Modeling of snrnp Motion in the Nucleoplasm
|
|
|
- Arthur Hodge
- 6 years ago
- Views:
Transcription
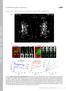
1 Excerpt from the Proceedings of the COMSOL Conference 008 Hannover Modeling of snrnp Motion in the Nucleoplasm Michaela Blažíková 1, Jan Malínský *,, David Staněk 3 and Petr Heřman 1 1 Charles University Prague, Faculty of Mathematics and Physics, Prague, Czech Republic, Institute of Experimental Medicine ASCR, v.v.i., Vídeňská 1083, 14 0 Prague 4, Czech Republic, 3 Institute of Molecular Genetics ASCR, v.v.i., Vídeňská 1083,14 0 Prague 4, Czech Republic *Corresponding author: malinsky@biomed.cas.cz Abstract: Small nuclear ribonucleoprotein particles (snrnps) are essential supramolecular complexes involved in pre-mrna splicing, the process of post-transcriptional RNA modifications. The particles undergo complex assembly steps inside the cell nucleus in a highly dynamic compartment called the Cajal body. We have previously shown that the free diffusion model does not fully describe the snrnp motion when tested for 1D fundamental solution of the diffusion equation. Numerical solutions of the diffusion equation were used instead to fit the experimental data obtained by fluorescence microscopy of a single cell in order to test the influence of geometry, dimensionality and boundary conditions. Finally, optimal characteristics of the model were determined. Keywords: snrnp, diffusion 1. Introduction Cajal bodies (CBs) are nuclear domains found in metabolically active cells [1,]. Somatic cells contain usually -4 round-shaped CBs µm in diameter [3]. The molecular factors found in CBs are involved in the pre-mrna splicing, the histone pre-mrna maturation, and in the pre-rrna processing [4,5,6]. The pre-mrna splicing is a two-step reaction catalyzed by spliceosomes. Spliceosomes are large complexes of hundreds of proteins containing small nuclear ribonucleoprotein particles (snrnps), namely U1, U, U4, U5 and U6 [7]. All spliceosomal snrnps except the U6 contain Sm proteins that make up an extremely stable Sm core of the snrnp, while the U6 snrnp interacts with similar Like-Sm (LSm) proteins [6]. Both the Sm and the LSm proteins assemble into snrnps during maturation and remain stably associated there. Cajal bodies were shown to be sites of the snrnps assembly [8,9,10]. The snrnps form an active spliceosome that enables the pre-mrna splicing to proceed. Spliceosomes are then inactivated and disassemble for further rounds of splicing. It has been recently shown that the snrnps reassemble in CBs after splicing [11]. Currently no model that would describe dynamics of the snrnps in vivo is available. A lack of metabolic energy necessary for an active movement of intra-nuclear proteins and RNA was reported in many studies [e.g. 1]. This strongly suggests that these molecules and complexes undergo a passive diffusive movement in the nucleus. Considering this knowledge, we have previously tested whether the simplest model based on a free diffusion of snrnps could describe movement of the particles in the nucleus [13]. An extended set of diffusion data describing spreading of the photoactivated protein complex was acquired at different locations in the nucleoplasm of 50 HeLa cell nuclei. A global fitting of the data to the fundamental solution of the diffusion equation in 1D indicated that the spreading of the complex has a diffusion character. However, the free diffusion model with a global and spatially invariant diffusion coefficient D was inconsistent with the data and could not explain the dynamics of the snrnp particles in the nucleoplasm. Individual analysis of the diffusion curves revealed that the D value increases with increasing radial distance from the photoactivated CB. In the first approximation D(x) depends linearly on the distance x, D( x) D0 D x, where D 0 = ( ) m /s and D = ( ) 10-8 m/s [13]. In order to eliminate a possible variability of environment in different cell nuclei, in this study we performed similar measurements and analyses on a single nucleus. We also examined influence of dimensionality of the solved problem on the obtained results. D was therefore
2 evaluated using 1-, - and 3-dimensional model of the nucleus. Finally, accuracy and applicability of the models was tested and discussed.. Methods.1 Live cell imaging HeLa cells were co-transfected with two fluorescently labeled proteins. The first one was SART3 tagged with the cyan fluorescent protein (SART3:CFP). SART3 is known to accumulate in Cajal bodies and was therefore used as a CBs marker to visualize and localize them in the nucleus. The second protein was SmB, one of the Sm proteins taking part in the snrnp complex formation. This protein carried a photoactivatable green fluorescent protein tag (PA-GFP) that can be converted from nonfluorescent to fluorescent form by ~400 nm laser light illumination [14]. It has been recently reported that SmB:PA-GFP proteins remain stably associated with the snrnp complex after its formation [11]. Spreading and distribution of the complex can be therefore followed by fluorescence after the local photoactivation of SmB:PA-GFP. The cells were imaged by the Zeiss LSM 510 confocal microscope equipped with a 63x/1. NA water immersion objective. Three images were acquired before the photoactivation. Then the SmB:PA-GFP localized in one selected CB was photoactivated by a short laser pulse at 405 nm and time series of fluorescence images was acquired for 5 minutes with 15s increments for each of the selected nuclei. For the fluorescence imaging the SmB:PA-GFP and SART3:CFP complexes were excited at 488 nm and 458 nm, respectively, and their emission was collected into two channels through the long-pass LP505 and the band-pass BP filters, respectively.. Image processing Images were analyzed using the Matlab software. Images from the cyan channel (SART3:CFP) were used to identify locations of Cajal bodies before the photoactivation. Fluorescence intensity of the photoactivated complexes (SmB:PA-GFP) was evaluated from the green channel images. After the photoactivation, tracking by means of the thresholding was used to identify and follow a slight motion of the photoactivated CB. Time course of the fluorescence intensity of SmB:PA-GFP was evaluated in the nucleoplasm at properly chosen distances from the photoactivated CB. 3. Numerical Model 3.1 Analytical fitting The method of analytical fitting was reported in detail in [13]. Briefly, the motion of snrnps was described by a diffusion equation with a spatially invariant diffusion coefficient D: c( x, t) t D c( x, t) x f, (1) where x is a space coordinate, t denotes time, and c is a concentration of diffusing particles. The source term f was set to zero. We used an initial condition c ( x,0) ( x xcb) to simulate the photoactivation at x CB, i.e. in the location of the selected Cajal body. The fundamental solution F of Eq.(1) can be written as [15]: ( x xcb ) k 4Dt F x xcb, t) e (, () Dt where k is a multiplicative parameter describing fluorescence brightness of SmB:PA-GFP in the cell. Eq. () was used for a non-linear least squares (NLS) fitting [16] of the experimental data. The fitting was performed in the Matlab software (function nlinfit). As a result we obtained optimized parameters D and k for each dataset. 3. Numerical fitting Solutions from the Diffusion Application Module (Comsol, Eq.1) were transferred to the Matlab and used for the NLS fitting of the measured intensity evolutions. In order to evaluate influence of dimensionality on the fitting results, we performed the analysis sequentially in 1D, D and 3D. Specific geometry was added into the model. In c( x,0) c particular, initial conditions 0 for
3 x CB and c(x,0) 0 elsewhere simulated the photoactivation in the CB area only. Neumann boundary conditions n. N 0, N D c were used to define impermeability of the nuclear membrane for the diffusing particles. The fitting performed in the Matlab software yielded optimized parameters D and c 0 as functions of distance from the photoactivation area. 4. Experimental Results Cajal bodies serve as spliceosomal snrnp assembly sites; the concentration of the snrnps in CBs is substantially higher than in the nucleoplasm [3]. A population of the snrnps confined to one selected Cajal body (CB) was photoactivated by a laser pulse. Movement of the photoactivated protein fraction was followed by the time lapse imaging. Arrangement of the experiment is shown in Fig. 1. fundamental solution of the diffusion equation in 1D (see methods). For the single cell we found a similar linear dependence of D(x) on the radial distance x from the photoactivated CB (Fig.3, analytical 1D fit) as when D was evaluated in different cell nuclei [13]. In this case the linear regression coefficients were D 0 = (0 1.5) m /s and D = ( ) 10-8 m/s. The dependence of D on the distance from the photoactivated CB was tested on other 10 cell nuclei with similar results (data not shown). In order to make modeling more realistic we applied a numerical solution of the diffusion model. On the contrary of the analytical solution, the numerical approach allows to include specific cell geometry and to add initial and boundary conditions to the model (see Methods). Geometries of the 1D and D models best corresponding to the real cell shape measured from Fig. 1 are schematically depicted in Fig.. Figure 1. Arrangement of the photoactivation experiment. Diffusion of the snrnp complexes inside the nucleoplasm delimited by the nuclear envelope (NE, green) was visualized by the SmB:PA-GFP fluorescent fusion protein. PA-GFP was photoactivated within the circular area (red circle) corresponding to one of the Cajal bodies (CB). Spreading of the photoactivated complexes (red arrow) was measured in the nucleoplasmic area (yellow circle). Diffusion coefficient was evaluated in one cell nucleus at different distances from the CB by analytical fitting of the data to the Figure. Model used for numerical 1D and D fitting. A) In 1D the cell nucleus was represented by a line segment of 1 µm length. Center of the CB (a line segment of 0.5 µm length) was located inside the nucleus, 8. µm from the lower NE end. The measurement points were located along the red line with 0.96 µm distance from each other. B) In the D model, the nucleus was represented by the ellipse (semi-axes of 10.5 µm and 5 µm) and the CB was modeled as a circle (with 0.5 µm radius) located on its major axis at 8. µm distance from the lower nuclear pole. The measurement points were located along the major axis with 0.96 µm distance from each other. The 3D model was created from the D one by formation of an ellipsoid with the third semi-
4 axis of.5 µm. This dimension could not be measured and it was taken from the literature according to an average size of HeLa cells [3]. Positions of the CB and the measurement points remained the same as for the D model. Results of the NLS fitting for 1-, - and 3-dimensional models are shown in Fig. 3. Figure 3. Evaluation of the diffusion coefficient in different distances from the photoactivated CB. Comparison of analytical fit in 1D with numerical fits in 1D, D and 3D. For all the numerical models the results are qualitatively similar. As expected, for longer diffusion distances they all differ from the analytical 1D fit: instead of the continuous growth, the numerical solutions exhibit a sigmoidal shape of the D(x) curve with an inflection point at approx. 7 µm. This is a consequence of the added geometry and boundary conditions to the model. Fits of an example data are shown in Fig.4. Figure 4. Example of experimental data measured in the distance of.8 μm from the photoactivated Cajal body (open circles) and numerical fits in 1D, D and 3D. From our simulations we can conclude that the model dimensionality does not significantly affect qualitative results obtained on the improved numerical model with the specific geometry and boundary conditions. Because the 1D model does not allow to properly describe and change the cell footprint, we decided to use the D model for further qualitative evaluation of model properties. In order to evaluate how accurately can be an irregular shape of the cell nucleus represented by the chosen ellipsoid, we tested a sensitivity of the recovered diffusion coefficients to changes in the model geometry, Fig. 5A. In particular, the minor axis of the ellipsoid was both elongated (I) and shortened (II). The major axis was elongated (III). Finally, we kept the original nuclear size unchanged and the CB position was moved closer to the nuclear membrane (IV). Results are summarized in Fig. 5. The elongation of the minor-axis induced a shift of the sigmoidal D(x) curve transition to a longer distance while shortening caused an opposite effect, Fig. 5B. The plateau of the curves remained in both cases unchanged. Diffusion coefficients at short distances were found to be slightly lower and higher for shorter and longer minor-axis, respectively. The elongation of the major-axis induced slower growth of D(x) and a dramatic shift of the transition point to even longer distances, Fig. 5C. The plateau of D(x) was not reached and it was probably shifted out of the analyzed area. The relocation of the CB with the linked measurement positions closer to the nuclear envelope caused a slight increase of the diffusion coefficient at short distances (similar to the case II). Moreover, a slower growth of the curve and a plateau elevation, similar to the case III, was observed, Fig. 5C. These results reveal a strong dependency of the calculated diffusion coefficient on the position of the CB within the nucleus. In summary, model geometry as well as positions of the measurement points in the nucleoplasm was found to have a considerable effect on the fitted values of the apparent diffusion coefficient. For quantitative evaluation of D within a cell nucleus it is therefore imperative to use a detailed shape of the particular nucleus and to accurately model position of the photoactivated Cajal body within the nucleus.
5 Figure 5. Effect of geometry changes on the apparent diffusion coefficient in the D model. A) Scheme of the geometry changes. Elongation of the minor axis from 10 m to 1 m (I) and shortening the axis from 10 m to 5 m (II). Elongation of the major axis from 1 m to 40 m (III) and change of the CB position within the nucleus (IV). The red line indicates the analysis area. B) Effect of the minor axis change (configurations I and II). C) Effect of the major axis elongation (III) and the CB position change (IV). 5. Discussion Consistently with previous results obtained on a set of cells [Blazikova et al., 008] the current diffusion analysis restricted to data from a single cell nucleus indicate that the value of the apparent diffusion coefficient increases with increasing radial distance from the photoactivated Cajal body. This qualitative observation was found to be independent of major model-geometry changes. Mechanisms underlying the observed spatial dependence of the diffusion coefficient remain unknown at present. We propose that the effect could result from interactions of the diffusing particles with other large macromolecular complexes in the nucleus. Such interactions should modulate the value of the apparent diffusion coefficient obtained at different locations in the nucleus. Construction of a model that includes such interactions is in progress. 6. Conclusions Our study of the snrnp dynamics within the single cell nucleus shows that the snrnp motion cannot be explained in the terms of free diffusion. The apparent diffusion coefficient of SmB:PA-GFP was found to show a significant dependence on the distance from the photoactivated Cajal body. Essentially the same result was obtained for 1D, D and 3D models. Dependency of the apparent diffusion coefficient on the model geometry revealed that a detailed knowledge of the nuclear shape is crucial for the accurate modeling. Our calculations indicate that data from different cell nuclei cannot be analyzed together by a common model based on an average nucleus geometry. Instead, each
6 nucleus has to be modeled and analyzed separately. 7. References 1. Boudonck, K., et al., Coiled body numbers in the Arabidopsis root epidermis are regulated by cell type, developmental stage and cell cycle parameters, Journal of Cell Science, 111, (1998). Pena, E., et al., Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons, The Journal of Comparative Neurology, 430, (001) 3. Klingauf, M., et al., Enhancement of U4/U6 Small Nuclear Ribonucleoprotein Particle Association in Cajal Bodies predicted by Mathematical Modeling, Molecular Biology of the Cell, 17, (006) 4. Gall, J. G., Cajal bodies: the first 100 years, Annual Review of Cell and Developmental Biology, 16, (000) 5. Matera, A. G. and K. B. Shpargel, Pumping RNA: nuclear bodybuilding along the RNP pipeline, Current opinion in cell biology, 18, (006) 6. Stanek, D. and Neugebauer K. M., The Cajal body: a meeting place for spliceosomal snrnps in the nuclear maze, Chromosoma, 115, (006) 7. Jurica, M. S. and M. J. Moore, Pre m-rna splicing: awash in sea of proteins, Molecular Cell, 1, 5-14 (003) 8. Schaffert, N., et al., RNAi knockdown of hprp31 leads to an accumulation of U4/U6 disnrnps in Cajal bodies, EMBO Journal, 3, (004) 9. Stanek, D. and Neugebauer K. M., Detection of snrnp assembly intermediates in Cajal bodies by fluorescence resonance energy transfer, The Journal of Cell Biology, 166, (004) 10. Nesic, D., et al., A role for Cajal bodies in the final steps of U snrnp biogenesis, Journal of Cell Science, 117, (004) 11. Stanek, D., et al., Spliceosomal snrnps Repeatedly Cycle through Cajal Bodies, Molecular Biology of the Cell, 19(6), (008) 1. Dundr, M., et al., In vivo kinetics of Cajal body components, The Journal of Cell Biology, 164, (004) 13. Blazikova M., et al., Diffusion Modeling of snrnp Dynamics, WDS'08 Proceedings of Contributed Papers: Part III - Physics, in press (008) 14. Patterson, G. H. and J. Lippincott-Schwarz, A Photoactivable GFP for Selective Photobleaching of Proteins and Cells, Science, 97, (00) 15. Itô S., Diffusion equations, American Mathematical Society (199) 16. Johnson, M. L., Use of Least-Squares Techniques in Biochemistry, Methods in Enzymology, 40, 1-36 (1994) 8. Acknowledgements This work was supported by the Grant Agency of the Czech Republic (04/07/0133), by the Grant Agency of Charles University (79508, to M.B.) and by the Ministry of Education, Youth and Sports of the Czech Republic (MSM , to P.H.).
Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes
 9 The Nucleus Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes Explain general structures of Nuclear Envelope, Nuclear Lamina, Nuclear Pore Complex Explain movement of proteins
9 The Nucleus Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes Explain general structures of Nuclear Envelope, Nuclear Lamina, Nuclear Pore Complex Explain movement of proteins
Nucleus. The nucleus is a membrane bound organelle that store, protect and express most of the genetic information(dna) found in the cell.
 Nucleus The nucleus is a membrane bound organelle that store, protect and express most of the genetic information(dna) found in the cell. Since regulation of gene expression takes place in the nucleus,
Nucleus The nucleus is a membrane bound organelle that store, protect and express most of the genetic information(dna) found in the cell. Since regulation of gene expression takes place in the nucleus,
GCD3033:Cell Biology. Transcription
 Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Transcription Transcription: DNA to RNA A) production of complementary strand of DNA B) RNA types C) transcription start/stop signals D) Initiation of eukaryotic gene expression E) transcription factors
Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus:
 m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
1. In most cases, genes code for and it is that
 Name Chapter 10 Reading Guide From DNA to Protein: Gene Expression Concept 10.1 Genetics Shows That Genes Code for Proteins 1. In most cases, genes code for and it is that determine. 2. Describe what Garrod
Name Chapter 10 Reading Guide From DNA to Protein: Gene Expression Concept 10.1 Genetics Shows That Genes Code for Proteins 1. In most cases, genes code for and it is that determine. 2. Describe what Garrod
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Analysis of Active Transport by Fluorescence Recovery after Photobleaching. MV Ciocanel, JA Kreiling, JA Gagnon, KL Mowry, B Sandstede
 Analysis of Active Transport by Fluorescence Recovery after Photobleaching MV Ciocanel, JA Kreiling, JA Gagnon, KL Mowry, B Sandstede Analysis of Active Transport by Fluorescence Recovery after Photobleaching
Analysis of Active Transport by Fluorescence Recovery after Photobleaching MV Ciocanel, JA Kreiling, JA Gagnon, KL Mowry, B Sandstede Analysis of Active Transport by Fluorescence Recovery after Photobleaching
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Eisner et al., http://www.jcb.org/cgi/content/full/jcb.201312066/dc1 Figure S1. Mitochondrial continuity in adult skeletal muscle. (A)
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Eisner et al., http://www.jcb.org/cgi/content/full/jcb.201312066/dc1 Figure S1. Mitochondrial continuity in adult skeletal muscle. (A)
Name: SBI 4U. Gene Expression Quiz. Overall Expectation:
 Gene Expression Quiz Overall Expectation: - Demonstrate an understanding of concepts related to molecular genetics, and how genetic modification is applied in industry and agriculture Specific Expectation(s):
Gene Expression Quiz Overall Expectation: - Demonstrate an understanding of concepts related to molecular genetics, and how genetic modification is applied in industry and agriculture Specific Expectation(s):
Suppression of Static Magnetic Field in Diffusion Measurements of Heterogeneous Materials
 PIERS ONLINE, VOL. 5, NO. 1, 2009 81 Suppression of Static Magnetic Field in Diffusion Measurements of Heterogeneous Materials Eva Gescheidtova 1 and Karel Bartusek 2 1 Faculty of Electrical Engineering
PIERS ONLINE, VOL. 5, NO. 1, 2009 81 Suppression of Static Magnetic Field in Diffusion Measurements of Heterogeneous Materials Eva Gescheidtova 1 and Karel Bartusek 2 1 Faculty of Electrical Engineering
Cell Division. OpenStax College. 1 Genomic DNA
 OpenStax-CNX module: m44459 1 Cell Division OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you will be
OpenStax-CNX module: m44459 1 Cell Division OpenStax College This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution License 3.0 By the end of this section, you will be
Modelling Biochemical Reaction Networks. Lecture 1: Overview of cell biology
 Modelling Biochemical Reaction Networks Lecture 1: Overview of cell biology Marc R. Roussel Department of Chemistry and Biochemistry Types of cells Prokaryotes: Cells without nuclei ( bacteria ) Very little
Modelling Biochemical Reaction Networks Lecture 1: Overview of cell biology Marc R. Roussel Department of Chemistry and Biochemistry Types of cells Prokaryotes: Cells without nuclei ( bacteria ) Very little
exotic anthelmintics have negatives charges (e.g. quisqualate and Kainite); however, these drugs are not used and need not be taken into account.
 Introduction This supplementary file has been created to present a study on the isolation condition of the drug within the drug well. All experiments were completed using the same assay setup described
Introduction This supplementary file has been created to present a study on the isolation condition of the drug within the drug well. All experiments were completed using the same assay setup described
Supplemental Information: Govindaraghavan, Lad and Osmani
 Supplemental Information: Govindaraghavan, Lad and Osmani Table S1: List of strains used in this study Strain Name Genotype (all strains also carry vea1) Source MG190 nima7; pyrg89; argb2; ndc80-cr::pyroa
Supplemental Information: Govindaraghavan, Lad and Osmani Table S1: List of strains used in this study Strain Name Genotype (all strains also carry vea1) Source MG190 nima7; pyrg89; argb2; ndc80-cr::pyroa
Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering
 Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering Lu Wei, Fanghao Hu, Yihui Shen, Zhixing Chen, Yong Yu, Chih-Chun Lin, Meng C. Wang, Wei Min * * To whom correspondence
Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering Lu Wei, Fanghao Hu, Yihui Shen, Zhixing Chen, Yong Yu, Chih-Chun Lin, Meng C. Wang, Wei Min * * To whom correspondence
Cover Page. The handle holds various files of this Leiden University dissertation
 Cover Page The handle http://hdl.handle.net/1887/41480 holds various files of this Leiden University dissertation Author: Tleis, Mohamed Title: Image analysis for gene expression based phenotype characterization
Cover Page The handle http://hdl.handle.net/1887/41480 holds various files of this Leiden University dissertation Author: Tleis, Mohamed Title: Image analysis for gene expression based phenotype characterization
Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry
 Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry Petr Pracna J. Heyrovský Institute of Physical Chemistry Academy of Sciences of the Czech Republic, Prague ZiF Cooperation
Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry Petr Pracna J. Heyrovský Institute of Physical Chemistry Academy of Sciences of the Czech Republic, Prague ZiF Cooperation
Spliceosome and Localization of Its Catalytic Core
 Molecular Cell, Volume 40 Supplemental Information 3D Cryo-EM Structure of an Active Step I Spliceosome and Localization of Its Catalytic Core Monika M. Golas, Bjoern Sander, Sergey Bessonov, Michael Grote,
Molecular Cell, Volume 40 Supplemental Information 3D Cryo-EM Structure of an Active Step I Spliceosome and Localization of Its Catalytic Core Monika M. Golas, Bjoern Sander, Sergey Bessonov, Michael Grote,
LIST of SUPPLEMENTARY MATERIALS
 LIST of SUPPLEMENTARY MATERIALS Mir et al., Dense Bicoid Hubs Accentuate Binding along the Morphogen Gradient Supplemental Movie S1 (Related to Figure 1). Movies corresponding to the still frames shown
LIST of SUPPLEMENTARY MATERIALS Mir et al., Dense Bicoid Hubs Accentuate Binding along the Morphogen Gradient Supplemental Movie S1 (Related to Figure 1). Movies corresponding to the still frames shown
Nature Protocols: doi: /nprot Supplementary Figure 1
 Supplementary Figure 1 Photographs of the 3D-MTC device and the confocal fluorescence microscopy. I: The system consists of a Leica SP8-Confocal microscope (with an option of STED), a confocal PC, a 3D-MTC
Supplementary Figure 1 Photographs of the 3D-MTC device and the confocal fluorescence microscopy. I: The system consists of a Leica SP8-Confocal microscope (with an option of STED), a confocal PC, a 3D-MTC
GENES AND CHROMOSOMES III. Lecture 5. Biology Department Concordia University. Dr. S. Azam BIOL 266/
 GENES AND CHROMOSOMES III Lecture 5 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION OPERONS Introduction All cells in a multi-cellular
GENES AND CHROMOSOMES III Lecture 5 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION OPERONS Introduction All cells in a multi-cellular
natural development from this collection of knowledge: it is more reliable to predict the property
 1 Chapter 1 Introduction As the basis of all life phenomena, the interaction of biomolecules has been under the scrutiny of scientists and cataloged meticulously [2]. The recent advent of systems biology
1 Chapter 1 Introduction As the basis of all life phenomena, the interaction of biomolecules has been under the scrutiny of scientists and cataloged meticulously [2]. The recent advent of systems biology
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
CHAPTER 3. Cell Structure and Genetic Control. Chapter 3 Outline
 CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
An intracellular phase transition drives nucleolar assembly and size control
 An intracellular phase transition drives nucleolar assembly and size control Steph Weber Department of Biology McGill University Membranes spatially organize eukaryotic cells Cells also contain membraneless
An intracellular phase transition drives nucleolar assembly and size control Steph Weber Department of Biology McGill University Membranes spatially organize eukaryotic cells Cells also contain membraneless
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Note. File Name: Peer Review File Description:
 File Name: Supplementary Information Description: Supplementary Figures and Supplementary Note File Name: Peer Revie File Description: Supplementary Fig.1. Complete set of ACFs extracted by FLCS analysis.
File Name: Supplementary Information Description: Supplementary Figures and Supplementary Note File Name: Peer Revie File Description: Supplementary Fig.1. Complete set of ACFs extracted by FLCS analysis.
The Discovery of the Cell
 The Discovery of the Cell The Discovery of the Cell Because there were no instruments to make cells visible, the existence of cells was unknown for most of human history. This changed with the invention
The Discovery of the Cell The Discovery of the Cell Because there were no instruments to make cells visible, the existence of cells was unknown for most of human history. This changed with the invention
nucleus: DNA & chromosomes
 nucleus: DNA & chromosomes chapter 5 nuclear organization nuclear structure nuclear envelope nucleoplasm nuclear matrix nucleolus nuclear envelope nucleolus nuclear matrix nucleoplasm nuclear pore nuclear
nucleus: DNA & chromosomes chapter 5 nuclear organization nuclear structure nuclear envelope nucleoplasm nuclear matrix nucleolus nuclear envelope nucleolus nuclear matrix nucleoplasm nuclear pore nuclear
AP Biology. Biology is the only subject in which multiplication is the same thing as division. The Cell Cycle: Cell Growth, Cell Division
 QuickTime and and a TIFF TIFF (Uncompressed) decompressor are are needed needed to to see see this this picture. picture. Biology is the only subject in which multiplication is the same thing as division
QuickTime and and a TIFF TIFF (Uncompressed) decompressor are are needed needed to to see see this this picture. picture. Biology is the only subject in which multiplication is the same thing as division
4.3 Intracellular calcium dynamics
 coupled equations: dv dv 2 dv 3 dv 4 = i + I e A + g,2 (V 2 V ) = i 2 + g 2,3 (V 3 V 2 )+g 2, (V V 2 )+g 2,4 (V 4 V 2 ) = i 3 + g 3,2 (V 2 V 3 ) = i 4 + g 4,2 (V 2 V 4 ) This can be written in matrix/vector
coupled equations: dv dv 2 dv 3 dv 4 = i + I e A + g,2 (V 2 V ) = i 2 + g 2,3 (V 3 V 2 )+g 2, (V V 2 )+g 2,4 (V 4 V 2 ) = i 3 + g 3,2 (V 2 V 3 ) = i 4 + g 4,2 (V 2 V 4 ) This can be written in matrix/vector
The Cell. C h a p t e r. PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas
 C h a p t e r 2 The Cell PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas Copyright 2009 Pearson Education, Inc., publishing as Pearson Benjamin Cummings Introduction
C h a p t e r 2 The Cell PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas Copyright 2009 Pearson Education, Inc., publishing as Pearson Benjamin Cummings Introduction
BMB Class 17, November 30, Single Molecule Biophysics (II)
 BMB 178 2018 Class 17, November 30, 2018 15. Single Molecule Biophysics (II) New Advances in Single Molecule Techniques Atomic Force Microscopy Single Molecule Manipulation - optical traps and tweezers
BMB 178 2018 Class 17, November 30, 2018 15. Single Molecule Biophysics (II) New Advances in Single Molecule Techniques Atomic Force Microscopy Single Molecule Manipulation - optical traps and tweezers
Yang et al.,
 JCB: SUPPLEMENTAL MATERIAL Yang et al., http://www.jcb.org/cgi/content/full/jcb.200605053/dc1 Supplemental results The NPC activity under improved transport conditions In earlier work where we demonstrated
JCB: SUPPLEMENTAL MATERIAL Yang et al., http://www.jcb.org/cgi/content/full/jcb.200605053/dc1 Supplemental results The NPC activity under improved transport conditions In earlier work where we demonstrated
Multiple Choice Identify the letter of the choice that best completes the statement or answers the question.
 chapter 7 Test Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. Who was one of the first people to identify and see cork cells? a. Anton van
chapter 7 Test Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. Who was one of the first people to identify and see cork cells? a. Anton van
Cell Growth and Division
 Cell Growth and Division Growth, Development, and Reproduction Q: How does a cell produce a new cell? 10.1 Why do cells divide? WHAT I KNOW SAMPLE ANSWER: Cells divide to produce more cells. WHAT I LEARNED
Cell Growth and Division Growth, Development, and Reproduction Q: How does a cell produce a new cell? 10.1 Why do cells divide? WHAT I KNOW SAMPLE ANSWER: Cells divide to produce more cells. WHAT I LEARNED
BIO 311C Spring 2010
 BIO 311C Spring 2010 Prokaryotic cells contain structures that are very similar to structures of the eukaryotic cytoskeleton. Prokaryotic cytoskeletal elements are required for cell division, maintaining
BIO 311C Spring 2010 Prokaryotic cells contain structures that are very similar to structures of the eukaryotic cytoskeleton. Prokaryotic cytoskeletal elements are required for cell division, maintaining
10.1 Cell Growth, Division, and Reproduction
 10.1 Cell Growth, Division, and Reproduction Lesson Objectives Explain the problems that growth causes for cells. Compare asexual and sexual reproduction. Lesson Summary Limits to Cell Size There are two
10.1 Cell Growth, Division, and Reproduction Lesson Objectives Explain the problems that growth causes for cells. Compare asexual and sexual reproduction. Lesson Summary Limits to Cell Size There are two
7.013 Spring 2005 Problem Set 4
 MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel NAME TA 7.013 Spring 2005 Problem Set 4 FRIDAY April 8th,
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel NAME TA 7.013 Spring 2005 Problem Set 4 FRIDAY April 8th,
Neurons and the membrane potential. N500 John Beggs 23 Aug, 2016
 Neurons and the membrane potential N500 John Beggs 23 Aug, 2016 My background, briefly Neurons Structural elements of a typical neuron Figure 1.2 Some nerve cell morphologies found in the human
Neurons and the membrane potential N500 John Beggs 23 Aug, 2016 My background, briefly Neurons Structural elements of a typical neuron Figure 1.2 Some nerve cell morphologies found in the human
Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding
 Supplementary information Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding Marcus Braun* 1,2,3, Zdenek Lansky* 1,2,3, Agata Szuba 1,2, Friedrich W. Schwarz 1,2, Aniruddha
Supplementary information Changes in microtubule overlap length regulate kinesin-14-driven microtubule sliding Marcus Braun* 1,2,3, Zdenek Lansky* 1,2,3, Agata Szuba 1,2, Friedrich W. Schwarz 1,2, Aniruddha
In order to confirm the contribution of diffusion to the FRAP recovery curves of
 Fonseca et al., Supplementary Information FRAP data analysis ) Contribution of Diffusion to the recovery curves In order to confirm the contribution of diffusion to the FRAP recovery curves of PH::GFP
Fonseca et al., Supplementary Information FRAP data analysis ) Contribution of Diffusion to the recovery curves In order to confirm the contribution of diffusion to the FRAP recovery curves of PH::GFP
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
BIOLOGY. Chapter 10 CELL REPRODUCTION PowerPoint Image Slideshow
 BIOLOGY Chapter 10 CELL REPRODUCTION PowerPoint Image Slideshow FIGURE 10.1 A sea urchin begins life as a single cell that (a) divides to form two cells, visible by scanning electron microscopy. After
BIOLOGY Chapter 10 CELL REPRODUCTION PowerPoint Image Slideshow FIGURE 10.1 A sea urchin begins life as a single cell that (a) divides to form two cells, visible by scanning electron microscopy. After
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
Richik N. Ghosh, Linnette Grove, and Oleg Lapets ASSAY and Drug Development Technologies 2004, 2:
 1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
Gene Control Mechanisms at Transcription and Translation Levels
 Gene Control Mechanisms at Transcription and Translation Levels Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by MHRD Page 1 of 9
Gene Control Mechanisms at Transcription and Translation Levels Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by MHRD Page 1 of 9
The Aβ40 and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary mixtures but cross-react during primary nucleation
 Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2015 The Aβ40 and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary
Electronic Supplementary Material (ESI) for Chemical Science. This journal is The Royal Society of Chemistry 2015 The Aβ40 and Aβ42 peptides self-assemble into separate homomolecular fibrils in binary
From gene to protein. Premedical biology
 From gene to protein Premedical biology Central dogma of Biology, Molecular Biology, Genetics transcription replication reverse transcription translation DNA RNA Protein RNA chemically similar to DNA,
From gene to protein Premedical biology Central dogma of Biology, Molecular Biology, Genetics transcription replication reverse transcription translation DNA RNA Protein RNA chemically similar to DNA,
BE/APh161: Physical Biology of the Cell Homework 2 Due Date: Wednesday, January 18, 2017
 BE/APh161: Physical Biology of the Cell Homework 2 Due Date: Wednesday, January 18, 2017 Doubt is the father of creation. - Galileo Galilei 1. Number of mrna in a cell. In this problem, we are going to
BE/APh161: Physical Biology of the Cell Homework 2 Due Date: Wednesday, January 18, 2017 Doubt is the father of creation. - Galileo Galilei 1. Number of mrna in a cell. In this problem, we are going to
What is the central dogma of biology?
 Bellringer What is the central dogma of biology? A. RNA DNA Protein B. DNA Protein Gene C. DNA Gene RNA D. DNA RNA Protein Review of DNA processes Replication (7.1) Transcription(7.2) Translation(7.3)
Bellringer What is the central dogma of biology? A. RNA DNA Protein B. DNA Protein Gene C. DNA Gene RNA D. DNA RNA Protein Review of DNA processes Replication (7.1) Transcription(7.2) Translation(7.3)
Chapter 11. Development: Differentiation and Determination
 KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
FROM LOCALIZATION TO INTERACTION
 EPFL SV PTBIOP FROM LOCALIZATION TO INTERACTION BIOP COURSE 2015 COLOCALIZATION TYPICAL EXAMPLE EPFL SV PTBIOP Vinculin Alexa568 Actin Alexa488 http://www.olympusconfocal.com/applications/colocalization.html
EPFL SV PTBIOP FROM LOCALIZATION TO INTERACTION BIOP COURSE 2015 COLOCALIZATION TYPICAL EXAMPLE EPFL SV PTBIOP Vinculin Alexa568 Actin Alexa488 http://www.olympusconfocal.com/applications/colocalization.html
Honors Biology Reading Guide Chapter 11
 Honors Biology Reading Guide Chapter 11 v Promoter a specific nucleotide sequence in DNA located near the start of a gene that is the binding site for RNA polymerase and the place where transcription begins
Honors Biology Reading Guide Chapter 11 v Promoter a specific nucleotide sequence in DNA located near the start of a gene that is the binding site for RNA polymerase and the place where transcription begins
Measuring Colocalization within Fluorescence Microscopy Images
 from photonics.com: 03/01/2007 http://www.photonics.com/article.aspx?aid=39341 Measuring Colocalization within Fluorescence Microscopy Images Two-color fluorescence-based methods are uncovering molecular
from photonics.com: 03/01/2007 http://www.photonics.com/article.aspx?aid=39341 Measuring Colocalization within Fluorescence Microscopy Images Two-color fluorescence-based methods are uncovering molecular
The Nervous System and the Sodium-Potassium Pump
 The Nervous System and the Sodium-Potassium Pump 1. Define the following terms: Ion: A Student Activity on Membrane Potentials Cation: Anion: Concentration gradient: Simple diffusion: Sodium-Potassium
The Nervous System and the Sodium-Potassium Pump 1. Define the following terms: Ion: A Student Activity on Membrane Potentials Cation: Anion: Concentration gradient: Simple diffusion: Sodium-Potassium
T H E J O U R N A L O F C E L L B I O L O G Y
 Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Multiple Choice Review- Eukaryotic Gene Expression
 Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
DIFFERENTIATION MORPHOGENESIS GROWTH HOW CAN AN IDENTICAL SET OF GENETIC INSTRUCTIONS PRODUCE DIFFERENT TYPES OF CELLS?
 DIFFERENTIATION HOW CAN AN IDENTICAL SET OF GENETIC INSTRUCTIONS PRODUCE DIFFERENT TYPES OF CELLS? MORPHOGENESIS HOW CAN CELLS FORM ORDERED STRUCTURES? GROWTH HOW DO OUR CELLS KNOW WHEN TO STOP DIVIDING
DIFFERENTIATION HOW CAN AN IDENTICAL SET OF GENETIC INSTRUCTIONS PRODUCE DIFFERENT TYPES OF CELLS? MORPHOGENESIS HOW CAN CELLS FORM ORDERED STRUCTURES? GROWTH HOW DO OUR CELLS KNOW WHEN TO STOP DIVIDING
monomer polymer polymeric network cell
 3.1 Motivation 3.2 Polymerization The structural stability of the cell is provided by the cytoskeleton. Assembling and disassembling dynamically, the cytoskeleton enables cell movement through a highly
3.1 Motivation 3.2 Polymerization The structural stability of the cell is provided by the cytoskeleton. Assembling and disassembling dynamically, the cytoskeleton enables cell movement through a highly
Eukaryotic vs. Prokaryotic genes
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 18: Eukaryotic genes http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Eukaryotic vs. Prokaryotic genes Like in prokaryotes,
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 18: Eukaryotic genes http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Eukaryotic vs. Prokaryotic genes Like in prokaryotes,
Name Chapter 10: Chromosomes, Mitosis, and Meiosis Mrs. Laux Take home test #7 DUE: MONDAY, NOVEMBER 16, 2009 MULTIPLE CHOICE QUESTIONS
 MULTIPLE CHOICE QUESTIONS 1. A bacterial chromosome consists of: A. a linear DNA molecule many times larger than the cell. B. a circular DNA molecule many times larger than the cell. C. a circular DNA
MULTIPLE CHOICE QUESTIONS 1. A bacterial chromosome consists of: A. a linear DNA molecule many times larger than the cell. B. a circular DNA molecule many times larger than the cell. C. a circular DNA
FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell
 FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell All media is on the Instructors Resource CD/DVD JPEG Resources Figures, Photos, and Tables PowerPoint Resources Chapter Outline with Figures Lecture
FREEMAN MEDIA INTEGRATION GUIDE Chapter 7: Inside the Cell All media is on the Instructors Resource CD/DVD JPEG Resources Figures, Photos, and Tables PowerPoint Resources Chapter Outline with Figures Lecture
APh161: Physical Biology of the Cell Homework 1 Due Date: Tuesday, January 27, 2009
 APh161: Physical Biology of the Cell Homework 1 Due Date: Tuesday, January 27, 2009 Chance only favours the mind which is prepared. Louis Pasteur In my view, homeworks are one of the primary tools at a
APh161: Physical Biology of the Cell Homework 1 Due Date: Tuesday, January 27, 2009 Chance only favours the mind which is prepared. Louis Pasteur In my view, homeworks are one of the primary tools at a
Energy Transformations in Z-Pinches
 Energy Transformations in Z-Pinches P. Kubeš, J. Kravárik Czech Technical University, Prague, Czech Republic M. Scholz, M. Paduch, K. Tomaszewski, L. Rić Institute of Plasma Physics and Laser Microfusion,
Energy Transformations in Z-Pinches P. Kubeš, J. Kravárik Czech Technical University, Prague, Czech Republic M. Scholz, M. Paduch, K. Tomaszewski, L. Rić Institute of Plasma Physics and Laser Microfusion,
Components of a functional cell. Boundary-membrane Cytoplasm: Cytosol (soluble components) & particulates DNA-information Ribosomes-protein synthesis
 Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
Guided Reading Activities
 Name Period Chapter 4: A Tour of the Cell Guided Reading Activities Big Idea: Introduction to the Cell Answer the following questions as you read Modules 4.1 4.4: 1. A(n) uses a beam of light to illuminate
Name Period Chapter 4: A Tour of the Cell Guided Reading Activities Big Idea: Introduction to the Cell Answer the following questions as you read Modules 4.1 4.4: 1. A(n) uses a beam of light to illuminate
Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry
 Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry Petr Pracna J. Heyrovský Institute of Physical Chemistry Academy of Sciences of the Czech Republic, Prague ZiF Cooperation
Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry Petr Pracna J. Heyrovský Institute of Physical Chemistry Academy of Sciences of the Czech Republic, Prague ZiF Cooperation
Chapter 6: A Tour of the Cell
 AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb2830 Untreated Latrunculin A t=15 min Latrunculin A t=30 min Latrunculin A t=45 min a Untreated Xpo6 t=15 min Xpo6 t=30 min Xpo6 t=45 min b 10 0 c 10 0 d Probability
SUPPLEMENTARY INFORMATION DOI: 10.1038/ncb2830 Untreated Latrunculin A t=15 min Latrunculin A t=30 min Latrunculin A t=45 min a Untreated Xpo6 t=15 min Xpo6 t=30 min Xpo6 t=45 min b 10 0 c 10 0 d Probability
Translation Part 2 of Protein Synthesis
 Translation Part 2 of Protein Synthesis IN: How is transcription like making a jello mold? (be specific) What process does this diagram represent? A. Mutation B. Replication C.Transcription D.Translation
Translation Part 2 of Protein Synthesis IN: How is transcription like making a jello mold? (be specific) What process does this diagram represent? A. Mutation B. Replication C.Transcription D.Translation
Fluorescence fluctuation microscopy: FRAP and FCS
 Fluorescence fluctuation microscopy: FRAP and FCS Confocal laser scanning fluorescence microscope for the mobility analysis in living cells Ensemble measurements of protein mobility and interactions pre
Fluorescence fluctuation microscopy: FRAP and FCS Confocal laser scanning fluorescence microscope for the mobility analysis in living cells Ensemble measurements of protein mobility and interactions pre
Advanced Higher Biology. Unit 1- Cells and Proteins 2c) Membrane Proteins
 Advanced Higher Biology Unit 1- Cells and Proteins 2c) Membrane Proteins Membrane Structure Phospholipid bilayer Transmembrane protein Integral protein Movement of Molecules Across Membranes Phospholipid
Advanced Higher Biology Unit 1- Cells and Proteins 2c) Membrane Proteins Membrane Structure Phospholipid bilayer Transmembrane protein Integral protein Movement of Molecules Across Membranes Phospholipid
Applications of Mathematics
 Applications of Mathematics Tomáš Cipra Exponential smoothing for irregular data Applications of Mathematics Vol. 5 2006) No. 6 597--604 Persistent URL: http://dml.cz/dmlcz/34655 Terms of use: Institute
Applications of Mathematics Tomáš Cipra Exponential smoothing for irregular data Applications of Mathematics Vol. 5 2006) No. 6 597--604 Persistent URL: http://dml.cz/dmlcz/34655 Terms of use: Institute
SECOND PUBLIC EXAMINATION. Honour School of Physics Part C: 4 Year Course. Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS
 2757 SECOND PUBLIC EXAMINATION Honour School of Physics Part C: 4 Year Course Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS TRINITY TERM 2013 Monday, 17 June, 2.30 pm 5.45 pm 15
2757 SECOND PUBLIC EXAMINATION Honour School of Physics Part C: 4 Year Course Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS TRINITY TERM 2013 Monday, 17 June, 2.30 pm 5.45 pm 15
Old FINAL EXAM BIO409/509 NAME. Please number your answers and write them on the attached, lined paper.
 Old FINAL EXAM BIO409/509 NAME Please number your answers and write them on the attached, lined paper. Gene expression can be regulated at several steps. Describe one example for each of the following:
Old FINAL EXAM BIO409/509 NAME Please number your answers and write them on the attached, lined paper. Gene expression can be regulated at several steps. Describe one example for each of the following:
Division Ave. High School AP Biology
 Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
CS-E5880 Modeling biological networks Gene regulatory networks
 CS-E5880 Modeling biological networks Gene regulatory networks Jukka Intosalmi (based on slides by Harri Lähdesmäki) Department of Computer Science Aalto University January 12, 2018 Outline Modeling gene
CS-E5880 Modeling biological networks Gene regulatory networks Jukka Intosalmi (based on slides by Harri Lähdesmäki) Department of Computer Science Aalto University January 12, 2018 Outline Modeling gene
Supporting Information
 Supporting Information Fleissner et al. 10.1073/pnas.0907039106 Fig. S1. (A) MAK-2-GFP localized to CATs tips is not bound by membrane. his-3::pccg1 mak-2-gfp; mak-2 strain labeled with membrane dye FM4
Supporting Information Fleissner et al. 10.1073/pnas.0907039106 Fig. S1. (A) MAK-2-GFP localized to CATs tips is not bound by membrane. his-3::pccg1 mak-2-gfp; mak-2 strain labeled with membrane dye FM4
General Knowledge on Cell Cycle & Cell Division General knowledge on Cell Cycle and Cell Division [ Contents: Cell Cycle,
 General Knowledge on Cell Cycle & Cell Division A complete General knowledge on Cell Cycle and Cell Division for you competitive examinations such as UPSC, IAS, Banking SBI PO, Railway Group-D, SSC, CGL
General Knowledge on Cell Cycle & Cell Division A complete General knowledge on Cell Cycle and Cell Division for you competitive examinations such as UPSC, IAS, Banking SBI PO, Railway Group-D, SSC, CGL
(Excerpt from S. Ji, Molecular Theory of the Living Cell: Concepts, Molecular Mechanisms, and Biomedical Applications, Springer, New York, 2012)
 2.2 The Franck-Condon Principle (FCP) 2.2.1 FCP and Born-Oppenheimer Approximation The Franck-Condon Principle originated in molecular spectroscopy in 1925 when J. Franck proposed (and later Condon provided
2.2 The Franck-Condon Principle (FCP) 2.2.1 FCP and Born-Oppenheimer Approximation The Franck-Condon Principle originated in molecular spectroscopy in 1925 when J. Franck proposed (and later Condon provided
2. Which of the following are NOT prokaryotes? A) eubacteria B) archaea C) viruses D) ancient bacteria
 1. Which of the following statements is FALSE? A) Errors in chromosome separation are rarely a problem for an organism. B) Errors in chromosome separation can result in a miscarriage. C) Errors in chromosome
1. Which of the following statements is FALSE? A) Errors in chromosome separation are rarely a problem for an organism. B) Errors in chromosome separation can result in a miscarriage. C) Errors in chromosome
Supplementary Information for. Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria
 Supplementary Information for Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria Isabella Santi 1 *, Neeraj Dhar 1, Djenet Bousbaine 1, Yuichi Wakamoto, John D.
Supplementary Information for Single-cell dynamics of the chromosome replication and cell division cycles in mycobacteria Isabella Santi 1 *, Neeraj Dhar 1, Djenet Bousbaine 1, Yuichi Wakamoto, John D.
BIOLOGY 111. CHAPTER 5: Chromosomes and Inheritance
 BIOLOGY 111 CHAPTER 5: Chromosomes and Inheritance Chromosomes and Inheritance Learning Outcomes 5.1 Differentiate between sexual and asexual reproduction in terms of the genetic variation of the offspring.
BIOLOGY 111 CHAPTER 5: Chromosomes and Inheritance Chromosomes and Inheritance Learning Outcomes 5.1 Differentiate between sexual and asexual reproduction in terms of the genetic variation of the offspring.
Introduction. Gene expression is the combined process of :
 1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
Protein Sorting. By: Jarod, Tyler, and Tu
 Protein Sorting By: Jarod, Tyler, and Tu Definition Organizing of proteins Organelles Nucleus Ribosomes Endoplasmic Reticulum Golgi Apparatus/Vesicles How do they know where to go? Amino Acid Sequence
Protein Sorting By: Jarod, Tyler, and Tu Definition Organizing of proteins Organelles Nucleus Ribosomes Endoplasmic Reticulum Golgi Apparatus/Vesicles How do they know where to go? Amino Acid Sequence
IncuCyte ZOOM NeuroTrack Fluorescent Processing
 IncuCyte ZOOM NeuroTrack Fluorescent Processing The NeuroTrack TM Software Module (Cat No 9600-0011) is used to measure the processes of neurons in monoculture or with fluorescent labeling in co-culture.
IncuCyte ZOOM NeuroTrack Fluorescent Processing The NeuroTrack TM Software Module (Cat No 9600-0011) is used to measure the processes of neurons in monoculture or with fluorescent labeling in co-culture.
Haveouts Guided Notes Pen/pencil No Privacy Folder DFAD
 Haveouts Guided Notes Pen/pencil No Privacy Folder DFAD Do First: Answer in your DFAD Answer the questions from your Guided Notes as a GROUP, be sure to record your answers in your DFAD. You have 5 mins
Haveouts Guided Notes Pen/pencil No Privacy Folder DFAD Do First: Answer in your DFAD Answer the questions from your Guided Notes as a GROUP, be sure to record your answers in your DFAD. You have 5 mins
Developmental Biology Lecture Outlines
 Developmental Biology Lecture Outlines Lecture 01: Introduction Course content Developmental Biology Obsolete hypotheses Current theory Lecture 02: Gametogenesis Spermatozoa Spermatozoon function Spermatozoon
Developmental Biology Lecture Outlines Lecture 01: Introduction Course content Developmental Biology Obsolete hypotheses Current theory Lecture 02: Gametogenesis Spermatozoa Spermatozoon function Spermatozoon
BME 5742 Biosystems Modeling and Control
 BME 5742 Biosystems Modeling and Control Lecture 24 Unregulated Gene Expression Model Dr. Zvi Roth (FAU) 1 The genetic material inside a cell, encoded in its DNA, governs the response of a cell to various
BME 5742 Biosystems Modeling and Control Lecture 24 Unregulated Gene Expression Model Dr. Zvi Roth (FAU) 1 The genetic material inside a cell, encoded in its DNA, governs the response of a cell to various
The wavelength reconstruction from toroidal spectrometer image data
 Computational Methods and Experimental Measurements XIII 165 The wavelength reconstruction from toroidal spectrometer image data J. Blazej 1, M. Tamas 1, L. Pina 1, A. Jancarek 1, S. Palinek 1, P. Vrba
Computational Methods and Experimental Measurements XIII 165 The wavelength reconstruction from toroidal spectrometer image data J. Blazej 1, M. Tamas 1, L. Pina 1, A. Jancarek 1, S. Palinek 1, P. Vrba
Lecture 8: Temporal programs and the global structure of transcription networks. Chap 5 of Alon. 5.1 Introduction
 Lecture 8: Temporal programs and the global structure of transcription networks Chap 5 of Alon 5. Introduction We will see in this chapter that sensory transcription networks are largely made of just four
Lecture 8: Temporal programs and the global structure of transcription networks Chap 5 of Alon 5. Introduction We will see in this chapter that sensory transcription networks are largely made of just four
Improved Holt Method for Irregular Time Series
 WDS'08 Proceedings of Contributed Papers, Part I, 62 67, 2008. ISBN 978-80-7378-065-4 MATFYZPRESS Improved Holt Method for Irregular Time Series T. Hanzák Charles University, Faculty of Mathematics and
WDS'08 Proceedings of Contributed Papers, Part I, 62 67, 2008. ISBN 978-80-7378-065-4 MATFYZPRESS Improved Holt Method for Irregular Time Series T. Hanzák Charles University, Faculty of Mathematics and
Nucleus and Mitosis. VIBS 443 and VIBS 602. Undergraduate Graduate Histology Lecture Series
 Nucleus and Mitosis VIBS 443 and VIBS 602 Undergraduate Graduate Histology Lecture Series Larry Johnson, Professor Veterinary Integrative Biosciences Texas A&M University College Station, TX 77843 Objectives
Nucleus and Mitosis VIBS 443 and VIBS 602 Undergraduate Graduate Histology Lecture Series Larry Johnson, Professor Veterinary Integrative Biosciences Texas A&M University College Station, TX 77843 Objectives
Reading Assignments. A. Genes and the Synthesis of Polypeptides. Lecture Series 7 From DNA to Protein: Genotype to Phenotype
 Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Ron et al SUPPLEMENTAL DATA
 Ron et al SUPPLEMENTAL DATA Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model Mily Ron, Kaisa Kajala,
Ron et al SUPPLEMENTAL DATA Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model Mily Ron, Kaisa Kajala,
298 Chapter 6 Electronic Structure and Periodic Properties of Elements
 98 Chapter 6 Electronic Structure and Periodic Properties of Elements 6. The Bohr Model By the end of this section, you will be able to: Describe the Bohr model of the hydrogen atom Use the Rydberg equation
98 Chapter 6 Electronic Structure and Periodic Properties of Elements 6. The Bohr Model By the end of this section, you will be able to: Describe the Bohr model of the hydrogen atom Use the Rydberg equation
Nuclear Functional Organization
 Lecture #4 The Cell as a Machine Nuclear Functional Organization Background readings from Chapters 4 of Alberts et al. Molecular Biology of the Cell (4 th Edition) Description of Functions by Biosystems
Lecture #4 The Cell as a Machine Nuclear Functional Organization Background readings from Chapters 4 of Alberts et al. Molecular Biology of the Cell (4 th Edition) Description of Functions by Biosystems
Supplementary Figure 1.
 Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
EQUIPMENT Beta spectrometer, vacuum pump, Cs-137 source, Geiger-Muller (G-M) tube, scalar
 Modern Physics Laboratory Beta Spectroscopy Experiment In this experiment, electrons emitted as a result of the radioactive beta decay of Cs-137 are measured as a function of their momentum by deflecting
Modern Physics Laboratory Beta Spectroscopy Experiment In this experiment, electrons emitted as a result of the radioactive beta decay of Cs-137 are measured as a function of their momentum by deflecting
Name: Class: Date: ID: A
 Class: Date: Ch 7 Review Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. Researchers use fluorescent labels and light microscopy to a. follow
Class: Date: Ch 7 Review Multiple Choice Identify the letter of the choice that best completes the statement or answers the question. 1. Researchers use fluorescent labels and light microscopy to a. follow
