Historical biogeography models with dispersal probability as a func7on of distance(s)
|
|
|
- Norman Riley
- 6 years ago
- Views:
Transcription
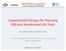
1 Historical biogeography models with dispersal probability as a func7on of distance(s) Nicholas J. Matzke, Postdoctoral Fellow, NIMBioS (Na6onal Ins6tute of Mathema6cal, SSE symposium: Fron6ers in Parametric Biogeography 10:45 am, Nobre Room, Guarujá, Brasil, June 30, 2015 Figure: Map of stochas6cally- mapped dispersal events BioGeoBEARS model: BAYAREALIKE- d- e+a+x+n Data from: Angiosperm megatree, Zanne et al. (2013, Nature)
2 Acknowledgements Ques6ons/comments/ collabora6ons at: (also: seeking a job) Thanks especially to: Jim Albert NIMBioS Brian O Meara Jeremy Beaulieu Ka?e Massana Michael Landis Ph.D. commicee John Huelsenbeck Tony Barnosky David Jablonski Roger Byrne Systema?c Biology editors & reviewers TRY IT YOURSELF AT: hmp://phylo.wikidot.com/biogeobears Funding: NIMBioS NSF Bivalves in Time and Space UC Berkeley Wang Fellowship UC Berkeley Tien Fellowship Google Summer of Code NIMBioS
3 Outline 1. Historical biogeography: What s the point? 2. Models morghulis 3. Models in BioGeoBEARS, and valida6on 4. Adding more realism with +x and +n 5. Es6ma6ng the global angiosperm macroevolu6onary dispersal kernel
4 Outline 1. Historical biogeography: What s the point? 2. Models morghulis 3. Models in BioGeoBEARS, and valida6on 4. Adding more realism with +x and +n 5. Es6ma6ng the global angiosperm macroevolu6onary dispersal kernel
5 Historical biogeography: What s the point? Tradi6onally, back to parsimony days, the point has been Ancestral Area Reconstruc6on e.g. Hawaiian Psychotria
6 Historical biogeography: What s the point? Sugges7on: let s replace Ancestral Area Reconstruc6on with Ancestral Range Es6ma6on (credit: Brian Moore) e.g. Hawaiian Psychotria
7 Historical biogeography: What s the point? Sugges7on: let s replace Ancestral Area Reconstruc6on with Ancestral Range Es6ma6on (credit: Brian Moore)
8 Historical biogeography: What s the point? Is Ancestral Range Es6ma6on the only point of historical biogeography? Not originally. The original hope was that by looking at many taxa, we could infer common pacerns and processes. (e.g.: General Area Cladograms, Lieberman- modified Brooks Parsimony Analysis)
9 Historical biogeography: What s the point? I think sta6s6cal model choice can bring process back into historical biogeography in a big way. We can do this by implemen6ng different models and seeing what probability they confer on the data (the likelihood). We don t expect this to be perfect, of course, but it is becer than just picking one model and not tes6ng it. - Already standard procedure in PCMs.
10 George Box All models are wrong, but some models are useful. This phrase is ubiquitous, but s6ll not kept in mind enough. Perhaps drama6za6on will help George E. P. Box ( )
11 For drama, look no further than HBO, 9 pm Sundays
12 In the free city of Braavos, the tradi7onal gree7ng is:
13 In the free city of Braavos, the tradi7onal gree7ng is:
14 In the free city of Braavos, the tradi7onal gree7ng is: The Faceless Man, Jaqen H'ghar
15 George E. P. Box ( )
16 George E. P. Box ( )
17 MODELS MODELS George E. P. Box ( ) MODELS MODELS
18 MODELS MODELS The Faceless Man Jaqen H'ghar George E. P. Box ( ) MODELS MODELS
19 MODELS MODELS The Faceless Man Jaqen H'ghar George E. P. Box ( ) MODELS MODELS
20 Models doeharis: All models must serve Let s make that happen with model comparison in BioGeoBEARS
21 Outline 1. Historical biogeography: What s the point? 2. Models morghulis 3. Models in BioGeoBEARS, and valida6on 4. Adding more realism with +x and +n 5. Es6ma6ng the global angiosperm macroevolu6onary dispersal kernel
22 Model comparison in BioGeoBEARS Example: DEC vs. DEC+J on Hawaiian Psychotria DEC LnL = DEC+J LnL = (tree & geog: Ree & Smith 2008)
23 Model comparison in BioGeoBEARS (for the +* model, set include_null_range=false) DEC* LnL = DEC*+J LnL = (tree & geog: Ree & Smith 2008)
24 Model comparison in BioGeoBEARS Example: DEC* vs. DEC*+J on Hawaiian Psychotria (for the * model, set include_null_range=false) LnL d e j DEC DEC+J DEC* (+) 0 DEC*+J (+) 0.12
25 Figure 1, Matzke 2013, Frontiers of Biogeography
26 Figure 1, Matzke 2013, Frontiers of Biogeography
27 Figure 1, Matzke 2013, Frontiers of Biogeography
28 Figure 1, Matzke 2013, Frontiers of Biogeography
29 Figure 1, Matzke 2013, Frontiers of Biogeography
30 Figure 1, Matzke 2013, Frontiers of Biogeography
31 Figure 1, Matzke 2013, Frontiers of Biogeography
32 Figure 1, Matzke 2013, Frontiers of Biogeography
33 Figure 1, Matzke 2013, Frontiers of Biogeography
34 Figure 1, Matzke 2013, Frontiers of Biogeography
35 Figure 1, Matzke 2013, Frontiers of Biogeography
36 Figure 1, Matzke 2013, Frontiers of Biogeography
37 Figure 1, Matzke 2013, Frontiers of Biogeography
38 DEC (LAGRANGE) Figure 1, Matzke 2013, Frontiers of Biogeography
39 DEC (LAGRANGE) Figure 1, Matzke 2013, Frontiers of Biogeography
40 DEC (LAGRANGE) Which model should we use? Figure 1, Matzke 2013, Frontiers of Biogeography
41 DEC (LAGRANGE) Figure 1, Matzke 2013, Frontiers of Biogeography
42 Model comparison in BioGeoBEARS DEC DEC+J
43 Model comparison in BioGeoBEARS DEC DEC+J DIVALIKE DIVALIKE+J
44 Model comparison in BioGeoBEARS DEC DEC+J DIVALIKE DIVALIKE+J BAYAREALIKE BAYAREALIKE+J
45 Model comparison in BioGeoBEARS DEC DEC+J DIVALIKE DIVALIKE+J BAYAREALIKE BAYAREALIKE+J DEC* DEC*+J DIVALIKE* DIVALIKE*+J BAYAREALIKE* BAYAREALIKE*+J
46 Model comparison in BioGeoBEARS AIC model weights across 14 datasets (assembled in Massana, Beaulieu, Matzke, O Meara, in prep.) (mostly island datasets from Matzke 2014) Across 14 datasets: Caecilians Cyrtandra Salamanders Leafhoppers Lonicera Drosophila Honeycreepers Megalagrion Orsonwelles Palpimanoid spiders Psychotria Plantago Scaptomyza Silverswords
47 What corresponds to the 3 models in RASP? AIC model weights across 14 datasets (assembled in Massana, Beaulieu, Matzke, O Meara, in prep.) (mostly island datasets from Matzke 2014) Across 14 datasets: Caecilians Cyrtandra Salamanders Leafhoppers Lonicera Drosophila Honeycreepers Megalagrion Orsonwelles Palpimanoid spiders Psychotria Plantago Scaptomyza Silverswords
48 What corresponds to the 3 models in RASP? AIC model weights across 14 datasets (assembled in Massana, Beaulieu, Matzke, O Meara, in prep.) (mostly island datasets from Matzke 2014) Across 14 datasets: Caecilians Cyrtandra Salamanders Leafhoppers Lonicera Drosophila Honeycreepers Megalagrion Orsonwelles Palpimanoid spiders Psychotria Plantago Scaptomyza Silverswords
49 Now we have 12 models. Is that the end? Nope. Models morghulis. Models dohaeris.
50 DEC (LAGRANGE) Figure 1, Matzke 2013, Frontiers of Biogeography
51 Anagene7c range- switching parameter, a In one sense, anagene6c range- switching is an absurd process (models morghulis) a But, it might be a decent approxima6on, if jump dispersal is the main dispersal mode, but many lineages are ex6nct or unsampled (models dohaeris) BAYAREALIKE*- d- e+a equals unordered character model (island model)
52 BAYAREALIKE*- d- e+a is a model we can use for valida7on BioGeoBEARS state probabili6es phytools state probabili6es (Revell)
53 Validate Biogeographical Stochas7c Mapping
54 BAYAREALIKE*- d- e+a is a model we can use for valida7on BioGeoBEARS, distribu6on of many Biogeographical Stochas7c Maps phytools state probabili6es (Revell)
55 Biogeographical Stochas7c Maps converge on state probabili7es BioGeoBEARS, distribu6on of many Biogeographical Stochas7c Maps DEC state probabili6es
56 DEC, DEC+J (BioGeoBEARS) vs. ClaSSE (diversitree) To make ClaSSE & DEC equivalent: DEC or DEC+J equals ClaSSE, *if*: - d and e control character change rates - all ex6nc6on rates set to zero - specia6on rate for each cladogenesis = (Yule rate) 6mes weight/sum(weights) - subtract equilibrium probabili6es at the root
57 DEC, DEC+J (BioGeoBEARS) vs. ClaSSE (diversitree) LnLs for: DEC- e = d DEC = e DEC+J = j
58 Outline 1. Historical biogeography: What s the point? 2. Models morghulis 3. Models in BioGeoBEARS, and valida6on 4. Adding more realism with +x and +n 5. Es6ma6ng the global angiosperm macroevolu6onary dispersal kernel
59 Standard approach: manual dispersal matrix One of the revolu6onary features of Lagrange was adding manual dispersal modifiers E.g., You might try a constrained model, where * dispersal from North America - > South America gets a mul6plier of 1 * dispersal from Africa - > South America gets a mul6plier of 0.1 * dispersal from Africa - > North America gets a mul6plier of 0.01
60 Standard approach: manual dispersal matrix These mul6pliers are subjec6ve, but I think such constrained models are useful. If you get improvement, it indicates some improved fit between the phylogene6c da6ng, distribu6ons, and observed geography at the 6ps If LnL gets worse, it indicates at least one of these is off (probably the da6ng really)
61 Distances Geographic distance = Great Circle Distance between area centroids (rescaled by dividing by min. observed distance) Environmental distance = Difference in absolute value of la7tude
62 New approach: es7mate dispersal matrix Let s try it. Phylogeny: Zanne et al. (2013), Nature, 15,000+ angiosperms Geography: median lat/long of species ranges Regions: Realms x Biomes (58 regions total) Assump7on: everything lives in 1 area
63 Realms x Biomes
64 Realms x Biomes
65 Realms x Biomes
66 Realms x Biomes
67 Realms x Biomes
68 Model comparison Base model: BAYAREALIKE-d-e +a means base rate of dispersal +x means dispersal prob. modified by distance^x +n means dispersal prob. modified by enviromental distance^n +J means weight of cladogenetic jump dispersal Param.LnL +a
69 Model comparison Base model: BAYAREALIKE-d-e +a means base rate of dispersal +x means dispersal prob. modified by distance^x +n means dispersal prob. modified by enviromental distance^n +J means weight of cladogenetic jump dispersal Param.LnL +a a+x
70 Model comparison Base model: BAYAREALIKE-d-e +a means base rate of dispersal +x means dispersal prob. modified by distance^x +n means dispersal prob. modified by enviromental distance^n +J means weight of cladogenetic jump dispersal Param.LnL +a a+x a+x+n
71 Model comparison Base model: BAYAREALIKE-d-e +a means base rate of dispersal +x means dispersal prob. modified by distance^x +n means dispersal prob. modified by enviromental distance^n +J means weight of cladogenetic jump dispersal Param.LnL a x n +a a+x a+x+n
72 Model comparison Base model: BAYAREALIKE-d-e +a means base rate of dispersal +x means dispersal prob. modified by distance^x +n means dispersal prob. modified by enviromental distance^n +J means weight of cladogenetic jump dispersal Param.LnL a x n +a a+x a+x+n J models -- no significant improvement (probably due to many missing species within genera?)
73 Model comparison Base model: BAYAREALIKE-d-e +a means base rate of dispersal +x means dispersal prob. modified by distance^x +n means dispersal prob. modified by enviromental distance^n +J means weight of cladogenetic jump dispersal Param.LnL a x n +a a+x a+x+n J models -- no significant improvement (probably due to many missing species within genera?) Both distance and environmental distance have big effects on angiosperm dispersal
74 Looking at the global angiosperm macroevolu7onary dispersal kernel
75 Looking at the global angiosperm macroevolu7onary dispersal kernel
76 Looking at the global angiosperm macroevolu7onary dispersal kernel
77 Looking at the global angiosperm macroevolu7onary dispersal kernel
78 Looking at the global angiosperm macroevolu7onary dispersal kernel
79 Looking at the global angiosperm macroevolu7onary dispersal kernel
80 Looking at the global angiosperm macroevolu7onary dispersal kernel
81 Looking at the global angiosperm macroevolu7onary dispersal kernel
82 Biogeographical stochas7c mapping
83 Biogeographical stochas7c mapping mapping
84 Biogeographical stochas7c mapping mapping
85 Biogeographical stochas7c mapping mapping
86 Outline 1. Historical biogeography: What s the point? ancestral ranges < learning about process 2. Models morghulis Models dohaeris 3. Models in BioGeoBEARS Test your hypotheses with model choice 4. Adding more realism with +x and +n Distance and environmental distance marer 5. Es6ma6ng the global angiosperm macroevolu6onary dispersal kernel
87 Acknowledgements Ques6ons/comments/ collabora6ons at: (also: seeking a job) Thanks especially to: Jim Albert NIMBioS Brian O Meara Jeremy Beaulieu Ka?e Massana Michael Landis Ph.D. commicee John Huelsenbeck Tony Barnosky David Jablonski Roger Byrne Systema?c Biology editors & reviewers TRY IT YOURSELF AT: hmp://phylo.wikidot.com/biogeobears Funding: NIMBioS NSF Bivalves in Time and Space UC Berkeley Wang Fellowship UC Berkeley Tien Fellowship Google Summer of Code NIMBioS
A primer on phylogenetic biogeography and DEC models. March 13, 2017 Michael Landis Bodega Bay Workshop Sunny California
 A primer on phylogenetic biogeography and DEC models March 13, 2017 Michael Landis Bodega Bay Workshop Sunny California Epidemiology Air travel communities H3N2 Influenza virus samples 2000 2007 CE Flu,
A primer on phylogenetic biogeography and DEC models March 13, 2017 Michael Landis Bodega Bay Workshop Sunny California Epidemiology Air travel communities H3N2 Influenza virus samples 2000 2007 CE Flu,
CS 6140: Machine Learning Spring What We Learned Last Week. Survey 2/26/16. VS. Model
 Logis@cs CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa@on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Assignment
Logis@cs CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa@on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Assignment
CS 6140: Machine Learning Spring 2016
 CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa?on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Logis?cs Assignment
CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa?on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Logis?cs Assignment
Model Selection in Historical Biogeography Reveals that Founder-Event Speciation Is a Crucial Process in Island Clades
 Syst. iol. 63(6):951 970, 2014 The uthor(s) 2014. Published by Oxford University Press, on behalf of the Society of Systematic iologists. ll rights reserved. For Permissions, please email: journals.permissions@oup.com
Syst. iol. 63(6):951 970, 2014 The uthor(s) 2014. Published by Oxford University Press, on behalf of the Society of Systematic iologists. ll rights reserved. For Permissions, please email: journals.permissions@oup.com
The Importance of Systema2cs and Rassenkreis. Reading: Willi Hennig
 The Importance of Systema2cs and Rassenkreis Reading: Throughout the class so far we seen that the distribu2on of an organism is the result of its biological history as well as geologic and clima4c history
The Importance of Systema2cs and Rassenkreis Reading: Throughout the class so far we seen that the distribu2on of an organism is the result of its biological history as well as geologic and clima4c history
Using Phylogenetic Comparative Methods To Understand Diversification and Geographic Range Evolution
 University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 5-2017 Using Phylogenetic Comparative Methods To Understand Diversification and
University of Tennessee, Knoxville Trace: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 5-2017 Using Phylogenetic Comparative Methods To Understand Diversification and
Some Review and Hypothesis Tes4ng. Friday, March 15, 13
 Some Review and Hypothesis Tes4ng Outline Discussing the homework ques4ons from Joey and Phoebe Review of Sta4s4cal Inference Proper4es of OLS under the normality assump4on Confidence Intervals, T test,
Some Review and Hypothesis Tes4ng Outline Discussing the homework ques4ons from Joey and Phoebe Review of Sta4s4cal Inference Proper4es of OLS under the normality assump4on Confidence Intervals, T test,
Sta$s$cal sequence recogni$on
 Sta$s$cal sequence recogni$on Determinis$c sequence recogni$on Last $me, temporal integra$on of local distances via DP Integrates local matches over $me Normalizes $me varia$ons For cts speech, segments
Sta$s$cal sequence recogni$on Determinis$c sequence recogni$on Last $me, temporal integra$on of local distances via DP Integrates local matches over $me Normalizes $me varia$ons For cts speech, segments
Maximum Likelihood Inference of Geographic Range Evolution by Dispersal, Local Extinction, and Cladogenesis
 Syst. Biol. 57(1):4 14, 2008 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150701883881 Maximum Likelihood Inference of Geographic Range Evolution
Syst. Biol. 57(1):4 14, 2008 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150701883881 Maximum Likelihood Inference of Geographic Range Evolution
CS 6140: Machine Learning Spring What We Learned Last Week 2/26/16
 Logis@cs CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa@on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Sign
Logis@cs CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa@on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Sign
Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2018 University of California, Berkeley
 Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2018 University of California, Berkeley B.D. Mishler Feb. 14, 2018. Phylogenetic trees VI: Dating in the 21st century: clocks, & calibrations;
Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2018 University of California, Berkeley B.D. Mishler Feb. 14, 2018. Phylogenetic trees VI: Dating in the 21st century: clocks, & calibrations;
Phylogene)cs. IMBB 2016 BecA- ILRI Hub, Nairobi May 9 20, Joyce Nzioki
 Phylogene)cs IMBB 2016 BecA- ILRI Hub, Nairobi May 9 20, 2016 Joyce Nzioki Phylogenetics The study of evolutionary relatedness of organisms. Derived from two Greek words:» Phle/Phylon: Tribe/Race» Genetikos:
Phylogene)cs IMBB 2016 BecA- ILRI Hub, Nairobi May 9 20, 2016 Joyce Nzioki Phylogenetics The study of evolutionary relatedness of organisms. Derived from two Greek words:» Phle/Phylon: Tribe/Race» Genetikos:
Greater host breadth still not associated with increased diversification rate in the Nymphalidae A response to Janz et al.
 doi:10.1111/evo.12914 Greater host breadth still not associated with increased diversification rate in the Nymphalidae A response to Janz et al. Christopher A. Hamm 1,2 and James A. Fordyce 3 1 Department
doi:10.1111/evo.12914 Greater host breadth still not associated with increased diversification rate in the Nymphalidae A response to Janz et al. Christopher A. Hamm 1,2 and James A. Fordyce 3 1 Department
Mul$ple Sequence Alignment Methods. Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu
 Mul$ple Sequence Alignment Methods Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu Species Tree Orangutan Gorilla Chimpanzee Human From the Tree of the Life
Mul$ple Sequence Alignment Methods Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu Species Tree Orangutan Gorilla Chimpanzee Human From the Tree of the Life
Sta$s$cal Significance Tes$ng In Theory and In Prac$ce
 Sta$s$cal Significance Tes$ng In Theory and In Prac$ce Ben Cartere8e University of Delaware h8p://ir.cis.udel.edu/ictir13tutorial Hypotheses and Experiments Hypothesis: Using an SVM for classifica$on will
Sta$s$cal Significance Tes$ng In Theory and In Prac$ce Ben Cartere8e University of Delaware h8p://ir.cis.udel.edu/ictir13tutorial Hypotheses and Experiments Hypothesis: Using an SVM for classifica$on will
SNPs versus sequences for phylogeography an explora:on using simula:ons and massively parallel sequencing in a non- model bird
 SNPs versus sequences for phylogeography an explora:on using simula:ons and massively parallel sequencing in a non- model bird Michael G. Harvey, Brian T. Smith, Brant C. Faircloth, Travis C. Glenn, and
SNPs versus sequences for phylogeography an explora:on using simula:ons and massively parallel sequencing in a non- model bird Michael G. Harvey, Brian T. Smith, Brant C. Faircloth, Travis C. Glenn, and
Reconstructing the history of lineages
 Reconstructing the history of lineages Class outline Systematics Phylogenetic systematics Phylogenetic trees and maps Class outline Definitions Systematics Phylogenetic systematics/cladistics Systematics
Reconstructing the history of lineages Class outline Systematics Phylogenetic systematics Phylogenetic trees and maps Class outline Definitions Systematics Phylogenetic systematics/cladistics Systematics
Algebra Exam. Solutions and Grading Guide
 Algebra Exam Solutions and Grading Guide You should use this grading guide to carefully grade your own exam, trying to be as objective as possible about what score the TAs would give your responses. Full
Algebra Exam Solutions and Grading Guide You should use this grading guide to carefully grade your own exam, trying to be as objective as possible about what score the TAs would give your responses. Full
Bayesian phylogenetics. the one true tree? Bayesian phylogenetics
 Bayesian phylogenetics the one true tree? the methods we ve learned so far try to get a single tree that best describes the data however, they admit that they don t search everywhere, and that it is difficult
Bayesian phylogenetics the one true tree? the methods we ve learned so far try to get a single tree that best describes the data however, they admit that they don t search everywhere, and that it is difficult
Historical Biogeography. Historical Biogeography. Historical Biogeography. Historical Biogeography
 "... that grand subject, that almost keystone of the laws of creation, Geographical Distribution" [Charles Darwin, 1845, in a letter to Joseph Dalton Hooker, the Director of the Royal Botanic Garden, Kew]
"... that grand subject, that almost keystone of the laws of creation, Geographical Distribution" [Charles Darwin, 1845, in a letter to Joseph Dalton Hooker, the Director of the Royal Botanic Garden, Kew]
An Introduc+on to Sta+s+cs and Machine Learning for Quan+ta+ve Biology. Anirvan Sengupta Dept. of Physics and Astronomy Rutgers University
 An Introduc+on to Sta+s+cs and Machine Learning for Quan+ta+ve Biology Anirvan Sengupta Dept. of Physics and Astronomy Rutgers University Why Do We Care? Necessity in today s labs Principled approach:
An Introduc+on to Sta+s+cs and Machine Learning for Quan+ta+ve Biology Anirvan Sengupta Dept. of Physics and Astronomy Rutgers University Why Do We Care? Necessity in today s labs Principled approach:
Test of neutral theory predic3ons for the BCI tree community informed by regional abundance data
 Test of neutral theory predic3ons for the BCI tree community informed by regional abundance data Anne%e Ostling Cody Weinberger Devin Riley Ecology and Evolu:onary Biology University of Michigan 1 Outline
Test of neutral theory predic3ons for the BCI tree community informed by regional abundance data Anne%e Ostling Cody Weinberger Devin Riley Ecology and Evolu:onary Biology University of Michigan 1 Outline
Machine Learning and Data Mining. Bayes Classifiers. Prof. Alexander Ihler
 + Machine Learning and Data Mining Bayes Classifiers Prof. Alexander Ihler A basic classifier Training data D={x (i),y (i) }, Classifier f(x ; D) Discrete feature vector x f(x ; D) is a con@ngency table
+ Machine Learning and Data Mining Bayes Classifiers Prof. Alexander Ihler A basic classifier Training data D={x (i),y (i) }, Classifier f(x ; D) Discrete feature vector x f(x ; D) is a con@ngency table
BIOGEOGRAPHY - BIOL 5010/ FALL 2017
 BIOGEOGRAPHY - BIOL 5010/6010 - FALL 2017 The study of the distribution of species and ecosystems in geographic space and through geological time MWF 2:30-3:20 pm, Biology/Natural Resources Building, room
BIOGEOGRAPHY - BIOL 5010/6010 - FALL 2017 The study of the distribution of species and ecosystems in geographic space and through geological time MWF 2:30-3:20 pm, Biology/Natural Resources Building, room
Methods to reconstruct phylogene1c networks accoun1ng for ILS
 Methods to reconstruct phylogene1c networks accoun1ng for ILS Céline Scornavacca some slides have been kindly provided by Fabio Pardi ISE-M, Equipe Phylogénie & Evolu1on Moléculaires Montpellier, France
Methods to reconstruct phylogene1c networks accoun1ng for ILS Céline Scornavacca some slides have been kindly provided by Fabio Pardi ISE-M, Equipe Phylogénie & Evolu1on Moléculaires Montpellier, France
CSE 473: Ar+ficial Intelligence. Hidden Markov Models. Bayes Nets. Two random variable at each +me step Hidden state, X i Observa+on, E i
 CSE 473: Ar+ficial Intelligence Bayes Nets Daniel Weld [Most slides were created by Dan Klein and Pieter Abbeel for CS188 Intro to AI at UC Berkeley. All CS188 materials are available at hnp://ai.berkeley.edu.]
CSE 473: Ar+ficial Intelligence Bayes Nets Daniel Weld [Most slides were created by Dan Klein and Pieter Abbeel for CS188 Intro to AI at UC Berkeley. All CS188 materials are available at hnp://ai.berkeley.edu.]
Harvesting and harnessing data for biogeographical research
 How do we know what grows where? Harvesting and harnessing data for biogeographical research A. Geography Tree B. Species Tree inventories and surveys natural areas, preserves, state forests, private properties
How do we know what grows where? Harvesting and harnessing data for biogeographical research A. Geography Tree B. Species Tree inventories and surveys natural areas, preserves, state forests, private properties
Big Bang, Black Holes, No Math
 ASTR/PHYS 109 Dr. David Toback Lecture 6 & 7 1 Prep For Today (is now due) L7 Reading: Required: BBBHNM Unit 2 (Chapters 5-9) Recommended Reading: See P3 of http://people.physics.tamu.edu/toback/109/syllabus.pdf
ASTR/PHYS 109 Dr. David Toback Lecture 6 & 7 1 Prep For Today (is now due) L7 Reading: Required: BBBHNM Unit 2 (Chapters 5-9) Recommended Reading: See P3 of http://people.physics.tamu.edu/toback/109/syllabus.pdf
CSE546: SVMs, Dual Formula5on, and Kernels Winter 2012
 CSE546: SVMs, Dual Formula5on, and Kernels Winter 2012 Luke ZeClemoyer Slides adapted from Carlos Guestrin Linear classifiers Which line is becer? w. = j w (j) x (j) Data Example i Pick the one with the
CSE546: SVMs, Dual Formula5on, and Kernels Winter 2012 Luke ZeClemoyer Slides adapted from Carlos Guestrin Linear classifiers Which line is becer? w. = j w (j) x (j) Data Example i Pick the one with the
Summary of Columbia REU Research. ATLAS: Summer 2014
 Summary of Columbia REU Research ATLAS: Summer 2014 ì 1 2 Outline ì Brief Analysis Summary ì Overview of Sta
Summary of Columbia REU Research ATLAS: Summer 2014 ì 1 2 Outline ì Brief Analysis Summary ì Overview of Sta
Historical Biogeography: Evolution in Time and Space
 Evo Edu Outreach (2012) 5:555 568 DOI 10.1007/s12052-012-0421-2 GEOGRAPHY AND EVOLUTION Historical Biogeography: Evolution in Time and Space Isabel Sanmartín Published online: 21 June 2012 # Springer Science+Business
Evo Edu Outreach (2012) 5:555 568 DOI 10.1007/s12052-012-0421-2 GEOGRAPHY AND EVOLUTION Historical Biogeography: Evolution in Time and Space Isabel Sanmartín Published online: 21 June 2012 # Springer Science+Business
Introduction to Particle Filters for Data Assimilation
 Introduction to Particle Filters for Data Assimilation Mike Dowd Dept of Mathematics & Statistics (and Dept of Oceanography Dalhousie University, Halifax, Canada STATMOS Summer School in Data Assimila5on,
Introduction to Particle Filters for Data Assimilation Mike Dowd Dept of Mathematics & Statistics (and Dept of Oceanography Dalhousie University, Halifax, Canada STATMOS Summer School in Data Assimila5on,
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200 Spring 2018 University of California, Berkeley
 "PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200 Spring 2018 University of California, Berkeley D.D. Ackerly Feb. 26, 2018 Maximum Likelihood Principles, and Applications to
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200 Spring 2018 University of California, Berkeley D.D. Ackerly Feb. 26, 2018 Maximum Likelihood Principles, and Applications to
CSCI1950 Z Computa3onal Methods for Biology* (*Working Title) Lecture 1. Ben Raphael January 21, Course Par3culars
 CSCI1950 Z Computa3onal Methods for Biology* (*Working Title) Lecture 1 Ben Raphael January 21, 2009 Course Par3culars Three major topics 1. Phylogeny: ~50% lectures 2. Func3onal Genomics: ~25% lectures
CSCI1950 Z Computa3onal Methods for Biology* (*Working Title) Lecture 1 Ben Raphael January 21, 2009 Course Par3culars Three major topics 1. Phylogeny: ~50% lectures 2. Func3onal Genomics: ~25% lectures
Appendix from L. J. Revell, On the Analysis of Evolutionary Change along Single Branches in a Phylogeny
 008 by The University of Chicago. All rights reserved.doi: 10.1086/588078 Appendix from L. J. Revell, On the Analysis of Evolutionary Change along Single Branches in a Phylogeny (Am. Nat., vol. 17, no.
008 by The University of Chicago. All rights reserved.doi: 10.1086/588078 Appendix from L. J. Revell, On the Analysis of Evolutionary Change along Single Branches in a Phylogeny (Am. Nat., vol. 17, no.
UoN, CAS, DBSC BIOL102 lecture notes by: Dr. Mustafa A. Mansi. The Phylogenetic Systematics (Phylogeny and Systematics)
 - Phylogeny? - Systematics? The Phylogenetic Systematics (Phylogeny and Systematics) - Phylogenetic systematics? Connection between phylogeny and classification. - Phylogenetic systematics informs the
- Phylogeny? - Systematics? The Phylogenetic Systematics (Phylogeny and Systematics) - Phylogenetic systematics? Connection between phylogeny and classification. - Phylogenetic systematics informs the
Biome evolution and biogeographical change through time
 thesis abstract Biome evolution and biogeographical change through time Christine D. Bacon ISSN 1948-6596 Department of Biological and Environmental Sciences, University of Gothenburg, Gothenburg, Sweden;
thesis abstract Biome evolution and biogeographical change through time Christine D. Bacon ISSN 1948-6596 Department of Biological and Environmental Sciences, University of Gothenburg, Gothenburg, Sweden;
Introduc)on to Ar)ficial Intelligence
 Introduc)on to Ar)ficial Intelligence Lecture 13 Approximate Inference CS/CNS/EE 154 Andreas Krause Bayesian networks! Compact representa)on of distribu)ons over large number of variables! (OQen) allows
Introduc)on to Ar)ficial Intelligence Lecture 13 Approximate Inference CS/CNS/EE 154 Andreas Krause Bayesian networks! Compact representa)on of distribu)ons over large number of variables! (OQen) allows
Big Bang, Black Holes, No Math
 ASTR/PHYS 109 Dr. David Toback Lecture 8 1 Prep For Today (is now due) L8 Reading: If you haven t already: Unit 2 (Chapters 5-9) Pre-Lecture Reading Questions: If you were misgraded, need help or an extension
ASTR/PHYS 109 Dr. David Toback Lecture 8 1 Prep For Today (is now due) L8 Reading: If you haven t already: Unit 2 (Chapters 5-9) Pre-Lecture Reading Questions: If you were misgraded, need help or an extension
Bayesian networks Lecture 18. David Sontag New York University
 Bayesian networks Lecture 18 David Sontag New York University Outline for today Modeling sequen&al data (e.g., =me series, speech processing) using hidden Markov models (HMMs) Bayesian networks Independence
Bayesian networks Lecture 18 David Sontag New York University Outline for today Modeling sequen&al data (e.g., =me series, speech processing) using hidden Markov models (HMMs) Bayesian networks Independence
X X (2) X Pr(X = x θ) (3)
 Notes for 848 lecture 6: A ML basis for compatibility and parsimony Notation θ Θ (1) Θ is the space of all possible trees (and model parameters) θ is a point in the parameter space = a particular tree
Notes for 848 lecture 6: A ML basis for compatibility and parsimony Notation θ Θ (1) Θ is the space of all possible trees (and model parameters) θ is a point in the parameter space = a particular tree
Introduction to Error Analysis
 Introduction to Error Analysis This is a brief and incomplete discussion of error analysis. It is incomplete out of necessity; there are many books devoted entirely to the subject, and we cannot hope to
Introduction to Error Analysis This is a brief and incomplete discussion of error analysis. It is incomplete out of necessity; there are many books devoted entirely to the subject, and we cannot hope to
CHAPTER 52: Ecology. Name: Question Set Define each of the following terms: a. ecology. b. biotic. c. abiotic. d. population. e.
 CHAPTER 52: Ecology 1. Define each of the following terms: a. ecology b. biotic c. abiotic d. population e. community f. ecosystem g. biosphere 2. What is dispersal? 3. What are the important factors that
CHAPTER 52: Ecology 1. Define each of the following terms: a. ecology b. biotic c. abiotic d. population e. community f. ecosystem g. biosphere 2. What is dispersal? 3. What are the important factors that
Natural Language Processing. Classification. Features. Some Definitions. Classification. Feature Vectors. Classification I. Dan Klein UC Berkeley
 Natural Language Processing Classification Classification I Dan Klein UC Berkeley Classification Automatically make a decision about inputs Example: document category Example: image of digit digit Example:
Natural Language Processing Classification Classification I Dan Klein UC Berkeley Classification Automatically make a decision about inputs Example: document category Example: image of digit digit Example:
The practice of naming and classifying organisms is called taxonomy.
 Chapter 18 Key Idea: Biologists use taxonomic systems to organize their knowledge of organisms. These systems attempt to provide consistent ways to name and categorize organisms. The practice of naming
Chapter 18 Key Idea: Biologists use taxonomic systems to organize their knowledge of organisms. These systems attempt to provide consistent ways to name and categorize organisms. The practice of naming
MA/CS 109 Lecture 7. Back To Exponen:al Growth Popula:on Models
 MA/CS 109 Lecture 7 Back To Exponen:al Growth Popula:on Models Homework this week 1. Due next Thursday (not Tuesday) 2. Do most of computa:ons in discussion next week 3. If possible, bring your laptop
MA/CS 109 Lecture 7 Back To Exponen:al Growth Popula:on Models Homework this week 1. Due next Thursday (not Tuesday) 2. Do most of computa:ons in discussion next week 3. If possible, bring your laptop
The statistical and informatics challenges posed by ascertainment biases in phylogenetic data collection
 The statistical and informatics challenges posed by ascertainment biases in phylogenetic data collection Mark T. Holder and Jordan M. Koch Department of Ecology and Evolutionary Biology, University of
The statistical and informatics challenges posed by ascertainment biases in phylogenetic data collection Mark T. Holder and Jordan M. Koch Department of Ecology and Evolutionary Biology, University of
Lecture 11 Friday, October 21, 2011
 Lecture 11 Friday, October 21, 2011 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean system
Lecture 11 Friday, October 21, 2011 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean system
Chaos, Complexity, and Inference (36-462)
 Chaos, Complexity, and Inference (36-462) Lecture 4 Cosma Shalizi 22 January 2009 Reconstruction Inferring the attractor from a time series; powerful in a weird way Using the reconstructed attractor to
Chaos, Complexity, and Inference (36-462) Lecture 4 Cosma Shalizi 22 January 2009 Reconstruction Inferring the attractor from a time series; powerful in a weird way Using the reconstructed attractor to
Chapter 27: Evolutionary Genetics
 Chapter 27: Evolutionary Genetics Student Learning Objectives Upon completion of this chapter you should be able to: 1. Understand what the term species means to biology. 2. Recognize the various patterns
Chapter 27: Evolutionary Genetics Student Learning Objectives Upon completion of this chapter you should be able to: 1. Understand what the term species means to biology. 2. Recognize the various patterns
Consistency Index (CI)
 Consistency Index (CI) minimum number of changes divided by the number required on the tree. CI=1 if there is no homoplasy negatively correlated with the number of species sampled Retention Index (RI)
Consistency Index (CI) minimum number of changes divided by the number required on the tree. CI=1 if there is no homoplasy negatively correlated with the number of species sampled Retention Index (RI)
CSE 473: Ar+ficial Intelligence
 CSE 473: Ar+ficial Intelligence Hidden Markov Models Luke Ze@lemoyer - University of Washington [These slides were created by Dan Klein and Pieter Abbeel for CS188 Intro to AI at UC Berkeley. All CS188
CSE 473: Ar+ficial Intelligence Hidden Markov Models Luke Ze@lemoyer - University of Washington [These slides were created by Dan Klein and Pieter Abbeel for CS188 Intro to AI at UC Berkeley. All CS188
Package OUwie. August 29, 2013
 Package OUwie August 29, 2013 Version 1.34 Date 2013-5-21 Title Analysis of evolutionary rates in an OU framework Author Jeremy M. Beaulieu , Brian O Meara Maintainer
Package OUwie August 29, 2013 Version 1.34 Date 2013-5-21 Title Analysis of evolutionary rates in an OU framework Author Jeremy M. Beaulieu , Brian O Meara Maintainer
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics
 POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2011 University of California, Berkeley
 "PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2011 University of California, Berkeley B.D. Mishler Feb. 1, 2011. Qualitative character evolution (cont.) - comparing
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2011 University of California, Berkeley B.D. Mishler Feb. 1, 2011. Qualitative character evolution (cont.) - comparing
CSCI1950 Z Computa4onal Methods for Biology Lecture 5
 CSCI1950 Z Computa4onal Methods for Biology Lecture 5 Ben Raphael February 6, 2009 hip://cs.brown.edu/courses/csci1950 z/ Alignment vs. Distance Matrix Mouse: ACAGTGACGCCACACACGT Gorilla: CCTGCGACGTAACAAACGC
CSCI1950 Z Computa4onal Methods for Biology Lecture 5 Ben Raphael February 6, 2009 hip://cs.brown.edu/courses/csci1950 z/ Alignment vs. Distance Matrix Mouse: ACAGTGACGCCACACACGT Gorilla: CCTGCGACGTAACAAACGC
Experimental Designs for Planning Efficient Accelerated Life Tests
 Experimental Designs for Planning Efficient Accelerated Life Tests Kangwon Seo and Rong Pan School of Compu@ng, Informa@cs, and Decision Systems Engineering Arizona State University ASTR 2015, Sep 9-11,
Experimental Designs for Planning Efficient Accelerated Life Tests Kangwon Seo and Rong Pan School of Compu@ng, Informa@cs, and Decision Systems Engineering Arizona State University ASTR 2015, Sep 9-11,
Chapter 22: Descent with Modification 1. BRIEFLY summarize the main points that Darwin made in The Origin of Species.
 AP Biology Chapter Packet 7- Evolution Name Chapter 22: Descent with Modification 1. BRIEFLY summarize the main points that Darwin made in The Origin of Species. 2. Define the following terms: a. Natural
AP Biology Chapter Packet 7- Evolution Name Chapter 22: Descent with Modification 1. BRIEFLY summarize the main points that Darwin made in The Origin of Species. 2. Define the following terms: a. Natural
Machine learning for Dynamic Social Network Analysis
 Machine learning for Dynamic Social Network Analysis Manuel Gomez Rodriguez Max Planck Ins7tute for So;ware Systems UC3M, MAY 2017 Interconnected World SOCIAL NETWORKS TRANSPORTATION NETWORKS WORLD WIDE
Machine learning for Dynamic Social Network Analysis Manuel Gomez Rodriguez Max Planck Ins7tute for So;ware Systems UC3M, MAY 2017 Interconnected World SOCIAL NETWORKS TRANSPORTATION NETWORKS WORLD WIDE
Phylogenetics. Applications of phylogenetics. Unrooted networks vs. rooted trees. Outline
 Phylogenetics Todd Vision iology 522 March 26, 2007 pplications of phylogenetics Studying organismal or biogeographic history Systematics ating events in the fossil record onservation biology Studying
Phylogenetics Todd Vision iology 522 March 26, 2007 pplications of phylogenetics Studying organismal or biogeographic history Systematics ating events in the fossil record onservation biology Studying
Panama and the Great American Interchange. Reading: Marshall et al. 1982
 Panama and the Great American Interchange Reading: Marshall et al. 1982 Lecture 6 Recap Lars Brundin Lars Brundin What about the general suitability of chironomid midges as indicators in biogeography?
Panama and the Great American Interchange Reading: Marshall et al. 1982 Lecture 6 Recap Lars Brundin Lars Brundin What about the general suitability of chironomid midges as indicators in biogeography?
Integrative Biology 200A "PRINCIPLES OF PHYLOGENETICS" Spring 2008
 Integrative Biology 200A "PRINCIPLES OF PHYLOGENETICS" Spring 2008 University of California, Berkeley B.D. Mishler March 18, 2008. Phylogenetic Trees I: Reconstruction; Models, Algorithms & Assumptions
Integrative Biology 200A "PRINCIPLES OF PHYLOGENETICS" Spring 2008 University of California, Berkeley B.D. Mishler March 18, 2008. Phylogenetic Trees I: Reconstruction; Models, Algorithms & Assumptions
Geography 3251: Mountain Geography Assignment II: Island Biogeography Theory Assigned: May 22, 2012 Due: May 29, 9 AM
 Names: Geography 3251: Mountain Geography Assignment II: Island Biogeography Theory Assigned: May 22, 2012 Due: May 29, 2012 @ 9 AM NOTE: This lab is a modified version of the Island Biogeography lab that
Names: Geography 3251: Mountain Geography Assignment II: Island Biogeography Theory Assigned: May 22, 2012 Due: May 29, 2012 @ 9 AM NOTE: This lab is a modified version of the Island Biogeography lab that
From Gene Trees to Species Trees. Tandy Warnow The University of Texas at Aus<n
 From Gene Trees to Species Trees Tandy Warnow The University of Texas at Aus
From Gene Trees to Species Trees Tandy Warnow The University of Texas at Aus
Lab 9: Maximum Likelihood and Modeltest
 Integrative Biology 200A University of California, Berkeley "PRINCIPLES OF PHYLOGENETICS" Spring 2010 Updated by Nick Matzke Lab 9: Maximum Likelihood and Modeltest In this lab we re going to use PAUP*
Integrative Biology 200A University of California, Berkeley "PRINCIPLES OF PHYLOGENETICS" Spring 2010 Updated by Nick Matzke Lab 9: Maximum Likelihood and Modeltest In this lab we re going to use PAUP*
Methods for Cross-Analyzing Radio and ray Time Series Data Fermi Marries Jansky
 Methods for Cross-Analyzing Radio and ray Time Series Data Fermi Marries Jansky Jeff Scargle NASA Ames Research Center Fermi Gamma Ray Space Telescope Special Thanks to Jim Chiang, Jay Norris, Brad Jackson,
Methods for Cross-Analyzing Radio and ray Time Series Data Fermi Marries Jansky Jeff Scargle NASA Ames Research Center Fermi Gamma Ray Space Telescope Special Thanks to Jim Chiang, Jay Norris, Brad Jackson,
Lecture V Phylogeny and Systematics Dr. Kopeny
 Delivered 1/30 and 2/1 Lecture V Phylogeny and Systematics Dr. Kopeny Lecture V How to Determine Evolutionary Relationships: Concepts in Phylogeny and Systematics Textbook Reading: pp 425-433, 435-437
Delivered 1/30 and 2/1 Lecture V Phylogeny and Systematics Dr. Kopeny Lecture V How to Determine Evolutionary Relationships: Concepts in Phylogeny and Systematics Textbook Reading: pp 425-433, 435-437
PCB6675C, BOT6935, ZOO6927 Evolutionary Biogeography Spring 2014
 PCB6675C, BOT6935, ZOO6927 Evolutionary Biogeography Spring 2014 Credits: 3 Schedule: Wednesdays and Fridays, 4 th & 5 th Period (10:40 am - 12:35 pm) Location: Carr 221 Instructors Dr. Nico Cellinese
PCB6675C, BOT6935, ZOO6927 Evolutionary Biogeography Spring 2014 Credits: 3 Schedule: Wednesdays and Fridays, 4 th & 5 th Period (10:40 am - 12:35 pm) Location: Carr 221 Instructors Dr. Nico Cellinese
HOW TO GET A GOOD GRADE ON THE MME 2273B FLUID MECHANICS 1 EXAM. Common mistakes made on the final exam and how to avoid them
 HOW TO GET A GOOD GRADE ON THE MME 2273B FLUID MECHANICS 1 EXAM Common mistakes made on the final exam and how to avoid them HOW TO GET A GOOD GRADE ON THE MME 2273B EXAM Introduction You now have a lot
HOW TO GET A GOOD GRADE ON THE MME 2273B FLUID MECHANICS 1 EXAM Common mistakes made on the final exam and how to avoid them HOW TO GET A GOOD GRADE ON THE MME 2273B EXAM Introduction You now have a lot
Estimating Evolutionary Trees. Phylogenetic Methods
 Estimating Evolutionary Trees v if the data are consistent with infinite sites then all methods should yield the same tree v it gets more complicated when there is homoplasy, i.e., parallel or convergent
Estimating Evolutionary Trees v if the data are consistent with infinite sites then all methods should yield the same tree v it gets more complicated when there is homoplasy, i.e., parallel or convergent
Using Phylogeny to Infer Vicariance. Reading: Lars Brundin
 Using Phylogeny to Infer Vicariance Reading: Lars Brundin Lecture 5 Recap Alfred Wegener Alfred Wegener In his book, Wegener described how the con(nents of SA and Africa fit together (not the first to
Using Phylogeny to Infer Vicariance Reading: Lars Brundin Lecture 5 Recap Alfred Wegener Alfred Wegener In his book, Wegener described how the con(nents of SA and Africa fit together (not the first to
Linear Regression and Correla/on. Correla/on and Regression Analysis. Three Ques/ons 9/14/14. Chapter 13. Dr. Richard Jerz
 Linear Regression and Correla/on Chapter 13 Dr. Richard Jerz 1 Correla/on and Regression Analysis Correla/on Analysis is the study of the rela/onship between variables. It is also defined as group of techniques
Linear Regression and Correla/on Chapter 13 Dr. Richard Jerz 1 Correla/on and Regression Analysis Correla/on Analysis is the study of the rela/onship between variables. It is also defined as group of techniques
Linear Regression and Correla/on
 Linear Regression and Correla/on Chapter 13 Dr. Richard Jerz 1 Correla/on and Regression Analysis Correla/on Analysis is the study of the rela/onship between variables. It is also defined as group of techniques
Linear Regression and Correla/on Chapter 13 Dr. Richard Jerz 1 Correla/on and Regression Analysis Correla/on Analysis is the study of the rela/onship between variables. It is also defined as group of techniques
CSCI1950 Z Computa4onal Methods for Biology Lecture 4. Ben Raphael February 2, hhp://cs.brown.edu/courses/csci1950 z/ Algorithm Summary
 CSCI1950 Z Computa4onal Methods for Biology Lecture 4 Ben Raphael February 2, 2009 hhp://cs.brown.edu/courses/csci1950 z/ Algorithm Summary Parsimony Probabilis4c Method Input Output Sankoff s & Fitch
CSCI1950 Z Computa4onal Methods for Biology Lecture 4 Ben Raphael February 2, 2009 hhp://cs.brown.edu/courses/csci1950 z/ Algorithm Summary Parsimony Probabilis4c Method Input Output Sankoff s & Fitch
Quan&fying Uncertainty. Sai Ravela Massachuse7s Ins&tute of Technology
 Quan&fying Uncertainty Sai Ravela Massachuse7s Ins&tute of Technology 1 the many sources of uncertainty! 2 Two days ago 3 Quan&fying Indefinite Delay 4 Finally 5 Quan&fying Indefinite Delay P(X=delay M=
Quan&fying Uncertainty Sai Ravela Massachuse7s Ins&tute of Technology 1 the many sources of uncertainty! 2 Two days ago 3 Quan&fying Indefinite Delay 4 Finally 5 Quan&fying Indefinite Delay P(X=delay M=
Adaptation. Evolution. What is evolution? What are the tools used by scientists to understand evolutionary time?
 Adaptation Evolution: The golden Thread Adaptation - Process where species acquire* traits that allow them to survive in their environments. Limited range of physiological modifications. Inheritance of
Adaptation Evolution: The golden Thread Adaptation - Process where species acquire* traits that allow them to survive in their environments. Limited range of physiological modifications. Inheritance of
BIOL 1010 Introduction to Biology: The Evolution and Diversity of Life. Spring 2011 Sections A & B
 BIOL 1010 Introduction to Biology: The Evolution and Diversity of Life. Spring 2011 Sections A & B Steve Thompson: stthompson@valdosta.edu http://www.bioinfo4u.net 1 ʻTree of Life,ʼ ʻprimitive,ʼ ʻprogressʼ
BIOL 1010 Introduction to Biology: The Evolution and Diversity of Life. Spring 2011 Sections A & B Steve Thompson: stthompson@valdosta.edu http://www.bioinfo4u.net 1 ʻTree of Life,ʼ ʻprimitive,ʼ ʻprogressʼ
The Tempo of Macroevolution: Patterns of Diversification and Extinction
 The Tempo of Macroevolution: Patterns of Diversification and Extinction During the semester we have been consider various aspects parameters associated with biodiversity. Current usage stems from 1980's
The Tempo of Macroevolution: Patterns of Diversification and Extinction During the semester we have been consider various aspects parameters associated with biodiversity. Current usage stems from 1980's
Geography of Evolution
 Geography of Evolution Biogeography - the study of the geographic distribution of organisms. The current distribution of organisms can be explained by historical events and current climatic patterns. Darwin
Geography of Evolution Biogeography - the study of the geographic distribution of organisms. The current distribution of organisms can be explained by historical events and current climatic patterns. Darwin
Machine Learning. Gaussian Mixture Models. Zhiyao Duan & Bryan Pardo, Machine Learning: EECS 349 Fall
 Machine Learning Gaussian Mixture Models Zhiyao Duan & Bryan Pardo, Machine Learning: EECS 349 Fall 2012 1 The Generative Model POV We think of the data as being generated from some process. We assume
Machine Learning Gaussian Mixture Models Zhiyao Duan & Bryan Pardo, Machine Learning: EECS 349 Fall 2012 1 The Generative Model POV We think of the data as being generated from some process. We assume
Gels and Trees. Jacob Landis
 Gels and Trees Jacob Landis Recap from Monday Pollination syndromes certain flower traits associated with unique kinds of pollinators DNA extractions leaf punch for material, Extraction buffer to get DNA
Gels and Trees Jacob Landis Recap from Monday Pollination syndromes certain flower traits associated with unique kinds of pollinators DNA extractions leaf punch for material, Extraction buffer to get DNA
A (short) introduction to phylogenetics
 A (short) introduction to phylogenetics Thibaut Jombart, Marie-Pauline Beugin MRC Centre for Outbreak Analysis and Modelling Imperial College London Genetic data analysis with PR Statistics, Millport Field
A (short) introduction to phylogenetics Thibaut Jombart, Marie-Pauline Beugin MRC Centre for Outbreak Analysis and Modelling Imperial College London Genetic data analysis with PR Statistics, Millport Field
Big Bang, Black Holes, No Math
 ASTR/PHYS 109 Dr. David Toback Lecture 5 1 Prep For Today (is now due) L5 Reading: No new reading Unit 2 reading assigned at the end of class Pre-Lecture Reading Questions: Unit 1: Grades have been posted
ASTR/PHYS 109 Dr. David Toback Lecture 5 1 Prep For Today (is now due) L5 Reading: No new reading Unit 2 reading assigned at the end of class Pre-Lecture Reading Questions: Unit 1: Grades have been posted
Biogeography. An ecological and evolutionary approach SEVENTH EDITION. C. Barry Cox MA, PhD, DSc and Peter D. Moore PhD
 Biogeography An ecological and evolutionary approach C. Barry Cox MA, PhD, DSc and Peter D. Moore PhD Division of Life Sciences, King's College London, Fmnklin-Wilkins Building, Stamford Street, London
Biogeography An ecological and evolutionary approach C. Barry Cox MA, PhD, DSc and Peter D. Moore PhD Division of Life Sciences, King's College London, Fmnklin-Wilkins Building, Stamford Street, London
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2009 University of California, Berkeley
 "PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2009 University of California, Berkeley B.D. Mishler Jan. 22, 2009. Trees I. Summary of previous lecture: Hennigian
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2009 University of California, Berkeley B.D. Mishler Jan. 22, 2009. Trees I. Summary of previous lecture: Hennigian
Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week:
 Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week: Course general information About the course Course objectives Comparative methods: An overview R as language: uses and
Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week: Course general information About the course Course objectives Comparative methods: An overview R as language: uses and
Concepts and Methods in Molecular Divergence Time Estimation
 Concepts and Methods in Molecular Divergence Time Estimation 26 November 2012 Prashant P. Sharma American Museum of Natural History Overview 1. Why do we date trees? 2. The molecular clock 3. Local clocks
Concepts and Methods in Molecular Divergence Time Estimation 26 November 2012 Prashant P. Sharma American Museum of Natural History Overview 1. Why do we date trees? 2. The molecular clock 3. Local clocks
Maximum Likelihood Tree Estimation. Carrie Tribble IB Feb 2018
 Maximum Likelihood Tree Estimation Carrie Tribble IB 200 9 Feb 2018 Outline 1. Tree building process under maximum likelihood 2. Key differences between maximum likelihood and parsimony 3. Some fancy extras
Maximum Likelihood Tree Estimation Carrie Tribble IB 200 9 Feb 2018 Outline 1. Tree building process under maximum likelihood 2. Key differences between maximum likelihood and parsimony 3. Some fancy extras
Natural Language Processing
 Natural Language Processing Info 159/259 Lecture 12: Features and hypothesis tests (Oct 3, 2017) David Bamman, UC Berkeley Announcements No office hours for DB this Friday (email if you d like to chat)
Natural Language Processing Info 159/259 Lecture 12: Features and hypothesis tests (Oct 3, 2017) David Bamman, UC Berkeley Announcements No office hours for DB this Friday (email if you d like to chat)
Last Lecture Recap UVA CS / Introduc8on to Machine Learning and Data Mining. Lecture 3: Linear Regression
 UVA CS 4501-001 / 6501 007 Introduc8on to Machine Learning and Data Mining Lecture 3: Linear Regression Yanjun Qi / Jane University of Virginia Department of Computer Science 1 Last Lecture Recap q Data
UVA CS 4501-001 / 6501 007 Introduc8on to Machine Learning and Data Mining Lecture 3: Linear Regression Yanjun Qi / Jane University of Virginia Department of Computer Science 1 Last Lecture Recap q Data
The California Hotspots Project: I.
 The California Hotspots Project: I. Identifying regions of rapid diversification of mammals Ed Davis, M. Koo, C. Conroy, J. Patton & C. Moritz Museum of Vertebrate Zoology, UC Berkeley *Funded by Resources
The California Hotspots Project: I. Identifying regions of rapid diversification of mammals Ed Davis, M. Koo, C. Conroy, J. Patton & C. Moritz Museum of Vertebrate Zoology, UC Berkeley *Funded by Resources
Bias/variance tradeoff, Model assessment and selec+on
 Applied induc+ve learning Bias/variance tradeoff, Model assessment and selec+on Pierre Geurts Department of Electrical Engineering and Computer Science University of Liège October 29, 2012 1 Supervised
Applied induc+ve learning Bias/variance tradeoff, Model assessment and selec+on Pierre Geurts Department of Electrical Engineering and Computer Science University of Liège October 29, 2012 1 Supervised
The Intersection of Chemistry and Biology: An Interview with Professor W. E. Moerner
 The Intersection of Chemistry and Biology: An Interview with Professor W. E. Moerner Joseph Nicolls Stanford University Professor W.E Moerner earned two B.S. degrees, in Physics and Electrical Engineering,
The Intersection of Chemistry and Biology: An Interview with Professor W. E. Moerner Joseph Nicolls Stanford University Professor W.E Moerner earned two B.S. degrees, in Physics and Electrical Engineering,
CHAPTER 26 PHYLOGENY AND THE TREE OF LIFE Connecting Classification to Phylogeny
 CHAPTER 26 PHYLOGENY AND THE TREE OF LIFE Connecting Classification to Phylogeny To trace phylogeny or the evolutionary history of life, biologists use evidence from paleontology, molecular data, comparative
CHAPTER 26 PHYLOGENY AND THE TREE OF LIFE Connecting Classification to Phylogeny To trace phylogeny or the evolutionary history of life, biologists use evidence from paleontology, molecular data, comparative
Some of these slides have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks!
 Some of these slides have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks! Paul has many great tools for teaching phylogenetics at his web site: http://hydrodictyon.eeb.uconn.edu/people/plewis
Some of these slides have been borrowed from Dr. Paul Lewis, Dr. Joe Felsenstein. Thanks! Paul has many great tools for teaching phylogenetics at his web site: http://hydrodictyon.eeb.uconn.edu/people/plewis
Measuring the physical proper2es of the Milky Way nuclear star cluster with 3D kinema2cs
 Measuring the physical proper2es of the Milky Way nuclear star cluster with 3D kinema2cs Tuan Do Dunlap Ins2tute, University of Toronto Dunlap Fellow Collaborators: A. Ghez (UCLA), J. Lu (IfA), G. Mar2nez
Measuring the physical proper2es of the Milky Way nuclear star cluster with 3D kinema2cs Tuan Do Dunlap Ins2tute, University of Toronto Dunlap Fellow Collaborators: A. Ghez (UCLA), J. Lu (IfA), G. Mar2nez
Estimating Phylogenies (Evolutionary Trees) II. Biol4230 Thurs, March 2, 2017 Bill Pearson Jordan 6-057
 Estimating Phylogenies (Evolutionary Trees) II Biol4230 Thurs, March 2, 2017 Bill Pearson wrp@virginia.edu 4-2818 Jordan 6-057 Tree estimation strategies: Parsimony?no model, simply count minimum number
Estimating Phylogenies (Evolutionary Trees) II Biol4230 Thurs, March 2, 2017 Bill Pearson wrp@virginia.edu 4-2818 Jordan 6-057 Tree estimation strategies: Parsimony?no model, simply count minimum number
The Importance of Time/Space in Diagnosing the Causality of Phylogenetic Events: Towards a Chronobiogeographical Paradigm?
 Syst. Biol. 50(3):391 407, 2001 The Importance of Time/Space in Diagnosing the Causality of Phylogenetic Events: Towards a Chronobiogeographical Paradigm? CRAIG ANDREW HUNN AND PAUL UPCHURCH Department
Syst. Biol. 50(3):391 407, 2001 The Importance of Time/Space in Diagnosing the Causality of Phylogenetic Events: Towards a Chronobiogeographical Paradigm? CRAIG ANDREW HUNN AND PAUL UPCHURCH Department
COMS 4771 Regression. Nakul Verma
 COMS 4771 Regression Nakul Verma Last time Support Vector Machines Maximum Margin formulation Constrained Optimization Lagrange Duality Theory Convex Optimization SVM dual and Interpretation How get the
COMS 4771 Regression Nakul Verma Last time Support Vector Machines Maximum Margin formulation Constrained Optimization Lagrange Duality Theory Convex Optimization SVM dual and Interpretation How get the
Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2016 University of California, Berkeley. Parsimony & Likelihood [draft]
![Integrative Biology 200 PRINCIPLES OF PHYLOGENETICS Spring 2016 University of California, Berkeley. Parsimony & Likelihood [draft] Integrative Biology 200 PRINCIPLES OF PHYLOGENETICS Spring 2016 University of California, Berkeley. Parsimony & Likelihood [draft]](/thumbs/83/87443456.jpg) Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2016 University of California, Berkeley K.W. Will Parsimony & Likelihood [draft] 1. Hennig and Parsimony: Hennig was not concerned with parsimony
Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2016 University of California, Berkeley K.W. Will Parsimony & Likelihood [draft] 1. Hennig and Parsimony: Hennig was not concerned with parsimony
