Spliceosome and Localization of Its Catalytic Core
|
|
|
- Bartholomew Sutton
- 5 years ago
- Views:
Transcription
1 Molecular Cell, Volume 40 Supplemental Information 3D Cryo-EM Structure of an Active Step I Spliceosome and Localization of Its Catalytic Core Monika M. Golas, Bjoern Sander, Sergey Bessonov, Michael Grote, Elmar Wolf, Berthold Kastner, Holger Stark, and Reinhard Lührmann This Supplemental Information contains: Figures S1 S4 Table S1 Supplemental References 1
2 Figure S1. Human Spliceosomal C Complexes Assembled on the PM5 or MINX GG Construct, Related to Figure 2 A. Schematic representation of the PM5 (top) and MINX GG (bottom) pre-mrna constructs. B. RNA and protein composition of the step I spliceosome assembled on the 2
3 PM5 pre-mrna (lanes 1 3) and MINX GG pre-mrna construct (lanes 4 6). The RNA composition was determined by denaturing PAGE (lanes 1, 2, 4, 5) and the protein composition by SDS-PAGE (lane 3 and 6). RNA was visualized by silver staining (lane 1 and 4) or by autoradiography (lane 2 and 5), and protein by staining with Coomassie blue (lane 3 and 6). Identities of RNAs (lanes 1, 2, 4 and 5) and the molecular weight marker (lane 3 and 6; in kda) are indicated on the right, respectively. C. Influence of GraFix on the structural integrity of the spliceosomal complexes. Shown is the gradient profile of the final ultracentrifugation step in the absence (purple curve) or presence i.e. GraFix (blue curve) of a low, increasing concentration of glutaraldehyde (0 0.1%). Gradients were fractionated from the bottom (i.e. fraction #1 corresponds to the bottom fraction of the gradient). Particles purified using the GraFix protocol exhibited a sharp peak. In contrast, particles purified in a parallel gradient lacking glutaraldehyde exhibited a broader peak that was shifted somewhat to lower molecular weight fractions. Although similar amounts of step I spliceosomes were loaded onto each gradient, less overall material was observed in the gradient without glutaraldehyde while more radioactivity was found on the walls of the gradient tube. Together, this suggests dissociation and/or aggregation of the particles during ultracentrifugation in the absence of glutaraldehyde. D. Gallery of 5 selected particle views of the spliceosomal C complex. The upper row shows band-pass filtered images of individual particles, and the second row shows the corresponding class averages, i.e. cluster centers in an MSA-based unsupervised image clustering. Upon Euler angle determination, row 3 shows the 3D structure under the same view as the two-dimensional projection views in row 2, and row 4 shows computed reprojections from the 3D structure, which are in excellent agreement with the class averages in row 2. The scale bars correspond to 10 nm. E. Fourier-Shell-Correlation (FSC) analysis of the refined cryo-em map of the step I spliceosome determined from unstained cryo-em images. The FSC of the cryo-em map of the step I spliceosome suggests a resolution of nm depending on the criteria used (i.e. 5σ, FSC, 0.5 FSC). F. 3D structure of the step I spliceosome assembled on the PM5 pre-mrna. The C complex has a size of ~36 nm 33.5 nm 27 nm with maximum dimensions of ~42 nm (diagonal as indicated by the arrowheads). The domain numbering is as in Figure 2; densities are colored in silver, purple, green, gold, turquoise, red and blue according to the domain identity. 3
4 Figure S2. Three-Dimensional Structure of the Salt-stable RNP Core of the Step I Spliceosome as Determined by Weighted Averaging of Random Conical Tilt 3D maps, Related to Figure 3 Subsequent to 3D alignment and weighted integration of the 3D RCT maps over all rotations, a weighted average 3D structure of the salt-stable RNP core complex of the C complex was determined. This structure was used as reference to align the 3D RCT maps as prerequisite for 3D classification of the maps shown in Figure 3D of the main manuscript. 4
5 Figure S3: Three-Dimensional Structure of the 35S U5 snrnp, Related to Figure 4 35S U5 snrnps were purified by anti-flag immuno-chromatography and exhibited essentially the same protein composition as found in our earlier study (Makarov et al., 5
6 2002), except that a few proteins such as S100A8/A9, KIAA0773 and FLJ10206, which were present in substoichiometric amounts in our earlier preparations, were absent from the preparations described here (in fact, these proteins have never been observed in native spliceosomal complexes indicating that they were contaminants). On the other hand, the proteins Morg1, Q9BRR8, hprp22 and Slu7, which were identified in the 35S U5 snrnp described here were absent from the previous preparations. These minor differences are likely due to the use of different antibodies during immunoaffinity chromatography of the 35S U5 snrnps. A. Gallery of 5 selected particle views of the 35S U5 snrnp. The upper row shows band-pass filtered images of individual particles, and the second row shows the corresponding class averages, i.e. cluster centers in an MSA-based unsupervised image clustering. Upon Euler angle determination, row 3 shows the 3D structure under the same view as the two-dimensional projection views in row 2, and row 4 shows computed re-projections from the 3D structure, which are in excellent agreement with the class averages in row 2. The scale bars correspond to 10 nm. B. Resolution of the refined cryo-em map of the 35S U5 snrnp determined from unstained cryo-em images. The Fourier-Shell-Correlation (FSC) of the cryo-em map of the 35S U5 snrnp suggests a resolution of nm, depending on the criteria used (i.e. 5σ, FSC, 0.5 FSC). C. 3D structure of the post-spliceosomal 35S U5 snrnp. The 35S U5 snrnp has a size of ~26.5 nm 26 nm 23 nm with maximum dimensions of ~28.5 nm (diagonal as indicated by the arrowheads). The domain numbering (i.e. domains 1, 2b and 3 5) is as in Fig. 7. 6
7 Figure S4. Comparison of the 3D Structures of Human Spliceosomal Complexes and the 70S Ribosome, Related to Figure 7 Shown are the 3D structures of the pre-catalytic BΔU1 complex (EMDB entry 1066; Boehringer et al., 2004), the C complex isolated under stringent conditions (EMDB entry 1062; Jurica et al., 2004), our catalytically active step I spliceosome (C complex) isolated under physiological conditions, our salt-stable RNP core of the C complex, our postspliceosomal 35S U5 snrnp and the bacterial 70S ribosome (PDB entries 2WRI, 2WRJ, 2WRK, 2WRL; Gao et al., 2009) filtered to slightly better than 3 nm resolution. The structures of the BΔU1 complex and of the C complex isolated under stringent conditions are shown in their (unfitted) standard views as given in the original publications. 7
8 Table S1. Details of the Electron Microscopic Data Sets Used for Image Processing, Related to Figure 2 Complex Type of Imaging Particle Numbers Used for PM5 step I spliceosome (NS) Salt-stable RNP core of the spliceosome (NS) Salt-stable RNP core of the spliceosome (Tilt pairs) Negative staining 4,618 2D image processing: class averaging, difference mapping Negative staining 29,382 2D image processing: class averaging, difference mapping Negative staining 63,045 (zero tilt) 6,331 (tilt pairs) 3D image processing: determination of a weighted average RCT structure and 3D class averages 35S U5 snrnp (NS) Negative staining 59,380 2D image processing: class averaging, difference mapping MINX GG step I spliceosome (Cryo) PM5 step I spliceosome (Cryo tilt pairs) 35S U5 snrnp (Cryo tilt pairs) PM5 step I spliceosome (Cryo refine) 35S U5 snrnp (Cryo refine) Anti-MBP labeled C complexes (label at 5 end of 5 exon) Unstained cryo in vitrified ice Unstained cryo tilt pairs in vitrified ice Unstained cryo tilt pairs in vitrified ice Unstained cryo in vitrified ice Unstained cryo in vitrified ice Negative staining ~300 (60 typical views) 56,202 2D image processing: class averaging 18,152 3D image processing: determination of a weighted average RCT structure 7,890 3D image processing: determination of a weighted average RCT structure 36,071 3D image processing: refinement 21,476 3D image processing: refinement 2D analysis: Localization of 5 exon end 8
9 Supplemental References Boehringer, D., Makarov, E.M., Sander, B., Makarova, O.V., Kastner, B., Lührmann, R., and Stark, H. (2004). Three-dimensional structure of a pre-catalytic human spliceosomal complex B. Nat Struct Mol Biol 11, Gao, Y.G., Selmer, M., Dunham, C.M., Weixlbaumer, A., Kelley, A.C., and Ramakrishnan, V. (2009). The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326, 694. Jurica, M.S., Sousa, D., Moore, M.J., and Grigorieff, N. (2004). Three-dimensional structure of C complex spliceosomes by electron microscopy. Nat Struct Mol Biol 11, Makarov, E.M., Makarova, O.V., Urlaub, H., Gentzel, M., Will, C.L., Wilm, M., and Lührmann, R. (2002). Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298,
GraFix: sample preparation for single-particle electron cryomicroscopy
 GraFix: sample preparation for single-particle electron cryomicroscopy Berthold Kastner, Niels Fischer, Monika Mariola Golas, Bjoern Sander, Prakash Dube, Daniel Boehringer, Klaus Hartmuth, Jochen Deckert,
GraFix: sample preparation for single-particle electron cryomicroscopy Berthold Kastner, Niels Fischer, Monika Mariola Golas, Bjoern Sander, Prakash Dube, Daniel Boehringer, Klaus Hartmuth, Jochen Deckert,
Sample Preparation. Holger Stark MPI for biophysical Chemistry Göttingen Germany
 Sample Preparation Holger Stark MPI for biophysical Chemistry Göttingen Germany Motivation PDB search results: s with min 4 different chains (complexes) make up only ~1% of the PDB s in contrast to the
Sample Preparation Holger Stark MPI for biophysical Chemistry Göttingen Germany Motivation PDB search results: s with min 4 different chains (complexes) make up only ~1% of the PDB s in contrast to the
Cryo-EM data collection, refinement and validation statistics
 1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/310/5753/1513/dc1 Supporting Online Material for Structural Roles for Human Translation Factor eif3 in Initiation of Protein Synthesis Bunpote Siridechadilok, Christopher
www.sciencemag.org/cgi/content/full/310/5753/1513/dc1 Supporting Online Material for Structural Roles for Human Translation Factor eif3 in Initiation of Protein Synthesis Bunpote Siridechadilok, Christopher
Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome
 Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome Ahmad Jomaa 1, Yu-Hsien Hwang Fu 2, Daniel Boehringer 1, Marc Leibundgut 1, Shu-ou Shan 2, and Nenad
Structure of the quaternary complex between SRP, SR, and translocon bound to the translating ribosome Ahmad Jomaa 1, Yu-Hsien Hwang Fu 2, Daniel Boehringer 1, Marc Leibundgut 1, Shu-ou Shan 2, and Nenad
Purification, SDS-PAGE and cryo-em characterization of the MCM hexamer and Cdt1 MCM heptamer samples.
 Supplementary Figure 1 Purification, SDS-PAGE and cryo-em characterization of the MCM hexamer and Cdt1 MCM heptamer samples. (a-b) SDS-PAGE analysis of the hexamer and heptamer samples. The eluted hexamer
Supplementary Figure 1 Purification, SDS-PAGE and cryo-em characterization of the MCM hexamer and Cdt1 MCM heptamer samples. (a-b) SDS-PAGE analysis of the hexamer and heptamer samples. The eluted hexamer
Center for Cell Imaging Department of Cell Biology
 Center for Cell Imaging Department of Cell Biology Contents Preparation of Colloidal Gold Conjugates Coupling the Protein A to the Gold Particles Purification of the protein A-gold. Storage Influence of
Center for Cell Imaging Department of Cell Biology Contents Preparation of Colloidal Gold Conjugates Coupling the Protein A to the Gold Particles Purification of the protein A-gold. Storage Influence of
Recognition of the amber UAG stop codon by release factor RF1
 Manuscript EMBO-2010-73984 Recognition of the amber UAG stop codon by release factor RF1 Andrei Korostelev, Jianyu Zhu, Haruichi Asahara and Harry F. Noller Corresponding author: Harry F. Noller, Univ.
Manuscript EMBO-2010-73984 Recognition of the amber UAG stop codon by release factor RF1 Andrei Korostelev, Jianyu Zhu, Haruichi Asahara and Harry F. Noller Corresponding author: Harry F. Noller, Univ.
Lecture CIMST Winter School. 1. What can you see by TEM?
 Lecture CIMST Winter School Cryo-electron microscopy and tomography of biological macromolecules 20.1.2011 9:00-9:45 in Y03G91 Dr. Takashi Ishikawa OFLB/005 Tel: 056 310 4217 e-mail: takashi.ishikawa@psi.ch
Lecture CIMST Winter School Cryo-electron microscopy and tomography of biological macromolecules 20.1.2011 9:00-9:45 in Y03G91 Dr. Takashi Ishikawa OFLB/005 Tel: 056 310 4217 e-mail: takashi.ishikawa@psi.ch
Reading the genetic code: The 3D version. Venki Ramakrishnan MRC Laboratory of Molecular Biology Cambridge, United Kingdom
 Reading the genetic code: The 3D version Venki Ramakrishnan MRC Laboratory of Molecular Biology Cambridge, United Kingdom DNA The Central Dogma The Central Dogma DNA mrna The Central Dogma DNA mrna Protein
Reading the genetic code: The 3D version Venki Ramakrishnan MRC Laboratory of Molecular Biology Cambridge, United Kingdom DNA The Central Dogma The Central Dogma DNA mrna The Central Dogma DNA mrna Protein
Preparing Colloidal Gold for Electron Microscopy
 Corporate Headquarters 400 Valley Road Warrington, PA 18976 1-800-523-2575 FAX 1-800-343-3291 Email: info@polysciences.com www.polysciences.com Europe - Germany Polysciences Europe GmbH Handelsstr. 3 D-69214
Corporate Headquarters 400 Valley Road Warrington, PA 18976 1-800-523-2575 FAX 1-800-343-3291 Email: info@polysciences.com www.polysciences.com Europe - Germany Polysciences Europe GmbH Handelsstr. 3 D-69214
Three-Dimensional Electron Microscopy of Macromolecular Assemblies
 Three-Dimensional Electron Microscopy of Macromolecular Assemblies Joachim Frank Wadsworth Center for Laboratories and Research State of New York Department of Health The Governor Nelson A. Rockefeller
Three-Dimensional Electron Microscopy of Macromolecular Assemblies Joachim Frank Wadsworth Center for Laboratories and Research State of New York Department of Health The Governor Nelson A. Rockefeller
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Cryo-EM structure and model of the C. thermophilum 90S preribosome. a, Gold standard FSC curve showing the average resolution of the 90S preribosome masked and unmasked (left). FSC
Supplementary Figure 1 Cryo-EM structure and model of the C. thermophilum 90S preribosome. a, Gold standard FSC curve showing the average resolution of the 90S preribosome masked and unmasked (left). FSC
SUPPLEMENTARY INFORMATION
 SUPPLMTARY IFORMATIO a doi:10.108/nature10402 b 100 nm 100 nm c SAXS Model d ulers assigned to reference- Back-projected free class averages class averages Refinement against single particles Reconstructed
SUPPLMTARY IFORMATIO a doi:10.108/nature10402 b 100 nm 100 nm c SAXS Model d ulers assigned to reference- Back-projected free class averages class averages Refinement against single particles Reconstructed
Model building and validation for cryo-em maps at low resolution
 International Workshop of Advanced Image Processing of Cryo-Electron Microscopy 2015 Tsinghua University, Beijing, China June 3-7, 2015 Model building and validation for cryo-em maps at low resolution
International Workshop of Advanced Image Processing of Cryo-Electron Microscopy 2015 Tsinghua University, Beijing, China June 3-7, 2015 Model building and validation for cryo-em maps at low resolution
 Supplemental Figure and Movie Legends Figure S1. Time course experiments on the thermal stability of apo AT cpn-α by native PAGE (related to Figure 2B). The samples were heated, respectively, to (A) 45
Supplemental Figure and Movie Legends Figure S1. Time course experiments on the thermal stability of apo AT cpn-α by native PAGE (related to Figure 2B). The samples were heated, respectively, to (A) 45
Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes
 9 The Nucleus Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes Explain general structures of Nuclear Envelope, Nuclear Lamina, Nuclear Pore Complex Explain movement of proteins
9 The Nucleus Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes Explain general structures of Nuclear Envelope, Nuclear Lamina, Nuclear Pore Complex Explain movement of proteins
Application examples of single particle 3D reconstruction. Ning Gao Tsinghua University
 Application examples of single particle 3D reconstruction Ning Gao Tsinghua University ninggao@tsinghua.edu.cn Electron Microscopes First electron microscope constructed by Ernst Ruska in 1930 s (1986
Application examples of single particle 3D reconstruction Ning Gao Tsinghua University ninggao@tsinghua.edu.cn Electron Microscopes First electron microscope constructed by Ernst Ruska in 1930 s (1986
Processing Heterogeneity
 New Challenges for Processing Heterogeneity Nikolaus Grigorieff Larson, The Far Side Heterogeneity and Biology Translocation, Brilot et al 2013 Glutamate receptor, Dürr et al 2014 Spliceosome, Wahl et
New Challenges for Processing Heterogeneity Nikolaus Grigorieff Larson, The Far Side Heterogeneity and Biology Translocation, Brilot et al 2013 Glutamate receptor, Dürr et al 2014 Spliceosome, Wahl et
Measurement of Dynamic trna Movement and Visualizing Ribosome Biogenesis Applications of Time resolved Electron Cryo microscopy
 2011 TIGP CBMB Seminar Measurement of Dynamic trna Movement and Visualizing Ribosome Biogenesis Applications of Time resolved Electron Cryo microscopy Ribosome dynamics and trna movement by time resolved
2011 TIGP CBMB Seminar Measurement of Dynamic trna Movement and Visualizing Ribosome Biogenesis Applications of Time resolved Electron Cryo microscopy Ribosome dynamics and trna movement by time resolved
ParM filament images were extracted and from the electron micrographs and
 Supplemental methods Outline of the EM reconstruction: ParM filament images were extracted and from the electron micrographs and straightened. The digitized images were corrected for the phase of the Contrast
Supplemental methods Outline of the EM reconstruction: ParM filament images were extracted and from the electron micrographs and straightened. The digitized images were corrected for the phase of the Contrast
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11991 Supplementary Figure 1 - Refinement strategy for PIC intermediate assemblies by negative stain EM. The cryo-negative stain structure of free Pol II 1 (a) was used as initial reference
doi:10.1038/nature11991 Supplementary Figure 1 - Refinement strategy for PIC intermediate assemblies by negative stain EM. The cryo-negative stain structure of free Pol II 1 (a) was used as initial reference
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Watanabe'et'al.' Supplementary'Figure'1'
 MW IN O 80 2 IN O 80 SW 3 R -C 3 MW SW R -C SW 1 R -C 2 IN O 80 1 Supplementary'Figure'1' Supplementary Figure 1. SDS-PAGE image of INO80-C and SWR-C. Three independent preparations of each TAP-tagged
MW IN O 80 2 IN O 80 SW 3 R -C 3 MW SW R -C SW 1 R -C 2 IN O 80 1 Supplementary'Figure'1' Supplementary Figure 1. SDS-PAGE image of INO80-C and SWR-C. Three independent preparations of each TAP-tagged
History of 3D Electron Microscopy and Helical Reconstruction
 T H E U N I V E R S I T Y of T E X A S S C H O O L O F H E A L T H I N F O R M A T I O N S C I E N C E S A T H O U S T O N History of 3D Electron Microscopy and Helical Reconstruction For students of HI
T H E U N I V E R S I T Y of T E X A S S C H O O L O F H E A L T H I N F O R M A T I O N S C I E N C E S A T H O U S T O N History of 3D Electron Microscopy and Helical Reconstruction For students of HI
Electron microscopy in molecular cell biology II
 Electron microscopy in molecular cell biology II Cryo-EM and image processing Werner Kühlbrandt Max Planck Institute of Biophysics Sample preparation for cryo-em Preparation laboratory Specimen preparation
Electron microscopy in molecular cell biology II Cryo-EM and image processing Werner Kühlbrandt Max Planck Institute of Biophysics Sample preparation for cryo-em Preparation laboratory Specimen preparation
Modeling of snrnp Motion in the Nucleoplasm
 Excerpt from the Proceedings of the COMSOL Conference 008 Hannover Modeling of snrnp Motion in the Nucleoplasm Michaela Blažíková 1, Jan Malínský *,, David Staněk 3 and Petr Heřman 1 1 Charles University
Excerpt from the Proceedings of the COMSOL Conference 008 Hannover Modeling of snrnp Motion in the Nucleoplasm Michaela Blažíková 1, Jan Malínský *,, David Staněk 3 and Petr Heřman 1 1 Charles University
GCD3033:Cell Biology. Transcription
 Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/6/e1700147/dc1 Supplementary Materials for Ribosome rearrangements at the onset of translational bypassing Xabier Agirrezabala, Ekaterina Samatova, Mariia Klimova,
advances.sciencemag.org/cgi/content/full/3/6/e1700147/dc1 Supplementary Materials for Ribosome rearrangements at the onset of translational bypassing Xabier Agirrezabala, Ekaterina Samatova, Mariia Klimova,
Part 1 X-ray Crystallography
 Part 1 X-ray Crystallography What happens to electron when it is hit by x-rays? 1. The electron starts vibrating with the same frequency as the x-ray beam 2. As a result, secondary beams will be scattered
Part 1 X-ray Crystallography What happens to electron when it is hit by x-rays? 1. The electron starts vibrating with the same frequency as the x-ray beam 2. As a result, secondary beams will be scattered
Post-class review: Glisovic et al. (2008) RNA-binding proteins and posttranscriptional gene regulation. FEBS letters 582:
 BMB 632 Probing structure and function of complex RNA-protein machines Instructor: Kristen W Lynch Co-Instructor: Jeremy Wilusz Tuesdays, 12-2 CRB 302 (and Anat-Chem 250/255) Summary: RNA-Protein complexes
BMB 632 Probing structure and function of complex RNA-protein machines Instructor: Kristen W Lynch Co-Instructor: Jeremy Wilusz Tuesdays, 12-2 CRB 302 (and Anat-Chem 250/255) Summary: RNA-Protein complexes
V19 Topology of Protein Complexes on Graphs
 V19 Topology of Protein Complexes on Graphs (1) CombDock (2) Nuclear Pore Complex 19. Lecture WS 2013/14 Bioinformatics III 1 Prediction Structures of Protein Complexes from Connectivities CombDock: automated
V19 Topology of Protein Complexes on Graphs (1) CombDock (2) Nuclear Pore Complex 19. Lecture WS 2013/14 Bioinformatics III 1 Prediction Structures of Protein Complexes from Connectivities CombDock: automated
Structure and RNA-binding properties. of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex
 Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Image Assessment San Diego, November 2005
 Image Assessment San Diego, November 005 Pawel A. Penczek The University of Texas Houston Medical School Department of Biochemistry and Molecular Biology 6431 Fannin, MSB6.18, Houston, TX 77030, USA phone:
Image Assessment San Diego, November 005 Pawel A. Penczek The University of Texas Houston Medical School Department of Biochemistry and Molecular Biology 6431 Fannin, MSB6.18, Houston, TX 77030, USA phone:
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing
 Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
Supplemental Data SUPPLEMENTAL FIGURES
 Supplemental Data CRYSTAL STRUCTURE OF THE MG.ADP-INHIBITED STATE OF THE YEAST F 1 C 10 ATP SYNTHASE Alain Dautant*, Jean Velours and Marie-France Giraud* From Université Bordeaux 2, CNRS; Institut de
Supplemental Data CRYSTAL STRUCTURE OF THE MG.ADP-INHIBITED STATE OF THE YEAST F 1 C 10 ATP SYNTHASE Alain Dautant*, Jean Velours and Marie-France Giraud* From Université Bordeaux 2, CNRS; Institut de
Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants. GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
 SUPPLEMENTAL DATA Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants Sense primer (5 to 3 ) Anti-sense primer (5 to 3 ) GAL1 mutants GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
SUPPLEMENTAL DATA Table S1. Primers used for the constructions of recombinant GAL1 and λ5 mutants Sense primer (5 to 3 ) Anti-sense primer (5 to 3 ) GAL1 mutants GAL1-E74A ccgagcagcgggcggctgtctttcc ggaaagacagccgcccgctgctcgg
SUPPLEMENTARY INFORMATION
 GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
Molecular Rainbow Dye Electrophoresis Student Materials
 Dye Electrophoresis Student Materials Introduction. 2 Pre-Lab Questions 4 Lab Protocol. 5 Data Collection Worksheet 6 Post-Lab Questions and Analysis.. 7 Last updated: 10/23/2017 Introduction Take just
Dye Electrophoresis Student Materials Introduction. 2 Pre-Lab Questions 4 Lab Protocol. 5 Data Collection Worksheet 6 Post-Lab Questions and Analysis.. 7 Last updated: 10/23/2017 Introduction Take just
Supplementary Information: Long range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Information: Long range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 Supplementary Information: Long range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs
Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition
 Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
Joachim Frank Wadsworth Center Empire State Plaza P.O. Box 509 Albany, New York Tel: (518)
 This material is provided for educational use only. The information in these slides including all data, images and related materials are the property of : Joachim Frank Wadsworth Center Empire State Plaza
This material is provided for educational use only. The information in these slides including all data, images and related materials are the property of : Joachim Frank Wadsworth Center Empire State Plaza
From gene to protein. Premedical biology
 From gene to protein Premedical biology Central dogma of Biology, Molecular Biology, Genetics transcription replication reverse transcription translation DNA RNA Protein RNA chemically similar to DNA,
From gene to protein Premedical biology Central dogma of Biology, Molecular Biology, Genetics transcription replication reverse transcription translation DNA RNA Protein RNA chemically similar to DNA,
Supplementary Information. Autocatalytic backbone N-methylation in a family of ribosomal peptide natural products
 Supplementary Information Autocatalytic backbone -methylation in a family of ribosomal peptide natural products iels S. van der Velden 1, oemi Kälin 1, Maximilian J. Helf 1, Jörn Piel 1, Michael F. Freeman
Supplementary Information Autocatalytic backbone -methylation in a family of ribosomal peptide natural products iels S. van der Velden 1, oemi Kälin 1, Maximilian J. Helf 1, Jörn Piel 1, Michael F. Freeman
Quantification Strategies Using the High Sensitivity Protein 250 Assay for the Agilent 2100 Bioanalyzer. Technical Note. Abstract
 Quantification Strategies Using the High Sensitivity Protein 25 Assay for the Agilent 21 Bioanalyzer Technical Note.. 1. 1. 1..1.1 1. 1. 1... Abstract The Agilent High Sensitivity Protein 25 assay for
Quantification Strategies Using the High Sensitivity Protein 25 Assay for the Agilent 21 Bioanalyzer Technical Note.. 1. 1. 1..1.1 1. 1. 1... Abstract The Agilent High Sensitivity Protein 25 assay for
Complete the table by stating the function associated with each organelle. contains the genetic material.... lysosome ribosome... Table 6.
 1 (a) Table 6.1 gives the functions of certain organelles in a eukaryotic cell. Complete the table by stating the function associated with each organelle. The first row has been completed for you. Organelle
1 (a) Table 6.1 gives the functions of certain organelles in a eukaryotic cell. Complete the table by stating the function associated with each organelle. The first row has been completed for you. Organelle
1. In most cases, genes code for and it is that
 Name Chapter 10 Reading Guide From DNA to Protein: Gene Expression Concept 10.1 Genetics Shows That Genes Code for Proteins 1. In most cases, genes code for and it is that determine. 2. Describe what Garrod
Name Chapter 10 Reading Guide From DNA to Protein: Gene Expression Concept 10.1 Genetics Shows That Genes Code for Proteins 1. In most cases, genes code for and it is that determine. 2. Describe what Garrod
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1. Api137-induced ribosome stalling at the end of the ORFs
 Supplementary Figure 1 Api137-induced ribosome stalling at the end of the ORFs Toeprinting analysis of translation arrest in the synthetic ORF RST2 (left) and the natural ORF ermbl (right) mediated by
Supplementary Figure 1 Api137-induced ribosome stalling at the end of the ORFs Toeprinting analysis of translation arrest in the synthetic ORF RST2 (left) and the natural ORF ermbl (right) mediated by
Three types of RNA polymerase in eukaryotic nuclei
 Three types of RNA polymerase in eukaryotic nuclei Type Location RNA synthesized Effect of α-amanitin I Nucleolus Pre-rRNA for 18,.8 and 8S rrnas Insensitive II Nucleoplasm Pre-mRNA, some snrnas Sensitive
Three types of RNA polymerase in eukaryotic nuclei Type Location RNA synthesized Effect of α-amanitin I Nucleolus Pre-rRNA for 18,.8 and 8S rrnas Insensitive II Nucleoplasm Pre-mRNA, some snrnas Sensitive
SI Text S1 Solution Scattering Data Collection and Analysis. SI references
 SI Text S1 Solution Scattering Data Collection and Analysis. The X-ray photon energy was set to 8 kev. The PILATUS hybrid pixel array detector (RIGAKU) was positioned at a distance of 606 mm from the sample.
SI Text S1 Solution Scattering Data Collection and Analysis. The X-ray photon energy was set to 8 kev. The PILATUS hybrid pixel array detector (RIGAKU) was positioned at a distance of 606 mm from the sample.
Supplementary Figure 1. Biochemical and sequence alignment analyses the
 Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Introduction to Cryo Microscopy. Nikolaus Grigorieff
 Introduction to Cryo Microscopy Nikolaus Grigorieff Early TEM and Biology 1906-1988 Bacterial culture in negative stain. Friedrich Krause, 1937 Nobel Prize in Physics 1986 Early design, 1931 Decades of
Introduction to Cryo Microscopy Nikolaus Grigorieff Early TEM and Biology 1906-1988 Bacterial culture in negative stain. Friedrich Krause, 1937 Nobel Prize in Physics 1986 Early design, 1931 Decades of
Visualizing the transfer-messenger RNA as the ribosome resumes translation
 Manuscript EMBO-2010-75355 Visualizing the transfer-messenger RNA as the ribosome resumes translation Jie Fu, Yaser Hashem, Iwona Wower, Jianlin Lei, Hstau Y. Liao, Christian Zwieb, Jacek Wower and Joachim
Manuscript EMBO-2010-75355 Visualizing the transfer-messenger RNA as the ribosome resumes translation Jie Fu, Yaser Hashem, Iwona Wower, Jianlin Lei, Hstau Y. Liao, Christian Zwieb, Jacek Wower and Joachim
Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr.
 Molecular Cell, Volume 30 Supplemental Data Coupling of Ribosomal L1 Stalk and trna Dynamics during Translation Elongation Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr. Supplemental
Molecular Cell, Volume 30 Supplemental Data Coupling of Ribosomal L1 Stalk and trna Dynamics during Translation Elongation Jingyi Fei, Pallav Kosuri, Daniel D. MacDougall, and Ruben L. Gonzalez, Jr. Supplemental
REVIEW SESSION. Wednesday, September 15 5:30 PM SHANTZ 242 E
 REVIEW SESSION Wednesday, September 15 5:30 PM SHANTZ 242 E Gene Regulation Gene Regulation Gene expression can be turned on, turned off, turned up or turned down! For example, as test time approaches,
REVIEW SESSION Wednesday, September 15 5:30 PM SHANTZ 242 E Gene Regulation Gene Regulation Gene expression can be turned on, turned off, turned up or turned down! For example, as test time approaches,
What can electron microscopy tell us about chaperoned protein folding? Helen R Saibil
 Review R45 What can electron microscopy tell us about chaperoned protein folding? Helen R Saibil Chaperonins form large, cage-like structures that act as protein folding machines. Recent developments in
Review R45 What can electron microscopy tell us about chaperoned protein folding? Helen R Saibil Chaperonins form large, cage-like structures that act as protein folding machines. Recent developments in
Studying conformational dynamics and molecular recognition using integrated structural biology in solution Michael Sattler
 Studying conformational dynamics and molecular recognition using integrated structural biology in solution Michael Sattler http://www.nmr.ch.tum.de http://www.helmholtz-muenchen.de/stb/ Outline Dynamics
Studying conformational dynamics and molecular recognition using integrated structural biology in solution Michael Sattler http://www.nmr.ch.tum.de http://www.helmholtz-muenchen.de/stb/ Outline Dynamics
SUPPLEMENTARY INFORMATION
 Fig. 1 Influences of crystal lattice contacts on Pol η structures. a. The dominant lattice contact between two hpol η molecules (silver and gold) in the type 1 crystals. b. A close-up view of the hydrophobic
Fig. 1 Influences of crystal lattice contacts on Pol η structures. a. The dominant lattice contact between two hpol η molecules (silver and gold) in the type 1 crystals. b. A close-up view of the hydrophobic
Biological Sciences 11 Spring Experiment 4. Protein crosslinking
 Biological Sciences 11 Spring 2000 Experiment 4. Protein crosslinking = C - CH 2 - CH 2 - CH 2 - C = H H GA Cl - H 2 N N H 2 Cl - C - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - C DMS CH 3 CH 3 N - - C -
Biological Sciences 11 Spring 2000 Experiment 4. Protein crosslinking = C - CH 2 - CH 2 - CH 2 - C = H H GA Cl - H 2 N N H 2 Cl - C - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - C DMS CH 3 CH 3 N - - C -
Motifs and Logos. Six Introduction to Bioinformatics. Importance and Abundance of Motifs. Getting the CDS. From DNA to Protein 6.1.
 Motifs and Logos Six Discovering Genomics, Proteomics, and Bioinformatics by A. Malcolm Campbell and Laurie J. Heyer Chapter 2 Genome Sequence Acquisition and Analysis Sami Khuri Department of Computer
Motifs and Logos Six Discovering Genomics, Proteomics, and Bioinformatics by A. Malcolm Campbell and Laurie J. Heyer Chapter 2 Genome Sequence Acquisition and Analysis Sami Khuri Department of Computer
Protein Interaction Mapping: Use of Osprey to map Survival of Motor Neuron Protein interactions
 Protein Interaction Mapping: Use of Osprey to map Survival of Motor Neuron Protein interactions Presented by: Meg Barnhart Computational Biosciences Arizona State University The Spinal Muscular Atrophy
Protein Interaction Mapping: Use of Osprey to map Survival of Motor Neuron Protein interactions Presented by: Meg Barnhart Computational Biosciences Arizona State University The Spinal Muscular Atrophy
for Molecular Biology and Neuroscience and Institute of Medical Microbiology, Rikshospitalet-Radiumhospitalet
 SUPPLEMENTARY INFORMATION TO Structural basis for enzymatic excision of N -methyladenine and N 3 -methylcytosine from DNA Ingar Leiros,5, Marivi P. Nabong 2,3,5, Kristin Grøsvik 3, Jeanette Ringvoll 2,
SUPPLEMENTARY INFORMATION TO Structural basis for enzymatic excision of N -methyladenine and N 3 -methylcytosine from DNA Ingar Leiros,5, Marivi P. Nabong 2,3,5, Kristin Grøsvik 3, Jeanette Ringvoll 2,
SUPPLEMENTARY INFORMATION
 In the format provided by the authors and unedited. DOI: 10.1038/NCHEM.2675 Real-time molecular scale observation of crystal formation Roy E. Schreiber, 1 Lothar Houben, 2 Sharon G. Wolf, 2 Gregory Leitus,
In the format provided by the authors and unedited. DOI: 10.1038/NCHEM.2675 Real-time molecular scale observation of crystal formation Roy E. Schreiber, 1 Lothar Houben, 2 Sharon G. Wolf, 2 Gregory Leitus,
Supplemental Information
 1 Supplemental Information Head swivel on the ribosome facilitates translocation via intra-subunit trna hybrid sites Andreas H. Ratje, Justus Loerke, Aleksandra Mikolajka, Matthias Brünner,Peter W. Hildebrand,
1 Supplemental Information Head swivel on the ribosome facilitates translocation via intra-subunit trna hybrid sites Andreas H. Ratje, Justus Loerke, Aleksandra Mikolajka, Matthias Brünner,Peter W. Hildebrand,
Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus:
 m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
SOCS3 binds specific receptor JAK complexes to control cytokine signaling by direct kinase inhibition SUPPLEMENTARY INFORMATION
 SOCS3 binds specific receptor JAK complexes to control cytokine signaling by direct kinase inhibition Nadia J. Kershaw 1,2, James M. Murphy 1,2, Nicholas P.D. Liau 1,2, Leila N. Varghese 1,2, Artem Laktyushin
SOCS3 binds specific receptor JAK complexes to control cytokine signaling by direct kinase inhibition Nadia J. Kershaw 1,2, James M. Murphy 1,2, Nicholas P.D. Liau 1,2, Leila N. Varghese 1,2, Artem Laktyushin
ENCODE DCC Antibody Validation Document
 ENCODE DCC Antibody Validation Document Date of Submission Name: Email: Lab Antibody Name: Target: Company/ Source: Catalog Number, database ID, laboratory Lot Number Antibody Description: Target Description:
ENCODE DCC Antibody Validation Document Date of Submission Name: Email: Lab Antibody Name: Target: Company/ Source: Catalog Number, database ID, laboratory Lot Number Antibody Description: Target Description:
Supplementary Figure 1. Fourier shell correlation curves for sub-tomogram averages and
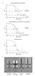 Supplementary Figure 1. Fourier shell correlation curves for sub-tomogram averages and comparisons to other published in situ T3SS structures. a, Resolution estimates after applying Fourier shell correlation
Supplementary Figure 1. Fourier shell correlation curves for sub-tomogram averages and comparisons to other published in situ T3SS structures. a, Resolution estimates after applying Fourier shell correlation
Improving cryo-em maps: focused 2D & 3D classifications and focused refinements
 Improving cryo-em maps: focused 2D & 3D classifications and focused refinements 3rd International Symposium on Cryo-3D Image Analysis 23.3.2018, Tahoe Bruno Klaholz Centre for Integrative Biology, IGBMC,
Improving cryo-em maps: focused 2D & 3D classifications and focused refinements 3rd International Symposium on Cryo-3D Image Analysis 23.3.2018, Tahoe Bruno Klaholz Centre for Integrative Biology, IGBMC,
Laboratory-Based Cryogenic Soft X-ray Tomography and Correlative Microscopy: 3D Visualization Inside the Cell
 Laboratory-Based Cryogenic Soft X-ray Tomography and Correlative Microscopy: 3D Visualization Inside the Cell David Carlson, Jeff Gelb, Vadim Palshin and James Evans Pacific Northwest National Laboratory
Laboratory-Based Cryogenic Soft X-ray Tomography and Correlative Microscopy: 3D Visualization Inside the Cell David Carlson, Jeff Gelb, Vadim Palshin and James Evans Pacific Northwest National Laboratory
Introduction to the Ribosome Overview of protein synthesis on the ribosome Prof. Anders Liljas
 Introduction to the Ribosome Molecular Biophysics Lund University 1 A B C D E F G H I J Genome Protein aa1 aa2 aa3 aa4 aa5 aa6 aa7 aa10 aa9 aa8 aa11 aa12 aa13 a a 14 How is a polypeptide synthesized? 2
Introduction to the Ribosome Molecular Biophysics Lund University 1 A B C D E F G H I J Genome Protein aa1 aa2 aa3 aa4 aa5 aa6 aa7 aa10 aa9 aa8 aa11 aa12 aa13 a a 14 How is a polypeptide synthesized? 2
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Structural characterization of NiV N 0 P in solution and in crystal.
 Supplementary Figure 1 Structural characterization of NiV N 0 P in solution and in crystal. (a) SAXS analysis of the N 32-383 0 -P 50 complex. The Guinier plot for complex concentrations of 0.55, 1.1,
Supplementary Figure 1 Structural characterization of NiV N 0 P in solution and in crystal. (a) SAXS analysis of the N 32-383 0 -P 50 complex. The Guinier plot for complex concentrations of 0.55, 1.1,
Supplementary Information
 Supplementary Information Supplementary Figure 1. X-ray diffraction patterns of (a) pure LDH, (b) AuCl 4 ion-exchanged LDH and (c) the Au/LDH hybrid catalyst. The refined cell parameters for pure, ion-exchanged,
Supplementary Information Supplementary Figure 1. X-ray diffraction patterns of (a) pure LDH, (b) AuCl 4 ion-exchanged LDH and (c) the Au/LDH hybrid catalyst. The refined cell parameters for pure, ion-exchanged,
GENE ACTIVITY Gene structure Transcription Transcript processing mrna transport mrna stability Translation Posttranslational modifications
 1 GENE ACTIVITY Gene structure Transcription Transcript processing mrna transport mrna stability Translation Posttranslational modifications 2 DNA Promoter Gene A Gene B Termination Signal Transcription
1 GENE ACTIVITY Gene structure Transcription Transcript processing mrna transport mrna stability Translation Posttranslational modifications 2 DNA Promoter Gene A Gene B Termination Signal Transcription
Supplementary materials. Crystal structure of the carboxyltransferase domain. of acetyl coenzyme A carboxylase. Department of Biological Sciences
 Supplementary materials Crystal structure of the carboxyltransferase domain of acetyl coenzyme A carboxylase Hailong Zhang, Zhiru Yang, 1 Yang Shen, 1 Liang Tong Department of Biological Sciences Columbia
Supplementary materials Crystal structure of the carboxyltransferase domain of acetyl coenzyme A carboxylase Hailong Zhang, Zhiru Yang, 1 Yang Shen, 1 Liang Tong Department of Biological Sciences Columbia
A recyclable supramolecular membrane for size-selective separation of nanoparticles
 A recyclable supramolecular membrane for size-selective separation of nanoparticles E. Krieg, E. Shirman, H. Weissman, E. Shimoni, S. G. Wolf, I. Pinkas, B. Rybtchinski J. Am. Chem. Soc. 2009, 131, 14365-14373.
A recyclable supramolecular membrane for size-selective separation of nanoparticles E. Krieg, E. Shirman, H. Weissman, E. Shimoni, S. G. Wolf, I. Pinkas, B. Rybtchinski J. Am. Chem. Soc. 2009, 131, 14365-14373.
Protein separation and characterization
 Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: tech@aladdin-e.com Email EU: eutech@aladdin-e.com Email Asia Pacific: cntech@aladdin-e.com Protein separation
Address:800 S Wineville Avenue, Ontario, CA 91761,USA Website:www.aladdin-e.com Email USA: tech@aladdin-e.com Email EU: eutech@aladdin-e.com Email Asia Pacific: cntech@aladdin-e.com Protein separation
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
A Visualized Mathematical Model of Atomic Structure
 A Visualized Mathematical Model of Atomic Structure Dr. Khalid Abdel Fattah Faculty of Engineering, Karary University, Khartoum, Sudan. E-mail: khdfattah@gmail.com Abstract: A model of atomic structure
A Visualized Mathematical Model of Atomic Structure Dr. Khalid Abdel Fattah Faculty of Engineering, Karary University, Khartoum, Sudan. E-mail: khdfattah@gmail.com Abstract: A model of atomic structure
Measurement of ice thickness on vitreous ice embedded cryo-em grids: investigation of optimizing condition for visualizing macromolecules
 Cho et al. Journal of Analytical Science and Technology 2013, 4:7 TECHNICAL NOTE Measurement of ice thickness on vitreous ice embedded cryo-em grids: investigation of optimizing condition for visualizing
Cho et al. Journal of Analytical Science and Technology 2013, 4:7 TECHNICAL NOTE Measurement of ice thickness on vitreous ice embedded cryo-em grids: investigation of optimizing condition for visualizing
Supplementary Information. Cryo-EM Studies of Drp1 Reveal Cardiolipin Interactions that Activate
 Supplementary Information Cryo-EM Studies of Drp1 Reveal Cardiolipin Interactions that Activate the Helical Oligomer Christopher A. Francy 1, 2, 3, Ryan W. Clinton 1, 2, 3, Chris Fröhlich 4, 5, Colleen
Supplementary Information Cryo-EM Studies of Drp1 Reveal Cardiolipin Interactions that Activate the Helical Oligomer Christopher A. Francy 1, 2, 3, Ryan W. Clinton 1, 2, 3, Chris Fröhlich 4, 5, Colleen
Protein Structures: Experiments and Modeling. Patrice Koehl
 Protein Structures: Experiments and Modeling Patrice Koehl Structural Bioinformatics: Proteins Proteins: Sources of Structure Information Proteins: Homology Modeling Proteins: Ab initio prediction Proteins:
Protein Structures: Experiments and Modeling Patrice Koehl Structural Bioinformatics: Proteins Proteins: Sources of Structure Information Proteins: Homology Modeling Proteins: Ab initio prediction Proteins:
Name: SBI 4U. Gene Expression Quiz. Overall Expectation:
 Gene Expression Quiz Overall Expectation: - Demonstrate an understanding of concepts related to molecular genetics, and how genetic modification is applied in industry and agriculture Specific Expectation(s):
Gene Expression Quiz Overall Expectation: - Demonstrate an understanding of concepts related to molecular genetics, and how genetic modification is applied in industry and agriculture Specific Expectation(s):
BCMP 201 Protein biochemistry
 BCMP 201 Protein biochemistry BCMP 201 Protein biochemistry with emphasis on the interrelated roles of protein structure, catalytic activity, and macromolecular interactions in biological processes. The
BCMP 201 Protein biochemistry BCMP 201 Protein biochemistry with emphasis on the interrelated roles of protein structure, catalytic activity, and macromolecular interactions in biological processes. The
Structural and functional aspects of gastric proton pump, H +,K + -ATPase
 Kazuhiro ABE, Ph. D. E-mail:kabe@cespi.nagoya-u.ac.jp Structural and functional aspects of gastric proton pump, H +,K + -ATPase In response to food intake, ph of our stomach reaches around 1. This highly
Kazuhiro ABE, Ph. D. E-mail:kabe@cespi.nagoya-u.ac.jp Structural and functional aspects of gastric proton pump, H +,K + -ATPase In response to food intake, ph of our stomach reaches around 1. This highly
Measuring quaternary structure similarity using global versus local measures.
 Supplementary Figure 1 Measuring quaternary structure similarity using global versus local measures. (a) Structural similarity of two protein complexes can be inferred from a global superposition, which
Supplementary Figure 1 Measuring quaternary structure similarity using global versus local measures. (a) Structural similarity of two protein complexes can be inferred from a global superposition, which
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
Supporting Information
 Copyright WILEY-VCH Verlag GmbH & Co. KGaA, 69469 Weinheim, Germany, 2013. Supporting Information for Adv. Mater., DOI: 10.1002/adma.201301472 Reconfigurable Infrared Camouflage Coatings from a Cephalopod
Copyright WILEY-VCH Verlag GmbH & Co. KGaA, 69469 Weinheim, Germany, 2013. Supporting Information for Adv. Mater., DOI: 10.1002/adma.201301472 Reconfigurable Infrared Camouflage Coatings from a Cephalopod
Translation Part 2 of Protein Synthesis
 Translation Part 2 of Protein Synthesis IN: How is transcription like making a jello mold? (be specific) What process does this diagram represent? A. Mutation B. Replication C.Transcription D.Translation
Translation Part 2 of Protein Synthesis IN: How is transcription like making a jello mold? (be specific) What process does this diagram represent? A. Mutation B. Replication C.Transcription D.Translation
Supporting Protocol This protocol describes the construction and the force-field parameters of the non-standard residue for the Ag + -site using CNS
 Supporting Protocol This protocol describes the construction and the force-field parameters of the non-standard residue for the Ag + -site using CNS CNS input file generatemetal.inp: remarks file generate/generatemetal.inp
Supporting Protocol This protocol describes the construction and the force-field parameters of the non-standard residue for the Ag + -site using CNS CNS input file generatemetal.inp: remarks file generate/generatemetal.inp
Supplementary Information. The protease GtgE from Salmonella exclusively targets. inactive Rab GTPases
 Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
7.06 Problem Set
 7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
Supplementary Figure 1
 Supplementary Figure 1 The correlation of n-score cutoff and FDR in both CID-only and CID-ETD fragmentation strategies. A bar diagram of different n-score thresholds applied in the search, plotted against
Supplementary Figure 1 The correlation of n-score cutoff and FDR in both CID-only and CID-ETD fragmentation strategies. A bar diagram of different n-score thresholds applied in the search, plotted against
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11539 Supplementary Figure 1 Schematic representation of plant (A) and mammalian (B) P 2B -ATPase domain organization. Actuator (A-), nucleotide binding (N-),
SUPPLEMENTARY INFORMATION doi:10.1038/nature11539 Supplementary Figure 1 Schematic representation of plant (A) and mammalian (B) P 2B -ATPase domain organization. Actuator (A-), nucleotide binding (N-),
