Crystallization and Stability of Membrane Proteins. David Drew
|
|
|
- Cuthbert Cross
- 5 years ago
- Views:
Transcription
1 Crystallization and Stability of Membrane Proteins David Drew
2 1. Produc)on, Purifica)on, Crystalliza)on (Talk 1) GFP- based E. coli pipeline - Bacterial proteins GFP- based S. cerevisiae pipeline - Eukaryo)c proteins Op)miza)on of membrane protein crystals based on detergent stability (Talk 2)
3 Whole-cell Drew et al., FEBS (2001) MPs tested rapidly for quantity and quality In-gel Drew et al., Nat Methods (2005), Newstead et al, PNAS (2007), Geertsma ER., PNAS (2008) FSEC Kawate T, Gouaux E. Structure (2006)
4 Plasmids and strains sent on request (no MTA)
5 Transporter structural summary? Crystallization success rate similar between proand eukaryotic transporters. Crystal optimization is the biggest difference. Sonoda Y et al. Structure. (2011)
6 The Emperical Triangle Y. Sonoda et al. Structure. (2011)
7 Micelle size is important Sonoda Y et al. FEBS (2010).
8 Micelle size correlates with dynamic radius Kunji ER. et al, Methods (2008)
9 Detergent (conformational) stability is important Serrano-Vega MJ, et al. PNAS (2008), Warne T et al. Nature (2008)
10 8Å 3Å DDM LDAO
11 How detergent stable does the MP need to be? Sonoda Y et al. Structure. (2011)
12 CPM assay to measure detergent MP stability DDM Different detergents Cys" CPM" 40 C Fluorescence Fluorescence CPM: the thiol-specific fluorochrome, N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide Alexandrov A., Mileni M., Chien E., Hanson M., Stevens RC., Structure (2008)
13
14
15 Include test-set of control membrane proteins Sonoda Y et al. Structure. (2011)
16 Benchmarking Stability for Optimizing Crystallization CPM assay Rela)ve fluorescence (R.F.U) LDAO OG 9M 10M C 12 E 9 / 12M Time (min) Measure unfolding rate at 40 C for 130 min in proteins all purified In 12M and diluted 150- fold into buffer containing detergent at 3 x cmc
17 Calculate a relative unfolding rate (t 1/2 ) based on cysteine accessibility over time Fraction of folded protein = 1 - (fluorescence t =x) (max fluorescence) Sonoda Y et al. Structure. (2011)
18 Bacterial MPs more stable than Eukaryotic MPs Prokaryotic Eukaryotic N = 16 N = 8 Y. Sonoda et al. Structure. (2011)
19 Mean membrane protein detergent stability correlates with micelle-size Y. Sonoda et al. Structure. (2011)
20 Hydrophobic mismatch Olaf Anderson and Roger Koepe (2007). Annu. Rev. Biopys. Biomol. Struct
21 Comparing t 1/2 values to monodispersity
22 Decreasing stability
23 Benchmarking Stability for Optimizing Crystallization LDAO Eukaryo)c Prokaryo)c stable 17 min not- stable
24 Benchmarking Stability for Optimizing Crystallization 12M > > Eukaryo)c Prokaryo)c stable 17 min
25 MPs stable in harsh detergents are also more in general LDAO vs 9M
26 MPs stable in harsh detergents are also more in general 9M vs 10M 12M vs C 12 E 9
27 Stable MPs are more likely to crystallize in small detergent
28 All our structure success are only the most stable MPs LDAO min
29 4 bacterial and 1 plant transporter crystals diffract well
30 How do we tackle less stable transporters? LDAO min 5?
31 Benchmarking Stability Lipid for addition Optimizing Crystallization Rat sugar transporter
32 Benchmarking Stability for Optimizing Crystallization? Solved
33 wt turkey β1-receptor ronn prediction (xenopus)
34 Wt turkey β1 (N- and C-terminal truncations) Δ30aa ICL3 Turkey β1 structure sequence
35 Mammalian proteins can also screen for more stable homologue
36
37 Mammalian transporter-fab complex
38 H1 structure Shimamura et al. Nature (2011)
39
40 Other strategies 3. LCP in combination with T4-L (Ray Stevens, Brian Kolbilka, Martin Caffrey) 4. Stabilization ligands/ mutagenesis (Chris Tate) e.g. human β2-receptor e.g. turkey β1-receptor Cherezov V et al., Science (2007) Warne et al. Nature (2010)
41 The Emperical Triangle Y. Sonoda et al. Structure. (2011)
42 Acknowledgements So Iwata s MPC/MPL/Kyoto/RaCH group Yo Sonoda, Simon Newstead, Nein-Jen Hu, Emmanuel Nji, Alex Cameron, Norimichi Nomura, Hae-Joo Kang Bernadette Byrne Yilmaz Alguel, James Lee My Current group Chiara Lee, Hae-Joo Kang, Nein-Jen Hu
GFP-based pipelines for the overexpression and purification of membrane proteins. David Drew
 GFP-based pipelines for the overexpression and purification of membrane proteins David Drew 1. Produc0on, Purifica0on, Crystalliza0on (Talk 1) GFP- based E. coli pipeline - Bacterial proteins GFP- based
GFP-based pipelines for the overexpression and purification of membrane proteins David Drew 1. Produc0on, Purifica0on, Crystalliza0on (Talk 1) GFP- based E. coli pipeline - Bacterial proteins GFP- based
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
Cell day 1.notebook September 01, Study the picture of a prokaryotic cell on page 162 in a textbook and the two eukaryotic cells on page 163.
 BellRinger: Log into a clicker! Study the picture of a prokaryotic cell on page 162 in a textbook and the two eukaryotic cells on page 163. Compare them and list similarities and differences. Sep 11 11:00
BellRinger: Log into a clicker! Study the picture of a prokaryotic cell on page 162 in a textbook and the two eukaryotic cells on page 163. Compare them and list similarities and differences. Sep 11 11:00
Supplementary information for:
 SUPPLEMETARY IFRMATI Supplementary information for: Structure of a β 1 -adrenergic G protein-coupled receptor Tony Warne, Maria J. Serrano-Vega, Jillian G. Baker#, Rouslan Moukhametzianov, Patricia C.
SUPPLEMETARY IFRMATI Supplementary information for: Structure of a β 1 -adrenergic G protein-coupled receptor Tony Warne, Maria J. Serrano-Vega, Jillian G. Baker#, Rouslan Moukhametzianov, Patricia C.
Supplementary Information. The protease GtgE from Salmonella exclusively targets. inactive Rab GTPases
 Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Structure and RNA-binding properties. of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex
 Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Sample preparation and characterization around SAXS
 Sample preparation and characterization around SAXS Experimental verification and validation? Rob Meijers EMBL Hamburg Garbage in? The right stuff Molecular weight Oligomerization state Monodispersity
Sample preparation and characterization around SAXS Experimental verification and validation? Rob Meijers EMBL Hamburg Garbage in? The right stuff Molecular weight Oligomerization state Monodispersity
Copyright Mark Brandt, Ph.D A third method, cryogenic electron microscopy has seen increasing use over the past few years.
 Structure Determination and Sequence Analysis The vast majority of the experimentally determined three-dimensional protein structures have been solved by one of two methods: X-ray diffraction and Nuclear
Structure Determination and Sequence Analysis The vast majority of the experimentally determined three-dimensional protein structures have been solved by one of two methods: X-ray diffraction and Nuclear
ml. ph 7.5 ph 6.5 ph 5.5 ph 4.5. β 2 AR-Gs complex + GDP β 2 AR-Gs complex + GTPγS
 a UV28 absorption (mau) 9 8 7 5 3 β 2 AR-Gs complex β 2 AR-Gs complex + GDP β 2 AR-Gs complex + GTPγS β 2 AR-Gs complex dissociated complex excess nucleotides b 9 8 7 5 3 β 2 AR-Gs complex β 2 AR-Gs complex
a UV28 absorption (mau) 9 8 7 5 3 β 2 AR-Gs complex β 2 AR-Gs complex + GDP β 2 AR-Gs complex + GTPγS β 2 AR-Gs complex dissociated complex excess nucleotides b 9 8 7 5 3 β 2 AR-Gs complex β 2 AR-Gs complex
Differential Scanning Fluorimetry: Detection of ligands and conditions that promote protein stability and crystallization
 Differential Scanning Fluorimetry: Detection of ligands and conditions that promote protein stability and crystallization Frank Niesen PX school 2008, Como, Italy Method introduction Topics Applications
Differential Scanning Fluorimetry: Detection of ligands and conditions that promote protein stability and crystallization Frank Niesen PX school 2008, Como, Italy Method introduction Topics Applications
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.108/nature1099 Supplementary Tables: Supplementary Table 1. Data collection and refinement statistics. Highest resolution shell is shown in parentheses. Structure Data
SUPPLEMENTARY INFORMATION doi:10.108/nature1099 Supplementary Tables: Supplementary Table 1. Data collection and refinement statistics. Highest resolution shell is shown in parentheses. Structure Data
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
Purinergic P2X receptors: structural and functional features depicted by X-ray and molecular modelling studies
 Purinergic P2X receptors: structural and functional features depicted by X-ray and molecular modelling studies Leanne Grimes and Mark T Young* School of Biosciences, Cardiff University, Museum Avenue,
Purinergic P2X receptors: structural and functional features depicted by X-ray and molecular modelling studies Leanne Grimes and Mark T Young* School of Biosciences, Cardiff University, Museum Avenue,
New G-protein-coupled receptor crystal structures: insights and limitations
 Opinion New G-protein-coupled receptor crystal structures: insights and limitations Brian Kobilka 1 and Gebhard F.X. Schertler 2 1 Departments of Molecular and Cellular Physiology and Medicine, Stanford
Opinion New G-protein-coupled receptor crystal structures: insights and limitations Brian Kobilka 1 and Gebhard F.X. Schertler 2 1 Departments of Molecular and Cellular Physiology and Medicine, Stanford
Structure Prediction of Membrane Proteins. Introduction. Secondary Structure Prediction and Transmembrane Segments Topology Prediction
 Review Structure Prediction of Membrane Proteins Chunlong Zhou 1, Yao Zheng 2, and Yan Zhou 1 * 1 Hangzhou Genomics Institute/James D. Watson Institute of Genome Sciences, Zhejiang University/Key Laboratory
Review Structure Prediction of Membrane Proteins Chunlong Zhou 1, Yao Zheng 2, and Yan Zhou 1 * 1 Hangzhou Genomics Institute/James D. Watson Institute of Genome Sciences, Zhejiang University/Key Laboratory
Biophysics Lectures Three and Four
 Biophysics Lectures Three and Four Kevin Cahill cahill@unm.edu http://dna.phys.unm.edu/ 1 The Atoms and Molecules of Life Cells are mostly made from the most abundant chemical elements, H, C, O, N, Ca,
Biophysics Lectures Three and Four Kevin Cahill cahill@unm.edu http://dna.phys.unm.edu/ 1 The Atoms and Molecules of Life Cells are mostly made from the most abundant chemical elements, H, C, O, N, Ca,
Membrane proteins Porins: FadL. Oriol Solà, Dimitri Ivancic, Daniel Folch, Marc Olivella
 Membrane proteins Porins: FadL Oriol Solà, Dimitri Ivancic, Daniel Folch, Marc Olivella INDEX 1. INTRODUCTION TO MEMBRANE PROTEINS 2. FADL: OUTER MEMBRANE TRANSPORT PROTEIN 3. MAIN FEATURES OF FADL STRUCTURE
Membrane proteins Porins: FadL Oriol Solà, Dimitri Ivancic, Daniel Folch, Marc Olivella INDEX 1. INTRODUCTION TO MEMBRANE PROTEINS 2. FADL: OUTER MEMBRANE TRANSPORT PROTEIN 3. MAIN FEATURES OF FADL STRUCTURE
Cell Structure and Function
 Cell Structure and Function Cell size comparison Animal cell Bacterial cell What jobs do cells have to do for an organism to live Gas exchange CO 2 & O 2 Eat (take in & digest food) Make energy ATP Build
Cell Structure and Function Cell size comparison Animal cell Bacterial cell What jobs do cells have to do for an organism to live Gas exchange CO 2 & O 2 Eat (take in & digest food) Make energy ATP Build
Ligand screening system using fusion proteins of G protein coupled receptors with G protein α subunits
 2 Ligand screening system using fusion proteins of G protein coupled receptors with G protein α subunits G protein coupled receptors A key player of signaling transduction. Cell membranes are packed with
2 Ligand screening system using fusion proteins of G protein coupled receptors with G protein α subunits G protein coupled receptors A key player of signaling transduction. Cell membranes are packed with
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Chemical structure of LPS and LPS biogenesis in Gram-negative bacteria. a. Chemical structure of LPS. LPS molecule consists of Lipid A, core oligosaccharide and O-antigen. The polar
Supplementary Figure 1 Chemical structure of LPS and LPS biogenesis in Gram-negative bacteria. a. Chemical structure of LPS. LPS molecule consists of Lipid A, core oligosaccharide and O-antigen. The polar
Reason... (2) Reason... (2) Reason... (2)
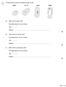 1 The figure below shows four different types of cell. (a) Which cell is a plant cell? Give one reason for your answer. Cell... Reason... (b) Which cell is an animal cell? Give one reason for your answer.
1 The figure below shows four different types of cell. (a) Which cell is a plant cell? Give one reason for your answer. Cell... Reason... (b) Which cell is an animal cell? Give one reason for your answer.
MCB 110. "Molecular Biology: Macromolecular Synthesis and Cellular Function" Spring, 2018
 MCB 110 "Molecular Biology: Macromolecular Synthesis and Cellular Function" Spring, 2018 Faculty Instructors: Prof. Jeremy Thorner Prof. Qiang Zhou Prof. Eva Nogales GSIs:!!!! Ms. Samantha Fernandez Mr.
MCB 110 "Molecular Biology: Macromolecular Synthesis and Cellular Function" Spring, 2018 Faculty Instructors: Prof. Jeremy Thorner Prof. Qiang Zhou Prof. Eva Nogales GSIs:!!!! Ms. Samantha Fernandez Mr.
Hydrogen/Deuterium Exchange Mass Spectrometry: A Mini-Tutorial
 Florida State University National High Magnetic Field Laboratory Tallahassee-Florida Hydrogen/euterium Exchange Mass Spectrometry: A Mini-Tutorial George Bou-Assaf 56 th ASMS Conference June 2 nd, 2008
Florida State University National High Magnetic Field Laboratory Tallahassee-Florida Hydrogen/euterium Exchange Mass Spectrometry: A Mini-Tutorial George Bou-Assaf 56 th ASMS Conference June 2 nd, 2008
Drug binding and subtype selectivity in G-protein-coupled receptors
 Drug binding and subtype selectivity in G-protein-coupled receptors Albert C. Pan The Aspect of Time in Drug Design Schloss Rauischholzhausen Marburg, Germany Thursday, March 27 th, 2014 D. E. Shaw Research
Drug binding and subtype selectivity in G-protein-coupled receptors Albert C. Pan The Aspect of Time in Drug Design Schloss Rauischholzhausen Marburg, Germany Thursday, March 27 th, 2014 D. E. Shaw Research
Biophysical Techniques in Drug Discovery
 Biophysical Techniques in Drug Discovery Angeles Canales Universidad Complutense de Madrid, Spain Drug Discovery Biophysical Techniques in Drug Discovery Edited by Angeles Canales Biophysical techniques
Biophysical Techniques in Drug Discovery Angeles Canales Universidad Complutense de Madrid, Spain Drug Discovery Biophysical Techniques in Drug Discovery Edited by Angeles Canales Biophysical techniques
S2004 Methods for characterization of biomolecular interactions - classical versus modern
 S2004 Methods for characterization of biomolecular interactions - classical versus modern Isothermal Titration Calorimetry (ITC) Eva Dubská email: eva.dubska@ceitec.cz Outline Calorimetry - history + a
S2004 Methods for characterization of biomolecular interactions - classical versus modern Isothermal Titration Calorimetry (ITC) Eva Dubská email: eva.dubska@ceitec.cz Outline Calorimetry - history + a
New Tools for Breaking Barriers to GPCR Expression in E. coli
 doi:10.1016/j.jmb.2011.03.032 J. Mol. Biol. (2011) 408, 597 598 Contents lists available at www.sciencedirect.com Journal of Molecular Biology journal homepage: http://ees.elsevier.com.jmb COMMENTARY New
doi:10.1016/j.jmb.2011.03.032 J. Mol. Biol. (2011) 408, 597 598 Contents lists available at www.sciencedirect.com Journal of Molecular Biology journal homepage: http://ees.elsevier.com.jmb COMMENTARY New
It took more than years for scientists to develop that would allow them to really study.
 CELLS NOTES All living things are made of! THE DISCOVERY OF CELLS The Scientist Who? When? What was discovered? Robert Hooke Anton van Leeuwenhoek Looked through a very simple at a thin slice of and saw
CELLS NOTES All living things are made of! THE DISCOVERY OF CELLS The Scientist Who? When? What was discovered? Robert Hooke Anton van Leeuwenhoek Looked through a very simple at a thin slice of and saw
Ch 7: Cell Structure and Functions. AP Biology
 Ch 7: Cell Structure and Functions AP Biology The Cell Theory 1. All living things are made of cells. 2. New cells come from existing cells. 3. Cells are the basic units of structure and function of living
Ch 7: Cell Structure and Functions AP Biology The Cell Theory 1. All living things are made of cells. 2. New cells come from existing cells. 3. Cells are the basic units of structure and function of living
Cells & Cell Organelles. Doing Life s Work
 Cells & Cell Organelles Doing Life s Work Types of cells bacteria cells Prokaryote Eukaryotes animal cells plant cells Cell size comparison Animal cell Bacterial cell most bacteria 1-10 microns eukaryotic
Cells & Cell Organelles Doing Life s Work Types of cells bacteria cells Prokaryote Eukaryotes animal cells plant cells Cell size comparison Animal cell Bacterial cell most bacteria 1-10 microns eukaryotic
Organisms: We will need to have some examples in mind for our spherical cows.
 Lecture 4: Structure and Composition (Sept. 15) 4.1 Reading Assignment for Lectures 3-4: Phillips, Kondev, Theriot (PKT), Chapter 2 Problem Set 1 (due Sept. 24) now posted on the website. Cellular materials:
Lecture 4: Structure and Composition (Sept. 15) 4.1 Reading Assignment for Lectures 3-4: Phillips, Kondev, Theriot (PKT), Chapter 2 Problem Set 1 (due Sept. 24) now posted on the website. Cellular materials:
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Welcome to BIOL 572: Recombinant DNA techniques
 Lecture 1: 1 Welcome to BIOL 572: Recombinant DNA techniques Agenda 1: Introduce yourselves Agenda 2: Course introduction Agenda 3: Some logistics for BIOL 572 Agenda 4: Q&A section Agenda 1: Introduce
Lecture 1: 1 Welcome to BIOL 572: Recombinant DNA techniques Agenda 1: Introduce yourselves Agenda 2: Course introduction Agenda 3: Some logistics for BIOL 572 Agenda 4: Q&A section Agenda 1: Introduce
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature12045 Supplementary Table 1 Data collection and refinement statistics. Native Pt-SAD X-ray source SSRF BL17U SPring-8 BL41XU Wavelength (Å) 0.97947 1.07171 Space group P2 1 2 1 2 1 P2
doi:10.1038/nature12045 Supplementary Table 1 Data collection and refinement statistics. Native Pt-SAD X-ray source SSRF BL17U SPring-8 BL41XU Wavelength (Å) 0.97947 1.07171 Space group P2 1 2 1 2 1 P2
Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea nd Semester
 Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea 2018. 2 nd Semester Absorbance Assay (280 nm) Considerations for use Absorbance assays are fast and
Kang, Lin-Woo, Ph.D. Professor Department of Biological Sciences Konkuk University Seoul, Korea 2018. 2 nd Semester Absorbance Assay (280 nm) Considerations for use Absorbance assays are fast and
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a
 Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Plasmid Relevant features Source. W18N_D20N and TrXE-W18N_D20N-anti
 Table S1. E. coli plasmids Plasmid Relevant features Source pdg680 T. reesei XynII AA 2-190 with C-terminal His 6 tag optimized for E. coli expression in pjexpress401 Wan et al. (in press) psbn44d psbn44h
Table S1. E. coli plasmids Plasmid Relevant features Source pdg680 T. reesei XynII AA 2-190 with C-terminal His 6 tag optimized for E. coli expression in pjexpress401 Wan et al. (in press) psbn44d psbn44h
Supporting online materials. For Zhou et al., LeuT-desipramine structure suggests. how antidepressants inhibit human neurotransmitter transporters
 Supporting online materials For Zhou et al., LeuT-desipramine structure suggests how antidepressants inhibit human neurotransmitter transporters Materials and methods LeuT overexpression and purification.
Supporting online materials For Zhou et al., LeuT-desipramine structure suggests how antidepressants inhibit human neurotransmitter transporters Materials and methods LeuT overexpression and purification.
Cell Organelles. Wednesday, October 22, 14
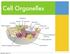 Cell Organelles Cell/Plasma Membrane ALL cells have a cell membrane It is the layer that surrounds the cell and controls what goes in and out Bacteria (Prokaryotic Cell) Cell/Plasma Membrane ALL cells
Cell Organelles Cell/Plasma Membrane ALL cells have a cell membrane It is the layer that surrounds the cell and controls what goes in and out Bacteria (Prokaryotic Cell) Cell/Plasma Membrane ALL cells
7.06 Problem Set
 7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
The Riboswitch is functionally separated into the ligand binding APTAMER and the decision-making EXPRESSION PLATFORM
 The Riboswitch is functionally separated into the ligand binding APTAMER and the decision-making EXPRESSION PLATFORM Purine riboswitch TPP riboswitch SAM riboswitch glms ribozyme In-line probing is used
The Riboswitch is functionally separated into the ligand binding APTAMER and the decision-making EXPRESSION PLATFORM Purine riboswitch TPP riboswitch SAM riboswitch glms ribozyme In-line probing is used
WHAT DO CELLS DO? CHALLENGE QUESTION. What are the functions of the structures inside of cells?
 WHAT DO CELLS DO? CHALLENGE QUESTION What are the functions of the structures inside of cells? WHAT DO CELLS DO? Understanding normal cell structures and their functions help scientists understand how
WHAT DO CELLS DO? CHALLENGE QUESTION What are the functions of the structures inside of cells? WHAT DO CELLS DO? Understanding normal cell structures and their functions help scientists understand how
Measuring S using an analytical ultracentrifuge. Moving boundary
 Measuring S using an analytical ultracentrifuge Moving boundary [C] t = 0 t 1 t 2 0 top r bottom 1 dr b r b (t) r b ω 2 = S ln = ω 2 S (t-t dt r b (t o ) o ) r b = boundary position velocity = dr b dt
Measuring S using an analytical ultracentrifuge Moving boundary [C] t = 0 t 1 t 2 0 top r bottom 1 dr b r b (t) r b ω 2 = S ln = ω 2 S (t-t dt r b (t o ) o ) r b = boundary position velocity = dr b dt
Unit 5: Cells Mr. Nagel Meade High School
 Unit 5: Cells Mr. Nagel Meade High School Hierarchy of Biology Organize the following from smallest to greatest: Tissue, atom, organism, organelle, organ, population, cell, community, ecosystem, organ
Unit 5: Cells Mr. Nagel Meade High School Hierarchy of Biology Organize the following from smallest to greatest: Tissue, atom, organism, organelle, organ, population, cell, community, ecosystem, organ
7.06 Cell Biology EXAM #3 KEY
 7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
The Plant Cell, Chapter 2
 Sugar maple, Acer saccharum Mitosis produces clones in Kalanchoe The Plant Cell, Chapter 2 hdp://maple.dnr.cornell.edu Chapter 2 The Plant Cell Macroscopic 4me- lapse video of plants The tree of life Proper4es
Sugar maple, Acer saccharum Mitosis produces clones in Kalanchoe The Plant Cell, Chapter 2 hdp://maple.dnr.cornell.edu Chapter 2 The Plant Cell Macroscopic 4me- lapse video of plants The tree of life Proper4es
THE CRYSTAL STRUCTURE OF THE SGT1-SKP1 COMPLEX: THE LINK BETWEEN
 THE CRYSTAL STRUCTURE OF THE SGT1-SKP1 COMPLEX: THE LINK BETWEEN HSP90 AND BOTH SCF E3 UBIQUITIN LIGASES AND KINETOCHORES Oliver Willhoft, Richard Kerr, Dipali Patel, Wenjuan Zhang, Caezar Al-Jassar, Tina
THE CRYSTAL STRUCTURE OF THE SGT1-SKP1 COMPLEX: THE LINK BETWEEN HSP90 AND BOTH SCF E3 UBIQUITIN LIGASES AND KINETOCHORES Oliver Willhoft, Richard Kerr, Dipali Patel, Wenjuan Zhang, Caezar Al-Jassar, Tina
Transcription Regulation And Gene Expression in Eukaryotes UPSTREAM TRANSCRIPTION FACTORS
 Transcription Regulation And Gene Expression in Eukaryotes UPSTREAM TRANSCRIPTION FACTORS RG. Clerc March 26. 2008 UPSTREAM TRANSCRIPTION FACTORS Experimental approaches DNA binding domains (DBD) Transcription
Transcription Regulation And Gene Expression in Eukaryotes UPSTREAM TRANSCRIPTION FACTORS RG. Clerc March 26. 2008 UPSTREAM TRANSCRIPTION FACTORS Experimental approaches DNA binding domains (DBD) Transcription
Development and Evaluation of Visual Biosensors for Rapid Detection of Salmonella spp. and Listeria monocytogenes
 Development and Evaluation of Visual Biosensors for Rapid Detection of Salmonella spp. and Listeria monocytogenes Lawrence D. Goodridge Department of Animal Sciences Colorado State University Lawrence.Goodridge@colostate.edu
Development and Evaluation of Visual Biosensors for Rapid Detection of Salmonella spp. and Listeria monocytogenes Lawrence D. Goodridge Department of Animal Sciences Colorado State University Lawrence.Goodridge@colostate.edu
Stochastic simulations
 Stochastic simulations Application to molecular networks Literature overview Noise in genetic networks Origins How to measure and distinguish between the two types of noise (intrinsic vs extrinsic)? What
Stochastic simulations Application to molecular networks Literature overview Noise in genetic networks Origins How to measure and distinguish between the two types of noise (intrinsic vs extrinsic)? What
Unlocking the eukaryotic membrane protein structural proteome John Kyongwon Lee and Robert Michael Stroud
 Available online at Unlocking the eukaryotic membrane protein structural proteome John Kyongwon Lee and Robert Michael Stroud Most of the 231 unique membrane protein structures (as of 3/2010) are of bacterial
Available online at Unlocking the eukaryotic membrane protein structural proteome John Kyongwon Lee and Robert Michael Stroud Most of the 231 unique membrane protein structures (as of 3/2010) are of bacterial
Membrane Protein Channels
 Membrane Protein Channels Potassium ions queuing up in the potassium channel Pumps: 1000 s -1 Channels: 1000000 s -1 Pumps & Channels The lipid bilayer of biological membranes is intrinsically impermeable
Membrane Protein Channels Potassium ions queuing up in the potassium channel Pumps: 1000 s -1 Channels: 1000000 s -1 Pumps & Channels The lipid bilayer of biological membranes is intrinsically impermeable
Supporting Information
 Supporting Information Micelle-Triggered b-hairpin to a-helix Transition in a 14-Residue Peptide from a Choline-Binding Repeat of the Pneumococcal Autolysin LytA HØctor Zamora-Carreras, [a] Beatriz Maestro,
Supporting Information Micelle-Triggered b-hairpin to a-helix Transition in a 14-Residue Peptide from a Choline-Binding Repeat of the Pneumococcal Autolysin LytA HØctor Zamora-Carreras, [a] Beatriz Maestro,
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1.
 Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
return in class, or Rm B
 Last lectures: Genetic Switches and Oscillators PS #2 due today bf before 3PM return in class, or Rm. 68 371B Naturally occurring: lambda lysis-lysogeny decision lactose operon in E. coli Engineered: genetic
Last lectures: Genetic Switches and Oscillators PS #2 due today bf before 3PM return in class, or Rm. 68 371B Naturally occurring: lambda lysis-lysogeny decision lactose operon in E. coli Engineered: genetic
The living cell is a society of molecules: molecules assembling and cooperating bring about life!
 The Computational Microscope Computational microscope views the cell 100-1,000,000 processors http://micro.magnet.fsu.edu/cells/animals/images/animalcellsfigure1.jpg The living cell is a society of molecules:
The Computational Microscope Computational microscope views the cell 100-1,000,000 processors http://micro.magnet.fsu.edu/cells/animals/images/animalcellsfigure1.jpg The living cell is a society of molecules:
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Microcalorimetry for the Life Sciences
 Microcalorimetry for the Life Sciences Why Microcalorimetry? Microcalorimetry is universal detector Heat is generated or absorbed in every chemical process In-solution No molecular weight limitations Label-free
Microcalorimetry for the Life Sciences Why Microcalorimetry? Microcalorimetry is universal detector Heat is generated or absorbed in every chemical process In-solution No molecular weight limitations Label-free
Building a Homology Model of the Transmembrane Domain of the Human Glycine α-1 Receptor
 Building a Homology Model of the Transmembrane Domain of the Human Glycine α-1 Receptor Presented by Stephanie Lee Research Mentor: Dr. Rob Coalson Glycine Alpha 1 Receptor (GlyRa1) Member of the superfamily
Building a Homology Model of the Transmembrane Domain of the Human Glycine α-1 Receptor Presented by Stephanie Lee Research Mentor: Dr. Rob Coalson Glycine Alpha 1 Receptor (GlyRa1) Member of the superfamily
Cells and the Stuff They re Made of. Indiana University P575 1
 Cells and the Stuff They re Made of Indiana University P575 1 Goal: Establish hierarchy of spatial and temporal scales, and governing processes at each scale of cellular function: o Where does emergent
Cells and the Stuff They re Made of Indiana University P575 1 Goal: Establish hierarchy of spatial and temporal scales, and governing processes at each scale of cellular function: o Where does emergent
Andriy Anishkin, Ph.D.
 Address Work Department of Biology, University of Maryland College Park, MD 20742 Tel: (310) 405-8378. Fax: (310) 314-9358 E-mail: anisan@umd.edu Andriy Anishkin, Ph.D. Curriculum Vitae Home 4005 Lawrence
Address Work Department of Biology, University of Maryland College Park, MD 20742 Tel: (310) 405-8378. Fax: (310) 314-9358 E-mail: anisan@umd.edu Andriy Anishkin, Ph.D. Curriculum Vitae Home 4005 Lawrence
Overview of Cells. Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory
 Overview of Cells Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory Prokaryotic Cells Archaea Bacteria Come in many different shapes and sizes.5 µm 2 µm, up to 60 µm long Have large
Overview of Cells Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory Prokaryotic Cells Archaea Bacteria Come in many different shapes and sizes.5 µm 2 µm, up to 60 µm long Have large
Introduction to Molecular and Cell Biology
 Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the molecular basis of disease? What
Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the molecular basis of disease? What
Biology Exam: Chapters 6 & 7
 Biology Exam: Chapters 6 & 7 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Which of the following compounds may be polymers? A) carbohydrates C) proteins
Biology Exam: Chapters 6 & 7 Multiple Choice Identify the choice that best completes the statement or answers the question. 1. Which of the following compounds may be polymers? A) carbohydrates C) proteins
The outer membrane of Borrelia 7/3/2014. LDA Conference Richard Bingham 1. The Outer Membrane of Borrelia; The Interface Between Them and Us
 The Outer Membrane of Borrelia; The Interface Between Them and Us Richard Bingham The University of Huddersfield Lecture Outline I will give an overview of the outer membrane of Borrelia I will present
The Outer Membrane of Borrelia; The Interface Between Them and Us Richard Bingham The University of Huddersfield Lecture Outline I will give an overview of the outer membrane of Borrelia I will present
2012 Univ Aguilera Lecture. Introduction to Molecular and Cell Biology
 2012 Univ. 1301 Aguilera Lecture Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the
2012 Univ. 1301 Aguilera Lecture Introduction to Molecular and Cell Biology Molecular biology seeks to understand the physical and chemical basis of life. and helps us answer the following? What is the
LIFE! (A BRIEF snapshot)
 LIFE! (A BRIEF snapshot) HTTP://WWW.PBS.ORG/WGBH/NOVA/EVOLUTION/ORIGINS-LIFE.HTML Atmospheric Stuff of Life- Coacervates When exactly (what criteria) do we obtain a living cell? Cellular Reproduction
LIFE! (A BRIEF snapshot) HTTP://WWW.PBS.ORG/WGBH/NOVA/EVOLUTION/ORIGINS-LIFE.HTML Atmospheric Stuff of Life- Coacervates When exactly (what criteria) do we obtain a living cell? Cellular Reproduction
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature17991 Supplementary Discussion Structural comparison with E. coli EmrE The DMT superfamily includes a wide variety of transporters with 4-10 TM segments 1. Since the subfamilies of the
doi:10.1038/nature17991 Supplementary Discussion Structural comparison with E. coli EmrE The DMT superfamily includes a wide variety of transporters with 4-10 TM segments 1. Since the subfamilies of the
Regulation and signaling. Overview. Control of gene expression. Cells need to regulate the amounts of different proteins they express, depending on
 Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
Translocation through the endoplasmic reticulum membrane. Institute of Biochemistry Benoît Kornmann
 Translocation through the endoplasmic reticulum membrane Institute of Biochemistry Benoît Kornmann Endomembrane system Protein sorting Permeable to proteins but not to ions IgG tetramer (16 nm) Fully hydrated
Translocation through the endoplasmic reticulum membrane Institute of Biochemistry Benoît Kornmann Endomembrane system Protein sorting Permeable to proteins but not to ions IgG tetramer (16 nm) Fully hydrated
Structure and function of AMPA receptors
 J Physiol 554.2 pp 249 253 249 Topical Review Structure and function of AMPA receptors Eric Gouaux Department of Biochemistry and Molecular Biophysics and Howard Hughes Medical Institute, Columbia University,
J Physiol 554.2 pp 249 253 249 Topical Review Structure and function of AMPA receptors Eric Gouaux Department of Biochemistry and Molecular Biophysics and Howard Hughes Medical Institute, Columbia University,
Review. Membrane proteins. Membrane transport
 Quiz 1 For problem set 11 Q1, you need the equation for the average lateral distance transversed (s) of a molecule in the membrane with respect to the diffusion constant (D) and time (t). s = (4 D t) 1/2
Quiz 1 For problem set 11 Q1, you need the equation for the average lateral distance transversed (s) of a molecule in the membrane with respect to the diffusion constant (D) and time (t). s = (4 D t) 1/2
REVIEW 2: CELLS & CELL COMMUNICATION. A. Top 10 If you learned anything from this unit, you should have learned:
 Name AP Biology REVIEW 2: CELLS & CELL COMMUNICATION A. Top 10 If you learned anything from this unit, you should have learned: 1. Prokaryotes vs. eukaryotes No internal membranes vs. membrane-bound organelles
Name AP Biology REVIEW 2: CELLS & CELL COMMUNICATION A. Top 10 If you learned anything from this unit, you should have learned: 1. Prokaryotes vs. eukaryotes No internal membranes vs. membrane-bound organelles
Enhancing hydrogen production of microalgae by redirecting electrons from photosystem I to hydrogenase
 Electronic Supplementary Material (ESI) for Energy & Environmental Science. This journal is The Royal Society of Chemistry 2014 Supplementary information for Enhancing hydrogen production of microalgae
Electronic Supplementary Material (ESI) for Energy & Environmental Science. This journal is The Royal Society of Chemistry 2014 Supplementary information for Enhancing hydrogen production of microalgae
Kingdom Monera(Archaebacteria & Eubacteria)
 Kingdom Monera(Archaebacteria & All bacteria are prokaryotes Characteristics: 1. No nucleus Eubacteria) 2. No membrane bound organelles 3. Smaller & less ribosomes 4. Most are smaller than eukaryotes 5.
Kingdom Monera(Archaebacteria & All bacteria are prokaryotes Characteristics: 1. No nucleus Eubacteria) 2. No membrane bound organelles 3. Smaller & less ribosomes 4. Most are smaller than eukaryotes 5.
Other Cells. Hormones. Viruses. Toxins. Cell. Bacteria
 Other Cells Hormones Viruses Toxins Cell Bacteria ΔH < 0 reaction is exothermic, tells us nothing about the spontaneity of the reaction Δ H > 0 reaction is endothermic, tells us nothing about the spontaneity
Other Cells Hormones Viruses Toxins Cell Bacteria ΔH < 0 reaction is exothermic, tells us nothing about the spontaneity of the reaction Δ H > 0 reaction is endothermic, tells us nothing about the spontaneity
Analysis of Escherichia coli amino acid transporters
 Ph.D thesis Analysis of Escherichia coli amino acid transporters Presented by Attila Szvetnik Supervisor: Dr. Miklós Kálmán Biology Ph.D School University of Szeged Bay Zoltán Foundation for Applied Research
Ph.D thesis Analysis of Escherichia coli amino acid transporters Presented by Attila Szvetnik Supervisor: Dr. Miklós Kálmán Biology Ph.D School University of Szeged Bay Zoltán Foundation for Applied Research
Structural properties determining low K + affinity of the selectivity filter in the TWIK1 K + channel
 Supporting Information Structural properties determining low K affinity of the selectivity filter in the TWIK1 K channel Hisao Tsukamoto 1, 2, Masahiro Higashi 3, Hideyoshi Motoki 3, Hiroki Watanabe 4,
Supporting Information Structural properties determining low K affinity of the selectivity filter in the TWIK1 K channel Hisao Tsukamoto 1, 2, Masahiro Higashi 3, Hideyoshi Motoki 3, Hiroki Watanabe 4,
Denaturation and renaturation of proteins
 Denaturation and renaturation of proteins Higher levels of protein structure are formed without covalent bonds. Therefore, they are not as stable as peptide covalent bonds which make protein primary structure
Denaturation and renaturation of proteins Higher levels of protein structure are formed without covalent bonds. Therefore, they are not as stable as peptide covalent bonds which make protein primary structure
Do First NO DFAD today
 Do First NO DFAD today Listen to instructions very carefully. Take out your cell phone and place it on the table. Remove your hands from your phone and make eye contact with me to let me know you have
Do First NO DFAD today Listen to instructions very carefully. Take out your cell phone and place it on the table. Remove your hands from your phone and make eye contact with me to let me know you have
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1)
 MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) 1) Which of the following statements about the atom A) It has 12 neutrons in its nucleus. B) It
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) 1) Which of the following statements about the atom A) It has 12 neutrons in its nucleus. B) It
Tellurite resistance protein/ethidium efflux transporter/ proflavin transporter. Putative inner membrane protein: function unknown
 Additional file 1. Table S1 and Figures S1-4 of Zhang et al. High-level production of membrane proteins in E. coli BL21(DE3) by omitting the inducer IPTG Table S1. Properties of the membrane proteins used
Additional file 1. Table S1 and Figures S1-4 of Zhang et al. High-level production of membrane proteins in E. coli BL21(DE3) by omitting the inducer IPTG Table S1. Properties of the membrane proteins used
!"#$%&!"&'(&%")(*(+& '4567,846/-&*,/69450.:&*,.;42/9450&<ᶫ.=097,.4.& #259,40& &95&523/0,--,.
 1 2 Contact Information Dr. Sonish Azam Office: SP 375.23 Tel: 514-848-2424 ex 3488 Email: sonish.azam@concordia.ca (Please mention BIOL 266 in subject) Office hours: after the class or by appointment
1 2 Contact Information Dr. Sonish Azam Office: SP 375.23 Tel: 514-848-2424 ex 3488 Email: sonish.azam@concordia.ca (Please mention BIOL 266 in subject) Office hours: after the class or by appointment
T i t l e o f t h e w o r k : L a M a r e a Y o k o h a m a. A r t i s t : M a r i a n o P e n s o t t i ( P l a y w r i g h t, D i r e c t o r )
 v e r. E N G O u t l i n e T i t l e o f t h e w o r k : L a M a r e a Y o k o h a m a A r t i s t : M a r i a n o P e n s o t t i ( P l a y w r i g h t, D i r e c t o r ) C o n t e n t s : T h i s w o
v e r. E N G O u t l i n e T i t l e o f t h e w o r k : L a M a r e a Y o k o h a m a A r t i s t : M a r i a n o P e n s o t t i ( P l a y w r i g h t, D i r e c t o r ) C o n t e n t s : T h i s w o
Gene regulation I Biochemistry 302. Bob Kelm February 25, 2005
 Gene regulation I Biochemistry 302 Bob Kelm February 25, 2005 Principles of gene regulation (cellular versus molecular level) Extracellular signals Chemical (e.g. hormones, growth factors) Environmental
Gene regulation I Biochemistry 302 Bob Kelm February 25, 2005 Principles of gene regulation (cellular versus molecular level) Extracellular signals Chemical (e.g. hormones, growth factors) Environmental
THE UNIVERSITY OF MANITOBA. PAPER NO: 409 LOCATION: Fr. Kennedy Gold Gym PAGE NO: 1 of 6 DEPARTMENT & COURSE NO: CHEM 4630 TIME: 3 HOURS
 PAPER NO: 409 LOCATION: Fr. Kennedy Gold Gym PAGE NO: 1 of 6 DEPARTMENT & COURSE NO: CHEM 4630 TIME: 3 HOURS EXAMINATION: Biochemistry of Proteins EXAMINER: J. O'Neil Section 1: You must answer all of
PAPER NO: 409 LOCATION: Fr. Kennedy Gold Gym PAGE NO: 1 of 6 DEPARTMENT & COURSE NO: CHEM 4630 TIME: 3 HOURS EXAMINATION: Biochemistry of Proteins EXAMINER: J. O'Neil Section 1: You must answer all of
1. Protein Data Bank (PDB) 1. Protein Data Bank (PDB)
 Protein structure databases; visualization; and classifications 1. Introduction to Protein Data Bank (PDB) 2. Free graphic software for 3D structure visualization 3. Hierarchical classification of protein
Protein structure databases; visualization; and classifications 1. Introduction to Protein Data Bank (PDB) 2. Free graphic software for 3D structure visualization 3. Hierarchical classification of protein
Chapter 4 Cell Structure and Function Sections 1-6
 Chapter 4 Cell Structure and Function Sections 1-6 4.1 Food For Thought E. coli O157:H7A, strain of bacteria that causes severe illness or death, occasionally contaminates foods such as ground beef and
Chapter 4 Cell Structure and Function Sections 1-6 4.1 Food For Thought E. coli O157:H7A, strain of bacteria that causes severe illness or death, occasionally contaminates foods such as ground beef and
Ribosome readthrough
 Ribosome readthrough Starting from the base PROTEIN SYNTHESIS Eukaryotic translation can be divided into four stages: Initiation, Elongation, Termination and Recycling During translation, the ribosome
Ribosome readthrough Starting from the base PROTEIN SYNTHESIS Eukaryotic translation can be divided into four stages: Initiation, Elongation, Termination and Recycling During translation, the ribosome
Eukaryotic cells are more complex than prokaryotic cells. They are identified by the presence of certain membrane-bound organelles.
 Eukaryotic Cells Eukaryotic cells are more complex than prokaryotic cells. They are identified by the presence of certain membrane-bound organelles. Prokaryotic cells have organelles too, but much fewer
Eukaryotic Cells Eukaryotic cells are more complex than prokaryotic cells. They are identified by the presence of certain membrane-bound organelles. Prokaryotic cells have organelles too, but much fewer
Full-length GlpG sequence was generated by PCR from E. coli genomic DNA. (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
 Supplementary Methods Protein expression and purification Full-length GlpG sequence was generated by PCR from E. coli genomic DNA (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
Supplementary Methods Protein expression and purification Full-length GlpG sequence was generated by PCR from E. coli genomic DNA (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
Secondary Structure. Bioch/BIMS 503 Lecture 2. Structure and Function of Proteins. Further Reading. Φ, Ψ angles alone determine protein structure
 Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Classifying Prokaryotes: Eubacteria Plasma Membrane. Ribosomes. Plasmid (DNA) Capsule. Cytoplasm. Outer Membrane DNA. Flagellum.
 Bacteria The yellow band surrounding this hot spring is sulfur, a waste product of extremophilic prokaryotes, probably of the Domain Archaea, Kingdom Archaebacteria. Bacteria are prokaryotic cells (no
Bacteria The yellow band surrounding this hot spring is sulfur, a waste product of extremophilic prokaryotes, probably of the Domain Archaea, Kingdom Archaebacteria. Bacteria are prokaryotic cells (no
DSC Characterization of the Structure/Function Relationship for Proteins
 DSC Characterization of the Structure/Function Relationship for Proteins Differential Scanning Calorimetry (DSC) DSC is recognized as Gold Std technique for measuring molecular thermal stability and structure
DSC Characterization of the Structure/Function Relationship for Proteins Differential Scanning Calorimetry (DSC) DSC is recognized as Gold Std technique for measuring molecular thermal stability and structure
NIH Public Access Author Manuscript Nat Rev Drug Discov. Author manuscript; available in PMC 2014 January 01.
 NIH Public Access Author Manuscript Published in final edited form as: Nat Rev Drug Discov. 2013 January ; 12(1): 25 34. doi:10.1038/nrd3859. GPCR Network: a large-scale collaboration on GPCR structure
NIH Public Access Author Manuscript Published in final edited form as: Nat Rev Drug Discov. 2013 January ; 12(1): 25 34. doi:10.1038/nrd3859. GPCR Network: a large-scale collaboration on GPCR structure
Machines with Moving Parts See How They Run.
 NRAMM Workshop, CIMBio, La Jolla CA November 8-13, 2009 Machines with Moving Parts See How They Run. Alasdair C. Steven Laboratory of Structural Biology, NIAMS-NIH Sources of Variability in Electron Micrographs
NRAMM Workshop, CIMBio, La Jolla CA November 8-13, 2009 Machines with Moving Parts See How They Run. Alasdair C. Steven Laboratory of Structural Biology, NIAMS-NIH Sources of Variability in Electron Micrographs
Macromolecular X-ray Crystallography
 Protein Structural Models for CHEM 641 Fall 07 Brian Bahnson Department of Chemistry & Biochemistry University of Delaware Macromolecular X-ray Crystallography Purified Protein X-ray Diffraction Data collection
Protein Structural Models for CHEM 641 Fall 07 Brian Bahnson Department of Chemistry & Biochemistry University of Delaware Macromolecular X-ray Crystallography Purified Protein X-ray Diffraction Data collection
BE/APh161 Physical Biology of the Cell. Rob Phillips Applied Physics and Bioengineering California Institute of Technology
 BE/APh161 Physical Biology of the Cell Rob Phillips Applied Physics and Bioengineering California Institute of Technology Cells Decide: Where to Go The Hunters of the Immune Response (Berman et al.) There
BE/APh161 Physical Biology of the Cell Rob Phillips Applied Physics and Bioengineering California Institute of Technology Cells Decide: Where to Go The Hunters of the Immune Response (Berman et al.) There
Warm Up. What are some examples of living things? Describe the characteristics of living things
 Warm Up What are some examples of living things? Describe the characteristics of living things Objectives Identify the levels of biological organization and explain their relationships Describe cell structure
Warm Up What are some examples of living things? Describe the characteristics of living things Objectives Identify the levels of biological organization and explain their relationships Describe cell structure
Structural investigation of single biomolecules
 Structural investigation of single biomolecules NMR spectroscopy and X-ray crystallography are currently the most common techniques capable of determining the structures of biological macromolecules like
Structural investigation of single biomolecules NMR spectroscopy and X-ray crystallography are currently the most common techniques capable of determining the structures of biological macromolecules like
