Table S1. 10μM_rfp_B1 2,074, ohr 10μM_rfp_C2 2,074, ohr
|
|
|
- Eustace McBride
- 6 years ago
- Views:
Transcription
1 Table S1 Selection without RpoD Insert Start Position Insert End Position Potential Tolerance Gene Candidates 0μM_rfp_A1 2,076, ohr 0μM_rfp_B1 2,082, ohr 0μM_rfp_C1 2,076, ohr 1μM_rfp_A1 2,040,459 2,045,404 pyrf: COG284 (PyrF) Orotidine-5'-phosphate decarboxylase [F] Pp_1816: COG2130 Putative NADP-dependent oxidoreductases [R] Pp_1817: COG1028 (FabG) Dehydrogenases with different specificities (related to short-chain alcohol dehydrogenases) [IQR] Pp_1818: hypothetical protein 1μM_rfp_C1 2,076, ohr 10μM_rfp_B1 2,074, ohr 10μM_rfp_C2 2,074, ohr 100μM_rfp_A1 2,084, ohr 100μM_rfp_C2 5,479,207 5,481,748 prma: COG2264 (PrmA) Ribosomal protein L11 methylase [J] Pp_4819: hypothetical protein Selection with RpoD Insert Start Position Insert End Position Potential Tolerance Gene Candidates 0μM_rpoD_A1 2,078, ohr 0μM_rpoD_B1 2,075, ohr 0μM_rpoD_C1 2,074, ohr 1μM_rpoD_A2 4,908,095 4,994,115 bkdb: COG508 (AceF) Pyruvate/2-oxoglutarate dehydrogenase complex, lpdv: COG1249 (Lpd) Pyruvate/2-oxoglutarate dehydrogenase complex,
2 dihydrolipoamide dehydrogenase (E3) component, and related enzymes [C] Pp_4405: COG2199 FOG: GGDEF domain [T] Pp_4312: hypothetical protein Pp_4313: hypothetical protein Pp_4314: COG3791 Uncharacterized conserved protein [S] Pp_4315: COG384 Predicted epimerase, PhzC/PhzF homolog [R] Pp_4316: COG1052 (LdhA) Lactate dehydrogenase and related dehydrogenases [CHR] Pp_4317: hypothetical protein Pp_4318: hypothetical protein Pp_4319: hypothetical protein Pp_4320: hypothetical protein (Approximate insert size: 4kb) 1μM_rpoD_B1 2,076, ohr 1μM_rpoD_C1 2,074, ohr 10μM_rpoD_A1 (referred to as 10µM_rpoD_A in the main text) 10μM_rpoD_B1 (referred to as 10µM_rpoD_B in the main text) 1,400,873 3,611,923 tola: biopolymer transport protein TolA tolb: COG823 (TolB) Periplasmic component of the Tol biopolymer transport system [U] oprl: COG2885 (OmpA) Outer membrane protein and related peptidoglycanassociated (lipo)proteins[m] Pp_1224: COG1729 Uncharacterized protein conserved in bacteria [S] Pp_1225: COG602 (NrdG) Organic radical activating enzymes [O] Pp_1226: COG603 Predicted PP-loop superfamily ATPase [R] Pp_1227: COG3965 Predicted Co/Zn/Cd cation transporters [P] Pp_3143: COG583 (LysR) Transcriptional regulator [K] Pp_3144: COG1760 (SdaA) L-serine deaminase [E] Pp_3145: conserved hypothetical protein Pp_3146: COG665 (DadA) Glycine/D-amino acid oxidases (deaminating) [E] Pp_3147: COG687 (PotD) Spermidine/putrescine-binding periplasmic protein [E] (Approximate insert size: 3kb) 264,579 3,325,971 Pp_0219: COG2011 (AbcD) ABC-type metal ion transport system, permease component [P] Pp_0220: COG1135 (AbcC) ABC-type metal ion transport system, ATPase component [P]
3 Pp_0221: COG1464 (NlpA) ABC-type metal ion transport system, periplasmic component/surface antigen [P] Pp_0222: COG2141 Coenzyme F420-dependent N5,N10-methylene tetrahydromethanopterin reductase and related flavin-dependent oxidoreductases [C] Pp_0223: COG1960 (CaiA) Acyl-CoA dehydrogenases [I] Pp_0224: COG1960 (CaiA) Acyl-CoA dehydrogenases [I] (Approximate insert size: 2.5kb) 10μM_rpoD_C1 254, ,579 Pp_0204: COG2188 (PhnF) Transcriptional regulators [K] Pp_0205: COG1053 (SdhA) Succinate dehydrogenase/fumarate reductase, flavoprotein subunit [C] Pp_0206: COG1146 Ferredoxin [C] Pp_0207: COG715 (TauA) ABC-type nitrate/sulfonate/bicarbonate transport systems, periplasmic components [P] Pp_0208: COG600 (TauC) ABC-type nitrate/sulfonate/bicarbonate transport system, permease component [P] Pp_0209: COG1116 (TauB) ABC-type nitrate/sulfonate/bicarbonate transport system, ATPase component [P] Pp_0210: COG1413 FOG: HEAT repeat [C] Pp_0211: COG3536 Uncharacterized protein conserved in bacteria [S] Pp_0212: hypothetical protein gabd: COG1012 (PutA) NAD-dependent aldehyde dehydrogenases [C] 10μM_rpoD_B2 2,074, ohr 10μM_rpoD_C3 405, ,791 Pp_0337: COG5001 Predicted signal transduction protein containing a membrane domain, an EAL and a GGDEF domain [T] acef: COG508 (AceF) Pyruvate/2-oxoglutarate dehydrogenase complex, 100μM_rpoD_B1 1,554,529 3,769,293 Pp_1362: COG469 (PykF) Pyruvate kinase [G] Pp_1363: conserved hypothetical protein Pp_1364: conserved hypothetical protein Pp_1365: COG2925 (SbcB) Exonuclease I [L] Pp_1366: transcriptional regulator Pp_1367: COG788 (PurU) Formyltetrahydrofolate hydrolase [F] Pp_1368: COG392 Predicted integral membrane protein [S]
4 Pp_3329: COG4325 Predicted membrane protein [S] Pp_3330: COG1629 (CirA) Outer membrane receptor proteins, mostly Fe transport [P] Pp_3331: conserved hypothetical protein (Approximate insert size: 3kb) 100μM_rpoD_A1 2,078, ohr 100μM_rpoD_B3 4,994,115 4,998,562 bkdb: COG508 (AceF) Pyruvate/2-oxoglutarate dehydrogenase complex, lpdv: COG1249 (Lpd) Pyruvate/2-oxoglutarate dehydrogenase complex, dihydrolipoamide dehydrogenase (E3) component, and related enzymes [C] Pp_4405: COG2199 FOG: GGDEF domain [T] 100μM_rpoD_C3 2,693,079 2,696,334 csuc: COG3121 (FimC) P pilus assembly protein, chaperone PapD [NU] csud: COG3188 (FimD) P pilus assembly protein, porin PapC [NU] 100μM_rpoD_A5 5,182,053 5,188,548 Pp_4561: conserved hypothetical protein Pp_4562: conserved hypothetical protein Pp_4563: COG2996 Uncharacterized protein conserved in bacteria [S] Pp_4564: conserved hypothetical protein acek: COG4579 Isocitrate dehydrogenase kinase/phosphatase [T] Pp_4566: membrane protein Pp_4567: hypothetical protein Pp_4568: membrane protein 1000μM_rpoD_A1 2,082, ohr 1000μM_rpoD_B1 2,086, ohr 1000μM_rpoD_C1 2,086, ohr
5 Table S2 Selection without RpoD Insert Start Position Insert End Position Known Tolerance Gene Candidates FosA1 2,041,821 2,082,192 ohr FosA2 2,041,821 2,082,192 ohr FosB1 3,594,543 2,096,380 ohr FosB2 3,594,543 2,096,594 ohr FosC1 2,069,717 2,111,474 ohr Sample with RpoD Insert Start Position Insert End Position rpod_fosa1 3,912,065 3,945,734 Pp_3456 rpod_fosa2 2,057,403 2,539,218 rpod_fosa3 1,584,206 1,544,143 ttgabc rpod_fosb1 2,438,045 2,399,033 rpod_fosb2 2,438,045 2,399,033 rpod_fosb3 2,438, rpod_fosc1 1,469,748 1,431,620 Pp_1271 rpod_fosc2 3,912,042 3,945,729 rpod_fosc3 2,075,733 2,040,789
Table 5. Genes unique to G. thermodenitrificans NG80-2 Gene ID Gene name Gene product COG functional category
 Table 5. Genes unique to G. thermodenitrificans NG80-2 GT0030 gt30 Methionine--tRNA ligase/methionyl-trna synthetase COG0143, Translation GT0033 gt33 Unknown GT0106 gt106 Ribosomal protein L3 COG0087,
Table 5. Genes unique to G. thermodenitrificans NG80-2 GT0030 gt30 Methionine--tRNA ligase/methionyl-trna synthetase COG0143, Translation GT0033 gt33 Unknown GT0106 gt106 Ribosomal protein L3 COG0087,
COG1487 Predicted nucleic acid binding protein, contains PIN domain R General function prediction only
 COG Function Functional Category COG2258 Uncharacterized protein conserved in bacteria S Function unknown COG4251 Bacteriophytochrome (light regulated signal transduction histidine kinase) T Signal transduction
COG Function Functional Category COG2258 Uncharacterized protein conserved in bacteria S Function unknown COG4251 Bacteriophytochrome (light regulated signal transduction histidine kinase) T Signal transduction
Table S1. 45 significantly up-regulated P. mirabilis HI4320 genes as determined by SAM during iron-limiting microarray a
 Table S1. 45 significantly up-regulated P. mirabilis HI4320 genes as determined by SAM during iron-limiting microarray a PMI no. Gene Description Log 2 fold-change b PMI0029 exbb biopolymer transport protein
Table S1. 45 significantly up-regulated P. mirabilis HI4320 genes as determined by SAM during iron-limiting microarray a PMI no. Gene Description Log 2 fold-change b PMI0029 exbb biopolymer transport protein
TCA Cycle. Voet Biochemistry 3e John Wiley & Sons, Inc.
 TCA Cycle Voet Biochemistry 3e Voet Biochemistry 3e The Electron Transport System (ETS) and Oxidative Phosphorylation (OxPhos) We have seen that glycolysis, the linking step, and TCA generate a large number
TCA Cycle Voet Biochemistry 3e Voet Biochemistry 3e The Electron Transport System (ETS) and Oxidative Phosphorylation (OxPhos) We have seen that glycolysis, the linking step, and TCA generate a large number
Genes for transport and metabolism of spermidine in Ruegeria pomeroyi DSS-3 and other marine bacteria
 The following supplement accompanies the article Genes for transport and metabolism of spermidine in Ruegeria pomeroyi DSS-3 and other marine bacteria Xiaozhen Mou 1,, Shulei Sun 2, Pratibha Rayapati 2,
The following supplement accompanies the article Genes for transport and metabolism of spermidine in Ruegeria pomeroyi DSS-3 and other marine bacteria Xiaozhen Mou 1,, Shulei Sun 2, Pratibha Rayapati 2,
Enzymes and Enzyme Kinetics I. Dr. Kevin Ahern
 Enzymes and Enzyme Kinetics I Dr. Kevin Ahern Enzymatic Reactions Enzymatic Reactions Enzymatically Catalyzed Reactions Background Substrates Bound at Active Site of the Methylene Tetrahydrofolate Reductase
Enzymes and Enzyme Kinetics I Dr. Kevin Ahern Enzymatic Reactions Enzymatic Reactions Enzymatically Catalyzed Reactions Background Substrates Bound at Active Site of the Methylene Tetrahydrofolate Reductase
What is an enzyme? Lecture 12: Enzymes & Kinetics I Introduction to Enzymes and Kinetics. Margaret A. Daugherty Fall 2004 KEY FEATURES OF ENZYMES
 Lecture 12: Enzymes & Kinetics I Introduction to Enzymes and Kinetics Margaret A. Daugherty Fall 2004 What is an enzyme? General Properties Mostly proteins, but some are actually RNAs Biological catalysts
Lecture 12: Enzymes & Kinetics I Introduction to Enzymes and Kinetics Margaret A. Daugherty Fall 2004 What is an enzyme? General Properties Mostly proteins, but some are actually RNAs Biological catalysts
GO ID GO term Number of members GO: translation 225 GO: nucleosome 50 GO: calcium ion binding 76 GO: structural
 GO ID GO term Number of members GO:0006412 translation 225 GO:0000786 nucleosome 50 GO:0005509 calcium ion binding 76 GO:0003735 structural constituent of ribosome 170 GO:0019861 flagellum 23 GO:0005840
GO ID GO term Number of members GO:0006412 translation 225 GO:0000786 nucleosome 50 GO:0005509 calcium ion binding 76 GO:0003735 structural constituent of ribosome 170 GO:0019861 flagellum 23 GO:0005840
What is an enzyme? Lecture 12: Enzymes & Kinetics I Introduction to Enzymes and Kinetics. Margaret A. Daugherty Fall General Properties
 Lecture 12: Enzymes & Kinetics I Introduction to Enzymes and Kinetics Margaret A. Daugherty Fall 2003 ENZYMES: Why, what, when, where, how? All but the who! What: proteins that exert kinetic control over
Lecture 12: Enzymes & Kinetics I Introduction to Enzymes and Kinetics Margaret A. Daugherty Fall 2003 ENZYMES: Why, what, when, where, how? All but the who! What: proteins that exert kinetic control over
Biologic catalysts 1. Shared properties with chemical catalysts a. Enzymes are neither consumed nor produced during the course of a reaction. b.
 Enzyme definition Enzymes are protein catalysts that increase the velocity of a chemical reaction and are not consumed during the reaction they catalyze. [Note: Some types of RNA can act like enzymes,
Enzyme definition Enzymes are protein catalysts that increase the velocity of a chemical reaction and are not consumed during the reaction they catalyze. [Note: Some types of RNA can act like enzymes,
Catalysis. Instructor: Dr. Tsung-Lin Li Genomics Research Center Academia Sinica
 Catalysis Instructor: Dr. Tsung-Lin Li Genomics Research Center Academia Sinica References: Biochemistry" by Donald Voet and Judith G. Voet Biochemistry" by Christopher K. Mathews, K. E. Van Hold and Kevin
Catalysis Instructor: Dr. Tsung-Lin Li Genomics Research Center Academia Sinica References: Biochemistry" by Donald Voet and Judith G. Voet Biochemistry" by Christopher K. Mathews, K. E. Van Hold and Kevin
Biological Process Term Enrichment
 Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
Biological Process Term Enrichment cellular protein localization cellular macromolecule localization intracellular protein transport intracellular transport generation of precursor metabolites and energy
(starvation). Description a. Predicted operon members b. Gene no. a. Relative change in expression (n-fold) mutant vs. wild type.
 1 Table S1. Genes whose expression differ in the phyr mutant 8402 and/or in the ecfg mutant 8404 compared with the wild type when grown to the mid-exponential phase (OD600 0.5-0.7) in rich medium (PSY)
1 Table S1. Genes whose expression differ in the phyr mutant 8402 and/or in the ecfg mutant 8404 compared with the wild type when grown to the mid-exponential phase (OD600 0.5-0.7) in rich medium (PSY)
Energy and Cellular Metabolism
 1 Chapter 4 About This Chapter Energy and Cellular Metabolism 2 Energy in biological systems Chemical reactions Enzymes Metabolism Figure 4.1 Energy transfer in the environment Table 4.1 Properties of
1 Chapter 4 About This Chapter Energy and Cellular Metabolism 2 Energy in biological systems Chemical reactions Enzymes Metabolism Figure 4.1 Energy transfer in the environment Table 4.1 Properties of
Transcriptome analysis of leaf tissue from Bermudagrass (Cynodon dactylon) using a normalised cdna library
 Ó CSIRO 2008 ISSN 1445-4408 10.1071/FP08133_AC Functional Plant Biology, 2008, 35, 585 594 Transcriptome analysis of leaf tissue from Bermudagrass (Cynodon dactylon) using a normalised cdna library Changsoo
Ó CSIRO 2008 ISSN 1445-4408 10.1071/FP08133_AC Functional Plant Biology, 2008, 35, 585 594 Transcriptome analysis of leaf tissue from Bermudagrass (Cynodon dactylon) using a normalised cdna library Changsoo
Photoperiodic effect on protein profiles of potato petiole
 Photoperiodic effect on protein profiles of potato petiole Shweta Shah 1, Young Jin-Lee 2, David J. Hannapel 3 and A. Gururaj Rao* 1 1 Department of Biochemistry, Biophysics and Molecular Biology, Iowa
Photoperiodic effect on protein profiles of potato petiole Shweta Shah 1, Young Jin-Lee 2, David J. Hannapel 3 and A. Gururaj Rao* 1 1 Department of Biochemistry, Biophysics and Molecular Biology, Iowa
Supplementary Figure 1. The proportion S. aureus CFU of the total CFU (S. aureus + E. faecalis CFU) per host in worms
 Supplementary Figure 1. The proportion S. aureus CFU of the total CFU (S. aureus + E. faecalis CFU) per host in worms alive or dead at 24 hours of exposure. Two sample t-test: t = 1.22, df = 10, P= 0.25.
Supplementary Figure 1. The proportion S. aureus CFU of the total CFU (S. aureus + E. faecalis CFU) per host in worms alive or dead at 24 hours of exposure. Two sample t-test: t = 1.22, df = 10, P= 0.25.
The srka gene encodes a protein member of CAMK serine- threonine protein kinase superfamily ARL Deletion of the srka gene.
 FIG. S1. The srka gene encodes a protein member of CAMK serine- threonine protein kinase superfamily. Protein alignment of A. nidulans SrkA, S. pombe Srk1, S. cerevisiae Rck, C. albicans Rck and homologs
FIG. S1. The srka gene encodes a protein member of CAMK serine- threonine protein kinase superfamily. Protein alignment of A. nidulans SrkA, S. pombe Srk1, S. cerevisiae Rck, C. albicans Rck and homologs
Division Ave. High School AP Biology
 Overview 10 reactions u convert () to pyruvate (3C) u produces: 4 & NADH u consumes: u net: & NADH C-C-C-C-C-C fructose-1,6bp P-C-C-C-C-C-C-P DHAP P-C-C-C G3P C-C-C-P H P i P i pyruvate C-C-C 4 4 NAD +
Overview 10 reactions u convert () to pyruvate (3C) u produces: 4 & NADH u consumes: u net: & NADH C-C-C-C-C-C fructose-1,6bp P-C-C-C-C-C-C-P DHAP P-C-C-C G3P C-C-C-P H P i P i pyruvate C-C-C 4 4 NAD +
MOLECULAR CELL BIOLOGY
 1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
1 Lodish Berk Kaiser Krieger scott Bretscher Ploegh Matsudaira MOLECULAR CELL BIOLOGY SEVENTH EDITION CHAPTER 13 Moving Proteins into Membranes and Organelles Copyright 2013 by W. H. Freeman and Company
Supplementary Table 3. Membrane/Signaling/Neural Genes of the DmSP. FBgn CG5265 acetyltransferase amino acid metabolism
 Supplementary Table 3 Membrane/Signaling/Neural Genes of the DmSP FlyBase ID Gene Name Molecular function summary Membrane Biological process summary FBgn0038486 CG5265 acetyltransferase amino acid metabolism
Supplementary Table 3 Membrane/Signaling/Neural Genes of the DmSP FlyBase ID Gene Name Molecular function summary Membrane Biological process summary FBgn0038486 CG5265 acetyltransferase amino acid metabolism
Transmembrane Domains (TMDs) of ABC transporters
 Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Transmembrane Domains (TMDs) of ABC transporters Most ABC transporters contain heterodimeric TMDs (e.g. HisMQ, MalFG) TMDs show only limited sequence homology (high diversity) High degree of conservation
Bio102 Problems Photosynthesis
 Bio102 Problems Photosynthesis 1. Why is it advantageous for chloroplasts to have a very large (in surface area) thylakoid membrane contained within the inner membrane? A. This limits the amount of stroma
Bio102 Problems Photosynthesis 1. Why is it advantageous for chloroplasts to have a very large (in surface area) thylakoid membrane contained within the inner membrane? A. This limits the amount of stroma
BBS2710 Microbial Physiology. Module 5 - Energy and Metabolism
 BBS2710 Microbial Physiology Module 5 - Energy and Metabolism Topics Energy production - an overview Fermentation Aerobic respiration Alternative approaches to respiration Photosynthesis Summary Introduction
BBS2710 Microbial Physiology Module 5 - Energy and Metabolism Topics Energy production - an overview Fermentation Aerobic respiration Alternative approaches to respiration Photosynthesis Summary Introduction
Electron Transport Chain (Respiratory Chain) - exercise - Vladimíra Kvasnicová
 Electron Transport Chain (Respiratory Chain) - exercise - Vladimíra Kvasnicová Respiratory chain (RCH) a) is found in all cells b) is located in a mitochondrion c) includes enzymes integrated in the inner
Electron Transport Chain (Respiratory Chain) - exercise - Vladimíra Kvasnicová Respiratory chain (RCH) a) is found in all cells b) is located in a mitochondrion c) includes enzymes integrated in the inner
Parkhill et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica
 Parkhill et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica Supplementary table 1: Genes unique to each Bordetella species
Parkhill et al. Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica Supplementary table 1: Genes unique to each Bordetella species
Gene expression profiles of Nitrosomonas europaea, an obligate chemolithotroph. Daniel Arp. Department of Botany and Plant Pathology.
 Gene expression profiles of Nitrosomonas europaea, an obligate chemolithotroph. Daniel Arp. Department of Botany and Plant Pathology. Oregon State University, Corvallis OR. 97331 Technical report for product
Gene expression profiles of Nitrosomonas europaea, an obligate chemolithotroph. Daniel Arp. Department of Botany and Plant Pathology. Oregon State University, Corvallis OR. 97331 Technical report for product
Characters related to higher starch accumulation in cassava storage roots
 Characters related to higher starch accumulation in cassava storage roots You-Zhi Li **1, Jian-Yu Zhao *1, San-Min Wu 1, Xian-Wei Fan 1, Xing-Lu Luo 1 & Bao-Shan Chen **1 1 State Key Laboratory for Conservation
Characters related to higher starch accumulation in cassava storage roots You-Zhi Li **1, Jian-Yu Zhao *1, San-Min Wu 1, Xian-Wei Fan 1, Xing-Lu Luo 1 & Bao-Shan Chen **1 1 State Key Laboratory for Conservation
Life 21 - Aerobic respiration Raven & Johnson Chapter 9 (parts)
 1 Life 21 - Aerobic respiration Raven & Johnson Chapter 9 (parts) Objectives 1: Describe the overall action of the Krebs cycle in generating ATP, NADH and FADH 2 from acetyl-coa 2: Understand the generation
1 Life 21 - Aerobic respiration Raven & Johnson Chapter 9 (parts) Objectives 1: Describe the overall action of the Krebs cycle in generating ATP, NADH and FADH 2 from acetyl-coa 2: Understand the generation
SUPPLEMENTAL MATERIAL
 SUPPLEMENTAL MATERIAL SUPPLEMENTAL RESULTS Microarray analysis of F. tularensis LVS within hepatocytes: As described in Materials and Methods, the global gene expression of LVS organisms grown in FL83B
SUPPLEMENTAL MATERIAL SUPPLEMENTAL RESULTS Microarray analysis of F. tularensis LVS within hepatocytes: As described in Materials and Methods, the global gene expression of LVS organisms grown in FL83B
Additional File 9. Gene families amplified in A. adeninivorans.
 Additional File. Gene families amplified in A. adeninivorans. Figure SA Duplication rate per gene per branch of the species tree leading to A. adeninivorans. Species tree showing the evolution of the species
Additional File. Gene families amplified in A. adeninivorans. Figure SA Duplication rate per gene per branch of the species tree leading to A. adeninivorans. Species tree showing the evolution of the species
RECONSTRUCTING GENE REGULATORY NETWORKS FROM FUNGAL TRANSCRIPTOMIC DATA USING BAYESIAN NETWORK
 RECONSTRUCTING GENE REGULATORY NETWORKS FROM FUNGAL TRANSCRIPTOMIC DATA USING BAYESIAN NETWORK Li Guo Fungal Comparative Genomics Laboratory Department of Biochemistry and Molecular biology University
RECONSTRUCTING GENE REGULATORY NETWORKS FROM FUNGAL TRANSCRIPTOMIC DATA USING BAYESIAN NETWORK Li Guo Fungal Comparative Genomics Laboratory Department of Biochemistry and Molecular biology University
Chapter 8 Metabolism: Energy, Enzymes, and Regulation
 Chapter 8 Metabolism: Energy, Enzymes, and Regulation Energy: Capacity to do work or cause a particular change. Thus, all physical and chemical processes are the result of the application or movement of
Chapter 8 Metabolism: Energy, Enzymes, and Regulation Energy: Capacity to do work or cause a particular change. Thus, all physical and chemical processes are the result of the application or movement of
Center for Academic Services & Advising
 March 2, 2017 Biology I CSI Worksheet 6 1. List the four components of cellular respiration, where it occurs in the cell, and list major products consumed and produced in each step. i. Hint: Think about
March 2, 2017 Biology I CSI Worksheet 6 1. List the four components of cellular respiration, where it occurs in the cell, and list major products consumed and produced in each step. i. Hint: Think about
Generation of new compounds through unbalanced transcription of landomycin A
 Applied Microbiology and Biotechnology Generation of new compounds through unbalanced transcription of landomycin A cluster Maksym Myronovskyi #1, Elke Brötz #1, Birgit Rosenkränzer 1, Niko Manderscheid
Applied Microbiology and Biotechnology Generation of new compounds through unbalanced transcription of landomycin A cluster Maksym Myronovskyi #1, Elke Brötz #1, Birgit Rosenkränzer 1, Niko Manderscheid
BIOCHEMISTRY. František Vácha. JKU, Linz.
 BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
Table S1: Domain based (meta)genome comparison of selected metagenomes of methanotrophs and genomes of selected methanogens*
 ANME-1-s ANME-1-m ANME-2a ANME-2d-h ANME-2d-a AM A M-1 M-2 M-3 H-1 H-2 H-3 H-4 S Table S1: Domain based (meta)genome comparison of selected metagenomes of methanotrophs and genomes of selected methanogens*
ANME-1-s ANME-1-m ANME-2a ANME-2d-h ANME-2d-a AM A M-1 M-2 M-3 H-1 H-2 H-3 H-4 S Table S1: Domain based (meta)genome comparison of selected metagenomes of methanotrophs and genomes of selected methanogens*
A. Reaction Mechanisms and Catalysis (1) proximity effect (2) acid-base catalysts (3) electrostatic (4) functional groups (5) structural flexibility
 (P&S Ch 5; Fer Ch 2, 9; Palm Ch 10,11; Zub Ch 9) A. Reaction Mechanisms and Catalysis (1) proximity effect (2) acid-base catalysts (3) electrostatic (4) functional groups (5) structural flexibility B.
(P&S Ch 5; Fer Ch 2, 9; Palm Ch 10,11; Zub Ch 9) A. Reaction Mechanisms and Catalysis (1) proximity effect (2) acid-base catalysts (3) electrostatic (4) functional groups (5) structural flexibility B.
20. Electron Transport and Oxidative Phosphorylation
 20. Electron Transport and Oxidative Phosphorylation 20.1 What Role Does Electron Transport Play in Metabolism? Electron transport - Role of oxygen in metabolism as final acceptor of electrons - In inner
20. Electron Transport and Oxidative Phosphorylation 20.1 What Role Does Electron Transport Play in Metabolism? Electron transport - Role of oxygen in metabolism as final acceptor of electrons - In inner
MITOCW watch?v=0xajihttcns
 MITOCW watch?v=0xajihttcns The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources for free. To
MITOCW watch?v=0xajihttcns The following content is provided under a Creative Commons license. Your support will help MIT OpenCourseWare continue to offer high quality educational resources for free. To
Population transcriptomics uncovers the regulation of gene. expression variation in adaptation to changing environment
 Supplementary information Population transcriptomics uncovers the regulation of gene expression variation in adaptation to changing environment Qin Xu 1, Caiyun Zhu 2,4, Yangyang Fan 1,4, Zhihong Song
Supplementary information Population transcriptomics uncovers the regulation of gene expression variation in adaptation to changing environment Qin Xu 1, Caiyun Zhu 2,4, Yangyang Fan 1,4, Zhihong Song
ENZYMES. by: Dr. Hadi Mozafari
 ENZYMES by: Dr. Hadi Mozafari 1 Specifications Often are Polymers Have a protein structures Enzymes are the biochemical reactions Katalyzers Enzymes are Simple & Complex compounds 2 Enzymatic Reactions
ENZYMES by: Dr. Hadi Mozafari 1 Specifications Often are Polymers Have a protein structures Enzymes are the biochemical reactions Katalyzers Enzymes are Simple & Complex compounds 2 Enzymatic Reactions
Oxidative Phosphorylation versus. Photophosphorylation
 Photosynthesis Oxidative Phosphorylation versus Photophosphorylation Oxidative Phosphorylation Electrons from the reduced cofactors NADH and FADH 2 are passed to proteins in the respiratory chain. In eukaryotes,
Photosynthesis Oxidative Phosphorylation versus Photophosphorylation Oxidative Phosphorylation Electrons from the reduced cofactors NADH and FADH 2 are passed to proteins in the respiratory chain. In eukaryotes,
Enzymes I. Dr. Mamoun Ahram Summer semester,
 Enzymes I Dr. Mamoun Ahram Summer semester, 2017-2018 Resources Mark's Basic Medical Biochemistry Other resources NCBI Bookshelf: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=books The Medical Biochemistry
Enzymes I Dr. Mamoun Ahram Summer semester, 2017-2018 Resources Mark's Basic Medical Biochemistry Other resources NCBI Bookshelf: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=books The Medical Biochemistry
Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Outer Glycolysis mitochondrial membrane Glucose ATP
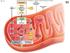 Fig. 7.5 uter Glycolysis mitochondrial membrane Glucose Intermembrane space xidation Mitochondrial matrix Acetyl-oA Krebs FAD e NAD + FAD Inner mitochondrial membrane e Electron e Transport hain hemiosmosis
Fig. 7.5 uter Glycolysis mitochondrial membrane Glucose Intermembrane space xidation Mitochondrial matrix Acetyl-oA Krebs FAD e NAD + FAD Inner mitochondrial membrane e Electron e Transport hain hemiosmosis
Genomic insights into high exopolysaccharide-producing dairy starter. bacterium Streptococcus thermophilus ASCC 1275
 Supplementary information of Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275 Qinglong Wu, Hein Min Tun, Frederick Chi-Ching Leung, Nagendra
Supplementary information of Genomic insights into high exopolysaccharide-producing dairy starter bacterium Streptococcus thermophilus ASCC 1275 Qinglong Wu, Hein Min Tun, Frederick Chi-Ching Leung, Nagendra
Patrick: An Introduction to Medicinal Chemistry 5e Chapter 04
 01) Which of the following statements is not true about receptors? a. Most receptors are proteins situated inside the cell. b. Receptors contain a hollow or cleft on their surface which is known as a binding
01) Which of the following statements is not true about receptors? a. Most receptors are proteins situated inside the cell. b. Receptors contain a hollow or cleft on their surface which is known as a binding
A Microfluidic Platform Enables Large-Scale Single-Cell Screening to Identify Genes Involved in Bacterial Persistence
 A Microfluidic Platform Enables Large-Scale Single-Cell Screening to Identify Genes Involved in Bacterial Persistence THÈSE N O 7641 (2017) PRÉSENTÉE LE 31 MARS 2017 À LA FACULTÉ DES SCIENCES DE LA VIE
A Microfluidic Platform Enables Large-Scale Single-Cell Screening to Identify Genes Involved in Bacterial Persistence THÈSE N O 7641 (2017) PRÉSENTÉE LE 31 MARS 2017 À LA FACULTÉ DES SCIENCES DE LA VIE
The roles of RelA/(p)ppGpp in glucose-starvation induced adaptive
 Supplementary Information The roles of RelA/(p)ppGpp in glucose-starvation induced adaptive response in the zoonotic Streptococcus suis Tengfei Zhang, 1, 2, a Jiawen Zhu, 1, a Shun Wei, 1 Qingping Luo,
Supplementary Information The roles of RelA/(p)ppGpp in glucose-starvation induced adaptive response in the zoonotic Streptococcus suis Tengfei Zhang, 1, 2, a Jiawen Zhu, 1, a Shun Wei, 1 Qingping Luo,
Subsystem: Succinate dehydrogenase
 Subsystem: Succinate dehydrogenase Olga Vassieva Fellowship for Interpretation of Genomes The super-macromolecular respiratory complex II (succinate:quinone oxidoreductase) couples the oxidation of succinate
Subsystem: Succinate dehydrogenase Olga Vassieva Fellowship for Interpretation of Genomes The super-macromolecular respiratory complex II (succinate:quinone oxidoreductase) couples the oxidation of succinate
M i t o c h o n d r i a
 M i t o c h o n d r i a Dr. Diala Abu-Hassan School of Medicine dr.abuhassand@gmail.com Mitochondria Function: generation of metabolic energy in eukaryotic cells Generation of ATP from the breakdown of
M i t o c h o n d r i a Dr. Diala Abu-Hassan School of Medicine dr.abuhassand@gmail.com Mitochondria Function: generation of metabolic energy in eukaryotic cells Generation of ATP from the breakdown of
number Done by Corrected by Doctor Nafeth Abu Tarboush
 number 6 Done by أنس القيشاوي Corrected by Zaid Emad Doctor Nafeth Abu Tarboush 1 P a g e In the previous lecture, we talked about redox reactions and the reduction potential briefly and how it can help
number 6 Done by أنس القيشاوي Corrected by Zaid Emad Doctor Nafeth Abu Tarboush 1 P a g e In the previous lecture, we talked about redox reactions and the reduction potential briefly and how it can help
Metabolism. Fermentation vs. Respiration. End products of fermentations are waste products and not fully.
 Outline: Metabolism Part I: Fermentations Part II: Respiration Part III: Metabolic Diversity Learning objectives are: Learn about respiratory metabolism, ATP generation by respiration linked (oxidative)
Outline: Metabolism Part I: Fermentations Part II: Respiration Part III: Metabolic Diversity Learning objectives are: Learn about respiratory metabolism, ATP generation by respiration linked (oxidative)
The Electron-Transfer Chain and Oxidative Phosphorylation
 The Electron-Transfer Chain and Oxidative Phosphorylation The Mitochondrion - Scene of the Action 1.1 The inner mitochondrial membrane compartmentalizes metabolic functions. Although mitochondria in different
The Electron-Transfer Chain and Oxidative Phosphorylation The Mitochondrion - Scene of the Action 1.1 The inner mitochondrial membrane compartmentalizes metabolic functions. Although mitochondria in different
Constraint-Based Workshops
 Constraint-Based Workshops 2. Reconstruction Databases November 29 th, 2007 Defining Metabolic Reactions ydbh hslj ldha 1st level: Primary metabolites LAC 2nd level: Neutral Formulas C 3 H 6 O 3 Charged
Constraint-Based Workshops 2. Reconstruction Databases November 29 th, 2007 Defining Metabolic Reactions ydbh hslj ldha 1st level: Primary metabolites LAC 2nd level: Neutral Formulas C 3 H 6 O 3 Charged
Supporting Information
 Supporting Information Proteomic Analyses of Cysteine Redox in High-fat-fed and Fasted Mouse Livers: Implications for Liver Metabolic Homeostasis Yixing Li 1#, Zupeng Luo 1#, Xilong Wu 2, Jun Zhu 2, Kai
Supporting Information Proteomic Analyses of Cysteine Redox in High-fat-fed and Fasted Mouse Livers: Implications for Liver Metabolic Homeostasis Yixing Li 1#, Zupeng Luo 1#, Xilong Wu 2, Jun Zhu 2, Kai
CSCE555 Bioinformatics. Protein Function Annotation
 CSCE555 Bioinformatics Protein Function Annotation Why we need to do function annotation? Fig from: Network-based prediction of protein function. Molecular Systems Biology 3:88. 2007 What s function? The
CSCE555 Bioinformatics Protein Function Annotation Why we need to do function annotation? Fig from: Network-based prediction of protein function. Molecular Systems Biology 3:88. 2007 What s function? The
Lecture 10. Proton Gradient-dependent ATP Synthesis. Oxidative. Photo-Phosphorylation
 Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation Model of the Electron Transport Chain (ETC) Glycerol-3-P Shuttle Outer Mitochondrial Membrane G3P DHAP
Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation Model of the Electron Transport Chain (ETC) Glycerol-3-P Shuttle Outer Mitochondrial Membrane G3P DHAP
Proteins. Department of Biochemistry Faculty of medicine University of Szeged
 Proteins Classification Function Structure Connecting peptide parts Synthesis Transcription - nucleus Translation - cytoplasma Posttranslation changes (folding) - endoplasmatic reticulum Amino acids Amino
Proteins Classification Function Structure Connecting peptide parts Synthesis Transcription - nucleus Translation - cytoplasma Posttranslation changes (folding) - endoplasmatic reticulum Amino acids Amino
Module organization and variance in protein-protein interaction networks
 Module organization and variance in protein-protein interaction networks Chun-Yu Lin 1, Tsai-Ling Lee 1, Yi-Yuan Chiu 1, Yi-Wei Lin 1, Yu-Shu Lo 1, Chih-Ta Lin 1, and Jinn-Moon Yang 1,2* 1 Institute of
Module organization and variance in protein-protein interaction networks Chun-Yu Lin 1, Tsai-Ling Lee 1, Yi-Yuan Chiu 1, Yi-Wei Lin 1, Yu-Shu Lo 1, Chih-Ta Lin 1, and Jinn-Moon Yang 1,2* 1 Institute of
Translation Part 2 of Protein Synthesis
 Translation Part 2 of Protein Synthesis IN: How is transcription like making a jello mold? (be specific) What process does this diagram represent? A. Mutation B. Replication C.Transcription D.Translation
Translation Part 2 of Protein Synthesis IN: How is transcription like making a jello mold? (be specific) What process does this diagram represent? A. Mutation B. Replication C.Transcription D.Translation
Basic Concepts of Enzyme Action. Enzymes. Rate Enhancement 9/17/2015. Stryer Short Course Chapter 6
 Basic Concepts of Enzyme Action Stryer Short Course Chapter 6 Enzymes Biocatalysts Active site Substrate and product Catalyzed rate Uncatalyzed rate Rate Enhancement Which is a better catalyst, carbonic
Basic Concepts of Enzyme Action Stryer Short Course Chapter 6 Enzymes Biocatalysts Active site Substrate and product Catalyzed rate Uncatalyzed rate Rate Enhancement Which is a better catalyst, carbonic
Ca. Liberibacter asiaticus Proteins Orthologous with psyma-encoded Proteins of Sinorhizobium meliloti: Hypothetical Roles in Plant Host Interaction
 Ca. Liberibacter asiaticus Proteins Orthologous with psyma-encoded Proteins of Sinorhizobium meliloti: Hypothetical Roles in Plant Host Interaction L. David Kuykendall, Jonathan Y. Shao, John S. Hartung*
Ca. Liberibacter asiaticus Proteins Orthologous with psyma-encoded Proteins of Sinorhizobium meliloti: Hypothetical Roles in Plant Host Interaction L. David Kuykendall, Jonathan Y. Shao, John S. Hartung*
Enzymes and Protein Structure
 Enzymes and Protein Structure Last Week PTM s We (Re)Learned About Primary Structure And Tertiary Structure S-Q-D-A-G-M-Q-Q-G-A-D-M-D-Q-V-S-A Secondary Structure Enzymes What are these crazy things called
Enzymes and Protein Structure Last Week PTM s We (Re)Learned About Primary Structure And Tertiary Structure S-Q-D-A-G-M-Q-Q-G-A-D-M-D-Q-V-S-A Secondary Structure Enzymes What are these crazy things called
Chapter 2 Energy in Biology Demand and Use
 Chapter 2 Energy in Biology Demand and Use A coupled energy source is a prerequisite of sustained dynamics in thermodynamically open systems. Abstract From the point of view of energy management in biological
Chapter 2 Energy in Biology Demand and Use A coupled energy source is a prerequisite of sustained dynamics in thermodynamically open systems. Abstract From the point of view of energy management in biological
Part II => PROTEINS and ENZYMES. 2.5 Enzyme Properties 2.5a Enzyme Nomenclature 2.5b Transition State Theory
 Part II => PROTEINS and ENZYMES 2.5 Enzyme Properties 2.5a Enzyme Nomenclature 2.5b Transition State Theory Section 2.5a: Enzyme Nomenclature Synopsis 2.5a - Enzymes are biological catalysts they are almost
Part II => PROTEINS and ENZYMES 2.5 Enzyme Properties 2.5a Enzyme Nomenclature 2.5b Transition State Theory Section 2.5a: Enzyme Nomenclature Synopsis 2.5a - Enzymes are biological catalysts they are almost
NUCLEOTIDE BINDING ENZYMES
 NUCLEOTIDE BINDING ENZYMES The Rossmann fold Relationship between sequence, structure and function. Anna Casas, Júlia Gasull and Nerea Vega Index 1. Introduction: Adenine nucleotides 1. The most commonly
NUCLEOTIDE BINDING ENZYMES The Rossmann fold Relationship between sequence, structure and function. Anna Casas, Júlia Gasull and Nerea Vega Index 1. Introduction: Adenine nucleotides 1. The most commonly
Rex-Family Repressor/NADH Complex
 Kasey Royer Michelle Lukosi Rex-Family Repressor/NADH Complex Part A The biological sensing protein that we selected is the Rex-family repressor/nadh complex. We chose this sensor because it is a calcium
Kasey Royer Michelle Lukosi Rex-Family Repressor/NADH Complex Part A The biological sensing protein that we selected is the Rex-family repressor/nadh complex. We chose this sensor because it is a calcium
A Novel Organotrophic Nitrate-reducing Fe(II)-oxidizing Bacterium. Isolated from a Paddy Soil and Draft Genome Sequencing
 Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2017 Supplementary information for A Novel Organotrophic Nitrate-reducing Fe(II)-oxidizing Bacterium
Electronic Supplementary Material (ESI) for RSC Advances. This journal is The Royal Society of Chemistry 2017 Supplementary information for A Novel Organotrophic Nitrate-reducing Fe(II)-oxidizing Bacterium
Cellular Neuroanatomy I The Prototypical Neuron: Soma. Reading: BCP Chapter 2
 Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Cellular Neuroanatomy I The Prototypical Neuron: Soma Reading: BCP Chapter 2 Functional Unit of the Nervous System The functional unit of the nervous system is the neuron. Neurons are cells specialized
Reconstruction and Analysis of Metabolic Networks
 1 Reconstruction and Analysis of Metabolic Networks 2 Outline What is a Reconstruction? Data Collection Interactions Between Network Components Special Considerations Applications Genome-scale Metabolic
1 Reconstruction and Analysis of Metabolic Networks 2 Outline What is a Reconstruction? Data Collection Interactions Between Network Components Special Considerations Applications Genome-scale Metabolic
A genome sequence based discriminator for vancomycin intermediate Staphyolococcus aureus Supplementary Methods
 1 Journal of Bacteriology Computational Biology Section 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 Supplementary Information for: A genome sequence based discriminator
1 Journal of Bacteriology Computational Biology Section 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 Supplementary Information for: A genome sequence based discriminator
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy
 Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
BCH 4054 Spring 2001 Chapter 21 Lecture Notes
 BCH 4054 Spring 2001 Chapter 21 Lecture Notes 1 Chapter 21 Electron Transport and Oxidative Phosphorylation 2 Overview Oxidation of NADH and CoQH 2 produced in TCA cycle by O 2 is very exergonic. Some
BCH 4054 Spring 2001 Chapter 21 Lecture Notes 1 Chapter 21 Electron Transport and Oxidative Phosphorylation 2 Overview Oxidation of NADH and CoQH 2 produced in TCA cycle by O 2 is very exergonic. Some
Warm-Up. Explain how a secondary messenger is activated, and how this affects gene expression. (LO 3.22)
 Warm-Up Explain how a secondary messenger is activated, and how this affects gene expression. (LO 3.22) Yesterday s Picture The first cell on Earth (approx. 3.5 billion years ago) was simple and prokaryotic,
Warm-Up Explain how a secondary messenger is activated, and how this affects gene expression. (LO 3.22) Yesterday s Picture The first cell on Earth (approx. 3.5 billion years ago) was simple and prokaryotic,
Enzymes and Enzyme Kinetics I. Dr.Nabil Bashir
 Enzymes and Enzyme Kinetics I Dr.Nabil Bashir Enzymes and Enzyme Kinetics I: Outlines Enzymes - Basic Concepts and Kinetics Enzymes as Catalysts Enzyme rate enhancement / Enzyme specificity Enzyme cofactors
Enzymes and Enzyme Kinetics I Dr.Nabil Bashir Enzymes and Enzyme Kinetics I: Outlines Enzymes - Basic Concepts and Kinetics Enzymes as Catalysts Enzyme rate enhancement / Enzyme specificity Enzyme cofactors
Quiz answers. Allele. BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 17: The Quiz (and back to Eukaryotic DNA)
 BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 17: The Quiz (and back to Eukaryotic DNA) http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Quiz answers Kinase: An enzyme
BIO 5099: Molecular Biology for Computer Scientists (et al) Lecture 17: The Quiz (and back to Eukaryotic DNA) http://compbio.uchsc.edu/hunter/bio5099 Larry.Hunter@uchsc.edu Quiz answers Kinase: An enzyme
Change to Office Hours this Friday and next Monday. Tomorrow (Abel): 8:30 10:30 am. Monday (Katrina): Cancelled (05/04)
 Change to Office Hours this Friday and next Monday Tomorrow (Abel): 8:30 10:30 am Monday (Katrina): Cancelled (05/04) Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation
Change to Office Hours this Friday and next Monday Tomorrow (Abel): 8:30 10:30 am Monday (Katrina): Cancelled (05/04) Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation
number Done by Corrected by Doctor Nafeth Abu Tarboush
 number 8 Done by Ali Yaghi Corrected by Mamoon Mohamad Alqtamin Doctor Nafeth Abu Tarboush 0 P a g e Oxidative phosphorylation Oxidative phosphorylation has 3 major aspects: 1. It involves flow of electrons
number 8 Done by Ali Yaghi Corrected by Mamoon Mohamad Alqtamin Doctor Nafeth Abu Tarboush 0 P a g e Oxidative phosphorylation Oxidative phosphorylation has 3 major aspects: 1. It involves flow of electrons
PHOTOSYNTHESIS: A BRIEF STORY!!!!!
 PHOTOSYNTHESIS: A BRIEF STORY!!!!! This is one of the most important biochemical processes in plants and is amongst the most expensive biochemical processes in plant in terms of investment. Photosynthesis
PHOTOSYNTHESIS: A BRIEF STORY!!!!! This is one of the most important biochemical processes in plants and is amongst the most expensive biochemical processes in plant in terms of investment. Photosynthesis
CHLOROPLASTS, CALVIN CYCLE, PHOTOSYNTHETIC ELECTRON TRANSFER AND PHOTOPHOSPHORYLATION (based on Chapter 19 and 20 of Stryer )
 CHLOROPLASTS, CALVIN CYCLE, PHOTOSYNTHETIC ELECTRON TRANSFER AND PHOTOPHOSPHORYLATION (based on Chapter 19 and 20 of Stryer ) Photosynthesis Photosynthesis Light driven transfer of electron across a membrane
CHLOROPLASTS, CALVIN CYCLE, PHOTOSYNTHETIC ELECTRON TRANSFER AND PHOTOPHOSPHORYLATION (based on Chapter 19 and 20 of Stryer ) Photosynthesis Photosynthesis Light driven transfer of electron across a membrane
NADH dehydrogenase in Corynebacterium glutamicum
 NADH dehydrogenase in Corynebacterium glutamicum Nawarat Nantapong 1, Hirohide Toyama, Kazunobu Matsushita Department of Biological Chemistry, Faculty of Agriculture, Yamaguchi University ( 1 Present address:
NADH dehydrogenase in Corynebacterium glutamicum Nawarat Nantapong 1, Hirohide Toyama, Kazunobu Matsushita Department of Biological Chemistry, Faculty of Agriculture, Yamaguchi University ( 1 Present address:
Lecture #8 9/21/01 Dr. Hirsh
 Lecture #8 9/21/01 Dr. Hirsh Types of Energy Kinetic = energy of motion - force x distance Potential = stored energy In bonds, concentration gradients, electrical potential gradients, torsional tension
Lecture #8 9/21/01 Dr. Hirsh Types of Energy Kinetic = energy of motion - force x distance Potential = stored energy In bonds, concentration gradients, electrical potential gradients, torsional tension
Bio-based route to the carbon-5 chemical glutaric acid and bio-polyamide PA6.5 using metabolically engineered Corynebacterium glutamicum
 Electronic Supplementary Material (ESI) for Green Chemistry. This journal is The Royal Society of Chemistry 2018 Online Supplementary Information to: Bio-based route to the carbon-5 chemical glutaric acid
Electronic Supplementary Material (ESI) for Green Chemistry. This journal is The Royal Society of Chemistry 2018 Online Supplementary Information to: Bio-based route to the carbon-5 chemical glutaric acid
Courtesy of Elsevier. Used with permission.
 Chemistry 5.07 2013 Problem Set 5 Answers Problem 1 Succinate dehydrogenase (SDH) is a heterotetramer enzyme complex that catalyzes the oxidation of succinate to fumarate with concomitant reduction of
Chemistry 5.07 2013 Problem Set 5 Answers Problem 1 Succinate dehydrogenase (SDH) is a heterotetramer enzyme complex that catalyzes the oxidation of succinate to fumarate with concomitant reduction of
Hybrid Quorum sensing in Vibrio harveyi- two component signalling
 Hybrid Quorum sensing in Vibrio harveyi- two component signalling Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by MHRD Page 1 of
Hybrid Quorum sensing in Vibrio harveyi- two component signalling Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by MHRD Page 1 of
The Mitochondrion. Definition Structure, ultrastructure Functions
 The Mitochondrion Definition Structure, ultrastructure Functions Organelle definition Etymology of the name Carl Benda (1903): (mitos) thread; (khondrion) granule. Light microscopy identification First
The Mitochondrion Definition Structure, ultrastructure Functions Organelle definition Etymology of the name Carl Benda (1903): (mitos) thread; (khondrion) granule. Light microscopy identification First
Bioinorganic Chemistry
 PRINCIPLES OF Bioinorganic Chemistry Stephen J. Lippard MASSACHUSETTS INSTITUTE OF TECHNOLOGY Jeremy M. Berg JOHNS HOPKINS SCHOOL OF MEDICINE f V University Science Books Mill Valley, California Preface
PRINCIPLES OF Bioinorganic Chemistry Stephen J. Lippard MASSACHUSETTS INSTITUTE OF TECHNOLOGY Jeremy M. Berg JOHNS HOPKINS SCHOOL OF MEDICINE f V University Science Books Mill Valley, California Preface
Chapter 6: Outline-2. Chapter 6: Outline Properties of Enzymes. Introduction. Activation Energy, E act. Activation Energy-2
 Chapter 6: Outline- Properties of Enzymes Classification of Enzymes Enzyme inetics Michaelis-Menten inetics Lineweaver-Burke Plots Enzyme Inhibition Catalysis Catalytic Mechanisms Cofactors Chapter 6:
Chapter 6: Outline- Properties of Enzymes Classification of Enzymes Enzyme inetics Michaelis-Menten inetics Lineweaver-Burke Plots Enzyme Inhibition Catalysis Catalytic Mechanisms Cofactors Chapter 6:
Introduction to Enzymes
 Introduction to Enzymes Lysozyme active site Chapter 8 Part 1 HIV-1 Protease with bound Inhibitor Dr. Ray How Enzymes Function What structural features allow an enzyme to have its unique biochemical function?
Introduction to Enzymes Lysozyme active site Chapter 8 Part 1 HIV-1 Protease with bound Inhibitor Dr. Ray How Enzymes Function What structural features allow an enzyme to have its unique biochemical function?
Chapter 8 Energy. I. Biological Energy. Part II Principles of Individual Cell Function
 Part II Principles of Individual Cell Function Chapter 8 This chapter discusses the mechanisms of mitochondrial respiration and photosynthesis by chloroplasts, which are processes involved in the production
Part II Principles of Individual Cell Function Chapter 8 This chapter discusses the mechanisms of mitochondrial respiration and photosynthesis by chloroplasts, which are processes involved in the production
Prokaryotic Regulation
 Prokaryotic Regulation Control of transcription initiation can be: Positive control increases transcription when activators bind DNA Negative control reduces transcription when repressors bind to DNA regulatory
Prokaryotic Regulation Control of transcription initiation can be: Positive control increases transcription when activators bind DNA Negative control reduces transcription when repressors bind to DNA regulatory
Name Synonym Product C4.3 SMD_ glna SMD_0103 glutamine synthetase glnb SMD_0104 nitrogen regulatory protein P-II - SMD_0155 glyoxalase -
 Name Synonym Product C4.3 SMD_0048 - glna SMD_0103 glutamine synthetase glnb SMD_0104 nitrogen regulatory protein P-II - SMD_0155 glyoxalase - SMD_0193 3-dehydroquinate dehydratase acea SMD_0200 Isocitrate
Name Synonym Product C4.3 SMD_0048 - glna SMD_0103 glutamine synthetase glnb SMD_0104 nitrogen regulatory protein P-II - SMD_0155 glyoxalase - SMD_0193 3-dehydroquinate dehydratase acea SMD_0200 Isocitrate
Supplementary Figure 1 The number of differentially expressed genes for uniparental males (green), uniparental females (yellow), biparental males
 Supplementary Figure 1 The number of differentially expressed genes for males (green), females (yellow), males (red), and females (blue) in caring vs. control comparisons in the caring gene set and the
Supplementary Figure 1 The number of differentially expressed genes for males (green), females (yellow), males (red), and females (blue) in caring vs. control comparisons in the caring gene set and the
Conceptofcolinearity: a continuous sequence of nucleotides in DNA encodes a continuous sequence of amino acids in a protein
 Translation Conceptofcolinearity: a continuous sequence of nucleotides in DNA encodes a continuous sequence of amino acids in a protein Para além do fenómeno do wobble, há que considerar Desvios ao código
Translation Conceptofcolinearity: a continuous sequence of nucleotides in DNA encodes a continuous sequence of amino acids in a protein Para além do fenómeno do wobble, há que considerar Desvios ao código
MITOCHONDRIAL LAB. We are alive because we make a lot of ATP and ATP makes (nonspontaneous) chemical reactions take place
 MITOCHONDRIAL LAB We are alive because we make a lot of ATP and ATP makes (nonspontaneous) chemical reactions take place We make about 95% of our ATP in the mitochondria We will isolate mitochondria, and
MITOCHONDRIAL LAB We are alive because we make a lot of ATP and ATP makes (nonspontaneous) chemical reactions take place We make about 95% of our ATP in the mitochondria We will isolate mitochondria, and
Chapter 12. Genes: Expression and Regulation
 Chapter 12 Genes: Expression and Regulation 1 DNA Transcription or RNA Synthesis produces three types of RNA trna carries amino acids during protein synthesis rrna component of ribosomes mrna directs protein
Chapter 12 Genes: Expression and Regulation 1 DNA Transcription or RNA Synthesis produces three types of RNA trna carries amino acids during protein synthesis rrna component of ribosomes mrna directs protein
Membrane-associated proteomics of chickpea identifies Sad1/UNC-84 protein (CaSUN1), a novel component of dehydration signaling
 Membrane-associated proteomics of chickpea identifies Sad/UNC-84 (CaSUN), a novel component of dehydration signaling Dinesh Kumar Jaiswal, Poonam Mishra, Pratigya Subba, Divya Rathi, Subhra Chakraborty*
Membrane-associated proteomics of chickpea identifies Sad/UNC-84 (CaSUN), a novel component of dehydration signaling Dinesh Kumar Jaiswal, Poonam Mishra, Pratigya Subba, Divya Rathi, Subhra Chakraborty*
Cellular Respiration Stage 4: Electron Transport Chain
 Cellular Respiration Stage 4: Electron Transport Chain 2006-2007 Cellular respiration What s the point? The point is to make ATP! ATP 2006-2007 ATP accounting so far Glycolysis 2 ATP Kreb s cycle 2 ATP
Cellular Respiration Stage 4: Electron Transport Chain 2006-2007 Cellular respiration What s the point? The point is to make ATP! ATP 2006-2007 ATP accounting so far Glycolysis 2 ATP Kreb s cycle 2 ATP
BME 5742 Biosystems Modeling and Control
 BME 5742 Biosystems Modeling and Control Lecture 24 Unregulated Gene Expression Model Dr. Zvi Roth (FAU) 1 The genetic material inside a cell, encoded in its DNA, governs the response of a cell to various
BME 5742 Biosystems Modeling and Control Lecture 24 Unregulated Gene Expression Model Dr. Zvi Roth (FAU) 1 The genetic material inside a cell, encoded in its DNA, governs the response of a cell to various
