Implementing Spherical Harmonics in the Poisson Boltzmann Equation for the Electrostatic-Potential. Alex Heitman Mentors: Dr. Madura Dr.
|
|
|
- Dwight Bruce
- 6 years ago
- Views:
Transcription
1 Implementing Spherical Harmonics in the Poisson Boltzmann Equation for the Electrostatic-Potential Alex Heitman Mentors: Dr. Madura Dr. Fleming
2 Background - Potential Electrostatic Potential why important? -electrostatic forces are derived from this -changes with each configuration change - clearly the potential is a vital yet complex calculation which needs to be performed optimally -solved with the Poisson Boltzmann equation
3 Background Poisson Boltzmann Equation The Poisson Boltzmann Equation (PBE) is a complex second order non-linear partial differential equation used the electrostatic potential. It takes the following form. The hyperbolic sine term can be linearized resulting in the Linearized PBE (LPBE)
4 Background- Molecular Case of the LPBE Discontinuous piecewise constant dielectric term Discrete individual point charges on molecule The LPBE splits into the following system
5 Background- Boundary Element Method Using Green s function we can convert the previous system of 2 nd order PDEs to a system of surface integrals of the following form This system is approximated using finite element methods creating a non sparse matrix
6 Project Idea Computational Speed and Efficiency -Matrix Compression : ACA algorithm Accuracy -Flexible approximation techniques: we can specify the order of approximating polynomial -Accurate functional representation: Spherical Harmonics
7 Project Idea part 1: Implementing ACA Algorithm Matrix manipulations are often the most computationally expensive Finite Element and Finite Difference Methods Sparse large matrices Boundary Element Method Smaller matrix but not sparse ACA compresses nearly singular portions of a surface integral matrix ACA programming and setting up the surface integral matrix consists of the bulk of the project
8 Project Idea part 2: Implementing Spherical Harmonics Clearly the solution of the PBE hinges on surface integrals To integrate over the surface we need a functional representation of the surface previously naive attempts were made fitting an ellipse over target molecule Spherical Harmonics can produce better shapes
9 Spherical Harmonics-What are they? an ortho-normal series of functions defined on the unit sphere (defined for all (phi,theta)) a generalization of the Fourier series to the sphere an infinite dimension basis for all continuous functions defined on the unit sphere
10 Spherical Harmonics-Why use them? we can express the radius of a star surface with a central point as a function of (phi,theta) this function can be then be written as an infinite sum of spherical harmonics we can specify the degree of accuracy by the adding harmonics for star shaped objects the spherical harmonics should converge to the shape
11 Spherical Harmonics- Coefficients For each function there exists a unique spherical harmonic coefficient for each harmonic The coefficients obey the following property The coefficients also minimize the following quantity
12 Spherical Harmonics- Process Start with a PDB file containing atomic coordinates of the nuclei of the molecule/protein Use Dr. Connolly s msroll program to generate a larger series of coordinates representing the dot surface of the molecule/protein Run spherical harmonics approximation of the desired degree
13 Results : Simple Amino Acids Alanine: only 13 atoms, a common residue
14 Results : Complex Amino Acids Tryptophan: 27 atoms
15 Results: Secondary Structure Beta Pleated Sheets ~ 15 residues
16 Results- Simple Proteins Due to slightly more regular shape these fit fairly well Crambin: only 46 residues
17 Results Not so simple protein Lysozyme: 129 Residues
18 Results - Summary Single Amino Acids: excellent fit Secondary Structure: way off because of poor shape Proteins: doesn t capture everything, but gives a pretty good general idea Overall fairly successful attempt
19 Thank You Slide I would like to thank my mentors Dr. Madura of Duquesne University Dr. Fleming of Duquesne University I would also like to thank Chase Smith of Duquesne for his work on the surface integral and the ACA algorithm I would also like to thank Dr. Michael L. Connolly for letting me use the Biohedron program to generate dot surfaces I Finally want to thank BBSI for the hands on introduction to Computational Biology
20 References Altman, Bardhan, Kuo, Tidor and White Fast Methods for Simulation of Biomolecule Electrostatics, IEEE Baker, Holst and Wang Adaptive Multilevel Finite Element Solution of the Poisson-Boltzmann Equation 1 Algorithms and Examples, Journal of Computational Chemistry, Boschitsch, Fenley and Zhou Fast Boundary Element Method for the Linear Poisson-Boltzmann Equation, Journal of Physical Chemistry, Connolly, Solvent-Accessible Surface of Proteins and Nucleic Acids, Science, Aug Getzoff and Max Spherical Harmonic Molecular Surfaces, IEEE Computer Graphics & Applications, July, Grandison, Penfold and Vanden-Broeck A rapid boundary integral equation technique for protein electrostatics, Journal of Computational Physics, Lee, Vouvakis and Zhao The Adaptive Cross Approximation Algorithm for Accelerated Method of Moments Computations of EMC Problems, IEEE 2005.
Numerical Optimization of a Walk-on-Spheres Solver for the Linear Poisson-Boltzmann Equation
 Commun. Comput. Phys. doi: 10.4208/cicp.220711.041011s Vol. 13, No. 1, pp. 195-206 January 2013 Numerical Optimization of a Walk-on-Spheres Solver for the Linear Poisson-Boltzmann Equation Travis Mackoy
Commun. Comput. Phys. doi: 10.4208/cicp.220711.041011s Vol. 13, No. 1, pp. 195-206 January 2013 Numerical Optimization of a Walk-on-Spheres Solver for the Linear Poisson-Boltzmann Equation Travis Mackoy
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
CMPS 6630: Introduction to Computational Biology and Bioinformatics. Tertiary Structure Prediction
 CMPS 6630: Introduction to Computational Biology and Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the
CMPS 6630: Introduction to Computational Biology and Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the
Orientational degeneracy in the presence of one alignment tensor.
 Orientational degeneracy in the presence of one alignment tensor. Rotation about the x, y and z axes can be performed in the aligned mode of the program to examine the four degenerate orientations of two
Orientational degeneracy in the presence of one alignment tensor. Rotation about the x, y and z axes can be performed in the aligned mode of the program to examine the four degenerate orientations of two
A SHORT NOTE COMPARING MULTIGRID AND DOMAIN DECOMPOSITION FOR PROTEIN MODELING EQUATIONS
 A SHORT NOTE COMPARING MULTIGRID AND DOMAIN DECOMPOSITION FOR PROTEIN MODELING EQUATIONS MICHAEL HOLST AND FAISAL SAIED Abstract. We consider multigrid and domain decomposition methods for the numerical
A SHORT NOTE COMPARING MULTIGRID AND DOMAIN DECOMPOSITION FOR PROTEIN MODELING EQUATIONS MICHAEL HOLST AND FAISAL SAIED Abstract. We consider multigrid and domain decomposition methods for the numerical
Can a continuum solvent model reproduce the free energy landscape of a β-hairpin folding in water?
 Can a continuum solvent model reproduce the free energy landscape of a β-hairpin folding in water? Ruhong Zhou 1 and Bruce J. Berne 2 1 IBM Thomas J. Watson Research Center; and 2 Department of Chemistry,
Can a continuum solvent model reproduce the free energy landscape of a β-hairpin folding in water? Ruhong Zhou 1 and Bruce J. Berne 2 1 IBM Thomas J. Watson Research Center; and 2 Department of Chemistry,
Physics 6303 Lecture 11 September 24, LAST TIME: Cylindrical coordinates, spherical coordinates, and Legendre s equation
 Physics 6303 Lecture September 24, 208 LAST TIME: Cylindrical coordinates, spherical coordinates, and Legendre s equation, l l l l l l. Consider problems that are no axisymmetric; i.e., the potential depends
Physics 6303 Lecture September 24, 208 LAST TIME: Cylindrical coordinates, spherical coordinates, and Legendre s equation, l l l l l l. Consider problems that are no axisymmetric; i.e., the potential depends
CMPS 3110: Bioinformatics. Tertiary Structure Prediction
 CMPS 3110: Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the laws of physics! Conformation space is finite
CMPS 3110: Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the laws of physics! Conformation space is finite
Modeling in Solution. Important Observations. Energy Concepts
 Modeling in Solution 1 Important Observations Various electrostatic effects for a solvated molecule are often less important than for an isolated gaseous molecule when the molecule is dissolved in a solvent
Modeling in Solution 1 Important Observations Various electrostatic effects for a solvated molecule are often less important than for an isolated gaseous molecule when the molecule is dissolved in a solvent
Laplacian-Centered Poisson Solvers and Multilevel Summation Algorithms
 Laplacian-Centered Poisson Solvers and Multilevel Summation Algorithms Dmitry Yershov 1 Stephen Bond 1 Robert Skeel 2 1 University of Illinois at Urbana-Champaign 2 Purdue University 2009 SIAM Annual Meeting
Laplacian-Centered Poisson Solvers and Multilevel Summation Algorithms Dmitry Yershov 1 Stephen Bond 1 Robert Skeel 2 1 University of Illinois at Urbana-Champaign 2 Purdue University 2009 SIAM Annual Meeting
Improved Solvation Models using Boundary Integral Equations
 Improved Solvation Models using Boundary Integral Equations Matthew Knepley and Jaydeep Bardhan Computational and Applied Mathematics Rice University SIAM Conference on the Life Sciences Minneapolis, MN
Improved Solvation Models using Boundary Integral Equations Matthew Knepley and Jaydeep Bardhan Computational and Applied Mathematics Rice University SIAM Conference on the Life Sciences Minneapolis, MN
Multigrid and Domain Decomposition Methods for Electrostatics Problems
 Multigrid and Domain Decomposition Methods for Electrostatics Problems Michael Holst and Faisal Saied Abstract. We consider multigrid and domain decomposition methods for the numerical solution of electrostatics
Multigrid and Domain Decomposition Methods for Electrostatics Problems Michael Holst and Faisal Saied Abstract. We consider multigrid and domain decomposition methods for the numerical solution of electrostatics
Molecular Basis of K + Conduction and Selectivity
 The Structure of the Potassium Channel: Molecular Basis of K + Conduction and Selectivity -Doyle, DA, et al. The structure of the potassium channel: molecular basis of K + conduction and selectivity. Science
The Structure of the Potassium Channel: Molecular Basis of K + Conduction and Selectivity -Doyle, DA, et al. The structure of the potassium channel: molecular basis of K + conduction and selectivity. Science
Junior-level Electrostatics Content Review
 Junior-level Electrostatics Content Review Please fill out the following exam to the best of your ability. This will not count towards your final grade in the course. Do your best to get to all the questions
Junior-level Electrostatics Content Review Please fill out the following exam to the best of your ability. This will not count towards your final grade in the course. Do your best to get to all the questions
All-atom Molecular Mechanics. Trent E. Balius AMS 535 / CHE /27/2010
 All-atom Molecular Mechanics Trent E. Balius AMS 535 / CHE 535 09/27/2010 Outline Molecular models Molecular mechanics Force Fields Potential energy function functional form parameters and parameterization
All-atom Molecular Mechanics Trent E. Balius AMS 535 / CHE 535 09/27/2010 Outline Molecular models Molecular mechanics Force Fields Potential energy function functional form parameters and parameterization
Computational Bioelectrostatics
 Computational Bioelectrostatics Matthew Knepley Computational and Applied Mathematics Rice University Scientific Computing Seminar University of Houston Houston, TX April 20, 2017 M. Knepley (CAAM) MGK
Computational Bioelectrostatics Matthew Knepley Computational and Applied Mathematics Rice University Scientific Computing Seminar University of Houston Houston, TX April 20, 2017 M. Knepley (CAAM) MGK
1. (3) Write Gauss Law in differential form. Explain the physical meaning.
 Electrodynamics I Midterm Exam - Part A - Closed Book KSU 204/0/23 Name Instructions: Use SI units. Where appropriate, define all variables or symbols you use, in words. Try to tell about the physics involved,
Electrodynamics I Midterm Exam - Part A - Closed Book KSU 204/0/23 Name Instructions: Use SI units. Where appropriate, define all variables or symbols you use, in words. Try to tell about the physics involved,
Applications of Molecular Dynamics
 June 4, 0 Molecular Modeling and Simulation Applications of Molecular Dynamics Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, The University of Tokyo Tohru
June 4, 0 Molecular Modeling and Simulation Applications of Molecular Dynamics Agricultural Bioinformatics Research Unit, Graduate School of Agricultural and Life Sciences, The University of Tokyo Tohru
ADVANCED ENGINEERING MATHEMATICS
 ADVANCED ENGINEERING MATHEMATICS DENNIS G. ZILL Loyola Marymount University MICHAEL R. CULLEN Loyola Marymount University PWS-KENT O I^7 3 PUBLISHING COMPANY E 9 U Boston CONTENTS Preface xiii Parti ORDINARY
ADVANCED ENGINEERING MATHEMATICS DENNIS G. ZILL Loyola Marymount University MICHAEL R. CULLEN Loyola Marymount University PWS-KENT O I^7 3 PUBLISHING COMPANY E 9 U Boston CONTENTS Preface xiii Parti ORDINARY
Solvation and Macromolecular Structure. The structure and dynamics of biological macromolecules are strongly influenced by water:
 Overview Solvation and Macromolecular Structure The structure and dynamics of biological macromolecules are strongly influenced by water: Electrostatic effects: charges are screened by water molecules
Overview Solvation and Macromolecular Structure The structure and dynamics of biological macromolecules are strongly influenced by water: Electrostatic effects: charges are screened by water molecules
The Select Command and Boolean Operators
 The Select Command and Boolean Operators Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingresources/jmol/jmoltraining/boolean.html
The Select Command and Boolean Operators Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingresources/jmol/jmoltraining/boolean.html
Modeling Biomolecular Systems II. BME 540 David Sept
 Modeling Biomolecular Systems II BME 540 David Sept Introduction Why do we perform simulations? What makes simulations possible? How do we perform simulations? What types of things/systems do we simulate?
Modeling Biomolecular Systems II BME 540 David Sept Introduction Why do we perform simulations? What makes simulations possible? How do we perform simulations? What types of things/systems do we simulate?
The Second Chilean Symposium on BOUNDARY ELEMENT METHODS
 The Second Chilean Symposium on BOUNDARY ELEMENT METHODS Friday, December 14, 2018 Universidad Técnica Federico Santa María Valparaíso, Chile http://bem2018.usm.cl/ 9:25 9:30 Welcome 9:30 10:30 Jaydeep
The Second Chilean Symposium on BOUNDARY ELEMENT METHODS Friday, December 14, 2018 Universidad Técnica Federico Santa María Valparaíso, Chile http://bem2018.usm.cl/ 9:25 9:30 Welcome 9:30 10:30 Jaydeep
NMR Spectroscopy of Polymers
 UNESCO/IUPAC Course 2005/2006 Jiri Brus NMR Spectroscopy of Polymers Brus J 1. part At the very beginning the phenomenon of nuclear spin resonance was studied predominantly by physicists and the application
UNESCO/IUPAC Course 2005/2006 Jiri Brus NMR Spectroscopy of Polymers Brus J 1. part At the very beginning the phenomenon of nuclear spin resonance was studied predominantly by physicists and the application
A Fast N-Body Solver for the Poisson(-Boltzmann) Equation
 A Fast N-Body Solver for the Poisson(-Boltzmann) Equation Robert D. Skeel Departments of Computer Science (and Mathematics) Purdue University http://bionum.cs.purdue.edu/2008december.pdf 1 Thesis Poisson(-Boltzmann)
A Fast N-Body Solver for the Poisson(-Boltzmann) Equation Robert D. Skeel Departments of Computer Science (and Mathematics) Purdue University http://bionum.cs.purdue.edu/2008december.pdf 1 Thesis Poisson(-Boltzmann)
Part 4 The Select Command and Boolean Operators
 Part 4 The Select Command and Boolean Operators http://cbm.msoe.edu/newwebsite/learntomodel Introduction By default, every command you enter into the Console affects the entire molecular structure. However,
Part 4 The Select Command and Boolean Operators http://cbm.msoe.edu/newwebsite/learntomodel Introduction By default, every command you enter into the Console affects the entire molecular structure. However,
2. TRIGONOMETRY 3. COORDINATEGEOMETRY: TWO DIMENSIONS
 1 TEACHERS RECRUITMENT BOARD, TRIPURA (TRBT) EDUCATION (SCHOOL) DEPARTMENT, GOVT. OF TRIPURA SYLLABUS: MATHEMATICS (MCQs OF 150 MARKS) SELECTION TEST FOR POST GRADUATE TEACHER(STPGT): 2016 1. ALGEBRA Sets:
1 TEACHERS RECRUITMENT BOARD, TRIPURA (TRBT) EDUCATION (SCHOOL) DEPARTMENT, GOVT. OF TRIPURA SYLLABUS: MATHEMATICS (MCQs OF 150 MARKS) SELECTION TEST FOR POST GRADUATE TEACHER(STPGT): 2016 1. ALGEBRA Sets:
Separation of Variables in Polar and Spherical Coordinates
 Separation of Variables in Polar and Spherical Coordinates Polar Coordinates Suppose we are given the potential on the inside surface of an infinitely long cylindrical cavity, and we want to find the potential
Separation of Variables in Polar and Spherical Coordinates Polar Coordinates Suppose we are given the potential on the inside surface of an infinitely long cylindrical cavity, and we want to find the potential
Development of Software Package for Determining Protein Titration Properties
 Development of Software Package for Determining Protein Titration Properties By Kaila Bennett, Amitoj Chopra, Jesse Johnson, Enrico Sagullo Bioengineering 175-Senior Design University of California Riverside
Development of Software Package for Determining Protein Titration Properties By Kaila Bennett, Amitoj Chopra, Jesse Johnson, Enrico Sagullo Bioengineering 175-Senior Design University of California Riverside
1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian. which is given in spherical coordinates by (2)
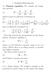 1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian operator (1) 2 = 2 x + 2 2 y + 2 2 z 2 which is given in spherical coordinates by (2) 2 = 1 ( r 2 ) + 1 r 2 r r r 2 sin θ θ and in
1. Poisson-Boltzmann 1.1. Poisson equation. We consider the Laplacian operator (1) 2 = 2 x + 2 2 y + 2 2 z 2 which is given in spherical coordinates by (2) 2 = 1 ( r 2 ) + 1 r 2 r r r 2 sin θ θ and in
Quintic beam closed form matrices (revised 2/21, 2/23/12) General elastic beam with an elastic foundation
 General elastic beam with an elastic foundation Figure 1 shows a beam-column on an elastic foundation. The beam is connected to a continuous series of foundation springs. The other end of the foundation
General elastic beam with an elastic foundation Figure 1 shows a beam-column on an elastic foundation. The beam is connected to a continuous series of foundation springs. The other end of the foundation
Molecular Visualization. Introduction
 Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Molecular Visualization Jeffry D. Madura Department of Chemistry & Biochemistry Center for Computational Sciences Duquesne University Introduction Assessments of change, dynamics, and cause and effect
Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche
 Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche The molecular structure of a protein can be broken down hierarchically. The primary structure of a protein is simply its
Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche The molecular structure of a protein can be broken down hierarchically. The primary structure of a protein is simply its
Partial differential equations
 Partial differential equations Many problems in science involve the evolution of quantities not only in time but also in space (this is the most common situation)! We will call partial differential equation
Partial differential equations Many problems in science involve the evolution of quantities not only in time but also in space (this is the most common situation)! We will call partial differential equation
FOURIER SERIES, TRANSFORMS, AND BOUNDARY VALUE PROBLEMS
 fc FOURIER SERIES, TRANSFORMS, AND BOUNDARY VALUE PROBLEMS Second Edition J. RAY HANNA Professor Emeritus University of Wyoming Laramie, Wyoming JOHN H. ROWLAND Department of Mathematics and Department
fc FOURIER SERIES, TRANSFORMS, AND BOUNDARY VALUE PROBLEMS Second Edition J. RAY HANNA Professor Emeritus University of Wyoming Laramie, Wyoming JOHN H. ROWLAND Department of Mathematics and Department
Region 16 Board of Education. Precalculus Curriculum
 Region 16 Board of Education Precalculus Curriculum 2008 1 Course Description This course offers students an opportunity to explore a variety of concepts designed to prepare them to go on to study calculus.
Region 16 Board of Education Precalculus Curriculum 2008 1 Course Description This course offers students an opportunity to explore a variety of concepts designed to prepare them to go on to study calculus.
Section II Understanding the Protein Data Bank
 Section II Understanding the Protein Data Bank The focus of Section II of the MSOE Center for BioMolecular Modeling Jmol Training Guide is to learn about the Protein Data Bank, the worldwide repository
Section II Understanding the Protein Data Bank The focus of Section II of the MSOE Center for BioMolecular Modeling Jmol Training Guide is to learn about the Protein Data Bank, the worldwide repository
UCLA and UC San Diego
 Home > News Center > Publications > EnVision Toward Computational Cell Biology NANOSTRUCTURES Contents Next PROJECT LEADERS Nathan A. Baker Michael J. Holst, UC San Diego PARTICIPANTS Burak Aksoylu Randolph
Home > News Center > Publications > EnVision Toward Computational Cell Biology NANOSTRUCTURES Contents Next PROJECT LEADERS Nathan A. Baker Michael J. Holst, UC San Diego PARTICIPANTS Burak Aksoylu Randolph
( nonlinear constraints)
 Wavelet Design & Applications Basic requirements: Admissibility (single constraint) Orthogonality ( nonlinear constraints) Sparse Representation Smooth functions well approx. by Fourier High-frequency
Wavelet Design & Applications Basic requirements: Admissibility (single constraint) Orthogonality ( nonlinear constraints) Sparse Representation Smooth functions well approx. by Fourier High-frequency
THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION
 THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION AND CALIBRATION Calculation of turn and beta intrinsic propensities. A statistical analysis of a protein structure
THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION AND CALIBRATION Calculation of turn and beta intrinsic propensities. A statistical analysis of a protein structure
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Name Final Exam December 7, 2015
 Name Final Exam December 7, 015 This test consists of five parts. Please note that in parts II through V, you can skip one question of those offered. Part I: Multiple Choice (mixed new and review questions)
Name Final Exam December 7, 015 This test consists of five parts. Please note that in parts II through V, you can skip one question of those offered. Part I: Multiple Choice (mixed new and review questions)
Protein structure (and biomolecular structure more generally) CS/CME/BioE/Biophys/BMI 279 Sept. 28 and Oct. 3, 2017 Ron Dror
 Protein structure (and biomolecular structure more generally) CS/CME/BioE/Biophys/BMI 279 Sept. 28 and Oct. 3, 2017 Ron Dror Please interrupt if you have questions, and especially if you re confused! Assignment
Protein structure (and biomolecular structure more generally) CS/CME/BioE/Biophys/BMI 279 Sept. 28 and Oct. 3, 2017 Ron Dror Please interrupt if you have questions, and especially if you re confused! Assignment
9/4/2018. Electric Field Models. Electric Field of a Point Charge. The Electric Field of Multiple Point Charges
 Electric Field Models One thing learned from last chapter was that sources determine the electric field. We can understand the essential physics on the basis of simplified models of the sources of electric
Electric Field Models One thing learned from last chapter was that sources determine the electric field. We can understand the essential physics on the basis of simplified models of the sources of electric
Electric Field Models
 Electric Field Models One thing learned from last chapter was that sources determine the electric field. We can understand the essential physics on the basis of simplified models of the sources of electric
Electric Field Models One thing learned from last chapter was that sources determine the electric field. We can understand the essential physics on the basis of simplified models of the sources of electric
Chapter 17: Fourier Series
 Section A Introduction to Fourier Series By the end of this section you will be able to recognise periodic functions sketch periodic functions determine the period of the given function Why are Fourier
Section A Introduction to Fourier Series By the end of this section you will be able to recognise periodic functions sketch periodic functions determine the period of the given function Why are Fourier
Rapid evaluation of electrostatic interactions in multi-phase media
 Rapid evaluation of electrostatic interactions in multi-phase media P.G. Martinsson, The University of Colorado at Boulder Acknowledgements: Some of the work presented is joint work with Mark Tygert and
Rapid evaluation of electrostatic interactions in multi-phase media P.G. Martinsson, The University of Colorado at Boulder Acknowledgements: Some of the work presented is joint work with Mark Tygert and
Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations
 Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations Alexandre V. Morozov, Tanja Kortemme, Kiril Tsemekhman, David Baker
Close agreement between the orientation dependence of hydrogen bonds observed in protein structures and quantum mechanical calculations Alexandre V. Morozov, Tanja Kortemme, Kiril Tsemekhman, David Baker
INTRODUCTION TO FINITE ELEMENT METHODS ON ELLIPTIC EQUATIONS LONG CHEN
 INTROUCTION TO FINITE ELEMENT METHOS ON ELLIPTIC EQUATIONS LONG CHEN CONTENTS 1. Poisson Equation 1 2. Outline of Topics 3 2.1. Finite ifference Method 3 2.2. Finite Element Method 3 2.3. Finite Volume
INTROUCTION TO FINITE ELEMENT METHOS ON ELLIPTIC EQUATIONS LONG CHEN CONTENTS 1. Poisson Equation 1 2. Outline of Topics 3 2.1. Finite ifference Method 3 2.2. Finite Element Method 3 2.3. Finite Volume
Pymol Practial Guide
 Pymol Practial Guide Pymol is a powerful visualizor very convenient to work with protein molecules. Its interface may seem complex at first, but you will see that with a little practice is simple and powerful
Pymol Practial Guide Pymol is a powerful visualizor very convenient to work with protein molecules. Its interface may seem complex at first, but you will see that with a little practice is simple and powerful
Dielectrics - III. Lecture 22: Electromagnetic Theory. Professor D. K. Ghosh, Physics Department, I.I.T., Bombay
 Dielectrics - III Lecture 22: Electromagnetic Theory Professor D. K. Ghosh, Physics Department, I.I.T., Bombay We continue with our discussion of dielectric medium. Example : Dielectric Sphere in a uniform
Dielectrics - III Lecture 22: Electromagnetic Theory Professor D. K. Ghosh, Physics Department, I.I.T., Bombay We continue with our discussion of dielectric medium. Example : Dielectric Sphere in a uniform
Research Statement. 1. Electrostatics Computation and Biomolecular Simulation. Weihua Geng Department of Mathematics, University of Michigan
 Research Statement Weihua Geng Department of Mathematics, University of Michigan I am interested in modeling problems in structural and systems biology using differential equations and integral equations,
Research Statement Weihua Geng Department of Mathematics, University of Michigan I am interested in modeling problems in structural and systems biology using differential equations and integral equations,
Indiana University Physics P331: Theory of Electromagnetism Review Problems #3
 Indiana University Physics P331: Theory of Electromagnetism Review Problems #3 Note: The final exam (Friday 1/14 8:00-10:00 AM will be comprehensive, covering lecture and homework material pertaining to
Indiana University Physics P331: Theory of Electromagnetism Review Problems #3 Note: The final exam (Friday 1/14 8:00-10:00 AM will be comprehensive, covering lecture and homework material pertaining to
Biochemistry 530: Introduction to Structural Biology. Autumn Quarter 2014 BIOC 530
 Biochemistry 530: Introduction to Structural Biology Autumn Quarter 2014 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic
Biochemistry 530: Introduction to Structural Biology Autumn Quarter 2014 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic
SUMMARY PHYSICS 707 Electrostatics. E(x) = 4πρ(x) and E(x) = 0 (1)
 SUMMARY PHYSICS 707 Electrostatics The basic differential equations of electrostatics are E(x) = 4πρ(x) and E(x) = 0 (1) where E(x) is the electric field and ρ(x) is the electric charge density. The field
SUMMARY PHYSICS 707 Electrostatics The basic differential equations of electrostatics are E(x) = 4πρ(x) and E(x) = 0 (1) where E(x) is the electric field and ρ(x) is the electric charge density. The field
Sunyia Hussain 06/15/2012 ChE210D final project. Hydration Dynamics at a Hydrophobic Surface. Abstract:
 Hydration Dynamics at a Hydrophobic Surface Sunyia Hussain 6/15/212 ChE21D final project Abstract: Water is the universal solvent of life, crucial to the function of all biomolecules. Proteins, membranes,
Hydration Dynamics at a Hydrophobic Surface Sunyia Hussain 6/15/212 ChE21D final project Abstract: Water is the universal solvent of life, crucial to the function of all biomolecules. Proteins, membranes,
UC Irvine UC Irvine Electronic Theses and Dissertations
 UC Irvine UC Irvine Electronic Theses and Dissertations Title Exploring Higher Accuracy Poisson-Boltzmann Methods for Biomolecular Simulations Permalink https://escholarship.org/uc/item/4988p8p8 Author
UC Irvine UC Irvine Electronic Theses and Dissertations Title Exploring Higher Accuracy Poisson-Boltzmann Methods for Biomolecular Simulations Permalink https://escholarship.org/uc/item/4988p8p8 Author
Bonds and Structural Supports
 Bonds and Structural Supports Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingresources/jmol/jmoltraining/struts.html
Bonds and Structural Supports Part of the Jmol Training Guide from the MSOE Center for BioMolecular Modeling Interactive version available at http://cbm.msoe.edu/teachingresources/jmol/jmoltraining/struts.html
General elastic beam with an elastic foundation
 General elastic beam with an elastic foundation Figure 1 shows a beam-column on an elastic foundation. The beam is connected to a continuous series of foundation springs. The other end of the foundation
General elastic beam with an elastic foundation Figure 1 shows a beam-column on an elastic foundation. The beam is connected to a continuous series of foundation springs. The other end of the foundation
Molecular Modeling -- Lecture 15 Surfaces and electrostatics
 Molecular Modeling -- Lecture 15 Surfaces and electrostatics Molecular surfaces The Hydrophobic Effect Electrostatics Poisson-Boltzmann Equation Electrostatic maps Electrostatic surfaces in MOE 15.1 The
Molecular Modeling -- Lecture 15 Surfaces and electrostatics Molecular surfaces The Hydrophobic Effect Electrostatics Poisson-Boltzmann Equation Electrostatic maps Electrostatic surfaces in MOE 15.1 The
Recent Progress in Numerical Methods for the Poisson- Boltzmann Equation in Biophysical Applications
 Recent Progress in Numerical Methods for the Poisson- Boltzmann Equation in Biophysical Applications B. Z. Lu 1,3,, Y. C. Zhou 1,2,3, M. J. Holst 2,3 and J. A. McCammon 1,3,4,5 1 Howard Hughes Medical
Recent Progress in Numerical Methods for the Poisson- Boltzmann Equation in Biophysical Applications B. Z. Lu 1,3,, Y. C. Zhou 1,2,3, M. J. Holst 2,3 and J. A. McCammon 1,3,4,5 1 Howard Hughes Medical
Lysozyme pka example - Software. APBS! >!Examples! >!pka calculations! >! Lysozyme pka example. Background
 Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Software Search this site Home Announcements An update on mailing lists APBS 1.2.0 released APBS 1.2.1 released APBS 1.3 released New APBS 1.3 Windows Installer PDB2PQR 1.7.1 released PDB2PQR 1.8 released
Chapter 22. Preview. Objectives Properties of the Nucleus Nuclear Stability Binding Energy Sample Problem. Section 1 The Nucleus
 Section 1 The Nucleus Preview Objectives Properties of the Nucleus Nuclear Stability Binding Energy Sample Problem Section 1 The Nucleus Objectives Identify the properties of the nucleus of an atom. Explain
Section 1 The Nucleus Preview Objectives Properties of the Nucleus Nuclear Stability Binding Energy Sample Problem Section 1 The Nucleus Objectives Identify the properties of the nucleus of an atom. Explain
Capacitors II. Physics 2415 Lecture 9. Michael Fowler, UVa
 Capacitors II Physics 2415 Lecture 9 Michael Fowler, UVa Today s Topics First, some review then Storing energy in a capacitor How energy is stored in the electric field Dielectrics: why they strengthen
Capacitors II Physics 2415 Lecture 9 Michael Fowler, UVa Today s Topics First, some review then Storing energy in a capacitor How energy is stored in the electric field Dielectrics: why they strengthen
Molecular dynamics simulation. CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror
 Molecular dynamics simulation CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror 1 Outline Molecular dynamics (MD): The basic idea Equations of motion Key properties of MD simulations Sample applications
Molecular dynamics simulation CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror 1 Outline Molecular dynamics (MD): The basic idea Equations of motion Key properties of MD simulations Sample applications
Copyright Mark Brandt, Ph.D A third method, cryogenic electron microscopy has seen increasing use over the past few years.
 Structure Determination and Sequence Analysis The vast majority of the experimentally determined three-dimensional protein structures have been solved by one of two methods: X-ray diffraction and Nuclear
Structure Determination and Sequence Analysis The vast majority of the experimentally determined three-dimensional protein structures have been solved by one of two methods: X-ray diffraction and Nuclear
Molecular Dynamics Simulation of HIV-1 Reverse. Transcriptase
 Molecular Dynamics Simulation of HIV-1 Reverse Transcriptase Abderrahmane Benghanem 1 Maria Kurnikova 2 1 Rensselaer Polytechnic Institute, Troy, NY 2 Carnegie Mellon University, Pittsburgh, PA June 16,
Molecular Dynamics Simulation of HIV-1 Reverse Transcriptase Abderrahmane Benghanem 1 Maria Kurnikova 2 1 Rensselaer Polytechnic Institute, Troy, NY 2 Carnegie Mellon University, Pittsburgh, PA June 16,
Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation
 Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation Ion:ion interactions What is the free energy of ion:ion interactions ΔG i-i? Consider an ion in a solution
Bchem 675 Lecture 9 Electrostatics-Lecture 2 Debye-Hückel: Continued Counter ion condensation Ion:ion interactions What is the free energy of ion:ion interactions ΔG i-i? Consider an ion in a solution
Lectures 11-13: Electrostatics of Salty Solutions
 Lectures 11-13: Electrostatics of Salty Solutions Lecturer: Brigita Urbanc Office: 1-909 (E-mail: brigita@drexel.edu) Course website: www.physics.drexel.edu/~brigita/courses/biophys_011-01/ 1 Water as
Lectures 11-13: Electrostatics of Salty Solutions Lecturer: Brigita Urbanc Office: 1-909 (E-mail: brigita@drexel.edu) Course website: www.physics.drexel.edu/~brigita/courses/biophys_011-01/ 1 Water as
Dielectrics. Lecture 20: Electromagnetic Theory. Professor D. K. Ghosh, Physics Department, I.I.T., Bombay
 What are dielectrics? Dielectrics Lecture 20: Electromagnetic Theory Professor D. K. Ghosh, Physics Department, I.I.T., Bombay So far we have been discussing electrostatics in either vacuum or in a conductor.
What are dielectrics? Dielectrics Lecture 20: Electromagnetic Theory Professor D. K. Ghosh, Physics Department, I.I.T., Bombay So far we have been discussing electrostatics in either vacuum or in a conductor.
arxiv: v1 [math.na] 10 Oct 2014
![arxiv: v1 [math.na] 10 Oct 2014 arxiv: v1 [math.na] 10 Oct 2014](/thumbs/82/87000254.jpg) Unconditionally stable time splitting methods for the electrostatic analysis of solvated biomolecules arxiv:1410.2788v1 [math.na] 10 Oct 2014 Leighton Wilson and Shan Zhao Department of Mathematics, University
Unconditionally stable time splitting methods for the electrostatic analysis of solvated biomolecules arxiv:1410.2788v1 [math.na] 10 Oct 2014 Leighton Wilson and Shan Zhao Department of Mathematics, University
Molecular Dynamics Flexible Fitting
 Molecular Dynamics Flexible Fitting Ryan McGreevy Research Programmer University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Molecular Dynamics Flexible
Molecular Dynamics Flexible Fitting Ryan McGreevy Research Programmer University of Illinois at Urbana-Champaign NIH Resource for Macromolecular Modeling and Bioinformatics Molecular Dynamics Flexible
The Discontinuous Galerkin Finite Element Method
 The Discontinuous Galerkin Finite Element Method Michael A. Saum msaum@math.utk.edu Department of Mathematics University of Tennessee, Knoxville The Discontinuous Galerkin Finite Element Method p.1/41
The Discontinuous Galerkin Finite Element Method Michael A. Saum msaum@math.utk.edu Department of Mathematics University of Tennessee, Knoxville The Discontinuous Galerkin Finite Element Method p.1/41
Physics 342 Lecture 23. Radial Separation. Lecture 23. Physics 342 Quantum Mechanics I
 Physics 342 Lecture 23 Radial Separation Lecture 23 Physics 342 Quantum Mechanics I Friday, March 26th, 2010 We begin our spherical solutions with the simplest possible case zero potential. Aside from
Physics 342 Lecture 23 Radial Separation Lecture 23 Physics 342 Quantum Mechanics I Friday, March 26th, 2010 We begin our spherical solutions with the simplest possible case zero potential. Aside from
Introduction to Biological Small Angle Scattering
 Introduction to Biological Small Angle Scattering Tom Grant, Ph.D. Staff Scientist BioXFEL Science and Technology Center Hauptman-Woodward Institute Buffalo, New York, USA tgrant@hwi.buffalo.edu SAXS Literature
Introduction to Biological Small Angle Scattering Tom Grant, Ph.D. Staff Scientist BioXFEL Science and Technology Center Hauptman-Woodward Institute Buffalo, New York, USA tgrant@hwi.buffalo.edu SAXS Literature
Chapter 1 Direct Modeling for Computational Fluid Dynamics
 Chapter 1 Direct Modeling for Computational Fluid Dynamics Computational fluid dynamics (CFD) is a scientific discipline, which aims to capture fluid motion in a discretized space. The description of the
Chapter 1 Direct Modeling for Computational Fluid Dynamics Computational fluid dynamics (CFD) is a scientific discipline, which aims to capture fluid motion in a discretized space. The description of the
Lysozyme pka example. Background
 Lysozyme pka example Background Hen egg white lysozyme (HEWL) is a very popular system for pka as it has a number of interesting values for its titratable residues. Early pka work on this enzyme is presented
Lysozyme pka example Background Hen egg white lysozyme (HEWL) is a very popular system for pka as it has a number of interesting values for its titratable residues. Early pka work on this enzyme is presented
PROTEIN STRUCTURE AMINO ACIDS H R. Zwitterion (dipolar ion) CO 2 H. PEPTIDES Formal reactions showing formation of peptide bond by dehydration:
 PTEI STUTUE ydrolysis of proteins with aqueous acid or base yields a mixture of free amino acids. Each type of protein yields a characteristic mixture of the ~ 20 amino acids. AMI AIDS Zwitterion (dipolar
PTEI STUTUE ydrolysis of proteins with aqueous acid or base yields a mixture of free amino acids. Each type of protein yields a characteristic mixture of the ~ 20 amino acids. AMI AIDS Zwitterion (dipolar
Non-bonded interactions
 speeding up the number-crunching continued Marcus Elstner and Tomáš Kubař December 3, 2013 why care? number of individual pair-wise interactions bonded interactions proportional to N: O(N) non-bonded interactions
speeding up the number-crunching continued Marcus Elstner and Tomáš Kubař December 3, 2013 why care? number of individual pair-wise interactions bonded interactions proportional to N: O(N) non-bonded interactions
Let s continue our discussion on the interaction between Fe(III) and 6,7-dihydroxynaphthalene-2- sulfonate.
 Chemistry 5995(133)-8990(013) Bioinorganic Chemistry: The Good, the Bad, and the Potential of Metals Assignment 2- Aqueous Speciation, Magnetism, Redox, UV-Vis Spectroscopy, and Pymol Let s continue our
Chemistry 5995(133)-8990(013) Bioinorganic Chemistry: The Good, the Bad, and the Potential of Metals Assignment 2- Aqueous Speciation, Magnetism, Redox, UV-Vis Spectroscopy, and Pymol Let s continue our
Ph.D. QUALIFYING EXAMINATION PART A. Tuesday, January 3, 2012, 1:00 5:00 P.M.
 PhD QUALIFYING EXAMINATION PART A Tuesday, January 3, 212, 1: 5: PM Work each problem on a separate sheet(s) of paper and put your identifying number on each page Do not use your name Each problem has
PhD QUALIFYING EXAMINATION PART A Tuesday, January 3, 212, 1: 5: PM Work each problem on a separate sheet(s) of paper and put your identifying number on each page Do not use your name Each problem has
Numerically Solving Partial Differential Equations
 Numerically Solving Partial Differential Equations Michael Lavell Department of Applied Mathematics and Statistics Abstract The physics describing the fundamental principles of fluid dynamics can be written
Numerically Solving Partial Differential Equations Michael Lavell Department of Applied Mathematics and Statistics Abstract The physics describing the fundamental principles of fluid dynamics can be written
The Molecular Dynamics Method
 H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
H-bond energy (kcal/mol) - 4.0 The Molecular Dynamics Method Fibronectin III_1, a mechanical protein that glues cells together in wound healing and in preventing tumor metastasis 0 ATPase, a molecular
Analysis of integral expressions for effective Born radii
 THE JOURNAL OF CHEMICAL PHYSICS 27, 850 2007 Analysis of integral expressions for effective Born radii John Mongan Bioinformatics Program, Medical Scientist Training Program, Center for Theoretical Biological
THE JOURNAL OF CHEMICAL PHYSICS 27, 850 2007 Analysis of integral expressions for effective Born radii John Mongan Bioinformatics Program, Medical Scientist Training Program, Center for Theoretical Biological
INTENSIVE COMPUTATION. Annalisa Massini
 INTENSIVE COMPUTATION Annalisa Massini 2015-2016 Course topics The course will cover topics that are in some sense related to intensive computation: Matlab (an introduction) GPU (an introduction) Sparse
INTENSIVE COMPUTATION Annalisa Massini 2015-2016 Course topics The course will cover topics that are in some sense related to intensive computation: Matlab (an introduction) GPU (an introduction) Sparse
PHYSICS. Course Syllabus. Section 1: Mathematical Physics. Subject Code: PH. Course Structure. Electromagnetic Theory
 PHYSICS Subject Code: PH Course Structure Sections/Units Topics Section 1 Section 2 Section 3 Section 4 Section 5 Section 6 Section 7 Section 8 Mathematical Physics Classical Mechanics Electromagnetic
PHYSICS Subject Code: PH Course Structure Sections/Units Topics Section 1 Section 2 Section 3 Section 4 Section 5 Section 6 Section 7 Section 8 Mathematical Physics Classical Mechanics Electromagnetic
Contents. 1 Basic Equations 1. Acknowledgment. 1.1 The Maxwell Equations Constitutive Relations 11
 Preface Foreword Acknowledgment xvi xviii xix 1 Basic Equations 1 1.1 The Maxwell Equations 1 1.1.1 Boundary Conditions at Interfaces 4 1.1.2 Energy Conservation and Poynting s Theorem 9 1.2 Constitutive
Preface Foreword Acknowledgment xvi xviii xix 1 Basic Equations 1 1.1 The Maxwell Equations 1 1.1.1 Boundary Conditions at Interfaces 4 1.1.2 Energy Conservation and Poynting s Theorem 9 1.2 Constitutive
A Generalization for Stable Mixed Finite Elements
 A Generalization for Stable Mixed Finite Elements Andrew Gillette joint work with Chandrajit Bajaj Department of Mathematics Institute of Computational Engineering and Sciences University of Texas at Austin,
A Generalization for Stable Mixed Finite Elements Andrew Gillette joint work with Chandrajit Bajaj Department of Mathematics Institute of Computational Engineering and Sciences University of Texas at Austin,
Molecular modeling with InsightII
 Molecular modeling with InsightII Yuk Sham Computational Biology/Biochemistry Consultant Phone: (612) 624 7427 (Walter Library) Phone: (612) 624 0783 (VWL) Email: shamy@msi.umn.edu How to run InsightII
Molecular modeling with InsightII Yuk Sham Computational Biology/Biochemistry Consultant Phone: (612) 624 7427 (Walter Library) Phone: (612) 624 0783 (VWL) Email: shamy@msi.umn.edu How to run InsightII
ENGINEERING MATHEMATICS I. CODE: 10 MAT 11 IA Marks: 25 Hrs/Week: 04 Exam Hrs: 03 PART-A
 ENGINEERING MATHEMATICS I CODE: 10 MAT 11 IA Marks: 25 Hrs/Week: 04 Exam Hrs: 03 Total Hrs: 52 Exam Marks:100 PART-A Unit-I: DIFFERENTIAL CALCULUS - 1 Determination of n th derivative of standard functions-illustrative
ENGINEERING MATHEMATICS I CODE: 10 MAT 11 IA Marks: 25 Hrs/Week: 04 Exam Hrs: 03 Total Hrs: 52 Exam Marks:100 PART-A Unit-I: DIFFERENTIAL CALCULUS - 1 Determination of n th derivative of standard functions-illustrative
Gauss s Law. Lecture 4. Chapter 27. Channel 61 (clicker) Physics II
 Lecture 4 Chapter 27 Physics II 01.30.2015 Gauss s Law 95.144 Course website: http://faculty.uml.edu/andriy_danylov/teaching/physicsii Lecture Capture: http://echo360.uml.edu/danylov201415/physics2spring.html
Lecture 4 Chapter 27 Physics II 01.30.2015 Gauss s Law 95.144 Course website: http://faculty.uml.edu/andriy_danylov/teaching/physicsii Lecture Capture: http://echo360.uml.edu/danylov201415/physics2spring.html
Recent Progress in Numerical Methods for the Poisson- Boltzmann Equation in Biophysical Applications
 COMMUNICATIONS IN COMPUTATIONAL PHYSICS Vol. 3, No. 5, pp. 973-1009 Commun. Comput. Phys. May 2008 REVIEW ARTICLE Recent Progress in Numerical Methods for the Poisson- Boltzmann Equation in Biophysical
COMMUNICATIONS IN COMPUTATIONAL PHYSICS Vol. 3, No. 5, pp. 973-1009 Commun. Comput. Phys. May 2008 REVIEW ARTICLE Recent Progress in Numerical Methods for the Poisson- Boltzmann Equation in Biophysical
lim = F F = F x x + F y y + F z
 Physics 361 Summary of Results from Lecture Physics 361 Derivatives of Scalar and Vector Fields The gradient of a scalar field f( r) is given by g = f. coordinates f g = ê x x + ê f y y + ê f z z Expressed
Physics 361 Summary of Results from Lecture Physics 361 Derivatives of Scalar and Vector Fields The gradient of a scalar field f( r) is given by g = f. coordinates f g = ê x x + ê f y y + ê f z z Expressed
Protein surface descriptors for binding sites comparison and ligand prediction
 Protein surface descriptors for binding sites comparison and ligand prediction Rayan Chikhi Internship report, Kihara Bioinformatics Laboratory, Purdue University, 2007 Abstract. Proteins molecular recognition
Protein surface descriptors for binding sites comparison and ligand prediction Rayan Chikhi Internship report, Kihara Bioinformatics Laboratory, Purdue University, 2007 Abstract. Proteins molecular recognition
Analytical Technologies in Biotechnology Prof. Dr. Ashwani K. Sharma Department of Biotechnology Indian Institute of Technology, Roorkee
 Analytical Technologies in Biotechnology Prof. Dr. Ashwani K. Sharma Department of Biotechnology Indian Institute of Technology, Roorkee Module - 4 Electrophoresis Lecture - 1 Basis Concept in Electrophoresis
Analytical Technologies in Biotechnology Prof. Dr. Ashwani K. Sharma Department of Biotechnology Indian Institute of Technology, Roorkee Module - 4 Electrophoresis Lecture - 1 Basis Concept in Electrophoresis
Non-bonded interactions
 speeding up the number-crunching Marcus Elstner and Tomáš Kubař May 8, 2015 why care? key to understand biomolecular structure and function binding of a ligand efficiency of a reaction color of a chromophore
speeding up the number-crunching Marcus Elstner and Tomáš Kubař May 8, 2015 why care? key to understand biomolecular structure and function binding of a ligand efficiency of a reaction color of a chromophore
Electrodynamics I Midterm - Part A - Closed Book KSU 2005/10/17 Electro Dynamic
 Electrodynamics I Midterm - Part A - Closed Book KSU 5//7 Name Electro Dynamic. () Write Gauss Law in differential form. E( r) =ρ( r)/ɛ, or D = ρ, E= electricfield,ρ=volume charge density, ɛ =permittivity
Electrodynamics I Midterm - Part A - Closed Book KSU 5//7 Name Electro Dynamic. () Write Gauss Law in differential form. E( r) =ρ( r)/ɛ, or D = ρ, E= electricfield,ρ=volume charge density, ɛ =permittivity
Computational Molecular Modeling
 Computational Molecular Modeling Lecture 1: Structure Models, Properties Chandrajit Bajaj Today s Outline Intro to atoms, bonds, structure, biomolecules, Geometry of Proteins, Nucleic Acids, Ribosomes,
Computational Molecular Modeling Lecture 1: Structure Models, Properties Chandrajit Bajaj Today s Outline Intro to atoms, bonds, structure, biomolecules, Geometry of Proteins, Nucleic Acids, Ribosomes,
CAP 5510 Lecture 3 Protein Structures
 CAP 5510 Lecture 3 Protein Structures Su-Shing Chen Bioinformatics CISE 8/19/2005 Su-Shing Chen, CISE 1 Protein Conformation 8/19/2005 Su-Shing Chen, CISE 2 Protein Conformational Structures Hydrophobicity
CAP 5510 Lecture 3 Protein Structures Su-Shing Chen Bioinformatics CISE 8/19/2005 Su-Shing Chen, CISE 1 Protein Conformation 8/19/2005 Su-Shing Chen, CISE 2 Protein Conformational Structures Hydrophobicity
Squigonometry (doing trig on a squircle)
 (doing trig on a squircle) http://www.southernct.edu/~fields/ abstract outline The point of radian measure (although this probably isn t apparent when we first learn it) is to give a parameterization of
(doing trig on a squircle) http://www.southernct.edu/~fields/ abstract outline The point of radian measure (although this probably isn t apparent when we first learn it) is to give a parameterization of
