Yeast ORFan Gene Project: Module 5 Guide
|
|
|
- Daisy Rich
- 5 years ago
- Views:
Transcription
1 Cellular Localization Data (Part 1) The tools described below will help you predict where your gene s product is most likely to be found in the cell, based on its sequence patterns. Each tool adds an additional layer of analysis to the others. Saccharomyces cerevisiae is a single-celled eukaryote; thus, it is important that you understand the structure of the cellular membranes including the options of membrane-bound organelles and the plasma membrane. Transmembrane Helices Hidden Markov Models (TMHMM) The TMHMM tool is used to assess the likelihood that some portion of a protein is embedded in a cellular membrane, and to predict the path that the amino acid chain follows if the protein is membrane-associated. TMHMM compares an amino acid sequence to a database of hidden Markov models or HMMs (you have already seen some of these in assessing conserved domains and regions in your predicted protein). These were created on the basis of known transmembrane (TM) helical sequences; that is, amino acid sequences demonstrated by experimental means to form α-helical secondary structures that cross cell membranes. If portions of a query sequence appear to be similar to any TMHMM s, then your gene product assuming it is a protein and not a functional RNA molecule may be localized to one of the cellular membranes. The search results will include a prediction of which segments of the protein are most likely to lie within the cytoplasm, to span a membrane, or to lie on the external side of the plasma membrane. Note that predictions of inside or outside location for a protein segment should be viewed cautiously, since it can be difficult to determine if a potential helical structure completely crosses a membrane without experimental evidence. Also be aware that these predictions are not valid at all if your protein sequence does not include any membrane-spanning helices.* Navigate to TMHMM at and enter your FASTA-formatted protein sequence in the search box. [ Recall this can be downloaded from the Protein tab on your gene page of SGD ] Leave the default Output Format as Extensive, with graphics and click the Submit button to begin your search. You will be presented with a graphic that shows the positions of amino acids in the primary sequence on the X-axis (these are numbered in the N-terminal to C-terminal direction) and the probability of being in a particular location on the Y-axis. Vertical red lines beneath the curve indicate portions of the sequence that match a Transmembrane Helix and are likely to enter or cross a membrane. The blue line represents the probability that a given portion of the sequence lies inside the cytoplasm and the pink line represents the probability of being external to the plasma membrane or outer membrane.* REMEMBER THAT THE INSIDE AND OUTSIDE PREDICTIONS SHOULD BE IGNORED IF THE SEQUENCE DOES NOT INCLUDE ANY REGIONS THAT ARE LIKELY TO CROSS A MEMBRANE COMPLETELY. Be alert for predicted transmembrane segments at the far N terminus of the protein. These could be signal peptides (see next section) rather than functional membrane-spanning regions of a mature protein. Record the number of predicted transmembrane helices in your Module 5 Worksheet. You may also want to record the amino acid sequence ranges covered by these TMHMMs. Save the graphic generated by TMHMM and upload it to the designated space in your Module 5 Worksheet. You will need to examine information obtained in the following sections before settling on a probable cellular location for your protein.
2 SignalP The signal peptide (SP) is an amino acid sequence found at the N terminus of newly-translated proteins that need to be secreted or become integrated into a membrane. The signal peptide directs the ribosome synthesizing the protein to a transport complex on the cytoplasmic face of the membrane, where the emerging polypeptide is threaded into or through the membrane. In most cases, the signal peptide is later cleaved from the protein as part of a maturation process; the enzyme responsible for this cleavage is called a signal peptidase. SignalP is a tool that attempts to predict SP regions on the basis of amino acid sequence similarity to known signal peptides.* Navigate to Enter the entire amino acid sequence in FASTA format. Select Eukaryotes under Organism group and click Submit. The graphical output from SignalP (euk networks) comprises three different scores: C, S and Y. Two additional scores, the S-mean and the D-score, are reported in the SignalP-4.1 output but these are only stated as numerical values. For each class of organism, two different networks are used: one for predicting the actual signal peptide and one for predicting the position of the signal peptidase I (SPase I) cleavage site. The S-score for the signal peptide prediction is reported for each individual amino acid position in the submitted sequence, with high scores indicating that the corresponding amino acid is likely to be part of a signal peptide and low scores indicating that the amino acid is part of a mature protein. The C-score is the cleavage site score. This is reported for each position in the submitted sequence and should only be significantly elevated at the cleavage site. Position numbering of the cleavage site can often be a source of confusion. When a cleavage position is shown as a single number, it refers to the first residue in the mature protein after removal of the signal peptide. [Reminder: the term residue refers to an amino acid that is part of a protein or peptide. Dehydration synthesis results in removal of a water molecule, and what remains of each amino acid is called an amino-acid residue]. If a cleavage site is reported as being between two amino acid residues e.g. amino acids this means that the mature protein begins with the second residue shown (number 27 in the example) and that the signal peptide terminates with the first. Y-max is a derivative of the C-score combined with the S-score, and is a better predictor of cleavage-site location than the raw C-score alone. This is because multiple high-peaking C-scores can be found in one sequence, even though only one of them is the actual cleavage site. The cleavage site is assigned from the Y-score where the slope of the S-score is steep and a significant C-score is found.* The S-mean is the average of the S-scores, ranging from the N-terminal amino acid to the amino acid that is assigned the highest Y-max score. Thus, the S-mean score is calculated for the entire length of the predicted signal peptide. The S-mean score was used in SignalP versions 1.0 and 2.0 as the criterion for discrimination of secretory and non-secretory proteins. The D-score was introduced with SignalP version 3.0 and is a simple average of the S-mean and Y-max score. The D-score is superior to the S-mean score in discriminating secretory from non-secretory polypeptides. Note that for non-secretory proteins, all the scores presented in the SignalP-4.1 output should be very low, at least in theory. * The hidden Markov model calculates the probability that the submitted amino acid sequence contains a signal peptide, and the cleavage site (if applicable) is also assigned on the basis of a probability score. If a probable signal peptide is identified, scores indicating the most likely locations of the n-region, h-region, and c-region are reported. The SignalP value is considered significant if it is >0.75. Upload the HMM graph to the Module 5 Worksheet. Report the Signal Peptide Probability in the Module 5 Worksheet.
3 PSORT II PSORT II is another useful tool for predicting the subcellular location of a protein. PSORT II predicts the subcellular localization sites of proteins based on their amino acid sequences. The method makes predictions based on both known sorting signal motifs and some correlative sequence features such as amino acid content. In this version of PSORT, parameters for analyzing yeast (or plant) sequences are the same with parameters for animal sequences. Yeast (and plant) have a candidate site, vacuole, instead of lysosome in animal. In yeast, the consensus sequence for ER-lumen retention is HDEL rather than KDEL in others.* Navigate to PSORT II at Enter the amino acid sequence in FASTA-format in the space provided (NOTE: this program requires the REMOVAL of the FASTA header which is the >genename). Select Yeast/Animal for source of input sequence, the click Submit to run the search. Scroll down the Results page to view the localization predictions. The scores will vary depending on the particular algorithm that has been run. Interpreting this data will take attention to detail, reading through the explanations below and determining how to deduce meaning from each component. As you go through each section paste the information in your Module 5 Worksheet and comment on how you have interpreted the data. Recognition of Signal Sequence In eukaryotes, proteins sorted through the so-called vesicular pathway (bulk flow) usually have a signal sequence (also called a leader peptide, and different in character from the signal peptide described above) in the N- terminus, which is cleaved off after the translocation through the ER membrane. Some N-terminal signal sequences are not cleaved off, remaining as transmembrane segments but it does not mean these proteins are retained in the ER; they can be further sorted and included in vesicles. * PSORT first predicts the presence of signal sequences, it considers the N-terminal positively-charged region (N-region) and the central hydrophobic region (H-region) of signal sequences. A discriminant score is calculated from the three values: length of H-region, peak value of H-region, and net charge of N-region. These results are summarized in "PSG". A large positive discriminant score means a high possibility to possess a signal sequence but it is unrelated to the possibility of its cleavage. * Next, PSORT applies a weight-matrix method and incorporates the information of consensus pattern around the cleavage sites (the (-3,-1)-rule) as well as the feature of the H-region. Thus it can be used to detect signal-anchor sequences. The output score of this "GvH" is the original weight-matrix score (for eukaryotes) subtracted by 3.5. A large positive output means a high possibility that it has a cleavable signal sequence. The position of possible cleavage site, i.e., the most C-terminal position of a signal sequence, is also reported. * Recognition of Transmembrane Segments PSORT II assumes that all integral membrane proteins have hydrophobic transmembrane segment(s) which are thought to be alpha- helices in membranes. PSORT detects potential transmembrane segments finding the most probable transmembrane segment from the average hydrophobicity value of 17-residue segments, if any. It predicts whether that segment is a transmembrane segment (INTEGRAL) or not (PERIPHERAL) comparing the discriminant score (reported as 'ALOM score') with a threshold parameter. For an integral membrane protein, position(s) of transmembrane segment(s) are also reported. Their length is fixed to 17 but their extension, i.e., the maximal range that satisfies the discriminant
4 criterion, is also given in parentheses. The item 'number of TMSs' is the number of predicted transmembrane segments. Specifically, PSORT first tentatively evaluates the number of TMSs using less stringent value (0.5). Then, it re-evaluates the number by using a more stringent threshold (-2.0). If it is still predicted to have at least one TMS, the former threshold value is used. Record the ALOM Score and number of TMs.* Recognition of Mitochondrial Proteins Although many proteins which engage in mitochondrial protein targeting have been characterized, their exact pathways have not been fully understood. Many proteins transported to mitochondria have a mitochondrial targeting signal on their N-terminus. Some seem to have internal signals but they may be recognized by a common cytosolic factor (MSF). PSORT employs a very simple method to recognize mitochondrial targeting signals: the discriminant analysis (called "MITDISC" ) whose variables are the amino acid composition of the N-terminal 20 residues. In this version, further discrimination of signals directing to the substructures of mitochondria, e.g., intermembrane space, is not attempted although proteins which are predicted to have both the mitochondrial targeting signal and transmembrane segment(s) are likely to be localized at the inner membrane. PSORT also reports some consensus patterns around the cleavage sites ("Gavel").* Note the score and data for this section but wait until completion of TargetP in the next module before deciding upon mitochondrial localization. Recognition of Nuclear Proteins Although it seems possible that a protein without its own nuclear localization signal (NLS) enters the nucleus via cotransport with a protein that has one, many nuclear proteins have their own NLSs. Presently, NLSs are classified into three categories. The classical type of NLSs is that of SV40 large T antigen. PSORT uses the following two rules to detect it: 4 residue pattern (called 'pat4') composed of 4 basic amino acids (K or R), or composed of three basic amino acids (K or R) and either H or P; the other (called 'pat7') is a pattern starting with P and followed within 3 residues by a basic segment containing 3 K/R residues out of 4. * Another type of NLS is the bipartite NLS, first found in Xenopus nucleoplasmin. The pattern (called 'bipartite') is: 2 basic residues, 10 residue spacer, and another basic region consisting of at least 3 basic residues out of 5 residues. The last category of NLS is the type of an N-terminal signal found in yeast protein, Mat alpha2. However, PSORT doesn't try to find it because this type of signal has not been well studied. Nor the knowledge of Nuclear Export Signals (NESs) has not been incorporated yet. * In the yeast genome, nuclear proteins occupy the majority. Since the precise discrimination of NLSs is presently difficult, the prediction of nuclear proteins affects much to the total prediction accuracy. Then, PSORT uses a heuristic that nuclear proteins are generally rich in basic residues: If the sum of K and R compositions are higher than 20%, then the protein is considered to have higher possibility of being nuclear than cytoplasmic. Moreover, a score (called "NNCN" ) which discriminates the tendency to be at either the nucleus or the cytoplasm is calculated based on the amino acid composition. Most scores mentioned above are combined by a discriminant function to give the 'NLS score'. In addition, PSORT examines the presence of RNP (ribonucleoprotein) consensus motif (called 'RNA-binding motif') because some RNPs are transported to the nucleus by signals existing in the bound RNAs. However, it is apparently insufficient for actual prediction. In this version, we classify ribosomal proteins as cytoplasmic proteins although some of them have NLSs and are once transported into the nucleus. *
5 Note the score and data for this section but wait until completion of NucPred in the next module before deciding upon nuclear localization. Recognition of ER (endoplasmic reticulum) Proteins PSORT postulates that the proteins with N-terminal signal sequence will be transported to the cell surface by default unless they have any other signals for specific retrieval, retention, or commitment; a luminal protein will be secreted constitutively to the extracellular space and a membrane protein will reside at the plasma membrane. The retrieval signal of ER luminal proteins from the bulk flow is the consensus motif, KDEL (HDEL in yeast), in the C-terminus. In addition, these proteins should have a cleavable signal sequence in their N-terminus but the existence of KDEL is often practically sufficient. Although PSORT only recognizes the (K/H)DEL pattern, it is known that some variations of this motif are allowed in some organisms and/or cell types. * The retrieval signals for ER membrane proteins appear more complex. Two kinds of signals are known; one is the di-lysine motif (the KKXX motif) which exist near the C-terminus of type Ia proteins and the other is the di-arginine motif (the XXRR motif) which exist near the C-terminus of type II proteins. Note that both of these motifs exist close to the terminus of the cytoplasmic tail. However, for the practical prediction, the existence of these motifs itself is neither necessary nor sufficient for the localization at the ER membrane. * Recognition of Peroxisomal Proteins Peroxisomes, sometimes called glyoxisomes, glycosomes, or microbodies, are organelles found in almost every eukaryotic cell. Several sorting signals into peroxisomes have been. As for peroxisomal-matrix targeting sequences (PTSs), two kinds of them are known. One is the tripeptide, (S/A/C)(K/R/H)L, at the C-terminus, known as PTS1 or the SKL-motif. PSORT calculates score of a given sequence empirically. The other is the N-terminal segment known as PTS2. The consensus patterns of known PTSs, (R/K)(L/I)xxxxx(H/Q)L is searched in the present version, although its importance has not been fully verified. The sorting signal of peroxisomal membrane proteins (mptss) is not well understood although the importance of a hydrophilic loop of 20 residues facing the matrix in Pmp47 is known. * Analysis of Proteins in Vesicular Pathway The signals that govern the protein sorting through the vesicle transport are complex because they are inter-related and they seem to be recognized in various situation. Moreover, there are redistribution processes via endocytosis and a pathway to lysosomes. As already exemplified above, many sorting signals of membrane proteins transported through these pathways exist in the cytoplasmic tail, a short terminal segment exposed to the cytosol in type Ia, Ib, and II proteins. For membrane proteins with (usually) a single transmembrane segment, PSORT tries to find the following motifs in the cytoplasmic tail: the YQRL motif which directs the transport from cell surface to Golgi; the tyrosine-containing motif and the dileucine motif for selective inclusion in clathrin-coated vesicles (endocytosis) and lysosomal targeting. * Lysosomal and Vacuolar Proteins Lysosomes are acidic organelles that contain numerous hydrolytic enzymes. In yeast and plant cells, similar activities are seen in vacuoles (lysosome-like vacuoles) and the protein sorting mechanisms for these organelles are conserved to some degree.* In mammalian lysosomes, one sorting signal of soluble (lumenal) proteins is a post-translational modification, addition of mannose-6-phosphate, but there is also a pathway which is independent of mannose-6-phosphate. Two kinds of
6 mannose-6-phosphate receptor are known but the substrate specificity of the enzyme which adds mannose-6-phosphate is not well understood although the importance of a specific conformation, a beta-hairpin motif, has been implicated. * Although yeast vacuoles have been studied as a model system of mammalian lysosomes, soluble proteins of yeast vacuole do not use the mannose-6-phosphate dependent pathway. Analyses of several yeast proteins suggest that the pro-peptides, which are exposed to the N-terminus after the cleavage of 'pre' signal sequence, work as a sorting signal. However, there are not apparently conserved motifs except for a weak motif, (T/I/K)LP(L/K/I), which PSORT searches for. The sorting mechanism of lysosomal membrane proteins seems different from that of lysosomal luminal proteins. The existence of the GY motif within 17 residues from the membrane boundary in the cytoplasmic tail seems to be important for some of them. * Lipid Anchors The protein modification reactions which bind lipid molecules to proteins are important because a linked lipid moiety can be integrated into various membranes and can anchor the bound protein. * For example, myristoylations occur at the consensus sequence in the N-terminal 9 residues. PSORT predicts its presence by the 'NMYR' program but note that the N-myristoylated proteins are not always anchored to the membrane. In contrast, all proteins linked to the glycosyl-phosphatidylinositol (GPI) molecules are thought to be anchored at the extracellular surface of the plasma membrane. In addition, GPI anchor plays some roles on the protein sorting in polarized cells. Although much is known about the biosynthesis of GPI-anchor, PSORT predicts GPI-anchored proteins by empirical knowledge that most of them are the type Ia membrane proteins with very short cytoplasmic tail (within 10 residues). * Lastly, there is a lipid modification known as (iso)prenylation ( i.e., farnesylation or geranylgeranylation.) This modification requires a CaaX motif in the C-terminus, where 'a' denotes an aliphatic amino acid. Prenylated proteins have been found in the plasma membrane and the nuclear envelope. * Miscellaneous Motifs Experimentally the knowledge of various protein functional motifs in the PROSITE database has been included. To discriminate cytoskeletal proteins, two motifs of actin-binding proteins were examined as well, these PROSITE motifs are experimentally introduced: 63 DNA binding motifs, which may be useful to distinguish nuclear proteins; 71 ribosomal protein motifs, which may be necessary because the sorting processes of ribosomal proteins are complex; 33 prokaryotic DNA binding motifs, which might be useful for the prediction of bacterial sequences. * Coiled-coil Structure Coiled-coil structures are found in some structural proteins, e.g., myosins, and in some DNA-binding proteins as the so-called leucine-zipper. In this structure, two alpha-helices bind each other making a coil, where these two alpha-helices show a 3.5-residue periodicity which is slightly different from the typical value, 3.6. Thus, the detection of coiled-coil structure by searching for 7-residue periodicity is relatively more accurate than usual secondary structure prediction. Although the presence of coiled-coil structure in a protein itself does not indicate its subcellular localization, such information might be useful for the people who try to characterize ORFs of unknown function. Currently, a classical detection algorithm is used and the sequences which are likely to be in a coiled-coil conformation are reported. *
7 Ignore the Results of the k -NN Prediction for now. Phobius Phobius combines the methods used by TMHMM and SignalP and generates a single graphical output. This tool is especially useful when TMHMM predicts that the N-terminal portion of the sequence forms a transmembrane helix, while SignalP predicts it is a signal peptide. Phobius will help you decide which of these predictions is most likely to be accurate. * Navigate to Enter the amino acid sequence in FASTA-format. Select Long with Graphics as your output format and submit the query. The prediction at the top of the output lists the most probable locations of transmembrane helices and a signal peptide (if any) in the sequence. The graphic plot shows the probability that a given amino acid residue is cytoplasmic, non-cytoplasmic, within a TM helix, or part of a signal peptide. Here one can see weak potential TM helices that fell below the prediction threshold and get an idea of the confidence level for each segment in the prediction. At the very bottom of the plot, in the range between 0 and on the Y axis, the overall predictions are represented in graphic form. * If the entire sequence is labeled as being cytoplasmic or non-cytoplasmic, the prediction is that it contains no transmembrane helices. AS WITH TMHMM, YOU SHOULD NOT INTERPRET THE STATED LOCATION AS ACCURATE IN THIS CASE SINCE THE LOCALIZATION MODEL IS ONLY VALID IF THE PROTEIN SPANS A MEMBRANE AT LEAST ONCE. Record the graph and probability data in the Module 5 Worksheet. One might assume that a protein predicted to contain transmembrane helices would include a signal peptide to direct it to the membrane. If this is not the case with your gene product, what might that say about the protein sequence? Note, that examples of internal signal sequences that are not cleaved and remain as transmembrane domains of mature proteins have been reported in both bacterial and eukaryotic cells. Hypothesis Where do you expect to find your protein? Take the results of all of the above analyses into consideration and make a final localization prediction. Did all the tools yield the same result? If one disagreed with the others, what might that tell you about the protein's function, based on the prediction method used in that tool? Record the final prediction in the Module 5 Worksheet. If the combined results of the analyses are inconclusive, enter Unknown. Feel free to consult one of the instructors if you re unsure about this. Note we will do another module on Cellular Localization so this preliminary hypothesis may be changed after further experimentation and addition of data.
Chapter 12: Intracellular sorting
 Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
CHAPTER 3. Cell Structure and Genetic Control. Chapter 3 Outline
 CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%%
 !"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
Transport between cytosol and nucleus
 of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
Intro Secondary structure Transmembrane proteins Function End. Last time. Domains Hidden Markov Models
 Last time Domains Hidden Markov Models Today Secondary structure Transmembrane proteins Structure prediction NAD-specific glutamate dehydrogenase Hard Easy >P24295 DHE2_CLOSY MSKYVDRVIAEVEKKYADEPEFVQTVEEVL
Last time Domains Hidden Markov Models Today Secondary structure Transmembrane proteins Structure prediction NAD-specific glutamate dehydrogenase Hard Easy >P24295 DHE2_CLOSY MSKYVDRVIAEVEKKYADEPEFVQTVEEVL
Today. Last time. Secondary structure Transmembrane proteins. Domains Hidden Markov Models. Structure prediction. Secondary structure
 Last time Today Domains Hidden Markov Models Structure prediction NAD-specific glutamate dehydrogenase Hard Easy >P24295 DHE2_CLOSY MSKYVDRVIAEVEKKYADEPEFVQTVEEVL SSLGPVVDAHPEYEEVALLERMVIPERVIE FRVPWEDDNGKVHVNTGYRVQFNGAIGPYK
Last time Today Domains Hidden Markov Models Structure prediction NAD-specific glutamate dehydrogenase Hard Easy >P24295 DHE2_CLOSY MSKYVDRVIAEVEKKYADEPEFVQTVEEVL SSLGPVVDAHPEYEEVALLERMVIPERVIE FRVPWEDDNGKVHVNTGYRVQFNGAIGPYK
Protein Sorting. By: Jarod, Tyler, and Tu
 Protein Sorting By: Jarod, Tyler, and Tu Definition Organizing of proteins Organelles Nucleus Ribosomes Endoplasmic Reticulum Golgi Apparatus/Vesicles How do they know where to go? Amino Acid Sequence
Protein Sorting By: Jarod, Tyler, and Tu Definition Organizing of proteins Organelles Nucleus Ribosomes Endoplasmic Reticulum Golgi Apparatus/Vesicles How do they know where to go? Amino Acid Sequence
Protein Sorting, Intracellular Trafficking, and Vesicular Transport
 Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
Chapter 1. DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d
 Chapter 1 1. Matching Questions DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d 2. Matching Questions : Unbranched polymer that, when folded into its three-dimensional shape,
Chapter 1 1. Matching Questions DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d 2. Matching Questions : Unbranched polymer that, when folded into its three-dimensional shape,
Chapter 7.2. Cell Structure
 Chapter 7.2 Cell Structure Daily Objectives Describe the structure and function of the cell nucleus. Describe the function and structure of membrane bound organelles found within the cell. Describe the
Chapter 7.2 Cell Structure Daily Objectives Describe the structure and function of the cell nucleus. Describe the function and structure of membrane bound organelles found within the cell. Describe the
The Discovery of Cells
 The Discovery of Cells Microscope observations! General Cell & Organelle Discovery 1600s Observations made by scientists using more powerful microscopes in the 1800s led to the formation of the cell theory.
The Discovery of Cells Microscope observations! General Cell & Organelle Discovery 1600s Observations made by scientists using more powerful microscopes in the 1800s led to the formation of the cell theory.
Genome Annotation Project Presentation
 Halogeometricum borinquense Genome Annotation Project Presentation Loci Hbor_05620 & Hbor_05470 Presented by: Mohammad Reza Najaf Tomaraei Hbor_05620 Basic Information DNA Coordinates: 527,512 528,261
Halogeometricum borinquense Genome Annotation Project Presentation Loci Hbor_05620 & Hbor_05470 Presented by: Mohammad Reza Najaf Tomaraei Hbor_05620 Basic Information DNA Coordinates: 527,512 528,261
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
Cellular basis of life History of cell Biology Year Name of the scientist Importance
 Cellular basis of life History of cell Biology Year Name of the scientist Importance 1590 Jansen 1650 Anton van Leeuwenhoek 1665 Robert Hooke 1831 Matthias Schleiden 1831 Theodore Schwann 1855 Rudolf Virchow
Cellular basis of life History of cell Biology Year Name of the scientist Importance 1590 Jansen 1650 Anton van Leeuwenhoek 1665 Robert Hooke 1831 Matthias Schleiden 1831 Theodore Schwann 1855 Rudolf Virchow
Class Work 31. Describe the function of the Golgi apparatus? 32. How do proteins travel from the E.R. to the Golgi apparatus? 33. After proteins are m
 Eukaryotes Class Work 1. What does the word eukaryote mean? 2. What is the one major difference between eukaryotes and prokaryotes? 3. List the different kingdoms of the eukaryote domain in the order in
Eukaryotes Class Work 1. What does the word eukaryote mean? 2. What is the one major difference between eukaryotes and prokaryotes? 3. List the different kingdoms of the eukaryote domain in the order in
Biology Midterm Review
 Biology Midterm Review Unit 1 Keystone Objectives: A.1.1, A.1.2, B.4.1.1 1.1 Biology explores life from the global to the microscopic level. Put the levels of organization in order, starting with subatomic
Biology Midterm Review Unit 1 Keystone Objectives: A.1.1, A.1.2, B.4.1.1 1.1 Biology explores life from the global to the microscopic level. Put the levels of organization in order, starting with subatomic
Introduction to Pattern Recognition. Sequence structure function
 Introduction to Pattern Recognition Sequence structure function Prediction in Bioinformatics What do we want to predict? Features from sequence Data mining How can we predict? Homology / Alignment Pattern
Introduction to Pattern Recognition Sequence structure function Prediction in Bioinformatics What do we want to predict? Features from sequence Data mining How can we predict? Homology / Alignment Pattern
Guided Reading Activities
 Name Period Chapter 4: A Tour of the Cell Guided Reading Activities Big Idea: Introduction to the Cell Answer the following questions as you read Modules 4.1 4.4: 1. A(n) uses a beam of light to illuminate
Name Period Chapter 4: A Tour of the Cell Guided Reading Activities Big Idea: Introduction to the Cell Answer the following questions as you read Modules 4.1 4.4: 1. A(n) uses a beam of light to illuminate
Cell Structure. Chapter 4. Cell Theory. Cells were discovered in 1665 by Robert Hooke.
 Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
2011 The Simple Homeschool Simple Days Unit Studies Cells
 1 We have a full line of high school biology units and courses at CurrClick and as online courses! Subscribe to our interactive unit study classroom and make science fun and exciting! 2 A cell is a small
1 We have a full line of high school biology units and courses at CurrClick and as online courses! Subscribe to our interactive unit study classroom and make science fun and exciting! 2 A cell is a small
CELL PART Expanded Definition Cell Structure Illustration Function Summary Location ALL CELLS DNA Common in Animals Uncommon in Plants Lysosome
 CELL PART Expanded Definition Cell Structure Illustration Function Summary Location is the material that contains the Carry genetic ALL CELLS information that determines material inherited characteristics.
CELL PART Expanded Definition Cell Structure Illustration Function Summary Location is the material that contains the Carry genetic ALL CELLS information that determines material inherited characteristics.
7-2 Eukaryotic Cell Structure
 1 of 49 Comparing the Cell to a Factory Eukaryotic Cell Structures Structures within a eukaryotic cell that perform important cellular functions are known as organelles. Cell biologists divide the eukaryotic
1 of 49 Comparing the Cell to a Factory Eukaryotic Cell Structures Structures within a eukaryotic cell that perform important cellular functions are known as organelles. Cell biologists divide the eukaryotic
It s a Small World After All
 It s a Small World After All Engage: Cities, factories, even your own home is a network of dependent and independent parts that make the whole function properly. Think of another network that has subunits
It s a Small World After All Engage: Cities, factories, even your own home is a network of dependent and independent parts that make the whole function properly. Think of another network that has subunits
Cell Organelles. Wednesday, October 22, 14
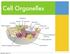 Cell Organelles Cell/Plasma Membrane ALL cells have a cell membrane It is the layer that surrounds the cell and controls what goes in and out Bacteria (Prokaryotic Cell) Cell/Plasma Membrane ALL cells
Cell Organelles Cell/Plasma Membrane ALL cells have a cell membrane It is the layer that surrounds the cell and controls what goes in and out Bacteria (Prokaryotic Cell) Cell/Plasma Membrane ALL cells
UNIT 3 CP BIOLOGY: Cell Structure
 UNIT 3 CP BIOLOGY: Cell Structure Page CP: CHAPTER 3, Sections 1-3; HN: CHAPTER 7, Sections 1-2 Standard B-2: The student will demonstrate an understanding of the structure and function of cells and their
UNIT 3 CP BIOLOGY: Cell Structure Page CP: CHAPTER 3, Sections 1-3; HN: CHAPTER 7, Sections 1-2 Standard B-2: The student will demonstrate an understanding of the structure and function of cells and their
Division Ave. High School AP Biology
 Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Cell Structure. Chapter 4
 Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Components of a functional cell. Boundary-membrane Cytoplasm: Cytosol (soluble components) & particulates DNA-information Ribosomes-protein synthesis
 Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
7.06 Cell Biology EXAM #3 KEY
 7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
Overview of Cells. Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory
 Overview of Cells Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory Prokaryotic Cells Archaea Bacteria Come in many different shapes and sizes.5 µm 2 µm, up to 60 µm long Have large
Overview of Cells Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory Prokaryotic Cells Archaea Bacteria Come in many different shapes and sizes.5 µm 2 µm, up to 60 µm long Have large
Topic 3: Cells Ch. 6. Microscopes pp Microscopes. Microscopes. Microscopes. Microscopes
 Topic 3: Cells Ch. 6 -All life is composed of cells and all cells have a plasma membrane, cytoplasm, and DNA. pp.105-107 - The development of the microscope was the key to understanding that all living
Topic 3: Cells Ch. 6 -All life is composed of cells and all cells have a plasma membrane, cytoplasm, and DNA. pp.105-107 - The development of the microscope was the key to understanding that all living
Cell Theory. Cell Structure. Chapter 4. Cell is basic unit of life. Cells discovered in 1665 by Robert Hooke
 Cell Structure Chapter 4 Cell is basic unit of life Cell Theory Cells discovered in 1665 by Robert Hooke Early cell studies conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden &
Cell Structure Chapter 4 Cell is basic unit of life Cell Theory Cells discovered in 1665 by Robert Hooke Early cell studies conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden &
Chapter 4. Table of Contents. Section 1 The History of Cell Biology. Section 2 Introduction to Cells. Section 3 Cell Organelles and Features
 Cell Structure and Function Table of Contents Section 1 The History of Cell Biology Section 2 Introduction to Cells Section 3 Cell Organelles and Features Section 4 Unique Features of Plant Cells Section
Cell Structure and Function Table of Contents Section 1 The History of Cell Biology Section 2 Introduction to Cells Section 3 Cell Organelles and Features Section 4 Unique Features of Plant Cells Section
9/2/17. Molecular and Cellular Biology. 3. The Cell From Genes to Proteins. key processes
 Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Cellular Transport. 1. Transport to and across the membrane 1a. Transport of small molecules and ions 1b. Transport of proteins
 Transport Processes Cellular Transport 1. Transport to and across the membrane 1a. Transport of small molecules and ions 1b. Transport of proteins 2. Vesicular transport 3. Transport through the nuclear
Transport Processes Cellular Transport 1. Transport to and across the membrane 1a. Transport of small molecules and ions 1b. Transport of proteins 2. Vesicular transport 3. Transport through the nuclear
Cell Organelles Tutorial
 1 Name: Cell Organelles Tutorial TEK 7.12D: Differentiate between structure and function in plant and animal cell organelles, including cell membrane, cell wall, nucleus, cytoplasm, mitochondrion, chloroplast,
1 Name: Cell Organelles Tutorial TEK 7.12D: Differentiate between structure and function in plant and animal cell organelles, including cell membrane, cell wall, nucleus, cytoplasm, mitochondrion, chloroplast,
9/11/18. Molecular and Cellular Biology. 3. The Cell From Genes to Proteins. key processes
 Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
122-Biology Guide-5thPass 12/06/14. Topic 1 An overview of the topic
 Topic 1 http://bioichiban.blogspot.com Cellular Functions 1.1 The eukaryotic cell* An overview of the topic Key idea 1: Cell Organelles Key idea 2: Plasma Membrane Key idea 3: Transport Across Membrane
Topic 1 http://bioichiban.blogspot.com Cellular Functions 1.1 The eukaryotic cell* An overview of the topic Key idea 1: Cell Organelles Key idea 2: Plasma Membrane Key idea 3: Transport Across Membrane
Why Bother? Predicting the Cellular Localization Sites of Proteins Using Bayesian Model Averaging. Yetian Chen
 Predicting the Cellular Localization Sites of Proteins Using Bayesian Model Averaging Yetian Chen 04-27-2010 Why Bother? 2008 Nobel Prize in Chemistry Roger Tsien Osamu Shimomura Martin Chalfie Green Fluorescent
Predicting the Cellular Localization Sites of Proteins Using Bayesian Model Averaging Yetian Chen 04-27-2010 Why Bother? 2008 Nobel Prize in Chemistry Roger Tsien Osamu Shimomura Martin Chalfie Green Fluorescent
Multiple Choice Review- Eukaryotic Gene Expression
 Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
Class IX: Biology Chapter 5: The fundamental unit of life. Chapter Notes. 1) In 1665, Robert Hooke first discovered and named the cells.
 Class IX: Biology Chapter 5: The fundamental unit of life. Key learnings: Chapter Notes 1) In 1665, Robert Hooke first discovered and named the cells. 2) Cell is the structural and functional unit of all
Class IX: Biology Chapter 5: The fundamental unit of life. Key learnings: Chapter Notes 1) In 1665, Robert Hooke first discovered and named the cells. 2) Cell is the structural and functional unit of all
Chapter 4: Cells: The Working Units of Life
 Name Period Chapter 4: Cells: The Working Units of Life 1. What are the three critical components of the cell theory? 2. What are the two important conceptual implications of the cell theory? 3. Which
Name Period Chapter 4: Cells: The Working Units of Life 1. What are the three critical components of the cell theory? 2. What are the two important conceptual implications of the cell theory? 3. Which
Takehome group assignment #1 - Individual worksheet NAME
 Takehome group assignment #1 - Individual worksheet NAME Answer the questions below, on your own (pp. 1-5). Then get together with your group, discuss your answers and complete the final group worksheet
Takehome group assignment #1 - Individual worksheet NAME Answer the questions below, on your own (pp. 1-5). Then get together with your group, discuss your answers and complete the final group worksheet
7.06 Problem Set
 7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
What is the central dogma of biology?
 Bellringer What is the central dogma of biology? A. RNA DNA Protein B. DNA Protein Gene C. DNA Gene RNA D. DNA RNA Protein Review of DNA processes Replication (7.1) Transcription(7.2) Translation(7.3)
Bellringer What is the central dogma of biology? A. RNA DNA Protein B. DNA Protein Gene C. DNA Gene RNA D. DNA RNA Protein Review of DNA processes Replication (7.1) Transcription(7.2) Translation(7.3)
FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell
 FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell All media is on the Instructors Resource CD/DVD JPEG Resources Figures, Photos, and Tables PowerPoint Resources Chapter Outline with Figures Lecture
FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell All media is on the Instructors Resource CD/DVD JPEG Resources Figures, Photos, and Tables PowerPoint Resources Chapter Outline with Figures Lecture
The Cell. What is a cell?
 The Cell What is a cell? The Cell What is a cell? Structure which makes up living organisms. The Cell Theory l All living things are composed of cells. l Cells are the basic unit of life. l Cells come
The Cell What is a cell? The Cell What is a cell? Structure which makes up living organisms. The Cell Theory l All living things are composed of cells. l Cells are the basic unit of life. l Cells come
Scale in the biological world
 Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Biology. 7-2 Eukaryotic Cell Structure 10/29/2013. Eukaryotic Cell Structures
 Biology Biology 1of 49 2of 49 Eukaryotic Cell Structures Eukaryotic Cell Structures Structures within a eukaryotic cell that perform important cellular functions are known as organelles. Cell biologists
Biology Biology 1of 49 2of 49 Eukaryotic Cell Structures Eukaryotic Cell Structures Structures within a eukaryotic cell that perform important cellular functions are known as organelles. Cell biologists
Biology. Mrs. Michaelsen. Types of cells. Cells & Cell Organelles. Cell size comparison. The Cell. Doing Life s Work. Hooke first viewed cork 1600 s
 Types of cells bacteria cells Prokaryote - no organelles Cells & Cell Organelles Doing Life s Work Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell Bacterial cell most
Types of cells bacteria cells Prokaryote - no organelles Cells & Cell Organelles Doing Life s Work Eukaryotes - organelles animal cells plant cells Cell size comparison Animal cell Bacterial cell most
Cell (Learning Objectives)
 Cell (Learning Objectives) 1. Understand & describe the basic components necessary for a functional cell. 2. Review the order of appearance of cells on earth and explain the endosymbiotic theory. 3. Compare
Cell (Learning Objectives) 1. Understand & describe the basic components necessary for a functional cell. 2. Review the order of appearance of cells on earth and explain the endosymbiotic theory. 3. Compare
Cell Organelles. a review of structure and function
 Cell Organelles a review of structure and function TEKS and Student Expectations (SE s) B.4 Science concepts. The student knows that cells are the basic structures of all living things with specialized
Cell Organelles a review of structure and function TEKS and Student Expectations (SE s) B.4 Science concepts. The student knows that cells are the basic structures of all living things with specialized
Cells & Cell Organelles. Doing Life s Work
 Cells & Cell Organelles Doing Life s Work Types of cells bacteria cells Prokaryote Eukaryotes animal cells plant cells Cell size comparison Animal cell Bacterial cell most bacteria 1-10 microns eukaryotic
Cells & Cell Organelles Doing Life s Work Types of cells bacteria cells Prokaryote Eukaryotes animal cells plant cells Cell size comparison Animal cell Bacterial cell most bacteria 1-10 microns eukaryotic
Unit 3: Cells. Objective: To be able to compare and contrast the differences between Prokaryotic and Eukaryotic Cells.
 Unit 3: Cells Objective: To be able to compare and contrast the differences between Prokaryotic and Eukaryotic Cells. The Cell Theory All living things are composed of cells (unicellular or multicellular).
Unit 3: Cells Objective: To be able to compare and contrast the differences between Prokaryotic and Eukaryotic Cells. The Cell Theory All living things are composed of cells (unicellular or multicellular).
Reading Assignments. A. Genes and the Synthesis of Polypeptides. Lecture Series 7 From DNA to Protein: Genotype to Phenotype
 Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus:
 m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
Tutorial 4 Protein Biochemistry 2 Genes to proteins: Protein synthesis, transport, targeting, and degradation
 IPAM Cells and Materials: At the Interface between Mathematics, Biology and Engineering Tutorial 4 Protein Biochemistry 2 Genes to proteins: Protein synthesis, transport, targeting, and degradation Dr.
IPAM Cells and Materials: At the Interface between Mathematics, Biology and Engineering Tutorial 4 Protein Biochemistry 2 Genes to proteins: Protein synthesis, transport, targeting, and degradation Dr.
Cell Structure: What cells are made of. Can you pick out the cells from this picture?
 Cell Structure: What cells are made of Can you pick out the cells from this picture? Review of the cell theory Microscope was developed 1610. Anton van Leeuwenhoek saw living things in pond water. 1677
Cell Structure: What cells are made of Can you pick out the cells from this picture? Review of the cell theory Microscope was developed 1610. Anton van Leeuwenhoek saw living things in pond water. 1677
Cell Alive Homeostasis Plants Animals Fungi Bacteria. Loose DNA DNA Nucleus Membrane-Bound Organelles Humans
 UNIT 3: The Cell DAYSHEET 45: Introduction to Cellular Organelles Name: Biology I Date: Bellringer: Place the words below into the correct space on the Venn Diagram: Cell Alive Homeostasis Plants Animals
UNIT 3: The Cell DAYSHEET 45: Introduction to Cellular Organelles Name: Biology I Date: Bellringer: Place the words below into the correct space on the Venn Diagram: Cell Alive Homeostasis Plants Animals
Review. Membrane proteins. Membrane transport
 Quiz 1 For problem set 11 Q1, you need the equation for the average lateral distance transversed (s) of a molecule in the membrane with respect to the diffusion constant (D) and time (t). s = (4 D t) 1/2
Quiz 1 For problem set 11 Q1, you need the equation for the average lateral distance transversed (s) of a molecule in the membrane with respect to the diffusion constant (D) and time (t). s = (4 D t) 1/2
Now starts the fun stuff Cell structure and function
 Now starts the fun stuff Cell structure and function Cell Theory The three statements of the cell theory are: All organisms are composed of one or more cells and the processes of life occur in these cells.
Now starts the fun stuff Cell structure and function Cell Theory The three statements of the cell theory are: All organisms are composed of one or more cells and the processes of life occur in these cells.
10/1/2014. Chapter Explain why the cell is considered to be the basic unit of life.
 Chapter 4 PSAT $ by October by October 11 Test 3- Tuesday October 14 over Chapter 4 and 5 DFA- Monday October 20 over everything covered so far (Chapters 1-5) Review on Thursday and Friday before 1. Explain
Chapter 4 PSAT $ by October by October 11 Test 3- Tuesday October 14 over Chapter 4 and 5 DFA- Monday October 20 over everything covered so far (Chapters 1-5) Review on Thursday and Friday before 1. Explain
and their organelles
 and their organelles Discovery Video: Cells REVIEW!!!! The Cell Theory 1. Every living organism is made of one or more cells. 2. The cell is the basic unit of structure and function. It is the smallest
and their organelles Discovery Video: Cells REVIEW!!!! The Cell Theory 1. Every living organism is made of one or more cells. 2. The cell is the basic unit of structure and function. It is the smallest
Cell Types. Prokaryotes
 Cell Types Prokaryotes before nucleus no membrane-bound nucleus only organelle present is the ribosome all other reactions occur in the cytoplasm not very efficient Ex.: bacteria 1 Cell Types Eukaryotes
Cell Types Prokaryotes before nucleus no membrane-bound nucleus only organelle present is the ribosome all other reactions occur in the cytoplasm not very efficient Ex.: bacteria 1 Cell Types Eukaryotes
O.k., Now Starts the Good Stuff (Part II) Eukaryotic Cell Structure and Function
 O.k., Now Starts the Good Stuff (Part II) Eukaryotic Cell Structure and Function Eukaryotic Cells These cells have membrane-bound structures called organelles. Cell processes occur in these organelles.
O.k., Now Starts the Good Stuff (Part II) Eukaryotic Cell Structure and Function Eukaryotic Cells These cells have membrane-bound structures called organelles. Cell processes occur in these organelles.
Cellular Neuroanatomy I The Prototypical Neuron: Soma. Reading: BCP Chapter 2
 Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Today s materials: Cell Structure and Function. 1. Prokaryote and Eukaryote 2. DNA as a blue print of life Prokaryote and Eukaryote. What is a cell?
 Today s materials: 1. Prokaryote and Eukaryote 2. DNA as a blue print of life Prokaryote and Eukaryote Achadiah Rachmawati What is a cell? Cell Structure and Function All living things are made of cells
Today s materials: 1. Prokaryote and Eukaryote 2. DNA as a blue print of life Prokaryote and Eukaryote Achadiah Rachmawati What is a cell? Cell Structure and Function All living things are made of cells
Chapter 7 Learning Targets Cell Structure & Function
 Name: Chapter 7 Learning Targets Cell Structure & Function a. Define the word cell: 1. I know the history of the cell: b. Who discovered the cell? What did he observe? 2. I can list the three parts of
Name: Chapter 7 Learning Targets Cell Structure & Function a. Define the word cell: 1. I know the history of the cell: b. Who discovered the cell? What did he observe? 2. I can list the three parts of
General A&P Cell Labs - Cellular Anatomy & Division (Mitosis) Pre-Lab Guide
 1 General A&P Cell Labs - Cellular Anatomy & Division (Mitosis) Pre-Lab AWalk-About@ Guide Have someone in your group read the following out loud, while the others read along: In this "Walk About", we
1 General A&P Cell Labs - Cellular Anatomy & Division (Mitosis) Pre-Lab AWalk-About@ Guide Have someone in your group read the following out loud, while the others read along: In this "Walk About", we
Chapter 6: A Tour of the Cell
 Chapter 6: A Tour of the Cell Concept 6.2 Eukaryotic cells have internal membranes that compartmentalize their functions 1. Which two domains consist of prokaryotic cells? 2. A major difference between
Chapter 6: A Tour of the Cell Concept 6.2 Eukaryotic cells have internal membranes that compartmentalize their functions 1. Which two domains consist of prokaryotic cells? 2. A major difference between
Introduction to Cells
 Life Science Introduction to Cells All life forms on our planet are made up of cells. In ALL organisms, cells have the same basic structure. The scientist Robert Hooke was the first to see cells under
Life Science Introduction to Cells All life forms on our planet are made up of cells. In ALL organisms, cells have the same basic structure. The scientist Robert Hooke was the first to see cells under
Biological Process Term Enrichment
 Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
Translocation through the endoplasmic reticulum membrane. Institute of Biochemistry Benoît Kornmann
 Translocation through the endoplasmic reticulum membrane Institute of Biochemistry Benoît Kornmann Endomembrane system Protein sorting Permeable to proteins but not to ions IgG tetramer (16 nm) Fully hydrated
Translocation through the endoplasmic reticulum membrane Institute of Biochemistry Benoît Kornmann Endomembrane system Protein sorting Permeable to proteins but not to ions IgG tetramer (16 nm) Fully hydrated
Gene Control Mechanisms at Transcription and Translation Levels
 Gene Control Mechanisms at Transcription and Translation Levels Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by MHRD Page 1 of 9
Gene Control Mechanisms at Transcription and Translation Levels Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by MHRD Page 1 of 9
4.1 Cells are the Fundamental Units of Life. Cell Structure. Cells. Fundamental units of life Cell theory. Except possibly viruses.
 Cells 4.1 Cells are the Fundamental Units of Life Fundamental units of life Cell theory All living things are composed of one or more cells. The cell is the most basic unit of life. All cells come from
Cells 4.1 Cells are the Fundamental Units of Life Fundamental units of life Cell theory All living things are composed of one or more cells. The cell is the most basic unit of life. All cells come from
Predicting protein subcellular localisation from amino acid sequence information Olof Emanuelsson Date received (in revised form): 10th September 2002
 Olof Emanuelsson works in close collaboration with Gunnar von Heijne on developing methods for predicting subcellular localisation of proteins. He is at the Stockholm Bioinformatics Center (SBC), a joint
Olof Emanuelsson works in close collaboration with Gunnar von Heijne on developing methods for predicting subcellular localisation of proteins. He is at the Stockholm Bioinformatics Center (SBC), a joint
CHARACTERISTICS OF LIFE ORGANIZATION OF LIFE CELL THEORY TIMELINE
 CHARACTERISTICS OF LIFE 1. composed of cells either uni/multi 2. reproduce sexual and/or asexual 3. contain DNA in cells 4. grow and develop 5. use material/energy in metabolic reactions 6. respond to
CHARACTERISTICS OF LIFE 1. composed of cells either uni/multi 2. reproduce sexual and/or asexual 3. contain DNA in cells 4. grow and develop 5. use material/energy in metabolic reactions 6. respond to
Chapter 6: A Tour of the Cell
 AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
Bio 111 Study Guide Chapter 6 Tour of the Cell
 Bio 111 Study Guide Chapter 6 Tour of the Cell BEFORE CLASS: Reading: Read the whole chapter from p. 93-121, mostly skimming Concept 6.1 on microscopy. Figure 6.8 on pp. 100-101 is really helpful in showing
Bio 111 Study Guide Chapter 6 Tour of the Cell BEFORE CLASS: Reading: Read the whole chapter from p. 93-121, mostly skimming Concept 6.1 on microscopy. Figure 6.8 on pp. 100-101 is really helpful in showing
Lecture 6 - Intracellular compartments and transport I
 01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
To study cells, biologists use microscopes and the tools of biochemistry [2].
![To study cells, biologists use microscopes and the tools of biochemistry [2]. To study cells, biologists use microscopes and the tools of biochemistry [2].](/thumbs/84/91204507.jpg) GUIDED READING - Ch. 6 - THE CELL NAME: Please print out these pages and HANDWRITE the answers directly on the printouts. Typed work or answers on separate sheets of paper will not be accepted. Importantly,
GUIDED READING - Ch. 6 - THE CELL NAME: Please print out these pages and HANDWRITE the answers directly on the printouts. Typed work or answers on separate sheets of paper will not be accepted. Importantly,
Cell Theory and Structure. Discoveries What are Cells? Cell Theory Cell Structures Organelles
 Cell Theory and Structure Discoveries What are Cells? Cell Theory Cell Structures Organelles Discoveries In 1665 Robert Hooke observed a thin slice of cork from an oak tree What he saw reminded him of
Cell Theory and Structure Discoveries What are Cells? Cell Theory Cell Structures Organelles Discoveries In 1665 Robert Hooke observed a thin slice of cork from an oak tree What he saw reminded him of
Biology: Life on Earth
 Teresa Audesirk Gerald Audesirk Bruce E. Byers Biology: Life on Earth Eighth Edition Lecture for Chapter 4 Cell Structure and Function Copyright 2008 Pearson Prentice Hall, Inc. Chapter 4 Outline 4.1 What
Teresa Audesirk Gerald Audesirk Bruce E. Byers Biology: Life on Earth Eighth Edition Lecture for Chapter 4 Cell Structure and Function Copyright 2008 Pearson Prentice Hall, Inc. Chapter 4 Outline 4.1 What
Function and Illustration. Nucleus. Nucleolus. Cell membrane. Cell wall. Capsule. Mitochondrion
 Intro to Organelles Name: Block: Organelles are small structures inside cells. They are often covered in membranes. Each organelle has a job to do in the cell. Their name means little organ. Just like
Intro to Organelles Name: Block: Organelles are small structures inside cells. They are often covered in membranes. Each organelle has a job to do in the cell. Their name means little organ. Just like
Honors Biology Fall Final Exam Study Guide
 Honors Biology Fall Final Exam Study Guide Helpful Information: Exam has 100 multiple choice questions. Be ready with pencils and a four-function calculator on the day of the test. Review ALL vocabulary,
Honors Biology Fall Final Exam Study Guide Helpful Information: Exam has 100 multiple choice questions. Be ready with pencils and a four-function calculator on the day of the test. Review ALL vocabulary,
Chapter 6 A Tour of the Cell
 Chapter 6 A Tour of the Cell The cell is the basic unit of life Although cells differ substantially from one another, they all share certain characteristics that reflect a common ancestry and remind us
Chapter 6 A Tour of the Cell The cell is the basic unit of life Although cells differ substantially from one another, they all share certain characteristics that reflect a common ancestry and remind us
Biology 1 Notebook. Review Answers Pages 17 -?
 Biology 1 Notebook Review Answers Pages 17 -? The History of Cell Studies 1. Robert Hook (1665) used a microscope to examine a thin slice of cork. The little boxes he observed reminded him of the small
Biology 1 Notebook Review Answers Pages 17 -? The History of Cell Studies 1. Robert Hook (1665) used a microscope to examine a thin slice of cork. The little boxes he observed reminded him of the small
Principles of Cellular Biology
 Principles of Cellular Biology آشنایی با مبانی اولیه سلول Biologists are interested in objects ranging in size from small molecules to the tallest trees: Cell Basic building blocks of life Understanding
Principles of Cellular Biology آشنایی با مبانی اولیه سلول Biologists are interested in objects ranging in size from small molecules to the tallest trees: Cell Basic building blocks of life Understanding
Biology Exam #1 Study Guide. True/False Indicate whether the statement is true or false. F 1. All living things are composed of many cells.
 Biology Exam #1 Study Guide True/False Indicate whether the statement is true or false. F 1. All living things are composed of many cells. T 2. Membranes are selectively permeable if they allow only certain
Biology Exam #1 Study Guide True/False Indicate whether the statement is true or false. F 1. All living things are composed of many cells. T 2. Membranes are selectively permeable if they allow only certain
7.L.1.2 Plant and Animal Cells. Plant and Animal Cells
 7.L.1.2 Plant and Animal Cells Plant and Animal Cells Clarifying Objective: 7.L.1.2 Compare the structures and functions of plant and animal cells; include major organelles (cell membrane, cell wall, nucleus,
7.L.1.2 Plant and Animal Cells Plant and Animal Cells Clarifying Objective: 7.L.1.2 Compare the structures and functions of plant and animal cells; include major organelles (cell membrane, cell wall, nucleus,
CELL : THE UNIT OF LIFE
 38 BIOLOGY, EXEMPLAR PROBLEMS CHAPTER 8 CELL : THE UNIT OF LIFE MULTIPLE CHOICE QUESTIONS 1. A common characteristic feature of plant sieve tube cells and most of mammalian erythrocytes is a. Absence of
38 BIOLOGY, EXEMPLAR PROBLEMS CHAPTER 8 CELL : THE UNIT OF LIFE MULTIPLE CHOICE QUESTIONS 1. A common characteristic feature of plant sieve tube cells and most of mammalian erythrocytes is a. Absence of
UNIT 1: WELLNESS & HOMEOSTASIS. Biology notes 1 Mr.Yeung
 UNIT 1: WELLNESS & HOMEOSTASIS Biology notes 1 Mr.Yeung WHAT IS UNIT 1 ABOUT? Wellness & Homeostasis 0. Wellness (Reading labels/homeostasis/ serving size) 1. Cell Biology (Ch.3) Cell organelles Cell types
UNIT 1: WELLNESS & HOMEOSTASIS Biology notes 1 Mr.Yeung WHAT IS UNIT 1 ABOUT? Wellness & Homeostasis 0. Wellness (Reading labels/homeostasis/ serving size) 1. Cell Biology (Ch.3) Cell organelles Cell types
Zimmerman AP Biology CBHS South Name Chapter 7&8 Guided Reading Assignment 1) What is resolving power and why is it important in biology?
 Zimmerman AP Biology CBHS South Name Chapter 7&8 Guided Reading Assignment 1) What is resolving power and why is it important in biology? 2) How does an electron microscope work and what is the difference
Zimmerman AP Biology CBHS South Name Chapter 7&8 Guided Reading Assignment 1) What is resolving power and why is it important in biology? 2) How does an electron microscope work and what is the difference
GCD3033:Cell Biology. Transcription
 Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
ST.JOSEPH S COLLEGE OF ARTS & SCIENCE (AUTONOMOUS) CUDDALORE 1 PG & RESEARCH DEPARTMENT OF BIOCHEMISTRY II-B.Sc Biochemistry QUESTION BANK
 1 ST.JOSEPH S COLLEGE OF ARTS & SCIENCE (AUTONOMOUS) CUDDALORE 1 PG & RESEARCH DEPARTMENT OF BIOCHEMISTRY II-B.Sc Biochemistry QUESTION BANK Subject: Cell Biology Subject code: BC102S Subject handled:
1 ST.JOSEPH S COLLEGE OF ARTS & SCIENCE (AUTONOMOUS) CUDDALORE 1 PG & RESEARCH DEPARTMENT OF BIOCHEMISTRY II-B.Sc Biochemistry QUESTION BANK Subject: Cell Biology Subject code: BC102S Subject handled:
Multiple Choice Identify the letter of the choice that best completes the statement or answers the question.
 Exam 1 Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. The smallest units of life in all living things are a. cells. c. cytoplasm. b. mitochondria.
Exam 1 Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. The smallest units of life in all living things are a. cells. c. cytoplasm. b. mitochondria.
Tuesday 9/6/2018 Mike Mueckler
 Tuesday 9/6/2018 Mike Mueckler mmueckler@wustl.edu Intracellular Targeting of Nascent Polypeptides Mitochondria are the Sites of Oxidative ATP Production Sugars Triglycerides Figure 14-10 Molecular Biology
Tuesday 9/6/2018 Mike Mueckler mmueckler@wustl.edu Intracellular Targeting of Nascent Polypeptides Mitochondria are the Sites of Oxidative ATP Production Sugars Triglycerides Figure 14-10 Molecular Biology
Introduction to Cells
 Life Science Introduction to Cells All life forms on our planet are made up of cells. In ALL organisms, cells have the same basic structure. The scientist Robert Hooke was the first to see cells under
Life Science Introduction to Cells All life forms on our planet are made up of cells. In ALL organisms, cells have the same basic structure. The scientist Robert Hooke was the first to see cells under
Cell Structure and Function
 Cell Structure and Function Cell size comparison Animal cell Bacterial cell What jobs do cells have to do for an organism to live Gas exchange CO 2 & O 2 Eat (take in & digest food) Make energy ATP Build
Cell Structure and Function Cell size comparison Animal cell Bacterial cell What jobs do cells have to do for an organism to live Gas exchange CO 2 & O 2 Eat (take in & digest food) Make energy ATP Build
