Supplemental Information. Expanded Coverage of the 26S Proteasome. Conformational Landscape Reveals. Mechanisms of Peptidase Gating
|
|
|
- Iris Snow
- 5 years ago
- Views:
Transcription
1 Cell Reports, Volume 24 Supplemental Information Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating Markus R. Eisele, Randi G. Reed, Till Rudack, Andreas Schweitzer, Florian Beck, Istvan Nagy, Günter Pfeifer, Jürgen M. Plitzko, Wolfgang Baumeister, Robert J. Tomko Jr., and Eri Sakata
2 Table S1 related to Main Figures 1-7: Yeast strains used in this study. Name RTY1 RTY432 RTY1168 RTY1171 RTY1173 RTY1179 RTY1301 RTY1321 RTY1372 RTY1504 RTY1506 RTY1664 RTY2013 RTY2033 RTY2091 RTY2099 RTY2112 RTY2123 RTY2166 RTY2135 RTY2137 YYS40 Genotype MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 (alias MHY500) MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 RPN5-6xGly-3xFLAG:hphMX4 rpt3δ::his3 [YCplac33-RPT3] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt1δ::his3 [pfl44-rpt1] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt4δ::his3 [YCplac33-RPT4] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt6δ::his3 [YCplac33-RPT6] MATα his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt6-δ1:natmx4 MATα his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpn6δ::natmx4 rpt6δ::his3 [prs316-rpn6; YCplac33-RPT6] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt2δ::his3 [prs316-rpt2] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpn7δ::natmx4 rpt2δ::his3 [prs316- RPN7; prs316-rpt2] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt3δ::his3 [YCplac33-RPT3] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt5δ::his3 [YCplac33-RPT5] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt1-δ1:kanmx6 MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 RPN5-6xGly-3xFLAG:hphMX4 rpt6δ::his3 [YCplac33-RPT6] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 RPN5-6xGly-3xFLAG:hphMX4 rpt2δ::his3 [prs316-rpt2] MATα his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 RPN7(D123C)-6xGly-V5:kanMX6 MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 RPT2(R407C):natMX4 MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 RPN7(D123C)-6xGly-V5:kanMX6 RPT2(R407C):natMX4 MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 rpt1-δ1:kanmx6 rpt6-δ1:natmx4 MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 pre8δ::his3 pre9δ::natmx4 [prs316-pre8] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 RPN7(D123C)-6xGly-V5:kanMX6 RPT2(R407C):natMX4 rpt3δ::his3 [YCplac33-RPT3] MATa his3-200 leu2-3,112 ura3-52 lys2-801 trp1-1 RPN7(D123C)-6xGly-V5:kanMX6 RPT2(R407C):natMX4 rpt6δ::his3 [YCplac33-RPT6] MATa ade2-1, ura3-1, his3-11, trp1-1, leu2-3,112, can1-100, rpn11::rpn11-3xflag- CYC1term-HIS3
3 Table S2 related to Main Figures 1-7: Plasmids used in this study. Plasmid Genotype Source prt357 prs314-rpt1 This study prt364 YCplac111-RPT3 This study prt702 YCplac111-RPT2 This study prt1122 pet42b-6his-cys-rpn12(c23s,d265a) (Tomko et al., 2015) prt1408 prs315-rpt5 This study prt1409 prs314-rpt1(e310q) This study prt1410 YCplac111-rpt2(E283Q) This study prt1411 YCplac111-rpt3(E273Q) This study prt1413 prs315-rpt5(e282q) This study prt1425 prs314-ha-rpn6 This study prt1496 YCplac111-RPT6 This study prt1497 YCplac111-rpt6(E249Q) This study prt1528 YCplac111-RPT4 This study prt1529 YCplac111-rpt4(E283Q) This study prt1780 prs314-ha-rpn6(t203c) This study prt1783 YCplac111-RPT2(R407C) This study prt1784 YCplac111-RPT6(G387C) This study prt1834 YCplac111-rpt2(E283Q, R407C) This study pes1 pet28a-5ub-dhfr-ur-his This study
4 A RPN7 rpn7-c RPN7 rpn7-c RPT2 RPT2 rpt2-c rpt2-c YPD, 2d, 30 o C YPD, 2d, 37 o C SC, 2d, 30 o C SC + 30 μm CdCl 2, 2d, 33 o C B Rpn6 Rpt6 s1: ~6 Å s2: ~16 Å s3: ~16 Å s4: ~10 Å C RPN6 rpn6-c RPN6 rpn6-c RPT6 RPT6 rpt6-c rpt6-c 30 o C, 2d D + DTT kda AMP- ATP ATP ATP PNP ATP minutes CuCl 2 * HA-Rpn6^Rpt6 50 HA-Rpn6 RPN6 RPT6 rpn6-c RPT6 RPN6 rpt6-c rpn6-c rpt6-c Figure S1
5 Figure S1 related to Figure 1. A, Cysteine substitutions do not impact cell health. The strains shown were plated as serial dilutions on YPD, SC, or SC+ 30 μm CdCl 2 plates and incubated at the temperatures shown. B, Rpn6-T203 and Rpt6-G387 serve as an alternative conformational reporter. Rpn6 is shown in gold, Rpt6 in blue, and the other five Rpt subunits are shown in grey. All other subunits are omitted for clarity. The T203 and G387C residues are shown as red spheres, and the distances between their α carbons is listed in Angstroms. C, No obvious growth impairment in cells harboring the T203C and G387C substitutions. Serial dilutions were performed as in (A) above. D, Disulfide crosslinking of Rpn6 and Rpt6 is dependent upon engineered cysteines and impacted by nucleotide. Crosslinking was induced in whole cell extracts (WCE) prepared in the presence of 2 mm of the indicated nucleotide with CuCl 2 for ten minutes. Proteins were resolved by non-denaturing SDS-PAGE and examined by anti-ha (Rpn6) immunoblotting. For the last lane, the WCE was incubated with 10 mm DTT for 10 minutes prior to gel loading.
6 FSC A B C s3 50 nm 50 nm s4 s5 s D E Resolution (1/Å) ATPγS 2 mm + substrate Ub Ub Ub Ub Ub DHFR UR ATPγS 2 mm - substrate 0% 20% 40% 60% 80% 100% s1 s3 s4 s5 F G s3 s4 H s5 vs. s1 s5 vs. s2 s5 vs. s3 s5 vs. s4 aligned to CP Å aligned to CP Å aligned to ATPase Å Figure S2
7 Figure S2 related to Figure 2. A, Double capped particles are predominantly visible on a typical micrograph. B, Particles were 2D sorted and only suitable reference-free 2D class averages were used for further 3D analysis. C, Combined particles of each state resulted in a resolution of 4.5 Å for s4, 4.9 Å for s5, 5.4 Å for s3 and 6.1 Å for s6 on the basis of the gold-standard FSC criterion (FSC 0.143). D, Schematic of linear ubiquitinated DHFR with an unstructured region at the C-terminus. E, State distribution of classified 2 mm ATPγS data set with and without model substrate. F and G, Cryo-EM reconstruction of classified s3 particles (F) and s4 particles (G). The 26S proteasome is colored according to Fig 2B. A clear density is visible in the center of the 20S in the s3 state and no density is visible in the s4 state. H, Residue-wise RMSD of the whole 26S proteasome, the 20S gate and the AAA+ ATPase (in Å) between the s5 vs. s1 state (left), s5 vs. s2 state, s5 vs. s3 state and s5 vs. s4 state, aligned to the CP or to the ATPase. For the gate RMSD plots, the compared structures (either s1, s2, s3 or s4) are shown. In all other panels, the measured RMSD is plotted on the s5 structure.
8 A RPT2 rpt2-eq YPD, 2d, 37 o C B C RP 2 CP RP 1 CP RP Lid RP 2 CP RP 1 CP Blm10-CP CP Intermediates Rpn12 Anti-Rpn12 (lid) blot Anti-20S (CP) blot Figure S3
9 Figure S3 related to Figure 3. A, No evidence of temperature sensitivity in rpt2-eq yeast. WT or rpt2-eq yeast were struck on YPD, and incubated at 37 o C for two days. B and C, Native gel immunoblot analysis of the indicated strains with anti-rpn12 (B) or anti-20s (C) antibodies reveal no obvious structural defects. CP assembly intermediates are indicated with a bracket.
10 s6 vs. s1 s6 vs. s2 s6 vs. s3 s6 vs. s4 s6 vs. s5 aligned to CP Å aligned to CP Å aligned to ATPase Å Figure S4
11 Figure S4 related to Figure 4. Residue-wise RMSD of the whole 26S proteasome, the 20S gate and the AAA+ ATPase (in Å) between the s6 vs. s1 state (left), s6 vs. s2 state, s6 vs. s3 state, s6 vs. s4 state and s6 vs. s5 state, aligned to the CP or to the ATPase. For the gate RMSD plots, the compared structures (either s1, s2, s3, s4 or s5) are shown. In all other panels, the RMSD is plotted on the s6 structure.
12 correlation score h (Å) A s1 s2 s3 s4 s5 s6 Rpt5 Rpt4 Rpt1 Rpt3 Rpt6 Rpt2 B Pocket distance s1 s2 s3 s4 s5 s Rpt3-Rpt6 Rpt6-Rpt2 Rpt2-Rpt1 Rpt1-Rpt5 Rpt5-Rpt4 Rpt4-Rpt3 C H10 H10 Phe nucleotide nucleotide cluster, engaged cluster, open Rpt3- Rpt6 Rpt6- Rpt2 Rpt2- Rpt1 Rpt1- Rpt5 Rpt5- Rpt4 Rpt4- Rpt3 s1 s2 s5 s3 s6 s Figure S5
13 Figure S5 related to Figure 5. A, Comparison of nucleotide bound states in all six states. A simulated map of the AAA+ ring model without nucleotides was subtracted from the experimental map. EMD-3534 and EMD-3535 were used for s1 and s2 (Wehmer et al., 2017). The difference maps always show six nucleotides for each state. B, The state of the pocket is determined by measuring the distance between the end of the H10 helix of one Rpt subunit and the tip of H6 helix of the adjacent clockwise subunit. Color coding of each state would corresponds to the one in Figure 6A. C, Assessment of the EM density of the nucleotide binding pocket. Volumes of all Rpt subunits interfaces were aligned and hierarchically clustered by their similarity. The aligned densities were divided into two large clusters; the average of the blue class is assigned to the engaged conformation whereas the gray is assigned to the open conformation. An overview of the clustering results of the Rpt interface is shown in right lower panel. The numbers in the columns correspond to the numbering of the clustering on the left.
14 A Rpt6 C-terminus Rpt1 C-terminus α3 α5 α2 α4 B C hsrpt1 mmrpt1 drrpt1 xlrpt1 dmrpt1 atrpt1 scrpt1 406 EKDFLEAVNKVIKSYAKFSATPRYMTYN 448 EKDFLEAVNKVIKSYAKFSATPRYMTYN 406 EKDFLEAVNKVIKSYAKFSATPRYMTYN 406 EKDFLEAVNKVIKSYAKFSATPRYMTYN 406 EKDFLEAVKKVIKSYAKFSATPRYMTYN 399 EKDFLDAVNKVIKGYQKFSATPKYMVYN 440 EKDFLKAVDKVISGYKKFSSTSRYMQYN hsrpt6 mmrpt6 drrpt6 xlrpt6 dmrpt6 atrpt6 scrpt6 380 QEDFEMAVAKVMQKDSEKNMSIKKLWK 380 QEDFEMAVAKVMQKDSEKNMSIKKLWK 380 QEDFEMAVAKVMQKDSEKNMSIKKLWK 388 QEDFEMAVAKVMQKDSEKNMSIKKLWK 379 QEDFEMAVAKVMQKDSEKNMSIKKLWK 378 QEDFEMAVSKVMKKEGESNMSLQRLWK 379 QEDFELAVGKVMNKNQETAISVAKLFK Figure S6
15 Figure S6 related to Figure 6. A, EM densities of the C-termini of Rpt6 (left) and Rpt1 (right). The Rpt6 C-termius is inserted between α2 and α3 and the Rpt1 C-terminus is inserted between α4 and α5. The EM density is depicted in gray. B, C, Boxshade alignments of the C-termini of Rpt1 (B) and Rpt6 (C) demonstrate high conservation of C- terminal amino acids.
16 A B C α1 - subunit S. cerevisiae 1 MSGAAAASAAGYDRHITIFSPEGRLYQVEYAFKATNQTNINSLAVRGKDCTVVISQKKVP 60 H. sapiens 1 ---MSRGSSAGFDRHITIFSPEGRLYQVEYAFKAINQGGLTSVAVRGKDCAVIVTQKKVP 57 M. musculus 1 ---MSRGSSAGFDRHITIFSPEGRLYQVEYAFKAINQGGLTSVAVRGKDCAVIVTQKKVP 57 A. thaliana 1 ---MSRGSGAGYDRHITIFSPEGRLFQVEYAFKAVKTAGITSIGVRGKDSVCVVTQKKVP 57 D. melanogaster 1 ---MSRGSSAGFDRHITIFSPEGRLYQVEYAFKAIAQENITTVALKSGDCAVVATQKKVT 57 M. jannaschii 1 GSHMQMVPPSAYDRAITVFSPEGRLYQVEYAREAVRRG-TTAIGIACKDGVVLAVDRRIT 60 S AV α1 α2 α3 α4 α5 α6 α7 S. cerevisiae YD P Y YSF P Y YD P Y YD P Y YD P Y YD P Y YD P Y H. sapiens FD P Y YSF P Y YD P Y YD P Y YD P Y YD P Y YD P Y M. musculus FD P Y YSF P Y YD P Y YD P Y YD P Y YD P Y YD P Y A. thaliana YD P F YSF P H YD P Y YD P Y YD P Y YD P Y YD P Y D. melanogaster FD P Y YSF P Y YD P Y YD P Y YD P Y YD P Y YD P Y M. jannaschii YD P Y YDR P Y YD P Y YD P Y YD P Y YD P Y YD P Y α6 α7 D α5 Rpt1 α4 α-helix (H0) canonical cluster Y6 Rpt5 Rpt2 α3 D9 α6 P15 Y26 Rpt6 α2 α7 α1 Rpt3 E closed (white) - open (color) ~3.5 Å P14 F Activated states (ATP cycle) Primed states α2 Ground state (Inactive) s4 s5 s7(?) s6 Rpt6 α3 s1 Substrate binding s2 s8(?) s3 s9(?) Figure S7
17 Figure S7 related to Figure 7. A, Sequence alignment between S. cerevisiae, H. sapiens, M. musculus, A. thaliana, D. melanogaster and M. jannaschii of the first 60 amino acids of α1. The canonical clusters are marked in blue and the first α-helix in green. B, Sequence alignment of the canonical cluster amino acids of all seven α-subunits from S. cerevisiae, H. sapiens, M. musculus, A. thaliana, D. melanogaster and M. jannaschii. All canonical cluster amino acids but those from α2 of M. jannaschii are highly conserved, which explains the non-canoncial cluster formed between α1- α2. C, Example of YD-P-Y motif between α6 and α7 in the s5 state. The EM density is shown as mesh. D, The α subunit N-termini of α2, α3 and α4 undergo large movements between the closed and open gate conformations. Each N-terminus is depicted in a different color in the s5 state: α1 light orange, α2 light blue, α3 dark green, α4 yellow, α5 dark blue, α6 dark orange, α7 light pink. The localization of the N-terminal extensions of α2, α3 and α4 in the s2 state are visible in white. The structures of s2 and s5 were aligned onto the complete α chains of the CP. E, A proline movement of ~3.5 Å in α2 can be identified between all open and closed structures, here shown in states s5 (colored, light blue and dark green) and s2 (white). The C-terminus of Rpt6 inserts into the α subunit pocket between α2 and α3 in s3 (closed) and all open gate states, s4, s5 and s6. F, Model for ATP hydrolysis by 26S proteasome. In the absence of substrate, the proteasome is present in the ground state (s1), which represents the lowest energy conformation amongst the conformational states. When the proteasome is activated, most likely by substrate binding (Lu et al., 2015), it undergoes a conformational change from the ground state to a primed state (s2 or s5). The translocation of substrate may be triggered by ATP hydrolysis by one of the initiating ATPases coupled with ATP binding by Rpt6, leading the proteasome to an activated state (s3, s4 and s6). ATP hydrolysis can occur in either the lowermost engaged and most likely ATP-bound subunit, as was suggested for Yme1 (Puchades et al., 2017), or the highest engaged subunit (Martin et al., 2008), and continues around the ring. ATP hydrolysis causes a major conformational change in the neighboring subunits, leading to a new nucleotide-pocket configuration and to the next proteasome state of the hydrolysis cycle. Additional states may exist (s7, s8, s9). It is probable that the cycle continues until substrate translocation is completed. The cycle could include reversion to s2, s5, or s1.
Supplementary Information: Long range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Information: Long range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Information: Long range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs
Supplemental Data SUPPLEMENTAL FIGURES
 Supplemental Data CRYSTAL STRUCTURE OF THE MG.ADP-INHIBITED STATE OF THE YEAST F 1 C 10 ATP SYNTHASE Alain Dautant*, Jean Velours and Marie-France Giraud* From Université Bordeaux 2, CNRS; Institut de
Supplemental Data CRYSTAL STRUCTURE OF THE MG.ADP-INHIBITED STATE OF THE YEAST F 1 C 10 ATP SYNTHASE Alain Dautant*, Jean Velours and Marie-France Giraud* From Université Bordeaux 2, CNRS; Institut de
Supplementary information Supplementary figures
 Supplementary information Supplementary figures Supplementary Figure 1. Quantitation by FCS of GFP-tagged proteasomes in supernatant of cell extract (a) In vitro assay for evaluation of the hydrodynamic
Supplementary information Supplementary figures Supplementary Figure 1. Quantitation by FCS of GFP-tagged proteasomes in supernatant of cell extract (a) In vitro assay for evaluation of the hydrodynamic
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Purification, SDS-PAGE and cryo-em characterization of the MCM hexamer and Cdt1 MCM heptamer samples.
 Supplementary Figure 1 Purification, SDS-PAGE and cryo-em characterization of the MCM hexamer and Cdt1 MCM heptamer samples. (a-b) SDS-PAGE analysis of the hexamer and heptamer samples. The eluted hexamer
Supplementary Figure 1 Purification, SDS-PAGE and cryo-em characterization of the MCM hexamer and Cdt1 MCM heptamer samples. (a-b) SDS-PAGE analysis of the hexamer and heptamer samples. The eluted hexamer
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase Cwc27
 Acta Cryst. (2014). D70, doi:10.1107/s1399004714021695 Supporting information Volume 70 (2014) Supporting information for article: Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase
Acta Cryst. (2014). D70, doi:10.1107/s1399004714021695 Supporting information Volume 70 (2014) Supporting information for article: Structure and evolution of the spliceosomal peptidyl-prolyl cistrans isomerase
Supplementary Figure 1. Aligned sequences of yeast IDH1 (top) and IDH2 (bottom) with isocitrate
 SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Aligned sequences of yeast IDH1 (top) and IDH2 (bottom) with isocitrate dehydrogenase from Escherichia coli [ICD, pdb 1PB1, Mesecar, A. D., and Koshland,
SUPPLEMENTARY FIGURE LEGENDS Supplementary Figure 1. Aligned sequences of yeast IDH1 (top) and IDH2 (bottom) with isocitrate dehydrogenase from Escherichia coli [ICD, pdb 1PB1, Mesecar, A. D., and Koshland,
Supporting Information
 Supporting Information Ottmann et al. 10.1073/pnas.0907587106 Fig. S1. Primary structure alignment of SBT3 with C5 peptidase from Streptococcus pyogenes. The Matchmaker tool in UCSF Chimera (http:// www.cgl.ucsf.edu/chimera)
Supporting Information Ottmann et al. 10.1073/pnas.0907587106 Fig. S1. Primary structure alignment of SBT3 with C5 peptidase from Streptococcus pyogenes. The Matchmaker tool in UCSF Chimera (http:// www.cgl.ucsf.edu/chimera)
Structure and RNA-binding properties. of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex
 Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
type GroEL-GroES complex. Crystals were grown in buffer D (100 mm HEPES, ph 7.5,
 Supplementary Material Supplementary Materials and Methods Structure Determination of SR1-GroES-ADP AlF x SR1-GroES-ADP AlF x was purified as described in Materials and Methods for the wild type GroEL-GroES
Supplementary Material Supplementary Materials and Methods Structure Determination of SR1-GroES-ADP AlF x SR1-GroES-ADP AlF x was purified as described in Materials and Methods for the wild type GroEL-GroES
Supplementary figure 1. Comparison of unbound ogm-csf and ogm-csf as captured in the GIF:GM-CSF complex. Alignment of two copies of unbound ovine
 Supplementary figure 1. Comparison of unbound and as captured in the GIF:GM-CSF complex. Alignment of two copies of unbound ovine GM-CSF (slate) with bound GM-CSF in the GIF:GM-CSF complex (GIF: green,
Supplementary figure 1. Comparison of unbound and as captured in the GIF:GM-CSF complex. Alignment of two copies of unbound ovine GM-CSF (slate) with bound GM-CSF in the GIF:GM-CSF complex (GIF: green,
SUPPLEMENTARY INFORMATION
 Supplementary Table S1 Kinetic Analyses of the AMSH-LP mutants AMSH-LP K M (μm) k cat x 10-3 (s -1 ) WT 71.8 ± 6.3 860 ± 65.4 T353A 76.8 ± 11.7 46.3 ± 3.7 F355A 58.9 ± 10.4 5.33 ± 0.30 proximal S358A 75.1
Supplementary Table S1 Kinetic Analyses of the AMSH-LP mutants AMSH-LP K M (μm) k cat x 10-3 (s -1 ) WT 71.8 ± 6.3 860 ± 65.4 T353A 76.8 ± 11.7 46.3 ± 3.7 F355A 58.9 ± 10.4 5.33 ± 0.30 proximal S358A 75.1
Nature Structural and Molecular Biology: doi: /nsmb.2938
 Supplementary Figure 1 Characterization of designed leucine-rich-repeat proteins. (a) Water-mediate hydrogen-bond network is frequently visible in the convex region of LRR crystal structures. Examples
Supplementary Figure 1 Characterization of designed leucine-rich-repeat proteins. (a) Water-mediate hydrogen-bond network is frequently visible in the convex region of LRR crystal structures. Examples
Cryo-EM data collection, refinement and validation statistics
 1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
SUPPLEMENTARY INFORMATION
 SUPPLMTARY IFORMATIO a doi:10.108/nature10402 b 100 nm 100 nm c SAXS Model d ulers assigned to reference- Back-projected free class averages class averages Refinement against single particles Reconstructed
SUPPLMTARY IFORMATIO a doi:10.108/nature10402 b 100 nm 100 nm c SAXS Model d ulers assigned to reference- Back-projected free class averages class averages Refinement against single particles Reconstructed
Bacterial protease uses distinct thermodynamic signatures for substrate recognition
 Bacterial protease uses distinct thermodynamic signatures for substrate recognition Gustavo Arruda Bezerra, Yuko Ohara-Nemoto, Irina Cornaciu, Sofiya Fedosyuk, Guillaume Hoffmann, Adam Round, José A. Márquez,
Bacterial protease uses distinct thermodynamic signatures for substrate recognition Gustavo Arruda Bezerra, Yuko Ohara-Nemoto, Irina Cornaciu, Sofiya Fedosyuk, Guillaume Hoffmann, Adam Round, José A. Márquez,
Heterohexameric Ring Arrangement of the Eukaryotic Proteasomal ATPases: Implications for Proteasome Structure and Assembly
 Article Heterohexameric Ring Arrangement of the Eukaryotic Proteasomal ATPases: Implications for Proteasome Structure and Assembly Robert J. Tomko, Jr., 1 Minoru Funakoshi, 1,2 Kyle Schneider, 1 Jimin
Article Heterohexameric Ring Arrangement of the Eukaryotic Proteasomal ATPases: Implications for Proteasome Structure and Assembly Robert J. Tomko, Jr., 1 Minoru Funakoshi, 1,2 Kyle Schneider, 1 Jimin
Viewing and Analyzing Proteins, Ligands and their Complexes 2
 2 Viewing and Analyzing Proteins, Ligands and their Complexes 2 Overview Viewing the accessible surface Analyzing the properties of proteins containing thousands of atoms is best accomplished by representing
2 Viewing and Analyzing Proteins, Ligands and their Complexes 2 Overview Viewing the accessible surface Analyzing the properties of proteins containing thousands of atoms is best accomplished by representing
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/6/e1700147/dc1 Supplementary Materials for Ribosome rearrangements at the onset of translational bypassing Xabier Agirrezabala, Ekaterina Samatova, Mariia Klimova,
advances.sciencemag.org/cgi/content/full/3/6/e1700147/dc1 Supplementary Materials for Ribosome rearrangements at the onset of translational bypassing Xabier Agirrezabala, Ekaterina Samatova, Mariia Klimova,
 Supplemental Figure and Movie Legends Figure S1. Time course experiments on the thermal stability of apo AT cpn-α by native PAGE (related to Figure 2B). The samples were heated, respectively, to (A) 45
Supplemental Figure and Movie Legends Figure S1. Time course experiments on the thermal stability of apo AT cpn-α by native PAGE (related to Figure 2B). The samples were heated, respectively, to (A) 45
Three-dimensional structure of a viral genome-delivery portal vertex
 Three-dimensional structure of a viral genome-delivery portal vertex Adam S. Olia 1, Peter E. Prevelige Jr. 2, John E. Johnson 3 and Gino Cingolani 4 1 Department of Biological Sciences, Purdue University,
Three-dimensional structure of a viral genome-delivery portal vertex Adam S. Olia 1, Peter E. Prevelige Jr. 2, John E. Johnson 3 and Gino Cingolani 4 1 Department of Biological Sciences, Purdue University,
Rho1 binding site PtdIns(4,5)P2 binding site Both sites
 localization Mutation site DMSO LatB WT F77A I115A I131A K134A Rho1 binding site PtdIns(4,5)P2 binding site Both sites E186A E199A N201A R84A-E186A-E199A L131A-K136A-E186A L131A-E186A-E199A K136A-E186A-E199A
localization Mutation site DMSO LatB WT F77A I115A I131A K134A Rho1 binding site PtdIns(4,5)P2 binding site Both sites E186A E199A N201A R84A-E186A-E199A L131A-K136A-E186A L131A-E186A-E199A K136A-E186A-E199A
Supplementary information
 Supplementary information The structural basis of modularity in ECF-type ABC transporters Guus B. Erkens 1,2, Ronnie P-A. Berntsson 1,2, Faizah Fulyani 1,2, Maria Majsnerowska 1,2, Andreja Vujičić-Žagar
Supplementary information The structural basis of modularity in ECF-type ABC transporters Guus B. Erkens 1,2, Ronnie P-A. Berntsson 1,2, Faizah Fulyani 1,2, Maria Majsnerowska 1,2, Andreja Vujičić-Žagar
SUPPLEMENTARY INFORMATION
 Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11744 Supplementary Table 1. Crystallographic data collection and refinement statistics. Wild-type Se-Met-BcsA-B SmCl 3 -soaked EMTS-soaked Data collection Space
SUPPLEMENTARY INFORMATION doi:10.1038/nature11744 Supplementary Table 1. Crystallographic data collection and refinement statistics. Wild-type Se-Met-BcsA-B SmCl 3 -soaked EMTS-soaked Data collection Space
Supplementary Information. Overlap between folding and functional energy landscapes for. adenylate kinase conformational change
 Supplementary Information Overlap between folding and functional energy landscapes for adenylate kinase conformational change by Ulrika Olsson & Magnus Wolf-Watz Contents: 1. Supplementary Note 2. Supplementary
Supplementary Information Overlap between folding and functional energy landscapes for adenylate kinase conformational change by Ulrika Olsson & Magnus Wolf-Watz Contents: 1. Supplementary Note 2. Supplementary
EST1 Homology Domain. 100 aa. hest1a / SMG6 PIN TPR TPR. Est1-like DBD? hest1b / SMG5. TPR-like TPR. a helical. hest1c / SMG7.
 hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
Introduction to Comparative Protein Modeling. Chapter 4 Part I
 Introduction to Comparative Protein Modeling Chapter 4 Part I 1 Information on Proteins Each modeling study depends on the quality of the known experimental data. Basis of the model Search in the literature
Introduction to Comparative Protein Modeling Chapter 4 Part I 1 Information on Proteins Each modeling study depends on the quality of the known experimental data. Basis of the model Search in the literature
Scale in the biological world
 Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
THE CRYSTAL STRUCTURE OF THE SGT1-SKP1 COMPLEX: THE LINK BETWEEN
 THE CRYSTAL STRUCTURE OF THE SGT1-SKP1 COMPLEX: THE LINK BETWEEN HSP90 AND BOTH SCF E3 UBIQUITIN LIGASES AND KINETOCHORES Oliver Willhoft, Richard Kerr, Dipali Patel, Wenjuan Zhang, Caezar Al-Jassar, Tina
THE CRYSTAL STRUCTURE OF THE SGT1-SKP1 COMPLEX: THE LINK BETWEEN HSP90 AND BOTH SCF E3 UBIQUITIN LIGASES AND KINETOCHORES Oliver Willhoft, Richard Kerr, Dipali Patel, Wenjuan Zhang, Caezar Al-Jassar, Tina
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10955 Supplementary Figures Supplementary Figure 1. Electron-density maps and crystallographic dimer structures of the motor domain. (a f) Stereo views of the final electron-density maps
doi:10.1038/nature10955 Supplementary Figures Supplementary Figure 1. Electron-density maps and crystallographic dimer structures of the motor domain. (a f) Stereo views of the final electron-density maps
Tracking Protein Allostery in Evolution
 Tracking Protein Allostery in Evolution Glycogen phosphorylase frees sugars to provide energy GP orthologs diverged 600,000,000 years can respond to transcription controls, metabolite concentrations and
Tracking Protein Allostery in Evolution Glycogen phosphorylase frees sugars to provide energy GP orthologs diverged 600,000,000 years can respond to transcription controls, metabolite concentrations and
Nature Structural and Molecular Biology: doi: /nsmb Supplementary Figure 1. Definition and assessment of ciap1 constructs.
 Supplementary Figure 1 Definition and assessment of ciap1 constructs. (a) ciap1 constructs used in this study are shown as primary structure schematics with domains colored as in the main text. Mutations
Supplementary Figure 1 Definition and assessment of ciap1 constructs. (a) ciap1 constructs used in this study are shown as primary structure schematics with domains colored as in the main text. Mutations
SUPPLEMENTARY INFORMATION
 doi:1.138/nature1737 Supplementary Table 1 variant Description FSEC - 2B12 a FSEC - 6A1 a K d (leucine) c Leucine uptake e K (wild-type like) K (Y18F) K (TS) K (TSY) K288A mutant, lipid facing side chain
doi:1.138/nature1737 Supplementary Table 1 variant Description FSEC - 2B12 a FSEC - 6A1 a K d (leucine) c Leucine uptake e K (wild-type like) K (Y18F) K (TS) K (TSY) K288A mutant, lipid facing side chain
SUPPLEMENTARY INFORMATION
 Supplementary Table 1: Amplitudes of three current levels. Level 0 (pa) Level 1 (pa) Level 2 (pa) TrkA- TrkH WT 200 K 0.01 ± 0.01 9.5 ± 0.01 18.7 ± 0.03 200 Na * 0.001 ± 0.01 3.9 ± 0.01 12.5 ± 0.03 200
Supplementary Table 1: Amplitudes of three current levels. Level 0 (pa) Level 1 (pa) Level 2 (pa) TrkA- TrkH WT 200 K 0.01 ± 0.01 9.5 ± 0.01 18.7 ± 0.03 200 Na * 0.001 ± 0.01 3.9 ± 0.01 12.5 ± 0.03 200
FW 1 CDR 1 FW 2 CDR 2
 Supplementary Figure 1 Supplementary Figure 1: Interface of the E9:Fas structure. The two interfaces formed by V H and V L of E9 with Fas are shown in stereo. The Fas receptor is represented as a surface
Supplementary Figure 1 Supplementary Figure 1: Interface of the E9:Fas structure. The two interfaces formed by V H and V L of E9 with Fas are shown in stereo. The Fas receptor is represented as a surface
Exam I Answer Key: Summer 2006, Semester C
 1. Which of the following tripeptides would migrate most rapidly towards the negative electrode if electrophoresis is carried out at ph 3.0? a. gly-gly-gly b. glu-glu-asp c. lys-glu-lys d. val-asn-lys
1. Which of the following tripeptides would migrate most rapidly towards the negative electrode if electrophoresis is carried out at ph 3.0? a. gly-gly-gly b. glu-glu-asp c. lys-glu-lys d. val-asn-lys
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11054 Supplementary Fig. 1 Sequence alignment of Na v Rh with NaChBac, Na v Ab, and eukaryotic Na v and Ca v homologs. Secondary structural elements of Na v Rh are indicated above the
doi:10.1038/nature11054 Supplementary Fig. 1 Sequence alignment of Na v Rh with NaChBac, Na v Ab, and eukaryotic Na v and Ca v homologs. Secondary structural elements of Na v Rh are indicated above the
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
Model Mélange. Physical Models of Peptides and Proteins
 Model Mélange Physical Models of Peptides and Proteins In the Model Mélange activity, you will visit four different stations each featuring a variety of different physical models of peptides or proteins.
Model Mélange Physical Models of Peptides and Proteins In the Model Mélange activity, you will visit four different stations each featuring a variety of different physical models of peptides or proteins.
RNA Polymerase I Contains a TFIIF-Related DNA-Binding Subcomplex
 Molecular Cell, Volume 39 Supplemental Information RNA Polymerase I Contains a TFIIFRelated DNABinding Subcomplex Sebastian R. Geiger, Kristina Lorenzen, Amelie Schreieck, Patrizia Hanecker, Dirk Kostrewa,
Molecular Cell, Volume 39 Supplemental Information RNA Polymerase I Contains a TFIIFRelated DNABinding Subcomplex Sebastian R. Geiger, Kristina Lorenzen, Amelie Schreieck, Patrizia Hanecker, Dirk Kostrewa,
Supporting information for
 Supporting information for Rewiring multi-domain protein switches: transforming a fluorescent Zn 2+ -sensor into a light-responsive Zn 2+ binding protein Stijn J.A. Aper and Maarten Merkx Laboratory of
Supporting information for Rewiring multi-domain protein switches: transforming a fluorescent Zn 2+ -sensor into a light-responsive Zn 2+ binding protein Stijn J.A. Aper and Maarten Merkx Laboratory of
SUPPLEMENTARY INFORMATION
 Fig. 1 Influences of crystal lattice contacts on Pol η structures. a. The dominant lattice contact between two hpol η molecules (silver and gold) in the type 1 crystals. b. A close-up view of the hydrophobic
Fig. 1 Influences of crystal lattice contacts on Pol η structures. a. The dominant lattice contact between two hpol η molecules (silver and gold) in the type 1 crystals. b. A close-up view of the hydrophobic
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10162 Architecture of the Mediator Head module Tsuyoshi Imasaki, Guillermo Calero, Gang Cai, Kuang-Lei Tsai, Kentaro Yamada, Francesco Cardelli, Hediye Erdjument-Bromage, Paul Tempst,
doi:10.1038/nature10162 Architecture of the Mediator Head module Tsuyoshi Imasaki, Guillermo Calero, Gang Cai, Kuang-Lei Tsai, Kentaro Yamada, Francesco Cardelli, Hediye Erdjument-Bromage, Paul Tempst,
cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work
 SUPPLEMENTARY INFORMATION Table S1. List of strains Strains Genotype Source B. subtilis PY79 Prototrophic derivative of B. subtilis 168 (60) RL1070 spovid::kan (12) HS176 cotz phs2 (cotz-gfp spc) (22)
SUPPLEMENTARY INFORMATION Table S1. List of strains Strains Genotype Source B. subtilis PY79 Prototrophic derivative of B. subtilis 168 (60) RL1070 spovid::kan (12) HS176 cotz phs2 (cotz-gfp spc) (22)
Supplementary Information. The protease GtgE from Salmonella exclusively targets. inactive Rab GTPases
 Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Protein Structure Determination from Pseudocontact Shifts Using ROSETTA
 Supporting Information Protein Structure Determination from Pseudocontact Shifts Using ROSETTA Christophe Schmitz, Robert Vernon, Gottfried Otting, David Baker and Thomas Huber Table S0. Biological Magnetic
Supporting Information Protein Structure Determination from Pseudocontact Shifts Using ROSETTA Christophe Schmitz, Robert Vernon, Gottfried Otting, David Baker and Thomas Huber Table S0. Biological Magnetic
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/4/5/eaar2740/dc1 Supplementary Materials for The fission yeast Stn1-Ten1 complex limits telomerase activity via its SUMO-interacting motif and promotes telomeres
advances.sciencemag.org/cgi/content/full/4/5/eaar2740/dc1 Supplementary Materials for The fission yeast Stn1-Ten1 complex limits telomerase activity via its SUMO-interacting motif and promotes telomeres
The AAA+ enzymes (ATPases associated with a variety of
 Unfolding the mechanism of the AAA+ unfoldase VAT by a combined cryo-em, solution NMR study Rui Huang a,b,c,1, Zev A. Ripstein c,d,1, Rafal Augustyniak a,b,c, Michal Lazniewski e,f, Krzysztof Ginalski
Unfolding the mechanism of the AAA+ unfoldase VAT by a combined cryo-em, solution NMR study Rui Huang a,b,c,1, Zev A. Ripstein c,d,1, Rafal Augustyniak a,b,c, Michal Lazniewski e,f, Krzysztof Ginalski
Supplementary Information. Structural basis for precursor protein-directed ribosomal peptide macrocyclization
 Supplementary Information Structural basis for precursor protein-directed ribosomal peptide macrocyclization Kunhua Li 1,3, Heather L. Condurso 1,3, Gengnan Li 1, Yousong Ding 2 and Steven D. Bruner 1*
Supplementary Information Structural basis for precursor protein-directed ribosomal peptide macrocyclization Kunhua Li 1,3, Heather L. Condurso 1,3, Gengnan Li 1, Yousong Ding 2 and Steven D. Bruner 1*
Structure, mechanism and ensemble formation of the Alkylhydroperoxide Reductase subunits. AhpC and AhpF from Escherichia coli
 Structure, mechanism and ensemble formation of the Alkylhydroperoxide Reductase subunits AhpC and AhpF from Escherichia coli Phat Vinh Dip 1,#, Neelagandan Kamariah 2,#, Malathy Sony Subramanian Manimekalai
Structure, mechanism and ensemble formation of the Alkylhydroperoxide Reductase subunits AhpC and AhpF from Escherichia coli Phat Vinh Dip 1,#, Neelagandan Kamariah 2,#, Malathy Sony Subramanian Manimekalai
Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition
 Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Zn 2+ -binding sites in USP18. (a) The two molecules of USP18 present in the asymmetric unit are shown. Chain A is shown in blue, chain B in green. Bound Zn 2+ ions are shown as
Supplementary Figure 1 Zn 2+ -binding sites in USP18. (a) The two molecules of USP18 present in the asymmetric unit are shown. Chain A is shown in blue, chain B in green. Bound Zn 2+ ions are shown as
Lipid Regulated Intramolecular Conformational Dynamics of SNARE-Protein Ykt6
 Supplementary Information for: Lipid Regulated Intramolecular Conformational Dynamics of SNARE-Protein Ykt6 Yawei Dai 1, 2, Markus Seeger 3, Jingwei Weng 4, Song Song 1, 2, Wenning Wang 4, Yan-Wen 1, 2,
Supplementary Information for: Lipid Regulated Intramolecular Conformational Dynamics of SNARE-Protein Ykt6 Yawei Dai 1, 2, Markus Seeger 3, Jingwei Weng 4, Song Song 1, 2, Wenning Wang 4, Yan-Wen 1, 2,
Analysis of nucleotide binding to p97 reveals the properties of a tandem AAA hexameric ATPase
 SUPPLEMENTARY INFORMATION Analysis of nucleotide binding to p97 reveals the properties of a tandem AAA hexameric ATPase Louise C Briggs, Geoff S Baldwin, Non Miyata, Hisao Kondo, Xiaodong Zhang, Paul S
SUPPLEMENTARY INFORMATION Analysis of nucleotide binding to p97 reveals the properties of a tandem AAA hexameric ATPase Louise C Briggs, Geoff S Baldwin, Non Miyata, Hisao Kondo, Xiaodong Zhang, Paul S
Detailed description of overall and active site architecture of PPDC- 3dThDP, PPDC-2HE3dThDP, PPDC-3dThDP-PPA and PPDC- 3dThDP-POVA
 Online Supplemental Results Detailed description of overall and active site architecture of PPDC- 3dThDP, PPDC-2HE3dThDP, PPDC-3dThDP-PPA and PPDC- 3dThDP-POVA Structure solution and overall architecture
Online Supplemental Results Detailed description of overall and active site architecture of PPDC- 3dThDP, PPDC-2HE3dThDP, PPDC-3dThDP-PPA and PPDC- 3dThDP-POVA Structure solution and overall architecture
SUPPLEMENTARY INFORMATION
 UPPEER ORO doi:10.1038/nature10753 D D D D P E ntracellular C1 W P P C EC1 D Q R H C D W D R C C2 D E D E C R Q Q W P W W R P P EC2 EC3 P C C P W P W W P C W H R C R E C3 P R R P P P C Extracellular embrane
UPPEER ORO doi:10.1038/nature10753 D D D D P E ntracellular C1 W P P C EC1 D Q R H C D W D R C C2 D E D E C R Q Q W P W W R P P EC2 EC3 P C C P W P W W P C W H R C R E C3 P R R P P P C Extracellular embrane
Supplementary Information
 Supplementary Information The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis Marta C. Marques a, Cristina Tapia b, Oscar Gutiérrez-Sanz b, Ana Raquel Ramos a, Kimberly L.
Supplementary Information The direct role of selenocysteine in [NiFeSe] hydrogenase maturation and catalysis Marta C. Marques a, Cristina Tapia b, Oscar Gutiérrez-Sanz b, Ana Raquel Ramos a, Kimberly L.
Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants. GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
 SUPPLEMENTAL DATA Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants Sense primer (5 to 3 ) Anti-sense primer (5 to 3 ) GAL1 mutants GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
SUPPLEMENTAL DATA Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants Sense primer (5 to 3 ) Anti-sense primer (5 to 3 ) GAL1 mutants GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
Supporting Information
 Supporting Information Micelle-Triggered b-hairpin to a-helix Transition in a 14-Residue Peptide from a Choline-Binding Repeat of the Pneumococcal Autolysin LytA HØctor Zamora-Carreras, [a] Beatriz Maestro,
Supporting Information Micelle-Triggered b-hairpin to a-helix Transition in a 14-Residue Peptide from a Choline-Binding Repeat of the Pneumococcal Autolysin LytA HØctor Zamora-Carreras, [a] Beatriz Maestro,
ml. ph 7.5 ph 6.5 ph 5.5 ph 4.5. β 2 AR-Gs complex + GDP β 2 AR-Gs complex + GTPγS
 a UV28 absorption (mau) 9 8 7 5 3 β 2 AR-Gs complex β 2 AR-Gs complex + GDP β 2 AR-Gs complex + GTPγS β 2 AR-Gs complex dissociated complex excess nucleotides b 9 8 7 5 3 β 2 AR-Gs complex β 2 AR-Gs complex
a UV28 absorption (mau) 9 8 7 5 3 β 2 AR-Gs complex β 2 AR-Gs complex + GDP β 2 AR-Gs complex + GTPγS β 2 AR-Gs complex dissociated complex excess nucleotides b 9 8 7 5 3 β 2 AR-Gs complex β 2 AR-Gs complex
Protein Data Bank Contents Guide: Atomic Coordinate Entry Format Description. Version Document Published by the wwpdb
 Protein Data Bank Contents Guide: Atomic Coordinate Entry Format Description Version 3.30 Document Published by the wwpdb This format complies with the PDB Exchange Dictionary (PDBx) http://mmcif.pdb.org/dictionaries/mmcif_pdbx.dic/index/index.html.
Protein Data Bank Contents Guide: Atomic Coordinate Entry Format Description Version 3.30 Document Published by the wwpdb This format complies with the PDB Exchange Dictionary (PDBx) http://mmcif.pdb.org/dictionaries/mmcif_pdbx.dic/index/index.html.
Transmembrane Domains (TMDs) of ABC transporters
 Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12045 Supplementary Table 1 Data collection and refinement statistics. Native Pt-SAD X-ray source SSRF BL17U SPring-8 BL41XU Wavelength (Å) 0.97947 1.07171 Space group P2 1 2 1 2 1 P2
doi:10.1038/nature12045 Supplementary Table 1 Data collection and refinement statistics. Native Pt-SAD X-ray source SSRF BL17U SPring-8 BL41XU Wavelength (Å) 0.97947 1.07171 Space group P2 1 2 1 2 1 P2
Protein Structure Basics
 Protein Structure Basics Presented by Alison Fraser, Christine Lee, Pradhuman Jhala, Corban Rivera Importance of Proteins Muscle structure depends on protein-protein interactions Transport across membranes
Protein Structure Basics Presented by Alison Fraser, Christine Lee, Pradhuman Jhala, Corban Rivera Importance of Proteins Muscle structure depends on protein-protein interactions Transport across membranes
Supplementary Figure 1. Fourier shell correlation curves for sub-tomogram averages and
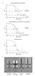 Supplementary Figure 1. Fourier shell correlation curves for sub-tomogram averages and comparisons to other published in situ T3SS structures. a, Resolution estimates after applying Fourier shell correlation
Supplementary Figure 1. Fourier shell correlation curves for sub-tomogram averages and comparisons to other published in situ T3SS structures. a, Resolution estimates after applying Fourier shell correlation
Structure of the SPRY domain of human DDX1 helicase, a putative interaction platform within a DEAD-box protein
 Supporting information Volume 71 (2015) Supporting information for article: Structure of the SPRY domain of human DDX1 helicase, a putative interaction platform within a DEAD-box protein Julian Kellner
Supporting information Volume 71 (2015) Supporting information for article: Structure of the SPRY domain of human DDX1 helicase, a putative interaction platform within a DEAD-box protein Julian Kellner
Lecture 10: Cyclins, cyclin kinases and cell division
 Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides Zaixing
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides Zaixing
Supporting information to: Time-resolved observation of protein allosteric communication. Sebastian Buchenberg, Florian Sittel and Gerhard Stock 1
 Supporting information to: Time-resolved observation of protein allosteric communication Sebastian Buchenberg, Florian Sittel and Gerhard Stock Biomolecular Dynamics, Institute of Physics, Albert Ludwigs
Supporting information to: Time-resolved observation of protein allosteric communication Sebastian Buchenberg, Florian Sittel and Gerhard Stock Biomolecular Dynamics, Institute of Physics, Albert Ludwigs
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a
 Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Structural insights into Aspergillus fumigatus lectin specificity - AFL binding sites are functionally non-equivalent
 Acta Cryst. (2015). D71, doi:10.1107/s1399004714026595 Supporting information Volume 71 (2015) Supporting information for article: Structural insights into Aspergillus fumigatus lectin specificity - AFL
Acta Cryst. (2015). D71, doi:10.1107/s1399004714026595 Supporting information Volume 71 (2015) Supporting information for article: Structural insights into Aspergillus fumigatus lectin specificity - AFL
V19 Topology of Protein Complexes on Graphs
 V19 Topology of Protein Complexes on Graphs (1) CombDock (2) Nuclear Pore Complex 19. Lecture WS 2013/14 Bioinformatics III 1 Prediction Structures of Protein Complexes from Connectivities CombDock: automated
V19 Topology of Protein Complexes on Graphs (1) CombDock (2) Nuclear Pore Complex 19. Lecture WS 2013/14 Bioinformatics III 1 Prediction Structures of Protein Complexes from Connectivities CombDock: automated
Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R)
 Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R) Shown in cyan and green are two adjacent tetramers from the crystallographic lattice of COP, forming the only unique inter-tetramer
Supplementary Figure 1 Crystal contacts in COP apo structure (PDB code 3S0R) Shown in cyan and green are two adjacent tetramers from the crystallographic lattice of COP, forming the only unique inter-tetramer
Conformational Analysis
 Conformational Analysis C01 3 C C 3 is the most stable by 0.9 kcal/mole C02 K eq = K 1-1 * K 2 = 0.45-1 * 0.048 = 0.11 C04 The intermediate in the reaction of 2 has an unfavorable syn-pentane interaction,
Conformational Analysis C01 3 C C 3 is the most stable by 0.9 kcal/mole C02 K eq = K 1-1 * K 2 = 0.45-1 * 0.048 = 0.11 C04 The intermediate in the reaction of 2 has an unfavorable syn-pentane interaction,
SUPPLEMENTARY INFORMATION
 Supplementary Table 1: Data collection, phasing and refinement statistics ChbC/Ta 6 Br 12 Native ChbC Data collection Space group P4 3 2 1 2 P4 3 2 1 2 Cell dimensions a, c (Å) 132.75, 453.57 132.81, 452.95
Supplementary Table 1: Data collection, phasing and refinement statistics ChbC/Ta 6 Br 12 Native ChbC Data collection Space group P4 3 2 1 2 P4 3 2 1 2 Cell dimensions a, c (Å) 132.75, 453.57 132.81, 452.95
Supporting Information for: Mechanism of Reversible Peptide-Bilayer. Attachment: Combined Simulation and Experimental Single-Molecule Study
 Supporting Information for: Mechanism of Reversible Peptide-Bilayer Attachment: Combined Simulation and Experimental Single-Molecule Study Nadine Schwierz a,, Stefanie Krysiak c, Thorsten Hugel c,d, Martin
Supporting Information for: Mechanism of Reversible Peptide-Bilayer Attachment: Combined Simulation and Experimental Single-Molecule Study Nadine Schwierz a,, Stefanie Krysiak c, Thorsten Hugel c,d, Martin
Supplementary Figure S1. Urea-mediated buffering mechanism of H. pylori. Gastric urea is funneled to a cytoplasmic urease that is presumably attached
 Supplementary Figure S1. Urea-mediated buffering mechanism of H. pylori. Gastric urea is funneled to a cytoplasmic urease that is presumably attached to HpUreI. Urea hydrolysis products 2NH 3 and 1CO 2
Supplementary Figure S1. Urea-mediated buffering mechanism of H. pylori. Gastric urea is funneled to a cytoplasmic urease that is presumably attached to HpUreI. Urea hydrolysis products 2NH 3 and 1CO 2
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1.
 Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Expanded View Figures
 The EMBO Journal Structure of a Dm peptide bound to the OT module Tobias Raisch et al Expanded View Figures A Hs Dm 262 297 685 8 HEAT HEAT MIF4G 9BD 1SHD 761 91 193 169 1152 1317 16 1376 1467 HEAT HEAT
The EMBO Journal Structure of a Dm peptide bound to the OT module Tobias Raisch et al Expanded View Figures A Hs Dm 262 297 685 8 HEAT HEAT MIF4G 9BD 1SHD 761 91 193 169 1152 1317 16 1376 1467 HEAT HEAT
2. Yeast two-hybrid system
 2. Yeast two-hybrid system I. Process workflow a. Mating of haploid two-hybrid strains on YPD plates b. Replica-plating of diploids on selective plates c. Two-hydrid experiment plating on selective plates
2. Yeast two-hybrid system I. Process workflow a. Mating of haploid two-hybrid strains on YPD plates b. Replica-plating of diploids on selective plates c. Two-hydrid experiment plating on selective plates
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1513/dc1 Supporting Online Material for Structural Roles for Human Translation Factor eif3 in Initiation of Protein Synthesis Bunpote Siridechadilok, Christopher
www.sciencemag.org/cgi/content/full/310/5753/1513/dc1 Supporting Online Material for Structural Roles for Human Translation Factor eif3 in Initiation of Protein Synthesis Bunpote Siridechadilok, Christopher
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
Secondary Structure. Bioch/BIMS 503 Lecture 2. Structure and Function of Proteins. Further Reading. Φ, Ψ angles alone determine protein structure
 Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Fig. 1. Stereo images showing (A) the best fit of the atomic model for F actin and the F actin map obtained by cryo-em and image analysis, and (B) goo
 Fig. 1. Stereo images showing (A) the best fit of the atomic model for F actin and the F actin map obtained by cryo-em and image analysis, and (B) good correspondence between the location of Cys374 and
Fig. 1. Stereo images showing (A) the best fit of the atomic model for F actin and the F actin map obtained by cryo-em and image analysis, and (B) good correspondence between the location of Cys374 and
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Cryo-EM structure and model of the C. thermophilum 90S preribosome. a, Gold standard FSC curve showing the average resolution of the 90S preribosome masked and unmasked (left). FSC
Supplementary Figure 1 Cryo-EM structure and model of the C. thermophilum 90S preribosome. a, Gold standard FSC curve showing the average resolution of the 90S preribosome masked and unmasked (left). FSC
Major Types of Association of Proteins with Cell Membranes. From Alberts et al
 Major Types of Association of Proteins with Cell Membranes From Alberts et al Proteins Are Polymers of Amino Acids Peptide Bond Formation Amino Acid central carbon atom to which are attached amino group
Major Types of Association of Proteins with Cell Membranes From Alberts et al Proteins Are Polymers of Amino Acids Peptide Bond Formation Amino Acid central carbon atom to which are attached amino group
A Push and Slide Mechanism Allows Sequence-Insensitive Translocation of Secretory Proteins by the SecA ATPase
 A Push and Slide Mechanism Allows Sequence-Insensitive Translocation of Secretory Proteins by the SecA ATPase Benedikt W. Bauer, 1,2 Tom Shemesh, 1,2 Yu Chen, 1,2 and Tom A. Rapoport 1,2, * 1 Howard Hughes
A Push and Slide Mechanism Allows Sequence-Insensitive Translocation of Secretory Proteins by the SecA ATPase Benedikt W. Bauer, 1,2 Tom Shemesh, 1,2 Yu Chen, 1,2 and Tom A. Rapoport 1,2, * 1 Howard Hughes
SUPPLEMENTARY INFORMATION
 Dph2 SeMet (iron-free) # Dph2 (iron-free) Dph2-[4Fe-4S] Data collection Space group P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 Cell dimensions a, b, c (Å) 58.26, 82.08, 160.42 58.74, 81.87, 160.01 55.70, 80.53,
Dph2 SeMet (iron-free) # Dph2 (iron-free) Dph2-[4Fe-4S] Data collection Space group P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 Cell dimensions a, b, c (Å) 58.26, 82.08, 160.42 58.74, 81.87, 160.01 55.70, 80.53,
Supplementary Information. Broad Spectrum Anti-Influenza Agents by Inhibiting Self- Association of Matrix Protein 1
 Supplementary Information Broad Spectrum Anti-Influenza Agents by Inhibiting Self- Association of Matrix Protein 1 Philip D. Mosier 1, Meng-Jung Chiang 2, Zhengshi Lin 2, Yamei Gao 2, Bashayer Althufairi
Supplementary Information Broad Spectrum Anti-Influenza Agents by Inhibiting Self- Association of Matrix Protein 1 Philip D. Mosier 1, Meng-Jung Chiang 2, Zhengshi Lin 2, Yamei Gao 2, Bashayer Althufairi
PURDUE UNIVERSITY GRADUATE SCHOOL Thesis/Dissertation Acceptance
 01 14 PURDUE UNIVERSITY GRADUATE SCHOOL Thesis/Dissertation Acceptance AISHWARYA RAMAMURTHY INVESTIGATING THE EARLY EVENTS IN PROTEASOME ASSEMBLY Master of Science Dr. ANDREW KUSMIERCZYK Dr. SIMON ATKINSON
01 14 PURDUE UNIVERSITY GRADUATE SCHOOL Thesis/Dissertation Acceptance AISHWARYA RAMAMURTHY INVESTIGATING THE EARLY EVENTS IN PROTEASOME ASSEMBLY Master of Science Dr. ANDREW KUSMIERCZYK Dr. SIMON ATKINSON
SUPPLEMENTARY INFORMATION
 Table of Contents Page Supplementary Table 1. Diffraction data collection statistics 2 Supplementary Table 2. Crystallographic refinement statistics 3 Supplementary Fig. 1. casic1mfc packing in the R3
Table of Contents Page Supplementary Table 1. Diffraction data collection statistics 2 Supplementary Table 2. Crystallographic refinement statistics 3 Supplementary Fig. 1. casic1mfc packing in the R3
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11991 Supplementary Figure 1 - Refinement strategy for PIC intermediate assemblies by negative stain EM. The cryo-negative stain structure of free Pol II 1 (a) was used as initial reference
doi:10.1038/nature11991 Supplementary Figure 1 - Refinement strategy for PIC intermediate assemblies by negative stain EM. The cryo-negative stain structure of free Pol II 1 (a) was used as initial reference
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature17991 Supplementary Discussion Structural comparison with E. coli EmrE The DMT superfamily includes a wide variety of transporters with 4-10 TM segments 1. Since the subfamilies of the
doi:10.1038/nature17991 Supplementary Discussion Structural comparison with E. coli EmrE The DMT superfamily includes a wide variety of transporters with 4-10 TM segments 1. Since the subfamilies of the
Supplementary Information
 Supplementary Information Resveratrol Serves as a Protein-Substrate Interaction Stabilizer in Human SIRT1 Activation Xuben Hou,, David Rooklin, Hao Fang *,,, Yingkai Zhang Department of Medicinal Chemistry
Supplementary Information Resveratrol Serves as a Protein-Substrate Interaction Stabilizer in Human SIRT1 Activation Xuben Hou,, David Rooklin, Hao Fang *,,, Yingkai Zhang Department of Medicinal Chemistry
Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08 Å resolution: comparison with the Azotobacter vinelandii MoFe protein
 Acta Cryst. (2015). D71, 274-282, doi:10.1107/s1399004714025243 Supporting information Volume 71 (2015) Supporting information for article: Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08
Acta Cryst. (2015). D71, 274-282, doi:10.1107/s1399004714025243 Supporting information Volume 71 (2015) Supporting information for article: Nitrogenase MoFe protein from Clostridium pasteurianum at 1.08
