Hands-on : Model Potential Molecular Dynamics
|
|
|
- Clinton McKinney
- 5 years ago
- Views:
Transcription
1 Hands-on : Model Potential Molecular Dynamics
2 OUTLINE 0. DL_POLY code introduction 0.a Input files 1. THF solvent molecule 1.a Geometry optimization 1.b NVE/NVT dynamics 2. Liquid THF 2.a Equilibration NPT 2.b Dipole moment calculation 2.c Static dielectric constant calculation
3 0. DL_POLY code introduction 0.a Input files DL_POLY code is a general purpose parallel molecular dynamics simulation package developed at Daresbury Laboratory by W. Smith and I.T. Todorov. It can be freely downloaded at ( upon registration, and its manual can be found at Copy the executable (DLPOLY.Z) from $WORKSHOP/dl_poly_4.04/execute/ into your working directory, and the manual (USRMAN4.04.pdf) from $WORKSHOP/dl_poly_4.04/manual. We need three files to start a DL_POLY MD simulation, named as CONFIG, CONTROL and FIELD: CONFIG contains the informations on the system geometry and the simulation cell FIELD contains the parameters of the force field which describes the molecular interactions (bond stetching, Van der Waals interactions, atomic partial charges etc.) CONTROL contains the simulation parameters (e.g. temperature, pressure, number of timesteps...)
4 CONFIG typical file structure Periodic boundary conditions key (see manual) Config file key (see manual) n. of atoms Simulation cell Title Atom name x,y,z coordinates Atom index
5 FIELD typical file structure title units name of first ms number of atoms per molecule number of molecular species (ms) number of molecules (of first ms) name,mass, charge of atoms bonded interactions (intramolecular) nonbonded (Van der Waals) interactions (for all ms) end of FIELD file end of first ms
6 FIELD typical file structure (2) details on the first molecular species details on the second molecular species details on the third molecular species nonbonded interactions unique section for all the molecular species If there are more than 1 molecular species, the FIELD file will look like this:
7 CONTROL typical file structure integration algorithm temperature (K) pressure (katm) thermodynamic ensemble Number of simulation timesteps I/O info timestep duration (ps) maximum allowed job time (s) cutoff for Coulomb and Van der Waals interactions (Å)
8 1. THF solvent molecule 1.a Geometry optimization 1.b NVE/NVT dynamics Edit CONTROL file by adding the highlighted row minimise force n f tells the code to perform a local optimization of the molecule every n step by minimizing the intramolecular forces with respect to the tolerance f, using conjugate gradient method. The minimization can be performed also on the energy or maximum displacement.
9 1. THF solvent molecule 1.a Geometry optimization 1.b NVE/NVT dynamics Run the calculation by executing DLPOLY.Z (it should be very fast) After the calculation, some new files have been created by the program: OUTPUT contains a summary of the simulation details, force field, system configuration at different timesteps etc. HISTORY contains the atomic coordinates at every timestep REVCON contains the final atomic coordinates, velocities and forces. It is used mainly as restart file. REVIVE binary file which is used as restart file STATIS contains a collection of statistical data, useful for post-processing, such as system temperature, energy, volume etc.
10 1. THF solvent molecule 1.a Geometry optimization 1.b NVE/NVT dynamics We can now perform a dynamic simulation, and we will verify that the theoretical assumptions on the NVE ensemble are correct. We can use the REVCON file generated from the geometry optimization as a restart file (rename this file as CONFIG), or use the CONFIG file in the folder. Edit the CONTROL file: choose the number of steps (5000 steps will take about 20 sec) comment the minimise line choose how often you want to collect data (in this example, all data are collected every 100 steps. See manual for details) RUN!!!
11 We have performed an NVE run, i.e. we should have constant number of particles, volume and energy. We can use the STATIS file to verify that everything is OK: potential energy total energy system temperature system volume this file contains also other useful data, we invite you to read the manual for details
12 TASKS Write a simple tool to extract the relevant data from the STATIS file Plot total energy, system temperature and system volume vs time Is average total energy conserved? What happens to temperature and volume?
13 If we want the system temperature to be conserved, we need to perform an NVT simulation. To do so, slightly modify the CONTROL file: change ensemble, choose the temperature, and rerun. You will also have to add two new keywords. These indicate that the code will scale the temperature every scale timesteps during the first equilibration steps (choose it to be about 10% of the total simulation time). Verify that the average system temperature is conserved now: NVE NVT Perform a NVT simulation at T=300K fot both the systems of one and two THF molecules (nvt/1mol/ and nvt/2mol/). Observe the trajectories with vmd (vmd dlpoly3hist HISTORY).
14 2. Liquid THF 2.a Equilibration NPT 2.b Dipole moment calculation 2.c Static dielectric constant calculation Since a liquid has no crystalline structure, i.e. it is disordered, we do not know where to put the molecules. We can start from any configuration (for example, a cubic crystal of arbitrary lattice constant), and then anneal the system to get a disordered, liquid-like structure. Annealing
15 2. Liquid THF 2.a Equilibration - NPT 2.b Dipole moment calculation 2.c Static dielectric constant calculation Any liquid has its own density at room temperature (g/cm³), we must reproduce it to have a good simulated liquid. The standard way to obtain it is to perform a constant pressure simulation (ensemble NPT): in this ensemble the volume is free to change in order to achieve the correct density. Ambient contitions: T=300 K, p=1 atm ensemble NPT Allows the cell density to vary during the equilibration Run the calculation and plot the volume and the potential energy. NB: this run will take about 1min for 1000 steps!
16 2. Liquid THF 2.a Equilibration - NPT 2.b Dipole moment calculation 2.c Static dielectric constant calculation If the volume converges too slowly or it oscillates too much, you may consider changing the relaxation constant of the barostat: high values provide smaller oscillations but slow convergence, and vice versa.
17 2. Liquid THF 2.a Equilibration NPT 2.b Dipole moment calculation 2.c Static dielectric constant calculation Once your volume and potential energy have reasonably converged, we can calculate the liquid density ρ and other thermodynamic properties, e.g. the vaporization enthalpy ΔHv E NB liq N mol M mol H v = RT = N mol V N mol number of molecules M mol molecular mass ( g/mol for THF) V system volume RT E NB liq gas constant times temperature ( kcal at 300 K) experimental values ρ g/cm³ ΔHv 7.65 kcal/mol nonbonding energy of the system (i.e. electrostatic+ VdW) NB: Very long simulations are required to obtain correct thermodynamic properties! Use the STATIS.ref file to extract the data
18 2. Liquid THF 2.a Equilibration NPT 2.b Dipole moment calculation 2.c Static dielectric constant calculation Let's now calculate the molecular dipole moment for the solvent THF. This will be useful to calculate the static dielectric permittivity of the liquid Na = q i r i=1 Where qi is the atom's charge and r is the atom-atom distance Since we have the atoms' coordinates (in r HISTORY file) and charges (in FIELD file), we can easily compute the value of the molecular dipole moment. Edit the dipole.f90 code in codes/step1/ to calculate the molecular dipole moment for THF, and compare it to the experimental value of 1.75 D. (NB: since charges are given in units of electron charge and the positions in Å, you will have to multiply the calculated value by to get the value in Debyes) Calculate the dipole moment for the system of 1 and 2 molecules (use the STATIS files in folders nvt/1mol/ and nvt/2mol/ and edit the codes in codes/step2/ ). Do you get the same results? Why?
19 2. Liquid THF 2.a Equilibration - NPT 2.b Static dielectric constant calculation The relative static dielectric permittivity can be calculated from the total dipole moment fluctuations (fluctuation-dissipation theorem) in a NPT equilibrium simulation r =1 4 2 M 2 M 3k B T V (M. Neumann, Mol.Phys. 1983) N mol = i M sum of all the molecular dipoles in the liquid i=1 V average system volume Modify the code for the single molecule dipole (as a function of time) to calculate the total dipole moment and hence the dielectric permittivity. Edit the code in codes/step3 and use the liquid/history.ref and liquid/statis.ref to extract the data.
20 2. Liquid THF 2.a Equilibration - NPT 2.b Static dielectric constant calculation r = M M 3k B T V NB: this formula is valid with 18 1 D=10 N mol in Debyes M statc cm= C cm = i M i=1 M 2 = M 2x M 2y M 2z 2 = M x 2 M y 2 M z 2 M Compare the calculated value with the experimental one εr=7.52 Does the two values compare well? Why?
21 A GENERAL RULE In Model Potential Molecular Dynamics the calculated properties strongly depend upon the choice of the parameters, such as bond strength, atomic charges etc. Furthermore, properties (such as the dielectric constant) which are related to electronic charge displacement cannot be properly taken into account with a simple fixed-charge model. Polarizable models have been developed and are available in Literature. Sec. 2.5 of the DL_POLY manual illustrates the ones which are implemented in this code.
22 Thanks for your attention!
Exercise 2: Solvating the Structure Before you continue, follow these steps: Setting up Periodic Boundary Conditions
 Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
Exercise 2: Solvating the Structure HyperChem lets you place a molecular system in a periodic box of water molecules to simulate behavior in aqueous solution, as in a biological system. In this exercise,
Molecular Dynamics Simulations. Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia
 Molecular Dynamics Simulations Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia 1 An Introduction to Molecular Dynamics Simulations Macroscopic properties
Molecular Dynamics Simulations Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia 1 An Introduction to Molecular Dynamics Simulations Macroscopic properties
Introduction to molecular dynamics
 1 Introduction to molecular dynamics Yves Lansac Université François Rabelais, Tours, France Visiting MSE, GIST for the summer Molecular Simulation 2 Molecular simulation is a computational experiment.
1 Introduction to molecular dynamics Yves Lansac Université François Rabelais, Tours, France Visiting MSE, GIST for the summer Molecular Simulation 2 Molecular simulation is a computational experiment.
 A Nobel Prize for Molecular Dynamics and QM/MM What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential
A Nobel Prize for Molecular Dynamics and QM/MM What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential
Free energy, electrostatics, and the hydrophobic effect
 Protein Physics 2016 Lecture 3, January 26 Free energy, electrostatics, and the hydrophobic effect Magnus Andersson magnus.andersson@scilifelab.se Theoretical & Computational Biophysics Recap Protein structure
Protein Physics 2016 Lecture 3, January 26 Free energy, electrostatics, and the hydrophobic effect Magnus Andersson magnus.andersson@scilifelab.se Theoretical & Computational Biophysics Recap Protein structure
The change in free energy on transferring an ion from a medium of low dielectric constantε1 to one of high dielectric constant ε2:
 The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
What is Classical Molecular Dynamics?
 What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
Introduction to CPMD 1
 Introduction to CPMD 1 CPMD program ab initio electronic structure (DFT) and molecular dynamics program plane wave basis set (PBC), pseudopotentials massively parallelized, linear scaling up to thousands
Introduction to CPMD 1 CPMD program ab initio electronic structure (DFT) and molecular dynamics program plane wave basis set (PBC), pseudopotentials massively parallelized, linear scaling up to thousands
Winmostar tutorial LAMMPS Melting point V X-Ability Co,. Ltd. 2017/8/17
 Winmostar tutorial LAMMPS Melting point V7.025 X-Ability Co,. Ltd. question@winmostar.com Contents Configure I. Build solid phase II. Equilibration of solid phase III. Equilibration of liquid phase IV.
Winmostar tutorial LAMMPS Melting point V7.025 X-Ability Co,. Ltd. question@winmostar.com Contents Configure I. Build solid phase II. Equilibration of solid phase III. Equilibration of liquid phase IV.
CPMD Tutorial Atosim/RFCT 2009/10
 These exercices were inspired by the CPMD Tutorial of Axel Kohlmeyer http://www.theochem.ruhruni-bochum.de/ axel.kohlmeyer/cpmd-tutor/index.html and by other tutorials. Here is a summary of what we will
These exercices were inspired by the CPMD Tutorial of Axel Kohlmeyer http://www.theochem.ruhruni-bochum.de/ axel.kohlmeyer/cpmd-tutor/index.html and by other tutorials. Here is a summary of what we will
2D Clausius Clapeyron equation
 2D Clausius Clapeyron equation 1/14 Aim: Verify the Clausius Clapeyron equation by simulations of a 2D model of matter Model: 8-4 type potential ( Lennard-Jones in 2D) u(r) = 1 r 8 1 r 4 Hard attractive
2D Clausius Clapeyron equation 1/14 Aim: Verify the Clausius Clapeyron equation by simulations of a 2D model of matter Model: 8-4 type potential ( Lennard-Jones in 2D) u(r) = 1 r 8 1 r 4 Hard attractive
Calculating Bond Enthalpies of the Hydrides
 Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
Proposed Exercise for the General Chemistry Section of the Teaching with Cache Workbook: Calculating Bond Enthalpies of the Hydrides Contributed by James Foresman, Rachel Fogle, and Jeremy Beck, York College
2D Clausius Clapeyron equation
 2D Clausius Clapeyron equation 1/14 Aim: Verify the Clausius Clapeyron equation by simulations of a 2D model of matter Model: 8-4 type potential ( Lennard-Jones in 2D) u(r) = 1 r 8 1 r 4 Hard attractive
2D Clausius Clapeyron equation 1/14 Aim: Verify the Clausius Clapeyron equation by simulations of a 2D model of matter Model: 8-4 type potential ( Lennard-Jones in 2D) u(r) = 1 r 8 1 r 4 Hard attractive
Molecular dynamics simulation of graphene formation on 6H SiC substrate via simulated annealing
 Molecular dynamics simulation of graphene formation on 6H SiC substrate via simulated annealing Yoon Tiem Leong @ Min Tjun Kit School of Physics Universiti Sains Malaysia 1 Aug 2012 Single layer graphene
Molecular dynamics simulation of graphene formation on 6H SiC substrate via simulated annealing Yoon Tiem Leong @ Min Tjun Kit School of Physics Universiti Sains Malaysia 1 Aug 2012 Single layer graphene
Hyeyoung Shin a, Tod A. Pascal ab, William A. Goddard III abc*, and Hyungjun Kim a* Korea
 The Scaled Effective Solvent Method for Predicting the Equilibrium Ensemble of Structures with Analysis of Thermodynamic Properties of Amorphous Polyethylene Glycol-Water Mixtures Hyeyoung Shin a, Tod
The Scaled Effective Solvent Method for Predicting the Equilibrium Ensemble of Structures with Analysis of Thermodynamic Properties of Amorphous Polyethylene Glycol-Water Mixtures Hyeyoung Shin a, Tod
Analysis of the simulation
 Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Analysis of the simulation Marcus Elstner and Tomáš Kubař January 7, 2014 Thermodynamic properties time averages of thermodynamic quantites correspond to ensemble averages (ergodic theorem) some quantities
Molecular Dynamics Simulation of a Nanoconfined Water Film
 Molecular Dynamics Simulation of a Nanoconfined Water Film Kyle Lindquist, Shu-Han Chao May 7, 2013 1 Introduction The behavior of water confined in nano-scale environment is of interest in many applications.
Molecular Dynamics Simulation of a Nanoconfined Water Film Kyle Lindquist, Shu-Han Chao May 7, 2013 1 Introduction The behavior of water confined in nano-scale environment is of interest in many applications.
Ch. 9 Liquids and Solids
 Intermolecular Forces I. A note about gases, liquids and gases. A. Gases: very disordered, particles move fast and are far apart. B. Liquid: disordered, particles are close together but can still move.
Intermolecular Forces I. A note about gases, liquids and gases. A. Gases: very disordered, particles move fast and are far apart. B. Liquid: disordered, particles are close together but can still move.
2008 Biowerkzeug Ltd.
 2008 Biowerkzeug Ltd. 1 Contents Summary...3 1 Simulation...4 1.1 Setup...4 1.2 Output...4 2 Settings...5 3 Analysis...9 3.1 Setup...9 3.2 Input options...9 3.3 Descriptions...10 Please note that we cannot
2008 Biowerkzeug Ltd. 1 Contents Summary...3 1 Simulation...4 1.1 Setup...4 1.2 Output...4 2 Settings...5 3 Analysis...9 3.1 Setup...9 3.2 Input options...9 3.3 Descriptions...10 Please note that we cannot
Computer simulation methods (2) Dr. Vania Calandrini
 Computer simulation methods (2) Dr. Vania Calandrini in the previous lecture: time average versus ensemble average MC versus MD simulations equipartition theorem (=> computing T) virial theorem (=> computing
Computer simulation methods (2) Dr. Vania Calandrini in the previous lecture: time average versus ensemble average MC versus MD simulations equipartition theorem (=> computing T) virial theorem (=> computing
CHAPTER 11: INTERMOLECULAR FORCES AND LIQUIDS AND SOLIDS. Chemistry 1411 Joanna Sabey
 CHAPTER 11: INTERMOLECULAR FORCES AND LIQUIDS AND SOLIDS Chemistry 1411 Joanna Sabey Forces Phase: homogeneous part of the system in contact with other parts of the system but separated from them by a
CHAPTER 11: INTERMOLECULAR FORCES AND LIQUIDS AND SOLIDS Chemistry 1411 Joanna Sabey Forces Phase: homogeneous part of the system in contact with other parts of the system but separated from them by a
Biomolecules are dynamic no single structure is a perfect model
 Molecular Dynamics Simulations of Biomolecules References: A. R. Leach Molecular Modeling Principles and Applications Prentice Hall, 2001. M. P. Allen and D. J. Tildesley "Computer Simulation of Liquids",
Molecular Dynamics Simulations of Biomolecules References: A. R. Leach Molecular Modeling Principles and Applications Prentice Hall, 2001. M. P. Allen and D. J. Tildesley "Computer Simulation of Liquids",
Potential Energy (hyper)surface
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = " d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = " d dx U(x) Conformation
The Molecular Dynamics Method
 The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
The Molecular Dynamics Method Thermal motion of a lipid bilayer Water permeation through channels Selective sugar transport Potential Energy (hyper)surface What is Force? Energy U(x) F = d dx U(x) Conformation
Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany
 Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals Phase Equilibria Phase diagrams and classical thermodynamics
Phase Equilibria and Molecular Solutions Jan G. Korvink and Evgenii Rudnyi IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals Phase Equilibria Phase diagrams and classical thermodynamics
The micro-properties of [hmpy+] [Tf 2 N-] Ionic liquid: a simulation. study. 1. Introduction
![The micro-properties of [hmpy+] [Tf 2 N-] Ionic liquid: a simulation. study. 1. Introduction The micro-properties of [hmpy+] [Tf 2 N-] Ionic liquid: a simulation. study. 1. Introduction](/thumbs/72/66941683.jpg) ISBN 978-1-84626-081-0 Proceedings of the 2010 International Conference on Application of Mathematics and Physics Volume 1: Advances on Space Weather, Meteorology and Applied Physics Nanjing, P. R. China,
ISBN 978-1-84626-081-0 Proceedings of the 2010 International Conference on Application of Mathematics and Physics Volume 1: Advances on Space Weather, Meteorology and Applied Physics Nanjing, P. R. China,
Michael W. Mahoney Department of Physics, Yale University, New Haven, Connecticut 06520
 JOURNAL OF CHEMICAL PHYSICS VOLUME 115, NUMBER 23 15 DECEMBER 2001 Quantum, intramolecular flexibility, and polarizability effects on the reproduction of the density anomaly of liquid water by simple potential
JOURNAL OF CHEMICAL PHYSICS VOLUME 115, NUMBER 23 15 DECEMBER 2001 Quantum, intramolecular flexibility, and polarizability effects on the reproduction of the density anomaly of liquid water by simple potential
Case study: molecular dynamics of solvent diffusion in polymers
 Course MP3 Lecture 11 29/11/2006 Case study: molecular dynamics of solvent diffusion in polymers A real-life research example to illustrate the use of molecular dynamics Dr James Elliott 11.1 Research
Course MP3 Lecture 11 29/11/2006 Case study: molecular dynamics of solvent diffusion in polymers A real-life research example to illustrate the use of molecular dynamics Dr James Elliott 11.1 Research
Chem 112 Dr. Kevin Moore
 Chem 112 Dr. Kevin Moore Gas Liquid Solid Polar Covalent Bond Partial Separation of Charge Electronegativity: H 2.1 Cl 3.0 H Cl δ + δ - Dipole Moment measure of the net polarity in a molecule Q Q magnitude
Chem 112 Dr. Kevin Moore Gas Liquid Solid Polar Covalent Bond Partial Separation of Charge Electronegativity: H 2.1 Cl 3.0 H Cl δ + δ - Dipole Moment measure of the net polarity in a molecule Q Q magnitude
Universal Repulsive Contribution to the. Solvent-Induced Interaction Between Sizable, Curved Hydrophobes: Supporting Information
 Universal Repulsive Contribution to the Solvent-Induced Interaction Between Sizable, Curved Hydrophobes: Supporting Information B. Shadrack Jabes, Dusan Bratko, and Alenka Luzar Department of Chemistry,
Universal Repulsive Contribution to the Solvent-Induced Interaction Between Sizable, Curved Hydrophobes: Supporting Information B. Shadrack Jabes, Dusan Bratko, and Alenka Luzar Department of Chemistry,
Chapter 10. Liquids and Solids
 Chapter 10 Liquids and Solids Chapter 10 Table of Contents 10.1 Intermolecular Forces 10.2 The Liquid State 10.3 An Introduction to Structures and Types of Solids 10.4 Structure and Bonding in Metals 10.5
Chapter 10 Liquids and Solids Chapter 10 Table of Contents 10.1 Intermolecular Forces 10.2 The Liquid State 10.3 An Introduction to Structures and Types of Solids 10.4 Structure and Bonding in Metals 10.5
Intermolecular Forces & Condensed Phases
 Intermolecular Forces & Condensed Phases CHEM 107 T. Hughbanks READING We will discuss some of Chapter 5 that we skipped earlier (Van der Waals equation, pp. 145-8), but this is just a segue into intermolecular
Intermolecular Forces & Condensed Phases CHEM 107 T. Hughbanks READING We will discuss some of Chapter 5 that we skipped earlier (Van der Waals equation, pp. 145-8), but this is just a segue into intermolecular
CE 530 Molecular Simulation
 1 CE 530 Molecular Simulation Lecture 14 Molecular Models David A. Kofke Department of Chemical Engineering SUNY Buffalo kofke@eng.buffalo.edu 2 Review Monte Carlo ensemble averaging, no dynamics easy
1 CE 530 Molecular Simulation Lecture 14 Molecular Models David A. Kofke Department of Chemical Engineering SUNY Buffalo kofke@eng.buffalo.edu 2 Review Monte Carlo ensemble averaging, no dynamics easy
CHEMISTRY. CHM202 Class #3 CHEMISTRY. Chapter 11. Chapter Outline for Class #3. Solutions: Properties and Behavior
 CHEMISTRY Fifth Edition Gilbert Kirss Foster Bretz Davies CHM202 Class #3 1 Chemistry, 5 th Edition Copyright 2017, W. W. Norton & Company CHEMISTRY Fifth Edition Gilbert Kirss Foster Bretz Davies Chapter
CHEMISTRY Fifth Edition Gilbert Kirss Foster Bretz Davies CHM202 Class #3 1 Chemistry, 5 th Edition Copyright 2017, W. W. Norton & Company CHEMISTRY Fifth Edition Gilbert Kirss Foster Bretz Davies Chapter
First Law of Thermodynamics
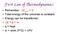 First Law of Thermodynamics Remember: ΔE univ = 0 Total energy of the universe is constant. Energy can be transferred: ΔE = q + w q = heat w = work (F*D) = ΔPV 1 st Law, review For constant volume process:
First Law of Thermodynamics Remember: ΔE univ = 0 Total energy of the universe is constant. Energy can be transferred: ΔE = q + w q = heat w = work (F*D) = ΔPV 1 st Law, review For constant volume process:
Why study protein dynamics?
 Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Diffusion of Water and Diatomic Oxygen in Poly(3-hexylthiophene) Melt: A Molecular Dynamics Simulation Study
 Diffusion of Water and Diatomic Oxygen in Poly(3-hexylthiophene) Melt: A Molecular Dynamics Simulation Study Julia Deitz, Yeneneh Yimer, and Mesfin Tsige Department of Polymer Science University of Akron
Diffusion of Water and Diatomic Oxygen in Poly(3-hexylthiophene) Melt: A Molecular Dynamics Simulation Study Julia Deitz, Yeneneh Yimer, and Mesfin Tsige Department of Polymer Science University of Akron
The broad topic of physical metallurgy provides a basis that links the structure of materials with their properties, focusing primarily on metals.
 Physical Metallurgy The broad topic of physical metallurgy provides a basis that links the structure of materials with their properties, focusing primarily on metals. Crystal Binding In our discussions
Physical Metallurgy The broad topic of physical metallurgy provides a basis that links the structure of materials with their properties, focusing primarily on metals. Crystal Binding In our discussions
CHE3935. Lecture 4 Quantum Mechanical Simulation Methods Continued
 CHE3935 Lecture 4 Quantum Mechanical Simulation Methods Continued 1 OUTLINE Review Introduction to CPMD MD and ensembles The functionals of density functional theory Return to ab initio methods Binding
CHE3935 Lecture 4 Quantum Mechanical Simulation Methods Continued 1 OUTLINE Review Introduction to CPMD MD and ensembles The functionals of density functional theory Return to ab initio methods Binding
Ionic Liquids simulations : obtention of structural and transport properties from molecular dynamics. C. J. F. Solano, D. Beljonne, R.
 Ionic Liquids simulations : obtention of structural and transport properties from molecular dynamics C. J. F. Solano, D. Beljonne, R. Lazzaroni Ionic Liquids simulations : obtention of structural and transport
Ionic Liquids simulations : obtention of structural and transport properties from molecular dynamics C. J. F. Solano, D. Beljonne, R. Lazzaroni Ionic Liquids simulations : obtention of structural and transport
Density dependence of dielectric permittivity of water and estimation of the electric field for the breakdown inception
 Journal of Physics: Conference Series PAPER OPEN ACCESS Density dependence of dielectric permittivity of water and estimation of the electric field for the breakdown inception To cite this article: D I
Journal of Physics: Conference Series PAPER OPEN ACCESS Density dependence of dielectric permittivity of water and estimation of the electric field for the breakdown inception To cite this article: D I
CHEMISTRY. Chapter 11 Intermolecular Forces Liquids and Solids
 CHEMISTRY The Central Science 8 th Edition Chapter 11 Liquids and Solids Kozet YAPSAKLI States of Matter difference between states of matter is the distance between particles. In the solid and liquid states
CHEMISTRY The Central Science 8 th Edition Chapter 11 Liquids and Solids Kozet YAPSAKLI States of Matter difference between states of matter is the distance between particles. In the solid and liquid states
Ch. 11: Liquids and Intermolecular Forces
 Ch. 11: Liquids and Intermolecular Forces Learning goals and key skills: Identify the intermolecular attractive interactions (dispersion, dipole-dipole, hydrogen bonding, ion-dipole) that exist between
Ch. 11: Liquids and Intermolecular Forces Learning goals and key skills: Identify the intermolecular attractive interactions (dispersion, dipole-dipole, hydrogen bonding, ion-dipole) that exist between
Solutions to Assignment #4 Getting Started with HyperChem
 Solutions to Assignment #4 Getting Started with HyperChem 1. This first exercise is meant to familiarize you with the different methods for visualizing molecules available in HyperChem. (a) Create a molecule
Solutions to Assignment #4 Getting Started with HyperChem 1. This first exercise is meant to familiarize you with the different methods for visualizing molecules available in HyperChem. (a) Create a molecule
This Answer/Solution Example is taken from a student s actual homework report. I thank him for permission to use it here. JG
 This Answer/Solution Example is taken from a student s actual homework report. I thank him for permission to use it here. JG Chem 8021 Spring 2005 Project II Calculation of Self-Diffusion Coeffecient of
This Answer/Solution Example is taken from a student s actual homework report. I thank him for permission to use it here. JG Chem 8021 Spring 2005 Project II Calculation of Self-Diffusion Coeffecient of
Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland
 Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland 1) Question. Two methods which are widely used for the optimization of molecular geometies are the Steepest descents and Newton-Raphson
Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland 1) Question. Two methods which are widely used for the optimization of molecular geometies are the Steepest descents and Newton-Raphson
arxiv: v1 [cond-mat.mtrl-sci] 18 Aug 2011
![arxiv: v1 [cond-mat.mtrl-sci] 18 Aug 2011 arxiv: v1 [cond-mat.mtrl-sci] 18 Aug 2011](/thumbs/78/78407616.jpg) Quantum path integral simulation of isotope effects in the melting temperature of ice Ih R. Ramírez and C. P. Herrero Instituto de Ciencia de Materiales de Madrid (ICMM), Consejo Superior de Investigaciones
Quantum path integral simulation of isotope effects in the melting temperature of ice Ih R. Ramírez and C. P. Herrero Instituto de Ciencia de Materiales de Madrid (ICMM), Consejo Superior de Investigaciones
Gear methods I + 1/18
 Gear methods I + 1/18 Predictor-corrector type: knowledge of history is used to predict an approximate solution, which is made more accurate in the following step we do not want (otherwise good) methods
Gear methods I + 1/18 Predictor-corrector type: knowledge of history is used to predict an approximate solution, which is made more accurate in the following step we do not want (otherwise good) methods
Supplementary Information for Atomistic Simulation of Spinodal Phase Separation Preceding Polymer Crystallization
 Supplementary Information for Atomistic Simulation of Spinodal Phase Separation Preceding Polymer Crystallization Richard H. Gee * Naida Lacevic and Laurence E. Fried University of California Lawrence
Supplementary Information for Atomistic Simulation of Spinodal Phase Separation Preceding Polymer Crystallization Richard H. Gee * Naida Lacevic and Laurence E. Fried University of California Lawrence
Chem 112 Exam 1 Version A Spring /16/ :00am/Odago, M. O.
 Chem 112 Exam 1 Version A Spring 2011 02/16/2011 10:00am/Odago, M. O. 1. The pressure of a certain gas is measured to be 25.1 mmhg. What is this pressure expressed in units of pascals? (1 atm=1.0125 x10
Chem 112 Exam 1 Version A Spring 2011 02/16/2011 10:00am/Odago, M. O. 1. The pressure of a certain gas is measured to be 25.1 mmhg. What is this pressure expressed in units of pascals? (1 atm=1.0125 x10
Introduction to model potential Molecular Dynamics A3hourcourseatICTP
 Introduction to model potential Molecular Dynamics A3hourcourseatICTP Alessandro Mattoni 1 1 Istituto Officina dei Materiali CNR-IOM Unità di Cagliari SLACS ICTP School on numerical methods for energy,
Introduction to model potential Molecular Dynamics A3hourcourseatICTP Alessandro Mattoni 1 1 Istituto Officina dei Materiali CNR-IOM Unità di Cagliari SLACS ICTP School on numerical methods for energy,
Solid to liquid. Liquid to gas. Gas to solid. Liquid to solid. Gas to liquid. +energy. -energy
 33 PHASE CHANGES - To understand solids and liquids at the molecular level, it will help to examine PHASE CHANGES in a little more detail. A quick review of the phase changes... Phase change Description
33 PHASE CHANGES - To understand solids and liquids at the molecular level, it will help to examine PHASE CHANGES in a little more detail. A quick review of the phase changes... Phase change Description
CHEM1100 Summary Notes Module 2
 CHEM1100 Summary Notes Module 2 Lecture 14 Introduction to Kinetic Theory & Ideal Gases What are Boyle s and Charles Laws? Boyle s Law the pressure of a given mass of an ideal gas is inversely proportional
CHEM1100 Summary Notes Module 2 Lecture 14 Introduction to Kinetic Theory & Ideal Gases What are Boyle s and Charles Laws? Boyle s Law the pressure of a given mass of an ideal gas is inversely proportional
Atoms can form stable units called molecules by sharing electrons.
 Atoms can form stable units called molecules by sharing electrons. The formation of molecules is the result of intramolecular bonding (within the molecule) e.g. ionic, covalent. Forces that cause the aggregation
Atoms can form stable units called molecules by sharing electrons. The formation of molecules is the result of intramolecular bonding (within the molecule) e.g. ionic, covalent. Forces that cause the aggregation
Theoretical models for the solvent effect
 Theoretical models for the solvent effect Benedetta Mennucci Dipartimento di Chimica e Chimica Industriale Web: http://benedetta.dcci.unipi.it Email: bene@dcci.unipi.it 8. Excited electronic states in
Theoretical models for the solvent effect Benedetta Mennucci Dipartimento di Chimica e Chimica Industriale Web: http://benedetta.dcci.unipi.it Email: bene@dcci.unipi.it 8. Excited electronic states in
Intermolecular and Intramolecular Forces. Introduction
 Intermolecular and Intramolecular Forces Introduction Atoms can form stable units called molecules by sharing electrons. The formation of molecules is the result of intramolecular bonding (within the molecule)
Intermolecular and Intramolecular Forces Introduction Atoms can form stable units called molecules by sharing electrons. The formation of molecules is the result of intramolecular bonding (within the molecule)
A Phase Transition in Ammonium Chloride. CHM 335 TA: David Robinson Office Hour: Wednesday, 11 am
 A Phase Transition in Ammonium Chloride CHM 335 TA: David Robinson Email: Drobinson@chm.uri.edu Office Hour: Wednesday, 11 am Purpose Determine the critical exponent for the order to disorder transition
A Phase Transition in Ammonium Chloride CHM 335 TA: David Robinson Email: Drobinson@chm.uri.edu Office Hour: Wednesday, 11 am Purpose Determine the critical exponent for the order to disorder transition
Introduction to Chemical Thermodynamics. (10 Lectures) Michaelmas Term
 Introduction to Chemical Thermodynamics Dr. D. E. Manolopoulos First Year (0 Lectures) Michaelmas Term Lecture Synopsis. Introduction & Background. Le Chatelier s Principle. Equations of state. Systems
Introduction to Chemical Thermodynamics Dr. D. E. Manolopoulos First Year (0 Lectures) Michaelmas Term Lecture Synopsis. Introduction & Background. Le Chatelier s Principle. Equations of state. Systems
16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit. Lecture 2 (9/11/17)
 16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit By Anthony Quintano - https://www.flickr.com/photos/quintanomedia/15071865580, CC BY 2.0, https://commons.wikimedia.org/w/index.php?curid=38538291
16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit By Anthony Quintano - https://www.flickr.com/photos/quintanomedia/15071865580, CC BY 2.0, https://commons.wikimedia.org/w/index.php?curid=38538291
FUNCTIONALISATION, HYDRATION & DYNAMICS
 FUNCTIONALISATION, HYDRATION & DYNAMICS MATERIALS STUDIO TUTORIAL WAMMBAT - DAY 4 Our main goal: MD simulation of glucose in hydrated & functionalised silica nanopore It is not a rocket science and you
FUNCTIONALISATION, HYDRATION & DYNAMICS MATERIALS STUDIO TUTORIAL WAMMBAT - DAY 4 Our main goal: MD simulation of glucose in hydrated & functionalised silica nanopore It is not a rocket science and you
Simulation of molecular systems by molecular dynamics
 Simulation of molecular systems by molecular dynamics Yohann Moreau yohann.moreau@ujf-grenoble.fr November 26, 2015 Yohann Moreau (UJF) Molecular Dynamics, Label RFCT 2015 November 26, 2015 1 / 35 Introduction
Simulation of molecular systems by molecular dynamics Yohann Moreau yohann.moreau@ujf-grenoble.fr November 26, 2015 Yohann Moreau (UJF) Molecular Dynamics, Label RFCT 2015 November 26, 2015 1 / 35 Introduction
Supplementary Information. Optimizing the Binding Energy of the Surfactant to. Iron-Oxide Yields Truly Monodisperse
 Supplementary Information Optimizing the Binding Energy of the Surfactant to Iron-Oxide Yields Truly Monodisperse Nanoparticles Hamed Sharifi Dehsari 1, Richard Anthony Harris 2, Anielen Halda Ribeiro
Supplementary Information Optimizing the Binding Energy of the Surfactant to Iron-Oxide Yields Truly Monodisperse Nanoparticles Hamed Sharifi Dehsari 1, Richard Anthony Harris 2, Anielen Halda Ribeiro
Superprotonic phase transition of CsHSO 4 : A molecular dynamics simulation study
 Superprotonic phase transition of CsHSO 4 : A molecular dynamics simulation study Calum R. I. Chisholm, 1 Yun Hee Jang, 2 Sossina M. Haile, 1 and William A. Goddard III 2, * 1 Department of Materials Science,
Superprotonic phase transition of CsHSO 4 : A molecular dynamics simulation study Calum R. I. Chisholm, 1 Yun Hee Jang, 2 Sossina M. Haile, 1 and William A. Goddard III 2, * 1 Department of Materials Science,
States of Matter. Intermolecular Forces. The States of Matter. Intermolecular Forces. Intermolecular Forces
 Intermolecular Forces Have studied INTRAmolecular forces the forces holding atoms together to form compounds. Now turn to forces between molecules INTERmolecular forces. Forces between molecules, between
Intermolecular Forces Have studied INTRAmolecular forces the forces holding atoms together to form compounds. Now turn to forces between molecules INTERmolecular forces. Forces between molecules, between
Supplementary Materials
 Supplementary Materials Atomistic Origin of Brittle Failure of Boron Carbide from Large Scale Reactive Dynamics Simulations; Suggestions toward Improved Ductility Qi An and William A. Goddard III * Materials
Supplementary Materials Atomistic Origin of Brittle Failure of Boron Carbide from Large Scale Reactive Dynamics Simulations; Suggestions toward Improved Ductility Qi An and William A. Goddard III * Materials
A Brief Guide to All Atom/Coarse Grain Simulations 1.1
 A Brief Guide to All Atom/Coarse Grain Simulations 1.1 Contents 1. Introduction 2. AACG simulations 2.1. Capabilities and limitations 2.2. AACG commands 2.3. Visualization and analysis 2.4. AACG simulation
A Brief Guide to All Atom/Coarse Grain Simulations 1.1 Contents 1. Introduction 2. AACG simulations 2.1. Capabilities and limitations 2.2. AACG commands 2.3. Visualization and analysis 2.4. AACG simulation
Sample Exercise 11.1 Identifying Substances That Can Form Hydrogen Bonds
 Sample Exercise 11.1 Identifying Substances That Can Form Hydrogen Bonds In which of these substances is hydrogen bonding likely to play an important role in determining physical properties: methane (CH
Sample Exercise 11.1 Identifying Substances That Can Form Hydrogen Bonds In which of these substances is hydrogen bonding likely to play an important role in determining physical properties: methane (CH
Determination of Kamlet-Taft parameters for selected solvate ionic liquids.
 Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2016 Determination of Kamlet-Taft parameters for selected solvate ionic liquids. Daniel
Electronic Supplementary Material (ESI) for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2016 Determination of Kamlet-Taft parameters for selected solvate ionic liquids. Daniel
T6.2 Molecular Mechanics
 T6.2 Molecular Mechanics We have seen that Benson group additivities are capable of giving heats of formation of molecules with accuracies comparable to those of the best ab initio procedures. However,
T6.2 Molecular Mechanics We have seen that Benson group additivities are capable of giving heats of formation of molecules with accuracies comparable to those of the best ab initio procedures. However,
Multiscale Coarse-Graining of Ionic Liquids
 3564 J. Phys. Chem. B 2006, 110, 3564-3575 Multiscale Coarse-Graining of Ionic Liquids Yanting Wang, Sergei Izvekov, Tianying Yan, and Gregory A. Voth* Center for Biophysical Modeling and Simulation and
3564 J. Phys. Chem. B 2006, 110, 3564-3575 Multiscale Coarse-Graining of Ionic Liquids Yanting Wang, Sergei Izvekov, Tianying Yan, and Gregory A. Voth* Center for Biophysical Modeling and Simulation and
Towards accurate calculations of Zn 2+ binding free energies in zinc finger proteins
 Towards accurate calculations of Zn 2+ binding free energies in zinc finger proteins UNDERGRADUATE HONORS RESEARCH THESIS Presented in Partial Fulfillment of the Requirements for the Bachelor of Science
Towards accurate calculations of Zn 2+ binding free energies in zinc finger proteins UNDERGRADUATE HONORS RESEARCH THESIS Presented in Partial Fulfillment of the Requirements for the Bachelor of Science
Free energy simulations
 Free energy simulations Marcus Elstner and Tomáš Kubař January 14, 2013 Motivation a physical quantity that is of most interest in chemistry? free energies Helmholtz F or Gibbs G holy grail of computational
Free energy simulations Marcus Elstner and Tomáš Kubař January 14, 2013 Motivation a physical quantity that is of most interest in chemistry? free energies Helmholtz F or Gibbs G holy grail of computational
Energy Barriers and Rates - Transition State Theory for Physicists
 Energy Barriers and Rates - Transition State Theory for Physicists Daniel C. Elton October 12, 2013 Useful relations 1 cal = 4.184 J 1 kcal mole 1 = 0.0434 ev per particle 1 kj mole 1 = 0.0104 ev per particle
Energy Barriers and Rates - Transition State Theory for Physicists Daniel C. Elton October 12, 2013 Useful relations 1 cal = 4.184 J 1 kcal mole 1 = 0.0434 ev per particle 1 kj mole 1 = 0.0104 ev per particle
Chapter 2. Dielectric Theories
 Chapter Dielectric Theories . Dielectric Theories 1.1. Introduction Measurements of dielectric properties of materials is very important because it provide vital information regarding the material characteristics,
Chapter Dielectric Theories . Dielectric Theories 1.1. Introduction Measurements of dielectric properties of materials is very important because it provide vital information regarding the material characteristics,
compared to gases. They are incompressible. Their density doesn t change with temperature. These similarities are due
 Liquids and solids They are similar compared to gases. They are incompressible. Their density doesn t change with temperature. These similarities are due to the molecules being close together in solids
Liquids and solids They are similar compared to gases. They are incompressible. Their density doesn t change with temperature. These similarities are due to the molecules being close together in solids
Methods of Computer Simulation. Molecular Dynamics and Monte Carlo
 Molecular Dynamics Time is of the essence in biological processes therefore how do we understand time-dependent processes at the molecular level? How do we do this experimentally? How do we do this computationally?
Molecular Dynamics Time is of the essence in biological processes therefore how do we understand time-dependent processes at the molecular level? How do we do this experimentally? How do we do this computationally?
Supporting Information
 Supporting Information Energies and 2 -hydroxyl group orientations of RNA backbone conformations. Benchmark CCSD(T)/CBS database, electronic analysis and assessment of DFT methods and MD simulations Arnošt
Supporting Information Energies and 2 -hydroxyl group orientations of RNA backbone conformations. Benchmark CCSD(T)/CBS database, electronic analysis and assessment of DFT methods and MD simulations Arnošt
Introduction to Computer Simulations of Soft Matter Methodologies and Applications Boulder July, 19-20, 2012
 Introduction to Computer Simulations of Soft Matter Methodologies and Applications Boulder July, 19-20, 2012 K. Kremer Max Planck Institute for Polymer Research, Mainz Overview Simulations, general considerations
Introduction to Computer Simulations of Soft Matter Methodologies and Applications Boulder July, 19-20, 2012 K. Kremer Max Planck Institute for Polymer Research, Mainz Overview Simulations, general considerations
Development of a simple, self-consistent polarizable model for liquid water
 University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2003 Development of a simple, self-consistent polarizable model for liquid water Haibo
University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2003 Development of a simple, self-consistent polarizable model for liquid water Haibo
Scientific Computing II
 Scientific Computing II Molecular Dynamics Simulation Michael Bader SCCS Summer Term 2015 Molecular Dynamics Simulation, Summer Term 2015 1 Continuum Mechanics for Fluid Mechanics? Molecular Dynamics the
Scientific Computing II Molecular Dynamics Simulation Michael Bader SCCS Summer Term 2015 Molecular Dynamics Simulation, Summer Term 2015 1 Continuum Mechanics for Fluid Mechanics? Molecular Dynamics the
Department of Chemical Engineering University of California, Santa Barbara Spring Exercise 3. Due: Thursday, 5/3/12
 Department of Chemical Engineering ChE 210D University of California, Santa Barbara Spring 2012 Exercise 3 Due: Thursday, 5/3/12 Objective: To learn how to write & compile Fortran libraries for Python,
Department of Chemical Engineering ChE 210D University of California, Santa Barbara Spring 2012 Exercise 3 Due: Thursday, 5/3/12 Objective: To learn how to write & compile Fortran libraries for Python,
Developing Monovalent Ion Parameters for the Optimal Point Charge (OPC) Water Model. John Dood Hope College
 Developing Monovalent Ion Parameters for the Optimal Point Charge (OPC) Water Model John Dood Hope College What are MD simulations? Model and predict the structure and dynamics of large macromolecules.
Developing Monovalent Ion Parameters for the Optimal Point Charge (OPC) Water Model John Dood Hope College What are MD simulations? Model and predict the structure and dynamics of large macromolecules.
Molecular dynamics simulation of Aquaporin-1. 4 nm
 Molecular dynamics simulation of Aquaporin-1 4 nm Molecular Dynamics Simulations Schrödinger equation i~@ t (r, R) =H (r, R) Born-Oppenheimer approximation H e e(r; R) =E e (R) e(r; R) Nucleic motion described
Molecular dynamics simulation of Aquaporin-1 4 nm Molecular Dynamics Simulations Schrödinger equation i~@ t (r, R) =H (r, R) Born-Oppenheimer approximation H e e(r; R) =E e (R) e(r; R) Nucleic motion described
CHEM 498Q / CHEM 630Q: Molecular Modeling of Proteins TUTORIAL #3a: Empirical force fields
 Department of Chemistry and Biochemistry, Concordia University! page 1 of 6 CHEM 498Q / CHEM 630Q: Molecular Modeling of Proteins TUTORIAL #3a: Empirical force fields INTRODUCTION The goal of this tutorial
Department of Chemistry and Biochemistry, Concordia University! page 1 of 6 CHEM 498Q / CHEM 630Q: Molecular Modeling of Proteins TUTORIAL #3a: Empirical force fields INTRODUCTION The goal of this tutorial
Supporting information
 Electronic Supplementary Material ESI for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2018 Supporting information Understanding three-body contributions to coarse-grained force
Electronic Supplementary Material ESI for Physical Chemistry Chemical Physics. This journal is the Owner Societies 2018 Supporting information Understanding three-body contributions to coarse-grained force
- intermolecular forces forces that exist between molecules
 Chapter 11: Intermolecular Forces, Liquids, and Solids - intermolecular forces forces that exist between molecules 11.1 A Molecular Comparison of Liquids and Solids - gases - average kinetic energy of
Chapter 11: Intermolecular Forces, Liquids, and Solids - intermolecular forces forces that exist between molecules 11.1 A Molecular Comparison of Liquids and Solids - gases - average kinetic energy of
Atomic and molecular interaction forces in biology
 Atomic and molecular interaction forces in biology 1 Outline Types of interactions relevant to biology Van der Waals interactions H-bond interactions Some properties of water Hydrophobic effect 2 Types
Atomic and molecular interaction forces in biology 1 Outline Types of interactions relevant to biology Van der Waals interactions H-bond interactions Some properties of water Hydrophobic effect 2 Types
Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar.
 Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar. 25, 2002 Molecular Dynamics: Introduction At physiological conditions, the
Why Proteins Fold? (Parts of this presentation are based on work of Ashok Kolaskar) CS490B: Introduction to Bioinformatics Mar. 25, 2002 Molecular Dynamics: Introduction At physiological conditions, the
SOLIDS AND LIQUIDS - Here's a brief review of the atomic picture or gases, liquids, and solids GASES
 30 SOLIDS AND LIQUIDS - Here's a brief review of the atomic picture or gases, liquids, and solids GASES * Gas molecules are small compared to the space between them. * Gas molecules move in straight lines
30 SOLIDS AND LIQUIDS - Here's a brief review of the atomic picture or gases, liquids, and solids GASES * Gas molecules are small compared to the space between them. * Gas molecules move in straight lines
Thermodynamics System Surrounding Boundary State, Property Process Quasi Actual Equilibrium English
 Session-1 Thermodynamics: An Overview System, Surrounding and Boundary State, Property and Process Quasi and Actual Equilibrium SI and English Units Thermodynamic Properties 1 Thermodynamics, An Overview
Session-1 Thermodynamics: An Overview System, Surrounding and Boundary State, Property and Process Quasi and Actual Equilibrium SI and English Units Thermodynamic Properties 1 Thermodynamics, An Overview
LONDON DISPERSION FORCES. - often called "London forces" for short. - London forces occur in all molecules, polar or nonpolar.
 43 LONDON DISPERSION FORCES - often called "London forces" for short. - occurs because electron density is - at any given point in time - likely to be uneven across a molecule due to the simple fact that
43 LONDON DISPERSION FORCES - often called "London forces" for short. - occurs because electron density is - at any given point in time - likely to be uneven across a molecule due to the simple fact that
Sodium, Na. Gallium, Ga CHEMISTRY Topic #2: The Chemical Alphabet Fall 2017 Dr. Susan Findlay See Exercises 9.2 to 9.7.
 Sodium, Na Gallium, Ga CHEMISTRY 1000 Topic #2: The Chemical Alphabet Fall 2017 Dr. Susan Findlay See Exercises 9.2 to 9.7 Forms of Carbon Kinetic Molecular Theory of Matter The kinetic-molecular theory
Sodium, Na Gallium, Ga CHEMISTRY 1000 Topic #2: The Chemical Alphabet Fall 2017 Dr. Susan Findlay See Exercises 9.2 to 9.7 Forms of Carbon Kinetic Molecular Theory of Matter The kinetic-molecular theory
Javier Junquera. Statistical mechanics
 Javier Junquera Statistical mechanics From the microscopic to the macroscopic level: the realm of statistical mechanics Computer simulations Thermodynamic state Generates information at the microscopic
Javier Junquera Statistical mechanics From the microscopic to the macroscopic level: the realm of statistical mechanics Computer simulations Thermodynamic state Generates information at the microscopic
Topic0990 Electrical Units In attempting to understand the properties of chemical substances, chemists divide chemistry into two parts.
 Topic99 Electrical Units In attempting to understand the properties of chemical substances, chemists divide chemistry into two parts. In one part, chemists are interested in understanding intramolecular
Topic99 Electrical Units In attempting to understand the properties of chemical substances, chemists divide chemistry into two parts. In one part, chemists are interested in understanding intramolecular
Chapters 11 and 12: Intermolecular Forces of Liquids and Solids
 1 Chapters 11 and 12: Intermolecular Forces of Liquids and Solids 11.1 A Molecular Comparison of Liquids and Solids The state of matter (Gas, liquid or solid) at a particular temperature and pressure depends
1 Chapters 11 and 12: Intermolecular Forces of Liquids and Solids 11.1 A Molecular Comparison of Liquids and Solids The state of matter (Gas, liquid or solid) at a particular temperature and pressure depends
Biomolecular modeling II
 2015, December 16 System boundary and the solvent Biomolecule in solution typical MD simulations molecular system in aqueous solution task make the system as small as possible (reduce cost) straightforward
2015, December 16 System boundary and the solvent Biomolecule in solution typical MD simulations molecular system in aqueous solution task make the system as small as possible (reduce cost) straightforward
Charge-on-spring polarizable water models revisited: From water clusters to liquid water to ice
 University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2004 Charge-on-spring polarizable water models revisited: From water clusters to liquid
University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2004 Charge-on-spring polarizable water models revisited: From water clusters to liquid
Winmostar tutorial Gromacs Interfacial Tension V X-Ability Co., Ltd. 2018/01/15
 Winmostar tutorial Gromacs Interfacial Tension V8.007 X-Ability Co., Ltd. question@winmostar.com Summary In this tutorial, we will calculate density distribution and interfacial tension between liquid-liquid
Winmostar tutorial Gromacs Interfacial Tension V8.007 X-Ability Co., Ltd. question@winmostar.com Summary In this tutorial, we will calculate density distribution and interfacial tension between liquid-liquid
Force Field for Water Based on Neural Network
 Force Field for Water Based on Neural Network Hao Wang Department of Chemistry, Duke University, Durham, NC 27708, USA Weitao Yang* Department of Chemistry, Duke University, Durham, NC 27708, USA Department
Force Field for Water Based on Neural Network Hao Wang Department of Chemistry, Duke University, Durham, NC 27708, USA Weitao Yang* Department of Chemistry, Duke University, Durham, NC 27708, USA Department
Winmostar tutorial LAMMPS Polymer modeling V X-Ability Co,. Ltd. 2017/7/6
 Winmostar tutorial LAMMPS Polymer modeling V7.021 X-Ability Co,. Ltd. question@winmostar.com 2017/7/6 Contents Configure I. Register a monomer II. Define a polymer III. Build a simulation cell IV. Execute
Winmostar tutorial LAMMPS Polymer modeling V7.021 X-Ability Co,. Ltd. question@winmostar.com 2017/7/6 Contents Configure I. Register a monomer II. Define a polymer III. Build a simulation cell IV. Execute
