Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion
|
|
|
- Justin Oliver
- 6 years ago
- Views:
Transcription
1 DOI /s Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion Sally L. Hanton Æ Laurent Chatre Æ Loren A. Matheson Æ Marika Rossi Æ Michael A. Held Æ Federica Brandizzi Received: 7 June 2007 / Accepted: 22 February 2008 Ó Springer Science+Business Media B.V Abstract In plants, differentiation of subdomains of the endoplasmic reticulum (ER) dedicated to protein export, the ER export sites (ERES), is influenced by the type of export-competent membrane cargo to be delivered to the Golgi. This raises a fundamental biological question: is the formation of transport intermediates at the ER for trafficking to the Golgi always regulated in the same manner? To test this, we followed the distribution and activity of two plant Sar1 isoforms. Sar1 is the small GTPase that regulates assembly of COPII (coat protein complex II) on carriers that transport secretory cargo from ER to Golgi. We show that, in contrast to a tobacco Sar1 isoform, the two Arabidopsis Sar1 GTPases were localised at ERES, independently of co-expression of Golgi-destined membrane cargo in tobacco cells. Although both isoforms labelled ERES, one was found to partition with the membrane fraction to a greater extent. The different distribution of fluorescent fusions of the two isoforms was influenced by the nature of an amino acid residue at the C-terminus of the protein, suggesting that the requirements for membrane Sally L. Hanton and Laurent Chatre have contributed equally to this work. Electronic supplementary material The online version of this article (doi: /s ) contains supplementary material, which is available to authorized users. S. L. Hanton L. Chatre L. A. Matheson M. Rossi F. Brandizzi Department of Biology, University of Saskatchewan, 112 Science Place, Saskatoon, SK, Canada S7N 5E2 M. Rossi M. A. Held F. Brandizzi (&) Department of Energy, Plant Research Laboratory, Michigan State University, East Lansing, MI 48824, USA brandizz@msu.edu association of the two GTPases are not equal. Furthermore, functional analyses based on the secretion of the bulk flow marker a-amylase indicated that over-expression of GTPrestricted mutants of the two isoforms caused different levels of ER export inhibition. These novel results indicate a functional heterogeneity among plant Sar1 isoforms. Keywords COPII Endoplasmic reticulum (ER) Golgi apparatus GTPase Sar1 Abbreviations ER Endoplasmic reticulum ERES ER export site COPII Coat protein complex II GEF Guanosine nucleotide exchange factor YFP Yellow fluorescent protein GFP Green fluorescent protein Introduction The export of proteins from the endoplasmic reticulum (ER) is mediated by the coat protein complex II (COPII) that assembles onto ER membranes (reviewed by Lee et al. 2004). The cytosolic COPII component proteins are the regulatory GTPase Sar1 and two structural heterodimeric complexes: Sec23/Sec24 and Sec13/31 (Barlowe et al. 1994; Matsuoka et al. 2001). Polymerisation of the COPII coat is initiated by activation of Sar1 by the guanosine nucleotide exchange factor (GEF) Sec12, an ER membrane protein. Activation of Sar1 is followed by the sequential recruitment of Sec23/24 and Sec13/31 to the ER membrane (Aridor et al. 2001;Matsuoka et al. 1998;Weissmanetal.2001). Recent evidence gathered in vitro and in mammalian cells indicates that Sar1 has additional functions beside the
2 recruitment of the COPII coat. These include cargo sorting, regulation of COPII coat dynamics, and membrane deformation during the formation of COPII carriers (Giraudo and Maccioni 2003; Lee et al. 2005; Sato and Nakano 2005). Despite the central role of Sar1 in protein export from the ER, our understanding of the functions of the isoforms of this GTPase in plants is limited. It is not yet known whether plant Sar1 isoforms have identical subcellular distributions, and whether they might perform different functions in the cell. Bioinformatic analyses have identified homologues of Sar1 in various plant species, including Arabidopsis thaliana (Bar-Peled and Raikhel 1997; d Enfert et al. 1992; Vernoud et al. 2003), Nicotiana tabacum (Andreeva et al. 1998; Takeuchi et al. 1998) and Brassica campestris (Kim et al. 1997). Dominant negative mutations that trap Sar1 in the GTP-restricted form have been instrumental in demonstrating that at least one of the tobacco and one of the Arabidopsis Sar1 isoforms are involved in ER export (Andreeva et al. 1998; dasilva et al. 2004; Phillipson et al. 2001; Takeuchi et al. 2000; Yang et al. 2005). Studies on the intracellular localisations of plant Sar1 isoforms suggest that Sar1 proteins may be functionally distinct in plant cells. For example, a tobacco Sar1 (termed NtSar1P) has been shown to localise predominantly in the cytosol of tobacco leaf epidermal cells, and to associate visibly with the ER membranes only under conditions of increased secretion of membrane cargo destined for the Golgi (dasilva et al. 2004). On the other hand, it has been demonstrated that half of the cellular pool of an Arabidopsis Sar1 (AtSARA1b) is associated with ER membranes in Arabidopsis (Bar-Peled and Raikhel 1997). The importance of the possibility that different plant Sar1 isoforms may have different cellular activities is mirrored by emerging findings on the functions of Sec24 in yeast and mammalian cells. It has been shown that different isoforms of the yeast and mammalian Sec24 are responsible for the selection of different cargo proteins for export via the recognition of different export motifs (Peng et al. 2000; Roberg et al. 1999; Shimoni et al. 2000; Wendeler et al. 2007). These findings suggest that the composition of the COPII coat may be heterogeneous, depending on the type of secretory cargo destined to leave the ER. It is not yet known whether Sar1 isoforms interact with different Sec24 proteins or with different cargo proteins, perhaps creating alternative COPII carriers. The different behaviour of plant Sar1 isoforms reported to date (Bar-Peled and Raikhel 1997; dasilva et al. 2004) may support this possibility. This aspect of ER export is particularly relevant in the study of formation of transport carriers at ERES in plants. We have shown recently that the plant ER can respond to a sudden need for secretion of export-competent membrane proteins by a signal-mediated recruitment of COPII coat proteins onto existing ERES, accompanied by the de novo generation of new ERES (Hanton et al. 2007). The extent of COPII coat recruitment to ERES and formation of ERES depended largely on the type of protein cargo to be exported (Hanton et al. 2007). These findings raise the important biological question as to whether all the regulatory mechanisms controlling the transport intermediates that carry proteins between the ER and Golgi act in an identical manner. In order to advance our knowledge of the regulation of COPII assembly in plant cells, we undertook a study to compare the subcellular distribution of different Sar1 isoforms, and their influence on export of proteins from the ER. To do so, we adopted two Arabidopsis Sar1 GTPases as model proteins. Our results show that despite the high similarity of their protein sequences, plant Sar1 proteins can behave differently in terms of subcellular distribution, and that maintenance of the different localisations of the two proteins is influenced by the nature of the amino acid residue at position 193 in the protein sequence. Finally, functional studies showed that Arabidopsis Sar1 mutants that are restricted to the GTP-bound form exhibited different levels of inhibition of ER export of the bulk flow marker a-amylase. Our results strongly suggest a functional heterogeneity of the plant Sar1 isoforms. Materials and methods Molecular cloning Standard molecular techniques were used as described by Sambrook et al. (1989). The fluorescent proteins used in this study were based on fusions with either mgfp5 (Haseloff et al. 1997) or EYFP (Clontech Inc., California, USA). The spectral properties of mgfp5 allow efficient spectral separation from YFP (Brandizzi et al. 2002a, b). AtSARA1a (At1g09180) and AtSARA1b (At1g56330) were amplified by PCR from ABRC cdna clones, and subcloned upstream of the EYFP sequence using the unique XbaI and SalI sites of the binary vector pvkh18en6, which contains the CaMV35S promoter sequence (Batoko et al. 2000; dasilva et al. 2004). Each of the mutants was subcloned in a similar manner. The primer sequences used for the cloning and mutagenesis are available upon request. Expression in leaves The confocal images produced for this work were obtained from transient transformation of tobacco leaves. Four week-old Nicotiana tabacum (cv Petit Havana) greenhouse
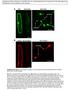
3 plants grown at 25 C were used for Agrobacterium tumefaciens (strain GV3101)-mediated DNA integration into the plant genome (Batoko et al. 2000). The bacterial optical density (OD 600 ) used for plant transformation was 0.05 for all Sar1 derivatives and 0.2 for ERD2-GFP. Transient expression in Arabidopsis leaves was performed using a biolistic method according to Seki et al. (1998). Sampling and imaging Transformed leaves were analysed 48 h after infection of the lower epidermis. Confocal imaging was performed using an inverted Zeiss LSM 510 META confocal microscope and a 639 water immersion objective. For imaging expression of GFP constructs, YFP constructs or both we used imaging settings as described in Brandizzi et al. (2002a, b) with a 3 lm optical slice, unless otherwise specified. Post-acquisition image processing was done with CorelDraw 12 software. Cell fractionation and western blot analysis Protoplasts were isolated from transiently transformed tobacco leaves as described by Hanton et al. (2005). Preparation of the soluble and membrane fractions was based on the method given in dasilva et al. (2004). About 200 ll of GFP extraction buffer (Hanton et al 2005) was added to the protoplast pellet and the soluble phase was extracted by sonication for 5 s, followed by centrifugation at 22,000 rcf for 15 min. The supernatant was carefully removed and a further 250 ll of extraction buffer added, followed by sonication for 5 s. 2% v/v Triton X-100 was added to each sample after extraction to ensure the release of proteins from the membrane. Extracts were diluted 50:50 with 2X SDS loading buffer (Crofts et al. 1999) and the samples analysed by western blot with anti-gfp serum (AbCam), as given in Hanton et al. (2005). Band intensities were quantified using ImageJ software. A rectangle of area 35 mm 2 was centred on each band and the intensity measured using the software tools. The total intensity for each sample was calculated by adding the intensities for the soluble and membrane fractions, and the intensity for each fraction was then calculated as a percentage of the total for the sample. This process was performed on each of three blots from three independent experiments. Electroporation and a-amylase assay For transient expression in protoplasts, Nicotiana tabacum (cv Petit Havana) were grown in MS medium (Murashige and Skoog 1962) supplemented with 2% sucrose, at 25 C with 16 h light/8 h dark regime at a light irradiance of 200 me m -2 s -1. Tobacco leaf protoplast preparation and subsequent DNA transfection via electroporation were performed as described by Phillipson et al. (2001). Plasmid concentrations used are given in Fig. 4. Extraction of protoplasts and a-amylase assays were performed as described by Crofts et al. (1999) and Phillipson et al. (2001). The secretion index was calculated as a ratio between the extracellular and intracellular enzyme activities. Results AtSARA1a and AtSARA1b have different intracellular localisations We have adopted two Arabidopsis Sar1 homologues termed AtSARA1a and AtSARA1b (Vernoud et al 2003), AGI numbers At1g09180 and At1g56330 respectively, as model proteins. It has been shown that about one-half of the cellular AtSARA1b pool is associated with the ER membranes (Bar-Peled and Raikhel 1997), which is markedly different from the reported predominantly cytosolic distribution of a tobacco Sar1 (NtSar1P) (dasilva et al. 2004). We wanted to investigate whether another Arabidopsis Sar1 isoform that is closely related to AtSARA1b would behave in the same manner as AtSARA1b, or whether its distribution might be more similar to that of NtSar1P. AtSARA1a and AtSARA1b share 93% sequence identity, and both have considerable sequence identity with NtSar1P (72% and 76% respectively, Fig. 1). To establish the intracellular localisations of AtSARA1a and AtSARA1b and to compare them with that reported for NtSar1P (dasilva et al. 2004), we subcloned AtSARA1a and AtSARA1b into the same binary vector with the C-terminal yellow fluorescent protein (YFP) cassette that was used for visualisation of NtSar1P in living cells (dasilva et al. 2004), by substituting NtSar1P with either AtSar1 isoform. This ensured that the expression system for the two proteins was identical to that of NtSar1P (dasilva et al. 2004). We then expressed each AtSar1 protein fusion in tobacco leaf epidermal cells using an Agrobacteriumbased protocol for DNA integration into the plant genome (Batoko et al. 2000; dasilva et al. 2004) and analysed the protein localisations by laser scanning confocal microscopy. We found that both isoforms were distributed at multiple punctate structures (Fig. 2a, i, arrowheads), at the ER and in the cytosol (Fig. 2a, i, arrows). We verified that a similar distribution occurred in Arabidopsis leaf epidermal cells (Supplementary Figure 1). Co-expression of either Sar1 isoform with the ER/Golgi marker ERD2-GFP (Boevink et al. 1998) revealed that the punctate structures were localised in the peri-golgi area (Fig. 2b d, j l, arrowheads), suggesting
4 Fig. 1 Alignment of the sequences of AtSARA1a, AtSARA1b and NtSar1P. All three sequences have a high level of similarity, emphasised by grey shading of residues that are identical between at least two of the protein sequences. Black arrowheads indicate the amino acids in positions 159 and 193 (164 and 198 for NtSar1P), in that they may correspond to ERES (dasilva et al. 2004; Hanton et al. 2007; Stefano et al. 2006). The distributions of the two AtSar1 isoforms therefore differ from that of NtSar1P- YFP, in that NtSar1P-YFP labels the cytosol and occasional bright punctate structures, and is visibly recruited to ERES upon over-expression of membrane cargo (dasilva et al. 2004). The distribution of both AtSar1 proteins at punctate structures resembled that previously reported for fluorescent fusions of AtSec23 and AtSec24 (Stefano et al. 2006). However, both AtSar1 proteins were also found in association with the ER, as shown by colocalisation experiments with the ER marker GFP-calnexin (Fig. 2f h, n p). This confocal analysis also suggested that, despite the fact that both AtSar1 isoforms labelled punctate structures, AtSARA1b-YFP had a slightly more reticular appearance in the areas of fluorescence that were not associated with ERES, compared with AtSARA1a-YFP. Given the very close association of ER and cytosol in plant cells, the relative distributions of the two isoforms could not be quantified reliably using live cell imaging. Therefore, to confirm our microscopy observations on the non-identical distribution of the two Sar1 isoforms, we adopted a biochemical method. We performed a cell fractionation based on that described by dasilva et al. (2004) to separate proteins in the soluble phase from those associated with membranes, using protoplasts prepared from tobacco leaves that had been transfected with AtSARA1a-YFP or AtSARA1b-YFP. The presence of the fluorescent tag allowed us to visualise the two nearly identical Sar1 proteins by western blotting with anti-gfp serum. Figure 3a shows that AtSARA1a-YFP was found in both the soluble which AtSARA1a and NtSar1P have identical amino acids while AtSARA1b differs. Open arrowheads indicate the histidine at position 74 that was mutated to create the GTP-restricted mutant. The GNKXD motif is underlined in AtSARA1a and AtSARA1b, but is not present in the sequence of NtSar1P and membrane fractions, though predominantly in the former. In comparison with AtSARA1a-YFP, AtSARA1b- YFP was more evenly distributed between the soluble and membrane fractions, indicating a higher level of membrane association than that observed for AtSARA1a-YFP. The amounts of proteins used in the experiment were similar (Fig. 3b), thereby excluding the possibility that different quantities of proteins used in the extraction could be responsible for the observed distributions. Quantification of the intensities of the bands shown in Fig. 3a (see Materials and methods ) confirmed that the differences in the intracellular partitioning of AtSARA1a-YFP and AtSARA1b-YFP were significant, with AtSARA1b-YFP being considerably more membrane-associated than AtSARA1a-YFP (Fig. 3c). The distribution of AtSARA1a and AtSARA1b is influenced by the amino acid composition of the C-terminus The different subcellular distributions of the AtSar1 isoforms were intriguing given the high sequence homology of the two proteins, and prompted us to investigate how the different distributions of the two proteins may be achieved. As NtSar1P-YFP was found in the cytosol and at ERES (dasilva et al. 2004) and AtSARA1a-YFP was predominantly distributed in the soluble fraction (Fig. 3a), we hypothesised that amino acids that appeared different only in AtSARA1b in a comparison between NtSar1P and the two AtSar1 isoforms might be responsible for the higher level of membrane association of AtSARA1b in comparison to AtSARA1a. Analysis of the Sar1 sequences
5 Fig. 2 Comparison of the intracellular localisations of AtSARA1a and AtSARA1b. (a) AtSARA1a-YFP labels the ER, cytosol (arrow) and punctate structures (arrowheads) (b d) Co-expression of At- SARA1a-YFP (b) with the Golgi marker ERD2-GFP (c) demonstrates that the punctate structures labelled by AtSARA1a-YFP are in the peri-golgi area (d, arrowheads). (e) AtSARA1a-YFP imaged using a pinhole of 1 Airy unit to give higher definition. The labelling pattern is similar to that using a wider pinhole. (f h) Co-expression of AtSARA1a-YFP (f) with the ER membrane marker GFP-calnexin (g) to allow distinction between labelling of the ER and cytosol (h, merge of f and g). Imaging parameters used were the same as in e. (i) AtSARA1b-YFP appears to be associated with the ER membranes (Fig. 1) showed that there are two residues that are conserved in NtSar1P and AtSARA1a but that differ in AtSARA1b, at positions 159 and 193 in the Arabidopsis proteins (Fig. 1, black arrowheads). The residues at position 159 in AtSARA1a and 164 in NtSar1P are both asparagines (N), while the corresponding residue in AtSARA1b is threonine (T). At position 198 in NtSar1P and 193 in AtSARA1a, the residues are both lysines (K), while in AtSARA1b residue 193 is asparagine (N). We postulated that the difference in the nature of one or both of the residues 159 and 193 in AtSARA1b could influence the association of the protein with membranes. We therefore (arrow) as well as labelling punctate structures (arrowheads) and cytosol. (j l) The punctate structures labelled by AtSARA1b-YFP (j) are found in the peri-golgi area, as shown by co-expression of ERD2- GFP (k, l, arrowheads). Possible ER staining is shown by both AtSARA1b-YFP and ERD2-GFP (arrows). (m) AtSARA1b-YFP imaged using a pinhole of 1 Airy unit to give higher definition. The labelling pattern is similar to that using a wider pinhole. (n p) Co-expression of AtSARA1b-YFP (n) with the ER membrane marker GFP-calnexin (o) to allow distinction between labelling of the ER and cytosol (p, merge of n and o). Imaging parameters used were the same as in e. Bars = 5 lm carried out mutagenesis of AtSARA1a and AtSARA1b so that the amino acid at position 159 was changed to that of the other isoform, giving AtSARA1a N159T and AtSARA1b T159N. A similar mutagenesis was performed to swap the residues at position 193, giving AtSARA1a K193N and AtSARA1b N193K. The rationale for this mutagenesis was based on the hypothesis that switching the residues at position 159 or 193 might reverse the subcellular distributions of the two proteins. We next analysed the subcellular localisations of fluorescent protein fusions of the Sar1 mutant proteins by livecell imaging. The mutations at position 159 did not seem to
6 have any effect on the localisations of either AtSARA1a or AtSARA1b mutants in comparison to the wild-type proteins (compare Fig. 2 and Supplementary Figure 2). Analysis of the distribution of the AtSar1 isoforms carrying a mutation in position 193 gave different results. For both AtSARA1a K193N -YFP and AtSARA1b N193K -YFP mutants, the defined ERES labelling at the peri-golgi area was clearly visible as confirmed by co-expression analyses with the ER/Golgi marker ERD2-GFP (Fig. 3e g, m o). However, expression of AtSARA1a K193N -YFP (Fig. 3d) appeared to give a slightly more reticular pattern compared to that of the wild-type (Fig. 2a), resembling the
7 b Fig. 3 The distributions of AtSARA1a and AtSARA1b depend on the nature of the C-terminal amino acid. (a) Protoplasts expressing AtSARA1a-YFP, AtSARA1a K193N -YFP, AtSARA1b-YFP or AtSARA1b N193K -YFP were subjected to a cell fractionation, separating proteins in the soluble phase (S) from those in the cell membranes (M). UT = untransformed protoplasts. Equal volumes of each extract were loaded onto SDS-PAGE gels and were subjected to western blotting with anti-gfp serum. AtSARA1a-YFP is found predominantly in the soluble phase, although a signal is also visible in the membrane fraction. In contrast, AtSARA1b-YFP is present at similar levels in both the soluble and membrane fractions, indicating a higher level of membrane association than that of AtSARA1a-YFP. On mutagenesis of K193 to N in AtSARA1a-YFP, the signal in the membrane fraction increased significantly, indicating an increased membrane association. However, on mutagenesis of N193 to K in AtSARA1b-YFP, the signal in the membrane fraction is reduced, indicating a lower level of membrane association, with a concomitant increase in the soluble fraction. (b) Total extracts of each sample demonstrate similar levels of expression for each protein fusion (10-fold diluted compared with those in panel a). Approximate molecular weights are given at left in kilodaltons. (c) Quantification of signal intensities from western blots showing soluble and membrane fractions for each of AtSARA1a-YFP, AtSARA1a K193N - YFP, AtSARA1b-YFP or AtSARA1b N193K -YFP. Error bars represent standard error of the mean for three independent experiments. (d) AtSARA1a K193N -YFP shows a more reticular appearance (arrow) in comparison with the wild-type protein. Cytosol and punctate structures (arrowheads) remain visible. (e g) The punctate structures labelled by AtSARA1a K193N -YFP (e) are in the peri-golgi area, shown by co-expression with ERD2-GFP (f, g, arrowheads), while the reticular pattern appears to colocalise more with the ER staining of ERD2-GFP (arrows) in comparison with the wild-type protein (compare with Fig. 2). (h) AtSARA1a K193N -YFP imaged using a pinhole of 1 Airy unit to give higher definition. (i k) Co-expression of AtSARA1a K193N -YFP (i) with the ER membrane marker GFPcalnexin (j) to allow distinction between labelling of the ER and cytosol (k, merge of i and j). Imaging parameters used were the same as in h. (l) AtSARA1b N193K YFP has a reduced level of membrane association compared with the wild-type protein, but punctate structures are still present (arrowheads) and the level of cytosolic staining appears to increase (arrow) in comparison with the wild-type protein (Fig. 2). (m o) AtSARA1b N193K -YFP-labelled punctate structures (m) are found in the peri-golgi area, shown by co-expression with ERD2-GFP (n, o, arrowheads). (p) AtSARA1b N193K -YFP imaged using a pinhole of 1 Airy unit to give higher definition. (q s) Co-expression of AtSARA1b N193K -YFP (q) with the ER membrane marker GFP-calnexin (r) to allow distinction between labelling of the ER and cytosol (s, merge of q and r). Imaging parameters used were the same as in h. Bars = 5 lm distribution of AtSARA1b-YFP (Fig. 2i). On the other hand, AtSARA1b N193K -YFP (Fig. 3l) appeared to be more cytosolic in comparison to the wild-type form (Fig. 2i), showing similarity to AtSARA1a-YFP (Fig. 2a). Co-expression of either mutant with GFP-calnexin (Fig. 3i k, q s), analysed with a smaller pinhole setting, indicated that both proteins display some ER association. However, this is not as clear as the distribution observed for AtSARA1b-YFP (Fig. 2m, n). Therefore, although both AtSARA1a K193N -YFP and AtSARA1b N193K -YFP showed membrane association and cytosolic distribution, suggesting that other parts of the proteins could also contribute to their distributions, these results indicate that the localisation of the two Sar1 isoforms was influenced by the amino acid residue at position 193. To ensure that the redistribution of these mutant proteins was quantitative, we carried out a cell fractionation experiment on protoplasts from leaves expressing the proteins carrying point mutations at position 193, in the same manner as for the wild-type proteins (Fig. 3a). We found that AtSARA1a K193N -YFP was redistributed from the soluble fraction to the membrane fraction (Fig. 3a). In contrast, AtSARA1b N193K -YFP was detected at much higher levels in the soluble fraction than the membrane fraction, indicating a reduced level of membrane association compared with its wild-type counterpart. An analysis of total cell extracts for each fusion protein confirmed that all the proteins were expressed at similar levels, excluding the possibility that the different fractionation of the proteins could be due to different expression levels of the proteins (Fig. 3b). Quantification of the intensities of the bands in Fig. 3a, carried out as for the wild-type proteins (Fig. 3c), confirmed that the differences observed for the AtSARA mutants and wild-type proteins using live cell imaging were significant. Although the effect of the mutation was not complete in either case, the combined confocal imaging and cell fractionation data supported our hypothesis that exchange of the 193 residues between AtSARA1a and AtSARA1b could influence the level of membrane association of the two proteins in comparison with their respective wild-type forms. The two Sar1 isoforms exhibit different effects on secretion We next wanted to test whether the different subcellular distributions of AtSARA1a and AtSARA1b could be reflected in different activities of these proteins in the regulation of ER protein export. To test this, we used a quantitative secretion assay based on the co-transformation of protoplasts with a-amylase and the AtSar1 isoforms as effector molecules (Phillipson et al. 2001). This assay would allow us to measure the relative influences of the two AtSar1 isoforms on ER protein export. We therefore co-expressed dilution series of AtSARA1a-YFP or AtSARA1b-YFP with a constant amount of a-amylase in tobacco leaf protoplasts, then measured the activity of the enzyme in the intracellular extracts and extracellular medium in order to calculate the secretion index (ratio of the extracellular and intracellular enzyme activities, (Phillipson et al. 2001, Fig. 4a). We found that although the secretion index was slightly reduced by co-expression of AtSARA1a-YFP, expression of AtSARA1b-YFP resulted in a significant inhibition of the secretion of a-amylase. To confirm these data further, we also wanted to test the
8 Fig. 4 AtSARA1a and AtSARA1b affect secretion to different extents. Tobacco leaf protoplasts were electroporated with a constant amount of DNA encoding a-amylase and increasing amounts of DNA encoding AtSARA1a-YFP, AtSARA1b-YFP, AtSARA1a H74L -YFP or AtSARA1b H74L -YFP. (a) The a-amylase activity was measured for cell extracts and extracellular medium, and the secretion index was calculated, being the ratio of activity in the medium to that in the cell extracts. Error bars represent standard deviation for three repetitions. (b) Equal quantities of protein extracts from the intracellular sample were subjected to SDS-PAGE and western blotting with anti-gfp serum (AbCam) to ensure similar levels of protein expression at each DNA concentration. DNA concentrations used were as given in the figure. Approximate molecular weights are given at left in kilodaltons effect of the GTP-restricted forms of the two Arabidopsis Sar1 GTPases. It has been shown that a GTP-restricted form of AtSARA1b causes a reduced level of secretion of a-amylase, possibly due to sequestration of other COPII components at the membrane by the GTPase (Phillipson et al. 2001). We wanted to see whether this effect would also be observed in AtSARA1a, and whether the extent of inhibition of secretion would be similar between the two isoforms. Given the results obtained with the wild-type forms, we expected that the AtSARA1b-GTP mutant would affect ER export to a larger extent than AtSARA1a- GTP. We therefore generated mutants of AtSARA1a and AtSARA1b in which the conserved histidine at position 74 (Fig. 1, white arrowhead) was changed to a leucine to obtain a GTP-restricted mutation (H74L, Phillipson et al. 2001) and fused them to YFP. We expressed a dilution series of each mutant protein with a-amylase and calculated the secretion index of the reporter enzyme, as described above. AtSARA1a H74L -YFP caused a reduction in the secretion index, but this was considerably less dramatic than the reduction caused by AtSARA1b H74L -YFP (Fig. 4a). Comparable expression levels of each of the Sar1 proteins were ensured by western blotting with anti-gfp (Fig. 4b); this confirms that even at low levels of expression, AtSARA1b H74L -YFP has approximately a 1.5-fold greater effect than AtSARA1a H74L -YFP. These results support our initial hypothesis of differential effects of the mutant AtSar1 isoforms on secretion, based on the data obtained with the wild-type counterparts of the two AtSar1 isoforms, and substantiate our suggestion that the two AtSar1 isoforms influence ER export to different extents. We next wanted to test whether swapping the residue at position 193 would cause the Sar1 isoforms to exhibit the opposite effect on secretion due to their altered localisation. We created a double mutant of AtSARA1b (AtSARA1b H74LN193K -YFP) and performed a similar experiment to that shown in Fig. 4. We hypothesised that this double mutant would have a lesser inhibitory effect on secretion than AtSARA1b H74L -YFP, but greater than AtSARA1a H74L -YFP, as the change in localisation caused by the 193 mutation was not complete. Figure 5a confirms that AtSARA1b H74LN193K -YFP causes a significant reduction in the level of secretion of a-amylase, but that it is less dramatic than that caused by AtSARA1b H74L -YFP. This indicates that the nature of the residue at position 193 influences the localisation of the GTPase and its regulation of secretion. Discussion Plant Sar1 isoforms have different activities Here we present evidence that plant Sar1 isoforms can have different cellular activities. In particular, we have shown that fluorescent protein fusions to two Arabidopsis Sar1 isoforms are visibly recruited to the ER membrane in tobacco and Arabidopsis leaves (Fig. 2 and Supplementary Figure 1). This recruitment occurs in cells where the constructs are expressed alone. The distributions of the two Arabidopsis isoforms differ from that of a fluorescent fusion to a tobacco Sar1 isoform (NtSar1P), which is largely distributed in the cytosol of tobacco leaves but is visibly recruited to ERES upon co-expression of membrane cargo destined for the Golgi (dasilva et al. 2004). Interestingly, comparison of the sequences of Arabidopsis and two other tobacco Sar1 isoforms (NtSar1A and NtSar1B) shows that the overall homology between these protein sequences is higher than that with NtSar1P (Supplementary Figure 3). NtSar1P does not contain a GNKXD motif that is present in the other Sar1 isoforms (Supplementary Figure 3, underlined). It is possible that this contributes to the more cytosolic
9 Fig. 5 AtSARA1b H74LN193K reduces secretion to a lesser extent than does AtSARA1b H74L. Tobacco leaf protoplasts were electroporated with a constant amount of DNA encoding a-amylase and increasing amounts of DNA encoding AtSARA1a H74L -YFP, AtSARA1b H74L - YFP or AtSARA1b H74LN193K -YFP. (a) The a-amylase activity was measured for cell extracts and extracellular medium, and the secretion index was calculated, being the ratio of activity in the medium to that in the cell extracts. Error bars represent standard deviation for two repetitions. (b) Equal quantities of protein extracts from the intracellular sample were subjected to SDS-PAGE and western blotting with anti-gfp serum to ensure similar levels of protein expression at each DNA concentration. DNA concentrations used were as given in the figure. Approximate molecular weights are given at left in kilodaltons distribution of NtSar1P compared with the Arabidopsis Sar1 isoforms. In this work, we have also verified that the subcellular distributions of the AtSar1 isoforms are not identical (Figs. 2, 3 and Supplementary Figure 1); AtSARA1b exhibits a higher level of membrane association than AtSARA1a. The distributions of the isoforms suggest that these Sar1 proteins may have different activities in plant cells. This possibility is further supported by the finding that over-expression of GTP-restricted mutants of each AtSar1 isoform led to different effects on secretion. Overexpression of either the wild-type or GTP-restricted mutant of AtSARA1b had a much stronger influence on secretion than AtSARA1a or its GTP-restricted mutant (Fig. 4). The residue at position 193 in both AtSar1 isoforms appears to play a role in the distribution and activity of the two proteins. By swapping this residue in AtSARA1a with that in AtSARA1b, the AtSARA1b mutant acquired a more cytosolic distribution than the wild-type (Fig. 3), which appears to be a property of AtSARA1a (this work) and NtSar1P (dasilva et al. 2004). In accordance with the hypothesis that the localisation of the protein has a bearing on its activity, we also found that a GTP-restricted mutant of AtSARA1b N193K had a less inhibitory effect on secretion than the GTP-restricted mutant of AtSARA1b (Fig. 5), although some inhibition was still observed. This corresponds to the finding that changing the nature of the residue at position 193 does not cause a complete exchange of membrane association properties between the two isoforms (Fig. 3). These data together also indicate that although the level of membrane association of the two isoforms influences the effect on secretion, other factors must also play a role in the activities of these proteins. Of all the proteins compared in Supplementary Figure 3, AtSARA1b is the only Sar1 isoform to possess an asparagine residue at the extreme C-terminus, all the others having a lysine residue. However, AtSARA1a has less homology to the other tobacco Sar1 isoforms (Supplementary Figure 3). A comparative analysis of the distributions and activities of the tobacco and Arabidopsis Sar1 proteins may contribute further to the understanding of the underlying mechanisms that control the distribution and function of Sar1 proteins across species. The analysis of the distribution and activity of the two Arabidopsis Sar1 isoforms in one expression system (tobacco leaf cells) has allowed us to identify differences between these GTPases. These findings prompt an important biological question: what is the reason for the existence of multiple isoforms of Sar1 in Arabidopsis? It seems unlikely that Sar1 GTPases are functionally redundant, as both the variation in localisation and difference in regulating secretion of soluble cargo suggest differences in functionality. A large body of evidence that is emerging from mammalian and yeast studies strongly indicates that the COPII coat composition may be heterogeneous (Peng et al. 2000; Roberg et al. 1999; Shimoni et al. 2000; Wendeler et al. 2007). By extrapolating these data to the plant system, we may speculate that different Sar1 isoforms could operate in plant cells to regulate the formation of COPII carriers with different coat compositions. If this were the case, the recruitment of Sec23/24 heterodimers to ERES could be dependent on the specific affinity of both cargo proteins and Sar1 isoforms for any given Sec23/24 heterodimer. On the other hand, it is also possible that different Sar1 isoforms recognise specific signals in cargo proteins. This would be consistent with the finding that a mammalian Sar1 isoform is able to associate with a cytoplasmic dibasic motif of a membrane cargo protein and may perform a selection function (Giraudo and Maccioni
10 2003). Therefore, similar to the mammalian Sec24 (Wendeler et al. 2007), multiple Sar1 isoforms could recognise different ER export signals. It cannot be ruled out that the Sar1 isoforms used in this study are expressed in different tissues in Arabidopsis, and that the different localisations and activities observed in this study are required to fulfil the secretory requirements of the tissue in which the protein is expressed. How is the different distribution of the two AtSar1 isoforms achieved? The distribution of AtSARA1b shown in this work is in agreement with that reported by Bar-Peled and Raikhel (1997) for the same protein, where a steady-state association of half of the cellular pool of the protein with ER membranes prepared from Arabidopsis tissue was determined. The distribution of AtSARA1a reported in this study is novel, showing more similarity to the localisation of NtSar1P (dasilva et al. 2004). However, both AtSARA1a and AtSARA1b are concentrated at ERES, similar to other COPII marker proteins (Hanton et al. 2007; Matheson et al. 2006; Stefano et al. 2006; Yang et al. 2005) but differing from the behaviour of NtSar1P. Possible explanations for the different steady-state partitioning of the AtSar1 isoforms between soluble and membrane phases may include different affinities of the two isoforms for membrane binding sites, or factors that relate to the GTP hydrolysis rate of the GTPases. Since cycles of activation and inactivation of Sar1 occur at the membrane, it is possible that different rates of GTP hydrolysis may influence the residence of the two AtSar1 isoforms at the ER. The fluorescent tag that allowed us to monitor the distribution of the two nearly identical protein isoforms and their mutants is unlikely to be the factor that determined the different distributions of AtSARA1a, AtSARA1b (this work) and NtSar1P (dasilva et al. 2004). The positioning of the tag at the C-terminus was not expected to alter the distribution of Sar1-YFP relative to untagged Sar1 (dasilva et al 2004), and preserves the integrity of the N-terminus of the GTPase, which is crucial for its membrane anchoring (Bi et al. 2002; Huang et al. 2001). We cannot exclude the possibility that the presence of the tag may affect the dynamics of the tagged Sar1 proteins compared with the endogenous proteins. dasilva et al. (2004) showed that the tag caused a slight reduction in the ability of a GTPrestricted mutant to inhibit secretion; however, the tag was an identical feature for all proteins used in this study, making the results comparable among isoforms. The experiments demonstrating the changed distribution of the Sar1 isoforms in response to altering the amino acid residue at position 193 (Fig. 3) provide further evidence that the YFP tag is unlikely to determine the relative distributions of the proteins. The involvement of residue 193 in the localisation of the Sar1 isoforms is particularly interesting, as previous studies have shown that the association of Sar1 with membranes is dependent on a hydrophobic region at the N-terminus of the protein, termed the STAR motif (Huang et al. 2001). The evidence that a residue at the C-terminus of the protein can also influence the distribution of Sar1 is important as it adds to our knowledge of mechanisms that Sar1-GTPases may use to target membranes. It is also interesting that we observed only a partial redistribution of the Sar1 proteins bearing mutations in position 193; the AtSARA1a K193N mutant did not have an identical distribution to AtSARA1b, and vice versa, the distribution of AtSARA1b K193N mutant was not exactly the same as that of AtSARA1a (Fig. 3). This suggests that the N- and C-termini may both contribute directly to the membrane association of the protein, or that each is involved in a separate step of the process. For example, the N-terminus may determine the initial association of Sar1 with ER membranes, while the C-terminus could be involved in recognition of factors that influence the distribution of the Sar1 proteins. For example, the membrane-associated GEF Sec12 is distributed throughout the ER membranes (dasilva et al. 2004). It is possible that AtSARA1b may have a stronger affinity for Sec12 than does AtSARA1a, or a slower rate of GTP hydrolysis, which would explain the different patterns of localisation of each of the AtSar1 proteins. The assembly of COPII coat proteins at ERES It has been shown previously that plant COPII proteins have a unique subcellular distribution. Unlike other systems, fluorescent protein fusions of several of the COPII coat proteins (Sec13, Sec23 and Sec24) have been found localised at ERES in the vicinity of mobile Golgi stacks (Hanton et al. 2007; Matheson et al. 2006; Stefano et al. 2006; Yang et al. 2005), while Sec12 is distributed uniformly over the ER (dasilva et al. 2004; Yang et al. 2005). The data provided in this study show that AtSar1 proteins can associate with ER membranes as well as localising at Golgi-associated ERES. This finding suggests that COPII may be assembled on areas of the ER besides the ERES at the ER/Golgi interface. However, the COPII coat proteins Sec24 and Sec23, which are involved in cargo selection (Aridor et al. 1998; Miller et al. 2003; Peng et al. 2000; Roberg et al. 1999; Shimoni et al. 2000; Votsmeier and Gallwitz 2001; Wendeler et al. 2007) and regulation of the GTPase activity of Sar1 (Yoshihisa et al. 1993), have not been found in association with plant ER membranes except at ERES (Hanton et al. 2007; Matheson et al. 2007; Stefano et al. 2006). A similar situation has been described
11 in mammalian cells, where Sar1 is localised homogeneously across the ER membrane with some accumulation at ERES (Kuge et al. 1994) and the COPII coat proteins Sec23/24 and Sec13/31 are exclusively localised to punctate structures at ERES (Forster et al. 2006; Ward et al. 2001), similar to plant cells. The spatial distribution of the mammalian and Arabidopsis Sar1 isoforms with respect to the COPII coat proteins suggests that the activation of Sar1 and its subsequent ER membrane association at non-eres areas may not provide sufficient stimulus for the formation of COPII carriers. On the other hand, the presence of cargo at ERES in conjunction with the presence of activated Sar1 might stimulate the recruitment of Sec23/24 to ERES, ultimately resulting in the formation of COPII carriers. This model would be consistent with a selective recruitment of COPII machinery that depends on the availability of cargo at ERES. In support of this, it has been shown that over-expression of membrane cargo proteins results in increased recruitment of Arabidopsis Sec24 and tobacco Sar1 isoforms to ERES in plants (dasilva et al. 2004; Hanton et al. 2007), and of Sar1 in mammalian cells (Guo and Linstedt 2006). These data support the model that the export machinery is available within the cytosol, and that its recruitment to ERES depends on the localised accumulation of export-competent cargo molecules at defined areas of the ER membrane. Acknowledgements This work was developed with grants awarded to F.B. from the Canada Foundation for Innovation (CFI), the Canada Research Chair (CRC) program, Natural Science and Engineering Research Council of Canada (NSERC) and Department of Energy, Michigan State University. L.A.M. is supported by an NSERC postdoctoral fellowship. References Andreeva AV, Kutuzov MA, Evans DE, Hawes CR (1998) Proteins involved in membrane transport between the ER and the Golgi apparatus: 21 putative plant homologues revealed by dbest searching. Cell Biol Int 22: Aridor M, Weissman J, Bannykh S, Nuoffer C, Balch WE (1998) Cargo selection by the COPII budding machinery during export from the ER. J Cell Biol 141:61 70 Aridor M, Fish KN, Bannykh S, Weissman J, Roberts TH, Lippincott- Schwartz J, Balch WE (2001) The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J Cell Biol 152: Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R (1994) COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77: Bar-Peled M, Raikhel NV (1997) Characterization of AtSEC12 and AtSAR1. Proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol 114: Batoko H, Zheng HQ, Hawes C, Moore I (2000) A rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12: Bi X, Corpina RA, Goldberg J (2002) Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature 419: Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C (1998) Stacks on tracks: the plant Golgi apparatus traffics on an actin/er network. Plant J 15: Brandizzi F, Frangne N, Marc-Martin S, Hawes C, Neuhaus JM, Paris N (2002a) The destination for single-pass membrane proteins is influenced markedly by the length of the hydrophobic domain. Plant Cell 14: Brandizzi F, Fricker M, Hawes C (2002b) A greener world: the revolution in plant bioimaging. Nat Rev Mol Cell Biol 3: Crofts AJ, Leborgne-Castel N, Hillmer S, Robinson DG, Phillipson B, Carlsson LE, Ashford DA, Denecke J (1999) Saturation of the endoplasmic reticulum retention machinery reveals anterograde bulk flow. Plant Cell 11: dasilva LL, Snapp EL, Denecke J, Lippincott-Schwartz J, Hawes C, Brandizzi F (2004) Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16: d Enfert C, Gensse M, Gaillardin C (1992) Fission yeast and a plant have functional homologues of the Sar1 and Sec12 proteins involved in ER to Golgi traffic in budding yeast. Embo J 11: Forster R, Weiss M, Zimmermann T, Reynaud EG, Verissimo F, Stephens DJ, Pepperkok R (2006) Secretory cargo regulates the turnover of COPII subunits at single ER exit sites. Curr Biol 16: Giraudo CG, Maccioni HJ (2003) Endoplasmic reticulum export of glycosyltransferases depends on interaction of a cytoplasmic dibasic motif with Sar1. Mol Biol Cell 14: Guo Y, Linstedt AD (2006) COPII-Golgi protein interactions regulate COPII coat assembly and Golgi size. J Cell Biol 174:53 63 Hanton SL, Renna L, Bortolotti LE, Chatre L, Stefano G, Brandizzi F (2005) Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. Plant Cell 17: Hanton SL, Chatre L, Renna L, Matheson LA, Brandizzi F (2007) De novo formation of plant endoplasmic reticulum export sites is membrane cargo-induced and signal-mediated. Plant Physiol 143: Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94: Huang M, Weissman JT, Beraud-Dufour S, Luan P, Wang C, Chen W, Aridor M, Wilson IA, Balch WE (2001) Crystal structure of Sar1-GDP at 1.7 A resolution and the role of the NH 2 terminus in ER export. J Cell Biol 155: Kim WY, Cheong NE, Je DY, Kim MG, Lim CO, Bahk JD, Cho MJ, Lee SY (1997) The presence of a Sar1 gene family in Brassica campestris that suppresses a yeast vesicular transport mutation Sec12-1. Plant Mol Biol 33: Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE (1994) Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol 125:51 65 Lee MC, Miller EA, Goldberg J, Orci L, Schekman R (2004) Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 20:87 Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R (2005) Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122:
12 Matheson LA, Hanton SL, Brandizzi F (2006) Traffic between the plant endoplasmic reticulum and Golgi apparatus: to the Golgi and beyond. Curr Opin Plant Biol 9: Matheson LA, Hanton SL, Rossi M, Latijnhouwers M, Stefano G, Renna L, Brandizzi F (2007) Multiple roles of ADP-ribosylation factor 1 in plant cells include spatially regulated recruitment of coatomer and elements of the Golgi matrix. Plant Physiol 143: Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T (1998) COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell 93: Matsuoka K, Schekman R, Orci L, Heuser JE (2001) Surface structure of the COPII-coated vesicle. Proc Natl Acad Sci USA 98: Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R (2003) Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114: Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: Peng R, De Antoni A, Gallwitz D (2000) Evidence for overlapping and distinct functions in protein transport of coat protein Sec24p family members. J Biol Chem 275: Phillipson BA, Pimpl P, dasilva LL, Crofts AJ, Taylor JP, Movafeghi A, Robinson DG, Denecke J (2001) Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell 13: Roberg KJ, Crotwell M, Espenshade P, Gimeno R, Kaiser CA (1999) LST1 is a SEC24 homologue used for selective export of the plasma membrane ATPase from the endoplasmic reticulum. J Cell Biol 145: Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbour Press, Cold Spring Harbour Sato K, Nakano A (2005) Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis. Nat Struct Mol Biol 12: Seki M, Iida A, Morikawa H (1998) Transient expression of foreign genes in tissues of Arabidopsis thaliana by bombardmentmediated transformation. Methods Mol Biol 82: Shimoni Y, Kurihara T, Ravazzola M, Amherdt M, Orci L, Schekman R (2000) Lst1p and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. J Cell Biol 151: Stefano G, Renna L, Chatre L, Hanton SL, Moreau P, Hawes C, Brandizzi F (2006) In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J 46: Takeuchi M, Tada M, Saito C, Yashiroda H, Nakano A (1998) Isolation of a tobacco cdna encoding Sar1 GTPase and analysis of its dominant mutations in vesicular traffic using a yeast complementation system. Plant Cell Physiol 39: Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A (2000) A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J 23: Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131: Votsmeier C, Gallwitz D (2001) An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. Embo J 20: Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott- Schwartz J (2001) Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol 155: Weissman JT, Aridor M, Balch WE (2001) Purification and properties of rat liver Sec23-Sec24 complex. Meth Enzymol 329: Wendeler MW, Paccaud JP, Hauri HP (2007) Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep 8: Yang YD, Elamawi R, Bubeck J, Pepperkok R, Ritzenthaler C, Robinson DG (2005) Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell 17: Yoshihisa T, Barlowe C, Schekman R (1993) Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science 259:
CELB40060 Membrane Trafficking in Animal Cells. Prof. Jeremy C. Simpson. Lecture 2 COPII and export from the ER
 CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
Correct Targeting of Plant ARF GTPases Relies on Distinct Protein Domains
 Traffic 2008; 9: 103 120 Blackwell Munksgaard # 2007 The Authors Journal compilation # 2007 Blackwell Publishing Ltd doi: 10.1111/j.1600-0854.2007.00671.x Correct Targeting of Plant ARF GTPases Relies
Traffic 2008; 9: 103 120 Blackwell Munksgaard # 2007 The Authors Journal compilation # 2007 Blackwell Publishing Ltd doi: 10.1111/j.1600-0854.2007.00671.x Correct Targeting of Plant ARF GTPases Relies
Secretory Cargo Regulates the Turnover of COPII Subunits at Single ER Exit Sites
 Current Biology 16, 173 179, January 24, 2006 ª2006 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2005.11.076 Secretory Cargo Regulates the Turnover of COPII Subunits at Single ER Exit Sites Report
Current Biology 16, 173 179, January 24, 2006 ª2006 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2005.11.076 Secretory Cargo Regulates the Turnover of COPII Subunits at Single ER Exit Sites Report
Diacidic Motifs Influence the Export of Transmembrane Proteins from the Endoplasmic Reticulum in Plant Cells W
 This article is published in The Plant Cell Online, The Plant Cell Preview Section, which publishes manuscripts accepted for publication after they have been edited and the authors have corrected proofs,
This article is published in The Plant Cell Online, The Plant Cell Preview Section, which publishes manuscripts accepted for publication after they have been edited and the authors have corrected proofs,
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
Wave line information sheet
 Wave line information sheet Niko Geldner May 200 Wave constructs and lines, as well as pnigel vector series were published: Rapid, combinatorial analysis of membrane compartments in intact plants with
Wave line information sheet Niko Geldner May 200 Wave constructs and lines, as well as pnigel vector series were published: Rapid, combinatorial analysis of membrane compartments in intact plants with
Secretory Bulk Flow of Soluble Proteins Is Efficient and COPII Dependent
 The Plant Cell, Vol. 13, 2005 2020, September 2001, www.plantcell.org 2001 American Society of Plant Biologists Secretory Bulk Flow of Soluble Proteins Is Efficient and COPII Dependent Belinda A. Phillipson,
The Plant Cell, Vol. 13, 2005 2020, September 2001, www.plantcell.org 2001 American Society of Plant Biologists Secretory Bulk Flow of Soluble Proteins Is Efficient and COPII Dependent Belinda A. Phillipson,
COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity to ER exit sites
 Journal of Cell Science 113, 2177-2185 (2000) Printed in Great Britain The Company of Biologists Limited 2000 JCS1368 2177 COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity
Journal of Cell Science 113, 2177-2185 (2000) Printed in Great Britain The Company of Biologists Limited 2000 JCS1368 2177 COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity
Protein Sorting, Intracellular Trafficking, and Vesicular Transport
 Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Supplemental Data. Gao et al. (2012). Plant Cell /tpc
 Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Supplemental Figure 1. Plant EMP Proteins. (A) The Accession numbers of the 12 EMP members from Arabidopsis. (B) Phylogenetic analysis of EMP proteins from Arabidopsis, human and yeast using the Mac Vector
Department of Cell Biology, Heidelberg Institute for Plant Sciences, University of Heidelberg, Heidelberg, Germany b
 The Plant Cell, Vol. 17, 1513 1531, May 2005, www.plantcell.org ª 2005 American Society of Plant Biologists Dynamics of COPII Vesicles and the Golgi Apparatus in Cultured Nicotiana tabacum BY-2 Cells Provides
The Plant Cell, Vol. 17, 1513 1531, May 2005, www.plantcell.org ª 2005 American Society of Plant Biologists Dynamics of COPII Vesicles and the Golgi Apparatus in Cultured Nicotiana tabacum BY-2 Cells Provides
Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar Compartment tothelyticvacuoleinvivo W
 The Plant Cell, Vol. 18, 2275 2293, September 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar
The Plant Cell, Vol. 18, 2275 2293, September 2006, www.plantcell.org ª 2006 American Society of Plant Biologists Overexpression of the Arabidopsis Syntaxin PEP12/SYP21 Inhibits Transport from the Prevacuolar
DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS. Scientific Background on the Nobel Prize in Medicine 2013
 DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS Scientific Background on the Nobel Prize in Medicine 2013 Daniela Scalet 6/12/2013 The Nobel Prize in Medicine
DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS Scientific Background on the Nobel Prize in Medicine 2013 Daniela Scalet 6/12/2013 The Nobel Prize in Medicine
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%%
 !"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
Predominant Golgi Residency of the Plant K/HDEL Receptor Is Essential for its Function in Mediating ER Retention
 Plant Cell Advance Publication. Published on August 2, 2018, doi:10.1105/tpc.18.00426 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41
Plant Cell Advance Publication. Published on August 2, 2018, doi:10.1105/tpc.18.00426 1 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41
A Missense Mutation in the Arabidopsis COPII Coat Protein Sec24A Induces the Formation of Clusters of the Endoplasmic Reticulum and Golgi Apparatus W
 The Plant Cell, Vol. 21: 3655 3671, November 2009, www.plantcell.org ã 2009 American Society of Plant Biologists A Missense Mutation in the Arabidopsis COPII Coat Protein Sec24A Induces the Formation of
The Plant Cell, Vol. 21: 3655 3671, November 2009, www.plantcell.org ã 2009 American Society of Plant Biologists A Missense Mutation in the Arabidopsis COPII Coat Protein Sec24A Induces the Formation of
THE ROLE OF SEC24 IN PROTEIN EXPORT FROM THE PLANT ENDOPLASMIC RETICULUM
 THE ROLE OF SEC24 IN PROTEIN EXPORT FROM THE PLANT ENDOPLASMIC RETICULUM A Thesis Submitted to the College of Graduate Studies and Research in Partial Fulfillment of the Requirements for the Degree in
THE ROLE OF SEC24 IN PROTEIN EXPORT FROM THE PLANT ENDOPLASMIC RETICULUM A Thesis Submitted to the College of Graduate Studies and Research in Partial Fulfillment of the Requirements for the Degree in
Signal Transduction. Dr. Chaidir, Apt
 Signal Transduction Dr. Chaidir, Apt Background Complex unicellular organisms existed on Earth for approximately 2.5 billion years before the first multicellular organisms appeared.this long period for
Signal Transduction Dr. Chaidir, Apt Background Complex unicellular organisms existed on Earth for approximately 2.5 billion years before the first multicellular organisms appeared.this long period for
A novel di-acidic motif facilitates ER export of the syntaxin SYP31
 Journal of Experimental Botany, Vol. 60, No. 11, pp. 3157 3165, 2009 doi:10.1093/jxb/erp155 Advance Access publication 10 June, 2009 This paper is available online free of all access charges (see http://jxb.oxfordjournals.org/open_access.html
Journal of Experimental Botany, Vol. 60, No. 11, pp. 3157 3165, 2009 doi:10.1093/jxb/erp155 Advance Access publication 10 June, 2009 This paper is available online free of all access charges (see http://jxb.oxfordjournals.org/open_access.html
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
www.sciencemag.org/cgi/content/full/1121356/dc1 Supporting Online Material for Polar PIN Localization Directs Auxin Flow in Plants Justyna Wiśniewska, Jian Xu, Daniela Seifertová, Philip B. Brewer, Kamil
Molecular Cell Biology 5068 In Class Exam 1 September 30, Please print your name:
 Molecular Cell Biology 5068 In Class Exam 1 September 30, 2014 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
Molecular Cell Biology 5068 In Class Exam 1 September 30, 2014 Exam Number: Please print your name: Instructions: Please write only on these pages, in the spaces allotted and not on the back. Write your
GTP hydrolysis by Sar1 mediates proof-reading for protein sorting into COPII vesicles
 JBC Papers in Press. Published on November 19, 2003 as Manuscript C300457200 GTP hydrolysis by Sar1 mediates proof-reading for protein sorting into COPII vesicles Ken Sato 1,2,4 and Akihiko Nakano 1,3
JBC Papers in Press. Published on November 19, 2003 as Manuscript C300457200 GTP hydrolysis by Sar1 mediates proof-reading for protein sorting into COPII vesicles Ken Sato 1,2,4 and Akihiko Nakano 1,3
Differential effects of a GTP-restricted mutant of Sar1p on segregation of cargo during export from the endoplasmic reticulum
 Research Article 3635 Differential effects of a GTP-restricted mutant of Sar1p on segregation of cargo during export from the endoplasmic reticulum David J. Stephens 1, * and Rainer Pepperkok 2 1 Department
Research Article 3635 Differential effects of a GTP-restricted mutant of Sar1p on segregation of cargo during export from the endoplasmic reticulum David J. Stephens 1, * and Rainer Pepperkok 2 1 Department
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells
 Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Looking for LOV: Location of LOV1 function in Nicotiana benthamiana cells By: Patrick Rutledge 1 Dr. Jennifer Lorang 2,3, Dr. Marc Curtis 2,3, Dr. Thomas Wolpert 2,3 BioResource Research 1, Botany and
Importance of Protein sorting. A clue from plastid development
 Importance of Protein sorting Cell organization depend on sorting proteins to their right destination. Cell functions depend on sorting proteins to their right destination. Examples: A. Energy production
Importance of Protein sorting Cell organization depend on sorting proteins to their right destination. Cell functions depend on sorting proteins to their right destination. Examples: A. Energy production
Transmembrane Domains (TMDs) of ABC transporters
 Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Ron et al SUPPLEMENTAL DATA
 Ron et al SUPPLEMENTAL DATA Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model Mily Ron, Kaisa Kajala,
Ron et al SUPPLEMENTAL DATA Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model Mily Ron, Kaisa Kajala,
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Lecture 6 - Intracellular compartments and transport I
 01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
A model for the self-organization of exit sites in the endoplasmic reticulum
 Research Article 55 A model for the self-organization of exit sites in the endoplasmic reticulum Stephan Heinzer 1, Stefan Wörz 2,3, Claudia Kalla 1, Karl Rohr 2,3 and Matthias Weiss 1, * 1 Cellular Biophysics
Research Article 55 A model for the self-organization of exit sites in the endoplasmic reticulum Stephan Heinzer 1, Stefan Wörz 2,3, Claudia Kalla 1, Karl Rohr 2,3 and Matthias Weiss 1, * 1 Cellular Biophysics
The neuron as a secretory cell
 The neuron as a secretory cell EXOCYTOSIS ENDOCYTOSIS The secretory pathway. Transport and sorting of proteins in the secretory pathway occur as they pass through the Golgi complex before reaching the
The neuron as a secretory cell EXOCYTOSIS ENDOCYTOSIS The secretory pathway. Transport and sorting of proteins in the secretory pathway occur as they pass through the Golgi complex before reaching the
Supplemental Data. Perrella et al. (2013). Plant Cell /tpc
 Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Intensity Intensity Intensity Intensity Intensity Intensity 150 50 150 0 10 20 50 C 150 0 10 20 50 D 0 10 20 Distance (μm) 50 20 40 E 50 F 0 10 20 50 0 15 30 Distance (μm) Supplemental Figure 1: Co-localization
Heather Currinn, Benjamin Guscott, Zita Balklava, Alice Rothnie and Thomas Wassmer*
 Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
7.06 Problem Set
 7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
Supplementary Information. The protease GtgE from Salmonella exclusively targets. inactive Rab GTPases
 Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Selective Targeting of ER Exit Sites Supports Axon Development
 Traffic 2009; 10: 1669 1684 2009 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2009.00974.x Selective Targeting of ER Exit Sites Supports Axon Development Meir Aridor 1 and Kenneth N. Fish 2, 1 Department
Traffic 2009; 10: 1669 1684 2009 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2009.00974.x Selective Targeting of ER Exit Sites Supports Axon Development Meir Aridor 1 and Kenneth N. Fish 2, 1 Department
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Identification and functional analysis of acidic signal motifs
 ER export of the plant K + channel KAT1: Identification and functional analysis of acidic signal motifs Vom Fachbereich Biologie der Technischen Universität Darmstadt zur Erlangung des akademischen Grades
ER export of the plant K + channel KAT1: Identification and functional analysis of acidic signal motifs Vom Fachbereich Biologie der Technischen Universität Darmstadt zur Erlangung des akademischen Grades
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
GFP WxTP-GFP WxTP-GFP. stromules. DsRed WxTP-DsRed WxTP-DsRed. stromules
 Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Elucidating the recycling mechanism of ER resident proteins with ERD2
 Elucidating the recycling mechanism of ER resident proteins with ERD2 Jing An Submitted in accordance with the requirements for the degree of Doctor of Philosophy The University of Leeds Faculty of Biological
Elucidating the recycling mechanism of ER resident proteins with ERD2 Jing An Submitted in accordance with the requirements for the degree of Doctor of Philosophy The University of Leeds Faculty of Biological
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Last time: Obtaining information from a cloned gene
 Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Last time: Obtaining information from a cloned gene Objectives: 1. What is the biochemical role of the gene? 2. Where and when is the gene expressed (transcribed)? 3. Where and when is the protein made?
Cell Biology Review. The key components of cells that concern us are as follows: 1. Nucleus
 Cell Biology Review Development involves the collective behavior and activities of cells, working together in a coordinated manner to construct an organism. As such, the regulation of development is intimately
Cell Biology Review Development involves the collective behavior and activities of cells, working together in a coordinated manner to construct an organism. As such, the regulation of development is intimately
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis
 Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis Ken Sato 1,2 & Akihiko Nakano 1,3 COPII coat proteins are required for direct capture of cargo and SNARE
Dissection of COPII subunit-cargo assembly and disassembly kinetics during Sar1p-GTP hydrolysis Ken Sato 1,2 & Akihiko Nakano 1,3 COPII coat proteins are required for direct capture of cargo and SNARE
The length of cargo-protein transmembrane segments drives secretory transport by facilitating cargo concentration in export domains
 JCS epress online publication date 12 May 2009 Research Article 1759 The length of cargo-protein transmembrane segments drives secretory transport by facilitating cargo concentration in export domains
JCS epress online publication date 12 May 2009 Research Article 1759 The length of cargo-protein transmembrane segments drives secretory transport by facilitating cargo concentration in export domains
Biochemical and Biophysical Research Communications 348 (2006)
 Biochemical and Biophysical Research Communications 348 (2006) 908 915 BBRC www.elsevier.com/locate/ybbrc An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg 2+ Yijian Rao
Biochemical and Biophysical Research Communications 348 (2006) 908 915 BBRC www.elsevier.com/locate/ybbrc An open conformation of switch I revealed by Sar1-GDP crystal structure at low Mg 2+ Yijian Rao
Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang Chen, Qiang Dong, Jiangqi Wen, Kirankumar S. Mysore and Jian Zhao
 New Phytologist Supporting Information Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bhlh transcription factor MtTT8 Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang
New Phytologist Supporting Information Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bhlh transcription factor MtTT8 Penghui Li, Beibei Chen, Gaoyang Zhang, Longxiang
Cellular Neuroanatomy I The Prototypical Neuron: Soma. Reading: BCP Chapter 2
 Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
targets. clustering show that different complex pathway
 Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Chapter 12: Intracellular sorting
 Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
Nature Genetics: doi: /ng Supplementary Figure 1. ssp mutant phenotypes in a functional SP background.
 Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Supplementary Figure 1 ssp mutant phenotypes in a functional SP background. (a,b) Statistical comparisons of primary and sympodial shoot flowering times as determined by mean values for leaf number on
Rab2 GTPase Regulates Vesicle Trafficking between the Endoplasmic Reticulum and the Golgi Bodies and Is Important to Pollen Tube Growth
 The Plant Cell, Vol. 14, 945 962, April 2002, www.plantcell.org 2002 American Society of Plant Biologists Rab2 GTPase Regulates Vesicle Trafficking between the Endoplasmic Reticulum and the Golgi Bodies
The Plant Cell, Vol. 14, 945 962, April 2002, www.plantcell.org 2002 American Society of Plant Biologists Rab2 GTPase Regulates Vesicle Trafficking between the Endoplasmic Reticulum and the Golgi Bodies
Regulation and signaling. Overview. Control of gene expression. Cells need to regulate the amounts of different proteins they express, depending on
 Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
AGD5 is a GTPase-activating protein at the trans-golgi network
 The Plant Journal (2010) 64, 790 799 doi: 10.1111/j.1365-313X.2010.04369.x AGD5 is a GTPase-activating protein at the trans-golgi network Giovanni Stefano 1,*,,, Luciana Renna 1,,, Marika Rossi 1, Elisa
The Plant Journal (2010) 64, 790 799 doi: 10.1111/j.1365-313X.2010.04369.x AGD5 is a GTPase-activating protein at the trans-golgi network Giovanni Stefano 1,*,,, Luciana Renna 1,,, Marika Rossi 1, Elisa
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
Effect of G15V and T20N Point Mutations of OsRacD from Oryza sativa on
 ISSN 100727626 CN 1123870ΠQ Chinese Journal of Biochemistry and Molecular Biology 2009 9 25 (9) :828 833 OsRacD G15 V T20 N 3,,,, (, 453007) OsRacD GTP Rho,,,., PCR, GTPase G15V T20N, GTP GDP OsRacD. CaMV35S
ISSN 100727626 CN 1123870ΠQ Chinese Journal of Biochemistry and Molecular Biology 2009 9 25 (9) :828 833 OsRacD G15 V T20 N 3,,,, (, 453007) OsRacD GTP Rho,,,., PCR, GTPase G15V T20N, GTP GDP OsRacD. CaMV35S
Division Ave. High School AP Biology
 Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Translocation through the endoplasmic reticulum membrane. Institute of Biochemistry Benoît Kornmann
 Translocation through the endoplasmic reticulum membrane Institute of Biochemistry Benoît Kornmann Endomembrane system Protein sorting Permeable to proteins but not to ions IgG tetramer (16 nm) Fully hydrated
Translocation through the endoplasmic reticulum membrane Institute of Biochemistry Benoît Kornmann Endomembrane system Protein sorting Permeable to proteins but not to ions IgG tetramer (16 nm) Fully hydrated
Name: TF: Section Time: LS1a ICE 5. Practice ICE Version B
 Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Supplemental material
 Supplemental material Table 1- Segregation analysis of sgt1a sgt1b double mutant plants Parental genotype No. of lines Normal seeds Phenotype Abnormal seeds Ratio of abnormal seeds (%) WT 3 171 2 1.16
Supplemental material Table 1- Segregation analysis of sgt1a sgt1b double mutant plants Parental genotype No. of lines Normal seeds Phenotype Abnormal seeds Ratio of abnormal seeds (%) WT 3 171 2 1.16
Activation of a receptor. Assembly of the complex
 Activation of a receptor ligand inactive, monomeric active, dimeric When activated by growth factor binding, the growth factor receptor tyrosine kinase phosphorylates the neighboring receptor. Assembly
Activation of a receptor ligand inactive, monomeric active, dimeric When activated by growth factor binding, the growth factor receptor tyrosine kinase phosphorylates the neighboring receptor. Assembly
Richik N. Ghosh, Linnette Grove, and Oleg Lapets ASSAY and Drug Development Technologies 2004, 2:
 1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
Stacks off tracks: a role for the golgin AtCASP in plant endoplasmic reticulum-golgi apparatus tethering
 Journal of Experimental Botany doi:10.1093/jxb/erx167 This paper is available online free of all access charges (see http://jxb.oxfordjournals.org/open_access.html for further details) RESEARCH PAPER Stacks
Journal of Experimental Botany doi:10.1093/jxb/erx167 This paper is available online free of all access charges (see http://jxb.oxfordjournals.org/open_access.html for further details) RESEARCH PAPER Stacks
Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition
 Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises
 Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
Supplementary Figure 1 Structure of the Orai channel. (a) The hexameric Drosophila Orai channel structure derived from crystallography 1 comprises six Orai subunits, each with identical amino acid sequences
Supplemental Materials Molecular Biology of the Cell
 Supplemental Materials Molecular iology of the Cell Figure S1 Krüger et al. Arabidopsis Plasmodium H. sapiens* 1) Xenopus* 1) Drosophila C.elegans S.cerevisae S.pombe L.major T.cruzi T.brucei DCP5 CITH
Supplemental Materials Molecular iology of the Cell Figure S1 Krüger et al. Arabidopsis Plasmodium H. sapiens* 1) Xenopus* 1) Drosophila C.elegans S.cerevisae S.pombe L.major T.cruzi T.brucei DCP5 CITH
Gene expression in prokaryotic and eukaryotic cells, Plasmids: types, maintenance and functions. Mitesh Shrestha
 Gene expression in prokaryotic and eukaryotic cells, Plasmids: types, maintenance and functions. Mitesh Shrestha Plasmids 1. Extrachromosomal DNA, usually circular-parasite 2. Usually encode ancillary
Gene expression in prokaryotic and eukaryotic cells, Plasmids: types, maintenance and functions. Mitesh Shrestha Plasmids 1. Extrachromosomal DNA, usually circular-parasite 2. Usually encode ancillary
Lecture 4. Protein Translocation & Nucleocytoplasmic Transport
 Lecture 4 Protein Translocation & Nucleocytoplasmic Transport Chapter 12 MBoC (5th Edition) Alberts et al. Reference paper: Tran and Wente, Cell 125, 1041-1053, 2006 2/8/2012 1 Page 713 Molecular Biology
Lecture 4 Protein Translocation & Nucleocytoplasmic Transport Chapter 12 MBoC (5th Edition) Alberts et al. Reference paper: Tran and Wente, Cell 125, 1041-1053, 2006 2/8/2012 1 Page 713 Molecular Biology
Golgi-Dependent Transport of Vacuolar Sorting Receptors Is Regulated by COPII, AP1, and AP4 Protein Complexes in Tobacco W OPEN
 The Plant Cell, Vol. 26: 1308 1329, March 2014, www.plantcell.org ã 2014 American Society of Plant Biologists. All rights reserved. Golgi-Dependent Transport of Vacuolar Sorting Receptors Is Regulated
The Plant Cell, Vol. 26: 1308 1329, March 2014, www.plantcell.org ã 2014 American Society of Plant Biologists. All rights reserved. Golgi-Dependent Transport of Vacuolar Sorting Receptors Is Regulated
COMPUTER SIMULATION OF DIFFERENTIAL KINETICS OF MAPK ACTIVATION UPON EGF RECEPTOR OVEREXPRESSION
 COMPUTER SIMULATION OF DIFFERENTIAL KINETICS OF MAPK ACTIVATION UPON EGF RECEPTOR OVEREXPRESSION I. Aksan 1, M. Sen 2, M. K. Araz 3, and M. L. Kurnaz 3 1 School of Biological Sciences, University of Manchester,
COMPUTER SIMULATION OF DIFFERENTIAL KINETICS OF MAPK ACTIVATION UPON EGF RECEPTOR OVEREXPRESSION I. Aksan 1, M. Sen 2, M. K. Araz 3, and M. L. Kurnaz 3 1 School of Biological Sciences, University of Manchester,
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
Stress Effects on Myosin Mutant Root Length in Arabidopsis thaliana
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2011 Stress Effects on Myosin
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange University of Tennessee Honors Thesis Projects University of Tennessee Honors Program 5-2011 Stress Effects on Myosin
7.06 Spring 2004 PS 6 KEY 1 of 14
 7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
Supplementary information
 Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature17991 Supplementary Discussion Structural comparison with E. coli EmrE The DMT superfamily includes a wide variety of transporters with 4-10 TM segments 1. Since the subfamilies of the
doi:10.1038/nature17991 Supplementary Discussion Structural comparison with E. coli EmrE The DMT superfamily includes a wide variety of transporters with 4-10 TM segments 1. Since the subfamilies of the
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis.
 Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice Supplementary Figure 2.
 Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Supplementary Figure 1. Markedly decreased numbers of marginal zone B cells in DOCK8 mutant mice. Percentage of marginal zone B cells in the spleen of wild-type mice (+/+), mice homozygous for cpm or pri
Chapter 1. DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d
 Chapter 1 1. Matching Questions DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d 2. Matching Questions : Unbranched polymer that, when folded into its three-dimensional shape,
Chapter 1 1. Matching Questions DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d 2. Matching Questions : Unbranched polymer that, when folded into its three-dimensional shape,
Analysis of correlated mutations in Ras G-domain
 www.bioinformation.net Volume 13(6) Hypothesis Analysis of correlated mutations in Ras G-domain Ekta Pathak * Bioinformatics Department, MMV, Banaras Hindu University. Ekta Pathak - E-mail: ektavpathak@gmail.com;
www.bioinformation.net Volume 13(6) Hypothesis Analysis of correlated mutations in Ras G-domain Ekta Pathak * Bioinformatics Department, MMV, Banaras Hindu University. Ekta Pathak - E-mail: ektavpathak@gmail.com;
Waithe et al Supplementary Figures
 Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Waithe et al Supplementary Figures Supplementary Figure 1 Expression and properties of WT and W391A mutant YFP- Ca V 2.2. A Immunoblot using Ca V 2.2 Ab for untransfected cells (UT, lane 1), YFP-Ca V 2.2
Nobel Lecture. The Principle of Membrane Fusion in the Cell. James E. Rothman Yale University. Karolinska Institutet Stockholm December 7, 2013
 Nobel Lecture James E. Rothman Yale University The Principle of Membrane Fusion in the Cell Karolinska Institutet Stockholm December 7, 2013 Stockholm 10:32 am October 7, 2013 Dr. Göran Hansson Manhattan
Nobel Lecture James E. Rothman Yale University The Principle of Membrane Fusion in the Cell Karolinska Institutet Stockholm December 7, 2013 Stockholm 10:32 am October 7, 2013 Dr. Göran Hansson Manhattan
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
CHAPTER 3. Cell Structure and Genetic Control. Chapter 3 Outline
 CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
CHAPTER 3 Cell Structure and Genetic Control Chapter 3 Outline Plasma Membrane Cytoplasm and Its Organelles Cell Nucleus and Gene Expression Protein Synthesis and Secretion DNA Synthesis and Cell Division
SUPPLEMENTARY INFORMATION
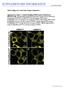 doi:10.1038/nature09606 Chen Li-Qing et al. A novel class of sugar transporters. Supplementary Figure 1. Confocal imaging of FRET sensor localization in HEK293T cells. The subcellular localization of the
doi:10.1038/nature09606 Chen Li-Qing et al. A novel class of sugar transporters. Supplementary Figure 1. Confocal imaging of FRET sensor localization in HEK293T cells. The subcellular localization of the
Chapter 6: A Tour of the Cell
 Chapter 6: A Tour of the Cell 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when Robert Hooke first looked at dead cells from an oak tree.
Chapter 6: A Tour of the Cell 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when Robert Hooke first looked at dead cells from an oak tree.
7.06 Cell Biology EXAM #3 KEY
 7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
Transport between cytosol and nucleus
 of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
SUPPLEMENTARY INFORMATION
 Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
Supplementary Figure 1 Sns and Duf co-localise in embryonic nephrocytes a-c, Wild-type stage 11 (a),14 (b) and 16 (c) embryos stained with anti-duf (green) and anti-sns (red). Higher magnification images
JCB Article. ARF-GAP mediated interaction between the ER-Golgi v-snares and the COPI coat
 JCB Article ARF-GAP mediated interaction between the ER-Golgi v-snares and the COPI coat Ulrike Rein, 1 Uwe Andag, 2 Rainer Duden, 3 Hans Dieter Schmitt, 2 and Anne Spang 1 1 Friedrich Miescher Laboratory,
JCB Article ARF-GAP mediated interaction between the ER-Golgi v-snares and the COPI coat Ulrike Rein, 1 Uwe Andag, 2 Rainer Duden, 3 Hans Dieter Schmitt, 2 and Anne Spang 1 1 Friedrich Miescher Laboratory,
Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures
 Nature Methods Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures Yannick Doyon, Thuy D Vo, Matthew C Mendel, Shon G Greenberg, Jianbin Wang, Danny F Xia, Jeffrey
Nature Methods Enhanced zinc-finger-nuclease activity with improved obligate heterodimeric architectures Yannick Doyon, Thuy D Vo, Matthew C Mendel, Shon G Greenberg, Jianbin Wang, Danny F Xia, Jeffrey
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a
 Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
