FURCA loci. Genetic control of trichome branch number in Arabidopsis: the roles of the. Danlin Luo and David G. Oppenheimer* SUMMARY
|
|
|
- Gloria Tucker
- 5 years ago
- Views:
Transcription
1 Development 126, (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV Genetic control of trichome branch number in Arabidopsis: the roles of the FURCA loci Danlin Luo and David G. Oppenheimer* Department of Biological Sciences and Coalition for BioMolecular Products, University of Alabama, Tuscaloosa, AL , USA *Author for correspondence ( Accepted 5 October; published on WWW 24 November 1999 SUMMARY We are using trichome (hair) morphogenesis as a model to study how plant cell shape is controlled. During a screen for new mutations that affect trichome branch initiation in Arabidopsis, we identified seven new mutants that show a reduction in trichome branch number from three branches to two. These mutations were named furca, after the Latin word for two-pronged fork. These seven recessive mutations were placed into four complementation groups that define four new genes: FURCA1, FURCA2, FURCA3 and FURCA4. The trichome branch number phenotype indicates that the FURCA genes encode positive regulators of trichome branch initiation. Analysis of double mutants suggests that primary and secondary branch initiation events are not genetically distinct, but rely on the levels of partially redundant groups of regulators of trichome branch initiation. Based on the analysis of both epistatic and genetic interactions between the FURCA genes and other genes that control trichome branch number, we propose a model that explains how these genes interact to control trichome branch initiation. This model successfully predicts the phenotypes of all the single and double mutants examined and suggests points of control of the trichome branch pathway. Key words: Arabidopsis thaliana, Trichome, Cell shape, Cell differentiation, Cell expansion INTRODUCTION A fundamental question in developmental biology is how is cell shape controlled. In plants, cell shape is important not only for the function of the individual cell, but also for its contribution to the generation of plant form. Because plant cells are surrounded by a cell wall any changes in cell shape must be coupled with appropriate expansion of the cell wall. Although much has been learned about the mechanisms of cell expansion (Cosgrove, 1997a,b; Giddings and Staehelin, 1991; McQueen-Mason, 1997; Nicol and Höfte, 1998; Pennell, 1998), the molecular events underlying the initiation of localized cell expansion events remain obscure. However, comparisons with fungal and algal model systems provide insights into the initiation of localized expansion events in higher plants (Fowler and Quatrano, 1997). First, a cortical site is chosen at the position of future cell expansion. Then, targeted secretion modifies the site of expansion. Finally, interactions with the cytoskeleton place the cell wall synthesis machinery at the selected site and localized expansion ensues. To uncover the mechanisms by which cell expansion events are initiated, we have studied the initiation of branches on trichomes (epidermal hairs) of Arabidopsis as a model. Arabidopsis trichomes are large, single cells that project from the epidermis of leaves, stems and sepals. Trichome development begins when a protodermal cell, in response to a complex interplay of tissue- and organ-specific regulators, adopts the trichome cell fate (Chien and Sussex, 1996; Larkin et al., 1999, 1996; Perazza et al., 1998; Schnittger et al., 1998; Szymanski et al., 1988; Telfer et al., 1997). Once the trichome fate is determined, the nascent trichome undergoes several rounds of endoreplication concomitant with cell enlargement and expansion out of the plane of the leaf blade (Hülskamp et al., 1994). When the developing trichome has expanded to the shape of a small cylinder, branching is initiated as a bulge on the distal face of the developing trichome not at the tip of the cylinder but approximately halfway between the base and the tip of the cylinder (Folkers et al., 1997; Hülskamp et al., 1994). The bulge expands to become the first trichome branch while the tip of the cylinder continues to expand to form a second branch. As the first branch continues to expand, a third branch is initiated at its base (Folkers et al., 1997; Hülskamp et al., 1994). Expansion continues until the trichome is 300 to 500 µm tall. The final phase of trichome differentiation is maturation. During maturation, the trichome cell wall thickens, accessory cells differentiate around the base of the trichome, and the surface of the trichome develops numerous papillae (Hülskamp et al., 1994; Marks et al., 1991). Previous studies of trichome development have identified a number of mutations that affect trichome branch number (Hülskamp et al., 1994; Folkers, et al., 1997; Perazza et al., 1999). Mutations in NOEK (NOK), TRIPTYCHON (TRY),
2 5548 D. Luo and D. G. Oppenheimer KAKTUS (KAK), POLYCHOME (PYM), RASTAFARI (RFI) and SPINDLY (SPY) lead to an increase in trichome branch number, whereas mutations in STICHEL (STI), ANGUSTAFOLIA (AN), ZWICHEL (ZWI), GLABRA3 (GL3) and STACHEL (STA) lead to trichomes with fewer than normal branches. The first genetic analysis of trichome branching was conducted by Folkers et al. (1997) who studied branching in single and double mutant combinations of an, zwi, sti, gl3, sta, nok and try. Based on their results, Folkers et al. (1997) proposed that trichome branching was controlled by trichome cell growth (or level of endoreplication). In addition, Folkers et al. (1997) proposed that primary and secondary branching were genetically distinct events. To further examine the genetic control of trichome branching, we screened for additional mutants that showed an altered trichome branch number. Seven new mutations that define four new genes were isolated; these mutations cause a decrease in trichome branch initiation resulting in trichomes with fewer than normal branches. Thus, these four new genes act as positive regulators of trichome branch initiation. The new mutations cannot be clearly assigned to roles in primary and secondary branching which suggests that this distinction is an oversimplification. Our data suggest instead that trichome branching is regulated by parallel, partially redundant pathways. Furthermore, our results from the analysis of double mutants suggest that cell growth and the control of trichome branch number are most likely independent processes. MATERIALS AND METHODS Plant strains and growth conditions The plant strains used in this study are summarized in Table 1. Mutants were backcrossed at least once to wild-type plants of their respective ecotypes. Plants were grown under constant illumination as previously described (Krishnakumar and Oppenheimer, 1999) and fertilized twice with a complete nutrient solution (Pollock and Oppenheimer, 1999). Genetic mapping A representative allele from each of the four furca (frc) complementation groups was mapped using classical and molecular markers. To determine the genetic map position of the FRC genes relative to classical phenotypic markers, the frc mutants were crossed to the following strains (obtained from the Arabidopsis Biological Resource Center, The Ohio State University; strain [marker scored]): cs124 (tt1-1), cs35 (cer5-1), cs30 (bp-1), cs34 (cer4-1), cs38 (cer7-1), cs85 (tt4-1), cs75 (ms1-1), cs137 (cer3-1) and Ler (er). Plants showing the frc phenotype were selected from the subsequent F 2 population and scored for the phenotype of the classical phenotypic marker. The frequency of recombination between the frc mutation and the classical marker was used to determine the approximate map position of each FRC locus. Map positions were confirmed by using molecular markers (Bell and Ecker, 1994; Konieczny and Ausubel, 1993). Construction of double mutants Double mutants were constructed by intercrossing the strains of interest and letting the F 1 plants self. Plants displaying only one of the parental phenotypes were collected from the F 2 population and allowed to self. Putative double mutants (plants with a novel phenotype) were selected from the F 3 population. Putative double mutants were confirmed by complementation tests with each of the original parents. In addition, double mutants were backcrossed to wild-type plants, and the subsequent F 2 population was examined for segregation of both of the original parental phenotypes. Physical characterization of the frc mutants Trichomes on wild-type and mutant plants were examined by scanning electron microscopy (SEM) as previously described (Oppenheimer et al., 1997). To quantify the trichome branch numbers for each mutant, all the trichome branches were counted on the adaxial side of either the third or fourth leaf from at least eight plants. RESULTS Isolation of new trichome branch number mutants To identify additional genes that control trichome branch initiation, we screened for Arabidopsis mutants that showed an altered trichome branch number. We isolated seven mutants that showed a reduction in the number of trichome branches (Table 1; Fig. 1). These mutants were named furca (frc). To determine if the mutations were dominant or recessive, we backcrossed the frc mutants to wild-type plants. All the crosses produced wild-type F 1 progeny which demonstrated that all the frc mutations were recessive. Furthermore, analysis of the segregation ratios of the frc plants to wild-type plants in the F 2 population from each backcross indicated that each of the frc mutations was monogenic. To determine if the frc mutations represented new trichome loci or previously identified loci, we performed complementation tests with sti, an, zwi, gl3, sta, nok and try mutants. We also crossed the frc mutants to each other to determine if any were allelic. The results of the pairwise complementation tests showed that the seven frc mutations represent four new genes that control trichome branch number. The new trichome branch number genes defined by the four complementation groups were named FRC1 (two alleles), FRC2 (three alleles), FRC3 (one allele) and FRC4 (one allele). In addition to the complementation tests, we determined the genetic map positions of each of the FRC genes to confirm that each of the FRC complementation groups represented independent loci. The FRC1 locus is located near the bottom of chromosome III, tightly linked to the classical genetic marker CER7 (we found no frc1-1 cer7 recombinants out of 680 frc1-1 plants scored). The FRC2 locus is approximately 12 Table 1. Name and origin of the trichome branch number mutants used in this study Strain Mutagen Ecotype Source (reference) frc1-1 EMS Col This study frc1-2 EMS Col This study frc2-1 EMS Col Joy Chien frc2-2 EMS Col This study frc2-3 Fast neutrons RLD This study frc3-1 Fast neutrons RLD This study frc4-1 Fast neutrons RLD This study sta-23 EMS Ler M. Hülskamp (Hülskamp et al., 1994) try-240 EMS Ler M. Hülskamp (Hülskamp et al., 1994) an-496 EMS RLD This study zwi-3 EMS Col Krisnakumar and Oppenheimer (1999) zwi Fast neutrons RLD Krisnakumar and Oppenheimer (1999) zwi-w2 EMS RLD Krisnakumar and Oppenheimer (1999) sti Fast neutrons RLD This study nok Fast neutrons RLD This study gl3 EMS Ler Koornneef et al. (1982)
3 Genetic control of trichome morphogenesis 5549 Table 2. Number of branches on wild-type and frc mutant trichomes Trichome branch points* Genotype (ecotype) Total Wild type (RLD) < Wild type (Col) frc1-1 (Col) frc2-1 (Col) frc2-3 (RLD) frc3-1 (RLD) frc4-1 (RLD) *% of trichomes having the indicated number of branch points (1 branch point indicates a trichome with two branches). Total number of trichomes counted. Fig. 1. Light micrographs of developing leaves of wild-type and frc mutants showing mature trichomes. (A) Fifth leaf of a Col wild-type plant showing mostly three-branched trichomes. (B) Fifth leaf of a frc1-1 plant. (C) Fifth leaf of a frc2-3 plant. (D) Fifth leaf of a frc3-1 plant and (E) Fifth leaf of a frc4-1 plant. cm south of classical marker cer5, and approximately 23 cm south of SSLP marker nga280 on chromosome I. The map position of the FRC3 locus is approximately 3 cm south of the molecular marker g4539 on chromosome IV. FRC4 is located on chromosome II, linked to the classical marker er (we found no frc4-1 er recombinants out of 232 frc4-1 plants scored). Because no known trichome branch mutants have been mapped to these positions, these results demonstrate that each of the FRC complementation groups represents a new gene. Phenotypic analysis of the frc mutants To determine if any of the frc mutations affect trichome number, we counted the number of trichomes on either the third or fourth leaf of at least 10 plants of each of the frc mutants. No significant difference was observed between the frc mutants and wild-type plants of the same ecotype (data not shown). This result suggests that the FRC genes play no role in cell fate decisions. Wild-type plants generally have three or four trichome branches whereas most of the trichomes on frc mutant plants have only two branches. All of the branched trichomes on the plants were affected by the frc mutations including trichomes on the abaxial sides of the leaves and the few branched trichomes found on the floral stem. The unbranched stem and sepal trichomes were unaffected by the frc mutations. To quantify the extent of the trichome branch number reduction, trichome branches were counted on leaves of frc mutants and wild-type plants (Table 2). The frc2 mutants have the weakest phenotype of the frc mutations. Generally, only about 60% of the trichomes on frc2 mutants have two branches whereas more than 90% of the trichomes on the other frc mutants have two branches (Table 2). However, all the frc mutants display a significant decrease in trichome branch number compared to wild-type plants, and their segregation can be easily followed in crosses. Because the frc mutations are likely to be loss-offunction mutations, this result suggests that the FRC genes act as positive regulators of trichome branch initiation. We also used SEM to examine the trichomes of the frc mutants (Fig. 2). Only trichome branch initiation appears to be affected in the frc mutants; trichome maturation is not affected. Variation in the size and/or morphology of the accessory cells in frc mutants is similar to that observed in wild-type plants. The stalk height of frc mutant trichomes was within the normal variation seen in wild-type plants. Plant size, growth rate, flowering time, fertility and general appearance of the frc mutants were also examined. Plants homozygous for either frc1 or frc3 are wild type in overall appearance, growth rate, flowering time and fertility. Plants homozygous for either the frc2-1 or the frc2-3 alleles, however, show decreased fertility compared to wild-type plants (data not shown). This decrease in fertility appears to be caused by a premature extension of the pistil from the unopened flower before the anthers mature. In outcrosses, frc2 plants showed no obvious decrease in fertility either when used as the female parent or as the male parent (data not shown). These results suggest that the fertility defect is a pleiotropic effect of the frc2 mutation and not a secondary mutation linked to the frc2 mutation. Plants homozygous for the frc4-1 mutation also show apparent pleiotropic effects. The frc4 mutant plants grow slower than wild-type plants, and produce abnormally short and bushy bolts at the time of flowering (data not shown). In addition, frc4 mutants also show a decrease in fertility (data not shown). However, because we isolated only one mutant allele of frc4, we cannot rule out tightly linked, secondary mutations as the cause of the pleiotropic effects of frc4-1. Double mutants Genetic interactions among the frc mutations To understand the genetic relationships among the frc mutations, pairwise combinations of frc double mutants were constructed.
4 5550 D. Luo and D. G. Oppenheimer Fig. 2. Scanning electron micrographs of mature leaf trichomes of wild-type, frc and other trichome branch number mutants. If the double mutant phenotype was similar to either one of the single mutants, then the interaction was interpreted as being epistatic. If the double mutant phenotype was more severe than either single mutant, then the interaction was interpreted as being. Mild effects were disregarded because they were indistinguishable from background effects. The frc1-1 frc3-1 and frc1-1 frc4-1 double mutants produce predominantly unbranched trichomes, and frc3-1 frc4-1 double mutants have exclusively unbranched trichomes (Table 3; Fig. 3). These results suggest that FRC1, FRC3 and FRC4 act independently in the control of trichome branch number because the effects of mutations in these three genes are. The phenotype of frc2-1 frc4-1 double mutants is the same as that of the frc4-1 single mutants (Table 3; Fig. 3); therefore, frc4-1 is epistatic to frc2-1. Although more than 60% of the trichomes on frc2-1 frc1-1 double mutants and the frc2-3 frc3-1 double mutants have two branches, at least 30% of the trichomes on these double mutants are unbranched (Table 3). Therefore, the effect of frc2-1 in combination with
5 Genetic control of trichome morphogenesis 5551 either frc1-1 or frc3-1 is. These results suggest that the FRC2 gene is likely to act independently of both the FRC1 and the FRC3 genes. Genetic interactions between the frc mutations and the other mutations that control trichome branch number To determine the genetic relationships between the frc mutations and the other mutations that control trichome branch number, we constructed pairwise combinations of double mutant strains. We uncovered epistatic relationships among several pairs of branch mutant alleles. These are described below. frc sti Plants homozygous for both the sti mutation and any of the frc mutations produced only unbranched trichomes (Table 3; Fig. 2). However, sti single mutants also produced exclusively unbranched trichomes. Therefore, effects could not be distinguished from epistatic effects (see Discussion). frc gl3 The gl3 mutation has pleiotropic effects on trichome development; gl3 mutants have aborted trichomes on the first leaf pair (D. G. O., unpublished results), a reduction in the
6 5552 D. Luo and D. G. Oppenheimer Table 3. Number of trichome branches on frc double mutants Trichome branch points* Genotype >4 Total frc1-1 frc frc1-1 frc frc1-1 frc frc2-3 frc frc2-1 frc frc3-1 frc sti frc1-1 sti frc2-1 sti frc3-1 sti frc4-1 sti gl frc1-1 gl frc2-1 gl frc3-1 gl frc4-1 gl nok frc1-1 nok < frc2-1 nok frc3-1 nok frc4-1 nok try frc1-1 try frc2-1 try frc3-1 try frc4-1 try an frc1-1 an frc2-1 an frc3-1 an frc4-1 an sta frc1-1 sta frc2-1 sta frc3-1 sta zwi frc1-1 zwi frc2-1 zwi frc3-1 zwi frc4-1 zwi *% of trichomes having the indicated number of branch points (1 branch point indicates a trichome with two branches). Total number of trichomes counted. number of trichome branches (Table 3 and Folkers et al., 1996), a reduction in the amount of nuclear DNA (Hülskamp et al., 1994), a decrease in apparent trichome cell size (Hülskamp et al., 1994), and an increase in trichome clusters (D. G. O., unpublished results). Double mutant strains containing gl3 and frc1 or frc3 produce predominantly (98%) unbranched trichomes (Table 3; Fig. 2). However, in these mutants, aborted trichomes are produced on the first leaf pair, and trichome clusters are present at the same frequency as seen in gl3 mutants (data not shown). All aspects of the gl3 phenotype appear to be epistatic to the frc1 or frc3 mutations except the trichome branch number reduction the effect of which was in combination with either frc1 or frc3. This result suggests that GL3 may independently control different aspects of trichome development, and that FRC1 and FRC3 contribute independently to the control of trichome branch number. The gl3 mutation is completely epistatic to the frc2 and frc4 mutations: the gl3 frc2-1 and gl3 frc4-1 double mutants have Fig. 3. Scanning electron micrographs of mature leaf trichomes of frc frc double mutants. approximately the same proportions of two-branched and unbranched trichomes as gl3 single mutants (Table 3; Fig. 2). This result suggests that GL3, FRC2 and FRC4 act in the same pathway to control trichome branch number (see Discussion). frc nok and frc try Plants homozygous for the nok mutation produce trichomes that have more branches than trichomes on wild-type plants (Folkers et al., 1997). Wild-type plants normally produce trichomes with three or four branches (very rarely five branches), whereas trichomes on nok mutants have up to eight branches (Folkers, et al., 1997; Tables 2, 3; Fig. 2). Thus, the NOK gene may act as a negative regulator of trichome branching. In addition to the effect on trichome branching, nok mutants also fail to complete trichome maturation; the trichomes on nok plants lack the wild-type number of papillae and appear glassy. To determine if any of the FRC genes might be negatively regulated by NOK, we constructed all the frc nok double mutants. The frc1-1 nok , frc2-1 nok and frc3-1 nok double mutants had more trichome branches than any of the frc single mutants, but fewer branches than in nok single mutants (Table 3; Fig. 2). This intermediate number of trichome branches demonstrates an effect of these frc mutations in combination with the nok mutation. In contrast to the frc1, frc2 and frc3 mutations, the frc4-1
7 Genetic control of trichome morphogenesis 5553 mutation was completely epistatic to the nok mutation with respect to trichome branch number. Approximately 92% of the trichomes on frc4-1 nok double mutants had two branches compared with 98% two-branched trichomes on frc4-1 single mutants (Table 3). However, the trichomes on all the frc nok double mutants have the same glassy phenotype as nok single mutants. This result shows that the effects of nok on maturation are genetically separable from the effects on branching. Mutations in TRY also have pleiotropic effects on trichome development: in addition to an increase in branch number, trichomes on try mutants develop in clusters and have an increased amount of nuclear DNA (Hülskamp et al., 1994). The results of the frc try double mutant analysis were similar to those of the frc nok double mutants. The frc1-1 try-240 and frc3-1 try-240 double mutants generally had fewer trichome branches than observed on try-240 single mutants, but more than observed on either frc1-1 or frc3-1 single mutants (Table 3; Fig. 2). For example, only 0.9% of the trichomes on frc1-1 plants have three branches compared with 20% on frc1-1 try- 240 double mutants, and 41.6% of the trichomes on try-240 single mutants have five branches compared with 0% of the trichomes on frc1-1 try-240 double mutants (Table 3). These results show an effect on trichome branching of either the frc1 or frc3 mutation in combination with the try mutation. Like the frc4-1 nok double mutant, the frc4-1 try- 240 double mutant produces mostly (92.3%) two-branched trichomes (Table 3; Fig. 2). Thus, the frc4-1 mutation is epistatic to the try-240 mutation with respect to trichome branch number. The frc2-1 mutation was weakly epistatic to the try-240 mutation; close to 40% of the trichomes on the frc2-1 try-240 double mutant had two branches compared with approximately 60% of the trichomes on frc2-1 plants (Table 3). All the frc try double mutants produced clusters of trichomes (Fig. 2) at the same frequency as try single mutants (data not shown). frc an Plants homozygous for the an mutation mostly produce trichomes with two branches instead of the wild-type number of three or four branches. All the frc an double mutants produced predominantly (82-100%) unbranched trichomes (Table 3; Fig. 2). Thus, the frc mutations have an negative effect on trichome branch number when in combination with the an mutation. frc sta and frc zwi Mutations in the STA gene lead to trichomes with mostly two branches (Table 9 and Fig. 9). The sta mutation was found to be epistatic to only the frc2 mutations; the frc2-1 sta-23 double mutants produced two- and three-branched trichomes in roughly the same proportions as in sta-23 single mutants (Table 3). The frc1 and frc3 mutations had an effect in combination with the sta-23 mutation. For example, both frc3-1 and sta-23 single mutants produced no unbranched trichomes, but nearly 60% of the trichomes on the frc3-1 sta- 23 double mutant were unbranched (Table 3). We were unable to isolate a fertile frc4-1 sta-23 double mutant both single mutants were small, slow growing plants with decreased fertility, and the putative frc4-1 sta-23 double mutant was severely stunted and infertile. However, we were able to identify several infertile, putative double mutants from which we estimated that they produced approximately 80% unbranched trichomes and 20% two-branched trichomes. Thus, sta-23 had an effect in combination with frc4-1. The results for the frc zwi double mutants were similar to those for the frc sta double mutants; zwi was epistatic only to frc2 (Table 3; Fig. 2). Whereas zwi single mutants produced approximately 60% two-branched trichomes, frc1-1 zwi , frc3-1 zwi and frc4-1 zwi double mutants had more than 93% unbranched trichomes (Table 3). DISCUSSION During a screen for mutants with altered trichome branch number, we discovered seven mutations that define four new genes involved in the control of trichome branch number. The discovery of these new mutants allowed us to further test a previously published model for the control of trichome branch number (Folkers et al., 1997). We characterized the trichome phenotypes of these new mutants, and determined the genetic relationships of the new genes to each other and to previously identified genes known to control trichome branch number. We found that the phenotypes of the single and double mutants deviated significantly from those predicted by the model presented by Folkers et al. (1997) which suggested a fundamentally different view of the mechanisms by which trichome branch number is controlled. The FURCA genes act as positive regulators of trichome branch formation Four new mutants (frc1, frc2, frc3 and frc4) that show a reduction in the number of trichome branches were described in this report. Because all of these mutations lead to fewer than the normal number of trichome branches, these results demonstrate that the products of the wild-type FRC genes play positive roles in trichome branch initiation. We attempted to determine whether primary or secondary branch initiation was affected in the frc mutants according to the criteria set by Folkers et al. (1997). However, the distinction between effects on primary or secondary branch initiation was ambiguous in our mutants, and we were unable to draw a clear distinction between them for the frc mutants. This issue is addressed in detail below. A new model for the control of trichome branch number None of the mutations examined in this study is epistatic to all others; this result suggests that parallel, partially redundant pathways exist for the control of trichome branch initiation. To construct a model for the control of trichome branch number we used the following rationale. If one branch mutation is epistatic to another then the two genes function in the same pathway; however, if the effects of two branch mutations are in the double mutant then the two genes function in separate pathways. Generally, this interpretation is applied to complete loss-of-function (null) alleles. None of the FRC genes have been cloned yet; therefore, we cannot determine if the mutant frc alleles are null alleles. However, because most induced mutations in Arabidopsis are loss-of-function
8 5554 D. Luo and D. G. Oppenheimer Col wildtype > sti gl3 > nok ª try-240 > > an-496 sta-23 > zwi ª frc1-1 frc1-1 frc2-1 > frc1-1 frc3-1 frc1-1 frc4-1 frc1-1 sti frc1-1 gl3 frc1-1 nok > > frc1-1 try-240 > frc1-1 an-496 frc1-1 sta-23 ª frc1-1 zwi frc2-1 > frc2-3 frc3-1 > frc2-1 frc4-1 frc2-1 sti frc2-1 gl3 frc2-1 nok > frc2-1 try-240 > WEAKLY frc2-1 an-496 frc2-1 sta-23 frc2-1 zwi > frc3-1 frc3-1 frc4-1 frc3-1 sti frc3-1 gl3 frc3-1 nok > frc3-1 try-240 ª frc3-1 an-496 frc3-1 sta-23 > frc3-1 zwi frc4-1 frc4-1 sti frc4-1 gl3 > frc4-1 nok frc4-1 try-240 frc4-1 an-496 frc4-1 zwi try-em1 sti-emu* try-em1 an-em1* try-em1 zwi-em1* try-em1 sta-23* try-em1 nok-122* 8-10 branches try-em1 gl3* > weakly nok-122 sti-emu* nok-122 an-em1* nok-122 zwi-em1* nok-122 sta-23* > nok-122 gl3* WEAKLY gl3 sti-emu* gl3 an-em1* > gl3 zwi-em1* gl3 sta-23* > sti-emu an-em1* sti-emu zwi-em1* sti-emu sta-23* zwi-em1 an-em1* zwi-em1 sta-23* > an-em1 sta-23* Fig. 4. Summary of the phenotypes of the double mutants. represents an unbranched trichome; represents a two-branched trichome; represents a three-branched trichome, and so on. The interpretation of the interaction for each double mutant combination is given in the third column. The data for the double mutant combinations markedwith a * were taken from Folkers et al. (1997). A > between two trichomes means that the proportion of trichomes of the first type is greater than the proportion of trichomes of the second type. A symbol between two trichomes means that both types of trichomes are present in approximately equal proportions. mutations, we will assume that the phenotypes of the frc mutants used in this study represent the phenotypes of strong loss-of-function alleles. A summary of the results of our genetic analysis is presented in Fig. 4. Applying the above rationale to the pairwise combinations of double mutants, we have developed a model for trichome branch initiation (Fig. 5). With this model, it is possible to predict the phenotypes of all the single and double mutants in this study as well as all the single and double mutants in the study conducted by Folkers et al. (1997). It is important to note that the model represents the genetic interactions between the genes controlling trichome branch initiation and not a developmental pathway per se. The model shows that trichome branch initiation is controlled by parallel, partially redundant pathways. Our double mutant analyses did not support the hypothesis that primary and secondary branching are genetically separate events. If primary and secondary branching is controlled by different sets of genes, then the mutations that produce two-
9 Genetic control of trichome morphogenesis 5555 NOK AN STI FRC1 ZWI Branch Initiation FRC2 TRY FRC3 STA FRC4 GL3 Fig. 5. A model for the genetic control of trichome branch number in Arabidopsis. Arrows indicate positive roles, blunt-ended lines indicate negative roles. The open arrow indicates the greater requirement for STI function during branching than the other branch number genes. The three-tailed arrow (under FRC2) indicates that each of the three genes (ZWI, STA and FRC4) must be present for FRC2 function. branched trichomes should fall into one of two classes: those that affect primary branching (like STA) and those that affect secondary branching (like AN). In double mutants containing combinations of primary branch mutations and secondary branch mutations, no trichome branches should be produced (as seen in an sta double mutants). Similarly, if primary and secondary branching are genetically distinct, then double mutant combinations of only primary branch mutations should produce branched trichomes. If double mutant combinations of two primary branch mutations produced unbranched trichomes, then this would indicate that both mutations affect both branching events, and thus are not genetically distinct. We found that all double mutant combinations of frc1, frc3 and frc4 produced predominantly unbranched trichomes (Table 3). Likewise, we found that all double mutant combinations of an, frc3 and frc4 (Table 3) produced unbranched trichomes. Therefore, we conclude that one or more of these genes must participate in both branching events even though the single mutants produce two-branched trichomes. Although, it is still possible that some components of branch initiation may be specific for one or the other of the branching events, the hypothesis that branch initiation is controlled by parallel, partially redundant pathways is sufficient to explain the observed phenotypes of the single and double mutants. Redundancy in pathways (both developmental and biochemical) is common in biology (Normanly and Bartel, 1999; Pickett and Meeks-Wagner, 1995; Thomas, 1993; Thomas et al., 1993). This makes sense, because redundancy in biological pathways increases the reliability of the pathway (McAdams and Arkin, 1999). Cell expansion is a complex process involving the cytoskeleton, cell wall degradation and synthesis enzymes and the proteins that are needed to target them to the site of expansion. Thus, it is likely that trichome branch formation involves multiple proteins acting together at the site of branch initiation, and that such a multi-protein complex could be used for both branching events. With this in mind, one may expect that the genetic analysis of branch initiation would identify many genes acting at the same level in the genetic pathway (such as ZWI, AN, FRC1, etc.). The relationship of STI to the FRC genes We found that the sti mutation appeared to be epistatic to all the frc mutations. However, because sti single mutants produce exclusively unbranched trichomes, we were unable to distinguish from epistatic interactions. Therefore, it was difficult to determine whether STI acts upstream or downstream of the other branch number genes, or in which pathways STI functions. Several possibilities exist. First, STI may act upstream of several of the other branch number genes. For example, the STI product could be a transcription factor that regulates several of the branch number genes. Thus, sti mutations would phenotypically resemble the double (or multiple) mutants of those genes STI regulates. Second, STI may function downstream of several of the other branch number genes. Again, STI would need to be regulated by more than one of the other branch number genes because all the single mutants produce two-branched trichomes, and sti mutants produce unbranched trichomes. A third possibility is that STI lies on a pathway independent from the other branch number genes but is absolutely required for trichome branch initiation. Previously, nok mutations were shown to rescue branch initiation in a sti mutant background (Folkers et al., 1997). This result suggests that NOK and STI are on separate pathways. We have provisionally placed STI downstream from TRY in a pathway independent from the other branch number genes. This placement was chosen in part because STI is epistatic to TRY (Folkers et al., 1997). Because nok sti double mutants can initiate trichome branches (Folkers et al., 1997 and D. G. O., unpublished results), this result suggests that an increase in AN function (due to the nok mutation) can partially substitute for decreased STI function. FRC2 function is partially redundant to both FRC4 and ZWI function At present, the frc2 mutations have a less severe effect on trichome branch number than any of the other trichome branch number mutations (see Tables 2 and 3). One explanation for this effect is that FRC2 function is redundant with some other branch number regulator. Both zwi and frc4 mutations are epistatic to frc2 mutations which suggests that FRC2 is in the same pathway as ZWI and FRC4. However, the effect of the zwi mutation is in combination with the frc4 mutation which suggests that ZWI and FRC4 function in separate pathways. The most parsimonious resolution is to place FRC2 downstream of, but redundant to, both FRC4 and ZWI. In this position, one would predict that frc2 mutations would be less severe than either frc4 or zwi mutations (which is the case), and that both frc4 and zwi mutations would be epistatic to frc2 mutations (which is also the case). This hypothesis is supported by other frc2 double mutants; the effects of frc2 mutations were in combination with the other branch number reduction mutations (except sta; see below). Therefore, we conclude that the function supplied by FRC2 is redundant to both FRC4 and ZWI. Relationship between GL3 and the FRC genes The gl3 mutation has a number of effects on trichome development including trichome clustering, reduction in trichome branching, reduction in trichome endoreplication, and failure of some trichomes to expand out from the surface of the leaf. Trichome clustering is thought to be the result of the
10 5556 D. Luo and D. G. Oppenheimer inability of the nascent trichome to inhibit its neighbors in the equivalence group from also adopting the trichome cell fate. This lateral inhibition occurs early during the trichome cell fate determination process. Likewise, expansion of the trichome out from the plane of the leaf blade precedes trichome branch initiation. Therefore, it is likely that GL3 acts upstream of the FRC genes. In a previous study of trichome branching (Folkers et al., 1997), mutations in GL3 were shown to further reduce trichome branching in combination with an, sta, nok, and zwi mutations. These effects of gl3 were interpreted as indicating that a minimum cell size (or level of endoreplication) was required before trichome branching could be initiated. However, we found that gl3 was epistatic to both frc2 and frc4, but when in combination with either frc1 or frc3 mutations. The simplest explanation for our results is that GL3 functions in the same pathway as FRC4 and FRC2, but in a pathway separate from FRC1 and FRC3. In addition, in all the double mutant combinations of frc with gl3, the trichome clustering phenotype of gl3 is still observed (Fig. 2 and data not shown). Therefore, we propose that GL3 controls several independent processes during trichome development, two of which are endoreplication and trichome branch initiation. Additional support of our hypothesis comes from the recent identification of GL3 as a member of the DRAT family of transcription activators (Payne et al., 1999). It is possible that the FRC4 gene is a target of GL3 along with genes that control the endoreplication cycle in trichomes. Thus, the processes of trichome branch initiation and endoreplication may be coupled, but one need not be dependent upon the other. Similarly, the try mutation also has pleiotropic effects on trichome development. These effects include increased trichome endoreplication, trichome clustering, and an increased number of trichome branches. It was proposed that the increase in trichome branch number is due to the increased cell growth brought about by the extra rounds of endoreplication that occur in try mutants (Folkers et al., 1997; Perazza et al., 1999). As proposed for GL3, the data are consistent with the hypothesis that TRY regulates several independent processes. The relationship of STA to the FRC genes Our double mutant analysis has shown that the sta mutation has effects in combination with all the frc mutations except frc2. These results suggest that STA functions in the same pathway as FRC2, but in a separate pathway from the other FRC genes. Because the sta mutation is more severe than the frc2 mutation (Tables 2 and 3), we hypothesize that STA function may be partially redundant to FRC2 function. The relationship between the negative regulators of trichome branching (NOK and TRY) and the FRC genes Mutations in either TRY or NOK produce trichomes with more than the wild-type number of branches (see Fig. 2; Tables 2 and 3). This phenotype suggests that both NOK and TRY act as negative regulators of trichome branching. We found that frc4 mutations were epistatic to both nok and try mutations. Folkers et al. (1997) previously reported that both an and zwi mutations were epistatic to both try and nok mutations. However, AN and ZWI do not function in a linear pathway because an zwi double mutants show an effect (Folkers et al., 1997; Fig. 4). When both the results of Folkers et al. (1997) and our results are considered together, the simplest explanation is that TRY and/or NOK negatively regulate FRC4, AN and ZWI. However, if this were the case, then in a nok an double mutant there should be an increase in ZWI function due to the loss of negative regulation by nok. But nok mutations do not rescue the reduction in branch number of an mutants; therefore, increased ZWI function cannot compensate for the loss of AN function. Therefore, we conclude that increased levels of ZWI and AN cannot substitute for one another in branch initiation: both must be active for branching to occur. This hypothesis is supported by an zwi double mutants which produce unbranched trichomes (Folkers et al., 1997; D. G. O., unpublished results). This same line of reasoning applies to the relationships between NOK and FRC4 as well as the relationships between TRY and FRC4, ZWI, and AN. Therefore, we conclude that the FRC4, AN and ZWI products function together in trichome branch initiation, and that an increase in activity of one product cannot compensate for the loss of another. In addition, because the results from the epistasis analysis suggest that NOK is on a pathway separate from FRC2, NOK need only regulate AN to account for all the phenotypes of the nok double mutants. Given that NOK and FRC2 function in separate pathways, then the rescue of the frc2 phenotype by nok mutations suggests that increased AN function (due to loss of negative regulation by nok) can compensate for loss of FRC2 function. The most likely explanation for this result is that FRC2 and AN perform similar functions, either as members of the same gene family, or as physically distinct proteins engaged in the same function. We also found that the two-branched phenotype of both the frc1 and frc3 mutations can be suppressed by either try or nok mutations (see Fig. 2 and Table 3). These results suggest that FRC1 and FRC3 are not regulated by either NOK or TRY, but function in a pathway distinct from NOK and TRY. We conclude that an increase in FRC4, ZWI, or AN function can compensate for a decrease in FRC3 or FRC1 function. Similarly, we predict that an increase in either FRC1 or FRC3 function can compensate for a decrease in ZWI, AN, or FRC4 function. Several other negative regulators of trichome branching have been identified recently (Perazza et al., 1999), and one or more of these may function to negatively regulate FRC1 and/or FRC3. In conclusion, we have isolated seven mutations that identify four new genes (FRC1-4) controlling trichome branch number in Arabidopsis. Based on genetic interactions among the trichome branch number mutations we have tested an earlier model of trichome branching. We have proposed a new model for trichome branch initiation in which parallel, partially redundant pathways regulate branch formation. In addition, we provide evidence that cell growth (or endoreplication) may be controlled independently from trichome branch initiation. Cloning of the FRC genes and biochemical analysis of the products will help unravel their role in the localized cell expansion events that give trichomes their distinctive shape. The authors gratefully acknowledge M. Hülskamp for supplying Arabidopsis trichome mutants and Jolanta Nunley for the scanning
11 Genetic control of trichome morphogenesis 5557 electron micrographs. The authors also thank John Larkin, Ed Stephenson, Janis O Donnell, Sujatha Krishnakumar and Kevin Redding for stimulating discussions and critical reading of the manuscript. The authors also thank Mary Pollock for careful editing of the manuscript. The authors also gratefully acknowledge support from the NIH (R29 GM ). D. L. was supported in part by the University of Alabama SARTC and a Graduate Council Research Fellowship. The authors thank the Arabidopsis stock center for supplying seed of classical marker strains. REFERENCES Bell, C. J. and Ecker, J. R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19, Chien, J. and Sussex, I. M. (1996). Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 111, Cosgrove, D. J. (1997a). Assembly and enlargement of the primary cell wall in plants. Annu. Rev. Cell Biol. 13, Cosgrove, D. J. (1997b). Relaxation in a high-stress environment: the molecular bases of extensible cell walls and cell enlargement. Plant Cell 9, Folkers, U., Berger, J. and Hülskamp, M. (1997). Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary branching by branch initiation regulators and cell growth. Development 124, Fowler, J. E. and Quatrano, R. S. (1997). Plant cell morphogenesis: Plasma membrane interactions with the cytoskeleton and cell wall. Ann. Rev. Cell Dev. Biol. 13, Giddings, T. H. J. and Staehelin, L. A. (1991). Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In The Cytoskeletal Basis of Plant Growth and Form (ed. C. W. Lloyd), pp New York: Academic Press. Hülskamp, M., Miséra, S. and Jürgens, G. (1994). Genetic dissection of trichome cell development in Arabidopsis. Cell 76, Konieczny, A. and Ausubel, F. M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, Koornneef, M., Dellaert, S. W. M. and van der Veen, J. H. (1982). EMSand radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L) Heynh. Mutation Research 93, Krishnakumar, S. and Oppenheimer, D. G. (1999). Extragenic suppressors of the Arabidopsis zwi-3 mutation identify new genes that function in trichome branch formation and pollen tube growth. Development 126, Larkin, J. C., Walker, J. D., Bolognesi-Winfield, A. C., Gray, J. C. and Walker, A. R. (1999). Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 151, Larkin, J. C., Young, N., Prigge, M. and Marks, M. D. (1996). The control of trichome spacing and number in Arabidopsis. Development 122, Marks, M. D., Esch, J., Herman, P., Sivakumaran, S. and Oppenheimer, D. (1991). A model for cell-type determination and differentiation in plants. In Molecular Biology of Plant Development (ed. G. Jenkins and W. Schuch), pp Cambridge: Company of Biologists Limited. McAdams, H. H. and Arkin, A. (1999). It s a noisy business! Genetic regulation at the nanomolar scale. Trends Genet. 15, McQueen-Mason, S. (1997). Plant cell walls and the control of growth. Biochem. Soc. Trans. 25, Nicol, F. and Höfte, H. (1998). Plant cell expansion: scaling the wall. Curr. Opin. Plant Biol. 1, Normanly, J. and Bartel, B. (1999). Redundancy as a way of life IAA metabolism. Curr. Opin. Plant Biol. 2, Oppenheimer, D. G., Pollock, M. A., Vacik, J., Szymanski, D. B., Ericson, B., Feldmann, K. and Marks, M. D. (1997). Essential role of a kinesinlike protein in Arabidopsis trichome morphogenesis. Proc. Natl. Acad. Sci. USA 94, Payne, T., Zhang, F. and Lloyd, A. (1999). GLABROUS3 is a bhlh transcription factor that interacts with other regulators of trichome development. 10th International Conference On Arabidopsis Research, 4 th - 8th July, Melbourne, Australia. Pennell, R. (1998). Cell walls: structures and signals. Curr. Opin. Plant Biol. 1, Perazza, D., Herzog, M., Hülskamp, M., Brown, S., Dorne, A.-M. and Bonneville, J.-M. (1999). Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152, Perazza, D., Vachon, G. and Herzog, M. (1998). Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 117, Pickett, F. B. and Meeks-Wagner, D. R. (1995). Seeing double: appreciating genetic redundancy. The Plant Cell 7, Pollock, M. A. and Oppenheimer, D. G. (1999). Inexpensive alternative to M&S medium for selection of Arabidopsis plants in culture. BioTechniques 26, Schnittger, A., Jürgens, G. and Hülskamp, M. (1998). Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON in Arabidopsis. Development 125, Szymanski, D. B., Klis, D. A., Larkin, J. C. and Marks, M. D. (1988). cot1: a regulator of Arabidopsis trichome initiation. Genetics 149, Telfer, A., Bollman, K. M. and Poethig, R. S. (1997). Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124, Thomas, J. H. (1993). Thinking about genetic redundancy. Trends Genet. 9, Thomas, J. H., Birnby, D. A. and Vowels, J. J. (1993). Evidence for parallel processing of sensory information controlling dauer formation in Caenorhabditis elegans. Genetics 134,
Ulrike Folkers 1, Jürgen Berger 2 and Martin Hülskamp 1, * SUMMARY
 Development 124, 3779-3786 (1997) Printed in Great Britain The Company of Biologists Limited 1997 DEV0088 3779 Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary
Development 124, 3779-3786 (1997) Printed in Great Britain The Company of Biologists Limited 1997 DEV0088 3779 Cell morphogenesis of trichomes in Arabidopsis: differential control of primary and secondary
The IRREGULAR TRICHOME BRANCH loci regulate trichome elongation in Arabidopsis
 Plant Cell Physiol. 46(9): 1549 1560 (2005) doi:10.1093/pcp/pci168, available online at www.pcp.oupjournals.org JSPP 2005 The IRREGULAR TRICHOME BRANCH loci regulate trichome elongation in Arabidopsis
Plant Cell Physiol. 46(9): 1549 1560 (2005) doi:10.1093/pcp/pci168, available online at www.pcp.oupjournals.org JSPP 2005 The IRREGULAR TRICHOME BRANCH loci regulate trichome elongation in Arabidopsis
TRICHOMES are epidermal hairs found on the ae- other hand, are predominantly unbranched (Marks
 Copyright 1999 by the Genetics Society of America Trichome Cell Growth in Arabidopsis thaliana Can Be Derepressed by Mutations in at Least Five Genes Daniel Perazza,*,1 Michel Herzog,* Martin Hülskamp,
Copyright 1999 by the Genetics Society of America Trichome Cell Growth in Arabidopsis thaliana Can Be Derepressed by Mutations in at Least Five Genes Daniel Perazza,*,1 Michel Herzog,* Martin Hülskamp,
A complementation test would be done by crossing the haploid strains and scoring the phenotype in the diploids.
 Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
Bypass and interaction suppressors; pathway analysis
 Bypass and interaction suppressors; pathway analysis The isolation of extragenic suppressors is a powerful tool for identifying genes that encode proteins that function in the same process as a gene of
Bypass and interaction suppressors; pathway analysis The isolation of extragenic suppressors is a powerful tool for identifying genes that encode proteins that function in the same process as a gene of
THE GENETICS OF CERTAIN COMMON VARIATIONS IN COLEUS 1
 THE GENETICS OF CERTAIN COMMON VARIATIONS IN COLEUS DAVID C. RIFE, The Ohio State University, Columbus, Ohio Coleus are characterized by great variations in leaf color, and to a lesser degree by variations
THE GENETICS OF CERTAIN COMMON VARIATIONS IN COLEUS DAVID C. RIFE, The Ohio State University, Columbus, Ohio Coleus are characterized by great variations in leaf color, and to a lesser degree by variations
Solutions to Problem Set 4
 Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
GLABRA1 and TRIPTYCHON in Arabidopsis
 Development 125, 2283-2289 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV0160 2283 Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON
Development 125, 2283-2289 (1998) Printed in Great Britain The Company of Biologists Limited 1998 DEV0160 2283 Tissue layer and organ specificity of trichome formation are regulated by GLABRA1 and TRIPTYCHON
The bhlh genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis
 Research article 291 The bhlh genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis Christine Bernhardt 1, Mingzhe Zhao 2,
Research article 291 The bhlh genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis Christine Bernhardt 1, Mingzhe Zhao 2,
The phenotype of this worm is wild type. When both genes are mutant: The phenotype of this worm is double mutant Dpy and Unc phenotype.
 Series 2: Cross Diagrams - Complementation There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome:
Series 2: Cross Diagrams - Complementation There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome:
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
GFP GAL bp 3964 bp
 Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Biological Roles of Cytokinins
 Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Genetic and Molecular Regulation by DELLA Proteins of Trichome Development in Arabidopsis 1[W][OA]
![Genetic and Molecular Regulation by DELLA Proteins of Trichome Development in Arabidopsis 1[W][OA] Genetic and Molecular Regulation by DELLA Proteins of Trichome Development in Arabidopsis 1[W][OA]](/thumbs/94/120779971.jpg) Genetic and Molecular Regulation by DELLA Proteins of Trichome Development in Arabidopsis 1[W][OA] Yinbo Gan, Hao Yu, Jinrong Peng, and Pierre Broun* Centre for Novel Agricultural Products, Department
Genetic and Molecular Regulation by DELLA Proteins of Trichome Development in Arabidopsis 1[W][OA] Yinbo Gan, Hao Yu, Jinrong Peng, and Pierre Broun* Centre for Novel Agricultural Products, Department
SUPPLEMENTARY INFORMATION
 Figure S1. Haploid plant produced by centromere-mediated genome elimination Chromosomes containing altered CENH3 in their centromeres (green dots) are eliminated after fertilization in a cross to wild
Figure S1. Haploid plant produced by centromere-mediated genome elimination Chromosomes containing altered CENH3 in their centromeres (green dots) are eliminated after fertilization in a cross to wild
Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section B and ONE question from Section C.
 UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
UNIVERSITY OF EAST ANGLIA School of Biological Sciences Main Series UG Examination 2017-2018 GENETICS BIO-5009A Time allowed: 2 hours Answer ALL questions in Section A, ALL PARTS of the question in Section
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL
 GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
Epidermal cell patterning and differentiation throughout the apical basal axis of the seedling
 Journal of Experimental Botany, Vol. 56, No. 418, pp. 1983 1989, August 2005 doi:10.1093/jxb/eri213 Advance Access publication 20 June, 2005 REVIEW ARTICLE Epidermal cell patterning and differentiation
Journal of Experimental Botany, Vol. 56, No. 418, pp. 1983 1989, August 2005 doi:10.1093/jxb/eri213 Advance Access publication 20 June, 2005 REVIEW ARTICLE Epidermal cell patterning and differentiation
2 Numbers in parentheses refer to literature cited.
 A Genetic Study of Monogerm and Multigerm Characters in Beets V. F. SAVITSKY 1 Introduction Monogerm beets were found in the variety Michigan Hybrid 18 in Oregon in 1948. Two of these monogerm plants,
A Genetic Study of Monogerm and Multigerm Characters in Beets V. F. SAVITSKY 1 Introduction Monogerm beets were found in the variety Michigan Hybrid 18 in Oregon in 1948. Two of these monogerm plants,
CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON
 PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
AP Biology Summer 2017
 Directions: Questions 1 and 2 are long free response questions that require about 22 minutes to answer and are worth 10 points each. Questions 3-6 are short free- response questions that require about
Directions: Questions 1 and 2 are long free response questions that require about 22 minutes to answer and are worth 10 points each. Questions 3-6 are short free- response questions that require about
Genetics 275 Notes Week 7
 Cytoplasmic Inheritance Genetics 275 Notes Week 7 Criteriafor recognition of cytoplasmic inheritance: 1. Reciprocal crosses give different results -mainly due to the fact that the female parent contributes
Cytoplasmic Inheritance Genetics 275 Notes Week 7 Criteriafor recognition of cytoplasmic inheritance: 1. Reciprocal crosses give different results -mainly due to the fact that the female parent contributes
Arabidopsis thaliana. Lucia Strader. Assistant Professor, Biology
 Arabidopsis thaliana Lucia Strader Assistant Professor, Biology Arabidopsis as a genetic model Easy to grow Small genome Short life cycle Self fertile Produces many progeny Easily transformed HIV E. coli
Arabidopsis thaliana Lucia Strader Assistant Professor, Biology Arabidopsis as a genetic model Easy to grow Small genome Short life cycle Self fertile Produces many progeny Easily transformed HIV E. coli
Reinforcement Unit 3 Resource Book. Meiosis and Mendel KEY CONCEPT Gametes have half the number of chromosomes that body cells have.
 6.1 CHROMOSOMES AND MEIOSIS KEY CONCEPT Gametes have half the number of chromosomes that body cells have. Your body is made of two basic cell types. One basic type are somatic cells, also called body cells,
6.1 CHROMOSOMES AND MEIOSIS KEY CONCEPT Gametes have half the number of chromosomes that body cells have. Your body is made of two basic cell types. One basic type are somatic cells, also called body cells,
You are required to know all terms defined in lecture. EXPLORE THE COURSE WEB SITE 1/6/2010 MENDEL AND MODELS
 1/6/2010 MENDEL AND MODELS!!! GENETIC TERMINOLOGY!!! Essential to the mastery of genetics is a thorough knowledge and understanding of the vocabulary of this science. New terms will be introduced and defined
1/6/2010 MENDEL AND MODELS!!! GENETIC TERMINOLOGY!!! Essential to the mastery of genetics is a thorough knowledge and understanding of the vocabulary of this science. New terms will be introduced and defined
Phase change and the regulation of trichome distribution in Arabidopsis
 Development 124, 645-654 (1997) Printed in Great Britain The Company of Biologists Limited 1997 DEV0093 645 Phase change and the regulation of trichome distribution in Arabidopsis thaliana Abby Telfer,
Development 124, 645-654 (1997) Printed in Great Britain The Company of Biologists Limited 1997 DEV0093 645 Phase change and the regulation of trichome distribution in Arabidopsis thaliana Abby Telfer,
CELL fate specification is a process of fundamental are selected from a field of developmentally equivalent
 Copyright 1999 by the Genetics Society of America Allele-Specific Interactions Between ttg and gl1 During Trichome Development in Arabidopsis thaliana John C. Larkin,* Jason D. Walker,* Agnese C. Bolognesi-Winfield,
Copyright 1999 by the Genetics Society of America Allele-Specific Interactions Between ttg and gl1 During Trichome Development in Arabidopsis thaliana John C. Larkin,* Jason D. Walker,* Agnese C. Bolognesi-Winfield,
Principles of QTL Mapping. M.Imtiaz
 Principles of QTL Mapping M.Imtiaz Introduction Definitions of terminology Reasons for QTL mapping Principles of QTL mapping Requirements For QTL Mapping Demonstration with experimental data Merit of QTL
Principles of QTL Mapping M.Imtiaz Introduction Definitions of terminology Reasons for QTL mapping Principles of QTL mapping Requirements For QTL Mapping Demonstration with experimental data Merit of QTL
The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis thaliana
 Development 126, 4763-477 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV248 4763 The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis
Development 126, 4763-477 (1999) Printed in Great Britain The Company of Biologists Limited 1999 DEV248 4763 The early-flowering mutant efs is involved in the autonomous promotion pathway of Arabidopsis
Utilizing Illumina high-throughput sequencing technology to gain insights into small RNA biogenesis and function
 Utilizing Illumina high-throughput sequencing technology to gain insights into small RNA biogenesis and function Brian D. Gregory Department of Biology Penn Genome Frontiers Institute University of Pennsylvania
Utilizing Illumina high-throughput sequencing technology to gain insights into small RNA biogenesis and function Brian D. Gregory Department of Biology Penn Genome Frontiers Institute University of Pennsylvania
Name Class Date. KEY CONCEPT Gametes have half the number of chromosomes that body cells have.
 Section 1: Chromosomes and Meiosis KEY CONCEPT Gametes have half the number of chromosomes that body cells have. VOCABULARY somatic cell autosome fertilization gamete sex chromosome diploid homologous
Section 1: Chromosomes and Meiosis KEY CONCEPT Gametes have half the number of chromosomes that body cells have. VOCABULARY somatic cell autosome fertilization gamete sex chromosome diploid homologous
Temperature-sensitive mutations that arrest Arabidopsis shoot development
 Development 122, 3799-3807 (1996) Printed in Great Britain The Company of Biologists Limited 1996 DEV0078 3799 Temperature-sensitive mutations that arrest Arabidopsis shoot development F. Bryan Pickett,
Development 122, 3799-3807 (1996) Printed in Great Britain The Company of Biologists Limited 1996 DEV0078 3799 Temperature-sensitive mutations that arrest Arabidopsis shoot development F. Bryan Pickett,
Evolution of phenotypic traits
 Quantitative genetics Evolution of phenotypic traits Very few phenotypic traits are controlled by one locus, as in our previous discussion of genetics and evolution Quantitative genetics considers characters
Quantitative genetics Evolution of phenotypic traits Very few phenotypic traits are controlled by one locus, as in our previous discussion of genetics and evolution Quantitative genetics considers characters
AP Biology Unit 6 Practice Test 1. A group of cells is assayed for DNA content immediately following mitosis and is found to have an average of 8
 AP Biology Unit 6 Practice Test Name: 1. A group of cells is assayed for DNA content immediately following mitosis and is found to have an average of 8 picograms of DNA per nucleus. How many picograms
AP Biology Unit 6 Practice Test Name: 1. A group of cells is assayed for DNA content immediately following mitosis and is found to have an average of 8 picograms of DNA per nucleus. How many picograms
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Chapter 2: Extensions to Mendel: Complexities in Relating Genotype to Phenotype.
 Chapter 2: Extensions to Mendel: Complexities in Relating Genotype to Phenotype. please read pages 38-47; 49-55;57-63. Slide 1 of Chapter 2 1 Extension sot Mendelian Behavior of Genes Single gene inheritance
Chapter 2: Extensions to Mendel: Complexities in Relating Genotype to Phenotype. please read pages 38-47; 49-55;57-63. Slide 1 of Chapter 2 1 Extension sot Mendelian Behavior of Genes Single gene inheritance
TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a WRKY Transcription Factor
 The Plant Cell, Vol. 14, 1359 1375, June 2002, www.plantcell.org 2002 American Society of Plant Biologists TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a
The Plant Cell, Vol. 14, 1359 1375, June 2002, www.plantcell.org 2002 American Society of Plant Biologists TRANSPARENT TESTA GLABRA2, a Trichome and Seed Coat Development Gene of Arabidopsis, Encodes a
When one gene is wild type and the other mutant:
 Series 2: Cross Diagrams Linkage Analysis There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome:
Series 2: Cross Diagrams Linkage Analysis There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome:
SUPPLEMENTARY INFORMATION
 Supplemental Methods Isolation and mapping of SPCH An EMS-mutagenized population of tmm-1 (Col);E1728 (an enhancer trap GFP marking guard cells) was created. M2 seeds were collected from M1 s planted in
Supplemental Methods Isolation and mapping of SPCH An EMS-mutagenized population of tmm-1 (Col);E1728 (an enhancer trap GFP marking guard cells) was created. M2 seeds were collected from M1 s planted in
The control of trichome spacing and number in Arabidopsis
 Development 1, 997-1 (1996) Printed in Great Britain The Company of Biologists Limited 1996 DEV6 997 The control of trichome spacing and number in Arabidopsis John C. Larkin 1, *, Nevin Young, Michael
Development 1, 997-1 (1996) Printed in Great Britain The Company of Biologists Limited 1996 DEV6 997 The control of trichome spacing and number in Arabidopsis John C. Larkin 1, *, Nevin Young, Michael
1 Mendel and His Peas
 CHAPTER 3 1 Mendel and His Peas SECTION Heredity BEFORE YOU READ After you read this section, you should be able to answer these questions: What is heredity? How did Gregor Mendel study heredity? National
CHAPTER 3 1 Mendel and His Peas SECTION Heredity BEFORE YOU READ After you read this section, you should be able to answer these questions: What is heredity? How did Gregor Mendel study heredity? National
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family
 Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
1. Ch 6. #17 In sweet peas, the synthesis of purple anthocyanin pigment in the petals is controlled by two genes, B and D.
 Day 11 Tutorial Answers for BIOL 234 section 921 summer 2014 students ONLY J. Klenz L. McDonnell Not for sale or duplication. Learning Goals: Given the functions of two or more gene products that control
Day 11 Tutorial Answers for BIOL 234 section 921 summer 2014 students ONLY J. Klenz L. McDonnell Not for sale or duplication. Learning Goals: Given the functions of two or more gene products that control
The Chromosomal Basis of Inheritance
 The Chromosomal Basis of Inheritance Mitosis and meiosis were first described in the late 800s. The chromosome theory of inheritance states: Mendelian genes have specific loci (positions) on chromosomes.
The Chromosomal Basis of Inheritance Mitosis and meiosis were first described in the late 800s. The chromosome theory of inheritance states: Mendelian genes have specific loci (positions) on chromosomes.
Exam 1 PBG430/
 1 Exam 1 PBG430/530 2014 1. You read that the genome size of maize is 2,300 Mb and that in this species 2n = 20. This means that there are 2,300 Mb of DNA in a cell that is a. n (e.g. gamete) b. 2n (e.g.
1 Exam 1 PBG430/530 2014 1. You read that the genome size of maize is 2,300 Mb and that in this species 2n = 20. This means that there are 2,300 Mb of DNA in a cell that is a. n (e.g. gamete) b. 2n (e.g.
Plant Structure, Growth, and Development
 Plant Structure, Growth, and Development Plant hierarchy: Cells Tissue: group of similar cells with similar function: Dermal, Ground, Vascular Organs: multiple kinds of tissue, very diverse function Organ
Plant Structure, Growth, and Development Plant hierarchy: Cells Tissue: group of similar cells with similar function: Dermal, Ground, Vascular Organs: multiple kinds of tissue, very diverse function Organ
Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation
 Erratum Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation Victor Kirik, Myeong Min Lee, Katja Wester, Ullrich Herrmann, Zhengui Zheng, David Oppenheimer, John Schiefelbein
Erratum Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation Victor Kirik, Myeong Min Lee, Katja Wester, Ullrich Herrmann, Zhengui Zheng, David Oppenheimer, John Schiefelbein
SAD2 in Arabidopsis Functions in Trichome Initiation through Mediating GL3 Function and Regulating GL1, TTG1 and GL2 Expression
 Journal of Integrative Plant Biology 2008, 50 (7): 906 917 SAD2 in Arabidopsis Functions in Trichome Initiation through Mediating GL3 Function and Regulating GL1, TTG1 and GL2 Expression Ying Gao 1,2,3,
Journal of Integrative Plant Biology 2008, 50 (7): 906 917 SAD2 in Arabidopsis Functions in Trichome Initiation through Mediating GL3 Function and Regulating GL1, TTG1 and GL2 Expression Ying Gao 1,2,3,
- interactions between alleles. - multiple phenotypic effects of one gene. - phenotypic variability in single genes. - interactions between genes
 Genotype Phenotype - interactions between alleles - dominance - multiple phenotypic effects of one gene - phenotypic variability in single genes - interactions between genes amount of product threshold
Genotype Phenotype - interactions between alleles - dominance - multiple phenotypic effects of one gene - phenotypic variability in single genes - interactions between genes amount of product threshold
Biology Chapter 11: Introduction to Genetics
 Biology Chapter 11: Introduction to Genetics Meiosis - The mechanism that halves the number of chromosomes in cells is a form of cell division called meiosis - Meiosis consists of two successive nuclear
Biology Chapter 11: Introduction to Genetics Meiosis - The mechanism that halves the number of chromosomes in cells is a form of cell division called meiosis - Meiosis consists of two successive nuclear
Bi Lecture 8 Genetic Pathways and Genetic Screens
 Bi190-2013 Lecture 8 Genetic Pathways and Genetic Screens WT A 2X:2A her-1 tra-1 1X:2A her-1 tra-1 Female body Male body Female body Male body her-1(lf) B 2X:2A her-1(lf) tra-1 1X:2A her-1(lf) tra-1 Female
Bi190-2013 Lecture 8 Genetic Pathways and Genetic Screens WT A 2X:2A her-1 tra-1 1X:2A her-1 tra-1 Female body Male body Female body Male body her-1(lf) B 2X:2A her-1(lf) tra-1 1X:2A her-1(lf) tra-1 Female
Chapter 18 Lecture. Concepts of Genetics. Tenth Edition. Developmental Genetics
 Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Reading: Chapter 5, pp ; Reference chapter D, pp Problem set F
 Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
DNA Structure and Function
 DNA Structure and Function Nucleotide Structure 1. 5-C sugar RNA ribose DNA deoxyribose 2. Nitrogenous Base N attaches to 1 C of sugar Double or single ring Four Bases Adenine, Guanine, Thymine, Cytosine
DNA Structure and Function Nucleotide Structure 1. 5-C sugar RNA ribose DNA deoxyribose 2. Nitrogenous Base N attaches to 1 C of sugar Double or single ring Four Bases Adenine, Guanine, Thymine, Cytosine
is the scientific study of. Gregor Mendel was an Austrian monk. He is considered the of genetics. Mendel carried out his work with ordinary garden.
 11-1 The 11-1 Work of Gregor Mendel The Work of Gregor Mendel is the scientific study of. Gregor Mendel was an Austrian monk. He is considered the of genetics. Mendel carried out his work with ordinary
11-1 The 11-1 Work of Gregor Mendel The Work of Gregor Mendel is the scientific study of. Gregor Mendel was an Austrian monk. He is considered the of genetics. Mendel carried out his work with ordinary
Introduction to Genetics
 Introduction to Genetics The Work of Gregor Mendel B.1.21, B.1.22, B.1.29 Genetic Inheritance Heredity: the transmission of characteristics from parent to offspring The study of heredity in biology is
Introduction to Genetics The Work of Gregor Mendel B.1.21, B.1.22, B.1.29 Genetic Inheritance Heredity: the transmission of characteristics from parent to offspring The study of heredity in biology is
Sexual Reproduction and Genetics
 Chapter Test A CHAPTER 10 Sexual Reproduction and Genetics Part A: Multiple Choice In the space at the left, write the letter of the term, number, or phrase that best answers each question. 1. How many
Chapter Test A CHAPTER 10 Sexual Reproduction and Genetics Part A: Multiple Choice In the space at the left, write the letter of the term, number, or phrase that best answers each question. 1. How many
Untitled Document. A. antibiotics B. cell structure C. DNA structure D. sterile procedures
 Name: Date: 1. The discovery of which of the following has most directly led to advances in the identification of suspects in criminal investigations and in the identification of genetic diseases? A. antibiotics
Name: Date: 1. The discovery of which of the following has most directly led to advances in the identification of suspects in criminal investigations and in the identification of genetic diseases? A. antibiotics
Major Plant Hormones 1.Auxins 2.Cytokinins 3.Gibberelins 4.Ethylene 5.Abscisic acid
 Plant Hormones Lecture 9: Control Systems in Plants What is a Plant Hormone? Compound produced by one part of an organism that is translocated to other parts where it triggers a response in target cells
Plant Hormones Lecture 9: Control Systems in Plants What is a Plant Hormone? Compound produced by one part of an organism that is translocated to other parts where it triggers a response in target cells
Mendel and the Gene Idea. Biology Exploring Life Section Modern Biology Section 9-1
 Mendel and the Gene Idea Biology Exploring Life Section 10.0-10.2 Modern Biology Section 9-1 Objectives Summarize the Blending Hypothesis and the problems associated with it. Describe the methods used
Mendel and the Gene Idea Biology Exploring Life Section 10.0-10.2 Modern Biology Section 9-1 Objectives Summarize the Blending Hypothesis and the problems associated with it. Describe the methods used
Reproduction, Seeds and Propagation
 Reproduction, Seeds and Propagation Diploid (2n) somatic cell Two diploid (2n) somatic cells Telophase Anaphase Metaphase Prophase I One pair of homologous chromosomes (homologues) II Homologues condense
Reproduction, Seeds and Propagation Diploid (2n) somatic cell Two diploid (2n) somatic cells Telophase Anaphase Metaphase Prophase I One pair of homologous chromosomes (homologues) II Homologues condense
A species of plants. In maize 20 genes for male sterility are listed by
 THE INTERRELATION OF PLASMAGENES AND CHROMO- GENES IN POLLEN PRODUCTION IN MAIZE DONALD F. JONES Connecticut Agricultural Experiment Station, New Haven, Connecticut Received January 24, 195 IV INHERITED
THE INTERRELATION OF PLASMAGENES AND CHROMO- GENES IN POLLEN PRODUCTION IN MAIZE DONALD F. JONES Connecticut Agricultural Experiment Station, New Haven, Connecticut Received January 24, 195 IV INHERITED
Anatomy of Plants Student Notes
 Directions: Fill in the blanks. Anatomy of Plants Student Notes Plant Cell Biology Segment 1. Plants Plants are organisms are incapable of movement produce food through 2. Animals Animals are multicellular
Directions: Fill in the blanks. Anatomy of Plants Student Notes Plant Cell Biology Segment 1. Plants Plants are organisms are incapable of movement produce food through 2. Animals Animals are multicellular
EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA
 EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA Item Type text; Electronic Thesis Authors Bergstrand, Lauren Janel Publisher The University of Arizona. Rights Copyright is held by the author. Digital
EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA Item Type text; Electronic Thesis Authors Bergstrand, Lauren Janel Publisher The University of Arizona. Rights Copyright is held by the author. Digital
Section 11 1 The Work of Gregor Mendel
 Chapter 11 Introduction to Genetics Section 11 1 The Work of Gregor Mendel (pages 263 266) What is the principle of dominance? What happens during segregation? Gregor Mendel s Peas (pages 263 264) 1. The
Chapter 11 Introduction to Genetics Section 11 1 The Work of Gregor Mendel (pages 263 266) What is the principle of dominance? What happens during segregation? Gregor Mendel s Peas (pages 263 264) 1. The
Outline. Leaf Development. Leaf Structure - Morphology. Leaf Structure - Morphology
 Outline 1. Leaf Structure: Morphology & Anatomy 2. Leaf Development A. Anatomy B. Sector analysis C. Leaf Development Leaf Structure - Morphology Leaf Structure - Morphology 1 Leaf Structure - Morphology
Outline 1. Leaf Structure: Morphology & Anatomy 2. Leaf Development A. Anatomy B. Sector analysis C. Leaf Development Leaf Structure - Morphology Leaf Structure - Morphology 1 Leaf Structure - Morphology
Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in Arabidopsis
 Zhang et al. BMC Plant Biology (2018) 18:63 https://doi.org/10.1186/s12870-018-1271-z RESEARCH ARTICLE Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in
Zhang et al. BMC Plant Biology (2018) 18:63 https://doi.org/10.1186/s12870-018-1271-z RESEARCH ARTICLE Genetic evidence suggests that GIS functions downstream of TCL1 to regulate trichome formation in
THE WORK OF GREGOR MENDEL
 GENETICS NOTES THE WORK OF GREGOR MENDEL Genetics-. - Austrian monk- the father of genetics- carried out his work on. Pea flowers are naturally, which means that sperm cells fertilize the egg cells in
GENETICS NOTES THE WORK OF GREGOR MENDEL Genetics-. - Austrian monk- the father of genetics- carried out his work on. Pea flowers are naturally, which means that sperm cells fertilize the egg cells in
Full file at CHAPTER 2 Genetics
 CHAPTER 2 Genetics MULTIPLE CHOICE 1. Chromosomes are a. small linear bodies. b. contained in cells. c. replicated during cell division. 2. A cross between true-breeding plants bearing yellow seeds produces
CHAPTER 2 Genetics MULTIPLE CHOICE 1. Chromosomes are a. small linear bodies. b. contained in cells. c. replicated during cell division. 2. A cross between true-breeding plants bearing yellow seeds produces
Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin
 Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin CHAPTER 1 1.2 The expected homozygosity, given allele
Solutions to Even-Numbered Exercises to accompany An Introduction to Population Genetics: Theory and Applications Rasmus Nielsen Montgomery Slatkin CHAPTER 1 1.2 The expected homozygosity, given allele
Ginkgo leaf. Ginkgo is dioecious, separate sexes: male and female plants are separate. Monoecious plants have both male and female parts.
 Ginkgo leaf Figure 22-30 Ginkgo tree. Ginkgo is dioecious, separate sexes: male and female plants are separate. Monoecious plants have both male and female parts. The vein pattern is dichotomous: Divided
Ginkgo leaf Figure 22-30 Ginkgo tree. Ginkgo is dioecious, separate sexes: male and female plants are separate. Monoecious plants have both male and female parts. The vein pattern is dichotomous: Divided
Labs 7 and 8: Mitosis, Meiosis, Gametes and Genetics
 Biology 107 General Biology Labs 7 and 8: Mitosis, Meiosis, Gametes and Genetics In Biology 107, our discussion of the cell has focused on the structure and function of subcellular organelles. The next
Biology 107 General Biology Labs 7 and 8: Mitosis, Meiosis, Gametes and Genetics In Biology 107, our discussion of the cell has focused on the structure and function of subcellular organelles. The next
The phenotype of this worm is wild type. When both genes are mutant: The phenotype of this worm is double mutant Dpy and Unc phenotype.
 Series 1: Cross Diagrams There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome: When both
Series 1: Cross Diagrams There are two alleles for each trait in a diploid organism In C. elegans gene symbols are ALWAYS italicized. To represent two different genes on the same chromosome: When both
Meiosis and Tetrad Analysis Lab
 Meiosis and Tetrad Analysis Lab Objectives: - Explain how meiosis and crossing over result in the different arrangements of ascospores within asci. - Learn how to calculate the map distance between a gene
Meiosis and Tetrad Analysis Lab Objectives: - Explain how meiosis and crossing over result in the different arrangements of ascospores within asci. - Learn how to calculate the map distance between a gene
early in short days 4, a mutation in Arabidopsis that causes early flowering
 Development 129, 5349-5361 2002 The Company of Biologists Ltd doi:10.1242/dev.00113 5349 early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mrna abundance of the
Development 129, 5349-5361 2002 The Company of Biologists Ltd doi:10.1242/dev.00113 5349 early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mrna abundance of the
Supplemental Data. Wang et al. (2014). Plant Cell /tpc
 Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Q2 (4.6) Put the following in order from biggest to smallest: Gene DNA Cell Chromosome Nucleus. Q8 (Biology) (4.6)
 Q1 (4.6) What is variation? Q2 (4.6) Put the following in order from biggest to smallest: Gene DNA Cell Chromosome Nucleus Q3 (4.6) What are genes? Q4 (4.6) What sort of reproduction produces genetically
Q1 (4.6) What is variation? Q2 (4.6) Put the following in order from biggest to smallest: Gene DNA Cell Chromosome Nucleus Q3 (4.6) What are genes? Q4 (4.6) What sort of reproduction produces genetically
Homework Assignment, Evolutionary Systems Biology, Spring Homework Part I: Phylogenetics:
 Homework Assignment, Evolutionary Systems Biology, Spring 2009. Homework Part I: Phylogenetics: Introduction. The objective of this assignment is to understand the basics of phylogenetic relationships
Homework Assignment, Evolutionary Systems Biology, Spring 2009. Homework Part I: Phylogenetics: Introduction. The objective of this assignment is to understand the basics of phylogenetic relationships
Cell Division and Genetics
 Name Date Cell Division and Genetics 1. Black fur is dominant over brown fur in a particular population of guinea pig. The genetic information that gives a guinea pig brown fur is described as having A.
Name Date Cell Division and Genetics 1. Black fur is dominant over brown fur in a particular population of guinea pig. The genetic information that gives a guinea pig brown fur is described as having A.
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Allele interactions: Terms used to specify interactions between alleles of the same gene: Dominant/recessive incompletely dominant codominant
 Biol 321 Feb 3, 2010 Allele interactions: Terms used to specify interactions between alleles of the same gene: Dominant/recessive incompletely dominant codominant Gene interactions: the collaborative efforts
Biol 321 Feb 3, 2010 Allele interactions: Terms used to specify interactions between alleles of the same gene: Dominant/recessive incompletely dominant codominant Gene interactions: the collaborative efforts
Introduction to Genetics
 Chapter 11 Introduction to Genetics Section 11 1 The Work of Gregor Mendel (pages 263 266) This section describes how Gregor Mendel studied the inheritance of traits in garden peas and what his conclusions
Chapter 11 Introduction to Genetics Section 11 1 The Work of Gregor Mendel (pages 263 266) This section describes how Gregor Mendel studied the inheritance of traits in garden peas and what his conclusions
GACE Biology Assessment Test I (026) Curriculum Crosswalk
 Subarea I. Cell Biology: Cell Structure and Function (50%) Objective 1: Understands the basic biochemistry and metabolism of living organisms A. Understands the chemical structures and properties of biologically
Subarea I. Cell Biology: Cell Structure and Function (50%) Objective 1: Understands the basic biochemistry and metabolism of living organisms A. Understands the chemical structures and properties of biologically
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/331/6019/876/dc1 Supporting Online Material for Synthetic Clonal Reproduction Through Seeds Mohan P. A. Marimuthu, Sylvie Jolivet, Maruthachalam Ravi, Lucie Pereira,
www.sciencemag.org/cgi/content/full/331/6019/876/dc1 Supporting Online Material for Synthetic Clonal Reproduction Through Seeds Mohan P. A. Marimuthu, Sylvie Jolivet, Maruthachalam Ravi, Lucie Pereira,
The distinctive roles of ve different ARC genes in the chloroplast division process in Arabidopsis
 The Plant Journal (1999) 18(6), 651±662 The distinctive roles of ve different ARC genes in the chloroplast division process in Arabidopsis Joanne L. Marrison 1, Stephen M. Rutherford 1,², Elizabeth J.
The Plant Journal (1999) 18(6), 651±662 The distinctive roles of ve different ARC genes in the chloroplast division process in Arabidopsis Joanne L. Marrison 1, Stephen M. Rutherford 1,², Elizabeth J.
Plant hormones: a. produced in many parts of the plant b. have many functions
 Plant hormones: a. produced in many parts of the plant b. have many functions Illustrated with 4 plant hormones: Gibberellins Auxin Cytokinins Ethylene Gibberellins Gibberellins illustrate how plant hormones
Plant hormones: a. produced in many parts of the plant b. have many functions Illustrated with 4 plant hormones: Gibberellins Auxin Cytokinins Ethylene Gibberellins Gibberellins illustrate how plant hormones
Chapter 6 Meiosis and Mendel
 UNIT 3 GENETICS Chapter 6 Meiosis and Mendel 1 hairy ears (hypertrichosis)- due to holandric gene. (Y chromosome)-only occurs in males. Appears in all sons. 2 Polydactyly- having extra fingers Wendy the
UNIT 3 GENETICS Chapter 6 Meiosis and Mendel 1 hairy ears (hypertrichosis)- due to holandric gene. (Y chromosome)-only occurs in males. Appears in all sons. 2 Polydactyly- having extra fingers Wendy the
A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development
 A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development Xuemei Chen Waksman Institute, Rutgers University, Piscataway, NJ 08854, USA. E-mail: xuemei@waksman.rutgers.edu Plant
A MicroRNA as a Translational Repressor of APETALA2 in Arabidopsis Flower Development Xuemei Chen Waksman Institute, Rutgers University, Piscataway, NJ 08854, USA. E-mail: xuemei@waksman.rutgers.edu Plant
Chapter Three. The effect of reduced DNA methylation on the flowering time and vernalization. response of Arabidopsis thaliana
 Chapter Three The effect of reduced DNA methylation on the flowering time and vernalization response of Arabidopsis thaliana 3.1 Introduction The time at which a plant flowers is influenced by environmental
Chapter Three The effect of reduced DNA methylation on the flowering time and vernalization response of Arabidopsis thaliana 3.1 Introduction The time at which a plant flowers is influenced by environmental
PRINCIPLES OF MENDELIAN GENETICS APPLICABLE IN FORESTRY. by Erich Steiner 1/
 PRINCIPLES OF MENDELIAN GENETICS APPLICABLE IN FORESTRY by Erich Steiner 1/ It is well known that the variation exhibited by living things has two components, one hereditary, the other environmental. One
PRINCIPLES OF MENDELIAN GENETICS APPLICABLE IN FORESTRY by Erich Steiner 1/ It is well known that the variation exhibited by living things has two components, one hereditary, the other environmental. One
1 Mendel and His Peas
 CHAPTER 5 1 Mendel and His Peas SECTION Heredity BEFORE YOU READ After you read this section, you should be able to answer these questions: What is heredity? How did Gregor Mendel study heredity? National
CHAPTER 5 1 Mendel and His Peas SECTION Heredity BEFORE YOU READ After you read this section, you should be able to answer these questions: What is heredity? How did Gregor Mendel study heredity? National
Problem Set 3 10:35 AM January 27, 2011
 BIO322: Genetics Douglas J. Burks Department of Biology Wilmington College of Ohio Problem Set 3 Due @ 10:35 AM January 27, 2011 Chapter 4: Problems 3, 5, 12, 23, 25, 31, 37, and 41. Chapter 5: Problems
BIO322: Genetics Douglas J. Burks Department of Biology Wilmington College of Ohio Problem Set 3 Due @ 10:35 AM January 27, 2011 Chapter 4: Problems 3, 5, 12, 23, 25, 31, 37, and 41. Chapter 5: Problems
Hairy s Inheritance: Investigating Variation, Selection, and Evolution with Wisconsin Fast Plants
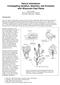 Introduction Hairy s Inheritance: Investigating Variation, Selection, and Evolution with Wisconsin Fast Plants Daniel Lauffer Wisconsin Fast Plants Program University of Wisconsin - Madison Since the dawn
Introduction Hairy s Inheritance: Investigating Variation, Selection, and Evolution with Wisconsin Fast Plants Daniel Lauffer Wisconsin Fast Plants Program University of Wisconsin - Madison Since the dawn
The Plant Cell, November. 2017, American Society of Plant Biologists. All rights reserved
 The Genetics of Floral Development Teaching Guide Overview The development of flowers in angiosperm plants provided a critical evolutionary advantage, allowing more options for pollen dispersal and seed
The Genetics of Floral Development Teaching Guide Overview The development of flowers in angiosperm plants provided a critical evolutionary advantage, allowing more options for pollen dispersal and seed
Name Class Date. Pearson Education, Inc., publishing as Pearson Prentice Hall. 33
 Chapter 11 Introduction to Genetics Chapter Vocabulary Review Matching On the lines provided, write the letter of the definition of each term. 1. genetics a. likelihood that something will happen 2. trait
Chapter 11 Introduction to Genetics Chapter Vocabulary Review Matching On the lines provided, write the letter of the definition of each term. 1. genetics a. likelihood that something will happen 2. trait
1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms.
 Practicing Biology BIG IDEA 3.A 1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms. 2. Using at least 2 well-known experiments, describe which features of DNA and RNA
Practicing Biology BIG IDEA 3.A 1. Draw, label and describe the structure of DNA and RNA including bonding mechanisms. 2. Using at least 2 well-known experiments, describe which features of DNA and RNA
GENE TRANSFER IN NICOTIANA RUSTICA BY MEANS OF IRRADIATED POLLEN. I. UNSELECTED PROGENIES
 Heredity (1981), 47(1), 17-26 1981. The Genetical Society of Great Britain 0018-067X/81/03290017$02.00 GENE TRANSFER IN NICOTIANA RUSTICA BY MEANS OF IRRADIATED POLLEN. I. UNSELECTED PROGENIES P. D. S.
Heredity (1981), 47(1), 17-26 1981. The Genetical Society of Great Britain 0018-067X/81/03290017$02.00 GENE TRANSFER IN NICOTIANA RUSTICA BY MEANS OF IRRADIATED POLLEN. I. UNSELECTED PROGENIES P. D. S.
Science Unit Learning Summary
 Learning Summary Inheritance, variation and evolution Content Sexual and asexual reproduction. Meiosis leads to non-identical cells being formed while mitosis leads to identical cells being formed. In
Learning Summary Inheritance, variation and evolution Content Sexual and asexual reproduction. Meiosis leads to non-identical cells being formed while mitosis leads to identical cells being formed. In
Lesson Overview Meiosis
 11.4 As geneticists in the early 1900s applied Mendel s laws, they wondered where genes might be located. They expected genes to be carried on structures inside the cell, but which structures? What cellular
11.4 As geneticists in the early 1900s applied Mendel s laws, they wondered where genes might be located. They expected genes to be carried on structures inside the cell, but which structures? What cellular
