Title the chloroplast accumulation respon. Takayuki; Wada, Masamitsu. Citation United States of America (2016), 11.
|
|
|
- Kimberly Turner
- 5 years ago
- Views:
Transcription
1 Title RPT2/NCH1 subfamily of NPH3-like pr the chloroplast accumulation respon Suetsugu, Noriyuki; Takemiya, Atsus Author(s) Higa, Takeshi; Komatsu, Aino; Shima Takayuki; Wada, Masamitsu Citation Proceedings of the National Academy United States of America (2016), 11 Issue Date URL Copyright 2016 National Academy o Right the published version. Please cite この論文は出版社版でありません 引用の際には出版社版をご確認ご利用ください Type Journal Article Textversion author Kyoto University
2 BIOLOGICAL SCIENCES: Plant Biology Short title: NPH3-like proteins mediate for chloroplast movement RPT2/NCH1 subfamily of NPH3-like proteins is essential for the chloroplast accumulation response in land plants Noriyuki Suetsugu a,b,3, Atsushi Takemiya a,1, Sam-Geun Kong a,2, Takeshi Higa a,3, Aino Komatsu b, Ken-ichiro Shimazaki a, Takayuki Kohchi b and Masamitsu Wada a,3,4 a Department of Biology, Faculty of Sciences, Kyushu University, Fukuoka , Japan b Graduate School of Biostudies, Kyoto University, Kyoto , Japan 1 Present address: Graduate School of Science and Technology for innovation, Yamaguchi University, Yamaguchi , Japan 2 Present address: Department of Biological Science, College of Natural Sciences, Kongju National University, Gongju, Chungnam 32588, Korea 3 Present address: Department of Biological Sciences, Graduate School of Science and Engineering, Tokyo Metropolitan University, Tokyo , Japan 4 To whom correspondence should be addressed. suetsugu@lif.kyoto-u.ac.jp or masamitsu.wada@gmail.com Corresponding author: Noriyuki Suetsugu 1
3 Graduate School of Biostudies, Kyoto University, Kyoto , Japan phone: fax: or Masamitsu Wada Department of Biological Sciences, Graduate School of Science and Engineering, Tokyo Metropolitan University, Tokyo , Japan phone: Key words: Arabidopsis, blue light, chloroplast movement, phototropin Manuscript information: Number of text pages (including references and figure legends): 27 Number of figures: 4 Number of supplementary figures: 10 Number of supplementary movies: 1 Number of words in abstract: 178 Total number of characters (including spaces) in the text (including abstract, references, notes, and figure legends):
4 Abstract In green plants, the blue-light receptor kinase phototropin mediates various photomovements and developmental responses, such as phototropism, chloroplast photorelocation movements (accumulation and avoidance), stomatal opening, and leaf flattening, which facilitate photosynthesis. In Arabidopsis, two phototropins (phot1 and phot2) redundantly mediate these responses. Two phototropin-interacting proteins NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3) and ROOT PHOTOTROPISM 2 (RPT2), which belong to the NPH3/RPT2-like (NRL) family of BTB domain proteins, mediate phototropism and leaf flattening. However, the roles of NRL proteins in chloroplast photorelocation movement remain to be determined. Here, we show that another phototropin-interacting NRL protein, NRL PROTEIN FOR CHLOROPLAST MOVEMENT 1 (NCH1), and RPT2 redundantly mediate the chloroplast accumulation response but not the avoidance response. NPH3, RPT2, and NCH1 are not involved in the chloroplast avoidance response or stomatal opening. In the liverwort Marchantia polymorpha, the NCH1 ortholog, MpNCH1, is essential for the chloroplast accumulation response but not the avoidance response, indicating that the regulation of the phototropin-mediated chloroplast accumulation response by RPT2/NCH1 is conserved in land plants. Thus, the NRL protein combination could determine the specificity of diverse phototropin-mediated responses. 3
5 Significance The photoreceptor phototropin mediates various blue-light-induced responses, including phototropism, chloroplast movement, stomatal opening, and leaf flattening. Two BTB/POZ proteins, NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3) and ROOT PHOTOTROPISM 2 (RPT2), were identified as early signaling components in phototropin-mediated phototropism and leaf flattening, and a phototropin substrate, BLUE LIGHT SIGNALING1 kinase, specifically mediates the phototropin-mediated stomatal opening. However, early signaling components in the chloroplast movement remain to be determined. We found that RPT2 and the NPH3/RPT2-like (NRL) protein NRL PROTEIN FOR CHLOROPLAST MOVEMENT 1 (NCH1) redundantly mediate the chloroplast accumulation response but not the avoidance response. Our findings indicate that phototropin-mediated phototropism, leaf flattening, and the chloroplast accumulation response, but not the chloroplast avoidance response and stomatal opening, are mediated by NRL proteins. 4
6 \body To adapt to a fluctuating light environment, plants evolved various light responses and photoreceptor systems. Phototropins (phot1 and phot2 in Arabidopsis thaliana) are blue-light receptor kinases (1) belonging to AGC kinase VIII family (named after the camp-dependent protein kinase A, cgmp-dependent protein kinase G and phospholipid-dependent protein kinase C) (2). phot1 and phot2 redundantly mediate hypocotyl phototropism, the chloroplast accumulation response, stomatal opening, leaf positioning, and leaf flattening, although functional differences between phot1 and phot2 exist (3-6). For example, phot2 is the main photoreceptor for the chloroplast avoidance response (7, 8) and palisade cell development (9). However, how phot1 and phot2 regulate these responses was still undetermined. NON-PHOTOTROPIC HYPOCOTYL 3 (NPH3) and ROOT PHOTOTROPISM 2 (RPT2) were identified through molecular genetic analyses of phototropism-deficient mutants (10, 11). NPH3 and RPT2 are founding members of the NPH3/RPT2-like (NRL) family, which consists of 30~ proteins in Arabidopsis (12). NRL proteins have an N-terminal BTB (broad complex, tramtrack, and bric à brac) domain and four conserved regions, I-IV (10) with region I partially overlapping the BTB domain (Fig. 1A and S1). NPH3 and RPT2 are localized on the plasma membrane, like phototropins, and interact with them (10, 13). The nph3 mutant is completely defective in hypocotyl and root phototropism (10, 11). The rpt2 mutant lacks root phototropism but retains partial hypocotyl phototropism under a low blue-light fluence rate (11, 13). The blue-light-induced dephosphorylation of NPH3, which is dependent on phot1, is thought to be an early event in the phototropic response (10, 14, 15). rpt2 mutants exhibit excessive levels of blue-light-induced NPH3 dephosphorylation and have an enhanced rate of NPH3 internalization, indicating that RPT2 mediates phototropism via the regulation of the phosphorylation status and localization of NPH3 (16). Both nph3 and 5
7 rpt2 mutants are also impaired in leaf flattening and positioning (6, 17, 18, 19). However, nph3 and rpt2 exhibit normal chloroplast photorelocation movements and stomatal opening (6, 17, 20). Thus, NPH3 and RPT2 likely mediate auxin-dependent processes among phototropin-mediated responses. Indeed, phot1 and NPH3 mediate auxin transport during blue-light-induced root phototropism via the regulation of the subcellular localization of an auxin efflux transporter PIN-FORMED 2 (21). Another phototropin-interacting protein family, PHYTOCHROME KINASE SUBSTRATE, is also necessary for leaf positioning and flattening, as well as phototropism, but not for chloroplast photorelocation movements and stomatal opening (17, 22, 23). Thus, signaling pathways for chloroplast photorelocation movements and stomatal opening may be very different from those for phototropin-mediated auxin responses. Chloroplasts move towards weak light (the accumulation response) and escape from strong light (the avoidance response). In land plants from liverwort to flowering plants, phototropin is the blue-light receptor for chloroplast photorelocation movement (24). Similar to Arabidopsis, the functional divergence between phototropins is found also in the fern Adiantum capillus-veneris and the moss Physcomitrella patens (25, 26). Importantly, in the liverwort Marchantia polymorpha, which belongs to the most basal land plant lineage, a single phototropin, Mpphot, mediates both the accumulation and avoidance responses (27). Thus, although the functional divergence of phototropins occurred during land plant evolution, phototropins intrinsically have the ability to mediate both the accumulation and avoidance responses. How one type of photoreceptor (i.e., phototropin) mediates different physiological responses is an important theme in understanding complex light-signaling pathways in plants. Here, we report that two phototropin-interacting NRL proteins, NRL PROTEIN FOR CHLOROPLAST MOVEMENT 1 (NCH1) and RPT2, are essential for the phototropin-mediated chloroplast accumulation response. Furthermore, NPH3, RPT2, 6
8 and NCH1 are not involved in the chloroplast avoidance response or stomatal opening. Thus, the combination of NRL proteins may determine the functional specificities among phototropin-mediated responses. 7
9 Results Identification of NCH1 as a phototropin signaling factor. Because NPH3 and RPT2 are early signaling components in phototropin-mediated responses, we hypothesized that NRL proteins, other than NPH3 and RPT2, are involved in chloroplast photorelocation movements. While analyzing the T-DNA insertion mutants of some NRL genes, we found that the T-DNA lines of the NRL gene At5g67385 were deficient in chloroplast photorelocation movements (see below in detail). Here, we named At5g67385 as NRL PROTEIN FOR CHLOROPLAST MOVEMENT 1 (NCH1), and the two T-DNA lines as nch1-1 (SALK_064178) and nch1-2 (GABI_326_A01) (Fig. 1B). Previously, NCH1 was identified as a plant immunity regulator gene, AtSR1 interaction protein 1 (SR1IP1) (28) and also named as NPH3/RPT2-Like 31 (NRL31), but hereafter we call At5g67385 as NCH1 in this paper. NCH1 showed a higher similarity to RPT2 than to NPH3 (Fig. S1). A phylogenetic analysis indicated that NRL proteins are classified into seven major clades and that NCH1 belonged to the same clade as RPT2 but a distinct clade from NPH3 (Fig. 1C and S2; 12, 29). At3g49970 (named as NCH1-LIKE 1, NCL1) was the closest paralog of NCH1 (Fig. 1B, S1 and S2; 12, 29). However, there is no evidence of its in vivo expression and we did not detect any defects of ncl1 mutant at least in chloroplast photorelocation movement and stomatal opening (see below), suggesting that the NCL1 in A. thaliana, if functional, has a role other than chloroplast photorelocation movement and stomatal opening (Supplemental Text, Fig. S1 and see below). To examine the subcellular localization of NCH1, we constructed transgenic nch1-1 plants expressing the NCH1-GFP fusion gene under the control of the native NCH1 promoter (NCH1pro:NCH1-GFP). NCH1-GFP fluorescence was detected in both the pavement and mesophyll cells. In both cell types, NCH1-GFP was localized on the plasma membrane (Fig. 2A). An immunoblot analysis using an NCH1 antiserum 8
10 revealed smeared bands around ~60 kda (frequently as prominent two bands) in wild-type, phot1phot2, and rpt2, but not in nch1 mutant plants (Fig. S3A). They were detected in the microsomal fraction, similar to phot1 (Fig. S3A and B). In an immunoprecipitation analysis of the microsomal fraction with an anti-gfp antibody, endogenous NCH1 was co-immunoprecipitated with phot1-gfp but not with GFP alone in a blue-light-independent manner (Fig. 2B; Fig. S3C). In vitro pull-down assay revealed that NCH1 directly interacted with both N- and C-terminal domains of phot1 (Fig. 2C and D). Similar to nph3 and rpt2 mutant plants (10, 11), the blue-light-induced autophosphorylation of phot1 was normal in nch1 mutant plants (Fig. S3A and D), indicating that NCH1 is downstream of the phototropins. Consistent with the interaction between NPH3 and RPT2 (13), yeast two hybrid analysis revealed the interactions between N- and C-terminal domains of NPH3, RPT2, NCH1 (Fig. 1A), although the interaction between RPT2 and NCH1 was much weaker (Fig. S4). Collectively, NCH1 is a phototropin-interactive protein similar to NPH3 and RPT2. NCH1 is not involved in the hypocotyl phototropic response and leaf flattening. We analyzed hypocotyl phototropic responses in wild-type, nch1 (nch1-1 and nch1-2), rpt2-4 (SAIL_140_D03; Fig. 1A), and rpt2nch1 (rpt2-4nch1-1 and rpt2-4nch1-2). Similar to other rpt2 mutant plants (11, 13, 20), rpt2-4 is defective in the phototropic response, especially at higher fluence rates (Fig. S5A). The nch1 mutant plants showed a slightly strong phototropic response compared with wild type, although the only differences between wild type and nch1-2 in phototropic response at 4 and 40 µmol m 2 s 1 were statistically significant (Fig. S5A; Student s t test, 0.01 < P < 0.05 for wild type vs. nch1-2 at 4 and 40 µmol m 2 s 1 ). rpt2nch1 mutant plants were similar to rpt2-4 (Fig. S5A; Student s t test, P > 0.1 for rpt2-4 vs. rpt2-4nch1-1 or rpt2-4nch1-2 at all fluence rates), indicating that NCH1 is not a positive regulator of phototropin-mediated 9
11 phototropic responses. When grown under the continuous white light, nch1-1 mutants exhibited normal leaf flattening, like wild type (Fig. S5B). Under our light conditions, the leaves of rpt2 mutant plants were slightly curled but leaf curling was not as severe as in the phot1phot2 double mutants (Fig. S5B). The leaf curling of rpt2nch1 double mutants was similar to that of rpt2 (Fig. S5B). Thus, NCH1 is not involved in leaf flattening. RPT2, NCH1, and NPH3 are not involved in stomatal opening. Recently, it was reported that rpt2 mutant and phot2rpt2 double mutant exhibit normal blue-light-induced stomatal opening (20), contrary to a previous study (13). We examined a possible redundancy among four NRL genes in stomatal opening using rpt2nch1, phot2rpt2, phot2rpt2nch1, and nph3rpt2nch1ncl1 mutant plants (Fig. S6). When the blue-light-induced stomatal opening in intact leaves was measured using an infrared thermograph, a blue-light-induced decrease in leaf temperature caused by stomatal opening was found in wild type but not phot1phot2 double mutants as described previously (Fig. S6A) (30). Consistently, blue-light-induced stomatal opening in isolated epidermal tissues was found in wild type but not in phot1phot2 (Fig. S6B). In both assays, normal blue-light-induced stomatal opening was detected in rpt2nch1, phot2rpt2, and phot2rpt2nch1 (Fig. S6A and B), indicating that RPT2 and NCH1 are dispensable for phototropin-mediated stomatal opening. Furthermore, nph3rpt2nch1ncl1 quadruple mutant plants exhibited normal blue-light-induced stomatal opening (Fig. S6A and B). Blue-light-induced proton pumping (Fig. S6C) and H + -ATPase phosphorylation (Fig. S6D), which are essential for phototropin-mediated stomatal opening (30), were also normal in the quadruple mutant plants (Fig. S6C and D). Thus, at least four NRL genes are not involved in phototropin-mediated stomatal opening. 10
12 RPT2 and NCH1 are essential for the chloroplast accumulation response but not the avoidance response. The chloroplast photorelocation movement was analyzed through the leaf transmittance of red light (31). In wild type (black in Fig. 3 and Fig. S7), weak blue light (3 µmol m 2 s 1 ) induces a decrease in leaf transmittance caused by the chloroplast accumulation response whereas strong blue light (20 and 50 µmol m 2 s 1 ) induces an increase in the leaf transmittance as a result of the avoidance response. After the strong blue light was turned off, a rapid decrease in leaf transmittance occurred (called the dark recovery response ). In the nch1 mutant plants (indigo in Fig. 3 and Fig. S7), the chloroplast accumulation response was slightly weaker compared with that of the wild type, although the difference was statistically insignificant in many experiments (Fig. 3A and B). However, the avoidance response was strongly enhanced in the nch1 mutants (Fig. 3A and B). Not only the speed, but also the amplitude of the avoidance response was strongly enhanced in nch1 mutants, especially at 20 µmol m 2 s 1 (Fig. 3A and B; Student s t test, P < for wild type vs. nch1). Furthermore, the dark recovery response was impaired in the nch1 mutants (Fig. 3A and B; Student s t test, P < 0.01 for wild type vs. nch1). The nch1ncl1 mutant showed a phenotype similar to that of nch1 single mutants (cyan in Fig. S7G and H; Student s t test, P > 0.2 for nch1-1 vs. nch1-1ncl1-1 at all fluence rates). However, in rpt2nch1 (orange in Fig. 3 and Fig. S7), the increase, but not the decrease, in leaf transmittance occurred even under the weak blue-light condition (Fig. 3A and B; Fig. S7A and B), indicating that rpt2nch1 is completely defective in the accumulation response. The enhanced avoidance response at 20 and 50 µmol m 2 s 1 observed in nch1 was suppressed in rpt2nch1. At 50 µmol m 2 s 1, the increase in the leaf transmittance in rpt2nch1 was much lower than that in wild type. The avoidance response may have been saturated by the preceding light condition (20 µmol m -2 s -1 ). Contrary to a previous study that detected no defects 11
13 in chloroplast photorelocation movements in the rpt2 mutant plants (13), we found that the rpt2 mutant plants, rpt2-3 (20) and rpt2-4, exhibited a weak accumulation response (red in Fig. 3A and B; Fig. S7A and B; Student s t test, P < 0.01 for wild type vs. rpt2 at 3 µmol m 2 s 1 ) but the avoidance response and the dark recovery response were normal (Fig. 3A and B and Fig. S7A and B; Student s t test, for wild type vs. rpt2, P > 0.05 in the avoidance response and P > 0.2 in the dark recovery response). Four rpt2nch1 (rpt2-3nch1-1, rpt2-3nch1-2, rpt2-4nch1-1, and rpt2-4nch1-2) mutants displayed the same defect in the accumulation response (Fig. 3A and B; Fig. S7A and B). Furthermore, the expression of NCH1-GFP driven by the NCH1 native promoter (NCH1pro:NCH1-GFP, abbreviated as CCG; cyan in Fig. S7C and D) rescued the accumulation rate in phot2rpt2nch1 (Fig. S7C and D; Student s t test, P < for phot2-1rpt2-4nch1-1 vs. CCG in phot2-1rpt2-4nch1-1 at 3 µmol m 2 s 1 ). NCH1pro:NCH1-GFP rescued nch1 defects in the accumulation response, although the initial accumulation rate was not different among the genotypes in the dark recovery response (Fig. S7E and F; Student s t test, P<0.05 for nch1-1 vs CCG lines). This partial rescue might be partly caused by lesser accumulation of NCH1-GFP in CCG lines (Fig. S8). Thus, NCH1 and RPT2 are necessary specifically for the accumulation response. Similar to rpt2nch1, the nph3rpt2nch1ncl1 quadruple mutant plants (orange in Fig. S7G and H) retained the avoidance response (Fig. S7G and H), indicating that NPH3, RPT2, NCH1, and NCL1 are not involved in the avoidance response. phot2rpt2nch1 showed no chloroplast photorelocation movement like phot1phot2. To reveal the relationships of RPT2 and NCH1 with phototropins, the chloroplast movement in double- and triple-mutant plants between nch1, rpt2, phot1, and phot2 was analyzed. phot1rpt2 (green in Fig. S7I and J) exhibited a weak accumulation response similar to the phot1 (cyan in Fig. S7I and J) and rpt2 single mutants (Fig. S7I and J; 12
14 Student s t test, P > 0.3 for phot1-5rpt2-4 vs. phot1-5 or rpt2-4 at 3 µmol m 2 s 1 ). The phot1nch1 (yellow green in Fig. S7I and J) displayed an enhanced avoidance response and an impaired dark recovery response similar to nch1 but showed a weak accumulation response similar to phot1 (Fig. S7I and J; Student s t test, P < 0.05 for nch1-1 vs. phot1-5nch1-1 at 3 µmol m 2 s 1 ). Both chloroplast accumulation and the avoidance responses in phot1rpt2nch1 (magenta in Fig. S7I and J) were similar to those of rpt2nch1 (Fig. S7I and J; Student s t test, P > 0.5 for rpt2-4nch1-1 vs. phot1-5rpt2-4nch1-1 at all fluence rates). phot2rpt2 (green in Fig. 3C and D) and phot2nch1 (yellow green in Fig. 3C and D) exhibited a severe impairment of the avoidance response, similar to that of the phot2 mutant (cyan in Fig. 3C and D), although they showed weaker accumulation responses than phot2, and similar to those of rpt2 and nch1 (Fig. 3C and D; Student s t test, P < 0.01 for phot2-1 vs. phot2-1rpt2-4 or phot2-1nch1-1 at 3 µmol m 2 s 1 ). Prominently, the phot2rpt2nch1 [magenta in Fig. 3C and D (and S7C and E)] completely lacked both the accumulation and avoidance responses (Fig. 3C and D), like the phot1phot2 double mutant. These results indicated that both phot1 and phot2 mediated the accumulation response through RPT2 and NCH1. The RPT2/NCH1 homolog mediates the chloroplast accumulation response in the liverwort M. polymorpha. To investigate the conserved role of NRL proteins in phototropin-mediated chloroplast movements in land plants, we searched and characterized RPT2/NCH1 homologous genes in the liverwort M. polymorpha. M. polymorpha has seven NRL genes and each one NRL gene in seven major clades (Fig. S2). MpNCH1 belongs to a clade that includes the Arabidopsis RPT2 and NCH1 genes (Fig. 1B and S2), and has the same intron position as Arabidopsis NCH1 (the third intron of MpNCH1 corresponds to the fourth intron of NCH1; Fig. 1A and 4A). The 13
15 amino acid sequence of MpNCH1 is similar to those of RPT2 and NCH1 (Fig. S9). We generated an MpNCH1 knockout line, Mpnch1 ko, using the homologous-recombination-based gene-targeting method (32; Fig. 4B). In the rim region of wild-type gemmalings (the early stage of thalli developing from gemmae) that were cultured under the white light conditions for 3 days, chloroplasts covered the upper surface of the cells as a result of the chloroplast accumulation response (Fig. 4C). However, in Mpnch1 ko, only a few chloroplasts were situated on the upper cell surface under the same conditions (Fig. 4C), and the distribution of chloroplasts was similar to that in the strong-light-irradiated wild-type cells where the chloroplast avoidance response was induced (27). When the chloroplasts that were localized on the anticlinal walls in Mpnch1 ko were irradiated with a strong blue light using a confocal laser scanning microscope, chloroplasts escaped from the irradiated area (Supplemental movie S1), indicating that Mpnch1 ko is deficient in the accumulation response but not in the avoidance response. The defects of Mpnch1 ko in the chloroplast distribution were completely rescued by the expression of wild-type MpNCH1 cdna or the NCH1-Citrine fusion gene driven under the control of the cauliflower mosaic virus 35S promoter (Fig. 4C and D). The functional NCH1-Citrine was predominantly localized on the plasma membrane (Fig. 4D). Thus, NCH1 is necessary for the phototropin-mediated accumulation response in both Arabidopsis and M. polymorpha, and phototropin-nch1 signaling on the plasma membrane is conserved in land plants. Discussion NRL proteins, such as NPH3 and NAKED PINS IN YUC MUTANTS 1 (NPY1), may mediate auxin-mediated responses via auxin transport and auxin signaling, together with AGC kinase VIIIs, such as phototropin and PINOID (33). The phototropin-nph3 module regulates phototropic responses and leaf flattening (6, 10, 11, 17), and the 14
16 PINOID-NPY1 module regulates auxin-mediated organogenesis and root gravitropic responses (29, 34, 35, 36). Phototropin-NPH3 and PINOID-NPY1 regulate the localization of auxin efflux carrier PIN-FORMED (PIN) proteins (21, 37) and result in the activation of AUXIN RESPONSE FACTOR transcription factors (38, 39). In this study, we found that two phototropin-interacting NRL proteins, RPT2 and NCH1, mediate the chloroplast accumulation response. Because chloroplast photorelocation movement is cell autonomous and occurs even in enucleated cells (40), RPT2 and NCH1 mediate the chloroplast accumulation response independently of auxin transport and auxin-mediated transcription. Thus, the AGC kinase-nrl module can regulate other molecular mechanisms, in addition to the auxin-related mechanism, via PIN regulation. NCH1 is specific to the chloroplast accumulation response and NPH3 is specific to phototropism and leaf flattening, but RPT2 mediates both responses. Because NCH1 and NPH3 can activate phototropin signaling pathways in the absence of RPT2 (i.e., rpt2 mutants), RPT2 could not simply function as a molecular hub for the phototropin signaling pathways. Although RPT2 is a modulator of NPH3, which is essential for phototropism (16), the relationship between RPT2 and NCH1 in the chloroplast accumulation response is redundant. Although blue light induces the changes in NPH3 localization pattern, i.e., the internalization from the plasma membrane to cytosol (16), we could not detect any clear changes of localization pattern of NCH1 at least during 10 min of blue light irradiation in both A. thaliana and M. polymorpha (Fig. S10). Thus, blue-light-activated phototropins might mediate chloroplast accumulation response through the mechanism other than the regulation of NCH1 localization. Previously, we identified J-domain protein required for chloroplast accumulation response 1 (jac1) mutants (41) that were defective in the accumulation response and the enhanced avoidance response, like rpt2nch1 (42), suggesting that JAC1 is a regulator 15
17 for the accumulation response, together with RPT2/NCH1. However, the phenotypes of both jac1 and rpt2nch1jac1 mutant plants were considerably different from that of rpt2nch1 (Supplemental Text, S7K and L), and the amount of NCH1 was normal in jac1 (Fig. S3D), suggesting that JAC1 has slightly different functions from RPT2 and NCH1. Nevertheless, it is important to elucidate the relationship between JAC1 and RPT2/NCH1. An RPT2/NCH1 ortholog, MpNCH1, mediates the chloroplast accumulation response and localizes on the plasma membrane in M. polymorpha, indicating that the signaling mechanism for the accumulation response is conserved in land plants. Phototropins and components for the motility system of chloroplast photorelocation movements, such as CHLOROPLAST UNUSUAL POSITIONING 1 (CHUP1) and KINESIN-LIKE PROTEIN FOR ACTIN-BASED CHLOROPLAST MOVEMENT (KAC) are highly conserved from liverwort to seed plants (25-27, 43-46). However, M. polymorpha and other non-seed plant genome database searches identified RPT2/NCH1, CHUP1 and KAC orthologs, but not those of JAC1 or some other genes, in non-seed plant genome sequences. Therefore, although the photoreceptor (phototropin), phototropin-interacting protein (RPT2/NCH1), and motility system (CHUP1 and KAC) are conserved, the signal transduction pathway might have developed during land plant evolution. In conclusion, the specificity of some phototropin-mediated signaling pathways was determined by the members and the combinations of phototropin-interacting NRL proteins. NPH3 is specific to phototropism and leaf flattening, and NCH1 is specific to chloroplast accumulation response. The involvement of NRL proteins in the chloroplast avoidance response is presently unknown. The phototropin-interacting kinase BLUE LIGHT SIGNALING1 mediates stomatal opening and is the direct substrate of phototropin (30). However, NRL proteins are not direct substrates of phototropin. In 16
18 addition to NPH3 and RPT2, we should analyze the phototropin-dependent signaling pathway taking into account NCH1. We identified one NPH3 ortholog, MpNPH3, in the M. polymorpha genome (Fig. S2), implying that the divergence of NRL proteins in phototropin-dependent signaling occurred early in land plant evolution and that AGC kinase-nrl modules are an early innovation during land plant evolution. 17
19 Materials and Methods For the plant materials, the analysis of phototropin-mediated responses, confocal laser scanning microscopy, immunoblotting, and other methods, please see SI Materials and Methods. ACKNOWLEDGEMENTS We thank Kimitsune Ishizaki for the database search of the MpNCH1 gene, Yoriko Matsuda for the generation of M. polymorpha Mpnch1 ko, and Yasuomi Tada for in vitro transcription/translation system. We also thank Mineko Shimizu, Atsuko Tsutsumi, and Mika Niwaki for assistance in plant culturing. The BAC clones and SALK T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center. This work was supported by the Grant-in-Aid for Scientific Research ( , 15KK0254 to N. S., and to A. T., to S.-G. K., to K. S., , , and to T. K., , , , and to M.W.) from the Japan Society for the Promotion of Science. 18
20 References 1. Christie JM (2007) Phototropin blue-light receptors. Annu Rev Plant Biol 58: Bögre L, Ökrész L, Henriques R, Anthony RG (2003) Growth signaling pathways in Arabidopsis and the AGC protein kinases. Trends Plant Sci 8(9): Suetsugu N, Wada M (2013) Evolution of three LOV blue light receptor families in green plants and photosynthetic stramenopiles: phototropin, ZTL/FKF1/LKP2 and aureochrome. Plant Cell Physiol 54(1): Sakai T, et al. (2001) Arabidopsis nph1 and npl1: Blue light receptors that mediate both phototropism and chloroplast relocation. Proc Natl Acad Sci USA 98(12): Kinoshita T, et al. (2001) phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414(6864): Inoue S, Kinoshita T, Takemiya A, Doi M, Shimazaki K (2008) Leaf positioning of Arabidopsis in response to blue light. Mol Plant 1(1): Kagawa T, et al. (2001) Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 291(5511): Jarillo JA, et al. (2001) Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410(6831): Kozuka T, Kong SG, Doi M, Shimazaki K, Nagatani A (2011) Tissue-autonomous promotion of palisade cell development by phototropin 2 in Arabidopsis. Plant Cell 23(10): Motchoulski A, Liscum E (1999) Arabidopsis NPH3: A NPH1 photoreceptor-interacting protein essential for phototropism. Science 286(5441): Sakai T, Wada T, Ishiguro S, Okada K (2000) RPT2: A signal transducer of the phototropic response in Arabidopsis. Plant Cell 12(2):
21 12. Gingerich DJ, Hanada K, Shiu SH, Vierstra RD (2007) Large-scale, lineage-specific expansion of a bric-a-brac/tramtrack/broad complex ubiquitin-ligase gene family in rice. Plant Cell 19(8): Inada S, Ohgishi M, Mayama T, Okada K, Sakai T (2004) RPT2 is a signal transducer involved in phototropic response and stomatal opening by association with phototropin 1 in Arabidopsis thaliana. Plant Cell 16(4): Pedmale UV, Liscum E (2007) Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3. J Biol Chem 282(27): Tsuchida-Mayama T, et al. (2008) Mapping of the phosphorylation sites on the phototropic signal transducer, NPH3. Plant Sci 174(6): Haga K, Tsuchida-Mayama T, Yamada M, Sakai T (2015) Arabidopsis ROOT PHOTOTROPISM2 contributes to the adaptation to high-intensity light in phototropic responses. Plant Cell 27(4): de Carbonnel M, et al. (2010) The Arabidopsis PHYTOCHROME KINASE SUBSTRATE2 protein is a phototropin signaling element that regulates leaf flattening and leaf positioning. Plant Physiol 152(3): Harada A, Takemiya A, Inoue S, Sakai T, Shimazaki K (2013) Role of RPT2 in leaf positioning and flattening and a possible inhibition of phot2 signaling by phot1. Plant Cell Physiol 54(1): Kozuka T, Suetsugu N, Wada M, Nagatani A (2013) Antagonistic regulation of leaf flattening by phytochrome B and phototropin in Arabidopsis thaliana. Plant Cell Physiol 54(1): Tsutsumi T, Takemiya A, Harada A, Shimazaki K (2013) Disruption of ROOT PHOTOTROPISM2 gene does not affect phototropin-mediated stomatal opening. Plant Sci :
22 21. Wan Y, et al. (2012) The signal transducer NPH3 integrates the phototropin1 photosensor with PIN2-based polar auxin transport in Arabidopsis root phototropism. Plant Cell 24(2): Lariguet P, et al. (2006) PHYTOCHROME KINASE SUBSTRATE 1 is a phototropin 1 binding protein required for phototropism. Proc Natl Acad Sci USA 103(26): Demarsy E, et al. (2012) Phytochrome Kinase Substrate 4 is phosphorylated by the phototropin 1 photoreceptor. EMBO J 31(16): Suetsugu N, Wada M (2013) Chloroplast photorelocation movement: A sophisticated strategy for chloroplasts to perform efficient photosynthesis. Advances in Photosynthesis - Fundamental Aspects, ed Najafpour MM (InTech, Rijeka, Croatia), pp Kagawa T, Kasahara M, Abe T, Yoshida S, Wada M (2004) Functional analysis of phototropin2 using fern mutants deficient in blue light-induced chloroplast avoidance movement. Plant Cell Physiol 45(4): Kasahara M, Kagawa T, Sato Y, Kiyosue T, Wada M (2004) Phototropins mediate blue and red light-induced chloroplast movements in Physcomitrella patens. Plant Physiol 135(3): Komatsu A, et al. (2014) Phototropin encoded by a single-copy gene mediates chloroplast photorelocation movements in the liverwort Marchantia polymorpha. Plant Physiol 166(1): Zhang L, Du L, Shen C, Yang Y, Poovaiah BW (2014) Regulation of plant immunity through ubiquitin-mediated modulation of Ca 2+ -calmodulin-atsr1/camta3 signaling. Plant J 78(2): Furutani M, et al. (2007) The gene MACCHI-BOU4/ENHANCER OF PINOID encodes a NPH3-like protein and reveals similarities between organogenesis and 21
23 phototropism at the molecular level. Development 134(21): Takemiya A, et al. (2013) Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat Commun 4: Wada M, Kong SG (2011) Analysis of chloroplast movement and relocation in Arabidopsis. Methods Mol Biol 774: Ishizaki K, Johzuka-Hisatomi Y, Ishida S, Iida S, Kohchi T (2013) Homologous recombination-mediated gene targeting in the liverwort Marchantia polymorpha L. Sci Rep 3: Barbosa ICR, Schwechheimer C (2014) Dynamic control of auxin transport-dependent growth by AGCVIII protein kinase. Curr Opin Plant Biol 22: Cheng Y, Qin G, Dai X, Zhao Y (2007) NPY1, a BTB-NPH3-like protein, plays a critical role in auxin-regulated organogenesis in Arabidopsis. Proc Natl Acad Sci USA 104(47): Cheng Y, Qin G, Dai X, Zhao Y (2008) NPY genes and AGC kinases define two key steps in auxin-mediated organogenesis in Arabidopsis. Proc Natl Acad Sci USA 105(52): Li Y, Cheng Y, Dai X, Zhao Y (2011) NPY genes play an essential role in root gravitropic responses in Arabidopsis. Mol Plant 4(1): Furutani M, Nakano Y, Tasaka M (2014) MAB4-induced auxin sink generates local auxin gradients in Arabidopsis organ formation. Proc Natl Acad Sci USA 111(3): Harper RM, et al. (2000) The NPH4 locus encodes the auxin response factor ARF7, a conditional regulator of differential growth in aerial Arabidopsis tissue. Plant Cell 12(5): Aida M, Vernoux T, Furutani M, Traas J, Tasaka M (2002) Roles of PIN-FORMED1 22
24 and MONOPTEROS in pattern formation of the apical region of the Arabidopsis embryo. Development 129(17): Wada M (1988) Chloroplast photoorientation in enucleated fern protonemata. Plant Cell Physiol 29(7): Suetsugu N, Kagawa T, Wada M (2005) An auxilin-like J-domain protein, JAC1, regulates phototropin-mediated chloroplast movement in Arabidopsis. Plant Physiol 139(1): Kodama Y, Suetsugu N, Kong SG, Wada M (2010) Two interacting coiled-coil proteins, WEB1 and PMI2, maintain the chloroplast photorelocation movement velocity in Arabidopsis. Proc Natl Acad Sci USA 107(45): Usami H, et al. (2012) CHUP1 mediates actin-based light-induced chloroplast avoidance movement in the moss Physcomitrella patens. Planta 236(6): Suetsugu N, et al. (2012) The KAC family of kinesin-like proteins is essential for the association of chloroplasts with the plasma membrane in land plants. Plant Cell Physiol 53(11): Oikawa K, et al. (2003) CHLOROPLAST UNUSUAL POSITIONING1 is essential for proper chloroplast positioning. Plant Cell 15(12): Suetsugu N, et al. (2010) Two kinesin-like proteins mediate actin-based chloroplast movement in Arabidopsis thaliana. Proc Natl Acad Sci USA 107(19): Ichikawa S, Yamada N, Suetsugu N, Wada M, Kadota A (2011) Red light, phot1, and JAC1 modulate phot2-dependent reorganization of chloroplast actin filaments and chloroplast avoidance movement. Plant Cell Physiol 52(8): Rosso MG, et al. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53(1-2): Huala T, et al. (1997) Arabidopsis NPH1: A protein kinase with a putative 23
25 redox-sensing domain. Science 278(5346): Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5): Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17(8): Suetsugu N, Higa T, Kong SG, Wada M (2015) PLASTID MOVEMENT IMPAIRED1 and PLASTID MOVEMENT IMPAIRED1-RELATED 1 mediate photorelocation movements of both chloroplasts and nuclei. Plant Physiol 169(2): Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile ppzp family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25(6): Suetsugu N, Kong SG, Kasahara M, Wada M (2013) Both LOV1 and LOV2 domains of phototropin2 function as the photosensory domain for hypocotyl phototropic responses in Arabidopsis thaliana (Brassicaceae). Am J Bot 100(1): Ueno K, Kinoshita T, Inoue S, Emi T, Shimazaki K (2005) Biochemical characterization of plasma membrane H + -ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol 46(6): Takemiya A, Yamauchi S, Yano T, Ariyoshi C, Shimazaki K (2013) Identification of a regulatory subunit of protein phosphatase 1 which mediates blue light signaling for stomatal opening. Plant Cell Physiol 54(1): Kinoshita T, Shimazaki K (1999) Blue light activates the plasma membrane H + -ATPase by phosphorylation of the C-terminus in stomatal guard cells. EMBO J 18(20): Kong SG, Arai Y, Suetsugu N, Yanagida T, Wada M (2013) Rapid severing and 24
26 motility of chloroplast-actin filaments are required for the chloroplast avoidance response in Arabidopsis. Plant Cell 25(2): Ishizaki K, et al. (2015) Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. Plos ONE 10(9):e Kubota A, Ishizaki K, Hosaka M, Kohchi T (2013) Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci Biotechnol Biochem 77(1):
27 FIGURE LEGENDS Fig. 1. Identification of NCH1 as a phototropin-signaling component. (A) Protein structure of NCH1. Four conserved regions I to IV (black boxes) and a BTB/POZ domain (black bar) are indicated. N- and C-terminal regions that are used in yeast two hybrid assay are indicated below. (B) Gene structure and mutation sites of RPT2, NCH1, and NCL1 genes. The rectangles and the intervening bars indicate exons and introns, respectively. The white rectangles and bars in the NCL1 gene indicate the predicted exon and intron sequences, respectively, which are similar to those of NCH1 but were deduced to be non-coding. The positions of the T-DNA insertion sites and the point mutation are indicated. (C) A RPT2/NCH1 clade in the NRL protein phylogenetic tree. The full NRL protein phylogenetic tree is presented in Fig. S2. The number indicates the branch support value of the RPT2/NCH1 branch. Bar = 0.5 substitutions per site. AT, Arabidopsis thaliana; AmTr, Amborella trichopoda; MA, Pinus abies; Smoe, Selaginella moellendorfii; Phpat, Physcomitrella patens; and Mapoly, Marchantia polymorpha. Fig. 2. (A) Plasma-membrane localization of NCH1-GFP. Images are false colored to show GFP (green) and chlorophyll (red) fluorescence. P: pavement cells, M: mesophyll cells. Bars = 10 µm. (B) Co-immunoprecipitation of NCH1 with phot1. Microsomal fraction of the proteins from 10-day-old light-grown seedlings of transgenic GFP and phot1-gfp lines probed with anti-nch1 or anti-phot1 antibodies. (C and D) Pull-down assay of phot1 and NCH1. FLAG- or His-tagged phot1 or NCH1 proteins were synthesized by in vitro transcription/translation reactions. The proteins bound to anti-flag beads were detected using anti-flag and anti-his. Fig. 3. Chloroplast photorelocation movements in rpt2 and nch1 mutant plants. (A D) 26
28 Analysis of chloroplast photorelocation movements through the measurement of leaf transmittance changes. (A and C) After a dark treatment for 10 min, the samples were sequentially irradiated with continuous blue light at 3, 20, and 50 µmol m 2 s 1 for 60, 40, and 40 min, indicated by white, light blue, and blue arrows, respectively. The light was turned off after 150 min (black arrow). Data are presented as means of three independent experiments, and the error bars indicate standard errors. (B and D) Changes in leaf transmittance rates during the 2 6 min after changes in the light fluence rates (3, 20, and 50 µmol m 2 s 1 or dark) are indicated as averages of transmittance changes for 1 min. Data are presented as means of three independent experiments and the error bars indicate standard errors. Fig. 4. MpNCH1 mediates the chloroplast accumulation response in Marchantia polymorpha. (A) Targeting of the MpNCH1 gene by homologous recombination. Rectangles indicate exons and the intervening bars indicate introns. A 210-bp region, including a part of the first exon, was replaced with the MpEF1a:HPTII cassette of pjhy-tmp1-mpnch1. The positions and directions of PCR primers used in B are indicated by lines and arrows, respectively. (B) Genotyping of the Mpnch1 ko line by PCR. The positions of the PCR primers used are shown in A. M: DNA molecular size marker, WT: wild type, KO: Mpnch1 ko.the size of the DNA marker is indicated at both sides of the gel. (C) Chloroplast distribution under the weak continuous white-light conditions (~50 µmol m 2 s 1 ). Gemmalings of wild type, Mpnch1 ko, and two complemented Mpnch1 ko lines were cultured under continuous white light for 3 days. (D) Plasma-membrane localization of MpNCH1-Citrine. Gemmalings of a complemented 35S:MpNCH1-Citrine Mpnch1 ko line (Line 3) were cultured under continuous white light for 3 days. Images are false-colored to show Citrine (green). Plant samples in the bright field and epi-fluorescent images are different. Bars = 20 µm. 27
29 A BTB/POZ NCH1 I II III IV N 265 C B RPT2 NCH1 rpt2-3 rpt2-4 G1436A nch1-2 nch1-1 ncl C NCL bp
30 Mesophyll Pavement B GFP phot1-gfp D BL D BL Input Pull-down (anti-flag) His-P1N His-P1C FLAG - NCH1 - NCH1 Immunoblot (anti-his) Immunoblot (anti-flag) Input IP (anti-gfp) Input Pull-down (anti-flag) His-NCH1 FLAG - P1N - P1C Immunoblot (anti-nch1) Immunoblot (anti-phot1) Immunoblot (anti-his) Immunoblot (anti-flag)
31 A Δ% Transmittance C Δ% Transmittance Wild type nch1-1 nch1-2 rpt2-4 rpt2-4nch1-1 rpt2-4nch Time (min) Wild type rpt2-4 nch1-1 rpt2-4nch1-1 phot2-1 phot2-1rpt2-4 phot2-1nch1-1 phot2-1rpt2-4nch Time (min) B Δ% Transmittance/min Δ% Transmittance/min D Dark Fluence rates of blue light (μmol m -2 s -1 ) Dark Fluence rates of blue light (μmol m -2 s -1 )
32 A c a b d MpNCH bp 3348 bp MpEF1a pro hpt II En/Spm pjhy-tmp1-mpnch1 a EF_R1 H1F b B a/b c/ef_r1 H1F/d M WT KO WT KO WT KO M ATG 1 k bp C Wild type Mpnch1 ko D 35S:MpNCH1 -Citrine (Mpnch1 ko ) Bright field Line 3 35S:MpNCH1 (Mpnch1 ko ) Line 1 Line 2 Citrine Fluorescence
33 RPT2/NCH1 subfamily of NPH3-like proteins is essential for the chloroplast accumulation response in land plants Noriyuki Suetsugu *, Atsushi Takemiya, Sam-Geun Kong, Takeshi Higa, Aino Komatsu, Ken-ichiro Shimazaki, Takayuki Kohchi and Masamitsu Wada * * To whom correspondence should be addressed. suetsugu@lif.kyoto-u.ac.jp or masamitsu.wada@gmail.com This PDF file includes: SI Text SI Materials and Methods Figs. S1 to S6 Movie 1 SI Text NCL1 is likely non-functional The chromosomal regions containing NCH1 and NCL1 are in an intragenome syntenic relationship (PLANT GENOME DUPLICATION DATABASE: NCH1/NCL1 gene duplication might have occurred in Brassicaceae, whereas the RPT2/NCH1 gene duplication seems to have occurred before the separation of Amborella and other angiosperm (Fig. 1C). However, among the NCL1 genes from some Brassicaceae species whose genome sequences are available, only the Arabidopsis NCL1 harbors a thymine insertion in the annotated second exon (Fig. S1B). This insertion produces the premature stop codon (specific to 1
34 Arabidopsis NCL1) if the start codon conserved in the Brassicaceae NCH1/NCL1 genes is used. Furthermore, there is no cdna or EST data for NCL1 in the TAIR database, while there are several cdna and EST clones for NCH1. Additionally, an ncl1 T-DNA line (Fig. 1B) showed the wild-type phenotype (cyan in Fig. S7G and H; Student s t test, P > 0.1 for wild type vs. ncl1-1 at all fluence rates) and the nch1ncl1 double mutant resembled the nch1 single mutant (cyan in Fig. S7G and H; Student s t test, P > 0.2 for nch1-1 vs. nch1-1ncl1-1 at all fluence rates). Furthermore, nph3rpt2nch1ncl1 quadruple mutant plants exhibited normal blue-light-induced stomatal opening (Fig. S6). Thus, NCL1 might regulate other responses if functional. The phenotypes of rpt2nch1 and jac1 mutants are similar but distinct The J-domain protein required for chloroplast accumulation response 1 (jac1) mutants (41) share common phenotypes, including the defective accumulation response, the enhanced avoidance response (the magnitude), and the impaired dark recovery response (42) with rpt2nch1, suggesting that RPT2 and NCH1 mediate the accumulation response through the regulation of JAC1. However, a pair-wise mutant analysis indicated that the jac1phenotypes (cyan in Fig. S7K and L) were somewhat different from those of rpt2nch1. Although the magnitude of the avoidance response increased in jac1 and rpt2nch1, the avoidance response was much stronger at 20 µmol m 2 s 1 in rpt2nch1 compared with jac1 (Fig. S7K and L; Student s t test, P < 0.01 for rpt2-4nch1-1 vs. jac1-1 at 20 µmol m 2 s 1 ). At 20 and 50 µmol m 2 s 1, the avoidance response was saturated in rpt2nch1 but not in jac1 (Fig. S7K and L). Furthermore, rpt2nch1jac1 (red in Fig. S7K and L) exhibited a weaker avoidance response than rpt2nch1 (Fig. S7K and L; Student s t test, P < for rpt2-4nch1-1 vs. rpt2-4nch1-1jac1-1 at 20 µmol m 2 s 1 ), consistent with jac1 being defective in the regulation of actin filaments under the high-light condition (47). We have never detected 2
35 the interaction between NCH1/RPT2 and JAC1 in yeast two hybrid assay. Our findings suggest that JAC1 has some functions that are independent of RPT2 and NCH1. SI Materials and Methods Arabidopsis lines and the growth condition The wild-type and mutant lines have a Columbia gl1 background. Arabidopsis cultures were performed as described previously (41). Briefly, seeds were sown on 0.8% agar medium containing 1/3 strength Murashige & Skoog inorganic salt and 1% sucrose, and cultured under white light at ca. ~100 μmol m 2 s 1 (16h) / dark (8h) cycle at 23 C in an incubator. SALK and SAIL T-DNA knockout lines, SALK_ (nch1-1), SALK_ (ncl1-1), and SAIL_140_D03 (rpt2-4), were provided by ABRC. nch1-2 is a GABI-Kat line, 326_A01 (48). phot1-5 (49), phot2-1 (7), nph3-6 (10), rpt2-3 (20), and jac1-1 (41) were described previously. Double, triple, and quadruple mutants were generated by genetic crossing. Transgenic GFP and phot1-gfp lines were described previously (30). Phylogenetic analyses of RPT2 and NCH1 Amino acid sequences of the full length NRL proteins were aligned using the MUSCLE program (50) implemented in Geneious software (version 6.1.8; Biomatters; with default parameters. Amino acid sequences for the conserved BTB/POZ domain and DVI regions were extracted and combined. Unaligned gaps were removed from the resulting alignment using Gblocks ( html). The Bayesian phylogenetic tree construction was performed using MrBayes (51). Generation of transgenic Arabidopsis plants To construct a vector for NCH1pro:NCH1-GFP, the NCH1 gene fragment including a 3
36 2,568-bp 5 sequence (before the start codon) and the gene body region that contained the open reading frame but lacked the stop codon, was cloned into the PstI and KpnI restriction enzyme sites of the ppzp221/35s:gfp-nost binary vector (52). The latter was constructed by cloning the GFP cdna into the ppzp221/35s-nost binary vector (53). The nch1-1 mutants were transformed with ppzp221/nch1pro:nch1-gfp-nost by the floral-dipping method using Agrobacterium tumefaciens (GV3101::pMP90). Confocal laser scanning microscopy To observe the subcellular localization of NCH1-GFP in A. thaliana and MpNCH1-Citrine in M. polymorpha, a confocal microscope (C2+; Nikon, Tokyo, Japan) was used. Fluorescence was observed at nm for fluorescent proteins and nm for chlorophyll autofluorescence. Immunoblot analysis An NCH1 rabbit polyclonal antibody was raised against a purified His-tag-NCH1 C-terminal domain ( ) fusion protein (eurofins genomics, Tokyo, Japan). The phot1 antibody was described previously (46). Seedlings were homogenized in extraction buffer [50 mm Mops-KOH (ph 7.5), 100 mm NaCl, 2.5 mm EDTA, 1 mm DTT, 1 mm PMSF, 10 µm leupeptin, and phosphatase inhibitor cocktail (Nacalai tesque, Kyoto, Japan)]. After centrifuging three times at 12,000 g, the supernatants (total protein fraction) were quantified using the Bio-Rad Protein Assay (Bio-Rad, CA, USA). Total protein homogenates were ultracentrifuged at 100,000 g for 1 h at 4 C. The resultant supernatants and pellets were used as soluble and microsomal protein fractions, respectively. An immunoprecipitation analysis with anti-gfp was performed as previously described (30). 4
37 In vitro pull-down assay In vitro pull-down assay was performed as previously described (30). FLAG- and His-tagged protein pair were synthesized using an in vitro transcription/translation system (BioSieg, Tokushima, Japan) and kept on ice for 10 min in a binding buffer containing 20 mm Tris-HCl (ph 7.4), 140 mm NaCl and 0.1% Triton X-100. After brief centrifugation, the supernatants were incubated with anti-flag Ms agarose (Sigma-Aldrich, MO, USA) for 1 h. The bound proteins were used for immunoblotting analysis using anti-flag (Sigma-Aldrich) and anti-his antibodies (GeneScript, NJ, USA). Yeast two hybrid assay The binding domain (bait) or the activation domain (prey) plasmids containing the genes for NCH1, RPT2, and NPH3 were constructed by subcloning the cdnas into the pgbkt7 or pgadt7 vectors, respectively (Matchmaker TM Yeast Two-Hybrid System; Takara Bio Inc., Kusatsu, Japan). The bait and prey plasmids were transformed into the yeast strain AH109 and Y187, respectively. The mated transformants were selected on SD agar media without Leu and Trp and spotted on to the indicated media. Analysis of hypocotyl phototropic responses The phototropic responses were analyzed as previously described with some modifications (54). The sterilized seeds were sown on 1/3-strength Murashige & Skoog agar plates without sucrose. After the cold treatment at 4 C in the darkness for 2 days, the seeds were irradiated with white light (~80 µmol m 2 s 1 ) for 3 h and then grown vertically in the dark for 54 h at 23ºC. The etiolated seedlings were irradiated for 12 h with unilateral blue light (at 0.04, 0.4, 4, or 40 µmol m 2 s 1 ) using blue light-emitting diodes. The hypocotyl curvature was measured using the Image J program (National 5
38 Institutes of Health, Bethesda, MD, USA; Measurement of stomatal responses to blue light Thermal imaging was carried out using TVS-8500 (NEC Avio Infrared Technologies, Tokyo, Japan) (30). Subtraction images were prepared using PE Professional software (NEC Avio Infrared Technologies, Tokyo, Japan). The stomatal aperture in the abaxial epidermis was measured as described previously (5). Briefly, epidermal strips were incubated in 5 mm MES-Bis-Tris propane (ph 6.5), 50 mm KCl and 0.1 mm CaCl 2 under red light (50 µmol m 2 s 1 ) superimposed by blue light (10 µmol m 2 s 1 ) for 2 h at 24 C. Blue light-dependent H + pumping from guard cell protoplasts was determined with a glass ph electrode (5). The reaction mixture (0.8 ml) contains mm MES-NaOH (ph 6.0), 1 mm CaCl 2, 0.4 M mannitol, 10 mm KCl, and guard cell protoplasts (50 µg proteins). Guard cell protoplasts were prepared enzymatically from leaves (55, 56). The phosphorylation of a penultimate Thr residue of the H + -ATPase was determined by protein blotting using GST (57). An immunoblot analysis was carried out using anti-h + -ATPase antibodies (57). Analyses of chloroplast photorelocation movements To detect chloroplast photorelocation movements in A. thaliana, the measurement of leaf transmittance changes was performed as described previously (31). Detached third leaves from 16-day-old seedlings were placed on 1% (w/v) gellan gum in a 96-well plate. Leaves were dark-adapted at least for 1 h before transmittance measurement. In M. polymorpha, chloroplast distribution and Citrine fluorescence were observed using a fluorescent microscopy (Axiophot; ZEISS, Germany). For the observation of the chloroplast avoidance response in Mpnch1 ko, a confocal microscope (SP5; Leica Microsystems, Wetzlar, Germany) was used as described previously (58). 6
39 Growth conditions of M. polymorpha The wild-type male accession of M. polymorpha, Takaragaike (Tak)-1, was used. Plants were cultured on one-half-strength Gamborg s B5 agar medium at 22 C under continuous white light (~50 µmol m 2 s 1 ). Targeted gene knockout and genetic complementation of MpNCH1 The 5 and 3 homology arms [ 3,313~+57 (3370 bp) and +268~+3,615 (3348 bp) from the start codon, respectively] of the MpNCH1 gene were cloned into the PacI and AscI sites of pjhy-tmp1 (32), respectively. Tak-1 Tak-2 F 1 spores were used for Agrobacterium-mediated transformation as described previously (32). The selection and PCR screening of MpNCH1 knockout plants were performed as described previously (32). Briefly, hygromycin-resistant plants were screened by PCR with the primer pair 5 -AAGGAGGAGGAACGAACCAT-3 (a) and 5 -CGAATCTATCCCCCACGAAT-3 (b). Putative Mpnch1 ko lines were confirmed using PCR with primers outside of the 5 - or 3 -homologous arms (c, 5 -CCCACTCACCTTTTCCACAT-3 or d, 5 -GTGTTCTTCTCAGGCGGAAG-3, respectively) and inside the MpEFpro::hptII cassette (EF_R1, 5 -GAAGGCTTCTGATTGAAGTTTCCTTTTCTG-3 for the 5 -arm and H1F, 5 -GTATAATGTATGCTATACGAAGTTATGTTT-3 for the 3 -arm). To generate the complementation construct, MpNCH1 cdna PCR fragments with or without a stop codon were cloned into pentr/d-topo (Life Technologies, MA, USA). The resulting MpNCH1 cassette was subcloned into a binary vector pmpgwb302 or pmpgwb306 (59) using LR clonase II (Life Technologies, MA, USA), resulting in the cauliflower mosaic virus pro35s:mpnch1 or pro35s:mpnch1-citrine binary vector, respectively. These binary vectors were introduced into regenerating thalli of Mpnch1 ko as described previously (60). 7
40 Supplementary Figure Legends Fig. S1. NRL protein sequence alignment. (A) The protein sequence alignment was performed using the MUSCLE program ( Regions colored red, blue, orange, and green indicate the four conserved sequence domains (I IV, respectively) defined by Motchoulski and Liscum (10). Upper red lines indicate the position of the BTB/POZ domain. A lower black line indicates the deletion in the conserved N-terminal region specific to Arabidopsis NCL1. NPH3, At5g64330; RPT2, At2g30520; NCH1, At5g67385; and NCL1, At3g (B) Alignment of Arabidopsis NCH1 and NCL1 gene sequences. The start codons are colored orange. The 5 region of the start codon of the Arabidopsis NCL1 gene is highly similar to NCH1 and Brassicaceae NCL1 exon1 and exon2 sequences but a thymine insertion (red) produces a premature stop codon (red bold). AlNCL1, Arabidopsis lyrata NCL1, XM_ ; EsNCL1, Eutrema salsugineum NCL1, XM_ ; CrNCL1, Capsella rubella NCL1, XM_ ; and BrNCL1, Brassica rapa NCL1, XM_ Fig. S2. The Bayesian phylogenetic tree of NRL proteins. The parameters used were as follows: preset amino acid model program, mixed (LG, blosum, and wag); length of set rates, inverse-gamma; number of gamma categories, 4; Markov chain Monte Carlo start tree, random; number of generation, 1,000,000; number of runs, 2; number of chains, 4; heated chain temperature, 0.2; sample frequency, 200; and burn-in fraction, klfl00053_0080 were used as the outgroup. The numbers at the nodes indicate posterior probabilities. Bars indicate substitution values per site. Both RPT2/NCH1 and NPH3 clades are indicated (red box). Seven Marchantia and four Arabidopsis NRL proteins (NCH1, RPT2, NCL1, and NPH3) are bold-faced. AT, Arabidopsis thaliana; AmTr, Amborella trichopoda; MA, Pinus abies; Smoe, Selaginella moellendorfii; Phpat, 8
41 Physcomitrella patens; Mapoly, Marchantia polymorpha; and kfl, Klebsormidium flaccidum. Fig. S3. An immunoblot analysis of NCH1 proteins. (A) Immunoblot analysis of phot1 and NCH1 proteins. Microsomal fraction of the proteins (corresponding to 50 µg total proteins) were used for an immunoblotting analysis with phot1 and NCH1 antibodies. The 13-day-old seedlings were dark adapted for 24 h (D) and then irradiated with 6.4 µmol m 2 s 1 of blue light for 1 h (BL). An asterisk indicates the position of a non-specific protein band. (B) Total protein extracts (T) were fractionated into soluble (S) and microsomal (M) fractions by ultracentrifugation (100,000 g, 1 h, 4 C). Immunoblotting was performed using phot1 and anti-nch1 antibodies. (C) Microsomal fraction of the proteins from 10-day-old light-grown seedlings of transgenic GFP, phot1-gfp, and nch1-1 mutant plants probed with anti-nch1 or anti- H + -ATPase antibodies. H + -ATPase is a marker for microsomal proteins. (D) Immunoblot analysis of phot1 and NCH1 proteins in jac1 mutant plants. Details are same as A. Fig. S4. Yeast two hybrid analysis. Yeast cells expressing the indicated bait and prey vectors were cultured on SD agar media supplemented with a mixture of the appropriate amino acids without Leu and Trp (-LT), His, Leu, and Trp (-HLT), or Ala, His, Leu, and Trp (-AHLT). Yeast cells were spotted with serial dilution (1, 1/10, and 1/100 from left). Yeast cells expressing BD-RPT2 C/AD-NPH3 N, BD-RPT2 C/AD-RPT2 N, or BD-NCH1 C/AD-NPH3 N could grow well on both HLT and AHLT media, indicating that RPT2 C-NPH3 N, RPT2 C-RPT2 N, or NCH1 C-NPH3 N interactions is strong. Conversely, yeast cells expressing BD-NCH1 C/AD-NCH1 N could grow well on both -HLT but very weakly on -AHLT media. Yeast cells expressing BD-RPT2 C/AD-NCH1 N could grow weakly on -HLT media. Thus, RPT2 C-NCH1 N or NCH1 9
42 C-NCH1 N interaction is weak. Yeast cells containing empty BD or AD vectors could not grow on both HLT and AHLT media, indicating that the interactions detected here are specific. Fig. S5. Hypocotyl phototropic response and leaf flattening in rpt2 and nch1 mutant backgrounds. (A) The phototropic response of the hypocotyls in etiolated seedlings. The hypocotyl curvature was induced by unilateral irradiation with blue light at the indicated fluence rates in the 3-day-old etiolated seedlings for 12 h. Data are presented as means of three independent experiments, and the error bars indicate standard errors. (B) Leaf-flattening response. After seeds were sown on vermiculite soil, plants were grown under continuous white-light conditions (~70 µmol m 2 s 1 ) for 25 days at 22 C. Bar = 1 cm. Fig. S6. nph3rpt2nch1ncl1 quadruple mutants are not impaired in the blue-light-induced stomatal opening. (A) Thermal image of a blue-light-dependent leaf temperature decrease. Arabidopsis wild-type and mutant (right panel) plants were illuminated with red light (RL: 80 µmol m 2 s 1 ) for 50 min and then a weak blue light (BL: 5 µmol m 2 s 1 ) was superimposed. Subtractive images (left panel) were obtained by subtracting an initial thermal image (prior to blue light exposure) from an image taken 15 min after blue light exposure. The bar represents 1 cm. (B) Light-dependent stomatal opening in the epidermis. Epidermal strips from dark-adapted plants were illuminated with red light (RL: 50 µmol m 2 s 1 ) and blue light (BL: 10 µmol m 2 s 1 ) for 2 h. Bars represent means ± standard deviations (n = 75, pooled from triplicate experiments). (C) Blue-light-dependent H + pumping in guard cell protoplasts from wild-type and nph3rpt2nch1ncl1 quadruple mutant plants. Guard cell protoplasts were illuminated by red light (600 µmol m 2 s 1 ) for 2 h, and a 10
43 pulse of blue light (100 µmol m 2 s 1, 30 s) was applied as indicated. (D) Blue light-dependent phosphorylation of the H + -ATPase in guard cell protoplasts. The phosphorylation of the H + -ATPase was determined by protein blotting using a GST protein. The total H + -ATPase proteins were detected by immunoblotting using an anti-h + -ATPase antibody. Fig. S7 Chloroplast photorelocation movements in rpt2 and nch1 mutant backgrounds. (A L) Analysis of chloroplast photorelocation movements through the measurement of leaf transmittance changes in the indicated lines. (A, C, E, G, I, and K) After dark treatment for 10 min, the samples were sequentially irradiated with continuous blue light at 3, 20, and 50 µmol m 2 s 1 for 60, 40, and 40 min, as indicated by white, light blue, and blue arrows, respectively. The light was turned off at 150 min (black arrow). Data are presented as means of three independent experiments, and the error bars indicate standard errors. (B, D, F, H, J, and L) Changes in leaf transmittance rates over 2 6 min after changes in the light fluence rate (3, 20, and 50 µmol m 2 s 1 or dark) are indicated as percentages of transmittance changes for 1 min. Data are presented as means of three independent experiments, and the error bars indicate standard errors. (A and B) Analysis in two rpt2 alleles. (C and D) The NCH1pro:NCH1-GFP transgene (CCG1-2) rescued the impaired chloroplast accumulation response in phot2-1rpt2-3nch1-1. (E and F) Partial rescue of the nch1 mutant phenotype in NCH1pro:NCH1-GFP transgenic lines (CCG1-2, CCG2-8, and CCG5-5). (G and H) No detectable involvement of four NRL proteins in the chloroplast avoidance response. (I and J) Genetic interactions between PHOT1 and RPT2/NCH1. (K and L) Comparison of phenotypes between rpt2nch1 and jac1 mutants. Data are presented as means of three independent experiments (five experiments in A and B, and four experiments in I and J) and the error bars indicate standard errors. 11
44 Fig. S8. An immunoblot analysis of NCH1-GFP proteins. The 50 µg total proteins extracted from 15-day-old seedlings were used for the immunoblotting analyses with NCH1 and phot1 antibodies. A star indicates full-length NCH1 in wild type. Black and white circles indicate full-length NCH1-GFP and the degradation products, respectively. Fig. S9. The sequence alignment of M. polymorpha and Arabidopsis RPT2/NCH1 subfamily proteins. The protein sequence alignment was performed using the MUSCLE program ( Regions colored red, blue, orange, and green indicate the four conserved sequence domains (I to IV, respectively) defined by Motchoulski and Liscum (10). Upper red lines indicate the positions of the BTB/POZ domains. A bottom black line indicates the deletion in the conserved N-terminal region specific to Arabidopsis NCL1. RPT2, At2g30520; NCH1, At5g67385; and NCL1, At3g Fig. S10. No apparent light-induced changes in NCH1 localization pattern. A part (indicated by white rectangles) of pavement (A) and mesophyll cells (B) of A. thaliana NCH1pro:NCH1-GFP line or cells of M. polymorpha 35S:MpNCH1-Citrine line (C) were irradiated with blue laser (488 nm, ca µmol m 2 s 1 ) for 10 min. ch: chloroplasts. Bars = 10 µm. Supplementary Movie Legends Movie 1. This time-lapse movie shows the chloroplast avoidance response induced by a blue laser in the M. polymorpha Mpnch1 ko line. A piece of weak light-adapted nch1 ko gemmaling was placed into a cuvette that was composed of two round cover slips that were supported by a ring-shaped silicon-rubber spacer. The time-lapse images 12
45 (maximized with three images in a 1.2-µm depth) were collected at approximately ~01:06 (mm:ss) intervals and played back at five frames per second (fps). The total elapsed time is 33:17 (mm:ss). The images are false-colored to indicate chlorophyll (red) fluorescence and bright transmittance field (gray). The region indicated by a blue rectangle (15 µm 50 µm) was irradiated using 458-nm laser scans during the intervals between the image acquisitions to induce the avoidance response. Scale bar = 25 µm. 13
46 A NPH3 RPT2 NCH1 NCL1 1 MMWESESDGGVGVGGGGGREYGDGVLSSNKHGGVKTDGFELRGQSWFVATDIPSDLLVKIGDMNFHLHKYPLLSRSGKMNRLIYE-SRDPDPTILILDDL MATEGKNPINMNSMSSSLA RTGQWVFSQDIPTDVVVEVGEANFSLHKFMLVAKSNYIRKLIME-SKDSDVTRINLSDI MSAKKKDLLSSAMK RTSEWISSQEVSSDVTVHVGEASFSLHKFPLMSKCGFIKKLVSESSKDSDSTVIKIPDI MLEKLSFLLHKFPLVSKCGFIKKLASESSNDSN--IIRIPDF NPH3 100 PGGPEAFELASKFCYGVPVDLTATNISGLRCAAEYLEMTEDLEEGNLIFKTEAFLSYVVLSSWRDSILVLKSCEKLSPWAENLQIVRRCSESIAWKACSN RPT2 78 PGGPEIFEKAAKFCYGVNFEITVQNVAALHCAAEFLQMTDKYCDNNLAGRTQDFLSQVALSSLSGAIVVLKSCEILLPISRDLGIVRRCVDVVGAKAC-- NCH1 74 PGGSEAFELAAKFCYGINFDMSTENIAMLRCAAEYLEMTEEHSVENLVVRAEAYLNEVALKSLSSSITVLHKSEKLLPIAERVKLVSRCIDAIAYMTC-- NCL1 41 PGGAEGFELVIKFCYDISFEINTENIAMLLCAAEYLEMTEEHSVENLVETIEVYLNEVILKSLSKSVKVLQKSQDLLPIAERVRLVDRCIDSIAYAIC-- NPH3 200 PKGIRWAYTGKAPSPSTTNFAGSSPRWNESKDSSFYCSPS RNTNSQPVPPDWWFEDVSILRIDHFVRVITAIKVKGMRFELLGAVIMHY PPDWWFEDVSILRIDHFVRVITAIKVKGMRFELLGAVIMHY RPT NEAMF PCRTP--PNWWTEELCILDVDFFSDVVSSMKQRGVKPSSLASAIITY P--PNWWTEELCILDVDFFSDVVSSMKQRGVKPSSLASAIITY NCH QESHFCSPSSSNSGNNEVVVQQQSKQPV-VDWWAEDLTVLRIDSFQRVLIAMMARGFKQYGLGPVLMLY V-VDWWAEDLTVLRIDSFQRVLIAMMARGFKQYGLGPVLMLY NCL QES QSNEDI-VDWWADDLAVLKIDMFRRVLVAMIARGFKRYSLGPVLKLY -VDWWADDLAVLKIDMFRRVLVAMIARGFKRYSLGPVLKLY NPH3 289 AGKWLPGLIKEGGVAIAPAMSSAIGGGLGLGGDEMSISCGSNSSGGSSGPDWKGGLHMVLSAGKTNGHQDSVACLAGLGISPKDQRMIVESLISIIPPQK DQRMIVESLISIIPPQK RPT2 226 TEKSLRDLVR DHSGRGVKY SDPGDNESDERS QQRDLVQSIVSLLPSDK NCH1 240 AQKSLRGL EIFGKGMK KIEPKQEH EKRVILETIVSLLPREK KRVILETIVSLLPREK NCL1 188 AEKALRGLVR--FLNFLTEQCDIFGKEAK KMEAEQEH EKRLILETIVSLLPRER KRLILETIVSLLPRER NPH3 389 DSVTCSFLLRLLRAANMLKVAPALITELEKRVGMQFEQATLQDLLIPGYNNKGE-TMYDVDLVQRLLEHFLVQEQTEGSSPSRMSPSPSQSMYADIPRGN RPT2 274 GLFPVNFLCSLLRCAVFLDTSLTCKNELEKRISVVLEHVSVDDLLIPSFTYDGE-RLLDLDSVRRIISAFVEKEKNVGV F-- NCH1 281 NAMSVSFLSMLLRAAIFLETTVACRLDLENRMGLQLGQAVLDDLLIPSYSFTGDHSMFDTDTVQRILMNYLEFE-VEGV RLS NCL1 240 NSVSVSFLSILLRAAIYLETTVACRLDLEKRMGLQLRQAVIDDLLIPYYSFNGDNTMLDVDTVQRILMNYLEFE-VEG NPH3 488 NNNGGGGGGNNQNAKMRVARLVDSYLTEVARDRNLPLTKFQVLAEALPESARTCDDGLYRAIDSYLKAHPTLSEHERKRLCRVMDCQKLSMDACMHAAQN RPT NGGDFNRGVCSVSLQRVAKTVDSYLAEIATYGDLTISKFNAIANLVPKSARKSDDDLYRAIDIFLKAHPNLDEIEREKVCSSMDPLKLSYDARLHASQN VAKTVDSYLAEIATYGDLTISKFNAIANLVPKSARKSDDDLYRAIDIFLKAHPNLDEIEREKVCSSMDPLKLSYDARLHASQN NCH1 362 NNGVDLAG-----DMERVGKLLENYMAEIASDRNVSLQKFIGLAELIPEQSRVTEDGMYRAVDIYLKAHPNMSDVERKKVCSLMDCQKLSREACAHAAQN VGKLLENYMAEIASDRNVSLQKFIGLAELIPEQSRVTEDGMYRAVDIYLKAHPNMSDVERKKVCSLMDCQKLSREACAHAAQNR NCL NSADFAS DIGELMETYLAEIASDRNINFAKFIGFAECIPKQSR-----MYRAIDIFLKTHPNISEVEKKKVCSLMDCKKLSRDVYAHAAQN NPH3 588 ERLPLRVVVQVLFSEQVKISNALANTSLKESTTLGEAMGTYQPMIPNRKTLIEATPQSFQ--EGWAAAKKDINTLKFELETVKTKYVELQNEMEVMQRQF RPT2 452 KRLPVNIVLHALYYDQLKLRSGVAEQEERAV VVLPEALKTRSQLQADTTLAKENEALRSELMKMKMYVSDMQKN-----KNG NCH1 457 DRLPVQTIVQVLYYEQQRLRGEVTNDSDSPA PPPPQPAAVLPPKLSSYT--DELSKLKRENQDLKLELLKMKMKLKEFEKESE--KKTS NCL1 403 DRFQ ENLSNSDSPA PATAEK-TLSPPELSSYK--NELSKLNRENQYLKLELLKVKMKFKELEKE-----KAF NPH3 686 EKTGKVKNTPSSSAWTSGWKKLSKLTKMSGQESHDISSGGEQAG VD----HPPPRKPRRWRNSIS RPT2 529 AGASSSNSSSLVSS------KKSKHTFFSSV-SKKLGKL-NPFKNGSKDTSHIDEDLGGVDITKPRRRRFSIS NCH1 542 SSTISTNPSSPISTASTGKPPLPRKSFINSV-SKKLGKL-NPFS ITPYNGRGRTKPPKDRRHSIS NCL1 467 EVMSGSDCSSSVSTASVAKPRLPRKSFINSV-SQKLGKLINPFG LK----QGQTKQPKSRRHSIS B AtNCH1 ATGTCAGCAAAGAAGAAAGATCTTTTGTCCTCAGCTATGAAAAGAACCAGTGAATG AtNCL1 ATGTCTGGAAAGAAGAAGGAGCTTTTGTCCTCAGCCATGAAGAGAACTAGCGAATG AlNCL1 ATGTCTAGAAAGAAG---GAGCTTCTGTCCTCAGCTATGGAGAGAACTAGTGAATG EsNCL1 ATGTCCGCAAAGCAG---GAGCTTATGTCCTTAGCCATGAAGAGAACTAGTGAATG CrNCL1 ATGTCTGGAAAGAAGAAGGAGCTTCTGTCCTCAGCCATGAAGAGAACTAGTGAATG BrNCL1 ATGTCCGCAAAGAGGAAGGAGCTTCTGTCCTTAGCCATGGAGAGAACTAGTGAATG (intron) AtNCH1 GATTTCTTCTCAAGAAGTATCTAGTGATGTCACCGTTCATGTAGGAGAAGCT-TCGTTTTCACTGCACAAG AtNCL1 GATTTCTTCTCAAGAAATTCCTAGTGACGTCACTGTTCATGTTGGAGAAACTTTCGTTTTTACTCCACAAG AlNCL1 GATTTCTTCTCAAAAAATTCCTAGTGACGTCACTGTTCATGTTGGAGAAACT-TCGTTTTCACTCCACAAG EsNCL1 GATATCTTCTCAGGAAATTCCTTGTGACATCGCTGTTCATATAGGAGAGACT-TCGTTTTCACTTCACAAG CrNCL1 GATTTATTCTCAAGAGATTCCAAGTGACATCACTGTTGATGTTGGAGAAACT-TTGTTTTCACTACACAAG BrNCL1 GATTTCATCTCAAGAGATTCCAAGTGATATCACTGTTTACATAGGAGACACG-TCCTTTTCCCTCCATAAG (intron) AtNCH1 TTTCCACTCATGTCAAAATGTGGGTTTATCAAGAAACTTGTGTCGGAATCAAGCAAAGATTCAGATT AtNCL1 TTTCCACTTGTTTCAAAATGTGGGTTTATAAAGAAACTTGCTTCTGAATCAAGCAATGATTCCAACA AlNCL1 TTTCCACTTGTTTCAAAATGTGGGTTTATAAAGAAACTTGCTTCCAAATCAAGCAACGATTCGAACA EsNCL1 TTTACAGTTGTGTCAAAATGTGGGTTTCTAAAGAAACTTGCTTCCGAAACAAGCAACGATTCCAACA CrNCL1 TTTCCACTTGTGTCAAAATGTGGGTTTATAAAGAAACTTGCTTCCGAAACAAGCAACGATTCGAAAG BrNCL1 ATT---CTTCTGTCAAAATGTGGGTTTATAAAGAAACTTACTTCCGAGTCAAGCAACGATTCGAACA
47 RPT2/NCH1 clade (Fig.1B) NPH3 clade
48 A Wild type D BL phot1-5 phot2-1 rpt2-4 nch1-1 D BL D BL D BL phot1 B T wild type S M T nch1-1 S M phot1 * NCH1 NCH1 C GFP phot1-gfp nch1 NCH1 H + -ATPase D wild type phot1-5 phot2-1 jac1-2 nch1-1 D BL D BL D BL D BL phot1 NCH1
49 Bait -LT -HLT -AHLT Prey Empty vector (pgadt7) Empty vector (pgbkt7) AD-NPH3 N (1-371) AD-RPT2 N (1-257) AD-NCH1 N (1-264) Empty vector (pgadt7) BD-RPT2 C ( ) AD-NPH3 N (1-371) AD-RPT2 N (1-257) AD-NCH1 N (1-264) Empty vector (pgadt7) BD-NCH1 C ( ) AD-NPH3 N (1-371) AD-RPT2 N (1-257) AD-NCH1 N (1-264)
50 A Wild type nch1-1 nch1-2 rpt2-4 rpt2-4nch1-1 rpt2-4nch Hypocotyl curvature (degree) B Fluence rates of blue light (μmol m -2 s -1 ) Wild type nch1-1 phot1-5phot2-1 rpt2-4 rpt2-4nch1-1
51 A Wild type rpt2-3 nch1-1 phot2-1 rpt2-3 nch1-1 C 1.0 Wild type rpt2-3 nch1-1 phot2-1 rpt2-3 nch1-1 phot1-5 phot2-1 phot2-1 rpt2-3 nph3-6 rpt2-3 nch1-1 ncl phot1-5 phot2-1 B H+ pumping (ph decrease) 0.0 C nph3-6 rpt2-3 nch1-1 ncl1-1 Dark RL+BL 3.0 Wild type phot1-5 phot2-1 rpt2-3 nch1-1 phot2-1 rpt2-3 phot2-1 rpt2-3 nch1-1 nph3-6 rpt2-3 nch1-1 ncl1-1 D BL 1 nmol H + Stomatal aperture (µm) 4.0 phot2-1 rpt2-3 3 min BL Wild type nph3-6 rpt2-3 nch1-1 ncl1-1 Wild type nph3-6 rpt2-3 nch1-1 ncl1-1 RL BL RL BL Protein blot (GST ) Immunoblot (anti-h+-atpase)
52 A Δ% Transmittance C Δ% Transmittance Wild type rpt2-3 rpt2-4 nch1-1 rpt2-3nch1-1 rpt2-4nch Time (min) Wild type phot2-1rpt2-4 phot2-1rpt2-4nch1-1 CCG1-2 in phot2-1rpt2-4nch1-1 B Δ% Transmittance/min Δ% Transmittance/min D Dark Fluence rates of blue light (μmol m -2 s -1 ) E Δ% Transmittance G Δ% Transmittance I Δ% Transmittance Wild type nch1-1 CCG1-2 CCG2-8 CCG5-5 Time (min) Time (min) Wild type nch1-1 ncl1-1 nch1-1ncl1-1 nph3-6rpt2-3 nch1-1ncl1-1 Time (min) Wild type rpt2-4 nch1-1 rpt2-4nch1-1 phot1-5 phot1-5rpt2-4 phot1-5nch1-1 phot1-5rpt2-4nch Time (min) F Δ% Transmittance/min H Δ% Transmittance/min Δ% Transmittance/min J Dark Fluence rates of blue light (μmol m -2 s -1 ) Dark Fluence rates of blue light (μmol m -2 s -1 ) Dark Fluence rates of blue light (μmol m -2 s -1 ) Dark Fluence rates of blue light (μmol m -2 s -1 ) K 0.3 L 0.03 Δ% Transmittance Wild type rpt2-4nch1-1 jac1-1 rpt2-4nch1-1jac1-1 Δ% Transmittance/min Time (min) Dark Fluence rates of blue light (μmol m -2 s -1 )
53 NCH1pro:NCH1-GFP (nch1-1) wild type nch NCH1 phot1
54 RPT2 MpNCH1 NCH1 NCL MATEGKNPINMNSMSSSLARTGQWVFSQDIPTDVVVEVGEANFSLHKFMLVAKSNYIRKLIMESK-DSDVTRINLSDIPGGPEIFEKAAKFCYGV PGGPEIFEKAAKFCYGV 1 MAVSAKPEKAAVVKRPFRTSFTMKRTNDWVLATDVPSDVVVIVGGVSFALHKFPLVSRCGRIRMLVVEAG-DSDVSQMELPDVPGGAEGFDMAARFCYGI VPGGAEGFDMAARFCYGI MSAKKKDLLSSAMKRTSEWISSQEVSSDVTVHVGEASFSLHKFPLMSKCGFIKKLVSESSKDSDSTVIKIPDIPGGSEAFELAAKFCYGI PGGSEAFELAAKFCYGI MLEKLSFLLHKFPLVSKCGFIKKLASESS--NDSNIIRIPDFPGGAEGFELVIKFCYDI PGGAEGFELVIKFCYDI RPT2 95 NFEITVQNVAALHCAAEFLQMTDKYCDNNLAGRTQDFLSQVALSSLSGAIVVLKSCEILLPISRDLGIVRRCVDVVGAKACN EAMF MpNCH1 100 NFEITTANVAVLRCAAEYLEMTEEYGEGNLVARTEAFLSEVVCQNLADSITVLHNCENLLPYAEELKIVSRTIDAIASKACREQIAQGMGESDYDISGRN NCH1 91 NFDMSTENIAMLRCAAEYLEMTEEHSVENLVVRAEAYLNEVALKSLSSSITVLHKSEKLLPIAERVKLVSRCIDAIAYMTCQ ESHFCSPSSS NCL1 58 SFEINTENIAMLLCAAEYLEMTEEHSVENLVETIEVYLNEVILKSLSKSVKVLQKSQDLLPIAERVRLVDRCIDSIAYAICQ ES RPT PCRTPP-NWWTEELCILDVDFFSDVVSSMKQRGVKPSSLASAIITYTEKSLRDLVRDHSGR----GVKYSDPGDNESDERSQQRDLVQS PP-NWWTEELCILDVDFFSDVVSSMKQRGVKPSSLASAIITYTEKSL QRDLVQS MpNCH1 200 DSNVKLSFSKGPQKVTPSDWWAEDLAVLRIDFYQRVLAAMRSRGLRYESIGGALMHYAHRSLKGLHRKQTGRDMYKGGQQKKVHDSTSAIEHEQRILVET PSDWWAEDLAVLRIDFYQRVLAAMRSRGLRYESIGGALMHYAHRSL QRILVET NCH1 183 NSGNNEVVVQQQSKQPVVDWWAEDLTVLRIDSFQRVLIAMMARGFKQYGLGPVLMLYAQKSLRGL EIFGKGMKKIEPKQEHEKRVILET VVDWWAEDLTVLRIDSFQRVLIAMMARGFKQYGLGPVLMLYAQKSL KRVILET NCL QSNEDIVDWWADDLAVLKIDMFRRVLVAMIARGFKRYSLGPVLKLYAEKALRGLVRFLNFLTEQCDIFGKEAKKMEAEQEHEKRLILET KRLILET RPT2 265 IVSLLPSDKGLFPVNFLCSLLRCAVFLDTSLTCKNELEKRISVVLEHVSVDDLLIPSFTYDGE-RLLDLDSVRRIISAFVEKEKNVGV------F----- MpNCH1 300 IVSLLPPEKNTASCSFLFGLLRTAIILDTTIACRLDLERRIGQQLEQATLDDLLIPSFSYTGD-TLFDVDIVQRIIVCFLQQTDNEDVHDPHSMYESDG- NCH1 272 IVSLLPREKNAMSVSFLSMLLRAAIFLETTVACRLDLENRMGLQLGQAVLDDLLIPSYSFTGDHSMFDTDTVQRILMNYLE FEVEGV NCL1 231 IVSLLPRERNSVSVSFLSILLRAAIYLETTVACRLDLEKRMGLQLRQAVIDDLLIPYYSFNGDNTMLDVDTVQRILMNYLE FEVEG- RPT NGGDFNRGVCSVSLQRVAKTVDSYLAEIATYGDLTISKFNAIANLVPKSARKSDDDLYRAIDIFLKAHPNLDEIEREKVCSSMDPLKLSYDARLHA VAKTVDSYLAEIATYGDLTISKFNAIANLVPKSARKSDDDLYRAIDIFLKAHPNLDEIEREKVCSSMDPLKLSYDARLHA MpNCH MGSPTQS-----ALMKVAKLLDSYLAEIAPDANLKLAKFISLAELLPEYARVVDDGLYRAIDIYLKAHPSLTEIDRKKLCKLMDVQKLSQEACTHA NCH1 359 RLSNNGVDLAG-----DMERVGKLLENYMAEIASDRNVSLQKFIGLAELIPEQSRVTEDGMYRAVDIYLKAHPNMSDVERKKVCSLMDCQKLSREACAHA VGKLLENYMAEIASDRNVSLQKFIGLAELIPEQSRVTEDGMYRAVDIYLKAHPNMSDVERKKVCSLMDCQKLSREACAHA NCL NSADFAS-----D---IGELMETYLAEIASDRNINFAKFIGFAECIPKQSR-----MYRAIDIFLKTHPNISEVEKKKVCSLMDCKKLSRDVYAHA RPT2 449 SQNKRLPVNIVLHALYYDQLKLR SGVAEQEERAVVVLPEALKT-RSQLQADTTLAKENEALRSELMKMKMYVSDMQKN KNG MpNCH1 489 AQNERLPVQIMVQVLYFEQLRLRTAMTGSLADVDHVGHNSQRISGAFSSAVSP-RDNY---ASVRRENRELKLEVARMRMRLTDLEKDHVCMKQDLEKGG NCH1 454 AQNDRLPVQTIVQVLYYEQQRLRGEVT---NDSDSPAPPPPQPAAVLPPKLSSYTDEL---SKLKRENQDLKLELLKMKMKLKEFEKESEKKTSSSTIST NCL1 400 AQNDRFQENL SNSDSPAPATAEKTLS-PPELSSYKNEL---SKLNRENQYLKLELLKVKMKFKELEKE---KAFEVMSGS RPT2 529 AGASSSNSSSLVSSKKSKHTFFSSVSKKLGKL-NPF-KNGSKDTSHIDEDLGGVDITKP RRRRFSIS MpNCH1 585 G QKTFLQSVSKKLTRL-NPFSKSGIKG GALDSKAPNTPDSRQGGSGGRRRRHSIS NCH1 548 NPSSPISTASTGKPPLPRKSFINSVSKKLGKL-NPF-SITPYN GRGRTKPP KDRRHSIS NCL1 473 DCSSSVSTASVAKPRLPRKSFINSVSQKLGKLINPF---GLKQ G--QTKQP KSRRHSIS
55 A A. thaliana pavement cells 0 min 10 min ch B A. thaliana mesophyll cells 0 min 10 min ch C M. polymorpha thallus cells 0 min 10 min
Figure 18.1 Blue-light stimulated phototropism Blue light Inhibits seedling hypocotyl elongation
 Blue Light and Photomorphogenesis Q: Figure 18.3 Blue light responses - phototropsim of growing Corn Coleoptile 1. How do we know plants respond to blue light? 2. What are the functions of multiple BL
Blue Light and Photomorphogenesis Q: Figure 18.3 Blue light responses - phototropsim of growing Corn Coleoptile 1. How do we know plants respond to blue light? 2. What are the functions of multiple BL
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
LECTURE 4: PHOTOTROPISM
 http://smtom.lecture.ub.ac.id/ Password: https://syukur16tom.wordpress.com/ LECTURE 4: PHOTOTROPISM LECTURE FLOW 1. 2. 3. 4. 5. INTRODUCTION DEFINITION INITIAL STUDY PHOTROPISM MECHANISM PHOTORECEPTORS
http://smtom.lecture.ub.ac.id/ Password: https://syukur16tom.wordpress.com/ LECTURE 4: PHOTOTROPISM LECTURE FLOW 1. 2. 3. 4. 5. INTRODUCTION DEFINITION INITIAL STUDY PHOTROPISM MECHANISM PHOTORECEPTORS
The C-terminal kinase fragment of Arabidopsis phototropin 2 triggers constitutive phototropin responses
 The Plant Journal (2007) 51, 862 873 doi: 10.1111/j.1365-313X.2007.03187.x The C-terminal kinase fragment of Arabidopsis phototropin 2 triggers constitutive phototropin responses Sam-Geun Kong 1,, Toshinori
The Plant Journal (2007) 51, 862 873 doi: 10.1111/j.1365-313X.2007.03187.x The C-terminal kinase fragment of Arabidopsis phototropin 2 triggers constitutive phototropin responses Sam-Geun Kong 1,, Toshinori
Leaf Positioning of Arabidopsis in Response to Blue Light
 Molecular Plant Volume 1 Number 1 Pages 15 26 January 2008 Leaf Positioning of Arabidopsis in Response to Blue Light Shin-ichiro Inoue a, Toshinori Kinoshita a,2, Atsushi Takemiya a, Michio Doi b and Ken-ichiro
Molecular Plant Volume 1 Number 1 Pages 15 26 January 2008 Leaf Positioning of Arabidopsis in Response to Blue Light Shin-ichiro Inoue a, Toshinori Kinoshita a,2, Atsushi Takemiya a, Michio Doi b and Ken-ichiro
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Supplemental Data. Chen and Thelen (2010). Plant Cell /tpc
 Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family
 Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering
 Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
The Role of a Protein in Stomatal Opening Mediated by PHOT2 in Arabidopsis W OA
 The Plant Cell, Vol. 24: 1114 1126, March 2012, www.plantcell.org ã 2012 American Society of Plant Biologists. All rights reserved. The Role of a 14-3-3 Protein in Stomatal Opening Mediated by PHOT2 in
The Plant Cell, Vol. 24: 1114 1126, March 2012, www.plantcell.org ã 2012 American Society of Plant Biologists. All rights reserved. The Role of a 14-3-3 Protein in Stomatal Opening Mediated by PHOT2 in
Cytokinin. Fig Cytokinin needed for growth of shoot apical meristem. F Cytokinin stimulates chloroplast development in the dark
 Cytokinin Abundant in young, dividing cells Shoot apical meristem Root apical meristem Synthesized in root tip, developing embryos, young leaves, fruits Transported passively via xylem into shoots from
Cytokinin Abundant in young, dividing cells Shoot apical meristem Root apical meristem Synthesized in root tip, developing embryos, young leaves, fruits Transported passively via xylem into shoots from
Analysis of regulatory function of circadian clock. on photoreceptor gene expression
 Thesis of Ph.D. dissertation Analysis of regulatory function of circadian clock on photoreceptor gene expression Tóth Réka Supervisor: Dr. Ferenc Nagy Biological Research Center of the Hungarian Academy
Thesis of Ph.D. dissertation Analysis of regulatory function of circadian clock on photoreceptor gene expression Tóth Réka Supervisor: Dr. Ferenc Nagy Biological Research Center of the Hungarian Academy
Chloroplast Photorelocation Movement
 Chloroplast Photorelocation Movement N. Suetsugu and M. Wada( *ü ) Abstract Chloroplast photorelocation movement is one of the best-characterized plant organelle movements and is found in various plant
Chloroplast Photorelocation Movement N. Suetsugu and M. Wada( *ü ) Abstract Chloroplast photorelocation movement is one of the best-characterized plant organelle movements and is found in various plant
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated
 Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
Figure 1. Identification of UGT74E2 as an IBA glycosyltransferase. (A) Relative conversion rates of different plant hormones to their glucosylated form by recombinant UGT74E2. The naturally occurring auxin
THE ROLE OF THE PHYTOCHROME B PHOTORECEPTOR IN THE REGULATION OF PHOTOPERIODIC FLOWERING. AnitaHajdu. Thesis of the Ph.D.
 THE ROLE OF THE PHYTOCHROME B PHOTORECEPTOR IN THE REGULATION OF PHOTOPERIODIC FLOWERING AnitaHajdu Thesis of the Ph.D. dissertation Supervisor: Dr. LászlóKozma-Bognár - senior research associate Doctoral
THE ROLE OF THE PHYTOCHROME B PHOTORECEPTOR IN THE REGULATION OF PHOTOPERIODIC FLOWERING AnitaHajdu Thesis of the Ph.D. dissertation Supervisor: Dr. LászlóKozma-Bognár - senior research associate Doctoral
Supplemental Data. Wang et al. (2014). Plant Cell /tpc
 Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Supplemental Figure1: Mock and NPA-treated tomato plants. (A) NPA treated tomato (cv. Moneymaker) developed a pin-like inflorescence (arrowhead). (B) Comparison of first and second leaves from mock and
Plant Growth and Development
 Plant Growth and Development Concept 26.1 Plants Develop in Response to the Environment Factors involved in regulating plant growth and development: 1. Environmental cues (e.g., day length) 2. Receptors
Plant Growth and Development Concept 26.1 Plants Develop in Response to the Environment Factors involved in regulating plant growth and development: 1. Environmental cues (e.g., day length) 2. Receptors
Supplemental Data. Perrella et al. (2013). Plant Cell /tpc
 Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Functional characterization of Arabidopsis phototropin 1 in the hypocotyl apex
 The Plant Journal (2016) 88, 907 920 doi: 10.1111/tpj.13313 Functional characterization of Arabidopsis phototropin 1 in the hypocotyl apex Stuart Sullivan 1, Atsushi Takemiya 2,, Eros Kharshiing 1,3, Catherine
The Plant Journal (2016) 88, 907 920 doi: 10.1111/tpj.13313 Functional characterization of Arabidopsis phototropin 1 in the hypocotyl apex Stuart Sullivan 1, Atsushi Takemiya 2,, Eros Kharshiing 1,3, Catherine
Molecular Genetic Analysis of Phototropism in Arabidopsis
 Molecular Genetic Analysis of Phototropism in Arabidopsis Tatsuya Sakai* and Ken Haga Graduate School of Science and Technology, Niigata University, Nishi-ku, Niigata, 950-2181 Japan *Corresponding author:
Molecular Genetic Analysis of Phototropism in Arabidopsis Tatsuya Sakai* and Ken Haga Graduate School of Science and Technology, Niigata University, Nishi-ku, Niigata, 950-2181 Japan *Corresponding author:
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011) /tpc
 Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Supplemental Data. Fernández-Calvo et al. Plant Cell. (2011). 10.1105/tpc.110.080788 Supplemental Figure S1. Phylogenetic tree of MYC2-related proteins from Arabidopsis and other plants. Phenogram representation
Biological Roles of Cytokinins
 Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
Direct Control of Shoot Meristem Activity by a Cytokinin-Activating Enzyme By Kurakawa et. Al. Published in Nature Presented by Boyana Grigorova Biological Roles of Cytokinins Cytokinins are positive regulators
Electromagenetic spectrum
 Light Controls of Plant Development 1 Electromagenetic spectrum 2 Light It is vital for photosynthesis and is also necessary to direct plant growth and development. It acts as a signal to initiate and
Light Controls of Plant Development 1 Electromagenetic spectrum 2 Light It is vital for photosynthesis and is also necessary to direct plant growth and development. It acts as a signal to initiate and
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Supplemental Data. Yamamoto et al. (2016). Plant Cell /tpc WT + HCO 3. slac1-4/δnc #5 + HCO 3 WT + ABA. slac1-4/δnc #5 + ABA
 I (pa) I (pa) A WT + HCO 3 B 2 V (mv) 15 1 5 5 2 slac14/δnc #5 + HCO 3 4 6 WT (n = 6) 8 WT + HCO 3 (n = 7) slac14/δnc #5 (n = 6) slac14/δnc #5 + HCO 3 (n = 1) C WT + ABA D 2 V (mv) 15 1 5 5 2 slac14/δnc
I (pa) I (pa) A WT + HCO 3 B 2 V (mv) 15 1 5 5 2 slac14/δnc #5 + HCO 3 4 6 WT (n = 6) 8 WT + HCO 3 (n = 7) slac14/δnc #5 (n = 6) slac14/δnc #5 + HCO 3 (n = 1) C WT + ABA D 2 V (mv) 15 1 5 5 2 slac14/δnc
Antagonistic Regulation of Leaf Flattening by Phytochrome B and Phototropin in Arabidopsis thaliana
 Antagonistic Regulation of Leaf Flattening by Phytochrome B and Phototropin in Arabidopsis thaliana Toshiaki Kozuka 1, Noriyuki Suetsugu 2, Masamitsu Wada 2 and Akira Nagatani 1, * 1 Department of Botany,
Antagonistic Regulation of Leaf Flattening by Phytochrome B and Phototropin in Arabidopsis thaliana Toshiaki Kozuka 1, Noriyuki Suetsugu 2, Masamitsu Wada 2 and Akira Nagatani 1, * 1 Department of Botany,
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL
 GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
GENETIC ANALYSES OF ROOT SYSTEM DEVELOPMENT IN THE TOMATO CROP MODEL Kelsey Hoth 1 Dr. Maria Ivanchenko 2 Bioresourse Research 1, Department of Botany and Plant Physiology 2, Oregon State University, Corvallis,
** * * * Col-0 cau1 CAU1. Actin2 CAS. Actin2. Supplemental Figure 1. CAU1 affects calcium accumulation.
 Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Ca 2+ ug g -1 DW Supplemental Data. Fu et al. Plant Cell. (213). 1.115/tpc.113.113886 A 5 4 3 * Col- cau1 B 4 3 2 Col- cau1 ** * * ** C 2 1 25 2 15 1 5 Shoots Roots *
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
Blue light signalling in chloroplast movements
 Journal of Experimental Botany, Vol. 63, No. 4, pp. 1559 1574, 2012 doi:10.1093/jxb/err429 Advance Access publication 6 February, 2012 REVIEW PAPER Blue light signalling in chloroplast movements Agnieszka
Journal of Experimental Botany, Vol. 63, No. 4, pp. 1559 1574, 2012 doi:10.1093/jxb/err429 Advance Access publication 6 February, 2012 REVIEW PAPER Blue light signalling in chloroplast movements Agnieszka
Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport
 Ph.D. thesis Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport Zsigmond Laura Supervisor: Dr. Szabados László Arabidopsis Molecular Genetic Group Institute of Plant
Ph.D. thesis Arabidopsis PPR40 connects abiotic stress responses to mitochondrial electron transport Zsigmond Laura Supervisor: Dr. Szabados László Arabidopsis Molecular Genetic Group Institute of Plant
Actions of auxin. Hormones: communicating with chemicals History: Discovery of a growth substance (hormone- auxin)
 Hormones: communicating with chemicals History- discovery of plant hormone. Auxin Concepts of hormones Auxin levels are regulated by synthesis/degradation, transport, compartmentation, conjugation. Polar
Hormones: communicating with chemicals History- discovery of plant hormone. Auxin Concepts of hormones Auxin levels are regulated by synthesis/degradation, transport, compartmentation, conjugation. Polar
Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang Chen, Qiang Dong, Jiangqi Wen, Kirankumar S. Mysore and Jian Zhao
 New Phytologist Supporting Information Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bhlh transcription factor MtTT8 Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang
New Phytologist Supporting Information Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bhlh transcription factor MtTT8 Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Plant evolution: AGC kinases tell the auxin tale
 Opinion TRENDS in Plant Science Vol.12 No.12 Plant evolution: AGC kinases tell the auxin tale Carlos S. Galván-Ampudia and Remko Offringa Leiden University, Institute of Biology, Molecular and Developmental
Opinion TRENDS in Plant Science Vol.12 No.12 Plant evolution: AGC kinases tell the auxin tale Carlos S. Galván-Ampudia and Remko Offringa Leiden University, Institute of Biology, Molecular and Developmental
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Ron et al SUPPLEMENTAL DATA
 Ron et al SUPPLEMENTAL DATA Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model Mily Ron, Kaisa Kajala,
Ron et al SUPPLEMENTAL DATA Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model Mily Ron, Kaisa Kajala,
Plant Physiology Preview. Published on June 3, 2016, as DOI: /pp
 Plant Physiology Preview. Published on June 3, 2016, as DOI:10.1104/pp.16.01581 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 RUNNING HEAD Blue light response of stomata requires AHA1 CORRESPONDING
Plant Physiology Preview. Published on June 3, 2016, as DOI:10.1104/pp.16.01581 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 RUNNING HEAD Blue light response of stomata requires AHA1 CORRESPONDING
Major Plant Hormones 1.Auxins 2.Cytokinins 3.Gibberelins 4.Ethylene 5.Abscisic acid
 Plant Hormones Lecture 9: Control Systems in Plants What is a Plant Hormone? Compound produced by one part of an organism that is translocated to other parts where it triggers a response in target cells
Plant Hormones Lecture 9: Control Systems in Plants What is a Plant Hormone? Compound produced by one part of an organism that is translocated to other parts where it triggers a response in target cells
Phytochrome A Regulates the Intracellular Distribution of Phototropin 1 Green Fluorescent Protein in Arabidopsis thaliana W
 The Plant Cell, Vol. 20: 2835 2847, October 2008, www.plantcell.org ã 2008 American Society of Plant Biologists Phytochrome A Regulates the Intracellular Distribution of Phototropin 1 Green Fluorescent
The Plant Cell, Vol. 20: 2835 2847, October 2008, www.plantcell.org ã 2008 American Society of Plant Biologists Phytochrome A Regulates the Intracellular Distribution of Phototropin 1 Green Fluorescent
CONTROL OF PLANT GROWTH AND DEVELOPMENT BI-2232 RIZKITA R E
 CONTROL OF PLANT GROWTH AND DEVELOPMENT BI-2232 RIZKITA R E The development of a plant the series of progressive changes that take place throughout its life is regulated in complex ways. Factors take part
CONTROL OF PLANT GROWTH AND DEVELOPMENT BI-2232 RIZKITA R E The development of a plant the series of progressive changes that take place throughout its life is regulated in complex ways. Factors take part
Supplemental Data. Perea-Resa et al. Plant Cell. (2012) /tpc
 Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Supplemental Data. Perea-Resa et al. Plant Cell. (22)..5/tpc.2.3697 Sm Sm2 Supplemental Figure. Sequence alignment of Arabidopsis LSM proteins. Alignment of the eleven Arabidopsis LSM proteins. Sm and
Author(s) Fujiwara, Yoshikazu; Fukukawa, Kenj. Citation Few-Body Systems (2013), 54(1-4): 4.
 Title Nucleon Deuteron Breakup Differenti Derived from the Quark-Model NN Int Author(s) Fujiwara, Yoshikazu; Fukukawa, Kenj Citation Few-Body Systems (), (-): Issue Date - URL http://hdl.handle.net//699
Title Nucleon Deuteron Breakup Differenti Derived from the Quark-Model NN Int Author(s) Fujiwara, Yoshikazu; Fukukawa, Kenj Citation Few-Body Systems (), (-): Issue Date - URL http://hdl.handle.net//699
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/9/452/ra106/dc1 Supplementary Materials for Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots Hyo-Jun Lee, Jun-Ho
www.sciencesignaling.org/cgi/content/full/9/452/ra106/dc1 Supplementary Materials for Stem-piped light activates phytochrome B to trigger light responses in Arabidopsis thaliana roots Hyo-Jun Lee, Jun-Ho
EST1 Homology Domain. 100 aa. hest1a / SMG6 PIN TPR TPR. Est1-like DBD? hest1b / SMG5. TPR-like TPR. a helical. hest1c / SMG7.
 hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
Nature Genetics: doi: /ng Supplementary Figure 1. ssp mutant phenotypes in a functional SP background.
 Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
CBMG688R. ADVANCED PLANT DEVELOPMENT AND PHYSIOLOGY II G. Deitzer Spring 2006 LECTURE
 1 CBMG688R. ADVANCED PLANT DEVELOPMENT AND PHYSIOLOGY II G. Deitzer Spring 2006 LECTURE Photomorphogenesis and Light Signaling Photoregulation 1. Light Quantity 2. Light Quality 3. Light Duration 4. Light
1 CBMG688R. ADVANCED PLANT DEVELOPMENT AND PHYSIOLOGY II G. Deitzer Spring 2006 LECTURE Photomorphogenesis and Light Signaling Photoregulation 1. Light Quantity 2. Light Quality 3. Light Duration 4. Light
POTASSIUM IN PLANT GROWTH AND YIELD. by Ismail Cakmak Sabanci University Istanbul, Turkey
 POTASSIUM IN PLANT GROWTH AND YIELD by Ismail Cakmak Sabanci University Istanbul, Turkey Low K High K High K Low K Low K High K Low K High K Control K Deficiency Cakmak et al., 1994, J. Experimental Bot.
POTASSIUM IN PLANT GROWTH AND YIELD by Ismail Cakmak Sabanci University Istanbul, Turkey Low K High K High K Low K Low K High K Low K High K Control K Deficiency Cakmak et al., 1994, J. Experimental Bot.
A copy can be downloaded for personal non-commercial research or study, without prior permission or charge
 Kaiserli, Eirini, Sullivan, Stuart, Jones, Matthew A., Feeney, Kevin A., and Christie, John M. (2009) Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation
Kaiserli, Eirini, Sullivan, Stuart, Jones, Matthew A., Feeney, Kevin A., and Christie, John M. (2009) Domain swapping to assess the mechanistic basis of Arabidopsis phototropin 1 receptor kinase activation
expression of AUX1, LAX1, LAX2 and LAX3 in etiolated hypocotyls.
 Table of Contents Supplementary Material and Methods Supplementary Figure S1: Phototropic response of various aux1lax mutants and expression of AUX1, LAX1, LAX2 and LAX3 in etiolated hypocotyls. Supplementary
Table of Contents Supplementary Material and Methods Supplementary Figure S1: Phototropic response of various aux1lax mutants and expression of AUX1, LAX1, LAX2 and LAX3 in etiolated hypocotyls. Supplementary
GFP GAL bp 3964 bp
 Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
Supplemental Data. Møller et al. (2009) Shoot Na + exclusion and increased salinity tolerance engineered by cell type-specific alteration of Na + transport in Arabidopsis Supplemental Figure 1. Salt-sensitive
HRS1 Acts as a Negative Regulator of Abscisic Acid Signaling to Promote Timely Germination of Arabidopsis Seeds
 HRS1 Acts as a Negative Regulator of Abscisic Acid Signaling to Promote Timely Germination of Arabidopsis Seeds Chongming Wu 1,2., Juanjuan Feng 1,2., Ran Wang 1,2, Hong Liu 1,2, Huixia Yang 1,2, Pedro
HRS1 Acts as a Negative Regulator of Abscisic Acid Signaling to Promote Timely Germination of Arabidopsis Seeds Chongming Wu 1,2., Juanjuan Feng 1,2., Ran Wang 1,2, Hong Liu 1,2, Huixia Yang 1,2, Pedro
Pairs of guard cells forming stomatal pores regulate gas
 Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba Atsushi Takemiya*, Toshinori Kinoshita*, Miwako Asanuma*, and Ken-ichiro Shimazaki* *Department of Biology,
Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba Atsushi Takemiya*, Toshinori Kinoshita*, Miwako Asanuma*, and Ken-ichiro Shimazaki* *Department of Biology,
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Life Science Journal 2014;11(9) Cryptochrome 2 negatively regulates ABA-dependent seed germination in Arabidopsis
 Cryptochrome 2 negatively regulates ABA-dependent seed germination in Arabidopsis Sung-Il Kim 1, Sang Ik Song 3, Hak Soo Seo 1, 2, 4 * 1 Department of Plant Science and Research Institute of Agriculture
Cryptochrome 2 negatively regulates ABA-dependent seed germination in Arabidopsis Sung-Il Kim 1, Sang Ik Song 3, Hak Soo Seo 1, 2, 4 * 1 Department of Plant Science and Research Institute of Agriculture
SUPPLEMENTARY INFORMATION
 Supplemental Methods Isolation and mapping of SPCH An EMS-mutagenized population of tmm-1 (Col);E1728 (an enhancer trap GFP marking guard cells) was created. M2 seeds were collected from M1 s planted in
Supplemental Methods Isolation and mapping of SPCH An EMS-mutagenized population of tmm-1 (Col);E1728 (an enhancer trap GFP marking guard cells) was created. M2 seeds were collected from M1 s planted in
GFP WxTP-GFP WxTP-GFP. stromules. DsRed WxTP-DsRed WxTP-DsRed. stromules
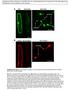 Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Lecture 13: PROTEIN SYNTHESIS II- TRANSLATION
 http://smtom.lecture.ub.ac.id/ Password: https://syukur16tom.wordpress.com/ Password: Lecture 13: PROTEIN SYNTHESIS II- TRANSLATION http://hyperphysics.phy-astr.gsu.edu/hbase/organic/imgorg/translation2.gif
http://smtom.lecture.ub.ac.id/ Password: https://syukur16tom.wordpress.com/ Password: Lecture 13: PROTEIN SYNTHESIS II- TRANSLATION http://hyperphysics.phy-astr.gsu.edu/hbase/organic/imgorg/translation2.gif
Last time: Obtaining information from a cloned gene
 Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Functional characterization of Ostreococcus tauri phototropin
 Research Functional characterization of Ostreococcus tauri phototropin Stuart Sullivan*, Jan Petersen*, Lisa Blackwood, Maria Papanatsiou and John M. Christie Institute of Molecular, Cell and Systems Biology,
Research Functional characterization of Ostreococcus tauri phototropin Stuart Sullivan*, Jan Petersen*, Lisa Blackwood, Maria Papanatsiou and John M. Christie Institute of Molecular, Cell and Systems Biology,
Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA
 Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Nature Genetics: doi: /ng Supplementary Figure 1. Icm/Dot secretion system region I in 41 Legionella species.
 Supplementary Figure 1 Icm/Dot secretion system region I in 41 Legionella species. Homologs of the effector-coding gene lega15 (orange) were found within Icm/Dot region I in 13 Legionella species. In four
Supplementary Figure 1 Icm/Dot secretion system region I in 41 Legionella species. Homologs of the effector-coding gene lega15 (orange) were found within Icm/Dot region I in 13 Legionella species. In four
Stress Effects on Myosin Mutant Root Length in Arabidopsis thaliana
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2011 Stress Effects on Myosin
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2011 Stress Effects on Myosin
Bio 1B Lecture Outline (please print and bring along) Fall, 2007
 Bio 1B Lecture Outline (please print and bring along) Fall, 2007 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #5 -- Molecular genetics and molecular evolution
Bio 1B Lecture Outline (please print and bring along) Fall, 2007 B.D. Mishler, Dept. of Integrative Biology 2-6810, bmishler@berkeley.edu Evolution lecture #5 -- Molecular genetics and molecular evolution
Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach
 Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach ABSTRACT SUBMITTED TO JAMIA MILLIA ISLAMIA NEW DELHI IN PARTIAL FULFILMENT OF
Characterisation of abiotic stress inducible plant promoters and bacterial genes for osmotolerance using transgenic approach ABSTRACT SUBMITTED TO JAMIA MILLIA ISLAMIA NEW DELHI IN PARTIAL FULFILMENT OF
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells
 Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Regulation of Phosphate Homeostasis by microrna in Plants
 Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Regulation of Phosphate Homeostasis by microrna in Plants Tzyy-Jen Chiou 1 *, Kyaw Aung 1,2, Shu-I Lin 1,3, Chia-Chune Wu 1, Su-Fen Chiang 1, and Chun-Lin Su 1 Abstract Upon phosphate (Pi) starvation,
Lecture 10: Cyclins, cyclin kinases and cell division
 Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA
 EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA Item Type text; Electronic Thesis Authors Bergstrand, Lauren Janel Publisher The University of Arizona. Rights Copyright is held by the author. Digital
EXPRESSION OF THE FIS2 PROMOTER IN ARABIDOPSIS THALIANA Item Type text; Electronic Thesis Authors Bergstrand, Lauren Janel Publisher The University of Arizona. Rights Copyright is held by the author. Digital
Ti plasmid derived plant vector systems: binary and co - integrative vectors transformation process; regeneration of the transformed lines
 Ti plasmid derived plant vector systems: binary and co - integrative vectors transformation process; regeneration of the transformed lines Mitesh Shrestha Constraints of Wild type Ti/Ri-plasmid Very large
Ti plasmid derived plant vector systems: binary and co - integrative vectors transformation process; regeneration of the transformed lines Mitesh Shrestha Constraints of Wild type Ti/Ri-plasmid Very large
Genetic interaction and phenotypic analysis of the Arabidopsis MAP kinase pathway mutations mekk1 and mpk4 suggests signaling pathway complexity
 Genetic interaction and phenotypic analysis of the Arabidopsis MAP kinase pathway mutations mekk1 and mpk4 suggests signaling pathway complexity Shih-Heng Su, Maria Cristina Suarez-Rodriguez, Patrick Krysan
Genetic interaction and phenotypic analysis of the Arabidopsis MAP kinase pathway mutations mekk1 and mpk4 suggests signaling pathway complexity Shih-Heng Su, Maria Cristina Suarez-Rodriguez, Patrick Krysan
SOMNUS, a CCCH-Type Zinc Finger Protein in Arabidopsis, Negatively Regulates Light-Dependent Seed Germination Downstream of PIL5 W
 The Plant Cell, Vol. 20: 1260 1277, May 2008, www.plantcell.org ª 2008 American Society of Plant Biologists SOMNUS, a CCCH-Type Zinc Finger Protein in Arabidopsis, Negatively Regulates Light-Dependent
The Plant Cell, Vol. 20: 1260 1277, May 2008, www.plantcell.org ª 2008 American Society of Plant Biologists SOMNUS, a CCCH-Type Zinc Finger Protein in Arabidopsis, Negatively Regulates Light-Dependent
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
SUPPLEMENTARY INFORMATION doi:10.1038/nature10534 Supplementary Fig. 1. Diagrammatic representation of the N-end rule pathway of targeted proteolysis (after Graciet and Wellmer 2010 9 ). Tertiary, secondary
Discovery of compounds that keep plants fresh ~ Controlling plant pore openings for drought tolerance and delay in leaf withering ~
 Discovery of compounds that keep plants fresh ~ Controlling plant pore openings for drought tolerance and delay in leaf withering ~ April 9, 2018 A team of scientists at Nagoya University has discovered
Discovery of compounds that keep plants fresh ~ Controlling plant pore openings for drought tolerance and delay in leaf withering ~ April 9, 2018 A team of scientists at Nagoya University has discovered
Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field.
 Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field. A, Phenotypes of 30-day old wild-type (WT) and abci8 mutant plants grown in a paddy field under normal sunny
Supplementary Figure 1 Characterization of wild type (WT) and abci8 mutant in the paddy field. A, Phenotypes of 30-day old wild-type (WT) and abci8 mutant plants grown in a paddy field under normal sunny
Chapter 18 Lecture. Concepts of Genetics. Tenth Edition. Developmental Genetics
 Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc
 OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner
 ABA represses the transcription of chloroplast genes Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner Supplementary data Supplementary tables Table
ABA represses the transcription of chloroplast genes Maria V. Yamburenko, Yan O. Zubo, Radomíra Vanková, Victor V. Kusnetsov, Olga N. Kulaeva, Thomas Börner Supplementary data Supplementary tables Table
Ph.D. thesis. Study of proline accumulation and transcriptional regulation of genes involved in this process in Arabidopsis thaliana
 Ph.D. thesis Study of proline accumulation and transcriptional regulation of genes involved in this process in Arabidopsis thaliana Written by: Edit Ábrahám Temesváriné Supervisors: Dr. László Szabados
Ph.D. thesis Study of proline accumulation and transcriptional regulation of genes involved in this process in Arabidopsis thaliana Written by: Edit Ábrahám Temesváriné Supervisors: Dr. László Szabados
Model plants and their Role in genetic manipulation. Mitesh Shrestha
 Model plants and their Role in genetic manipulation Mitesh Shrestha Definition of Model Organism Specific species or organism Extensively studied in research laboratories Advance our understanding of Cellular
Model plants and their Role in genetic manipulation Mitesh Shrestha Definition of Model Organism Specific species or organism Extensively studied in research laboratories Advance our understanding of Cellular
SMALL AUXIN UP RNA (SAUR) genes are the largest class of primary auxin (growth
 Abstract SMALL AUXIN UP RNA (SAUR) genes are the largest class of primary auxin (growth hormone) responsive genes in all land plants, many of which are expressed in actively growing plant tissues. This
Abstract SMALL AUXIN UP RNA (SAUR) genes are the largest class of primary auxin (growth hormone) responsive genes in all land plants, many of which are expressed in actively growing plant tissues. This
Supporting Online Material
 1 Stomatal Patterning and Differentiation by Synergistic Interactions of Receptor Kinases Elena D. Shpak, Jessica Messmer McAbee, Lynn Jo Pillitteri, and Keiko U. Torii Supporting Online Material Material
1 Stomatal Patterning and Differentiation by Synergistic Interactions of Receptor Kinases Elena D. Shpak, Jessica Messmer McAbee, Lynn Jo Pillitteri, and Keiko U. Torii Supporting Online Material Material
DOWNLOAD OR READ : CHLOROPLASTS PDF EBOOK EPUB MOBI
 DOWNLOAD OR READ : CHLOROPLASTS PDF EBOOK EPUB MOBI Page 1 Page 2 chloroplasts chloroplasts pdf chloroplasts Light has been shown to be a requirement for chloroplast division. Chloroplasts can grow and
DOWNLOAD OR READ : CHLOROPLASTS PDF EBOOK EPUB MOBI Page 1 Page 2 chloroplasts chloroplasts pdf chloroplasts Light has been shown to be a requirement for chloroplast division. Chloroplasts can grow and
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
downstream (0.8 kb) homologous sequences to the genomic locus of DIC. A DIC mutant strain (ro- 6
 A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
7.06 Problem Set
 7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
Reorganized actin filaments anchor chloroplasts along the anticlinal walls of Vallisneria epidermal cells under high-intensity blue light
 Planta (2005) 221: 823 830 DOI 10.1007/s00425-005-1493-9 ORIGINAL ARTICLE Yuuki Sakai Æ Shingo Takagi Reorganized actin filaments anchor chloroplasts along the anticlinal walls of Vallisneria epidermal
Planta (2005) 221: 823 830 DOI 10.1007/s00425-005-1493-9 ORIGINAL ARTICLE Yuuki Sakai Æ Shingo Takagi Reorganized actin filaments anchor chloroplasts along the anticlinal walls of Vallisneria epidermal
Arabidopsis thaliana. Lucia Strader. Assistant Professor, Biology
 Arabidopsis thaliana Lucia Strader Assistant Professor, Biology Arabidopsis as a genetic model Easy to grow Small genome Short life cycle Self fertile Produces many progeny Easily transformed HIV E. coli
Arabidopsis thaliana Lucia Strader Assistant Professor, Biology Arabidopsis as a genetic model Easy to grow Small genome Short life cycle Self fertile Produces many progeny Easily transformed HIV E. coli
Suetsugu, Noriyuki; Takami, Tsuneak. original author and source are cred
 Title Guard Cell Chloroplasts Are Dependent Stomatal Opening in Essenti Arabi Suetsugu, Noriyuki; Takami, Tsuneak Author(s) Watanabe, Harutaka; Iiboshi, Chihok Shimazaki, Ken-ichiro Citation PLoS ONE (2014),
Title Guard Cell Chloroplasts Are Dependent Stomatal Opening in Essenti Arabi Suetsugu, Noriyuki; Takami, Tsuneak Author(s) Watanabe, Harutaka; Iiboshi, Chihok Shimazaki, Ken-ichiro Citation PLoS ONE (2014),
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed
 Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
Supplementary Figure S1. Amino acid alignment of selected monocot FT-like and TFL-like sequences. Sequences were aligned using ClustalW and analyzed using the Geneious software. Accession numbers of the
(17) CYCLANILIDE: MECHANISM OF ACTION AND USES AS A PLANT GROWTH REGULATOR IN COTTON
 (17) CYCLANILIDE: MECHANISM OF ACTION AND USES AS A PLANT GROWTH REGULATOR IN COTTON Jim Burton 1 and Marianne Pedersen Abstract. Cyclanilide [1-(2,4-dichlorophenylaminocarbonyl)-cyclopropane carboxylic
(17) CYCLANILIDE: MECHANISM OF ACTION AND USES AS A PLANT GROWTH REGULATOR IN COTTON Jim Burton 1 and Marianne Pedersen Abstract. Cyclanilide [1-(2,4-dichlorophenylaminocarbonyl)-cyclopropane carboxylic
Genetic Characterization and Functional Analysis of the GID1 Gibberellin Receptors in Arabidopsis W
 The Plant Cell, Vol. 18, 3399 3414, December 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Genetic Characterization and Functional Analysis of the GID1 Gibberellin Receptors in Arabidopsis
The Plant Cell, Vol. 18, 3399 3414, December 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Genetic Characterization and Functional Analysis of the GID1 Gibberellin Receptors in Arabidopsis
Computational methods for predicting protein-protein interactions
 Computational methods for predicting protein-protein interactions Tomi Peltola T-61.6070 Special course in bioinformatics I 3.4.2008 Outline Biological background Protein-protein interactions Computational
Computational methods for predicting protein-protein interactions Tomi Peltola T-61.6070 Special course in bioinformatics I 3.4.2008 Outline Biological background Protein-protein interactions Computational
AND MOLECULAR CHARACT~RIZATION. By DEBORAH MARIE LONG,
 NITRATE REDUCTA3E IN MAIZE ROOTS: LOCALIZATION AND MOLECULAR CHARACT~RIZATION By DEBORAH MARIE LONG, B.Sc. A Thesis Submitted to the School of Graduate Studies in Partial Fulfilment of the Requirements
NITRATE REDUCTA3E IN MAIZE ROOTS: LOCALIZATION AND MOLECULAR CHARACT~RIZATION By DEBORAH MARIE LONG, B.Sc. A Thesis Submitted to the School of Graduate Studies in Partial Fulfilment of the Requirements
