Improved Solvation Models using Boundary Integral Equations
|
|
|
- Antonia Wilson
- 5 years ago
- Views:
Transcription
1 Improved Solvation Models using Boundary Integral Equations Matthew Knepley and Jaydeep Bardhan Computational and Applied Mathematics Rice University SIAM Conference on the Life Sciences Minneapolis, MN August 9, 2018 M. Knepley (Rice) Solvation LS18 1 / 52
2 Collaborators Amir Molvai Tabrizi (postdoc, NE) Tom Klotz (grad student, Rice) Spencer Goossens (grad student, NE) Ali Rahimi (grad student, NE) M. Knepley (Rice) Solvation LS18 3 / 52
3 Main Point Solvation computation can benefit from non-poisson models. M. Knepley (Rice) Solvation LS18 4 / 52
4 Bioelectrostatics The Natural World Induced Surface Charge on Lysozyme M. Knepley (Rice) Solvation LS18 5 / 52
5 Bioelectrostatics Physical Model Electrostatic Potential φ Region I: protein Region II: solvent Surface M. Knepley (Rice) Solvation LS18 6 / 52
6 Bioelectrostatics Mathematical Model We can write a Boundary Integral Equation (BIE) for the induced surface charge σ, σ( r) + ˆɛ Γ σ( r )d 2 r Q n( r) 4π r r = ˆɛ q k n( r) 4π r r k (I + ˆɛD ) σ( r) = k=1 where we define ˆɛ = 2 ɛ I ɛ II ɛ I + ɛ II < 0 M. Knepley (Rice) Solvation LS18 7 / 52
7 Bioelectrostatics Mathematical Model This is equivalent to a PDE model for the potentials Φ I,II in the two regions, and boundary conditions at the solute surface: ɛ I Φ I = ɛ II Φ II = 0 Q q k δ( r r k ) k=1 Φ I r=b = Φ II r=b ɛ I Φ I r r=b = ɛ II Φ II r r=b M. Knepley (Rice) Solvation LS18 8 / 52
8 Bioelectrostatics Mathematical Model The reaction potential is given by φ R σ( r )d 2 r ( r) = 4πɛ 1 r r = Cσ Γ which defines G es, the electrostatic part of the solvation free energy G es = 1 q, φ R 2 = 1 q, Lq 2 = 1 q, CA 1 Bq 2 where Bq = ˆɛ Ω n( r) Aσ = I + ˆɛD q( r )d 3 r 4π r r M. Knepley (Rice) Solvation LS18 9 / 52
9 Outline Some History 1 Some History 2 Improving the Poisson Operator M. Knepley (Rice) Solvation LS18 10 / 52
10 Some History Generalized Born Approximation The pairwise energy between charges is defined by the Still equation: Ges ij = 1 ( 1 1 ) N 8π ɛ II ɛ I i,j where the effective Born radius is R i = 1 ( 1 1 ) 1 8π ɛ II ɛ I E i q i q j r 2 ij + R i R j e r 2 ij /4R i R j where E i is the self-energy of the charge q i, the electrostatic energy when atom i has unit charge and all others are neutral. M. Knepley (Rice) Solvation LS18 11 / 52
11 GB Problems Some History No global potential solution, only energy No analysis of the error For example, Salsbury 2006 consists of parameter tuning No path for systematic improvement For example, Sigalov 2006 changes the model The same atoms have different radii in different molecules, solvents temperatures LOTS of parameters Nina, Beglov, Roux 1997 M. Knepley (Rice) Solvation LS18 12 / 52
12 GB Problems Some History No global potential solution, only energy No analysis of the error For example, Salsbury 2006 consists of parameter tuning No path for systematic improvement For example, Sigalov 2006 changes the model The same atoms have different radii in different molecules, solvents temperatures LOTS of parameters Nina, Beglov, Roux 1997 M. Knepley (Rice) Solvation LS18 12 / 52
13 Some History Implicit Solvent Models State-of-the-art solvation models still use the same variation in radii Biomolecular electrostatics I want your solvation (model), J. Bardhan, Comp. Sci. & Disc., 5(1), (2012) M. Knepley (Rice) Solvation LS18 13 / 52
14 Outline Improving the Poisson Operator 1 Some History 2 Improving the Poisson Operator M. Knepley (Rice) Solvation LS18 14 / 52
15 Improving the Poisson Operator Origins of Electrostatic Asymmetry Explicit solvent molecular dynamics FEP Standard Maxwell boundary condition Proposed nonlinear boundary condition 1.4 / σ E n Coul E n Coul M. Knepley (Rice) Solvation LS18 15 / 52
16 Improving the Poisson Operator Origins of Electrostatic Asymmetry M. Knepley (Rice) Solvation LS18 16 / 52
17 Improving the Poisson Operator Origins of Electrostatic Asymmetry 10 Deeply buried charge The nonzero slope at q=0 is the static potential 0 G charging (kcal/mol) Solvent-exposed charge Symbols: explicit-solvent freeenergy perturbation calculations Lines: piecewise-linear model plus static potential q Asymmetry for a deeply buried charge is exclusively due to the static potential Asymmetry for a solvent-exposed charge results from both the static potential and the hydrogen-oxygen size difference M. Knepley (Rice) Solvation LS18 17 / 52
18 Main Idea Improving the Poisson Operator Maxwell Boundary Condition Assume the model and energy, and derive the radii, Φ I ɛ I n = ɛ Φ II II n M. Knepley (Rice) Solvation LS18 18 / 52
19 Main Idea Improving the Poisson Operator Solvation-Layer Interface Condition (SLIC) Assume the energy and radii, and derive the model. (ɛ I ɛh(e n )) Φ I n = (ɛ II ɛh(e n )) Φ II n M. Knepley (Rice) Solvation LS18 18 / 52
20 Main Idea Improving the Poisson Operator Using our correspondence with the BIE form, ( I + h(e n ) + ˆɛ ( 12 )) I + D σ = ˆɛ Q k=1 G n where h is a diagonal nonlinear integral operator. M. Knepley (Rice) Solvation LS18 18 / 52
21 SLIC Boundary Perturbation Improving the Poisson Operator h(e n ) = α tanh (βe n γ) + µ where α Size of the asymmetry β Width of the transition region γ The transition field strength µ Assures h(0) = 0, so µ = α tanh( γ) M. Knepley (Rice) Solvation LS18 19 / 52
22 Improving the Poisson Operator Accuracy of SLIC Residues 0 20 Charging free energy (kcal/mol) MD FEP Poisson, Roux radii NLBC, extrapolated NLBC, 4 vertices/a 2 NLBC, 2 vertices/a 2 ARG ASP CYS GLU HIS LYS TYR M. Knepley (Rice) Solvation LS18 20 / 52
23 Improving the Poisson Operator Accuracy of SLIC Protonation Symmetric LPB NLBC LPB G prot,salt (kcal/mol) ARG ASP CYS GLU HIS LYS TYR M. Knepley (Rice) Solvation LS18 21 / 52
24 Improving the Poisson Operator Accuracy of SLIC Synthetic Molecules 20 Hydration Free Energy (kcal/mol) Negative (MD, Mobley et al) Positive (MD, Mobley et al) Negative (NLBC, this work) Positive (NLBC, this work) Problem Index M. Knepley (Rice) Solvation LS18 22 / 52
25 Improving the Poisson Operator Accuracy of SLIC Synthetic Molecules Asymmetry in Electrostatic Free Energy (kcal/mol) MD, Mobley et al NLBC, this work Number of Atoms in Ring M. Knepley (Rice) Solvation LS18 23 / 52
26 Improving the Poisson Operator Accuracy of SLIC Synthetic Molecules 0 Electrostatic Solvation Free Energy (kcal/mol) Negative (MD, Mobley et al) 60 Positive (MD, Mobley et al) Negative (NLBC, this work) Positive (NLBC, this work) Number of Atoms in Ring M. Knepley (Rice) Solvation LS18 24 / 52
27 Improving the Poisson Operator Thermodynamics The parameters show linear temperature dependence α (Angstroms) W 0.6 MeOH F 15 AN DMF M. Knepley (Rice) Solvation LS18 25 / AN F
28 Improving the Poisson Operator Model Validation Courtesy A. Molvai Tabrizi Model validation and verification using experiment Water H 2 O Formamide CH 3 NO Dimethyl sulfoxide C 2 H 6 OS Methanol CH 3 OH Acetonitrile C 2 H 3 N Nitromethane CH 3 NO 2 Ethanol C 2 H 5 OH Dimethyl formamide C 3 H 7 NO Propylene carbonate CH 3 C 2 H 3 O 2 CO Northeastern University Mechanical and Industrial Engineering Bardhan Lab M. Knepley (Rice) Solvation LS18 26 / 52
29 Improving the Poisson Operator Model Validation Courtesy A. Molvai Tabrizi M. Knepley (Rice) Solvation LS18 27 / 52
30 Improving the Poisson Operator Model Validation Courtesy A. Molvai Tabrizi M. Knepley (Rice) Solvation LS18 28 / 52
31 Improving the Poisson Operator Model Validation Courtesy A. Molvai Tabrizi Model validation and verification using experiment Dimethyl 25 o C C 3 H 7 NO Computed Experimental Northeastern University Mechanical and Industrial Engineering Bardhan Lab M. Knepley (Rice) Solvation LS18 29 / 52
32 Improving the Poisson Operator Model Validation Courtesy A. Molvai Tabrizi A. Molavi Tabrizi, M.G. Knepley, and J.P. Bardhan, Generalising the mean spherical approximation as a multiscale, nonlinear boundary condition at the solute-solvent interface, Molecular Physics (2016). M. Knepley (Rice) Solvation LS18 30 / 52
33 Improving the Poisson Operator Thermodynamic Predictions Courtesy A. Molvai Tabrizi Experimental Data in Parentheses M. Knepley (Rice) Solvation LS18 31 / 52
34 Improving the Poisson Operator Thermodynamic Predictions Courtesy A. Molvai Tabrizi A. Molavi Tabrizi, S. Goossens, A. Rahimi, M.G. Knepley, and J.P. Bardhan, Predicting solvation free energies and thermodynamics in polar solvents and mixtures using a solvation-layer interface condition. Journal of Chemical Physical (2017). M. Knepley (Rice) Solvation LS18 32 / 52
35 Improving the Poisson Operator Main Successes of SLIC Accurate charging free energy using crystal radii (no fitting/temp dep) for (de-)protonation for individual atoms for mixtures M. Knepley (Rice) Solvation LS18 33 / 52
36 Improving the Poisson Operator Main Successes of SLIC Accurate charging free energy using crystal radii (no fitting/temp dep) for (de-)protonation for individual atoms for mixtures M. Knepley (Rice) Solvation LS18 33 / 52
37 Improving the Poisson Operator Main Successes of SLIC Accurate charging free energy using crystal radii (no fitting/temp dep) for (de-)protonation for individual atoms for mixtures M. Knepley (Rice) Solvation LS18 33 / 52
38 Improving the Poisson Operator Main Successes of SLIC Accurate charging free energy using crystal radii (no fitting/temp dep) for (de-)protonation for individual atoms for mixtures M. Knepley (Rice) Solvation LS18 33 / 52
39 Improving the Poisson Operator Main Successes of SLIC Accurate charging free energy using crystal radii (no fitting/temp dep) for (de-)protonation for individual atoms for mixtures M. Knepley (Rice) Solvation LS18 33 / 52
40 Improving the Poisson Operator Main Successes of SLIC Accurate transfer free energy for water-octanol system on par with explicit-solvent MD Reinterpretation of Mean Spherical Approximation Explains temperature dependence of model coefficients M. Knepley (Rice) Solvation LS18 34 / 52
41 Improving the Poisson Operator Main Successes of SLIC Accurate transfer free energy for water-octanol system on par with explicit-solvent MD Reinterpretation of Mean Spherical Approximation Explains temperature dependence of model coefficients M. Knepley (Rice) Solvation LS18 34 / 52
42 Improving the Poisson Operator What is missing from SLIC? Large packing fraction No charge oscillation or overcharging Classical DFT? (Gillespie, Microfluidics and Nanofluidics, 2015) No dielectric saturation Possible with different condition No long range correlations Use nonlocal dielectric M. Knepley (Rice) Solvation LS18 35 / 52
43 Future Work Future Work More complex solutes Better nonlinear solvers Mixtures Preliminary work is accurate Integration into community code Psi4, QChem, APBS M. Knepley (Rice) Solvation LS18 36 / 52
44 Thank You!
45 Outline Future Work Approximate Boundary Conditions Approximate Boundary Conditions M. Knepley (Rice) Solvation LS18 38 / 52
46 Bioelectrostatics Physical Model Future Work Approximate Boundary Conditions Electrostatic Potential φ Region I: protein Region II: solvent Surface M. Knepley (Rice) Solvation LS18 39 / 52
47 Future Work Kirkwood s Solution (1934) Approximate Boundary Conditions The potential inside Region I is given by Φ I = Q k=1 q k ɛ 1 r r k + ψ, and the potential in Region II is given by Φ II = n n=0 m= n C nm r n+1 Pm n (cos θ)e imφ. M. Knepley (Rice) Solvation LS18 40 / 52
48 Future Work Kirkwood s Solution (1934) Approximate Boundary Conditions The reaction potential ψ is expanded in a series n ψ = B nm r n Pn m (cos θ)e imφ. n=0 m= n and the source distribution is also expanded Q k=1 q k ɛ 1 r r k = n n=0 m= n E nm ɛ 1 r n+1 Pm n (cos θ)e imφ. M. Knepley (Rice) Solvation LS18 41 / 52
49 Future Work Kirkwood s Solution (1934) Approximate Boundary Conditions By applying the boundary conditions, letting the sphere have radius b, Φ I r=b = Φ II r=b ɛ I Φ I r r=b = ɛ II Φ II r r=b we can eliminate C nm, and determine the reaction potential coefficients in terms of the source distribution, B nm = 1 (ɛ I ɛ II )(n + 1) ɛ I b 2n+1 ɛ I n + ɛ II (n + 1) E nm. M. Knepley (Rice) Solvation LS18 42 / 52
50 Future Work Approximate Boundary Conditions Approximate Boundary Conditions Theorem: The BIBEE boundary integral operator approximations ( A CFA = I 1 + ˆɛ ) 2 A P = I have an equivalent PDE formulation, Q ɛ I Φ CFA,P = q k δ( r r k ) ɛ II Φ CFA,P = 0 k=1 ɛ I Φ C I ɛ II r r=b = Φ II r or ψ CFA r=b r Φ I r=b = Φ II r=b 3ɛ I ɛ II ɛ I + ɛ II Φ C I r r=b = Φ II r ψ P r r=b, where Φ C 1 is the Coulomb field due to interior charges. M. Knepley (Rice) Solvation LS18 43 / 52
51 Future Work Approximate Boundary Conditions Approximate Boundary Conditions Theorem: For spherical solute, the BIBEE boundary integral operator approximations have eigenspaces are identical to that of the original operator. BEM eigenvector e i e j BIBEE/P eigenvector M. Knepley (Rice) Solvation LS18 43 / 52
52 Future Work Proof of PDE Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these PDEs are equivalent to the original BIEs, Start with the fundamental solution to Laplace s equation G(r, r ) Note that Γ G(r, r )σ(r )dγ satisfies the bulk equation and decay at infinity Insertion into the approximate BC gives the BIBEE boundary integral approximation M. Knepley (Rice) Solvation LS18 44 / 52
53 Future Work Proof of PDE Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these PDEs are equivalent to the original BIEs, Start with the fundamental solution to Laplace s equation G(r, r ) Note that Γ G(r, r )σ(r )dγ satisfies the bulk equation and decay at infinity Insertion into the approximate BC gives the BIBEE boundary integral approximation M. Knepley (Rice) Solvation LS18 44 / 52
54 Future Work Proof of PDE Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these PDEs are equivalent to the original BIEs, Start with the fundamental solution to Laplace s equation G(r, r ) Note that Γ G(r, r )σ(r )dγ satisfies the bulk equation and decay at infinity Insertion into the approximate BC gives the BIBEE boundary integral approximation M. Knepley (Rice) Solvation LS18 44 / 52
55 Future Work Proof of PDE Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these PDEs are equivalent to the original BIEs, Start with the fundamental solution to Laplace s equation G(r, r ) Note that Γ G(r, r )σ(r )dγ satisfies the bulk equation and decay at infinity Insertion into the approximate BC gives the BIBEE boundary integral approximation M. Knepley (Rice) Solvation LS18 44 / 52
56 Future Work Proof of Eigenspace Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these integral operators share a common eigenbasis, Note that, for a spherical boundary, D is compact and has a pure point spectrum Examine the effect of the operator on a unit spherical harmonic charge distribution Use completeness of the spherical harmonic basis M. Knepley (Rice) Solvation LS18 45 / 52
57 Future Work Proof of Eigenspace Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these integral operators share a common eigenbasis, Note that, for a spherical boundary, D is compact and has a pure point spectrum Examine the effect of the operator on a unit spherical harmonic charge distribution Use completeness of the spherical harmonic basis M. Knepley (Rice) Solvation LS18 45 / 52
58 Future Work Proof of Eigenspace Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these integral operators share a common eigenbasis, Note that, for a spherical boundary, D is compact and has a pure point spectrum Examine the effect of the operator on a unit spherical harmonic charge distribution Use completeness of the spherical harmonic basis M. Knepley (Rice) Solvation LS18 45 / 52
59 Future Work Proof of Eigenspace Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these integral operators share a common eigenbasis, Note that, for a spherical boundary, D is compact and has a pure point spectrum Examine the effect of the operator on a unit spherical harmonic charge distribution Use completeness of the spherical harmonic basis M. Knepley (Rice) Solvation LS18 45 / 52
60 Future Work Proof of Eigenspace Equivalence Approximate Boundary Conditions Proof: Bardhan and Knepley, JCP, 135(12), In order to show that these integral operators share a common eigenbasis, Note that, for a spherical boundary, D is compact and has a pure point spectrum Examine the effect of the operator on a unit spherical harmonic charge distribution Use completeness of the spherical harmonic basis The result does not hold for general boundaries. M. Knepley (Rice) Solvation LS18 45 / 52
61 Series Solutions Future Work Approximate Boundary Conditions Note that the approximate solutions are separable: B nm = 1 ɛ 1 n + ɛ 2 (n + 1) γ nm B CFA nm = 1 ɛ 2 1 2n + 1 γ nm B P nm = 1 1 ɛ 1 + ɛ 2 n + 1 γ nm. 2 If ɛ I = ɛ II = ɛ, both approximations are exact: B nm = B CFA nm = B P nm = 1 ɛ(2n + 1) γ nm. M. Knepley (Rice) Solvation LS18 46 / 52
62 Series Solutions Future Work Approximate Boundary Conditions Note that the approximate solutions are separable: B nm = 1 ɛ 1 n + ɛ 2 (n + 1) γ nm B CFA nm = 1 ɛ 2 1 2n + 1 γ nm B P nm = 1 1 ɛ 1 + ɛ 2 n + 1 γ nm. 2 If ɛ I = ɛ II = ɛ, both approximations are exact: B nm = B CFA nm = B P nm = 1 ɛ(2n + 1) γ nm. M. Knepley (Rice) Solvation LS18 46 / 52
63 Asymptotics Future Work Approximate Boundary Conditions BIBEE/CFA is exact for the n = 0 mode, B 00 = B CFA 00 = γ 00 ɛ 2, whereas BIBEE/P approaches the exact response in the limit n : lim B nm = lim B P n n nm = 1 (ɛ 1 + ɛ 2 )n γ nm. M. Knepley (Rice) Solvation LS18 47 / 52
64 Asymptotics Future Work Approximate Boundary Conditions BIBEE/CFA is exact for the n = 0 mode, B 00 = B CFA 00 = γ 00 ɛ 2, whereas BIBEE/P approaches the exact response in the limit n : lim B nm = lim B P n n nm = 1 (ɛ 1 + ɛ 2 )n γ nm. M. Knepley (Rice) Solvation LS18 47 / 52
65 Asymptotics Future Work Approximate Boundary Conditions In the limit ɛ 1 /ɛ 2 0, lim B nm = ɛ 1 /ɛ 2 0 lim ɛ 1 /ɛ 2 0 BCFA nm = lim ɛ 1 /ɛ 2 0 BP nm = γ nm ɛ 2 (n + 1) γ nm ɛ 2 (2n + 1), γ nm ɛ 2 ( n so that the approximation ratios are given by ), B CFA nm B nm = n + 1 2n + 1, Bnm P = n + 1 B nm n M. Knepley (Rice) Solvation LS18 48 / 52
66 Improved Accuracy Future Work Approximate Boundary Conditions BIBEE/I interpolates between BIBEE/CFA and BIBEE/P Bardhan, Knepley, JCP, M. Knepley (Rice) Solvation LS18 49 / 52
67 Basis Augmentation Future Work Approximate Boundary Conditions We examined the more complex problem of protein-ligand binding using trypsin and bovine pancreatic trypsin inhibitor (BPTI), using electrostatic component analysis to identify residue contributions to binding and molecular recognition. M. Knepley (Rice) Solvation LS18 50 / 52
68 Basis Augmentation Future Work Approximate Boundary Conditions Looking at an ensemble of synthetic proteins, we can see that BIBEE/CFA becomes more accurate as the monopole moment increases, and BIBEE/P more accurate as it decreases. BIBEE/I is accurate for spheres, but must be extended for ellipses. M. Knepley (Rice) Solvation LS18 51 / 52
69 Basis Augmentation Future Work Approximate Boundary Conditions For ellipses, we add a few low order multipole moments, up to the octopole, to recover 5% accuracy for all synthetic proteins tested. M. Knepley (Rice) Solvation LS18 52 / 52
Computational Bioelectrostatics
 Computational Bioelectrostatics Matthew Knepley Computational and Applied Mathematics Rice University Scientific Computing Seminar University of Houston Houston, TX April 20, 2017 M. Knepley (CAAM) MGK
Computational Bioelectrostatics Matthew Knepley Computational and Applied Mathematics Rice University Scientific Computing Seminar University of Houston Houston, TX April 20, 2017 M. Knepley (CAAM) MGK
Computational Bioelectrostatics
 Computational Bioelectrostatics Matthew Knepley Computation Institute University of Chicago Numerical Analysis Seminar Texas A&M University College Station, TX January 27, 2015 M. Knepley (UC) MGK TAMU
Computational Bioelectrostatics Matthew Knepley Computation Institute University of Chicago Numerical Analysis Seminar Texas A&M University College Station, TX January 27, 2015 M. Knepley (UC) MGK TAMU
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany
 Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals Phase Equilibria Phase diagrams and classical thermodynamics
Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals Phase Equilibria Phase diagrams and classical thermodynamics
Potential Energy (hyper)surface
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = " d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = " d dx U(x) Conformation
Laplacian-Centered Poisson Solvers and Multilevel Summation Algorithms
 Laplacian-Centered Poisson Solvers and Multilevel Summation Algorithms Dmitry Yershov 1 Stephen Bond 1 Robert Skeel 2 1 University of Illinois at Urbana-Champaign 2 Purdue University 2009 SIAM Annual Meeting
Laplacian-Centered Poisson Solvers and Multilevel Summation Algorithms Dmitry Yershov 1 Stephen Bond 1 Robert Skeel 2 1 University of Illinois at Urbana-Champaign 2 Purdue University 2009 SIAM Annual Meeting
All-atom Molecular Mechanics. Trent E. Balius AMS 535 / CHE /27/2010
 All-atom Molecular Mechanics Trent E. Balius AMS 535 / CHE 535 09/27/2010 Outline Molecular models Molecular mechanics Force Fields Potential energy function functional form parameters and parameterization
All-atom Molecular Mechanics Trent E. Balius AMS 535 / CHE 535 09/27/2010 Outline Molecular models Molecular mechanics Force Fields Potential energy function functional form parameters and parameterization
Rapid evaluation of electrostatic interactions in multi-phase media
 Rapid evaluation of electrostatic interactions in multi-phase media P.G. Martinsson, The University of Colorado at Boulder Acknowledgements: Some of the work presented is joint work with Mark Tygert and
Rapid evaluation of electrostatic interactions in multi-phase media P.G. Martinsson, The University of Colorado at Boulder Acknowledgements: Some of the work presented is joint work with Mark Tygert and
Research Statement. Shenggao Zhou. November 3, 2014
 Shenggao Zhou November 3, My research focuses on: () Scientific computing and numerical analysis (numerical PDEs, numerical optimization, computational fluid dynamics, and level-set method for interface
Shenggao Zhou November 3, My research focuses on: () Scientific computing and numerical analysis (numerical PDEs, numerical optimization, computational fluid dynamics, and level-set method for interface
arxiv: v1 [physics.chem-ph] 25 Sep 2014
![arxiv: v1 [physics.chem-ph] 25 Sep 2014 arxiv: v1 [physics.chem-ph] 25 Sep 2014](/thumbs/89/99600310.jpg) Modeling Charge-Sign Asymmetric Solvation Free Energies With Nonlinear Boundary Conditions Jaydeep P. Bardhan 1 and Matthew G. Knepley 2 1 Dept. of Electrical and Computer Enginering, Northeastern University,
Modeling Charge-Sign Asymmetric Solvation Free Energies With Nonlinear Boundary Conditions Jaydeep P. Bardhan 1 and Matthew G. Knepley 2 1 Dept. of Electrical and Computer Enginering, Northeastern University,
1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian. which is given in spherical coordinates by (2)
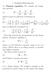 1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian operator (1) 2 = 2 x + 2 2 y + 2 2 z 2 which is given in spherical coordinates by (2) 2 = 1 ( r 2 ) + 1 r 2 r r r 2 sin θ θ and in
1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian operator (1) 2 = 2 x + 2 2 y + 2 2 z 2 which is given in spherical coordinates by (2) 2 = 1 ( r 2 ) + 1 r 2 r r r 2 sin θ θ and in
Solvation and Macromolecular Structure. The structure and dynamics of biological macromolecules are strongly influenced by water:
 Overview Solvation and Macromolecular Structure The structure and dynamics of biological macromolecules are strongly influenced by water: Electrostatic effects: charges are screened by water molecules
Overview Solvation and Macromolecular Structure The structure and dynamics of biological macromolecules are strongly influenced by water: Electrostatic effects: charges are screened by water molecules
The change in free energy on transferring an ion from a medium of low dielectric constantε1 to one of high dielectric constant ε2:
 The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
ψ s a ˆn a s b ˆn b ψ Hint: Because the state is spherically symmetric the answer can depend only on the angle between the two directions.
 1. Quantum Mechanics (Fall 2004) Two spin-half particles are in a state with total spin zero. Let ˆn a and ˆn b be unit vectors in two arbitrary directions. Calculate the expectation value of the product
1. Quantum Mechanics (Fall 2004) Two spin-half particles are in a state with total spin zero. Let ˆn a and ˆn b be unit vectors in two arbitrary directions. Calculate the expectation value of the product
Molecular Simulation III
 Molecular Simulation III Quantum Chemistry Classical Mechanics E = Ψ H Ψ ΨΨ U = E bond +E angle +E torsion +E non-bond Molecular Dynamics Jeffry D. Madura Department of Chemistry & Biochemistry Center
Molecular Simulation III Quantum Chemistry Classical Mechanics E = Ψ H Ψ ΨΨ U = E bond +E angle +E torsion +E non-bond Molecular Dynamics Jeffry D. Madura Department of Chemistry & Biochemistry Center
Solutions and Non-Covalent Binding Forces
 Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes
 THE JOURNAL OF CHEMICAL PHYSICS 122, 124706 2005 A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes Seiichiro Tanizaki
THE JOURNAL OF CHEMICAL PHYSICS 122, 124706 2005 A generalized Born formalism for heterogeneous dielectric environments: Application to the implicit modeling of biological membranes Seiichiro Tanizaki
Alchemical free energy calculations in OpenMM
 Alchemical free energy calculations in OpenMM Lee-Ping Wang Stanford Department of Chemistry OpenMM Workshop, Stanford University September 7, 2012 Special thanks to: John Chodera, Morgan Lawrenz Outline
Alchemical free energy calculations in OpenMM Lee-Ping Wang Stanford Department of Chemistry OpenMM Workshop, Stanford University September 7, 2012 Special thanks to: John Chodera, Morgan Lawrenz Outline
Alek Marenich Cramer -Truhlar Group. University of Minnesota
 and Computational Electrochemistry Alek Marenich Cramer -Truhlar Group University of Minnesota Outline Solvation and charge models Energetics and dynamics in solution Computational electrochemistry Future
and Computational Electrochemistry Alek Marenich Cramer -Truhlar Group University of Minnesota Outline Solvation and charge models Energetics and dynamics in solution Computational electrochemistry Future
Accounting for Solvation in Quantum Chemistry. Comp chem spring school 2014 CSC, Finland
 Accounting for Solvation in Quantum Chemistry Comp chem spring school 2014 CSC, Finland In solution or in a vacuum? Solvent description is important when: Polar solvent: electrostatic stabilization Solvent
Accounting for Solvation in Quantum Chemistry Comp chem spring school 2014 CSC, Finland In solution or in a vacuum? Solvent description is important when: Polar solvent: electrostatic stabilization Solvent
Biochemistry 675, Lecture 8 Electrostatic Interactions
 Biochemistry 675, Lecture 8 Electrostatic Interactions Previous Classes Hydrophobic Effect Hydrogen Bonds Today: Electrostatic Interactions Reading: Handout:P.R. Bergethon & E.R. Simons, Biophysical Chemistry,
Biochemistry 675, Lecture 8 Electrostatic Interactions Previous Classes Hydrophobic Effect Hydrogen Bonds Today: Electrostatic Interactions Reading: Handout:P.R. Bergethon & E.R. Simons, Biophysical Chemistry,
Variational Implicit Solvation of Biomolecules: From Theory to Numerical Computations
 Variational Implicit Solvation of Biomolecules: From Theory to Numerical Computations Bo Li Department of Mathematics and Center for Theoretical Biological Physics UC San Diego CECAM Workshop: New Perspectives
Variational Implicit Solvation of Biomolecules: From Theory to Numerical Computations Bo Li Department of Mathematics and Center for Theoretical Biological Physics UC San Diego CECAM Workshop: New Perspectives
Figure 1. Molecules geometries of 5021 and Each neutral group in CHARMM topology was grouped in dash circle.
 Project I Chemistry 8021, Spring 2005/2/23 This document was turned in by a student as a homework paper. 1. Methods First, the cartesian coordinates of 5021 and 8021 molecules (Fig. 1) are generated, in
Project I Chemistry 8021, Spring 2005/2/23 This document was turned in by a student as a homework paper. 1. Methods First, the cartesian coordinates of 5021 and 8021 molecules (Fig. 1) are generated, in
Computer simulation methods (2) Dr. Vania Calandrini
 Computer simulation methods (2) Dr. Vania Calandrini in the previous lecture: time average versus ensemble average MC versus MD simulations equipartition theorem (=> computing T) virial theorem (=> computing
Computer simulation methods (2) Dr. Vania Calandrini in the previous lecture: time average versus ensemble average MC versus MD simulations equipartition theorem (=> computing T) virial theorem (=> computing
Indiana University Physics P331: Theory of Electromagnetism Review Problems #3
 Indiana University Physics P331: Theory of Electromagnetism Review Problems #3 Note: The final exam (Friday 1/14 8:00-10:00 AM will be comprehensive, covering lecture and homework material pertaining to
Indiana University Physics P331: Theory of Electromagnetism Review Problems #3 Note: The final exam (Friday 1/14 8:00-10:00 AM will be comprehensive, covering lecture and homework material pertaining to
Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects
 Statistical Thermodynamics Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects Dr. Ronald M. Levy ronlevy@temple.edu Bare Molecular Mechanics Atomistic Force Fields: torsion
Statistical Thermodynamics Lecture 12: Solvation Models: Molecular Mechanics Modeling of Hydration Effects Dr. Ronald M. Levy ronlevy@temple.edu Bare Molecular Mechanics Atomistic Force Fields: torsion
PROTEIN-PROTEIN DOCKING REFINEMENT USING RESTRAINT MOLECULAR DYNAMICS SIMULATIONS
 TASKQUARTERLYvol.20,No4,2016,pp.353 360 PROTEIN-PROTEIN DOCKING REFINEMENT USING RESTRAINT MOLECULAR DYNAMICS SIMULATIONS MARTIN ZACHARIAS Physics Department T38, Technical University of Munich James-Franck-Str.
TASKQUARTERLYvol.20,No4,2016,pp.353 360 PROTEIN-PROTEIN DOCKING REFINEMENT USING RESTRAINT MOLECULAR DYNAMICS SIMULATIONS MARTIN ZACHARIAS Physics Department T38, Technical University of Munich James-Franck-Str.
q 2 Da This looks very similar to Coulomb doesn t it? The only difference to Coulomb is the factor of 1/2.
 Born Lets now think about a somewhat different, but related problem that is associated with the name Born. Using a name (usually the name of the guy who first discovered or solved the problem) to describe
Born Lets now think about a somewhat different, but related problem that is associated with the name Born. Using a name (usually the name of the guy who first discovered or solved the problem) to describe
PDEs in Spherical and Circular Coordinates
 Introduction to Partial Differential Equations part of EM, Scalar and Vector Fields module (PHY2064) This lecture Laplacian in spherical & circular polar coordinates Laplace s PDE in electrostatics Schrödinger
Introduction to Partial Differential Equations part of EM, Scalar and Vector Fields module (PHY2064) This lecture Laplacian in spherical & circular polar coordinates Laplace s PDE in electrostatics Schrödinger
INTRODUCTION TO FINITE ELEMENT METHODS ON ELLIPTIC EQUATIONS LONG CHEN
 INTROUCTION TO FINITE ELEMENT METHOS ON ELLIPTIC EQUATIONS LONG CHEN CONTENTS 1. Poisson Equation 1 2. Outline of Topics 3 2.1. Finite ifference Method 3 2.2. Finite Element Method 3 2.3. Finite Volume
INTROUCTION TO FINITE ELEMENT METHOS ON ELLIPTIC EQUATIONS LONG CHEN CONTENTS 1. Poisson Equation 1 2. Outline of Topics 3 2.1. Finite ifference Method 3 2.2. Finite Element Method 3 2.3. Finite Volume
Secondary Structure. Bioch/BIMS 503 Lecture 2. Structure and Function of Proteins. Further Reading. Φ, Ψ angles alone determine protein structure
 Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Amino Acids and Proteins at ZnO-water Interfaces in Molecular Dynamics Simulations: Electronic Supplementary Information
 Amino Acids and Proteins at ZnO-water Interfaces in Molecular Dynamics Simulations: Electronic Supplementary Information Grzegorz Nawrocki and Marek Cieplak Institute of Physics, Polish Academy of Sciences,
Amino Acids and Proteins at ZnO-water Interfaces in Molecular Dynamics Simulations: Electronic Supplementary Information Grzegorz Nawrocki and Marek Cieplak Institute of Physics, Polish Academy of Sciences,
Applications of Molecular Dynamics
 June 4, 0 Molecular Modeling and Simulation Applications of Molecular Dynamics Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, The University of Tokyo Tohru
June 4, 0 Molecular Modeling and Simulation Applications of Molecular Dynamics Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, The University of Tokyo Tohru
Dielectrics. Lecture 20: Electromagnetic Theory. Professor D. K. Ghosh, Physics Department, I.I.T., Bombay
 What are dielectrics? Dielectrics Lecture 20: Electromagnetic Theory Professor D. K. Ghosh, Physics Department, I.I.T., Bombay So far we have been discussing electrostatics in either vacuum or in a conductor.
What are dielectrics? Dielectrics Lecture 20: Electromagnetic Theory Professor D. K. Ghosh, Physics Department, I.I.T., Bombay So far we have been discussing electrostatics in either vacuum or in a conductor.
Implementing Spherical Harmonics in the Poisson Boltzmann Equation for the Electrostatic-Potential. Alex Heitman Mentors: Dr. Madura Dr.
 Implementing Spherical Harmonics in the Poisson Boltzmann Equation for the Electrostatic-Potential Alex Heitman Mentors: Dr. Madura Dr. Fleming Background - Potential Electrostatic Potential why important?
Implementing Spherical Harmonics in the Poisson Boltzmann Equation for the Electrostatic-Potential Alex Heitman Mentors: Dr. Madura Dr. Fleming Background - Potential Electrostatic Potential why important?
Protein Folding & Stability. Lecture 11: Margaret A. Daugherty. Fall How do we go from an unfolded polypeptide chain to a
 Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2004 How do we go from an unfolded polypeptide chain to a compact folded protein? (Folding of thioredoxin, F. Richards) Structure - Function
Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2004 How do we go from an unfolded polypeptide chain to a compact folded protein? (Folding of thioredoxin, F. Richards) Structure - Function
Multipole moments. November 9, 2015
 Multipole moments November 9, 5 The far field expansion Suppose we have a localized charge distribution, confined to a region near the origin with r < R. Then for values of r > R, the electric field must
Multipole moments November 9, 5 The far field expansion Suppose we have a localized charge distribution, confined to a region near the origin with r < R. Then for values of r > R, the electric field must
Profiling Charge Complementarity and Selectivity for Binding at the Protein Surface
 Biophysical Journal Volume 84 May 2003 2883 2896 2883 Profiling Charge Complementarity and Selectivity for Binding at the Protein Surface Traian Sulea and Enrico O. Purisima Biotechnology Research Institute,
Biophysical Journal Volume 84 May 2003 2883 2896 2883 Profiling Charge Complementarity and Selectivity for Binding at the Protein Surface Traian Sulea and Enrico O. Purisima Biotechnology Research Institute,
GENERALIZED BORN MODELS OF MACROMOLECULARSOLVATION EFFECTS
 Annu. Rev. Phys. Chem. 2000. 51:129 52 Copyright c 2000 by Annual Reviews. All rights reserved GENERALIZED BORN MODELS OF MACROMOLECULARSOLVATION EFFECTS Donald Bashford and David A. Case Department of
Annu. Rev. Phys. Chem. 2000. 51:129 52 Copyright c 2000 by Annual Reviews. All rights reserved GENERALIZED BORN MODELS OF MACROMOLECULARSOLVATION EFFECTS Donald Bashford and David A. Case Department of
Dielectric Boundary in Biomolecular Solvation Bo Li Department of Mathematics and Center for Theoretical Biological Physics UC San Diego
 Dielectric Boundary in Biomolecular Solvation Bo Li Department of Mathematics and Center for Theoretical Biological Physics UC San Diego International Conference on Free Boundary Problems Newton Institute,
Dielectric Boundary in Biomolecular Solvation Bo Li Department of Mathematics and Center for Theoretical Biological Physics UC San Diego International Conference on Free Boundary Problems Newton Institute,
An introduction to Molecular Dynamics. EMBO, June 2016
 An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
Dispersion Interactions from the Exchange-Hole Dipole Moment
 Dispersion Interactions from the Exchange-Hole Dipole Moment Erin R. Johnson and Alberto Otero-de-la-Roza Chemistry and Chemical Biology, University of California, Merced E. R. Johnson (UC Merced) Dispersion
Dispersion Interactions from the Exchange-Hole Dipole Moment Erin R. Johnson and Alberto Otero-de-la-Roza Chemistry and Chemical Biology, University of California, Merced E. R. Johnson (UC Merced) Dispersion
COSMO-RS Theory. The Basics
 Theory The Basics From µ to properties Property µ 1 µ 2 activity coefficient vapor pressure Infinite dilution Gas phase Pure compound Pure bulk compound Partition coefficient Phase 1 Phase 2 Liquid-liquid
Theory The Basics From µ to properties Property µ 1 µ 2 activity coefficient vapor pressure Infinite dilution Gas phase Pure compound Pure bulk compound Partition coefficient Phase 1 Phase 2 Liquid-liquid
Speeding up path integral simulations
 Speeding up path integral simulations Thomas Markland and David Manolopoulos Department of Chemistry University of Oxford Funded by the US Office of Naval Research and the UK EPSRC Outline 1. Ring polymer
Speeding up path integral simulations Thomas Markland and David Manolopoulos Department of Chemistry University of Oxford Funded by the US Office of Naval Research and the UK EPSRC Outline 1. Ring polymer
Lecture 4 Quantum mechanics in more than one-dimension
 Lecture 4 Quantum mechanics in more than one-dimension Background Previously, we have addressed quantum mechanics of 1d systems and explored bound and unbound (scattering) states. Although general concepts
Lecture 4 Quantum mechanics in more than one-dimension Background Previously, we have addressed quantum mechanics of 1d systems and explored bound and unbound (scattering) states. Although general concepts
Biochemistry 530: Introduction to Structural Biology. Autumn Quarter 2014 BIOC 530
 Biochemistry 530: Introduction to Structural Biology Autumn Quarter 2014 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic
Biochemistry 530: Introduction to Structural Biology Autumn Quarter 2014 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic
Earth Solid Earth Rocks Minerals Atoms. How to make a mineral from the start of atoms?
 Earth Solid Earth Rocks Minerals Atoms How to make a mineral from the start of atoms? Formation of ions Ions excess or deficit of electrons relative to protons Anions net negative charge Cations net
Earth Solid Earth Rocks Minerals Atoms How to make a mineral from the start of atoms? Formation of ions Ions excess or deficit of electrons relative to protons Anions net negative charge Cations net
Structural and mechanistic insight into the substrate. binding from the conformational dynamics in apo. and substrate-bound DapE enzyme
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 215 Structural and mechanistic insight into the substrate binding from the conformational
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 215 Structural and mechanistic insight into the substrate binding from the conformational
V. Electrostatics Lecture 24: Diffuse Charge in Electrolytes
 V. Electrostatics Lecture 24: Diffuse Charge in Electrolytes MIT Student 1. Poisson-Nernst-Planck Equations The Nernst-Planck Equation is a conservation of mass equation that describes the influence of
V. Electrostatics Lecture 24: Diffuse Charge in Electrolytes MIT Student 1. Poisson-Nernst-Planck Equations The Nernst-Planck Equation is a conservation of mass equation that describes the influence of
Performing pka Calculations in Proteins Using Free Energy Perturbation Adiabatic Charging (FEP/AC)
 Performing pka Calculations in Proteins Using Free Energy Perturbation Adiabatic Charging (FEP/AC) Lynn Kamerlin Uppsala University Purpose of the workshop: The aim of this workshop will be to provide
Performing pka Calculations in Proteins Using Free Energy Perturbation Adiabatic Charging (FEP/AC) Lynn Kamerlin Uppsala University Purpose of the workshop: The aim of this workshop will be to provide
G pol = 1 2 where ρ( x) is the charge density at position x and the reaction electrostatic potential
 EFFICIENT AND ACCURATE HIGHER-ORDER FAST MULTIPOLE BOUNDARY ELEMENT METHOD FOR POISSON BOLTZMANN ELECTROSTATICS CHANDRAJIT BAJAJ SHUN-CHUAN CHEN Abstract. The Poisson-Boltzmann equation is a partial differential
EFFICIENT AND ACCURATE HIGHER-ORDER FAST MULTIPOLE BOUNDARY ELEMENT METHOD FOR POISSON BOLTZMANN ELECTROSTATICS CHANDRAJIT BAJAJ SHUN-CHUAN CHEN Abstract. The Poisson-Boltzmann equation is a partial differential
J07M.1 - Ball on a Turntable
 Part I - Mechanics J07M.1 - Ball on a Turntable J07M.1 - Ball on a Turntable ẑ Ω A spherically symmetric ball of mass m, moment of inertia I about any axis through its center, and radius a, rolls without
Part I - Mechanics J07M.1 - Ball on a Turntable J07M.1 - Ball on a Turntable ẑ Ω A spherically symmetric ball of mass m, moment of inertia I about any axis through its center, and radius a, rolls without
Free Energy of Solvation, Interaction, and Binding of Arbitrary Charge Distributions Imbedded in a Dielectric Continuum
 J. Phys. Chem. 1994, 98, 5113-5111 5773 Free Energy of Solvation, Interaction, and Binding of Arbitrary Charge Distributions Imbedded in a Dielectric Continuum B. Jayaram Department of Chemistry, Indian
J. Phys. Chem. 1994, 98, 5113-5111 5773 Free Energy of Solvation, Interaction, and Binding of Arbitrary Charge Distributions Imbedded in a Dielectric Continuum B. Jayaram Department of Chemistry, Indian
Fluctuating Hydrodynamics for Transport in Neutral and Charged Fluid Mixtures
 Fluctuating Hydrodynamics for Transport in Neutral and Charged Fluid Mixtures Alejandro L. Garcia San Jose State University Lawrence Berkeley Nat. Lab. Hydrodynamic Fluctuations in Soft Matter Simulations
Fluctuating Hydrodynamics for Transport in Neutral and Charged Fluid Mixtures Alejandro L. Garcia San Jose State University Lawrence Berkeley Nat. Lab. Hydrodynamic Fluctuations in Soft Matter Simulations
A half submerged metal sphere (UIC comprehensive
 Problem 1. exam) A half submerged metal sphere (UIC comprehensive A very light neutral hollow metal spherical shell of mass m and radius a is slightly submerged by a distance b a below the surface of a
Problem 1. exam) A half submerged metal sphere (UIC comprehensive A very light neutral hollow metal spherical shell of mass m and radius a is slightly submerged by a distance b a below the surface of a
Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation
 Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation Ion:ion interactions What is the free energy of ion:ion interactions ΔG i-i? Consider an ion in a solution
Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation Ion:ion interactions What is the free energy of ion:ion interactions ΔG i-i? Consider an ion in a solution
The Molecular Dynamics Method
 H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
A cylinder in a magnetic field (Jackson)
 Problem 1. A cylinder in a magnetic field (Jackson) A very long hollow cylinder of inner radius a and outer radius b of permeability µ is placed in an initially uniform magnetic field B o at right angles
Problem 1. A cylinder in a magnetic field (Jackson) A very long hollow cylinder of inner radius a and outer radius b of permeability µ is placed in an initially uniform magnetic field B o at right angles
On the adaptive finite element analysis of the Kohn-Sham equations
 On the adaptive finite element analysis of the Kohn-Sham equations Denis Davydov, Toby Young, Paul Steinmann Denis Davydov, LTM, Erlangen, Germany August 2015 Denis Davydov, LTM, Erlangen, Germany College
On the adaptive finite element analysis of the Kohn-Sham equations Denis Davydov, Toby Young, Paul Steinmann Denis Davydov, LTM, Erlangen, Germany August 2015 Denis Davydov, LTM, Erlangen, Germany College
It s the amino acids!
 Catalytic Mechanisms HOW do enzymes do their job? Reducing activation energy sure, but HOW does an enzyme catalysis reduce the energy barrier ΔG? Remember: The rate of a chemical reaction of substrate
Catalytic Mechanisms HOW do enzymes do their job? Reducing activation energy sure, but HOW does an enzyme catalysis reduce the energy barrier ΔG? Remember: The rate of a chemical reaction of substrate
Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions. Let's get together.
 Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions Let's get together. Most gases are NOT ideal except at very low pressures: Z=1 for ideal gases Intermolecular interactions come
Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions Let's get together. Most gases are NOT ideal except at very low pressures: Z=1 for ideal gases Intermolecular interactions come
Sunyia Hussain 06/15/2012 ChE210D final project. Hydration Dynamics at a Hydrophobic Surface. Abstract:
 Hydration Dynamics at a Hydrophobic Surface Sunyia Hussain 6/15/212 ChE21D final project Abstract: Water is the universal solvent of life, crucial to the function of all biomolecules. Proteins, membranes,
Hydration Dynamics at a Hydrophobic Surface Sunyia Hussain 6/15/212 ChE21D final project Abstract: Water is the universal solvent of life, crucial to the function of all biomolecules. Proteins, membranes,
Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry
 Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry Petr Pracna J. Heyrovský Institute of Physical Chemistry Academy of Sciences of the Czech Republic, Prague ZiF Cooperation
Finite Ring Geometries and Role of Coupling in Molecular Dynamics and Chemistry Petr Pracna J. Heyrovský Institute of Physical Chemistry Academy of Sciences of the Czech Republic, Prague ZiF Cooperation
lim = F F = F x x + F y y + F z
 Physics 361 Summary of Results from Lecture Physics 361 Derivatives of Scalar and Vector Fields The gradient of a scalar field f( r) is given by g = f. coordinates f g = ê x x + ê f y y + ê f z z Expressed
Physics 361 Summary of Results from Lecture Physics 361 Derivatives of Scalar and Vector Fields The gradient of a scalar field f( r) is given by g = f. coordinates f g = ê x x + ê f y y + ê f z z Expressed
10 Chemical reactions in aqueous solutions
 The Physics and Chemistry of Water 10 Chemical reactions in aqueous solutions Effects of water in reactions Hydration of reactants cause steric barriers Increases attraction between nonpolar reactants
The Physics and Chemistry of Water 10 Chemical reactions in aqueous solutions Effects of water in reactions Hydration of reactants cause steric barriers Increases attraction between nonpolar reactants
Chapter 15: Enyzmatic Catalysis
 Chapter 15: Enyzmatic Catalysis Voet & Voet: Pages 496-508 Slide 1 Catalytic Mechanisms Catalysis is a process that increases the rate at which a reaction approaches equilibrium Rate enhancement depends
Chapter 15: Enyzmatic Catalysis Voet & Voet: Pages 496-508 Slide 1 Catalytic Mechanisms Catalysis is a process that increases the rate at which a reaction approaches equilibrium Rate enhancement depends
PHYSICS. Course Syllabus. Section 1: Mathematical Physics. Subject Code: PH. Course Structure. Electromagnetic Theory
 PHYSICS Subject Code: PH Course Structure Sections/Units Topics Section 1 Section 2 Section 3 Section 4 Section 5 Section 6 Section 7 Section 8 Mathematical Physics Classical Mechanics Electromagnetic
PHYSICS Subject Code: PH Course Structure Sections/Units Topics Section 1 Section 2 Section 3 Section 4 Section 5 Section 6 Section 7 Section 8 Mathematical Physics Classical Mechanics Electromagnetic
Level-Set Variational Solvation Coupling Solute Molecular Mechanics with Continuum Solvent
 Level-Set Variational Solvation Coupling Solute Molecular Mechanics with Continuum Solvent Bo Li Department of Mathematics and Center for Theoretical Biological Physics (CTBP) University of California,
Level-Set Variational Solvation Coupling Solute Molecular Mechanics with Continuum Solvent Bo Li Department of Mathematics and Center for Theoretical Biological Physics (CTBP) University of California,
MM-PBSA Validation Study. Trent E. Balius Department of Applied Mathematics and Statistics AMS
 MM-PBSA Validation Study Trent. Balius Department of Applied Mathematics and Statistics AMS 535 11-26-2008 Overview MM-PBSA Introduction MD ensembles one snap-shots relaxed structures nrichment Computational
MM-PBSA Validation Study Trent. Balius Department of Applied Mathematics and Statistics AMS 535 11-26-2008 Overview MM-PBSA Introduction MD ensembles one snap-shots relaxed structures nrichment Computational
NH 2. Biochemistry I, Fall Term Sept 9, Lecture 5: Amino Acids & Peptides Assigned reading in Campbell: Chapter
 Biochemistry I, Fall Term Sept 9, 2005 Lecture 5: Amino Acids & Peptides Assigned reading in Campbell: Chapter 3.1-3.4. Key Terms: ptical Activity, Chirality Peptide bond Condensation reaction ydrolysis
Biochemistry I, Fall Term Sept 9, 2005 Lecture 5: Amino Acids & Peptides Assigned reading in Campbell: Chapter 3.1-3.4. Key Terms: ptical Activity, Chirality Peptide bond Condensation reaction ydrolysis
Molecular dynamics simulation of Aquaporin-1. 4 nm
 Molecular dynamics simulation of Aquaporin-1 4 nm Molecular Dynamics Simulations Schrödinger equation i~@ t (r, R) =H (r, R) Born-Oppenheimer approximation H e e(r; R) =E e (R) e(r; R) Nucleic motion described
Molecular dynamics simulation of Aquaporin-1 4 nm Molecular Dynamics Simulations Schrödinger equation i~@ t (r, R) =H (r, R) Born-Oppenheimer approximation H e e(r; R) =E e (R) e(r; R) Nucleic motion described
Classical Field Theory: Electrostatics-Magnetostatics
 Classical Field Theory: Electrostatics-Magnetostatics April 27, 2010 1 1 J.D.Jackson, Classical Electrodynamics, 2nd Edition, Section 1-5 Electrostatics The behavior of an electrostatic field can be described
Classical Field Theory: Electrostatics-Magnetostatics April 27, 2010 1 1 J.D.Jackson, Classical Electrodynamics, 2nd Edition, Section 1-5 Electrostatics The behavior of an electrostatic field can be described
NYU Physics Preliminary Examination in Electricity & Magnetism Fall 2011
 This is a closed-book exam. No reference materials of any sort are permitted. Full credit will be given for complete solutions to the following five questions. 1. An impenetrable sphere of radius a carries
This is a closed-book exam. No reference materials of any sort are permitted. Full credit will be given for complete solutions to the following five questions. 1. An impenetrable sphere of radius a carries
PHY 407 QUANTUM MECHANICS Fall 05 Problem set 1 Due Sep
 Problem set 1 Due Sep 15 2005 1. Let V be the set of all complex valued functions of a real variable θ, that are periodic with period 2π. That is u(θ + 2π) = u(θ), for all u V. (1) (i) Show that this V
Problem set 1 Due Sep 15 2005 1. Let V be the set of all complex valued functions of a real variable θ, that are periodic with period 2π. That is u(θ + 2π) = u(θ), for all u V. (1) (i) Show that this V
Notes: Most of the material presented in this chapter is taken from Jackson, Chap. 2, 3, and 4, and Di Bartolo, Chap. 2. 2π nx i a. ( ) = G n.
 Chapter. Electrostatic II Notes: Most of the material presented in this chapter is taken from Jackson, Chap.,, and 4, and Di Bartolo, Chap... Mathematical Considerations.. The Fourier series and the Fourier
Chapter. Electrostatic II Notes: Most of the material presented in this chapter is taken from Jackson, Chap.,, and 4, and Di Bartolo, Chap... Mathematical Considerations.. The Fourier series and the Fourier
Homology modeling. Dinesh Gupta ICGEB, New Delhi 1/27/2010 5:59 PM
 Homology modeling Dinesh Gupta ICGEB, New Delhi Protein structure prediction Methods: Homology (comparative) modelling Threading Ab-initio Protein Homology modeling Homology modeling is an extrapolation
Homology modeling Dinesh Gupta ICGEB, New Delhi Protein structure prediction Methods: Homology (comparative) modelling Threading Ab-initio Protein Homology modeling Homology modeling is an extrapolation
Oxygen Binding in Hemocyanin
 Supporting Information for Quantum Mechanics/Molecular Mechanics Study of Oxygen Binding in Hemocyanin Toru Saito and Walter Thiel* Max-Planck-Institut für Kohlenforschung, Kaiser-Wilhelm-Platz 1, D-45470
Supporting Information for Quantum Mechanics/Molecular Mechanics Study of Oxygen Binding in Hemocyanin Toru Saito and Walter Thiel* Max-Planck-Institut für Kohlenforschung, Kaiser-Wilhelm-Platz 1, D-45470
Towards accurate calculations of Zn 2+ binding free energies in zinc finger proteins
 Towards accurate calculations of Zn 2+ binding free energies in zinc finger proteins UNDERGRADUATE HONORS RESEARCH THESIS Presented in Partial Fulfillment of the Requirements for the Bachelor of Science
Towards accurate calculations of Zn 2+ binding free energies in zinc finger proteins UNDERGRADUATE HONORS RESEARCH THESIS Presented in Partial Fulfillment of the Requirements for the Bachelor of Science
Free energy simulations
 Free energy simulations Marcus Elstner and Tomáš Kubař January 14, 2013 Motivation a physical quantity that is of most interest in chemistry? free energies Helmholtz F or Gibbs G holy grail of computational
Free energy simulations Marcus Elstner and Tomáš Kubař January 14, 2013 Motivation a physical quantity that is of most interest in chemistry? free energies Helmholtz F or Gibbs G holy grail of computational
Coupling the Level-Set Method with Variational Implicit Solvent Modeling of Molecular Solvation
 Coupling the Level-Set Method with Variational Implicit Solvent Modeling of Molecular Solvation Bo Li Math Dept & CTBP, UCSD Li-Tien Cheng (Math, UCSD) Zhongming Wang (Math & Biochem, UCSD) Yang Xie (MAE,
Coupling the Level-Set Method with Variational Implicit Solvent Modeling of Molecular Solvation Bo Li Math Dept & CTBP, UCSD Li-Tien Cheng (Math, UCSD) Zhongming Wang (Math & Biochem, UCSD) Yang Xie (MAE,
The Second Chilean Symposium on BOUNDARY ELEMENT METHODS
 The Second Chilean Symposium on BOUNDARY ELEMENT METHODS Friday, December 14, 2018 Universidad Técnica Federico Santa María Valparaíso, Chile http://bem2018.usm.cl/ 9:25 9:30 Welcome 9:30 10:30 Jaydeep
The Second Chilean Symposium on BOUNDARY ELEMENT METHODS Friday, December 14, 2018 Universidad Técnica Federico Santa María Valparaíso, Chile http://bem2018.usm.cl/ 9:25 9:30 Welcome 9:30 10:30 Jaydeep
Physics 222, Spring 2010 Quiz 3, Form: A
 Physics 222, Spring 2010 Quiz 3, Form: A Name: Date: Instructions You must sketch correct pictures and vectors, you must show all calculations, and you must explain all answers for full credit. Neatness
Physics 222, Spring 2010 Quiz 3, Form: A Name: Date: Instructions You must sketch correct pictures and vectors, you must show all calculations, and you must explain all answers for full credit. Neatness
CHAPTER 3 POTENTIALS 10/13/2016. Outlines. 1. Laplace s equation. 2. The Method of Images. 3. Separation of Variables. 4. Multipole Expansion
 CHAPTER 3 POTENTIALS Lee Chow Department of Physics University of Central Florida Orlando, FL 32816 Outlines 1. Laplace s equation 2. The Method of Images 3. Separation of Variables 4. Multipole Expansion
CHAPTER 3 POTENTIALS Lee Chow Department of Physics University of Central Florida Orlando, FL 32816 Outlines 1. Laplace s equation 2. The Method of Images 3. Separation of Variables 4. Multipole Expansion
Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine. JAK2 Selective Mechanism Combined Molecular Dynamics Simulation
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supporting Information Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supporting Information Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine
Chapter 9 Ionic and Covalent Bonding
 Chem 1045 Prof George W.J. Kenney, Jr General Chemistry by Ebbing and Gammon, 8th Edition Last Update: 06-April-2009 Chapter 9 Ionic and Covalent Bonding These Notes are to SUPPLIMENT the Text, They do
Chem 1045 Prof George W.J. Kenney, Jr General Chemistry by Ebbing and Gammon, 8th Edition Last Update: 06-April-2009 Chapter 9 Ionic and Covalent Bonding These Notes are to SUPPLIMENT the Text, They do
Molecular Dynamics Simulations
 Molecular Dynamics Simulations Dr. Kasra Momeni www.knanosys.com Outline Long-range Interactions Ewald Sum Fast Multipole Method Spherically Truncated Coulombic Potential Speeding up Calculations SPaSM
Molecular Dynamics Simulations Dr. Kasra Momeni www.knanosys.com Outline Long-range Interactions Ewald Sum Fast Multipole Method Spherically Truncated Coulombic Potential Speeding up Calculations SPaSM
RESEARCH ARTICLES. Effective Energy Function for Proteins in Solution
 PROTEINS: Structure, Function, and Genetics 35:133 152 (1999) RESEARCH ARTICLES Effective Energy Function for Proteins in Solution Themis Lazaridis 1 and Martin Karplus 1,2 * 1 Department of Chemistry
PROTEINS: Structure, Function, and Genetics 35:133 152 (1999) RESEARCH ARTICLES Effective Energy Function for Proteins in Solution Themis Lazaridis 1 and Martin Karplus 1,2 * 1 Department of Chemistry
Solving PDEs with Multigrid Methods p.1
 Solving PDEs with Multigrid Methods Scott MacLachlan maclachl@colorado.edu Department of Applied Mathematics, University of Colorado at Boulder Solving PDEs with Multigrid Methods p.1 Support and Collaboration
Solving PDEs with Multigrid Methods Scott MacLachlan maclachl@colorado.edu Department of Applied Mathematics, University of Colorado at Boulder Solving PDEs with Multigrid Methods p.1 Support and Collaboration
l=0 The expansion coefficients can be determined, for example, by finding the potential on the z-axis and expanding that result in z.
 Electrodynamics I Exam - Part A - Closed Book KSU 15/11/6 Name Electrodynamic Score = 14 / 14 points Instructions: Use SI units. Where appropriate, define all variables or symbols you use, in words. Try
Electrodynamics I Exam - Part A - Closed Book KSU 15/11/6 Name Electrodynamic Score = 14 / 14 points Instructions: Use SI units. Where appropriate, define all variables or symbols you use, in words. Try
Chapter 4. Glutamic Acid in Solution - Correlations
 Chapter 4 Glutamic Acid in Solution - Correlations 4. Introduction Glutamic acid crystallises from aqueous solution, therefore the study of these molecules in an aqueous environment is necessary to understand
Chapter 4 Glutamic Acid in Solution - Correlations 4. Introduction Glutamic acid crystallises from aqueous solution, therefore the study of these molecules in an aqueous environment is necessary to understand
Structural Bioinformatics (C3210) Molecular Mechanics
 Structural Bioinformatics (C3210) Molecular Mechanics How to Calculate Energies Calculation of molecular energies is of key importance in protein folding, molecular modelling etc. There are two main computational
Structural Bioinformatics (C3210) Molecular Mechanics How to Calculate Energies Calculation of molecular energies is of key importance in protein folding, molecular modelling etc. There are two main computational
Lecture 11: Potential Energy Functions
 Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Lecture 11: Potential Energy Functions Dr. Ronald M. Levy ronlevy@temple.edu Originally contributed by Lauren Wickstrom (2011) Microscopic/Macroscopic Connection The connection between microscopic interactions
Advanced Molecular Molecular Dynamics
 Advanced Molecular Molecular Dynamics Technical details May 12, 2014 Integration of harmonic oscillator r m period = 2 k k and the temperature T determine the sampling of x (here T is related with v 0
Advanced Molecular Molecular Dynamics Technical details May 12, 2014 Integration of harmonic oscillator r m period = 2 k k and the temperature T determine the sampling of x (here T is related with v 0
The AGBNP2 Implicit Solvation Model
 2544 J. Chem. Theory Comput. 2009, 5, 2544 2564 The AGBNP2 Implicit Solvation Model Emilio Gallicchio,* Kristina Paris, and Ronald M. Levy Department of Chemistry and Chemical Biology and BioMaPS Institute
2544 J. Chem. Theory Comput. 2009, 5, 2544 2564 The AGBNP2 Implicit Solvation Model Emilio Gallicchio,* Kristina Paris, and Ronald M. Levy Department of Chemistry and Chemical Biology and BioMaPS Institute
Lecture 6 - Bonding in Crystals
 Lecture 6 onding in Crystals inding in Crystals (Kittel Ch. 3) inding of atoms to form crystals A crystal is a repeated array of atoms Why do they form? What are characteristic bonding mechanisms? How
Lecture 6 onding in Crystals inding in Crystals (Kittel Ch. 3) inding of atoms to form crystals A crystal is a repeated array of atoms Why do they form? What are characteristic bonding mechanisms? How
Advanced in silico drug design
 Advanced in silico drug design RNDr. Martin Lepšík, Ph.D. Lecture: Advanced scoring Palacky University, Olomouc 2016 1 Outline 1. Scoring Definition, Types 2. Physics-based Scoring: Master Equation Terms
Advanced in silico drug design RNDr. Martin Lepšík, Ph.D. Lecture: Advanced scoring Palacky University, Olomouc 2016 1 Outline 1. Scoring Definition, Types 2. Physics-based Scoring: Master Equation Terms
Cooperativity and Specificity of Cys 2 His 2 Zinc Finger Protein-DNA Interactions: A Molecular Dynamics Simulation Study
 7662 J. Phys. Chem. B 2010, 114, 7662 7671 Cooperativity and Specificity of Cys 2 His 2 Zinc Finger Protein-DNA Interactions: A Molecular Dynamics Simulation Study Juyong Lee, Jin-Soo Kim, and Chaok Seok*
7662 J. Phys. Chem. B 2010, 114, 7662 7671 Cooperativity and Specificity of Cys 2 His 2 Zinc Finger Protein-DNA Interactions: A Molecular Dynamics Simulation Study Juyong Lee, Jin-Soo Kim, and Chaok Seok*
Lecture 11: Protein Folding & Stability
 Structure - Function Protein Folding: What we know Lecture 11: Protein Folding & Stability 1). Amino acid sequence dictates structure. 2). The native structure represents the lowest energy state for a
Structure - Function Protein Folding: What we know Lecture 11: Protein Folding & Stability 1). Amino acid sequence dictates structure. 2). The native structure represents the lowest energy state for a
Protein Folding & Stability. Lecture 11: Margaret A. Daugherty. Fall Protein Folding: What we know. Protein Folding
 Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2003 Structure - Function Protein Folding: What we know 1). Amino acid sequence dictates structure. 2). The native structure represents
Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2003 Structure - Function Protein Folding: What we know 1). Amino acid sequence dictates structure. 2). The native structure represents
Lecture 4 Quantum mechanics in more than one-dimension
 Lecture 4 Quantum mechanics in more than one-dimension Background Previously, we have addressed quantum mechanics of 1d systems and explored bound and unbound (scattering) states. Although general concepts
Lecture 4 Quantum mechanics in more than one-dimension Background Previously, we have addressed quantum mechanics of 1d systems and explored bound and unbound (scattering) states. Although general concepts
