Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis
|
|
|
- Gary Wheeler
- 5 years ago
- Views:
Transcription
1 RESEARCH ARTICLE 5039 Development 138, (2011) doi: /dev Published by The Company of Biologists Ltd Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis Rachappa Balkunde*, Daniel Bouyer*, and Martin Hülskamp SUMMARY Trichome patterning on Arabidopsis leaves is one of the best-studied model systems for two-dimensional de novo patterning. In addition to an activator-inhibitor-related mechanism, we previously proposed a depletion mechanism to operate during this process such that GLABRA3 (GL3) traps the trichome-promoting factor TRANSPARENT TESTA GLABRA1 (TTG1) in trichomes that, in turn, results in a depletion of TTG1 in trichome neighbouring cells. In this manuscript we analyze the molecular basis underlying this trapping mechanism. We demonstrate the ability of GL3 to regulate TTG1 mobility by expressing TTG1 and GL3 in different tissue layers in different combinations. We further show that TTG1 trapping by GL3 is based on direct interaction between both proteins and recruitment in the nucleus. KEY WORDS: TTG1, GL3, Nuclear trapping INTRODUCTION The regular distribution of trichomes on the leaf surface of Arabidopsis is particularly suited to study two-dimensional de novo patterning processes (Hulskamp, 2004; Ishida et al., 2008; Pesch and Hulskamp, 2009). At the basal part of young leaves, trichome cells are singled out from apparently equivalent protodermal cells. Clonal analysis rendered it unlikely that a cell lineage mechanism is responsible for trichome spacing but rather relies on cellular interactions of initially equivalent cells (Larkin et al., 1996; Schnittger et al., 1999). The genetic and molecular analysis of trichome patterning has suggested two distinct patterning processes that are likely to operate in parallel as the key regulators are involved in both of them (Pesch and Hulskamp, 2009). The first mechanism is similar to an activator-inhibitor-based model (for a general introduction, see Meinhardt and Gierer, 2000). According to this model, trichome-promoting factors turn on their own inhibitors that can move into neighbouring cells to mediate lateral inhibition. Three activators, the WD40 protein TRANSPARENT TESTA GLABRA1 (TTG1) (Koornneef, 1981; Galway et al., 1994; Walker et al., 1999), the R2R3 MYB related transcription factor GLABRA1 (GL1) (Oppenheimer et al., 1991) and the helix-loophelix (bhlh)-like transcription factor GLABRA3 (GL3) (Koornneef et al., 1982; Hulskamp et al., 1994; Payne et al., 2000) act together by forming a trimeric transcriptional activator complex (Payne et al., 2000). They trigger the expression of the R3 singlerepeat MYB inhibitor TRIPTYCHON (TRY) and probably also five redundantly acting homologs (Wada et al., 1997; Schellmann et al., 2002; Kirik et al., 2004a; Kirik et al., 2004b; Digiuni et al., 2008; Tominaga et al., 2008; Wang et al., 2008; Wester et al., 2009). Some of the inhibitors were shown to move between cells University of Cologne, Botanical Institute III, Zülpicher Strasse 47b, Köln, Germany. *These authors contributed equally to this work Present address: Institut de Biologie Moléculaire des Plantes du CNRS, BMP-CNRS UPR2357, 12 rue du Général Zimmer, Strasbourg, France Author for correspondence (martin.huelskamp@uni-koeln.de) Accepted 16 September 2011 and can repress the function of the activators by competitive complex formation (Esch et al., 2003; Kurata et al., 2005; Wang et al., 2007; Digiuni et al., 2008; Wester et al., 2009). A second model explains de novo trichome patterning by a depletion of the activator TTG1 in non-trichome cells that is brought about through trapping of TTG1 by GL3 in trichome initials, causing lateral inhibition (Bouyer et al., 2008; Pesch and Hulskamp, 2009). This model is based on several findings. First, TTG1 protein was found to be able to move between cells. As TTG1 movement results in a co-movement of marker molecules that are immobile on their own, a plasmodesmatal transport is postulated (Bouyer et al., 2008). Translational TTG1-YFP fusion protein distribution was shown to be markedly reduced in cells next to trichome initials, though the initial expression pattern exhibited the same level of expression in all epidermal cells of the trichomepatterning zone (Bouyer et al., 2008). This depletion of TTG1 in trichome neighbouring cells was lost in gl3 mutants, suggesting that TTG1 is trapped in trichomes where GL3 is strongly expressed (Bouyer et al., 2008). Most aspects of the activator-depletion model, however, remain speculative. In particular, it is not known whether a direct interaction between GL3 and TTG1 is important or whether depletion is indirectly dependent on GL3. Here, we have analysed the molecular details of the proposed trapping mechanism. We demonstrate that GL3 can regulate TTG1 mobility by expressing TTG1 and GL3 in different tissue layers in different mutant combinations. We further show that GL3 mediates the nuclear localization of TTG1 by direct interaction between GL3 and TTG1, indicating that TTG1 trapping is mediated by intracellular compartmentalization. MATERIALS AND METHODS Constructs TTG1-YFPpEN was described previously (Bouyer et al., 2008). TTG1 C26-YFPpEN and C26TTG1-YFPpEN constructs were created by inverse PCR using primers flanking the deletion region and TTG1- YFPpEN as a template. To create CFP-GL3pEN, the full-length coding sequence of GL3 was amplified by PCR introducing SalI restriction sites and was cloned in pbluescript vector. The SalI fragment was then cloned into pen1a vector digested with SalI and XhoI restriction enzymes to obtain GL3pEN. CFP was cloned at the XmnI site N-terminally to GL3 to create CFP-GL3pEN. CFP-GL3 78pEN and CFP-GL3 NLSpEN were
2 5040 RESEARCH ARTICLE Development 138 (22) created by inverse PCR using primers flanking the deletion region and CFP-GL3pEN as a template. An internal 78 amino acid fragment ( amino acids) from GL3 was amplified by PCR using both forward and reverse primers attached with NcoI restriction sites. The PCR product was cloned N-terminally to GUS at the NcoI restriction site in GUSpEN (Invitrogen) to obtain 78GL3-GUSpEN. TTG1pAMPAT-GW, RbcS2b/RBCpAMPAT-GW and AtML1-GW binary vectors were described previously (Bouyer et al., 2008; Wester et al., 2008). 35S:GL3, 35S:TTG1-YFP, RBC:TTG1-YFP, RBC:GFP-GL3, AtML1:GFP-GL3, 35S:CFP-GL3, 35S:CFP-GL3 78, 35S:CFP- GL3 NLS, 35S:NLS-TTG1-YFP, TTG1:NLS-TTG1-YFP, RBC:NLS- TTG1-YFP, 35S:TTG1 26-YFP and 35S:C26TTG1-YFP constructs were created by gateway LR reaction system (Invitrogen). Plant materials and growth conditions The mutant lines used in this study, ttg1-1, gl3-1, gl3 egl3, gl3 ttg1 (Ler background), ttg1-13 (RLD background) and gl3 egl3 tt8 have been described previously (Koornneef et al., 1982; Walker et al., 1999; Larkin et al., 1999; Payne et al., 2000; Zhang et al., 2003; Bouyer et al., 2008). ttg1 TTG1:TTG1-YFP has been described previously (Bouyer et al., 2008). The ttg1 RBC:TTG1-YFP, gl3 RBC:GFP-GL3, ttg1 gl3 RBC:TTG1-YFP, ttg1 gl3 RBC:GFP-GL3, ttg1 gl3 AtML1:GFP-GL3, Ler 35S:GL3, gl3 egl3 TTG1:TTG1-YFP, gl3 egl3 tt8 TTG1:TTG1-YFP, ttg1 TTG1:NLS- TTG1-YFP and ttg1 RBC:NLS-TTG1-YFP lines were generated by Agrobacterium-mediated transformation in the corresponding backgrounds using the floral dip method described previously (Clough and Bent, 1998). Transformants were selected in the T1 generation on soil using 0.1% BASTA solution. Homozygous lines were used to make the genetic crossings and F1 plants were selfed to identify the progeny with the desired background among the segregating F2 population. gl3 TTG1:TTG1-YFP, 35S:GL3 TTG1:TTG1-YFP, ttg1 gl3 RBC:TTG1-YFP RBC:GFP-GL3 and ttg1 gl3 RBC:TTG1YFP AtML1:GFP-GL3 were created by genetic crossings. Arabidopsis plants were grown on soil at 22 C in continuous light conditions. Yeast two hybrid assay The Saccharomyces cerevisiae strain AH109 was used for the yeast twohybrid assays. Yeast transformation was performed as described before (Gietz et al., 1995). pc-act2 and pas2 plasmids (Clontech) were used for the fusion with GAL4 activation domain and GAL4 DNA-binding domain, respectively. GL3, GL3 78, GL3 NLS, 78GL3-GUS, EGL3, TT8 and GUS were fused to the GAL4 activation domain, and TTG1, TTG1 C26, C26TTG1, 78GL3-GUS and NLS-TTG1 were fused to the DNA-binding domain by the gateway LR reaction system (Invitrogen). Yeast were grown on synthetic dropout media lacking leucine, tryptophan and histidine (Leu /Trp /His ) and supplemented with 5 mm 3-amino- 1,2,4-triazole (3-AT) to analyze the protein-protein interactions. Nuclear transportation trap (NTT) assay Plasmid vectors pnh2 (NES-LexAD) and pnh3 (NES-LexAD-NLS) and pns (modified pnh2) have been described previously (Ueki et al., 1998). pns-ttg1 and pns-ttg1 C26 were constructed by cloning the SalI/XhoI fragments of TTG1 and TTG1 C26 into the SalI restriction site in the pns vector. pns-gl3 and pns-gl3 78 were constructed by cloning the SalI fragment of GL3 and GL3 78 into the SalI restriction site of the pns vector. SalI/Ecl36I and SalI/PvuII fragments of GL3 and GL3 78, respectively, were cloned into the XhoI/PvuII-digested pvt-u vector to obtain pvt-u-gl3 and pvt-u-gl3 78. The nuclear transportation trap (NTT) assay was carried out as described previously (Ueki et al., 1998) using the yeast strain EGY 48 (Clontech). NES-LexAD [pnh2], NES-LexAD-NLS [pnh3], NES-LexAD-TTG1 [pns-ttg1], NES-LexAD-TTG1 C26 [pns-ttg1 C26], NES-LexAD- GL3[pNS-GL3] and NES-LexAD-GL3 78 [pns-gl3 78] plasmids were transformed individually and grown on synthetic dropout media lacking leucine and histidine (Leu /His ) for 4-7 days at 30 C to detect the expression of the LEU2 reporter gene expression for the transport of the fusion protein into the nucleus. Similarly, pns-ttg1 and pns-ttg1 C26 were separately co-transformed with either pvt-u-gl3 or pvt-u-gl3 78. Transient expression The biolistic PDS 1000/He system (Bio-Rad) was used for the transient expression studies. Gold particles (1.0 m) were coated with 300 ng of each DNA and were co-bombarded into onion or Arabidopsis cotyledon epidermal cells with 900 psi rupture discs under a vacuum of 26 inches of Hg. Fluorescence was analyzed hours after the bombardment. Microscopy and quantification of YFP signal Fluorescent images were captured using the Leica TCS-SP2 confocal microscope equipped with the LCS software. Images were made using 40 water immersion objective. The z-stack images were obtained and merged to one plane. Raw images were used for quantifying the YFP fluorescence using the histogram quantification tool of the LCS software. Young rosette leaves were stained with 5 g/ml of propidium iodide (PI) for 1-2 minutes to mark the cell walls. Fluorescent pictures of onion epidermal cells were captured using the LEICA-DMRE microscope equipped with a high-resolution KY-F70 3-CCD JVC camera and DISKUS software. Images were processed with Adobe Photoshop CS2. Scanning Electron Microscopy (SEM) Twelve to 15-day-old seedlings were mounted on stubs with silver adhesive (Electron Microscopy Sciences) and kept cold on ice until viewing in a Hitachi S-3500N environmental scanning electron microscope (ESEM) using high vacuum mode. Images were taken quickly (within 15 minutes) to minimize damage by the electron beam on the live sample, using an accelerating voltage of 5.0 kv and a working distance of mm. RESULTS GL3 counteracts TTG1 mobility The previous finding that TTG1 protein depletion is lost in gl3 mutants led to the hypothesis that TTG1 binding to GL3 leads to a trapping of TTG1 in trichomes due to elevated GL3 levels in these cells (Bouyer et al., 2008; Zhao et al., 2008). One prediction of this hypothesis is that mobility of TTG1 can be altered depending on the presence or absence of GL3. We took advantage of the previous finding that TTG1 can rescue the ttg1 mutant trichome phenotype when expressed in the sub-epidermis. If the hypothesis were correct, one would expect that tissue-specific GL3 expression modulates the rescue efficiency of sub-epidermal expressed TTG1. The experiments were designed to test this in two directions, by either providing GL3 exclusively in the epidermis or in the subepidermis. Does epidermal GL3 promote the rescue by trapping TTG1 in the upper layer? In order to address this question, we tested the rescue ability of sub-epidermal TTG1 in the absence or abundance of epidermal GL3. As shown before using the Table 1. Mobility between cell layers Number of trichomes Genotype on leaves 3 and 4 ±s.d. n Ler 86± gl3 37± ttg ttg1 gl ttg1 RBC:TTG1-YFP 70± ttg1 gl3 RBC:TTG1-YFP 2± gl3 RBC:GFP-GL3 36± ttg1 gl3 RBC:GFP-GL ttg1 gl3 RBC:GFP-GL3 RBC:TTG1-YFP 0 30 ttg1 gl3 AtML1:GFP-GL3 RBC:TTG1-YFP > ttg1 gl3 AtML1:GFP-GL3 70± s.d., standard deviation; n, number of plants analyzed.
3 TTG1 trapping by GL3 RESEARCH ARTICLE 5041 Fig. 1. GL3 controls TTG1 movement. (A- C,F,G,J,K) Scanning electron microscopy micrographs of rosette leaves. (D,E,H,I,L) Confocal images. (A) ttg1 gl3. No trichomes are found. (B) ttg1 RBC:TTG1-YFP. Trichome phenotype is restored. (C) ttg1 gl3 RBC:TTG1-YFP. Weak rescue of trichome phenotype. Arrows indicate trichomes. (D) ttg1 RBC:TTG1-YFP. YFP fluorescence is found in the sub-epidermis, as well as in the epidermis. (E) ttg1 gl3 RBC:TTG1-YFP. YFP fluorescence is found in the sub-epidermis, as well as in the epidermis. (F) ttg1 gl3 RBC:GFP-GL3. No trichomes are found. (G) ttg1 gl3 RBC:GFP-GL3 RBC:TTG1-YFP. No trichomes are found. (H) ttg1 gl3 RBC:GFP-GL3. GFP fluorescence is found exclusively in the sub-epidermis. (I) ttg1 gl3 RBC:GFP-GL3 RBC:TTG1-YFP. No YFP fluorescence is found in the epidermis. (J) ttg1 gl3 AtML1:GFP-GL3. Moderate trichome rescue mostly at the leaf margin. (K) ttg1 gl3 AtML1:GFP-GL3 RBC:TTG1-YFP. Strong over-production of trichomes. (L) ttg1 gl3 AtML1:GFP-GL3. GFP fluorescence is exclusively found in the epidermis. heterologous PPCA1 promoter, sub-epidermal TTG1 expression driven by the widely used Rubisco small subunit 2B promoter (RBC) also rescues the ttg1 mutant trichome phenotype (Table 1; Fig. 1A,B). In these lines, TTG1-YFP is clearly found in both cell layers, the sub-epidermis as well as the epidermis, demonstrating the movement between cell layers (Fig. 1D). The influence of GL3 in the epidermis on TTG1-YFP movement from the sub-epidermis was assayed by studying the rescue ability of sub-epidermal TTG1 in ttg1 gl3 double mutants. If GL3 does not interfere with TTG1 mobility, then ttg1 gl3 double mutants expressing sub-epidermal TTG1 should show the same number of trichomes as seen in gl3 single mutants. However, the rescue of the ttg1 gl3 double mutant by RBC:TTG1-YFP was much less effective. In spite of this, the intercellular transport of TTG1-YFP per se is not affected as seen by the presence of TTG1-YFP fluorescence in all cell layers comparable with the situation in the ttg1 single mutant (Table 1; Fig. 1C,E). Although gl3 mutants show on average 37 trichomes on leaves 3 and 4, the corresponding RBC:TTG1-YFP lines display on average only two trichomes. This suggests that GL3 strongly promotes the rescue efficiency of sub-epidermal TTG1. In order to distinguish between the possibilities that GL3 promotes the rescue either through co-movement from the sub-epidermis and/or by modification of TTG1 function, or by the proposed trapping mechanism, we created several lines in which GFP-GL3 is expressed in the sub-epidermis or the epidermis. GFP-GL3 was found exclusively in the layers in which it was expressed, indicating that it cannot move between the layers in either direction in the leaf. This result also confirms the tissue specificity of the used promoters (Fig. 1H,L). Both, the comovement and modification hypotheses are ruled out by the findings that no rescue is observed in gl3 RBC:GFP-GL3, ttg1 gl3 RBC:GFP-GL3 and ttg1gl3 RBC:TTG1-YFP RBC:GFP- GL3 lines (Table 1; Fig. 1F,G). In fact, when crossing low efficiently rescuing RBC:TTG1-YFP in the gl3 ttg1 double homozygous background together with RBC:GFP-GL3 in the same background, trichome development is completely abolished (Table 1). As the fluorescence of GFP/YFP is exclusively detected in the nuclei of the sub-epidermis (Fig. 1I), this strongly suggests that the presence of GL3 restricts the mobility of TTG1 rather than modifying its function or being co-transported. In a complementary experiment, we increased the epidermal GL3 levels using an epidermis-specific AtML1:GFP-GL3 line. As GL3 is able to partially complement ttg1, these lines show restoration of trichomes mainly at the margin of the leaves (Table 1; Fig. 1J). However, these trichomes show strong distortions as previously described for lines overexpressing GL3 in a ttg1 background (Fig. 1J) (Zhang et al., 2003). When crossing this line with the ttg1 gl3 RBC:TTG1-YFP line, we observed a drastic increase in trichome number, suggesting that GL3 promotes TTG1-triggered sub-epidermal rescue by capturing TTG1 in the epidermis (Table 1; Fig. 1K). Together, these data demonstrate that GL3 modulates TTG1 mobility in a manner consistent with the hypothesis that it traps TTG1 in cells with high GL3 expression. GL3 modulates the intracellular localization of TTG1 In wild-type leaves, TTG1 is typically found in the cytoplasm and the nucleus of epidermal cells (Fig. 2A) (Bouyer et al., 2008; Zhao et al., 2008). This intracellular localization appears to be altered in gl3 mutants or in plants overexpressing GL3. In gl3 mutants, the TTG1-YFP distribution appears to be more diffuse, indicating a shift of the protein from the nucleus to the cytoplasm (Fig. 2C). By contrast, TTG1-YFP appears to be localized mainly into the nucleus in the 35S:GL3 overexpression background (Fig. 2B). A quantitative analysis using confocal laser scanning microscopy (CLSM) confirmed this impression. The intracellular distribution pattern was found to be significantly different with respect to the level of GL3 (Table 2). Whereas, in wild type, 69% of the TTG1- YFP fluorescence was found in the nucleus, gl3 mutants show a statistical significant (P ) reduction to 56% nuclear
4 5042 RESEARCH ARTICLE Development 138 (22) Fig. 2. Cellular distribution pattern of TTG1-YFP fusion protein in the trichome patterning zone in the leaf epidermis. (A-E) The upper row shows TTG-YFP in yellow and propidium iodide staining the cell wall in blue. Trichome cells are marked by red arrowheads. (F-J) The bottom row shows intensity projection images of confocal stacks of TTG1:TTG1-YFP lines. The fluorescence intensity is indicated by size of the peaks. (A,F) ttg1 TTG1:TTG1-YFP. The YFP fluorescence intensity is much weaker in the nuclei of trichome neighbouring cells. (B,G) 35S:GL3 TTG1:TTG1- YFP. YFP fluorescence intensity in trichome neighbouring cells is comparable with that in trichome cells. (C,H) gl3 TTG1:TTG1-YFP. The YFP signal is also found in the cytoplasm and shows similar strength in all cells. (D,I) gl3 egl3 TTG1:TTG1-YFP. YFP signal distribution is similar to that in gl3 (C,H). (E,J) gl3 egl3 tt8 TTG1:TTG1-YFP. YFP signal distribution is similar to that in gl3 (C,H). Scale bars: 10 m. distribution and in 35S:GL3 lines the nuclear localized TTG1-YFP is increased to 89% (Table 2). During the course of these experiments, we noted that 35S:GL3 lines showed a subtle but significant depletion of the TTG1-YFP signal. We found 77% of the trichome fluorescence in the first tier of cells around the trichome initials, 85% in the second and 90% in the third with a significant difference between the first and the other two tiers (ttest, P<0.05) (Fig. 2F-H). Our TTG1 misexpression data in different cell layers indicated that GL3 plays an important role in TTG1 trapping. In order to test the relevance of other bhlh homologs of GL3 in TTG1-YFP localization, we tested also gl3 egl3 double mutants and gl3 egl3 tt8 triple mutants as GL3 and EGL3 act in a partially redundant manner and differ in their expression pattern in leaves; furthermore, TT8 is required for the development of leaf marginal trichomes (Zhang et al., 2003; Zhao et al., 2008). We saw a significant change in the gl3 egl3 double mutant compared with the gl3 single mutant, but the additional removal of TT8 (gl3 egl3 tt8) had no additional effect (Table 2; Fig. 2D,E,I,J). Taken together, we conclude that the intracellular localization of TTG1 depends largely on the bhlh proteins GL3 and EGL3 in a partially redundant manner, which is reflected in the mutant phenotype of single and double mutants of gl3 egl3 (Zhang et al., 2003). GL3 promotes the nuclear transport of TTG1 The regulation of nuclear targeting of GL3 and TTG1 was analysed using a heterologous yeast-based nuclear transportation trap (NTT) assay (Ueki et al., 1998). In this system the protein of interest is expressed as a translational fusion to an artificial transactivator LexAD (consisting of LexA DNA-binding domain and GAL4AD transactivation domain) fused to a nuclear export signal (NES) from HIV Rev protein (NES-LexAD- Protein-ofinterest ). Owing to the presence of the NES, non-nuclear-targeted proteins that lack a functional nuclear localization signal (NLS) are excluded from the nucleus. However, nuclear-targeted proteins can overcome the NES-mediated nuclear export and enter the nucleus, thereby activating the LexAD-responsive LEUCINE2 Table 2. Quantification of nuclear concentration of TTG1-YFP expressed under TTG1 promoter in wild-type, gl3, gl3 egl3, gl3 egl3 tt8 and 35S:GL3 lines Wild type p35s:gl3* gl3 gl3 egl3, gl3 egl3 tt8 #, ** Percentage of total YFP fluorescence in nucleus ±s.d. 69±6.9 89±4.4 56±6.9 48±6.8 49±6.8 n WT, wild type; s.d., standard deviation; n, number of single cells in the patterning zone used for measurement. *The values for 35S:GL3 are statistically significantly different from wild type (P ). The values for gl3 are statistically significantly different from wild type (P ). The values for gl3 egl3 are statistically significantly different from wild type (P ). The values for gl3 egl3 are statistically significantly different from gl3 (P ). # The values for gl3 egl3 tt8 are statistically significantly different from wild type (P ). **The values for gl3 egl3 tt8 are statistically not significantly different from gl3 egl3 (P 0.056). Nuclear YFP fluorescence is expressed as a percentage of the total YFP fluorescence in the cell.
5 TTG1 trapping by GL3 RESEARCH ARTICLE 5043 Fig. 3. Nuclear targeting of TTG1 by GL3. (A,B) Nuclear transportation trap (NTT) assay in yeast. When fusion protein (NES- LexAD- gene-of-interest ) is in the nucleus, the reporter gene is expressed, which allows growth on the medium lacking leucine. (A) NES-LexAD-SV40NLS (2), NES-LexAD-GL3 (3) and NES-LexAD- GL3 78 (4) activates the reporter gene, allowing growth on medium lacking leucine, whereas yeasts containing NES-LexAD (1), NES-LexAD- TTG1 (5) or NES-LexAD-TTG1 C26 (6) are unable to activate the reporter and cannot grow on medium lacking leucine. (B) When coexpressed with GL3, NES-LexAD-TTG1 allows growth on medium lacking leucine (7), whereas co-expression of GL3 78 with NES-LexAD- TTG1 (8), GL3 with NES-LexAD-TTG1 C26 (9) and GL3 78 with NES- LexAD-TTG1 C26 (10) did not result in the activation of reporter and cannot grow on medium lacking leucine. SD-LH, synthetic dropout medium lacking leucine and histidine; SD-LHU, synthetic dropout medium lacking leucine, histidine and uracil. (C-O) Transient expression in Arabidopsis cotyledon epidermal cells. (C-F) Subcellular localization of CFP-GL3 (C), CFP-GL3 78 (D), CFP-GL3 NLS (E) and TTG1-YFP (F). Deletion of TTG1 interaction domain in GL3 (GL3 78) does not affect its subcellular localization; deletion of NLS from GL3 shifts GL3 NLS protein completely to cytoplasm. (G-I) Co-expression of TTG1-YFP (G) and CFP-GL3 (H). The bulk of the TTG1-YFP is accumulated in the nucleus. (J-L) Co-expression of TTG1-YFP (J) and CFP-GL3 78 (K). Subcellular distribution of TTG1-YFP is similar to the situation when TTG1-YFP is expressed alone. (M-O) Co-expression of TTG1-YFP (M) and CFP-GL3 NLS (N). The bulk of the TTG1-YFP is in the cytoplasm and is co-localized with CFP-GL3. Insets in C-E show higher magnification of the nucleus. Scale bars: 50 m. Arrows indicate the position of the nucleus. (LEU2) reporter gene. In this assay, we found that TTG1 fused to the NES did not enter the nucleus, suggesting that it is not actively transported into the nucleus (Fig. 3A). By contrast, GL3 behaved as a nuclear protein when expressed as a fusion to NES in this assay (Fig. 3A). We extended the NTT assay using co-expression of NES-fused proteins and its interaction partners without NES attachment to test their interaction with respect to intracellular distribution. In this experiment, we found that co-expression of NES-LexAD-TTG1 fusion protein with GL3 protein resulted in a re-localization of TTG1 to the nucleus (Fig. 3B). This indicates that the nuclear localization of TTG1 is mediated by its direct interaction with GL3. To demonstrate that the specific interaction between TTG1 and GL3 is responsible for nuclear targeting, we created TTG1 and GL3 variants that disturb their interaction. A TTG1 variant lacking the C-terminal 26 amino acids has been shown to lose the interaction with GL3 (Payne et al., 2000) and can no longer be targeted by GL3 to the nucleus in the yeast NTT assay (Fig. 3B). In the case of GL3, we used a deletion corresponding to an EGL3 deletion previously shown not to interact with TTG1 in yeast two hybrid screens (I. Zimmermann, PhD Thesis, University of Cologne, 2003). The resulting GL3 protein (hereafter referred as GL3 78) has an internal 78 amino acid deletion between amino acids and showed no interaction with TTG1 in yeast twohybrid analysis (Table 3). In the next experiment, we fused this 78 amino acid fragment from GL3 (hereafter referred as 78GL3) N- terminally to GUS in order to ensure stability and found binding to TTG1 in a yeast two-hybrid system. Control experiments revealed no binding of this fragment to GL3 itself, showing that the TTG1- GL3 interaction domain can be separated from the GL3-selfdimerization domain (Table 3) (Payne et al., 2000). Although the GL3 78 variant localizes to the nucleus, it is unable to trigger TTG1 nuclear localization in co-expression assays in yeast (Fig. 3B). These findings were confirmed in plants using transient expression analysis following biolistic transformation of Arabidopsis epidermal cells. In one set of experiments, we coexpressed TTG1-YFP and CFP-GL3 in Arabidopsis cotyledon epidermal cells. While TTG1-YFP alone is localized in the nucleus and the cytoplasm (Fig. 3F), co-expression with CFP-GL3 (Fig. 3G) causes a nuclear localization of the majority of TTG1-YFP (Fig. 3H). CFP-GL3 78 was still found in the nucleus (Fig. 3D,J), but had no influence on the subcellular distribution of TTG1-YFP (Fig. 3K). Here, TTG1-YFP localization was similar to TTG-YFP expressed alone. In order to test whether targeting is mediated by the GL3 NLS signal, we also tested a CFP-marked GL3 variant lacking the NLS nuclear targeting sequence. GL3 NLS still interacts with TTG1 in
6 5044 RESEARCH ARTICLE Development 138 (22) Table 3. Yeast two hybrid interaction between TTG1 and GL3 mutant proteins BD-TTG1 BD-TTG1 C26 BD-78GL3-GUS BD-NLS-TTG1 BD-C26TTG1 AD-GL3 + + AD-GL3 78 n.d. n.d. n.d. AD-78GL3-GUS + n.d. n.d. n.d. AD-GUS n.d. n.d. n.d. AD-GL3 NLS + n.d. n.d. n.d. AD-EGL3 + n.d. + n.d. AD-TT8 + n.d. + n.d. +, positive interactions;, no interaction; BD, GAL4 DNA-binding domain; AD, GAL4 DNA activation domain; n.d., not determined. yeast-two-hybrid assays (Table 3). The CFP-GL3 NLS was found in the cytoplasm (Fig. 3E,M) and co-expression experiments revealed that TTG1-YFP is not targeted to the nucleus anymore (Fig. 3N). The co-expression of TTG1-YFP with CFP-GL3 variants in onion epidermal cells showed the same results (supplementary material Fig. S1). As deletions may cause aberrant protein functions, we aimed to demonstrate the relevance of TTG1-GL3 binding by an aptamer approach (Fig. 4). The aptamer approach uses a small protein fragment to compete with the binding of the two proteins under consideration (Rudolph et al., 2003). For this experiment, we used the 78 amino acids of the GL3 protein known to be relevant for the interaction with TTG1 (Table 3). To ensure the stability of the protein fragment and to enable its visualization, we fused GUS to the C terminus and RFP to the N terminus. This RFP-78GL3-GUS protein was co-expressed with CFP-GL3 and TTG-YFP in onion epidermal cells as described above and the RFP-78GL3-GUS fluorescence was observed in the cytoplasm as expected (Fig. 4B). TTG1-YFP was observed both in the cytoplasm and the nucleus (Fig. 4C), and comparatively more signal was observed in the cytoplasm when compared with experiments carried out using RFP-GUS without the 78GL3 aptamer (Fig. 4D-F). This result suggests that RFP-78GL3- GUS interfered with the ability of GL3 to recruit TTG1-YFP to the nucleus (Fig. 4C). In order to demonstrate that this also occurs in trichomes, we analyzed the distribution of a TTG1-YFP variant not binding to GL3 (Table 3), TTG1 C26-YFP. As expected, TTG1 C26-YFP localization was shifted to the cytoplasm. This redistribution, however, was much more pronounced than in gl3 egl3 double mutants, indicating additional regulatory complexity for the intracellular localization of TTG1 (Fig. 5A). Attempts to show that the deleted 26 amino acids alone can bind to GL3 failed (Table 3; supplementary material Fig. S2) NLS-TTG1 is able to move and does not affect TTG1 depletion In a next step, we aimed to analyze whether nuclear targeting affects the intercellular mobility of TTG1. Towards this end, we created NLS-TTG1-YFP construct. In transient expression assays, the NLS efficiently targeted the fusion protein to the nucleus (Fig. 6A). Protein-protein interactions of NLS-TTG1-YFP with GL3 are indistinguishable from TTG1-YFP in yeast two hybrid interaction assay (Table 3). When this fusion protein was expressed under the TTG1 promoter, ttg1 mutants were rescued, indicating that the NLS fusion does not interfere with TTG1 function (Table 4). To test whether TTG1 mobility between cells is affected by NLS-mediated nuclear targeting, we expressed the NLS-TTG1-YFP fusion protein under the RBC promoter in the sub-epidermis of ttg1 mutants. These plants showed a rescue of the trichome phenotype, indicating that sub-epidermal nuclear-targeted TTG1 can move from the subepidermis into the epidermis. Fig. 4. GL3 fragment (78GL3) competes with GL3 for binding to TTG1 in aptamer approach. (A-C) Co-expression of TTG1-YFP, RFP-78GL3- GUS and CFP-GL3. (A) Localization of CFP-GL3 in the nucleus. (B) Localization of RFP-78GL3-GUS in the cytoplasm and weakly in the nucleus. (C) TTG1-YFP is localized in the cytoplasm and in the nucleus. (D-F) Co-expression of TTG1-YFP, RFP-GUS and CFP-GL3. (D) CFP-GL3 is localized in the nucleus. (E) RFP-GUS is localized in the cytoplasm and weakly in the nucleus. (F) Bulk of the TTG1-YFP is trapped in the nucleus. Inset in B and E shows a higher magnification of the nucleus.
7 TTG1 trapping by GL3 RESEARCH ARTICLE 5045 Fig. 5. TTG1 C26YFP shifts to the cytoplasm. (A) TTG1:TTG1 C26-YFP. FYP fluorescence in the nucleus is drastically reduced as a result of the deletion of the GL3 interaction domain from TTG1. (B) TTG1:TTG1-YFP. YFP fluorescence is observed both in the nucleus and the cytoplasm. If nuclear targeting does not change the intercellular mobility, one would expect that NLS-TTG1-YFP distribution should not be changed when compared with TTG1-YFP in a wild-type background. Consistent with this, we found a depletion around trichome in TTG1:NLS-TTG1-YFP lines (Fig. 6C-F). The first tier of cells around the trichomes exhibited 45% of the YFP fluorescence found in trichomes, the second tier 62% and the third tier 74%. Statistical analysis showed that these values were highly significant (t-test, P<0.001) similar to TTG1-YFP without NLS fusion. Together, these data show that the nuclear localization of TTG1 by GL3 is not as important as the trapping of TTG1 in the nucleus. DISCUSSION The TTG1 depletion model was initially derived from three observations (Bouyer et al., 2008): first, TTG1 protein can move between cells; second, the depletion of TTG1 in trichome neighbouring cells is not found in gl3 mutants; and, third, GL3 can bind to TTG1. Because GL3 is expressed in trichomes, it was proposed that TTG1 movement is regulated by GL3 such that the elevated GL3 levels in trichomes result in an accumulation of TTG1. However, the molecular nature that triggers TTG1 depletion by GL3 have not been elucidated so far and here we show that the direct interaction between TTG1 and GL3 give rise to nuclear translocation and trapping in trichomes, which is a prerequisite in the computational modelling that we proposed previously (Bouyer et al., 2008). Is TTG1 movement controlled by GL3? The depletion model predicts that TTG1 can freely move between cells and that its movement is retained by GL3 in trichomes. If this were the case, one would expect that GL3 should also be able to modulate TTG1 movement when expressed in a cell-type-specific manner. We tested this by analyzing the movement behaviour of TTG1-YFP between cell layers. This assay is based on the observation that TTG1-YFP can complement the epidermal trichome phenotype of ttg1 mutants when expressed in the subepidermis (Bouyer et al., 2008). This rescue ability depends strongly on the presence of GL3 as sub-epidermally provided TTG1 is no longer able to compensate for the loss of endogenous TTG1 in a gl3 ttg1 double mutant. Two additional findings are particularly relevant: first, the observation that exclusive expression of GL3 in the sub-epidermis further precludes the rescue and prevents TTG1-YFP mobility from this layer; and, second, exclusive expression of GL3 in the epidermis conversely enhances the RBC:TTG1-YFP-mediated trichome production dramatically. Notably, GL3 seems to differ with respect to its intercellular mobility in the leaf and in the root. In contrast to leaves (Digiuni et al., 2008; Zhao et al., 2008), it has been suggested that GL3 moves in the root as the protein is found in atrichoblasts, whereas its RNA expression pattern is confined to trichoblasts in the root epidermis (Bernhardt et al., 2005). GL3 itself has no obvious influence on the mobility of TTG1 between the sub-epidermis and the epidermal layer as the TTG1- YFP signal can clearly be detected in the L1 in gl3 mutants when expressed from the L2. It is, however, possible that a redundant function of EGL3 masks a requirement of GL3 for TTG1 mobility. Yet, GL3 is essential to prevent further movement owing to recruitment of TTG1 to the nucleus and its binding within a immobile protein complex in a trichome initial. This is demonstrated by the retention of TTG1-YFP in the sub-epidermis when GL3 is co-expressed there (RBC:GFP-GL3). Together, these data demonstrate that the mobility of TTG1 is controlled by GL3. Intracellular localization of TTG1 and its regulation by GL3 Whether TTG1 is present and/or required predominantly in the nucleus or in the cytoplasm is not clear from the current data. On the one hand, cell fractionation studies showed that the ortholog of TTG1 from Petunia, AN11, is cytoplasmic (Vetten et al., 1997). On the other hand, molecular data indicate that TTG1, together with GL3, directly controls the expression of a common set of target genes and that GL3 protein forms subnuclear speckles in the absence of TTG1, indicating a role for TTG1 in the nucleus (Gonzalez et al., 2008; Zhao et al., 2008; Yoshida et al., 2009). As TTG1 does not contain any obvious NLS motif, an active nuclear translocation seems unlikely (Walker et al., 1999). A direct assessment of its localization in Arabidopsis using TTG1-YFP fusions revealed nuclear and cytoplasmic fluorescence (Bouyer et al., 2008; Zhao et al., 2008). Because GL3 was shown to control the distribution of TTG1-YFP in the epidermis, we speculated that the intracellular Table 4. Trichome rescue efficiency of nuclear-targeted TTG1-YFP Genotype Number of trichomes ±s.d. n ttg1-13 TTG1:NLS-TTG1-YFP 128±18 26 ttg1-13 RBC:NLS-TTG1-YFP 124±30 26 ttg1-13 TTG1:TTG1-YFP 127±20 26 ttg1-13 RBC:TTG1-YFP 98 1 Trichome number is the average number of trichomes on leaves 3 and 4 in T1 plants; s.d., standard deviation; n, number of plants analyzed.
8 5046 RESEARCH ARTICLE Development 138 (22) 2002), suggesting an evolutionary conservation of this mechanism but offering the possibility that different nuclear and cytoplasmic functions might exist because AN11 has been shown to be cytoplasmic in petunia (Vetten et al., 1997). Fig. 6. Cellular distribution of nuclear targeted TTG1-YFP. (A,B) Transient expression of NLS-TTG1-YFP(A) and TTG1-YFP (B) in Arabidopsis cotyledon epidermal cells. (C-F) Distribution of NLS-TTG1- YFP in the epidermal cells of the trichome patterning zone on leaves in lines carrying TTG1:NLS-TTG1-YFP. (C) NLS-TTG1-YFP distribution. (E) Cell walls counterstained with propidium iodide. (E) Overlay image of B and C. (F) Intensity projection images of confocal stacks of TTG1:NLS- TTG1-YFP lines. The fluorescence intensity is indicated by size of the peaks. Arrowheads indicate the nucleus. Scale bars: 10 m. localization of TTG1 is also controlled by GL3 such that nuclear localized GL3 targets TTG1 to the nucleus. We confirmed this hypothesis in several experiments. First, in planta analysis of TTG1- YFP expressed under the TTG1 promoter in wild-type, gl3 mutant and 35S:GL3 lines demonstrated that the fraction of nuclear TTG1- YFP depends on the amount of GL3. Second, co-expression of CFP- GL3 and TTG1-YFP in Arabidopsis cotyledon epidermal cells, as well as in onion epidermal cells, showed that TTG1-YFP is targeted by GL3 to the nucleus and that GL3 mutations interfering with the GL3-TTG1 interaction cannot trigger nuclear targeting anymore. Third, yeast nuclear targeting assays showed that TTG1 has no functional nuclear localization sites. Instead, GL3 recruits TTG1 into the nucleus and this depends on the direct interaction between GL3 and TTG1. These data are in line with the observation that the TTG1 homolog PFWD in Perilla frutescens accumulates in the nucleus when co-expressed with bhlh gene MYC-RP (Sompornpailin et al., bhlh redundancy in the regulation of TTG1 nuclear localization The bhlh genes involved in TTG1-regulated pathways have partial functional redundancy. TT8 regulates seed coat mucilage production, seed coat pigment production, anthocyanin biosynthesis and trichome development (Zhang et al., 2003; Baudry et al., 2004; Maes et al., 2008). EGL3 controls seed coat pigmentation, seed coat mucilage production, anthocyanin biosynthesis, trichome and root hair development, and GL3 is involved in anthocyanin biosynthesis, trichome and root hair development (Nesi et al., 2000; Bernhardt et al., 2003; Zhang et al., 2003; Baudry et al., 2004; Bernhardt et al., 2005; Baudry et al., 2006; Gonzalez et al., 2008; Maes et al., 2008). All three bhlh proteins interact with TTG1 in yeast two hybrid assays and it is therefore conceivable that they all can sequester TTG1 in the nucleus. Our analysis of double and triple mutants revealed that GL3 has a major role and that EGL3 acts redundantly in this respect, whereas TT8 cannot further shift the TTG1-YFP nuclear/cytoplasm balance in the gl3 egl3 tt8 triple mutant. A comparison of the TTG1-YFP nuclear/cytoplasm balance between the gl3 egl3 tt8 triple mutant and the TTG1 C26-YFP protein revealed a noteworthy discrepancy. About 50% of the TTG1-YFP is found in the cytoplasm in the triple mutant. By contrast the TTG1 C26-YFP protein that does not interact with the bhlh proteins anymore is exclusively found in the cytoplasm. One possibility to explain this is the involvement of additional factors in the regulation of TTG1 localization. This may be caused by a loss of interaction between TTG1 C26-YFP and further nuclear translocation regulators or by promoting its interaction with components that direct its cytoplasmic distribution, or a combination of both. As GFP on its own or even as double- and triple-fusion still appears both nuclear and cytoplasmic (Kim et al., 2005), a passive mechanism is unlikely to account for the exclusively cytoplasmic compartmentalization of TTG1 C26-YFP. The second explanation would introduce the additional assumption that movement into the nucleus is actively mediated by the deleted C-terminal 26 amino acids. However, such an active nuclear transport mechanism seems unlikely, as TTG1 does not seem to contain any NLS in the yeast NTT assay (Fig. 3A). Nuclear trapping and intercellular transport In most experimental systems, nuclear targeting and intercellular transport are interrelated. Mutations or modification in CAPRICE (CPC), KNOTTED1 (KN1) and SHORT ROOT (SHR) proteins that reduce their nuclear localization also affect the mobility between cells (Lucas et al., 1995; Prochiantz and Joliot, 2003; Gallagher et al., 2004; Kurata et al., 2005). Whereas these transcription factors have been shown to require a nuclear localization signal for intercellular transport, nuclear targeting of GFP partially reduced its passive transport ability (Crawford and Zambryski, 2000). These examples suggest that nuclear targeting can prevent, but might also be required for, the movement of proteins into neighbouring cells. A more detailed analysis of SHR protein movement indicated that nuclear as well as cytoplasmic localization are relevant for intercellular movement (Gallagher and Benfey, 2009). By contrast, steady cytoplasmic localization of TTG1 does not seem to be relevant for intercellular protein movement. This is suggested by the
9 TTG1 trapping by GL3 RESEARCH ARTICLE 5047 finding that plants expressing a NLS-TTG1-YFP fusion protein exhibit exclusively a nuclear localization signal and when expressed in the sub-epidermis can fully rescue the epidermal trichome phenotype (Table 4; Fig. 6). Principles of protein movement regulation during patterning The intercellular movement of transcription factors has been shown to play an important role in various patterning processes in plants, including cortex/endodermis specification, root hair and trichome patterning. In such cases, protein movement provides positional information to other cells, which makes it extremely important to control their distribution in the tissue. This can, in principle, be achieved in two ways. One principle is the local production of a protein that in turn can move into the surrounding tissue thereby forming a gradient. This is thought to be realized during trichome patterning where the R3 single repeat inhibitors are produced in trichomes from where they can move into the neighbouring cells (Kurata et al., 2005; Digiuni et al., 2008; Wester et al., 2009). A second principle is the trapping of moving proteins. This has been elegantly demonstrated for the SHR protein, which is sequestered by SCARECROW (SCR) in the nucleus (Cui et al., 2007). In this system, SHR is produced in the stele and its movement into outer tissue layers is restricted by SCR trapping. Thus, trichome patterning relies on a similar molecular mechanism, although the systems in which these different patterning processes take place are strikingly different. Here, trapping is thought to create a depletion pattern from an initial ubiquitous distribution, whereas root patterning is strongly based on positional cues that give rise to a highly regular radial and epidermal distribution of distinct cell types. Here, we could show that the protein interaction between TTG1 and GL3 is necessary for the intracellular and epidermal distribution pattern of TTG1, which in mathematical simulations is required and sufficient to generate a spacing pattern (Bouyer et al., 2008). Acknowledgements We sincerely thank Jae Yean Kim and David Jackson laboratory for the RUBISCO SMALL SUBUNIT 2b promoter; and Alan Lloyd s laboratory for the AtML1 promoter and the gl3 egl3 tt8 triple mutant seeds. We are grateful to Klaus Harter for providing the yeast vectors pnh2, pnh3 and pns; and to Joachim Uhrig for the yeast vector pvt-u. We thank Martina Pesch for the pc- ACT2-EGL3 and pc-act2-tt8 constructs, and for fruitful discussions. Funding This project was supported by the Deutsche Forschungsgemeinschaf priority program [SFB 572]. R.B. was funded by the International Graduate School in Genetic and Functional Genomics (IGS-GFG), University of Cologne. Competing interests statement The authors declare no competing financial interests. Supplementary material Supplementary material available online at References Baudry, A., Heim, M. A., Dubreucq, B., Caboche, M., Weisshaar, B. and Lepiniec, L. (2004). TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 39, Baudry, A., Caboche, M. and Lepiniec, L. (2006). TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bhlh factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 46, Bernhardt, C., Lee, M. M., Gonzalez, A., Zhang, F., Lloyd, A. and Schiefelbein, J. (2003). The bhlh genes GLABRA3 (GL3) andenhancer OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130, Bernhardt, C., Zhao, M., Gonzalez, A., Lloyd, A. and Schiefelbein, J. (2005). The bhlh genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132, Bouyer, D., Geier, F., Kragler, F., Schnittger, A., Pesch, M., Wester, K., Balkunde, R., Timmer, J., Fleck, C. and Hulskamp, M. (2008). Twodimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol. 6, e141. Clough, S. and Bent, A. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, Crawford, K. M. and Zambryski, P. C. (2000). Subcellular localization determines the availability of non-targeted proteins to plasmodesmatal transport. Curr. Biol. 10, Cui, H., Levesque, M. P., Vernoux, T., Jung, J. W., Paquette, A. J., Gallagher, K. L., Wang, J. Y. I., Blilou Scheres, B. and Benfey, P. N. (2007). An evolutionary conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316, Digiuni, S., Schellmann, S., Geier, F., Greese, B., Pesch, M., Wester, K., Dartan, B., Mach, V., Srinivas, B. P., Timmer, J. et al. (2008). A competitive complex formation mechanism underlies trichome patterning on Arabidopsis leaves. Mol. Syst. Biol. 4, 217. Esch, J. J., Chen, M., Sanders, M., Hillestad, M., Ndkium, S., Idelkope, B., Neizer, J. and Marks, M. D. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130, Gallagher, K. L. and Benfey, P. N. (2009). Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J. 57, Gallagher, K. L., Paquette, A. J., Nakajima, K. and Benfey, P. N. (2004). Mechanisms regulating SHORT-ROOT intercellular movement. Curr. Biol. 14, Galway, M. E., Masucci, J. D., Lloyd, A. M., Walbot, V., Davis, R. W. and Schiefelbein, J. W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166, Gietz, R. D., Schiestl, R. H., Willems, A. R. and Woods, R. A. (1995). Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, Gonzalez, A., Zhao, M., Leavitt, J. M. and Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, Hulskamp, M. (2004). Plant trichomes: a model for cell differentiation. Nat. Rev. Mol. Cell Biol. 5, Hulskamp, M., Misera, S. and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, Ishida, T., Kurata, T., Okada, K. and Wada, T. (2008). A genetic regulatory network in the development of trichomes and root hairs. Annu. Rev. Plant Biol. 59, Kim, I., Kobayashi, K., Cho, E. and Zambryski, P. C. (2005). Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc. Natl. Acad. Sci. USA 102, Kirik, V., Simon, M., Hulskamp, M. and Schiefelbein, J. (2004a). The ENHANCER OF TRY AND CPC1 (ETC1) gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 268, Kirik, V., Simon, M., Wester, K., Schiefelbein, J. and Hulskamp, M. (2004b). ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 55, Koornneef, M. (1981). The complex syndrome of ttg mutants. Arabid. Inf. Serv. 18, Koornneef, M., Dellaert, L. W. M. and VanderVeen, J. H. (1982). EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana. Mutat. Res. 93, Kurata, T., Ishida, T., Kawabata-Awai, C., Noguchi, M., Hattori, S., Sano, R., Nagasaka, R., Tominaga, R., Koshino-Kimura, Y., Kato, T. et al. (2005). Cellto-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132, Larkin, J. C., Young, N., Prigge, M. and Marks, M. D. (1996). The control of trichome spacing and number in Arabidopsis. Development 122, Lucas, W. J., Bouche-Pillon, S., Jackson, D. P., Nguyen, L., Baker, L., Ding, B. and Hake, S. (1995). Selective trafficking of KNOTTED1 homeodomain protein and its mrna through plasmodesmata. Science 270, Maes, L., Inze, D. and Goossens, A. (2008). Functional specialization of TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 148, Nesi, N., Debeaujon, I., Jond, C., Pelletier, G., Caboche, M. and Lepiniec, L. (2000). The TT8 gene encodes a basic helix-loop-helix domain protein required
Epidermal cell patterning and differentiation throughout the apical basal axis of the seedling
 Journal of Experimental Botany, Vol. 56, No. 418, pp. 1983 1989, August 2005 doi:10.1093/jxb/eri213 Advance Access publication 20 June, 2005 REVIEW ARTICLE Epidermal cell patterning and differentiation
Journal of Experimental Botany, Vol. 56, No. 418, pp. 1983 1989, August 2005 doi:10.1093/jxb/eri213 Advance Access publication 20 June, 2005 REVIEW ARTICLE Epidermal cell patterning and differentiation
The bhlh genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis
 Research article 291 The bhlh genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis Christine Bernhardt 1, Mingzhe Zhao 2,
Research article 291 The bhlh genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis Christine Bernhardt 1, Mingzhe Zhao 2,
The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci
 Access the Development most First recent posted version epress online at http://dev.biologists.org/lookup/doi/10.1242/dev.016873 on online 23 April publication 2008 as 10.1242/dev.016873 date 23 April
Access the Development most First recent posted version epress online at http://dev.biologists.org/lookup/doi/10.1242/dev.016873 on online 23 April publication 2008 as 10.1242/dev.016873 date 23 April
Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in Arabidopsis
 Zhang et al. BMC Plant Biology (2018) 18:63 https://doi.org/10.1186/s12870-018-1271-z RESEARCH ARTICLE Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in
Zhang et al. BMC Plant Biology (2018) 18:63 https://doi.org/10.1186/s12870-018-1271-z RESEARCH ARTICLE Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in
Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang Chen, Qiang Dong, Jiangqi Wen, Kirankumar S. Mysore and Jian Zhao
 New Phytologist Supporting Information Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bhlh transcription factor MtTT8 Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang
New Phytologist Supporting Information Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bhlh transcription factor MtTT8 Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang
A Mutual Support Mechanism through Intercellular Movement of CAPRICE and GLABRA3 Can Pattern the Arabidopsis Root Epidermis
 A Mutual Support Mechanism through Intercellular Movement of CAPRICE and GLABRA3 Can Pattern the Arabidopsis Root Epidermis PLoS BIOLOGY Natasha Saint Savage 1, Tom Walker 2, Yana Wieckowski 3, John Schiefelbein
A Mutual Support Mechanism through Intercellular Movement of CAPRICE and GLABRA3 Can Pattern the Arabidopsis Root Epidermis PLoS BIOLOGY Natasha Saint Savage 1, Tom Walker 2, Yana Wieckowski 3, John Schiefelbein
Nature Genetics: doi: /ng Supplementary Figure 1. ssp mutant phenotypes in a functional SP background.
 Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation
 Erratum Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation Victor Kirik, Myeong Min Lee, Katja Wester, Ullrich Herrmann, Zhengui Zheng, David Oppenheimer, John Schiefelbein
Erratum Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation Victor Kirik, Myeong Min Lee, Katja Wester, Ullrich Herrmann, Zhengui Zheng, David Oppenheimer, John Schiefelbein
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplemental Data. Perrella et al. (2013). Plant Cell /tpc
 Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
A novel regulatory circuit specifies cell fate in the Arabidopsis root epidermis
 Physiologia Plantarum 126: 503 510. 2006 Copyright ß Physiologia Plantarum 2006, ISSN 0031-9317 REVIEW A novel regulatory circuit specifies cell fate in the Arabidopsis root epidermis John Schiefelbein
Physiologia Plantarum 126: 503 510. 2006 Copyright ß Physiologia Plantarum 2006, ISSN 0031-9317 REVIEW A novel regulatory circuit specifies cell fate in the Arabidopsis root epidermis John Schiefelbein
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
SAD2 in Arabidopsis Functions in Trichome Initiation through Mediating GL3 Function and Regulating GL1, TTG1 and GL2 Expression
 Journal of Integrative Plant Biology 2008, 50 (7): 906 917 SAD2 in Arabidopsis Functions in Trichome Initiation through Mediating GL3 Function and Regulating GL1, TTG1 and GL2 Expression Ying Gao 1,2,3,
Journal of Integrative Plant Biology 2008, 50 (7): 906 917 SAD2 in Arabidopsis Functions in Trichome Initiation through Mediating GL3 Function and Regulating GL1, TTG1 and GL2 Expression Ying Gao 1,2,3,
Supplemental Data. Wang et al. (2014). Plant Cell /tpc
 Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Development 143: doi: /dev : Supplementary information
 Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
Supplementary Materials and Methods Plant materials The mutants and transgenic plants used in the present study were as follows: E361 (from Alex Webb s laboratory); tmm-1, ptmm::tmm-gfp and flp-1 (from
Lecture 4: Radial Patterning and Intercellular Communication.
 Lecture 4: Radial Patterning and Intercellular Communication. Summary: Description of the structure of plasmodesmata, and the demonstration of selective movement of solutes and large molecules between
Lecture 4: Radial Patterning and Intercellular Communication. Summary: Description of the structure of plasmodesmata, and the demonstration of selective movement of solutes and large molecules between
Supplemental Data. Gao et al. (2012). Plant Cell /tpc
 Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
GFP GAL bp 3964 bp
 Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL
 GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
Non-linear signaling for pattern formation? Ben Scheres
 412 Non-linear signaling for pattern formation? Ben Scheres Developmental pathways are often simplified as linear cascades of gene activation caused by initial signaling events. Recent data, however, support
412 Non-linear signaling for pattern formation? Ben Scheres Developmental pathways are often simplified as linear cascades of gene activation caused by initial signaling events. Recent data, however, support
HOW DO CELLS KNOW WHAT THEY WANT TO
 Annu. Rev. Plant Biol. 2003. 54:403 30 doi: 10.1146/annurev.arplant.54.031902.134823 Copyright c 2003 by Annual Reviews. All rights reserved HOW DO CELLS KNOW WHAT THEY WANT TO BE WHEN THEY GROW UP? Lessons
Annu. Rev. Plant Biol. 2003. 54:403 30 doi: 10.1146/annurev.arplant.54.031902.134823 Copyright c 2003 by Annual Reviews. All rights reserved HOW DO CELLS KNOW WHAT THEY WANT TO BE WHEN THEY GROW UP? Lessons
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
GLABRA1 and TRIPTYCHON in Arabidopsis
 Development 125, 2283-2289 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV0160 2283 Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON
Development 125, 2283-2289 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV0160 2283 Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON
TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a WRKY Transcription Factor
 The Plant Cell, Vol. 14, 1359 1375, June 2002, www.plantcell.org 2002 American Society of Plant Biologists TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a
The Plant Cell, Vol. 14, 1359 1375, June 2002, www.plantcell.org 2002 American Society of Plant Biologists TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a
Arabidopsis thaliana Myosin XI Is Necessary for Cell Fate Determination in Root Epidermis
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2010 Arabidopsis thaliana Myosin
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2010 Arabidopsis thaliana Myosin
Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus:
 m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
m Eukaryotic mrna processing Newly made RNA is called primary transcript and is modified in three ways before leaving the nucleus: Cap structure a modified guanine base is added to the 5 end. Poly-A tail
purpose of this Chapter is to highlight some problems that will likely provide new
 119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
SUPPLEMENTARY INFORMATION
 Supplemental Methods Isolation and mapping of SPCH An EMS-mutagenized population of tmm-1 (Col);E1728 (an enhancer trap GFP marking guard cells) was created. M2 seeds were collected from M1 s planted in
Supplemental Methods Isolation and mapping of SPCH An EMS-mutagenized population of tmm-1 (Col);E1728 (an enhancer trap GFP marking guard cells) was created. M2 seeds were collected from M1 s planted in
2. Yeast two-hybrid system
 2. Yeast two-hybrid system I. Process workflow a. Mating of haploid two-hybrid strains on YPD plates b. Replica-plating of diploids on selective plates c. Two-hydrid experiment plating on selective plates
2. Yeast two-hybrid system I. Process workflow a. Mating of haploid two-hybrid strains on YPD plates b. Replica-plating of diploids on selective plates c. Two-hydrid experiment plating on selective plates
Genetic analysis of seed coat development in Arabidopsis
 Review TRENDS in Plant Science Vol.0 No.0 October 00 Genetic analysis of seed coat development in Arabidopsis George Haughn and Abed Chaudhury Department of Botany, University of British Columbia, 670
Review TRENDS in Plant Science Vol.0 No.0 October 00 Genetic analysis of seed coat development in Arabidopsis George Haughn and Abed Chaudhury Department of Botany, University of British Columbia, 670
Ron et al SUPPLEMENTAL DATA
 Ron et al SUPPLEMENTAL DATA Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model Mily Ron, Kaisa Kajala,
Ron et al SUPPLEMENTAL DATA Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model Mily Ron, Kaisa Kajala,
Last time: Obtaining information from a cloned gene
 Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
The Plant Cell, November. 2017, American Society of Plant Biologists. All rights reserved
 The Genetics of Floral Development Teaching Guide Overview The development of flowers in angiosperm plants provided a critical evolutionary advantage, allowing more options for pollen dispersal and seed
The Genetics of Floral Development Teaching Guide Overview The development of flowers in angiosperm plants provided a critical evolutionary advantage, allowing more options for pollen dispersal and seed
Chapter 4 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays.
 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Biological Roles of Cytokinins
 Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA
 EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA Item Type text; Electronic Thesis Authors Bergstrand, Lauren Janel Publisher The University of Arizona. Rights Copyright is held by the author. Digital
EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA Item Type text; Electronic Thesis Authors Bergstrand, Lauren Janel Publisher The University of Arizona. Rights Copyright is held by the author. Digital
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering
 Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON
 PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
Supplemental Data. Chen and Thelen (2010). Plant Cell /tpc
 Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings
 The Plant Journal (2008) 53, 814 827 doi: 10.1111/j.1365-313X.2007.03373.x Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings Antonio
The Plant Journal (2008) 53, 814 827 doi: 10.1111/j.1365-313X.2007.03373.x Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings Antonio
Reading Assignments. A. Genes and the Synthesis of Polypeptides. Lecture Series 7 From DNA to Protein: Genotype to Phenotype
 Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Supplemental material
 Supplemental material Table 1- Segregation analysis of sgt1a sgt1b double mutant plants Parental genotype No. of lines Normal seeds Phenotype Abnormal seeds Ratio of abnormal seeds (%) WT 3 171 2 1.16
Supplemental material Table 1- Segregation analysis of sgt1a sgt1b double mutant plants Parental genotype No. of lines Normal seeds Phenotype Abnormal seeds Ratio of abnormal seeds (%) WT 3 171 2 1.16
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family
 Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
FURCA loci. Genetic control of trichome branch number in Arabidopsis: the roles of the. Danlin Luo and David G. Oppenheimer* SUMMARY
 Development 126, 5547-5557 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV0265 5547 Genetic control of trichome branch number in Arabidopsis: the roles of the FURCA loci Danlin
Development 126, 5547-5557 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV0265 5547 Genetic control of trichome branch number in Arabidopsis: the roles of the FURCA loci Danlin
Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach
 Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach ABSTRACT SUBMITTED TO JAMIA MILLIA ISLAMIA NEW DELHI IN PARTIAL FULFILMENT OF
Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach ABSTRACT SUBMITTED TO JAMIA MILLIA ISLAMIA NEW DELHI IN PARTIAL FULFILMENT OF
Actions of auxin. Hormones: communicating with chemicals History: Discovery of a growth substance (hormone- auxin)
 Hormones: communicating with chemicals History- discovery of plant hormone. Auxin Concepts of hormones Auxin levels are regulated by synthesis/degradation, transport, compartmentation, conjugation. Polar
Hormones: communicating with chemicals History- discovery of plant hormone. Auxin Concepts of hormones Auxin levels are regulated by synthesis/degradation, transport, compartmentation, conjugation. Polar
Regulation of Phosphate Homeostasis by microrna in Plants
 Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Outline. Leaf Development. Leaf Structure - Morphology. Leaf Structure - Morphology
 Outline 1. Leaf Structure: Morphology & Anatomy 2. Leaf Development A. Anatomy B. Sector analysis C. Leaf Development Leaf Structure - Morphology Leaf Structure - Morphology 1 Leaf Structure - Morphology
Outline 1. Leaf Structure: Morphology & Anatomy 2. Leaf Development A. Anatomy B. Sector analysis C. Leaf Development Leaf Structure - Morphology Leaf Structure - Morphology 1 Leaf Structure - Morphology
Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA
 Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
Arabidopsis CAPRICE (MYB) and GLABRA3 (bhlh) Control Tomato (Solanum lycopersicum) Anthocyanin Biosynthesis
 Arabidopsis CAPRICE (MYB) and GLABRA3 (bhlh) Control Tomato (Solanum lycopersicum) Anthocyanin Biosynthesis Takuji Wada 1, Asuka Kunihiro 2, Rumi Tominaga-Wada 1 * 1 Graduate School of Biosphere Sciences,
Arabidopsis CAPRICE (MYB) and GLABRA3 (bhlh) Control Tomato (Solanum lycopersicum) Anthocyanin Biosynthesis Takuji Wada 1, Asuka Kunihiro 2, Rumi Tominaga-Wada 1 * 1 Graduate School of Biosphere Sciences,
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells
 Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
1. Contains the sugar ribose instead of deoxyribose. 2. Single-stranded instead of double stranded. 3. Contains uracil in place of thymine.
 Protein Synthesis & Mutations RNA 1. Contains the sugar ribose instead of deoxyribose. 2. Single-stranded instead of double stranded. 3. Contains uracil in place of thymine. RNA Contains: 1. Adenine 2.
Protein Synthesis & Mutations RNA 1. Contains the sugar ribose instead of deoxyribose. 2. Single-stranded instead of double stranded. 3. Contains uracil in place of thymine. RNA Contains: 1. Adenine 2.
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Analysis of regulatory function of circadian clock. on photoreceptor gene expression
 Thesis of Ph.D. dissertation Analysis of regulatory function of circadian clock on photoreceptor gene expression Tóth Réka Supervisor: Dr. Ferenc Nagy Biological Research Center of the Hungarian Academy
Thesis of Ph.D. dissertation Analysis of regulatory function of circadian clock on photoreceptor gene expression Tóth Réka Supervisor: Dr. Ferenc Nagy Biological Research Center of the Hungarian Academy
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011) /tpc
 Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
GENES AND CHROMOSOMES III. Lecture 5. Biology Department Concordia University. Dr. S. Azam BIOL 266/
 GENES AND CHROMOSOMES III Lecture 5 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION OPERONS Introduction All cells in a multi-cellular
GENES AND CHROMOSOMES III Lecture 5 BIOL 266/4 2014-15 Dr. S. Azam Biology Department Concordia University CELL NUCLEUS AND THE CONTROL OF GENE EXPRESSION OPERONS Introduction All cells in a multi-cellular
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Stress Effects on Myosin Mutant Root Length in Arabidopsis thaliana
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2011 Stress Effects on Myosin
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2011 Stress Effects on Myosin
Bi 1x Spring 2014: LacI Titration
 Bi 1x Spring 2014: LacI Titration 1 Overview In this experiment, you will measure the effect of various mutated LacI repressor ribosome binding sites in an E. coli cell by measuring the expression of a
Bi 1x Spring 2014: LacI Titration 1 Overview In this experiment, you will measure the effect of various mutated LacI repressor ribosome binding sites in an E. coli cell by measuring the expression of a
ENDODERMIS & POLARITY
 https://en.wikipedia.org/wiki/casparian_strip ENDODERMIS & POLARITY Niloufar Pirayesh 13.01.2016 PCDU SEMINAR 2 What is Endodermis? It helps with Regulate the movement of water ions and hormones. (in and
https://en.wikipedia.org/wiki/casparian_strip ENDODERMIS & POLARITY Niloufar Pirayesh 13.01.2016 PCDU SEMINAR 2 What is Endodermis? It helps with Regulate the movement of water ions and hormones. (in and
Welcome to Class 21!
 Welcome to Class 21! Introductory Biochemistry! Lecture 21: Outline and Objectives l Regulation of Gene Expression in Prokaryotes! l transcriptional regulation! l principles! l lac operon! l trp attenuation!
Welcome to Class 21! Introductory Biochemistry! Lecture 21: Outline and Objectives l Regulation of Gene Expression in Prokaryotes! l transcriptional regulation! l principles! l lac operon! l trp attenuation!
Supporting Information
 Supporting Information Fleissner et al. 10.1073/pnas.0907039106 Fig. S1. (A) MAK-2-GFP localized to CATs tips is not bound by membrane. his-3::pccg1 mak-2-gfp; mak-2 strain labeled with membrane dye FM4
Supporting Information Fleissner et al. 10.1073/pnas.0907039106 Fig. S1. (A) MAK-2-GFP localized to CATs tips is not bound by membrane. his-3::pccg1 mak-2-gfp; mak-2 strain labeled with membrane dye FM4
Temporal Control of Trichome Distribution by MicroRNA156-Targeted SPL Genes in Arabidopsis thaliana W OA
 This article is a Plant Cell Advance Online Publication. The date of its first appearance online is the official date of publication. The article has been edited and the authors have corrected proofs,
This article is a Plant Cell Advance Online Publication. The date of its first appearance online is the official date of publication. The article has been edited and the authors have corrected proofs,
Factors involved in asymmetric cell division in the Arabidopsis root meristem
 Factors involved in asymmetric cell division in the Arabidopsis root meristem Bachelor thesis biology Utrecht University Martinus Schneijderberg Student number 3252469 Instructor: Dr. I. Blilou Labwork
Factors involved in asymmetric cell division in the Arabidopsis root meristem Bachelor thesis biology Utrecht University Martinus Schneijderberg Student number 3252469 Instructor: Dr. I. Blilou Labwork
** * * * Col-0 cau1 CAU1. Actin2 CAS. Actin2. Supplemental Figure 1. CAU1 affects calcium accumulation.
 Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
Genetic Characterization and Functional Analysis of the GID1 Gibberellin Receptors in Arabidopsis W
 The Plant Cell, Vol. 18, 3399 3414, December 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Genetic Characterization and Functional Analysis of the GID1 Gibberellin Receptors in Arabidopsis
The Plant Cell, Vol. 18, 3399 3414, December 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Genetic Characterization and Functional Analysis of the GID1 Gibberellin Receptors in Arabidopsis
Stochastic simulations
 Stochastic simulations Application to molecular networks Literature overview Noise in genetic networks Origins How to measure and distinguish between the two types of noise (intrinsic vs extrinsic)? What
Stochastic simulations Application to molecular networks Literature overview Noise in genetic networks Origins How to measure and distinguish between the two types of noise (intrinsic vs extrinsic)? What
cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work cote cote-yfp spc This work
 SUPPLEMENTARY INFORMATION Table S1. List of strains Strains Genotype Source B. subtilis PY79 Prototrophic derivative of B. subtilis 168 (60) RL1070 spovid::kan (12) HS176 cotz phs2 (cotz-gfp spc) (22)
SUPPLEMENTARY INFORMATION Table S1. List of strains Strains Genotype Source B. subtilis PY79 Prototrophic derivative of B. subtilis 168 (60) RL1070 spovid::kan (12) HS176 cotz phs2 (cotz-gfp spc) (22)
Novel Functions of Plant Cyclin-Dependent Kinase Inhibitors, ICK1/KRP1, Can Act Non-Cell-Autonomously and Inhibit Entry into Mitosis W
 This article is published in The Plant Cell Online, The Plant Cell Preview Section, which publishes manuscripts accepted for publication after they have been edited and the authors have corrected proofs,
This article is published in The Plant Cell Online, The Plant Cell Preview Section, which publishes manuscripts accepted for publication after they have been edited and the authors have corrected proofs,
DNA Structure and Function
 DNA Structure and Function Nucleotide Structure 1. 5-C sugar RNA ribose DNA deoxyribose 2. Nitrogenous Base N attaches to 1 C of sugar Double or single ring Four Bases Adenine, Guanine, Thymine, Cytosine
DNA Structure and Function Nucleotide Structure 1. 5-C sugar RNA ribose DNA deoxyribose 2. Nitrogenous Base N attaches to 1 C of sugar Double or single ring Four Bases Adenine, Guanine, Thymine, Cytosine
Variability in the Control of Cell Division Underlies Sepal Epidermal Patterning in Arabidopsis thaliana
 Variability in the Control of Cell Division Underlies Sepal Epidermal Patterning in Arabidopsis thaliana Adrienne H. K. Roeder 1,2., Vijay Chickarmane 1., Alexandre Cunha 2,3, Boguslaw Obara 4,B.S. Manjunath
Variability in the Control of Cell Division Underlies Sepal Epidermal Patterning in Arabidopsis thaliana Adrienne H. K. Roeder 1,2., Vijay Chickarmane 1., Alexandre Cunha 2,3, Boguslaw Obara 4,B.S. Manjunath
3% three branches, n 661) compared to the corresponding wild-type WS (0% unbranched, 10% two branches, Correspondence: M. Hülskamp
 Brief Communication 1891 CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein V. Kirik*, D. Bouyer*, U. Schöbinger*, N. Bechtold, M. Herzog, J.-M. Bonneville
Brief Communication 1891 CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein V. Kirik*, D. Bouyer*, U. Schöbinger*, N. Bechtold, M. Herzog, J.-M. Bonneville
GFP WxTP-GFP WxTP-GFP. stromules. DsRed WxTP-DsRed WxTP-DsRed. stromules
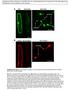 Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Lecture 10: Cyclins, cyclin kinases and cell division
 Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Supp- Figure 2 Confocal micrograph of N. benthamiana tissues transiently expressing 35S:YFP-PDCB1. PDCB1 was targeted to plasmodesmata (twin punctate
 Supplemental Data. Simpson et al. (009). n rabidopsis GPI-anchor plasmodesmal neck protein with callosebinding activity and potential to regulate cell-to-cell trafficking. 5 0 stack Supp- Figure Confocal
Supplemental Data. Simpson et al. (009). n rabidopsis GPI-anchor plasmodesmal neck protein with callosebinding activity and potential to regulate cell-to-cell trafficking. 5 0 stack Supp- Figure Confocal
Plant Molecular and Cellular Biology Lecture 8: Mechanisms of Cell Cycle Control and DNA Synthesis Gary Peter
 Plant Molecular and Cellular Biology Lecture 8: Mechanisms of Cell Cycle Control and DNA Synthesis Gary Peter 9/10/2008 1 Learning Objectives Explain why a cell cycle was selected for during evolution
Plant Molecular and Cellular Biology Lecture 8: Mechanisms of Cell Cycle Control and DNA Synthesis Gary Peter 9/10/2008 1 Learning Objectives Explain why a cell cycle was selected for during evolution
Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner
 ABA represses the transcription of chloroplast genes Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner Supplementary data Supplementary tables Table
ABA represses the transcription of chloroplast genes Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner Supplementary data Supplementary tables Table
Genetic transcription and regulation
 Genetic transcription and regulation Central dogma of biology DNA codes for DNA DNA codes for RNA RNA codes for proteins not surprisingly, many points for regulation of the process DNA codes for DNA replication
Genetic transcription and regulation Central dogma of biology DNA codes for DNA DNA codes for RNA RNA codes for proteins not surprisingly, many points for regulation of the process DNA codes for DNA replication
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
Richik N. Ghosh, Linnette Grove, and Oleg Lapets ASSAY and Drug Development Technologies 2004, 2:
 1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
Epigenetics and Flowering Any potentially stable and heritable change in gene expression that occurs without a change in DNA sequence
 Epigenetics and Flowering Any potentially stable and heritable change in gene expression that occurs without a change in DNA sequence www.plantcell.org/cgi/doi/10.1105/tpc.110.tt0110 Epigenetics Usually
Epigenetics and Flowering Any potentially stable and heritable change in gene expression that occurs without a change in DNA sequence www.plantcell.org/cgi/doi/10.1105/tpc.110.tt0110 Epigenetics Usually
Hormones Act Downstream of TTG and GL2 to Pmmote Root Hair Outgmwth during Epidermis Development in the Arabidopsis Root
 The Plant Cell, Vol. 8, 1505-1517, September 1996 0 1996 American Society of Plant Physiologists Hormones Act Downstream of TTG and GL2 to Pmmote Root Hair Outgmwth during Epidermis Development in the
The Plant Cell, Vol. 8, 1505-1517, September 1996 0 1996 American Society of Plant Physiologists Hormones Act Downstream of TTG and GL2 to Pmmote Root Hair Outgmwth during Epidermis Development in the
Making Holes in Leaves: Promoting Cell State Transitions in Stomatal Development
 The Plant Cell, Vol. 19: 1140 1143, April 2007, www.plantcell.org ª 2007 American Society of Plant Biologists Making Holes in Leaves: Promoting Cell State Transitions in Stomatal Development The leaves
The Plant Cell, Vol. 19: 1140 1143, April 2007, www.plantcell.org ª 2007 American Society of Plant Biologists Making Holes in Leaves: Promoting Cell State Transitions in Stomatal Development The leaves
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its
 Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its transcriptional activity in wild-type embryo. A gradient of canonical
Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its transcriptional activity in wild-type embryo. A gradient of canonical
The geneticist s questions. Deleting yeast genes. Functional genomics. From Wikipedia, the free encyclopedia
 From Wikipedia, the free encyclopedia Functional genomics..is a field of molecular biology that attempts to make use of the vast wealth of data produced by genomic projects (such as genome sequencing projects)
From Wikipedia, the free encyclopedia Functional genomics..is a field of molecular biology that attempts to make use of the vast wealth of data produced by genomic projects (such as genome sequencing projects)
Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct
 SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed
 Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
Host-Pathogen Interaction. PN Sharma Department of Plant Pathology CSK HPKV, Palampur
 Host-Pathogen Interaction PN Sharma Department of Plant Pathology CSK HPKV, Palampur-176062 PATHOGEN DEFENCE IN PLANTS A BIOLOGICAL AND MOLECULAR VIEW Two types of plant resistance response to potential
Host-Pathogen Interaction PN Sharma Department of Plant Pathology CSK HPKV, Palampur-176062 PATHOGEN DEFENCE IN PLANTS A BIOLOGICAL AND MOLECULAR VIEW Two types of plant resistance response to potential
3.B.1 Gene Regulation. Gene regulation results in differential gene expression, leading to cell specialization.
 3.B.1 Gene Regulation Gene regulation results in differential gene expression, leading to cell specialization. We will focus on gene regulation in prokaryotes first. Gene regulation accounts for some of
3.B.1 Gene Regulation Gene regulation results in differential gene expression, leading to cell specialization. We will focus on gene regulation in prokaryotes first. Gene regulation accounts for some of
Ulrike Folkers 1, Jürgen Berger 2 and Martin Hülskamp 1, * SUMMARY
 Development 124, 3779-3786 (1997) Printed in Great Britain The Company of Biologists Limited 1997 DEV0088 3779 Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary
Development 124, 3779-3786 (1997) Printed in Great Britain The Company of Biologists Limited 1997 DEV0088 3779 Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary
A developmental geneticist s guide to roots Find out about the hidden half of plants
 the Centre for Plant Integrative Biology A developmental geneticist s guide to roots Find out about the hidden half of plants What do roots look like from the inside? How do roots form? Can we improve
the Centre for Plant Integrative Biology A developmental geneticist s guide to roots Find out about the hidden half of plants What do roots look like from the inside? How do roots form? Can we improve
A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development
 A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development Xuemei Chen Waksman Institute, Rutgers University, Piscataway, NJ 08854, USA. E-mail: xuemei@waksman.rutgers.edu Plant
A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development Xuemei Chen Waksman Institute, Rutgers University, Piscataway, NJ 08854, USA. E-mail: xuemei@waksman.rutgers.edu Plant
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
P-Glycoprotein4 Displays Auxin Efflux Transporter Like Action in Arabidopsis Root Hair Cells and Tobacco Cells W OA
 This article is published in The Plant Cell Online, The Plant Cell Preview Section, which publishes manuscripts accepted for publication after they have been edited and the authors have corrected proofs,
This article is published in The Plant Cell Online, The Plant Cell Preview Section, which publishes manuscripts accepted for publication after they have been edited and the authors have corrected proofs,
Genetically Engineering Yeast to Understand Molecular Modes of Speciation
 Genetically Engineering Yeast to Understand Molecular Modes of Speciation Mark Umbarger Biophysics 242 May 6, 2004 Abstract: An understanding of the molecular mechanisms of speciation (reproductive isolation)
Genetically Engineering Yeast to Understand Molecular Modes of Speciation Mark Umbarger Biophysics 242 May 6, 2004 Abstract: An understanding of the molecular mechanisms of speciation (reproductive isolation)
SUPPLEMENTARY INFORMATION
 PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis Momoko Ikeuchi 1 *, Akira Iwase 1 *, Bart Rymen 1, Hirofumi Harashima 1, Michitaro Shibata 1, Mariko Ohnuma 1, Christian Breuer 1,
PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis Momoko Ikeuchi 1 *, Akira Iwase 1 *, Bart Rymen 1, Hirofumi Harashima 1, Michitaro Shibata 1, Mariko Ohnuma 1, Christian Breuer 1,
