Trafficking of Phosphatidylinositol 3-Phosphate from the trans-golgi Network to the Lumen of the Central Vacuole in Plant Cells
|
|
|
- Violet Richardson
- 6 years ago
- Views:
Transcription
1 The Plant Cell, Vol. 13, , February 2001, American Society of Plant Physiologists Trafficking of Phosphatidylinositol 3-Phosphate from the trans-golgi Network to the Lumen of the Central Vacuole in Plant Cells Dae Heon Kim, a Young-Jae Eu, b Cheol Min Yoo, b Yong-Woo Kim, b Kyeong Tae Pih, b Jing Bo Jin, a Soo Jin Kim, b Harald Stenmark, c and Inhwan Hwang b,1 a Department of Molecular Biology, Gyeongsang National University, Chinju, , Korea b Division of Molecular and Life Science and Center for Plant Intracellular Trafficking, Pohang University of Science and Technology, Pohang, , Korea c Department of Biochemistry, Norwegian Radium Hospital, Montebello, N-0310 Oslo, Norway Very limited information is available on the role of phosphatidylinositol 3-phosphate (PI[3]P) in vesicle trafficking in plant cells. To investigate the role of PI(3)P during the vesicle trafficking in plant cells, we exploited the PI(3)P-specific binding property of the endosome binding domain (EBD) (amino acids 1257 to 1411) of human early endosome antigen 1, which is involved in endosome fusion. When expressed transiently in Arabidopsis protoplasts, a green fluorescent protein (GFP):EBD fusion protein exhibited PI(3)P-dependent localization to various compartments such as the trans- Golgi network, the prevacuolar compartment, the tonoplasts, and the vesicles in the vacuolar lumen that varied with time. The internalized GFP:EBD eventually disappeared from the lumen. Deletion experiments revealed that the PI(3)Pdependent localization required the Rab5 binding motif in addition to the zinc finger motif. Overexpression of GFP:EBD inhibited vacuolar trafficking of sporamin but not trafficking of H -ATPase to the plasma membrane. On the basis of these results, we propose that the trafficking of GFP:EBD reflects that of PI(3)P and that PI(3)P synthesized at the trans-golgi network is transported to the vacuole through the prevacuolar compartment for degradation in plant cells. INTRODUCTION Evidence suggests that phosphoinositides play a regulatory role in vesicle trafficking (Camilli et al., 1996; Corvera and Czech, 1998; Gary et al., 1998; Cremona et al., 1999; Leevers et al., 1999; Roth, 1999). Phosphatidylinositol 3-phosphate (PI[3]P) has been implicated in this process (Whitman et al., 1998; Corvera et al., 1999). Direct evidence for the role of PI(3)P in vesicle trafficking was obtained when the yeast VPS34 gene, one of the genes involved in the vesicular protein sorting in yeast, was found to encode a PI3-kinase (Schu et al., 1993). Almost all PI3-kinase activity in yeast can be attributed to Vps34p (Stack et al., 1995; Wurmser and Emr, 1998). However, Vps34p requires a protein kinase, Vps15p, for its activation and membrane association (Stack et al., 1995). Also, a mammalian protein, p110, which is homologous to yeast Vps34p, has PI3-kinase activity and can transduce signals from tyrosine-phosphorylated receptors into a variety of intracellular responses (Volinia et al., 1995). However, this mammalian PI3-kinase, in association with an adapter protein p85, is able to phosphorylate other PI compounds such as PI(4)P and PI(4,5)P 2 at the D3 position of PI 1 To whom correspondence should be addressed. ihhwang@ postech.ac.kr; fax (Volinia et al., 1995; Panaretou et al., 1997). It is now clear that PI3-kinases play critical roles in various trafficking events, such as endocytosis of transferrin (Li et al., 1995), endosome fusion (Jones et al., 1998), vacuolar trafficking in yeast (Peterson et al., 1999), vesicle formation at the trans- Golgi network (TGN) (Hickinson et al., 1997; Jones and Howell, 1997; Jones et al., 1998), vacuole morphogenesis in Schizosaccharomyces pombe (Takegawa et al., 1995), and multivesicular body formation (Fernandez-Borja et al., 1999). PI(3)P is likely to act in the regulation of various steps of vesicle trafficking through its binding proteins. In mammalian cells, early endosome antigen1 (EEA1) has been well characterized as a PI(3)P binding protein (Stenmark et al., 1996; Simonsen et al., 1998; Christoforidis et al., 1999; McBride et al., 1999). EEA1 binds specifically to PI(3)P via a Zn 2 -coordinating finger at its C terminus (Patki et al., 1997; Burd and Emr, 1998; Gaullier et al., 1998). A similar motif has been found in other proteins that bind to PI(3)P. These include Vps27p (Misra and Hurley, 1999), Vac1p (Tall et al., 1999), and Fab1p (Gary et al., 1998), all of which are implicated in intracellular trafficking. In addition, Hrs, a tyrosine kinase substrate, has the motif and binds to PI(3)P with high affinity and has been implicated in vesicular trafficking via early endosomes (Komada and Soriano, 1999). This motif has
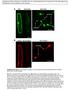
2 288 The Plant Cell Figure 1. PI(3)P-Dependent Targeting of GFP:EBD in Plant Cells.
3 Trafficking of PI(3)P in Plant Cells 289 been dubbed a FYVE finger (Stenmark and Aasland, 1999). In EEA1, the FYVE motif is sufficient for binding to PI(3)P; however, it appears that the cooperation of the neighboring Rab binding motif is required for PI(3)P-dependent targeting of the protein to the endosome in vivo (Li et al., 1995; Callaghan et al., 1999; Pfeffer, 1999; Lawe et al., 2000). In plant cells, Vps34p homologs have been isolated from Arabidopsis (Welters et al., 1994) and soybean (Hong and Verma, 1994). Transgenic Arabidopsis that expressed the AtVPS34 gene in the antisense orientation showed abnormal development, which suggested that PI(3)P plays an important role in plants. Evidence for the role of PI(3)P in vesicle trafficking in plant cells was obtained by inhibition studies using wortmannin in BY2 cells (Matsuoka et al., 1995). In this study, we addressed the question of whether PI(3)P plays a role in intracellular trafficking in plant cells. We used the endosome binding domain (amino acids 1257 to 1411) of EEA1 as an in vivo probe for PI(3)P in plant cells because the C-terminal endosome binding domain (EBD; amino acids 1257 to 1411) of human EEA1 was shown to be sufficient for binding of PI(3)P in vitro and could be directed to the endosome in a PI(3)P-dependent manner. Our evidence shows that PI(3)P is present in various compartments of plant cells and is transported to vacuoles for degradation and that PI(3)P plays a role in vacuolar trafficking of sporamin in Arabidopsis. RESULTS EBD of Human EEA1 Exhibits PI(3)P-Dependent Localization in Plant Cells To investigate whether PI(3)P plays a role in intracellular trafficking in plant cells, we used the PI(3)P-specific binding property of the FYVE domain as a molecular probe for PI(3)P. In this study, we used the C-terminal region of EEA1 (amino acids 1257 to 1411), that is, the EBD, consisting of the Rab5 binding and FYVE domains, because this region is sufficient for PI(3)P-dependent localization of the fused green fluorescent protein (GFP) to the endosome in vivo in animal cells (Stenmark et al., 1996). To visualize the EBD in plant cells, we constructed a fusion protein gene, GFP:EBD (Figure 1A). As a control, we constructed a similar GFP fusion protein gene that contained a mutant form of EBD (EBDC1358S) that does not bind to PI(3)P due to the mutation (C1358S) at the FYVE domain (Stenmark et al., 1996; Burd and Emr, 1998; Gaullier et al., 1998). These and soluble modified GFP (smgfp) (Davis and Vierstra, 1998) constructs were introduced into protoplasts prepared from 1- to 3-week-old whole Arabidopsis seedlings as described previously (Kang et al., 1998), and the fluorescence of transiently expressed GFP:EBD was examined 12 to 24 hr after transformation. As shown in Figure 1B, most of the green fluorescent signal from GFP:EBD was associated with the tonoplasts and within the central vacuoles (center), whereas in the control (GFP:EBDC1358S), only one or a few green fluorescent spots were observed in the cytoplasm (right). This is in sharp contrast to the pattern seen in animal cells, in which the mutant form was distributed uniformly in the cytosol. One possible explanation for this difference is that the mutation might have introduced a new localization signal in the molecule. The green fluorescent signal of smgfp was distributed uniformly in the cytosol as expected (left). To confirm that the fluorescent staining pattern of GFP:EBD was due to binding to PI(3)P at the FYVE domain of EBD in plant cells, we treated the protoplasts transformed with the GFP:EBD and GFP:EBDC1358S constructs with wortmannin (Ui et al., 1995) and LY (Vlahos et al., 1994), which have been shown to be inhibitors for PI3-kinases in animal cells. As shown in Figure 1C, in the presence of both inhibitors, the fluorescence signals from GFP:EBD were distributed uniformly in the cytosol, whereas the inhibitors did Figure 1. (continued). (A) Scheme of various EBD constructs. EBD is the C-terminal region of EEA1 from amino acid residues 1257 to 1411, which contains the FYVE and Rab5 binding domains. (a) GFP:EBD; (b) GFP:EBDC1358S; (c) GFP. The 35S cauliflower mosaic virus promoter was used for expression of these constructs. (B) Patterns of green fluorescent signals of proteins fused with GFP. Protoplasts prepared from whole Arabidopsis seedlings were transformed and examined 12 to 24 hr later. The red fluorescent signals of the chloroplasts are due to autofluorescence of chlorophyll. Yellow indicates overlap of red and green fluorescent signals. CH, chloroplasts; VM, vacuolar membrane. (C) The effect of PI3-kinase inhibitors on the pattern of GFP signals. Protoplasts were incubated in the presence of wortmannin (10 M) and LY (100 M) at room temperature, and the fluorescent pattern was examined 12 to 24 hr after transformation. The images are representative of at least three independent transformations for each construct and each condition. Note that some protoplasts do not contain chloroplasts because they originated from the root cells. CH, chloroplasts. Bars 20 m. (D) Formation of similar types of vesicles in BY2 cells and tobacco leaf cells. The BY2 cells were incubated with FM4-64 (40 M) for 5 min and then washed twice with Murashige and Skoog (1962) (MS) medium. The cells were examined under a fluorescent microscope at 20 to 30 hr after washing ([a] and [b]). Tobacco leaf tissues were bombarded using gold particles coated with GFP:EBD and incubated for 20 to 30 hr on MS plates after the bombardment. The cells were then examined under a fluorescent microscope ([c] and [d]). (a) and (c) show bright field images; (b) and (d) show fluorescent images. N, nucleus. Arrowheads indicate vesicles formed from the central vacuole. Bars 20 m.
4 290 The Plant Cell Figure 2. Temporal Expression Pattern of GFP:EBD. Transformed protoplasts were examined at various times, as indicated. The whole population contained a mixture of protoplasts with different GFP patterns, especially between 6 to 12 hr after transformation. Each image represents the fluorescent pattern of the majority ( 20 to 30%) of transformed protoplasts in the whole population at each time, from at least three independent transformation experiments. CH, chloroplasts; VM, vacuolar membrane. not affect the fluorescence pattern of GFP:EBDC1358S. The behavior of GFP:EBD was very similar to that observed in animal cells in the presence of wortmannin, implying that these inhibitors affected production of PI(3)P in Arabidopsis protoplasts either directly or indirectly. These results strongly suggest that the FYVE domain of EEA1 may bind specifically to PI(3)P in plant cells, as has been shown in animal cells (Patki et al., 1997). Next, we performed two experiments to exclude the possibility that the vacuolar targeting and vesicle formation at the central vacuole were caused artificially by expression of an animal protein (EBD). It has been shown that the lipophilic dye FM4-64 can be taken up efficiently by endocytosis and transported to the central vacuole in yeast cells (Vida and Emr, 1995). Therefore, we used FM4-64 to determine whether the dye can be used to visualize the tonoplast and the internalization process from the tonoplast into the lumen. When BY2 cells and Arabidopsis protoplasts were incubated with FM4-64, the dye was internalized into the lumen of the central vacuole by vesicles similar to those shown in Figure 1D (panels a and b) (data not shown for Arabidopsis protoplasts). In the second experiment, we introduced GFP:EBD into tobacco leaf cells by the particle bombardment method (Takeuchi et al., 1992) and examined the targeting of GFP:EBD in the leaf cells. As shown in Figure 1D (panels c and d), green fluorescent signals were accumulated at the central vacuole and vesicles similar to those seen in the protoplasts were formed from the central vacuole. Thus, these results strongly argue against the possibility of artifacts caused by experimental conditions. PI(3)P-Dependent Trafficking of GFP:EBD to the Central Vacuole in Plant Cells Protoplasts expressing GFP:EBD showed several different patterns of fluorescence that changed with time. Therefore, we examined in more detail the patterns of the fluorescent signals at various times after transformation. Green fluorescent signals were observed 4 to 6 hr after transformation. Initially, fluorescence was observed in the cytosol of protoplasts (Figure 2, top left). This pattern was replaced with numerous small punctate stains (Figure 2, top middle). Soon, the majority of protoplasts contained large fluorescent spots near the central vacuoles (Figure 2, top right). This was followed by localization of the GFP signal to the tonoplasts of the central vacuole, where the fusion protein was internalized into the vacuoles by numerous vesicles (Figure 2, bot-
5 Trafficking of PI(3)P in Plant Cells 291 tom). The green fluorescent vesicles present in the lumen disappeared with time. The internalization of the fusion protein by numerous vesicles is depicted more clearly in Figure 3. The z-sections of protoplasts show the numerous vesicles that were formed within the vacuole. Often, the vesicles contained few speckles. The disappearance of the fluorescent signals may be due to the degradation of the GFP:EBD protein by vacuolar proteases. The observed behavior of GFP:EBD in plant cells closely resembled the proposed turnover mechanism of PI(3)P in yeast, in which PI(3)P is transported to the vacuole by vesicle trafficking that requires Vam3p and Ypt7p and is then degraded in the vacuole lumen by lipases and phosphatases (Wurmser and Emr, 1998; Wurmser et al., 1999). Therefore, the results strongly suggest that PI(3)P is turned over in plant cells by transportation from the cytosol to the vacuoles. The GFP:EBD Fusion Protein Is Initially Targeted to the TGN The green fluorescent signal was present as numerous punctate stains in the cytosol 4 to 6 hr after transformation. One explanation for such localization is that GFP:EBD may be targeted to an organelle in which PI(3)P is being synthesized by a PI3-kinase. In yeast, PI(3)P has been shown to be synthesized in the Golgi and/or endosome (Stack et al., 1995). Also, at least one isoform of PI3-kinase is localized to the TGN in animal cells (Domin et al., 2000). To examine the localization of GFP:EBD, we attempted colocalization with the Golgi marker rat sialyltransferase (Munro, 1995; Wee et al., 1998) fused to red fluorescent protein (DsRed). First, we confirmed the localization of rat sialyltransferase:red fluorescent protein (ST:RFP) to the Golgi apparatus by cotransforming Figure 3. Internalization of GFP:EBD into the Lumen of Vacuoles by Numerous Vesicles. Serial sections were obtained from the middle to the bottom of a protoplast through the z axis at 2- m intervals. (1) and (9) show the middle and bottom views of the protoplast, respectively. PM, plasma membrane; VM, vacuolar membrane. Bar 20 m.
6 292 The Plant Cell Figure 4. The GFP:EBD Fusion Protein Is Localized to the TGN Early after Transformation. (A) Colocalization of GFP:EBD with ST. Protoplasts were cotransformed with ST:RFP and BiP:GFP, and the localization of ST and BiP was examined in the absence ([a] to [c]) or presence ([d] to [f]) of brefeldin A (30 g/ml). Note the relocation and complete overlap of red fluorescent signals with green fluorescent signals. Protoplasts were cotransformed with ST:RFP and GFP:EBD, and the colocalization of red and green fluorescent signals was examined ([g] to [i]). The red fluorescent signals from the chlorophyll and red fluorescent protein are quite different from
7 Trafficking of PI(3)P in Plant Cells 293 the protoplasts with a fusion (BiP:GFP) between GFP and portions of the chaperone binding protein (BiP). BiP is an endoplasmic reticulum resident, and it is widely used as a marker of this compartment. The BiP:GFP fusion gives the typical punctate pattern indicative of the endoplasmic reticulum (Figure 4A, first and second rows). ST:RFP gave the pattern of punctate staining that was expected if the marker protein was localized at the Golgi apparatus (panel b) (Boevink et al., 1998; Wee et al., 1998). In the presence of brefeldin A, an agent that disrupts the Golgi apparatus (Fujiwara et al., 1988), the punctate staining pattern disappeared (panel e), and red fluorescent signals overlapped with the green fluorescent signals of BiP:GFP, thus giving yellow fluorescent signals (panel f). These results suggest that ST:RFP was localized at the Golgi apparatus, as shown previously (Munro, 1995; Wee et al., 1998). Next, we examined the localization of GFP:EBD in the protoplasts cotransformed with ST:RFP (Figure 4A, bottom row). Most of the green fluorescent signal of GFP:EBD overlapped with the red fluorescent signal of ST:RFP (panel i). Also, some signals were juxtaposed to each other (panel i), thus raising the possibility that GFP:EBD may be localized to the Golgi apparatus or to an organelle that is closely associated with the Golgi apparatus, such as the TGN. To obtain additional evidence for the localization, we performed colocalization of RFP:EBD with AtVTI1a:GFP, a fusion protein between GFP and AtVTI1a, a t-snare that is localized primarily to the TGN with a minor portion localized to the prevacuolar compartment in plant cells (Zheng et al., 1999). RFP:EBD was cotransformed with AtVTI1a:GFP into protoplasts. Most of the red and green fluorescent signals clearly overlapped each other (Figure 4B), which further supported the notion that EBD is targeted to the TGN 4 to 6 hr after transformation in plant cells. At 1 to 2 hr later, when the small punctate stains were visible, we started to observe protoplasts with several large fluorescent spots or a group of small punctate stains connected to each other (Figure 5A). The protoplasts with these structures were 20 to 30% at 8 to 10 hr after transformation. At 9 hr after transformation, some of the protoplasts with this structure showed a weak fluorescent signal at the tonoplast, indicating that GFP:EBD started to accumulate at the tonoplast (Figure 2, top right). The latter structures (indicated by arrows in Figure 5A, panels 1 and 2) were clearly different from the small punctate stains seen at earlier times, indicating the possibility of another organelle. Therefore, we attempted colocalization of GFP:EBD with a marker protein for the prevacuolar compartment. We tagged a fusion protein, 491 (aleurain without the targeting signal motif, NPIR, fused to the C terminus of BP80), with DsRed (491:RFP) to use as a marker for the prevacuolar/multivesicular compartment (Jiang and Rogers, 1998). As shown in Figure 5B, the green fluorescent signals clearly overlapped with the red fluorescent signals of 491:RFP (arrowheads), indicating that the green fluorescent signals of the large spots are associated with the prevacuolar compartment. These results suggested that GFP:EBD was transported to the prevacuolar compartment from the TGN. PI4-Kinase Is Involved in the PI(3)P-Dependent Trafficking to the Central Vacuoles To enhance our understanding of the mechanism of the PI(3)P-dependent trafficking of GFP:EBD, we set out to determine whether PI4-kinase was involved in the trafficking in plant cells, because PI4-kinases have been proposed to play a role in intracellular trafficking in yeast and animal cells (Wiedemann et al., 1996; Hama et al., 1999). Also, Arabidopsis PI4-kinase has been proposed to play a role in intracellular trafficking (Xue et al., 1999). We constructed various deletion mutants of the PI4-kinase gene of Arabidopsis and cotransformed them with the GFP:EBD construct into protoplasts to determine whether any of these deletion mutants had a dominant negative effect on the transportation of GFP:EBD. As a control, a full-length construct was included in the experiment. After cotransformation, we compared the pattern of fluorescence among the population of transformed protoplasts at various times. Only the deletion mutant, LKU(PI4-K ), caused a significant retardation of the vacuolar trafficking of GFP:EBD (Figures 6A and 6B). The inhibition was pronounced early in the time course; however, the effect was lost eventually. These results suggest that PI4-kinase may be involved in the PI(3)P-dependent trafficking of GFP:EBD to the vacuole membranes. The GFP:EBD Fusion Protein Inhibits Trafficking of Sporamin to the Central Vacuoles Overexpression of the FYVE domain may deplete endogenous PI(3)P, or at least compete with an endogenous PI(3)P binding protein, which in turn may result in the inhibition of Figure 4. (continued). each other morphologically and can be distinguished easily. CH, chloroplasts; OV, fluorescent signals of chlorophyll that were detected by the GFP filter due to overexposure. (B) Protoplasts were cotransformed with AtVTI1a:GFP and RFP:EBD and examined at various times after transformation. The photograph is representative of protoplasts at 4 to 6 hr after transformation. At least three independent transformation experiments were performed for each construct and each condition.
8 294 The Plant Cell Figure 5. GFP:EBD Was Temporally Localized to the Prevacuolar Compartment. (A) Protoplasts transformed with GFP:EBD had structures that appeared to be formed by the fusion of multiple vesicles (left ). Enlarged images of boxes 1 and 2, respectively, are shown in the middle and at right. (B) Protoplasts were cotransformed with GFP:EBD and 491:RFP. Most of the green fluorescent signals clearly overlapped with the red fluorescent signals of 491 (arrowheads). CH, chloroplasts. PI(3)P-dependent trafficking in plant cells. To examine this possibility, an EBD construct was cotransformed with a reporter construct, sporamin:gfp, that is normally targeted to the large central vacuole (Figure 7A, panel d) (Matsuoka et al., 1995). Vacuolar localization of sporamin:gfp was monitored at various times, and the efficiency of targeting was calculated as the number of protoplasts containing a central vacuole with green fluorescent signals among total protoplasts expressing sporamin:gfp (Figure 7B). The targeting efficiency reached nearly 80% at 70 hr after transformation with sporamin:gfp alone or together with the EBDC1358S. In contrast to the control experiments, the efficiency of targeting was 38%, when an EBD construct was cotransformed into protoplasts. To determine whether this inhibition was specific to the vacuolar trafficking of sporamin, we examined the effect of overexpression of EBD on the trafficking of a plasma membrane type H -ATPase (DeWitt et al., 1996) as a GFP fusion protein. As shown in Figure 7A, in the presence of EBD there was no inhibition of trafficking of H -ATPase:GFP to the plasma membrane (Figure 7A, panel b), which indicates that the inhibition was specific to the trafficking of sporamin. This finding strongly suggests that PI(3)P is involved in the trafficking of sporamin to the central vacuole. However, it has been shown that sporamin is not missorted by wortmannin in BY2 cells (Matsuoka et al., 1995). Thus, we wanted to examine whether vacuolar trafficking of sporamin:gfp is inhibited by wortmannin in Arabidopsis protoplasts. In the presence of wortmannin, the green fluorescent signal of sporamin:gfp was accumulated in the cytosol as punctate stains instead of being localized at the central vacuole. Only less than 5% of transformed protoplasts showed targeting of sporamin:gfp to the central vacuole at 40 hr after transformation (Figure 7C). Another PI3-kinase inhibitor, LY294002, gave a similar result with wortmannin (data not shown). Furthermore, an AtVPS34 mutant that has a small deletion (30 amino acids) at the C-terminal kinase domain resulted in 30 to 40% inhibition of sporamin:gfp targeting when cotransformed into
9 Trafficking of PI(3)P in Plant Cells 295 protoplasts (D.H. Kim and I. Hwang, unpublished data). Thus, these results further support the notion that PI(3)P may be involved in the vacuolar targeting of sporamin in Arabidopsis cells. The Rab5 Binding Domain Is Required for the PI(3)P-Dependent Localization and Trafficking of GFP:EBD in Plant Cells The C-terminal domain (amino acids 1257 to 1411) of EEA1, which was used as a molecular probe for PI(3)P, also contained the IQ (calmodulin binding) and Rab5 binding domains in addition to the zinc finger domain. The Rab5 binding domain is necessary for PI(3)P-dependent localization of EEA1 to the early endosome in animal cells (Lawe et al., 2000). Therefore, we investigated whether these neighboring domains contributed to the PI(3)P-dependent localization and transportation of EBD in plant cells. We generated two deletion constructs, EBD1306 (amino acids 1306 to 1411) and EBD1336 (amino acids 1336 to 1411), and fused them with GFP (Figure 8A). The EBD1306 construct contained the zinc finger domain and the neighboring Rab5 binding domain, but not the IQ domain, whereas the EBD1336 construct contained only the zinc finger domain. As shown in Figure 8B, the green fluorescent signal was distributed uniformly in the protoplasts transformed with GFP:EBD1336, whereas the protoplasts transformed with GFP:EBD1306 showed a pattern of fluorescence identical to that of the original construct, GFP:EBD. These results indicate that in addition to Figure 6. Overexpression of the LKU Domain of PI4-Kinase Inhibits Transport of GFP:EBD to Vacuoles. Protoplasts were transformed with GFP:EBD either alone or together with Full(PI4-K ) or LKU(PI4-K ), and the targeting efficiency of GFP:EBD to the vacuole was examined at various times. (A) Typical GFP patterns of protoplasts with GFP:EBD targeted to vacuoles (a) and not targeted to vacuoles (b). CH, chloroplasts. Bars 20 m. (B) The number of protoplasts with the GFP pattern shown in (A) were counted from a whole population to give the efficiency of targeting. Also, protoplasts with punctate staining together with the vacuolar staining were counted as GFP:EBD targeted to the vacuoles. Three independent transformation experiments were performed, and each time 200 transformed protoplasts were counted. The first, second, and third columns at each time point indicate protoplasts transformed with GFP:EBD alone, GFP:EBD plus Full(PI4-K ), and GFP:EBD plus LKU(PI4-K ), respectively. The numbers indicate the percentage of protoplasts with GFP:EBD targeted to the vacuoles. Error bars indicate SD.
10 296 The Plant Cell Figure 7. Inhibition of Vacuolar Targeting of Sporamin. (A) Protoplasts were transformed with H -ATPase:GFP alone (a), with 35S-EBD (b), or with 35S-EBDC1358S (c), and examined 24 hr later. Also, protoplasts were transformed with sporamin:gfp (SPO:GFP) alone (d), with 35S-EBD (e), or with 35S-EBDC1358S (f), and examined 40 and 70 hr later. Protoplasts with the GFP pattern observed in (e) and (f) were counted as sporamin:gfp not targeted to vacuoles and targeted to vacuoles, respectively. CH, chloroplasts; PM, plasma membrane. Bars 20 m. (B) Efficiency of vacuolar targeting of sporamin:gfp. Protoplasts with vacuolar staining together with punctate staining were considered as sporamin:gfp targeted to the vacuoles. At least three independent transformation experiments were performed, and more than 200 transformed protoplasts were counted each time. The numbers indicate the percentage of protoplasts with sporamin:gfp targeted to the vacuoles. (C) Inhibition of vacuolar targeting of sporamin:gfp by wortmannin. Protoplasts transformed with sporamin:gfp were incubated with 5 M wortmannin (SPO:GFP WM) at room temperature, and targeting efficiency was measured at various times. At least three independent transformation experiments were performed, and more than 200 transformed protoplasts were counted each time. Criterion for the inhibition was the accumulation of fluorescent punctate stains in the cytosol versus the accumulation of the green fluorescent signals in the central vacuole. The numbers indicate the percentage of protoplasts with sporamin:gfp targeted to the vacuoles. Error bars indicate SD.
11 Trafficking of PI(3)P in Plant Cells 297 the zinc finger domain, the Rab5 binding domain is necessary for the PI(3)P-dependent localization and transportation of GFP:EBD in plant cells, as has been reported for animal cells (Lawe et al., 2000). DISCUSSION PI(3)P-Dependent Binding of GFP:EBD in Plant Cells The FYVE domain alone has been shown to be sufficient for PI(3)P-dependent binding in vitro and also can be targeted to the endosome in vivo in a PI(3)P-dependent manner when introduced into cells as a GFP fusion protein (Stenmark et al., 1996; Burd and Emr, 1998; Lawe et al., 2000). In this study, we constructed a similar GFP:EBD fusion protein using the endosome binding domain of human EEA1 and introduced the construct into protoplasts of Arabidopsis. We observed three different fluorescent patterns of the GFP:EBD fusion protein in protoplasts: green fluorescent signals were observed as numerous punctate stains in the cytosol, at the tonoplasts, and at vesicles within the central vacuoles. Inhibition studies using wortmannin and LY294002, which have been shown to be PI3-kinase inhibitors in animal cells, suggest that these staining patterns may be due to the binding of GFP:EBD to PI(3)P. In addition, the fact that GFP:EBDC1358S showed a completely different staining pattern supports the notion that the localization of GFP:EBD is dependent on PI(3)P as in animal cells. Interestingly, experiments with the deletion constructs EBD1306 and EBD1336 indicate that the neighboring Rab5 binding domain, but not the IQ domain, is necessary for PI(3)P-dependent localization of the fusion protein, as shown in animal cells (Lawe et al., 2000). Together, these results strongly suggest that PI(3)P is present in various organelles in plant cells and that the targeting mechanism of a protein with the EBD domain may be highly conserved in yeast, animal, and plant cells. In addition, these results imply that an endogenous Rab5 homolog of Arabidopsis may play a role in the localization of the fusion protein. Transport of PI(3)P to the Vacuole for Degradation The pattern of the green fluorescent signal of GFP:EBD that was expressed transiently in protoplasts changed with time. The signal was observed initially at the TGN and then at the prevacuolar compartment. The signal was then localized to the membrane of the central vacuole and finally in the lumen Figure 8. The Rab5 Binding Motif Is Necessary for PI(3)P-Dependent Targeting and Transport of GFP:EBD. (A) Scheme of deletion constructs of EBD. FYVE and Rab5 indicate the zinc finger and Rab5 binding domains, respectively. (B) Protoplasts were transformed with GFP:EBD (denoted as GFP:1257), GFP:EBD1306 (GFP:1306), or GFP:EBD1336 (GFP:1336). CH, chloroplasts. Bars 20 m.
12 298 The Plant Cell of the central vacuole. Because GFP:EBD localization is dependent on the presence of PI(3)P, the differential localization of GFP:EBD likely reflects the localization of PI(3)P in plant cells. In yeast, PI(3)P is synthesized at the Golgi/endosome by Vps34p (Schu et al., 1993; Wurmser and Emr, 1998), which is recruited to the Golgi/endosome and activated by Vps15p (Stack et al., 1995). In Arabidopsis, AtVPS34, a homolog of yeast Vps34p, has been isolated and shown to play a critical role in plant development (Welters et al., 1994). However, it is not known whether AtVPS34 is responsible for biosynthesis of PI(3)P, which was identified by GFP:EBD. In the protoplasts transformed with GFP:EBD, one of the earliest patterns of fluorescence was the punctate stains at the TGN. Initial localization of GFP:EBD at the TGN may indicate that PI(3)P is synthesized by a PI3-kinase at the TGN in plant cells, as occurs in animal cells (Domin et al., 2000). Of course, it is also possible that PI(3)P synthesized at other places was transported to the TGN. However, we believe the first interpretation to be correct because this is the mechanism reported for other cells (Wurmser and Emr, 1998). To prove unequivocally where PI(3)P is synthesized in plant cells, it is necessary to identify and localize the PI3- kinase involved. Once PI(3)P is synthesized at the Golgi/endosome, it is transported for turnover to vacuoles by a vesicletrafficking process that requires Vam3p and Ypt7p in yeast (Wurmser and Emr, 1998). Degradation of PI(3)P in the vacuole is the primary turnover mechanism of PI(3)P in yeast, because blocking of vacuolar transport causes a several-fold increase in the PI(3)P level. Also, Fab1p, a PI(3)P 5-kinase localized at the endosome and the vacuolar membrane, is involved in the turnover of PI(3)P by phosphorylating the D5 position of the inositol ring (Gary et al., 1998; Odorizzi et al., 1998). In plant cells, the fluorescent signal of GFP:EBD that was initially localized to the TGN was subsequently observed to be associated with the prevacuolar compartment and with the central vacuole. These results suggest that the complex of GFP:EBD and PI(3)P was transported from the TGN through the prevacuolar compartment to the central vacuole, and finally internalized into the central vacuole by vesicles, as depicted in our model (Figure 9). The behavior of GFP:EBD was very similar to the proposed turnover mechanism of PI(3)P in yeast (Wurmser and Emr, 1998; Wurmser et al., 1999), which suggests that the turnover mechanism may be highly conserved in yeast and plant cells. However, it is not known whether plant cells possess a homolog of yeast Fab1p (Wurmser et al., 1999) that downregulates the level of PI(3)P. Interestingly, the expression of LKU(PI4-K ), a deletion mutant of PI4-kinase, causes retardation of the PI(3)P-dependent trafficking of GFP:EBD to the tonoplasts of the central vacuole, suggesting that PI(4)P may play a role in the trafficking. PI(3)P-Dependent Vesicle Trafficking in Plant Cells It is highly likely that overexpressed GFP:EBD may bind to a significant amount of PI(3)P, which results in inhibition of a process that requires PI(3)P in plant cells. The vacuolar trafficking of sporamin:gfp was inhibited by 50% when EBD Figure 9. A Model for the PI(3)P-Dependent Trafficking of EBD of EEA1 in Plant Cells. The EBD binds to PI(3)P at the TGN, and the EBD:PI(3)P complex is transported to the tonoplasts through the prevacuolar compartment by a PI4-kinase-dependent mechanism. The complex is then internalized into the lumen of the central vacuole for degradation by vacuolar hydrolyases. PVC, prevacuolar compartment.
13 Trafficking of PI(3)P in Plant Cells 299 was overexpressed in the protoplasts. Also, this inhibition was specific to the trafficking of sporamin, because targeting of H -ATPase:GFP to the plasma membrane was not affected under the same conditions. Interestingly, targeting of sporamin:gfp in the Arabidopsis protoplasts was also inhibited by wortmannin, which is in stark contrast to the previous results seen in the BY2 cells (Matsuoka et al., 1995), implying that there may be a difference between cell types in the inhibition. One possible explanation for the inhibition is competition for PI(3)P between the FYVE domain and the endogenous PI(3)P binding protein. However, we cannot exclude the possibility that other mechanisms, such as depletion of a Rab5 homolog by EBD, may be responsible for the inhibition. In fact, these two possibilities are not mutually exclusive, because Rab5 and PI(3)P work together to form a complex involved in endosome fusion in animal cells. EBD inhibits early endosome fusion by interfering with normal EEA1/SNARE complex formation (McBride et al., 1999). Therefore, EBD may also interfere with the formation of a similar complex containing PI(3)P and a Rab5 homolog in plant cells. Interestingly, analysis of Arabidopsis genomic sequences revealed a gene that potentially codes for a protein with the FYVE domain. Also, there are several Rab5 homologs in the Arabidopsis genome. To generate a chimeric fusion construct of GFP:EBD, DNA that encoded the C-terminal fragment of human early endosome antigen 1 (EEA1; amino acid residues 1257 to 1411) was amplified by polymerase chain reaction (PCR) using primers 5 -GAATTCGTGGCA- ATCTAGTCAACGG-3 and 5 -CTAATGTTAGTGTAATATTAC-3, sequenced to confirm the nucleotide sequence, and ligated in frame to the C terminus of the green fluorescent protein (GFP) coding region without the terminator codon. The FYVEC1358S mutant (Stenmark et al., 1996) was also fused to soluble modified GFP (smgfp) to give GFP:EBDC1358S. To construct GFP:EBD1306 and GFP:EBD1336, EBD1306 and EBD1336 were amplified by PCR using primers (1306, 5 -ATAACAATGCTTCAAACCAAAGTATTAGAA-3 and 5 -ATAACA- ATGATCAAACATACACAAGCG-3 ; 1336, 5 -CTAATGTTAGTGTAATAT- TAC-3 and 5 -CTAATGTTAGTGTAATATTAC-3 ) and ligated to the 3 end of the gene encoding smgfp. To construct the reporter fusion genes, AtVTI1a:GFP, 491:RFP, H -ATPase:GFP, sporamin:gfp, and ST:RFP, DsRed1-1 or GFP were ligated in frame to DNA that encoded the C termini of AtVTI1a, 491, AHA2, sporamin B, and sialyltransferase, respectively. To construct BiP:GFP, the Arabidopsis chaperone binding protein (BiP) gene (GenBank accession number D82817) was amplified by PCR from total cdna using specific primers BiP5 (5 -TACGCAAAAGTTTCC-GAT-3 ) and BiP3 (5 -CTA- GAGCTCATCGTGAGA-3 ). The N-terminal leader sequence (44 amino acid residues) and the C-terminal region (79 amino acid residues with the retrieval signal HDEL) were fused in frame to the N- and C-termini, respectively, of smgfp without the termination codon. All the chimeric GFP fusion constructs were placed under the control of the 35S promoter in a puc vector for expression in protoplasts. Plasmids were purified using Qiagen (Valencia, CA) columns according to the manufacturer s protocol. The fusion constructs were introduced into Arabidopsis protoplasts prepared from whole seedlings by polyethylene glycol mediated transformation (Kang et al., 1998). Expression of the fusion constructs was monitored at various times after transformation, and images were captured with a cooled charge-coupled device camera using a Zeiss (Jena, Germany) Axioplan fluorescence microscope. The filter sets used were XF116 (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23) and XF137 (exciter, 540AF30; dichroic, 570DRLP; emitter, 585ALP) (Omega Optical, Brattleboro, VT) for green and red fluorescent proteins, respectively. Images were then processed using Adobe Photoshop (Mountain View, CA), and the images are in pseudocolor. ACKNOWLEDGMENTS METHODS Growth of Plants Arabidopsis thaliana (ecotype Columbia) was grown on Murashige and Skoog (1962) plates at 20 C in a culture room with a 16-hr-light/ 8-hr-dark cycle. Arabidopsis seedlings used for preparation of protoplasts were grown in MS liquid medium with constant shaking (160 rpm) at 20 C under a 16-hr-light/8-hr-dark cycle. We thank Dr. M.S. Yang (Chunbook National University, Chunjoo, Korea) for the sporamin plasmids. We also thank Dr. J.C. Rogers (Washington State University, Pullman) for the clone 491 (aleurain:bp80 construct), a prevacuolar marker. This work was supported by a grant from the National Creative Research Initiatives. Received September 5, 2000; accepted November 17, REFERENCES In Vivo Targeting of Fusion Proteins Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus trafficks on an actin/er network. Plant J. 15, Burd, C.G., and Emr, S.D. (1998). Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to Ring FYVE domains. Mol. Cell 2, Callaghan, J., Nixon, S., Bucci, C., Toh, B.H., and Stenmark, H. (1999). Direct interaction of EEA1 with Rab5b. Eur. J. Biochem. 265, Camilli, P.D., Emr, S.D., McPherson, P.S., and Novick, P. (1996). Phosphoinositides as regulators in membrane traffic. Science 271, Christoforidis, S., McBride, H.M., Burgoyne, R.D., and Zerial, M. (1999). The Rab5 effector EEA1 is a core component of endosome docking. Nature 397, Corvera, S., and Czech, M.P. (1998). Direct targets of phosphoinositide 3-kinase products in membrane traffic and signal transduction. Trends Cell Biol. 8,
14 300 The Plant Cell Corvera, S., D Arrigo, A., and Stenmark, H. (1999). Phosphoinositides in membrane traffic. Curr. Opin. Cell Biol. 11, Cremona, O., Paolo, G.D., Wenk, M.R., Luthi, A., Kim, W.T., Takei, K., Daniell, L., Nemoto, Y., Shears, S.B., Flavell, R.A., McCormick, D.A., and Camilli, P.D. (1999). Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 99, Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, DeWitt, N.D., Hong, B., Sussman, M.R., and Harper, J.F. (1996). Targeting of two Arabidopsis H ( ) -ATPase isoforms to the plasma membrane. Plant Physiol. 112, Domin, J., Gaidarov, I., Smith, M.E., Keen, J.H., and Waterfield, M.D. (2000). The class II phosphoinositide 3-kinase PI3K C2alpha is concentrated in the trans-golgi network and present in clathrincoated vesicles. J. Biol. Chem. 275, Fernandez-Borja, M., Wubbolts, R., Calafat, J., Janssen, H., Divecha, N., Dusseljee, S., and Neefjes, J. (1999). Multivesicular body morphogenesis requires phosphatidylinositol 3-kinase activity. Curr. Biol. 9, Fujiwara, T., Oda, K., Yokota, S., Takatsuki, A., and Ikehara, Y. (1988). Brefeldin A causes disassembly of the Golgi complex and accumulation of secretory proteins in the endoplasmic reticulum. J. Biol.Chem. 263, Gary, J.D., Wurmser, A.E., Bonangelino, C.J., Weisman, L.S., and Emr, S.D. (1998). Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143, Gaullier, J.-M., Simonsen, A., D Arrigo, A., Bremnes, B., Stenmark, H., and Aasland, R. (1998). FYVE fingers bind PtdIns(3)P. Nature 394, Hama, H., Schnieders, E.A., Thorner, J., Takemoto, J.Y., and DeWald, D.B. (1999). Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 274, Hickinson, D.M., Lucocq, J.M., Towler, M.C., Clough, S., James, J., James, S.R., Downes, C.P., and Ponnambalam, S. (1997). Association of a phosphatidylinositol-specific 3-kinase with a human trans-golgi network resident protein. Curr. Biol. 7, Hong, Z., and Verma, D.P.S. (1994). A phosphatidylinositol 3-kinase is induced during soybean nodule organogenesis and is associated with membrane proliferation. Proc. Natl. Acad. Sci. USA 91, Jiang, L., and Rogers, J.C. (1998). Integral membrane protein sorting to vacuoles in plant cells: Evidence for two pathways. J. Cell Biol. 143, Jones, A.T., Mills, I.G., Scheidig, A.J., Alexandrov, K., and Clague, M.J. (1998). Inhibition of endosome fusion by wortmannin persists in the presence of activated Rab5. Mol. Biol. Cell 9, Jones, S.M., and Howell, K.E. (1997). Phosphatidylinositol 3-kinase is required for the formation of constitutive transport vesicles from the TGN. J. Cell Biol. 139, Kang, S.G., Jin, J.B., Piao, H.L., Pih, K.T., Jang, H.J., Lim, J.H., and Hwang, I. (1998). Molecular cloning of an Arabidopsis cdna encoding a dynamin-like protein that is localized to plastids. Plant Mol. Biol. 38, Komada, M., and Soriano, P. (1999). Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 13, Lawe, D.C., Patki, V., Heller-Harrison, R., Lambright, D., and Corvera, S. (2000). The FYVE domain of early endosome antigen 1 is required for both phosphatidylinositol 3-phosphate and Rab5 binding. Critical role of this dual interaction for endosomal localization. J. Biol. Chem. 275, Leevers, S.J., Vanhaesebroeck, B., and Waterfield, M.D. (1999). Signaling through phosphoinositide 3-kinases: The lipids take centre stage. Curr. Opin. Cell Biol. 11, Li, G., D Souza-Schorey, C., Barbieri, M.A., Roberts, R., Klippel, A., Williams, L.T., and Stahl, P.D. (1995). Evidence for phosphatidylinositol 3-kinase as a regulator of endocytosis via activation of Rab5. Proc. Natl. Acad. Sci. USA 92, Matsuoka, K., Bassham, D.C., Raikhel, N.V., and Nakamura, K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130, McBride, H.M., Rybin, V., Murphy, C., Giner, A., Teasdale, R., and Zerial, M. (1999). Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell 98, Misra, S., and Hurley, J.H. (1999). Crystal structure of a phosphatidylinositol 3-phosphate-specific membrane-targeting motif, the FYVE domain of Vps27p. Cell 97, Munro, S. (1995). An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14, Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, Odorizzi, G., Babst, M., and Emr, S.D. (1998). Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell 95, Panaretou, C., Domin, J., Cockcroft, S., and Waterfield, M.D. (1997). Characterization of p150, an adaptor protein for the human phosphatidylinositol (PtdIns) 3-kinase: Substrate presentation by phosphatidylinositol transfer protein to the p150 Ptdins 3-kinase complex. J. Biol. Chem. 272, Patki, V., Virbasius, J., Lane, W.S., Toh, B.H., Shpetner, H.S., and Corvera, S. (1997). Identification of an early endosomal protein regulated by phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 94, Peterson, M.R., Burd, C.G., and Emr, S.D. (1999). Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45pdependent vesicle docking/fusion at the endosome. Curr. Biol. 9, Pfeffer, S.R. (1999). Motivating endosome motility. Nat. Cell Biol. 1, E145 E147. Roth, M.G. (1999). Lipid regulators of membrane traffic through the Golgi complex. Trends Cell Biol. 9, Schu, P.V., Takegawa, K., Fry, M.J., Stack, J.H., Waterfield, M.D., and Emr, S.D. (1993). Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260,
15 Trafficking of PI(3)P in Plant Cells 301 Simonsen, A., Lippe, R., Christoforidis, S., Gaullier, J.-M., Brech, A., Callaghan, J., Toh, B.-H., Murphy, C., Zerial, M., and Stenmark, H. (1998). EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394, Stack, J.H., DeWald, D.B., Takegawa, K., and Emr, S.D. (1995). Vesicle-mediated protein transport: Regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129, Stenmark, H., and Aasland, R. (1999). FYVE-finger proteins Effectors of an inositol lipid. J. Cell Science 112, Stenmark, H., Aasland, R., Toh, B.H., and D Arrigo, A. (1996). Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 271, Takegawa, K., DeWald, D.B., and Emr, S.D. (1995). Schizosaccharomyces pombe Vps34p, a phosphatidylinositol-specific PI 3-kinase essential for normal cell growth and vacuole morphology. J. Cell Sci. 108, Takeuchi, Y., Dotson, M., and Keen, N.T. (1992). Plant transformation: A simple particle bombardment device based on flowing helium. Plant Mol. Biol. 18, Tall, G.G., Hama, H., DeWald, D.B., and Horazdovsky, B.F. (1999). The phosphatidylinositol 3-phosphate binding protein Vac1p interacts with a Rab GTPase and a Sec1p homologue to facilitate vesicle-mediated vacuolar protein sorting. Mol. Biol. Cell 10, Ui, M., Okada, T., Hazeki, K., and Haxeki, O. (1995). Wortmannin as a unique probe for an intracellular signaling protein, phosphoinositide 3-kinase. TIBS 20, Vida, T.A., and Emr, S.D. (1995). A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, Vlahos, C., Matter, W.F., Hui, K., and Brown, R.F. (1994). A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)- 8-phenyl-4H 1-benzopyran-4-one (LY294002). J. Biol. Chem. 269, Volinia, S., Dhand, R., Vanhaesebroeck, B., MacDougall, L.K., Stein, R., Zvelebil, M.J., Domin, J., Panaretou, C., and Waterfield, M.D. (1995). A human phosphatidylinositol 3-kinase complex related to the yeast Vps34p-Vps15p protein sorting system. EMBO J. 14, Wee, E.G.-T., Sherrier, D.J., Prime, T.A., and Dupree, P. (1998). Targeting of active sialyltransferase to the plant Golgi apparatus. Plant Cell 10, Welters, P., Takegawa, K., Emr, S.D., and Chrispeels, M.J. (1994). AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calciumdependent lipid binding domain. Proc. Natl. Acad. Sci. USA 91, Whitman, M., Downes, C.P., Keeler, M., Keller, T., and Cantley, L. (1998). Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature 332, Wiedemann, C., Schafer, T., and Burger, M.M. (1996). Chromaffin granule associated phosphatidylinositol 4-kinase activity is required for stimulated secretion. EMBO J. 15, Wurmser, A.E., and Emr, S.D. (1998). Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 17, Wurmser, A.E., Gary, J.D., and Emr, S.D. (1999). Phosphoinositide 3-kinases and their FYVE domain-containing effectors as regulators of vacuolar/lysosomal membrane trafficking pathways. J. Biol. Chem. 274, Xue, H.W., Pical, C., Brearley, C., Elge, S., and Muller-Rober, B. (1999). A plant 126-kDa phosphatidylinositol 4-kinase with a novel repeat structure. Cloning and functional expression in baculovirus-infected insect cells. J. Biol. Chem. 274, Zheng, H., von Mollard, G.F., Kovaleva, V., Stevens, T.H., and Raikhel, N.V. (1999). The plant vesicle Associated SNARE AtVTI1a likely mediates vesicle transport from the trans-golgi network to the prevacuolar compartment. Mol. Biol. Cell 10,
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
Wave line information sheet
 Wave line information sheet Niko Geldner May 200 Wave constructs and lines, as well as pnigel vector series were published: Rapid, combinatorial analysis of membrane compartments in intact plants with
Wave line information sheet Niko Geldner May 200 Wave constructs and lines, as well as pnigel vector series were published: Rapid, combinatorial analysis of membrane compartments in intact plants with
Protein Sorting, Intracellular Trafficking, and Vesicular Transport
 Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS. Scientific Background on the Nobel Prize in Medicine 2013
 DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS Scientific Background on the Nobel Prize in Medicine 2013 Daniela Scalet 6/12/2013 The Nobel Prize in Medicine
DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS Scientific Background on the Nobel Prize in Medicine 2013 Daniela Scalet 6/12/2013 The Nobel Prize in Medicine
A New Dynamin-Like Protein, ADL6, Is Involved in Trafficking from the trans-golgi Network to the Central Vacuole in Arabidopsis
 The Plant Cell, Vol. 13, 1511 1525, July 2001, www.plantcell.org 2001 American Society of Plant Biologists RESEARCH ARTICLE A New Dynamin-Like Protein, ADL6, Is Involved in Trafficking from the trans-golgi
The Plant Cell, Vol. 13, 1511 1525, July 2001, www.plantcell.org 2001 American Society of Plant Biologists RESEARCH ARTICLE A New Dynamin-Like Protein, ADL6, Is Involved in Trafficking from the trans-golgi
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%%
 !"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar Compartment tothelyticvacuoleinvivo W
 The Plant Cell, Vol. 18, 2275 2293, September 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar
The Plant Cell, Vol. 18, 2275 2293, September 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar
Vacuole Biogenesis in Saccharomyces cerevisiae: Protein Transport Pathways to the Yeast Vacuole
 MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Mar. 1998, p. 230 247 Vol. 62, No. 1 1092-2172/98/$04.00 0 Copyright 1998, American Society for Microbiology Vacuole Biogenesis in Saccharomyces cerevisiae:
MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Mar. 1998, p. 230 247 Vol. 62, No. 1 1092-2172/98/$04.00 0 Copyright 1998, American Society for Microbiology Vacuole Biogenesis in Saccharomyces cerevisiae:
Signal Transduction. Dr. Chaidir, Apt
 Signal Transduction Dr. Chaidir, Apt Background Complex unicellular organisms existed on Earth for approximately 2.5 billion years before the first multicellular organisms appeared.this long period for
Signal Transduction Dr. Chaidir, Apt Background Complex unicellular organisms existed on Earth for approximately 2.5 billion years before the first multicellular organisms appeared.this long period for
CELB40060 Membrane Trafficking in Animal Cells. Prof. Jeremy C. Simpson. Lecture 2 COPII and export from the ER
 CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
GFP WxTP-GFP WxTP-GFP. stromules. DsRed WxTP-DsRed WxTP-DsRed. stromules
 Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Heather Currinn, Benjamin Guscott, Zita Balklava, Alice Rothnie and Thomas Wassmer*
 Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Supplemental Data. Gao et al. (2012). Plant Cell /tpc
 Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Identification of Multivesicular Bodies as Prevacuolar Compartments in Nicotiana tabacum BY-2 Cells W
 The Plant Cell, Vol. 16, 672 693, March 2004, www.plantcell.org ª 2004 American Society of Plant Biologists Identification of Multivesicular Bodies as Prevacuolar Compartments in Nicotiana tabacum BY-2
The Plant Cell, Vol. 16, 672 693, March 2004, www.plantcell.org ª 2004 American Society of Plant Biologists Identification of Multivesicular Bodies as Prevacuolar Compartments in Nicotiana tabacum BY-2
The Plant Cell, Vol. 15, , October 2003, American Society of Plant Biologists
 , Vol. 15, 2357 2369, October 2003, www.plantcell.org 2003 American Society of Plant Biologists The Arabidopsis Dynamin-Like Proteins ADL1C and ADL1E Play a Critical Role in Mitochondrial Morphogenesis
, Vol. 15, 2357 2369, October 2003, www.plantcell.org 2003 American Society of Plant Biologists The Arabidopsis Dynamin-Like Proteins ADL1C and ADL1E Play a Critical Role in Mitochondrial Morphogenesis
Cell Biology Review. The key components of cells that concern us are as follows: 1. Nucleus
 Cell Biology Review Development involves the collective behavior and activities of cells, working together in a coordinated manner to construct an organism. As such, the regulation of development is intimately
Cell Biology Review Development involves the collective behavior and activities of cells, working together in a coordinated manner to construct an organism. As such, the regulation of development is intimately
Transport between cytosol and nucleus
 of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
The Arabidopsis Dynamin-Like Proteins ADL1C and ADL1E Play a Critical Role in Mitochondrial Morphogenesis
 This article is published in Online, Preview Section, which publishes manuscripts accepted for publication after they have been edited and the authors have corrected proofs, but before the final, complete
This article is published in Online, Preview Section, which publishes manuscripts accepted for publication after they have been edited and the authors have corrected proofs, but before the final, complete
Lecture 6 - Intracellular compartments and transport I
 01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
The neuron as a secretory cell
 The neuron as a secretory cell EXOCYTOSIS ENDOCYTOSIS The secretory pathway. Transport and sorting of proteins in the secretory pathway occur as they pass through the Golgi complex before reaching the
The neuron as a secretory cell EXOCYTOSIS ENDOCYTOSIS The secretory pathway. Transport and sorting of proteins in the secretory pathway occur as they pass through the Golgi complex before reaching the
Membrane tethering in intracellular transport M Gerard Waters* and Suzanne R Pfeffer
 453 Membrane tethering in intracellular transport M Gerard Waters* and Suzanne R Pfeffer Studies of various membrane trafficking steps over the past year indicate that membranes are tethered together prior
453 Membrane tethering in intracellular transport M Gerard Waters* and Suzanne R Pfeffer Studies of various membrane trafficking steps over the past year indicate that membranes are tethered together prior
OsGAP1 Functions as a Positive Regulator of OsRab11-mediated TGN to PM or Vacuole Trafficking
 Plant Cell Physiol. 46(12): 2005 2018 (2005) doi:10.1093/pcp/pci215, available online at www.pcp.oupjournals.org JSPP 2005 OsGAP1 Functions as a Positive Regulator of OsRab11-mediated TGN to PM or Vacuole
Plant Cell Physiol. 46(12): 2005 2018 (2005) doi:10.1093/pcp/pci215, available online at www.pcp.oupjournals.org JSPP 2005 OsGAP1 Functions as a Positive Regulator of OsRab11-mediated TGN to PM or Vacuole
Supplementary information
 Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
Lecture 4. Protein Translocation & Nucleocytoplasmic Transport
 Lecture 4 Protein Translocation & Nucleocytoplasmic Transport Chapter 12 MBoC (5th Edition) Alberts et al. Reference paper: Tran and Wente, Cell 125, 1041-1053, 2006 2/8/2012 1 Page 713 Molecular Biology
Lecture 4 Protein Translocation & Nucleocytoplasmic Transport Chapter 12 MBoC (5th Edition) Alberts et al. Reference paper: Tran and Wente, Cell 125, 1041-1053, 2006 2/8/2012 1 Page 713 Molecular Biology
GGA proteins: new players in the sorting game
 COMMENTARY 3413 GGA proteins: new players in the sorting game Annette L. Boman Department of Biochemistry and Molecular Biology, University of Minnesota, School of Medicine Duluth, 10 University Drive,
COMMENTARY 3413 GGA proteins: new players in the sorting game Annette L. Boman Department of Biochemistry and Molecular Biology, University of Minnesota, School of Medicine Duluth, 10 University Drive,
targets. clustering show that different complex pathway
 Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Correct Targeting of Plant ARF GTPases Relies on Distinct Protein Domains
 Traffic 2008; 9: 103 120 Blackwell Munksgaard # 2007 The Authors Journal compilation # 2007 Blackwell Publishing Ltd doi: 10.1111/j.1600-0854.2007.00671.x Correct Targeting of Plant ARF GTPases Relies
Traffic 2008; 9: 103 120 Blackwell Munksgaard # 2007 The Authors Journal compilation # 2007 Blackwell Publishing Ltd doi: 10.1111/j.1600-0854.2007.00671.x Correct Targeting of Plant ARF GTPases Relies
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Chapter 12: Intracellular sorting
 Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
CHAPTER 3. Cell Structure and Genetic Control. Chapter 3 Outline
 CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis.
 Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
The Where, When, and How of Organelle Acidification by the Yeast Vacuolar H -ATPase
 MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Mar. 2006, p. 177 191 Vol. 70, No. 1 1092-2172/06/$08.00 0 doi:10.1128/mmbr.70.1.177 191.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved.
MICROBIOLOGY AND MOLECULAR BIOLOGY REVIEWS, Mar. 2006, p. 177 191 Vol. 70, No. 1 1092-2172/06/$08.00 0 doi:10.1128/mmbr.70.1.177 191.2006 Copyright 2006, American Society for Microbiology. All Rights Reserved.
Richik N. Ghosh, Linnette Grove, and Oleg Lapets ASSAY and Drug Development Technologies 2004, 2:
 1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
Importance of Protein sorting. A clue from plastid development
 Importance of Protein sorting Cell organization depend on sorting proteins to their right destination. Cell functions depend on sorting proteins to their right destination. Examples: A. Energy production
Importance of Protein sorting Cell organization depend on sorting proteins to their right destination. Cell functions depend on sorting proteins to their right destination. Examples: A. Energy production
7.06 Spring 2004 PS 6 KEY 1 of 14
 7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
A Genome-Wide Analysis of Arabidopsis Rop-Interactive CRIB Motif Containing Proteins That Act as Rop GTPase Targets
 The Plant Cell, Vol. 13, 2841 2856, December 2001, www.plantcell.org 2001 American Society of Plant Biologists A Genome-Wide Analysis of Arabidopsis Rop-Interactive CRIB Motif Containing Proteins That
The Plant Cell, Vol. 13, 2841 2856, December 2001, www.plantcell.org 2001 American Society of Plant Biologists A Genome-Wide Analysis of Arabidopsis Rop-Interactive CRIB Motif Containing Proteins That
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
7.06 Problem Set
 7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
Last time: Obtaining information from a cloned gene
 Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Phosphatidylinositol 3- and 4-Phosphate Are Required for Normal Stomatal Movements
 The Plant Cell, Vol. 14, 2399 2412, October 2002, www.plantcell.org 2002 American Society of Plant Biologists Phosphatidylinositol 3- and 4-Phosphate Are Required for Normal Stomatal Movements Ji-Yul Jung,
The Plant Cell, Vol. 14, 2399 2412, October 2002, www.plantcell.org 2002 American Society of Plant Biologists Phosphatidylinositol 3- and 4-Phosphate Are Required for Normal Stomatal Movements Ji-Yul Jung,
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and
 Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Division Ave. High School AP Biology
 Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Cellular Neuroanatomy I The Prototypical Neuron: Soma. Reading: BCP Chapter 2
 Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Transmembrane Domains (TMDs) of ABC transporters
 Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Overexpression of Arabidopsis Sorting Nexin AtSNX2b Inhibits Endocytic Trafficking to the Vacuole
 Molecular Plant Volume 1 Number 6 Pages 961 976 November 2008 RESEARCH ARTICLE Overexpression of Arabidopsis Sorting Nexin AtSNX2b Inhibits Endocytic Trafficking to the Vacuole Nguyen Q. Phan a,b, Sang-Jin
Molecular Plant Volume 1 Number 6 Pages 961 976 November 2008 RESEARCH ARTICLE Overexpression of Arabidopsis Sorting Nexin AtSNX2b Inhibits Endocytic Trafficking to the Vacuole Nguyen Q. Phan a,b, Sang-Jin
Secretory Bulk Flow of Soluble Proteins Is Efficient and COPII Dependent
 The Plant Cell, Vol. 13, 2005 2020, September 2001, www.plantcell.org 2001 American Society of Plant Biologists Secretory Bulk Flow of Soluble Proteins Is Efficient and COPII Dependent Belinda A. Phillipson,
The Plant Cell, Vol. 13, 2005 2020, September 2001, www.plantcell.org 2001 American Society of Plant Biologists Secretory Bulk Flow of Soluble Proteins Is Efficient and COPII Dependent Belinda A. Phillipson,
Endocytosis and Endocytic Network in Plants. Communication in Plants: Neuronal Aspects of Plant Life
 Endocytosis and Endocytic Network in Plants Communication in Plants: Neuronal Aspects of Plant Life Bonn, 29 November 2006 WHAT IS ENDOCYTOSIS? Endocytosis Endo (within) and Cytosis (cell) A process in
Endocytosis and Endocytic Network in Plants Communication in Plants: Neuronal Aspects of Plant Life Bonn, 29 November 2006 WHAT IS ENDOCYTOSIS? Endocytosis Endo (within) and Cytosis (cell) A process in
Biology Teach Yourself Series Topic 2: Cells
 Biology Teach Yourself Series Topic 2: Cells A: Level 14, 474 Flinders Street Melbourne VIC 3000 T: 1300 134 518 W: tssm.com.au E: info@tssm.com.au TSSM 2013 Page 1 of 14 Contents Cells... 3 Prokaryotic
Biology Teach Yourself Series Topic 2: Cells A: Level 14, 474 Flinders Street Melbourne VIC 3000 T: 1300 134 518 W: tssm.com.au E: info@tssm.com.au TSSM 2013 Page 1 of 14 Contents Cells... 3 Prokaryotic
Multivesicular Bodies Mature from the Trans-Golgi Network/Early Endosome in Arabidopsis W
 This article is a Plant Cell Advance Online Publication. The date of its first appearance online is the official date of publication. The article has been edited and the authors have corrected proofs,
This article is a Plant Cell Advance Online Publication. The date of its first appearance online is the official date of publication. The article has been edited and the authors have corrected proofs,
Proportion of Arabidopsis genes in different functions. 5% of genome consists of transporters (>1000 genes)
 Membranes are crucial to the life of the cell 1. PM define boundary of cell, and maintains difference between cytosol and external environment. Inside, organelles are distinct from one another. 2. Membrane
Membranes are crucial to the life of the cell 1. PM define boundary of cell, and maintains difference between cytosol and external environment. Inside, organelles are distinct from one another. 2. Membrane
Chapter 1. DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d
 Chapter 1 1. Matching Questions DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d 2. Matching Questions : Unbranched polymer that, when folded into its three-dimensional shape,
Chapter 1 1. Matching Questions DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d 2. Matching Questions : Unbranched polymer that, when folded into its three-dimensional shape,
Cytokinin. Fig Cytokinin needed for growth of shoot apical meristem. F Cytokinin stimulates chloroplast development in the dark
 Cytokinin Abundant in young, dividing cells Shoot apical meristem Root apical meristem Synthesized in root tip, developing embryos, young leaves, fruits Transported passively via xylem into shoots from
Cytokinin Abundant in young, dividing cells Shoot apical meristem Root apical meristem Synthesized in root tip, developing embryos, young leaves, fruits Transported passively via xylem into shoots from
CELL : THE UNIT OF LIFE
 38 BIOLOGY, EXEMPLAR PROBLEMS CHAPTER 8 CELL : THE UNIT OF LIFE MULTIPLE CHOICE QUESTIONS 1. A common characteristic feature of plant sieve tube cells and most of mammalian erythrocytes is a. Absence of
38 BIOLOGY, EXEMPLAR PROBLEMS CHAPTER 8 CELL : THE UNIT OF LIFE MULTIPLE CHOICE QUESTIONS 1. A common characteristic feature of plant sieve tube cells and most of mammalian erythrocytes is a. Absence of
Class IX: Biology Chapter 5: The fundamental unit of life. Chapter Notes. 1) In 1665, Robert Hooke first discovered and named the cells.
 Class IX: Biology Chapter 5: The fundamental unit of life. Key learnings: Chapter Notes 1) In 1665, Robert Hooke first discovered and named the cells. 2) Cell is the structural and functional unit of all
Class IX: Biology Chapter 5: The fundamental unit of life. Key learnings: Chapter Notes 1) In 1665, Robert Hooke first discovered and named the cells. 2) Cell is the structural and functional unit of all
CELL PART Expanded Definition Cell Structure Illustration Function Summary Location ALL CELLS DNA Common in Animals Uncommon in Plants Lysosome
 CELL PART Expanded Definition Cell Structure Illustration Function Summary Location is the material that contains the Carry genetic ALL CELLS information that determines material inherited characteristics.
CELL PART Expanded Definition Cell Structure Illustration Function Summary Location is the material that contains the Carry genetic ALL CELLS information that determines material inherited characteristics.
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
Molecular Characterization of Plant Vacuolar Sorting Receptor (VSR) Proteins
 Functional Plant Science and Biotechnology 2007 Global Science Books Molecular Characterization of Plant Vacuolar Sorting Receptor (VSR) Proteins Yansong Miao 1 Minghui Yang 1 Kwun Yee Li 1 Pui Kit Suen
Functional Plant Science and Biotechnology 2007 Global Science Books Molecular Characterization of Plant Vacuolar Sorting Receptor (VSR) Proteins Yansong Miao 1 Minghui Yang 1 Kwun Yee Li 1 Pui Kit Suen
Name: TF: Section Time: LS1a ICE 5. Practice ICE Version B
 Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Outline. Introduction to membrane fusion Experimental methods Results Discussion and conclusion
 Three-dimensional Structure of the rsly1 N-terminal Domain Reveals a Conformational Change Induced by Binding to Syntaxin 5 Demet Arac, Irina Dulubova, Jimin Pei, Iryna Huryeva, Nick V. Grishin and Josep
Three-dimensional Structure of the rsly1 N-terminal Domain Reveals a Conformational Change Induced by Binding to Syntaxin 5 Demet Arac, Irina Dulubova, Jimin Pei, Iryna Huryeva, Nick V. Grishin and Josep
Explain how cell size and shape affect the overall rate of nutrient intake and the rate of waste elimination. [LO 2.7, SP 6.2]
![Explain how cell size and shape affect the overall rate of nutrient intake and the rate of waste elimination. [LO 2.7, SP 6.2] Explain how cell size and shape affect the overall rate of nutrient intake and the rate of waste elimination. [LO 2.7, SP 6.2]](/thumbs/77/76271213.jpg) Cells Learning Objectives Use calculated surface area-to-volume ratios to predict which cell(s) might eliminate wastes or procure nutrients faster by diffusion. [LO 2.6, SP 2.2] Explain how cell size and
Cells Learning Objectives Use calculated surface area-to-volume ratios to predict which cell(s) might eliminate wastes or procure nutrients faster by diffusion. [LO 2.6, SP 2.2] Explain how cell size and
Lecture 10: Cyclins, cyclin kinases and cell division
 Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Biochemistry: A Review and Introduction
 Biochemistry: A Review and Introduction CHAPTER 1 Chem 40/ Chem 35/ Fundamentals of 1 Outline: I. Essence of Biochemistry II. Essential Elements for Living Systems III. Classes of Organic Compounds IV.
Biochemistry: A Review and Introduction CHAPTER 1 Chem 40/ Chem 35/ Fundamentals of 1 Outline: I. Essence of Biochemistry II. Essential Elements for Living Systems III. Classes of Organic Compounds IV.
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
CHAPTER 1 INTRODUCTION TO CELLS 2009 Garland Science Publishing 3 rd Edition
 Unity and Diversity of Cells 1-1 The smallest unit of life is a(n) (a) DNA molecule. (b) cell. (c) organelle. (d) virus. (e) protein. CHAPTER 1 INTRODUCTION TO CELLS 2009 Garland Science Publishing 3 rd
Unity and Diversity of Cells 1-1 The smallest unit of life is a(n) (a) DNA molecule. (b) cell. (c) organelle. (d) virus. (e) protein. CHAPTER 1 INTRODUCTION TO CELLS 2009 Garland Science Publishing 3 rd
Biology. 7-2 Eukaryotic Cell Structure 10/29/2013. Eukaryotic Cell Structures
 Biology Biology 1of 49 2of 49 Eukaryotic Cell Structures Eukaryotic Cell Structures Structures within a eukaryotic cell that perform important cellular functions are known as organelles. Cell biologists
Biology Biology 1of 49 2of 49 Eukaryotic Cell Structures Eukaryotic Cell Structures Structures within a eukaryotic cell that perform important cellular functions are known as organelles. Cell biologists
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Molecular Cell Biology 5068 In Class Exam 2 November 8, 2016
 Molecular Cell Biology 5068 In Class Exam 2 November 8, 2016 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Molecular Cell Biology 5068 In Class Exam 2 November 8, 2016 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Scale in the biological world
 Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
The Ultrastructure of Cells (1.2) IB Diploma Biology
 The Ultrastructure of Cells (1.2) IB Diploma Biology Explain why cells with different functions have different structures. Cells have different organelles depending on the primary function of the cell
The Ultrastructure of Cells (1.2) IB Diploma Biology Explain why cells with different functions have different structures. Cells have different organelles depending on the primary function of the cell
GO ID GO term Number of members GO: translation 225 GO: nucleosome 50 GO: calcium ion binding 76 GO: structural
 GO ID GO term Number of members GO:0006412 translation 225 GO:0000786 nucleosome 50 GO:0005509 calcium ion binding 76 GO:0003735 structural constituent of ribosome 170 GO:0019861 flagellum 23 GO:0005840
GO ID GO term Number of members GO:0006412 translation 225 GO:0000786 nucleosome 50 GO:0005509 calcium ion binding 76 GO:0003735 structural constituent of ribosome 170 GO:0019861 flagellum 23 GO:0005840
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Chapter 7.2. Cell Structure
 Chapter 7.2 Cell Structure Daily Objectives Describe the structure and function of the cell nucleus. Describe the function and structure of membrane bound organelles found within the cell. Describe the
Chapter 7.2 Cell Structure Daily Objectives Describe the structure and function of the cell nucleus. Describe the function and structure of membrane bound organelles found within the cell. Describe the
The Discovery of Cells
 The Discovery of Cells Microscope observations! General Cell & Organelle Discovery 1600s Observations made by scientists using more powerful microscopes in the 1800s led to the formation of the cell theory.
The Discovery of Cells Microscope observations! General Cell & Organelle Discovery 1600s Observations made by scientists using more powerful microscopes in the 1800s led to the formation of the cell theory.
Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition
 Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family
 Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Introduction to cells
 Almen Cellebiologi Introduction to cells 1. Unity and diversity of cells 2. Microscopes and visualization of cells 3. Prokaryotic cells, eubacteria and archaea 4. Eucaryotic cells, nucleus, mitochondria
Almen Cellebiologi Introduction to cells 1. Unity and diversity of cells 2. Microscopes and visualization of cells 3. Prokaryotic cells, eubacteria and archaea 4. Eucaryotic cells, nucleus, mitochondria
Cell (Learning Objectives)
 Cell (Learning Objectives) 1. Understand & describe the basic components necessary for a functional cell. 2. Review the order of appearance of cells on earth and explain the endosymbiotic theory. 3. Compare
Cell (Learning Objectives) 1. Understand & describe the basic components necessary for a functional cell. 2. Review the order of appearance of cells on earth and explain the endosymbiotic theory. 3. Compare
Cell Review. 1. The diagram below represents levels of organization in living things.
 Cell Review 1. The diagram below represents levels of organization in living things. Which term would best represent X? 1) human 2) tissue 3) stomach 4) chloroplast 2. Which statement is not a part of
Cell Review 1. The diagram below represents levels of organization in living things. Which term would best represent X? 1) human 2) tissue 3) stomach 4) chloroplast 2. Which statement is not a part of
Components of a functional cell. Boundary-membrane Cytoplasm: Cytosol (soluble components) & particulates DNA-information Ribosomes-protein synthesis
 Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
Cell (Outline) - Components of a functional cell - Major Events in the History of Earth: abiotic and biotic phases; anaerobic and aerobic atmosphere - Prokaryotic cells impact on the biosphere - Origin
Biological Process Term Enrichment
 Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
Biology: Life on Earth
 Teresa Audesirk Gerald Audesirk Bruce E. Byers Biology: Life on Earth Eighth Edition Lecture for Chapter 4 Cell Structure and Function Copyright 2008 Pearson Prentice Hall, Inc. Chapter 4 Outline 4.1 What
Teresa Audesirk Gerald Audesirk Bruce E. Byers Biology: Life on Earth Eighth Edition Lecture for Chapter 4 Cell Structure and Function Copyright 2008 Pearson Prentice Hall, Inc. Chapter 4 Outline 4.1 What
Host-Pathogen interaction-ii. Pl Path 604 PN Sharma Department of Plant Pathology CSK HPKV, Palampur
 Host-Pathogen interaction-ii Pl Path 604 PN Sharma Department of Plant Pathology CSK HPKV, Palampur-176062 It was originally believed that gene-for-gene resistance was conferred by a direct interaction
Host-Pathogen interaction-ii Pl Path 604 PN Sharma Department of Plant Pathology CSK HPKV, Palampur-176062 It was originally believed that gene-for-gene resistance was conferred by a direct interaction
16 The Cell Cycle. Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization
 The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
AS Biology Summer Work 2015
 AS Biology Summer Work 2015 You will be following the OCR Biology A course and in preparation for this you are required to do the following for September 2015: Activity to complete Date done Purchased
AS Biology Summer Work 2015 You will be following the OCR Biology A course and in preparation for this you are required to do the following for September 2015: Activity to complete Date done Purchased
7.06 Cell Biology EXAM #3 KEY
 7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
Chapter 4 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays.
 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Molecular Cell Biology 5068 In Class Exam 1 September 30, Please print your name:
 Molecular Cell Biology 5068 In Class Exam 1 September 30, 2014 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Molecular Cell Biology 5068 In Class Exam 1 September 30, 2014 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Demonstration in Yeast of the Function of BP-80, a Putative Plant Vacuolar Sorting Receptor
 The Plant Cell, Vol. 13, 781 792, April 2001, www.plantcell.org 2001 American Society of Plant Physiologists Demonstration in Yeast of the Function of BP-80, a Putative Plant Vacuolar Sorting Receptor
The Plant Cell, Vol. 13, 781 792, April 2001, www.plantcell.org 2001 American Society of Plant Physiologists Demonstration in Yeast of the Function of BP-80, a Putative Plant Vacuolar Sorting Receptor
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice Supplementary Figure 2.
 Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Biology Exam #1 Study Guide. True/False Indicate whether the statement is true or false. F 1. All living things are composed of many cells.
 Biology Exam #1 Study Guide True/False Indicate whether the statement is true or false. F 1. All living things are composed of many cells. T 2. Membranes are selectively permeable if they allow only certain
Biology Exam #1 Study Guide True/False Indicate whether the statement is true or false. F 1. All living things are composed of many cells. T 2. Membranes are selectively permeable if they allow only certain
Which row in the chart correctly identifies the functions of structures A, B, and C? A) 1 B) 2 C) 3 D) 4
 1. What is a similarity between all bacteria and plants? A) They both have a nucleus B) They are both composed of cells C) They both have chloroplasts D) They both lack a cell wall 2. Which statement is
1. What is a similarity between all bacteria and plants? A) They both have a nucleus B) They are both composed of cells C) They both have chloroplasts D) They both lack a cell wall 2. Which statement is
Parenchyma Cell. Magnification 2375X
 Parenchyma Cell The large size of parenchyma cells is due in part to their relatively large vacuole (V) and in part also to the large number of chloroplasts (Cp) they contain. From a crimson clover, Trifolium
Parenchyma Cell The large size of parenchyma cells is due in part to their relatively large vacuole (V) and in part also to the large number of chloroplasts (Cp) they contain. From a crimson clover, Trifolium
Predominant Golgi Residency of the Plant K/HDEL Receptor Is Essential for its Function in Mediating ER Retention
 Plant Cell Advance Publication. Published on August 2, 2018, doi:10.1105/tpc.18.00426 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41
Plant Cell Advance Publication. Published on August 2, 2018, doi:10.1105/tpc.18.00426 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41
SUPPLEMENTARY INFORMATION
 GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
Multiple Choice Review- Eukaryotic Gene Expression
 Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
Multiple Choice Review- Eukaryotic Gene Expression 1. Which of the following is the Central Dogma of cell biology? a. DNA Nucleic Acid Protein Amino Acid b. Prokaryote Bacteria - Eukaryote c. Atom Molecule
The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar. compartment in Arabidopsis
 MBC in Press, published on November 18, 2002 as 10.1091/mbc.E02-08-0509 The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis Running Title: Tonoplast
MBC in Press, published on November 18, 2002 as 10.1091/mbc.E02-08-0509 The AtC-VPS protein complex is localized to the tonoplast and the prevacuolar compartment in Arabidopsis Running Title: Tonoplast
Supplemental Data. Chen and Thelen (2010). Plant Cell /tpc
 Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Supplemental Data. Chen and Thelen (2010). Plant Cell 10.1105/tpc.109.071837 1 C Total 5 kg 20 kg 100 kg Transmission Image 100 kg soluble pdtpi-gfp Plastid (PDH-alpha) Mito (PDH-alpha) GFP Image vector
Introduction to Botany. Lecture 3
 Introduction to Botany. Lecture 3 Alexey Shipunov Minot State University August 30th, 2010 Taxonomic pyramid Monday test Name at least one function of proteins in plasma membrane Monday test What will
Introduction to Botany. Lecture 3 Alexey Shipunov Minot State University August 30th, 2010 Taxonomic pyramid Monday test Name at least one function of proteins in plasma membrane Monday test What will
Base your answers to questions 1 and 2 on the diagram below which represents a typical green plant cell and on your knowledge of biology.
 Base your answers to questions 1 and 2 on the diagram below which represents a typical green plant cell and on your knowledge of biology. 5. Which letter corresponds to that of the endoplasmic reticulum?
Base your answers to questions 1 and 2 on the diagram below which represents a typical green plant cell and on your knowledge of biology. 5. Which letter corresponds to that of the endoplasmic reticulum?
