Winter School in Mathematical & Computational Biology
|
|
|
- Delilah Kelley
- 5 years ago
- Views:
Transcription
1 1
2 Network reconstruction, topology and feasible solution space
3 From component to systems biology Component biology Systems biology Component view Systems view Needed homeostasis Function S+E X E+P Reaction network Time-dependent concentration Steady state flux map HT analytical chemistry Integrative analysis Compute flux Calculate C Calculate k Palsson (2000) Nat Biotech 20:649 3
4 E. coli on glycerol Objective Constraints Max growth rate on glycerol Network topology Steady state Maximum rates Ibarra et al, Nature 420, (2002) 4
5 Permanence of networks The permanent feature of life is networks Interconnections or links between components define the essence of a living process Components have finite turn-over time Metabolites: ~1 min mrna: ~2 hour Cells: a few years (in human) Yet, organisms remain essentially identical (if older) Kinetics is secondary Networks are relatively insensitive to kinetics Kinetics can evolve rapidly to realise network potential 5
6 Systems biology Components Gen- Transcript- Prote- Metabol- Systemic annotation Reconstruction of biochemical network (unique!) In silico modelling Topology Constraints Dynamics Sensitivity Noise Hypothesis generation & testing Phenotypic space is essentially infinite 6
7 Reconstruction All cellular networks Metabolic Regulatory Signaling are (bio)chemical The chemical nature is important Defines a stoichiometric relationship between components (invariable, integer) Defines fundamental constraints for the systems Thermodynamics: irreversibility and relative rates, maximum concentrations Mass transfer: maximum rates Spatial constraints: maximum concentrations, maximum rates Chemical networks are readily described by a stoichiometric matrix 7
8 Systemic (2D) annotation Palsson (2004) Nat Biotech 22:1218 8
9 Reconstruction process Genome sequence ORF prediction Gene annotation BLAST, Phylogeny, context Pathway reconstruction Synthesis of all biomass components Missing genes Functional validation Historical data, phenotype arrays Metabolomics Additional information Regulation, e.g., array analysis 9
10 Annotation workflow 10
11 Automated model compilation KEGG: Mus musculus release 46 Primary: pathway maps GENE & RN files plus COORD files Covers 50% of genes in mmu-genome LST file Secondary: global files mmu-enzyme LST file Less specific EC entries Reaction attributes: LIGAND Reaction & reaction-name LST Compound names and ID Reaction-mapformula: reversibility UniProtKB: Localisation Default: cytoplasm 11
12 Manual curation Technical (KEGG issues) Inconsistent compound or reaction labels (network gaps) Reactions violating atom conservation E.g., DNA + nucleotide = DNA Generic molecules: R H 2 O, H +, redox (difficult to pick up) Lumped reactions (e.g., PDH) Connectivity Membrane transporters Biomass drains 12
13 Network gaps Visual inspection of KEGG maps Good for synthesis pathways 6 essential reactions identified w/o known gene association All found in human GSM Linear programming Test generation of each biomass component 3 reactions w/o gene association found (all had irreversible counterparts in opposite direction) 5 reactions mapped to mitochondria but needed in cytosol (3 cytosolic in human GSM) Literature data 43 reactions w/o gene association added 21 subcellular localisations corrected 13
14 Model overview Unique gene-reaction associations 4617 Number of genes 1399 Mapped reactions 1757 Other reactions Reactions w/o gene association Membrane transporters Biomass reactions Autocatalytic Total number of reactions Rxns in mitochondria Metabolites
15 Topology S [m,n] Rows for m metabolites Columns for reactions
16 glc-d g6p f6p fdp dhap g3p 13dpg 3pg 2pg pep pyr lac-l atp adp pi h2o nadh nad h glc-d[e] lac-d[e] h[e] Enzyme Protein EC # Reaction HEX Hexokinase hk glc-d + atp -> g6p + adp + h v1 PGI1 Phosphoglucose isomerase pgi g6p <-> f6p v2 PFKA Phosphofructokinase pfka f6p + atp -> fdp + adp + h v3 FBA Fructose-1,6-bisphosphatate aldolase fba fdp <-> dhap + g3p v4 TPIA Triosphosphate Isomerase tpia dhap <-> g3p v5 GAPA G3P dehydrogenase-a complex gapa g3p + pi + nad <-> 13dpg + nadh + h v6 PGK Phosphoglycerate kinase pgk dpg + adp <-> 3pg + atp v7 GPMA Phosphoglycerate mutase 1 gpma pg <-> 2pg v8 ENO Enolase eno pg <-> pep + h2o v9 PYKF Pyruvate Kinase I pykf pep + adp + h -> pyr + atp v10 LDH_L L-Lactate dehydrogenase Ldh pyr + nadh + h <-> lac-l + nad v11 ATP hydrolysis atp + h2o -> adp + pi + h v12 GLCt glucose exchange glc-d[e] <-> glc-d b1 L-LAC-t lactate transport lac-l + h <-> h[e] + lac-l[e] b2 16
17 glc-d g6p f6p fdp dhap g3p Reaction S' glc-d + atp -> g6p + adp + h v g6p <-> f6p v2-1 1 f6p + atp -> fdp + adp + h v fdp <-> dhap + g3p v dhap <-> g3p v5-1 1 g3p + pi + nad <-> 13dpg + nadh + h v dpg + adp <-> 3pg + atp v pg <-> 2pg v pg <-> pep + h2o v pep + adp + h -> pyr + atp v pyr + nadh + h <-> lac-l + nad v atp + h2o -> adp + pi + h v glc-d[e] <-> glc-d b1 1-1 lac-l + h <-> h[e] + lac-l[e] b dpg 3pg 2pg pep pyr lac-l atp adp pi h2o nadh nad h glc-d[e] lac-d[e] h[e] 17
18 S v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 b1 b2 glc-d -1 1 g6p 1-1 f6p 1-1 fdp 1-1 dhap 1-1 g3p dpg 1-1 3pg 1-1 2pg 1-1 pep 1-1 pyr 1-1 lac-l 1-1 atp adp pi -1 1 h2o 1-1 nadh 1-1 nad -1 1 h glc-d[e] -1 lac-d[e] 1 h[e] 1 18
19 S tot, S exch, S int Internal fluxes Exchange Secondary Primary External S tot 19
20 S tot v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 b1 b2 glc-d -1 1 g6p 1-1 f6p 1-1 fdp 1-1 dhap 1-1 g3p dpg 1-1 3pg 1-1 2pg 1-1 pep 1-1 pyr 1-1 lac-l 1-1 atp adp pi -1 1 h2o 1-1 nadh 1-1 nad -1 1 h glc-d[e] -1 lac-l[e] 1 h[e] 1 dhap glc-d[e] glc-d g6p f6p fdp g3p 13dpg 3pg 2pg pep pyr lac-l lac-l[e]
21 S tot, S exch, S int Internal fluxes Exchange Primary Secondary S exch External S tot 21
22 S exch v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 b1 b2 glc-d -1 1 g6p 1-1 f6p 1-1 fdp 1-1 dhap 1-1 g3p dpg 1-1 3pg 1-1 2pg 1-1 pep 1-1 pyr 1-1 lac-l 1-1 atp adp pi -1 1 h2o 1-1 nadh 1-1 nad -1 1 h glc-d[e] -1 lac-l[e] 1 h[e] 1 dhap glc-d[e] glc-d g6p f6p fdp g3p 13dpg 3pg 2pg pep pyr lac-l lac-l[e]
23 S tot, S exch, S int Internal fluxes Exchange Primary Secondary S int S exch External S tot 23
24 S int v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 b1 b2 glc-d -1 1 g6p 1-1 f6p 1-1 fdp 1-1 dhap 1-1 g3p dpg 1-1 3pg 1-1 2pg 1-1 pep 1-1 pyr 1-1 lac-l 1-1 atp adp pi -1 1 h2o 1-1 nadh 1-1 nad -1 1 h glc-d[e] -1 lac-d[e] 1 h[e] 1 dhap glc-d[e] glc-d g6p f6p fdp g3p 13dpg 3pg 2pg pep pyr lac-l lac-l[e]
25 Reaction map: S Metabolites are nodes For reaction draw edge between substrates (- entry) to products (+entry) Highly non-linear map Participation: 4 is typical 25
26 Compound map: -S T Reaction as nodes Compounds as links Connectivity number 2 is common High for ATP etc Soft link metabolites flowing through reactions not fixed by stoichiometry 26
27 Open or closed systems A B v 1 C S int A B v 1 C b 1 A b v 1 b 1 A 3 C C S b v 1 2 exch b 3 B b 2 B A e B e b 1 A b v b 1 A 1 3 C b C C e v 2 S tot 1 b 3 B B e b 2 B A e C e 27
28 Binary S sˆ Sˆ : s ˆ ij ij = 0 = 1 if if s s ij ij = 0 0 Binary S S v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 b1 b2 Sb v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 b1 b2 glc-d -1 1 glc-d g6p 1-1 g6p f6p 1-1 f6p fdp 1-1 fdp dhap 1-1 dhap g3p g3p dpg dpg pg 1-1 3pg pg 1-1 2pg pep 1-1 pep pyr 1-1 pyr lac-l 1-1 lac-l atp atp adp adp pi -1 1 pi h2o 1-1 h2o nadh 1-1 nadh nad -1 1 nad h h glc-d[e] -1 glc-d[e] lac-l[e] 1 lac-l[e] h[e] 1 h[e]
29 Reaction adjacency matrix, A v = ˆ T S Sˆ diag( A v Av=Sb'Sb v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 b1 b2 v v v v v v v v v v v v b b ) i ˆ ˆ T = i i = 2 s s ski = k k ˆ sˆ ki = π π i = participation number for reaction i, i.e., number of compounds participating in reaction i. Off-diagonal elements indicates how many compounds two reactions i and j have in common i
30 Compound adjacency matrix, A x = SS ˆ ˆ T ( a x ) ii glc-d g6p ˆ f6p fdp dhap Ax=SbSb' glc-d g6p f6p fdp dhap g3p dpg pg pg pep pyr lac-l atp adp pi h2o nadh nad h glc-d[e] lac-l[e] h[e] = 2 sik = k k g3p sˆ ik 13dpg = 3pg ρ 2pg i pep pyr lac-l ρ i = connectivity number for compound i, i.e., number of reactions in which compound i participates. Off-diagonal elements indicates how many reactions both compounds i and j participate in. atp adp pi h2o nadh nad h glc-d[e] lac-l[e] h[e]
31 Singletons 31
32 Network topology 32
33 Active, essential and zero-flux reactions 950 singletons of 2104 metabolites Minor biomass components (e.g., spermidine) C-unconnected, e.g., xenobiotics Annotation errors 987 reactions linked to singletons (dead-end metabolites) 1050 active (i.e., non-zero flux) reactions Approximately 270 essential 409 degrees of freedom 33
34 S as a linear transformation v = dx dt i dx dt Range T ( v v K v ) x = ( x x K x ) = = v R 1 n k Sv s 2 ik v k S dx [ m, n] R dt Domain n (Dynamic mass balance) m 1 2 m T 34
35 Four fundamental spaces S [m,n] Row(S) R n R m Col(S) v dyn v dx/dt v ss Null(S) Left null(s) 35
36 Dimensions of subspaces r = Rank(S) = # linearly, independent relationships between compounds and reactions dim(col(s)) = dim(row(s)) = r dim(right null(s)) = n r dim(left null(s)) = m r 36
37 (Right) null space v = v ss + v dyn, where Sv ss =0 v ss is in (right) null space of S Null(S) contains all allowable steady-state flux distributions. 37
38 Row space Together v ss + v dyn span R n v dyn orthogonal to null space, i.e., in row(s) row(s) contains all dynamic flux distributions, i.e., the thermodynamic driving forces that changes state 38
39 Column space dx/dt = s 1 v 1 + s 2 v 2 + s n v n, where s i is the i th column in S Hence, dx/dt is in the column space of S Contains all allowable time derivatives of the concentrations vector and hence how the thermodynamic driving forces move concentration state of network 39
40 Left column space Together Left null(s) and Row(S) span R m Dim(Left null(s)) + Dim(Col(S)) = m Vectors in the left null space of S are orthogonal to Col(S) Left null(s) contains all the conservation relationships, i.e., time invariants, defined by the network. This defines conserved metabolic pools as combinations of metabolites. 40
41 Four fundamental spaces S [m,n] Row(S) R n R m Col(S) v dyn v dx/dt v ss Null(S) Left null(s) 41
42 Basis for vector spaces A basis for a space is a set of vectors that can be used to span the space, e.g., b 1 =(1,0,0), b 2 =(0,1,0) and b 3 =(0,0,1) Any vector v R (3) can be decomposed as v = w 1 b 1 + w 2 b 2 + w 3 b 3 So [b 1,b 2,b 3 ] is a basis for R (3) Many types of bases Linear basis Orthonormal (linear) basis for linear spaces Convex basis for finite linear spaces b 3 z b 1 b 2 (w 1,w 2,w 3 ) y x 42
43 Singular value decomposition S = U Σ V T [ m, n] [ m, m] [ m, n] [ n, n] U and V are orthonormal matrices U T U = I (mxm) and V T V = I (nxn) U T = U -1 and V T = V -1 Σ = diag(σ 1, σ 2,, σ r ), where σ 1 σ 2,, σ r >0 43
44 Orthonormal bases for subspaces S U Σ V T = rxr Row(S) mxn Null(S) Col(S) Left null(s) Columns in U defines orthonormal basis for Col(S) and Left null(s) Columns in V defines orthonormal basis for Row(S) and Null(S) 44
45 U -1.20E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E-01 Col(S) Left Null(S) 45
46 Σ r = 13, i.e., 13 linearly, independent relationships between compounds and reactions 46
47 V 4.56E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E E-10-2E E E E E E E E-10 Row(S) Null(S) 47
48 Eigen-reaction dx T T = Sv = UΣV v uk x = uk1x1 + uk 2x2 + L+ ukmxm ( pool) dt T v T dx k v = vk1v1 + vk 2v2 + L+ vknvn pathway T T U = U UΣV v dt Eigen reaction : ( T d U x) Σ( T = V v) vkjv j for vkj > 0 dt for k r : u kixi ukixi ( T d u x) for uki < 0 for uki > 0 ( T v v) k = σ vkjv j k k for vkj < 0 dt ( ) 48
49 1 st Mode (σ = 4.060) v8 2.12E-02 v6 2.28E-01 v E-01 dhap v1 4.56E-01 2pg lac-l v3 4.57E-01 pi pyr nadh g3p pep pg fdp nad v9-1.32e-03 g6p h2o v5-1.58e-02 13dpg glc-d b1-2.94e-02 adp f6p v4-5.08e-02 h atp v2-6.30e-02 b2-1.39e-01 v e-01 v7-3.06e-01 v e h adp atp u 2 v v 3 v 1 v 10 v
50 Mode 1 50
51 Mode 2 51
52 Null spaces glc-d + atp -> g6p + adp + h v g6p <-> f6p v f6p + atp -> fdp + adp + h v fdp <-> dhap + g3p v dhap <-> g3p v g3p + pi + nad <-> 13dpg + nadh + h v dpg + adp <-> 3pg + atp v pg <-> 2pg v pg <-> pep + h2o v pep + adp + h -> pyr + atp v pyr + nadh + h <-> lac-l + nad v atp + h2o -> adp + pi + h v glc-d[e] <-> glc-d b lac-l + h <-> h[e] + lac-l[e] b Conserved metabolite pools Steady state flux balance 52
53 0 v 1 10 A 0 v v 3 8 dx dt = Sv da dt = v ( 1 1 1) v 2 v3 1 S = = ()( ) 0 Eigen reaction : u v T 1 T 1 x = 1 A ( pool) v = v v v 3 ( pathway) v v v A 53
54 Sv v SS 0 SS = w 1 + w 2 v 10 0 v 6 0 v 8 1 = Orthonormal basis for Null(S) 0 v 1 10 A 0 v v 3 8 vss = α 1 + α 2 v 10 0 v v 3 8 Convex basis for Null(S) 54
55 vss = α 1 + α 2 v 10 0 v v 3 8 Convex basis for Null(S) Since all v i 0 and all basis elements positive: α i 0 0 v 10 : 0 α1 + α v 6 : 0 α1 2 0 v 8: 0 α
56 α 2 6 v α v v 2 56
57 Linear vs convex spaces Linear space Described by linear equations Vector space defined by set of linearly independent basis vectors (b i ) v = Σw i b i - < w i <+ Every point uniquely described by linear combination of b i Number of basis vectors equals dim(null(s)) Infinite number of bases can span space Convex space Described by linear equations and inequalities Convex polyhedral cone defined by conically independent generating vectors (p i ) v = Σα i p i 0 α i <+ Every point described by nonnegative, linear combination of p i (non-unique) Number of generation vectors may exceed dim(null(s)) Unique set of generating vectors 57
58 Extreme pathways v ss = Σ α i p i 0 α i < α i,max P i s are unique & correspond to edges in (n-r)- dimensional cone; α i s not unique Correspond to pathways on a flux map Termed extreme pathways, since they define edges of bounded null space in its conical representation 58
59 ExPa classification p 1 p n Internal fluxes v 1 Type I Primary pathways Type II Futile cycles Type III Internal cycles Exchange fluxes Currency Primary b 1 c =0 =0 0 =0 Type I Type II Type III 59
60 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 b1 b2 b3 b4 c1 c2 c3 c4 c5 c6 c7 glc-d g6p b1 f6p glc-d fdp v1 dhap g6p g3p v2 v3 3pg f6p pep v4 pyr fdp accoa v5 v6 acp dhap g3p lac v7/v8 v9/v10 etoh pg ac v11/v12 for pep atp v13 adp pyr lac pi v14 v17/v18 b2 nadh accoa etoh nad v15/16 v19/v20 b3 coa acp ac v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 b1 b2 b3 b4 c1 c2 c3 c4 c5 c6 c7 v21/v22 b4 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 b1 b2 b3 b4 c1 c2 c3 c4 c5 c6 c7 Type Type Type Type Type Type glc-lac Type Type lac-etoh glc-etoh Type ac-etoh etoh-ac lac-ac glc-ac
61 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 b1 b2 b3 b4 c1 c2 c3 c4 c5 c6 c7 glc-d b1 g6p glc-d f6p v1 fdp g6p dhap v2 v3 g3p f6p 3pg v4 pep fdp pyr v5 v6 accoa dhap g3p acp v7/v8 v9/v10 lac pg etoh v11/v12 ac pep for v13 atp pyr lac adp v14 v17/v18 b2 pi accoa etoh nadh v15 v19/v20 b3 nad acp ac coa v21/v22 b4 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 b1 b2 b3 b4 c1 c2 c3 c4 c5 c6 c7 v16 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 b1 b2 b3 b4 c1 c2 c3 c4 c5 c6 c7 Type Type Type Type Type Type glc-lac ac-etoh Type lac-etoh glc-etoh Type Futile cycle etoh-ac lac-ac glc-ac glc-etoh/ac
62 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 v23 b1 b2 b3 b4 b5 glc-d b1 g6p glc-d f6p v1 fdp g6p dhap v2 v3 g3p f6p 3pg v4 pep fdp pyr v5 v6 accoa dhap g3p acp v7/v8 v9/v10 lac pg etoh v11/v12 ac pep for v13 atp pyr lac adp v14 v17/v18 b2 pi accoa etoh nadh v15 v19/v20 b3 nad acp ac coa v21/v22 b4 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 v23 b1 b2 b3 b4 b5 v16 t t t t t t t t glc-lac glc-lac lac-etoh/ac lac-etoh/ac glc-etoh/ac glc-etoh/ac v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 v23 b1 b2 b3 b4 b5
63 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 v23 b1 b2 b3 b4 b5 glc-d g6p b1 f6p glc-d fdp v1 dhap g6p g3p v2 v3 3pg f6p pep v4 pyr fdp accoa v5 v6 acp dhap g3p lac v7/v8 v9/v10 etoh pg ac v11/v12 for pep atp v13 adp pyr lac pi v14 v17/v18 b2 nadh accoa etoh nad v15 v19/v20 b3 coa acp ac v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 v23 b1 b2 b3 b4 b5 v21/v22 b4 v16 v1 v2 v3 v4 v5 v6 v7 v8 v9 v10 v11 v12 v13 v14 v15 v16 v17 v18 v19 v20 v21 v22 v23 b1 b2 b3 b4 b
64 Sv = 0 Stoichiometry Linear algebra v R (n) Eigenreactions v subspace of R (n) v 0 Reaction direction Convex analysis ExPa v = Σα i p i, α i 0 convex cone 64
65 Pathway matrix, P, and Binary P ( p p p ) = ( ) P = 2 3 ˆ pˆ ij = 0 if p P : pˆ ij = 1 if p P LM = 1 K ) T P ) P ij ij = 0 0 Binary P Pathway length matrix Diagonal elements = # reactions in each ExPa Off-diagonal elements = # shared reactions between two ExPa R PM ˆ ˆ T = PP Reaction participation matrix Diagonal elements = # ExPa in which a given reaction is found Off-diagonal elements = # ExPa that contains given pair of reactions 65
66 JAK-STAT signaling network Input 15 receptors with ligands Output 7 STAT homo- and heterodimers 297 reactions 216 internal 81 irreversible exchange 147 extreme pathways 66
67 Reaction participation 168 reactions participate in only one extreme pathway Very specific function, i.e., ideal drug targets 67
68 Cross-talk Cross-talk 147 ExPAs 10,731 pairwise comparisons Observations All pathways have single output 99.8% disjoint output 0.2% identical output 63.9% deterministic 14.8% classical cross talk
69 Correlated reaction sets 69
70 Sv = 0 Stoichiometry Linear algebra v R (n) Eigenreactions v subspace of R (n) v 0 Reaction direction Convex analysis ExPa v = Σα i p i, α i 0 convex cone 70
71 COBRA = Constraint-based reconstruction and analysis of metabolic and regulatory networks
72 Sv = 0 Stoichiometry Linear algebra v R (n) Eigenreactions v subspace of R (n) v 0 Reaction direction Convex analysis ExPa v = Σα i p i, α i 0 convex cone 72
73 Link to real world Defines feasible solution space in terms of balanced pathways Thermodynamic, regulatory, kinetic constraints Delete unfeasible ExPas to see true solution space Gene KO and upregulation Removes all ExPas that use particular gene Identify KO candidates among genes not required in desired ExPas Identify upregulation candidates among genes with high coefficients or that are unique in desired ExPas Max yields Yields calculated for individual ExPa(s) Trade-off between biomass vs product 73
74 Sv = 0 Stoichiometry Linear algebra v R (n) Eigenreactions v subspace of R (n) v 0 Reaction direction Convex analysis ExPa Union of convex subsets Regulation Thermo Kinetics v = Σα i p i, α i 0 convex cone 74
75 Link to real world Defines feasible solution space in terms of balanced pathways Thermodynamic, regulatory, kinetic constraints Delete unfeasible ExPas to see true solution space Gene KO and upregulation Removes all ExPas that use particular gene Identify KO candidates among genes not required in desired ExPas Identify upregulation candidates among genes with high coefficients or that are unique in desired ExPas Max yields Yields calculated for individual ExPa(s) Trade-off between biomass vs product 75
76 In silico E. coli model Network: 71 reactions - 21 reversible 55 internal metabolites 13 external metabolites Captures: - Substrate uptake - Glycolysis - PPP, EDP, TCA-Cycle - Anaplerosis - Respiration -Fermentation - Biomass formation - 4 different pathways for product formation 76
77 4 natural pathways for 3HP production (KEGG) 3HP 77
78 Effect on product formation 3-HP Yield (aerobic) 3-HP Yield (anaerobic) (anaerobic, aerobic) Left (1) Right (2) Center(3) Center-right (4) 95.8 % 96.6 % 84.2 % 100 % ** 100 % 100 % * (50%) Yields are given in C mol / C mol [%] 85.7 % 100 % ** In general the yields are higher under anaerobic conditions The second pathway is strongly dependent on a reversible acetate kinase. The third pathway underperforms the other in both scenarios. Two unknown enzymes in the fourth pathway 78
79 In silico analysis of β-alanine pathway for the production of 3-HP in E. coli #product synthesis left pathway R54 : OAA + GLU = ASP + 2 OXO R55 : ASP = bala + CO2 R56 : bala + 2 OXO = 3 OXOPRO + GLU R57 : 3 OXOPRO + NADH = 3 HPA + NAD 3HP 79
80 Elementary mode analysis Carbon yield for biomass elementary modes 2292 make the desired product Mode #11048 Max P with X>0 Carbon yield for 3-HP Anaerobic Does not form acetate Does not require PEP-carboxylase and GPI Highly dependent on malic enzyme (NADPH) 80
81 Knock-out of GPI, PEP-C and Acetate kinase Anaerobic conditions 464 elementary modes 126 make the desired product Carbon yield for biomass Carbon yield for 3-HP 81
82 Sv = 0 Stoichiometry Linear algebra v R (n) Eigenreactions v subspace of R (n) Capacity constraints v v max v = Σα i p i, α i 0 Bounded convex cone 0 α i α i,max v 0 ExPa Reaction direction Convex analysis Union of convex subsets Regulation Thermo Kinetics v = Σα i p i, α i 0 convex cone 82
83 Linear programming maximize subject to and Z = w v = wivi Sv = 0 v v v, i = 1, i, min i i,max K, n Objectives Linear Max growth (μ) Max product (π) Min substrate (σ) Max w 1 μ+w 2 π Nonlinear Min v 2 (QP) 83
84 LP solutions Unique Shadow price (dual) Increase in objective function for a unit increase in a constraint Degenerate Reduced cost Increase in objective function for unit increase in flux Unbounded 84
85 ExPa vs LP ExPa Define all feasible points Define all extreme points in unbounded problem Degenerate solutions are linear combinations of ExPas with identical objective function Does not define all points in bounded problem Length constrained by capacities Computationally challenging LP Computationally inexpensive even for large problems Give one extreme solution point, reconstructing all more difficult Many approaches developed 85
86 Mixed integer programming maximize subject to and Z = w y v A y v = b v v v i,min 0 y i y i i n j i,max, j = 1, K, m integer (binary), i = 1, K, n 86
87 87
88 PhPP 88
89 89
90 Phenotypic phase plane Ibarra et al, Nature 420, (2002) 90
91 AraGEM Gene-reaction-association entries 5253 ORFs (unique) 1419 Metabolites 1748 Unique reactions 1567 Cytosolic reactions 1265 Mitochondrial reactions 60 Plastidic reactions 159 Peroxisomal reactions 98 Modified reactions 36 Biomass drains and transporters 148 Biomass drains 47 Transporters (Intercellular) 18 Transporters (Inter-organelle) 83 Gaps (unique reactions ID) filled by manual curation 75 Singleton metabolites
92 92
93 Assumptions Inputs, outputs and constraints Case 1 Case 2 Photons uptake (free flux) + + Glutamine transporter (mitochondria) - - Glutamate transporter(mitochondria) - - Glutamine transporter (plastid) - - Glutamate transporter (plastid) + + RuBisCO; EC (carboxylation:oxygenation; 3:1) Fd-GOGAT ; EC (plastid) + + NADH-GOGAT; EC (plastid) - - Optimization: minimize uptake of Photons Photons Biomass rate (estimated and fixed) Leaf Leaf 93
94 RuBisCO 3:1 C:O: ~40% increase in photon requirements (litt 30-50%) 94
Constraint-Based Workshops
 Constraint-Based Workshops 2. Reconstruction Databases November 29 th, 2007 Defining Metabolic Reactions ydbh hslj ldha 1st level: Primary metabolites LAC 2nd level: Neutral Formulas C 3 H 6 O 3 Charged
Constraint-Based Workshops 2. Reconstruction Databases November 29 th, 2007 Defining Metabolic Reactions ydbh hslj ldha 1st level: Primary metabolites LAC 2nd level: Neutral Formulas C 3 H 6 O 3 Charged
V15 Flux Balance Analysis Extreme Pathways
 V15 Flux Balance Analysis Extreme Pathways Stoichiometric matrix S: m n matrix with stochiometries of the n reactions as columns and participations of m metabolites as rows. The stochiometric matrix is
V15 Flux Balance Analysis Extreme Pathways Stoichiometric matrix S: m n matrix with stochiometries of the n reactions as columns and participations of m metabolites as rows. The stochiometric matrix is
V19 Metabolic Networks - Overview
 V19 Metabolic Networks - Overview There exist different levels of computational methods for describing metabolic networks: - stoichiometry/kinetics of classical biochemical pathways (glycolysis, TCA cycle,...
V19 Metabolic Networks - Overview There exist different levels of computational methods for describing metabolic networks: - stoichiometry/kinetics of classical biochemical pathways (glycolysis, TCA cycle,...
Chapter 7: Metabolic Networks
 Chapter 7: Metabolic Networks 7.1 Introduction Prof. Yechiam Yemini (YY) Computer Science epartment Columbia University Introduction Metabolic flux analysis Applications Overview 2 1 Introduction 3 Metabolism:
Chapter 7: Metabolic Networks 7.1 Introduction Prof. Yechiam Yemini (YY) Computer Science epartment Columbia University Introduction Metabolic flux analysis Applications Overview 2 1 Introduction 3 Metabolism:
V14 extreme pathways
 V14 extreme pathways A torch is directed at an open door and shines into a dark room... What area is lighted? Instead of marking all lighted points individually, it would be sufficient to characterize
V14 extreme pathways A torch is directed at an open door and shines into a dark room... What area is lighted? Instead of marking all lighted points individually, it would be sufficient to characterize
Stoichiometric vector and matrix
 Stoichiometric vector and matrix The stoichiometric coefficients of a reaction are collected to a vector s r In s r there is a one position for each metabolite in the metabolic system, and the stoichiometric
Stoichiometric vector and matrix The stoichiometric coefficients of a reaction are collected to a vector s r In s r there is a one position for each metabolite in the metabolic system, and the stoichiometric
Predicting rice (Oryza sativa) metabolism
 Predicting rice (Oryza sativa) metabolism Sudip Kundu Department of Biophysics, Molecular Biology & Bioinformatics, University of Calcutta, WB, India. skbmbg@caluniv.ac.in Collaborators: Mark G Poolman
Predicting rice (Oryza sativa) metabolism Sudip Kundu Department of Biophysics, Molecular Biology & Bioinformatics, University of Calcutta, WB, India. skbmbg@caluniv.ac.in Collaborators: Mark G Poolman
Metabolic Network Analysis
 Metabolic Network Analysis Christoph Flamm Institute for Theoretical Chemistry University Vienna Harvest Network Course, Brno, April 1-6, 2011 The Systems Biology Modelling Space Network Models Level of
Metabolic Network Analysis Christoph Flamm Institute for Theoretical Chemistry University Vienna Harvest Network Course, Brno, April 1-6, 2011 The Systems Biology Modelling Space Network Models Level of
Metabolic modelling. Metabolic networks, reconstruction and analysis. Esa Pitkänen Computational Methods for Systems Biology 1 December 2009
 Metabolic modelling Metabolic networks, reconstruction and analysis Esa Pitkänen Computational Methods for Systems Biology 1 December 2009 Department of Computer Science, University of Helsinki Metabolic
Metabolic modelling Metabolic networks, reconstruction and analysis Esa Pitkänen Computational Methods for Systems Biology 1 December 2009 Department of Computer Science, University of Helsinki Metabolic
Description of the algorithm for computing elementary flux modes
 Description of the algorithm for computing elementary flux modes by Stefan Schuster, Thomas Dandekar,, and David Fell Department of Bioinformatics, Max Delbrück Centre for Molecular Medicine D-9 Berlin-Buch,
Description of the algorithm for computing elementary flux modes by Stefan Schuster, Thomas Dandekar,, and David Fell Department of Bioinformatics, Max Delbrück Centre for Molecular Medicine D-9 Berlin-Buch,
Reconstruction and Analysis of Metabolic Networks
 1 Reconstruction and Analysis of Metabolic Networks 2 Outline What is a Reconstruction? Data Collection Interactions Between Network Components Special Considerations Applications Genome-scale Metabolic
1 Reconstruction and Analysis of Metabolic Networks 2 Outline What is a Reconstruction? Data Collection Interactions Between Network Components Special Considerations Applications Genome-scale Metabolic
Giving you the energy you need!
 Giving you the energy you need! Use your dominant hand Open and close the pin (with your thumb and forefinger) as many times as you can for 20 seconds while holding the other fingers straight out! Repeat
Giving you the energy you need! Use your dominant hand Open and close the pin (with your thumb and forefinger) as many times as you can for 20 seconds while holding the other fingers straight out! Repeat
Review Questions - Lecture 5: Metabolism, Part 1
 Review Questions - Lecture 5: Metabolism, Part 1 Questions: 1. What is metabolism? 2. What does it mean to say that a cell has emergent properties? 3. Define metabolic pathway. 4. What is the difference
Review Questions - Lecture 5: Metabolism, Part 1 Questions: 1. What is metabolism? 2. What does it mean to say that a cell has emergent properties? 3. Define metabolic pathway. 4. What is the difference
Stoichiometric analysis of metabolic networks
 1 SGN-6156 Computational systems biology II Antti Larjo antti.larjo@tut.fi Computational Systems Biology group 30.4.2008 30.4.2008 2 Contents Networks in cells Metabolism Metabolic networks and pathways
1 SGN-6156 Computational systems biology II Antti Larjo antti.larjo@tut.fi Computational Systems Biology group 30.4.2008 30.4.2008 2 Contents Networks in cells Metabolism Metabolic networks and pathways
Integrated Knowledge-based Reverse Engineering of Metabolic Pathways
 Integrated Knowledge-based Reverse Engineering of Metabolic Pathways Shuo-Huan Hsu, Priyan R. Patkar, Santhoi Katare, John A. Morgan and Venkat Venkatasubramanian School of Chemical Engineering, Purdue
Integrated Knowledge-based Reverse Engineering of Metabolic Pathways Shuo-Huan Hsu, Priyan R. Patkar, Santhoi Katare, John A. Morgan and Venkat Venkatasubramanian School of Chemical Engineering, Purdue
Systems II. Metabolic Cycles
 Systems II Metabolic Cycles Overview Metabolism is central to cellular functioning Maintenance of mass and energy requirements for the cell Breakdown of toxic compounds Consists of a number of major pathways
Systems II Metabolic Cycles Overview Metabolism is central to cellular functioning Maintenance of mass and energy requirements for the cell Breakdown of toxic compounds Consists of a number of major pathways
Structural Analysis of Expanding Metabolic Networks
 Genome Informatics 15(1): 35 45 (24) 35 Structural Analysis of Expanding Metabolic Networks Oliver Ebenhöh oliver.ebenhoeh@rz.hu-berlin.de Reinhart Heinrich reinhart.heinrich@rz.hu-berlin.de Thomas Handorf
Genome Informatics 15(1): 35 45 (24) 35 Structural Analysis of Expanding Metabolic Networks Oliver Ebenhöh oliver.ebenhoeh@rz.hu-berlin.de Reinhart Heinrich reinhart.heinrich@rz.hu-berlin.de Thomas Handorf
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy
 Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
Stoichiometric network analysis
 Stoichiometric network analysis In stoichiometric analysis of metabolic networks, one concerns the effect of the network structure on the behaviour and capabilities of metabolism. Questions that can be
Stoichiometric network analysis In stoichiometric analysis of metabolic networks, one concerns the effect of the network structure on the behaviour and capabilities of metabolism. Questions that can be
Genomics, Computing, Economics & Society
 Genomics, Computing, Economics & Society 10 AM Thu 27-Oct 2005 week 6 of 14 MIT-OCW Health Sciences & Technology 508/510 Harvard Biophysics 101 Economics, Public Policy, Business, Health Policy Class outline
Genomics, Computing, Economics & Society 10 AM Thu 27-Oct 2005 week 6 of 14 MIT-OCW Health Sciences & Technology 508/510 Harvard Biophysics 101 Economics, Public Policy, Business, Health Policy Class outline
Bringing Genomes to Life: The Use of Genome-Scale In Silico Models
 2 Bringing Genomes to Life: The Use of Genome-Scale In Silico Models Ines Thiele and Bernhard Ø. Palsson Summary Metabolic network reconstruction has become an established procedure that allows the integration
2 Bringing Genomes to Life: The Use of Genome-Scale In Silico Models Ines Thiele and Bernhard Ø. Palsson Summary Metabolic network reconstruction has become an established procedure that allows the integration
Oxidative Phosphorylation versus. Photophosphorylation
 Photosynthesis Oxidative Phosphorylation versus Photophosphorylation Oxidative Phosphorylation Electrons from the reduced cofactors NADH and FADH 2 are passed to proteins in the respiratory chain. In eukaryotes,
Photosynthesis Oxidative Phosphorylation versus Photophosphorylation Oxidative Phosphorylation Electrons from the reduced cofactors NADH and FADH 2 are passed to proteins in the respiratory chain. In eukaryotes,
Cellular Metabolic Models
 Cellular Metabolic Models. Cellular metabolism. Modeling cellular metabolism. Flux balance model of yeast glycolysis 4. Kinetic model of yeast glycolysis Cellular Metabolic Models Cellular Metabolism Basic
Cellular Metabolic Models. Cellular metabolism. Modeling cellular metabolism. Flux balance model of yeast glycolysis 4. Kinetic model of yeast glycolysis Cellular Metabolic Models Cellular Metabolism Basic
Bioinformatics: Network Analysis
 Bioinformatics: Network Analysis Flux Balance Analysis and Metabolic Control Analysis COMP 572 (BIOS 572 / BIOE 564) - Fall 2013 Luay Nakhleh, Rice University 1 Flux Balance Analysis (FBA) Flux balance
Bioinformatics: Network Analysis Flux Balance Analysis and Metabolic Control Analysis COMP 572 (BIOS 572 / BIOE 564) - Fall 2013 Luay Nakhleh, Rice University 1 Flux Balance Analysis (FBA) Flux balance
Energy and Cellular Metabolism
 1 Chapter 4 About This Chapter Energy and Cellular Metabolism 2 Energy in biological systems Chemical reactions Enzymes Metabolism Figure 4.1 Energy transfer in the environment Table 4.1 Properties of
1 Chapter 4 About This Chapter Energy and Cellular Metabolism 2 Energy in biological systems Chemical reactions Enzymes Metabolism Figure 4.1 Energy transfer in the environment Table 4.1 Properties of
Biochemistry 3300 Problems (and Solutions) Metabolism I
 (1) Provide a reasonable systematic name for an enzyme that catalyzes the following reaction: fructose + ATP > fructose-1 phosphate + ADP (2) The IUBMB has a developed a set of rules for classifying enzymes
(1) Provide a reasonable systematic name for an enzyme that catalyzes the following reaction: fructose + ATP > fructose-1 phosphate + ADP (2) The IUBMB has a developed a set of rules for classifying enzymes
Gene Regulatory Network Identification
 Gene Regulatory Network Identification Korkut Uygun and Yinlun Huang Department of Chemical Engineering and Materials Science Wayne State University AIChE Annual National Meeting San Francisco November
Gene Regulatory Network Identification Korkut Uygun and Yinlun Huang Department of Chemical Engineering and Materials Science Wayne State University AIChE Annual National Meeting San Francisco November
Transformation of Energy! Energy is the ability to do work.! Thermodynamics is the study of the flow and transformation of energy in the universe.
 Section 1 How Organisms Obtain Energy Transformation of Energy! Energy is the ability to do work.! Thermodynamics is the study of the flow and transformation of energy in the universe. Section 1 How Organisms
Section 1 How Organisms Obtain Energy Transformation of Energy! Energy is the ability to do work.! Thermodynamics is the study of the flow and transformation of energy in the universe. Section 1 How Organisms
Written Exam 15 December Course name: Introduction to Systems Biology Course no
 Technical University of Denmark Written Exam 15 December 2008 Course name: Introduction to Systems Biology Course no. 27041 Aids allowed: Open book exam Provide your answers and calculations on separate
Technical University of Denmark Written Exam 15 December 2008 Course name: Introduction to Systems Biology Course no. 27041 Aids allowed: Open book exam Provide your answers and calculations on separate
Grundlagen der Systembiologie und der Modellierung epigenetischer Prozesse
 Grundlagen der Systembiologie und der Modellierung epigenetischer Prozesse Sonja J. Prohaska Bioinformatics Group Institute of Computer Science University of Leipzig October 25, 2010 Genome-scale in silico
Grundlagen der Systembiologie und der Modellierung epigenetischer Prozesse Sonja J. Prohaska Bioinformatics Group Institute of Computer Science University of Leipzig October 25, 2010 Genome-scale in silico
Flux balance analysis in metabolic networks
 Flux balance analysis in metabolic networks Lecture notes by Eran Eden, https://systemsbiology.usu.edu/documents/flux%20balance%20analysis%20overview%20-%202016.pdf additional slides by B. Palsson from
Flux balance analysis in metabolic networks Lecture notes by Eran Eden, https://systemsbiology.usu.edu/documents/flux%20balance%20analysis%20overview%20-%202016.pdf additional slides by B. Palsson from
V14 Graph connectivity Metabolic networks
 V14 Graph connectivity Metabolic networks In the first half of this lecture section, we use the theory of network flows to give constructive proofs of Menger s theorem. These proofs lead directly to algorithms
V14 Graph connectivity Metabolic networks In the first half of this lecture section, we use the theory of network flows to give constructive proofs of Menger s theorem. These proofs lead directly to algorithms
Introduction to Systems Biology. James Gomes KSBS, IITD
 Introduction to Systems Biology James Gomes KSBS, IITD Inner Life of a Cell Cells Exhibit a High Degree of Complexity What are the different types of cells? Is Life an emergent property? What is Systems
Introduction to Systems Biology James Gomes KSBS, IITD Inner Life of a Cell Cells Exhibit a High Degree of Complexity What are the different types of cells? Is Life an emergent property? What is Systems
2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of October
 Name: Class: _ Date: _ 2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of 19-23 October Multiple Choice Identify the choice that best completes the statement or answers the question. 1) Which
Name: Class: _ Date: _ 2015 AP Biology PRETEST Unit 3: Cellular Energetics Week of 19-23 October Multiple Choice Identify the choice that best completes the statement or answers the question. 1) Which
Thermodynamic principles governing metabolic operation : inference, analysis, and prediction Niebel, Bastian
 University of Groningen Thermodynamic principles governing metabolic operation : inference, analysis, and prediction Niebel, Bastian IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's
University of Groningen Thermodynamic principles governing metabolic operation : inference, analysis, and prediction Niebel, Bastian IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's
Bio102 Problems Photosynthesis
 Bio102 Problems Photosynthesis 1. Why is it advantageous for chloroplasts to have a very large (in surface area) thylakoid membrane contained within the inner membrane? A. This limits the amount of stroma
Bio102 Problems Photosynthesis 1. Why is it advantageous for chloroplasts to have a very large (in surface area) thylakoid membrane contained within the inner membrane? A. This limits the amount of stroma
Exam 4 April 15, 2005 CHEM 3511 Print Name: KEY Signature
 1) (8 pts) General Properties of Enzymes. Give four properties of enzymaticallycatalyzed reactions. The answers should indicate how enzymatic reactions differ from non-enzymatic reactions. Write four only
1) (8 pts) General Properties of Enzymes. Give four properties of enzymaticallycatalyzed reactions. The answers should indicate how enzymatic reactions differ from non-enzymatic reactions. Write four only
13. PG3 + ATP + NADH => GA3P + ADP + Pi + NAD +
 Additional file 2. 1.1 Stochiometric model for P. pastoris containing some additional reactions from the 13 C model (section 1.2) Methanol metabolism 1. Metoh => Form 2. Form => FOR + NADH 3. FOR + NAD
Additional file 2. 1.1 Stochiometric model for P. pastoris containing some additional reactions from the 13 C model (section 1.2) Methanol metabolism 1. Metoh => Form 2. Form => FOR + NADH 3. FOR + NAD
Cellular Energy. How Organisms Obtain Energy Section 2: Photosynthesis Section 3: Cellular Respiration. Click on a lesson name to select.
 Section 1: How Organisms Obtain Energy Section 2: Photosynthesis Section 3: Cellular Respiration Click on a lesson name to select. Section 1 How Organisms Obtain Energy Transformation of Energy Energy
Section 1: How Organisms Obtain Energy Section 2: Photosynthesis Section 3: Cellular Respiration Click on a lesson name to select. Section 1 How Organisms Obtain Energy Transformation of Energy Energy
BBS2710 Microbial Physiology. Module 5 - Energy and Metabolism
 BBS2710 Microbial Physiology Module 5 - Energy and Metabolism Topics Energy production - an overview Fermentation Aerobic respiration Alternative approaches to respiration Photosynthesis Summary Introduction
BBS2710 Microbial Physiology Module 5 - Energy and Metabolism Topics Energy production - an overview Fermentation Aerobic respiration Alternative approaches to respiration Photosynthesis Summary Introduction
ATLAS of Biochemistry
 ATLAS of Biochemistry USER GUIDE http://lcsb-databases.epfl.ch/atlas/ CONTENT 1 2 3 GET STARTED Create your user account NAVIGATE Curated KEGG reactions ATLAS reactions Pathways Maps USE IT! Fill a gap
ATLAS of Biochemistry USER GUIDE http://lcsb-databases.epfl.ch/atlas/ CONTENT 1 2 3 GET STARTED Create your user account NAVIGATE Curated KEGG reactions ATLAS reactions Pathways Maps USE IT! Fill a gap
Center for Academic Services & Advising
 March 2, 2017 Biology I CSI Worksheet 6 1. List the four components of cellular respiration, where it occurs in the cell, and list major products consumed and produced in each step. i. Hint: Think about
March 2, 2017 Biology I CSI Worksheet 6 1. List the four components of cellular respiration, where it occurs in the cell, and list major products consumed and produced in each step. i. Hint: Think about
An introduction to biochemical reaction networks
 Chapter 1 An introduction to biochemical reaction networks (This part is discussed in the slides). Biochemical reaction network distinguish themselves from general chemical reaction networks in that the
Chapter 1 An introduction to biochemical reaction networks (This part is discussed in the slides). Biochemical reaction network distinguish themselves from general chemical reaction networks in that the
Lectures by Kathleen Fitzpatrick
 Chapter 10 Chemotrophic Energy Metabolism: Aerobic Respiration Lectures by Kathleen Fitzpatrick Simon Fraser University Figure 10-1 Figure 10-6 Conversion of pyruvate The conversion of pyruvate to acetyl
Chapter 10 Chemotrophic Energy Metabolism: Aerobic Respiration Lectures by Kathleen Fitzpatrick Simon Fraser University Figure 10-1 Figure 10-6 Conversion of pyruvate The conversion of pyruvate to acetyl
Chapter 15 part 2. Biochemistry I Introduction to Metabolism Bioenergetics: Thermodynamics in Biochemistry. ATP 4- + H 2 O ADP 3- + P i + H +
 Biochemistry I Introduction to Metabolism Bioenergetics: Thermodynamics in Biochemistry ATP 4- + 2 ADP 3- + P i 2- + + Chapter 15 part 2 Dr. Ray 1 Energy flow in biological systems: Energy Transformations
Biochemistry I Introduction to Metabolism Bioenergetics: Thermodynamics in Biochemistry ATP 4- + 2 ADP 3- + P i 2- + + Chapter 15 part 2 Dr. Ray 1 Energy flow in biological systems: Energy Transformations
Lecture 10. Proton Gradient-dependent ATP Synthesis. Oxidative. Photo-Phosphorylation
 Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation Model of the Electron Transport Chain (ETC) Glycerol-3-P Shuttle Outer Mitochondrial Membrane G3P DHAP
Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation Model of the Electron Transport Chain (ETC) Glycerol-3-P Shuttle Outer Mitochondrial Membrane G3P DHAP
Biology A: Chapter 5 Annotating Notes
 Name: Pd: Biology A: Chapter 5 Annotating Notes -As you read your textbook, please fill out these notes. -Read each paragraph state the big/main idea on the left side. - On the right side you should take
Name: Pd: Biology A: Chapter 5 Annotating Notes -As you read your textbook, please fill out these notes. -Read each paragraph state the big/main idea on the left side. - On the right side you should take
AP Bio-Ms.Bell Unit#3 Cellular Energies Name
 AP Bio-Ms.Bell Unit#3 Cellular Energies Name 1. Base your answer to the following question on the image below. 7. Base your answer to the following question on Which of the following choices correctly
AP Bio-Ms.Bell Unit#3 Cellular Energies Name 1. Base your answer to the following question on the image below. 7. Base your answer to the following question on Which of the following choices correctly
Systems Biology Lecture 1 history, introduction and definitions. Pawan Dhar
 Systems Biology Lecture 1 history, introduction and definitions Pawan Dhar Historical context 1900 1950 2000 Dominant approach Physiology Molecular biology Focus of study Paradigmatic discovery Functioning
Systems Biology Lecture 1 history, introduction and definitions Pawan Dhar Historical context 1900 1950 2000 Dominant approach Physiology Molecular biology Focus of study Paradigmatic discovery Functioning
Photosynthesis and Cellular Respiration
 Photosynthesis and Cellular Respiration Photosynthesis and Cellular Respiration All cellular activities require energy. Directly or indirectly nearly all energy for life comes from the sun. Autotrophs:
Photosynthesis and Cellular Respiration Photosynthesis and Cellular Respiration All cellular activities require energy. Directly or indirectly nearly all energy for life comes from the sun. Autotrophs:
Photosynthesis and cellular respirations
 The Introduction of Biology Defining of life Basic chemistry, the chemistry of organic molecules Classification of living things History of cells and Cells structures and functions Photosynthesis and cellular
The Introduction of Biology Defining of life Basic chemistry, the chemistry of organic molecules Classification of living things History of cells and Cells structures and functions Photosynthesis and cellular
GENOME-SCALE MODELS OF MICROBIAL CELLS: EVALUATING THE CONSEQUENCES OF CONSTRAINTS
 GENOME-SCALE MODELS OF MICROBIAL CELLS: EVALUATING THE CONSEQUENCES OF CONSTRAINTS Nathan D. Price, Jennifer L. Reed and Bernhard Ø. Palsson Abstract Microbial cells operate under governing constraints
GENOME-SCALE MODELS OF MICROBIAL CELLS: EVALUATING THE CONSEQUENCES OF CONSTRAINTS Nathan D. Price, Jennifer L. Reed and Bernhard Ø. Palsson Abstract Microbial cells operate under governing constraints
Division Ave. High School AP Biology
 Overview 10 reactions u convert () to pyruvate (3C) u produces: 4 & NADH u consumes: u net: & NADH C-C-C-C-C-C fructose-1,6bp P-C-C-C-C-C-C-P DHAP P-C-C-C G3P C-C-C-P H P i P i pyruvate C-C-C 4 4 NAD +
Overview 10 reactions u convert () to pyruvate (3C) u produces: 4 & NADH u consumes: u net: & NADH C-C-C-C-C-C fructose-1,6bp P-C-C-C-C-C-C-P DHAP P-C-C-C G3P C-C-C-P H P i P i pyruvate C-C-C 4 4 NAD +
Respiration and Photosynthesis
 Respiration and Photosynthesis Cellular Respiration Glycolysis The Krebs Cycle Electron Transport Chains Anabolic Pathway Photosynthesis Calvin Cycle Flow of Energy Energy is needed to support all forms
Respiration and Photosynthesis Cellular Respiration Glycolysis The Krebs Cycle Electron Transport Chains Anabolic Pathway Photosynthesis Calvin Cycle Flow of Energy Energy is needed to support all forms
State state describe
 Warm-Up State the products of the light-dependent reaction of photosynthesis, state which product has chemical energy, and describe how that product is made. KREBS ETC FADH 2 Glucose Pyruvate H 2 O NADH
Warm-Up State the products of the light-dependent reaction of photosynthesis, state which product has chemical energy, and describe how that product is made. KREBS ETC FADH 2 Glucose Pyruvate H 2 O NADH
Biological Pathways Representation by Petri Nets and extension
 Biological Pathways Representation by and extensions December 6, 2006 Biological Pathways Representation by and extension 1 The cell Pathways 2 Definitions 3 4 Biological Pathways Representation by and
Biological Pathways Representation by and extensions December 6, 2006 Biological Pathways Representation by and extension 1 The cell Pathways 2 Definitions 3 4 Biological Pathways Representation by and
Pathways that Harvest and Store Chemical Energy
 6 Pathways that Harvest and Store Chemical Energy Energy is stored in chemical bonds and can be released and transformed by metabolic pathways. Chemical energy available to do work is termed free energy
6 Pathways that Harvest and Store Chemical Energy Energy is stored in chemical bonds and can be released and transformed by metabolic pathways. Chemical energy available to do work is termed free energy
Metabolism Review. A. Top 10
 A. Top 10 Metabolism Review 1. Energy production through chemiosmosis a. pumping of H+ ions onto one side of a membrane through protein pumps in an Electron Transport Chain (ETC) b. flow of H+ ions across
A. Top 10 Metabolism Review 1. Energy production through chemiosmosis a. pumping of H+ ions onto one side of a membrane through protein pumps in an Electron Transport Chain (ETC) b. flow of H+ ions across
RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES. Bio 107 Week 6
 RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES Bio 107 Week 6 Procedure 7.2 Label test tubes well, including group name 1) Add solutions listed to small test tubes 2)
RESPIRATION AND FERMENTATION: AEROBIC AND ANAEROBIC OXIDATION OF ORGANIC MOLECULES Bio 107 Week 6 Procedure 7.2 Label test tubes well, including group name 1) Add solutions listed to small test tubes 2)
CP Biology Unit 5 Cell Energy Study Guide. Electron Carriers Electron Transport Chain Fermentation Glycolysis Krebs cycle Light-Dependent Reactions
 Name: KEY CP Biology Unit 5 Cell Energy Study Guide Vocabulary to know: ATP ADP Aerobic Anaerobic ATP Synthases Cellular Respiration Chlorophyll Chloroplast Electron Carriers Electron Transport Chain Fermentation
Name: KEY CP Biology Unit 5 Cell Energy Study Guide Vocabulary to know: ATP ADP Aerobic Anaerobic ATP Synthases Cellular Respiration Chlorophyll Chloroplast Electron Carriers Electron Transport Chain Fermentation
Biological Chemistry and Metabolic Pathways
 Biological Chemistry and Metabolic Pathways 1. Reaction a. Thermodynamics b. Kinetics 2. Enzyme a. Structure and Function b. Regulation of Activity c. Kinetics d. Inhibition 3. Metabolic Pathways a. REDOX
Biological Chemistry and Metabolic Pathways 1. Reaction a. Thermodynamics b. Kinetics 2. Enzyme a. Structure and Function b. Regulation of Activity c. Kinetics d. Inhibition 3. Metabolic Pathways a. REDOX
Quantifying thermodynamic bottlenecks of metabolic pathways. Elad Noor TAU, October 2012
 Quantifying thermodynamic bottlenecks of metabolic pathways Elad Noor TAU, October 2012 Hulda S. Haraldsdóttir Ronan M. T. Fleming Wolfram Liebermeister Ron Milo Arren Bar-Even Avi Flamholz Why are there
Quantifying thermodynamic bottlenecks of metabolic pathways Elad Noor TAU, October 2012 Hulda S. Haraldsdóttir Ronan M. T. Fleming Wolfram Liebermeister Ron Milo Arren Bar-Even Avi Flamholz Why are there
Cellular Respiration: Harvesting Chemical Energy. 9.1 Catabolic pathways yield energy by oxidizing organic fuels
 Cellular Respiration: Harvesting Chemical Energy 9.1 Catabolic pathways yield energy by oxidizing organic fuels 9.2 Glycolysis harvests chemical energy by oxidizing glucose to pyruvate 9.3 The citric acid
Cellular Respiration: Harvesting Chemical Energy 9.1 Catabolic pathways yield energy by oxidizing organic fuels 9.2 Glycolysis harvests chemical energy by oxidizing glucose to pyruvate 9.3 The citric acid
CHAPTER 15 Metabolism: Basic Concepts and Design
 CHAPTER 15 Metabolism: Basic Concepts and Design Chapter 15 An overview of Metabolism Metabolism is the sum of cellular reactions - Metabolism the entire network of chemical reactions carried out by living
CHAPTER 15 Metabolism: Basic Concepts and Design Chapter 15 An overview of Metabolism Metabolism is the sum of cellular reactions - Metabolism the entire network of chemical reactions carried out by living
1) What is the one letter and the three letter abbreviations for the poly peptide shown
 Lecture 1 Short problems 1) What is the one letter and the three letter abbreviations for the poly peptide shown 2) How many peptide bonds does this polypeptide possess? 3) Circle the peptide bonds 4)
Lecture 1 Short problems 1) What is the one letter and the three letter abbreviations for the poly peptide shown 2) How many peptide bonds does this polypeptide possess? 3) Circle the peptide bonds 4)
Hypergraphs, Metabolic Networks, Bioreaction Systems. G. Bastin
 Hypergraphs, Metabolic Networks, Bioreaction Systems. G. Bastin PART 1 : Metabolic flux analysis and minimal bioreaction modelling PART 2 : Dynamic metabolic flux analysis of underdetermined networks 2
Hypergraphs, Metabolic Networks, Bioreaction Systems. G. Bastin PART 1 : Metabolic flux analysis and minimal bioreaction modelling PART 2 : Dynamic metabolic flux analysis of underdetermined networks 2
Extreme Pathways in the Post-Genome Era
 Extreme Pathways in the Post-Genome Era Bernhard Palsson Lecture #8 September 6, 003 ) Overview of extreme pathways Outline ) Extreme pathways of human red blood cell metabolism 3) Extreme pathway analysis
Extreme Pathways in the Post-Genome Era Bernhard Palsson Lecture #8 September 6, 003 ) Overview of extreme pathways Outline ) Extreme pathways of human red blood cell metabolism 3) Extreme pathway analysis
Be sure to understand:
 Learning Targets & Focus Questions for Unit 6: Bioenergetics Chapter 8: Thermodynamics Chapter 9: Cell Resp Focus Q Ch. 10: Photosynthesis Chapter 8 (141-150) 1. I can explain how living systems adhere
Learning Targets & Focus Questions for Unit 6: Bioenergetics Chapter 8: Thermodynamics Chapter 9: Cell Resp Focus Q Ch. 10: Photosynthesis Chapter 8 (141-150) 1. I can explain how living systems adhere
International Journal of Scientific & Engineering Research, Volume 6, Issue 2, February ISSN
 International Journal of Scientific & Engineering Research, Volume 6, Issue 2, February-2015 273 Analogizing And Investigating Some Applications of Metabolic Pathway Analysis Methods Gourav Mukherjee 1,
International Journal of Scientific & Engineering Research, Volume 6, Issue 2, February-2015 273 Analogizing And Investigating Some Applications of Metabolic Pathway Analysis Methods Gourav Mukherjee 1,
Study Guide A. Answer Key. Cells and Energy
 Cells and Energy Answer Key SECTION 1. CHEMICAL ENERGY AND ATP 1. molecule; food molecules 2. high-energy; lower-energy 3. phosphate group 4. a; d; b; c 5. b; e 6. c; d 7. a; f 8. chemical energy; light
Cells and Energy Answer Key SECTION 1. CHEMICAL ENERGY AND ATP 1. molecule; food molecules 2. high-energy; lower-energy 3. phosphate group 4. a; d; b; c 5. b; e 6. c; d 7. a; f 8. chemical energy; light
Change to Office Hours this Friday and next Monday. Tomorrow (Abel): 8:30 10:30 am. Monday (Katrina): Cancelled (05/04)
 Change to Office Hours this Friday and next Monday Tomorrow (Abel): 8:30 10:30 am Monday (Katrina): Cancelled (05/04) Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation
Change to Office Hours this Friday and next Monday Tomorrow (Abel): 8:30 10:30 am Monday (Katrina): Cancelled (05/04) Lecture 10 Proton Gradient-dependent ATP Synthesis Oxidative Phosphorylation Photo-Phosphorylation
Biological networks CS449 BIOINFORMATICS
 CS449 BIOINFORMATICS Biological networks Programming today is a race between software engineers striving to build bigger and better idiot-proof programs, and the Universe trying to produce bigger and better
CS449 BIOINFORMATICS Biological networks Programming today is a race between software engineers striving to build bigger and better idiot-proof programs, and the Universe trying to produce bigger and better
Photosynthesis in Higher Plants
 Photosynthesis in Higher Plants Very Short Answers Questions: 1. Name the processes which take place in the grana and stroma regions of chloroplasts? A: Grana Light reactions. Trapping light energy, synthesizing
Photosynthesis in Higher Plants Very Short Answers Questions: 1. Name the processes which take place in the grana and stroma regions of chloroplasts? A: Grana Light reactions. Trapping light energy, synthesizing
Photosynthesis and Cellular Respiration
 Photosynthesis and Cellular Respiration What you will learn: GPS Standard SB3a Explain the cycling of energy through the processes of photosynthesis and respiration. IN OTHER WORDS Photosynthesis and Cellular
Photosynthesis and Cellular Respiration What you will learn: GPS Standard SB3a Explain the cycling of energy through the processes of photosynthesis and respiration. IN OTHER WORDS Photosynthesis and Cellular
Cellular Energy: Respiration. Goals: Anaerobic respiration
 Cellular Energy: Respiration Anaerobic respiration Goals: Define and describe the 3 sets of chemical reactions that comprise aerobic cellular respiration Describe the types of anaerobic respiration Compare
Cellular Energy: Respiration Anaerobic respiration Goals: Define and describe the 3 sets of chemical reactions that comprise aerobic cellular respiration Describe the types of anaerobic respiration Compare
Metabolism. Fermentation vs. Respiration. End products of fermentations are waste products and not fully.
 Outline: Metabolism Part I: Fermentations Part II: Respiration Part III: Metabolic Diversity Learning objectives are: Learn about respiratory metabolism, ATP generation by respiration linked (oxidative)
Outline: Metabolism Part I: Fermentations Part II: Respiration Part III: Metabolic Diversity Learning objectives are: Learn about respiratory metabolism, ATP generation by respiration linked (oxidative)
Photosynthesis and Cellular Respiration Unit
 Photosynthesis and Cellular Respiration Unit All cellular activities require energy. Directly or indirectly nearly all energy for life comes from the sun. Autotrophs: organisms that can make their own
Photosynthesis and Cellular Respiration Unit All cellular activities require energy. Directly or indirectly nearly all energy for life comes from the sun. Autotrophs: organisms that can make their own
Energy Transformation. Metabolism = total chemical reactions in cells.
 Energy Transformation Metabolism = total chemical reactions in cells. metabole = change Metabolism is concerned with managing the material and energy resources of the cell -Catabolism -Anabolism -Catabolism
Energy Transformation Metabolism = total chemical reactions in cells. metabole = change Metabolism is concerned with managing the material and energy resources of the cell -Catabolism -Anabolism -Catabolism
4.1 Chemical Energy and ATP. KEY CONCEPT All cells need chemical energy.
 4.1 Chemical Energy and ATP KEY CONCEPT All cells need chemical energy. 4.1 Chemical Energy and ATP The chemical energy used for most cell processes is carried by ATP. Molecules in food store chemical
4.1 Chemical Energy and ATP KEY CONCEPT All cells need chemical energy. 4.1 Chemical Energy and ATP The chemical energy used for most cell processes is carried by ATP. Molecules in food store chemical
Resource allocation in metabolic networks: kinetic optimization and approximations by FBA
 Resource allocation in metabolic networks: kinetic optimization and approximations by FBA Stefan Müller *1, Georg Regensburger *, Ralf Steuer + * Johann Radon Institute for Computational and Applied Mathematics,
Resource allocation in metabolic networks: kinetic optimization and approximations by FBA Stefan Müller *1, Georg Regensburger *, Ralf Steuer + * Johann Radon Institute for Computational and Applied Mathematics,
MOLECULAR ACTIVITIES OF PLANT CELLS
 MOLECULAR ACTIVITIES OF PLANT CELLS An introduction to plant biochemistry JOHN W. ANDERSON BAgrSc, PhD Reader, Botany Department, School of Biological Sciences, La Trobe University, Bundoora, Victoria,
MOLECULAR ACTIVITIES OF PLANT CELLS An introduction to plant biochemistry JOHN W. ANDERSON BAgrSc, PhD Reader, Botany Department, School of Biological Sciences, La Trobe University, Bundoora, Victoria,
Copyright The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Outer Glycolysis mitochondrial membrane Glucose ATP
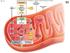 Fig. 7.5 uter Glycolysis mitochondrial membrane Glucose Intermembrane space xidation Mitochondrial matrix Acetyl-oA Krebs FAD e NAD + FAD Inner mitochondrial membrane e Electron e Transport hain hemiosmosis
Fig. 7.5 uter Glycolysis mitochondrial membrane Glucose Intermembrane space xidation Mitochondrial matrix Acetyl-oA Krebs FAD e NAD + FAD Inner mitochondrial membrane e Electron e Transport hain hemiosmosis
Chapter 8 Metabolism: Energy, Enzymes, and Regulation
 Chapter 8 Metabolism: Energy, Enzymes, and Regulation Energy: Capacity to do work or cause a particular change. Thus, all physical and chemical processes are the result of the application or movement of
Chapter 8 Metabolism: Energy, Enzymes, and Regulation Energy: Capacity to do work or cause a particular change. Thus, all physical and chemical processes are the result of the application or movement of
Modeling (bio-)chemical reaction networks
 Mathematical iology: Metabolic Network nalysis Interdisciplinary lecture series for ioinformatics, Mathematics, iology and/or Life Science & Technology dr. Sander Hille shille@math.leidenuniv.nl http://pub.math.leidenuniv.nl/~hillesc
Mathematical iology: Metabolic Network nalysis Interdisciplinary lecture series for ioinformatics, Mathematics, iology and/or Life Science & Technology dr. Sander Hille shille@math.leidenuniv.nl http://pub.math.leidenuniv.nl/~hillesc
A. Structures of PS. Site of PS in plants: mostly in leaves in chloroplasts. Leaf cross section. Vein. Mesophyll CO 2 O 2. Stomata
 PS Lecture Outline I. Introduction A. Structures B. Net Reaction II. Overview of PS A. Rxns in the chloroplast B. pigments III. Closer looks A. LD Rxns B. LI Rxns 1. non-cyclic e- flow 2. cyclic e- flow
PS Lecture Outline I. Introduction A. Structures B. Net Reaction II. Overview of PS A. Rxns in the chloroplast B. pigments III. Closer looks A. LD Rxns B. LI Rxns 1. non-cyclic e- flow 2. cyclic e- flow
2. In regards to the fluid mosaic model, which of the following is TRUE?
 General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
Exam 3 Review (4/12/2011) Lecture note excerpt covering lectures (Exam 3 topics: Chapters 8, 12, 14 & 15)
 Exam 3 Review (4/12/2011) Lecture note excerpt covering lectures 17-23 (Exam 3 topics: Chapters 8, 12, 14 & 15) Enzyme Kinetics, Inhibition, and Regulation Chapter 12 Enzyme Kinetics When the concentration
Exam 3 Review (4/12/2011) Lecture note excerpt covering lectures 17-23 (Exam 3 topics: Chapters 8, 12, 14 & 15) Enzyme Kinetics, Inhibition, and Regulation Chapter 12 Enzyme Kinetics When the concentration
MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 AP Exam Chapters 9 and 10; Photosynthesis and Respiration Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Carbon dioxide (CO2) is released
AP Exam Chapters 9 and 10; Photosynthesis and Respiration Name MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) Carbon dioxide (CO2) is released
AP Biology Day 16. Monday, September 26, 2016
 AP Biology Day 16 Monday, September 26, 2016 CW/HW Assignments 1. Ch. 9 Guided Reading 2. Ch. 9 Video Cornell Notes (2) PLANNER 1. Ch. 9 Video Cornell Notes (weebly) 2. Study & schedule test retake! unit
AP Biology Day 16 Monday, September 26, 2016 CW/HW Assignments 1. Ch. 9 Guided Reading 2. Ch. 9 Video Cornell Notes (2) PLANNER 1. Ch. 9 Video Cornell Notes (weebly) 2. Study & schedule test retake! unit
Biology Reading Assignments:
 Biology 205 5.13.08 Reading Assignments: Chapter 3 Energy, Catalysis and Biosynthesis pgs. 83-94; 106-116 (Note the various roles of nucleotide based carrier molecules); work questions 3-2 and 3-3 Chapter
Biology 205 5.13.08 Reading Assignments: Chapter 3 Energy, Catalysis and Biosynthesis pgs. 83-94; 106-116 (Note the various roles of nucleotide based carrier molecules); work questions 3-2 and 3-3 Chapter
Flux Balance Analysis
 Lecture 10: Flux Balance Analysis Tue 7 March 2006 with the collaboration of Luna De Ferrari 1 Images and text from: E. Klipp, Systems Biology in Practice, Wiley-VCH, 2005 Ch 5 Edwards JS, Palsson BO,
Lecture 10: Flux Balance Analysis Tue 7 March 2006 with the collaboration of Luna De Ferrari 1 Images and text from: E. Klipp, Systems Biology in Practice, Wiley-VCH, 2005 Ch 5 Edwards JS, Palsson BO,
Name Date Class. Photosynthesis and Respiration
 Concept Mapping Photosynthesis and Respiration Complete the Venn diagram about photosynthesis and respiration. These terms may be used more than once: absorbs, Calvin cycle, chlorophyll, CO 2, H 2 O, Krebs
Concept Mapping Photosynthesis and Respiration Complete the Venn diagram about photosynthesis and respiration. These terms may be used more than once: absorbs, Calvin cycle, chlorophyll, CO 2, H 2 O, Krebs
Enumerating constrained elementary flux vectors of metabolic networks
 Enumerating constrained elementary flux vectors of metabolic networks Robert Urbanczik Institute of Physiology, University of Bern Bühlplatz 5, 3012-Bern, Switzerland Abstract I generalize the concept
Enumerating constrained elementary flux vectors of metabolic networks Robert Urbanczik Institute of Physiology, University of Bern Bühlplatz 5, 3012-Bern, Switzerland Abstract I generalize the concept
2) This is a learning problem. You will learn something. You will appreciate this information. If not soon, than someday. Ready?
 Problem Set 2 Hello Class. This is a Big Round problem set. Lots of word problems. I do this because science is a language. Much of it, and medicine too, is transacted as spoken word: seminars, grand rounds,
Problem Set 2 Hello Class. This is a Big Round problem set. Lots of word problems. I do this because science is a language. Much of it, and medicine too, is transacted as spoken word: seminars, grand rounds,
Biology Reading Assignment: Chapter 9 in textbook
 Biology 205 5.10.06 Reading Assignment: Chapter 9 in textbook HTTP://WUNMR.WUSTL.EDU/EDUDEV/LABTUTORIALS/CYTOCHROMES/CYTOCHROMES.HTML What does a cell need to do? propagate itself (and its genetic program)
Biology 205 5.10.06 Reading Assignment: Chapter 9 in textbook HTTP://WUNMR.WUSTL.EDU/EDUDEV/LABTUTORIALS/CYTOCHROMES/CYTOCHROMES.HTML What does a cell need to do? propagate itself (and its genetic program)
Cell Energy: The Big Picture. So, What Exactly is ATP. Adenosine Triphosphate. Your turn to Practice converting ATP to ADP:
 Understanding How Living Things Obtain and Use Energy. Cell Energy: The Big Picture Most Autotrophs produce food (sugar) using light energy during Photosynthesis. Then, both Autotrophs and Heterotroph
Understanding How Living Things Obtain and Use Energy. Cell Energy: The Big Picture Most Autotrophs produce food (sugar) using light energy during Photosynthesis. Then, both Autotrophs and Heterotroph
Cellular Energetics Review
 Cellular Energetics Review 1. What two molecules are formed when a phosphate is removed from ATP? 2. Describe how photosynthesis and cellular respiration are reverse processes. 3. What is the function
Cellular Energetics Review 1. What two molecules are formed when a phosphate is removed from ATP? 2. Describe how photosynthesis and cellular respiration are reverse processes. 3. What is the function
Cells in silico: a Holistic Approach
 Cells in silico: a Holistic Approach Pierpaolo Degano Dipartimento di Informatica, Università di Pisa, Italia joint work with a lot of nice BISCA people :-) Bertinoro, 7th June 2007 SFM 2008 Bertinoro
Cells in silico: a Holistic Approach Pierpaolo Degano Dipartimento di Informatica, Università di Pisa, Italia joint work with a lot of nice BISCA people :-) Bertinoro, 7th June 2007 SFM 2008 Bertinoro
BIOLOGY. Monday 14 Dec 2015
 BIOLOGY Monday 14 Dec 2015 Entry Task An ATP molecule is made of what 3 components? What happens when ATP becomes ADP? Explain. Agenda Entry Task Housekeeping Chapter 8 Essential Question Section 8.2 (Photosynthesis:
BIOLOGY Monday 14 Dec 2015 Entry Task An ATP molecule is made of what 3 components? What happens when ATP becomes ADP? Explain. Agenda Entry Task Housekeeping Chapter 8 Essential Question Section 8.2 (Photosynthesis:
Table 1. ODEs used in the model based on mass balances.
 Table. ODEs used in the model based on mass balances. ODE s _ = _ 6 = + _ 6 = + + = +! 3 = +2!!$ = +!$ %&' ( = _ + %&' ) *+ = + _, - +2 ),$.(/0 = +,!%!-.(023 = + - $!. 56..(023_ = +!. 56. 2,3 8.3(20 =
Table. ODEs used in the model based on mass balances. ODE s _ = _ 6 = + _ 6 = + + = +! 3 = +2!!$ = +!$ %&' ( = _ + %&' ) *+ = + _, - +2 ),$.(/0 = +,!%!-.(023 = + - $!. 56..(023_ = +!. 56. 2,3 8.3(20 =
