Prediction of N-terminal protein sorting signals Manuel G Claros, Søren Brunak and Gunnar von Heijne
|
|
|
- Candice Clare Webster
- 6 years ago
- Views:
Transcription
1 394 Prediction of N-terminal protein sorting signals Manuel G Claros, Søren Brunak and Gunnar von Heijne Recently, neural networks have been applied to a widening range of problems in molecular biology. An area particularly suited to neural-network methods is the identification of protein sorting signals and the prediction of their cleavage sites, as these functional units are encoded by local, linear sequences of amino acids rather than global 3D structures. Addresses Laboratorio de Bioqumica y Biologa Molecular, Facultad de Ciencias, Universidad de Málaga, E Málaga, Spain Department of Chemistry, Centre for Biological Sequence Analysis, Technical University of Denmark, DK-2800 Lyngby, Denmark Department of Biochemistry, Arrhenius Laboratory, Stockholm University, S Stockholm, Sweden Current Opinion in Structural Biology 1997, 7: Current Biology Ltd ISSN X Abbreviations c-region C-terminal region ctp chloroplast transit peptide h-region hydrophobic core mtp mitochondrial targeting peptide n-region N-terminal region SP signal peptide Introduction Many proteins that are synthesized in the cytoplasm are ultimately found in noncytoplasmic locations. This requires an efficient and specific addressing system. The address labels are encoded by a specific sequence or structure in the protein [1 ], and various methods to detect different classes of targeting signals in protein sequences have been devised over the years. In this review, we will briefly discuss methods designed to identify signals that target proteins to the secretory pathway (signal peptides) and to mitochondria and chloroplasts. Our emphasis will be on the most recent work in this area, which is based mostly on the use of neural networks. Signal peptide characteristics Proteins destined for the endoplasmic reticulum, the Golgi compartment, the lysosome, the plasma membrane, and the exterior of the cell are initially transported across the endoplasmic reticulum membrane [2]. These proteins contain an N-terminal signal peptide (SP) of about residues. SPs consist of a short, positively charged N-terminal region (n-region), a 7 15 residue hydrophobic core (h-region), and a more polar 3 7 residue C-terminal region (c-region) leading up to the signal peptidase cleavage site between the SP and the mature protein [3]. Although minor differences in composition and length exist, the characteristics of eukaryotic SPs are essentially shared by the SPs of prokaryotic and archaeal proteins destined to the periplasm [4 ] (H Nielsen, S Brunak, von Heijne, unpublished data). Prokaryotic lipoproteins are cleaved by a distinct signal peptidase and their SPs have a different cleavage-site pattern [3]. Mitochondrial targeting peptide characteristics Most mitochondrial targeting peptides (mtps) consist of an N-terminal presequence of about residues, although several mitochondrial proteins are synthesized without a cleavable extension [5]. mtps are enriched in arginine, leucine, serine and alanine, contain none or a few acidic residues and are believed to form amphiphilic α-helices [6,7]. A loosely conserved cleavage-site motif has been proposed [8]. Proteins located in the inner mitochondrial membrane or the intermembrane space are often targeted by means of a bipartite mtp [9]. Although not related specifically to the mtps, it may be noted that mitochondrially targeted proteins have a high isoelectric point [10] and do not present physical constraints such as highly hydrophobic transmembrane segments that obstruct their import [11 ]. Chloroplast transit peptide characteristics Chloroplast transit peptides (ctps) are mainly enriched in serine and other hydroxylated residues. Because of a rather low content of acidic residues, their net charge is often positive. Their length varies from 20 to more than 100 residues. ctps seem to be organized in three domains [7]: an N-terminal part, which is around ten amino acids long, uncharged, poor in glycine and proline, and which has an alanine following the initial methionine in most cases; a central part, which is enriched in serine and contains few, if any, acidic residues; and a C-terminal part, which is enriched in arginine, and which may form an amphiphilic β strand. ctps that target proteins to the thylakoid are bipartite [12]. Two kinds of cleavage-site consensus motifs have been proposed: one for thylakoidal proteins, which is similar to the signal peptidase motif found in SPs [13], and another, less conserved motif for stromal proteins [14]. Neural networks Neural networks consist of a number of simple, nonlinear computational units that operate in parallel. The technique was originally developed with the purpose of modeling brain function, and neurons and synapses in biological neural networks, but it has so far had its greatest impact in practical pattern-recognition applications [15,16]. In the most popular network architecture, the units are organized into layers: an input layer, which receives signals from external sources, such as the numerical encoding of sequence data; one or more hidden layers; and an output layer, which sends analog signals to the environment for later interpretation and assignment of
2 Prediction of N-terminal protein sorting signals Claros, Brunak and von Heijne 395 functional categories. When only input and output layers are present, the architecture is often called a Perceptron, and it has the computational ability to resolve only linear classification tasks. A more powerful pattern-recognition device results when hidden layers are present. These layers are termed hidden because the activation of their units is not directly observable from the outside; they receive signals from the input units and relay weighted signals to the output units. Hidden units give the network the capability of handling nonlinear classification tasks. The optimal numbers of hidden layers and units are normally determined empirically, although tools originating from a more general Bayesian framework can be used for estimating a reasonable number of weight parameters [15 17]. One of the main advantages of neural networks is that a preconceived model is not required. The weights in a network (i.e. the strengths of the connections between units in adjacent layers) are determined automatically by so called learning procedures that use sequence data to adjust the parameter values. Information from both positive and negative cases is taken into account, such that residues and positions that are important will be weighted accordingly. Thus, no a priori relationships exist between the chosen parameters, leading to essentially unbiased, data-driven results [17]. Importantly, the trained network can detect nonlinear correlations between the residues in the input window and exploit them for improving the quality of the predictions. As expected, larger, nonhomologous, data sets increase the accuracy of neural-network methods [18]. A special benefit is the possibility of interpreting the weights resulting from training in a biochemically meaningful way, possibly leading to new insights into the recognition that takes place in the cellular environment. Below, we discuss the conventional and neural-network methods that have been developed for predicting protein targeting signals (Table 1). Prediction of signal peptides Statistical methods For a full characterization, one needs to predict both the existence of a particular sorting signal in a protein sequence and the cleavage site between the signal and the mature protein. A procedure to do the former was proposed early on by McGeoch [19], who built a discriminant function based on the net charge and length of the n-region, and the length and hydrophobicity of the h-region. Weight matrices were used early on to identify signal peptidase cleavage sites [20], with 75 80% predictions of cleavage sites in SPs outside the training set reported to be correct. These matrices have been widely used in several software applications such as SIGSEQ [21] for UNIX and MS-DOS operating systems, the PLOT.A/SIG program included in MacProt for Apple Macintosh computers (P Markiewicz, unpublished), SIGNAL [22] for UNIX, and Signalase (N Matei, unpublished) for Apple Macintosh computers. The most complete software that uses the original SP weight matrices is SIGCLEAVE in the extended pack of programs for GCG (EGCG) for VMS and UNIX machines [23]. SIGCLEAVE has been modified following the recommendations in [24], although the reported success rate is similar to the original method. Whereas most of the early methods focused only on cleavage-site predictions, Folz and Gordon [25] have combined two different algorithms to detect SPs: one algorithm (SIGSEQ1) is based on a series of empirically derived rules to generate a probability score for each position, and the second algorithm (SIGSEQ2) applies the statistical weight matrix approach. By combining the two Table 1 Software for predicting N-terminal sorting signals. Sorting signal Programs Internet addresses Reference Signal peptides SIGSED ftp://ftp.ebi.ac.uk/pub/software/ [21] SIGSEQ1, SIGSEQ2 folz@wums.bitnet [25] PLOT.A/SIG ftp://ftp/ebi.ac.uk/pub/software/mac (a) SIGNAL ftp://ftp/ebi.ac.uk/pub/software/unix/ (b) Signalase ftp://ftp/ebi.ac.uk/pub/software/mac/ (c) SIGCLEAVE ftp://ftp/ebi.ac.uk/pub/software/ [23] PROFI wrede@zedat.fu-berlin.de [29] PROSA, PROMIS wrede@zedat.fu-berlin.de [32] SIGNALP [41] signalp@cbs.dtu.dk ( server) Mitochondrial targeting peptides MitoProt II ftp://ftp.ebi.ac.uk/pub/software/ [36 ] mitoprot@biologie.ens.fr ( server) PROSA, PROMIS wrede@zedat.fu-berlin.de [32] Subcellular localizations PSORT [40] This published address does not exist. (a) P Marckiewicz, personal communication. (b) R Colgrove, personal communication. (c) N Matei, personal communication.
3 396 Sequences and topology methods, 80% of the SP cleavage sites in a collection of eukaryotic proteins was correctly predicted. Neural network approaches Neural networks were first applied to SP predictions in combination with weight matrices [26]. A multilayer architecture that self-adjusts to the data was trained to detect 20 N-terminal residues belonging to SPs. 82% and 74% of the SPs were correctly identified in the training and test set, respectively. When the network was applied to sequences first selected with the weight matrices, the success rate increased to 95%. This network failed to outperform the discrimination ability of the weight matrix method, even though a larger training set was used. An unsupervised neural network based on a self-organizing Kohonen feature map has unexpectedly been found to be able to extract SP motifs from insulin cdna sequences [27]. How well this method performs on more general data sets is unclear. Using a training set of only 24 proteins, a neural network, trained by a genetic algorithm and representing each amino acid by four physicochemical properties, has been able to correctly predict all SPs in the training set [28]. It uses an asymmetric window of 10 to +2 residues with a Perceptron-type neural network. This method, PROFI, has been improved by using a three-layer network and by dividing the same 24 proteins into training and test sets: the optimal input layer employs 4 7 physicochemical amino acid properties for the sequence description, one hidden layer for the feature extraction, and a single output layer for classification [29]. The success rate for predicting the cleavage sites is 99% for the training set and 91% for the test set. The network has been further used to design amino acid sequences, by using simulated molecular evolution, that should have ideal E. coli leader peptidase cleavage sites [30]. Two SP cleavage sites designed in this way have been studied in vivo, and the experimentally observed processing of the protein confirms the predictions made by the neural network [31]. A similar strategy of using a genetic neural network, but using a larger set of proteins (65 from eubacteria and 95 from eukaryotes), has enabled the generalization of E. coli SP characteristics to other organisms. Both the characteristics of the SP and the cleavage site have been studied [32]. The statistical program PROSA is used to extract rules that describe SPs and is able to predict 80% of the cleavage sites in both training and test sets. The neural network PROMIS has been used to predict SPs and their cleavage sites. PROMIS can predict 80% of the SP cleavage sites in the training and test sets. The most efficient method currently available to identify SPs and their cleavage sites is SIGNALP [33 ], which uses neural networks trained on very large sets of nonhomologous prokaryotic (266 sequences from Gram-negative and 141 from Gram-positive bacteria) and eukaryotic (1011 sequences) SPs [34 ]. SIGNALP combines two networks, one to recognize the cleavage site, and another to distinguish between SPs and non-sps. The best results have been obtained with networks that have zero or one hidden layer with up to four hidden units. SIGNALP uses asymmetric windows (including more positions upstream than downstream of the cleavage site) for predicting the SP, and a symmetric or nearly symmetric window to predict the cleavage site. Cleavage-site prediction on the test sets correctly identified 78% of the eukaryotic and 89% of the prokaryotic signal peptides. This is lower than the accuracy reported for the original weight matrix method (see above), but when new weight matrices were constructed based on the same training set as used for SIGNALP, the performance was much worse than in the original publication. This suggests that signal peptides are somewhat more variable than was apparent from the early, rather small collections, and that neural networks can extract useful information beyond what can be captured with weight matrices. It should be noted that the prediction performances reported for SIGNALP correspond to minimal values due to the low similarity between the sequences in the test set. Thus, the prediction accuracy will generally be higher when the method is applied to an unbiased sample of new sequences, as these will often be similar to sequences used in the training set. Interestingly, the difficulty of SP prediction has been found to increase in the order Gram-negative bacteria < eukaryotes < Gram-positive bacteria. This neural network is also, to some limited extent, able to discriminate among SPs and uncleaved signal anchor sequences. Prediction of mitochondrial and chloroplast sorting signals Statistical methods The program MitoProt for Apple Macintosh computers [35 ] represents an early attempt to help identifying mtps. It calculates a number of characteristics thought to describe mtps but leaves the final decision to the user. In the search for a more objective way to identify mtps, discriminant analysis turns out to be very useful [36 ]. An analysis based on 47 physicochemical parameters and a large set of mitochondrial and nonmitochondrial proteins has reached a success rate of 80% for the training set and 75% for the test set when predicting the existence of an mtp. Many of the mitochondrial proteins not detected by the method have been found to possess internal, noncleavable mtps or to be localized in the outer mitochondrial membrane. Among matrix and inner membrane proteins, only 7% of the mtps were missed. The method has been implemented in the program MitoProt II for Macintosh and UNIX machines. A discriminant function to separate both mitochondrial and chloroplast proteins from non-organellar proteins has also been included in the last release of MitoProt II
4 Prediction of N-terminal protein sorting signals Claros, Brunak and von Heijne 397 (MG Claros, P Vincens, unpublished). The crossvalidated success rate predicting mtps and ctps (77% for both the training and test sets) is slightly lower than for only mitochondrial proteins. The discrimination between mtps and ctps is performed with the equation described in [7], based on the observation that the frequencies of arginine and serine are very different in the two kinds of signals. Thylakoid lumen proteins have bipartite presequences that are cleaved off by a prokaryotic-like signal peptidase, and a weight matrix similar to that used for cleavage sites in bacterial SPs has been derived for such proteins [13]. The prediction accuracy is not known in this case, but it is probably higher than for Gram-negative SPs as the substrate specificity of the thylakoidal protease is more restricted than for the E. coli signal peptidase [37]. Neural network approaches Neural networks were first applied to a small set of proteins to infer mtps and ctps by Schneider et al. [28], who reported a success rate of 75% for the test set. Cleavage-site predictions based on the PROSA and PROMIS programs, however, have so far not been very successful [32]. The main problem in both cases has been overtraining and an inability to extract significant properties describing transit peptides. Using Neurospora crassa mitochondrial precursors, scanned with asymmetric windows and utilizing 13 Perceptron-type networks [38], 81% of the mtp cleavage sites have been correctly predicted. Recently, self-organizing Kohonen networks have been trained on mtps (with each residue represented by its size and hydrophobicity values) from a range of different organisms and have been found to be able to cluster these sequences according to three previously known cleavage-site motifs (G Schneider et al., unpublished data). Prediction of subcellular localization by expert systems Expert systems are programs that use domain-specific knowledge and heuristics to solve problems in a narrow and realistic problem area. The difference between an expert system and the algorithms described above is that the expert system applies a large number of different sorting signal detection algorithms at the same time, utilizing a collection of if then rules as a knowledge base to interpret their respective outputs. In the first application of expert systems to the protein sorting problem, four different locations (cytoplasm, inner membrane, periplasm, and outer membrane) in Gramnegative bacteria have been considered [39]. The basic if then rules are intended to simulate the actual sorting pathways, and the system uses the SP-detection methods described above as well as empirical rules relating to the amino acid composition in the mature part of the protein. It has been able to correctly distribute 83% of 106 nonhomologous prokaryotic proteins among the four localization sites. A similar expert system (PSORT) has been developed for predicting protein locations in eukaryotic cells [40]. 401 proteins from 14 different subcellular sites (cytoplasm, nucleus, four mitochondrial sites, peroxisome, two endoplasmic reticulum sites, Golgi, two lysosome sites, vacuole, two plasma membrane sites, and the extracellular space; and for plant cells, three more sites corresponding to chloroplast proteins) have been collected. Many of the rules are based on discriminant functions calculated on the overall amino acid composition of the protein. The organization of the if then rules follows a reasoning that tries to emulate the real pathway of sorting in vivo. Around 65% of all proteins can be localized to the correct compartment by the method, whereas a random guess would result in less than 10% accuracy. 66% of the mitochondrial proteins and 86% of the chloroplast proteins have been correctly localized. For proteins targeted by SPs to the secretory pathway, the accuracy is similar to that obtained using the non-neural network algorithms discussed above. Conclusions In recent years, workers trying to predict protein sorting signals have turned increasingly to the use of neural networks. By and large, this approach appears to give slight but significant improvements over previous methods, although the larger size and higher quality of the more recent training and test sets may be equally important. SPs, mtps, and ctps can now be quite reliably identified, but cleavage sites can so far only be predicted with an acceptable degree of confidence for SPs. Much of the development in the coming years will probably be an adaptation of the methods to the analysis of data from genome-sequencing projects. A particularly interesting (and difficult) problem in this context is the identification of sorting signals in incomplete sequences (such as ESTs). In addition, slight improvements will probably be obtained by developing species-specific prediction methods. References and recommended reading Papers of particular interest, published within the annual period of review, have been highlighted as: of special interest of outstanding interest 1. Zheng N, Gierasch LM: Signal sequences: the same yet different. Cell 1996, 86: Different targeting signals are compared, inferring evolutionary and functional relationships. Special emphasis is put on the fact that most of them fold into helical structures.
5 398 Sequences and topology 2. Rapoport TA, Jungnickel B, Kutay U: Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem 1996, 65: Von Heijne G: The signal peptide. J Membr Biol 1990, 115: Rusch SL, Kendall DA: Protein transport via amino-terminal targeting sequences: common themes in diverse systems. Mol Membr Biol 1995, 12: This review describes the sequence characteristics of N-terminal targeting signals from different organisms and sorting pathways. 5. Schwarz E, Neupert W: Mitochondrial protein import: mechanisms, components and energetics. Biochim Biophys Acta 1994, 1187: Von Heijne G: Mitochondrial targeting sequences may form amphiphilic helices. EMBO J 1986, 5: Von Heijne G, Steppuhn J, Herrmann RG: Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem 1989, 180: Gavel Y, Von Heijne G: Cleavage-site motifs in mitochondrial targeting peptides. Protein Eng 1990, 4: Stuart RA, Neupert W: Topogenesis of inner membrane proteins of mitochondria. Trends Biochem Sci 1996, 21: Jaussi R: Homologous nuclear-encoded mitochondrial and cytosolic isoproteins. A review of structure, biosynthesis and genes. Eur J Biochem 1995, 228: Claros MG, Perea J, Shu Y, Samatey FA, Popot JL, Jacq C: Limitations of the in vivo import of hydrophobic proteins into yeast mitochondria. The case of cytoplasmically synthesized apocytochrome b. Eur J Biochem 1995, 228: Using different parts of the cytochrome b gene, the authors demonstrate that imported mitochondrial proteins should not contain too many hydrophobic segments. A new parameter called mesohydrophobicity is developed to quantify a part of the constraints on hydrophobicity. 12. Robinson C, Klösgen RB: Targeting of proteins into and across the thylakoid membrane a multitude of mechanisms. Plant Mol Biol 1994, 26: Howe CJ, Wallace TP: Prediction of leader peptide cleavage sites for polypeptides of the thylakoid lumen. Nucleic Acids Res 1990, 18: Gavel Y, Von Heijne G: A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett 1990, 261: Presnell SR, Cohen FE: Artificial neural networks for pattern recognition in biochemical sequences. Annu Rev Biophys Biomol Struct 1993, 22: Bishop C: Neural Networks for Pattern Recognition. Oxford: Clarendon Press; Hirst JD: Prediction of structural, functional features of protein and nucleic acid sequences by artificial neural networks. Biochemistry 1992, 31: Chadonia JM, Karplus M: Importance of larger data sets for protein secondary structure prediction with neural networks. Protein Sci 1996, 5: McGeoch DJ: On the predictive recognition of signal peptide sequences. Virus Res 1985, 3: Von Heijne G: A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 1986, 14: Popowicz AM, Dash PF: SIGSEQ: a computer program for predicting signal sequence cleavage sites. Comput Appl Biosci 1988, 4: Colgrove R: SIGNAL for UNIX [PhD Thesis]. San Francisco: University of California, San Francisco; Rice P, López R, Doelz R, Leunissen J: EGCG 8.0. Embnet news 1995, 2: Von Heijne G: Sequence Analysis in Molecular Biology: Treasure Trove or Trivial Pursuit? London: Academic Press; Folz RJ, Gordon JI: Computers-assisted predictions of signal peptidase processing sites. Biochem Biophys Res Comm 1987, 146: Ladunga I, Czako F, Csabai I, Geszti T: Improving signal peptide prediction accuracy by simulated neural network. Comput Appl Biosci 1991, 7: Arrigo P, Giuliano F, Scalia F, Rapallo A, Damiani G: Identification of a new motif on nucleic acid sequence data using Kohonen s self-organizing map. Comput Appl Biosci 1991, 7: Schneider G, Röhlk S, Wrede P: Analysis of cleavage-site patterns in protein precursor sequences with a perceptrontype neural network. Biochem Biophys Res Comm 1993, 194: Schneider G, Wrede P: Development of artificial neural filters for pattern recognition in protein sequences. J Mol Evol 1993, 36: Schneider G, Wrede P: The rational design of amino acid sequences by artificial neural networks and simulated molecular evolution: de novo design of an idealized leader peptidase cleavage site. Biophys J 1994, 66: Schneider G, Hahn U, Fatemi A, Müller G, Wrede P: Peptide design in machina: artificial signal peptidase I cleavage sites are processed in vivo. Minim Invas Med 1995, 6: Schneider G, Wrede P: Signal analysis of protein targeting sequences. Protein Seq Data Anal 1993, 5: Nielsen H, Engelbrecht J, Brunak S, Von Heijne G: Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 1997, 10:1 6. Neural networks are trained to identify signal peptides in prokaryotic and eukaryotic proteins. Sequence logos are calculated for signal peptides from different groups of organisms. 34. Nielsen H, Engelbrecht J, Von Heijne G, Brunak S: Defining a similarity threshold for a functional protein sequence pattern: the signal peptide cleavage site. Proteins 1996, 24: A method to exclude redundant or homologous entries from sets of sequences that contain a common, well defined feature, such as a proteolytic cleavage site is presented. 35. Claros MG: MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci 1995, 11: Software that calculates and presents all the characteristics proposed to be important for mitochondrial targeting sequences is described. 36. Claros MG, Vincens P: Computational method to predict mitochondrially imported proteins and their transit peptides. Eur J Biochem 1996, 241: A discriminant analysis is used to identify mitochondrial proteins. The final function is implemented in a software that can also be used to identify the targeting sequence. The advantage of this approach is the large number of parameters and sequences considered to elaborate the discriminant function, and the rules to infer the mitochondrial targeting sequence. 37. Shackleton JB, Robinson C: Transport of proteins into chloroplasts the thylakoidal processing peptidase is a signaltype peptidase with stringent substrate requirements at the 3 position and 1 position. J Biol Chem 1991, 266: Schneider G, Schuchhardt J, Wrede P: Peptide design in machina: development of artificial mitochondrial protein precursor cleavage sites by simulated molecular evolution. Biophys J 1995, 68: Nakai K, Kanehisa M: Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins 1991, 11: Nakai K, Kanehisa M: A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 1992, 14: Nielsen H, Engelbrecht J, Brunak S, Von Heijne G: A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 1997, in press.
A NEURAL NETWORK METHOD FOR IDENTIFICATION OF PROKARYOTIC AND EUKARYOTIC SIGNAL PEPTIDES AND PREDICTION OF THEIR CLEAVAGE SITES
 International Journal of Neural Systems, Vol. 8, Nos. 5 & 6 (October/December, 1997) 581 599 c World Scientific Publishing Company A NEURAL NETWORK METHOD FOR IDENTIFICATION OF PROKARYOTIC AND EUKARYOTIC
International Journal of Neural Systems, Vol. 8, Nos. 5 & 6 (October/December, 1997) 581 599 c World Scientific Publishing Company A NEURAL NETWORK METHOD FOR IDENTIFICATION OF PROKARYOTIC AND EUKARYOTIC
ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites
 Protein Science ~1999!, 8:978 984. Cambridge University Press. Printed in the USA. Copyright 1999 The Protein Society ChloroP, a neural network-based method for predicting chloroplast transit peptides
Protein Science ~1999!, 8:978 984. Cambridge University Press. Printed in the USA. Copyright 1999 The Protein Society ChloroP, a neural network-based method for predicting chloroplast transit peptides
Prediction of signal peptides and signal anchors by a hidden Markov model
 In J. Glasgow et al., eds., Proc. Sixth Int. Conf. on Intelligent Systems for Molecular Biology, 122-13. AAAI Press, 1998. 1 Prediction of signal peptides and signal anchors by a hidden Markov model Henrik
In J. Glasgow et al., eds., Proc. Sixth Int. Conf. on Intelligent Systems for Molecular Biology, 122-13. AAAI Press, 1998. 1 Prediction of signal peptides and signal anchors by a hidden Markov model Henrik
Improved Prediction of Signal Peptides: SignalP 3.0
 doi:10.1016/j.jmb.2004.05.028 J. Mol. Biol. (2004) 340, 783 795 Improved Prediction of Signal Peptides: SignalP 3.0 Jannick Dyrløv Bendtsen 1, Henrik Nielsen 1, Gunnar von Heijne 2 and Søren Brunak 1 *
doi:10.1016/j.jmb.2004.05.028 J. Mol. Biol. (2004) 340, 783 795 Improved Prediction of Signal Peptides: SignalP 3.0 Jannick Dyrløv Bendtsen 1, Henrik Nielsen 1, Gunnar von Heijne 2 and Søren Brunak 1 *
Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites
 Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites Henrik Nielsen, Jacob Engelbrecht, øren Brunak, and unnar von Heijne y Center for Biological equence
Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites Henrik Nielsen, Jacob Engelbrecht, øren Brunak, and unnar von Heijne y Center for Biological equence
Why Bother? Predicting the Cellular Localization Sites of Proteins Using Bayesian Model Averaging. Yetian Chen
 Predicting the Cellular Localization Sites of Proteins Using Bayesian Model Averaging Yetian Chen 04-27-2010 Why Bother? 2008 Nobel Prize in Chemistry Roger Tsien Osamu Shimomura Martin Chalfie Green Fluorescent
Predicting the Cellular Localization Sites of Proteins Using Bayesian Model Averaging Yetian Chen 04-27-2010 Why Bother? 2008 Nobel Prize in Chemistry Roger Tsien Osamu Shimomura Martin Chalfie Green Fluorescent
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
Yeast ORFan Gene Project: Module 5 Guide
 Cellular Localization Data (Part 1) The tools described below will help you predict where your gene s product is most likely to be found in the cell, based on its sequence patterns. Each tool adds an additional
Cellular Localization Data (Part 1) The tools described below will help you predict where your gene s product is most likely to be found in the cell, based on its sequence patterns. Each tool adds an additional
Using AdaBoost for the prediction of subcellular location of prokaryotic and eukaryotic proteins
 Mol Divers (2008) 12:41 45 DOI 10.1007/s11030-008-9073-0 FULL LENGTH PAPER Using AdaBoost for the prediction of subcellular location of prokaryotic and eukaryotic proteins Bing Niu Yu-Huan Jin Kai-Yan
Mol Divers (2008) 12:41 45 DOI 10.1007/s11030-008-9073-0 FULL LENGTH PAPER Using AdaBoost for the prediction of subcellular location of prokaryotic and eukaryotic proteins Bing Niu Yu-Huan Jin Kai-Yan
Protein Bioinformatics. Rickard Sandberg Dept. of Cell and Molecular Biology Karolinska Institutet sandberg.cmb.ki.
 Protein Bioinformatics Rickard Sandberg Dept. of Cell and Molecular Biology Karolinska Institutet rickard.sandberg@ki.se sandberg.cmb.ki.se Outline Protein features motifs patterns profiles signals 2 Protein
Protein Bioinformatics Rickard Sandberg Dept. of Cell and Molecular Biology Karolinska Institutet rickard.sandberg@ki.se sandberg.cmb.ki.se Outline Protein features motifs patterns profiles signals 2 Protein
A Probabilistic Classification System for Predicting the Cellular Localization Sites of Proteins
 From: ISMB-96 Proceedings. Copyright 1996, AAAI (www.aaai.org). All rights reserved. A Probabilistic Classification System for Predicting the Cellular Localization Sites of Proteins Paul Horton Computer
From: ISMB-96 Proceedings. Copyright 1996, AAAI (www.aaai.org). All rights reserved. A Probabilistic Classification System for Predicting the Cellular Localization Sites of Proteins Paul Horton Computer
Chapter 12: Intracellular sorting
 Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
Chapter 12: Intracellular sorting Principles of intracellular sorting Principles of intracellular sorting Cells have many distinct compartments (What are they? What do they do?) Specific mechanisms are
PROTEIN SUBCELLULAR LOCALIZATION PREDICTION BASED ON COMPARTMENT-SPECIFIC BIOLOGICAL FEATURES
 3251 PROTEIN SUBCELLULAR LOCALIZATION PREDICTION BASED ON COMPARTMENT-SPECIFIC BIOLOGICAL FEATURES Chia-Yu Su 1,2, Allan Lo 1,3, Hua-Sheng Chiu 4, Ting-Yi Sung 4, Wen-Lian Hsu 4,* 1 Bioinformatics Program,
3251 PROTEIN SUBCELLULAR LOCALIZATION PREDICTION BASED ON COMPARTMENT-SPECIFIC BIOLOGICAL FEATURES Chia-Yu Su 1,2, Allan Lo 1,3, Hua-Sheng Chiu 4, Ting-Yi Sung 4, Wen-Lian Hsu 4,* 1 Bioinformatics Program,
Intro Secondary structure Transmembrane proteins Function End. Last time. Domains Hidden Markov Models
 Last time Domains Hidden Markov Models Today Secondary structure Transmembrane proteins Structure prediction NAD-specific glutamate dehydrogenase Hard Easy >P24295 DHE2_CLOSY MSKYVDRVIAEVEKKYADEPEFVQTVEEVL
Last time Domains Hidden Markov Models Today Secondary structure Transmembrane proteins Structure prediction NAD-specific glutamate dehydrogenase Hard Easy >P24295 DHE2_CLOSY MSKYVDRVIAEVEKKYADEPEFVQTVEEVL
Today. Last time. Secondary structure Transmembrane proteins. Domains Hidden Markov Models. Structure prediction. Secondary structure
 Last time Today Domains Hidden Markov Models Structure prediction NAD-specific glutamate dehydrogenase Hard Easy >P24295 DHE2_CLOSY MSKYVDRVIAEVEKKYADEPEFVQTVEEVL SSLGPVVDAHPEYEEVALLERMVIPERVIE FRVPWEDDNGKVHVNTGYRVQFNGAIGPYK
Last time Today Domains Hidden Markov Models Structure prediction NAD-specific glutamate dehydrogenase Hard Easy >P24295 DHE2_CLOSY MSKYVDRVIAEVEKKYADEPEFVQTVEEVL SSLGPVVDAHPEYEEVALLERMVIPERVIE FRVPWEDDNGKVHVNTGYRVQFNGAIGPYK
Signal peptides and protein localization prediction
 Downloaded from orbit.dtu.dk on: Jun 30, 2018 Signal peptides and protein localization prediction Nielsen, Henrik Published in: Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics Publication
Downloaded from orbit.dtu.dk on: Jun 30, 2018 Signal peptides and protein localization prediction Nielsen, Henrik Published in: Encyclopedia of Genetics, Genomics, Proteomics and Bioinformatics Publication
SVM Kernel Optimization: An Example in Yeast Protein Subcellular Localization Prediction
 SVM Kernel Optimization: An Example in Yeast Protein Subcellular Localization Prediction Ṭaráz E. Buck Computational Biology Program tebuck@andrew.cmu.edu Bin Zhang School of Public Policy and Management
SVM Kernel Optimization: An Example in Yeast Protein Subcellular Localization Prediction Ṭaráz E. Buck Computational Biology Program tebuck@andrew.cmu.edu Bin Zhang School of Public Policy and Management
SUB-CELLULAR LOCALIZATION PREDICTION USING MACHINE LEARNING APPROACH
 SUB-CELLULAR LOCALIZATION PREDICTION USING MACHINE LEARNING APPROACH Ashutosh Kumar Singh 1, S S Sahu 2, Ankita Mishra 3 1,2,3 Birla Institute of Technology, Mesra, Ranchi Email: 1 ashutosh.4kumar.4singh@gmail.com,
SUB-CELLULAR LOCALIZATION PREDICTION USING MACHINE LEARNING APPROACH Ashutosh Kumar Singh 1, S S Sahu 2, Ankita Mishra 3 1,2,3 Birla Institute of Technology, Mesra, Ranchi Email: 1 ashutosh.4kumar.4singh@gmail.com,
Meta-Learning for Escherichia Coli Bacteria Patterns Classification
 Meta-Learning for Escherichia Coli Bacteria Patterns Classification Hafida Bouziane, Belhadri Messabih, and Abdallah Chouarfia MB University, BP 1505 El M Naouer 3100 Oran Algeria e-mail: (h_bouziane,messabih,chouarfia)@univ-usto.dz
Meta-Learning for Escherichia Coli Bacteria Patterns Classification Hafida Bouziane, Belhadri Messabih, and Abdallah Chouarfia MB University, BP 1505 El M Naouer 3100 Oran Algeria e-mail: (h_bouziane,messabih,chouarfia)@univ-usto.dz
Chapter 6: A Tour of the Cell
 AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
 Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry 1. The study of cells has been limited by their small size, and so they were
Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry 1. The study of cells has been limited by their small size, and so they were
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%%
 !"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
Structure of mitochondria
 Structure of mitochondria Subcompartments of mitochondria Outer membrane (OM) Inner membrane (IM) Matrix Intermembrane space (IMS) Cristae membrane Inner boundary membrane Cristae compartment Molecular
Structure of mitochondria Subcompartments of mitochondria Outer membrane (OM) Inner membrane (IM) Matrix Intermembrane space (IMS) Cristae membrane Inner boundary membrane Cristae compartment Molecular
Chapter 6: A Tour of the Cell
 Chapter 6: A Tour of the Cell Concept 6.2 Eukaryotic cells have internal membranes that compartmentalize their functions 1. Which two domains consist of prokaryotic cells? 2. A major difference between
Chapter 6: A Tour of the Cell Concept 6.2 Eukaryotic cells have internal membranes that compartmentalize their functions 1. Which two domains consist of prokaryotic cells? 2. A major difference between
Introduction to Pattern Recognition. Sequence structure function
 Introduction to Pattern Recognition Sequence structure function Prediction in Bioinformatics What do we want to predict? Features from sequence Data mining How can we predict? Homology / Alignment Pattern
Introduction to Pattern Recognition Sequence structure function Prediction in Bioinformatics What do we want to predict? Features from sequence Data mining How can we predict? Homology / Alignment Pattern
Predicting protein subcellular localisation from amino acid sequence information Olof Emanuelsson Date received (in revised form): 10th September 2002
 Olof Emanuelsson works in close collaboration with Gunnar von Heijne on developing methods for predicting subcellular localisation of proteins. He is at the Stockholm Bioinformatics Center (SBC), a joint
Olof Emanuelsson works in close collaboration with Gunnar von Heijne on developing methods for predicting subcellular localisation of proteins. He is at the Stockholm Bioinformatics Center (SBC), a joint
Scale in the biological world
 Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Analysis of N-terminal Acetylation data with Kernel-Based Clustering
 Analysis of N-terminal Acetylation data with Kernel-Based Clustering Ying Liu Department of Computational Biology, School of Medicine University of Pittsburgh yil43@pitt.edu 1 Introduction N-terminal acetylation
Analysis of N-terminal Acetylation data with Kernel-Based Clustering Ying Liu Department of Computational Biology, School of Medicine University of Pittsburgh yil43@pitt.edu 1 Introduction N-terminal acetylation
Chapter 4 Active Reading Guide A Tour of the Cell
 Name: AP Biology Mr. Croft Chapter 4 Active Reading Guide A Tour of the Cell Section 1 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when
Name: AP Biology Mr. Croft Chapter 4 Active Reading Guide A Tour of the Cell Section 1 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when
Supervised Ensembles of Prediction Methods for Subcellular Localization
 In Proc. of the 6th Asia-Pacific Bioinformatics Conference (APBC 2008), Kyoto, Japan, pp. 29-38 1 Supervised Ensembles of Prediction Methods for Subcellular Localization Johannes Aßfalg, Jing Gong, Hans-Peter
In Proc. of the 6th Asia-Pacific Bioinformatics Conference (APBC 2008), Kyoto, Japan, pp. 29-38 1 Supervised Ensembles of Prediction Methods for Subcellular Localization Johannes Aßfalg, Jing Gong, Hans-Peter
Cell Organelles. a review of structure and function
 Cell Organelles a review of structure and function TEKS and Student Expectations (SE s) B.4 Science concepts. The student knows that cells are the basic structures of all living things with specialized
Cell Organelles a review of structure and function TEKS and Student Expectations (SE s) B.4 Science concepts. The student knows that cells are the basic structures of all living things with specialized
Chapter 4: Cells: The Working Units of Life
 Name Period Chapter 4: Cells: The Working Units of Life 1. What are the three critical components of the cell theory? 2. What are the two important conceptual implications of the cell theory? 3. Which
Name Period Chapter 4: Cells: The Working Units of Life 1. What are the three critical components of the cell theory? 2. What are the two important conceptual implications of the cell theory? 3. Which
Cell Alive Homeostasis Plants Animals Fungi Bacteria. Loose DNA DNA Nucleus Membrane-Bound Organelles Humans
 UNIT 3: The Cell DAYSHEET 45: Introduction to Cellular Organelles Name: Biology I Date: Bellringer: Place the words below into the correct space on the Venn Diagram: Cell Alive Homeostasis Plants Animals
UNIT 3: The Cell DAYSHEET 45: Introduction to Cellular Organelles Name: Biology I Date: Bellringer: Place the words below into the correct space on the Venn Diagram: Cell Alive Homeostasis Plants Animals
Chapter 1. DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d
 Chapter 1 1. Matching Questions DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d 2. Matching Questions : Unbranched polymer that, when folded into its three-dimensional shape,
Chapter 1 1. Matching Questions DNA is made from the building blocks adenine, guanine, cytosine, and. Answer: d 2. Matching Questions : Unbranched polymer that, when folded into its three-dimensional shape,
Overview of Cells. Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory
 Overview of Cells Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory Prokaryotic Cells Archaea Bacteria Come in many different shapes and sizes.5 µm 2 µm, up to 60 µm long Have large
Overview of Cells Prokaryotes vs Eukaryotes The Cell Organelles The Endosymbiotic Theory Prokaryotic Cells Archaea Bacteria Come in many different shapes and sizes.5 µm 2 µm, up to 60 µm long Have large
SCOP. all-β class. all-α class, 3 different folds. T4 endonuclease V. 4-helical cytokines. Globin-like
 SCOP all-β class 4-helical cytokines T4 endonuclease V all-α class, 3 different folds Globin-like TIM-barrel fold α/β class Profilin-like fold α+β class http://scop.mrc-lmb.cam.ac.uk/scop CATH Class, Architecture,
SCOP all-β class 4-helical cytokines T4 endonuclease V all-α class, 3 different folds Globin-like TIM-barrel fold α/β class Profilin-like fold α+β class http://scop.mrc-lmb.cam.ac.uk/scop CATH Class, Architecture,
Chapter 6: A Tour of the Cell
 Chapter 6: A Tour of the Cell 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when Robert Hooke first looked at dead cells from an oak tree.
Chapter 6: A Tour of the Cell 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when Robert Hooke first looked at dead cells from an oak tree.
Protein Secondary Structure Prediction
 part of Bioinformatik von RNA- und Proteinstrukturen Computational EvoDevo University Leipzig Leipzig, SS 2011 the goal is the prediction of the secondary structure conformation which is local each amino
part of Bioinformatik von RNA- und Proteinstrukturen Computational EvoDevo University Leipzig Leipzig, SS 2011 the goal is the prediction of the secondary structure conformation which is local each amino
122-Biology Guide-5thPass 12/06/14. Topic 1 An overview of the topic
 Topic 1 http://bioichiban.blogspot.com Cellular Functions 1.1 The eukaryotic cell* An overview of the topic Key idea 1: Cell Organelles Key idea 2: Plasma Membrane Key idea 3: Transport Across Membrane
Topic 1 http://bioichiban.blogspot.com Cellular Functions 1.1 The eukaryotic cell* An overview of the topic Key idea 1: Cell Organelles Key idea 2: Plasma Membrane Key idea 3: Transport Across Membrane
Reconstructing Mitochondrial Evolution?? Morphological Diversity. Mitochondrial Diversity??? What is your definition of a mitochondrion??
 Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial
Reconstructing Mitochondrial Evolution?? What is your definition of a mitochondrion?? Morphological Diversity Mitochondria as we all know them: Suprarenal gland Liver cell Plasma cell Adrenal cortex Mitochondrial
Importance of Protein sorting. A clue from plastid development
 Importance of Protein sorting Cell organization depend on sorting proteins to their right destination. Cell functions depend on sorting proteins to their right destination. Examples: A. Energy production
Importance of Protein sorting Cell organization depend on sorting proteins to their right destination. Cell functions depend on sorting proteins to their right destination. Examples: A. Energy production
Cell Structure. Chapter 4
 Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Genome Annotation Project Presentation
 Halogeometricum borinquense Genome Annotation Project Presentation Loci Hbor_05620 & Hbor_05470 Presented by: Mohammad Reza Najaf Tomaraei Hbor_05620 Basic Information DNA Coordinates: 527,512 528,261
Halogeometricum borinquense Genome Annotation Project Presentation Loci Hbor_05620 & Hbor_05470 Presented by: Mohammad Reza Najaf Tomaraei Hbor_05620 Basic Information DNA Coordinates: 527,512 528,261
Supplementary Materials for mplr-loc Web-server
 Supplementary Materials for mplr-loc Web-server Shibiao Wan and Man-Wai Mak email: shibiao.wan@connect.polyu.hk, enmwmak@polyu.edu.hk June 2014 Back to mplr-loc Server Contents 1 Introduction to mplr-loc
Supplementary Materials for mplr-loc Web-server Shibiao Wan and Man-Wai Mak email: shibiao.wan@connect.polyu.hk, enmwmak@polyu.edu.hk June 2014 Back to mplr-loc Server Contents 1 Introduction to mplr-loc
Public Database 의이용 (1) - SignalP (version 4.1)
 Public Database 의이용 (1) - SignalP (version 4.1) 2015. 8. KIST 이철주 Secretion pathway prediction ProteinCenter (Proxeon Bioinformatics, Odense, Denmark; http://www.cbs.dtu.dk/services) SignalP (version 4.1)
Public Database 의이용 (1) - SignalP (version 4.1) 2015. 8. KIST 이철주 Secretion pathway prediction ProteinCenter (Proxeon Bioinformatics, Odense, Denmark; http://www.cbs.dtu.dk/services) SignalP (version 4.1)
Cells. Structural and functional units of living organisms
 Cells Structural and functional units of living organisms Eukaryotic ( true nucleus ) vs. Prokaryotic ( before nucleus ) cells Proks Eukaryotic ( true nucleus ) vs. Prokaryotic ( before nucleus ) cells
Cells Structural and functional units of living organisms Eukaryotic ( true nucleus ) vs. Prokaryotic ( before nucleus ) cells Proks Eukaryotic ( true nucleus ) vs. Prokaryotic ( before nucleus ) cells
FUSION OF CONDITIONAL RANDOM FIELD AND SIGNALP FOR PROTEIN CLEAVAGE SITE PREDICTION
 FUSION OF CONDITIONAL RANDOM FIELD AND SIGNALP FOR PROTEIN CLEAVAGE SITE PREDICTION Man-Wai Mak and Wei Wang Dept. of Electronic and Information Engineering The Hong Kong Polytechnic University, Hong Kong
FUSION OF CONDITIONAL RANDOM FIELD AND SIGNALP FOR PROTEIN CLEAVAGE SITE PREDICTION Man-Wai Mak and Wei Wang Dept. of Electronic and Information Engineering The Hong Kong Polytechnic University, Hong Kong
Artificial Intelligence (AI) Common AI Methods. Training. Signals to Perceptrons. Artificial Neural Networks (ANN) Artificial Intelligence
 Artificial Intelligence (AI) Artificial Intelligence AI is an attempt to reproduce intelligent reasoning using machines * * H. M. Cartwright, Applications of Artificial Intelligence in Chemistry, 1993,
Artificial Intelligence (AI) Artificial Intelligence AI is an attempt to reproduce intelligent reasoning using machines * * H. M. Cartwright, Applications of Artificial Intelligence in Chemistry, 1993,
A Bayesian System Integrating Expression Data with Sequence Patterns for Localizing Proteins: Comprehensive Application to the Yeast Genome
 A Bayesian System Integrating Expression Data with Sequence Patterns for Localizing Proteins: Comprehensive Application to the Yeast Genome Amar Drawid 1 & Mark Gerstein 1,2 * Departments of (1) Molecular
A Bayesian System Integrating Expression Data with Sequence Patterns for Localizing Proteins: Comprehensive Application to the Yeast Genome Amar Drawid 1 & Mark Gerstein 1,2 * Departments of (1) Molecular
Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network
 Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network Marko Tscherepanow and Franz Kummert Applied Computer Science, Faculty of Technology, Bielefeld
Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network Marko Tscherepanow and Franz Kummert Applied Computer Science, Faculty of Technology, Bielefeld
Supplementary Materials for R3P-Loc Web-server
 Supplementary Materials for R3P-Loc Web-server Shibiao Wan and Man-Wai Mak email: shibiao.wan@connect.polyu.hk, enmwmak@polyu.edu.hk June 2014 Back to R3P-Loc Server Contents 1 Introduction to R3P-Loc
Supplementary Materials for R3P-Loc Web-server Shibiao Wan and Man-Wai Mak email: shibiao.wan@connect.polyu.hk, enmwmak@polyu.edu.hk June 2014 Back to R3P-Loc Server Contents 1 Introduction to R3P-Loc
Common ground for protein translocation: access control for mitochondria and chloroplasts
 Common ground for protein translocation: access control for mitochondria and chloroplasts Enrico Schleiff*and Thomas Becker Abstract Mitochondria and chloroplasts import the vast majority of their proteins
Common ground for protein translocation: access control for mitochondria and chloroplasts Enrico Schleiff*and Thomas Becker Abstract Mitochondria and chloroplasts import the vast majority of their proteins
9/11/18. Molecular and Cellular Biology. 3. The Cell From Genes to Proteins. key processes
 Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Structure-function studies of a protein-transporting transmembrane channel
 University of Oxford Department of Biochemistry D.Phil. studentship in Molecular ell Biology Structure-function studies of a protein-transporting transmembrane channel Periplasm Tat TatB TatA ytoplasm
University of Oxford Department of Biochemistry D.Phil. studentship in Molecular ell Biology Structure-function studies of a protein-transporting transmembrane channel Periplasm Tat TatB TatA ytoplasm
Protein structure alignments
 Protein structure alignments Proteins that fold in the same way, i.e. have the same fold are often homologs. Structure evolves slower than sequence Sequence is less conserved than structure If BLAST gives
Protein structure alignments Proteins that fold in the same way, i.e. have the same fold are often homologs. Structure evolves slower than sequence Sequence is less conserved than structure If BLAST gives
It s a Small World After All
 It s a Small World After All Engage: Cities, factories, even your own home is a network of dependent and independent parts that make the whole function properly. Think of another network that has subunits
It s a Small World After All Engage: Cities, factories, even your own home is a network of dependent and independent parts that make the whole function properly. Think of another network that has subunits
Cell Structure. Chapter 4. Cell Theory. Cells were discovered in 1665 by Robert Hooke.
 Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Cell Structure Chapter 4 Cell Theory Cells were discovered in 1665 by Robert Hooke. Early studies of cells were conducted by - Mathias Schleiden (1838) - Theodor Schwann (1839) Schleiden and Schwann proposed
Bioinformatics Chapter 1. Introduction
 Bioinformatics Chapter 1. Introduction Outline! Biological Data in Digital Symbol Sequences! Genomes Diversity, Size, and Structure! Proteins and Proteomes! On the Information Content of Biological Sequences!
Bioinformatics Chapter 1. Introduction Outline! Biological Data in Digital Symbol Sequences! Genomes Diversity, Size, and Structure! Proteins and Proteomes! On the Information Content of Biological Sequences!
Transport between cytosol and nucleus
 of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
1-D Predictions. Prediction of local features: Secondary structure & surface exposure
 1-D Predictions Prediction of local features: Secondary structure & surface exposure 1 Learning Objectives After today s session you should be able to: Explain the meaning and usage of the following local
1-D Predictions Prediction of local features: Secondary structure & surface exposure 1 Learning Objectives After today s session you should be able to: Explain the meaning and usage of the following local
Biochemistry: A Review and Introduction
 Biochemistry: A Review and Introduction CHAPTER 1 Chem 40/ Chem 35/ Fundamentals of 1 Outline: I. Essence of Biochemistry II. Essential Elements for Living Systems III. Classes of Organic Compounds IV.
Biochemistry: A Review and Introduction CHAPTER 1 Chem 40/ Chem 35/ Fundamentals of 1 Outline: I. Essence of Biochemistry II. Essential Elements for Living Systems III. Classes of Organic Compounds IV.
Neural Networks for Protein Structure Prediction Brown, JMB CS 466 Saurabh Sinha
 Neural Networks for Protein Structure Prediction Brown, JMB 1999 CS 466 Saurabh Sinha Outline Goal is to predict secondary structure of a protein from its sequence Artificial Neural Network used for this
Neural Networks for Protein Structure Prediction Brown, JMB 1999 CS 466 Saurabh Sinha Outline Goal is to predict secondary structure of a protein from its sequence Artificial Neural Network used for this
Translocation through the endoplasmic reticulum membrane. Institute of Biochemistry Benoît Kornmann
 Translocation through the endoplasmic reticulum membrane Institute of Biochemistry Benoît Kornmann Endomembrane system Protein sorting Permeable to proteins but not to ions IgG tetramer (16 nm) Fully hydrated
Translocation through the endoplasmic reticulum membrane Institute of Biochemistry Benoît Kornmann Endomembrane system Protein sorting Permeable to proteins but not to ions IgG tetramer (16 nm) Fully hydrated
10/1/2014. Chapter Explain why the cell is considered to be the basic unit of life.
 Chapter 4 PSAT $ by October by October 11 Test 3- Tuesday October 14 over Chapter 4 and 5 DFA- Monday October 20 over everything covered so far (Chapters 1-5) Review on Thursday and Friday before 1. Explain
Chapter 4 PSAT $ by October by October 11 Test 3- Tuesday October 14 over Chapter 4 and 5 DFA- Monday October 20 over everything covered so far (Chapters 1-5) Review on Thursday and Friday before 1. Explain
Identifying Extracellular Plant Proteins Based on Frequent Subsequences
 Identifying Extracellular Plant Proteins Based on Frequent Subsequences Yang Wang, Osmar R. Zaïane, Randy Goebel and Gregory Taylor University of Alberta {wyang, zaiane, goebel}@cs.ualberta.ca Abstract
Identifying Extracellular Plant Proteins Based on Frequent Subsequences Yang Wang, Osmar R. Zaïane, Randy Goebel and Gregory Taylor University of Alberta {wyang, zaiane, goebel}@cs.ualberta.ca Abstract
9/2/17. Molecular and Cellular Biology. 3. The Cell From Genes to Proteins. key processes
 Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Molecular and Cellular Biology Animal Cell ((eukaryotic cell) -----> compare with prokaryotic cell) ENDOPLASMIC RETICULUM (ER) Rough ER Smooth ER Flagellum Nuclear envelope Nucleolus NUCLEUS Chromatin
Protein Sorting. By: Jarod, Tyler, and Tu
 Protein Sorting By: Jarod, Tyler, and Tu Definition Organizing of proteins Organelles Nucleus Ribosomes Endoplasmic Reticulum Golgi Apparatus/Vesicles How do they know where to go? Amino Acid Sequence
Protein Sorting By: Jarod, Tyler, and Tu Definition Organizing of proteins Organelles Nucleus Ribosomes Endoplasmic Reticulum Golgi Apparatus/Vesicles How do they know where to go? Amino Acid Sequence
Reading Assignments. A. Genes and the Synthesis of Polypeptides. Lecture Series 7 From DNA to Protein: Genotype to Phenotype
 Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
Lecture Series 7 From DNA to Protein: Genotype to Phenotype Reading Assignments Read Chapter 7 From DNA to Protein A. Genes and the Synthesis of Polypeptides Genes are made up of DNA and are expressed
BL1102 Essay. The Cells Behind The Cells
 BL1102 Essay The Cells Behind The Cells Matriculation Number: 120019783 19 April 2013 1 The Cells Behind The Cells For the first 3,000 million years on the early planet, bacteria were largely dominant.
BL1102 Essay The Cells Behind The Cells Matriculation Number: 120019783 19 April 2013 1 The Cells Behind The Cells For the first 3,000 million years on the early planet, bacteria were largely dominant.
Cell Organelles. Wednesday, October 22, 14
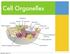 Cell Organelles Cell/Plasma Membrane ALL cells have a cell membrane It is the layer that surrounds the cell and controls what goes in and out Bacteria (Prokaryotic Cell) Cell/Plasma Membrane ALL cells
Cell Organelles Cell/Plasma Membrane ALL cells have a cell membrane It is the layer that surrounds the cell and controls what goes in and out Bacteria (Prokaryotic Cell) Cell/Plasma Membrane ALL cells
Protein Structure Prediction Using Multiple Artificial Neural Network Classifier *
 Protein Structure Prediction Using Multiple Artificial Neural Network Classifier * Hemashree Bordoloi and Kandarpa Kumar Sarma Abstract. Protein secondary structure prediction is the method of extracting
Protein Structure Prediction Using Multiple Artificial Neural Network Classifier * Hemashree Bordoloi and Kandarpa Kumar Sarma Abstract. Protein secondary structure prediction is the method of extracting
2011 The Simple Homeschool Simple Days Unit Studies Cells
 1 We have a full line of high school biology units and courses at CurrClick and as online courses! Subscribe to our interactive unit study classroom and make science fun and exciting! 2 A cell is a small
1 We have a full line of high school biology units and courses at CurrClick and as online courses! Subscribe to our interactive unit study classroom and make science fun and exciting! 2 A cell is a small
Topic 3: Cells Ch. 6. Microscopes pp Microscopes. Microscopes. Microscopes. Microscopes
 Topic 3: Cells Ch. 6 -All life is composed of cells and all cells have a plasma membrane, cytoplasm, and DNA. pp.105-107 - The development of the microscope was the key to understanding that all living
Topic 3: Cells Ch. 6 -All life is composed of cells and all cells have a plasma membrane, cytoplasm, and DNA. pp.105-107 - The development of the microscope was the key to understanding that all living
Biology Summer Assignments
 Biology Summer Assignments Welcome Mustangs! The following summer assignments are to assist you in obtaining background information for topics we will be learning during 1 st quarter, in Biology. Please
Biology Summer Assignments Welcome Mustangs! The following summer assignments are to assist you in obtaining background information for topics we will be learning during 1 st quarter, in Biology. Please
John F. Allen. Cell Biology and Developmental Genetics. Lectures by. jfallen.org
 Cell Biology and Developmental Genetics Lectures by John F. Allen School of Biological and Chemical Sciences, Queen Mary, University of London jfallen.org 1 Cell Biology and Developmental Genetics Lectures
Cell Biology and Developmental Genetics Lectures by John F. Allen School of Biological and Chemical Sciences, Queen Mary, University of London jfallen.org 1 Cell Biology and Developmental Genetics Lectures
Complete the table by stating the function associated with each organelle. contains the genetic material.... lysosome ribosome... Table 6.
 1 (a) Table 6.1 gives the functions of certain organelles in a eukaryotic cell. Complete the table by stating the function associated with each organelle. The first row has been completed for you. Organelle
1 (a) Table 6.1 gives the functions of certain organelles in a eukaryotic cell. Complete the table by stating the function associated with each organelle. The first row has been completed for you. Organelle
CHAPTER 9 PROTEIN SUBCELLULAR LOCALIZATION PREDICTION
 CHAPTER 9 PROTEIN SUBCELLULAR LOCALIZATION PREDICTION Paul Horton National Institute of Industrial Science and Technology horton-p@aist.go.jp Yuri Mukai National Institute of Industrial Science and Technology
CHAPTER 9 PROTEIN SUBCELLULAR LOCALIZATION PREDICTION Paul Horton National Institute of Industrial Science and Technology horton-p@aist.go.jp Yuri Mukai National Institute of Industrial Science and Technology
Cell Organelles Tutorial
 1 Name: Cell Organelles Tutorial TEK 7.12D: Differentiate between structure and function in plant and animal cell organelles, including cell membrane, cell wall, nucleus, cytoplasm, mitochondrion, chloroplast,
1 Name: Cell Organelles Tutorial TEK 7.12D: Differentiate between structure and function in plant and animal cell organelles, including cell membrane, cell wall, nucleus, cytoplasm, mitochondrion, chloroplast,
Organisms: We will need to have some examples in mind for our spherical cows.
 Lecture 4: Structure and Composition (Sept. 15) 4.1 Reading Assignment for Lectures 3-4: Phillips, Kondev, Theriot (PKT), Chapter 2 Problem Set 1 (due Sept. 24) now posted on the website. Cellular materials:
Lecture 4: Structure and Composition (Sept. 15) 4.1 Reading Assignment for Lectures 3-4: Phillips, Kondev, Theriot (PKT), Chapter 2 Problem Set 1 (due Sept. 24) now posted on the website. Cellular materials:
Organelles in Eukaryotic Cells
 Why? Organelles in Eukaryotic Cells What are the functions of different organelles in a cell? The cell is the basic unit and building block of all living things. Organisms rely on their cells to perform
Why? Organelles in Eukaryotic Cells What are the functions of different organelles in a cell? The cell is the basic unit and building block of all living things. Organisms rely on their cells to perform
Unicellular Marine Organisms. Chapter 4
 Unicellular Marine Organisms Chapter 4 The Cellular Structure of Life: Review Cell wall: firm, fairly rigid structure located outside the plasma membrane of plants, fungi, most bacteria, and some protists;
Unicellular Marine Organisms Chapter 4 The Cellular Structure of Life: Review Cell wall: firm, fairly rigid structure located outside the plasma membrane of plants, fungi, most bacteria, and some protists;
Cell Structure and Function
 Cell Structure and Function Cell size comparison Animal cell Bacterial cell What jobs do cells have to do for an organism to live Gas exchange CO 2 & O 2 Eat (take in & digest food) Make energy ATP Build
Cell Structure and Function Cell size comparison Animal cell Bacterial cell What jobs do cells have to do for an organism to live Gas exchange CO 2 & O 2 Eat (take in & digest food) Make energy ATP Build
A hidden Markov model for predicting transmembrane helices in protein sequences
 Procedings of ISMB 6, 1998, pages 175-182 A hidden Markov model for predicting transmembrane helices in protein sequences Erik L.L. Sonnhammer National Center for Biotechnology Information Building 38A,
Procedings of ISMB 6, 1998, pages 175-182 A hidden Markov model for predicting transmembrane helices in protein sequences Erik L.L. Sonnhammer National Center for Biotechnology Information Building 38A,
Biological Networks: Comparison, Conservation, and Evolution via Relative Description Length By: Tamir Tuller & Benny Chor
 Biological Networks:,, and via Relative Description Length By: Tamir Tuller & Benny Chor Presented by: Noga Grebla Content of the presentation Presenting the goals of the research Reviewing basic terms
Biological Networks:,, and via Relative Description Length By: Tamir Tuller & Benny Chor Presented by: Noga Grebla Content of the presentation Presenting the goals of the research Reviewing basic terms
2. Cellular and Molecular Biology
 2. Cellular and Molecular Biology 2.1 Cell Structure 2.2 Transport Across Cell Membranes 2.3 Cellular Metabolism 2.4 DNA Replication 2.5 Cell Division 2.6 Biosynthesis 2.1 Cell Structure What is a cell?
2. Cellular and Molecular Biology 2.1 Cell Structure 2.2 Transport Across Cell Membranes 2.3 Cellular Metabolism 2.4 DNA Replication 2.5 Cell Division 2.6 Biosynthesis 2.1 Cell Structure What is a cell?
Introduction to Cells- Stations Lab
 Introduction to Cells- Stations Lab Station 1: Microscopes allow scientists to study cells. Microscopes: How do light microscopes differ from electron microscopes? (How does each work? How much can each
Introduction to Cells- Stations Lab Station 1: Microscopes allow scientists to study cells. Microscopes: How do light microscopes differ from electron microscopes? (How does each work? How much can each
CHAPTER 1 INTRODUCTION TO CELLS 2009 Garland Science Publishing 3 rd Edition
 Unity and Diversity of Cells 1-1 The smallest unit of life is a(n) (a) DNA molecule. (b) cell. (c) organelle. (d) virus. (e) protein. CHAPTER 1 INTRODUCTION TO CELLS 2009 Garland Science Publishing 3 rd
Unity and Diversity of Cells 1-1 The smallest unit of life is a(n) (a) DNA molecule. (b) cell. (c) organelle. (d) virus. (e) protein. CHAPTER 1 INTRODUCTION TO CELLS 2009 Garland Science Publishing 3 rd
11. What are the four most abundant elements in a human body? A) C, N, O, H, P B) C, N, O, P C) C, S, O, H D) C, Na, O, H E) C, H, O, Fe
 48017 omework#1 on VVP Chapter 1: and in the provided answer template on Monday 4/10/17 @ 1:00pm; Answers on this document will not be graded! Matching A) Phylogenetic B) negative C) 2 D) Δ E) TS F) halobacteria
48017 omework#1 on VVP Chapter 1: and in the provided answer template on Monday 4/10/17 @ 1:00pm; Answers on this document will not be graded! Matching A) Phylogenetic B) negative C) 2 D) Δ E) TS F) halobacteria
Machine Learning in Action
 Machine Learning in Action Tatyana Goldberg (goldberg@rostlab.org) August 16, 2016 @ Machine Learning in Biology Beijing Genomics Institute in Shenzhen, China June 2014 GenBank 1 173,353,076 DNA sequences
Machine Learning in Action Tatyana Goldberg (goldberg@rostlab.org) August 16, 2016 @ Machine Learning in Biology Beijing Genomics Institute in Shenzhen, China June 2014 GenBank 1 173,353,076 DNA sequences
Part 2. The Basics of Biology:
 Part 2 The Basics of Biology: An Engineer s Perspective Chapter 2 An Overview of Biological Basics 21 2.1 Cells 2.2 Cell Construction 2.3 Cell Nutrient 2.1 Are all cells the same? Cells Basic unit of living
Part 2 The Basics of Biology: An Engineer s Perspective Chapter 2 An Overview of Biological Basics 21 2.1 Cells 2.2 Cell Construction 2.3 Cell Nutrient 2.1 Are all cells the same? Cells Basic unit of living
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis.
 Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Computational Genomics and Molecular Biology, Fall
 Computational Genomics and Molecular Biology, Fall 2014 1 HMM Lecture Notes Dannie Durand and Rose Hoberman November 6th Introduction In the last few lectures, we have focused on three problems related
Computational Genomics and Molecular Biology, Fall 2014 1 HMM Lecture Notes Dannie Durand and Rose Hoberman November 6th Introduction In the last few lectures, we have focused on three problems related
A. The Cell: The Basic Unit of Life. B. Prokaryotic Cells. D. Organelles that Process Information. E. Organelles that Process Energy
 The Organization of Cells A. The Cell: The Basic Unit of Life Lecture Series 4 The Organization of Cells B. Prokaryotic Cells C. Eukaryotic Cells D. Organelles that Process Information E. Organelles that
The Organization of Cells A. The Cell: The Basic Unit of Life Lecture Series 4 The Organization of Cells B. Prokaryotic Cells C. Eukaryotic Cells D. Organelles that Process Information E. Organelles that
Zimmerman AP Biology CBHS South Name Chapter 7&8 Guided Reading Assignment 1) What is resolving power and why is it important in biology?
 Zimmerman AP Biology CBHS South Name Chapter 7&8 Guided Reading Assignment 1) What is resolving power and why is it important in biology? 2) How does an electron microscope work and what is the difference
Zimmerman AP Biology CBHS South Name Chapter 7&8 Guided Reading Assignment 1) What is resolving power and why is it important in biology? 2) How does an electron microscope work and what is the difference
Chapter 4. Table of Contents. Section 1 The History of Cell Biology. Section 2 Introduction to Cells. Section 3 Cell Organelles and Features
 Cell Structure and Function Table of Contents Section 1 The History of Cell Biology Section 2 Introduction to Cells Section 3 Cell Organelles and Features Section 4 Unique Features of Plant Cells Section
Cell Structure and Function Table of Contents Section 1 The History of Cell Biology Section 2 Introduction to Cells Section 3 Cell Organelles and Features Section 4 Unique Features of Plant Cells Section
Using N-terminal targeting sequences, amino acid composition, and sequence motifs for predicting protein subcellular localization
 Using N-terminal targeting sequences, amino acid composition, and sequence motifs for predicting protein subcellular localization Annette Höglund, Pierre Dönnes, Torsten Blum, Hans-Werner Adolph, and Oliver
Using N-terminal targeting sequences, amino acid composition, and sequence motifs for predicting protein subcellular localization Annette Höglund, Pierre Dönnes, Torsten Blum, Hans-Werner Adolph, and Oliver
The Cell. The basic unit of all living things
 The Cell The basic unit of all living things 1 Robert Hooke was the first to name the cell (1665) 2 The Cell Theory The cell is the unit of Structure of all living things. The cell is the unit of Function
The Cell The basic unit of all living things 1 Robert Hooke was the first to name the cell (1665) 2 The Cell Theory The cell is the unit of Structure of all living things. The cell is the unit of Function
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
We may not be able to do any great thing, but if each of us will do something, however small it may be, a good deal will be accomplished.
 Chapter 4 Initial Experiments with Localization Methods We may not be able to do any great thing, but if each of us will do something, however small it may be, a good deal will be accomplished. D.L. Moody
Chapter 4 Initial Experiments with Localization Methods We may not be able to do any great thing, but if each of us will do something, however small it may be, a good deal will be accomplished. D.L. Moody
CMPS 6630: Introduction to Computational Biology and Bioinformatics. Structure Comparison
 CMPS 6630: Introduction to Computational Biology and Bioinformatics Structure Comparison Protein Structure Comparison Motivation Understand sequence and structure variability Understand Domain architecture
CMPS 6630: Introduction to Computational Biology and Bioinformatics Structure Comparison Protein Structure Comparison Motivation Understand sequence and structure variability Understand Domain architecture
