Neurotransmitter 2014; 1: e388. doi: /nt.388; 2014 by Takashi Hayashi
|
|
|
- Beatrice Pope
- 6 years ago
- Views:
Transcription
1 BRIEF REPORT Evolutionarily conserved palmitoylation-dependent regulation of ionotropic glutamate receptors in vertebrates Takashi Hayashi 1, 2 1 Section of Cellular Biochemistry, Department of Biochemistry and Cellular Biology, National Institute of Neuroscience, National Center of Neurology and Psychiatry (NCNP) Ogawa-Higashi, Kodaira, Tokyo , Japan 2 Department of Molecular Neurobiology and Pharmacology, Graduate School of Medicine, The University of Tokyo Hongo, Bunkyo-ku, Tokyo , Japan Correspondence: T. H. thayashi@ncnp.go.jp Received: October 26, 2014 Published online: December 01, 2014 In the vertebrate central nervous system, glutamate is the major excitatory neurotransmitter. Ionotoropic glutamate receptors (iglurs) are responsible for the glutamate-mediated postsynaptic excitation of neurons. Regulation of glutamatergic synapses in vertebrate central nervous system is critical for higher brain functions such as neural communication, memory formation, learning, emotion and behavior. Previous studies showed that post-translational protein palmitoylation, the only reversible covalent attachment of lipid to protein, regulates synaptic expression, intracellular localization and membrane trafficking of iglurs in mammalian neurons. Here, I further focus on conservation of palmitoylation sites found in iglur vertebrate orthologs. Analysis of databases shows that every palmitoylation sites of iglurs have been completely conserved during evolution in the vertebrate lineage, in spite of the divergence of iglurs amino acid sequences. Namely, palmitoylated cysteine residues of AMPA receptors, NMDA receptors and KA receptors, are evolutionarily conserved against mutation pressure throughout vertebrate species without exception. This finding suggests that the dynamic regulation of glutamatergic synapses made possible by this reversible palmitoylation of iglurs is critical for the vertebrate-specific refined functions of complex nervous systems. Keywords: palmitoylation; AMPA receptor; NMDA receptor; KA receptor; excitatory synapses; vertebrate To cite this article: Takashi Hayashi. Evolutionarily conserved palmitoylation-dependent regulation of ionotropic glutamate receptors in vertebrates. Neurotransmitter 2014; 1: e388. doi: /nt.388. Introduction Glutamate is the major excitatory neurotransmitter in the vertebrate central nervous system (CNS). The ionotropic glutamate receptors (iglurs) are classified into four groups, namely, -amino-3-hydroxy-5-methyl-4-isoxazole propionate (AMPA)-type, kainate (KA)-type, (delta)-type and N-methyl-D-aspartate (NMDA)-type receptors. Of these iglurs, AMPA receptors mediate the majority of fast excitatory synaptic transmission in the mammalian brain [1, 2]. The expression level of post-synaptic AMPA receptors is closely linked to excitatory synaptic strength. Quantitative control of synaptic AMPA receptor amount is thus not only critical for basal synaptic transmission, but also for mammalian synaptic plasticity and higher brain function [3-5]. AMPA receptors trafficking to and from synapses is dynamically regulated by post-translational protein modification, both of receptors themselves, and of scaffold proteins, which bind receptors and direct them to specific subcellular locations [6-9]. NMDA receptors are the main triggers for the induction of synaptic plasticity, synaptogenesis and excitotoxicity [5, 10-12]. KA receptors play roles distinct from other iglur families by changing neural Page 1 of 10
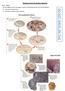
2 Figure 1. Vertebrate iglurs palmitoylation sites are evolutionarily conserved. Schematic of iglurs, showing location of palmitoylation sites (left). Summary of evolutionary subclasses and the presence or absence of iglur palmitoylation sites (right). +: Cys for palmitoylation site; : substituted residues at corresponding sites;?: residues not determined. Species numbers in each subclass are shown. (A) AMPA receptor subunits GluA1-4, (B) NMDA receptor subunits GluN2A and GluN2B, (C) KA receptor subunit GluK2. circuit activity through modulation of excitatory and inhibitory transmission and neuronal excitability [13]. One key modification of mammalian iglurs is the reversible addition of the lipid palmitate to intracellular cysteine residues. This process, post-translational protein Page 2 of 10
3 Table 1. The BLAST alignments of vertebrate iglurs. (A) GluA1 (AMPA receptor subunit), (B) GluN2A (NMDA receptor subunit) and (C) GluK2 (KA receptor subunit). Percent identity among orthologs across any two species were obtained by performing BLAST search (with BLOSUM62) with full length amino acid sequences of vertebrate iglur orthologs. Homo sapiens (human), Mus musculus (mouse), Gallus gallus (chicken), Chrysemys picta bellii (painted turtle), Xenopus laevis (African clawed frog), Danio rerio (zebrafish) are compared as representative of each vertebrate class. palmitoylation, acts as a sticky tag that can direct receptors to specific regions of the plasma membrane, or to specific intracellular membranes or vesicles [14-16]. Genetic evidence strongly links impaired palmitoylation to abnormal mammalian brain development and/or function, including human neuropsychiatric disorders [17-22]. We have previously shown that palmitoylation regulate the synaptic expression and localization of AMPA receptors and NMDA receptors [23-26]. All four subunits of the mammalian AMPA receptor, GluA1-4 (also known as GluR1-4, GluRA-D or GluR 1-4) and the regulatory subunits of the mammalian NMDA receptor, GluN2A and GluN2B (also known as NR2A, NR2B or GluR 1, GluR 2) are palmitoylated at their two distinct sites. Palmitoylation of one site causes AMPA receptors and NMDA receptors to accumulate in the Golgi apparatus. Palmitoylation thus appears to act as a quality control mechanism to ensure correct receptor maturation. In contrast, second palmitoylation sites in AMPA receptors and NMDA receptors are implicated in quantitative regulation of synaptic receptor numbers, which relate to complex neuronal events such as synaptic plasticity and higher brain function. In addition, surface insertion and stabilization of KA receptor subunit GluK2 (also known as GluR6 or GluR 2) also occurs in its palmitoylation-dependent manner [27, 28]. Summarizing the above, post-translational protein palmitoylation appears uniquely suited to create dynamic controls of the synaptic expression and intracellular trafficking of mammalian iglurs in the CNS. Many neurotransmitter receptors, including iglurs, Page 3 of 10
4 appear to be evolutionarily conserved; orthologs with identical domains and transmembrane topology are found in organisms from worms to man [29]. Here, I focused on the palmitoylation sites of iglurs in the vertebrate lineage and sequence analysis of databases provides evidence for the complete conservation of palmitoylation mechanism in iglur regulations during vertebrate evolution. Materials and methods For analysis of the iglur orthologs, available cdna sequences, protein sequences, expressed sequence tags (ESTs) and genomic sequences are obtained by searching the NCBI databases, Genbank, EST banks, elephant shark genome project ( Joint Genome Institute ( and the Ensembl database ( by sequence homologies. Results Completely conserved palmitoylation sites in iglurs Recent progress in genome analyses revealed that many animal species possess orthologs of mammalian iglurs [29-32]. Our previous examination of the evolutionary conservation of the AMPA receptor palmitoylated cysteines revealed that the first palmitoylation site, which commonly lies in GluA1-4 transmembrane domain (TMD) 2, is present in every animal species examined except for nematodes [23, 33]. In contrast, the second palmitoylation site, located in the GluA1-4 C-terminal region, present only in all vertebrate AMPA receptor orthologs, whereas this site is absent in the majority of invertebrate AMPA receptor homologs; the only exception being a subset of insect species [23, 33]. The emergence and conservation of the second palmitoylation site in vertebrates suggests that AMPA receptor C-terminal palmitoylation may have evolved to allow additional regulation of synaptic AMPA receptor trafficking that may be required for higher brain function (Fig. 1A). Furthermore, palmitoylation sites in both Cys cluster I and Cys cluster II of NMDA receptor subunits GluN2A and GluN2B are completely conserved in all reported vertebrate orthologs (Fig. 1B). Only vertebrates GluN2A and GluN2B have the long intracellular C-terminal domain, carrying both palmitoylation sites, and they are lost in invertebrates [34]. While palmitoylation at Cys cluster I regulates synaptic expression of NMDA receptors, palmitoylation at Cys cluster II accumulates the receptors in the Golgi apparatus, like AMPA receptors [25]. Additional analysis of sequence databases shows that palmitoylation sites in GluK2 (Cys 858, 871), which affects surface localization of KA receptors in neurons, are also completely conserved in every vertebrate species (Fig. 1C). See supplementary information for full details of data sources. Divergence of amino acid sequence in iglurs full length The homology comparison of full length amino acid sequences of AMPA receptor orthologs show ~98% identity among mammalian species, ~93% identity between mammals and birds, ~93% identity between mammals and reptiles, ~88% identity between mammals and amphibians, ~74% identity between mammals and fishes (Table 1A). NMDA receptor orthologs show ~95% identity among mammalian species, ~82% identity between mammals and birds, ~80% identity between mammals and reptiles, ~74% identity between mammals and amphibians, ~67% identity between mammals and fishes (Table 1B). Higher homology scores are obtained among KA receptor orthologs, ~99 identity among mammalian species, ~98% identity between mammals and birds, ~98% identity between mammals and reptiles, ~94% identity between mammals and amphibians, ~91% identity between mammals and fishes (Table 1C). In conclusion, random mutations are observed all over vertebrate iglur sequences during vertebrate evolution. Discussion Generally, structurally or functionally important amino acid residues are conserved during molecular evolution against mutation pressure. Despite divergences in amino acid sequences among iglurs, this study reveal that palmitoylation sites in AMPA receptors, NMDA receptors and KA receptors are completely conserved throughout vertebrate species without exception, which strongly suggest critical roles of the multi-layer regulation of vertebrate glutamatergic excitatory synapses by palmitoylation of iglurs. In other words, vertebrates commonly use palmitoylation in excitatory synapses to control their CNS. Especially, synaptic palmitoylation site in the AMPA receptor C-termini are completely conserved in the vertebrate lineage from the Agnatha superclass (jawless fishes, such as hagfish and lamprey) to the Gnathostomata (jawed vertebrate) superclass. This modification site is readily acquired in the vertebrate common ancestor by simple gain-of-function mutations. In contrast, invertebrate species have non-cysteine residues at the corresponding sites of AMPA receptor homologs presumably because of random mutations in molecular evolution [33]. These facts suggest that the palmitoylation-dependent regulation of the AMPA receptor was established only in the common vertebrate ancestor around 500 million years ago in the late Cambrian to the early Ordovician periods. The vertebrate specific palmitoylation mechanism has been completely maintained up to the present time. On the other hand, the origin of Page 4 of 10
5 evolutionary acquirements of vertebrate NMDA receptors and KA receptors palmitoylation still remains unclear, because sequence information of primitive receptor orthologs of jawless fishes and species in the Urochordata subphylum (ascidians or sea squirts) or in the Cephalochordata subphylum (amphioxus and lancelet) is currently unavailable. Future genome analyses will answer detailed processes of acquisition of the post-translational protein palmitoylation in vertebrate glutamatergic excitatory synapses. Conflict of Interest The author declares that he has no conflicting interests. Acknowledgements This work was supported in part by the Grants-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (MEXT), the Takeda Science Foundation, the Mitsubishi Foundation and the Brain Science Foundation. References 1. Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci 1994; 17: Seeburg PH. The TINS/TiPS Lecture. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci 1993; 16: Kessels HW, Malinow R. Synaptic AMPA receptor plasticity and behavior. Neuron 2009; 61: Shepherd JD, Huganir RL. The cell biology of synaptic plasticity: AMPA receptor trafficking. Annu Rev Cell Dev Biol 2007; 23: Collingridge GL, Isaac JT, Wang YT. Receptor trafficking and synaptic plasticity. Nat Rev Neurosci 2004; 5: Anggono V, Huganir RL. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr Opin Neurobiol 2012; 22: Lu W, Roche KW. Posttranslational regulation of AMPA receptor trafficking and function. Curr Opin Neurobiol 2012; 22: Thomas GM, Hayashi T, Chiu SL, Chen CM, Huganir RL. Palmitoylation by DHHC5/8 targets GRIP1 to dendritic endosomes to regulate AMPA-R trafficking. Neuron 2012; 73: Thomas GM, Hayashi T, Huganir RL, Linden DJ. DHHC8-dependent PICK1 palmitoylation is required for induction of cerebellar long-term synaptic depression. J Neurosci 2013; 33: Mori H, Mishina M. Structure and function of the NMDA receptor channel. Neuropharmacology 1995; 34: Lau CG, Zukin RS. NMDA receptor trafficking in synaptic plasticity and neuropsychiatric disorders. Nat Rev Neurosci 2007; 8: Mori H, Mishina M. Roles of diverse glutamate receptors in brain functions elucidated by subunit-specific and region-specific gene targeting. Life Sci 2003; 74: Contractor A, Mulle C, Swanson GT. Kainate receptors coming of age: milestones of two decades of research. Trends Neurosci 2011; 34: Fukata Y, Fukata M. Protein palmitoylation in neuronal development and synaptic plasticity. Nat Rev Neurosci 2010; 11: Linder ME, Deschenes RJ. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol 2007; 8: Nadolski MJ, Linder ME. Protein lipidation. FEBS J 2007; 274: Fallin MD, Lasseter VK, Wolyniec PS, McGrath JA, Nestadt G, Valle D, et al. Genomewide linkage scan for bipolar-disorder susceptibility loci among Ashkenazi Jewish families. Am J Hum Genet 2004; 75: Mansouri MR, Marklund L, Gustavsson P, Davey E, Carlsson B, Larsson C, et al. Loss of ZDHHC15 expression in a woman with a balanced translocation t(x;15)(q13.3;cen) and severe mental retardation. Eur J Hum Genet 2005; 13: Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, MacDermott AB, et al. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci 2008; 11: Otani K, Ujike H, Tanaka Y, Morita Y, Kishimoto M, Morio A, et al. The ZDHHC8 gene did not associate with bipolar disorder or schizophrenia. Neurosci Lett 2005; 390: Raymond FL, Tarpey PS, Edkins S, Tofts C, O'Meara S, Teague J, et al. Mutations in ZDHHC9, which encodes a palmitoyltransferase of NRAS and HRAS, cause X-linked mental retardation associated with a Marfanoid habitus. Am J Hum Genet 2007; 80: Young FB, Butland SL, Sanders SS, Sutton LM, Hayden MR. Putting proteins in their place: palmitoylation in Huntington disease and other neuropsychiatric diseases. Prog Neurobiol 2012; 97: Hayashi T, Rumbaugh G, Huganir RL. Differential regulation of AMPA receptor subunit trafficking by palmitoylation of two distinct sites. Neuron 2005; 47: Lin DT, Makino Y, Sharma K, Hayashi T, Neve R, Takamiya K, et al. Regulation of AMPA receptor extrasynaptic insertion by 4.1N, phosphorylation and palmitoylation. Nat Neurosci 2009; 12: Hayashi T, Thomas GM, Huganir RL. Dual palmitoylation of NR2 subunits regulates NMDA receptor trafficking. Neuron 2009; 64: Mattison HA, Hayashi T, Barria A. Palmitoylation at two cysteine clusters on the C-terminus of GluN2A and GluN2B differentially control synaptic targeting of NMDA receptors. PLoS One 2012; 7:e Pickering DS, Taverna FA, Salter MW, Hampson DR. Palmitoylation of the GluR6 kainate receptor. Proc Natl Acad Sci U S A 1995; 92: Page 5 of 10
6 28. Copits BA, Swanson GT. Kainate receptor post-translational modifications differentially regulate association with 4.1N to control activity-dependent receptor endocytosis. J Biol Chem 2013; 288: Okamura Y, Nishino A, Murata Y, Nakajo K, Iwasaki H, Ohtsuka Y, et al. Comprehensive analysis of the ascidian genome reveals novel insights into the molecular evolution of ion channel genes. Physiol Genomics 2005; 22: Chiu J, DeSalle R, Lam HM, Meisel L, Coruzzi G. Molecular evolution of glutamate receptors: a primitive signaling mechanism that existed before plants and animals diverged. Mol Biol Evol 1999; 16: Sprengel R, Aronoff R, Volkner M, Schmitt B, Mosbach R, Kuner T. Glutamate receptor channel signatures. Trends Pharmacol Sci 2001; 22: Chen YC, Kung SS, Chen BY, Hung CC, Chen CC, Wang TY, et al. Identifications, classification, and evolution of the vertebrate alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptor subunit genes. J Mol Evol 2001; 53: Thomas GM, Hayashi T. Smarter neuronal signaling complexes from existing components: how regulatory modifications were acquired during animal evolution: evolution of palmitoylation-dependent regulation of AMPA-type ionotropic glutamate receptors. Bioessays 2013; 35: Ryan TJ, Emes RD, Grant SG, Komiyama NH. Evolution of NMDA receptor cytoplasmic interaction domains: implications for organisation of synaptic signalling complexes. BMC Neurosci 2008; 9:6. Page 6 of 10
7 Supplementary information Figure 1. Vertebrate iglurs palmitoylation sites are evolutionarily conserved. The Vertebrata subphylum consists of two superclasses, Gnathostomata (jawed vertebrate) and Agnatha (jawless fish). This figure represents sequence data from the following vertebrate species. (A) All cloned and predicted AMPA receptor othologs in each class are shown below. GluA1-4 subunits have two distinct palmitoylation sites in their C-termini and TMD2 region: GluA1 Cys811, Cys585; GluA2 Cys836, Cys610; GluA3 Cys841, Cys615; GluA4 Cys817, Cys611 (amino acid residues in the mouse GluA sequence as representative). Gnathostome AMPA receptor subunits In the Mammalia class: Homo sapiens (human), Pan paniscus (bonobo), Pan troglodytes (chimpanzee), Gorilla gorilla (western lowland gorilla), Pongo abelii (Sumatran orangutan), Nomascus leucogenys (northern white-cheeked gibbon), Macaca fascicularis (crab-eating macaque), Macaca mulatta (rhesus monkey), Papio anubis (olive baboon), Callithrix jacchus (white-tufted-ear marmoset), Saimiri boliviensis (black-capped squirrel monkey), Tarsius syrichta (tarsier), Microcebus murinus (mouse lemur), Otolemur garnettii (northern greater galago), Tupaia belangeri (northern treeshrew), Spermophilus tridecemlineatus / Ictidomys tridecemlineatus (thirteen-lined ground squirrel), Mesocricetus auratus (golden hamster), Rattus norvegicus (Norway rat), Mus musculus (house mouse), Dipodomys ordii (kangaroo rat), Cavia porcellus (guinea pig), Octodon degus (degu), Heterocephalus glaber (naked mole rat), Oryctolagus cuniculus (rabbit), Ochotona princeps (American pika), Erinaceus europaeus (European hedgehog), Sorex araneus (European shrew), Pteropus vampyrus (megabat), Pteropus alecto (black flying fox), Myotis lucifugus (little brown bat), Myotis brandtii (Brandt's bat), Myotis davidii (mouse-eared bat), Felis catus (cat), Canis lupus familiaris (dog), Mustela putorius furo (ferret), Ailuropoda melanoleuca (giant panda), Leptonychotes weddellii (Weddell seal), Equus caballus (horse), Ceratotherium simum (white rhinoceros), Bos taurus (cattle), Ovis aries (sheep), Capra hircus (goat), Pantholops hodgsonii (chiru/tibetan antelope), Tursiops truncates (dolphin), Orcinus orca (killer whale), Sus scrofa (wild boar), Camelus ferus (wild Bactrian camel), Vicugna pacos (alpaca), Choloepus hoffmanni (two-toed sloth), Dasypus novemcinctus (armadillo), Loxodonta africana (African elephant), Trichechus manatus latirostris (Florida manatee), Procavia capensis (rock hyrax), Echinops telfairi (lesser hedgehog tenrec), Chrysochloris asiatica (Cape golden mole), Elephantulus edwardii (Cape elephant shrew), Macropus eugenii (wallaby), Sarcophilus harrisii (Tasmanian devil), Monodelphis domestica (gray short-tailed opossum), Ornithorhynchus anatinus (platypus). In the Aves class (birds): Gallus gallus (chicken), Meleagris gallopavo (turkey), Anas platyrhynchos (mallard/duck), Columba livia (domestic pigeon), Falco peregrinus (peregrine falcon), Falco cherrug (saker falcon), Pseudopodoces humilis (ground tit), Ficedula albicollis (collared flycatcher), Taeniopygia guttata (zebra finch), Geospiza fortis (medium ground-finch), Zonotrichia albicollis (white-throated sparrow), Melopsittacus undulatus (budgerigar). In the Reptilia class: Trachemys scripta (red-eared slider turtle), Chrysemys picta bellii (painted turtle), Pelodiscus sinensis (Chinese softshell turtle), Chelonia mydas (green sea turtle), Anolis carolinensis (green anole), Alligator mississippiensis (American alligator), Alligator sinensis (Chinese alligator). In the Amphibia class: Rana catesbeiana (bullfrog), Xenopus tropicalis (western clawed frog), Xenopus laevis (African clawed frog). In the Actinopterygii class (ray-finned fishes); Fugu rubripes (Japanese pufferfish), Tetraodon fluviatilis (green spotted pufferfish), Tetraodon nigroviridis (a similar brackish species of green spotted pufferfish), Gadus morhua (Atlantic cod), Morone chrysops x Morone saxatilis (white bass x striped sea-bass), Gasterosteus aculeatus (stickleback), Oreochromis mossambicus (Mozambique tilapia), Oreochromis niloticus (Nile tilapia), Danio rerio (zebrafish), Carassius auratus (goldfish), Carassius carassius (crucian carp), Oryzias latipes (Japanese medaka), Astyanax mexicanus (Mexican tetra/blind cave fish), Poecilia formosa (Amazon molly), Haplochromis burtoni (Burton's mouthbrooder), Pundamilia nyererei (cichlid), Xiphophorus maculatusi (platyfish), Lepisosteus oculatusi (spotted gar). In the Sarcopterygii class (lobe-finned fishes): Latimeria chalumnae (coelacanth). In the Chondrichthyes class (cartilaginous fishes): Squalus acanthias (spiny dogfish shark), Callorhinchus milii (elephant shark). Agnatha AMPA receptor subunits In the Myxini class: Paramyxine yangi (brown hagfish). In the Petromyzontida class: Petromyzon marinus (sea Page 7 of 10
8 lamprey). (B) All cloned and predicted NMDA receptor regulatory subunits GluN2 othologs in each class are shown below. GluN2A and GluN2B subunits have two distinct palmitoylation sites in their C-termini: GluN2A Cys cluster I (Cys 848, 853, 870), GluN2A Cys cluster II (Cys1214, 1217, 1236, 1239) and GluN2B Cys cluster I (Cys 849, 854, 871), GluN2B Cys cluster II (Cys1215, 1218, 1239, 1242, 1245) (amino acid residues in the mouse GluN2A and GluN2B sequence as representative). Gnathostome NMDA receptor subunits In the Mammalia class: Homo sapiens (human), Pan paniscus (bonobo), Pan troglodytes (chimpanzee), Gorilla gorilla (western lowland gorilla), Pongo pygmaeus (Borneo orangutan), Pongo abelii (Sumatran orangutan), Hylobates muelleri (Müller's Bornean gibbon), Nomascus leucogenys (northern white-cheeked gibbon), Cercopithecus ascanius (red-tailed monkey), Cercopithecus diana (Diana monkey), Cercopithecus wolfi (Wolf's mona monkey), Allenopithecus nigroviridis (Allen's swamp monkey), Miopithecus talapoin (Angolan talapoin), Macaca fascicularis (crab-eating macaque), Macaca mulatta (rhesus monkey), Macaca fuscata (Japanese macaque), Chlorocebus sabaeus (green monkey), Papio hamadryas (hamadryas baboon), Papio anubis (olive baboon), Colobus angolensis (Angola colobus), Trachypithecus cristatus (silvery lutung), Trachypithecus vetulus (purple-faced langur), Pygathrix nemaeus (douc langur), Nasalis larvatus (proboscis monkey), Callithrix jacchus (white-tufted-ear marmoset), Callithrix pygmaea (pygmy marmoset), Saimiri boliviensis (black-capped squirrel monkey), Alouatta caraya (black howler monkey), Ateles fusciceps (black-headed spider monkey), Lagothrix lagotricha (brown woolly monkey), Callicebus moloch (dusky titi), Tarsius syrichta (tarsier), Microcebus murinus (mouse lemur), Lemur catta (ring-tailed lemur), Varecia rubra (red ruffed lemur), Otolemur garnettii (northern greater galago), Galeopterus variegatus (Sunda flying lemur), Tupaia belangeri (northern treeshrew), Tupaia chinensis (Chinese treeshrew), Spermophilus tridecemlineatus / Ictidomys tridecemlineatus (thirteen-lined ground squirrel), Cricetulus griseus (Chinese hamster), Mesocricetus auratus (golden hamster), Microtus ochrogaster (prairie vole), Rattus norvegicus (Norway rat), Mus musculus (house mouse), Jaculus jaculus (lesser Egyptian jerboa), Peromyscus maniculatus bairdii (deer mouse), Chinchilla lanigera (chinchilla), Dipodomys ordii (kangaroo rat), Cavia porcellus (guinea pig), Octodon degus (degu), Heterocephalus glaber (naked mole rat), Oryctolagus cuniculus (rabbit), Ochotona princeps (American pika), Erinaceus europaeus (European hedgehog), Sorex araneus (European shrew), Condylura cristata (star-nosed mole), Eptesicus fuscus (big brown bat), Pteropus vampyrus (megabat), Pteropus alecto (black flying fox), Myotis lucifugus (little brown bat), Myotis brandtii (Brandt's bat), Myotis davidii (mouse-eared bat), Felis catus (cat), Panthera tigris altaica (Amur tiger), Canis lupus familiaris (dog), Mustela putorius furo (ferret), Ursus maritimus (polar bear), Ailuropoda melanoleuca (giant panda), Leptonychotes weddellii (Weddell seal), Odobenus rosmarus divergens (walrus), Equus caballus (horse), Equus przewalskii (Przewalski's horse), Ceratotherium simum (white rhinoceros), Bos taurus (cattle), Bos mutus (yak), Bubalus bubalis (water buffalo), Ovis aries (sheep), Capra hircus (goat), Moschus moschiferus (Siberian musk deer), Elaphurus davidianus (Père David's deer), Pantholops hodgsonii (chiru/tibetan antelope), Hippopotamus amphibius (hippopotamus), Tursiops truncates (dolphin), Orcinus orca (killer whale), Kogia sima (dwarf sperm whale), Physeter catodon (sperm whale), Mesoplodon densirostris (Blainville's beaked whale), Platanista gangetica (river dolphin), Lipotes vexillifer (baiji), Delphinus capensis (common dolphin), Delphinapterus leucas (beluga whale), Stenella attenuata (pantropical spotted dolphin), Sousa chinensis (Chinese white dolphin), Grampus griseus (Risso's dolphin), Lipotes vexillifer (Yangtze River dolphin), Balaenoptera acutorostrata (minke whale), Balaenoptera omurai (Omura's whale), Neophocaena phocaenoides (finless porpoise), Sus scrofa (wild boar), Sus scrofa domesticus (pig), Camelus bactrianus (Bactrian camel), Camelus ferus (wild Bactrian camel), Vicugna pacos (alpaca), Choloepus hoffmanni (two-toed sloth), Dasypus novemcinctus (armadillo), Loxodonta africana (African elephant), Trichechus manatus latirostris (Florida manatee), Procavia capensis (rock hyrax), Echinops telfairi (lesser hedgehog tenrec), Chrysochloris asiatica (Cape golden mole), Elephantulus edwardii (Cape elephant shrew), Orycteropus afer (aardvark), Macropus eugenii (wallaby), Sarcophilus harrisii (Tasmanian devil), Monodelphis domestica (gray short-tailed opossum), Ornithorhynchus anatinus (platypus). In the Aves class (birds): Gallus gallus (chicken), Meleagris gallopavo (turkey), Anas platyrhynchos (mallard/duck), Columba livia (domestic pigeon), Falco peregrinus (peregrine falcon), Falco cherrug (saker falcon), Corvus brachyrhynchos (American crow), Pseudopodoces humilis (ground tit), Ficedula albicollis (collared flycatcher), Taeniopygia guttata (zebra finch), Geospiza fortis (medium ground-finch), Zonotrichia albicollis (white-throated sparrow), Melopsittacus undulatus (budgerigar), Calypte anna (Anna's hummingbird). Page 8 of 10
9 In the Reptilia class: Chrysemys picta bellii (painted turtle), Pelodiscus sinensis (Chinese softshell turtle), Chelonia mydas (green sea turtle), Anolis carolinensis (green anole), Alligator mississippiensis (American alligator), Alligator sinensis (Chinese alligator), Python bivittatus (Burmese python), Ophiophagus hannah (king cobra). In the Amphibia class: Xenopus tropicalis (western clawed frog), Xenopus laevis (African clawed frog). In the Actinopterygii class (ray-finned fishes); Fugu rubripes (Japanese pufferfish), Tetraodon nigroviridis (a similar brackish species of green spotted pufferfish), Gadus morhua (Atlantic cod), Gasterosteus aculeatus (stickleback), Oreochromis niloticus (Nile tilapia), Danio rerio (zebrafish), Carassius auratus (goldfish), Oryzias latipes (Japanese medaka), Poecilia reticulata (guppy), Astyanax mexicanus (Mexican tetra/blind cave fish), Maylandia zebra (zebra mbuna), Poecilia formosa (Amazon molly), Haplochromis burtoni (Burton's mouthbrooder), Pundamilia nyererei (cichlid), Xiphophorus maculatusi (platyfish), Apteronotus leptorhynchus (brown ghost knifefish), Lepisosteus oculatusi (spotted gar), Cynoglossus semilaevis (tongue sole). In the Sarcopterygii class (lobe-finned fishes): Latimeria chalumnae (coelacanth). In the Chondrichthyes class (cartilaginous fishes): Callorhinchus milii (elephant shark). Agnatha NMDA receptor subunits In the Petromyzontida class: Petromyzon marinus (sea lamprey), the corresponding residues of palimitoyation sites at Cys cluster II have not been determined. (C) All cloned and predicted KA receptor subunit GluK2 orthologs in each class are shown below. GluK2 subunit has two distinct palmitoylation sites in their C-termini: GluK2 Cys858, Cys671 (amino acid residues in the mouse GluK2 sequence as representative). Gnathostome KA receptor subunit GluK2 In the Mammalia class: Homo sapiens (human), Pan paniscus (bonobo), Pan troglodytes (chimpanzee), Gorilla gorilla (western lowland gorilla), Pongo abelii (Sumatran orangutan), Nomascus leucogenys (northern white-cheeked gibbon), Macaca fascicularis (crab-eating macaque), Macaca mulatta (rhesus monkey), Chlorocebus sabaeus (green monkey), Papio anubis (olive baboon), Callithrix jacchus (white-tufted-ear marmoset), Saimiri boliviensis (black-capped squirrel monkey), Tarsius syrichta (tarsier), Otolemur garnettii (northern greater galago), Galeopterus variegatus (Sunda flying lemur), Tupaia belangeri (northern treeshrew), Tupaia chinensis (Chinese treeshrew), Spermophilus tridecemlineatus / Ictidomys tridecemlineatus (thirteen-lined ground squirrel), Cricetulus griseus (Chinese hamster), Mesocricetus auratus (golden hamster), Microtus ochrogaster (prairie vole), Rattus norvegicus (Norway rat), Mus musculus (house mouse), Jaculus jaculus (lesser Egyptian jerboa), Peromyscus maniculatus bairdii (deer mouse), Chinchilla lanigera (chinchilla), Dipodomys ordii (kangaroo rat), Cavia porcellus (guinea pig), Octodon degus (degu), Heterocephalus glaber (naked mole rat), Oryctolagus cuniculus (rabbit), Ochotona princeps (American pika), Erinaceus europaeus (European hedgehog), Sorex araneus (European shrew), Condylura cristata (star-nosed mole), Eptesicus fuscus (big brown bat), Pteropus vampyrus (megabat), Pteropus alecto (black flying fox), Myotis lucifugus (little brown bat), Myotis brandtii (Brandt's bat), Myotis davidii (mouse-eared bat), Felis catus (cat), Panthera tigris altaica (Amur tiger), Canis lupus familiaris (dog), Mustela putorius furo (ferret), Ursus maritimus (polar bear), Ailuropoda melanoleuca (giant panda), Leptonychotes weddellii (Weddell seal), Odobenus rosmarus divergens (walrus), Equus caballus (horse), Equus przewalskii (Przewalski's horse), Ceratotherium simum (white rhinoceros), Bos taurus (cattle), Bos mutus (yak), Bubalus bubalis (water buffalo), Ovis aries (sheep), Capra hircus (goat), Pantholops hodgsonii (chiru/tibetan antelope), Tursiops truncates (dolphin), Orcinus orca (killer whale), Physeter catodon (sperm whale), Lipotes vexillifer (baiji), Lipotes vexillifer (Yangtze River dolphin), Balaenoptera acutorostrata (minke whale), Sus scrofa (wild boar), Camelus ferus (wild Bactrian camel), Vicugna pacos (alpaca), Choloepus hoffmanni (two-toed sloth), Dasypus novemcinctus (armadillo), Loxodonta africana (African elephant), Trichechus manatus latirostris (Florida manatee), Procavia capensis (rock hyrax), Echinops telfairi (lesser hedgehog tenrec), Chrysochloris asiatica (Cape golden mole), Elephantulus edwardii (Cape elephant shrew), Orycteropus afer (aardvark), Macropus eugenii (wallaby), Sarcophilus harrisii (Tasmanian devil), Monodelphis domestica (gray short-tailed opossum), Ornithorhynchus anatinus (platypus). In the Aves class (birds): Gallus gallus (chicken), Meleagris gallopavo (turkey), Anas platyrhynchos (mallard/duck), Columba livia (domestic pigeon), Falco peregrinus (peregrine falcon), Falco cherrug (saker falcon), Corvus brachyrhynchos (American crow), Pseudopodoces humilis (ground tit), Ficedula albicollis (collared flycatcher), Taeniopygia guttata (zebra finch), Geospiza fortis (medium ground-finch), Zonotrichia albicollis (white-throated sparrow), Melopsittacus undulatus (budgerigar), Calypte anna (Anna's hummingbird). Page 9 of 10
10 In the Reptilia class: Chrysemys picta bellii (painted turtle), Pelodiscus sinensis (Chinese softshell turtle), Chelonia mydas (green sea turtle), Anolis carolinensis (green anole), Alligator mississippiensis (American alligator), Alligator sinensis (Chinese alligator), Python bivittatus (Burmese python), Ophiophagus hannah (king cobra). In the Amphibia class: Xenopus tropicalis (western clawed frog), Xenopus laevis (African clawed frog). In the Actinopterygii class (ray-finned fishes); Fugu rubripes (Japanese pufferfish), Tetraodon nigroviridis (a similar brackish species of green spotted pufferfish), Gadus morhua (Atlantic cod), Gasterosteus aculeatus (stickleback), Oreochromis niloticus (Nile tilapia), Danio rerio (zebrafish), Oryzias latipes (Japanese medaka), Poecilia reticulata (guppy), Astyanax mexicanus (Mexican tetra/blind cave fish), Maylandia zebra (zebra mbuna), Poecilia formosa (Amazon molly), Haplochromis burtoni (Burton's mouthbrooder), Pundamilia nyererei (cichlid), Xiphophorus maculatusi (platyfish), Lepisosteus oculatusi (spotted gar), Cynoglossus semilaevis (tongue sole). In the Sarcopterygii class (lobe-finned fishes): Latimeria chalumnae (coelacanth). In the Chondrichthyes class (cartilaginous fishes): Callorhinchus milii (elephant shark). Agnatha KA receptor subunit GluK2 In the Myxini class: Paramyxine yangi (brown hagfish), the corresponding residues of palimitoyation sites have not been determined. In the Petromyzontida class: Petromyzon marinus (sea lamprey), the corresponding residues of palimitoyation sites have not been determined. Page 10 of 10
Supplemental Figure 1.
 Supplemental Material: Annu. Rev. Genet. 2015. 49:213 42 doi: 10.1146/annurev-genet-120213-092023 A Uniform System for the Annotation of Vertebrate microrna Genes and the Evolution of the Human micrornaome
Supplemental Material: Annu. Rev. Genet. 2015. 49:213 42 doi: 10.1146/annurev-genet-120213-092023 A Uniform System for the Annotation of Vertebrate microrna Genes and the Evolution of the Human micrornaome
Comparing Genomes! Homologies and Families! Sequence Alignments!
 Comparing Genomes! Homologies and Families! Sequence Alignments! Allows us to achieve a greater understanding of vertebrate evolution! Tells us what is common and what is unique between different species
Comparing Genomes! Homologies and Families! Sequence Alignments! Allows us to achieve a greater understanding of vertebrate evolution! Tells us what is common and what is unique between different species
Mammalogy: the study of the evolution, ecology, physiology, and anatomy of members of the Class Mammalia (Chordata, Vertebrata).
 Mammalogy: the study of the evolution, ecology, physiology, and anatomy of members of the Class Mammalia (Chordata, Vertebrata). Mammalogy has been of practical interest to humans since our ancestors evolved
Mammalogy: the study of the evolution, ecology, physiology, and anatomy of members of the Class Mammalia (Chordata, Vertebrata). Mammalogy has been of practical interest to humans since our ancestors evolved
Combination of X-ray crystallography, SAXS and DEER to obtain the structure of the FnIII-3,4 domains of integrin α6β4
 Acta Cryst. (2015). D71, 969-985, doi:10.1107/s1399004715002485 Supporting information Volume 71 (2015) Supporting information for article: Combination of X-ray crystallography, SAXS and DEER to obtain
Acta Cryst. (2015). D71, 969-985, doi:10.1107/s1399004715002485 Supporting information Volume 71 (2015) Supporting information for article: Combination of X-ray crystallography, SAXS and DEER to obtain
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/7/e1602878/dc1 Supplementary Materials for Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades Michael J. Gaudry,
advances.sciencemag.org/cgi/content/full/3/7/e1602878/dc1 Supplementary Materials for Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades Michael J. Gaudry,
Expanded View Figures
 Marie-Thérèse El-aher et al non-polyq fragments and Huntington s disease The EMO Journal Expanded View Figures HTT-Q TEV rec + + + + αtub 15 5 37 sh-htt cells 167/586 167/586 TEV-Q -Q23 Particle volume
Marie-Thérèse El-aher et al non-polyq fragments and Huntington s disease The EMO Journal Expanded View Figures HTT-Q TEV rec + + + + αtub 15 5 37 sh-htt cells 167/586 167/586 TEV-Q -Q23 Particle volume
Master Biomedizin ) UCSC & UniProt 2) Homology 3) MSA 4) Phylogeny. Pablo Mier
 Master Biomedizin 2018 1) UCSC & UniProt 2) Homology 3) MSA 4) 1 12 a. All of the sequences in file1.fasta (https://cbdm.uni-mainz.de/mb18/) are homologs. How many groups of orthologs would you say there
Master Biomedizin 2018 1) UCSC & UniProt 2) Homology 3) MSA 4) 1 12 a. All of the sequences in file1.fasta (https://cbdm.uni-mainz.de/mb18/) are homologs. How many groups of orthologs would you say there
Phylogenetic tree construction based on amino acid composition and nucleotide content of complete vertebrate mitochondrial genomes
 IOSR Journal Of Pharmacy (e)-issn: 2250-3013, (p)-issn: 2319-4219 Www.Iosrphr.Org Volume 3, Issue 6 (July 2013), Pp 51-60 Phylogenetic tree construction based on amino acid composition and nucleotide content
IOSR Journal Of Pharmacy (e)-issn: 2250-3013, (p)-issn: 2319-4219 Www.Iosrphr.Org Volume 3, Issue 6 (July 2013), Pp 51-60 Phylogenetic tree construction based on amino acid composition and nucleotide content
Phylogenetic analysis of uroporphyrinogen III synthase (UROS) gene
 www.bioinformation.net Hypothesis Volume 8(25) Phylogenetic analysis of uroporphyrinogen III synthase (UROS) gene Abjal Pasha Shaik 1,$, *, Abbas H Alsaeed 1$ & Asma Sultana 2$ 1Department of Clinical
www.bioinformation.net Hypothesis Volume 8(25) Phylogenetic analysis of uroporphyrinogen III synthase (UROS) gene Abjal Pasha Shaik 1,$, *, Abbas H Alsaeed 1$ & Asma Sultana 2$ 1Department of Clinical
Biased amino acid composition in warm-blooded animals
 Biased amino acid composition in warm-blooded animals Guang-Zhong Wang and Martin J. Lercher Bioinformatics group, Heinrich-Heine-University, Düsseldorf, Germany Among eubacteria and archeabacteria, amino
Biased amino acid composition in warm-blooded animals Guang-Zhong Wang and Martin J. Lercher Bioinformatics group, Heinrich-Heine-University, Düsseldorf, Germany Among eubacteria and archeabacteria, amino
Genome Visualization in Space
 Genome Visualization in Space Leandro S. Marcolino 1, Brá ulio R. G. M. Couto 2 and Marcos A. dos Santos 1 Departamento de Ciência da Computação, Universidade Federal de Minas Gerais / UFMG 1 ; Programa
Genome Visualization in Space Leandro S. Marcolino 1, Brá ulio R. G. M. Couto 2 and Marcos A. dos Santos 1 Departamento de Ciência da Computação, Universidade Federal de Minas Gerais / UFMG 1 ; Programa
Bioinformatics Report Branchiostoma lanceolatum dopamine D 1 / receptor protein phylogenetic analysis. Alanna Lewis
 Bioinformatics Report Branchiostoma lanceolatum dopamine D 1 / receptor protein phylogenetic analysis Alanna Lewis 0 Abstract: Dopamine is an essential neurotransmitter for many species of chordates. The
Bioinformatics Report Branchiostoma lanceolatum dopamine D 1 / receptor protein phylogenetic analysis Alanna Lewis 0 Abstract: Dopamine is an essential neurotransmitter for many species of chordates. The
Marine medaka ATP-binding cassette (ABC) superfamily and new insight into teleost Abch nomenclature
 Supplementary file for: Marine medaka ATP-binding cassette (ABC) superfamily and new insight into teleost Abch nomenclature Chang-Bum Jeong a,b,#, Bo-Mi Kim a,#, Hye-Min Kang a, Ik-Young Choi c, Jae-Sung
Supplementary file for: Marine medaka ATP-binding cassette (ABC) superfamily and new insight into teleost Abch nomenclature Chang-Bum Jeong a,b,#, Bo-Mi Kim a,#, Hye-Min Kang a, Ik-Young Choi c, Jae-Sung
Rapid birth-and-death evolution of the xenobiotic metabolizing NAT gene family in vertebrates with evidence of adaptive selection
 Sabbagh et al. BMC Evolutionary Biology 2013, 13:62 RESEARCH ARTICLE Open Access Rapid birth-and-death evolution of the xenobiotic metabolizing NAT gene family in vertebrates with evidence of adaptive
Sabbagh et al. BMC Evolutionary Biology 2013, 13:62 RESEARCH ARTICLE Open Access Rapid birth-and-death evolution of the xenobiotic metabolizing NAT gene family in vertebrates with evidence of adaptive
Orthologous loci for phylogenomics from raw NGS data
 Orthologous loci for phylogenomics from raw NS data Rachel Schwartz The Biodesign Institute Arizona State University Rachel.Schwartz@asu.edu May 2, 205 Big data for phylogenetics Phylogenomics requires
Orthologous loci for phylogenomics from raw NS data Rachel Schwartz The Biodesign Institute Arizona State University Rachel.Schwartz@asu.edu May 2, 205 Big data for phylogenetics Phylogenomics requires
Cubic Spline Interpolation Reveals Different Evolutionary Trends of Various Species
 Cubic Spline Interpolation Reveals Different Evolutionary Trends of Various Species Zhiqiang Li 1 and Peter Z. Revesz 1,a 1 Department of Computer Science, University of Nebraska-Lincoln, Lincoln, NE,
Cubic Spline Interpolation Reveals Different Evolutionary Trends of Various Species Zhiqiang Li 1 and Peter Z. Revesz 1,a 1 Department of Computer Science, University of Nebraska-Lincoln, Lincoln, NE,
Sheet1. Page 1. protein
 build 1 version 1 Ab initio model Ab initio GPIPE/6085/1.1/gnomon_prot build 1 version 1 Build GPIPE/6085/1.1/ Acyrthosiphon pisum build 2 version 1 Build GPIPE/7029/2.1/ Acyrthosiphon pisum build 2.1
build 1 version 1 Ab initio model Ab initio GPIPE/6085/1.1/gnomon_prot build 1 version 1 Build GPIPE/6085/1.1/ Acyrthosiphon pisum build 2 version 1 Build GPIPE/7029/2.1/ Acyrthosiphon pisum build 2.1
Mechanisms of Evolution Darwinian Evolution
 Mechanisms of Evolution Darwinian Evolution Descent with modification by means of natural selection All life has descended from a common ancestor The mechanism of modification is natural selection Concept
Mechanisms of Evolution Darwinian Evolution Descent with modification by means of natural selection All life has descended from a common ancestor The mechanism of modification is natural selection Concept
Complex evolutionary history of the vertebrate sweet/umami taste receptor genes
 Article SPECIAL ISSUE Adaptive Evolution and Conservation Ecology of Wild Animals doi: 10.1007/s11434-013-5811-5 Complex evolutionary history of the vertebrate sweet/umami taste receptor genes FENG Ping
Article SPECIAL ISSUE Adaptive Evolution and Conservation Ecology of Wild Animals doi: 10.1007/s11434-013-5811-5 Complex evolutionary history of the vertebrate sweet/umami taste receptor genes FENG Ping
TRANSPOSABLE ELEMENTS DYNAMICS IN TAXA WITH DIFFERENT REPRODUCTIVE STRATEGIES OR SPECIATION RATE
 Alma Mater Studiorum Università di Bologna DOTTORATO DI RICERCA IN BIODIVERSITÀ ED EVOLUZIONE Ciclo XXV Settore scientifico-disciplinare di afferenza: BIO-05 Zoologia Settore concorsuale di afferenza:
Alma Mater Studiorum Università di Bologna DOTTORATO DI RICERCA IN BIODIVERSITÀ ED EVOLUZIONE Ciclo XXV Settore scientifico-disciplinare di afferenza: BIO-05 Zoologia Settore concorsuale di afferenza:
Biology Keystone (PA Core) Quiz Theory of Evolution - (BIO.B ) Theory Of Evolution, (BIO.B ) Scientific Terms
 Biology Keystone (PA Core) Quiz Theory of Evolution - (BIO.B.3.2.1 ) Theory Of Evolution, (BIO.B.3.3.1 ) Scientific Terms Student Name: Teacher Name: Jared George Date: Score: 1) Evidence for evolution
Biology Keystone (PA Core) Quiz Theory of Evolution - (BIO.B.3.2.1 ) Theory Of Evolution, (BIO.B.3.3.1 ) Scientific Terms Student Name: Teacher Name: Jared George Date: Score: 1) Evidence for evolution
Supplementary Material
 Evolution of substrate specificity in the Nucleobase-Ascorbate Transporter (NAT) protein family Anezia Kourkoulou, Alexandros A. Pittis & George Diallinas Supplementary Material Supplementary Figure S1.
Evolution of substrate specificity in the Nucleobase-Ascorbate Transporter (NAT) protein family Anezia Kourkoulou, Alexandros A. Pittis & George Diallinas Supplementary Material Supplementary Figure S1.
Supporting Information
 Supporting Information Chen et al. 1.173/pnas.11379113 Fig. S1. Life cycle of the social amoeba D. discoideum. D. discoideum amoebae propagate vegetatively as a unicellular organism when food (naturally
Supporting Information Chen et al. 1.173/pnas.11379113 Fig. S1. Life cycle of the social amoeba D. discoideum. D. discoideum amoebae propagate vegetatively as a unicellular organism when food (naturally
Research Article Placenta-Specific Protein 1 Is Conserved throughout the Placentalia under Purifying Selection
 e Scientific World Journal, Article ID 537356, 5 pages http://dx.doi.org/10.1155/2014/537356 Research Article Placenta-Specific Protein 1 Is Conserved throughout the Placentalia under Purifying Selection
e Scientific World Journal, Article ID 537356, 5 pages http://dx.doi.org/10.1155/2014/537356 Research Article Placenta-Specific Protein 1 Is Conserved throughout the Placentalia under Purifying Selection
Major Gene Families in Humans and Their Evolutionary History Prof. Yoshihito Niimura Prof. Masatoshi Nei
 Major Gene Families in Humans Yoshihito Niimura Tokyo Medical and Dental University and Masatoshi Nei Pennsylvania State University 1 1. Multigene family Contents 2. Olfactory receptors (ORs) 3. OR genes
Major Gene Families in Humans Yoshihito Niimura Tokyo Medical and Dental University and Masatoshi Nei Pennsylvania State University 1 1. Multigene family Contents 2. Olfactory receptors (ORs) 3. OR genes
CONSTRUCTION OF PHYLOGENETIC TREE FROM MULTIPLE GENE TREES USING PRINCIPAL COMPONENT ANALYSIS
 INTERNATIONAL JOURNAL OF ELECTRONICS AND COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) Proceedings of the International Conference on Emerging Trends in Engineering and Management (ICETEM14) ISSN 0976
INTERNATIONAL JOURNAL OF ELECTRONICS AND COMMUNICATION ENGINEERING & TECHNOLOGY (IJECET) Proceedings of the International Conference on Emerging Trends in Engineering and Management (ICETEM14) ISSN 0976
Cladistics and Bioinformatics Questions 2013
 AP Biology Name Cladistics and Bioinformatics Questions 2013 1. The following table shows the percentage similarity in sequences of nucleotides from a homologous gene derived from five different species
AP Biology Name Cladistics and Bioinformatics Questions 2013 1. The following table shows the percentage similarity in sequences of nucleotides from a homologous gene derived from five different species
AP Biology. Evolution is "so overwhelmingly established that it has become irrational to call it a theory." Evidence of Evolution by Natural Selection
 Evidence of Evolution by Natural Selection Evolution is "so overwhelmingly established that it has become irrational to call it a theory." -- Ernst Mayr What Evolution Is 2001 Professor Emeritus, Evolutionary
Evidence of Evolution by Natural Selection Evolution is "so overwhelmingly established that it has become irrational to call it a theory." -- Ernst Mayr What Evolution Is 2001 Professor Emeritus, Evolutionary
Bayesian models for molecular evolution Rates, rates and traits
 Bayesian models for molecular evolution Rates, rates and traits Nicolas Lartillot March 20, 2012 Nicolas Lartillot (Universite de Montréal) BCM2004: Molecular evolution March 20, 2012 1 / 38 1 Molecular
Bayesian models for molecular evolution Rates, rates and traits Nicolas Lartillot March 20, 2012 Nicolas Lartillot (Universite de Montréal) BCM2004: Molecular evolution March 20, 2012 1 / 38 1 Molecular
Cyclin-Dependent Kinase
 Cyclin-Dependent Kinase (At a glance) Cory Camasta Cyclin-Dependent Kinase Family E.C: 2.7.11.22 2 = Transferase 2.7 = Transfer of phosphate group 2.7.11 = Serine/Threonine Kinase 2.7.11.22 = Cyclin-dependent
Cyclin-Dependent Kinase (At a glance) Cory Camasta Cyclin-Dependent Kinase Family E.C: 2.7.11.22 2 = Transferase 2.7 = Transfer of phosphate group 2.7.11 = Serine/Threonine Kinase 2.7.11.22 = Cyclin-dependent
Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. [version 2; referees: 3 approved]
![Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. [version 2; referees: 3 approved] Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. [version 2; referees: 3 approved]](/thumbs/92/111047392.jpg) OBSERVATION ARTICLE Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. [version 2; referees: 3 approved] Leos Kral Department of Biology, University
OBSERVATION ARTICLE Possible identification of CENP-C in fish and the presence of the CENP-C motif in M18BP1 of vertebrates. [version 2; referees: 3 approved] Leos Kral Department of Biology, University
Evidence of Evolution by Natural Selection. Dodo bird
 Evidence of Evolution by Natural Selection Dodo bird 2007-2008 Evidence supporting evolution Fossil record transition species Anatomical record homologous & vestigial structures embryology & development
Evidence of Evolution by Natural Selection Dodo bird 2007-2008 Evidence supporting evolution Fossil record transition species Anatomical record homologous & vestigial structures embryology & development
CLASSIFICATION. Finding Order in Diversity
 CLASSIFICATION Finding Order in Diversity WHAT IS TAXONOMY? Discipline of classifying organisms and assigning each organism a universally accepted name. WHY CLASSIFY? To study the diversity of life, biologists
CLASSIFICATION Finding Order in Diversity WHAT IS TAXONOMY? Discipline of classifying organisms and assigning each organism a universally accepted name. WHY CLASSIFY? To study the diversity of life, biologists
Face area (cm 2 ) Brain surface area (cm 2 ) Cranial capacity (cm 3 ) 1, Jaw Angle ( º )
 Honors Biology Test : Evolution GOOD LUCK! You ve learned so much! Multiple Choice: Identify the choice that best completes the statement or answers the question. (2 pts each) 1. As we move through the
Honors Biology Test : Evolution GOOD LUCK! You ve learned so much! Multiple Choice: Identify the choice that best completes the statement or answers the question. (2 pts each) 1. As we move through the
Phylogenetic Trees. How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species?
 Why? Phylogenetic Trees How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species? The saying Don t judge a book by its cover. could be applied
Why? Phylogenetic Trees How do the changes in gene sequences allow us to reconstruct the evolutionary relationships between related species? The saying Don t judge a book by its cover. could be applied
Evolutionary analysis of the genomic region of FOXP3 in vertebrates
 University of Arkansas, Fayetteville ScholarWorks@UARK Biological Sciences Undergraduate Honors Theses Biological Sciences 5-2015 Evolutionary analysis of the genomic region of FOXP3 in vertebrates Ethan
University of Arkansas, Fayetteville ScholarWorks@UARK Biological Sciences Undergraduate Honors Theses Biological Sciences 5-2015 Evolutionary analysis of the genomic region of FOXP3 in vertebrates Ethan
Genomic organization and evolution of ruminant lysozyme c genes
 ZOOLOGICAL RESEARCH Genomic organization and evolution of ruminant lysozyme c genes David M. IRWIN 1,2,* 1 Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Canada 2 Banting
ZOOLOGICAL RESEARCH Genomic organization and evolution of ruminant lysozyme c genes David M. IRWIN 1,2,* 1 Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Canada 2 Banting
Evidence for Evolution by Natural Selection Regents Biology
 Evidence for Evolution by Natural Selection Objective: Determine the different types of evidence for proving evolution Evidence supporting evolution Fossil record shows change over time Comparative Anatomy
Evidence for Evolution by Natural Selection Objective: Determine the different types of evidence for proving evolution Evidence supporting evolution Fossil record shows change over time Comparative Anatomy
Phylogeny 9/8/2014. Evolutionary Relationships. Data Supporting Phylogeny. Chapter 26
 Phylogeny Chapter 26 Taxonomy Taxonomy: ordered division of organisms into categories based on a set of characteristics used to assess similarities and differences Carolus Linnaeus developed binomial nomenclature,
Phylogeny Chapter 26 Taxonomy Taxonomy: ordered division of organisms into categories based on a set of characteristics used to assess similarities and differences Carolus Linnaeus developed binomial nomenclature,
Vertebrate genome sequencing: building a backbone for comparative genomics
 104 Forum Web Watch Vertebrate genome sequencing: building a backbone for comparative genomics James W. Thomas and Jeffrey W. Touchman The human genome sequence provides a reference point from which we
104 Forum Web Watch Vertebrate genome sequencing: building a backbone for comparative genomics James W. Thomas and Jeffrey W. Touchman The human genome sequence provides a reference point from which we
Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST
 Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST Introduction Bioinformatics is a powerful tool which can be used to determine evolutionary relationships and
Investigation 3: Comparing DNA Sequences to Understand Evolutionary Relationships with BLAST Introduction Bioinformatics is a powerful tool which can be used to determine evolutionary relationships and
DO NOT WRITE ON THIS. Evidence from Evolution Activity. The Fossilization Process. Types of Fossils
 Evidence from Evolution Activity Part 1 - Fossils Use the diagrams on the next page to answer the following questions IN YOUR NOTEBOOK. 1. Describe how fossils form. 2. Describe the different types of
Evidence from Evolution Activity Part 1 - Fossils Use the diagrams on the next page to answer the following questions IN YOUR NOTEBOOK. 1. Describe how fossils form. 2. Describe the different types of
Anthro 101: Human Biological Evolution. Lecture 7: Taxonomy/Primate Adaptations. Prof. Kenneth Feldmeier
 Anthro 101: Human Biological Evolution Lecture 7: Taxonomy/Primate Adaptations Prof. Kenneth Feldmeier Here is the PLAN Listen to this lecture and read about Taxonomy in the text I will ask you a question(s)
Anthro 101: Human Biological Evolution Lecture 7: Taxonomy/Primate Adaptations Prof. Kenneth Feldmeier Here is the PLAN Listen to this lecture and read about Taxonomy in the text I will ask you a question(s)
Dynamic evolution of the GnRH receptor gene family in vertebrates
 Williams et al. BMC Evolutionary Biology 2014, 14:215 RESEARCH ARTICLE Open Access Dynamic evolution of the GnRH receptor gene family in vertebrates Barry L Williams 1,2, Yasuhisa Akazome 3, Yoshitaka
Williams et al. BMC Evolutionary Biology 2014, 14:215 RESEARCH ARTICLE Open Access Dynamic evolution of the GnRH receptor gene family in vertebrates Barry L Williams 1,2, Yasuhisa Akazome 3, Yoshitaka
An Evolutionary Trend Discovery Algorithm Based on Cubic Spline Interpolation
 An Evolutionary Trend Discovery Algorithm Based on Cubic Spline Interpolation ZHIQIANG LI and PETER Z. REVESZ Department of Computer Science and Engineering University of Nebraska-Lincoln Lincoln, NE 68588-0115
An Evolutionary Trend Discovery Algorithm Based on Cubic Spline Interpolation ZHIQIANG LI and PETER Z. REVESZ Department of Computer Science and Engineering University of Nebraska-Lincoln Lincoln, NE 68588-0115
Exploring evolution of brain genes involved in microcephaly through phylogeny and synteny analysis
 Rauf and Mir Theoretical Biology and Medical Modelling 2013, 10:61 RESEARCH Open Access Exploring evolution of brain genes involved in microcephaly through phylogeny and synteny analysis Sobiah Rauf and
Rauf and Mir Theoretical Biology and Medical Modelling 2013, 10:61 RESEARCH Open Access Exploring evolution of brain genes involved in microcephaly through phylogeny and synteny analysis Sobiah Rauf and
Lecture 16: Again on Regression
 Lecture 16: Again on Regression S. Massa, Department of Statistics, University of Oxford 10 February 2016 The Normality Assumption Body weights (Kg) and brain weights (Kg) of 62 mammals. Species Body weight
Lecture 16: Again on Regression S. Massa, Department of Statistics, University of Oxford 10 February 2016 The Normality Assumption Body weights (Kg) and brain weights (Kg) of 62 mammals. Species Body weight
The sex-determining locus in the tiger pufferfish, Takifugu rubripes. Kiyoshi Asahina and Yuzuru Suzuki
 The sex-determining locus in the tiger pufferfish, Takifugu rubripes Kiyoshi Kikuchi*, Wataru Kai*, Ayumi Hosokawa*, Naoki Mizuno, Hiroaki Suetake, Kiyoshi Asahina and Yuzuru Suzuki Fisheries Laboratory,
The sex-determining locus in the tiger pufferfish, Takifugu rubripes Kiyoshi Kikuchi*, Wataru Kai*, Ayumi Hosokawa*, Naoki Mizuno, Hiroaki Suetake, Kiyoshi Asahina and Yuzuru Suzuki Fisheries Laboratory,
7 th Grade SCIENCE FINAL REVIEW Ecology, Evolution, Classification
 7 th Grade SCIENCE FINAL REVIEW Ecology, Evolution, Classification ECOLOGY Students will be able to: Define species, population, community and ecosystem. species organisms that can mate and produce fertile
7 th Grade SCIENCE FINAL REVIEW Ecology, Evolution, Classification ECOLOGY Students will be able to: Define species, population, community and ecosystem. species organisms that can mate and produce fertile
Group activities: Making animal model of human behaviors e.g. Wine preference model in mice
 Lecture schedule 3/30 Natural selection of genes and behaviors 4/01 Mouse genetic approaches to behavior 4/06 Gene-knockout and Transgenic technology 4/08 Experimental methods for measuring behaviors 4/13
Lecture schedule 3/30 Natural selection of genes and behaviors 4/01 Mouse genetic approaches to behavior 4/06 Gene-knockout and Transgenic technology 4/08 Experimental methods for measuring behaviors 4/13
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/312/5780/1653/dc1 Supporting Online Material for The Xist RNA Gene Evolved in Eutherians by Pseudogenization of a Protein-Coding Gene Laurent Duret,* Corinne Chureau,
www.sciencemag.org/cgi/content/full/312/5780/1653/dc1 Supporting Online Material for The Xist RNA Gene Evolved in Eutherians by Pseudogenization of a Protein-Coding Gene Laurent Duret,* Corinne Chureau,
Anthro 101: Human Biological Evolution. Lecture 7: Taxonomy/Primate Adaptations. Prof. Kenneth Feldmeier
 Anthro 101: Human Biological Evolution Lecture 7: Taxonomy/Primate Adaptations Prof. Kenneth Feldmeier Here is the deal, read though the lecture and hopefully the audio works on youtube Classifying species
Anthro 101: Human Biological Evolution Lecture 7: Taxonomy/Primate Adaptations Prof. Kenneth Feldmeier Here is the deal, read though the lecture and hopefully the audio works on youtube Classifying species
Bayesian models for macroevolutionary studies
 Bayesian models for macroevolutionary studies Nicolas Lartillot February 28, 2012 Nicolas Lartillot (Universite de Montréal) Integrative models of macroevolution February 28, 2012 1 / 47 The molecular
Bayesian models for macroevolutionary studies Nicolas Lartillot February 28, 2012 Nicolas Lartillot (Universite de Montréal) Integrative models of macroevolution February 28, 2012 1 / 47 The molecular
GATA family of transcription factors of vertebrates: phylogenetics and chromosomal synteny
 Phylogenetics and chromosomal synteny of the GATAs 1273 GATA family of transcription factors of vertebrates: phylogenetics and chromosomal synteny CHUNJIANG HE, HANHUA CHENG* and RONGJIA ZHOU* Department
Phylogenetics and chromosomal synteny of the GATAs 1273 GATA family of transcription factors of vertebrates: phylogenetics and chromosomal synteny CHUNJIANG HE, HANHUA CHENG* and RONGJIA ZHOU* Department
Evidence of Evolution by Natural Selection. Evidence supporting evolution. Fossil record. Fossil record. Anatomical record.
 Evidence of Evolution by Natural Selection Dodo bird Evidence supporting evolution Fossil record transition species Anatomical record homologous & vestigial structures embryology & development Molecular
Evidence of Evolution by Natural Selection Dodo bird Evidence supporting evolution Fossil record transition species Anatomical record homologous & vestigial structures embryology & development Molecular
Evolution and divergence of the mammalian SAMD9/SAMD9L gene family
 Lemos de Matos et al. BMC Evolutionary Biology 2013, 13:121 RESEARCH ARTICLE Evolution and divergence of the mammalian SAMD9/SAMD9L gene family Ana Lemos de Matos 1,2,3, Jia Liu 3, Grant McFadden 3 and
Lemos de Matos et al. BMC Evolutionary Biology 2013, 13:121 RESEARCH ARTICLE Evolution and divergence of the mammalian SAMD9/SAMD9L gene family Ana Lemos de Matos 1,2,3, Jia Liu 3, Grant McFadden 3 and
Preliminary Assessment Report of an Archaeofauna from Eyri, lsafjord, NW Iceland
 Preliminary Assessment Report of an Archaeofauna from Eyri, lsafjord, NW Iceland Yekaterina Thomas H. McGovern CUNY Northern Science and Education Center NORSEC CUNY Doctoral Program in Anthropology Brooklyn
Preliminary Assessment Report of an Archaeofauna from Eyri, lsafjord, NW Iceland Yekaterina Thomas H. McGovern CUNY Northern Science and Education Center NORSEC CUNY Doctoral Program in Anthropology Brooklyn
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Zn 2+ -binding sites in USP18. (a) The two molecules of USP18 present in the asymmetric unit are shown. Chain A is shown in blue, chain B in green. Bound Zn 2+ ions are shown as
Supplementary Figure 1 Zn 2+ -binding sites in USP18. (a) The two molecules of USP18 present in the asymmetric unit are shown. Chain A is shown in blue, chain B in green. Bound Zn 2+ ions are shown as
Gene expression differences in human and chimpanzee cerebral cortex
 Evolution of the human genome by natural selection What you will learn in this lecture (1) What are the human genome and positive selection? (2) How do we analyze positive selection? (3) How is positive
Evolution of the human genome by natural selection What you will learn in this lecture (1) What are the human genome and positive selection? (2) How do we analyze positive selection? (3) How is positive
Introduction to Molecular Evolution (BIOL 3046) Course website:
 Introduction to Molecular Evolution (BIOL 3046) Course website: www.biol3046.info What is evolution? Evolution: from Latin evolvere, to unfold broad sense, to change e.g., stellar evolution, cultural evolution,
Introduction to Molecular Evolution (BIOL 3046) Course website: www.biol3046.info What is evolution? Evolution: from Latin evolvere, to unfold broad sense, to change e.g., stellar evolution, cultural evolution,
1 ATGGGTCTC 2 ATGAGTCTC
 We need an optimality criterion to choose a best estimate (tree) Other optimality criteria used to choose a best estimate (tree) Parsimony: begins with the assumption that the simplest hypothesis that
We need an optimality criterion to choose a best estimate (tree) Other optimality criteria used to choose a best estimate (tree) Parsimony: begins with the assumption that the simplest hypothesis that
Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are:
 Comparative genomics and proteomics Species available Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are: Vertebrates: human, chimpanzee, mouse, rat,
Comparative genomics and proteomics Species available Ensembl focuses on metazoan (animal) genomes. The genomes currently available at the Ensembl site are: Vertebrates: human, chimpanzee, mouse, rat,
b. In Table 1 (question #2 on the Answer Sheet describe the function of each set of bones and answer the question.)
 Biology EVIDENCE OF EVOLUTION INTRODUCTION: Evidence has been found to indicate that living things have changed gradually during their natural history. The study of fossils as well as embryology, biochemistry,
Biology EVIDENCE OF EVOLUTION INTRODUCTION: Evidence has been found to indicate that living things have changed gradually during their natural history. The study of fossils as well as embryology, biochemistry,
Application of new distance matrix to phylogenetic tree construction
 Application of new distance matrix to phylogenetic tree construction P.V.Lakshmi Computer Science & Engg Dept GITAM Institute of Technology GITAM University Andhra Pradesh India Allam Appa Rao Jawaharlal
Application of new distance matrix to phylogenetic tree construction P.V.Lakshmi Computer Science & Engg Dept GITAM Institute of Technology GITAM University Andhra Pradesh India Allam Appa Rao Jawaharlal
Evolution. Darwin s Voyage
 Evolution Darwin s Voyage Charles Darwin Explorer on an observation trip to the Galapagos Islands. He set sail on the HMS Beagle in 1858 from England on a 5 year trip. He was a naturalist (a person who
Evolution Darwin s Voyage Charles Darwin Explorer on an observation trip to the Galapagos Islands. He set sail on the HMS Beagle in 1858 from England on a 5 year trip. He was a naturalist (a person who
Station 1: Evidence from Current Examples
 Station 1: Evidence from Current Examples Go to the website below: http://www.pbs.org/wgbh/evolution/educators/lessons/lesson6/act1.html Watch the video segment called Why does evolution matter now? After
Station 1: Evidence from Current Examples Go to the website below: http://www.pbs.org/wgbh/evolution/educators/lessons/lesson6/act1.html Watch the video segment called Why does evolution matter now? After
Evidence of Common Ancestry Stations
 Stations Scientists have long wondered where organisms came from and how they evolved. One of the main sources of evidence for the evolution of organisms comes from the fossil record. Thousands of layers
Stations Scientists have long wondered where organisms came from and how they evolved. One of the main sources of evidence for the evolution of organisms comes from the fossil record. Thousands of layers
Evidence for Evolution
 Evidence for Evolution Evolution Biological evolution is descent with modification. It is important to remember that: Humans did not evolve from chimpanzees. Humans and chimpanzees are evolutionary cousins
Evidence for Evolution Evolution Biological evolution is descent with modification. It is important to remember that: Humans did not evolve from chimpanzees. Humans and chimpanzees are evolutionary cousins
Evolution of the Sry gene within the African pygmy mice Nannomys
 Evolution of the Sry gene within the African pygmy mice Nannomys Subgenus of the genus Mus Widespread in Sub Saharan Africa ~ 20 species Mus minutoides Very high proportion (> 75%) of fertile sex reversed
Evolution of the Sry gene within the African pygmy mice Nannomys Subgenus of the genus Mus Widespread in Sub Saharan Africa ~ 20 species Mus minutoides Very high proportion (> 75%) of fertile sex reversed
Fixation of Deleterious Mutations at Critical Positions in Human Proteins
 Fixation of Deleterious Mutations at Critical Positions in Human Proteins Author Sankarasubramanian, Sankar Published 2011 Journal Title Molecular Biology and Evolution DOI https://doi.org/10.1093/molbev/msr097
Fixation of Deleterious Mutations at Critical Positions in Human Proteins Author Sankarasubramanian, Sankar Published 2011 Journal Title Molecular Biology and Evolution DOI https://doi.org/10.1093/molbev/msr097
Small RNA in rice genome
 Vol. 45 No. 5 SCIENCE IN CHINA (Series C) October 2002 Small RNA in rice genome WANG Kai ( 1, ZHU Xiaopeng ( 2, ZHONG Lan ( 1,3 & CHEN Runsheng ( 1,2 1. Beijing Genomics Institute/Center of Genomics and
Vol. 45 No. 5 SCIENCE IN CHINA (Series C) October 2002 Small RNA in rice genome WANG Kai ( 1, ZHU Xiaopeng ( 2, ZHONG Lan ( 1,3 & CHEN Runsheng ( 1,2 1. Beijing Genomics Institute/Center of Genomics and
Vertebrates can discriminate among thousands of different odor
 Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods Yoshihito Niimura* and Masatoshi Nei Institute of Molecular Evolutionary Genetics and Department of Biology, Pennsylvania State
Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods Yoshihito Niimura* and Masatoshi Nei Institute of Molecular Evolutionary Genetics and Department of Biology, Pennsylvania State
Investigating Evolutionary Relationships between Species through the Light of Graph Theory based on the Multiplet Structure of the Genetic Code
 07 IEEE 7th International Advance Computing Conference Investigating Evolutionary Relationships between Species through the Light of Graph Theory based on the Multiplet Structure of the Genetic Code Antara
07 IEEE 7th International Advance Computing Conference Investigating Evolutionary Relationships between Species through the Light of Graph Theory based on the Multiplet Structure of the Genetic Code Antara
Chapter 26 Phylogeny and the Tree of Life
 Chapter 26 Phylogeny and the Tree of Life Biologists estimate that there are about 5 to 100 million species of organisms living on Earth today. Evidence from morphological, biochemical, and gene sequence
Chapter 26 Phylogeny and the Tree of Life Biologists estimate that there are about 5 to 100 million species of organisms living on Earth today. Evidence from morphological, biochemical, and gene sequence
Genomes and Their Evolution
 Chapter 21 Genomes and Their Evolution PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Chapter 21 Genomes and Their Evolution PowerPoint Lecture Presentations for Biology Eighth Edition Neil Campbell and Jane Reece Lectures by Chris Romero, updated by Erin Barley with contributions from
Classification. Rules of Classification
 ification Taxonomy=the science of classifying living things Hierarchical system of ification Carl Linnaeus (1750 s) Father of ification Botanist Binomial names (Scientific Names) Latin Carl Linnaeus Rules
ification Taxonomy=the science of classifying living things Hierarchical system of ification Carl Linnaeus (1750 s) Father of ification Botanist Binomial names (Scientific Names) Latin Carl Linnaeus Rules
Drosophila melanogaster and D. simulans, two fruit fly species that are nearly
 Comparative Genomics: Human versus chimpanzee 1. Introduction The chimpanzee is the closest living relative to humans. The two species are nearly identical in DNA sequence (>98% identity), yet vastly different
Comparative Genomics: Human versus chimpanzee 1. Introduction The chimpanzee is the closest living relative to humans. The two species are nearly identical in DNA sequence (>98% identity), yet vastly different
Molecular evolution. Joe Felsenstein. GENOME 453, Autumn Molecular evolution p.1/49
 Molecular evolution Joe Felsenstein GENOME 453, utumn 2009 Molecular evolution p.1/49 data example for phylogeny inference Five DN sequences, for some gene in an imaginary group of species whose names
Molecular evolution Joe Felsenstein GENOME 453, utumn 2009 Molecular evolution p.1/49 data example for phylogeny inference Five DN sequences, for some gene in an imaginary group of species whose names
Science in Motion Ursinus College
 Science in Motion Ursinus College NAME EVIDENCE FOR EVOLUTION LAB INTRODUCTION: Evolution is not just a historical process; it is occurring at this moment. Populations constantly adapt in response to changes
Science in Motion Ursinus College NAME EVIDENCE FOR EVOLUTION LAB INTRODUCTION: Evolution is not just a historical process; it is occurring at this moment. Populations constantly adapt in response to changes
Introduction Principles of Signaling and Organization p. 3 Signaling in Simple Neuronal Circuits p. 4 Organization of the Retina p.
 Introduction Principles of Signaling and Organization p. 3 Signaling in Simple Neuronal Circuits p. 4 Organization of the Retina p. 5 Signaling in Nerve Cells p. 9 Cellular and Molecular Biology of Neurons
Introduction Principles of Signaling and Organization p. 3 Signaling in Simple Neuronal Circuits p. 4 Organization of the Retina p. 5 Signaling in Nerve Cells p. 9 Cellular and Molecular Biology of Neurons
Evidence of Species Change
 Evidence of Species Change Evidence of Evolution What is evolution? Evolution is change over time Scientific theory of evolution explains how living things descended from earlier organisms Evidence of
Evidence of Species Change Evidence of Evolution What is evolution? Evolution is change over time Scientific theory of evolution explains how living things descended from earlier organisms Evidence of
A comprehensive analysis of teleost MHC class I sequences
 Grimholt et al. BMC Evolutionary Biology (2015) 15:32 DOI 10.1186/s12862-015-0309-1 RESEARCH ARTICLE Open Access A comprehensive analysis of teleost MHC class I sequences Unni Grimholt 1*, Kentaro Tsukamoto
Grimholt et al. BMC Evolutionary Biology (2015) 15:32 DOI 10.1186/s12862-015-0309-1 RESEARCH ARTICLE Open Access A comprehensive analysis of teleost MHC class I sequences Unni Grimholt 1*, Kentaro Tsukamoto
Evidence for Evolution by Natural Selection. Raven Chapters 1 & 22
 Evidence for Evolution by Natural Selection Raven Chapters 1 & 22 2006-2007 Science happens within a culture What was the doctrine of the time? TINTORETTO The Creation of the Animals 1550 Then along comes
Evidence for Evolution by Natural Selection Raven Chapters 1 & 22 2006-2007 Science happens within a culture What was the doctrine of the time? TINTORETTO The Creation of the Animals 1550 Then along comes
Evolution. 1. The figure below shows the classification of several types of prairie dogs.
 Name: Date: 1. The figure below shows the classification of several types of prairie dogs. 3. Which statement describes the best evidence that two species share a recent common ancestor? A. The species
Name: Date: 1. The figure below shows the classification of several types of prairie dogs. 3. Which statement describes the best evidence that two species share a recent common ancestor? A. The species
HUMAN EVOLUTION. Where did we come from?
 HUMAN EVOLUTION Where did we come from? www.christs.cam.ac.uk/darwin200 Darwin & Human evolution Darwin was very aware of the implications his theory had for humans. He saw monkeys during the Beagle voyage
HUMAN EVOLUTION Where did we come from? www.christs.cam.ac.uk/darwin200 Darwin & Human evolution Darwin was very aware of the implications his theory had for humans. He saw monkeys during the Beagle voyage
Emergence of Xin Demarcates a Key Innovation in Heart Evolution
 Emergence of Xin Demarcates a Key Innovation in Heart Evolution Shaun E. Grosskurth, Debashish Bhattacharya, Qinchuan Wang, Jim Jung-Ching Lin* Department of Biology, University of Iowa, Iowa City, Iowa,
Emergence of Xin Demarcates a Key Innovation in Heart Evolution Shaun E. Grosskurth, Debashish Bhattacharya, Qinchuan Wang, Jim Jung-Ching Lin* Department of Biology, University of Iowa, Iowa City, Iowa,
Primate Diversity & Human Evolution (Outline)
 Primate Diversity & Human Evolution (Outline) 1. Source of evidence for evolutionary relatedness of organisms 2. Primates features and function 3. Classification of primates and representative species
Primate Diversity & Human Evolution (Outline) 1. Source of evidence for evolutionary relatedness of organisms 2. Primates features and function 3. Classification of primates and representative species
Chapter 16: Reconstructing and Using Phylogenies
 Chapter Review 1. Use the phylogenetic tree shown at the right to complete the following. a. Explain how many clades are indicated: Three: (1) chimpanzee/human, (2) chimpanzee/ human/gorilla, and (3)chimpanzee/human/
Chapter Review 1. Use the phylogenetic tree shown at the right to complete the following. a. Explain how many clades are indicated: Three: (1) chimpanzee/human, (2) chimpanzee/ human/gorilla, and (3)chimpanzee/human/
Signatures of Natural Selection in a Primate Bitter Taste Receptor
 J Mol Evol (2011) 73:257 265 DOI 10.1007/s00239-011-9481-0 Signatures of Natural Selection in a Primate Bitter Taste Receptor Stephen Wooding Received: 10 August 2011 / Accepted: 14 December 2011 / Published
J Mol Evol (2011) 73:257 265 DOI 10.1007/s00239-011-9481-0 Signatures of Natural Selection in a Primate Bitter Taste Receptor Stephen Wooding Received: 10 August 2011 / Accepted: 14 December 2011 / Published
Evidence of EVOLUTION
 Evidence of EVOLUTION Evolution: Genetic change in a population through time Charles Darwin On his journey around the world, Darwin found evidence of GRADUAL CHANGE (evolution) He cited evidences he found
Evidence of EVOLUTION Evolution: Genetic change in a population through time Charles Darwin On his journey around the world, Darwin found evidence of GRADUAL CHANGE (evolution) He cited evidences he found
Phylogenetic Analyses Reveal Ancient Duplication of Estrogen Receptor Isoforms
 J Mol Evol (1999) 49:609 614 Springer-Verlag New York Inc. 1999 Phylogenetic Analyses Reveal Ancient Duplication of Estrogen Receptor Isoforms Scott T. Kelley, 1, * Varykina G. Thackray 2, * 1 Department
J Mol Evol (1999) 49:609 614 Springer-Verlag New York Inc. 1999 Phylogenetic Analyses Reveal Ancient Duplication of Estrogen Receptor Isoforms Scott T. Kelley, 1, * Varykina G. Thackray 2, * 1 Department
Hands-On Nine The PAX6 Gene and Protein
 Hands-On Nine The PAX6 Gene and Protein Main Purpose of Hands-On Activity: Using bioinformatics tools to examine the sequences, homology, and disease relevance of the Pax6: a master gene of eye formation.
Hands-On Nine The PAX6 Gene and Protein Main Purpose of Hands-On Activity: Using bioinformatics tools to examine the sequences, homology, and disease relevance of the Pax6: a master gene of eye formation.
Supplementary information
 Supplementary information Superoxide dismutase 1 is positively selected in great apes to minimize protein misfolding Pouria Dasmeh 1, and Kasper P. Kepp* 2 1 Harvard University, Department of Chemistry
Supplementary information Superoxide dismutase 1 is positively selected in great apes to minimize protein misfolding Pouria Dasmeh 1, and Kasper P. Kepp* 2 1 Harvard University, Department of Chemistry
ISSN (Online) ISSN (Print) *Corresponding author Padma Saxena
 Scholars Academic Journal of Biosciences (SAJB) Sch. Acad. J. Biosci., 2014; 2(4): 290-294 Scholars Academic and Scientific Publisher (An International Publisher for Academic and Scientific Resources)
Scholars Academic Journal of Biosciences (SAJB) Sch. Acad. J. Biosci., 2014; 2(4): 290-294 Scholars Academic and Scientific Publisher (An International Publisher for Academic and Scientific Resources)
General Patterns in Evolution
 General Patterns in Evolution Uses of Phylogenetic Analysis Allows mapping order of character state changes Documents evolutionary trends in development Reveals that Homoplasy is common Can attempt to
General Patterns in Evolution Uses of Phylogenetic Analysis Allows mapping order of character state changes Documents evolutionary trends in development Reveals that Homoplasy is common Can attempt to
Sequence motif analysis
 Sequence motif analysis Alan Moses Associate Professor and Canada Research Chair in Computational Biology Departments of Cell & Systems Biology, Computer Science, and Ecology & Evolutionary Biology Director,
Sequence motif analysis Alan Moses Associate Professor and Canada Research Chair in Computational Biology Departments of Cell & Systems Biology, Computer Science, and Ecology & Evolutionary Biology Director,
Open Access. Kenji Sorimachi 1,2,* and Teiji Okayasu 3
 Send Orders for Reprints to reprints@benthamscience.ae Current Chemical Genomics and Translational Medicine, 2015, 9, 1-5 1 Open Access Evidence for Natural Selection in Nucleotide Content Relationships
Send Orders for Reprints to reprints@benthamscience.ae Current Chemical Genomics and Translational Medicine, 2015, 9, 1-5 1 Open Access Evidence for Natural Selection in Nucleotide Content Relationships
Name: Period: Evidence for Evolution Part I. Introduction
 Name: Evidence for Evolution Period: Part I. Introduction Two of the most important ideas in evolution are as follows: 1) Every species is a modified (changed) descendant of a species that existed before.
Name: Evidence for Evolution Period: Part I. Introduction Two of the most important ideas in evolution are as follows: 1) Every species is a modified (changed) descendant of a species that existed before.
In the path of organic evolution, gene and genome
 Pakistan J. Zool., vol. 48(5), pp. 1331-1335, 2016 Transcription of Dynein Heavy Chain 9 (DNAH9) in Paralichthys olivaceus: Insights into the Evolution of the DNAH9 Gene Zhenwei Wang, Conghui Liu, Jingjing
Pakistan J. Zool., vol. 48(5), pp. 1331-1335, 2016 Transcription of Dynein Heavy Chain 9 (DNAH9) in Paralichthys olivaceus: Insights into the Evolution of the DNAH9 Gene Zhenwei Wang, Conghui Liu, Jingjing
