Molecular Simulation III
|
|
|
- Kerry Goodwin
- 5 years ago
- Views:
Transcription
1 Molecular Simulation III Quantum Chemistry Classical Mechanics E = Ψ H Ψ ΨΨ U = E bond +E angle +E torsion +E non-bond Molecular Dynamics Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Crystal-Phospholipid Bilayer Interactions Solvated DMPC Bilayer in Absence and Presence of CPPD Crystal Pseudogout (human inflamatory disease) caused by presence of in vivo crystals of calcium pyrophosphate dihydrate (CPPD). Molecular aspect of in vivo crystal induced inflammation is unknown Rupture of the lysosome phospholipid membrane is a commonly accepted mechanism of inflammation. Important to elucidate the nature of crystal-phospholipid bilayer interactions The knowledge will aid in developing inhibitors to diminish the adhesion of CPPD to membranes 00 ps 400 ps 600 ps 1
2 MD Review r( t) Molecular dynamics is a numerical integration of the classical equations of motion d x F = ma = m dt assuming conservative forces. F = U the integrated equations of motion become r t t r t r t t 1 F t t m ( + δ ) = ( ) ( δ ) + () δ r( t+ δ t) r( t+ nδ t) VMWARE VMWARE Molecular Dynamics of BPTI BPTI: Bovine Pancreatic Trypsin Inhibitor Small protein of 58 amino acid residues Protein used in first MD simulations
3 Dynamics Output AVER DYN: Step Time TOTEner TOTKe ENERgy TEMPerature AVER PROP: GRMS HFCTote HFCKe EHFCor VIRKe AVER INTERN: BONDs ANGLes UREY-b DIHEdrals IMPRopers AVER CROSS: CMAPs AVER EXTERN: VDWaals ELEC HBONds ASP USER AVER PRESS: VIRE VIRI PRESSE PRESSI VOLUme AVER> AVER PROP> AVER INTERN> AVER CROSS> AVER EXTERN> AVER PRESS> Docking Ligand-Receptor Docking Deals with identification of suitable ( best ) ligands for specific receptors in proteins. Ligands can act either as activators or as inhibitors of the biological function of the protein in the cell. Artificial ligands (i.e. drugs) can be used to up-regulate or down-regulate the activity of proteins that are associated with specific diseases. To the left, HIV-1 Protease complexed with an efficient inhibitor, TL Docking Three-dimensional molecular structure is one of the foundations of structure-based drug design. Often, data are available for the shape of a protein and a drug separately, but not for the two together. Docking is the process by which two molecules fit together in 3D space. 3
4 Docking Two general classes Unbiased Autodock Direct DOCK LUDI Goals Robust and accurate Computationally feasible Receptor Substrate - Ligand-Receptor Docking Approach: Challenges Must screen millions of possible compounds that fit a particular receptor. Must specifically select those ligands that show a high affinity. The set of ligands selected can then be screened further by more involved computational techniques, such as freeenergy perturbation theory (ΔG bind ) We would like an automated, standard protocol to find the best Ligand-Receptor fit. Docking Docking Terms to consider in docking Shape complementarity Interaction specificity Solvation/desolvation Hydrophobic Hydrogen bonding Terms considered in MOE-Dock (Autodock) Van der Waals Hydrogen bonding electrostatics Energy evaluation Based on a Grid approach Search engine Simulated Annealing (SA) Autodock MOE-Dock Genetic Algorithms (GA) Autodock 3.0 4
5 MOE-Dock Application We will look at a docking example of a TIBO-like inhibitor to HIV-1 Reverse Transcriptase (HIV-RT). Crystal structure to be used: HIV-RT with TIBO-R MOE-Dock Application Setting up the calculation. Prepare the protein. Color the ligand, receptor, and metal ions distinctly. Add H atoms to the X-ray structure if none are given MOE Edit Add Hydrogens Select ForceField. MOE Window Potential Control Minimize. MOE Compute Energy Min. Here you can turn on solvation model; Place partial charges on atoms MOE-Dock Application MOE Compute Simulations Dock MOE-Dock Application Docking Results Examine the docked structures compared to the crystal structure of the ligand and its receptor. In this database, columns contain the total energy of the complex, the electrostatic (U_ele) and van der Waals energies (U_vdw) between the protein and the ligand, and the energy of the (flexible) ligand (U_ligand). The docking box appears around the ligand. Graphic shows HIV-RT (red) and its ligand TIBO-R
6 MOE-Dock Application To find the best (lowest energy) docked structure, you will sort the database in ascending order with respect to the total energy (U_total) Poisson Boltzmann Electrostatics Application Areas of Electrostatics Electrostatic Energies Electrostatic Forces Electrostatic Binding Free Energy Electrostatic Solvation Free Energy pka Shifts Protein Stability Conformational ph Dependence Redox Electrostatic Steering in Enzyme/Substrate Encounters Electrostatic Forces Coupled to Molecular Mechanics/Dynamics Explicit vs. Continuum Solvent Model Based on a suggestion by Born, the explicit solvent model may be very crudely approximated by a structureless continuum. In this continuum picture the solvent is represented by a dielectric constant, ε sol, and the effect of ions by, κ. The solute is a set of embedded charges inside a cavity with a dielectric constant of, ε in. 6
7 Continuum Solvent Model ΔG = ΔG + ΔG solv np elec ΔG np γ SA ΔG elec N 1 atoms s v = q φ φ i= 1 i c i i h Poisson-Boltzmann Model of Molecular Electrostatics permittivity a f 4 f ε φ = πρ κ φλ electrostatic potential fixed charge density masking function inverse Debye length 8πeNI A κ = 1000εkT B Solving the FDPB Equation In practice, one knows the charge density (ρ) from the fixed charges in the receptor and substrate. the permittivity (dielectric constant). Kappa (κ), which is related to the ionic strength. Make a guess at the potential. Solve the equation for a new potential. Continue to solve until the change in potential is small. Poisson-Boltzmann Electrostatic Forces f = F + F + F + F Coul RF DBF IBF F Coul is the Coulombic force which is the interaction of all the solute atoms with each other and is referred to as the qe force. F RF is the reaction field force, F RF = qe RF where E RF is the solvent reaction field acting at an atom. F DBF is the dielectric boundary force. This is due to the tendency of high dielectric medium to reduce the field energy by moving into regions of low-dielectric constant. F IBF is the ionic boundary force and is generally small in comparison with the other forces in the system. This force results from the tendency of mobile ions to reduce the field energy by moving into regions of zero ionic strength (i. e. the molecular interior). 7
8 mass m dxt dt Langevin Dynamics position Fx m dx t = γ + Rt dt Random flucuations due to interactions with the solvent af a f af a f Force which depends upon the position of the particle relative to the other particles Force due to the motion through the solvent γ = k T B md diffusion constant Dichloroethane Summary of simulation parameters ε i = 1 ε s = 80 γ = 6.5 ps -1 dt = ps T = 1000 K grid spacing = 0.5 to 1. A Atom Type Charg e (e ) Radius (Å ) C l CH Trans conformer dominates in the gas phase Increased gauche conformer in liquid phase Summary of simulation results Dichloroethane Alanine dipeptide Summary of simulation parameters Normalized Prob. Density (1/deg) Reference Gas Phase Results ns stochastic dynamics simulation Dihedral Angle (degree) ε i = 1 ε s = 80 γ = 6.5 ps -1 dt = ps T = 1000 K grid spacing = 0.7 to 1.7 A Normalized Prob. Density (1/deg) Reference (PB solvation energies) ns stochastic dynamics simulation 15 x 15 x 15 grid) ns stochastic dynamics simulation 10 x 10 x 10 grid) ns stochastic dynamics simulation 10 x 10 x 10, no solvent boundary forces) Dihedral Angle (degree) Conclusions Good equilibration Good agreement with other computational models Weak sensititvity to grid spacing No heating from numerical forces 8
9 in vacuo Alanine dipeptide Thermodynamic Treatment of Ion- Solvent Interactions: The Born Model Ion-Solvent interaction: Consists of solvent dipoles interacting with the electric field of the ion. Two cases to consider for the solvent: A structure-less continuum of dielectric ε ( The Born Model ) Discrete molecules with dipoles, polarizability, etc. Aqueous Reference ns stochastic dynamics The Born Model Consider: Continuum model of ion solvation. If medium 1 is a vacuum, ΔG born. is just the free energy of solvation. We will calculate the free energy of transfer of an ion from medium 1 (ε 1 ) to medium (ε ). This will be called ΔG born. The path for ΔG born refers to: First discharging the ion in medium 1 (ΔG o 1 ) Transferring the ion from medium 1 to medium (G o ) Recharging the ion in solvent (ΔG o 3 ) 9
10 The Charging Process Energies of charging/discharging: computed by a model where infinitesimal pieces of charge are brought from infinity, and placed on the surface of the ion until the final charge is obtained The Charging Process What is the energy of bringing a charge dq from infinity and placing it on the surface of a sphere with radius a? dg = Φdq The Charging Process Knowing the potential (Φ) of a point charge, we have, The Charging Process Therefore, For ΔG o 1, ΔG o, and ΔGo born we have (Next Slide) Integrating this from 0 to the final charge on the ion, Ze (where Z is the valence)..(next Slide) 10
11 The Charging Process If ε < ε 1, then ΔG o >0 It takes work to move an ion from water to a less polar solvent (such as vacuum or hydrocarbon) Free Energy of Solvation Consider: Transferring an ion from a vacuum to a medium of ε. Assume ΔG o = 0. (No interaction between solvent and discharged ion). Two points to note: 1. ΔG < 0 if ε > 1. ΔG increases as ionic Radius increases. Why? The field and the potential At the ion surface becomes Less. Generalized Born Widely used to represent the electrostatic contribution to the free energy of solvation Model is comprised of a system of particles with radii a i and charges q i The total electrostatic free energy is given by the sum of the Coulomb energy and the Born free energy of solvation in a medium of relative permittivity ε. G elec qq 1 1 = 1 εr ε N N N i j i i= 1 j= i+ 1 ij i= 1 q a i Generalized Born The previous equation can be re-written into the generalized Born equation N N 1 1 qq i j Δ Gelec = 1 ε = 1 = 1 f r, a ( ) i j ij ij f(r ij,a ij ) depends upon the interparticle distances r ij and the Born radii a i. (, ) f r a = r + a e ij ij ij ij ij i j D a = aa D= r ij ( aij ) 11
12 Generalized Born Note the following When i=j the equation returns the Born expression When r ij << a i and a j the expression is close to the Onsager result (I.e. a dipole) When r ij >> a i and a j the result is very close to the sum of the Coulomb and Born expression A major advantage to this formulation is that the expression can be differentiate analytically, thereby enabling the solvation term to be included in gradient-based optimization methods MacroModel GB/SA Solvation Model Accounts for solvation effects, especially in complex systems. Generalized Born/Surface Area (GB/SA) approach (continuum). increases the speed of the calculation avoids convergence problems, apparent in explicit models, where longer simulations or different solvent starting geometries yield different final energies. The GB/SA model can be used to calculate absolute free energies of solvation. Application of GB/SA Solvation Model Hall group applied the GB/SA continuum solvation model to RNA hairpins with much success. Simulations of the UUCG tetraloop give average structures within 1. Å of the initial NMR model, in agreement with an explicit solvent simulation (Williams, D. J., Hall, K. B Biophys J. 76: ). Electrostatic Free Energy of Solvation Calculation In this calculation one computes the electrostatic energy difference between the molecule in the aqueous phase and in vacuum. This is equivalent to computing the work in moving a charge from a low dielectric to a high dielectric. This work is equivalent to a change in the free energy. MOE-Electrostatics can be used by performing two calculations Compute the electrostatic energy with both dielectric constants set to 1 Compute the electrostatic energy with the interior dielectric set to 1 and the exterior dielectric set to 80. 1
Molecular Simulation III
 Molecular Simulation III Quantum Chemistry Classical Mechanics E = Ψ H Ψ ΨΨ U = E bond +E angle +E torsion +E non-bond Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences
Molecular Simulation III Quantum Chemistry Classical Mechanics E = Ψ H Ψ ΨΨ U = E bond +E angle +E torsion +E non-bond Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences
Molecular Modeling -- Lecture 15 Surfaces and electrostatics
 Molecular Modeling -- Lecture 15 Surfaces and electrostatics Molecular surfaces The Hydrophobic Effect Electrostatics Poisson-Boltzmann Equation Electrostatic maps Electrostatic surfaces in MOE 15.1 The
Molecular Modeling -- Lecture 15 Surfaces and electrostatics Molecular surfaces The Hydrophobic Effect Electrostatics Poisson-Boltzmann Equation Electrostatic maps Electrostatic surfaces in MOE 15.1 The
All-atom Molecular Mechanics. Trent E. Balius AMS 535 / CHE /27/2010
 All-atom Molecular Mechanics Trent E. Balius AMS 535 / CHE 535 09/27/2010 Outline Molecular models Molecular mechanics Force Fields Potential energy function functional form parameters and parameterization
All-atom Molecular Mechanics Trent E. Balius AMS 535 / CHE 535 09/27/2010 Outline Molecular models Molecular mechanics Force Fields Potential energy function functional form parameters and parameterization
Potential Energy (hyper)surface
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = " d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = " d dx U(x) Conformation
MM-GBSA for Calculating Binding Affinity A rank-ordering study for the lead optimization of Fxa and COX-2 inhibitors
 MM-GBSA for Calculating Binding Affinity A rank-ordering study for the lead optimization of Fxa and COX-2 inhibitors Thomas Steinbrecher Senior Application Scientist Typical Docking Workflow Databases
MM-GBSA for Calculating Binding Affinity A rank-ordering study for the lead optimization of Fxa and COX-2 inhibitors Thomas Steinbrecher Senior Application Scientist Typical Docking Workflow Databases
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
The Molecular Dynamics Method
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
Free energy, electrostatics, and the hydrophobic effect
 Protein Physics 2016 Lecture 3, January 26 Free energy, electrostatics, and the hydrophobic effect Magnus Andersson magnus.andersson@scilifelab.se Theoretical & Computational Biophysics Recap Protein structure
Protein Physics 2016 Lecture 3, January 26 Free energy, electrostatics, and the hydrophobic effect Magnus Andersson magnus.andersson@scilifelab.se Theoretical & Computational Biophysics Recap Protein structure
Solvation and Macromolecular Structure. The structure and dynamics of biological macromolecules are strongly influenced by water:
 Overview Solvation and Macromolecular Structure The structure and dynamics of biological macromolecules are strongly influenced by water: Electrostatic effects: charges are screened by water molecules
Overview Solvation and Macromolecular Structure The structure and dynamics of biological macromolecules are strongly influenced by water: Electrostatic effects: charges are screened by water molecules
INTERMOLECULAR AND SURFACE FORCES
 INTERMOLECULAR AND SURFACE FORCES SECOND EDITION JACOB N. ISRAELACHVILI Department of Chemical & Nuclear Engineering and Materials Department University of California, Santa Barbara California, USA ACADEMIC
INTERMOLECULAR AND SURFACE FORCES SECOND EDITION JACOB N. ISRAELACHVILI Department of Chemical & Nuclear Engineering and Materials Department University of California, Santa Barbara California, USA ACADEMIC
Molecular Simulation III
 Molecular Simulation III Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Time and Length Scales Tamar Schlick s Biomolecular Structure and
Molecular Simulation III Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Time and Length Scales Tamar Schlick s Biomolecular Structure and
Docking. GBCB 5874: Problem Solving in GBCB
 Docking Benzamidine Docking to Trypsin Relationship to Drug Design Ligand-based design QSAR Pharmacophore modeling Can be done without 3-D structure of protein Receptor/Structure-based design Molecular
Docking Benzamidine Docking to Trypsin Relationship to Drug Design Ligand-based design QSAR Pharmacophore modeling Can be done without 3-D structure of protein Receptor/Structure-based design Molecular
Can a continuum solvent model reproduce the free energy landscape of a β-hairpin folding in water?
 Can a continuum solvent model reproduce the free energy landscape of a β-hairpin folding in water? Ruhong Zhou 1 and Bruce J. Berne 2 1 IBM Thomas J. Watson Research Center; and 2 Department of Chemistry,
Can a continuum solvent model reproduce the free energy landscape of a β-hairpin folding in water? Ruhong Zhou 1 and Bruce J. Berne 2 1 IBM Thomas J. Watson Research Center; and 2 Department of Chemistry,
Molecular Mechanics, Dynamics & Docking
 Molecular Mechanics, Dynamics & Docking Lawrence Hunter, Ph.D. Director, Computational Bioscience Program University of Colorado School of Medicine Larry.Hunter@uchsc.edu http://compbio.uchsc.edu/hunter
Molecular Mechanics, Dynamics & Docking Lawrence Hunter, Ph.D. Director, Computational Bioscience Program University of Colorado School of Medicine Larry.Hunter@uchsc.edu http://compbio.uchsc.edu/hunter
Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects
 Statistical Thermodynamics Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects Dr. Ronald M. Levy ronlevy@temple.edu Bare Molecular Mechanics Atomistic Force Fields: torsion
Statistical Thermodynamics Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects Dr. Ronald M. Levy ronlevy@temple.edu Bare Molecular Mechanics Atomistic Force Fields: torsion
ONETEP PB/SA: Application to G-Quadruplex DNA Stability. Danny Cole
 ONETEP PB/SA: Application to G-Quadruplex DNA Stability Danny Cole Introduction Historical overview of structure and free energy calculation of complex molecules using molecular mechanics and continuum
ONETEP PB/SA: Application to G-Quadruplex DNA Stability Danny Cole Introduction Historical overview of structure and free energy calculation of complex molecules using molecular mechanics and continuum
Polypeptide Folding Using Monte Carlo Sampling, Concerted Rotation, and Continuum Solvation
 Polypeptide Folding Using Monte Carlo Sampling, Concerted Rotation, and Continuum Solvation Jakob P. Ulmschneider and William L. Jorgensen J.A.C.S. 2004, 126, 1849-1857 Presented by Laura L. Thomas and
Polypeptide Folding Using Monte Carlo Sampling, Concerted Rotation, and Continuum Solvation Jakob P. Ulmschneider and William L. Jorgensen J.A.C.S. 2004, 126, 1849-1857 Presented by Laura L. Thomas and
Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany
 Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals Phase Equilibria Phase diagrams and classical thermodynamics
Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals Phase Equilibria Phase diagrams and classical thermodynamics
Biochemistry 530: Introduction to Structural Biology. Autumn Quarter 2014 BIOC 530
 Biochemistry 530: Introduction to Structural Biology Autumn Quarter 2014 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic
Biochemistry 530: Introduction to Structural Biology Autumn Quarter 2014 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic
Homology modeling. Dinesh Gupta ICGEB, New Delhi 1/27/2010 5:59 PM
 Homology modeling Dinesh Gupta ICGEB, New Delhi Protein structure prediction Methods: Homology (comparative) modelling Threading Ab-initio Protein Homology modeling Homology modeling is an extrapolation
Homology modeling Dinesh Gupta ICGEB, New Delhi Protein structure prediction Methods: Homology (comparative) modelling Threading Ab-initio Protein Homology modeling Homology modeling is an extrapolation
GENERALIZED BORN MODELS OF MACROMOLECULARSOLVATION EFFECTS
 Annu. Rev. Phys. Chem. 2000. 51:129 52 Copyright c 2000 by Annual Reviews. All rights reserved GENERALIZED BORN MODELS OF MACROMOLECULARSOLVATION EFFECTS Donald Bashford and David A. Case Department of
Annu. Rev. Phys. Chem. 2000. 51:129 52 Copyright c 2000 by Annual Reviews. All rights reserved GENERALIZED BORN MODELS OF MACROMOLECULARSOLVATION EFFECTS Donald Bashford and David A. Case Department of
Solutions and Non-Covalent Binding Forces
 Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
Biochemistry 675, Lecture 8 Electrostatic Interactions
 Biochemistry 675, Lecture 8 Electrostatic Interactions Previous Classes Hydrophobic Effect Hydrogen Bonds Today: Electrostatic Interactions Reading: Handout:P.R. Bergethon & E.R. Simons, Biophysical Chemistry,
Biochemistry 675, Lecture 8 Electrostatic Interactions Previous Classes Hydrophobic Effect Hydrogen Bonds Today: Electrostatic Interactions Reading: Handout:P.R. Bergethon & E.R. Simons, Biophysical Chemistry,
Using AutoDock 4 with ADT: A Tutorial
 Using AutoDock 4 with ADT: A Tutorial Ruth Huey Sargis Dallakyan Alex Perryman David S. Goodsell (Garrett Morris) 9/2/08 Using AutoDock 4 with ADT 1 What is Docking? Predicting the best ways two molecules
Using AutoDock 4 with ADT: A Tutorial Ruth Huey Sargis Dallakyan Alex Perryman David S. Goodsell (Garrett Morris) 9/2/08 Using AutoDock 4 with ADT 1 What is Docking? Predicting the best ways two molecules
3. Solutions W = N!/(N A!N B!) (3.1) Using Stirling s approximation ln(n!) = NlnN N: ΔS mix = k (N A lnn + N B lnn N A lnn A N B lnn B ) (3.
 3. Solutions Many biological processes occur between molecules in aqueous solution. In addition, many protein and nucleic acid molecules adopt three-dimensional structure ( fold ) in aqueous solution.
3. Solutions Many biological processes occur between molecules in aqueous solution. In addition, many protein and nucleic acid molecules adopt three-dimensional structure ( fold ) in aqueous solution.
Protein Folding & Stability. Lecture 11: Margaret A. Daugherty. Fall How do we go from an unfolded polypeptide chain to a
 Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2004 How do we go from an unfolded polypeptide chain to a compact folded protein? (Folding of thioredoxin, F. Richards) Structure - Function
Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2004 How do we go from an unfolded polypeptide chain to a compact folded protein? (Folding of thioredoxin, F. Richards) Structure - Function
Applications of Molecular Dynamics
 June 4, 0 Molecular Modeling and Simulation Applications of Molecular Dynamics Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, The University of Tokyo Tohru
June 4, 0 Molecular Modeling and Simulation Applications of Molecular Dynamics Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, The University of Tokyo Tohru
BIOC 530 Fall, 2011 BIOC 530
 Fall, 2011 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic acids, control of enzymatic reactions. Please see the course
Fall, 2011 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic acids, control of enzymatic reactions. Please see the course
The change in free energy on transferring an ion from a medium of low dielectric constantε1 to one of high dielectric constant ε2:
 The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar.
 Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar. 25, 2002 Molecular Dynamics: Introduction At physiological conditions, the
Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar. 25, 2002 Molecular Dynamics: Introduction At physiological conditions, the
An introduction to Molecular Dynamics. EMBO, June 2016
 An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
Fluorine in Peptide and Protein Engineering
 Fluorine in Peptide and Protein Engineering Rita Fernandes Porto, February 11 th 2016 Supervisor: Prof. Dr. Beate Koksch 1 Fluorine a unique element for molecule design The most abundant halogen in earth
Fluorine in Peptide and Protein Engineering Rita Fernandes Porto, February 11 th 2016 Supervisor: Prof. Dr. Beate Koksch 1 Fluorine a unique element for molecule design The most abundant halogen in earth
MM-PBSA Validation Study. Trent E. Balius Department of Applied Mathematics and Statistics AMS
 MM-PBSA Validation Study Trent. Balius Department of Applied Mathematics and Statistics AMS 535 11-26-2008 Overview MM-PBSA Introduction MD ensembles one snap-shots relaxed structures nrichment Computational
MM-PBSA Validation Study Trent. Balius Department of Applied Mathematics and Statistics AMS 535 11-26-2008 Overview MM-PBSA Introduction MD ensembles one snap-shots relaxed structures nrichment Computational
Peptide folding in non-aqueous environments investigated with molecular dynamics simulations Soto Becerra, Patricia
 University of Groningen Peptide folding in non-aqueous environments investigated with molecular dynamics simulations Soto Becerra, Patricia IMPORTANT NOTE: You are advised to consult the publisher's version
University of Groningen Peptide folding in non-aqueous environments investigated with molecular dynamics simulations Soto Becerra, Patricia IMPORTANT NOTE: You are advised to consult the publisher's version
Lecture 11: Protein Folding & Stability
 Structure - Function Protein Folding: What we know Lecture 11: Protein Folding & Stability 1). Amino acid sequence dictates structure. 2). The native structure represents the lowest energy state for a
Structure - Function Protein Folding: What we know Lecture 11: Protein Folding & Stability 1). Amino acid sequence dictates structure. 2). The native structure represents the lowest energy state for a
Protein Folding & Stability. Lecture 11: Margaret A. Daugherty. Fall Protein Folding: What we know. Protein Folding
 Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2003 Structure - Function Protein Folding: What we know 1). Amino acid sequence dictates structure. 2). The native structure represents
Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2003 Structure - Function Protein Folding: What we know 1). Amino acid sequence dictates structure. 2). The native structure represents
What is Protein-Ligand Docking?
 MOLECULAR DOCKING Definition: What is Protein-Ligand Docking? Computationally predict the structures of protein-ligand complexes from their conformations and orientations. The orientation that maximizes
MOLECULAR DOCKING Definition: What is Protein-Ligand Docking? Computationally predict the structures of protein-ligand complexes from their conformations and orientations. The orientation that maximizes
Bioengineering 215. An Introduction to Molecular Dynamics for Biomolecules
 Bioengineering 215 An Introduction to Molecular Dynamics for Biomolecules David Parker May 18, 2007 ntroduction A principal tool to study biological molecules is molecular dynamics simulations (MD). MD
Bioengineering 215 An Introduction to Molecular Dynamics for Biomolecules David Parker May 18, 2007 ntroduction A principal tool to study biological molecules is molecular dynamics simulations (MD). MD
Kd = koff/kon = [R][L]/[RL]
![Kd = koff/kon = [R][L]/[RL] Kd = koff/kon = [R][L]/[RL]](/thumbs/96/127564193.jpg) Taller de docking y cribado virtual: Uso de herramientas computacionales en el diseño de fármacos Docking program GLIDE El programa de docking GLIDE Sonsoles Martín-Santamaría Shrödinger is a scientific
Taller de docking y cribado virtual: Uso de herramientas computacionales en el diseño de fármacos Docking program GLIDE El programa de docking GLIDE Sonsoles Martín-Santamaría Shrödinger is a scientific
Structural Bioinformatics (C3210) Molecular Mechanics
 Structural Bioinformatics (C3210) Molecular Mechanics How to Calculate Energies Calculation of molecular energies is of key importance in protein folding, molecular modelling etc. There are two main computational
Structural Bioinformatics (C3210) Molecular Mechanics How to Calculate Energies Calculation of molecular energies is of key importance in protein folding, molecular modelling etc. There are two main computational
Complicated, short range. þq 1 Q 2 /4p3 0 r (Coulomb energy) Q 2 u 2 /6(4p3 0 ) 2 ktr 4. u 2 1 u2 2 =3ð4p3 0Þ 2 ktr 6 ðkeesom energyþ
 Bonding ¼ Type of interaction Interaction energy w(r) Covalent, metallic Complicated, short range Charge charge þq 1 Q 2 /4p3 0 r (Coulomb energy) Charge dipole Qu cos q/4p3 0 r 2 Q 2 u 2 /6(4p3 0 ) 2
Bonding ¼ Type of interaction Interaction energy w(r) Covalent, metallic Complicated, short range Charge charge þq 1 Q 2 /4p3 0 r (Coulomb energy) Charge dipole Qu cos q/4p3 0 r 2 Q 2 u 2 /6(4p3 0 ) 2
Dihedral Angles. Homayoun Valafar. Department of Computer Science and Engineering, USC 02/03/10 CSCE 769
 Dihedral Angles Homayoun Valafar Department of Computer Science and Engineering, USC The precise definition of a dihedral or torsion angle can be found in spatial geometry Angle between to planes Dihedral
Dihedral Angles Homayoun Valafar Department of Computer Science and Engineering, USC The precise definition of a dihedral or torsion angle can be found in spatial geometry Angle between to planes Dihedral
Dielectric Boundary in Biomolecular Solvation Bo Li Department of Mathematics and Center for Theoretical Biological Physics UC San Diego
 Dielectric Boundary in Biomolecular Solvation Bo Li Department of Mathematics and Center for Theoretical Biological Physics UC San Diego International Conference on Free Boundary Problems Newton Institute,
Dielectric Boundary in Biomolecular Solvation Bo Li Department of Mathematics and Center for Theoretical Biological Physics UC San Diego International Conference on Free Boundary Problems Newton Institute,
Molecular Dynamics, Monte Carlo and Docking. Lecture 21. Introduction to Bioinformatics MNW2
 Molecular Dynamics, Monte Carlo and Docking Lecture 21 Introduction to Bioinformatics MNW2 Allowed phi-psi angles Red areas are preferred, yellow areas are allowed, and white is avoided 2.3a Hamiltonian
Molecular Dynamics, Monte Carlo and Docking Lecture 21 Introduction to Bioinformatics MNW2 Allowed phi-psi angles Red areas are preferred, yellow areas are allowed, and white is avoided 2.3a Hamiltonian
Protein Dynamics. The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron.
 Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Protein Dynamics The space-filling structures of myoglobin and hemoglobin show that there are no pathways for O 2 to reach the heme iron. Below is myoglobin hydrated with 350 water molecules. Only a small
Bioengineering & Bioinformatics Summer Institute, Dept. Computational Biology, University of Pittsburgh, PGH, PA
 Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Softwares for Molecular Docking. Lokesh P. Tripathi NCBS 17 December 2007
 Softwares for Molecular Docking Lokesh P. Tripathi NCBS 17 December 2007 Molecular Docking Attempt to predict structures of an intermolecular complex between two or more molecules Receptor-ligand (or drug)
Softwares for Molecular Docking Lokesh P. Tripathi NCBS 17 December 2007 Molecular Docking Attempt to predict structures of an intermolecular complex between two or more molecules Receptor-ligand (or drug)
Studying complex macromolecules
 Studying complex macromolecules M2 SERP-Chem Fabien Cailliez LCP Bât 349 fabien.cailliez@u-psud.fr Outline I. Classical forcefields II. III. IV. Molecular simulations and biomolecules One typical example:
Studying complex macromolecules M2 SERP-Chem Fabien Cailliez LCP Bât 349 fabien.cailliez@u-psud.fr Outline I. Classical forcefields II. III. IV. Molecular simulations and biomolecules One typical example:
OpenDiscovery: Automated Docking of Ligands to Proteins and Molecular Simulation
 OpenDiscovery: Automated Docking of Ligands to Proteins and Molecular Simulation Gareth Price Computational MiniProject OpenDiscovery Aims + Achievements Produce a high-throughput protocol to screen a
OpenDiscovery: Automated Docking of Ligands to Proteins and Molecular Simulation Gareth Price Computational MiniProject OpenDiscovery Aims + Achievements Produce a high-throughput protocol to screen a
Intermolecular and Surface Forces
 Intermolecular and Surface Forces ThirH FHitinn '' I I 111 \J& LM* КтЛ I Km I W I 1 Jacob N. Israelachvili UNIVERSITY OF CALIFORNIA SANTA BARBARA, CALIFORNIA, USA AMSTERDAM BOSTON HEIDELBERG LONDON NEW
Intermolecular and Surface Forces ThirH FHitinn '' I I 111 \J& LM* КтЛ I Km I W I 1 Jacob N. Israelachvili UNIVERSITY OF CALIFORNIA SANTA BARBARA, CALIFORNIA, USA AMSTERDAM BOSTON HEIDELBERG LONDON NEW
Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation
 Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation Ion:ion interactions What is the free energy of ion:ion interactions ΔG i-i? Consider an ion in a solution
Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation Ion:ion interactions What is the free energy of ion:ion interactions ΔG i-i? Consider an ion in a solution
Why study protein dynamics?
 Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Concept review: Binding equilibria
 Concept review: Binding equilibria 1 Binding equilibria and association/dissociation constants 2 The binding of a protein to a ligand at equilibrium can be written as: P + L PL And so the equilibrium constant
Concept review: Binding equilibria 1 Binding equilibria and association/dissociation constants 2 The binding of a protein to a ligand at equilibrium can be written as: P + L PL And so the equilibrium constant
High Throughput In-Silico Screening Against Flexible Protein Receptors
 John von Neumann Institute for Computing High Throughput In-Silico Screening Against Flexible Protein Receptors H. Sánchez, B. Fischer, H. Merlitz, W. Wenzel published in From Computational Biophysics
John von Neumann Institute for Computing High Throughput In-Silico Screening Against Flexible Protein Receptors H. Sánchez, B. Fischer, H. Merlitz, W. Wenzel published in From Computational Biophysics
Fondamenti di Chimica Farmaceutica. Computer Chemistry in Drug Research: Introduction
 Fondamenti di Chimica Farmaceutica Computer Chemistry in Drug Research: Introduction Introduction Introduction Introduction Computer Chemistry in Drug Design Drug Discovery: Target identification Lead
Fondamenti di Chimica Farmaceutica Computer Chemistry in Drug Research: Introduction Introduction Introduction Introduction Computer Chemistry in Drug Design Drug Discovery: Target identification Lead
BCB410 Protein-Ligand Docking Exercise Set Shirin Shahsavand December 11, 2011
 BCB410 Protein-Ligand Docking Exercise Set Shirin Shahsavand December 11, 2011 1. Describe the search algorithm(s) AutoDock uses for solving protein-ligand docking problems. AutoDock uses 3 different approaches
BCB410 Protein-Ligand Docking Exercise Set Shirin Shahsavand December 11, 2011 1. Describe the search algorithm(s) AutoDock uses for solving protein-ligand docking problems. AutoDock uses 3 different approaches
Modeling in Solution. Important Observations. Energy Concepts
 Modeling in Solution 1 Important Observations Various electrostatic effects for a solvated molecule are often less important than for an isolated gaseous molecule when the molecule is dissolved in a solvent
Modeling in Solution 1 Important Observations Various electrostatic effects for a solvated molecule are often less important than for an isolated gaseous molecule when the molecule is dissolved in a solvent
Principles of Physical Biochemistry
 Principles of Physical Biochemistry Kensal E. van Hold e W. Curtis Johnso n P. Shing Ho Preface x i PART 1 MACROMOLECULAR STRUCTURE AND DYNAMICS 1 1 Biological Macromolecules 2 1.1 General Principles
Principles of Physical Biochemistry Kensal E. van Hold e W. Curtis Johnso n P. Shing Ho Preface x i PART 1 MACROMOLECULAR STRUCTURE AND DYNAMICS 1 1 Biological Macromolecules 2 1.1 General Principles
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
schematic diagram; EGF binding, dimerization, phosphorylation, Grb2 binding, etc.
 Lecture 1: Noncovalent Biomolecular Interactions Bioengineering and Modeling of biological processes -e.g. tissue engineering, cancer, autoimmune disease Example: RTK signaling, e.g. EGFR Growth responses
Lecture 1: Noncovalent Biomolecular Interactions Bioengineering and Modeling of biological processes -e.g. tissue engineering, cancer, autoimmune disease Example: RTK signaling, e.g. EGFR Growth responses
Molecular Dynamics Simulation of HIV-1 Reverse. Transcriptase
 Molecular Dynamics Simulation of HIV-1 Reverse Transcriptase Abderrahmane Benghanem 1 Maria Kurnikova 2 1 Rensselaer Polytechnic Institute, Troy, NY 2 Carnegie Mellon University, Pittsburgh, PA June 16,
Molecular Dynamics Simulation of HIV-1 Reverse Transcriptase Abderrahmane Benghanem 1 Maria Kurnikova 2 1 Rensselaer Polytechnic Institute, Troy, NY 2 Carnegie Mellon University, Pittsburgh, PA June 16,
Free Energy Landscapes of Encounter Complexes in Protein-Protein Association
 1166 Biophysical Journal Volume 76 March 1999 1166 1178 Free Energy Landscapes of Encounter Complexes in Protein-Protein Association Carlos J. Camacho, Zhiping Weng, Sandor Vajda, and Charles DeLisi Department
1166 Biophysical Journal Volume 76 March 1999 1166 1178 Free Energy Landscapes of Encounter Complexes in Protein-Protein Association Carlos J. Camacho, Zhiping Weng, Sandor Vajda, and Charles DeLisi Department
Lecture 11: Potential Energy Functions
 Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Improved Solvation Models using Boundary Integral Equations
 Improved Solvation Models using Boundary Integral Equations Matthew Knepley and Jaydeep Bardhan Computational and Applied Mathematics Rice University SIAM Conference on the Life Sciences Minneapolis, MN
Improved Solvation Models using Boundary Integral Equations Matthew Knepley and Jaydeep Bardhan Computational and Applied Mathematics Rice University SIAM Conference on the Life Sciences Minneapolis, MN
Conformational sampling of macrocycles in solution and in the solid state
 Conformational sampling of macrocycles in solution and in the solid state Paul Hawkins, Ph.D. Head of Scientific Solutions Stanislaw Wlodek, Ph.D. Senior Scientific Developer 6/6/2018 https://berkonomics.com/?p=2437
Conformational sampling of macrocycles in solution and in the solid state Paul Hawkins, Ph.D. Head of Scientific Solutions Stanislaw Wlodek, Ph.D. Senior Scientific Developer 6/6/2018 https://berkonomics.com/?p=2437
Structural Bioinformatics (C3210) Molecular Docking
 Structural Bioinformatics (C3210) Molecular Docking Molecular Recognition, Molecular Docking Molecular recognition is the ability of biomolecules to recognize other biomolecules and selectively interact
Structural Bioinformatics (C3210) Molecular Docking Molecular Recognition, Molecular Docking Molecular recognition is the ability of biomolecules to recognize other biomolecules and selectively interact
Level-Set Variational Solvation Coupling Solute Molecular Mechanics with Continuum Solvent
 Level-Set Variational Solvation Coupling Solute Molecular Mechanics with Continuum Solvent Bo Li Department of Mathematics and Center for Theoretical Biological Physics (CTBP) University of California,
Level-Set Variational Solvation Coupling Solute Molecular Mechanics with Continuum Solvent Bo Li Department of Mathematics and Center for Theoretical Biological Physics (CTBP) University of California,
1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian. which is given in spherical coordinates by (2)
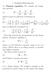 1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian operator (1) 2 = 2 x + 2 2 y + 2 2 z 2 which is given in spherical coordinates by (2) 2 = 1 ( r 2 ) + 1 r 2 r r r 2 sin θ θ and in
1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian operator (1) 2 = 2 x + 2 2 y + 2 2 z 2 which is given in spherical coordinates by (2) 2 = 1 ( r 2 ) + 1 r 2 r r r 2 sin θ θ and in
Molecular Interactions F14NMI. Lecture 4: worked answers to practice questions
 Molecular Interactions F14NMI Lecture 4: worked answers to practice questions http://comp.chem.nottingham.ac.uk/teaching/f14nmi jonathan.hirst@nottingham.ac.uk (1) (a) Describe the Monte Carlo algorithm
Molecular Interactions F14NMI Lecture 4: worked answers to practice questions http://comp.chem.nottingham.ac.uk/teaching/f14nmi jonathan.hirst@nottingham.ac.uk (1) (a) Describe the Monte Carlo algorithm
MOLECULAR RECOGNITION DOCKING ALGORITHMS. Natasja Brooijmans 1 and Irwin D. Kuntz 2
 Annu. Rev. Biophys. Biomol. Struct. 2003. 32:335 73 doi: 10.1146/annurev.biophys.32.110601.142532 Copyright c 2003 by Annual Reviews. All rights reserved First published online as a Review in Advance on
Annu. Rev. Biophys. Biomol. Struct. 2003. 32:335 73 doi: 10.1146/annurev.biophys.32.110601.142532 Copyright c 2003 by Annual Reviews. All rights reserved First published online as a Review in Advance on
APBS electrostatics in VMD - Software. APBS! >!Examples! >!Visualization! >! Contents
 Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Advanced in silico drug design
 Advanced in silico drug design RNDr. Martin Lepšík, Ph.D. Lecture: Advanced scoring Palacky University, Olomouc 2016 1 Outline 1. Scoring Definition, Types 2. Physics-based Scoring: Master Equation Terms
Advanced in silico drug design RNDr. Martin Lepšík, Ph.D. Lecture: Advanced scoring Palacky University, Olomouc 2016 1 Outline 1. Scoring Definition, Types 2. Physics-based Scoring: Master Equation Terms
Other Cells. Hormones. Viruses. Toxins. Cell. Bacteria
 Other Cells Hormones Viruses Toxins Cell Bacteria ΔH < 0 reaction is exothermic, tells us nothing about the spontaneity of the reaction Δ H > 0 reaction is endothermic, tells us nothing about the spontaneity
Other Cells Hormones Viruses Toxins Cell Bacteria ΔH < 0 reaction is exothermic, tells us nothing about the spontaneity of the reaction Δ H > 0 reaction is endothermic, tells us nothing about the spontaneity
Supplementary Figure 1 Preparation of PDA nanoparticles derived from self-assembly of PCDA. (a)
 Supplementary Figure 1 Preparation of PDA nanoparticles derived from self-assembly of PCDA. (a) Computer simulation on the self-assembly of PCDAs. Two kinds of totally different initial conformations were
Supplementary Figure 1 Preparation of PDA nanoparticles derived from self-assembly of PCDA. (a) Computer simulation on the self-assembly of PCDAs. Two kinds of totally different initial conformations were
Lectures 11-13: Electrostatics of Salty Solutions
 Lectures 11-13: Electrostatics of Salty Solutions Lecturer: Brigita Urbanc Office: 1-909 (E-mail: brigita@drexel.edu) Course website: www.physics.drexel.edu/~brigita/courses/biophys_011-01/ 1 Water as
Lectures 11-13: Electrostatics of Salty Solutions Lecturer: Brigita Urbanc Office: 1-909 (E-mail: brigita@drexel.edu) Course website: www.physics.drexel.edu/~brigita/courses/biophys_011-01/ 1 Water as
Dr. Sander B. Nabuurs. Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre
 Dr. Sander B. Nabuurs Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre The road to new drugs. How to find new hits? High Throughput
Dr. Sander B. Nabuurs Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre The road to new drugs. How to find new hits? High Throughput
Protein-Ligand Docking Methods
 Review Goal: Given a protein structure, predict its ligand bindings Protein-Ligand Docking Methods Applications: Function prediction Drug discovery etc. Thomas Funkhouser Princeton University S597A, Fall
Review Goal: Given a protein structure, predict its ligand bindings Protein-Ligand Docking Methods Applications: Function prediction Drug discovery etc. Thomas Funkhouser Princeton University S597A, Fall
Cheminformatics platform for drug discovery application
 EGI-InSPIRE Cheminformatics platform for drug discovery application Hsi-Kai, Wang Academic Sinica Grid Computing EGI User Forum, 13, April, 2011 1 Introduction to drug discovery Computing requirement of
EGI-InSPIRE Cheminformatics platform for drug discovery application Hsi-Kai, Wang Academic Sinica Grid Computing EGI User Forum, 13, April, 2011 1 Introduction to drug discovery Computing requirement of
Today s lecture. Molecular Mechanics and docking. Lecture 22. Introduction to Bioinformatics Docking - ZDOCK. Protein-protein docking
 C N F O N G A V B O N F O M A C S V U Molecular Mechanics and docking Lecture 22 ntroduction to Bioinformatics 2007 oday s lecture 1. Protein interaction and docking a) Zdock method 2. Molecular motion
C N F O N G A V B O N F O M A C S V U Molecular Mechanics and docking Lecture 22 ntroduction to Bioinformatics 2007 oday s lecture 1. Protein interaction and docking a) Zdock method 2. Molecular motion
Don t forget to bring your MD tutorial. Potential Energy (hyper)surface
 Don t forget to bring your MD tutorial Lab session starts at 1pm You will have to finish an MD/SMD exercise on α-conotoxin in oxidized and reduced forms Potential Energy (hyper)surface What is Force? Energy
Don t forget to bring your MD tutorial Lab session starts at 1pm You will have to finish an MD/SMD exercise on α-conotoxin in oxidized and reduced forms Potential Energy (hyper)surface What is Force? Energy
Aqueous solutions. Solubility of different compounds in water
 Aqueous solutions Solubility of different compounds in water The dissolution of molecules into water (in any solvent actually) causes a volume change of the solution; the size of this volume change is
Aqueous solutions Solubility of different compounds in water The dissolution of molecules into water (in any solvent actually) causes a volume change of the solution; the size of this volume change is
Biophysics II. Hydrophobic Bio-molecules. Key points to be covered. Molecular Interactions in Bio-molecular Structures - van der Waals Interaction
 Biophysics II Key points to be covered By A/Prof. Xiang Yang Liu Biophysics & Micro/nanostructures Lab Department of Physics, NUS 1. van der Waals Interaction 2. Hydrogen bond 3. Hydrophilic vs hydrophobic
Biophysics II Key points to be covered By A/Prof. Xiang Yang Liu Biophysics & Micro/nanostructures Lab Department of Physics, NUS 1. van der Waals Interaction 2. Hydrogen bond 3. Hydrophilic vs hydrophobic
ABSTRACT. Professor. Richard C. Stewart. Dept. of Cell Biology & Molecular Genetics.
 ABSTRACT Title of Document: PARAMETERIZATION OF AN ENERGY MODEL FOR SCORING OF ANTI-HIV DRUGS AND A COMPUTATIONAL METHOD OF LEAD COMPOUND OPTIMIZATION FOR DRUG DISCOVERY Himan Mookherjee, Master of Science,
ABSTRACT Title of Document: PARAMETERIZATION OF AN ENERGY MODEL FOR SCORING OF ANTI-HIV DRUGS AND A COMPUTATIONAL METHOD OF LEAD COMPOUND OPTIMIZATION FOR DRUG DISCOVERY Himan Mookherjee, Master of Science,
A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes
 THE JOURNAL OF CHEMICAL PHYSICS 122, 124706 2005 A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes Seiichiro Tanizaki
THE JOURNAL OF CHEMICAL PHYSICS 122, 124706 2005 A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes Seiichiro Tanizaki
Supplementary Methods
 Supplementary Methods MMPBSA Free energy calculation Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) has been widely used to calculate binding free energy for protein-ligand systems (1-7).
Supplementary Methods MMPBSA Free energy calculation Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) has been widely used to calculate binding free energy for protein-ligand systems (1-7).
A Gentle Introduction to (or Review of ) Fundamentals of Chemistry and Organic Chemistry
 Wright State University CORE Scholar Computer Science and Engineering Faculty Publications Computer Science and Engineering 2003 A Gentle Introduction to (or Review of ) Fundamentals of Chemistry and Organic
Wright State University CORE Scholar Computer Science and Engineering Faculty Publications Computer Science and Engineering 2003 A Gentle Introduction to (or Review of ) Fundamentals of Chemistry and Organic
A primer on pharmacology pharmacodynamics
 A primer on pharmacology pharmacodynamics Drug binding & effect Universidade do Algarve Faro 2017 by Ferdi Engels, Ph.D. 1 Pharmacodynamics Relation with pharmacokinetics? dosage plasma concentration site
A primer on pharmacology pharmacodynamics Drug binding & effect Universidade do Algarve Faro 2017 by Ferdi Engels, Ph.D. 1 Pharmacodynamics Relation with pharmacokinetics? dosage plasma concentration site
Computational modeling of G-Protein Coupled Receptors (GPCRs) has recently become
 Homology Modeling and Docking of Melatonin Receptors Andrew Kohlway, UMBC Jeffry D. Madura, Duquesne University 6/18/04 INTRODUCTION Computational modeling of G-Protein Coupled Receptors (GPCRs) has recently
Homology Modeling and Docking of Melatonin Receptors Andrew Kohlway, UMBC Jeffry D. Madura, Duquesne University 6/18/04 INTRODUCTION Computational modeling of G-Protein Coupled Receptors (GPCRs) has recently
Protein Folding experiments and theory
 Protein Folding experiments and theory 1, 2,and 3 Protein Structure Fig. 3-16 from Lehninger Biochemistry, 4 th ed. The 3D structure is not encoded at the single aa level Hydrogen Bonding Shared H atom
Protein Folding experiments and theory 1, 2,and 3 Protein Structure Fig. 3-16 from Lehninger Biochemistry, 4 th ed. The 3D structure is not encoded at the single aa level Hydrogen Bonding Shared H atom
The Molecular Dynamics Method
 H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations
 Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations Alexandre V. Morozov, Tanja Kortemme, Kiril Tsemekhman, David Baker
Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations Alexandre V. Morozov, Tanja Kortemme, Kiril Tsemekhman, David Baker
Molecular Forces in Biological Systems - Electrostatic Interactions; - Shielding of charged objects in solution
 Molecular Forces in Biological Systems - Electrostatic Interactions; - Shielding of charged objects in solution Electrostatic self-energy, effects of size and dielectric constant q q r r ε ε 1 ε 2? δq
Molecular Forces in Biological Systems - Electrostatic Interactions; - Shielding of charged objects in solution Electrostatic self-energy, effects of size and dielectric constant q q r r ε ε 1 ε 2? δq
Hands-on : Model Potential Molecular Dynamics
 Hands-on : Model Potential Molecular Dynamics OUTLINE 0. DL_POLY code introduction 0.a Input files 1. THF solvent molecule 1.a Geometry optimization 1.b NVE/NVT dynamics 2. Liquid THF 2.a Equilibration
Hands-on : Model Potential Molecular Dynamics OUTLINE 0. DL_POLY code introduction 0.a Input files 1. THF solvent molecule 1.a Geometry optimization 1.b NVE/NVT dynamics 2. Liquid THF 2.a Equilibration
Molecular dynamics simulation. CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror
 Molecular dynamics simulation CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror 1 Outline Molecular dynamics (MD): The basic idea Equations of motion Key properties of MD simulations Sample applications
Molecular dynamics simulation CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror 1 Outline Molecular dynamics (MD): The basic idea Equations of motion Key properties of MD simulations Sample applications
Scuola di Chimica Computazionale
 Societa Chimica Italiana Gruppo Interdivisionale di Chimica Computazionale Scuola di Chimica Computazionale Introduzione, per Esercizi, all Uso del Calcolatore in Chimica Organica e Biologica Modellistica
Societa Chimica Italiana Gruppo Interdivisionale di Chimica Computazionale Scuola di Chimica Computazionale Introduzione, per Esercizi, all Uso del Calcolatore in Chimica Organica e Biologica Modellistica
Principles of Drug Design
 (16:663:502) Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu For more current information, please check WebCT at https://webct.rutgers.edu
(16:663:502) Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu For more current information, please check WebCT at https://webct.rutgers.edu
Carbohydrate- Protein interac;ons are Cri;cal in Life and Death. Other Cells. Hormones. Viruses. Toxins. Cell. Bacteria
 ther Cells Carbohydrate- Protein interac;ons are Cri;cal in Life and Death ormones Viruses Toxins Cell Bacteria ow to Model Protein- ligand interac;ons? Protein Protein Protein DNA/RNA Protein Carbohydrate
ther Cells Carbohydrate- Protein interac;ons are Cri;cal in Life and Death ormones Viruses Toxins Cell Bacteria ow to Model Protein- ligand interac;ons? Protein Protein Protein DNA/RNA Protein Carbohydrate
Introduction to" Protein Structure
 Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
Introduction to" Protein Structure Function, evolution & experimental methods Thomas Blicher, Center for Biological Sequence Analysis Learning Objectives Outline the basic levels of protein structure.
User Guide for LeDock
 User Guide for LeDock Hongtao Zhao, PhD Email: htzhao@lephar.com Website: www.lephar.com Copyright 2017 Hongtao Zhao. All rights reserved. Introduction LeDock is flexible small-molecule docking software,
User Guide for LeDock Hongtao Zhao, PhD Email: htzhao@lephar.com Website: www.lephar.com Copyright 2017 Hongtao Zhao. All rights reserved. Introduction LeDock is flexible small-molecule docking software,
Computational methods of studying the binding of toxins from venomous animals to biological ion channels: theory and applications
 Computational methods of studying the binding of toxins from venomous animals to biological ion channels: theory and applications Dan Gordon, Rong Chen and Shin-Ho Chung * Research School of Biology, The
Computational methods of studying the binding of toxins from venomous animals to biological ion channels: theory and applications Dan Gordon, Rong Chen and Shin-Ho Chung * Research School of Biology, The
