SUPPLEMENTARY INFORMATION
|
|
|
- Bartholomew Fowler
- 5 years ago
- Views:
Transcription
1 SUPPLEMENTARY METHODS Mef2 primary screen. RNAi hairpins from the VDRC collection were crossed to Mef2-GAL4 at 27 C. After 2 weeks lethality rate and stage was scored, and if possible males containing Mef2-GAL4 and UAS-IR were sorted and incubated for another 7 days at 27ºC, at which time adult viability, wing posture and flight were scored as previously described 1. Every positive line was retested once or twice, blind to both the genotype and the outcome of the initial assay. Scores were averaged. Only lines with a s 19 score 1 better than 0.5 were scored in this work to avoid unspecific phenotypes (Supplementary Fig. 7). For a gene with two UAS-IR lines, the stronger phenotype was used to assign the phenotypic class. Larval assay. RNAi hairpins were crossed to Mef2-GAL4, ZCL0663, a GFP trap in CG6416 labelling the Z-line 7, and progeny assayed after 2 5 days at 27ºC, depending on the lethality stage observed in the initial screen. Larvae were immobilised by placing them into 65ºC water for about 1 sec, and then mounted in 50% glycerol. Images were acquired on a Zeiss Axiophot or Zeiss AxioImagerZ1 at 10x and 20x and analysed with Metamorph software. Embryo and larval staining. Embryos were stained as described 28 with mouse anti- 3e8 (1:100; ref. 29) and rat anti-projectin MAC150 (1:100) (Babraham Institute). For larval stainings larva-filets were prepared as described 30 with the modification that dissections were done in relaxing solution (20mM phosphate buffer, ph 7.0; 5 mm MgCl 2 ; 5 mm EGTA, 5 mm ATP). Samples stained with rat anti-how (1:100; ref. 31) were fixed with 4% paraformaldehyde (PFA), samples stained with rat anti-α- in MAK276 (1:3; ref. 32) (Brabraham Institute) were fixed with ice cold MeOH. All incubations were performed in PBST (PBS + 0,2% Tx100) instead of PBSTween. Mouse anti- 3e8 was used (1:30; ref. 29). Images were taken with a Leica SP2 with a 40x objective and processed with ImageJ and Photoshop. 1
2 Adult muscle assay. Hemi-thoraces from 7 10 day old Mef2-GAL4 UAS-IR adults were prepared by removing head, legs and abdomen with scissors, fixing the thorax in 4% paraformaldehyde (PFA) in relaxing solution (20mM phosphate buffer, ph 7.0; 5 mm MgCl 2 ; 5 mm EGTA) for 5-10 min and bisecting the thoraces sagittally with a sharp microtome blade. Hemi-thoraces were incubated for min in relaxing solution, fixed for 10 min in 4% PFA in relaxing solution, washed 2x in PBST (PBS + 0,1% Tween) and incubated with rhodamine phalloidin (Molecular Probes) for 30 min (1:500 in PBST). After washing 2x for 10 min in PBST, samples were mounted in 50% glycerol and imaged with a Zeiss LSM 510 or Leica SP2 with 10x and 100x objectives to analyze muscle fibers and myofibrils, respectively. For immunohistochemistry hemi-thoraces were prepared as above, apart from incubating the thorax in relaxing solution supplemented with 3 % normal goat serum for 20 min before fixation. Hemi-thoraces were incubated with primary and secondary antibodies for 1 h each in PBS with 0.2 % Triton-X100 and mounted in Vectashield. Mouse anti- 3e8 was used (1:50; ref. 29), rabbit anti-flii sc were used 1:50 (Santa Cruz), rabbit anti-unc-89 (1:500; ref. 33), rat anti-α-in MAK276 (1:3; ref. 32, Brabraham Institute) and guinea pig anti-tmod (1:100; ref 34). Microarrays. IFM from 50 flies were dissected with fine forceps in PBS and the RNA was isolated using TriPure (Roche). RNA was labelled and hybridized to Agilent chips according to the manufacturer (Agilent). Only transcripts expressed above log threshold 8 were analysed. All experiments were performed in biological duplicates. Bioinformatics. Hypergeometric distribution was used to assess the probability for a particular term to be enriched or depleted from a gene set compared with the reference set of all screened genes. A heatmap representation of scores is shown. To evaluate if the evolutionary conservation within a gene set is higher or lower than expected, we score the incidence of finding predicted orthologs for Flybase genes in a wide taxonomic range of species (At- Arabidopsis thaliana, Sc- Saccharomyces cerevisiae, Mm- Mus musculus, Hs- Homo sapiens, Ce- Caenorhabditis elegans, Am- Apis mellifera, Ag- Anopheles gambiae). The ortholog resources used are Compara (v49), Inparanoid (v6.1), Orthomcl (v2), Homologene and eggnog. For each gene 2
3 set potential functional conservation to human was evaluated using phenotype location data provided for disease-related OMIM records in their Clinical Synopsis ( Orthology information as above was used to establish links between Drosophila genes and human disease genes. To define the muscle expression set of genes we searched the BDGP database with the terms 'larval muscle', 'body muscle' and 'visceral muscle'. References. 28. Schnorrer, F., Kalchhauser, I. & Dickson, B.J. The transmembrane protein Kon-tiki couples to Dgrip to mediate myotube targeting in Drosophila. Dev Cell 12, (2007). 29. Saide, J.D. et al. Characterization of components of Z-bands in the fibrillar flight muscle of Drosophila melanogaster. J Cell Biol 109, (1989). 30. Schmid, A. & Sigrist, S.J. Analysis of neuromuscular junctions: histology and in vivo imaging. Methods Mol Biol 420, (2008). 31. Nabel-Rosen, H., Dorevitch, N., Reuveny, A. & Volk, T. The balance between two isoforms of the Drosophila RNA-binding protein how controls tendon cell differentiation. Mol Cell 4, (1999). 32. Lakey, A. et al. Identification and localization of high molecular weight proteins in insect flight and leg muscle. EMBO J 9, (1990). 33. Burkart, C. et al. Modular proteins from the Drosophila sallimus (sls) gene and their expression in muscles with different extensibility. J Mol Biol 367, (2007). 34. Dye, C.A. et al. The Drosophila sanpodo gene controls sibling cell fate and encodes a tropomodulin homolog, an actin/tropomyosin-associated protein. Development 125, (1998). 3
4 Screen overview 17,759 RNAi lines 10,461 genes Muscle knock-down Viability, flight, locomotion, posture 3,909 RNAi lines 2,785 genes 563 RNAi lines 436 genes (early lethals) 405 RNAi lines 328 genes (flightless) Muscle knock-down larval body wall muscles: patterning, sarcomere structure Muscle knock-down adult flight muscles: patterning, myofibril structure, sarcomere structure 190 genes 6 phenotypic classes 189 genes 9 phenotypic classes Supplementary Figure 1 Screen overview Overview of primary and secondary Mef2-GAL4 screens. Genes identified in the primary screen were assigned to specific classes according to viability, flight, locomotion and wing posture, and selected genes assayed in secondary screens according to their phenotype. 4
5 doi: /nature08799 a Projectin b ov UAS-mhc-IR c UAS-bt-IR d Actn e Actn UAS-Actn-IR f How g How UAS-how-IR Supplementary Figure 2 Larval muscle protein knock-down. 2 Body muscles of stage Supplementary 17 embryos stained Figure with anti- in red and anti-projectin in green in wild type (a), UAS-mhc-IR (TF7164) (b), and UAS-bt-IR (TF46253) (c); scale bar, 50 µm. Body muscles of L3 larvae stained with anti- in red and antiin (d, e) or anti-how (f, g) in green in wild type (d, f), UAS-actn-IR (TF7760) (e), and UAS-how-IR (TF13756) (g); larvae in (d) and (e) were fixed in methanol, scale bar, 25 µm. 5
6 doi: /nature08799 a FliI b FliI UAS-fliI-IR c Unc-89 d Unc-89 UAS-unc-89-IR Supplementary Figure 3 Supplementary IFM protein knock-down. Figure 3 Myofibrils from adult flight muscles stained in with phalloidin in red, anti-α-flii (a,b) in red or anti-unc-89 (c,d) in green in wild type (a,c), UAS-fliI-IR (TF39528) (b), UAS-unc89-IR (TF29412) (d). Scale bar, 10 µm. 6
7 doi: /nature08799 a Actn b UAS-fliI-IR c UAS-CG8578-IR d UAS-coro-IR e UAS-cora-IR f Tmod g UAS-hts-IR Supplementary Figure 4 Supplementary Figure 4 Myofibril morphology phenotypes. (a-e) Myofibrils from adult flight muscles stained in with phalloidin in blue, anti-αin in red and anti- in green in wild type (a), UAS-fliI-IR (TF39528) (b), UAS-CG8578-IR (TF35968) (c), UAS-coro-IR (TF44671) (d), and UAS-cora-IR (TF9787) (e). Myofibrils stained from wild type (f) and UAS-hts-IR (g) stained with phalloidin in red, anti-tmod in green and anti- in blue. Scale bar, 10 µm. 7
8 doi: /nature08799 a b UAS-unc-89-IR c UAS-eag-IR f UAS-pdp1-IR Supplementary Figure 5 No distinct M phenotype. Supplementary Figure 5 Myofibrils from adult flight muscles stained with phalloidin in red and anti- in green in wild type (a), UAS-unc89-IR (TF29412) (b), UAS-eag-IR (TF9127) (c), and UAS-pdp1-IR (TF37769) (d). Scale bar, 10 µm. 8
9 UAS-CG8578-IR CG mer off-target flii 16mer off-target UAS-fliI-IR UAS-pdp1-IR pdp1 16mer off-target unc89 16mer off-target UAS-unc89-IR Supplementary Figure 6 RNA levels in isolated IFMs. Supplementary Figure 6 Genome-wide expression profiles of dissected IFM fibers from wild type adults compared to CG8578, flii, unc89 and pdp1 knock-down. The log value of the fold change in mrna levels compared to wild-type controls was determined for each gene expressed above a log threshold of 8. Genes were ranked according to the foldchange, and the rankings of the on-target gene and all predicted off-targets are shown. Off-targets were defined as having at least one exact 16mer match to the hairpin. Offtarget genes show no enrichment for knock-down, whereas on-target mrna levels are consistently reduced. 9
10 5 4!"#$%&" "n!i*+,"n- (/0#1) 3 2 all /0si34e lethal flightless l0:0;030< s 19 (bin) Supplementary Figure 7 Choice of s 19 cut off. Number of RNAi lines scored positive in the Mef2 screen, and in the library as a whole, binned according to s 19 scores. Only lines with s 19 > 0.5 were used in this study. 10
11 For Supplementary Tables 1-12, see separate Supplementary Information files. Supplementary Table 1 Phenotypic classes in primary screen. Phenotypes in primary screen (Fig. 1b) listed by gene and transgenic RNAi line. Supplementary Table 2 Lethal stages in primary screen. Lethality stages in primary screen (Fig. 1c) listed by gene and transgenic RNAi line. Supplementary Table 3 Positive controls. Phenotypes of positive control genes in the primary Mef2-GAL4 screen, listed by gene and RNAi line. Supplementary Table 4 Negative controls. Phenotypes of negative control genes in the primary Mef2-GAL4 screen. Supplementary Table 5 Gene validation. List of genes tested with a second generation RNAi library. Supplementary Table 6 Muscle morphology phenotypes in larval screen. Muscle morphology defects observed in the secondary screen of larval body wall muscles (Fig. 2a), as well as a list of all transformant lines tested. Supplementary Table 7 Sarcomere morphology phenotypes in larval screen. Sarcomere morphology defects observed in the secondary screen of larval body wall muscles (Fig. 2b). Supplementary Table 8 Muscle morphology phenotypes in IFM screen. Muscle morphology defects observed in the secondary screen of adult IFMs (Fig. 3a), as well as a list of all transformant lines tested. Supplementary Table 9 Myofibril morphology phenotypes in IFM screen. Myofibril morphology defects observed in the secondary screen of adult IFMs (Fig. 3b),, including a list of genes with actin blobs. 11
12 Supplementary Table 10 Sarcomere morphology phenotypes in IFM screen. Sarcomere morphology defects observed in the secondary screen of adult IFMs (Fig. 3c). Supplementary Table 11 Overlap of Mef2 positives with Mef2 targets and muscle expression. List of genes that are predicted Mef2 target genes, expressed in muscles and positive in the Mef2 RNAi screen. Supplementary Table 12 Overlap of Mef2 positives with late muscle cluster or genes expressed in adult muscle precursors. List of genes positive in the Mef2 RNAi screen and found in the late muscle cluster or to be expressed in adult muscle precursors in wing discs. 12
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
Chapter 4 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays.
 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Supplementary Information
 Supplementary Information Supplementary Figure 1. JAK/STAT in early wing development (a-f) Wing primordia of second instar larvae of the indicated genotypes labeled to visualize expression of upd mrna
Supplementary Information Supplementary Figure 1. JAK/STAT in early wing development (a-f) Wing primordia of second instar larvae of the indicated genotypes labeled to visualize expression of upd mrna
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1244624/dc1 Supplementary Materials for Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein Sougata Roy, Hai Huang,
www.sciencemag.org/cgi/content/full/science.1244624/dc1 Supplementary Materials for Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein Sougata Roy, Hai Huang,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
Supplemental Data. Perea-Resa et al. Plant Cell. (2012) /tpc
 Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplementary Figure 1. Nature Genetics: doi: /ng.3848
 Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Original Article Overexpression of ter94, Drosophila VCP, improves motor neuron degeneration induced by knockdown of TBPH, Drosophila TDP-43
 Am J Neurodegener Dis 2018;7(1):11-31 www.ajnd.us /ISSN:2165-591X/AJND0069303 Original Article Overexpression of ter94, Drosophila VCP, improves motor neuron degeneration induced by knockdown of TBPH,
Am J Neurodegener Dis 2018;7(1):11-31 www.ajnd.us /ISSN:2165-591X/AJND0069303 Original Article Overexpression of ter94, Drosophila VCP, improves motor neuron degeneration induced by knockdown of TBPH,
Correspondence of D. melanogaster and C. elegans developmental stages revealed by alternative splicing characteristics of conserved exons
 Gao and Li BMC Genomics (2017) 18:234 DOI 10.1186/s12864-017-3600-2 RESEARCH ARTICLE Open Access Correspondence of D. melanogaster and C. elegans developmental stages revealed by alternative splicing characteristics
Gao and Li BMC Genomics (2017) 18:234 DOI 10.1186/s12864-017-3600-2 RESEARCH ARTICLE Open Access Correspondence of D. melanogaster and C. elegans developmental stages revealed by alternative splicing characteristics
Chapter 11. Development: Differentiation and Determination
 KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
Procedure to Create NCBI KOGS
 Procedure to Create NCBI KOGS full details in: Tatusov et al (2003) BMC Bioinformatics 4:41. 1. Detect and mask typical repetitive domains Reason: masking prevents spurious lumping of non-orthologs based
Procedure to Create NCBI KOGS full details in: Tatusov et al (2003) BMC Bioinformatics 4:41. 1. Detect and mask typical repetitive domains Reason: masking prevents spurious lumping of non-orthologs based
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Chapter 18 Lecture. Concepts of Genetics. Tenth Edition. Developmental Genetics
 Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Midterm 1. Average score: 74.4 Median score: 77
 Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Related Courses He who asks is a fool for five minutes, but he who does not ask remains a fool forever.
 CSE 527 Computational Biology http://www.cs.washington.edu/527 Lecture 1: Overview & Bio Review Autumn 2004 Larry Ruzzo Related Courses He who asks is a fool for five minutes, but he who does not ask remains
CSE 527 Computational Biology http://www.cs.washington.edu/527 Lecture 1: Overview & Bio Review Autumn 2004 Larry Ruzzo Related Courses He who asks is a fool for five minutes, but he who does not ask remains
Lecture 7. Development of the Fruit Fly Drosophila
 BIOLOGY 205/SECTION 7 DEVELOPMENT- LILJEGREN Lecture 7 Development of the Fruit Fly Drosophila 1. The fruit fly- a highly successful, specialized organism a. Quick life cycle includes three larval stages
BIOLOGY 205/SECTION 7 DEVELOPMENT- LILJEGREN Lecture 7 Development of the Fruit Fly Drosophila 1. The fruit fly- a highly successful, specialized organism a. Quick life cycle includes three larval stages
Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are:
 Comparative genomics and proteomics Species available Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are: Vertebrates: human, chimpanzee, mouse, rat,
Comparative genomics and proteomics Species available Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are: Vertebrates: human, chimpanzee, mouse, rat,
SALS, a WH2-Domain-Containing Protein, Promotes Sarcomeric Actin Filament Elongation from Pointed Ends during Drosophila Muscle Growth
 Article SALS, a WH2-Domain-Containing Protein, Promotes Sarcomeric Actin Filament Elongation from Pointed Ends during Drosophila Muscle Growth Jianwu Bai, 1, * John H. Hartwig, 3 and Norbert Perrimon 1,2,
Article SALS, a WH2-Domain-Containing Protein, Promotes Sarcomeric Actin Filament Elongation from Pointed Ends during Drosophila Muscle Growth Jianwu Bai, 1, * John H. Hartwig, 3 and Norbert Perrimon 1,2,
Computational Structural Bioinformatics
 Computational Structural Bioinformatics ECS129 Instructor: Patrice Koehl http://koehllab.genomecenter.ucdavis.edu/teaching/ecs129 koehl@cs.ucdavis.edu Learning curve Math / CS Biology/ Chemistry Pre-requisite
Computational Structural Bioinformatics ECS129 Instructor: Patrice Koehl http://koehllab.genomecenter.ucdavis.edu/teaching/ecs129 koehl@cs.ucdavis.edu Learning curve Math / CS Biology/ Chemistry Pre-requisite
18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis
 18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis An organism arises from a fertilized egg cell as the result of three interrelated processes: cell division, cell
18.4 Embryonic development involves cell division, cell differentiation, and morphogenesis An organism arises from a fertilized egg cell as the result of three interrelated processes: cell division, cell
Genes, Development, and Evolution
 14 Genes, Development, and Evolution Chapter 14 Genes, Development, and Evolution Key Concepts 14.1 Development Involves Distinct but Overlapping Processes 14.2 Changes in Gene Expression Underlie Cell
14 Genes, Development, and Evolution Chapter 14 Genes, Development, and Evolution Key Concepts 14.1 Development Involves Distinct but Overlapping Processes 14.2 Changes in Gene Expression Underlie Cell
Predicting Protein Functions and Domain Interactions from Protein Interactions
 Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/5/e1602426/dc1 Supplementary Materials for In vivo mapping of tissue- and subcellular-specific proteomes in Caenorhabditis elegans The PDF file includes: Aaron
advances.sciencemag.org/cgi/content/full/3/5/e1602426/dc1 Supplementary Materials for In vivo mapping of tissue- and subcellular-specific proteomes in Caenorhabditis elegans The PDF file includes: Aaron
Genomics and bioinformatics summary. Finding genes -- computer searches
 Genomics and bioinformatics summary 1. Gene finding: computer searches, cdnas, ESTs, 2. Microarrays 3. Use BLAST to find homologous sequences 4. Multiple sequence alignments (MSAs) 5. Trees quantify sequence
Genomics and bioinformatics summary 1. Gene finding: computer searches, cdnas, ESTs, 2. Microarrays 3. Use BLAST to find homologous sequences 4. Multiple sequence alignments (MSAs) 5. Trees quantify sequence
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1
 Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Figure 1: To test the role of mir-17~92 in orthologous genetic model of ADPKD, we generated Ksp/Cre;Pkd1 F/F (Pkd1-KO) and Ksp/Cre;Pkd1 F/F ;mir-17~92 F/F (Pkd1-miR-17~92KO) mice. (A) Q-PCR
Supplementary Information. Recognition of the pre-mirna structure by Drosophila Dicer-1
 Supplementary Information Recognition of the pre-mirna structure by Drosophila Dicer-1 Akihisa Tsutsumi 1,2, Tomoko Kawamata 1, Natsuko Izumi 1 Hervé Seitz 3,4 and Yukihide Tomari 1,2 Nature Structural
Supplementary Information Recognition of the pre-mirna structure by Drosophila Dicer-1 Akihisa Tsutsumi 1,2, Tomoko Kawamata 1, Natsuko Izumi 1 Hervé Seitz 3,4 and Yukihide Tomari 1,2 Nature Structural
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network
 Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network Sohyun Hwang 1, Seung Y Rhee 2, Edward M Marcotte 3,4 & Insuk Lee 1 protocol 1 Department of
Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network Sohyun Hwang 1, Seung Y Rhee 2, Edward M Marcotte 3,4 & Insuk Lee 1 protocol 1 Department of
Introduction to Bioinformatics
 Introduction to Bioinformatics Lecture : p he biological problem p lobal alignment p Local alignment p Multiple alignment 6 Background: comparative genomics p Basic question in biology: what properties
Introduction to Bioinformatics Lecture : p he biological problem p lobal alignment p Local alignment p Multiple alignment 6 Background: comparative genomics p Basic question in biology: what properties
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and
 Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Genomes and Their Evolution
 Chapter 21 Genomes and Their Evolution PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Chapter 21 Genomes and Their Evolution PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Germline stem cell differentiation entails regional control of cell fate. regulator GLD-1 in Caenorhabditis elegans
 Genetics: Early Online, published on January 12, 216 as 1.1534/genetics.115.185678 Germline stem cell differentiation entails regional control of cell fate regulator GLD-1 in Caenorhabditis elegans John
Genetics: Early Online, published on January 12, 216 as 1.1534/genetics.115.185678 Germline stem cell differentiation entails regional control of cell fate regulator GLD-1 in Caenorhabditis elegans John
SUPPLEMENTARY INFORMATION
 Supplementary Notes Downregulation of atlastin does not affect secretory traffic We investigated whether Atlastin might play a role in secretory traffic. Traffic impairment results in disruption of Golgi
Supplementary Notes Downregulation of atlastin does not affect secretory traffic We investigated whether Atlastin might play a role in secretory traffic. Traffic impairment results in disruption of Golgi
Caenorhabditis elegans
 Caenorhabditis elegans Why C. elegans? Sea urchins have told us much about embryogenesis. They are suited well for study in the lab; however, they do not tell us much about the genetics involved in embryogenesis.
Caenorhabditis elegans Why C. elegans? Sea urchins have told us much about embryogenesis. They are suited well for study in the lab; however, they do not tell us much about the genetics involved in embryogenesis.
Taxonomy. Content. How to determine & classify a species. Phylogeny and evolution
 Taxonomy Content Why Taxonomy? How to determine & classify a species Domains versus Kingdoms Phylogeny and evolution Why Taxonomy? Classification Arrangement in groups or taxa (taxon = group) Nomenclature
Taxonomy Content Why Taxonomy? How to determine & classify a species Domains versus Kingdoms Phylogeny and evolution Why Taxonomy? Classification Arrangement in groups or taxa (taxon = group) Nomenclature
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A)
 Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Supporting Information
 Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
Supplementary text for the section Interactions conserved across species: can one select the conserved interactions?
 1 Supporting Information: What Evidence is There for the Homology of Protein-Protein Interactions? Anna C. F. Lewis, Nick S. Jones, Mason A. Porter, Charlotte M. Deane Supplementary text for the section
1 Supporting Information: What Evidence is There for the Homology of Protein-Protein Interactions? Anna C. F. Lewis, Nick S. Jones, Mason A. Porter, Charlotte M. Deane Supplementary text for the section
Geert Geeven. April 14, 2010
 iction of Gene Regulatory Interactions NDNS+ Workshop April 14, 2010 Today s talk - Outline Outline Biological Background Construction of Predictors The main aim of my project is to better understand the
iction of Gene Regulatory Interactions NDNS+ Workshop April 14, 2010 Today s talk - Outline Outline Biological Background Construction of Predictors The main aim of my project is to better understand the
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Proteomics. Yeast two hybrid. Proteomics - PAGE techniques. Data obtained. What is it?
 Proteomics What is it? Reveal protein interactions Protein profiling in a sample Yeast two hybrid screening High throughput 2D PAGE Automatic analysis of 2D Page Yeast two hybrid Use two mating strains
Proteomics What is it? Reveal protein interactions Protein profiling in a sample Yeast two hybrid screening High throughput 2D PAGE Automatic analysis of 2D Page Yeast two hybrid Use two mating strains
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila
 Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
Development 143: doi: /dev : Supplementary information
 Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Bioinformatics 2. Yeast two hybrid. Proteomics. Proteomics
 GENOME Bioinformatics 2 Proteomics protein-gene PROTEOME protein-protein METABOLISM Slide from http://www.nd.edu/~networks/ Citrate Cycle Bio-chemical reactions What is it? Proteomics Reveal protein Protein
GENOME Bioinformatics 2 Proteomics protein-gene PROTEOME protein-protein METABOLISM Slide from http://www.nd.edu/~networks/ Citrate Cycle Bio-chemical reactions What is it? Proteomics Reveal protein Protein
T H E J O U R N A L O F C E L L B I O L O G Y
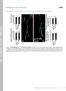 Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Supplemental material Kasprowicz et al., http://www.jcb.org/cgi/content/full/jcb.201310090/dc1 T H E J O U R N A L O F C E L L B I O L O G Y Figure S1. NMJ morphology of shi 12-12B ; shi-4c not treated
Comparing Genomes! Homologies and Families! Sequence Alignments!
 Comparing Genomes! Homologies and Families! Sequence Alignments! Allows us to achieve a greater understanding of vertebrate evolution! Tells us what is common and what is unique between different species
Comparing Genomes! Homologies and Families! Sequence Alignments! Allows us to achieve a greater understanding of vertebrate evolution! Tells us what is common and what is unique between different species
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10923 Supplementary Figure 1 Ten-a and Ten-m antibody and cell type specificities. a c, Representative single confocal sections of a Drosophila NMJ stained with antibodies to Ten-a (red),
doi:10.1038/nature10923 Supplementary Figure 1 Ten-a and Ten-m antibody and cell type specificities. a c, Representative single confocal sections of a Drosophila NMJ stained with antibodies to Ten-a (red),
Hox genes and evolution of body plan
 Hox genes and evolution of body plan Prof. LS Shashidhara Indian Institute of Science Education and Research (IISER), Pune ls.shashidhara@iiserpune.ac.in 2009 marks 150 years since Darwin and Wallace proposed
Hox genes and evolution of body plan Prof. LS Shashidhara Indian Institute of Science Education and Research (IISER), Pune ls.shashidhara@iiserpune.ac.in 2009 marks 150 years since Darwin and Wallace proposed
Lesson Overview. Gene Regulation and Expression. Lesson Overview Gene Regulation and Expression
 13.4 Gene Regulation and Expression THINK ABOUT IT Think of a library filled with how-to books. Would you ever need to use all of those books at the same time? Of course not. Now picture a tiny bacterium
13.4 Gene Regulation and Expression THINK ABOUT IT Think of a library filled with how-to books. Would you ever need to use all of those books at the same time? Of course not. Now picture a tiny bacterium
Small RNA in rice genome
 Vol. 45 No. 5 SCIENCE IN CHINA (Series C) October 2002 Small RNA in rice genome WANG Kai ( 1, ZHU Xiaopeng ( 2, ZHONG Lan ( 1,3 & CHEN Runsheng ( 1,2 1. Beijing Genomics Institute/Center of Genomics and
Vol. 45 No. 5 SCIENCE IN CHINA (Series C) October 2002 Small RNA in rice genome WANG Kai ( 1, ZHU Xiaopeng ( 2, ZHONG Lan ( 1,3 & CHEN Runsheng ( 1,2 1. Beijing Genomics Institute/Center of Genomics and
GFP GAL bp 3964 bp
 Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Identification and functional analysis of novel genes involved in Drosophila germ cell development
 Identification and functional analysis of novel genes involved in Drosophila germ cell development Summary of the Ph.D. thesis László Dániel Henn Supervisor: Dr. Miklós Erdélyi Biological Research Centre,
Identification and functional analysis of novel genes involved in Drosophila germ cell development Summary of the Ph.D. thesis László Dániel Henn Supervisor: Dr. Miklós Erdélyi Biological Research Centre,
Supplemental Figures S1 S5
 Beyond reduction of atherosclerosis: PON2 provides apoptosis resistance and stabilizes tumor cells Ines Witte (1), Sebastian Altenhöfer (1), Petra Wilgenbus (1), Julianna Amort (1), Albrecht M. Clement
Beyond reduction of atherosclerosis: PON2 provides apoptosis resistance and stabilizes tumor cells Ines Witte (1), Sebastian Altenhöfer (1), Petra Wilgenbus (1), Julianna Amort (1), Albrecht M. Clement
Network Biology-part II
 Network Biology-part II Jun Zhu, Ph. D. Professor of Genomics and Genetic Sciences Icahn Institute of Genomics and Multi-scale Biology The Tisch Cancer Institute Icahn Medical School at Mount Sinai New
Network Biology-part II Jun Zhu, Ph. D. Professor of Genomics and Genetic Sciences Icahn Institute of Genomics and Multi-scale Biology The Tisch Cancer Institute Icahn Medical School at Mount Sinai New
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools. Evaluation. Course Homepage.
 CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools Giri Narasimhan ECS 389; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs06.html 1/12/06 CAP5510/CGS5166 1 Evaluation
CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools Giri Narasimhan ECS 389; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs06.html 1/12/06 CAP5510/CGS5166 1 Evaluation
10-810: Advanced Algorithms and Models for Computational Biology. microrna and Whole Genome Comparison
 10-810: Advanced Algorithms and Models for Computational Biology microrna and Whole Genome Comparison Central Dogma: 90s Transcription factors DNA transcription mrna translation Proteins Central Dogma:
10-810: Advanced Algorithms and Models for Computational Biology microrna and Whole Genome Comparison Central Dogma: 90s Transcription factors DNA transcription mrna translation Proteins Central Dogma:
BIOINFORMATICS LAB AP BIOLOGY
 BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
According to the diagram, which of the following is NOT true?
 Instructions: Review Chapter 44 on muscular-skeletal systems and locomotion, and then complete the following Blackboard activity. This activity will introduce topics that will be covered in the next few
Instructions: Review Chapter 44 on muscular-skeletal systems and locomotion, and then complete the following Blackboard activity. This activity will introduce topics that will be covered in the next few
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplemental Data. Wang et al. (2014). Plant Cell /tpc
 Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
The Emergence of Modularity in Biological Systems
 The Emergence of Modularity in Biological Systems Zhenyu Wang Dec. 2007 Abstract: Modularity is a ubiquitous phenomenon in various biological systems, both in genotype and in phenotype. Biological modules,
The Emergence of Modularity in Biological Systems Zhenyu Wang Dec. 2007 Abstract: Modularity is a ubiquitous phenomenon in various biological systems, both in genotype and in phenotype. Biological modules,
Deciphering regulatory networks by promoter sequence analysis
 Bioinformatics Workshop 2009 Interpreting Gene Lists from -omics Studies Deciphering regulatory networks by promoter sequence analysis Elodie Portales-Casamar University of British Columbia www.cisreg.ca
Bioinformatics Workshop 2009 Interpreting Gene Lists from -omics Studies Deciphering regulatory networks by promoter sequence analysis Elodie Portales-Casamar University of British Columbia www.cisreg.ca
Exam 1 ID#: October 4, 2007
 Biology 4361 Name: KEY Exam 1 ID#: October 4, 2007 Multiple choice (one point each) (1-25) 1. The process of cells forming tissues and organs is called a. morphogenesis. b. differentiation. c. allometry.
Biology 4361 Name: KEY Exam 1 ID#: October 4, 2007 Multiple choice (one point each) (1-25) 1. The process of cells forming tissues and organs is called a. morphogenesis. b. differentiation. c. allometry.
bi lecture 17 integration of genetic interaction data
 bi190 2013 lecture 17 integration of genetic interaction data Genes interact to specify phenotype 20,000 genes 200,000,000 potential binary interactions (2 locus physiological epistasis) How will we efficiently
bi190 2013 lecture 17 integration of genetic interaction data Genes interact to specify phenotype 20,000 genes 200,000,000 potential binary interactions (2 locus physiological epistasis) How will we efficiently
Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290
 Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290 Question (from Introduction): How does svb control the
Shavenbaby Couples Patterning to Epidermal Cell Shape Control. Chanut-Delalande H, Fernandes I, Roch F, Payre F, Plaza S (2006) PLoS Biol 4(9): e290 Question (from Introduction): How does svb control the
11/24/13. Science, then, and now. Computational Structural Bioinformatics. Learning curve. ECS129 Instructor: Patrice Koehl
 Computational Structural Bioinformatics ECS129 Instructor: Patrice Koehl http://www.cs.ucdavis.edu/~koehl/teaching/ecs129/index.html koehl@cs.ucdavis.edu Learning curve Math / CS Biology/ Chemistry Pre-requisite
Computational Structural Bioinformatics ECS129 Instructor: Patrice Koehl http://www.cs.ucdavis.edu/~koehl/teaching/ecs129/index.html koehl@cs.ucdavis.edu Learning curve Math / CS Biology/ Chemistry Pre-requisite
Measuring TF-DNA interactions
 Measuring TF-DNA interactions How is Biological Complexity Achieved? Mediated by Transcription Factors (TFs) 2 Regulation of Gene Expression by Transcription Factors TF trans-acting factors TF TF TF TF
Measuring TF-DNA interactions How is Biological Complexity Achieved? Mediated by Transcription Factors (TFs) 2 Regulation of Gene Expression by Transcription Factors TF trans-acting factors TF TF TF TF
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
** * * * Col-0 cau1 CAU1. Actin2 CAS. Actin2. Supplemental Figure 1. CAU1 affects calcium accumulation.
 Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo
 Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Role of Mitochondrial Remodeling in Programmed Cell Death in
 Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Supplementary Information
 Supplementary Information Supplementary figures % Occupancy 90 80 70 60 50 40 30 20 Wt tol-1(nr2033) Figure S1. Avoidance behavior to S. enterica was not observed in wild-type or tol-1(nr2033) mutant nematodes.
Supplementary Information Supplementary figures % Occupancy 90 80 70 60 50 40 30 20 Wt tol-1(nr2033) Figure S1. Avoidance behavior to S. enterica was not observed in wild-type or tol-1(nr2033) mutant nematodes.
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Drosophila Life Cycle
 Drosophila Life Cycle 1 Early Drosophila Cleavage Nuclei migrate to periphery after 10 nuclear divisions. Cellularization occurs when plasma membrane folds in to divide nuclei into cells. Drosophila Superficial
Drosophila Life Cycle 1 Early Drosophila Cleavage Nuclei migrate to periphery after 10 nuclear divisions. Cellularization occurs when plasma membrane folds in to divide nuclei into cells. Drosophila Superficial
Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain
 Development 127, 367-379 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV5342 367 Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain Wei Du
Development 127, 367-379 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV5342 367 Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain Wei Du
Graph Alignment and Biological Networks
 Graph Alignment and Biological Networks Johannes Berg http://www.uni-koeln.de/ berg Institute for Theoretical Physics University of Cologne Germany p.1/12 Networks in molecular biology New large-scale
Graph Alignment and Biological Networks Johannes Berg http://www.uni-koeln.de/ berg Institute for Theoretical Physics University of Cologne Germany p.1/12 Networks in molecular biology New large-scale
Solutions to Problem Set 4
 Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated
 Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
Modulation of central pattern generator output by peripheral sensory cells in Drosophila larvae. BioNB4910 Cornell University.
 Modulation of central pattern generator output by peripheral sensory cells in Drosophila larvae BioNB4910 Cornell University Goals 1) Observe the behavioral effects of remotely activating different populations
Modulation of central pattern generator output by peripheral sensory cells in Drosophila larvae BioNB4910 Cornell University Goals 1) Observe the behavioral effects of remotely activating different populations
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST
 Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Big Idea 1 Evolution INVESTIGATION 3 COMPARING DNA SEQUENCES TO UNDERSTAND EVOLUTIONARY RELATIONSHIPS WITH BLAST How can bioinformatics be used as a tool to determine evolutionary relationships and to
Genetic Engineering for Species Conservation Applications in Hawai'i
 Genetic Engineering for Species Conservation Applications in Hawai'i Floyd A. Reed Department of Biology, University of Hawai'i at Mānoa, Honolulu, HI, USA http://www.hawaiireedlab.com/presentations Culex
Genetic Engineering for Species Conservation Applications in Hawai'i Floyd A. Reed Department of Biology, University of Hawai'i at Mānoa, Honolulu, HI, USA http://www.hawaiireedlab.com/presentations Culex
Welcome to BIOL 572: Recombinant DNA techniques
 Lecture 1: 1 Welcome to BIOL 572: Recombinant DNA techniques Agenda 1: Introduce yourselves Agenda 2: Course introduction Agenda 3: Some logistics for BIOL 572 Agenda 4: Q&A section Agenda 1: Introduce
Lecture 1: 1 Welcome to BIOL 572: Recombinant DNA techniques Agenda 1: Introduce yourselves Agenda 2: Course introduction Agenda 3: Some logistics for BIOL 572 Agenda 4: Q&A section Agenda 1: Introduce
BIS &003 Answers to Assigned Problems May 23, Week /18.6 How would you distinguish between an enhancer and a promoter?
 Week 9 Study Questions from the textbook: 6 th Edition: Chapter 19-19.6, 19.7, 19.15, 19.17 OR 7 th Edition: Chapter 18-18.6 18.7, 18.15, 18.17 19.6/18.6 How would you distinguish between an enhancer and
Week 9 Study Questions from the textbook: 6 th Edition: Chapter 19-19.6, 19.7, 19.15, 19.17 OR 7 th Edition: Chapter 18-18.6 18.7, 18.15, 18.17 19.6/18.6 How would you distinguish between an enhancer and
Gene Ontology and Functional Enrichment. Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein
 Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
Gene Ontology and Functional Enrichment Genome 559: Introduction to Statistical and Computational Genomics Elhanan Borenstein The parsimony principle: A quick review Find the tree that requires the fewest
robustness: revisting the significance of mirna-mediated regulation
 : revisting the significance of mirna-mediated regulation Hervé Seitz IGH (CNRS), Montpellier, France October 13, 2012 microrna target identification .. microrna target identification mir: target: 2 7
: revisting the significance of mirna-mediated regulation Hervé Seitz IGH (CNRS), Montpellier, France October 13, 2012 microrna target identification .. microrna target identification mir: target: 2 7
Proteomics. Areas of Interest
 Introduction to BioMEMS & Medical Microdevices Proteomics and Protein Microarrays Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
Introduction to BioMEMS & Medical Microdevices Proteomics and Protein Microarrays Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
CRISPRseek Workshop Design of target-specific guide RNAs in CRISPR-Cas9 genome-editing systems
 April 2008 CRISPRseek Workshop Design of target-specific guide RNAs in CRISPR-Cas9 genome-editing systems Lihua Julie Zhu Sept 10th 2014 Michael Brodsky Jianhong Ou INSTALLATION First install R 3.1.0 Windows:
April 2008 CRISPRseek Workshop Design of target-specific guide RNAs in CRISPR-Cas9 genome-editing systems Lihua Julie Zhu Sept 10th 2014 Michael Brodsky Jianhong Ou INSTALLATION First install R 3.1.0 Windows:
Evolution and Development Evo-Devo
 Evolution and Development Evo-Devo Darwin wrote a book on barnacles. Plate 1 (Lepas), from A monograph on the sub-class Cirripedia, by Charles Darwin. Comparative embryology There is an obvious similarity
Evolution and Development Evo-Devo Darwin wrote a book on barnacles. Plate 1 (Lepas), from A monograph on the sub-class Cirripedia, by Charles Darwin. Comparative embryology There is an obvious similarity
Cryo-EM data collection, refinement and validation statistics
 1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
