SPGB) and Genetic Resources Programme Second Meeting of the ECPGR Working Group on Leafy Vegetables 8 9 October, Ljubljana, Slovenia Vladimir MEGLIČ, Jelka ŠUŠTAR VOZLIČ Slovene
Slovene Plant Gene Bank (SPGB) and Genetic Resources Programme Second Meeting of the ECPGR Working Group on Leafy Vegetables 8 9 October, Ljubljana, Slovenia Vladimir MEGLIČ, Jelka ŠUŠTAR VOZLIČ Slovene
