Sta$s$cal Physics, Inference and Applica$ons to Biology
|
|
|
- Ashlie Leonard
- 5 years ago
- Views:
Transcription
1 Sta$s$cal Physics, Inference and Applica$ons to Biology Physics Department, Ecole Normale Superieure, Paris, France. Simona Cocco Office:GH301
2 Deriving Protein Structure and Func$on from Sequence Amino- Acid Sequence WW protein? FuncLon Structure One of the major unsolved problems in biology.
3 Deriving Protein Structure and Func$on from Sequence Amino- Acid Sequence A WW protein mutalon? FuncLon Structure MutaLons can be: neutral: no effect detrimental: func$onality decreases, e.g. gives a cancer beneficial: func$onality increases. Dangerous for us if the organism is a bacterium e.g. mutalon confers resistance to anlbiolcs
4 Mul$- sequence alignements (MSA) InformaLon on: - Structure - EvoluLonary Dynamics Big data bases! PFAM 15,000 protein families thousands of sequences for each Model Probability for Sequences in the MSA P(A 1,...,A N )?
5 State of the art on Protein Sequences GeneLc code P L P CCUCUUCCGCCA.
6 State of the art on Protein Sequences Theory Classify sequences corresponding to different proteins (bioinformalcs) (Uniprot,PFAM,genbank) 1995 ~2000 Extract structural informalon from ~2010 protein sequence data (stalslcal physics) 2015 CCUCUUCCGCCA. Sequencing Cost of sequencing ($ per base): 1 Cost of sequencing ($ per base): 10-6 Nb. of protein sequences in databases ~ 10 8 Non- enzymes Enzymes G.Petsko, D.Ringe
7 Evolu$on reflects func$onal and structural constraints Residue contacts induce residue co-evolution contact in 3D co-evolution statistical analysis R I D H R L K N T D H F L N G R L R D T D H H E R Q E T G E L K H K Y R T R L T D L D H R R A M E V G N L K H T Q K E E L A N L K H K Q Q S E V E N A K H R L N Q R A D D L D H correlation Inverse question: S : conservalon Are sequence correlations indicative for residue-residue contacts? [Göbel et al. 94, Neher 94, Ranganathan et al. 99]
8 Evolu$on reflects func$onal and structural constraints Residue contacts induce residue co-evolution contact in 3D co-evolution statistical analysis R I D H R L K N T D H F L N G R L R D T D H H E R Q E T G E L K H K Y R T R L T D L D H R R A M E V G N L K H T Q K E E L A N L K H K Q Q S E V E N A K H R L N Q R A D D L D H correlation Inverse question: S : conservalon Are sequence correlations indicative for residue-residue contacts? [Göbel et al. 94, Neher 94, Ranganathan et al. 99]
9 Evolu$on reflects func$onal and structural constraints Residue contacts induce residue co-evolution contact in 3D co-evolution statistical analysis R I D H R L K N T D H F L N G R L R D T D H H E R Q E T G E L K H K Y R T R L T D L D H R R A M E V G N L K H T Q K E E L A N L K H K Q Q S E V E N A K H R L N Q R A D D L D H correlation Inverse question: S : conservalon Are sequence correlations indicative for residue-residue contacts? [Göbel et al. 94, Neher 94, Ranganathan et al. 99]
10 Sta$s$cal Couplings are beker es$mators of contacts than correla$ons Trypsin inhibitor (small protein, N=52 residues, contact if distance < 0.8 nm) Trypsin inhibitor Correlations vs. residue contacts Trypsin inhibitor Couplings vs. residue contacts - contact - no contact - contact - no contact Wednesday, January 30, 2013 Wednesday, January 30, 2013 [Gobel et al., Proteins 1994] [Weigt et al. PNAS 2009, Structural prediclon: Hopf et al. Cell 2012, Nugent, Jones 2012 (psicov) Protein- protein interaclons.. ] c For Gaussian variables J=- C - 1 ij = J ij J ik J kj k J ik J kl J lj k,l but C is affected by noise, need for regularizalon [S.C., Weigt, Monasson PloS Comp. Biol. 2013]
11 Inference of func$onal interac$ons from the spiking ac$vity of a neural popula$on Raster Plot neurons Small Lme window: bin Lme: 1hour, spikes ReLna, Cortex, Correla=ons Interac=ons Aim: to find an Ising model P(s 1 s N ) which reproduces and interprets the data
12 Mul$- sequence alignements InformaLon on: - Structure - FuncLon Big data bases! PFAM 15,000 protein families thousands of sequences for each Aim: to find a Pons model P(A 1 A N ) which reproduces and interpret the data
13 A 20+1 possible state model: Pons model
14 Broad Outline: Applica$ons Pons model to understand structure of proteins Pons model to decode relalonship between protein s sequence and funclon (genotype- fenotype mapping): - Design new proteins with the same structure and funclon of the natural ones. - ForecasLng Viral EvoluLon - PredicLng anlbiolc resistance
15 Plan Theory : The coupled 20- lener model for Proteins: The Pons Model Pseudo- Likelihood Method ApplicaLons: Analysis of MulLple Sequence Alignment in Proteins for: Contacts prediclons Structural prediclon Design new funclonal proteins the case of a small protein domain WW Predict viral evolulon for HIV virus Predict anlbiolc resistance of TEM 1
16 Example 4: Multi-sequence alignements InformaLon on: - Structure - FuncLon Big data bases! PFAM 15,000 protein families thousands of sequences for each
17 Inference of couplings and fields for PoKs model {A im i=1,..,n ;m=1,..m} 1 M 1 M M Σ δ = f i (A) m=1 M A,Ai Σ δ δ A,Ai m=1 B,Aj = f ij (A,B) f i (A) f J (B) f ij (A,B)
18 Inference of couplings and fields for PoKs model {A im i=1,..,n ;m=1,..m} Σ P(A 1,...,A N ) δ {A} A,Ai Σ P(A 1,...,A N ) δ δ {A} A,Ai = f i (A) B,Aj = f ij (A,B) f i (A) f ij (A,B) f J (B) PoKs Model : P(A 1,...,A N ) = e Σ J ij (A i,a j ) + Σ h i (A i ) i<j Z[{J ij (A,B),h i (A)}] i 21 N+21 2 N (N- 1)/2 Coupled equalons to solve!!
19 Inference of couplings and fields for PoKs model {A im i=1,..,n ;m=1,..m} Σ P(A 1,...,A N ) δ {A} A,Ai Σ P(A 1,...,A N ) δ δ {A} A,Ai = f i (A) B,Ai = f ij (A,B) f i (A) f ij (A,B) f J (B) PoKs Model : P(A 1,...,A N ) = e Σ J ij (A i,a j ) + Σ h i (A i ) i<j Z[{J ij (A i,a j ),h i (A i )}] i 21 N+21 2 N (N- 1)/2 Coupled equalons to solve!! Problem rewrinen as the MinimizaLon of the Cross Entropy S = log Z[{J ij (A,B),h i (A)}] - Σ J ij (A,B) f ij (A,B) - Σ h i (A) f i (A) i<j,a,b i,a
20 Inference of couplings and fields for PoKs model {A im i=1,..,n ;m=1,..m} Σ P(A 1,...,A N ) δ {A} A,Ai Σ P(A 1,...,A N ) δ δ {A} A,Ai = f i (A) B,Ai = f ij (A,B) f i (A) f ij (A,B) f J (B) PoKs Model : P(A 1,...,A N ) = e Σ J ij (A i,a j ) + Σ h i (A i ) i<j Z[{J ij (A i,a j ),h i (A i )}] i 21 N+21 2 N (N- 1)/2 Coupled equalons to solve!! Problem rewrinen as the MinimizaLon of the Cross Entropy S = log Z[{J ij (A,B),h i (A)}] - Σ J ij (A,B) f ij (A,B) - Σ h i (A) f i (A) RegolarizaLon is needed! i<j,a,b i,a
21 1. The Pons model is a generalizalon of the Ising model (binary variables) for any given number of category or Pons states in each site (21- a.a.) 2. Gauge invariance and overparametrizalon It is possible to arbitrarly fix h i (c i ) =0, J ij (c i,b)=0 or other gauges without changing the model probability distribulon 3. Minimising the Cross Entropy allows to find the parameters to reproduce frequencies and correlalons 4. Equivalence between minimising the Cross Entropy and maximising the Log- Likelihood of the data given the model 5. Pseudo- Likelihood model
22 Pseudo- likelihood method Pseudo log-likelihood of the node 0: l 0 =- pseudo cross entropy: S 0 k J0k 0 1 B B S 0 = log [ exp( J 0j (A,A j ) + h 0 (A))] - [h 0 (A) p 0 (A) - J 0j (A,A j ) p ij (A,A j )] τ=1 A,j τ Idea: avoid calculation of partition function using the sampled configurations Total Pseudo- log likelihood: Sum of contribulons of each node A j,aj Prior: increase signal/noise ratio by exploiting the sparsity of J ij cost function ({J}) = pseudo-cross-entropy + { Γ J ij (A,B) ij Γ J ij2 (A,B) ij
23 Biblyography P. Ravikumar, M. J. Wainwright, and J. D. Lafferty (2010). High- dimensional Ising model selec=on using l1- regularized logis=c regression. J. Ann Stat. 38, M. Ekeberg, C. Lovkvist, Y. Lan, M. Weigt, E. Aurell (2013). Improved contact predic=on in proteins: Using pseudolikelihood to infer PoKs models. Phys. Rev. E 87,
24 State of the art in protein structure predic$on
25 State of the art in protein structure predic$on True PosiLve rates: (L prediclons, 8 Å) PconsC MaxEnt alone 0.37 CorrelaLons 0.09 plmdca: Ekeberg, Aurell (2014)
26 PredicLon of Contacts by Pseudo- Likelihood in CASP 2015 Marcin. Skwark (4th posilon on contact prediclo ) Ekeberg et al Phys Rev E 2013,Monastryskyy et al CASP 2015
27 PredicLon of Contacts in the Allosteric HSP70 protein Maliverni D,.., De Los Rios P. HSP70 dimerizalon predicted by Sequence VariaLon Plos Comp Bio (2015).
28 Contact Predic$on and Protein Folding D. Marks T.Hopf, C. Sanders Protein Structure PredicLon Via Sequence varialon Nat. Biotechnology Combine Contact PredicLon and classical Molecular Dynamic simulalons or Monte Carlo Methods with effeclve PotenLals: structures with very high resolulon in the range of 2-6 Å Classical Methods: - All atom PotenLal: ComputaLonally very expensive! - EffecLve PotenLal difficult: find right level of descriplon, take into account context dependence.. - Blind prediclon for transmembranar proteins that are hard to crystallize
29 Contact Predic$on and RNA Folding RNA + Rosena Rosena (Baker group): Monte Carlo MinimizaLon Of EffecLve InteracLon potenlal between amino acids. The bond angles are chosen among a library of known and possible structures. + Lennard Jones anraclve PotenLals for pairs in contact Riboswitch: 2gdi (RF00059) De Leonardis et al. NAR (2015) Rosena alone RMSD 16 Å Rosena + First 25 Coplings:7.5 Å Rosena+ All contacts: 6.3 Å
Supporting Information
 Supporting Information I. INFERRING THE ENERGY FUNCTION We downloaded a multiple sequence alignment (MSA) for the HIV-1 clade B Protease protein from the Los Alamos National Laboratory HIV database (http://www.hiv.lanl.gov).
Supporting Information I. INFERRING THE ENERGY FUNCTION We downloaded a multiple sequence alignment (MSA) for the HIV-1 clade B Protease protein from the Los Alamos National Laboratory HIV database (http://www.hiv.lanl.gov).
RecitaLon CB Lecture #10 RNA Secondary Structure
 RecitaLon 3-19 CB Lecture #10 RNA Secondary Structure 1 Announcements 2 Exam 1 grades and answer key will be posted Friday a=ernoon We will try to make exams available for pickup Friday a=ernoon (probably
RecitaLon 3-19 CB Lecture #10 RNA Secondary Structure 1 Announcements 2 Exam 1 grades and answer key will be posted Friday a=ernoon We will try to make exams available for pickup Friday a=ernoon (probably
Explore SNP polymorphism data. A. Dereeper, Y. Hueber
 Explore SNP polymorphism data A. Dereeper, Y. Hueber Bioinformatics trainings, Supagro, February, 2016 Tablet Graphical tool to visualize assemblies Accept many formats ACE, SAM, BAM GATK (Genome Analysis
Explore SNP polymorphism data A. Dereeper, Y. Hueber Bioinformatics trainings, Supagro, February, 2016 Tablet Graphical tool to visualize assemblies Accept many formats ACE, SAM, BAM GATK (Genome Analysis
Protein Contact Prediction by Integrating Joint Evolutionary Coupling Analysis and Supervised Learning
 Protein Contact Prediction by Integrating Joint Evolutionary Coupling Analysis and Supervised Learning Abstract Jianzhu Ma Sheng Wang Zhiyong Wang Jinbo Xu Toyota Technological Institute at Chicago {majianzhu,
Protein Contact Prediction by Integrating Joint Evolutionary Coupling Analysis and Supervised Learning Abstract Jianzhu Ma Sheng Wang Zhiyong Wang Jinbo Xu Toyota Technological Institute at Chicago {majianzhu,
Potts Models and Protein Covariation. Allan Haldane Ron Levy Group
 Temple University Structural Bioinformatics II Potts Models and Protein Covariation Allan Haldane Ron Levy Group Outline The Evolutionary Origins of Protein Sequence Variation Contents of the Pfam database
Temple University Structural Bioinformatics II Potts Models and Protein Covariation Allan Haldane Ron Levy Group Outline The Evolutionary Origins of Protein Sequence Variation Contents of the Pfam database
Linear Regression. Machine Learning Seyoung Kim. Many of these slides are derived from Tom Mitchell. Thanks!
 Linear Regression Machine Learning 10-601 Seyoung Kim Many of these slides are derived from Tom Mitchell. Thanks! Regression So far, we ve been interested in learning P(Y X) where Y has discrete values
Linear Regression Machine Learning 10-601 Seyoung Kim Many of these slides are derived from Tom Mitchell. Thanks! Regression So far, we ve been interested in learning P(Y X) where Y has discrete values
arxiv: v1 [q-bio.qm] 20 Jan 2014
![arxiv: v1 [q-bio.qm] 20 Jan 2014 arxiv: v1 [q-bio.qm] 20 Jan 2014](/thumbs/82/85310868.jpg) Fast pseudolikelihood maximization for direct-coupling analysis of protein structure from many homologous amino-acid sequences arxiv:1401.4832v1 [q-bio.qm] 20 Jan 2014 Magnus Ekeberg a,b,1,, Tuomo Hartonen
Fast pseudolikelihood maximization for direct-coupling analysis of protein structure from many homologous amino-acid sequences arxiv:1401.4832v1 [q-bio.qm] 20 Jan 2014 Magnus Ekeberg a,b,1,, Tuomo Hartonen
The Evolutionary Origins of Protein Sequence Variation
 Temple University Structural Bioinformatics II The Evolutionary Origins of Protein Sequence Variation Protein Evolution (tour of concepts & current ideas) Protein Fitness, Marginal Stability, Compensatory
Temple University Structural Bioinformatics II The Evolutionary Origins of Protein Sequence Variation Protein Evolution (tour of concepts & current ideas) Protein Fitness, Marginal Stability, Compensatory
From Principal Component to Direct Coupling Analysis of Coevolution in Proteins: Low-Eigenvalue Modes are Needed for Structure Prediction
 From Principal Component to Direct Coupling Analysis of Coevolution in Proteins: Low-Eigenvalue Modes are Needed for Structure Prediction 1 2 Simona Cocco, Remi Monasson, Martin Weigt 3,4 * 1 Laboratoire
From Principal Component to Direct Coupling Analysis of Coevolution in Proteins: Low-Eigenvalue Modes are Needed for Structure Prediction 1 2 Simona Cocco, Remi Monasson, Martin Weigt 3,4 * 1 Laboratoire
Jeremy Chang Identifying protein protein interactions with statistical coupling analysis
 Jeremy Chang Identifying protein protein interactions with statistical coupling analysis Abstract: We used an algorithm known as statistical coupling analysis (SCA) 1 to create a set of features for building
Jeremy Chang Identifying protein protein interactions with statistical coupling analysis Abstract: We used an algorithm known as statistical coupling analysis (SCA) 1 to create a set of features for building
Influence of Multiple Sequence Alignment Depth on Potts Statistical Models of Protein Covariation
 Influence of Multiple Sequence Alignment Depth on Potts Statistical Models of Protein Covariation Allan Haldane Center for Biophysics and Computational Biology, Department of Physics, and Institute for
Influence of Multiple Sequence Alignment Depth on Potts Statistical Models of Protein Covariation Allan Haldane Center for Biophysics and Computational Biology, Department of Physics, and Institute for
Learning the collective dynamics of complex biological systems. from neurons to animal groups. Thierry Mora
 Learning the collective dynamics of complex biological systems from neurons to animal groups Thierry Mora Università Sapienza Rome A. Cavagna I. Giardina O. Pohl E. Silvestri M. Viale Aberdeen University
Learning the collective dynamics of complex biological systems from neurons to animal groups Thierry Mora Università Sapienza Rome A. Cavagna I. Giardina O. Pohl E. Silvestri M. Viale Aberdeen University
CS 365 Introduc/on to Scien/fic Modeling Lecture 2: Cellular Automata
 CS 365 Introduc/on to Scien/fic Modeling Lecture 2: Cellular Automata Stephanie Forrest Dept. of Computer Science Univ. of New Mexico Fall Semester, 2014 Reading Assignment Mitchell, Ch. 10, 2 Wolfram,
CS 365 Introduc/on to Scien/fic Modeling Lecture 2: Cellular Automata Stephanie Forrest Dept. of Computer Science Univ. of New Mexico Fall Semester, 2014 Reading Assignment Mitchell, Ch. 10, 2 Wolfram,
arxiv: v2 [q-bio.bm] 5 Mar 2014
![arxiv: v2 [q-bio.bm] 5 Mar 2014 arxiv: v2 [q-bio.bm] 5 Mar 2014](/thumbs/88/115401288.jpg) 1 arxiv:1403.0379v2 [q-bio.bm] 5 Mar 2014 Improving contact prediction along three dimensions Christoph Feinauer 1,, Marcin J. Skwark 3,4,, Andrea Pagnani 1,2, Erik Aurell 3,4,5, 1 DISAT and Center for
1 arxiv:1403.0379v2 [q-bio.bm] 5 Mar 2014 Improving contact prediction along three dimensions Christoph Feinauer 1,, Marcin J. Skwark 3,4,, Andrea Pagnani 1,2, Erik Aurell 3,4,5, 1 DISAT and Center for
CISC 636 Computational Biology & Bioinformatics (Fall 2016)
 CISC 636 Computational Biology & Bioinformatics (Fall 2016) Predicting Protein-Protein Interactions CISC636, F16, Lec22, Liao 1 Background Proteins do not function as isolated entities. Protein-Protein
CISC 636 Computational Biology & Bioinformatics (Fall 2016) Predicting Protein-Protein Interactions CISC636, F16, Lec22, Liao 1 Background Proteins do not function as isolated entities. Protein-Protein
Gürol M. Süel, Steve W. Lockless, Mark A. Wall, and Rama Ra
 Gürol M. Süel, Steve W. Lockless, Mark A. Wall, and Rama Ranganathan, Evolutionarily conserved networks of residues mediate allosteric communication in proteins, Nature Structural Biology, vol. 10, no.
Gürol M. Süel, Steve W. Lockless, Mark A. Wall, and Rama Ranganathan, Evolutionarily conserved networks of residues mediate allosteric communication in proteins, Nature Structural Biology, vol. 10, no.
CS395T: Structured Models for NLP Lecture 5: Sequence Models II
 CS395T: Structured Models for NLP Lecture 5: Sequence Models II Greg Durrett Some slides adapted from Dan Klein, UC Berkeley Recall: HMMs Input x =(x 1,...,x n ) Output y =(y 1,...,y n ) y 1 y 2 y n P
CS395T: Structured Models for NLP Lecture 5: Sequence Models II Greg Durrett Some slides adapted from Dan Klein, UC Berkeley Recall: HMMs Input x =(x 1,...,x n ) Output y =(y 1,...,y n ) y 1 y 2 y n P
arxiv: v1 [physics.bio-ph] 13 Oct 2017
![arxiv: v1 [physics.bio-ph] 13 Oct 2017 arxiv: v1 [physics.bio-ph] 13 Oct 2017](/thumbs/89/99405972.jpg) Correlation-Compressed Direct Coupling Analysis Chen-Yi Gao 1,3, Hai-Jun Zhou 1,3,5, and Erik Aurell 2,4 1 Key Laboratory of Theoretical Physics, Institute of Theoretical Physics, Chinese Academy of Sciences,
Correlation-Compressed Direct Coupling Analysis Chen-Yi Gao 1,3, Hai-Jun Zhou 1,3,5, and Erik Aurell 2,4 1 Key Laboratory of Theoretical Physics, Institute of Theoretical Physics, Chinese Academy of Sciences,
CS395T: Structured Models for NLP Lecture 2: Machine Learning Review
 CS395T: Structured Models for NLP Lecture 2: Machine Learning Review Greg Durrett Some slides adapted from Vivek Srikumar, University of Utah Administrivia Course enrollment Lecture slides posted on website
CS395T: Structured Models for NLP Lecture 2: Machine Learning Review Greg Durrett Some slides adapted from Vivek Srikumar, University of Utah Administrivia Course enrollment Lecture slides posted on website
Neural Networks for Protein Structure Prediction Brown, JMB CS 466 Saurabh Sinha
 Neural Networks for Protein Structure Prediction Brown, JMB 1999 CS 466 Saurabh Sinha Outline Goal is to predict secondary structure of a protein from its sequence Artificial Neural Network used for this
Neural Networks for Protein Structure Prediction Brown, JMB 1999 CS 466 Saurabh Sinha Outline Goal is to predict secondary structure of a protein from its sequence Artificial Neural Network used for this
Analyzing large-scale spike trains data with spatio-temporal constraints
 Author manuscript, published in "NeuroComp/KEOpS'12 workshop beyond the retina: from computational models to outcomes in bioengineering. Focus on architecture and dynamics sustaining information flows
Author manuscript, published in "NeuroComp/KEOpS'12 workshop beyond the retina: from computational models to outcomes in bioengineering. Focus on architecture and dynamics sustaining information flows
Bayesian Inference of Interactions and Associations
 Bayesian Inference of Interactions and Associations Jun Liu Department of Statistics Harvard University http://www.fas.harvard.edu/~junliu Based on collaborations with Yu Zhang, Jing Zhang, Yuan Yuan,
Bayesian Inference of Interactions and Associations Jun Liu Department of Statistics Harvard University http://www.fas.harvard.edu/~junliu Based on collaborations with Yu Zhang, Jing Zhang, Yuan Yuan,
Ising models for neural activity inferred via Selective Cluster Expansion: structural and coding properties
 Ising models for neural activity inferred via Selective Cluster Expansion: structural and coding properties John Barton,, and Simona Cocco Department of Physics, Rutgers University, Piscataway, NJ 8854
Ising models for neural activity inferred via Selective Cluster Expansion: structural and coding properties John Barton,, and Simona Cocco Department of Physics, Rutgers University, Piscataway, NJ 8854
How pairwise coevolutionary models capture the collective residue variability in proteins
 How pairwise coevolutionary models capture the collective residue variability in proteins Matteo Figliuzzi, Pierre Barrat-Charlaix, and Martin Weigt arxiv:1801.04184v1 [q-bio.qm] 12 Jan 2018 Abstract Global
How pairwise coevolutionary models capture the collective residue variability in proteins Matteo Figliuzzi, Pierre Barrat-Charlaix, and Martin Weigt arxiv:1801.04184v1 [q-bio.qm] 12 Jan 2018 Abstract Global
Alpha-helical Topology and Tertiary Structure Prediction of Globular Proteins Scott R. McAllister Christodoulos A. Floudas Princeton University
 Alpha-helical Topology and Tertiary Structure Prediction of Globular Proteins Scott R. McAllister Christodoulos A. Floudas Princeton University Department of Chemical Engineering Program of Applied and
Alpha-helical Topology and Tertiary Structure Prediction of Globular Proteins Scott R. McAllister Christodoulos A. Floudas Princeton University Department of Chemical Engineering Program of Applied and
Improving De novo Protein Structure Prediction using Contact Maps Information
 CIBCB 2017 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology Improving De novo Protein Structure Prediction using Contact Maps Information Karina Baptista dos Santos
CIBCB 2017 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology Improving De novo Protein Structure Prediction using Contact Maps Information Karina Baptista dos Santos
Advanced Quantum Chemistry III: Part 6
 Advanced Quantum Chemistry III: Part 6 Norio Yoshida Kyushu University Last updated 14-1-15 2013 Winter Term 1 Quantum Chemistry for Condensed Phase Liquid phase Solid phase Biological systems 2 Divide
Advanced Quantum Chemistry III: Part 6 Norio Yoshida Kyushu University Last updated 14-1-15 2013 Winter Term 1 Quantum Chemistry for Condensed Phase Liquid phase Solid phase Biological systems 2 Divide
Learning the dynamics of biological networks
 Learning the dynamics of biological networks Thierry Mora ENS Paris A.Walczak Università Sapienza Rome A. Cavagna I. Giardina O. Pohl E. Silvestri M.Viale Aberdeen University F. Ginelli Laboratoire de
Learning the dynamics of biological networks Thierry Mora ENS Paris A.Walczak Università Sapienza Rome A. Cavagna I. Giardina O. Pohl E. Silvestri M.Viale Aberdeen University F. Ginelli Laboratoire de
Analyzing large-scale spike trains data with spatio-temporal constraints
 Analyzing large-scale spike trains data with spatio-temporal constraints Hassan Nasser, Olivier Marre, Selim Kraria, Thierry Viéville, Bruno Cessac To cite this version: Hassan Nasser, Olivier Marre, Selim
Analyzing large-scale spike trains data with spatio-temporal constraints Hassan Nasser, Olivier Marre, Selim Kraria, Thierry Viéville, Bruno Cessac To cite this version: Hassan Nasser, Olivier Marre, Selim
Design of a Novel Globular Protein Fold with Atomic-Level Accuracy
 Design of a Novel Globular Protein Fold with Atomic-Level Accuracy Brian Kuhlman, Gautam Dantas, Gregory C. Ireton, Gabriele Varani, Barry L. Stoddard, David Baker Presented by Kate Stafford 4 May 05 Protein
Design of a Novel Globular Protein Fold with Atomic-Level Accuracy Brian Kuhlman, Gautam Dantas, Gregory C. Ireton, Gabriele Varani, Barry L. Stoddard, David Baker Presented by Kate Stafford 4 May 05 Protein
Generalized Exponential Random Graph Models: Inference for Weighted Graphs
 Generalized Exponential Random Graph Models: Inference for Weighted Graphs James D. Wilson University of North Carolina at Chapel Hill June 18th, 2015 Political Networks, 2015 James D. Wilson GERGMs for
Generalized Exponential Random Graph Models: Inference for Weighted Graphs James D. Wilson University of North Carolina at Chapel Hill June 18th, 2015 Political Networks, 2015 James D. Wilson GERGMs for
Hamiltonians with focus on Anisotropic interac4ons (Chemical Shi8 Anisotropy and Dipolar Interac4ons) The (solid- state NMR) world is anisotropic
 Hamiltonians with focus on Anisotropic interac4ons (Chemical Shi8 Anisotropy and Dipolar Interac4ons) The (solid- state NMR) world is anisotropic! Munich School November 2014!! Beat H. Meier beme@ethz.ch
Hamiltonians with focus on Anisotropic interac4ons (Chemical Shi8 Anisotropy and Dipolar Interac4ons) The (solid- state NMR) world is anisotropic! Munich School November 2014!! Beat H. Meier beme@ethz.ch
arxiv: v2 [q-bio.bm] 14 Jun 2017
![arxiv: v2 [q-bio.bm] 14 Jun 2017 arxiv: v2 [q-bio.bm] 14 Jun 2017](/thumbs/86/94115461.jpg) Monte Carlo simulation of a statistical mechanical model of multiple protein sequence alignment Akira R. Kinjo Institute for Protein Research, Osaka University 3-2 Yamadaoka, Suita, Osaka, 565-0871, Japan.
Monte Carlo simulation of a statistical mechanical model of multiple protein sequence alignment Akira R. Kinjo Institute for Protein Research, Osaka University 3-2 Yamadaoka, Suita, Osaka, 565-0871, Japan.
Evolu&on of Cellular Interac&on Networks. Pedro Beltrao Krogan and Lim UCSF
 Evolu&on of Cellular Interac&on Networks Pedro Beltrao Krogan and Lim Labs @ UCSF Point muta&ons Recombina&on Duplica&ons Mutants Mutants Point muta&ons Recombina&on Duplica&ons Changes in protein- protein,
Evolu&on of Cellular Interac&on Networks Pedro Beltrao Krogan and Lim Labs @ UCSF Point muta&ons Recombina&on Duplica&ons Mutants Mutants Point muta&ons Recombina&on Duplica&ons Changes in protein- protein,
Variational Methods in Bayesian Deconvolution
 PHYSTAT, SLAC, Stanford, California, September 8-, Variational Methods in Bayesian Deconvolution K. Zarb Adami Cavendish Laboratory, University of Cambridge, UK This paper gives an introduction to the
PHYSTAT, SLAC, Stanford, California, September 8-, Variational Methods in Bayesian Deconvolution K. Zarb Adami Cavendish Laboratory, University of Cambridge, UK This paper gives an introduction to the
Contact map guided ab initio structure prediction
 Contact map guided ab initio structure prediction S M Golam Mortuza Postdoctoral Research Fellow I-TASSER Workshop 2017 North Carolina A&T State University, Greensboro, NC Outline Ab initio structure prediction:
Contact map guided ab initio structure prediction S M Golam Mortuza Postdoctoral Research Fellow I-TASSER Workshop 2017 North Carolina A&T State University, Greensboro, NC Outline Ab initio structure prediction:
Conditional Graphical Models
 PhD Thesis Proposal Conditional Graphical Models for Protein Structure Prediction Yan Liu Language Technologies Institute University Thesis Committee Jaime Carbonell (Chair) John Lafferty Eric P. Xing
PhD Thesis Proposal Conditional Graphical Models for Protein Structure Prediction Yan Liu Language Technologies Institute University Thesis Committee Jaime Carbonell (Chair) John Lafferty Eric P. Xing
Statistical Machine Learning Methods for Bioinformatics IV. Neural Network & Deep Learning Applications in Bioinformatics
 Statistical Machine Learning Methods for Bioinformatics IV. Neural Network & Deep Learning Applications in Bioinformatics Jianlin Cheng, PhD Department of Computer Science University of Missouri, Columbia
Statistical Machine Learning Methods for Bioinformatics IV. Neural Network & Deep Learning Applications in Bioinformatics Jianlin Cheng, PhD Department of Computer Science University of Missouri, Columbia
COMP 562: Introduction to Machine Learning
 COMP 562: Introduction to Machine Learning Lecture 20 : Support Vector Machines, Kernels Mahmoud Mostapha 1 Department of Computer Science University of North Carolina at Chapel Hill mahmoudm@cs.unc.edu
COMP 562: Introduction to Machine Learning Lecture 20 : Support Vector Machines, Kernels Mahmoud Mostapha 1 Department of Computer Science University of North Carolina at Chapel Hill mahmoudm@cs.unc.edu
On the Entropy of Protein Families
 On the Entropy of Protein Families The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Barton, John P.,
On the Entropy of Protein Families The MIT Faculty has made this article openly available. Please share how this access benefits you. Your story matters. Citation As Published Publisher Barton, John P.,
Can protein model accuracy be. identified? NO! CBS, BioCentrum, Morten Nielsen, DTU
 Can protein model accuracy be identified? Morten Nielsen, CBS, BioCentrum, DTU NO! Identification of Protein-model accuracy Why is it important? What is accuracy RMSD, fraction correct, Protein model correctness/quality
Can protein model accuracy be identified? Morten Nielsen, CBS, BioCentrum, DTU NO! Identification of Protein-model accuracy Why is it important? What is accuracy RMSD, fraction correct, Protein model correctness/quality
Supporting Online Material for
 www.sciencemag.org/cgi/content/full/309/5742/1868/dc1 Supporting Online Material for Toward High-Resolution de Novo Structure Prediction for Small Proteins Philip Bradley, Kira M. S. Misura, David Baker*
www.sciencemag.org/cgi/content/full/309/5742/1868/dc1 Supporting Online Material for Toward High-Resolution de Novo Structure Prediction for Small Proteins Philip Bradley, Kira M. S. Misura, David Baker*
arxiv: v2 [cond-mat.stat-mech] 17 Jun 2018
![arxiv: v2 [cond-mat.stat-mech] 17 Jun 2018 arxiv: v2 [cond-mat.stat-mech] 17 Jun 2018](/thumbs/89/99405865.jpg) Inverse Ising inference by combining Ornstein-Zernike theory with deep learning Soma Turi and Alpha A. Lee Cavendish Laboratory, University of Cambridge, Cambridge CB3 HE, United Kingdom arxiv:76.8466v
Inverse Ising inference by combining Ornstein-Zernike theory with deep learning Soma Turi and Alpha A. Lee Cavendish Laboratory, University of Cambridge, Cambridge CB3 HE, United Kingdom arxiv:76.8466v
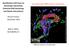
Island Biogeography 1
 Island Biogeography 1 Island Biogeography 2 Island Biogeography Islands of Petran subalpine coniferous forest (black areas) in the American Southwest. Contour lines are the lower edge of Petran montane
Island Biogeography 1 Island Biogeography 2 Island Biogeography Islands of Petran subalpine coniferous forest (black areas) in the American Southwest. Contour lines are the lower edge of Petran montane
Presentation Outline. Prediction of Protein Secondary Structure using Neural Networks at Better than 70% Accuracy
 Prediction of Protein Secondary Structure using Neural Networks at Better than 70% Accuracy Burkhard Rost and Chris Sander By Kalyan C. Gopavarapu 1 Presentation Outline Major Terminology Problem Method
Prediction of Protein Secondary Structure using Neural Networks at Better than 70% Accuracy Burkhard Rost and Chris Sander By Kalyan C. Gopavarapu 1 Presentation Outline Major Terminology Problem Method
Gene Regulatory Networks II Computa.onal Genomics Seyoung Kim
 Gene Regulatory Networks II 02-710 Computa.onal Genomics Seyoung Kim Goal: Discover Structure and Func;on of Complex systems in the Cell Identify the different regulators and their target genes that are
Gene Regulatory Networks II 02-710 Computa.onal Genomics Seyoung Kim Goal: Discover Structure and Func;on of Complex systems in the Cell Identify the different regulators and their target genes that are
Probabilistic Graphical Models
 School of Computer Science Probabilistic Graphical Models Gaussian graphical models and Ising models: modeling networks Eric Xing Lecture 0, February 7, 04 Reading: See class website Eric Xing @ CMU, 005-04
School of Computer Science Probabilistic Graphical Models Gaussian graphical models and Ising models: modeling networks Eric Xing Lecture 0, February 7, 04 Reading: See class website Eric Xing @ CMU, 005-04
High dimensional Ising model selection
 High dimensional Ising model selection Pradeep Ravikumar UT Austin (based on work with John Lafferty, Martin Wainwright) Sparse Ising model US Senate 109th Congress Banerjee et al, 2008 Estimate a sparse
High dimensional Ising model selection Pradeep Ravikumar UT Austin (based on work with John Lafferty, Martin Wainwright) Sparse Ising model US Senate 109th Congress Banerjee et al, 2008 Estimate a sparse
Ab-initio protein structure prediction
 Ab-initio protein structure prediction Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center, Cornell University Ithaca, NY USA Methods for predicting protein structure 1. Homology
Ab-initio protein structure prediction Jaroslaw Pillardy Computational Biology Service Unit Cornell Theory Center, Cornell University Ithaca, NY USA Methods for predicting protein structure 1. Homology
Bias/variance tradeoff, Model assessment and selec+on
 Applied induc+ve learning Bias/variance tradeoff, Model assessment and selec+on Pierre Geurts Department of Electrical Engineering and Computer Science University of Liège October 29, 2012 1 Supervised
Applied induc+ve learning Bias/variance tradeoff, Model assessment and selec+on Pierre Geurts Department of Electrical Engineering and Computer Science University of Liège October 29, 2012 1 Supervised
Predicting Protein Functions and Domain Interactions from Protein Interactions
 Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Priors in Dependency network learning
 Priors in Dependency network learning Sushmita Roy sroy@biostat.wisc.edu Computa:onal Network Biology Biosta2s2cs & Medical Informa2cs 826 Computer Sciences 838 hbps://compnetbiocourse.discovery.wisc.edu
Priors in Dependency network learning Sushmita Roy sroy@biostat.wisc.edu Computa:onal Network Biology Biosta2s2cs & Medical Informa2cs 826 Computer Sciences 838 hbps://compnetbiocourse.discovery.wisc.edu
CMPS 6630: Introduction to Computational Biology and Bioinformatics. Tertiary Structure Prediction
 CMPS 6630: Introduction to Computational Biology and Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the
CMPS 6630: Introduction to Computational Biology and Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the
Homology Modeling. Roberto Lins EPFL - summer semester 2005
 Homology Modeling Roberto Lins EPFL - summer semester 2005 Disclaimer: course material is mainly taken from: P.E. Bourne & H Weissig, Structural Bioinformatics; C.A. Orengo, D.T. Jones & J.M. Thornton,
Homology Modeling Roberto Lins EPFL - summer semester 2005 Disclaimer: course material is mainly taken from: P.E. Bourne & H Weissig, Structural Bioinformatics; C.A. Orengo, D.T. Jones & J.M. Thornton,
Sequence Analysis, '18 -- lecture 9. Families and superfamilies. Sequence weights. Profiles. Logos. Building a representative model for a gene.
 Sequence Analysis, '18 -- lecture 9 Families and superfamilies. Sequence weights. Profiles. Logos. Building a representative model for a gene. How can I represent thousands of homolog sequences in a compact
Sequence Analysis, '18 -- lecture 9 Families and superfamilies. Sequence weights. Profiles. Logos. Building a representative model for a gene. How can I represent thousands of homolog sequences in a compact
Structural biomathematics: an overview of molecular simulations and protein structure prediction
 : an overview of molecular simulations and protein structure prediction Figure: Parc de Recerca Biomèdica de Barcelona (PRBB). Contents 1 A Glance at Structural Biology 2 3 1 A Glance at Structural Biology
: an overview of molecular simulations and protein structure prediction Figure: Parc de Recerca Biomèdica de Barcelona (PRBB). Contents 1 A Glance at Structural Biology 2 3 1 A Glance at Structural Biology
High-dimensional graphical model selection: Practical and information-theoretic limits
 1 High-dimensional graphical model selection: Practical and information-theoretic limits Martin Wainwright Departments of Statistics, and EECS UC Berkeley, California, USA Based on joint work with: John
1 High-dimensional graphical model selection: Practical and information-theoretic limits Martin Wainwright Departments of Statistics, and EECS UC Berkeley, California, USA Based on joint work with: John
Detecting unfolded regions in protein sequences. Anne Poupon Génomique Structurale de la Levure IBBMC Université Paris-Sud / CNRS France
 Detecting unfolded regions in protein sequences Anne Poupon Génomique Structurale de la Levure IBBMC Université Paris-Sud / CNRS France Large proteins and complexes: a domain approach Structural studies
Detecting unfolded regions in protein sequences Anne Poupon Génomique Structurale de la Levure IBBMC Université Paris-Sud / CNRS France Large proteins and complexes: a domain approach Structural studies
Improved Protein Secondary Structure Prediction
 Improved Protein Secondary Structure Prediction Secondary Structure Prediction! Given a protein sequence a 1 a 2 a N, secondary structure prediction aims at defining the state of each amino acid ai as
Improved Protein Secondary Structure Prediction Secondary Structure Prediction! Given a protein sequence a 1 a 2 a N, secondary structure prediction aims at defining the state of each amino acid ai as
Information Theory. Mark van Rossum. January 24, School of Informatics, University of Edinburgh 1 / 35
 1 / 35 Information Theory Mark van Rossum School of Informatics, University of Edinburgh January 24, 2018 0 Version: January 24, 2018 Why information theory 2 / 35 Understanding the neural code. Encoding
1 / 35 Information Theory Mark van Rossum School of Informatics, University of Edinburgh January 24, 2018 0 Version: January 24, 2018 Why information theory 2 / 35 Understanding the neural code. Encoding
Parameter Estimation for Spatio-Temporal Maximum Entropy Distributions: Application to Neural Spike Trains
 Entropy 214, 16, 2244-2277; doi:1.339/e1642244 OPEN ACCESS entropy ISSN 199-43 www.mdpi.com/journal/entropy Article Parameter Estimation for Spatio-Temporal Maximum Entropy Distributions: Application to
Entropy 214, 16, 2244-2277; doi:1.339/e1642244 OPEN ACCESS entropy ISSN 199-43 www.mdpi.com/journal/entropy Article Parameter Estimation for Spatio-Temporal Maximum Entropy Distributions: Application to
Variational inference
 Simon Leglaive Télécom ParisTech, CNRS LTCI, Université Paris Saclay November 18, 2016, Télécom ParisTech, Paris, France. Outline Introduction Probabilistic model Problem Log-likelihood decomposition EM
Simon Leglaive Télécom ParisTech, CNRS LTCI, Université Paris Saclay November 18, 2016, Télécom ParisTech, Paris, France. Outline Introduction Probabilistic model Problem Log-likelihood decomposition EM
arxiv: v3 [cond-mat.dis-nn] 18 May 2011
![arxiv: v3 [cond-mat.dis-nn] 18 May 2011 arxiv: v3 [cond-mat.dis-nn] 18 May 2011](/thumbs/84/91004413.jpg) Exact mean field inference in asymmetric kinetic Ising systems M. Mézard and J. Sakellariou Laboratoire de Physique Théorique et Modèles Statistiques, CNRS and Université Paris-Sud, Bât 100, 91405 Orsay
Exact mean field inference in asymmetric kinetic Ising systems M. Mézard and J. Sakellariou Laboratoire de Physique Théorique et Modèles Statistiques, CNRS and Université Paris-Sud, Bât 100, 91405 Orsay
Goals. Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions
 Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions Jamie Duke 1,2 and Carlos Camacho 3 1 Bioengineering and Bioinformatics Summer Institute,
Structural Analysis of the EGR Family of Transcription Factors: Templates for Predicting Protein DNA Interactions Jamie Duke 1,2 and Carlos Camacho 3 1 Bioengineering and Bioinformatics Summer Institute,
CSE446: Linear Regression Regulariza5on Bias / Variance Tradeoff Winter 2015
 CSE446: Linear Regression Regulariza5on Bias / Variance Tradeoff Winter 2015 Luke ZeElemoyer Slides adapted from Carlos Guestrin Predic5on of con5nuous variables Billionaire says: Wait, that s not what
CSE446: Linear Regression Regulariza5on Bias / Variance Tradeoff Winter 2015 Luke ZeElemoyer Slides adapted from Carlos Guestrin Predic5on of con5nuous variables Billionaire says: Wait, that s not what
High-dimensional graphical model selection: Practical and information-theoretic limits
 1 High-dimensional graphical model selection: Practical and information-theoretic limits Martin Wainwright Departments of Statistics, and EECS UC Berkeley, California, USA Based on joint work with: John
1 High-dimensional graphical model selection: Practical and information-theoretic limits Martin Wainwright Departments of Statistics, and EECS UC Berkeley, California, USA Based on joint work with: John
Chapter 4 Dynamic Bayesian Networks Fall Jin Gu, Michael Zhang
 Chapter 4 Dynamic Bayesian Networks 2016 Fall Jin Gu, Michael Zhang Reviews: BN Representation Basic steps for BN representations Define variables Define the preliminary relations between variables Check
Chapter 4 Dynamic Bayesian Networks 2016 Fall Jin Gu, Michael Zhang Reviews: BN Representation Basic steps for BN representations Define variables Define the preliminary relations between variables Check
Inferring Transcriptional Regulatory Networks from Gene Expression Data II
 Inferring Transcriptional Regulatory Networks from Gene Expression Data II Lectures 9 Oct 26, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday
Inferring Transcriptional Regulatory Networks from Gene Expression Data II Lectures 9 Oct 26, 2011 CSE 527 Computational Biology, Fall 2011 Instructor: Su-In Lee TA: Christopher Miles Monday & Wednesday
Secondary Structure. Bioch/BIMS 503 Lecture 2. Structure and Function of Proteins. Further Reading. Φ, Ψ angles alone determine protein structure
 Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
Bioch/BIMS 503 Lecture 2 Structure and Function of Proteins August 28, 2008 Robert Nakamoto rkn3c@virginia.edu 2-0279 Secondary Structure Φ Ψ angles determine protein structure Φ Ψ angles are restricted
W/Z + Jets in ATLAS. Standard LHC Workshop April 12, 2012 Copenhagen, Denmark
 W/Z Jets in Standard Model @ LHC Workshop April, 0 Copenhagen, Denmark Imai Jen- La Plante (University of Chicago) on behalf of the CollaboraLon QCD Studies with W and Z Bosons Studies of produc8on in
W/Z Jets in Standard Model @ LHC Workshop April, 0 Copenhagen, Denmark Imai Jen- La Plante (University of Chicago) on behalf of the CollaboraLon QCD Studies with W and Z Bosons Studies of produc8on in
CS 6140: Machine Learning Spring What We Learned Last Week. Survey 2/26/16. VS. Model
 Logis@cs CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa@on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Assignment
Logis@cs CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa@on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Assignment
CMPS 6630: Introduction to Computational Biology and Bioinformatics. Structure Comparison
 CMPS 6630: Introduction to Computational Biology and Bioinformatics Structure Comparison Protein Structure Comparison Motivation Understand sequence and structure variability Understand Domain architecture
CMPS 6630: Introduction to Computational Biology and Bioinformatics Structure Comparison Protein Structure Comparison Motivation Understand sequence and structure variability Understand Domain architecture
CS 6140: Machine Learning Spring 2016
 CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa?on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Logis?cs Assignment
CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa?on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Logis?cs Assignment
Computational methods for predicting protein-protein interactions
 Computational methods for predicting protein-protein interactions Tomi Peltola T-61.6070 Special course in bioinformatics I 3.4.2008 Outline Biological background Protein-protein interactions Computational
Computational methods for predicting protein-protein interactions Tomi Peltola T-61.6070 Special course in bioinformatics I 3.4.2008 Outline Biological background Protein-protein interactions Computational
Biol478/ August
 Biol478/595 29 August # Day Inst. Topic Hwk Reading August 1 M 25 MG Introduction 2 W 27 MG Sequences and Evolution Handouts 3 F 29 MG Sequences and Evolution September M 1 Labor Day 4 W 3 MG Database
Biol478/595 29 August # Day Inst. Topic Hwk Reading August 1 M 25 MG Introduction 2 W 27 MG Sequences and Evolution Handouts 3 F 29 MG Sequences and Evolution September M 1 Labor Day 4 W 3 MG Database
Some recent results on the inverse Ising problem. Federico Ricci-Tersenghi Physics Department Sapienza University, Roma
 Some recent results on the inverse Ising problem Federico Ricci-Tersenghi Physics Department Sapienza University, Roma Outline of the talk Bethe approx. for inverse Ising problem Comparison among several
Some recent results on the inverse Ising problem Federico Ricci-Tersenghi Physics Department Sapienza University, Roma Outline of the talk Bethe approx. for inverse Ising problem Comparison among several
Graphical Model Selection
 May 6, 2013 Trevor Hastie, Stanford Statistics 1 Graphical Model Selection Trevor Hastie Stanford University joint work with Jerome Friedman, Rob Tibshirani, Rahul Mazumder and Jason Lee May 6, 2013 Trevor
May 6, 2013 Trevor Hastie, Stanford Statistics 1 Graphical Model Selection Trevor Hastie Stanford University joint work with Jerome Friedman, Rob Tibshirani, Rahul Mazumder and Jason Lee May 6, 2013 Trevor
Adaptive Crowdsourcing via EM with Prior
 Adaptive Crowdsourcing via EM with Prior Peter Maginnis and Tanmay Gupta May, 205 In this work, we make two primary contributions: derivation of the EM update for the shifted and rescaled beta prior and
Adaptive Crowdsourcing via EM with Prior Peter Maginnis and Tanmay Gupta May, 205 In this work, we make two primary contributions: derivation of the EM update for the shifted and rescaled beta prior and
Giri Narasimhan. CAP 5510: Introduction to Bioinformatics. ECS 254; Phone: x3748
 CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 2/15/07 CAP5510 1 EM Algorithm Goal: Find θ, Z that maximize Pr
CAP 5510: Introduction to Bioinformatics Giri Narasimhan ECS 254; Phone: x3748 giri@cis.fiu.edu www.cis.fiu.edu/~giri/teach/bioinfs07.html 2/15/07 CAP5510 1 EM Algorithm Goal: Find θ, Z that maximize Pr
CMPS 3110: Bioinformatics. Tertiary Structure Prediction
 CMPS 3110: Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the laws of physics! Conformation space is finite
CMPS 3110: Bioinformatics Tertiary Structure Prediction Tertiary Structure Prediction Why Should Tertiary Structure Prediction Be Possible? Molecules obey the laws of physics! Conformation space is finite
Protein Folding Prof. Eugene Shakhnovich
 Protein Folding Eugene Shakhnovich Department of Chemistry and Chemical Biology Harvard University 1 Proteins are folded on various scales As of now we know hundreds of thousands of sequences (Swissprot)
Protein Folding Eugene Shakhnovich Department of Chemistry and Chemical Biology Harvard University 1 Proteins are folded on various scales As of now we know hundreds of thousands of sequences (Swissprot)
Time- varying signals: cross- and auto- correla5on, correlograms. NEU 466M Instructor: Professor Ila R. Fiete Spring 2016
 Time- varying signals: cross- and auto- correla5on, correlograms NEU 466M Instructor: Professor Ila R. Fiete Spring 2016 Sta5s5cal measures We first considered simple sta5s5cal measures for single variables
Time- varying signals: cross- and auto- correla5on, correlograms NEU 466M Instructor: Professor Ila R. Fiete Spring 2016 Sta5s5cal measures We first considered simple sta5s5cal measures for single variables
Template Free Protein Structure Modeling Jianlin Cheng, PhD
 Template Free Protein Structure Modeling Jianlin Cheng, PhD Associate Professor Computer Science Department Informatics Institute University of Missouri, Columbia 2013 Protein Energy Landscape & Free Sampling
Template Free Protein Structure Modeling Jianlin Cheng, PhD Associate Professor Computer Science Department Informatics Institute University of Missouri, Columbia 2013 Protein Energy Landscape & Free Sampling
Machine Learning I Continuous Reinforcement Learning
 Machine Learning I Continuous Reinforcement Learning Thomas Rückstieß Technische Universität München January 7/8, 2010 RL Problem Statement (reminder) state s t+1 ENVIRONMENT reward r t+1 new step r t
Machine Learning I Continuous Reinforcement Learning Thomas Rückstieß Technische Universität München January 7/8, 2010 RL Problem Statement (reminder) state s t+1 ENVIRONMENT reward r t+1 new step r t
Logis&c Regression. Robot Image Credit: Viktoriya Sukhanova 123RF.com
 Logis&c Regression These slides were assembled by Eric Eaton, with grateful acknowledgement of the many others who made their course materials freely available online. Feel free to reuse or adapt these
Logis&c Regression These slides were assembled by Eric Eaton, with grateful acknowledgement of the many others who made their course materials freely available online. Feel free to reuse or adapt these
Politecnico di Torino. Porto Institutional Repository
 Politecnico di Torino Porto Institutional Repository [Doctoral thesis] The Statistical Mechanics Approach to Protein Sequence Data: Beyond Contact Prediction Original Citation: Feinauer, Christoph (2016).
Politecnico di Torino Porto Institutional Repository [Doctoral thesis] The Statistical Mechanics Approach to Protein Sequence Data: Beyond Contact Prediction Original Citation: Feinauer, Christoph (2016).
Machine Learning. Lecture Slides for. ETHEM ALPAYDIN The MIT Press, h1p://www.cmpe.boun.edu.
 Lecture Slides for INTRODUCTION TO Machine Learning ETHEM ALPAYDIN The MIT Press, 2010 alpaydin@boun.edu.tr h1p://www.cmpe.boun.edu.tr/~ethem/i2ml2e CHAPTER 19: Design and Analysis of Machine Learning
Lecture Slides for INTRODUCTION TO Machine Learning ETHEM ALPAYDIN The MIT Press, 2010 alpaydin@boun.edu.tr h1p://www.cmpe.boun.edu.tr/~ethem/i2ml2e CHAPTER 19: Design and Analysis of Machine Learning
Minimum Message Length Inference and Mixture Modelling of Inverse Gaussian Distributions
 Minimum Message Length Inference and Mixture Modelling of Inverse Gaussian Distributions Daniel F. Schmidt Enes Makalic Centre for Molecular, Environmental, Genetic & Analytic (MEGA) Epidemiology School
Minimum Message Length Inference and Mixture Modelling of Inverse Gaussian Distributions Daniel F. Schmidt Enes Makalic Centre for Molecular, Environmental, Genetic & Analytic (MEGA) Epidemiology School
Protein Structure Prediction
 Page 1 Protein Structure Prediction Russ B. Altman BMI 214 CS 274 Protein Folding is different from structure prediction --Folding is concerned with the process of taking the 3D shape, usually based on
Page 1 Protein Structure Prediction Russ B. Altman BMI 214 CS 274 Protein Folding is different from structure prediction --Folding is concerned with the process of taking the 3D shape, usually based on
+ + ( + ) = Linear recurrent networks. Simpler, much more amenable to analytic treatment E.g. by choosing
 Linear recurrent networks Simpler, much more amenable to analytic treatment E.g. by choosing + ( + ) = Firing rates can be negative Approximates dynamics around fixed point Approximation often reasonable
Linear recurrent networks Simpler, much more amenable to analytic treatment E.g. by choosing + ( + ) = Firing rates can be negative Approximates dynamics around fixed point Approximation often reasonable
Mul$ple Sequence Alignment Methods. Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu
 Mul$ple Sequence Alignment Methods Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu Species Tree Orangutan Gorilla Chimpanzee Human From the Tree of the Life
Mul$ple Sequence Alignment Methods Tandy Warnow Departments of Bioengineering and Computer Science h?p://tandy.cs.illinois.edu Species Tree Orangutan Gorilla Chimpanzee Human From the Tree of the Life
LEAP: Liquefac.on Experiments and Analysis Projects
 LEAP: Liquefac.on Experiments and Analysis Projects Bruce Ku(er and Trevor Carey, UC Davis Majid Manzari, George Washington University Mourad Zeghal, RPI 1 How accurate are numerical methods that are use
LEAP: Liquefac.on Experiments and Analysis Projects Bruce Ku(er and Trevor Carey, UC Davis Majid Manzari, George Washington University Mourad Zeghal, RPI 1 How accurate are numerical methods that are use
Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche
 Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche The molecular structure of a protein can be broken down hierarchically. The primary structure of a protein is simply its
Protein Structure Prediction II Lecturer: Serafim Batzoglou Scribe: Samy Hamdouche The molecular structure of a protein can be broken down hierarchically. The primary structure of a protein is simply its
Controlled healing of graphene nanopore
 Controlled healing of graphene nanopore Konstantin Zakharchenko Alexander Balatsky Zakharchenko K.V., Balatsky A.V. Controlled healing of graphene nanopore. Carbon (80), December 2014, pp. 12 18. http://dx.doi.org/10.1016/j.carbon.2014.07.085
Controlled healing of graphene nanopore Konstantin Zakharchenko Alexander Balatsky Zakharchenko K.V., Balatsky A.V. Controlled healing of graphene nanopore. Carbon (80), December 2014, pp. 12 18. http://dx.doi.org/10.1016/j.carbon.2014.07.085
CS388: Natural Language Processing Lecture 9: CNNs, Neural CRFs. CNNs. Administrivia. This Lecture. Greg Durrett. Project 1 due today at 5pm
 CS388: Natural Language Processing Lecture 9: CNNs, Neural CRFs Project 1 due today at 5pm Administrivia Mini 2 out tonight, due in two weeks Greg Durrett This Lecture CNNs CNNs for SenLment Neural CRFs
CS388: Natural Language Processing Lecture 9: CNNs, Neural CRFs Project 1 due today at 5pm Administrivia Mini 2 out tonight, due in two weeks Greg Durrett This Lecture CNNs CNNs for SenLment Neural CRFs
Variable Selection in High Dimensional Convex Regression
 Sensing and Analysis of High-Dimensional Data UCL-Duke Workshop September 4 & 5, 2014 Variable Selection in High Dimensional Convex Regression John Lafferty Department of Statistics & Department of Computer
Sensing and Analysis of High-Dimensional Data UCL-Duke Workshop September 4 & 5, 2014 Variable Selection in High Dimensional Convex Regression John Lafferty Department of Statistics & Department of Computer
Proteomics and Variable Selection
 Proteomics and Variable Selection p. 1/55 Proteomics and Variable Selection Alex Lewin With thanks to Paul Kirk for some graphs Department of Epidemiology and Biostatistics, School of Public Health, Imperial
Proteomics and Variable Selection p. 1/55 Proteomics and Variable Selection Alex Lewin With thanks to Paul Kirk for some graphs Department of Epidemiology and Biostatistics, School of Public Health, Imperial
CS 6140: Machine Learning Spring What We Learned Last Week 2/26/16
 Logis@cs CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa@on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Sign
Logis@cs CS 6140: Machine Learning Spring 2016 Instructor: Lu Wang College of Computer and Informa@on Science Northeastern University Webpage: www.ccs.neu.edu/home/luwang Email: luwang@ccs.neu.edu Sign
Deepening Ques+ons about Electron Deep Orbits of the Hydrogen Atom
 Deepening Ques+ons about Electron Deep Orbits of the Hydrogen Atom J. L. Paillet 1, A. Meulenberg 2 1 Aix- Marseille University, France, jean- luc.paillet@club- internet.fr 2 Science for Humanity Trust,
Deepening Ques+ons about Electron Deep Orbits of the Hydrogen Atom J. L. Paillet 1, A. Meulenberg 2 1 Aix- Marseille University, France, jean- luc.paillet@club- internet.fr 2 Science for Humanity Trust,
Scalable Inference for Neuronal Connectivity from Calcium Imaging
 Scalable Inference for Neuronal Connectivity from Calcium Imaging Alyson K. Fletcher Sundeep Rangan Abstract Fluorescent calcium imaging provides a potentially powerful tool for inferring connectivity
Scalable Inference for Neuronal Connectivity from Calcium Imaging Alyson K. Fletcher Sundeep Rangan Abstract Fluorescent calcium imaging provides a potentially powerful tool for inferring connectivity
