CRISPRseek Workshop Design of target-specific guide RNAs in CRISPR-Cas9 genome-editing systems
|
|
|
- Pierce Perkins
- 5 years ago
- Views:
Transcription
1 April 2008 CRISPRseek Workshop Design of target-specific guide RNAs in CRISPR-Cas9 genome-editing systems Lihua Julie Zhu Sept 10th 2014 Michael Brodsky Jianhong Ou
2 INSTALLATION First install R Windows: Mac OS X: Source (Linux): Then obtain Bioconductor 2.14 by starting R and entering the commands source(" bioclite() To install additional package, e.g., CRISPRseek, type: bioclite("crisprseek") Installed at ghpcc06 cluster and rstudio.umassmed.edu
3 AVAILABLE IN RSTUDIO SERVER AT UMASS CRISPRseek package has been installed in the Rstudio server at Umass. To access it, please send an to the following list. Also need to sign up for a HPCC account at Once your access is granted, please validate that you indeed can access the Rstudio by login at rstudio.umassmed.edu using your username and password in a web browser.
4 RUN CRISPRSEEK IN YOUR OWN INSTALLATION source(" bioclite("crisprseek") The human annotation packages need to be installed to run the example code in the vignette. bioclite("bsgenome.hsapiens.ucsc.hg19") bioclite("txdb.hsapiens.ucsc.hg19.knowngene") bioclite("org.hs.eg.db")
5 GENOME SEQUENCE PACKAGES (68) BSgenome BSgenome BSgenome.Hsapiens.UCSC.hg19 BSgenome.Hsapiens.UCSC.hg19.masked BSgenome.Mmusculus.UCSC.mm10 BSgenome.Mmusculus.UCSC.mm10.masked BSgenome.Dmelanogaster.UCSC.dm3 BSgenome.Dmelanogaster.UCSC.dm3.masked BSgenome.Rnorvegicus.UCSC.rn5 BSgenome.Rnorvegicus.UCSC.rn5.masked BSgenome.Drerio.UCSC.danRer7 BSgenome.Drerio.UCSC.danRer7.masked BSgenome.Celegans.UCSC.ce10 BSgenome.Athaliana.TAIR.TAIR9 BSgenome.Scerevisiae.UCSC.sacCer3 Description Full genome sequences for Homo sapiens (UCSC version hg19) Full masked genome sequences for Homo sapiens (UCSC version hg19) Full genome sequences for Mus musculus (UCSC version mm10) Full masked genome sequences for Mus musculus (UCSC version mm10) Full genome sequences for Drosophila melanogaster (UCSC version dm3) Full masked genome sequences for Drosophila melanogaster (UCSC version dm3) Full genome sequences for Rattus norvegicus (UCSC version rn5) Full masked genome sequences for Rattus norvegicus (UCSC version rn5) Full genome sequences for Danio rerio (UCSC version danrer7) Full masked genome sequences for Danio rerio (UCSC version danrer7) Full genome sequences for Caenorhabditis elegans (UCSC version ce10) Full genome sequences for Arabidopsis thaliana (TAIR9) Saccharomyces cerevisiae (Yeast) full genome (UCSC version saccer3)
6 TRANSCRIPT PACKAGES (18) AnnotationData txdb Description TxDb.Athaliana.BioMart.plantsmart21 Athaliana genes TxDb.Celegans.UCSC.ce6.ensGene Worm genes TxDb.Dmelanogaster.UCSC.dm3.ensGene Fly genes TxDb.Hsapiens.UCSC.hg18.knownGene Human hg18 genes TxDb.Hsapiens.UCSC.hg19.knownGene Human hg19 genes Human hg19 TxDb.Hsapiens.UCSC.hg19.lincRNAsTranscripts lincrnas TxDb.Mmusculus.UCSC.mm10.knownGene Mouse mm10 genes TxDb.Mmusculus.UCSC.mm9.knownGene Mouse mm9 genes TxDb.Rnorvegicus.UCSC.rn4.ensGene Rat rn4 TxDb.Rnorvegicus.UCSC.rn5.refGene Rat rn5 TxDb.Scerevisiae.UCSC.sacCer2.sgdGene Yeast Cer2 TxDb.Scerevisiae.UCSC.sacCer3.sgdGene Yeaset Cer3 To create additional TxDb object maketranscriptdbfromucsc and maketranscriptdbfromgff
7 Genome Annotation Packages (19) BiocViews.html# OrgDb Genome Annotation Package Genome organn org.at.tair.db Arabidopsis org.at.tairsymbol org.bt.eg.db Bovine org.bt.egsymbol org.ce.eg.db Worm org.ce.egsymbol org.cf.eg.db Canine org.cf.egsymbol org.dm.eg.db Fly org.dm.egflybase2eg org.dr.eg.db Zebrafish org.dr.egsymbol org.gg.eg.db Chicken org.gg.egsymbol org.hs.eg.db Human org.hs.egsymbol org.mm.eg.db Mouse org.mm.egsymbol org.mmu.eg.db Rhesus org.mmu.egsymbol org.pt.eg.db Chimp org.pt.egsymbol org.rn.eg.db Rat org.rn.egsymbol org.sc.sgd.db Yeast org.sc.sgdgenename org.ss.eg.db Pig org.ss.egsymbol org.xl.eg.db Xenopus org.xl.egsymbol
8 INSTALL COMMON ANNOTATION PACKAGES IN YOUR OWN INSTALLATION source(" bioclite("bsgenome.hsapiens.ucsc.hg19") bioclite("bsgenome.mmusculus.ucsc.mm10") bioclite("bsgenome.rnorvegicus.ucsc.rn5") bioclite("bsgenome.drerio.ucsc.danrer7") bioclite("bsgenome.dmelanogaster.ucsc.dm3") bioclite("bsgenome.celegans.ucsc.ce6 ) bioclite("txdb.hsapiens.ucsc.hg19.knowngene") bioclite("txdb.mmusculus.ucsc.mm10.knowngene ) bioclite("txdb.rnorvegicus.ucsc.rn5.refgene ) bioclite("txdb.dmelanogaster.ucsc.dm3.ensgene ) bioclite("txdb.celegans.ucsc.ce6.ensgene") - bioclite("org.hs.eg.db") - bioclite("org.mm.eg.db") - bioclite("org.dm.eg.db") - bioclite("org.ce.eg.db ) - bioclite("org.dr.eg.db") - bioclite("org.rn.eg.db")
9 REQUEST TO INSTALL ADDITIONAL PACKAGES IN UMASS SERVERS rstudio.umassmed.edu ghpcc06
10 MAIN FUNCTIONS OF CRISPRSEEK offtargetanalysis workflow grna searching and off-target analysis for one or a set of input sequences compare2sequences workflow Identify grnas that specifically target one of the two input sequences or both
11 offtargetanalysis Input Sequence (fasta or fastq) Find grnas Input Genome (BSgenome) Filter grnas Search for off-targets txdb organn annotateexon = TRUE Annotate target and off-targets Calculate cleavage score for off-targets Filter offtargets Fetch flanking sequences Generate report
12 DEFAULT PARAMETERS OFFTARGETANALYSIS offtargetanalysis(inputfilepath, format = "fasta", findgrnas = TRUE, exportallgrnas = c("all", "fasta", "genbank", "no"), findgrnaswithrecutonly = TRUE, REpatternFile, minrepatternsize = 6, overlap.grna.positions = c(17, 18), findpairedgrnaonly = TRUE, min.gap = 0, max.gap = 20, grna.name.prefix = "grna", PAM.size = 3, grna.size = 20, PAM = "NGG", BSgenomeName, chromtosearch = "all", max.mismatch = 4, PAM.pattern = "N[A G]G$", grna.pattern = "", min.score = 0.5, topn = 100, topn.offtargettotalscore = 10, annotateexon = TRUE, txdb, organn, outputdir, fetchsequence = TRUE, upstream = 200, downstream = 200, weights = c(0, 0, 0.014, 0, 0, 0.395, 0.317, 0, 0.389, 0.079, 0.445, 0.508, 0.613, 0.851, 0.732, 0.828, 0.615, 0.804, 0.685, 0.583), overwrite = FALSE) Default setting for CRISPR-cas9 system in S. pyogenes guide sequence /grna in CRISPRseek CCACTGTGTGCACTTCATCCTGG PAM sequence
13 OUTPUT FILES OF OFFTARGETANALYSIS grnas (genbank, fasta) Restriction Site Overlap (tab delimited) Paired Sites (tab delimited) Off-Target sites (tab delimited) Summary (tab delimited)
14 compare2sequences: Identify grnas that specifically target one of the two sequences or both Input Sequence 1 (fasta or fastq) Find grnas Input Sequence 2 (fasta or fastq) grnas for Sequence 1 Filter grnas grnas for Sequence 2 Search for off-targets in sequence 2 Search for off-targets in sequence 1 Calculate cleavage score for off-targets Generate report
15 DEFAULT PARAMETERS COMPARE2SEQUENCES compare2sequences(inputfile1path, inputfile2path, format = "fasta", findgrnaswithrecutonly = FALSE, REpatternFile, minrepatternsize = 6, overlap.grna.positions = c(17, 18), findpairedgrnaonly = FALSE, min.gap = 0, max.gap = 20, grna.name.prefix = "grna", PAM.size = 3, grna.size = 20, PAM = "NGG", PAM.pattern = "N[A G]G$", max.mismatch =4, outputdir, weights = c(0, 0, 0.014, 0, 0, 0.395, 0.317, 0, 0.389, 0.079, 0.445, 0.508, 0.613, 0.851, 0.732, 0.828, 0.615, 0.804, 0.685, 0.583), overwrite = FALSE) args(compare2sequences)
16 OUTPUT FILES Scores For 2 Input Sequences (tab delimited) grnas (genbank, fasta) Restriction Site Overlap (tab delimited) Paired Sites (tab delimited)
17 compare2sequences might output no match or more than one match to the alternative input sequence for each grnas identified for each input sequence depending on max.mismatch (default is 4 mismatches allowed) Solution Tuning of Maximum Mismatch Allowed Sort the output by grnapluspam and scorediff to examine possible multiple off-target sites in the alternative sequence, if you aim to identify grnas to target one of the two input sequences only. Tune max.mismatch
18 REFERENCE AND HELP CRISPRseek.pdf In a R session browsevignettes( CRISPRseek )?offtargetanalysis?compare2sequences Zhu LJ*, Holmes BR, Aronin N and Brodsky MH*. (2010) [* denotes cocorresponding author] CRISPRseek: a Bioconductor package to identify targetspecific guide RNAs for CRISPR-Cas9 genome-editing systems. PloS One (In press) bioconductor bioconductor@stat.math.ethz.ch
19 DEMO & EXERCISE
Procedure to Create NCBI KOGS
 Procedure to Create NCBI KOGS full details in: Tatusov et al (2003) BMC Bioinformatics 4:41. 1. Detect and mask typical repetitive domains Reason: masking prevents spurious lumping of non-orthologs based
Procedure to Create NCBI KOGS full details in: Tatusov et al (2003) BMC Bioinformatics 4:41. 1. Detect and mask typical repetitive domains Reason: masking prevents spurious lumping of non-orthologs based
The MANTiS Manual. Contents. MANTiS Version 1.1
 The MANTiS Manual MANTiS Version 1.1 Contents Connection to the MANTiS database... 2 Memory settings... 2 Main functionalities... 2 Character Mapping View... 4 Genome content View... 5 Biological processes
The MANTiS Manual MANTiS Version 1.1 Contents Connection to the MANTiS database... 2 Memory settings... 2 Main functionalities... 2 Character Mapping View... 4 Genome content View... 5 Biological processes
Computational Structural Bioinformatics
 Computational Structural Bioinformatics ECS129 Instructor: Patrice Koehl http://koehllab.genomecenter.ucdavis.edu/teaching/ecs129 koehl@cs.ucdavis.edu Learning curve Math / CS Biology/ Chemistry Pre-requisite
Computational Structural Bioinformatics ECS129 Instructor: Patrice Koehl http://koehllab.genomecenter.ucdavis.edu/teaching/ecs129 koehl@cs.ucdavis.edu Learning curve Math / CS Biology/ Chemistry Pre-requisite
Master Biomedizin ) UCSC & UniProt 2) Homology 3) MSA 4) Phylogeny. Pablo Mier
 Master Biomedizin 2018 1) UCSC & UniProt 2) Homology 3) MSA 4) 1 12 a. All of the sequences in file1.fasta (https://cbdm.uni-mainz.de/mb18/) are homologs. How many groups of orthologs would you say there
Master Biomedizin 2018 1) UCSC & UniProt 2) Homology 3) MSA 4) 1 12 a. All of the sequences in file1.fasta (https://cbdm.uni-mainz.de/mb18/) are homologs. How many groups of orthologs would you say there
Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are:
 Comparative genomics and proteomics Species available Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are: Vertebrates: human, chimpanzee, mouse, rat,
Comparative genomics and proteomics Species available Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are: Vertebrates: human, chimpanzee, mouse, rat,
Unit 5: Cell Division and Development Guided Reading Questions (45 pts total)
 Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 12 The Cell Cycle Unit 5: Cell Division and Development Guided
Name: AP Biology Biology, Campbell and Reece, 7th Edition Adapted from chapter reading guides originally created by Lynn Miriello Chapter 12 The Cell Cycle Unit 5: Cell Division and Development Guided
RNA- seq read mapping
 RNA- seq read mapping Pär Engström SciLifeLab RNA- seq workshop October 216 IniDal steps in RNA- seq data processing 1. Quality checks on reads 2. Trim 3' adapters (opdonal (for species with a reference
RNA- seq read mapping Pär Engström SciLifeLab RNA- seq workshop October 216 IniDal steps in RNA- seq data processing 1. Quality checks on reads 2. Trim 3' adapters (opdonal (for species with a reference
Multiple Sequence Alignments
 Multiple Sequence Alignments...... Elements of Bioinformatics Spring, 2003 Tom Carter http://astarte.csustan.edu/ tom/ March, 2003 1 Sequence Alignments Often, we would like to make direct comparisons
Multiple Sequence Alignments...... Elements of Bioinformatics Spring, 2003 Tom Carter http://astarte.csustan.edu/ tom/ March, 2003 1 Sequence Alignments Often, we would like to make direct comparisons
11/24/13. Science, then, and now. Computational Structural Bioinformatics. Learning curve. ECS129 Instructor: Patrice Koehl
 Computational Structural Bioinformatics ECS129 Instructor: Patrice Koehl http://www.cs.ucdavis.edu/~koehl/teaching/ecs129/index.html koehl@cs.ucdavis.edu Learning curve Math / CS Biology/ Chemistry Pre-requisite
Computational Structural Bioinformatics ECS129 Instructor: Patrice Koehl http://www.cs.ucdavis.edu/~koehl/teaching/ecs129/index.html koehl@cs.ucdavis.edu Learning curve Math / CS Biology/ Chemistry Pre-requisite
ECOL/MCB 320 and 320H Genetics
 ECOL/MCB 320 and 320H Genetics Instructors Dr. C. William Birky, Jr. Dept. of Ecology and Evolutionary Biology Lecturing on Molecular genetics Transmission genetics Population and evolutionary genetics
ECOL/MCB 320 and 320H Genetics Instructors Dr. C. William Birky, Jr. Dept. of Ecology and Evolutionary Biology Lecturing on Molecular genetics Transmission genetics Population and evolutionary genetics
Pyrobayes: an improved base caller for SNP discovery in pyrosequences
 Pyrobayes: an improved base caller for SNP discovery in pyrosequences Aaron R Quinlan, Donald A Stewart, Michael P Strömberg & Gábor T Marth Supplementary figures and text: Supplementary Figure 1. The
Pyrobayes: an improved base caller for SNP discovery in pyrosequences Aaron R Quinlan, Donald A Stewart, Michael P Strömberg & Gábor T Marth Supplementary figures and text: Supplementary Figure 1. The
Comparing Genomes! Homologies and Families! Sequence Alignments!
 Comparing Genomes! Homologies and Families! Sequence Alignments! Allows us to achieve a greater understanding of vertebrate evolution! Tells us what is common and what is unique between different species
Comparing Genomes! Homologies and Families! Sequence Alignments! Allows us to achieve a greater understanding of vertebrate evolution! Tells us what is common and what is unique between different species
Advanced Cell Biology. Lecture 2
 Advanced Cell Biology. Lecture 2 Alexey Shipunov Minot State University January 13, 2012 Outline Questions and answers Microscopy Prokaryotic and eukaryotic cells Outline Questions and answers Microscopy
Advanced Cell Biology. Lecture 2 Alexey Shipunov Minot State University January 13, 2012 Outline Questions and answers Microscopy Prokaryotic and eukaryotic cells Outline Questions and answers Microscopy
CGS 5991 (2 Credits) Bioinformatics Tools
 CAP 5991 (3 Credits) Introduction to Bioinformatics CGS 5991 (2 Credits) Bioinformatics Tools Giri Narasimhan 8/26/03 CAP/CGS 5991: Lecture 1 1 Course Schedules CAP 5991 (3 credit) will meet every Tue
CAP 5991 (3 Credits) Introduction to Bioinformatics CGS 5991 (2 Credits) Bioinformatics Tools Giri Narasimhan 8/26/03 CAP/CGS 5991: Lecture 1 1 Course Schedules CAP 5991 (3 credit) will meet every Tue
Gene mention normalization in full texts using GNAT and LINNAEUS
 Gene mention normalization in full texts using GNAT and LINNAEUS Illés Solt 1,2, Martin Gerner 3, Philippe Thomas 2, Goran Nenadic 4, Casey M. Bergman 3, Ulf Leser 2, Jörg Hakenberg 5 1 Department of Telecommunications
Gene mention normalization in full texts using GNAT and LINNAEUS Illés Solt 1,2, Martin Gerner 3, Philippe Thomas 2, Goran Nenadic 4, Casey M. Bergman 3, Ulf Leser 2, Jörg Hakenberg 5 1 Department of Telecommunications
Small RNA in rice genome
 Vol. 45 No. 5 SCIENCE IN CHINA (Series C) October 2002 Small RNA in rice genome WANG Kai ( 1, ZHU Xiaopeng ( 2, ZHONG Lan ( 1,3 & CHEN Runsheng ( 1,2 1. Beijing Genomics Institute/Center of Genomics and
Vol. 45 No. 5 SCIENCE IN CHINA (Series C) October 2002 Small RNA in rice genome WANG Kai ( 1, ZHU Xiaopeng ( 2, ZHONG Lan ( 1,3 & CHEN Runsheng ( 1,2 1. Beijing Genomics Institute/Center of Genomics and
Proteins: Structure & Function. Ulf Leser
 Proteins: Structure & Function Ulf Leser This Lecture Proteins Structure Function Databases Predicting Protein Secondary Structure Many figures from Zvelebil, M. and Baum, J. O. (2008). "Understanding
Proteins: Structure & Function Ulf Leser This Lecture Proteins Structure Function Databases Predicting Protein Secondary Structure Many figures from Zvelebil, M. and Baum, J. O. (2008). "Understanding
Heuristic Methods. Heuristic methods for alignment Sequence databases Multiple alignment Gene and protein prediction
 Heuristic methods for alignment Sequence databases Multiple alignment Gene and protein prediction Armstrong, 2010 Heuristic Methods! FASTA! BLAST! Gapped BLAST! PSI-BLAST Armstrong, 2010 1 Assumptions
Heuristic methods for alignment Sequence databases Multiple alignment Gene and protein prediction Armstrong, 2010 Heuristic Methods! FASTA! BLAST! Gapped BLAST! PSI-BLAST Armstrong, 2010 1 Assumptions
GEP Annotation Report
 GEP Annotation Report Note: For each gene described in this annotation report, you should also prepare the corresponding GFF, transcript and peptide sequence files as part of your submission. Student name:
GEP Annotation Report Note: For each gene described in this annotation report, you should also prepare the corresponding GFF, transcript and peptide sequence files as part of your submission. Student name:
A Browser for Pig Genome Data
 A Browser for Pig Genome Data Thomas Mailund January 2, 2004 This report briefly describe the blast and alignment data available at http://www.daimi.au.dk/ mailund/pig-genome/ hits.html. The report describes
A Browser for Pig Genome Data Thomas Mailund January 2, 2004 This report briefly describe the blast and alignment data available at http://www.daimi.au.dk/ mailund/pig-genome/ hits.html. The report describes
Supplemental Figure 1.
 Supplemental Material: Annu. Rev. Genet. 2015. 49:213 42 doi: 10.1146/annurev-genet-120213-092023 A Uniform System for the Annotation of Vertebrate microrna Genes and the Evolution of the Human micrornaome
Supplemental Material: Annu. Rev. Genet. 2015. 49:213 42 doi: 10.1146/annurev-genet-120213-092023 A Uniform System for the Annotation of Vertebrate microrna Genes and the Evolution of the Human micrornaome
Neural Network Prediction of Translation Initiation Sites in Eukaryotes: Perspectives for EST and Genome analysis.
 Neural Network Prediction of Translation Initiation Sites in Eukaryotes: Perspectives for EST and Genome analysis. Anders Gorm Pedersen and Henrik Nielsen Center for Biological Sequence Analysis The Technical
Neural Network Prediction of Translation Initiation Sites in Eukaryotes: Perspectives for EST and Genome analysis. Anders Gorm Pedersen and Henrik Nielsen Center for Biological Sequence Analysis The Technical
MegAlign Pro Pairwise Alignment Tutorials
 MegAlign Pro Pairwise Alignment Tutorials All demo data for the following tutorials can be found in the MegAlignProAlignments.zip archive here. Tutorial 1: Multiple versus pairwise alignments 1. Extract
MegAlign Pro Pairwise Alignment Tutorials All demo data for the following tutorials can be found in the MegAlignProAlignments.zip archive here. Tutorial 1: Multiple versus pairwise alignments 1. Extract
GATA family of transcription factors of vertebrates: phylogenetics and chromosomal synteny
 Phylogenetics and chromosomal synteny of the GATAs 1273 GATA family of transcription factors of vertebrates: phylogenetics and chromosomal synteny CHUNJIANG HE, HANHUA CHENG* and RONGJIA ZHOU* Department
Phylogenetics and chromosomal synteny of the GATAs 1273 GATA family of transcription factors of vertebrates: phylogenetics and chromosomal synteny CHUNJIANG HE, HANHUA CHENG* and RONGJIA ZHOU* Department
Reassessing Domain Architecture Evolution of Metazoan Proteins: Major Impact of Gene Prediction Errors
 Genes 2011, 2, 449-501; doi:10.3390/genes2030449 Article OPEN ACCESS genes ISSN 2073-4425 www.mdpi.com/journal/genes Reassessing Domain Architecture Evolution of Metazoan Proteins: Major Impact of Gene
Genes 2011, 2, 449-501; doi:10.3390/genes2030449 Article OPEN ACCESS genes ISSN 2073-4425 www.mdpi.com/journal/genes Reassessing Domain Architecture Evolution of Metazoan Proteins: Major Impact of Gene
ST-Links. SpatialKit. Version 3.0.x. For ArcMap. ArcMap Extension for Directly Connecting to Spatial Databases. ST-Links Corporation.
 ST-Links SpatialKit For ArcMap Version 3.0.x ArcMap Extension for Directly Connecting to Spatial Databases ST-Links Corporation www.st-links.com 2012 Contents Introduction... 3 Installation... 3 Database
ST-Links SpatialKit For ArcMap Version 3.0.x ArcMap Extension for Directly Connecting to Spatial Databases ST-Links Corporation www.st-links.com 2012 Contents Introduction... 3 Installation... 3 Database
ATLAS of Biochemistry
 ATLAS of Biochemistry USER GUIDE http://lcsb-databases.epfl.ch/atlas/ CONTENT 1 2 3 GET STARTED Create your user account NAVIGATE Curated KEGG reactions ATLAS reactions Pathways Maps USE IT! Fill a gap
ATLAS of Biochemistry USER GUIDE http://lcsb-databases.epfl.ch/atlas/ CONTENT 1 2 3 GET STARTED Create your user account NAVIGATE Curated KEGG reactions ATLAS reactions Pathways Maps USE IT! Fill a gap
Hands-On Nine The PAX6 Gene and Protein
 Hands-On Nine The PAX6 Gene and Protein Main Purpose of Hands-On Activity: Using bioinformatics tools to examine the sequences, homology, and disease relevance of the Pax6: a master gene of eye formation.
Hands-On Nine The PAX6 Gene and Protein Main Purpose of Hands-On Activity: Using bioinformatics tools to examine the sequences, homology, and disease relevance of the Pax6: a master gene of eye formation.
Genome Sequencing & DNA Sequence Analysis
 7.91 / 7.36 / BE.490 Lecture #1 Feb. 24, 2004 Genome Sequencing & DNA Sequence Analysis Chris Burge What is a Genome? A genome is NOT a bag of proteins What s in the Human Genome? Outline of Unit II: DNA/RNA
7.91 / 7.36 / BE.490 Lecture #1 Feb. 24, 2004 Genome Sequencing & DNA Sequence Analysis Chris Burge What is a Genome? A genome is NOT a bag of proteins What s in the Human Genome? Outline of Unit II: DNA/RNA
Introduction to protein alignments
 Introduction to protein alignments Comparative Analysis of Proteins Experimental evidence from one or more proteins can be used to infer function of related protein(s). Gene A Gene X Protein A compare
Introduction to protein alignments Comparative Analysis of Proteins Experimental evidence from one or more proteins can be used to infer function of related protein(s). Gene A Gene X Protein A compare
Bio2. Heuristics, Databases ; Multiple Sequence Alignment ; Gene Finding. Biological Databases (sequences) Armstrong, 2007 Bioinformatics 2
 Bio2 Heuristics, Databases ; Multiple Sequence Alignment ; Gene Finding Biological Databases (sequences) 1 Biological Databases Introduction to Sequence Databases Overview of primary query tools and the
Bio2 Heuristics, Databases ; Multiple Sequence Alignment ; Gene Finding Biological Databases (sequences) 1 Biological Databases Introduction to Sequence Databases Overview of primary query tools and the
Account Setup. STEP 1: Create Enhanced View Account
 SpyMeSatGov Access Guide - Android DigitalGlobe Imagery Enhanced View How to setup, search and download imagery from DigitalGlobe utilizing NGA s Enhanced View license Account Setup SpyMeSatGov uses a
SpyMeSatGov Access Guide - Android DigitalGlobe Imagery Enhanced View How to setup, search and download imagery from DigitalGlobe utilizing NGA s Enhanced View license Account Setup SpyMeSatGov uses a
19/10/2015. Response to environmental manipulation. Its not as simple as it seems!
 Jackie Mitchell Department of Basic and Clinical Neuroscience - IoPPN Why do we need model genetic systems? What does it take to be a good model? Common in vivo model species Genetic manipulations (Lecture
Jackie Mitchell Department of Basic and Clinical Neuroscience - IoPPN Why do we need model genetic systems? What does it take to be a good model? Common in vivo model species Genetic manipulations (Lecture
Genome-Wide Computational Prediction and Analysis of Core Promoter Elements across Plant Monocots and Dicots
 Genome-Wide Computational Prediction and Analysis of Core Promoter Elements across Plant Monocots and Dicots Sunita Kumari 1, Doreen Ware 1,2 * 1 Cold Spring Harbor Laboratory, Cold Spring Harbor, New
Genome-Wide Computational Prediction and Analysis of Core Promoter Elements across Plant Monocots and Dicots Sunita Kumari 1, Doreen Ware 1,2 * 1 Cold Spring Harbor Laboratory, Cold Spring Harbor, New
Inparanoid: a comprehensive database of eukaryotic orthologs
 D476 D480 Nucleic Acids Research, 2005, Vol. 33, Database issue doi:10.1093/nar/gki107 Inparanoid: a comprehensive database of eukaryotic orthologs Kevin P. O Brien, Maido Remm 1 and Erik L. L. Sonnhammer*
D476 D480 Nucleic Acids Research, 2005, Vol. 33, Database issue doi:10.1093/nar/gki107 Inparanoid: a comprehensive database of eukaryotic orthologs Kevin P. O Brien, Maido Remm 1 and Erik L. L. Sonnhammer*
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools. Evaluation. Course Homepage.
 CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools Giri Narasimhan ECS 389; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs06.html 1/12/06 CAP5510/CGS5166 1 Evaluation
CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools Giri Narasimhan ECS 389; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs06.html 1/12/06 CAP5510/CGS5166 1 Evaluation
Comparing whole genomes
 BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
10-810: Advanced Algorithms and Models for Computational Biology. microrna and Whole Genome Comparison
 10-810: Advanced Algorithms and Models for Computational Biology microrna and Whole Genome Comparison Central Dogma: 90s Transcription factors DNA transcription mrna translation Proteins Central Dogma:
10-810: Advanced Algorithms and Models for Computational Biology microrna and Whole Genome Comparison Central Dogma: 90s Transcription factors DNA transcription mrna translation Proteins Central Dogma:
New Universal Rules of Eukaryotic Translation Initiation Fidelity
 New Universal Rules of Eukaryotic Translation Initiation Fidelity Hadas Zur 1, Tamir Tuller 2,3 * 1 The Blavatnik School of Computer Science, Tel Aviv University, Tel-Aviv, Israel, 2 Department of Biomedical
New Universal Rules of Eukaryotic Translation Initiation Fidelity Hadas Zur 1, Tamir Tuller 2,3 * 1 The Blavatnik School of Computer Science, Tel Aviv University, Tel-Aviv, Israel, 2 Department of Biomedical
Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development
 Supplementary Information: Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development Krishanpal Karmodiya 1, Krishanpal Anamika 1,2, Vijaykumar
Supplementary Information: Camello, a novel family of Histone Acetyltransferases that acetylate histone H4 and is essential for zebrafish development Krishanpal Karmodiya 1, Krishanpal Anamika 1,2, Vijaykumar
Synteny Portal Documentation
 Synteny Portal Documentation Synteny Portal is a web application portal for visualizing, browsing, searching and building synteny blocks. Synteny Portal provides four main web applications: SynCircos,
Synteny Portal Documentation Synteny Portal is a web application portal for visualizing, browsing, searching and building synteny blocks. Synteny Portal provides four main web applications: SynCircos,
Graph Alignment and Biological Networks
 Graph Alignment and Biological Networks Johannes Berg http://www.uni-koeln.de/ berg Institute for Theoretical Physics University of Cologne Germany p.1/12 Networks in molecular biology New large-scale
Graph Alignment and Biological Networks Johannes Berg http://www.uni-koeln.de/ berg Institute for Theoretical Physics University of Cologne Germany p.1/12 Networks in molecular biology New large-scale
RELATIONSHIPS BETWEEN GENES/PROTEINS HOMOLOGUES
 Molecular Biology-2018 1 Definitions: RELATIONSHIPS BETWEEN GENES/PROTEINS HOMOLOGUES Heterologues: Genes or proteins that possess different sequences and activities. Homologues: Genes or proteins that
Molecular Biology-2018 1 Definitions: RELATIONSHIPS BETWEEN GENES/PROTEINS HOMOLOGUES Heterologues: Genes or proteins that possess different sequences and activities. Homologues: Genes or proteins that
CSE P 590 A Spring : MLE, EM
 CSE P 590 A Spring 2013 4: MLE, EM 1 Outline HW#2 Discussion MLE: Maximum Likelihood Estimators EM: the Expectation Maximization Algorithm Next: Motif description & discovery 2 HW # 2 Discussion Species
CSE P 590 A Spring 2013 4: MLE, EM 1 Outline HW#2 Discussion MLE: Maximum Likelihood Estimators EM: the Expectation Maximization Algorithm Next: Motif description & discovery 2 HW # 2 Discussion Species
Quantitative Measurement of Genome-wide Protein Domain Co-occurrence of Transcription Factors
 Quantitative Measurement of Genome-wide Protein Domain Co-occurrence of Transcription Factors Arli Parikesit, Peter F. Stadler, Sonja J. Prohaska Bioinformatics Group Institute of Computer Science University
Quantitative Measurement of Genome-wide Protein Domain Co-occurrence of Transcription Factors Arli Parikesit, Peter F. Stadler, Sonja J. Prohaska Bioinformatics Group Institute of Computer Science University
UNT FOLIOTEK FOR FACULTY FALL Alyssa Strong Foliotek Administrator
 UNT FOLIOTEK FOR FACULTY FALL 2018 Alyssa Strong Foliotek Administrator Alyssa.Strong@unt.edu 940-369-5157 Agenda What is Foliotek? Courses and Rubrics in Foliotek Best Practices Logging in Canvas/Blackboard
UNT FOLIOTEK FOR FACULTY FALL 2018 Alyssa Strong Foliotek Administrator Alyssa.Strong@unt.edu 940-369-5157 Agenda What is Foliotek? Courses and Rubrics in Foliotek Best Practices Logging in Canvas/Blackboard
Cryo-EM data collection, refinement and validation statistics
 1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
1 Table S1 Cryo-EM data collection, refinement and validation statistics Data collection and processing CPSF-160 WDR33 (EMDB-7114) (PDB 6BM0) CPSF-160 WDR33 (EMDB-7113) (PDB 6BLY) CPSF-160 WDR33 CPSF-30
microrna Studies Chen-Hanson Ting SVFIG June 23, 2018
 microrna Studies Chen-Hanson Ting SVFIG June 23, 2018 Summary MicroRNA (mirna) Species and organisms studied mirna in mitocondria Huge genome files mirna in human Chromosome 1 mirna in bacteria Tools used
microrna Studies Chen-Hanson Ting SVFIG June 23, 2018 Summary MicroRNA (mirna) Species and organisms studied mirna in mitocondria Huge genome files mirna in human Chromosome 1 mirna in bacteria Tools used
Introduction. SAMStat. QualiMap. Conclusions
 Introduction SAMStat QualiMap Conclusions Introduction SAMStat QualiMap Conclusions Where are we? Why QC on mapped sequences Acknowledgment: Fernando García Alcalde The reads may look OK in QC analyses
Introduction SAMStat QualiMap Conclusions Introduction SAMStat QualiMap Conclusions Where are we? Why QC on mapped sequences Acknowledgment: Fernando García Alcalde The reads may look OK in QC analyses
PNmerger: a Cytoscape plugin to merge biological pathways and protein interaction networks
 PNmerger: a Cytoscape plugin to merge biological pathways and protein interaction networks http://www.hupo.org.cn/pnmerger Fuchu He E-mail: hefc@nic.bmi.ac.cn Tel: 86-10-68171208 FAX: 86-10-68214653 Yunping
PNmerger: a Cytoscape plugin to merge biological pathways and protein interaction networks http://www.hupo.org.cn/pnmerger Fuchu He E-mail: hefc@nic.bmi.ac.cn Tel: 86-10-68171208 FAX: 86-10-68214653 Yunping
Browsing Genomic Information with Ensembl Plants
 Browsing Genomic Information with Ensembl Plants Etienne de Villiers, PhD (Adapted from slides by Bert Overduin EMBL-EBI) Outline of workshop Brief introduction to Ensembl Plants History Content Tutorial
Browsing Genomic Information with Ensembl Plants Etienne de Villiers, PhD (Adapted from slides by Bert Overduin EMBL-EBI) Outline of workshop Brief introduction to Ensembl Plants History Content Tutorial
Advanced Algorithms and Models for Computational Biology
 Advanced Algorithms and Models for Computational Biology Introduction to cell biology genomics development and probability Eric ing Lecture January 3 006 Reading: Chap. DTM book Introduction to cell biology
Advanced Algorithms and Models for Computational Biology Introduction to cell biology genomics development and probability Eric ing Lecture January 3 006 Reading: Chap. DTM book Introduction to cell biology
Yes, the Library will be accessible via the new PULSE and the existing desktop version of PULSE.
 F R E Q U E N T L Y A S K E D Q U E S T I O N S THE LIBRARY GENERAL W H A T I S T H E L I B R A R Y? The Library is the new, shorter, simpler name for the Business Development (Biz Dev) Library. It s your
F R E Q U E N T L Y A S K E D Q U E S T I O N S THE LIBRARY GENERAL W H A T I S T H E L I B R A R Y? The Library is the new, shorter, simpler name for the Business Development (Biz Dev) Library. It s your
Using Base Pairing Probabilities for MiRNA Recognition. Yet Another SVM for MiRNA Recognition: yasmir
 0. Using Base Pairing Probabilities for MiRNA Recognition Yet Another SVM for MiRNA Recognition: yasmir Daniel Pasailă, Irina Mohorianu, Liviu Ciortuz Department of Computer Science Al. I. Cuza University,
0. Using Base Pairing Probabilities for MiRNA Recognition Yet Another SVM for MiRNA Recognition: yasmir Daniel Pasailă, Irina Mohorianu, Liviu Ciortuz Department of Computer Science Al. I. Cuza University,
Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week:
 Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week: Course general information About the course Course objectives Comparative methods: An overview R as language: uses and
Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week: Course general information About the course Course objectives Comparative methods: An overview R as language: uses and
CSE 427 Comp Bio. Sequence Alignment
 CSE 427 Comp Bio Sequence Alignment 1 Sequence Alignment What Why A Dynamic Programming Algorithm 2 Sequence Alignment Goal: position characters in two strings to best line up identical/similar ones with
CSE 427 Comp Bio Sequence Alignment 1 Sequence Alignment What Why A Dynamic Programming Algorithm 2 Sequence Alignment Goal: position characters in two strings to best line up identical/similar ones with
Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network
 Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network Sohyun Hwang 1, Seung Y Rhee 2, Edward M Marcotte 3,4 & Insuk Lee 1 protocol 1 Department of
Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network Sohyun Hwang 1, Seung Y Rhee 2, Edward M Marcotte 3,4 & Insuk Lee 1 protocol 1 Department of
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
CAP 5510: Introduction to Bioinformatics CGS 5166: Bioinformatics Tools
 CAP 5510: to : Tools ECS 254A / EC 2474; Phone x3748; Email: giri@cis.fiu.edu My Homepage: http://www.cs.fiu.edu/~giri http://www.cs.fiu.edu/~giri/teach/bioinfs15.html Office ECS 254 (and EC 2474); Phone:
CAP 5510: to : Tools ECS 254A / EC 2474; Phone x3748; Email: giri@cis.fiu.edu My Homepage: http://www.cs.fiu.edu/~giri http://www.cs.fiu.edu/~giri/teach/bioinfs15.html Office ECS 254 (and EC 2474); Phone:
BLAST Database Searching. BME 110: CompBio Tools Todd Lowe April 8, 2010
 BLAST Database Searching BME 110: CompBio Tools Todd Lowe April 8, 2010 Admin Reading: Read chapter 7, and the NCBI Blast Guide and tutorial http://www.ncbi.nlm.nih.gov/blast/why.shtml Read Chapter 8 for
BLAST Database Searching BME 110: CompBio Tools Todd Lowe April 8, 2010 Admin Reading: Read chapter 7, and the NCBI Blast Guide and tutorial http://www.ncbi.nlm.nih.gov/blast/why.shtml Read Chapter 8 for
Hereditary Hemochromatosis
 Hereditary Hemochromatosis The HFE gene Becky Reese What is hereditary hemochromatosis? Recessively inherited Iron overload disorder Inability to regulate iron absorption No cure http://www.consultant360.com/article/genetics-gastroenterology-what-you-need-know-part-1
Hereditary Hemochromatosis The HFE gene Becky Reese What is hereditary hemochromatosis? Recessively inherited Iron overload disorder Inability to regulate iron absorption No cure http://www.consultant360.com/article/genetics-gastroenterology-what-you-need-know-part-1
You are required to know all terms defined in lecture. EXPLORE THE COURSE WEB SITE 1/6/2010 MENDEL AND MODELS
 1/6/2010 MENDEL AND MODELS!!! GENETIC TERMINOLOGY!!! Essential to the mastery of genetics is a thorough knowledge and understanding of the vocabulary of this science. New terms will be introduced and defined
1/6/2010 MENDEL AND MODELS!!! GENETIC TERMINOLOGY!!! Essential to the mastery of genetics is a thorough knowledge and understanding of the vocabulary of this science. New terms will be introduced and defined
Extensive identification and analysis of conserved small ORFs in animals
 Mackowiak et al. Genome Biology (2015) 16:179 DOI 10.1186/s13059-015-0742-x RESEARCH Open Access Extensive identification and analysis of conserved small ORFs in animals Sebastian D. Mackowiak 1, Henrik
Mackowiak et al. Genome Biology (2015) 16:179 DOI 10.1186/s13059-015-0742-x RESEARCH Open Access Extensive identification and analysis of conserved small ORFs in animals Sebastian D. Mackowiak 1, Henrik
MCB142/ICB163 Overview. Instructors:
 MCB142/ICB163 Overview Instructors: Professor Sharon Amacher Professor Abby Dernburg Professor Monty Slatkin GSIs: Kristin Camfield Nat Hallihan Hana Lee Christine Preston Richard Price Kate Smallenburg
MCB142/ICB163 Overview Instructors: Professor Sharon Amacher Professor Abby Dernburg Professor Monty Slatkin GSIs: Kristin Camfield Nat Hallihan Hana Lee Christine Preston Richard Price Kate Smallenburg
AN ALGORITHM FOR IDENTIFYING CLUSTERS OF FUNCTIONALLY RELATED GENES IN GENOMES. A Thesis GANG MAN YI
 AN ALGORITHM FOR IDENTIFYING CLUSTERS OF FUNCTIONALLY RELATED GENES IN GENOMES A Thesis by GANG MAN YI Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the
AN ALGORITHM FOR IDENTIFYING CLUSTERS OF FUNCTIONALLY RELATED GENES IN GENOMES A Thesis by GANG MAN YI Submitted to the Office of Graduate Studies of Texas A&M University in partial fulfillment of the
Related Courses He who asks is a fool for five minutes, but he who does not ask remains a fool forever.
 CSE 527 Computational Biology http://www.cs.washington.edu/527 Lecture 1: Overview & Bio Review Autumn 2004 Larry Ruzzo Related Courses He who asks is a fool for five minutes, but he who does not ask remains
CSE 527 Computational Biology http://www.cs.washington.edu/527 Lecture 1: Overview & Bio Review Autumn 2004 Larry Ruzzo Related Courses He who asks is a fool for five minutes, but he who does not ask remains
ChemmineR: A Compound Mining Toolkit for Chemical Genomics in R
 ChemmineR: A Compound Mining Toolkit for Chemical Genomics in R Yiqun Cao Anna Charisi Thomas Girke October 28, 2009 Introduction The ChemmineR package includes functions for calculating atom pair descriptors
ChemmineR: A Compound Mining Toolkit for Chemical Genomics in R Yiqun Cao Anna Charisi Thomas Girke October 28, 2009 Introduction The ChemmineR package includes functions for calculating atom pair descriptors
AMS 132: Discussion Section 2
 Prof. David Draper Department of Applied Mathematics and Statistics University of California, Santa Cruz AMS 132: Discussion Section 2 All computer operations in this course will be described for the Windows
Prof. David Draper Department of Applied Mathematics and Statistics University of California, Santa Cruz AMS 132: Discussion Section 2 All computer operations in this course will be described for the Windows
Purpose. Process. Students will locate the species listed below on the infographic and write down the domain to which they belong:
 Purpose This activity gives students practice with interpreting infographics and also supports student understanding of the similarities and differences between humans and other species. Process Students
Purpose This activity gives students practice with interpreting infographics and also supports student understanding of the similarities and differences between humans and other species. Process Students
BSC 4934: QʼBIC Capstone Workshop" Giri Narasimhan. ECS 254A; Phone: x3748
 BSC 4934: QʼBIC Capstone Workshop" Giri Narasimhan ECS 254A; Phone: x3748 giri@cs.fiu.edu http://www.cs.fiu.edu/~giri/teach/bsc4934_su10.html July 2010 7/12/10 Q'BIC Bioinformatics 1 Overview of Course"
BSC 4934: QʼBIC Capstone Workshop" Giri Narasimhan ECS 254A; Phone: x3748 giri@cs.fiu.edu http://www.cs.fiu.edu/~giri/teach/bsc4934_su10.html July 2010 7/12/10 Q'BIC Bioinformatics 1 Overview of Course"
Quantitative and qualitative analyses. of in-paralogs
 Quantitative and qualitative analyses of in-paralogs Dissertation zur Erlangung des naturwissentschaflichen Doktorgrades der Bayerischen Julius-Maximilians Universität Würzburg vorgelegt von Stanislav
Quantitative and qualitative analyses of in-paralogs Dissertation zur Erlangung des naturwissentschaflichen Doktorgrades der Bayerischen Julius-Maximilians Universität Würzburg vorgelegt von Stanislav
General Chemistry Lab Molecular Modeling
 PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
PURPOSE The objectives of this experiment are PROCEDURE General Chemistry Lab Molecular Modeling To learn how to use molecular modeling software, a commonly used tool in chemical research and industry.
Mathangi Thiagarajan Rice Genome Annotation Workshop May 23rd, 2007
 -2 Transcript Alignment Assembly and Automated Gene Structure Improvements Using PASA-2 Mathangi Thiagarajan mathangi@jcvi.org Rice Genome Annotation Workshop May 23rd, 2007 About PASA PASA is an open
-2 Transcript Alignment Assembly and Automated Gene Structure Improvements Using PASA-2 Mathangi Thiagarajan mathangi@jcvi.org Rice Genome Annotation Workshop May 23rd, 2007 About PASA PASA is an open
Standards-Based Quantification in DTSA-II Part II
 Standards-Based Quantification in DTSA-II Part II Nicholas W.M. Ritchie National Institute of Standards and Technology, Gaithersburg, MD 20899-8371 nicholas.ritchie@nist.gov Introduction This article is
Standards-Based Quantification in DTSA-II Part II Nicholas W.M. Ritchie National Institute of Standards and Technology, Gaithersburg, MD 20899-8371 nicholas.ritchie@nist.gov Introduction This article is
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY METHODS Mef2 primary screen. RNAi hairpins from the VDRC collection were crossed to Mef2-GAL4 at 27 C. After 2 weeks lethality rate and stage was scored, and if possible 20-30 males containing
SUPPLEMENTARY METHODS Mef2 primary screen. RNAi hairpins from the VDRC collection were crossed to Mef2-GAL4 at 27 C. After 2 weeks lethality rate and stage was scored, and if possible 20-30 males containing
Model plants and their Role in genetic manipulation. Mitesh Shrestha
 Model plants and their Role in genetic manipulation Mitesh Shrestha Definition of Model Organism Specific species or organism Extensively studied in research laboratories Advance our understanding of Cellular
Model plants and their Role in genetic manipulation Mitesh Shrestha Definition of Model Organism Specific species or organism Extensively studied in research laboratories Advance our understanding of Cellular
CSE 427 Computational Biology Winter Sequence Alignment. Sequence Alignment. Sequence Similarity: What
 CSE 427 Computational Biology Winter 2008 Sequence Alignment; DNA Replication Sequence Alignment Part I Motivation, dynamic programming, global alignment 1 3 Sequence Alignment Sequence Similarity: What
CSE 427 Computational Biology Winter 2008 Sequence Alignment; DNA Replication Sequence Alignment Part I Motivation, dynamic programming, global alignment 1 3 Sequence Alignment Sequence Similarity: What
Towards a Comprehensive Annotation of Structured RNAs in Drosophila
 Towards a Comprehensive Annotation of Structured RNAs in Drosophila Rebecca Kirsch 31st TBI Winterseminar, Bled 20/02/2016 Studying Non-Coding RNAs in Drosophila Why Drosophila? especially for novel molecules
Towards a Comprehensive Annotation of Structured RNAs in Drosophila Rebecca Kirsch 31st TBI Winterseminar, Bled 20/02/2016 Studying Non-Coding RNAs in Drosophila Why Drosophila? especially for novel molecules
G C T G U A. template strand
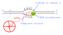 A 5 w 3 3 c 5 mrna RNA polymerase T G C A G C C T G U A coding or sense st AUG?? template strand The triplet code 3 bases = 1 amino acid Punctuation: sta rt: sto p: AUGA CUU CAGUAAC AU U AA C C AUG = methionine,
A 5 w 3 3 c 5 mrna RNA polymerase T G C A G C C T G U A coding or sense st AUG?? template strand The triplet code 3 bases = 1 amino acid Punctuation: sta rt: sto p: AUGA CUU CAGUAAC AU U AA C C AUG = methionine,
Review Article From Ontology to Semantic Similarity: Calculation of Ontology-Based Semantic Similarity
 Hindawi Publishing Corporation The Scientific World Journal Volume 2013, Article ID 793091, 11 pages http://dx.doi.org/10.1155/2013/793091 Review Article From Ontology to Semantic Similarity: Calculation
Hindawi Publishing Corporation The Scientific World Journal Volume 2013, Article ID 793091, 11 pages http://dx.doi.org/10.1155/2013/793091 Review Article From Ontology to Semantic Similarity: Calculation
Molecular Coevolution of the Vertebrate Cytochrome c 1 and Rieske Iron Sulfur Protein in the Cytochrome bc 1 Complex
 Molecular Coevolution of the Vertebrate Cytochrome c 1 and Rieske Iron Sulfur Protein in the Cytochrome bc 1 Complex Kimberly Baer *, David McClellan Department of Integrative Biology, Brigham Young University,
Molecular Coevolution of the Vertebrate Cytochrome c 1 and Rieske Iron Sulfur Protein in the Cytochrome bc 1 Complex Kimberly Baer *, David McClellan Department of Integrative Biology, Brigham Young University,
new interface and features
 Web version of SciFinder : new interface and features Bhawat Ruangying, CAS representative Updated at 22 Dec 2009 www.cas.org SciFinder web interface Technical aspects of SciFinder Web SciFinder URL :
Web version of SciFinder : new interface and features Bhawat Ruangying, CAS representative Updated at 22 Dec 2009 www.cas.org SciFinder web interface Technical aspects of SciFinder Web SciFinder URL :
The following steps will help you to record your work and save and submit it successfully.
 MATH 22AL Lab # 4 1 Objectives In this LAB you will explore the following topics using MATLAB. Properties of invertible matrices. Inverse of a Matrix Explore LU Factorization 2 Recording and submitting
MATH 22AL Lab # 4 1 Objectives In this LAB you will explore the following topics using MATLAB. Properties of invertible matrices. Inverse of a Matrix Explore LU Factorization 2 Recording and submitting
Genome Evolution Greg Lang, Department of Biological Sciences
 Genome Evolution Greg Lang, Department of Biological Sciences BioS 010: Bioscience in the 21st Century Mechanisms of genome evolution Gene Duplication Genome Rearrangement Whole Genome Duplication Gene
Genome Evolution Greg Lang, Department of Biological Sciences BioS 010: Bioscience in the 21st Century Mechanisms of genome evolution Gene Duplication Genome Rearrangement Whole Genome Duplication Gene
Improving Hox Protein Classification across the Major Model Organisms
 Improving Hox Protein Classification across the Major Model Organisms Stefanie D. Hueber, Georg F. Weiller*, Michael A. Djordjevic, Tancred Frickey Genomic Interactions Group, Research School of Biology,
Improving Hox Protein Classification across the Major Model Organisms Stefanie D. Hueber, Georg F. Weiller*, Michael A. Djordjevic, Tancred Frickey Genomic Interactions Group, Research School of Biology,
Homolog. Orthologue. Comparative Genomics. Paralog. What is Comparative Genomics. What is Comparative Genomics
 Orthologue Orthologs are genes in different species that evolved from a common ancestral gene by speciation. Normally, orthologs retain the same function in the course of evolution. Identification of orthologs
Orthologue Orthologs are genes in different species that evolved from a common ancestral gene by speciation. Normally, orthologs retain the same function in the course of evolution. Identification of orthologs
Biased amino acid composition in warm-blooded animals
 Biased amino acid composition in warm-blooded animals Guang-Zhong Wang and Martin J. Lercher Bioinformatics group, Heinrich-Heine-University, Düsseldorf, Germany Among eubacteria and archeabacteria, amino
Biased amino acid composition in warm-blooded animals Guang-Zhong Wang and Martin J. Lercher Bioinformatics group, Heinrich-Heine-University, Düsseldorf, Germany Among eubacteria and archeabacteria, amino
PRIDE Cluster: building the consensus of proteomics data
 Supplementary Materials PRIDE Cluster: building the consensus of proteomics data Johannes Griss, Joseph Michael Foster, Henning Hermjakob and Juan Antonio Vizcaíno EMBL-European Bioinformatics Institute,
Supplementary Materials PRIDE Cluster: building the consensus of proteomics data Johannes Griss, Joseph Michael Foster, Henning Hermjakob and Juan Antonio Vizcaíno EMBL-European Bioinformatics Institute,
SUPPLEMENTARY INFORMATION
 Fig. 1 Influences of crystal lattice contacts on Pol η structures. a. The dominant lattice contact between two hpol η molecules (silver and gold) in the type 1 crystals. b. A close-up view of the hydrophobic
Fig. 1 Influences of crystal lattice contacts on Pol η structures. a. The dominant lattice contact between two hpol η molecules (silver and gold) in the type 1 crystals. b. A close-up view of the hydrophobic
BIOINFORMATICS LAB AP BIOLOGY
 BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
BIOINFORMATICS LAB AP BIOLOGY Bioinformatics is the science of collecting and analyzing complex biological data. Bioinformatics combines computer science, statistics and biology to allow scientists to
CHEMICAL INVENTORY ENTRY GUIDE
 CHEMICAL INVENTORY ENTRY GUIDE Version Date Comments 1 October 2013 Initial A. SUMMARY All chemicals located in research and instructional laboratories at George Mason University are required to be input
CHEMICAL INVENTORY ENTRY GUIDE Version Date Comments 1 October 2013 Initial A. SUMMARY All chemicals located in research and instructional laboratories at George Mason University are required to be input
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/312/5780/1653/dc1 Supporting Online Material for The Xist RNA Gene Evolved in Eutherians by Pseudogenization of a Protein-Coding Gene Laurent Duret,* Corinne Chureau,
www.sciencemag.org/cgi/content/full/312/5780/1653/dc1 Supporting Online Material for The Xist RNA Gene Evolved in Eutherians by Pseudogenization of a Protein-Coding Gene Laurent Duret,* Corinne Chureau,
changes in gene expression, we developed and tested several models. Each model was
 Additional Files Additional File 1 File format: PDF Title: Experimental design and linear models Description: This additional file describes in detail the experimental design and linear models used to
Additional Files Additional File 1 File format: PDF Title: Experimental design and linear models Description: This additional file describes in detail the experimental design and linear models used to
Software BioScout-Calibrator June 2013
 SARAD GmbH BioScout -Calibrator 1 Manual Software BioScout-Calibrator June 2013 SARAD GmbH Tel.: ++49 (0)351 / 6580712 Wiesbadener Straße 10 FAX: ++49 (0)351 / 6580718 D-01159 Dresden email: support@sarad.de
SARAD GmbH BioScout -Calibrator 1 Manual Software BioScout-Calibrator June 2013 SARAD GmbH Tel.: ++49 (0)351 / 6580712 Wiesbadener Straße 10 FAX: ++49 (0)351 / 6580718 D-01159 Dresden email: support@sarad.de
Visualising very long data vectors with the Hilbert curve Description of the Bioconductor packages HilbertVis and HilbertVisGUI.
 Visualising very long data vectors with the Hilbert curve Description of the Bioconductor packages HilbertVis and HilbertVisGUI. Simon Anders European Bioinformatics Institute, Hinxton, Cambridge, UK sanders@fs.tum.de
Visualising very long data vectors with the Hilbert curve Description of the Bioconductor packages HilbertVis and HilbertVisGUI. Simon Anders European Bioinformatics Institute, Hinxton, Cambridge, UK sanders@fs.tum.de
Cascade Support Vector Machines applied to the Translation Initiation Site prediction problem
 Cascade Support Vector Machines applied to the Translation Initiation Site prediction problem Wallison W. Guimarães 1, Cristiano L. N. Pinto 2, Cristiane N. Nobre 1, Luis E. Zárate 1 1 Pontifical Catholic
Cascade Support Vector Machines applied to the Translation Initiation Site prediction problem Wallison W. Guimarães 1, Cristiano L. N. Pinto 2, Cristiane N. Nobre 1, Luis E. Zárate 1 1 Pontifical Catholic
GENE ONTOLOGY (GO) Wilver Martínez Martínez Giovanny Silva Rincón
 GENE ONTOLOGY (GO) Wilver Martínez Martínez Giovanny Silva Rincón What is GO? The Gene Ontology (GO) project is a collaborative effort to address the need for consistent descriptions of gene products in
GENE ONTOLOGY (GO) Wilver Martínez Martínez Giovanny Silva Rincón What is GO? The Gene Ontology (GO) project is a collaborative effort to address the need for consistent descriptions of gene products in
Genetic Engineering for Species Conservation Applications in Hawai'i
 Genetic Engineering for Species Conservation Applications in Hawai'i Floyd A. Reed Department of Biology, University of Hawai'i at Mānoa, Honolulu, HI, USA http://www.hawaiireedlab.com/presentations Culex
Genetic Engineering for Species Conservation Applications in Hawai'i Floyd A. Reed Department of Biology, University of Hawai'i at Mānoa, Honolulu, HI, USA http://www.hawaiireedlab.com/presentations Culex
Isoform discovery and quantification from RNA-Seq data
 Isoform discovery and quantification from RNA-Seq data C. Toffano-Nioche, T. Dayris, Y. Boursin, M. Deloger November 2016 C. Toffano-Nioche, T. Dayris, Y. Boursin, M. Isoform Deloger discovery and quantification
Isoform discovery and quantification from RNA-Seq data C. Toffano-Nioche, T. Dayris, Y. Boursin, M. Deloger November 2016 C. Toffano-Nioche, T. Dayris, Y. Boursin, M. Isoform Deloger discovery and quantification
Comparative studies on sequence characteristics around translation initiation codon in four eukaryotes
 Indian Academy of Sciences RESEARCH NOTE Comparative studies on sequence characteristics around translation initiation codon in four eukaryotes QINGPO LIU and QINGZHONG XUE* Department of Agronomy, College
Indian Academy of Sciences RESEARCH NOTE Comparative studies on sequence characteristics around translation initiation codon in four eukaryotes QINGPO LIU and QINGZHONG XUE* Department of Agronomy, College
