Received 7 March 2010/Accepted 16 June 2010
|
|
|
- Gregory Johnson
- 5 years ago
- Views:
Transcription
1 APPLIED AND ENVIRONMENTAL MICROBIOLOGY, Aug. 2010, p Vol. 76, No /10/$12.00 doi: /aem Copyright 2010, American Society for Microbiology. All Rights Reserved. The Genome Sequence of the Crenarchaeon Acidilobus saccharovorans Supports a New Order, Acidilobales, and Suggests an Important Ecological Role in Terrestrial Acidic Hot Springs Andrey V. Mardanov, 1 Vitali A. Svetlitchnyi, 1 Alexey V. Beletsky, 1 Maria I. Prokofeva, 2 Elizaveta A. Bonch-Osmolovskaya, 2 Nikolai V. Ravin, 1 * and Konstantin G. Skryabin 1 * Centre Bioengineering, Russian Academy of Sciences, Moscow , Russia, 1 and Winogradsky Institute of Microbiology, Russian Academy of Sciences, Moscow , Russia 2 Received 7 March 2010/Accepted 16 June 2010 Acidilobus saccharovorans is an anaerobic, organotrophic, thermoacidophilic crenarchaeon isolated from a terrestrial hot spring. We report the complete genome sequence of A. saccharovorans, which has permitted the prediction of genes for Embden-Meyerhof and Entner-Doudoroff pathways and genes associated with the oxidative tricarboxylic acid cycle. The electron transfer chain is branched with two sites of proton translocation and is linked to the reduction of elemental sulfur and thiosulfate. The genomic data suggest an important role of the order Acidilobales in thermoacidophilic ecosystems whereby its members can perform a complete oxidation of organic substrates, closing the anaerobic carbon cycle. Acidophilic microorganisms are widely dispersed in natural acidic environments, including volcanic hot springs, and are, in the majority, aerobes (14). However, such anoxic, high-temperature, acidic environments are inhabited by metabolically versatile anaerobic thermoacidophiles of the archaeal phylum Crenarchaeota. Lithoautotrophic thermoacidophiles oxidize molecular hydrogen in the course of elemental sulfur (S 0 ) respiration. Organotrophs couple the oxidation of organic substrates to the reduction of S 0 or thiosulfate. They all belong to the genus Acidilobus in the family Acidilobaceae and to the genus Caldisphaera in the family Caldisphaeraceae (4, 13, 22, 24). Acidilobaceae and Caldisphaeraceae form the crenarchaeal order Acidilobales (24). Acidilobus saccharovorans was isolated from an acidic hot spring of Uzon Caldera, Kamchatka, Russia (24). It is an obligately anaerobic acidophile with a range of growth from ph 2.5 to 5.8 (optimum at ph 3.5 to 4) and a temperature range from 60 to 90 C (optimum at 80 to 85 C). It utilizes a wide range of proteinaceous and carbohydrate substrates and cannot grow lithoautotrophically on H 2 and CO 2 (24). S 0 and thiosulfate stimulate growth and are reduced to H 2 S. Protons cannot serve as electron acceptors, since no H 2 is produced during growth in the absence of S 0 (24). Genomic sequences of aerobic, thermoacidophilic euryarchaea Thermoplasma acidophilum (26) and Picrophilus torridus (8) give an insight into the thermoacidophilic survival strategy. However, no genomes of obligately anaerobic, thermoacidophilic archaea were available until now. Here we present the genome of * Corresponding author. Mailing address: Centre Bioengineering, RAS, Prospekt 60-let Oktyabrya, Bldg. 7-1, Moscow , Russia. Phone for N. V. Ravin: (7) Fax: (7) nravin@biengi.ac.ru. Phone for K. G. Skryabin: (7) Fax: (7) skryabin@biengi.ac.ru. Present address: Direvo Industrial Biotechnology GmbH, D Cologne, Germany. Supplemental material for this article may be found at Published ahead of print on 25 June A. saccharovorans and show that it encodes numerous hydrolytic enzymes and metabolic pathways necessary for the utilization and complete mineralization of organic substrates in its natural habitat, acidic hot springs. Genome sequencing and annotation. A. saccharovorans T (DSM 16705) was obtained from the culture collection of the Laboratory of Hyperthermophilic Microbial Communities, the Winogradsky Institute of Microbiology of the Russian Academy of Sciences, and grown as described previously (24). The A. saccharovorans genome was sequenced on a Roche GS FLX genome sequencer by the standard protocol for a shotgun genome library. The GS FLX run (1/4 plate) resulted in the generation of about 22 Mb of sequences with an average read length of 230 bp. The GS FLX reads were assembled into 11 contigs by a GS De Novo Assembler (Roche). The contigs were oriented into scaffolds, and the complete genome sequence was obtained upon the generation and sequencing of appropriate PCR fragments. The rrna genes were identified with RNAmmer (17). trna genes were located with trnascan-se (19). Protein-encoding genes were identified with the GLIMMER gene finder (5), followed by a round of manual curation. Genome annotation was performed with the AutoFACT tool (16). Protein similarity was analyzed by BLASTP search (1). Signal peptides were predicted with SignalP version 3.0 (3). General features of the genome. A. saccharovorans has a single circular chromosome of 1,496,453 bp with no extrachromosomal elements (see Fig. S1 in the supplemental material). There is a single copy of the 16S-23S rrna operon and a single distantly located 5S rrna gene. A total of 45 trna genes coding for all 20 amino acids are scattered over the genome. The cumulative GC skew profile (9, 18) displays two local minima that likely correspond to the DNA replication origins (see Fig. S1 in the supplemental material). Genome annotation reveals 1,499 potential protein-encoding genes, 972 (65%) of which can be functionally assigned. The functions of the remaining 527 genes cannot be predicted from the deduced 5652
2 VOL. 76, 2010 ACIDILOBUS SACCHAROVORANS GENOME SEQUENCE 5653 FIG. 1. Comparisons of proteomes of A. saccharovorans and other archaea. (A) The numbers of A. saccharovorans protein-encoding genes present in the genomes of other archaea. Genes were considered present in a pair of genomes if the region of similarity covered 70% of the corresponding protein with an E value of Abbreviations: Asac, A. saccharovorans; Aper, Aeropyrum pernix; Dkam, Desulfurococcus kamchatkensis; Hbut, Hyperthermus butylicus; Ihos, Ignicoccus hospitalis; Smar, Staphylothermus marinus; Ssol, Sulfolobus solfataricus; Sacid, Sulfolobus acidocaldarius; Stok, Sulfolobus tokodaii; Msed, Metallosphaera sedula; Cmaq, Caldivirga maquilingensis; Tpen, Thermofilum pendens; Pisl, Pyrobaculum islandicum; Tneut, Thermoproteus neutrophilus; Ptor, Picrophilus torridus; Tacid, Thermoplasma acidophilum; Pfur, Pyrococcus furiosus; Aful, Archaeoglobus fulgidus. (B) The best BLASTP hits of A. saccharovorans proteins. Significant BLASTP hits were detected for 1,218 of 1,499 proteins found in the A. saccharovorans genome. Proteins unique to A. saccharovorans were excluded from this analysis. amino acid sequences; 246 of them are unique to A. saccharovorans. The numbers of predicted protein-encoding genes of A. saccharovorans that have homologs in other archaeal genomes are shown in Fig. 1A. The closest relative of A. saccharovorans is the aerobic, neutrophilic crenarchaeon Aeropyrum pernix; they share about half of their proteomes ( 740 proteins). Fewer A. saccharovorans proteins have homologs in the genomes of other members of the crenarchaeal orders Desulfurococcales and Thermoproteales (Fig. 1B). Notably, A. saccharovorans shares higher numbers of proteins (661 to 687, 16% of best BLASTP hits) with the phylogenetically distant aerobic thermoacidophiles of the order Sulfolobales (24). Predicted functions of most of the 199 proteins of A. saccharovorans showing the best BLASTP hits in Sulfolobales proteomes are related to transport (54 proteins) or are unknown (86 proteins). A. saccharovorans and Sulfolobales often coexist in acidic hot springs, and lateral gene transfer may be responsible for the relatively high similarity between their proteomes. Extensive lateral gene transfer was suggested to explain the high fraction of genes shared by Sulfolobales and aerobic euryarchaeal thermoacidophiles P. torridus and T. acidophilum (8, 26). However, it is unlikely that extensive gene transfer occurred between A. saccharovorans and euryarchaeal thermoacidophiles, because the numbers of proteins shared with P. torridus (428) and T. acidophilum (459), as well as with nonacidophilic euryarchaea Pyrococcus furiosus (440) and Archaeoglobus fulgidus (427), are similar. Metabolism of proteins, carbohydrates, and lipids. Growth of A. saccharovorans on proteins and peptides may be enabled by the function of several extracellular proteolytic enzymes. The imported peptides can be fermented with concomitant substrate-level phosphorylation to generate ATP (Fig. 2; see Table S1 in the supplemental material). The presence of genes for extra- and intracellular glycoside hydrolases and sugar transporters (Fig. 2 and Table S1) correlates with the growth of A. saccharovorans on carbohydrates (24). Notably, the A. saccharovorans genome encodes at least two primary ABC-type sugar transport systems (Table S1) and more than 20 secondary transporters. A similar high ratio of secondary transport systems to ATP-consuming primary transport systems was found in the thermoacidophile P. torridus (8), suggesting that the high external proton concentration is extensively used by A. saccharovorans for transport processes. Glucose in archaea is metabolized to pyruvate via either the Embden-Meyerhof (EM) pathway or the Entner-Doudoroff (ED) pathway. Both modified EM and ED pathways are encoded by A. saccharovorans genome (Fig. 2), suggesting their operation in parallel as found in Thermoproteus tenax (32). The ED pathway can be beneficial at the upper temperature range of growth, since it avoids the most heat-labile intermediates (31). The EM pathway may account for the synthesis of fructose-6-phosphate used in the ribulose monophosphate pathway (Fig. 2). The genome contains 14 genes encoding esterases and genes encoding a complete -oxidation pathway, suggesting the ability of A. saccharovorans to utilize triacylglycerides and longchain fatty acids available from the environment (Fig. 2 and Table S1). To our knowledge, besides A. saccharovorans, A. fulgidus is the only archaeon to encode the enzymes of -oxidation (15). Conservation of energy. A. saccharovorans may conserve energy by substrate-level phosphorylation as well as by oxidative phosphorylation. During growth in the presence of S 0 as a terminal electron acceptor, H 2 S is produced and acetate is formed (24). It is likely that in the presence of S 0, acetyl coenzyme A (acetyl-coa) generated from the oxidation of substrates is concomitantly converted to acetate by fermenta-
3 5654 MARDANOV ET AL. APPL. ENVIRON. MICROBIOL. tion pathway as well as oxidized to CO2 by anaerobic respiration (Fig. 2). The A. saccharovorans genome encodes all eight enzymes of the oxidative tricarboxylic acid (TCA) cycle (Fig. 2 and Table S1). This cycle operates in T. tenax and Pyrobaculum islandicum and was proposed to function in both oxidative and reductive directions, enabling the complete oxidation of or- ganic substrates to CO2 and H2S, as well as CO2 fixation during autotrophic growth (12, 30, 31, 32). In A. saccharovorans, which cannot grow autotrophically (24), the TCA cycle appears to operate only in the oxidative direction. Indeed, citrate lyase, the key enzyme of the reductive TCA cycle, is not encoded. FIG. 2. Overview of catabolic pathways encoded by the A. saccharovorans genome. Substrates utilized are in boldface, enzymes and proteins encoded by genes identified on the genome are in blue, and energy-rich intermediate compounds are in red. Panels: A, utilization of carbohydrates; B, utilization of proteins; C, glycolysis (EM and ED pathways); D, pyruvate degradation; E, pentose phosphate synthesis; F, -oxidation of long-chain fatty acids; G, formation of proton motive force coupled with ATP generation. TCA, oxidative tricarboxylic acid cycle. Abbreviations: POR, pyruvate:ferredoxin oxidoreductase; VOR, 2-ketoisovalerate:ferredoxin oxidoreductase; IOR, indolepyruvate:ferredoxin oxidoreductase; KGOR, 2-ketoglutarate:ferredoxin oxidoreductase; ACS, acetyl-coa synthetase; SCS, succinyl-coa synthetase; AOR, aldehyde:ferredoxin oxidoreductases; HK, ADP-dependent hexokinase; PGI, phosphoglucose isomerase; PFK, ADP-dependent phosphofructokinase; FBA, fructose-1,6-bisphosphate aldolase; GAPOR, glyceraldehyde-3-phosphate:ferredoxin oxidoreductase; GAPN, nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase; PGM, phosphoglycerate mutase; PYK, pyruvate kinase; PPDK, pyruvate phosphate dikinase; PEP, phosphoenolpyruvate; GDH, glucose dehydrogenase; GAD, gluconate dehydratase; KDGK, 2-keto-3-deoxy-gluconate kinase; KD(P)GA, 2-keto-3-deoxy-(6-phospho)gluconate aldolase; GK, glycerate kinase; PHI, 6-phospho-3-hexuloisomerase; HPS, 3-hexulose-6-phosphate synthase; RPI, ribose-5-phosphate isomerase; PPS, phosphoribosyl pyrophosphate synthase; FOR, formaldehyde:ferredoxin oxidoreductase; Fd(ox), oxidized ferredoxin; Fd(red), reduced ferredoxin; FNOR, ferredoxin:nad(p) oxidoreductase complex; NSR, NAD(P)H:elemental sulfur oxidoreductase; PPase, H -translocating pyrophosphatase; SQOR, succinate:quinone oxidoreductase complex; SR, sulfur reductase complex; Etf, electron transfer flavoprotein; Etf-QOR, electron transfer protein-quinone oxidoreductase; CoASH, coenzyme A; FAD, flavin adenine dinucleotide; FADH2, reduced FAD.
4 VOL. 76, 2010 ACIDILOBUS SACCHAROVORANS GENOME SEQUENCE 5655 During growth of A. saccharovorans by anaerobic respiration in the presence of S 0, reduced ferredoxin, NAD(P)H, and H produced in the TCA cycle can be oxidized by a concerted action of a set of membrane-bound protein complexes and cytoplasmic proteins, resulting in the generation of transmembrane proton gradient. The genome analysis reveals a putative membrane-bound protein complex (ASAC_0373 to ASAC_0383), which is partially homologous to bacterial NADH:quinone oxidoreductase (respiratory complex I). From the 14 subunits (NuoA to NuoN) present in Escherichia coli complex I (7), this putative A. saccharovorans complex contains 11 subunits (NuoA to NuoD and NuoH to NuoN), while three subunits (NuoEFG) known to form the dehydrogenase domain involved in NADH binding and oxidation (28) are missing. Thus, the A. saccharovorans complex most likely accepts electrons from a donor other than NADH. ASAC_0380 is also homologous to the MbxL subunit of the membrane-bound oxidoreductase (MBX) complex from P. furiosus (29). It has been shown that in the MBX complex, the oxidation of ferredoxin and the reduction of NAD(P) are coupled to the generation of a proton motive force in the presence of S 0 (29). A similar function of the MBX complex in the crenarchaea D. kamchatkensis (25) and S. marinus (2), as well as in the euryarchaeon T. sibiricus (20), has been proposed. Based on these data, we propose that ASAC_0373-ASAC_0383 encodes a proton-translocating ferredoxin:nad(p) oxidoreductase complex (FNOR) which accepts electrons from reduced ferredoxin and transfers them to NAD(P), thereby translocating protons and establishing a proton gradient (Fig. 2). NAD(P)H is then oxidized by the encoded NAD(P)H:elemental sulfur oxidoreductase (NSR) (ASAC_1028), forming H 2 S, or enters other processes linked to NAD(P)H reoxidation (e.g., thiosulfate reduction). In addition, the NuoB (Nqo6) subunit, which transfers electrons to menaquinone in Thermus thermophilus complex I (27), is present in the A. saccharovorans FNOR complex (ASAC_0382), suggesting the possibility of electron transfer not only to NAD(P) but also to quinones in the respiratory chain and further to membrane-bound sulfur reductase (SR; ASAC_1394 to ASAC_1397) (Fig. 2; see Table S1 in the supplemental material). The transmembrane proton gradient could also be generated by the function of the predicted integral membrane protein H -translocating pyrophosphatase (PPase; ASAC_1013) closely related to functionally characterized enzyme from Pyrobaculum aerophilum (6). The pyrophosphatase may work in concert with H -ATP synthase to scavenge energy from biosynthetic waste pyrophosphate in order to maintain the proton gradient, especially under energy-limiting conditions. Besides involving the proposed FNOR complex, the respiratory chain of A. saccharovorans presumably involves the succinate:quinone oxidoreductase complex (SQOR; ASAC_1440 to ASAC_1443), which is the membrane-bound component of the oxidative TCA cycle. Electrons from the oxidation of succinate in the TCA cycle would be delivered by SQOR to the quinones in the membrane. The genome contains a gene cluster (corresponding to ASAC_0264 to ASAC_0267) encoding two subunits of electron transfer flavoprotein (EtfAB; ASAC_0264 and ASAC_0265, respectively), a ferredoxin-like protein (ASAC_0266), and an electron transfer flavoprotein-quinone oxidoreductase (Etf- QOR; ASAC_0267). In analogy to the mitochondrial enzymes, ASAC_0264 to ASAC_0267 might be involved in the oxidation of fatty acids and some amino acids (10). These substrates would be oxidized by acyl-coa dehydrogenases (FadE) with the Etf as an acceptor, which would pass the electrons to Etf-QOR and further to quinone in the respiratory chain of A. saccharovorans. The genome analysis also suggests a second site of S 0 reduction in A. saccharovorans. It involves a predicted membranebound SR complex, which would function as the terminal oxidase of S 0 -dependent respiration. Based on the sequence analyses, we propose that the SR is composed of the hydrophilic catalytic subunit ASAC_1397, the electron transfer subunit ASAC_1396, and the membrane-bound subunits ASAC_1395 and ASAC_1394. Summarizing the data on the electron transfer in A. saccharovorans, we propose that its respiratory chain functioning during the growth with S 0 is branched (Fig. 2). First, the electrons from reduced ferredoxin could be utilized by FNOR and NSR. It would be linked to proton translocation and the first site of S 0 reduction. Second, a part of the electrons from FNOR, as well as the electrons from SQOR and Etf-QOR, would be transferred to the quinone pool. Oxidation of quinol by the SR would also be linked to proton translocation and the second site of reduction of S 0. During growth in the absence of S 0, the TCA cycle is not functional and the respiratory chain involving the SQOR complex, quinones, and SR seems not to operate. However, a proton gradient still could be established by the oxidation of reduced ferredoxin in the reaction of the proposed FNOR complex linked to reoxidation of NAD(P)H in reactions yielding reduced fermentation products. Some anaerobic Crenarchaeota were reported to use in addition to S 0 other terminal electron acceptors, including sulfate, thiosulfate, and nitrate. The presence of thiosulfate stimulated the growth of A. saccharovorans and led to production of H 2 S (24). We have predicted the function of ASAC_1397 as a component of membrane-bound SR complex (see above). Nevertheless, in analogy with thiosulfate reductase of Salmonella enterica involved also in the reduction of S 0 (11), the dual function of ASAC_1397 in the reduction of S 0 as well as of thiosulfate can be suggested. The established proton gradient can be used by ATP synthase for the synthesis of ATP. The genome of A. saccharovorans encodes a single A 1 A o ATP synthase composed of nine subunits (Table S1). The subunit K (c) (ASAC_0393) of the membrane-embedded motor does not contain conserved residues involved in Na binding in Na -ATP synthases (21), strongly suggesting that H is the coupling ion in the A 1 A o ATP synthase. During growth of A. saccharovorans in the absence of S 0, the required proton gradient could be established only partially by the function of the FNOR complex and H - translocating PPase. An additional proton gradient required for maintenance of normal intracellular ph could be generated by the reverse activity of A 1 A o ATP synthase. Possible ecological role of Acidilobales. Organic substances reaching terrestrial acidic hot springs from surrounding areas as well as produced by lithoautotrophic microorganisms serve as substrates for organotrophic thermoacidophiles. Metabolic activity of these organisms is supported by fast anaerobic oxidation of [ 14 C]sucrose to CO 2 in acidic hot springs (23). Cat-
5 5656 MARDANOV ET AL. APPL. ENVIRON. MICROBIOL. abolic pathways and energy generation mechanisms predicted from the genome of A. saccharovorans explain its growth on proteins, carbohydrates, and lipids (Fig. 2). Though phylogenetically Acidilobales are most closely related to Desulfurococcales, they metabolically resemble Thermoproteales. Similarly to T. tenax, A. saccharovorans possesses both the modified EM and ED pathways of glucose catabolism. Another feature common between A. saccharovorans and Thermoproteales is the presence of the TCA cycle, which enables a complete oxidation of organic substances in the presence of external anaerobic electron acceptors. Adaptation of A. saccharovorans to acidic conditions is reflected by a high ratio of secondary transporters to ATP-dependent primary transporters, the suggested reversibility of H -ATP synthase, the function of the encoded H - translocating PPase, and the proposed two proton translocation sites in the anaerobic respiratory chain. Physiological functions predicted by A. saccharovorans genome analysis (Fig. 2) and the wide distribution of Acidilobus spp. in acidic hot springs (23, 24) suggest an important role of Acidilobales in microbial communities of such habitats, closing the anaerobic carbon cycle through complete mineralization of organic substrates via S 0 respiration. Contrary to mesophilic microbial communities, in which the process of anaerobic destruction of organic substances consists of several stages performed by different groups of microorganisms (hydrolysis, fermentation, syntrophic reactions, and terminal oxidation), in thermal environments this could be achieved by a single group of organisms, Thermoproteales in neutral hot springs and Acidilobales in acidic ones. Nucleotide sequence accession number. The annotated genome sequence of A. saccharovorans T has been deposited in the GenBank database under accession number CP This work was supported by the Federal Agency for Science and Innovations of Russia (contract ). The work of M.I.P. and E.A.B.-O. on the analysis of growth characteristics of A. saccharovorans was supported by the Molecular and Cellular Biology program of RAS. REFERENCES 1. Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: Anderson, I. J., L. Dharmarajan, J. Rodriguez, S. Hooper, I. Porat, L. E. Ulrich, J. G. Elkins, K. Mavromatis, H. Sun, M. Land, A. Lapidus, S. Lucas, K. Barry, H. Huber, I. B. Zhulin, W. B. Whitman, B. Mukhopadhyay, C. Woese, J. Bristow, and N. Kyrpides The complete genome sequence of Staphylothermus marinus reveals differences in sulfur metabolism among heterotrophic Crenarchaeota. BMC Genomics 10: Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. J. Brunak Improved prediction of signal peptides: SignalP 3.0. Mol. Biol. 340: Boyd, E. S., R. A. Jackson, G. Encarnasion, J. A. Zahn, T. Beard, W. D. Leavitt, Y. Pi, C. L. Zhang, A. Pearson, and G. G. Geesey Isolation, characterization, and ecology of sulfur-respiring crenarchaea inhabiting acidsulfate-chloride-containing geothermal spring in Yellowstone National Park. Appl. Environ. Microbiol. 73: Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27: Drozdowicz, Y. M., Y. P. Lu, V. Patel, S. Fitz-Gibbon, J. H. Miller, and P. A. Rea A thermostable vacuolar-type membrane pyrophosphatase from the archaeon Pyrobaculum aerophilum: implications for the origins of pyrophosphate-energized pumps. FEBS Lett. 460: Friedrich, T., and D. Scheide The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 479: Fütterer, O., A. Angelov, H. Liesegang, G. Gottschalk, C. Schleper, B. Schepers, C. Dock, G. Antranikian, and W. Liebl Genome sequence of Picrophilus torridus and its implications for life around ph 0. Proc. Natl. Acad. Sci. U. S. A. 101: Grigoriev, A Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26: Herrmann, G., E. Jayamani, G. Mai, and W. Buckel Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J. Bacteriol. 190: Hinsley, A. P., and B. C. Berks Specificity of respiratory pathways involved in the reduction of sulfur compounds by Salmonella enterica. Microbiology 148: Hu, Y., and J. F. Holden Citric acid cycle in the hyperthermophilic archaeon Pyrobaculum islandicum grown autotrophically, heterotrophically, and mixotrophically with acetate. J. Bacteriol. 188: Itoh, T., K. Suzuki, P. C. Sanchez, and T. Nakase Caldisphaera lagunensis gen. nov., sp. nov., a novel thermoacidophilic crenarchaeote isolated from a hot spring at Mt Maquiling, Philippines. Int. J. Syst. Evol. Microbiol. 53: Johnson, D. B., and K. B. Hallberg Carbon, iron and sulfur metabolism in acidophilic microorganisms. Adv. Microb. Physiol. 54: Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, S. Peterson, C. I. Reich, L. K. McNeil, J. H. Badger, A. Glodek, L. Zhou, R. Overbeek, J. D. Gocayne, J. F. Weidman, L. McDonald, T. Utterback, M. D. Cotton, T. Spriggs, P. Artiach, B. P. Kaine, S. M. Sykes, P. W. Sadow, K. P. D Andrea, C. Bowman, C. Fujii, S. A. Garland, T. M. Mason, G. J. Olsen, C. M. Fraser, H. O. Smith, C. R. Woese, and J. C. Venter The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390: Koski, L. B., M. W. Gray, B. F. Langi, and G. Burger AutoFACT: an automatic functional annotation and classification tool. BMC Bioinformatics 6: Lagesen, K., P. Hallin, E. A. Rødland, H. H. Staerfeldt, T. Rognes, and D. W. Ussery RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35: Lobry, J. R Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13: Lowe, T., and S. R. Eddy trnascan-se: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: Mardanov, A. V., N. V. Ravin, V. A. Svetlitchnyi, A. V. Beletsky, M. L. Miroshnichenko, E. A. Bonch-Osmolovskaya, and K. G. Skryabin Metabolic versatility and indigenous origin of the archaeon Thermococcus sibiricus, isolated from a Siberian oil reservoir, as revealed by genome analysis. Appl. Environ. Microbiol. 75: Pisa, K. Y., H. Huber, M. Thomm, and V. Müller A sodium iondependent A 1 A O ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 274: Prokofeva, M. I., M. L. Miroshnichenko, N. A. Kostrikina, N. A. Chernyh, B. B. Kuznetsov, T. P. Tourova, and E. A. Bonch-Osmolovskaya Acidilobus aceticus gen. nov., sp. nov., a novel anaerobic thermoacidophilic archaeon from continental hot vents in Kamchatka. Int. J. Syst. Evol. Microbiol. 50: Prokofeva, M. I., I. I. Rusanov, N. V. Pimenov, and E. A. Bonch-Osmolovskaya Organotrophic activity in Kamchatka hot springs with low ph. Mikrobiologiia 75: Prokofeva, M. I., N. A. Kostrikina, T. V. Kolganova, T. P. Tourova, A. M. Lysenko, A. V. Lebedinsky, and E. A. Bonch-Osmolovskaya Isolation of the anaerobic thermoacidophilic crenarchaeote Acidilobus saccharovorans sp. nov. and proposal of Acidilobales ord. nov., including Acidilobaceae fam. nov. and Caldisphaeraceae fam. nov. Int. J. Syst. Evol. Microbiol. 59: Ravin, N. V., A. V. Mardanov, A. V. Beletsky, I. V. Kublanov, T. V. Kolganova, A. V. Lebedinsky, N. A. Chernyh, E. A. Bonch-Osmolovskaya, and K. G. Skryabin Complete genome sequence of the anaerobic, peptidefermenting hyperthermophilic crenarchaeon Desulfurococcus kamchatkensis. J. Bacteriol. 191: Ruepp, A., W. Graml, M. L. Santos-Martinez, K. K. Koretke, C. Volker, H. W. Mewes, D. Frishman, S. Stocker, A. N. Lupas, and W. Baumeister The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407: Sazanov, L. A., and P. Hinchliffe Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311: Sazanov, L. A Respiratory complex I: mechanistic and structural insights provided by the crystal structure of the hydrophilic domain. Biochemistry 46: Schut, G. J., S. L. Bridger, and M. W. Adams Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococ-
6 VOL. 76, 2010 ACIDILOBUS SACCHAROVORANS GENOME SEQUENCE 5657 cus furiosus: characterization of a coenzyme A-dependent NAD(P)H sulfur oxidoreductase. J. Bacteriol. 189: Selig, M., and P. Schönheit Oxidation of organic compounds to CO 2 with sulfur or thiosulfate as electron acceptor in the anaerobic hyperthermophilic archaea Thermoproteus tenax and Pyrobaculum islandicum proceeds via the citric acid cycle. Arch. Microbiol. 162: Siebers, B., B. Tjaden, K. Michalke, C. Dörr, H. Ahmed, M. Zaparty, P. Gordon, C. W. Sensen, A. Zibat, H. P. Klenk, S. C. Schuster, and R. Hensel Reconstruction of the central carbohydrate metabolism of Thermoproteus tenax by use of genomic and biochemical data. J. Bacteriol. 186: Zaparty, M., B. Tjaden, R. Hensel, and B. Siebers The central carbohydrate metabolism of the hyperthermophilic crenarchaeote Thermoproteus tenax: pathways and insights into their regulation. Arch. Microbiol. 190:
BBS2710 Microbial Physiology. Module 5 - Energy and Metabolism
 BBS2710 Microbial Physiology Module 5 - Energy and Metabolism Topics Energy production - an overview Fermentation Aerobic respiration Alternative approaches to respiration Photosynthesis Summary Introduction
BBS2710 Microbial Physiology Module 5 - Energy and Metabolism Topics Energy production - an overview Fermentation Aerobic respiration Alternative approaches to respiration Photosynthesis Summary Introduction
LIFE AT THE LIMITS - EXTREME ENVIRONMENTS "HOW CAN THEY SURVIVE?"
 COMPARATIVE MICROBIAL GENOMICS ANALYSIS LIFE AT THE LIMITS - EXTREME ENVIRONMENTS "HOW CAN THEY SURVIVE?" Group 5 Kanokwan Inban Lakkhana Khanhayuwa Parwin Tantayapirak Introduction 2 Source: http://pathmicro.med.sc.edu/fox/evol.gif
COMPARATIVE MICROBIAL GENOMICS ANALYSIS LIFE AT THE LIMITS - EXTREME ENVIRONMENTS "HOW CAN THEY SURVIVE?" Group 5 Kanokwan Inban Lakkhana Khanhayuwa Parwin Tantayapirak Introduction 2 Source: http://pathmicro.med.sc.edu/fox/evol.gif
Metabolism. Fermentation vs. Respiration. End products of fermentations are waste products and not fully.
 Outline: Metabolism Part I: Fermentations Part II: Respiration Part III: Metabolic Diversity Learning objectives are: Learn about respiratory metabolism, ATP generation by respiration linked (oxidative)
Outline: Metabolism Part I: Fermentations Part II: Respiration Part III: Metabolic Diversity Learning objectives are: Learn about respiratory metabolism, ATP generation by respiration linked (oxidative)
Center for Academic Services & Advising
 March 2, 2017 Biology I CSI Worksheet 6 1. List the four components of cellular respiration, where it occurs in the cell, and list major products consumed and produced in each step. i. Hint: Think about
March 2, 2017 Biology I CSI Worksheet 6 1. List the four components of cellular respiration, where it occurs in the cell, and list major products consumed and produced in each step. i. Hint: Think about
METABOLISM CHAPTER 04 BIO 211: ANATOMY & PHYSIOLOGY I. Dr. Lawrence G. Altman Some illustrations are courtesy of McGraw-Hill.
 BIO 211: ANATOMY & PHYSIOLOGY I CHAPTER 04 1 Please wait 20 seconds before starting slide show. Mouse click or Arrow keys to navigate. Hit ESCAPE Key to exit. CELLULAR METABOLISM Dr. Lawrence G. Altman
BIO 211: ANATOMY & PHYSIOLOGY I CHAPTER 04 1 Please wait 20 seconds before starting slide show. Mouse click or Arrow keys to navigate. Hit ESCAPE Key to exit. CELLULAR METABOLISM Dr. Lawrence G. Altman
CELL METABOLISM OVERVIEW Keep the big picture in mind as we discuss the particulars!
 BIO 211: ANATOMY & PHYSIOLOGY I CHAPTER 04 CELLULAR METABOLISM 1 Please wait 20 seconds before starting slide show. Mouse click or Arrow keys to navigate. Hit ESCAPE Key to exit. Dr. Lawrence G. Altman
BIO 211: ANATOMY & PHYSIOLOGY I CHAPTER 04 CELLULAR METABOLISM 1 Please wait 20 seconds before starting slide show. Mouse click or Arrow keys to navigate. Hit ESCAPE Key to exit. Dr. Lawrence G. Altman
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy
 Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of October
 Name: Class: _ Date: _ 2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of 19-23 October Multiple Choice Identify the choice that best completes the statement or answers the question. 1) Which
Name: Class: _ Date: _ 2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of 19-23 October Multiple Choice Identify the choice that best completes the statement or answers the question. 1) Which
Life 21 - Aerobic respiration Raven & Johnson Chapter 9 (parts)
 1 Life 21 - Aerobic respiration Raven & Johnson Chapter 9 (parts) Objectives 1: Describe the overall action of the Krebs cycle in generating ATP, NADH and FADH 2 from acetyl-coa 2: Understand the generation
1 Life 21 - Aerobic respiration Raven & Johnson Chapter 9 (parts) Objectives 1: Describe the overall action of the Krebs cycle in generating ATP, NADH and FADH 2 from acetyl-coa 2: Understand the generation
Oxidative Phosphorylation versus. Photophosphorylation
 Photosynthesis Oxidative Phosphorylation versus Photophosphorylation Oxidative Phosphorylation Electrons from the reduced cofactors NADH and FADH 2 are passed to proteins in the respiratory chain. In eukaryotes,
Photosynthesis Oxidative Phosphorylation versus Photophosphorylation Oxidative Phosphorylation Electrons from the reduced cofactors NADH and FADH 2 are passed to proteins in the respiratory chain. In eukaryotes,
Cellular Respiration: Harvesting Chemical Energy. 9.1 Catabolic pathways yield energy by oxidizing organic fuels
 Cellular Respiration: Harvesting Chemical Energy 9.1 Catabolic pathways yield energy by oxidizing organic fuels 9.2 Glycolysis harvests chemical energy by oxidizing glucose to pyruvate 9.3 The citric acid
Cellular Respiration: Harvesting Chemical Energy 9.1 Catabolic pathways yield energy by oxidizing organic fuels 9.2 Glycolysis harvests chemical energy by oxidizing glucose to pyruvate 9.3 The citric acid
Lectures by Kathleen Fitzpatrick
 Chapter 10 Chemotrophic Energy Metabolism: Aerobic Respiration Lectures by Kathleen Fitzpatrick Simon Fraser University Figure 10-1 Figure 10-6 Conversion of pyruvate The conversion of pyruvate to acetyl
Chapter 10 Chemotrophic Energy Metabolism: Aerobic Respiration Lectures by Kathleen Fitzpatrick Simon Fraser University Figure 10-1 Figure 10-6 Conversion of pyruvate The conversion of pyruvate to acetyl
Cell and Molecular Biology
 Cell and Molecular Biology (3000719): academic year 2013 Content & Objective :Cell Chemistry and Biosynthesis 3 rd Edition, 1994, pp. 41-88. 4 th Edition, 2002, pp. 47-127. 5 th Edition, 2008, pp. 45-124.
Cell and Molecular Biology (3000719): academic year 2013 Content & Objective :Cell Chemistry and Biosynthesis 3 rd Edition, 1994, pp. 41-88. 4 th Edition, 2002, pp. 47-127. 5 th Edition, 2008, pp. 45-124.
Giving you the energy you need!
 Giving you the energy you need! Use your dominant hand Open and close the pin (with your thumb and forefinger) as many times as you can for 20 seconds while holding the other fingers straight out! Repeat
Giving you the energy you need! Use your dominant hand Open and close the pin (with your thumb and forefinger) as many times as you can for 20 seconds while holding the other fingers straight out! Repeat
BIOCHEMISTRY. František Vácha. JKU, Linz.
 BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
Photosynthesis and Cellular Respiration
 Photosynthesis and Cellular Respiration Photosynthesis and Cellular Respiration All cellular activities require energy. Directly or indirectly nearly all energy for life comes from the sun. Autotrophs:
Photosynthesis and Cellular Respiration Photosynthesis and Cellular Respiration All cellular activities require energy. Directly or indirectly nearly all energy for life comes from the sun. Autotrophs:
Biochemical bases for energy transformations. Biochemical bases for energy transformations. Nutrition 202 Animal Energetics R. D.
 Biochemical bases for energy transformations Biochemical bases for energy transformations Nutrition 202 Animal Energetics R. D. Sainz Lecture 02 Energy originally from radiant sun energy Captured in chemical
Biochemical bases for energy transformations Biochemical bases for energy transformations Nutrition 202 Animal Energetics R. D. Sainz Lecture 02 Energy originally from radiant sun energy Captured in chemical
Reconstruction of the Central Carbohydrate Metabolism of Thermoproteus tenax by Use of Genomic and Biochemical Data
 JOURNAL OF BACTERIOLOGY, Apr. 2004, p. 2179 2194 Vol. 186, No. 7 0021-9193/04/$08.00 0 DOI: 10.1128/JB.186.7.2179 2194.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Reconstruction
JOURNAL OF BACTERIOLOGY, Apr. 2004, p. 2179 2194 Vol. 186, No. 7 0021-9193/04/$08.00 0 DOI: 10.1128/JB.186.7.2179 2194.2004 Copyright 2004, American Society for Microbiology. All Rights Reserved. Reconstruction
Physiological diversity
 Physiological diversity Principles Energetic considerations Biochemical pathways Organisms Ecological relevance Physiological diversity Sulfate- and nitrate reducers (5. Nov.) Methanogens and homoacetogens
Physiological diversity Principles Energetic considerations Biochemical pathways Organisms Ecological relevance Physiological diversity Sulfate- and nitrate reducers (5. Nov.) Methanogens and homoacetogens
Energy for biological processes
 1 Energy transfer When you have finished revising this topic, you should: be able to explain the difference between catabolic and anabolic reactions be able to describe the part played by in cell metabolism
1 Energy transfer When you have finished revising this topic, you should: be able to explain the difference between catabolic and anabolic reactions be able to describe the part played by in cell metabolism
Electron Transport Chain (Respiratory Chain) - exercise - Vladimíra Kvasnicová
 Electron Transport Chain (Respiratory Chain) - exercise - Vladimíra Kvasnicová Respiratory chain (RCH) a) is found in all cells b) is located in a mitochondrion c) includes enzymes integrated in the inner
Electron Transport Chain (Respiratory Chain) - exercise - Vladimíra Kvasnicová Respiratory chain (RCH) a) is found in all cells b) is located in a mitochondrion c) includes enzymes integrated in the inner
AQA Biology A-level Topic 5: Energy transfers in and between organisms
 AQA Biology A-level Topic 5: Energy transfers in and between organisms Notes Photosynthesis Photosynthesis is a reaction in which light energy is used to produce glucose in plants. The process requires
AQA Biology A-level Topic 5: Energy transfers in and between organisms Notes Photosynthesis Photosynthesis is a reaction in which light energy is used to produce glucose in plants. The process requires
Chapter 5. The Chloroplast. 5.1 Matter and Energy Pathways in Living Systems. Photosynthesis & Cellular Respiration
 Chapter 5 Photosynthesis & Cellular Respiration 5.1 Matter and Energy Pathways in Living Systems Both cellular respiration and photosynthesis are examples of biological processes that involve matter &
Chapter 5 Photosynthesis & Cellular Respiration 5.1 Matter and Energy Pathways in Living Systems Both cellular respiration and photosynthesis are examples of biological processes that involve matter &
Chapter 8 Metabolism: Energy, Enzymes, and Regulation
 Chapter 8 Metabolism: Energy, Enzymes, and Regulation Energy: Capacity to do work or cause a particular change. Thus, all physical and chemical processes are the result of the application or movement of
Chapter 8 Metabolism: Energy, Enzymes, and Regulation Energy: Capacity to do work or cause a particular change. Thus, all physical and chemical processes are the result of the application or movement of
Microbiology Helmut Pospiech.
 Microbiology 28.03.2018 Helmut Pospiech http://www.thescientificcartoonist.com/?p=107 Energy metabolism of Microorganisms Fermentation ADP +Pi Motility ATP Active transport (nutrient uptake) Lactic Acid
Microbiology 28.03.2018 Helmut Pospiech http://www.thescientificcartoonist.com/?p=107 Energy metabolism of Microorganisms Fermentation ADP +Pi Motility ATP Active transport (nutrient uptake) Lactic Acid
2. In regards to the fluid mosaic model, which of the following is TRUE?
 General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
METABOLISM. What is metabolism? Categories of metabolic reactions. Total of all chemical reactions occurring within the body
 METABOLISM What is metabolism? METABOLISM Total of all chemical reactions occurring within the body Categories of metabolic reactions Catabolic reactions Degradation pathways Anabolic reactions Synthesis
METABOLISM What is metabolism? METABOLISM Total of all chemical reactions occurring within the body Categories of metabolic reactions Catabolic reactions Degradation pathways Anabolic reactions Synthesis
Division Ave. High School AP Biology
 Overview 10 reactions u convert () to pyruvate (3C) u produces: 4 & NADH u consumes: u net: & NADH C-C-C-C-C-C fructose-1,6bp P-C-C-C-C-C-C-P DHAP P-C-C-C G3P C-C-C-P H P i P i pyruvate C-C-C 4 4 NAD +
Overview 10 reactions u convert () to pyruvate (3C) u produces: 4 & NADH u consumes: u net: & NADH C-C-C-C-C-C fructose-1,6bp P-C-C-C-C-C-C-P DHAP P-C-C-C G3P C-C-C-P H P i P i pyruvate C-C-C 4 4 NAD +
Review Quizzes Chapters 6-10
 Review Quizzes Chapters 6-10 1. The product(s) of the light reactions of photosynthesis is/are a. pyruvate b. glucose c. ATP and NADPH d. CO 2 and H 2 O e. ribulose bisphosphate (RuBP) 1. The product(s)
Review Quizzes Chapters 6-10 1. The product(s) of the light reactions of photosynthesis is/are a. pyruvate b. glucose c. ATP and NADPH d. CO 2 and H 2 O e. ribulose bisphosphate (RuBP) 1. The product(s)
Energy in the World of Life
 Cellular Energy Energy in the World of Life Sustaining life s organization requires ongoing energy inputs Assembly of the molecules of life starts with energy input into living cells Energy Conversion
Cellular Energy Energy in the World of Life Sustaining life s organization requires ongoing energy inputs Assembly of the molecules of life starts with energy input into living cells Energy Conversion
Energy for Life 12/11/14. Light Absorption in Chloroplasts
 Energy for Life Biochemical pathways A series of reactions where the products of one reaction is used in the next reaction Light Absorption in Chloroplasts Chloroplasts Two membranes Grana- layered stacks
Energy for Life Biochemical pathways A series of reactions where the products of one reaction is used in the next reaction Light Absorption in Chloroplasts Chloroplasts Two membranes Grana- layered stacks
Edexcel (B) Biology A-level
 Edexcel (B) Biology A-level Topic 5: Energy for Biological Processes Notes Aerobic Respiration Aerobic respiration as splitting of the respiratory substrate, to release carbon dioxide as a waste product
Edexcel (B) Biology A-level Topic 5: Energy for Biological Processes Notes Aerobic Respiration Aerobic respiration as splitting of the respiratory substrate, to release carbon dioxide as a waste product
The Life of a Cell. The Chemistry of Life. A View of the Cell. Cellular Transport and the Cell Cycle. Energy in a Cell
 The Life of a Cell The Chemistry of Life A View of the Cell Cellular Transport and the Cell Cycle Energy in a Cell Chapter 9 Energy in a Cell 9.1: The Need for Energy 9.1: Section Check 9.2: Photosynthesis:
The Life of a Cell The Chemistry of Life A View of the Cell Cellular Transport and the Cell Cycle Energy in a Cell Chapter 9 Energy in a Cell 9.1: The Need for Energy 9.1: Section Check 9.2: Photosynthesis:
2.A.2- Capture and Storage of Free Energy
 2.A.2- Capture and Storage of Free Energy Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce, and to maintain dynamic homeostasis. EU 2.A- Growth, reproduction
2.A.2- Capture and Storage of Free Energy Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce, and to maintain dynamic homeostasis. EU 2.A- Growth, reproduction
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 AP Exam Chapters 9 and 10; Photosynthesis and Respiration Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Carbon dioxide (CO2) is released
AP Exam Chapters 9 and 10; Photosynthesis and Respiration Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Carbon dioxide (CO2) is released
OBJECTIVES OUTCOMES FORMATIVE ASSESSMENT SUMMATIVE ASSESSMENT
 GOAL: A great diversity of metabolic and biochemical are seen among the Bacteria, Archaea, and Eukarya yet some common metabolic pathways exist across all domains. OBJECTIVES OUTCOMES FORMATIVE ASSESSMENT
GOAL: A great diversity of metabolic and biochemical are seen among the Bacteria, Archaea, and Eukarya yet some common metabolic pathways exist across all domains. OBJECTIVES OUTCOMES FORMATIVE ASSESSMENT
Chapter 5: Photosynthesis: The Energy of Life pg : Pathways of Photosynthesis pg
 UNIT 2: Metabolic Processes Chapter 5: Photosynthesis: The Energy of Life pg. 210-240 5.2: Pathways of Photosynthesis pg. 220-228 Light Dependent Reactions Photosystem II and I are the two light capturing
UNIT 2: Metabolic Processes Chapter 5: Photosynthesis: The Energy of Life pg. 210-240 5.2: Pathways of Photosynthesis pg. 220-228 Light Dependent Reactions Photosystem II and I are the two light capturing
Bio102 Problems Photosynthesis
 Bio102 Problems Photosynthesis 1. Why is it advantageous for chloroplasts to have a very large (in surface area) thylakoid membrane contained within the inner membrane? A. This limits the amount of stroma
Bio102 Problems Photosynthesis 1. Why is it advantageous for chloroplasts to have a very large (in surface area) thylakoid membrane contained within the inner membrane? A. This limits the amount of stroma
Lecture 10. Proton Gradient-dependent ATP Synthesis. Oxidative. Photo-Phosphorylation
 Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation Model of the Electron Transport Chain (ETC) Glycerol-3-P Shuttle Outer Mitochondrial Membrane G3P DHAP
Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation Model of the Electron Transport Chain (ETC) Glycerol-3-P Shuttle Outer Mitochondrial Membrane G3P DHAP
A + B = C C + D = E E + F = A
 Photosynthesis - Plants obtain energy directly from the sun - Organisms that do this are autotrophs (make their own food from inorganic forms) - Photosynthesis is a series of chemical reactions where the
Photosynthesis - Plants obtain energy directly from the sun - Organisms that do this are autotrophs (make their own food from inorganic forms) - Photosynthesis is a series of chemical reactions where the
Unit 5 Cellular Energy
 Unit 5 Cellular Energy I. Enzymes (159) 1.Are CATALYSTS: Speed up chemical reactions that would otherwise happen too slowly to support life. Catalysts DO NOT make reactions happen that couldn t happen
Unit 5 Cellular Energy I. Enzymes (159) 1.Are CATALYSTS: Speed up chemical reactions that would otherwise happen too slowly to support life. Catalysts DO NOT make reactions happen that couldn t happen
Metabolism Review. A. Top 10
 A. Top 10 Metabolism Review 1. Energy production through chemiosmosis a. pumping of H+ ions onto one side of a membrane through protein pumps in an Electron Transport Chain (ETC) b. flow of H+ ions across
A. Top 10 Metabolism Review 1. Energy production through chemiosmosis a. pumping of H+ ions onto one side of a membrane through protein pumps in an Electron Transport Chain (ETC) b. flow of H+ ions across
Chapter 5. Table of Contents. Section 1 Energy and Living Things. Section 2 Photosynthesis. Section 3 Cellular Respiration
 Photosynthesis and Cellular Respiration Table of Contents Section 1 Energy and Living Things Section 2 Photosynthesis Section 3 Cellular Respiration Section 1 Energy and Living Things Objectives Analyze
Photosynthesis and Cellular Respiration Table of Contents Section 1 Energy and Living Things Section 2 Photosynthesis Section 3 Cellular Respiration Section 1 Energy and Living Things Objectives Analyze
Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Outer Glycolysis mitochondrial membrane Glucose ATP
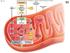 Fig. 7.5 uter Glycolysis mitochondrial membrane Glucose Intermembrane space xidation Mitochondrial matrix Acetyl-oA Krebs FAD e NAD + FAD Inner mitochondrial membrane e Electron e Transport hain hemiosmosis
Fig. 7.5 uter Glycolysis mitochondrial membrane Glucose Intermembrane space xidation Mitochondrial matrix Acetyl-oA Krebs FAD e NAD + FAD Inner mitochondrial membrane e Electron e Transport hain hemiosmosis
Remember, Earth s atmosphere was devoid of oxygen until about one billion years following the appearance of the first life forms.
 Chapter 8 (Strickberger 4 th ed.) Origins of Cells and the First Organisms Early organisms required energy to carry out life s functions. However this need for energy could only be satisfied once a system
Chapter 8 (Strickberger 4 th ed.) Origins of Cells and the First Organisms Early organisms required energy to carry out life s functions. However this need for energy could only be satisfied once a system
Bis2A 5.6: Oxidative Phosphorylation and the Electron Transport Chain *
 OpenStax-CNX module: m59707 1 Bis2A 5.6: Oxidative Phosphorylation and the Electron Transport Chain * The BIS2A Team This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution
OpenStax-CNX module: m59707 1 Bis2A 5.6: Oxidative Phosphorylation and the Electron Transport Chain * The BIS2A Team This work is produced by OpenStax-CNX and licensed under the Creative Commons Attribution
RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES. Bio 107 Week 6
 RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES Bio 107 Week 6 Procedure 7.2 Label test tubes well, including group name 1) Add solutions listed to small test tubes 2)
RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES Bio 107 Week 6 Procedure 7.2 Label test tubes well, including group name 1) Add solutions listed to small test tubes 2)
AP Bio-Ms.Bell Unit#3 Cellular Energies Name
 AP Bio-Ms.Bell Unit#3 Cellular Energies Name 1. Base your answer to the following question on the image below. 7. Base your answer to the following question on Which of the following choices correctly
AP Bio-Ms.Bell Unit#3 Cellular Energies Name 1. Base your answer to the following question on the image below. 7. Base your answer to the following question on Which of the following choices correctly
MICROBIOLOGIA GENERALE. The Archaea
 MICROBIOLOGIA GENERALE The Archaea The Archaea s traits 1. The cell wall of Archaea: pseudopeptidoglycan, polysaccharide, glycoprotein 2. The cytoplasmic membrane of Archaea: ether linkage, glycerol
MICROBIOLOGIA GENERALE The Archaea The Archaea s traits 1. The cell wall of Archaea: pseudopeptidoglycan, polysaccharide, glycoprotein 2. The cytoplasmic membrane of Archaea: ether linkage, glycerol
Energy and Cellular Metabolism
 1 Chapter 4 About This Chapter Energy and Cellular Metabolism 2 Energy in biological systems Chemical reactions Enzymes Metabolism Figure 4.1 Energy transfer in the environment Table 4.1 Properties of
1 Chapter 4 About This Chapter Energy and Cellular Metabolism 2 Energy in biological systems Chemical reactions Enzymes Metabolism Figure 4.1 Energy transfer in the environment Table 4.1 Properties of
Physiological diversity. Recommended text books. Physiological diversity. Sulfate and nitrate reducers. ! Principles. ! Energetic considerations
 Physiological diversity Recommended text books! Principles! Energetic considerations! Biochemical pathways! Organisms! Ecological relevance Physiological diversity! Sulfate- and nitrate reducers (11. Nov.)!
Physiological diversity Recommended text books! Principles! Energetic considerations! Biochemical pathways! Organisms! Ecological relevance Physiological diversity! Sulfate- and nitrate reducers (11. Nov.)!
Located in the thylakoid membranes. Chlorophyll have Mg + in the center. Chlorophyll pigments harvest energy (photons) by absorbing certain
 a review Located in the thylakoid membranes. Chlorophyll have Mg + in the center. Chlorophyll pigments harvest energy (photons) by absorbing certain wavelengths (blue-420 nm and red-660 nm are most important).
a review Located in the thylakoid membranes. Chlorophyll have Mg + in the center. Chlorophyll pigments harvest energy (photons) by absorbing certain wavelengths (blue-420 nm and red-660 nm are most important).
ΔG o' = ηf ΔΕ o' = (#e ( V mol) ΔΕ acceptor
 Reading: Sec. 19.1 Electron-Transfer Reactions in Mitochondria (listed subsections only) 19.1.1 Electrons are Funneled to Universal Electron Acceptors p. 692/709 19.1.2 Electrons Pass through a Series
Reading: Sec. 19.1 Electron-Transfer Reactions in Mitochondria (listed subsections only) 19.1.1 Electrons are Funneled to Universal Electron Acceptors p. 692/709 19.1.2 Electrons Pass through a Series
CHAPTER 15 Metabolism: Basic Concepts and Design
 CHAPTER 15 Metabolism: Basic Concepts and Design Chapter 15 An overview of Metabolism Metabolism is the sum of cellular reactions - Metabolism the entire network of chemical reactions carried out by living
CHAPTER 15 Metabolism: Basic Concepts and Design Chapter 15 An overview of Metabolism Metabolism is the sum of cellular reactions - Metabolism the entire network of chemical reactions carried out by living
Mechanism of CO 2 Fixation: Rubisco Step 1
 dark means light-indendent! The Dark Reaction: Carbon Fixation Requires Rubisco: Ribulose-1,5-bisphosphate carboxylase RuBPi + C 6 x 3PG MW = 550, 000 g/mol Subunits eight small (14,000 g/mol) eight large
dark means light-indendent! The Dark Reaction: Carbon Fixation Requires Rubisco: Ribulose-1,5-bisphosphate carboxylase RuBPi + C 6 x 3PG MW = 550, 000 g/mol Subunits eight small (14,000 g/mol) eight large
Photosynthesis
 Student Expectations: Cellular Energy Understand that cellular energy is temporarily stored in the nucleotide ATP (adenosine triphosphate) Describe how energy is released by ATP When the outer phosphate
Student Expectations: Cellular Energy Understand that cellular energy is temporarily stored in the nucleotide ATP (adenosine triphosphate) Describe how energy is released by ATP When the outer phosphate
Lecture 2 Carbon and Energy Transformations
 1.018/7.30J Fall 2003 Fundamentals of Ecology Lecture 2 Carbon and Energy Transformations READINGS FOR NEXT LECTURE: Krebs Chapter 25: Ecosystem Metabolism I: Primary Productivity Luria. 1975. Overview
1.018/7.30J Fall 2003 Fundamentals of Ecology Lecture 2 Carbon and Energy Transformations READINGS FOR NEXT LECTURE: Krebs Chapter 25: Ecosystem Metabolism I: Primary Productivity Luria. 1975. Overview
Cellular Energy. How Organisms Obtain Energy Section 2: Photosynthesis Section 3: Cellular Respiration. Click on a lesson name to select.
 Section 1: How Organisms Obtain Energy Section 2: Photosynthesis Section 3: Cellular Respiration Click on a lesson name to select. Section 1 How Organisms Obtain Energy Transformation of Energy Energy
Section 1: How Organisms Obtain Energy Section 2: Photosynthesis Section 3: Cellular Respiration Click on a lesson name to select. Section 1 How Organisms Obtain Energy Transformation of Energy Energy
TCA Cycle. Voet Biochemistry 3e John Wiley & Sons, Inc.
 TCA Cycle Voet Biochemistry 3e Voet Biochemistry 3e The Electron Transport System (ETS) and Oxidative Phosphorylation (OxPhos) We have seen that glycolysis, the linking step, and TCA generate a large number
TCA Cycle Voet Biochemistry 3e Voet Biochemistry 3e The Electron Transport System (ETS) and Oxidative Phosphorylation (OxPhos) We have seen that glycolysis, the linking step, and TCA generate a large number
BIOLOGY 345 Midterm II - 15 November 2010 PART I. Multiple choice questions (4 points each, 32 points total).
 BIOLOGY 345 Name Midterm II - 15 November 2010 PART I. Multiple choice questions (4 points each, 32 points total). 1. Considering the multitude of potential metabolic processes available to Bacteria and
BIOLOGY 345 Name Midterm II - 15 November 2010 PART I. Multiple choice questions (4 points each, 32 points total). 1. Considering the multitude of potential metabolic processes available to Bacteria and
CHLOROPLASTS, CALVIN CYCLE, PHOTOSYNTHETIC ELECTRON TRANSFER AND PHOTOPHOSPHORYLATION (based on Chapter 19 and 20 of Stryer )
 CHLOROPLASTS, CALVIN CYCLE, PHOTOSYNTHETIC ELECTRON TRANSFER AND PHOTOPHOSPHORYLATION (based on Chapter 19 and 20 of Stryer ) Photosynthesis Photosynthesis Light driven transfer of electron across a membrane
CHLOROPLASTS, CALVIN CYCLE, PHOTOSYNTHETIC ELECTRON TRANSFER AND PHOTOPHOSPHORYLATION (based on Chapter 19 and 20 of Stryer ) Photosynthesis Photosynthesis Light driven transfer of electron across a membrane
20. Electron Transport and Oxidative Phosphorylation
 20. Electron Transport and Oxidative Phosphorylation 20.1 What Role Does Electron Transport Play in Metabolism? Electron transport - Role of oxygen in metabolism as final acceptor of electrons - In inner
20. Electron Transport and Oxidative Phosphorylation 20.1 What Role Does Electron Transport Play in Metabolism? Electron transport - Role of oxygen in metabolism as final acceptor of electrons - In inner
Review Questions - Lecture 5: Metabolism, Part 1
 Review Questions - Lecture 5: Metabolism, Part 1 Questions: 1. What is metabolism? 2. What does it mean to say that a cell has emergent properties? 3. Define metabolic pathway. 4. What is the difference
Review Questions - Lecture 5: Metabolism, Part 1 Questions: 1. What is metabolism? 2. What does it mean to say that a cell has emergent properties? 3. Define metabolic pathway. 4. What is the difference
Cellular respiration. How do living things stay alive? Cellular Respiration Burning. Photosynthesis. Cellular Respiration
 How do living things stay alive? Cellular Respiration Burning Happens in ALL living things inside cells and has the main goal of producing ATP the fuel of life It does not matter whether the organisms
How do living things stay alive? Cellular Respiration Burning Happens in ALL living things inside cells and has the main goal of producing ATP the fuel of life It does not matter whether the organisms
Cellular Respiration. The mechanism of creating cellular energy. Thursday, 11 October, 12
 Cellular Respiration The mechanism of creating cellular energy What do we know?? What do we know?? Grade 5 - Food --> Energy What do we know?? Grade 5 - Food --> Energy Grade 10 - glu. + O2 --> CO2 + H20
Cellular Respiration The mechanism of creating cellular energy What do we know?? What do we know?? Grade 5 - Food --> Energy What do we know?? Grade 5 - Food --> Energy Grade 10 - glu. + O2 --> CO2 + H20
Be sure to understand:
 Learning Targets & Focus Questions for Unit 6: Bioenergetics Chapter 8: Thermodynamics Chapter 9: Cell Resp Focus Q Ch. 10: Photosynthesis Chapter 8 (141-150) 1. I can explain how living systems adhere
Learning Targets & Focus Questions for Unit 6: Bioenergetics Chapter 8: Thermodynamics Chapter 9: Cell Resp Focus Q Ch. 10: Photosynthesis Chapter 8 (141-150) 1. I can explain how living systems adhere
Enzymes I. Dr. Mamoun Ahram Summer semester,
 Enzymes I Dr. Mamoun Ahram Summer semester, 2017-2018 Resources Mark's Basic Medical Biochemistry Other resources NCBI Bookshelf: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=books The Medical Biochemistry
Enzymes I Dr. Mamoun Ahram Summer semester, 2017-2018 Resources Mark's Basic Medical Biochemistry Other resources NCBI Bookshelf: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=books The Medical Biochemistry
Photosynthesis: The Calvin Cycle
 Whoops! Wrong Calvin 1950s 1961 Photosynthesis: The Calvin Cycle Remember what it means to be a plant Need to produce all organic molecules necessary for growth carbohydrates, lipids, proteins, nucleic
Whoops! Wrong Calvin 1950s 1961 Photosynthesis: The Calvin Cycle Remember what it means to be a plant Need to produce all organic molecules necessary for growth carbohydrates, lipids, proteins, nucleic
Biological Process Term Enrichment
 Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
CHAPTER XI PHOTOSYNTHESIS. DMA: Chapter 11 Hartmann's 1
 CHAPTER XI PHOTOSYNTHESIS DMA: Chapter 11 Hartmann's 1 The nature of light The sun's energy travels through space to the earth as electromagnetic radiation waves at the speed of light, about 300,000 Km/s.
CHAPTER XI PHOTOSYNTHESIS DMA: Chapter 11 Hartmann's 1 The nature of light The sun's energy travels through space to the earth as electromagnetic radiation waves at the speed of light, about 300,000 Km/s.
Photosynthesis and Cellular Respiration
 Photosynthesis and Cellular Respiration Outline I. Energy and Carbon Cycle II. Photosynthesis A. Introduction B. Reactions II. Cellular Respiration A. Introduction B. Reactions Carbon Cycle All organisms
Photosynthesis and Cellular Respiration Outline I. Energy and Carbon Cycle II. Photosynthesis A. Introduction B. Reactions II. Cellular Respiration A. Introduction B. Reactions Carbon Cycle All organisms
MOLECULAR ACTIVITIES OF PLANT CELLS
 MOLECULAR ACTIVITIES OF PLANT CELLS An introduction to plant biochemistry JOHN W. ANDERSON BAgrSc, PhD Reader, Botany Department, School of Biological Sciences, La Trobe University, Bundoora, Victoria,
MOLECULAR ACTIVITIES OF PLANT CELLS An introduction to plant biochemistry JOHN W. ANDERSON BAgrSc, PhD Reader, Botany Department, School of Biological Sciences, La Trobe University, Bundoora, Victoria,
Name Date Class. Photosynthesis and Respiration
 Concept Mapping Photosynthesis and Respiration Complete the Venn diagram about photosynthesis and respiration. These terms may be used more than once: absorbs, Calvin cycle, chlorophyll, CO 2, H 2 O, Krebs
Concept Mapping Photosynthesis and Respiration Complete the Venn diagram about photosynthesis and respiration. These terms may be used more than once: absorbs, Calvin cycle, chlorophyll, CO 2, H 2 O, Krebs
Energy Metabolism exergonic reaction endergonic reaction Energy of activation
 Metabolism Energy Living things require energy to grow and reproduce Most energy used originates from the sun Plants capture 2% of solar energy Some captured energy is lost as metabolic heat All energy
Metabolism Energy Living things require energy to grow and reproduce Most energy used originates from the sun Plants capture 2% of solar energy Some captured energy is lost as metabolic heat All energy
1. Which of the following species have strains that are capable of undergoing the process of conjugation?
 Biology 3340 Summer 2005 Second Examination Version A Name Be sure to put your name on the mark-sense sheet as well Directions: Write your name in the correct space on the mark-sense sheet and the exam
Biology 3340 Summer 2005 Second Examination Version A Name Be sure to put your name on the mark-sense sheet as well Directions: Write your name in the correct space on the mark-sense sheet and the exam
GO ID GO term Number of members GO: translation 225 GO: nucleosome 50 GO: calcium ion binding 76 GO: structural
 GO ID GO term Number of members GO:0006412 translation 225 GO:0000786 nucleosome 50 GO:0005509 calcium ion binding 76 GO:0003735 structural constituent of ribosome 170 GO:0019861 flagellum 23 GO:0005840
GO ID GO term Number of members GO:0006412 translation 225 GO:0000786 nucleosome 50 GO:0005509 calcium ion binding 76 GO:0003735 structural constituent of ribosome 170 GO:0019861 flagellum 23 GO:0005840
Respiration and Photosynthesis
 Respiration and Photosynthesis Cellular Respiration Glycolysis The Krebs Cycle Electron Transport Chains Anabolic Pathway Photosynthesis Calvin Cycle Flow of Energy Energy is needed to support all forms
Respiration and Photosynthesis Cellular Respiration Glycolysis The Krebs Cycle Electron Transport Chains Anabolic Pathway Photosynthesis Calvin Cycle Flow of Energy Energy is needed to support all forms
All organisms require a constant expenditure of energy to maintain the living state - "LIFE".
 CELLULAR RESPIRATION All organisms require a constant expenditure of energy to maintain the living state - "LIFE". Where does the energy come from and how is it made available for life? With rare exception,
CELLULAR RESPIRATION All organisms require a constant expenditure of energy to maintain the living state - "LIFE". Where does the energy come from and how is it made available for life? With rare exception,
The products have more enthalpy and are more ordered than the reactants.
 hapters 7 & 10 Bioenergetics To live, organisms must obtain energy from their environment and use it to do the work of building and organizing cell components such as proteins, enzymes, nucleic acids,
hapters 7 & 10 Bioenergetics To live, organisms must obtain energy from their environment and use it to do the work of building and organizing cell components such as proteins, enzymes, nucleic acids,
Change to Office Hours this Friday and next Monday. Tomorrow (Abel): 8:30 10:30 am. Monday (Katrina): Cancelled (05/04)
 Change to Office Hours this Friday and next Monday Tomorrow (Abel): 8:30 10:30 am Monday (Katrina): Cancelled (05/04) Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation
Change to Office Hours this Friday and next Monday Tomorrow (Abel): 8:30 10:30 am Monday (Katrina): Cancelled (05/04) Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation
Catalysis. Instructor: Dr. Tsung-Lin Li Genomics Research Center Academia Sinica
 Catalysis Instructor: Dr. Tsung-Lin Li Genomics Research Center Academia Sinica References: Biochemistry" by Donald Voet and Judith G. Voet Biochemistry" by Christopher K. Mathews, K. E. Van Hold and Kevin
Catalysis Instructor: Dr. Tsung-Lin Li Genomics Research Center Academia Sinica References: Biochemistry" by Donald Voet and Judith G. Voet Biochemistry" by Christopher K. Mathews, K. E. Van Hold and Kevin
Dr. Mallery Biology 150 Workshop Fall Semester ENERGY and METABOLISM ANSWERS
 Dr. Mallery Biology 150 Workshop Fall Semester ENERGY and METABOLISM ANSWERS IN THE GRAND SCHEME The purpose of this workshop is to get the Learning Communities to begin thinking about why certain chemical
Dr. Mallery Biology 150 Workshop Fall Semester ENERGY and METABOLISM ANSWERS IN THE GRAND SCHEME The purpose of this workshop is to get the Learning Communities to begin thinking about why certain chemical
Forms of stored energy in cells
 Forms of stored energy in cells Electrochemical gradients Covalent bonds (ATP) Reducing power (NADH) During photosynthesis, respiration and glycolysis these forms of energy are converted from one to another
Forms of stored energy in cells Electrochemical gradients Covalent bonds (ATP) Reducing power (NADH) During photosynthesis, respiration and glycolysis these forms of energy are converted from one to another
Subsystem: TCA Cycle. List of Functional roles. Olga Vassieva 1 and Rick Stevens 2 1. FIG, 2 Argonne National Laboratory and University of Chicago
 Subsystem: TCA Cycle Olga Vassieva 1 and Rick Stevens 2 1 FIG, 2 Argonne National Laboratory and University of Chicago List of Functional roles Tricarboxylic acid cycle (TCA) oxidizes acetyl-coa to CO
Subsystem: TCA Cycle Olga Vassieva 1 and Rick Stevens 2 1 FIG, 2 Argonne National Laboratory and University of Chicago List of Functional roles Tricarboxylic acid cycle (TCA) oxidizes acetyl-coa to CO
PHOTOSYNTHESIS: A BRIEF STORY!!!!!
 PHOTOSYNTHESIS: A BRIEF STORY!!!!! This is one of the most important biochemical processes in plants and is amongst the most expensive biochemical processes in plant in terms of investment. Photosynthesis
PHOTOSYNTHESIS: A BRIEF STORY!!!!! This is one of the most important biochemical processes in plants and is amongst the most expensive biochemical processes in plant in terms of investment. Photosynthesis
Transformation of Energy! Energy is the ability to do work.! Thermodynamics is the study of the flow and transformation of energy in the universe.
 Section 1 How Organisms Obtain Energy Transformation of Energy! Energy is the ability to do work.! Thermodynamics is the study of the flow and transformation of energy in the universe. Section 1 How Organisms
Section 1 How Organisms Obtain Energy Transformation of Energy! Energy is the ability to do work.! Thermodynamics is the study of the flow and transformation of energy in the universe. Section 1 How Organisms
6CO 2 + 6H 2 O C 6 H 12 O 6 + 6O 2. sun. Occurs in chloroplasts ATP. enzymes CO 2 O 2 H 2 O. sugars
 4.2 8.2 Overview Photosynthesis: of Photosynthesis An Overview Photosynthesis process by which plants make food using energy from the sun Plants are autotrophs that make their own source of chemical energy.
4.2 8.2 Overview Photosynthesis: of Photosynthesis An Overview Photosynthesis process by which plants make food using energy from the sun Plants are autotrophs that make their own source of chemical energy.
Photosynthesis and Cellular Respiration Note-taking Guide
 Photosynthesis and Cellular Respiration Note-taking Guide Preview to Photosynthesis glucose, reactions, light-dependent, Calvin cycle, thylakoid, photosystem II, oxygen, light-harvesting, two, chloroplasts,
Photosynthesis and Cellular Respiration Note-taking Guide Preview to Photosynthesis glucose, reactions, light-dependent, Calvin cycle, thylakoid, photosystem II, oxygen, light-harvesting, two, chloroplasts,
Ch. 10 Photosynthesis: The Calvin Cycle Life from Air
 Ch. 10 Photosynthesis: The Calvin Cycle Life from Air 2007-2008 Whoops! Wrong Calvin The Calvin Cycle 1950s 1961 Remember what it means to be a plant Need to produce all organic molecules necessary for
Ch. 10 Photosynthesis: The Calvin Cycle Life from Air 2007-2008 Whoops! Wrong Calvin The Calvin Cycle 1950s 1961 Remember what it means to be a plant Need to produce all organic molecules necessary for
NAME ONE THING we have in common with plants. If
 Cellular Respiration NAME ONE THING we have in common with plants. If you said cellular respiration, you are right. That is one thing we have in common with plants, slugs, slime mold, and spiders. Living
Cellular Respiration NAME ONE THING we have in common with plants. If you said cellular respiration, you are right. That is one thing we have in common with plants, slugs, slime mold, and spiders. Living
f) Adding an enzyme does not change the Gibbs free energy. It only increases the rate of the reaction by lowering the activation energy.
 Problem Set 2-Answer Key BILD1 SP16 1) How does an enzyme catalyze a chemical reaction? Define the terms and substrate and active site. An enzyme lowers the energy of activation so the reaction proceeds
Problem Set 2-Answer Key BILD1 SP16 1) How does an enzyme catalyze a chemical reaction? Define the terms and substrate and active site. An enzyme lowers the energy of activation so the reaction proceeds
Photosynthesis and Life
 7-1 Chapter 7 Photosynthesis and Life During photosynthesis Organisms use the energy of light to build highenergy organic molecules. Plants, algae, and some bacteria can do this. Can make their own food
7-1 Chapter 7 Photosynthesis and Life During photosynthesis Organisms use the energy of light to build highenergy organic molecules. Plants, algae, and some bacteria can do this. Can make their own food
Courtesy of Elsevier. Used with permission.
 Chemistry 5.07 2013 Problem Set 5 Answers Problem 1 Succinate dehydrogenase (SDH) is a heterotetramer enzyme complex that catalyzes the oxidation of succinate to fumarate with concomitant reduction of
Chemistry 5.07 2013 Problem Set 5 Answers Problem 1 Succinate dehydrogenase (SDH) is a heterotetramer enzyme complex that catalyzes the oxidation of succinate to fumarate with concomitant reduction of
Biochemical Pathways
 Biochemical Pathways Living organisms can be divided into two large groups according to the chemical form in which they obtain carbon from the environment. Autotrophs can use carbon dioxide from the atmosphere
Biochemical Pathways Living organisms can be divided into two large groups according to the chemical form in which they obtain carbon from the environment. Autotrophs can use carbon dioxide from the atmosphere
CARBON FIXATION IN NITRIFIERS
 CARBON FIXATION IN NITRIFIERS Logan Hodgskiss, MSc PhD student Division of Archaea Biology and Ecogenomics Department of Ecogenomics and Systems Biology University of Vienna, Austria October 4 th, 2017
CARBON FIXATION IN NITRIFIERS Logan Hodgskiss, MSc PhD student Division of Archaea Biology and Ecogenomics Department of Ecogenomics and Systems Biology University of Vienna, Austria October 4 th, 2017
Subsystem: Succinate dehydrogenase
 Subsystem: Succinate dehydrogenase Olga Vassieva Fellowship for Interpretation of Genomes The super-macromolecular respiratory complex II (succinate:quinone oxidoreductase) couples the oxidation of succinate
Subsystem: Succinate dehydrogenase Olga Vassieva Fellowship for Interpretation of Genomes The super-macromolecular respiratory complex II (succinate:quinone oxidoreductase) couples the oxidation of succinate
Principles of Cellular Biology
 Principles of Cellular Biology آشنایی با مبانی اولیه سلول Biologists are interested in objects ranging in size from small molecules to the tallest trees: Cell Basic building blocks of life Understanding
Principles of Cellular Biology آشنایی با مبانی اولیه سلول Biologists are interested in objects ranging in size from small molecules to the tallest trees: Cell Basic building blocks of life Understanding
2. Cellular and Molecular Biology
 2. Cellular and Molecular Biology 2.1 Cell Structure 2.2 Transport Across Cell Membranes 2.3 Cellular Metabolism 2.4 DNA Replication 2.5 Cell Division 2.6 Biosynthesis 2.1 Cell Structure What is a cell?
2. Cellular and Molecular Biology 2.1 Cell Structure 2.2 Transport Across Cell Membranes 2.3 Cellular Metabolism 2.4 DNA Replication 2.5 Cell Division 2.6 Biosynthesis 2.1 Cell Structure What is a cell?
Hydrogen Metabolism and the Evolution of Biological Respiration
 Hydrogen Metabolism and the Evolution of Biological Respiration Two separate families of enzymes that oxidize hydrogen and also produce it arose through convergent evolution Eric S. Boyd, Gerrit J. Schut,
Hydrogen Metabolism and the Evolution of Biological Respiration Two separate families of enzymes that oxidize hydrogen and also produce it arose through convergent evolution Eric S. Boyd, Gerrit J. Schut,
Photosynthetic autotrophs use the energy of sunlight to convert low-g CO 2 and H 2 O into energy-rich complex sugar molecules.
 Chapters 7 & 10 Bioenergetics To live, organisms must obtain energy from their environment and use it to do the work of building and organizing cell components such as proteins, enzymes, nucleic acids,
Chapters 7 & 10 Bioenergetics To live, organisms must obtain energy from their environment and use it to do the work of building and organizing cell components such as proteins, enzymes, nucleic acids,
