Utah School of Medicine, Salt Lake City, UT Tel.: ; Fax: ;
|
|
|
- Bruno Rogers
- 6 years ago
- Views:
Transcription
1 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 281, NO. 25, pp , June 23, by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Dimeric Dnm1-G385D Interacts with Mdv1 on Mitochondria and Can Be Stimulated to Assemble into Fission Complexes Containing Mdv1 and Fis1 * Received for publication, December 20, 2005, and in revised form, February 27, 2006 Published, JBC Papers in Press, April 6, 2006, DOI /jbc.M Debjani Bhar, Mary Anne Karren, Markus Babst, and Janet M. Shaw 1 From the Biochemistry Department, University of Utah School of Medicine, Salt Lake City, Utah and Biology Department, University of Utah, Salt Lake City, Utah Interactions between yeast Dnm1p, Mdv1p, and Fis1p are required to form fission complexes that catalyze division of the mitochondrial compartment. During the formation of mitochondrial fission complexes, the Dnm1p GTPase self-assembles into large multimeric complexes on the outer mitochondrial membrane that are visualized as punctate structures by fluorescent labeling. Although it is clear that Fis1p Mdv1p complexes on mitochondria are required for the initial recruitment of Dnm1p, it is not clear whether Dnm1p puncta assemble before or after this recruitment step. Here we show that the minimum oligomeric form of cytoplasmic Dnm1p is a dimer. The middle domain mutant protein Dnm1 G385D p forms dimers in vivo but fails to assemble into punctate structures. However, this dimeric mutant stably interacts with Mdv1p on the outer mitochondrial membrane, demonstrating that assembly of stable Dnm1p multimers is not required for Dnm1p- Mdv1p association or for mitochondrial recruitment of Dnm1p. Dnm1 G385D p is reported to be a terminal dimer in vitro. We describe conditions that allow assembly of Dnm1 G385D p into functional fission complexes on mitochondria in vivo. Using these conditions, we demonstrate that multimerization of Dnm1p is required to promote reorganization of Mdv1p from a uniform mitochondrial localization into punctate fission complexes. Our studies also reveal that Fis1p is present in these assembled fission complexes. Based on our results, we propose that Dnm1p dimers are initially recruited to the membrane via interaction with Mdv1p Fis1p complexes. These dimers then assemble into multimers that subsequently promote the reorganization of Mdv1p into punctate fission complexes. Mitochondrial fission controls organelle shape, copy number, and function in most eukaryotic cells. Recent studies in mammalian cells highlight the importance of mitochondrial fission in programmed cell death (1 5), calcium homeostasis (6, 7), and neuronal development and function (8). Although the molecular machinery regulating mitochondrial fission is conserved from fungi to mammals, a mechanistic understanding of distinct steps in mitochondrial fission complex assembly has come mainly from studies in the budding yeast, Saccharomyces cerevisiae. Three proteins are known to be essential for mitochondrial fission in yeast. Dnm1p is a conserved GTPase related to dynamin and contains * This work was funded by National Institutes of Health Grant GM53466 (to J. M. S.) and American Heart Association Grant N (to M. B.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact. 1 To whom correspondence should be addressed: Dept. of Biochemistry, University of Utah School of Medicine, Salt Lake City, UT Tel.: ; Fax: ; shaw@biochem.utah.edu. an N-terminal GTPase domain, a middle domain, and an -helical/ GTPase effector domain (9 12). Fis1p is a conserved tail-anchored outer mitochondrial membrane protein with a single tetratricopeptide repeat-like fold facing the cytoplasm (13, 14). Mdv1p contains an N-terminal extension, a domain predicted to form coiled-coils, and seven WD repeats predicted to form a -propeller (15 17). A paralog of Mdv1p called Caf4p also binds both Dnm1p and Fis1p but is not essential for fission (18). Thus, Caf4p either plays a regulatory role in fission or is only required under specific, as yet unidentified physiological conditions. Interactions between these three proteins are required to form fission complexes that catalyze mitochondrial division. Fis1p initially recruits Mdv1p to the membrane, and this Fis1p Mdv1p complex mediates recruitment of Dnm1p (13, 15, 17 19). By fluorescence microscopy, Dnm1-GFP can be visualized as punctate complexes at discrete sites along mitochondria (9, 11, 20). In this study we refer to these punctate structures as Dnm1p multimers, although we do not exclude the possibility that cells may contain additional multimeric Dnm1p structures that are too small to visualize. In addition, large Dnm1-GFP punctate structures can be observed moving rapidly in the cytoplasm (21 23). Sometime after Dnm1p multimers appear on mitochondria, Mdv1p accumulates at these sites followed by mitochondrial fission (15, 17, 23). We refer to punctate structures on mitochondria containing both Mdv1p and Dnm1p as mitochondrial fission complexes. Interestingly, Fis1p has not been visualized in punctate fission complexes along with Dnm1p and Mdv1p, although it is thought to mediate post-assembly steps in mitochondrial fission (13, 19, 24). This finding raises the possibility that Fis1p physically interacts with the fission complex components before but not after their assembly into punctate fission complexes. Dnm1p molecules are proposed to cycle between small oligomeric subunits in the cytoplasm and the large, multimeric fission complexes that are found at mitochondrial constriction sites during fission (11). In support of this idea, purified Dnm1p forms a dimer that can further assemble into large, multimeric rings and spirals (25). The minimum oligomeric state of cytoplasmic Dnm1p in vivo is not known. However, the predominant cytoplasmic form of both dynamin (26) and Drp1 (27), the mammalian Dnm1p homolog, is reported to be tetramer. Although significant progress has been made, several key steps in fission complex assembly are not understood. First, it is not clear whether Dnm1p recruitment to mitochondria occurs before or after formation of large Dnm1p multimers. Second, the specific event required to stimulate Mdv1p reorganization from a uniform to a punctate appearance in fission complexes has not yet been defined. Third, the physical presence of Fis1p in the assembled fission complex has not been demonstrated. In this study we show that the minimum oligomeric form of cytoplasmic Dnm1p is a dimer. Using the dimeric middle domain mutant JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 281 NUMBER 25 JUNE 23, 2006
2 Dnm1 G385D p, we demonstrate that Dnm1p dimers are effectively recruited to mitochondria and stably interact with Mdv1p. Although Dnm1 G385D p is reported to be a terminal dimer in vitro (25), we describe conditions that promote assembly of Dnm1 G385D p into complexes containing Mdv1p and Fis1p, some of which catalyze fission. Thus, we have defined a role for the middle domain in stabilization of Dnm1p selfinteractions that lead to the formation of Dnm1p multimers. By controlling the assembly state of Dnm1 G385D p, we provide the first direct demonstration that Mdv1p reorganization into punctate fission complexes is stimulated by Dnm1p multimerization on the mitochondrial membrane. Our results support a model of fission complex assembly in which Dnm1p dimers are initially recruited to Fis1p Mdv1p complexes on mitochondria, where they self-assemble further into multimeric structures. These structures then promote the reorganization of Mdv1p into punctate fission complexes that also contain Fis1p. EXPERIMENTAL PROCEDURES Strain and Plasmid Construction Yeast strains used in this study were derived from the FY genetic background (28). Standard methods were used for growth, transformation, and genetic manipulation of S. cerevisiae (29, 30) and Escherichia coli (31). To construct prs425 3MYC-DNM1, an XhoI/SacII fragment of prs316 3MYC-DNM1 (provided by R. Jensen and H. Sesaki) was ligated into prs425 digested with XhoI/SacII. prs426 DNM1 3HA was constructed as described (10). prs415 DNM1 G385D, prs425 3MYC-DNM1 G385D, and prs426 DNM1 G385D -3HA were created by site-directed mutagenesis (QuikChange site-directed mutagenesis kit, Stratagene) using plasmids prs415 DNM1, prs425 3MYC-DNM1, and prs426 DNM1 3HA (10) as template. prs416 DNM1 G385D -GFP was created by sitedirected mutagenesis (Stratagene) using prs416 DNM1-GFP as template. Integration of DNM1 G385D at the DNM1 locus was carried out using a two-step replacement technique. First, the DNM1 open reading frame was replaced with URA3 using standard homologous recombination techniques (32). Second, URA3 was replaced with DNM1 G385D by homologous recombination, and integrants were selected by growth on 5-fluoroorotic acid. Mutations, disruptions, and replacements were confirmed by PCR, DNA sequencing, and Western blotting (where appropriate). Fluorescence Microscopy and Quantification of Mitochondrial Phenotypes Mitochondrial morphology was analyzed and quantified in cells expressing a fast folding mitochondrial matrix-targeted form of red fluorescent protein (mt-rfp) 2 from p414gpd-mtrfpff (19) or a mitochondrial targeted form of green fluorescent protein (mt-gfp) from pvt100u-gfp2 plasmid (provided by B. Westermann). For in vivo localization of Dnm1p, Dnm1-GFP was expressed from prs416 (13). For in vivo localization of Mdv1p, GFP-Mdv1p was expressed from prs415-met25 (19, 33). Expression of GFP-Mdv1p from the un-induced MET25 promoter produced sufficient GFP-Mdv1p (2 5-fold increase over endogenous Mdv1p levels) for visualization with no adverse effects on mitochondrial morphology or fission protein localization. Before quantification, strains were grown overnight at 30 C, diluted to 0.5 A 600, and grown for an additional 2 3 h into log phase. For experiments using sodium azide (NaN 3 ) and sodium fluoride (NaF), cells were grown in the appropriate synthetic media to log phase and incubated with 10 mm NaN 3 and 10 mm NaF at 30 C for 15 min before observation. Mitochondrial morphology and fission protein localization were visualized using a Zeiss Axioplan 2 Imaging microscope (Carl 2 The abbreviations used are: mt-rfp, mitochondrial matrix-targeted red fluorescent protein; GFP, green fluorescent protein; IP, immunoprecipitation; HA, hemagglutinin; DSP, dithiobis(succinimidyl propionate). Zeiss) (34). Images were captured, processed by deconvolution, and assembled as described previously (34). For each strain, 3 independent experiments of 100 cells each were performed, the average and S.D. of which are reported. Co-immunoprecipitation Analysis Co-immunoprecipitation (co-ip) assays were performed from the indicated yeast strains grown in the appropriate synthetic media to a density of A 600. For Dnm1p- Dnm1p co-ips, dnm1 cells were transformed with the following plasmids: prs426 DNM1 3HA (10), prs425 3MYC-DNM1, prs426 DNM1 G385D -3HA, and prs425 3MYC-DNM1 G385D. Co-IPs were performed from whole cell lysates as described previously (10) using anti-ha-agarose-conjugated beads (Sigma). For Dnm1p-Fis1p co-ips with cross-linking, yeast strains expressing Dnm1p (DNM1) or Dnm1 G385D p(dnm1 G385D ) from the genomic locus were transformed with either prs415met25 9MYC-FIS1 (13) or prs415met25 plasmids. For Dnm1p-Mdv1p co-ips with cross-linking, dnm1 cells expressing endogenous Mdv1p were transformed with either prs425 3MYC-DNM1 or prs425 MYC-DNM1 G385D plasmids. Dnm1p-Fis1p co-ips and Dnm1p-Mdv1p co-ips were performed as described previously (19). Briefly, log phase cells growing in synthetic dextrose media were harvested at 4200 g for 5 min and incubated in buffer TD (100 mm Tris-SO 4, ph 9.4, 10 mm dithiothreitol) for 15 min at 30 C. After washing twice with buffer SP (1.2 M sorbitol, 20 mm potassium phosphate, ph 7.4), cells were incubated with zymolyase (6 mg/ml) for 40 min at 30 C. Spheroplasts were treated with 2.5 mm dithiobis(succinimidyl propionate) (DSP) (Pierce) for 30 min, incubated with 50 mm glycine for 15 min to quench the cross-linking reaction, and homogenized in buffer SH (1.2 M sorbitol, 40 mm HEPES-KOH, ph 7.4). 50 A 600 units of homogenized cells were centrifuged at 12,500 g for 10 min. The pellet was solubilized with IP buffer (50 mm Tris-HCl, ph 7.5, 100 mm NaCl, 5 mm EDTA, 0.07% Triton X-100) containing 1:500 protein inhibitor mixture (Calbiochem) for 15 min, and the supernatant was incubated with anti-c-myc conjugated agarose beads for 1 h. Where indicated, spheroplasts were treated with 10 mm NaN 3 /NaF for 15 min at 30 C before dithiobis(succinimidyl propionate) cross-linking. Co-IP samples were analyzed by 10% SDS-PAGE and Western blotting using anti-ha (University of Utah), anti-myc (Santa Cruz Biotechnology), anti-porin (Molecular Probes, Inc.), anti-mdv1p (J. Nunnari), and affinity-purified anti-dnm1p (11) antibodies as indicated. Gel Filtration Analysis 100 A 600 of each indicated yeast strain in log phase was spheroplasted and lysed in phosphate-buffered saline (137 mm NaCl, 2.7 mm KCl, 4.3 mm Na 2 HPO 4, 1.4 mm KH 2 PO 4, ph 7.4) containing 1:100 protease inhibitor mixture (Sigma) as described previously (35). Cleared whole cell lysates were centrifuged at 100,000 g for 30 min, and the resulting supernatant ( 3 mg/ml) was loaded on a Sephacryl-S300 column (16/60; Amersham Biosciences) and separated in the presence of phosphate-buffered saline (35). Proteins in collected fractions were precipitated in 10% trichloroacetic acid (TCA), washed with cold acetone to remove residual TCA, resuspended in sample buffer, separated by 10% SDS-PAGE, and analyzed by Western blotting using affinity-purified anti-dnm1p antibody. The molecular mass of Dnm1p or Dnm1 G385D p was calculated based on the elution profile of high molecular weight gel filtration standards (Amersham Biosciences) run separately. RESULTS Mutation of a Conserved Glycine in the Dnm1p Middle Domain Blocks Protein Function and Causes Dominant Fission Defects The middle domain of Dnm1p contains a glycine residue (Gly-385) that is conserved among members of the dynamin family (Fig. 1A). In genetic JUNE 23, 2006 VOLUME 281 NUMBER 25 JOURNAL OF BIOLOGICAL CHEMISTRY 17313
3 FIGURE 2. Dnm1 G385D p forms a complex with itself in a co-immunoprecipitation assay. Myc-tagged and HA-tagged forms of Dnm1p or Dnm1 G385D p were co-expressed in dnm1 cells. HA-tagged Dnm1 proteins were immunoprecipitated using anti-ha agarose beads. Co-IP of Myc-tagged Dnm1 proteins was detected by immunoblot using anti-myc antibodies (top panel). Immunoprecipitation of HA-tagged Dnm1p or Dnm1 G385D p was detected with anti-ha antibodies (middle panel). Western blot shows total loaded (L), flow-through (FT), and eluted (E) proteins. The mitochondrial outer membrane protein porin is provided as a loading control (bottom panel). The asterisk denotes nonspecific binding of 3Myc-Dnm1p to beads. FIGURE 1. Mutation of the conserved Gly-385 residue of Dnm1p causes dominant localization and fission defects. A, alignment of a middle domain fragment of Dnm1p with Dynamin family members from yeast to humans. Conserved glycine residue Gly- 385 is shaded gray. S.c., S. cerevisiae; S.p., Saccharomyces pombe; H.s., Homo sapiens; C.e., Caenorhabditis elegans; A.t., Arabidopsis thaliana. B, quantification of mitochondrial morphology in DNM1 and dnm1 strains expressing Dnm1p or Dnm1 G385D p. C, GFP localization in cells expressing Dnm1-GFP or Dnm1 G385D -GFP. Differential interference contrast (DIC; left panel), GFP (middle panel), and GFP merged with mt-rfp (right panel) images are shown. The percentage of cells in a population that display visible punctate GFP structures co-localized with mitochondria is indicated at the far right. In B and C, three independent experiments were performed, and 100 cells were counted per experiment (S.D. is indicated). Genes indicated are genomic unless preceded by p, which denotes plasmid-borne. screens, our laboratory detected the substitution of glycine 385 to aspartate (G385D) 144 independent times, indicating the importance of this residue for middle domain function. Although the DNM1 G385D mutation is reported to cause dominant mitochondrial fission defects (22, 23, 25), a detailed characterization of the activity of the Dnm1 G385D protein in vivo has not been performed. Wild-type yeast cells contain tubular mitochondrial networks that are maintained by balanced fission and fusion events (12, 22, 36 43). When fission is blocked, ongoing fusion converts mitochondria into interconnected nets, which often collapse to one side of the cell (9, 11, 12). Using a mitochondrial-matrix-targeted form of red fluorescent protein (mt-rfp), we determined the effect of Dnm1 G385D ponmitochondrial morphology in the presence of wild-type Dnm1p. Cells containing branched and tubular mitochondria were scored as fission competent, whereas cells with net-like or collapsed mitochondria were scored as fission-incompetent. As shown in Fig. 1B, the majority of cells expressing Dnm1p from the genomic locus or a low copy plasmid contained fission-competent mitochondria. By contrast, only 9% of cells expressing Dnm1 G385D p in the presence of wild-type Dnm1p had fission competent mitochondria, indicating that Dnm1 G385D p causes severe dominant fission defects. In a dnm1 background, Dnm1 G385D p displayed very little fission activity, suggesting that this protein likely blocks activity of the native fission machinery. The inability of Dnm1 G385D p to support fission was not due to reduced protein levels, as Western blot analyses of whole cell extracts from these strains confirmed that similar levels of Dnm1p and Dnm1 G385D p were expressed at steady state (data not shown). Unlike wild-type Dnm1-GFP, which assembled into multimeric punctate structures on the outer mitochondrial membrane (Fig. 1C), the Dnm1 G385D -GFP protein localized diffusely in the cytoplasm. Thus, residue Gly-385 in the middle domain is critical for stable assembly of Dnm1p on mitochondria. However, the G385D mutation does not prevent Dnm1 G385D p from interacting with wild-type Dnm1p. In cells expressing both wild-type Dnm1-GFP and mutant Dnm1 G385D p, the Dnm1-GFP protein was restricted to the cytoplasm in 60% of the cells (data not shown), suggesting that Dnm1 G385D p mediates dominant effects by binding to and sequestering the wild-type protein in nonfunctional complexes. Dnm1p and Dnm1 G385D p Self-interact and Form Cytoplasmic Dimers at Steady State In a previous study, we demonstrated that Dnm1p forms complexes with itself in vivo (10). In this study, co-ip studies were used to determine whether the G385D mutation affected Dnm1p complex formation with itself. Fig. 2 shows that co-ip of 3Myc-Dnm1 G385D p with Dnm1 G385D -3HAp (Fig. 2, lane 6, -Myc) was only slightly less efficient than co-ip of 3Myc-Dnm1p with Dnm1 3HAp (Fig. 2 lane 3, -Myc), indicating that Dnm1 G385D proteins are able to form complexes in vivo. These interactions were specific, as we did not observe significant co-ip of porin (an outer membrane protein) under these conditions (Fig. 2, lanes 3, 6, 9, 12, -porin) or of 3Myc-tagged Dnm1p when an HA-tagged protein was absent (Fig. 2, lane 12, -Myc). In separate experiments, mutant 3Myc-Dnm1 G385D p co-immunoprecipitated efficiently with wild-type Dnm1 3HAp (data not shown), consistent with the idea that Dnm1 G385D p is dominant negative because it binds to and interferes with the function of the wild-type Dnm1 protein. Although Dnm1 G385D p formed complexes in the co-immunoprecipitation assay, we were unable to detect Dnm1 G385D -GFP assembled into punctate structures. A recent in vitro study reported that MGRS-10His-HA-Dnm1p purified from insect cells formed dimers that further assembled into rings and spirals in low salt buffer (25). These ring-like structures are presumed to represent the punctate JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 281 NUMBER 25 JUNE 23, 2006
4 mitochondrial fission complexes observed in vivo. In the same study, mutant MGRS-10His-HA-Dnm1 G385D p formed dimers but could not assemble further. In independent studies, we examined the oligomeric state of untagged, cytoplasmic Dnm1p and Dnm1 G385D p using gel filtration analysis of cytoplasmic extracts. In wild-type extracts, the majority of Dnm1p reproducibly eluted at a fraction corresponding to the 157-kDa molecular mass standard (Fig. 3A, top panel, fraction 6, arrow). The predicted molecular mass of monomeric Dnm1p is 85 kda (Fig. 3A, top panel, fraction 8, arrowhead). Assuming that Dnm1p is globular, this elution profile suggests that the majority of Dnm1p exists as a dimer in the cytosol. This Dnm1p elution profile was unaltered in cells lacking the two other fission proteins, Mdv1p and Fis1p, indicating that these proteins are not required for Dnm1p dimer formation in vivo (Fig. 3A, middle and bottom panels). Importantly, Dnm1 G385D p was also able to dimerize FIGURE 3. Dnm1p and Dnm1 G385D p behave as homodimers in gel-filtration assays. Cytoplasmic extracts from the indicated strains were fractionated on a Sephacryl-300 column and analyzed by Western blotting using affinity-purified anti-dnm1p antibodies. Dnm1p monomers are expected to elute in fraction 8 (arrowhead). Dnm1p dimers are expected to elute in fraction 6 (157kD, arrow). A, Dnm1p elution profile from wild-type (WT; top panel), mdv1 (middle panel), and fis1 (bottom panel) strains expressing endogenous Dnm1p. B, Dnm1 G385D p elution profile from dnm1 (top panel), dnm1 mdv1 (middle panel), and dnm1 fis1 (bottom panel) strains expressing plasmid-borne Dnm1 G385D p from the endogenous promoter. The asterisk denotes full-length protein. The filled circle denotes breakdown product. and eluted in the 157-kDa fraction even in the absence of Mdv1p and Fis1p (Fig. 3B). Relative to the Dnm1p profile, a larger portion of Dnm1 G385D p eluted in early fractions corresponding to a slightly larger molecular weight in the dnm1 strain background (Fig. 3B, top panel, fractions 5 and 6). Although the physiological relevance of this observation is currently unclear, these results demonstrate that the minimal dimeric form of Dnm1p is not disrupted by the G385D middle domain substitution. It is important to note that the S300 column used in this gel filtration experiment only resolves small Dnm1p cytoplasmic oligomers and not large complexes. Therefore, this experiment does not address the oligomeric state of the large, rapidly moving punctate structures visualized by Dnm1p-GFP in live cells. Sodium Azide/Sodium Fluoride Treatment Promotes Assembly of Dnm1 G385D -GFP into Mitochondrial Fission Complexes Although Dnm1 G385D p proteins interact to form cytosolic dimers, these dimers do not form visible punctate complexes. It is possible that Dnm1 G385D p dimers never interact further to form multimers. Alternatively, Dnm1 G385D p may form multimers that are unstable and rapidly dissociate, shifting the equilibrium to the dimeric form. In vitro, dynamin family members including Dnm1p assemble into stable rings and spirals in the absence of nucleotide or the presence of GDP and disassemble upon GTP hydrolysis (25, 44, 45). We reasoned that if Dnm1 G385D p forms unstable complexes, then depleting cells of GTP might stabilize these complexes, allowing their visualization. To test this idea, cells were treated with sodium azide and sodium fluoride (NaN 3 /NaF), which inhibits the cytochrome oxidase complex and enzymes of the tricarboxylic acid cycle, respectively. As a consequence, ATP production is rapidly abolished, and ATP pools required for GTP production are depleted (46 50). We examined the organization and distribution of Dnm1-GFP or Dnm1 G385D -GFP in cells depleted of GTP by a brief 10 mm NaN 3 /NaF treatment. As shown in Fig. 4, Dnm1-GFP localized to punctate fission complexes on mt-rfp-labeled mitochondria in 100% of the cells in the population (Fig. 4, top row), and NaN 3 /NaF treatment did not alter the punctate appearance or mitochondrial localization of wild-type Dnm1- GFP (Fig. 4, second row). By contrast, the Dnm1 G385D -GFP staining FIGURE 4. Dnm1 G385D -GFP forms punctate structures after treatment with NaN 3 /NaF. GFP localization in cells expressing Dnm1-GFP or Dnm1 G385D -GFP and treated (indicated by ) or not treated (indicated by ) with 10 mm NaN 3 /NaF for 15 min. Differential interference contrast (DIC; left panel), mt-rfp (second panel), GFP (third panel), and GFP merged with mt-rfp (right panel) are shown. The percentage of cells in a population that display visible punctate GFP structures co-localized with mitochondria is indicated at the far right (three independent experiments were performed, and 100 cells were counted per experiment, S.D. is indicated). Genes indicated are genomic unless preceded by p, which denotes plasmid-borne. JUNE 23, 2006 VOLUME 281 NUMBER 25 JOURNAL OF BIOLOGICAL CHEMISTRY 17315
5 FIGURE 6. Mdv1p forms a complex with Dnm1 G385D p in a co-immunoprecipitation assay. Myc-tagged Dnm1 proteins were immunoprecipitated using anti-myc-agarose-conjugated beads. Co-IP of Mdv1p was detected by immunoblot using native Mdv1p antibodies (top panel). Western blot shows total loaded (L) and eluted (E) proteins. Immunoprecipitation of Myc-tagged Dnm1p or Dnm1 G385D p was detected with anti-myc antibodies (middle panel). The mitochondrial protein porin is provided as a loading control (bottom panel). FIGURE 5. Dnm1 G385D p mediates mitochondrial fission in fzo1 dnm1 cells after treatment with NaN 3 /NaF. Mitochondrial morphology in fzo1 dnm1 cells carrying prs415-dnm1, prs415-dnm1 G385D, or prs415 and treated (indicated by ) ornot treated (indicated by ) with 10 mm NaN 3 /NaF for 15 min. Differential interference contrast (DIC; left panel) and mt-gfp (right panel) images are shown. Percentage of cells in a population that display fragmented mitochondria are indicated at the far right (three independent experiments were performed, and 100 cells were counted per experiment; S.D. is indicated). Genes indicated are genomic unless preceded by p, which denotes plasmid-borne. pattern was converted from diffuse cytoplasmic (Fig. 4, third row) to discrete puncta after NaN 3 /NaF treatment (Fig. 4, fourth row). The formation of Dnm1 G385D -GFP puncta was very rapid, occurring within 5 minofnan 3 /NaF treatment, and was rapidly reversed upon NaN 3 /NaF removal (data not shown). Quantification revealed that greater than 80% of these Dnm1 G385D -GFP-containing puncta stably co-localized with mt-rfp -labeled mitochondria in individual cells, suggesting that they are bona fide fission complexes. These findings are consistent with the idea that Dnm1 G385D p dimers form unstable multimeric complexes in vivo that rapidly disassemble unless they are stabilized by GTP depletion. In wild-type cells, NaN 3 treatment stimulates Dnm1p-dependent fission, leading to mitochondrial fragmentation (Fig. 4, second row, mt- RFP, and Fekkes et al. 16). If Dnm1 G385D p multimers that form after treatment with NaN 3 /NaF are physiologically relevant, they should also stimulate fission. However, in dnm1 cells expressing Dnm1 G385D p, we did not observe significant mitochondrial fragmentation after treatment with NaN 3 /NaF (Fig. 4, fourth row, mt-rfp). We reasoned that fission events might be difficult to detect in cells already containing mitochondrial nets. To test this possibility, we performed the same experiments in fzo1 dnm1 cells, which have tubular mitochondria. The lack of fusion, which prevents net formation, enhances detection of fission events in these cells. As shown in Fig. 5, mitochondria fragmented in the majority of fzo1 dnm1 cells expressing wild-type Dnm1p both in the presence and absence of NaN 3 /NaF (Fig. 5, first and second row). Significantly, when fzo1 dnm1 cells expressing Dnm1 G385D p were treated with 10 mm NaN 3 /NaF, mitochondria fragmented in 41.3% of cells. By contrast, only 4.6% of untreated cells contained fragmented mitochondria (Fig. 5, fifth and sixth row). Cells carrying the empty vector did not show any increase in mitochondrial fragmentation when treated with NaN 3 /NaF, indicating that stimulation of mitochondrial fission under these conditions is Dnm1p-dependent (Fig. 5, third and fourth row) (16). These results indicate that the Dnm1 G385D p punctate structures formed in the presence of NaN 3 /NaF mediate fission. Assembly of Dnm1 G385D p into Stable Multimers Is Not Required for Interaction with Mitochondrial Mdv1p Both co-ip (21) and yeast twohybrid (17, 18, 21) studies suggest that Dnm1p interacts with Mdv1p. We performed co-ip assays from membrane-enriched fractions to examine the possibility that defects in these Dnm1p-Mdv1p interactions were responsible for the inability of Dnm1 G385D p to form stable mitochondrial fission complexes in vivo. As shown in Fig. 6, a fraction of Mdv1p from chemically cross-linked membrane fractions reproducibly co-immunoprecipitated with 3Myc-Dnm1p (Fig. 6, lane 2, -Mdv1p). Importantly, membrane-bound Mdv1p also co-immunoprecipitated with 3Myc-Dnm1 G385D p (Fig. 6 lane 4, -Mdv1p) at levels similar to wild type. It is important to note that the fractions used for these experiments were generated from cells containing dimeric Dnm1 G385D p but lacking Dnm1 G385D p stable multimers. The ability of mitochondrial Mdv1p to interact with Dnm1 G385D p suggests that 1) stable assembly of Dnm1 G385D p into multimeric complexes is not a prerequisite for Mdv1p interaction, 2) dimeric Dnm1 G385D p is efficiently recruited to mitochondria, and 3) the instability of Dnm1 G385D p multimers is not due to a Dnm1 G385D p-mdv1p interaction defect. Assembly of Dnm1 G385D p into Stable Multimers Is Essential for Reorganization of GFP-Mdv1p into Punctate Structures In wild-type cells, Fis1p Mdv1p complexes are uniformly distributed on the cytoplasmic face of the mitochondrial outer membrane (15, 17, 19). GFP-Mdv1p is also found in punctate structures that co-localize with Dnm1p (15, 17). However, GFP-Mdv1p fails to form visible punctate structures in dnm1 cells, suggesting that Dnm1p plays an essential role in their formation. However, the specific step that promotes Mdv1p reorganization from a uniform mitochondrial distribution into punctate structures is unknown. We hypothesized that Dnm1p multimerization drives JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 281 NUMBER 25 JUNE 23, 2006
6 FIGURE 7. GFP-Mdv1p reorganizesintopunctate structures on mitochondria after Dnm1 G385D p multimersarestabilizedbynan 3 /NaFtreatment. GFP-Mdv1p localization in cells expressing Dnm1p or Dnm1 G385D p and treated (indicated by )ornot treated(indicatedby )with10mmnan 3 /NaFfor15 min. Differential interference contrast (DIC; left panel), mt-rfp (second panel), GFP-Mdv1p (third panel), and GFP-Mdv1p merged with mt-rfp (right panel) are shown. The percentage of cells in a population that display visible punctate GFP-Mdv1p structures co-localized with mitochondria is indicated at the far right (three independent experiments were performed, and 100 cells counted per experiment; S.D. is indicated). Genes indicated are genomic unless preceded by p, which denotes plasmid-borne. Mdv1p reorganization into punctate structures. To test this idea, we examined the localization pattern of GFP-Mdv1p in Dnm1 G385D p-expressing cells incubated in the absence or presence of NaN 3 /NaF. In both the absence and presence of NaN 3 /NaF treatment, GFP- Mdv1p formed mitochondrial puncta in cells expressing wild-type Dnm1p (Fig. 7, first and second rows). The NaN 3 /NaF treatment had no detectable effect on the size or shape of these puncta. By contrast, a uniform mitochondrial GFP-Mdv1p localization was observed in cells lacking Dnm1p whether or not NaN 3 /NaF was added (Fig. 7, third and fourth rows). Importantly, GFP-Mdv1p puncta were not observed in the majority of untreated cells expressing Dnm1 G385D p (Fig. 7, fifth row). Because Dnm1 G385D p forms dimers under these conditions that are able to bind and co-ip with Mdv1p, we conclude that interaction of dimeric Dnm1 G385D p with Mdv1p is not sufficient to promote reorganization of Mdv1p into puncta. Significantly, GFP-Mdv1p puncta were restored in 63% of cells expressing Dnm1 G385D p after treatment with NaN 3 /NaF (Fig. 7, sixth row). As described above, dimeric Dnm1 G385D p assembles into multimeric complexes after NaN 3 /NaF treatment. Thus, stabilizing Dnm1 G385D p multimeric complex formation stimulates the production of GFP-Mdv1p punctate structures. This GFP-Mdv1p puncta formation depends on assembly of Dnm1p or Dnm1 G385D p, as similar structures are not present in dnm1 cells or in untreated DNM1 G385D cells. Together, our co-ip and localization results provide a direct demonstration that stable assembly of Dnm1p into multimeric complexes is required to reorganize Mdv1p into punctate structures. Fis1p Is Present in Assembled Fission Complexes on Mitochondria Fis1p is essential for Dnm1p recruitment to mitochondria (13 17) and is also thought to play an important role after assembly of the fission complex (13, 19, 24). However, unlike Mdv1p and Dnm1p, fluorescently tagged Fis1p does not form visible puncta associated with fission complexes. This observation raises the possibility that Fis1p is required to initially recruit fission complex components but does not associate with these components after they form punctate structures. Alternatively, Fis1p may be an integral component of assembled complexes but is present in low stoichiometric amounts compared with Mdv1p and Dnm1p, making visualization by fluorescence microscopy difficult. We described conditions in which mitochondrial Mdv1p and Dnm1 G385D p are reorganized into punctate structures after NaN 3 /NaF treatment. To test whether Fis1p is present in these complexes, we performed co-ip experiments between Fis1p and Dnm1 G385D p from DSP cross-linked membrane fractions in the presence or absence of NaN 3 /NaF. A fraction of Dnm1p coimmunoprecipitated with 9Myc-Fis1p both in the absence and presence of NaN 3 /NaF (Fig. 8, lanes 2 and 4, -Dnm1p). This interaction was specific as Dnm1p failed to precipitate from cells lacking 9Myc- Fis1p (Fig. 8, lane 10, -Dnm1p). We did not detect Dnm1 G385D p JUNE 23, 2006 VOLUME 281 NUMBER 25 JOURNAL OF BIOLOGICAL CHEMISTRY 17317
7 FIGURE 8. Dnm1 G385D p-fis1p complexes are only detected in a co-ip assay after treatment with NaN 3 /NaF. Western blot showing Dnm1p and Dnm1 G385D p co-immunoprecipitation with Fis1p before (indicated by ) and after (indicated by ) treatment of intact cells with 10 mm NaN 3 /NaF. Myc-tagged Fis1p was immunoprecipitated using anti-myc-agarose-conjugated beads. Co-IP of Dnm1 proteins was detected by immunoblot using native Dnm1p antibodies (top panel). Western blot shows total loaded (L) and eluted (E) proteins. Immunoprecipitation of Myc-tagged Fis1p was detected with anti- Myc antibodies (middle panel). The mitochondrial protein porin is provided as a loading control (bottom panel). Genes indicated are genomic unless preceded by p, which denotes plasmid-borne. co-immunoprecipitation with 9Myc-Fis1p (Fig. 8, lane 6, -Dnm1p), suggesting that any amount of Dnm1p associated with Fis1p is below the level of detection under these conditions. However, Dnm1 G385D p co-immunoprecipitated efficiently with Fis1p in the presence of NaN 3 /NaF, indicating that Fis1p is a stable component of assembled fission complexes on mitochondria. DISCUSSION The results described here advance our understanding of fission complex assembly in several important ways. First, we demonstrate that the minimum cytoplasmic form of Dnm1p is a dimer. Second, our experiments support a role for the Dnm1p middle domain in the stabilization of Dnm1p multimers during or after assembly. Third, our data suggest that assembly of Dnm1p into stable multimers is not required for Dnm1p-Mdv1p interaction or mitochondrial recruitment of Dnm1p. Fourth, we demonstrate that reorganization of Mdv1p into punctate structures requires assembly of Dnm1p into multimers on mitochondria. Finally, we show that Fis1p is physically present in assembled fission complexes with Mdv1p and Dnm1p (25). Each of these points is discussed in detail below. Cytosolic Dnm1p Is a Dimer Our gel filtration assays demonstrate that the minimum soluble form of yeast Dnm1p under physiological conditions is a dimer. Moreover, we show that Dnm1p dimerization is not dependent upon the presence of Mdv1p or Fis1p. These results are consistent with an in vitro study reporting that yeast Dnm1p purified from insect cells forms a dimer in solution (25). However, the mammalian homolog of Dnm1p, Drp1, migrates as a tetramer in gel filtration studies (27). We suggest that the differences in oligomeric state may reflect variations in the assembly pathway of mitochondrial fission complexes between species. This view is supported by the fact that whereas Mdv1p is an important component of mitochondrial fission in yeast, its mammalian homolog has not yet been identified. The Middle Domain Is Required to Stabilize Multimeric Dnm1p Complexes We demonstrated previously that the Dnm1p middle domain interacts with itself and the GTPase effector domain/assembly domain (10). Although these findings suggest that the middle domain is important for Dnm1p self-assembly, this idea has never been rigorously tested in vivo. In this study we demonstrate that Dnm1 G385D p dimerizes to the same extent as the wild-type Dnm1 protein. However, this amino acid substitution compromises the ability of Dnm1p to form stable multimers. Dnm1 G385D p might fail to form multimeric complexes because the G385D substitution blocks dimer-dimer interactions required for multimer assembly. Alternatively, Dnm1 G385D p may form multimers that disassemble more rapidly, preventing visualization of stable fission complexes. By treating cells with NaN 3 /NaF, we demonstrated that dimeric Dnm1 G385D p is capable of assembling into functional fission complexes containing both Mdv1p and Fis1p. We suggest that the primary effect of the G385D middle domain mutation is to enhance disassembly of Dnm1p complexes, shifting the equilibrium to the dimeric state. The molecular basis for this enhanced disassembly is currently unclear. One possibility is that the G385D substitution alters the Dnm1p GTPase cycle in vivo by disrupting Dnm1p intramolecular interactions, resulting in premature GTP hydrolysis and rapid disassembly of Dnm1 G385D p-containing multimers. Dynamin and Dnm1p multimers assemble in the absence of nucleotide or the presence of GDP in vitro (25, 44, 45). In this study we show that Dnm1 G385D p can form multimeric complexes on mitochondria after treatment to deplete cellular GTP. Furthermore, mitochondrial fission is partially restored under these conditions. The simplest interpretation of this result is that Dnm1 G385D p self-interactions are stabilized when Dnm1 G385D p is in the GDP-bound or empty state, resulting in the formation of visible multimeric structures that can catalyze fission. A recent study reported that Dnm1 G385D p is a terminal dimer in vitro (25). It is possible that Dnm1 G385D p multimers also assemble in vitro under conditions not yet tested. Alternatively, the ability of Dnm1 G385D p multimers to assemble upon NaN 3 /NaF treatment may require the presence of additional proteins or factors that promote assembly in vivo but are absent in vitro. Assembly of Dnm1p into Multimers Is Not Essential for Dnm1-Mdv1p Interactions or Dnm1p Recruitment to Mitochondria Mdv1p interacts with Dnm1p in mitochondrial fission complexes (17, 19, 21). It has been proposed that Mdv1p only interacts with multimeric Dnm1p (23). This model was based on the observations that Mdv1p fails to interact with Dnm1 G385D p by the yeast two-hybrid assay and that Mdv1p fails to form punctate structures on mitochondria in cells expressing Dnm1 G385D p. We show here that dimeric Dnm1 G385D p forms a complex with mitochondrial Mdv1p in a co-ip assay. Thus, assembly of Dnm1p into stable multimers is not required for Dnm1p-Mdv1p interaction. Moreover, although Mdv1p can interact with dimeric Dnm1 G385D p, this interaction does not stabilize Dnm1 G385D p multimer formation. It is important to note that we failed to detect interactions between Dnm1 G385D p and Mdv1p using a sensitive yeast two-hybrid protocol (data not shown), consistent with published results (23). However, co-ip assays reproducibly detected this interaction, indicating that the two-hybrid assay may not always be a reliable means of assessing physiologically relevant interactions between these proteins. Membrane-bound Mdv1p is required to recruit Dnm1p to mitochondrial fission complexes (18, 19). We and others (21 23) have observed Dnm1p multimeric structures moving rapidly in the cytoplasm in live cells, leading to the hypothesis that Dnm1p multimers first assemble in the cytoplasm and are subsequently recruited to Mdv1p on mitochondria. However, these rapidly moving puncta have never been shown to stably bind mitochondria and subsequently become functional fission complexes. An alternative interpretation is that these cytoplasmic Dnm1p multimers are the products of completed fission reactions or simply Dnm1p storage units. Here we show that dimeric Dnm1 G385D p is efficiently recruited to mitochondrial Mdv1p, consistent with the idea that multimerization occurs after recruitment of dimeric Dnm1p to the membrane JOURNAL OF BIOLOGICAL CHEMISTRY VOLUME 281 NUMBER 25 JUNE 23, 2006
8 Mdv1p Reorganization into Punctate Structures on Mitochondria Requires Stable Assembly of Dnm1p Multimeric Complexes Previous studies established that Mdv1p Fis1p complexes are distributed uniformly on mitochondrial tubules (15, 17 19). In addition, Mdv1p forms punctate structures that co-localize with Dnm1p-containing fission complexes (15, 17). When Dnm1p is absent, Mdv1p remains uniformly distributed on mitochondria and fails to form punctate structures (15, 17), suggesting that Dnm1p is required for the reorganization of Mdv1p into visible puncta. Here we demonstrate that Mdv1p fails to form mitochondrial puncta even when it is associated with dimeric Dnm1 G385D p. However, when Dnm1 G385D p multimeric complexes are stabilized by NaN 3 /NaF treatment, Mdv1p forms visible mitochondrial puncta, providing a direct demonstration that assembly of Dnm1p into multimeric complexes promotes the reorganization of Mdv1p from a uniform to a punctate mitochondrial localization. Consistent with these observations, in time-lapse studies, GFP-Mdv1p puncta appear after Dnm1p- GFP puncta become visible on mitochondria (23). Exactly how Mdv1p is reorganized during this process is not yet clear. Mdv1p puncta formation may reflect a change in the stoichiometry of Dnm1p and Mdv1p in the fission complex due to Mdv1p self-assembly. In support of this model, Mdv1p has been shown to form a complex with itself in co-ip experiments (18). Further studies are required to determine the nature of Mdv1p reorganization into these punctate fission complexes. Fis1p Is Present in Assembled Fission Complexes Fis1p is essential for Dnm1p recruitment to mitochondria (13, 15 17) and also plays a role after assembly of the fission complex (13, 19, 24). Despite the apparent stability of the fission complexes visualized in intact cells, the presence of Fis1p in these structures has been difficult to demonstrate. In the co-ip studies described here we did not detect Fis1p in a complex with dimeric Dnm1 G385D p even after chemical cross-linking. However, this interaction was detected after NaN 3 /NaF treatment, which stabilizes Dnm1p G385D multimers. These results indicate that Fis1p remains associated with fission complexes after assembly. Given our results, it is surprising that GFP-Fis1p does not form visible puncta similar to those observed for Dnm1p-GFP and GFP-Mdv1p in fission complexes. It is possible that there are fewer Fis1p molecules compared with Dnm1p and Mdv1p in an assembled fission complex. Stepwise Assembly of Mitochondrial Fission Complexes This study provides new insights regarding the spatial and temporal dynamics of mitochondrial fission complex assembly (Fig. 9). We propose that fission complex assembly begins when Dnm1p dimers are recruited to pre-existing Mdv1p Fis1p complexes on mitochondria (Fig. 9, step 1). These mitochondrial dimers, which are in a dynamic equilibrium with the cytoplasmic pool of Dnm1p dimers, further assemble to form multimeric complexes (Fig. 9, step 2). Dnm1p multimerization then drives the reorganization of Mdv1p from a uniform mitochondrial distribution into punctate fission complexes that also contain Fis1p (Fig. 9, step 3). In later steps, these fission complexes are activated, leading to division of the mitochondrial compartment. How these three proteins work together in the assembled complex to achieve fission is the focus of our ongoing studies. FIGURE 9. Stepwise model for the assembly of fission complexes on mitochondria. Dnm1p dimers are recruited to Mdv1p Fis1p pre-existing complexes on mitochondria. In the next step multimerization of Dnm1p on mitochondria occurs. Dnm1p multimers then promote the reorganization of Mdv1p into fission complexes. These fission complexes also contain Fis1p. C, cytoplasm; M, outer mitochondrial membrane; IMS, intermembrane space. Note that large, rapidly moving cytoplasmic Dnm1p complexes visualized in vivo are not depicted in this model. Acknowledgments We appreciate stimulating discussions and technical assistance from members of Shaw laboratory, especially Noriko Okamoto. We thank J. Nunnari for providing anti-mdv1p antibody. The Utah health sciences DNA/peptide and sequencing facilities are supported by a National Cancer Institute Grant 5-P30CA REFERENCES 1. Frank, S., Gaume, B., Bergmann-Leitner, E. S., Leitner, W. W., Robert, E. G., Catez, F., Smith, C. L., and Youle, R. J. (2001) Dev. Cell 1, Jagasia, R., Grote, P., Westermann, B., and Conradt, B. (2005) Nature 433, James, D. I., Parone, P. A., Mattenberger, Y., and Martinou, J. C. (2003) J. Biol. Chem. 278, Karbowski, M., and Youle, R. J. (2003) Cell Death Differ. 10, Lee, Y. J., Jeong, S. Y., Karbowski, M., Smith, C. L., and Youle, R. J. (2004) Mol. Biol. Cell 15, Scorrano, L. (2003) Cell Death Differ. 10, Szabadkai, G., Simoni, A. M., Chami, M., Wieckowski, M. R., Youle, R. J., and Rizzuto, R. (2004) Mol. Cell. 16, Li, Z., Okamoto, K., Hayashi, Y., and Sheng, M. (2004) Cell 119, Bleazard, W., McCaffery, J. M., King, E. J., Bale, S., Mozdy, A., Tieu, Q., Nunnari, J., and Shaw, J. M. (1999) Nat. Cell Biol. 1, Fukushima, N. H., Brisch, E., Keegan, B. R., Bleazard, W., and Shaw, J. M. (2001) Mol. Biol. Cell 12, Otsuga, D., Keegan, B. R., Brisch, E., Thatcher, J. W., Hermann, G. J., Bleazard, W., and Shaw, J. M. (1998) J. Cell Biol. 143, Sesaki, H., and Jensen, R. E. (1999) J. Cell Biol. 147, Mozdy, A. D., McCaffery, J. M., and Shaw, J. M. (2000) J. Cell Biol. 151, Suzuki, M., Neutzner, A., Tjandra, N., and Youle, R. J. (2005) J. Biol. Chem. 280, Cerveny, K. L., McCaffery, J. M., and Jensen, R. E. (2001) Mol. Biol. Cell 12, Fekkes, P., Shepard, K. A., and Yaffe, M. P. (2000) J. Cell Biol. 151, Tieu, Q., and Nunnari, J. (2000) J. Cell Biol. 151, Griffin, E. E., Graumann, J., and Chan, D. C. (2005) J. Cell Biol. 170, Karren, M. A., Coonrod, E. M., Anderson, T. K., and Shaw, J. M. (2005) J. Cell Biol. 171, Sesaki, H., Southard, S. M., Yaffe, M. P., and Jensen, R. E. (2003) Mol. Biol. Cell 14, Cerveny, K. L., and Jensen, R. E. (2003) Mol. Biol. Cell 14, Jensen, R. E., Hobbs, A. E., Cerveny, K. L., and Sesaki, H. (2000) Microsc. Res. Tech. 51, Naylor, K., Ingerman, E., Okreglak, V., Marino, M., Hinshaw, J. E., and Nunnari, J. (2006) J. Biol. Chem. 281, Tieu, Q., Okreglak, V., Naylor, K., and Nunnari, J. (2002) J. Cell Biol. 158, Ingerman, E., Perkins, E. M., Marino, M., Mears, J. A., McCaffery, J. M., Hinshaw, J. E., and Nunnari, J. (2005) J. Cell Biol. 170, Okamoto, P. M., Tripet, B., Litowski, J., Hodges, R. S., and Vallee, R. B. (1999) J. Biol. Chem. 274, Zhu, P. P., Patterson, A., Stadler, J., Seeburg, D. P., Sheng, M., and Blackstone, C. (2004) J. Biol. Chem. 279, Winston, F., Dollard, C., and Ricupero-Hovasse, S. L. (1995) Yeast 11, Guthrie, C., and Fink, G. (1991) in Yeast Genetics and Molecular Biology (Abelson, J. N., and Simon, M. I., eds) Methods in Enzymology, pp. 12, 186, Academic Press, Inc., San Diego, CA 30. Sherman, F., Fink, G. R., and Hicks, J. B. (1986) Methods in Yeast Genetics, p. 121, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY 31. Maniatis, T., Fritsch, E. F., and Sambrook, J. (1982) Molecular Cloning: A Laboratory Manual, p. 1.21, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 32. Rothstein, R. (1991) Methods Enzymol. 194, Frederick, R. L., McCaffery, J. M., Cunningham, K. W., Okamoto, K., and Shaw, J. M. (2004) J. Cell Biol. 167, Kondo-Okamoto, N., Shaw, J. M., and Okamoto, K. (2003) J. Biol. Chem. 278, Babst, M., Katzmann, D. J., Snyder, W. B., Wendland, B., and Emr, S. D. (2002) Dev. Cell 3, Hermann, G. J., and Shaw, J. M. (1998) Annu. Rev. Cell Dev. Biol. 14, JUNE 23, 2006 VOLUME 281 NUMBER 25 JOURNAL OF BIOLOGICAL CHEMISTRY 17319
D The WD40 Protein Caf4p is a Component of the Mitochondrial Fission Machinery and Recruits Dnm1p to Mitochondria
 153 D The WD4 Protein Caf4p is a Component of the Mitochondrial Fission Machinery and Recruits Dnm1p to Mitochondria This chapter represents a further example of the use of MudPIT to analyze the polypeptide
153 D The WD4 Protein Caf4p is a Component of the Mitochondrial Fission Machinery and Recruits Dnm1p to Mitochondria This chapter represents a further example of the use of MudPIT to analyze the polypeptide
Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc
 OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
Supplemental Information. The Mitochondrial Fission Receptor MiD51. Requires ADP as a Cofactor
 Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
Structure, Volume 22 Supplemental Information The Mitochondrial Fission Receptor MiD51 Requires ADP as a Cofactor Oliver C. Losón, Raymond Liu, Michael E. Rome, Shuxia Meng, Jens T. Kaiser, Shu-ou Shan,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hfis1
 Research Article 4141 Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hfis1 Tianzheng Yu 1, Randall J. Fox 1, Lindsay S. Burwell 2 and Yisang Yoon 1, * 1 Department
Research Article 4141 Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hfis1 Tianzheng Yu 1, Randall J. Fox 1, Lindsay S. Burwell 2 and Yisang Yoon 1, * 1 Department
Stress-Induced Nuclear-to-Cytoplasmic Translocation of Cyclin C Promotes Mitochondrial Fission in Yeast
 Article Stress-Induced Nuclear-to-Cytoplasmic Translocation of Cyclin C Promotes Mitochondrial Fission in Yeast Katrina F. Cooper, 1 Svetlana Khakhina, 1 Stephen K. Kim, 1 and Randy Strich 1, * 1 Department
Article Stress-Induced Nuclear-to-Cytoplasmic Translocation of Cyclin C Promotes Mitochondrial Fission in Yeast Katrina F. Cooper, 1 Svetlana Khakhina, 1 Stephen K. Kim, 1 and Randy Strich 1, * 1 Department
Supplementary Figure 1.
 Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Supplementary Figure 1. Characterisation of IHG-1 overexpressing and knockdown cell lines. (A) Total cellular RNA was prepared from HeLa cells stably overexpressing IHG-1 or mts-ihg-1. IHG-1 mrna was quantified
Supplementary Information
 Supplementary Information MAP2/Hoechst Hyp.-AP ph 6.5 Hyp.-SD ph 7.2 Norm.-SD ph 7.2 Supplementary Figure 1. Mitochondrial elongation in cortical neurons by acidosis. Representative images of neuronal
Supplementary Information MAP2/Hoechst Hyp.-AP ph 6.5 Hyp.-SD ph 7.2 Norm.-SD ph 7.2 Supplementary Figure 1. Mitochondrial elongation in cortical neurons by acidosis. Representative images of neuronal
Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity
 Research Article 6535 Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity Naotada Ishihara, Yuka Eura and Katsuyoshi Mihara* Department of Molecular Biology, Graduate
Research Article 6535 Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity Naotada Ishihara, Yuka Eura and Katsuyoshi Mihara* Department of Molecular Biology, Graduate
The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria
 JCB: ARTICLE The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria Erik E. Griffin, Johannes Graumann, and David C. Chan Division of Biology, California
JCB: ARTICLE The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria Erik E. Griffin, Johannes Graumann, and David C. Chan Division of Biology, California
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
Lecture 10: Cyclins, cyclin kinases and cell division
 Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Full-length GlpG sequence was generated by PCR from E. coli genomic DNA. (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
 Supplementary Methods Protein expression and purification Full-length GlpG sequence was generated by PCR from E. coli genomic DNA (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
Supplementary Methods Protein expression and purification Full-length GlpG sequence was generated by PCR from E. coli genomic DNA (with two sequence variations, D51E/L52V, from the gene bank entry aac28166),
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p
 Research Article 2005 Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p Stefan Jakobs 1, *,, Nadia Martini 1, *, Astrid C. Schauss 1, Alexander Egner 1, Benedikt
Research Article 2005 Spatial and temporal dynamics of budding yeast mitochondria lacking the division component Fis1p Stefan Jakobs 1, *,, Nadia Martini 1, *, Astrid C. Schauss 1, Alexander Egner 1, Benedikt
7.06 Problem Set
 7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
7.06 Problem Set 5 -- 2006 1. In the first half of the course, we encountered many examples of proteins that entered the nucleus in response to the activation of a cell-signaling pathway. One example of
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL Figure S1. Mitochondrial morphology in Fis1-null, Mff-null and Fis1/Mff-null MEF cells. (A) Western blotting of lysates from Fis1-null, Mff-null and Fis1/Mff-null cells. Lysates were
SUPPLEMENTAL MATERIAL Figure S1. Mitochondrial morphology in Fis1-null, Mff-null and Fis1/Mff-null MEF cells. (A) Western blotting of lysates from Fis1-null, Mff-null and Fis1/Mff-null cells. Lysates were
Plant mitochondrial dynamics
 Plant mitochondrial dynamics Peroxisome Chloroplast Nucleus Mitochondria from Alberts et al. 1994 Living Arabidopsis leaf 10 µm Logan & Leaver (2000) Journal of Experimental Botany, 51: 865-871 5 µm S.
Plant mitochondrial dynamics Peroxisome Chloroplast Nucleus Mitochondria from Alberts et al. 1994 Living Arabidopsis leaf 10 µm Logan & Leaver (2000) Journal of Experimental Botany, 51: 865-871 5 µm S.
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing
 Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
EST1 Homology Domain. 100 aa. hest1a / SMG6 PIN TPR TPR. Est1-like DBD? hest1b / SMG5. TPR-like TPR. a helical. hest1c / SMG7.
 hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
hest1a / SMG6 EST1 Homology Domain 100 aa 853 695 761 780 1206 hest1 / SMG5 -like? -like 109 145 214 237 497 165 239 1016 114 207 212 381 583 hest1c / SMG7 a helical 1091 Sc 57 185 267 284 699 Figure S1:
Lipid transfer proteins confer resistance to trichothecenes
 Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
Lipid transfer proteins confer resistance to trichothecenes John McLaughlin and Anwar Bin-Umer Tumer Laboratory National Fusarium Head Blight Forum December 6th, 2012 FY09-11: Identify trichothecene resistance
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles
 Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Illegitimate translation causes unexpected gene expression from on-target out-of-frame alleles created by CRISPR-Cas9 Shigeru Makino, Ryutaro Fukumura, Yoichi Gondo* Mutagenesis and Genomics Team, RIKEN
Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology
 Research Article 1201 Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology Diana Stojanovski 1, Olga S. Koutsopoulos 1, Koji Okamoto 2 and Michael T. Ryan 1, * 1 Department
Research Article 1201 Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology Diana Stojanovski 1, Olga S. Koutsopoulos 1, Koji Okamoto 2 and Michael T. Ryan 1, * 1 Department
Three types of RNA polymerase in eukaryotic nuclei
 Three types of RNA polymerase in eukaryotic nuclei Type Location RNA synthesized Effect of α-amanitin I Nucleolus Pre-rRNA for 18,.8 and 8S rrnas Insensitive II Nucleoplasm Pre-mRNA, some snrnas Sensitive
Three types of RNA polymerase in eukaryotic nuclei Type Location RNA synthesized Effect of α-amanitin I Nucleolus Pre-rRNA for 18,.8 and 8S rrnas Insensitive II Nucleoplasm Pre-mRNA, some snrnas Sensitive
Supplementary Figure 1. Biochemical and sequence alignment analyses the
 Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Name: TF: Section Time: LS1a ICE 5. Practice ICE Version B
 Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces cerevisiae.
 Genetics: Published Articles Ahead of Print, published on July 30, 2012 as 10.1534/genetics.112.143818 A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces
Genetics: Published Articles Ahead of Print, published on July 30, 2012 as 10.1534/genetics.112.143818 A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
DOI: 10.1038/ncb2362 Figure S1 CYLD and CASPASE 8 genes are co-regulated. Analysis of gene expression across 79 tissues was carried out as described previously [Ref: PMID: 18636086]. Briefly, microarray
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
doi:10.1038/nature10244 a O07391_MYCAV/127-243 NLPC_HAEIN/80-181 SPR_SHIFL/79-183 P74160_SYNY3/112-245 O24914_HELPY/301-437 Q51835_PORGI/68-178 DPP6_BACSH/163-263 YKFC_BACSU/185-292 YDHO_ECOLI/153-263
7.06 Cell Biology EXAM #3 KEY
 7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 KEY May 2, 2006 This is an OPEN BOOK exam, and you are allowed access to books, a calculator, and notes BUT NOT computers or any other types of electronic devices. Please write
Scale in the biological world
 Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Importance of Mitochondrial Dynamics During Meiosis and Sporulation
 Molecular Biology of the Cell Vol. 15, 4369 4381, October 2004 Importance of Mitochondrial Dynamics During Meiosis and Sporulation Steven W. Gorsich* and Janet M. Shaw Department of Biology, University
Molecular Biology of the Cell Vol. 15, 4369 4381, October 2004 Importance of Mitochondrial Dynamics During Meiosis and Sporulation Steven W. Gorsich* and Janet M. Shaw Department of Biology, University
Structure and RNA-binding properties. of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex
 Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Structure and RNA-binding properties of the Not1 Not2 Not5 module of the yeast Ccr4 Not complex Varun Bhaskar 1, Vladimir Roudko 2,3, Jerome Basquin 1, Kundan Sharma 4, Henning Urlaub 4, Bertrand Seraphin
Dynamin-related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells
 Vol. 12, 2245 2256, August 2001 Dynamin-related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells Elena Smirnova, Lorena Griparic, Dixie-Lee Shurland, and Alexander M. van der Bliek*
Vol. 12, 2245 2256, August 2001 Dynamin-related Protein Drp1 Is Required for Mitochondrial Division in Mammalian Cells Elena Smirnova, Lorena Griparic, Dixie-Lee Shurland, and Alexander M. van der Bliek*
Types of biological networks. I. Intra-cellurar networks
 Types of biological networks I. Intra-cellurar networks 1 Some intra-cellular networks: 1. Metabolic networks 2. Transcriptional regulation networks 3. Cell signalling networks 4. Protein-protein interaction
Types of biological networks I. Intra-cellurar networks 1 Some intra-cellular networks: 1. Metabolic networks 2. Transcriptional regulation networks 3. Cell signalling networks 4. Protein-protein interaction
CELB40060 Membrane Trafficking in Animal Cells. Prof. Jeremy C. Simpson. Lecture 2 COPII and export from the ER
 CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
Supplementary Figure 1
 Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary Figure 1 Supplementary Figure 1. HSP21 expression in 35S:HSP21 and hsp21 knockdown plants. (a) Since no T- DNA insertion line for HSP21 is available in the publicly available T-DNA collections,
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a
 Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplementary materials. Crystal structure of the carboxyltransferase domain. of acetyl coenzyme A carboxylase. Department of Biological Sciences
 Supplementary materials Crystal structure of the carboxyltransferase domain of acetyl coenzyme A carboxylase Hailong Zhang, Zhiru Yang, 1 Yang Shen, 1 Liang Tong Department of Biological Sciences Columbia
Supplementary materials Crystal structure of the carboxyltransferase domain of acetyl coenzyme A carboxylase Hailong Zhang, Zhiru Yang, 1 Yang Shen, 1 Liang Tong Department of Biological Sciences Columbia
ydci GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC
 Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Table S1. DNA primers used in this study. Name ydci P1ydcIkd3 Sequence GTC TGT TTG AAC GCG GGC GAC TGG GCG CGC AAT TAA CGG TGT GTA GGC TGG AGC TGC TTC Kd3ydcIp2 lacz fusion YdcIendP1 YdcItrgP2 GAC AGC
Supporting Information
 Supporting Information Wang et al. 10.1073/pnas.0804871105 SI Materials and Methods Cell Culture and Transfection. Human neuroblastoma M17 cells were grown as described before (1). Transfection was performed
Supporting Information Wang et al. 10.1073/pnas.0804871105 SI Materials and Methods Cell Culture and Transfection. Human neuroblastoma M17 cells were grown as described before (1). Transfection was performed
SUPPLEMENTARY INFORMATION
 DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
DOI:.38/ncb97 P ( μm, hours) 1 2 4 P DMSO Figure S1 unningham et al. E 97 65 27 MFN1 GFP-Parkin Opa1 ctin GPDH HEK293 GFP-Parkin 19 115 97 65 27 Mitochondrial Fraction SH-SY5Y GFP-Parkin Mito DMSO Mito
Supporting Online Material. On-Chip Dielectrophoretic Co-Assembly of Live Cells and. Particles into Responsive Biomaterials
 Supporting Online Material On-Chip Dielectrophoretic Co-Assembly of Live Cells and Particles into esponsive Biomaterials Shalini Gupta, ossitza G. Alargova, Peter K. Kilpatrick and Orlin D. Velev* Description
Supporting Online Material On-Chip Dielectrophoretic Co-Assembly of Live Cells and Particles into esponsive Biomaterials Shalini Gupta, ossitza G. Alargova, Peter K. Kilpatrick and Orlin D. Velev* Description
Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway
 Published Online: 11 October, 2004 Supp Info: http://doi.org/10.1083/jcb.200405100 Downloaded from jcb.rupress.org on July 13, 2018 JCB: ARTICLE Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology
Published Online: 11 October, 2004 Supp Info: http://doi.org/10.1083/jcb.200405100 Downloaded from jcb.rupress.org on July 13, 2018 JCB: ARTICLE Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology
Biological Sciences 11 Spring Experiment 4. Protein crosslinking
 Biological Sciences 11 Spring 2000 Experiment 4. Protein crosslinking = C - CH 2 - CH 2 - CH 2 - C = H H GA Cl - H 2 N N H 2 Cl - C - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - C DMS CH 3 CH 3 N - - C -
Biological Sciences 11 Spring 2000 Experiment 4. Protein crosslinking = C - CH 2 - CH 2 - CH 2 - C = H H GA Cl - H 2 N N H 2 Cl - C - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - C DMS CH 3 CH 3 N - - C -
Supporting Information
 Supporting Information Self-Assembly of Glutathione S-transferases into Nanowires Wei Zhang, a Quan Luo,* a Lu Miao, a Yushi Bai, a Zeyuan Dong, a Jiayun Xu, a and Junqiu Liu* a a State Key Laboratory
Supporting Information Self-Assembly of Glutathione S-transferases into Nanowires Wei Zhang, a Quan Luo,* a Lu Miao, a Yushi Bai, a Zeyuan Dong, a Jiayun Xu, a and Junqiu Liu* a a State Key Laboratory
7.06 Spring 2004 PS 6 KEY 1 of 14
 7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition
 Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
The Outer Envelope Protein PDV1, Together with its Paralogue PDV2, Mediates
 The Outer Envelope Protein PDV1, Together with its Paralogue PDV2, Mediates Recruitment of the Dynamin-Related Protein ARC5 to the Plastid Division Site in Arabidopsis Shin-ya Miyagishima a, c, John E.
The Outer Envelope Protein PDV1, Together with its Paralogue PDV2, Mediates Recruitment of the Dynamin-Related Protein ARC5 to the Plastid Division Site in Arabidopsis Shin-ya Miyagishima a, c, John E.
Division of Mitochondria Requires a Novel DNM1- interacting Protein, Net2p
 Molecular Biology of the Cell Vol. 12, 309 321, February 2001 Division of Mitochondria Requires a Novel DNM1- interacting Protein, Net2p Kara L. Cerveny,* J. Michael McCaffery, and Robert E. Jensen* *Department
Molecular Biology of the Cell Vol. 12, 309 321, February 2001 Division of Mitochondria Requires a Novel DNM1- interacting Protein, Net2p Kara L. Cerveny,* J. Michael McCaffery, and Robert E. Jensen* *Department
SEPTEMBER 10, 2010 VOLUME 285 NUMBER 37 JOURNAL OF BIOLOGICAL CHEMISTRY 28749
 THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 285, NO. 37, pp. 28749 28763, September 10, 2010 2010 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Bcl-2 and Bax Interact
THE JOURNAL OF BIOLOGICAL CHEMISTRY VOL. 285, NO. 37, pp. 28749 28763, September 10, 2010 2010 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in the U.S.A. Bcl-2 and Bax Interact
P. syringae and E. coli
 CHAPTER 6 A comparison of the recd mutant phenotypes of P. syringae and E. coli 6.1 INTRODUCTION The RecBCD complex is essential for recombination mediated repair of double strand breaks (DSBs) of DNA
CHAPTER 6 A comparison of the recd mutant phenotypes of P. syringae and E. coli 6.1 INTRODUCTION The RecBCD complex is essential for recombination mediated repair of double strand breaks (DSBs) of DNA
Supporting online material
 Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Supporting online material Materials and Methods Target proteins All predicted ORFs in the E. coli genome (1) were downloaded from the Colibri data base (2) (http://genolist.pasteur.fr/colibri/). 737 proteins
Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes
 9 The Nucleus Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes Explain general structures of Nuclear Envelope, Nuclear Lamina, Nuclear Pore Complex Explain movement of proteins
9 The Nucleus Student Learning Outcomes: Nucleus distinguishes Eukaryotes from Prokaryotes Explain general structures of Nuclear Envelope, Nuclear Lamina, Nuclear Pore Complex Explain movement of proteins
Analysis of Escherichia coli amino acid transporters
 Ph.D thesis Analysis of Escherichia coli amino acid transporters Presented by Attila Szvetnik Supervisor: Dr. Miklós Kálmán Biology Ph.D School University of Szeged Bay Zoltán Foundation for Applied Research
Ph.D thesis Analysis of Escherichia coli amino acid transporters Presented by Attila Szvetnik Supervisor: Dr. Miklós Kálmán Biology Ph.D School University of Szeged Bay Zoltán Foundation for Applied Research
Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p
 Research Article 1633 Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p Alison M. Motley, Gemma P. Ward and Ewald H. Hettema* Department of Molecular Biology and Biotechnology, University
Research Article 1633 Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p Alison M. Motley, Gemma P. Ward and Ewald H. Hettema* Department of Molecular Biology and Biotechnology, University
Lecture 7 Cell Biolog y ٢٢٢ ١
 Lecture 7 ١ Mitochondria ٢ Mitochondria Mitochondria are the energy factories of the cells. The energy currency for the work that animals must do is the energy-rich molecule adenosine triphosphate (ATP).
Lecture 7 ١ Mitochondria ٢ Mitochondria Mitochondria are the energy factories of the cells. The energy currency for the work that animals must do is the energy-rich molecule adenosine triphosphate (ATP).
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
Supplementary Figures Supplementary Figure 1. Purification of yeast CKM. (a) Silver-stained SDS-PAGE analysis of CKM purified through a TAP-tag engineered into the Cdk8 C-terminus. (b) Kinase activity
T H E J O U R N A L O F C E L L B I O L O G Y
 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Breker et al., http://www.jcb.org/cgi/content/full/jcb.201301120/dc1 Figure S1. Single-cell proteomics of stress responses. (a) Using
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Breker et al., http://www.jcb.org/cgi/content/full/jcb.201301120/dc1 Figure S1. Single-cell proteomics of stress responses. (a) Using
Gene regulation I Biochemistry 302. Bob Kelm February 25, 2005
 Gene regulation I Biochemistry 302 Bob Kelm February 25, 2005 Principles of gene regulation (cellular versus molecular level) Extracellular signals Chemical (e.g. hormones, growth factors) Environmental
Gene regulation I Biochemistry 302 Bob Kelm February 25, 2005 Principles of gene regulation (cellular versus molecular level) Extracellular signals Chemical (e.g. hormones, growth factors) Environmental
Mitochondria associate with the cytoskeleton. Figure 14-5 Molecular Biology of the Cell ( Garland Science 2008)
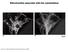 Mitochondria associate with the cytoskeleton Figure 14-5 Molecular Biology of the Cell ( Garland Science 2008) Special arrangements of mitochondria Figure 14-6 Molecular Biology of the Cell ( Garland Science
Mitochondria associate with the cytoskeleton Figure 14-5 Molecular Biology of the Cell ( Garland Science 2008) Special arrangements of mitochondria Figure 14-6 Molecular Biology of the Cell ( Garland Science
RNA Synthesis and Processing
 RNA Synthesis and Processing Introduction Regulation of gene expression allows cells to adapt to environmental changes and is responsible for the distinct activities of the differentiated cell types that
RNA Synthesis and Processing Introduction Regulation of gene expression allows cells to adapt to environmental changes and is responsible for the distinct activities of the differentiated cell types that
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
Activation of a receptor. Assembly of the complex
 Activation of a receptor ligand inactive, monomeric active, dimeric When activated by growth factor binding, the growth factor receptor tyrosine kinase phosphorylates the neighboring receptor. Assembly
Activation of a receptor ligand inactive, monomeric active, dimeric When activated by growth factor binding, the growth factor receptor tyrosine kinase phosphorylates the neighboring receptor. Assembly
Transmembrane Domains (TMDs) of ABC transporters
 Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON
 PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
PROKARYOTE GENES: E. COLI LAC OPERON CHAPTER 13 CHAPTER 13 PROKARYOTE GENES: E. COLI LAC OPERON Figure 1. Electron micrograph of growing E. coli. Some show the constriction at the location where daughter
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Supplementary Figure 1 Identification of the ScDcp2 minimal region interacting with both ScDcp1 and the ScEdc3 LSm domain. Pull-down experiment of untagged ScEdc3 LSm with various ScDcp1-Dcp2-His 6 fragments.
Lecture 6 - Intracellular compartments and transport I
 01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
01.26.11 Lecture 6 - Intracellular compartments and transport I Intracellular transport and compartments 1. Protein sorting: How proteins get to their appropriate destinations within the cell 2. Vesicular
Lecture 26: More on Gel Filtration Chromatography and the Trypsin Resurrection Experiment
 Biological Chemistry Laboratory Biology 3515/Chemistry 3515 Spring 2018 Lecture 26: More on Gel Filtration Chromatography and the Trypsin Resurrection Experiment 12 April 2018 c David P. Goldenberg University
Biological Chemistry Laboratory Biology 3515/Chemistry 3515 Spring 2018 Lecture 26: More on Gel Filtration Chromatography and the Trypsin Resurrection Experiment 12 April 2018 c David P. Goldenberg University
camp Direct Immunoassay Kit
 camp Direct Immunoassay Kit Catalog Number KA0886 100 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
camp Direct Immunoassay Kit Catalog Number KA0886 100 assays Version: 05 Intended for research use only www.abnova.com Table of Contents Introduction... 3 Background... 3 General Information... 4 Materials
Molecular Biology (9)
 Molecular Biology (9) Translation Mamoun Ahram, PhD Second semester, 2017-2018 1 Resources This lecture Cooper, Ch. 8 (297-319) 2 General information Protein synthesis involves interactions between three
Molecular Biology (9) Translation Mamoun Ahram, PhD Second semester, 2017-2018 1 Resources This lecture Cooper, Ch. 8 (297-319) 2 General information Protein synthesis involves interactions between three
Direct Interaction between Survivin and Smac/DIABLO Is Essential for the Anti-apoptotic Activity of Survivin during Taxol-induced Apoptosis*
 THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 278, No. 25, Issue of June 20, pp. 23130 23140, 2003 2003 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Direct Interaction
THE JOURNAL OF BIOLOGICAL CHEMISTRY Vol. 278, No. 25, Issue of June 20, pp. 23130 23140, 2003 2003 by The American Society for Biochemistry and Molecular Biology, Inc. Printed in U.S.A. Direct Interaction
Map of AP-Aligned Bio-Rad Kits with Learning Objectives
 Map of AP-Aligned Bio-Rad Kits with Learning Objectives Cover more than one AP Biology Big Idea with these AP-aligned Bio-Rad kits. Big Idea 1 Big Idea 2 Big Idea 3 Big Idea 4 ThINQ! pglo Transformation
Map of AP-Aligned Bio-Rad Kits with Learning Objectives Cover more than one AP Biology Big Idea with these AP-aligned Bio-Rad kits. Big Idea 1 Big Idea 2 Big Idea 3 Big Idea 4 ThINQ! pglo Transformation
Muscle regulation and Actin Topics: Tropomyosin and Troponin, Actin Assembly, Actin-dependent Movement
 1 Muscle regulation and Actin Topics: Tropomyosin and Troponin, Actin Assembly, Actin-dependent Movement In the last lecture, we saw that a repeating alternation between chemical (ATP hydrolysis) and vectorial
1 Muscle regulation and Actin Topics: Tropomyosin and Troponin, Actin Assembly, Actin-dependent Movement In the last lecture, we saw that a repeating alternation between chemical (ATP hydrolysis) and vectorial
Supplementary Information. The protease GtgE from Salmonella exclusively targets. inactive Rab GTPases
 Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
Supplementary Information The protease GtgE from Salmonella exclusively targets inactive Rab GTPases Table of Contents Supplementary Figures... 2 Supplementary Figure 1... 2 Supplementary Figure 2... 3
The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression
 Traffic 2010; 11: 827 842 2010 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2010.01055.x The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression Danming Tang, Hebao Yuan and Yanzhuang
Traffic 2010; 11: 827 842 2010 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2010.01055.x The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression Danming Tang, Hebao Yuan and Yanzhuang
cgmp ELISA Kit (Direct Competitive) Based on Monoclonal Anti-cGMP Antibody
 (FOR RESEARCH USE ONLY. DO NOT USE IT IN CLINICAL DIAGNOSIS!) cgmp ELISA Kit (Direct Competitive) Based on Monoclonal Anti-cGMP Antibody Catalog No: E-EL-DS02 96T This manual must be read attentively and
(FOR RESEARCH USE ONLY. DO NOT USE IT IN CLINICAL DIAGNOSIS!) cgmp ELISA Kit (Direct Competitive) Based on Monoclonal Anti-cGMP Antibody Catalog No: E-EL-DS02 96T This manual must be read attentively and
Malachite Green Phosphate Detection Kit Catalog Number: DY996
 Malachite Green Phosphate Detection Kit Catalog Number: DY996 This Malachite Green Phosphate Detection Kit employs a simple, sensitive, reproducible, and non-radioactive method for measuring inorganic
Malachite Green Phosphate Detection Kit Catalog Number: DY996 This Malachite Green Phosphate Detection Kit employs a simple, sensitive, reproducible, and non-radioactive method for measuring inorganic
DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS. Scientific Background on the Nobel Prize in Medicine 2013
 DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS Scientific Background on the Nobel Prize in Medicine 2013 Daniela Scalet 6/12/2013 The Nobel Prize in Medicine
DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS Scientific Background on the Nobel Prize in Medicine 2013 Daniela Scalet 6/12/2013 The Nobel Prize in Medicine
Quantification of Protein Half-Lives in the Budding Yeast Proteome
 Supporting Methods Quantification of Protein Half-Lives in the Budding Yeast Proteome 1 Cell Growth and Cycloheximide Treatment Three parallel cultures (17 ml) of each TAP-tagged strain were grown in separate
Supporting Methods Quantification of Protein Half-Lives in the Budding Yeast Proteome 1 Cell Growth and Cycloheximide Treatment Three parallel cultures (17 ml) of each TAP-tagged strain were grown in separate
NAD + /NADH Assay [Colorimetric]
![NAD + /NADH Assay [Colorimetric] NAD + /NADH Assay [Colorimetric]](/thumbs/91/105496846.jpg) G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name NAD + /NADH Assay [Colorimetric] (Cat. #786 1539, 786 1540) think proteins! think G-Biosciences
G-Biosciences 1-800-628-7730 1-314-991-6034 technical@gbiosciences.com A Geno Technology, Inc. (USA) brand name NAD + /NADH Assay [Colorimetric] (Cat. #786 1539, 786 1540) think proteins! think G-Biosciences
23-. Shoot and root development depend on ratio of IAA/CK
 Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Balance of Hormones regulate growth and development Environmental factors regulate hormone levels light- e.g. phototropism gravity- e.g. gravitropism temperature Mode of action of each hormone 1. Signal
Chapter 5. Partial purification of granule bound Pi-fA synthase
 Chapter 5 Partial purification of granule bound Pi-fA synthase 5.1 INTRODUCTION The enzyme PHA synthase occurs inside the bacterial cells both, as soluble and granule bound form (Haywood et al., 1989).
Chapter 5 Partial purification of granule bound Pi-fA synthase 5.1 INTRODUCTION The enzyme PHA synthase occurs inside the bacterial cells both, as soluble and granule bound form (Haywood et al., 1989).
Bis sulfone Reagents. Figure 1.
 Bis sulfone Reagents An intact IgG molecule has four accessible inter chain disulfide bonds that can be reduced to form eight free cysteine thiols, which can serve as sites for conjugation. The reaction
Bis sulfone Reagents An intact IgG molecule has four accessible inter chain disulfide bonds that can be reduced to form eight free cysteine thiols, which can serve as sites for conjugation. The reaction
Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis
 Molecular Biology of the Cell Vol. 15, 5001 5011, November 2004 Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis Yang-ja Lee,* Seon-Yong Jeong,* Mariusz
Molecular Biology of the Cell Vol. 15, 5001 5011, November 2004 Roles of the Mammalian Mitochondrial Fission and Fusion Mediators Fis1, Drp1, and Opa1 in Apoptosis Yang-ja Lee,* Seon-Yong Jeong,* Mariusz
Data Sheet. Azide Cy5 RNA T7 Transcription Kit
 Cat. No. Size 1. Description PP-501-Cy5 10 reactions à 40 µl For in vitro use only Quality guaranteed for 12 months Store all components at -20 C. Avoid freeze and thaw cycles. DBCO-Sulfo-Cy5 must be stored
Cat. No. Size 1. Description PP-501-Cy5 10 reactions à 40 µl For in vitro use only Quality guaranteed for 12 months Store all components at -20 C. Avoid freeze and thaw cycles. DBCO-Sulfo-Cy5 must be stored
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering
 Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Big Idea 1: The process of evolution drives the diversity and unity of life.
 Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
HEAVY METAL-INDUCED PROTEINS IN CHLAMYDOMONAS REINHARDTII AND THALASSIOSIRA WEISSFLOGII CELLS
 Proceedings of the 14 th International Conference on Environmental Science and Technology Rhodes, Greece, 3-5 September 2015 HEAVY METAL-INDUCED PROTEINS IN CHLAMYDOMONAS REINHARDTII AND THALASSIOSIRA
Proceedings of the 14 th International Conference on Environmental Science and Technology Rhodes, Greece, 3-5 September 2015 HEAVY METAL-INDUCED PROTEINS IN CHLAMYDOMONAS REINHARDTII AND THALASSIOSIRA
An intracellular phase transition drives nucleolar assembly and size control
 An intracellular phase transition drives nucleolar assembly and size control Steph Weber Department of Biology McGill University Membranes spatially organize eukaryotic cells Cells also contain membraneless
An intracellular phase transition drives nucleolar assembly and size control Steph Weber Department of Biology McGill University Membranes spatially organize eukaryotic cells Cells also contain membraneless
Prof. Fahd M. Nasr. Lebanese university Faculty of sciences I Department of Natural Sciences.
 Prof. Fahd M. Nasr Lebanese university Faculty of sciences I Department of Natural Sciences fnasr@ul.edu.lb B3206 Microbial Genetics Eukaryotic M. G. The yeast Saccharomyces cerevisiae as a genetic model
Prof. Fahd M. Nasr Lebanese university Faculty of sciences I Department of Natural Sciences fnasr@ul.edu.lb B3206 Microbial Genetics Eukaryotic M. G. The yeast Saccharomyces cerevisiae as a genetic model
16 The Cell Cycle. Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization
 The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
A A A A B B1
 LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
JCB Article. The inner membrane protein Mdm33 controls mitochondrial morphology in yeast. The Journal of Cell Biology
 JCB Article Published February 18, 2003 The inner membrane protein Mdm33 controls mitochondrial morphology in yeast Marlies Messerschmitt, 1 Stefan Jakobs, 2 Frank Vogel, 3 Stefan Fritz, 1 Kai Stefan Dimmer,
JCB Article Published February 18, 2003 The inner membrane protein Mdm33 controls mitochondrial morphology in yeast Marlies Messerschmitt, 1 Stefan Jakobs, 2 Frank Vogel, 3 Stefan Fritz, 1 Kai Stefan Dimmer,
Initiation of translation in eukaryotic cells:connecting the head and tail
 Initiation of translation in eukaryotic cells:connecting the head and tail GCCRCCAUGG 1: Multiple initiation factors with distinct biochemical roles (linking, tethering, recruiting, and scanning) 2: 5
Initiation of translation in eukaryotic cells:connecting the head and tail GCCRCCAUGG 1: Multiple initiation factors with distinct biochemical roles (linking, tethering, recruiting, and scanning) 2: 5
Immunoassay Kit (Colorimetric)
 RayBio cgmp Direct Immunoassay Kit (Colorimetric) User Manual Version 1.0 May 25, 2014 RayBio cgmp Direct Immunoassay Kit (Colorimetric) Protocol (Cat#: 68AT-cGMP-S100) RayBiotech, Inc. We Provide You
RayBio cgmp Direct Immunoassay Kit (Colorimetric) User Manual Version 1.0 May 25, 2014 RayBio cgmp Direct Immunoassay Kit (Colorimetric) Protocol (Cat#: 68AT-cGMP-S100) RayBiotech, Inc. We Provide You
Quantification Strategies Using the High Sensitivity Protein 250 Assay for the Agilent 2100 Bioanalyzer. Technical Note. Abstract
 Quantification Strategies Using the High Sensitivity Protein 25 Assay for the Agilent 21 Bioanalyzer Technical Note.. 1. 1. 1..1.1 1. 1. 1... Abstract The Agilent High Sensitivity Protein 25 assay for
Quantification Strategies Using the High Sensitivity Protein 25 Assay for the Agilent 21 Bioanalyzer Technical Note.. 1. 1. 1..1.1 1. 1. 1... Abstract The Agilent High Sensitivity Protein 25 assay for
