Ising models for neural activity inferred via Selective Cluster Expansion: structural and coding properties
|
|
|
- Kelley Johnston
- 5 years ago
- Views:
Transcription
1 Ising models for neural activity inferred via Selective Cluster Expansion: structural and coding properties John Barton,, and Simona Cocco Department of Physics, Rutgers University, Piscataway, NJ 8854 USA CNRS-Laboratoire de Physique Statistique de l ENS, 4 rue Lhomond, 755 Paris, France Present address: Department of Chemical Engineering, MIT, Cambridge, MA 39 Present address: Ragon Institute of Massachusetts General Hospital, Massachusetts Institute of Technology and Harvard University, Boston, MA 9 jpbarton@mit.edu, cocco@lps.ens.fr Abstract. We describe the Selective Cluster Expansion (SCE) of the entropy, a method for inferring an Ising model which describes the correlated activity of populations of neurons [, ]. We re-analyze data obtained from multielectrode recordings performed in vitro on the retina and in vivo on the prefrontal cortex. Recorded population sizes N range from N = 37 to N = 7 neurons. We compare the SCE method with the simplest mean field methods (corresponding to a Gaussian model) and with regularizations which favor sparse networks (L norm) or penalize large couplings (L norm). The network of the strongest interactions inferred via mean field methods generally agree with those obtained from SCE. Reconstruction of the sampled moments of the distributions, corresponding to neuron spiking frequencies and pairwise correlations, and the prediction of higher moments including three-cell correlations and multi-neuron firing frequencies, is more difficult than determining the large-scale structure of the interaction network, and apart from a cortical recording in which the measured correlation indices are small, these goals are achieved with the SCE but not with mean field approaches. We also find differences in the inferred structure of retinal and cortical networks: inferred interactions tend to be more irregular and sparse for cortical data than for retinal data. This result may reflect the structure of the recording. As a consequence the SCE is more effective for retinal data when expanding the entropy with respect to a mean field reference S S MF, while expansions of the entropy S alone perform better for cortical data. Keywords: statistical inference, neuronal networks
2 Ising models for neural activity inferred via Selective Cluster Expansion. Introduction Recent years have seen the advance of in vivo [3] and in vitro [4] recording of populations of neurons. The quantitative analysis of these data presents a formidable computational challenge. Bialek and coworkers [5] and Chichilnisky [6] and coworkers have introduced the idea of using the Ising model to describe patterns of correlated neural activity. The Ising model is the maximum entropy model, i.e. the least constrained model [7], capable of reproducing observed spiking frequencies and pairwise correlations in the neural population. This model has proved to be a good model for the activity, in the sense that even though one only constrains the model using the first and second moments, one can predict features of the activity such as third moments and multi-neuron firing patterns. The inverse Ising approach, consisting of the inference of an effective Ising model from experimental data, goes beyond the cross-correlation analysis of multielectrode recordings by disentangling the network of direct interactions from correlations. The inverse Ising approach can then be used to extract structural information on effective connections, such as how they change with the distance, time scale, and different types of cells. One can also explore how effective connections change during activity due to adaptation, short term memory, and long term learning. Another key feature of the Inverse Ising approach is the ability to take into account that measured correlations are affected by finite sampling noise. Experiments are limited in time and their ability to thoroughly sample the experimental system, therefore it is important to avoid overfitting of data. Overfitting can have drastic effects on the structure of the inferred interaction network. One way to avoid overfitting is to regularize the inference problem by introducing a priori information about the model parameters in the log-likelihood of the Ising model giving the data. However, inferring an Ising model from a set of experimental data is a challenging computational problem with no straightforward solution. Typical methods of solving the inference problem include the Boltzmann learning method, which involves iterative Monte Carlo simulations followed by small updates to the interaction parameters [8]. This method can be very slow for large systems, though recent advances have notably improved the speed, and this method has proved to be effective in the analysis of neural data [9 ]. Other methods such as iterative scaling algorithms [, 3], pseudolikelihood approximation [4, 5], various perturbative expansions [6 9] and mean field (or Gaussian model) approximations [] have also been developed which attempt to solve the Inverse Ising problem in certain limits. Such approximations are often computationally simple, but suffer from a limited range of validity. One method that has recently proven to effectively infer the parameters of spin glass models is the method of minimum probability flow []. Here we review a statistical mechanical approach [, ] based on a selective cluster expansion which improves upon mean field methods. Moreover it avoids overfitting by
3 Ising models for neural activity inferred via Selective Cluster Expansion 3 selecting only clusters with significant contributions to the inverse problem, minimizing the impact of finite sampling noise. We compare this approach with the mean field or Gaussian approximation and we test different forms of regularization, in particular the norm L regularization, which preferentially penalizes small but nonzero couplings, and norm L regularization, which penalizes large couplings in absolute value more heavily. Our aim is to compare the methods using the structure and sparsity of the inferred interaction network, and the ability of the inferred Ising model to reproduce the multineuron recorded activity. These two features correspond respectively to the capacity to give structural information about the system which has generated the data and the ability of the Ising model to encode neural activity. The outline of this paper is as follows. In the remainder of the introduction we give a description of the neural data, then detail two methods for analyzing the data: crosscorrelation histogram analysis, which focuses on correlations between pairs of cells, and the inverse Ising approach, which attempts to infer an effective network of interactions characterizing the whole system. In Section we give an overview of the effects of finite sampling noise in the data on the Ising model inference problem. This includes, in particular, quantifying the expected fluctuations of the empirical correlations and the inferred Ising model couplings and fields, as well as introducing methods of regularizing the inference procedure to minimize problems due to finite sampling. Section 3 gives a discussion of the difficulties of the inverse Ising problem. In Sections 4 and 5, we describe two methods for solving the inverse problem: the mean field or Gaussian approximation, and the Selective Cluster Expansion (SCE). Here we discuss regularization conventions for both the mean field and SCE, as well as practical computational methods and questions of convergence for the SCE. Beginning with Section 6 we focus on applications to real data. In this Section we assess the performance of the SCE algorithm on retinal and cortical data. We then describe the properties of the inferred Ising models following the analysis performed previously in [7] on retinal data, showing that the inferred models reconstruct the empirical spiking frequencies and pairwise correlations, and evaluating their ability to predict higher order correlations and multi-neuron firing frequencies. Structural properties of the interaction networks in retinal and cortical data, including maps of inferred couplings and the reliability of the inferred positive and negative couplings are also discussed. We illustrate the importance of network effects by comparing the couplings obtained through SCE with the those obtained from the correlation indices, which only depend on properties of pairs of cells rather than on the entire system. We also compare couplings inferred with SCE on a subset of each experimental system with those for the full system. In Section 7 we explore the performance of the mean field approximation using a variety of regularization methods and strengths. Couplings inferred via the mean field approximation and those obtained from SCE are also compared. Finally in Section 8 we discuss the limitations of the algorithm and some possible extensions of the static inverse Ising approach in treating neural data.
4 Ising models for neural activity inferred via Selective Cluster Expansion Dark N= Flicker N=5 4 3 Natural Movie N=4 4 3 Cortex N= Cortex N= time (sec) Figure. Raster plot of spike train data from recordings of neurons populations: in the retina in dark conditions (N = 6 cells) and with a random flickering stimulus (N = 5 cells), data by M. Meister []; in the retina with a natural movie stimulus (N = 4 cells), data by M. Berry [5]; in the prefrontal cortex of a rat (N = 3 cells), data by A. Peyrache and F. Battaglia [3]; in the medial prefrontal cortex of a rat (N = 7 cells), data by G. Buzsáki and S. Fujisawa [4]. Recordings last for 3 minutes hour, here we plot only s of the recording. One can translate the continuous time data in the raster plot into binary patterns of activity by arranging the time interval into bins of size t and recording whether or not each neuron spikes within each time bin. The probability p i that a neuron i spikes in a time window of size t is the number of windows in which the neuron i is active divided by the total number of time windows. The probability p ij that two neurons i, j are active in the same time window is given by the number of time windows in which both the neurons are active divided by the total number of time windows... Description of neural data We begin by briefly describing the in vitro and in vivo multielectrode recordings (see Fig. ) of neural activity used in our analysis. The first set of recording is done in vitro on the retina [5,, 5]. Here the retina is extracted from the eye of an animal typically a salamander, guinea pig, or primate and placed on a multielectrode array with all five layers of cells (photoreceptors, horizontal, bipolar, amacrine, and ganglion cells) responsible for detecting and preprocessing visual signals. This allows for the simultaneous recording of tens to hundreds of cells in the last layer consisting of ganglion cells while the retina is subjected to various visual stimuli. The neural network of the retina is relatively simple: it has a feed-forward architecture and preprocesses visual information, which is then transmitted through the optical nerve to the visual cortex [6, 6]. The retina thus presents an ideal environment for studying how a stimulus is encoded in the ganglion cell activity and how information is processed [7]. The first experiments on the retina [8] defined the concept of receptive fields, which are regions in the visual plane to which a ganglion cell is sensitive. Light which is applied to the receptive field of a ganglion cell stimulates a response from the cell. Early studies also identified different ganglion cell types [7], denoted as ON and OFF. ON cells respond when light is switched on in the center of the cell s receptive field, while OFF cells respond when light in the receptive field is switched off. In the experimental work of Arnett [9] and Mastronarde [3], the correlated
5 Ising models for neural activity inferred via Selective Cluster Expansion 5 activity of ganglion cells has been studied via the simultaneous recording of pairs of ganglion cells. Neighboring cells tend to spike in synchrony if they are of the same type and tend to be de-synchronized if they are of different types. Cross-correlation histograms, which describe the correlated firing of pairs of neurons [7], have been particularly useful for identifying circuits of connections between ganglion cells in the retina and for determining the distance dependence of functional connections between ganglions [4, 6, 3]. The second set of recordings is done in vivo on rats. These recordings are obtained by implanting tetrodes or silicon probes which simultaneously record neural activity on several layers of the prefrontal cortex [3]. Probes can be implanted for several weeks in a rat, and allowing for the study of mechanisms of memory formation. Rats are trained in a working memory task and recordings are typically performed before, during, and after the learning of the task [4, 3]. In the work of Peyrache, Battaglia and collaborators, recordings are performed on the medial prefrontal cortex (prelimbic and infralimbic area) with 6 tetrodes, each of which consists of 4 electrodes. The analysis of cross-correlations between cells through Principal Component Analysis has shown that during sleep neural patterns of activity appearing in the previous waking experiences are replayed. This could be a mechanism for consolidating memory formation [3, 3, 33]. In the work of Fujisawa, Buzsáki and collaborators, recordings are performed with silicon probes which record from either the superficial (layers and 3) or deep (layer 5) layers of the medial prefrontal cortex. Cross-correlation analysis of these recordings has identified neurons which differentiate between different trajectories of the rat in a maze, as well as signatures of short term plasticity in the working memory task. In all cases, raw data is obtained in the form of voltage measurements from each electrode in the experimental apparatus. The raw data is then analyzed to determine the number of neurons being recorded and to assign each voltage spike to a particular neuron, in a process known as spike sorting [5, 34, 35]. The end result is a set of spike trains a list of all of the times at which a particular neuron fired for each neuron observed in the experiment... Definition of correlation index Spatial and temporal correlations in the data can be described in terms of a crosscorrelation histogram, which examines the correlations in the firing of pairs of neurons over time. Cross-correlation analysis has been an important analytical tool in the study of neural activity. Let us denote the set of recorded times of each spike event for a neuron labeled by i by {t i,,..., t i,ni }, with N i the total number of times that the neuron spikes during the total recording time T. This data can be extracted from the spike trains depicted in Fig.. One can then determine the cross-correlation histogram of spiking
6 Ising models for neural activity inferred via Selective Cluster Expansion 6 H 3 Da 5-7 H H τ(ms) τ(ms) Fli -6 CB τ(ms) 4 Figure. Examples of cross-correlation histograms between two cells with an inferred positive coupling (see also discussion in Section 6), from recordings of 6 cells in retina in dark (Da 5-7), and 5 cells in retina with a random flickering stimulus (Fl -6); 7 cells in medial prefrontal cortex of a rat (CB 6-83). A characteristic correlation time of approximately ms corresponding to the central peak is visible. delays between each pair of cells i, j, H ij (τ, t) = T N i N j t N i N j a= b= θ τ, t (t i,a, t j,b ) () Here t is the bin width of the histogram, and θ τ, t (t i,a, t j,b ) is an indicator function which is equal to one if τ + t i,a t j,b < t/ and zero otherwise. The cross-correlation histogram can be interprered as the probability that a cell j spikes in the interval τ ± t/, conditioned on the cell i spiking at time zero, and normalized by the probability the cell j spikes in some time window t. The cross-correlation histogram approaches one at very long times τ when the firing of the cell j is independent from the fact that the cell i has fired at time zero.
7 Ising models for neural activity inferred via Selective Cluster Expansion 7 H H Fli 3-8 Da -6 τ(ms) -4-4 τ(ms) H Fli τ(ms) Figure 3. Examples of cross-correlation histograms between two cells with an inferred negative coupling (see also discussion in Section 6), from recordings of 5 cells in retina with a random flickering stimulus, (Fl 3-8) shows no large anticorrelation bump, (Fl -) shows a correlation peak delayed by about 5 ms; 6 cells in retina in dark, (Da -6) displays an anticorrelation bump with the characteristic time scale of ms. The correlation index is a measure of synchrony between two cells, and is defined as the function H ij at its central peak (τ = ), with bin size t CI ij ( t) = H ij (, t) p ij ( t) p i ( t) p j ( t). () C i,j ( t) is the number of spikes emitted by the cells i and j with a delay smaller than t, normalized by the number of times the two cells would spike in the same time window if they were independent. For t small with respect to the typical interval between subsequent spikes of a single cell, there are never two spikes of the same cell in the same time bin and therefore the correlation index is exactly the probability that two cells are active in the same bin p ij ( t) divided by the probability p i ( t) p j ( t) they would be active in the same time window assuming no correlation between them. Some cross-correlation histograms for the data sets we consider are shown in Figs. 4. The time bin t is a parameter in the correlation index analysis which defines
8 Ising models for neural activity inferred via Selective Cluster Expansion 8 H τ(ms) CB 6-83 H τ(ms) Da - H τ(ms) CB Figure 4. Examples of cross-correlation histograms between two cells at long time scales, from recordings of 6 cells in retina in dark, (Da -) corresponds to a positive coupling and no long time effects; 7 cells in medial prefrontal cortex of a rat, (CB 6-83) corresponds to a positive coupling and no long time effects, (CB 35-78) displays nonstationary effects because cell 78 is active only in the first half of the recording. the relevant time scale for correlations. A characteristic time scale of some tens of milliseconds is shown in Fig. [36]. However, in retinal recordings long time patterns appear especially in the presence of stimulus, and in vivo cortical recordings show long time correlations which can be due, for example, to different firing rhythms or trajectory-dependent activation of neurons (see Fig. 4) ), which are observed in the cross-correlations obtained from cortical recordings by Fujisawa and collaborators. Despite its usefulness as an analytical tool, cross-correlation analysis is unable to account for network effects in a controlled way. That is, simply by treating each pair independently it is not possible to determine whether the correlated activity of a single pair of neurons is due to a direct interaction between them, or whether it results from indirect interactions mediated by other neurons in the network. In the following section
9 Ising models for neural activity inferred via Selective Cluster Expansion 9 we introduce an artificial Ising network, inferred using the spiking probabilities p i ( t) and pairwise correlations p ij ( t) on a fixed time scale t, which goes in this direction by reproducing neural activity observed in the data beyond that which was used to infer the model. This approach was introduced to analyze retinal data in [5, 6]. In Section 7 we clarify the relationship between the couplings J ij inferred by the Ising approach and the two-cell approximation J () ij derived from cross-correlation analysis. The Ising model we apply in the following aims to go beyond cross-correlation analysis in the sense it that disentangles direct couplings from correlations, correcting for network effects..3. Ising model encoding of the activity Neural activity represented in the form of spike trains can also be encoded in a binary form. We first divide the total time interval of a recording of the activity into small time bins of size t. The activity is then represented by set of binary variables s τ i, hereafter called spins, where τ =,..., B labels the time bin and i =,..., N is a label which refers to a particular neuron. In the data we consider N spans a few decades (3 to ). If in bin τ neuron i has spiked at least once then we set s τ i =, otherwise s τ i =. We write the probability of an N-spin configuration P N [s] in the data is the fraction of bins carrying that configuration s = {s, s,..., s N }, and define the empirical Hamiltonian H N [s] as minus the logarithm of P N. By definition the Gibbs measure corresponding to this Hamiltonian reproduces the data exactly. As the spins are binary variables H N can be expanded in full generality as N H N [s] = J (k) i,i,...,i p s i s i... s ip, (3) i <i <...<i p k= where J () is simply a constant ensuring the normalization of P N. Notice that, given an experimental multielectrode recording, P N and H N depend on the binning interval t. Knowledge of the N coefficients J in (3) is equivalent to specifying the probability of each one of the spin configurations. The model defined by (3) provides a complete, but flawed, representation of the experimental data. First, it gives a poor compression of the data, requiring a number of variables which grows exponentially with the system size to exhaustively encode the probability of every configuration of the system. As a predictive model of the future behavior of the experimental system, it will also likely perform poorly compared to other reasonable models due to the overfitting of the data. It is tempting then to look for simpler approximate expressions of H N that retain most of the statistical structure of the spin configurations. A sensible approximation to the true interaction model should at least reproduce the values of the empirical oneand two-point correlation functions, p i = B s τ i, p ij = B s τ i s τ j. (4) B B τ= τ=
10 Ising models for neural activity inferred via Selective Cluster Expansion These constraints can be satisfied by the Ising model defined by the Hamiltonian N H [s] = h i s i J ij s i s j. (5) i= i<j The corresponding probability of a configuration in the Ising model is given by the standard equilibrium Gibbs measure, P [s] = e H [s] Z[{h i }, {J ij }], (6) where the partition function Z[{h i }, {J ij }] = s e H [s] ensures that s P [s] =. The local fields {h i } and pairwise couplings {J ij } are the natural parameters for encoding the one- and two-point correlations because they are the conjugate thermodynamical variables to these correlations. Indeed the N (N + )/ coupled equations (4) can be solved by finding the couplings and fields which minimize the cross entropy between the inferred Ising model and the data S [{h i }, {J ij }] = log Z[{h i }, {J ij }] i h i p i i<j J ij p ij. (7) This can be verified by computing the derivatives of S [{h i }, {J i,j }] with respect to the couplings and fields, S [{h i }, {J ij }] = s i e H[s] p i, (8) h i Z s S [{h i }, {J ij }] = s i s j e H[s] p ij. (9) J ij Z s At the minimum of (7) the derivatives must vanish, implying that the conditions (4) are satisfied. Moreover the minimal cross entropy S [{p i }, {p ij }] = min {hi },{J ij } S [{h i }, {J i,j }] is the Legendre transform of the free energy F = log Z[{h i }, {J ij }] of the Ising model. It can be shown (see []) that the Ising model (6) is the probabilistic model with the maximum entropy S[P ] = P [s] ln P [s] () s which satisfies the constraints that the average value of the one- and two-point correlations coincide with the observed data (4) and that the probability distribution P [s] is normalized such that all probabilities sum to one..4. Previous results of the inverse Ising approach It was put forward in [5, 37] that the Ising model defined by (6) not only reproduces the one- and two-point firing correlations extracted from the data, it also captures other features of the data such as higher order correlations and multi-neuron firing frequencies. Thus the energy H provides a good approximation to the whole energy H N ; interactions involving three or more spins in (3) are not quantitatively important, and it suffices to keep in the expansion local fields and pair interactions only. Further simplification,
11 Ising models for neural activity inferred via Selective Cluster Expansion however, is not generally possible. Hamiltonians H with no multi-spin interactions, corresponding to setting all J ij = in (5) with the fields h i chosen to reproduce the average neural activity, provide a very poor approximation to (3). Such independent neuron approximations fail to reproduce, for example, the probability that k cells spike together in the same bin for k =,..., N. The claim in [5, 37] is based on the following points: Information-theoretical arguments: call S the entropy of the independent-neuron model H, S the entropy of the Ising model (5), and S N the entropy of the experimental distribution P N. Then the reduction in entropy of spin configurations coming from pair correlations, I = S S, explains much of the reduction in entropy coming from correlations at all orders (or multi-information), I N = S S N : I /I N 9% [5]. Hence higher order correlations do not contribute much to the multi-information, implying that most of the correlation observed in the data is explained simply through pairwise interactions. The multi-neuron firing frequencies, i.e. the probability of k neurons spiking together in the same bin, predicted by the Ising model with pairwise interactions are much closer to the experimental values than those of its independent-cell counterpart. Higher order correlations (three spin and higher correlations) predicted by the Ising model are in good agreement with the experimental data.. Statistical effects of finite sampling Finite sampling of the experimental systems we consider here introduces fluctuations which complicate the inverse problem. In this Section we discuss the effects of finite sampling noise on the empirical one- and two-point correlations, consequences for the inference procedure, and statistical errors on the inferred couplings and fields... Statistical error on empirical correlations The inverse Ising approach requires measurements of the probability that each cell spikes in a time bin p i ( t) and that each pair of cells spike in the same time bin p ij ( t). These probabilities can be obtained from the spike train data by counting the number of time windows in which a cell, or two cells for the pair correlations, are active divided by the total number of time windows B. It is important to notice that the number of sampled configurations, while large (typically the total recording time is of the order of one hour, which with a time bin of ms corresponds to roughly 6 configurations), is limited. Therefore the empirical correlations {p i }, {p ij } we obtain will differ from the ones which would be obtained with an infinite sampling {p true i }, {p true ij the order of {δp i }, {δp ij } which we estimate in the following. In our model, the spins are stochastic variables with Gibbs average s i = p true i } by typical fluctuations of ()
12 Ising models for neural activity inferred via Selective Cluster Expansion and with variance σ i = s i s i = p true i ( p true i ). () Their average p i over B independent samples is also a stochastic variable with the same average as above, but with the standard deviation p true i ( p true i ) pi ( p i ) δp i =, (3) B B where in the last expression of (3) we have replaced the Gibbs average p true i with the empirical average, neglecting terms of the order /B. Similarly the pair correlations are have the average value and variance s i s j = p true ij, (4) σ ij = p true ij ( p true ij ), (5) therefore their sampled average over B samples p ij has a standard deviation pij ( p ij ) δp ij. (6) B.. The Hessian of the cross entropy: The importance of regularization terms Because of finite sampling problems the fields and couplings obtained as minimizers of (7) are not necessarily well defined. As a simple example consider a single neuron (N = ) with an average spiking rate r. The probability that the corresponding spin in the binary representation is zero is exp( r t) r t, which is close to one for small bin widths t. If the number B of bins is much smaller than /(r t) the spin is likely to be equal to zero in all configurations. Hence p = and the field solution of the inverse problem is h =. In practice the number B of available data is large enough to avoid this difficulty as far as single site probabilities are concerned. However, when two or more spins are considered and higher-order correlations are taken into account incomplete sampling cannot be avoided; some groups of cells are never active simultaneously, which lead to infinite couplings. This problem is easily cured with a Bayesian approach. We now start with a set of data consisting of B configurations of spins {s τ }, τ =,..., B, and assume that the data come from the Ising model with Hamiltonian (5). The probability or likelihood of a spin configuration is given by (6). The likelihood of the whole set of data is therefore B P like [{s τ } {h i, J kl }] = P [s τ {h i, J kl }] = exp ( B S [{h i }, {J ij }]), (7) τ= where the cross entropy S is defined in (7). In order to calculate the a posteriori probability of the couplings and fields we need to introduce a prior probability over those parameters. In the absence of any information
13 Ising models for neural activity inferred via Selective Cluster Expansion 3 (data) let us assume that the couplings have a Gaussian distribution, with zero mean and variance σ, [ ( )] P prior [{h i }, {J ij }] = (πσ ) N(N+)/ exp J σ ij. (8) So far the variance is an unknown parameter. We expect it to be of the order of unity, that is, much smaller than the number of configurations B in the data set. Multiplying (7) and (8) leads to the Bayesian a posteriori probability for the fields and couplings given the data, P post [{h i }, {J ij } {s τ }] = Pprior [{h i }, {J ij }] P like [{s τ } {h i }, {J kl }] P[{s τ }] i<j, (9) where P[{s τ }] is a normalization constant which is independent of the couplings and fields. The most likely values for the fields and couplings are obtained by maximizing (9), that is, by minimizing the modified cross entropy, ( ) S [{h i }, {J ij }, γ] = S [{h i }, {J ij }] + γ Jij, () where γ = B σ. () The value of the variance σ can be determined, once again, using a Bayesian criterion. P[{s τ }] in (9) is the a priori probability of the data set over all possible Ising models. The optimal variance is the one maximizing this quantity. The approach is illustrated in detail in the simplest case of a single neuron, then extended to the case of interacting neurons in Appendix A. The precise form of the regularization term is somewhat arbitrary. For convenience in the calculations we use the regularization γ i<j p i ( p i )p j ( p j ) J ij. () This choice corresponds to a Gaussian prior for the interactions in a Bayesian framework. Another regularization term we use is based on L norm rather than L, and favors sparse coupling networks, i.e. with many couplings equal to zero, γ p i ( p i )p j ( p j ) J ij, (3) i<j which corresponds to a Laplacian prior distribution. In Section 7 we describe the properties of the L and L norm penalties in more detail, and in Section 4. we motivate the p i, p i j dependence of the terms in the regularization. When the number of samples B in the data set becomes large, () shows that the regularization strength γ ; the couplings and fields determined by minimizing (7) always coincide with Bayes predictions in the perfect sampling limit. For finite B the i<j
14 Ising models for neural activity inferred via Selective Cluster Expansion 4 presence of a quadratic contribution in () ensures that S grows rapidly far from the origin along any direction in the N(N+) -dimensional space of fields and couplings. Thanks to the regularization there exists a unique and finite minimizer of the cross entropy. The Hessian matrix χ of the cross entropy S, also called the Fisher information matrix, is given by ) χ = = ( S ( h i h i S h i J kl S h i J k l S J kl J k l s i s i s i s i s i s k s l s i s k s l s i s k s l s i s k s l s k s l s k s l s k s l s k s l ), (4) where denotes the Gibbs average with measure (6), and i =,..., N, k < l N. The addition of a regularization term with a quadratic penalty on the couplings and fields adds γ times the identity matrix to the Hessian, and as the unregularized χ is a covariance matrix and hence nonnegative, this assures that the susceptibility of the regularized model is positive definite. Thus S [{h i }, {J ij }, γ] is strictly convex. This proves the existence and uniqueness of the solution to the inverse problem..3. Statistical error on couplings and fields In this Section we compute the statistical fluctuations of the inferred couplings and fields due to finite sampling of the experimental system. As in Section., let us assume that data are not extracted from experiments but rather generated from the Ising model (5) with the fields {h i } and couplings {Jij} which minimize the cross entropy (7). From Section. the probability of infering a set of fields {h i } and couplings {J ij } is proportional to exp( B S [{h i }, {J ij }]). When B is very large this probability is tightly concentrated around the minimum of S, that is, {h i }, {Jij}. The difference between the inferred and the true fields and couplings is encoded in the N(N+) -dimensional vector h,j of components {h i h i }, {J ij Jij}. The distribution of this vector is asymptotically Gaussian, P[ det χ h,j ] exp ( B ) (πb) h,j χ N(N+)/4 h,j. (5) Statistical fluctuations of the couplings and fields are therefore characterized by the standard deviations δh i = B (χ ) i,i, δj ij = B (χ ) ij,ij. (6) Let v = ( x, y) where x = (x, x,..., x N ) and y = (y, y 3,..., y N,N ) are, respectively N- and N(N ) -dimensional vectors. Then for any v. [ v ( H v = x i si s i ) + ( y kl sk s l s k s l )] i k<l
15 Ising models for neural activity inferred via Selective Cluster Expansion 5 Note that (3) and (6) can be also directly obtained from the covariance matrix χ. Indeed linear response theory tells us that p,c = χ h,j. (7) We deduce from (5) that p,c obeys a Gaussian distribution as h,j, with the Hessian matrix χ replaced with its inverse matrix in (5). The typical uncertainties of the oneand two-point correlations are given by δp i = (χ) B i,i and δp ij = (χ) B ij,ij, which correspond exactly with (3) and (6). 3. The difficulty of the inverse Ising problem The inverse problem can be subdivided in three different problems which are of increasing difficulty []. Reconstruction of the interaction graph It is possible to reproduce qualitative features of the underlying interaction graph (i.e. the set of interactions which would be inferred if the inverse problem was solved exactly), such as the number of connections to a given cell or the range of interactions, without determining the values of the couplings precisely. Knowing the rank of the couplings in absolute value, for instance, gives an idea of the structure of the interaction graph. We will see in Section 7 that mean field approximations are only useful for finding precisely the network of interactions in the case of weak interactions and not, as in the majority of the analyzed neural data, for a dilute network with large couplings. These approximations generally give, then, wrong values for the couplings but respect the ranking of large couplings by magnitude therefore allowing for the accurate recovery of the structure of the interaction graph of large couplings. Reconstruction of the interaction graph and values of the couplings It can be of interest to infer the values of the couplings more precisely, and to identify reliable couplings, which are different from zero even when taking into account statistical fluctuations due to finite sampling (6), and unreliable couplings which are compatible with zero. A precise inference of the value of the couplings compared with the error bar will be helpful to characterize changes in the values of couplings, which can occur for example in a memory task [4]. Also it would be of interest to characterize reliable negative couplings with the aim of identifying inhibitory connections, which are particularly susceptible to sampling noise and thus difficult to assign reliably. It has been pointed out in [] that the difficulty of the inference problem depends on the inverse of the Fisher information matrix χ (4). Whereas the susceptibility χ characterizes the response of the Ising model to a small change in the couplings or fields, the inverse susceptibility χ gives the change in the inferred couplings and fields due to a perturbation of the observed frequencies or correlations. If data are generated by sparse networks and if the sampling is good, then χ is localized, i.e. it decays fast with the length of the interaction path between spins in the interaction network. This
16 Ising models for neural activity inferred via Selective Cluster Expansion 6 property holds even in the presence of long-range correlations and makes the inverse problem not intrinsically hard. It is important to notice that the sampling fluctuations in frequencies (3) and correlations (6) do not come from a sparse network structure and thus χ can lose its localized properties because of sampling noise. In other words, overfitting can generate very complicated and densely connected networks which will have a large couplingssusceptibility. A way to filter the data must then be found to efficiently solve the inverse problem in presence of sampling noise. The selective cluster expansion, which we introduce in Section 5, approaches this problem by including methods of regularization and by building up a solution to the inference problem in a progressive way, including only terms which contribute significantly to the inference so as to avoid the overfitting of noise. Reconstruction of the empirical correlations The most difficult inference task is to find the local fields and the network of couplings which truly satisfy the minimum of (7), and which therefore reproduce the frequencies and correlations obtained from data. One can use such an inferred Ising model to then predict, for example, the observed higher moments of the distributions (three body or higher order correlations) or multi-neuron firing frequencies, as discussed in Section.4. The Ising model could also be used to predict the response to a local perturbation, which could be due to a stimulation, or the ablation of a connection. The reconstruction problem is difficult because a small change in the inferred parameters can result in large changes in the correlations and frequencies given by the inferred model. In other words, the susceptibility matrix may be dense, rather than sparse, with many large entries. Let again p,c denote the N(N+) -dimensional vector whose components are the differences between the predicted and true values of the frequencies and correlations. Typically inference errors come from the approximations done in the resolution of the inverse problem. Using the linear response theory of (7) we can estimate the order of magnitude of the reconstruction error due to an inference error: p,c χ h,j, (8) where χ is the norm of the matrix defined as the sum over the rows of the maximum element over the columns, χ = i max χ i,j. (9) j Let us imagine that a parameter h i has been inferred with an inference error of the order h., which is a typical order of magnitude of the expected statistical fluctuations in the inferred parameters. This inference error can drastically change the reconstructed p i. For example for the recording of N = 4 neurons in the retina subject to a natural movie stimulus [5] χ =.6, and the consequent δ p.6 is of the same order of magnitude as the p i, which is much larger than the statistical errors we expect
17 Ising models for neural activity inferred via Selective Cluster Expansion 7 on these variables. We discuss in Section 6 how the reconstruction behaves as a function of the inference precision in the cluster expansion. This example explains why even if the network is reconstructed at the precision corresponding to the statistical fluctuations (6) we expect from the finite sampling, and therefore the inverse problem is quite accurately solved, the statistical properties of the data on the inferred model can be much harder to reproduce and may require sophisticated expansion and fine tuning of some parameters. Note that equation (8) gives only an order of magnitude, the more precise matrix dependence of the response in (7) tells us that the modes which correspond to large eigenvalues of the Hessian matrix χ have to be more precisely determined to solve the reconstruction problem well, while the others, which correspond to small eigenvalues of the Hessian matrix, have less impact on the reconstruction problem. 4. High temperature expansions and the mean field entropy S MF The main problem of the minimization of the cross entropy S (7) is the calculation of the partition function Z which, if done exactly, requires the sum over all N possible configurations of the system of N spins. Because this sum becomes prohibitive for systems with more than spins some approximate solution of the inverse problem must be found. High temperature expansions [6, 38 4] of the Legendre transform of the free energy are useful when the pairs of variables interact through weak couplings. The Ising model with weak (of the order of N / ) interactions is the so-called Sherrington- Kirkpatrick model. In this case the entropy S[{p i }, {p ij }] coincides asymptotically for large N with where S MF [{p i }, {p ij }] = S ind [{p i }] + log det M[{p i}, {p ij }], (3) S ind [{p i }] = i [ p i log p i ( p i ) log( p i )] (3) is the entropy of independent spin variables with averages {p i }, and p ij p i p j M ij [{p i }, {p ij }] = pi ( p i )p j ( p j ), (3) which can be calculated in O(N 3 ) time [6, 4], and is consistent with the so-called TAP equations [4]. It is important to underline that what we call the mean field entropy S MF corresponds to the entropy of a Gaussian model of continous spin variables with averages p i and variances p i ( p i ). The derivatives of S MF with respect to the {p ij } and {p i } give the value of the couplings and fields, (J MF ) ij = S MF p ij (M ) ij = pi ( p i )p j ( p j ),
18 Ising models for neural activity inferred via Selective Cluster Expansion 8 (h MF ) i = S MF = p i (J MF ) ij (c ) ij p i p i ( p i ) p j, (33) j( i) where c ij = p ij p i p j is the connected correlation. It is possible to get an idea of the goodness of the high temperature expansion from the size of the correlation indices CI ij = p ij /(p i p j ), which are the expansion parameters of the high temperature series [6]. We note that in neural data, even if the connected correlations are small, the correlation indices CI ij can be large because the probabilities p i, p j that individual cells are active in a time bin are also generally small, and CI ij is of order one. Histograms of the correlation indices for the different data sets we analyze are shown in Figs. 3 and 4, and show that the correlation indices are small in absolute value ( CI ij.) only in the data sets of in vivo recordings of the medial prefrontal cortex of rats with tetrodes [3, 3]. We therefore expect the high temperature expansion and in particular S MF to be a good approximation to the inverse problem only for this data set. 4.. L -Regularized mean field entropy A regularized version of the mean field entropy can be computed analytically []. The derivation is as follows. First we use the mean field expression for the cross-entropy at fixed couplings J ij and frequencies p i, see [39], to rewrite S Ising [{p i }, {J ij }] = S ind [{p i }] log det (Id J ) i<j J ij (p ij p i p j ),(34) where J ij = J ij pi ( p i )p j ( p j ) and Id denotes the N-dimensional identity matrix. We consider the L -norm regularization (). The entropy at fixed data {p i }, {p ij } is [ S L MF [{p i}, {p ij }] = S ind [{p i }] + min S Ising [{p i }, {J ij }] {J ij } + γ ] Jij p i ( p i )p j ( p j ) i<j [ = S ind [{p i }] + min {J ij } log det (Id J ) tr (J M[{p i }, {p ij }]) + γ tr (J ) ], (35) where M[{p i }, {p ij }] is defined in (3). The optimal interaction matrix J is the root of the equation (Id J ) M[{p i }, {p ij }] + γ J =. (36) Hence, J has the same eigenvectors as M[{p i }, {p ij }], a consequence of the dependence on p i we have chosen for the quadratic regularization term in (). Let j q denote the q th eigenvalue of J. Then S L MF [{p i}, {p ij }, γ] = S ind [{p i }] + N (log ˆm q + ˆm q ), (37) q=
19 Ising models for neural activity inferred via Selective Cluster Expansion 9 where ˆm q is the largest root of ˆm q ˆm q (m q γ) = γ, and m q is the q th eigenvalue of M[{p i }, {p ij }]. Note that ˆm q = m q when γ =, as expected. 4.. L -Regularized mean field entropy With the choice of an L -norm regularization, the entropy at fixed data p becomes [ S L MF [{p i}, {p ij }, γ] = S ind [{p i }] + min {J ij } log det (Id J ) tr (J M[{p i }, {p ij }]) + γ i<j ] J ij. (38) No analytical expression exists for the optimal J. However, it can be found in a polynomial time using convex optimization techniques. The minimization of (38) is known in the statistics literature as the estimate of the precision matrix with a constraint of sparsity. Several numerical procedures to compute J efficiently are available [43]. 5. Selective Cluster Expansion When an exact calculation of the partition function is out of reach, and the amplitudes of the correlation indices are large, an accurate estimate of the interactions which solve the inverse Ising problem can be obtained through cluster expansions. Cluster expansions have a rich history in statistical mechanics, e.g. the virial expansion in the theory of liquids or cluster variational methods [44]. The selective cluster expansion algorithm (SCE) which we have recently proposed [, ] makes use of an assumption about the properties of the inverse susceptibility χ to efficiently generate an approximate solution to the inverse Ising inference problem. χ is typically much sparser and shorter range than χ, implying that most interactions inferred from a given set of data depend strongly on only a small set of correlations. Thus an estimate of the interactions for the entire system can be constructed by solving the inference problem on small subsets (clusters) of spins and combining the results. The SCE gives such an estimate by recursively solving the inverse Ising problem on small clusters of spins, selecting the clusters which give significant information about the underlying interaction graph according to their contribution to the entropy S of the inferred Ising model, and building from these a new set of clusters to analyze. At the end of this cluster expansion procedure, an estimate of the entropy and of the interactions for the full system is produced based on the values inferred on each cluster. Let S Γ denote the entropy of the Ising model defined just on a subset Γ = {i, i,...} of the full set of spins, which reproduces the one- and two-point correlations obtained from data for this subset. The entropy S of the inferred Ising model on the full system of N spins can then be expanded formally as a sum of individual contributions from each of the N nonempty subsets, S = Γ S Γ, S Γ = S Γ Γ Γ S Γ. (39)
20 Ising models for neural activity inferred via Selective Cluster Expansion The cluster entropy S Γ, defined recursively in (39), measures the contribution of the cluster Γ to the total entropy. It is calculated by substracting the cluster entropies of all possible smaller subsets of Γ from the subset entropy S Γ. Each cluster entropy depends only upon the correlations between the spins in that cluster, and for small clusters it is easy to compute numerically. The contribution of each cluster to the interactions, which we denote J Γ, is defined analogously and computed at the same time as the cluster entropy. The elements of J Γ are the couplings and fields which minimize the cross entropy S restricted to the cluster Γ. Note that in applications to real data, we employ some form of regularization (), (3), to ensure that the Hessian χ is positive definite and to reduce the effects of sampling noise. As an example, the entropy of a single-spin cluster is, using (39) with N =, S (i) [p i ] S ind [p i ] = p i log p i ( p i ) log( p i ). (4) The contribution to the field of a single-spin cluster is h () i h () i S (i) = log (p i /( p i )). (4) p i The entropy of a two spin subsystem {i, j} is S () [p i, p j, p ij ] = p ij log p ij (p i p ij ) log (p i p ij ) (p j p ij ) log (p j p ij ) ( p i p j + p ij ) log ( p i p j + p ij ) Using again (39) with N =, we obtain S (i,j) [p i, p j, p ij ] = S (i,j) [p i, p j, p ij ] S (i) [p i ] S (j) [p j ], which measures the loss in entropy when imposing the constraint s i s j = p ij to a system of two spins with fixed magnetizations, s i = p i, s j = p j. The contribution to the field of the two spin cluster is h () i = log ((p i p ij )/( p i p j + p ij )) h () i, (4) and the contribution to the coupling is J () ij J () ij = log p ij log (p i p ij ) log (p j p ij ) + log ( p i p j + p ij )(43). Note that the two-cell couplings, which do not take into account network effects, reduce to the logarithm of the correlation index for p ij p i, which is typically the case when the time bin is much smaller than the typical interval between spikes. In this case p i t and p ij t, thus J () ij log p ij p i p j. (44) It is difficult to write the entropy analytically for clusters of more than two spins, but S Γ can be computed numerically as the minimum of the cross entropy (7). This involves calculating Z, a sum over an exponential number of spin configurations. In practice this limits the size of the clusters which we can consider to those with spins. A recursive use of (39), or the Möbius inversion formula, allows us to obtain S Γ for larger and larger clusters Γ (see also the pseudocode of Algorithm ). It is important
+ + ( + ) = Linear recurrent networks. Simpler, much more amenable to analytic treatment E.g. by choosing
 Linear recurrent networks Simpler, much more amenable to analytic treatment E.g. by choosing + ( + ) = Firing rates can be negative Approximates dynamics around fixed point Approximation often reasonable
Linear recurrent networks Simpler, much more amenable to analytic treatment E.g. by choosing + ( + ) = Firing rates can be negative Approximates dynamics around fixed point Approximation often reasonable
THE retina in general consists of three layers: photoreceptors
 CS229 MACHINE LEARNING, STANFORD UNIVERSITY, DECEMBER 2016 1 Models of Neuron Coding in Retinal Ganglion Cells and Clustering by Receptive Field Kevin Fegelis, SUID: 005996192, Claire Hebert, SUID: 006122438,
CS229 MACHINE LEARNING, STANFORD UNIVERSITY, DECEMBER 2016 1 Models of Neuron Coding in Retinal Ganglion Cells and Clustering by Receptive Field Kevin Fegelis, SUID: 005996192, Claire Hebert, SUID: 006122438,
Exercises. Chapter 1. of τ approx that produces the most accurate estimate for this firing pattern.
 1 Exercises Chapter 1 1. Generate spike sequences with a constant firing rate r 0 using a Poisson spike generator. Then, add a refractory period to the model by allowing the firing rate r(t) to depend
1 Exercises Chapter 1 1. Generate spike sequences with a constant firing rate r 0 using a Poisson spike generator. Then, add a refractory period to the model by allowing the firing rate r(t) to depend
encoding and estimation bottleneck and limits to visual fidelity
 Retina Light Optic Nerve photoreceptors encoding and estimation bottleneck and limits to visual fidelity interneurons ganglion cells light The Neural Coding Problem s(t) {t i } Central goals for today:
Retina Light Optic Nerve photoreceptors encoding and estimation bottleneck and limits to visual fidelity interneurons ganglion cells light The Neural Coding Problem s(t) {t i } Central goals for today:
Analyzing large-scale spike trains data with spatio-temporal constraints
 Author manuscript, published in "NeuroComp/KEOpS'12 workshop beyond the retina: from computational models to outcomes in bioengineering. Focus on architecture and dynamics sustaining information flows
Author manuscript, published in "NeuroComp/KEOpS'12 workshop beyond the retina: from computational models to outcomes in bioengineering. Focus on architecture and dynamics sustaining information flows
Mid Year Project Report: Statistical models of visual neurons
 Mid Year Project Report: Statistical models of visual neurons Anna Sotnikova asotniko@math.umd.edu Project Advisor: Prof. Daniel A. Butts dab@umd.edu Department of Biology Abstract Studying visual neurons
Mid Year Project Report: Statistical models of visual neurons Anna Sotnikova asotniko@math.umd.edu Project Advisor: Prof. Daniel A. Butts dab@umd.edu Department of Biology Abstract Studying visual neurons
ECE 521. Lecture 11 (not on midterm material) 13 February K-means clustering, Dimensionality reduction
 ECE 521 Lecture 11 (not on midterm material) 13 February 2017 K-means clustering, Dimensionality reduction With thanks to Ruslan Salakhutdinov for an earlier version of the slides Overview K-means clustering
ECE 521 Lecture 11 (not on midterm material) 13 February 2017 K-means clustering, Dimensionality reduction With thanks to Ruslan Salakhutdinov for an earlier version of the slides Overview K-means clustering
Neural Coding: Integrate-and-Fire Models of Single and Multi-Neuron Responses
 Neural Coding: Integrate-and-Fire Models of Single and Multi-Neuron Responses Jonathan Pillow HHMI and NYU http://www.cns.nyu.edu/~pillow Oct 5, Course lecture: Computational Modeling of Neuronal Systems
Neural Coding: Integrate-and-Fire Models of Single and Multi-Neuron Responses Jonathan Pillow HHMI and NYU http://www.cns.nyu.edu/~pillow Oct 5, Course lecture: Computational Modeling of Neuronal Systems
Deep learning / Ian Goodfellow, Yoshua Bengio and Aaron Courville. - Cambridge, MA ; London, Spis treści
 Deep learning / Ian Goodfellow, Yoshua Bengio and Aaron Courville. - Cambridge, MA ; London, 2017 Spis treści Website Acknowledgments Notation xiii xv xix 1 Introduction 1 1.1 Who Should Read This Book?
Deep learning / Ian Goodfellow, Yoshua Bengio and Aaron Courville. - Cambridge, MA ; London, 2017 Spis treści Website Acknowledgments Notation xiii xv xix 1 Introduction 1 1.1 Who Should Read This Book?
Gaussian processes. Chuong B. Do (updated by Honglak Lee) November 22, 2008
 Gaussian processes Chuong B Do (updated by Honglak Lee) November 22, 2008 Many of the classical machine learning algorithms that we talked about during the first half of this course fit the following pattern:
Gaussian processes Chuong B Do (updated by Honglak Lee) November 22, 2008 Many of the classical machine learning algorithms that we talked about during the first half of this course fit the following pattern:
Parameter Estimation for Spatio-Temporal Maximum Entropy Distributions: Application to Neural Spike Trains
 Entropy 214, 16, 2244-2277; doi:1.339/e1642244 OPEN ACCESS entropy ISSN 199-43 www.mdpi.com/journal/entropy Article Parameter Estimation for Spatio-Temporal Maximum Entropy Distributions: Application to
Entropy 214, 16, 2244-2277; doi:1.339/e1642244 OPEN ACCESS entropy ISSN 199-43 www.mdpi.com/journal/entropy Article Parameter Estimation for Spatio-Temporal Maximum Entropy Distributions: Application to
Adaptation in the Neural Code of the Retina
 Adaptation in the Neural Code of the Retina Lens Retina Fovea Optic Nerve Optic Nerve Bottleneck Neurons Information Receptors: 108 95% Optic Nerve 106 5% After Polyak 1941 Visual Cortex ~1010 Mean Intensity
Adaptation in the Neural Code of the Retina Lens Retina Fovea Optic Nerve Optic Nerve Bottleneck Neurons Information Receptors: 108 95% Optic Nerve 106 5% After Polyak 1941 Visual Cortex ~1010 Mean Intensity
Characterization of Nonlinear Neuron Responses
 Characterization of Nonlinear Neuron Responses Final Report Matt Whiteway Department of Applied Mathematics and Scientific Computing whit822@umd.edu Advisor Dr. Daniel A. Butts Neuroscience and Cognitive
Characterization of Nonlinear Neuron Responses Final Report Matt Whiteway Department of Applied Mathematics and Scientific Computing whit822@umd.edu Advisor Dr. Daniel A. Butts Neuroscience and Cognitive
Variational Methods in Bayesian Deconvolution
 PHYSTAT, SLAC, Stanford, California, September 8-, Variational Methods in Bayesian Deconvolution K. Zarb Adami Cavendish Laboratory, University of Cambridge, UK This paper gives an introduction to the
PHYSTAT, SLAC, Stanford, California, September 8-, Variational Methods in Bayesian Deconvolution K. Zarb Adami Cavendish Laboratory, University of Cambridge, UK This paper gives an introduction to the
11. Learning graphical models
 Learning graphical models 11-1 11. Learning graphical models Maximum likelihood Parameter learning Structural learning Learning partially observed graphical models Learning graphical models 11-2 statistical
Learning graphical models 11-1 11. Learning graphical models Maximum likelihood Parameter learning Structural learning Learning partially observed graphical models Learning graphical models 11-2 statistical
ECE521 week 3: 23/26 January 2017
 ECE521 week 3: 23/26 January 2017 Outline Probabilistic interpretation of linear regression - Maximum likelihood estimation (MLE) - Maximum a posteriori (MAP) estimation Bias-variance trade-off Linear
ECE521 week 3: 23/26 January 2017 Outline Probabilistic interpretation of linear regression - Maximum likelihood estimation (MLE) - Maximum a posteriori (MAP) estimation Bias-variance trade-off Linear
Variational Principal Components
 Variational Principal Components Christopher M. Bishop Microsoft Research 7 J. J. Thomson Avenue, Cambridge, CB3 0FB, U.K. cmbishop@microsoft.com http://research.microsoft.com/ cmbishop In Proceedings
Variational Principal Components Christopher M. Bishop Microsoft Research 7 J. J. Thomson Avenue, Cambridge, CB3 0FB, U.K. cmbishop@microsoft.com http://research.microsoft.com/ cmbishop In Proceedings
The Bayesian Brain. Robert Jacobs Department of Brain & Cognitive Sciences University of Rochester. May 11, 2017
 The Bayesian Brain Robert Jacobs Department of Brain & Cognitive Sciences University of Rochester May 11, 2017 Bayesian Brain How do neurons represent the states of the world? How do neurons represent
The Bayesian Brain Robert Jacobs Department of Brain & Cognitive Sciences University of Rochester May 11, 2017 Bayesian Brain How do neurons represent the states of the world? How do neurons represent
Linear & nonlinear classifiers
 Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1396 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1396 1 / 44 Table
Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1396 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1396 1 / 44 Table
Analyzing large-scale spike trains data with spatio-temporal constraints
 Analyzing large-scale spike trains data with spatio-temporal constraints Hassan Nasser, Olivier Marre, Selim Kraria, Thierry Viéville, Bruno Cessac To cite this version: Hassan Nasser, Olivier Marre, Selim
Analyzing large-scale spike trains data with spatio-temporal constraints Hassan Nasser, Olivier Marre, Selim Kraria, Thierry Viéville, Bruno Cessac To cite this version: Hassan Nasser, Olivier Marre, Selim
Neural coding Ecological approach to sensory coding: efficient adaptation to the natural environment
 Neural coding Ecological approach to sensory coding: efficient adaptation to the natural environment Jean-Pierre Nadal CNRS & EHESS Laboratoire de Physique Statistique (LPS, UMR 8550 CNRS - ENS UPMC Univ.
Neural coding Ecological approach to sensory coding: efficient adaptation to the natural environment Jean-Pierre Nadal CNRS & EHESS Laboratoire de Physique Statistique (LPS, UMR 8550 CNRS - ENS UPMC Univ.
Limulus. The Neural Code. Response of Visual Neurons 9/21/2011
 Crab cam (Barlow et al., 2001) self inhibition recurrent inhibition lateral inhibition - L16. Neural processing in Linear Systems: Temporal and Spatial Filtering C. D. Hopkins Sept. 21, 2011 The Neural
Crab cam (Barlow et al., 2001) self inhibition recurrent inhibition lateral inhibition - L16. Neural processing in Linear Systems: Temporal and Spatial Filtering C. D. Hopkins Sept. 21, 2011 The Neural
Learning the collective dynamics of complex biological systems. from neurons to animal groups. Thierry Mora
 Learning the collective dynamics of complex biological systems from neurons to animal groups Thierry Mora Università Sapienza Rome A. Cavagna I. Giardina O. Pohl E. Silvestri M. Viale Aberdeen University
Learning the collective dynamics of complex biological systems from neurons to animal groups Thierry Mora Università Sapienza Rome A. Cavagna I. Giardina O. Pohl E. Silvestri M. Viale Aberdeen University
1/12/2017. Computational neuroscience. Neurotechnology.
 Computational neuroscience Neurotechnology https://devblogs.nvidia.com/parallelforall/deep-learning-nutshell-core-concepts/ 1 Neurotechnology http://www.lce.hut.fi/research/cogntech/neurophysiology Recording
Computational neuroscience Neurotechnology https://devblogs.nvidia.com/parallelforall/deep-learning-nutshell-core-concepts/ 1 Neurotechnology http://www.lce.hut.fi/research/cogntech/neurophysiology Recording
Bayesian Modeling and Classification of Neural Signals
 Bayesian Modeling and Classification of Neural Signals Michael S. Lewicki Computation and Neural Systems Program California Institute of Technology 216-76 Pasadena, CA 91125 lewickiocns.caltech.edu Abstract
Bayesian Modeling and Classification of Neural Signals Michael S. Lewicki Computation and Neural Systems Program California Institute of Technology 216-76 Pasadena, CA 91125 lewickiocns.caltech.edu Abstract
Lecture 11: Non-Equilibrium Statistical Physics
 Massachusetts Institute of Technology MITES 2018 Physics III Lecture 11: Non-Equilibrium Statistical Physics In the previous notes, we studied how systems in thermal equilibrium can change in time even
Massachusetts Institute of Technology MITES 2018 Physics III Lecture 11: Non-Equilibrium Statistical Physics In the previous notes, we studied how systems in thermal equilibrium can change in time even
Need for Deep Networks Perceptron. Can only model linear functions. Kernel Machines. Non-linearity provided by kernels
 Need for Deep Networks Perceptron Can only model linear functions Kernel Machines Non-linearity provided by kernels Need to design appropriate kernels (possibly selecting from a set, i.e. kernel learning)
Need for Deep Networks Perceptron Can only model linear functions Kernel Machines Non-linearity provided by kernels Need to design appropriate kernels (possibly selecting from a set, i.e. kernel learning)
Estimation of information-theoretic quantities
 Estimation of information-theoretic quantities Liam Paninski Gatsby Computational Neuroscience Unit University College London http://www.gatsby.ucl.ac.uk/ liam liam@gatsby.ucl.ac.uk November 16, 2004 Some
Estimation of information-theoretic quantities Liam Paninski Gatsby Computational Neuroscience Unit University College London http://www.gatsby.ucl.ac.uk/ liam liam@gatsby.ucl.ac.uk November 16, 2004 Some
Bayesian probability theory and generative models
 Bayesian probability theory and generative models Bruno A. Olshausen November 8, 2006 Abstract Bayesian probability theory provides a mathematical framework for peforming inference, or reasoning, using
Bayesian probability theory and generative models Bruno A. Olshausen November 8, 2006 Abstract Bayesian probability theory provides a mathematical framework for peforming inference, or reasoning, using
Simultaneous Multi-frame MAP Super-Resolution Video Enhancement using Spatio-temporal Priors
 Simultaneous Multi-frame MAP Super-Resolution Video Enhancement using Spatio-temporal Priors Sean Borman and Robert L. Stevenson Department of Electrical Engineering, University of Notre Dame Notre Dame,
Simultaneous Multi-frame MAP Super-Resolution Video Enhancement using Spatio-temporal Priors Sean Borman and Robert L. Stevenson Department of Electrical Engineering, University of Notre Dame Notre Dame,
Finding a Basis for the Neural State
 Finding a Basis for the Neural State Chris Cueva ccueva@stanford.edu I. INTRODUCTION How is information represented in the brain? For example, consider arm movement. Neurons in dorsal premotor cortex (PMd)
Finding a Basis for the Neural State Chris Cueva ccueva@stanford.edu I. INTRODUCTION How is information represented in the brain? For example, consider arm movement. Neurons in dorsal premotor cortex (PMd)
arxiv: v1 [q-bio.nc] 26 Oct 2017
![arxiv: v1 [q-bio.nc] 26 Oct 2017 arxiv: v1 [q-bio.nc] 26 Oct 2017](/thumbs/79/78949314.jpg) Pairwise Ising model analysis of human cortical neuron recordings Trang-Anh Nghiem 1, Olivier Marre 2 Alain Destexhe 1, and Ulisse Ferrari 23 arxiv:1710.09929v1 [q-bio.nc] 26 Oct 2017 1 Laboratory of Computational
Pairwise Ising model analysis of human cortical neuron recordings Trang-Anh Nghiem 1, Olivier Marre 2 Alain Destexhe 1, and Ulisse Ferrari 23 arxiv:1710.09929v1 [q-bio.nc] 26 Oct 2017 1 Laboratory of Computational
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2013
 UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2013 Exam policy: This exam allows two one-page, two-sided cheat sheets; No other materials. Time: 2 hours. Be sure to write your name and
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2013 Exam policy: This exam allows two one-page, two-sided cheat sheets; No other materials. Time: 2 hours. Be sure to write your name and
Markov Chain Monte Carlo The Metropolis-Hastings Algorithm
 Markov Chain Monte Carlo The Metropolis-Hastings Algorithm Anthony Trubiano April 11th, 2018 1 Introduction Markov Chain Monte Carlo (MCMC) methods are a class of algorithms for sampling from a probability
Markov Chain Monte Carlo The Metropolis-Hastings Algorithm Anthony Trubiano April 11th, 2018 1 Introduction Markov Chain Monte Carlo (MCMC) methods are a class of algorithms for sampling from a probability
The homogeneous Poisson process
 The homogeneous Poisson process during very short time interval Δt there is a fixed probability of an event (spike) occurring independent of what happened previously if r is the rate of the Poisson process,
The homogeneous Poisson process during very short time interval Δt there is a fixed probability of an event (spike) occurring independent of what happened previously if r is the rate of the Poisson process,
Statistical models for neural encoding, decoding, information estimation, and optimal on-line stimulus design
 Statistical models for neural encoding, decoding, information estimation, and optimal on-line stimulus design Liam Paninski Department of Statistics and Center for Theoretical Neuroscience Columbia University
Statistical models for neural encoding, decoding, information estimation, and optimal on-line stimulus design Liam Paninski Department of Statistics and Center for Theoretical Neuroscience Columbia University
State-Space Methods for Inferring Spike Trains from Calcium Imaging
 State-Space Methods for Inferring Spike Trains from Calcium Imaging Joshua Vogelstein Johns Hopkins April 23, 2009 Joshua Vogelstein (Johns Hopkins) State-Space Calcium Imaging April 23, 2009 1 / 78 Outline
State-Space Methods for Inferring Spike Trains from Calcium Imaging Joshua Vogelstein Johns Hopkins April 23, 2009 Joshua Vogelstein (Johns Hopkins) State-Space Calcium Imaging April 23, 2009 1 / 78 Outline
Neural networks: Unsupervised learning
 Neural networks: Unsupervised learning 1 Previously The supervised learning paradigm: given example inputs x and target outputs t learning the mapping between them the trained network is supposed to give
Neural networks: Unsupervised learning 1 Previously The supervised learning paradigm: given example inputs x and target outputs t learning the mapping between them the trained network is supposed to give
Bayesian Feature Selection with Strongly Regularizing Priors Maps to the Ising Model
 LETTER Communicated by Ilya M. Nemenman Bayesian Feature Selection with Strongly Regularizing Priors Maps to the Ising Model Charles K. Fisher charleskennethfisher@gmail.com Pankaj Mehta pankajm@bu.edu
LETTER Communicated by Ilya M. Nemenman Bayesian Feature Selection with Strongly Regularizing Priors Maps to the Ising Model Charles K. Fisher charleskennethfisher@gmail.com Pankaj Mehta pankajm@bu.edu
Bayesian Learning in Undirected Graphical Models
 Bayesian Learning in Undirected Graphical Models Zoubin Ghahramani Gatsby Computational Neuroscience Unit University College London, UK http://www.gatsby.ucl.ac.uk/ Work with: Iain Murray and Hyun-Chul
Bayesian Learning in Undirected Graphical Models Zoubin Ghahramani Gatsby Computational Neuroscience Unit University College London, UK http://www.gatsby.ucl.ac.uk/ Work with: Iain Murray and Hyun-Chul
5. Discriminant analysis
 5. Discriminant analysis We continue from Bayes s rule presented in Section 3 on p. 85 (5.1) where c i is a class, x isap-dimensional vector (data case) and we use class conditional probability (density
5. Discriminant analysis We continue from Bayes s rule presented in Section 3 on p. 85 (5.1) where c i is a class, x isap-dimensional vector (data case) and we use class conditional probability (density
Learning the dynamics of biological networks
 Learning the dynamics of biological networks Thierry Mora ENS Paris A.Walczak Università Sapienza Rome A. Cavagna I. Giardina O. Pohl E. Silvestri M.Viale Aberdeen University F. Ginelli Laboratoire de
Learning the dynamics of biological networks Thierry Mora ENS Paris A.Walczak Università Sapienza Rome A. Cavagna I. Giardina O. Pohl E. Silvestri M.Viale Aberdeen University F. Ginelli Laboratoire de
Discriminative Direction for Kernel Classifiers
 Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Evolution of the Average Synaptic Update Rule
 Supporting Text Evolution of the Average Synaptic Update Rule In this appendix we evaluate the derivative of Eq. 9 in the main text, i.e., we need to calculate log P (yk Y k, X k ) γ log P (yk Y k ). ()
Supporting Text Evolution of the Average Synaptic Update Rule In this appendix we evaluate the derivative of Eq. 9 in the main text, i.e., we need to calculate log P (yk Y k, X k ) γ log P (yk Y k ). ()
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie
 A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
Inference in kinetic Ising models: mean field and Bayes estimators
 Inference in kinetic Ising models: mean field and Bayes estimators Ludovica Bachschmid-Romano, Manfred Opper Artificial Intelligence group, Computer Science, TU Berlin, Germany October 23, 2015 L. Bachschmid-Romano,
Inference in kinetic Ising models: mean field and Bayes estimators Ludovica Bachschmid-Romano, Manfred Opper Artificial Intelligence group, Computer Science, TU Berlin, Germany October 23, 2015 L. Bachschmid-Romano,
Efficient coding of natural images with a population of noisy Linear-Nonlinear neurons
 Efficient coding of natural images with a population of noisy Linear-Nonlinear neurons Yan Karklin and Eero P. Simoncelli NYU Overview Efficient coding is a well-known objective for the evaluation and
Efficient coding of natural images with a population of noisy Linear-Nonlinear neurons Yan Karklin and Eero P. Simoncelli NYU Overview Efficient coding is a well-known objective for the evaluation and
Linear & nonlinear classifiers
 Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1394 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1394 1 / 34 Table
Linear & nonlinear classifiers Machine Learning Hamid Beigy Sharif University of Technology Fall 1394 Hamid Beigy (Sharif University of Technology) Linear & nonlinear classifiers Fall 1394 1 / 34 Table
How to do backpropagation in a brain
 How to do backpropagation in a brain Geoffrey Hinton Canadian Institute for Advanced Research & University of Toronto & Google Inc. Prelude I will start with three slides explaining a popular type of deep
How to do backpropagation in a brain Geoffrey Hinton Canadian Institute for Advanced Research & University of Toronto & Google Inc. Prelude I will start with three slides explaining a popular type of deep
When Do Microcircuits Produce Beyond-Pairwise Correlations?
 When Do Microcircuits Produce Beyond-Pairwise Correlations? Andrea K. Barreiro,4,, Julijana Gjorgjieva 3,5, Fred Rieke 2, and Eric Shea-Brown Department of Applied Mathematics, University of Washington
When Do Microcircuits Produce Beyond-Pairwise Correlations? Andrea K. Barreiro,4,, Julijana Gjorgjieva 3,5, Fred Rieke 2, and Eric Shea-Brown Department of Applied Mathematics, University of Washington
(Feed-Forward) Neural Networks Dr. Hajira Jabeen, Prof. Jens Lehmann
 (Feed-Forward) Neural Networks 2016-12-06 Dr. Hajira Jabeen, Prof. Jens Lehmann Outline In the previous lectures we have learned about tensors and factorization methods. RESCAL is a bilinear model for
(Feed-Forward) Neural Networks 2016-12-06 Dr. Hajira Jabeen, Prof. Jens Lehmann Outline In the previous lectures we have learned about tensors and factorization methods. RESCAL is a bilinear model for
Tensor network simulations of strongly correlated quantum systems
 CENTRE FOR QUANTUM TECHNOLOGIES NATIONAL UNIVERSITY OF SINGAPORE AND CLARENDON LABORATORY UNIVERSITY OF OXFORD Tensor network simulations of strongly correlated quantum systems Stephen Clark LXXT[[[GSQPEFS\EGYOEGXMZMXMIWUYERXYQGSYVWI
CENTRE FOR QUANTUM TECHNOLOGIES NATIONAL UNIVERSITY OF SINGAPORE AND CLARENDON LABORATORY UNIVERSITY OF OXFORD Tensor network simulations of strongly correlated quantum systems Stephen Clark LXXT[[[GSQPEFS\EGYOEGXMZMXMIWUYERXYQGSYVWI
STA 414/2104: Lecture 8
 STA 414/2104: Lecture 8 6-7 March 2017: Continuous Latent Variable Models, Neural networks With thanks to Russ Salakhutdinov, Jimmy Ba and others Outline Continuous latent variable models Background PCA
STA 414/2104: Lecture 8 6-7 March 2017: Continuous Latent Variable Models, Neural networks With thanks to Russ Salakhutdinov, Jimmy Ba and others Outline Continuous latent variable models Background PCA
Neural Networks. Mark van Rossum. January 15, School of Informatics, University of Edinburgh 1 / 28
 1 / 28 Neural Networks Mark van Rossum School of Informatics, University of Edinburgh January 15, 2018 2 / 28 Goals: Understand how (recurrent) networks behave Find a way to teach networks to do a certain
1 / 28 Neural Networks Mark van Rossum School of Informatics, University of Edinburgh January 15, 2018 2 / 28 Goals: Understand how (recurrent) networks behave Find a way to teach networks to do a certain
Does the Wake-sleep Algorithm Produce Good Density Estimators?
 Does the Wake-sleep Algorithm Produce Good Density Estimators? Brendan J. Frey, Geoffrey E. Hinton Peter Dayan Department of Computer Science Department of Brain and Cognitive Sciences University of Toronto
Does the Wake-sleep Algorithm Produce Good Density Estimators? Brendan J. Frey, Geoffrey E. Hinton Peter Dayan Department of Computer Science Department of Brain and Cognitive Sciences University of Toronto
Statistical Rock Physics
 Statistical - Introduction Book review 3.1-3.3 Min Sun March. 13, 2009 Outline. What is Statistical. Why we need Statistical. How Statistical works Statistical Rock physics Information theory Statistics
Statistical - Introduction Book review 3.1-3.3 Min Sun March. 13, 2009 Outline. What is Statistical. Why we need Statistical. How Statistical works Statistical Rock physics Information theory Statistics
7 Rate-Based Recurrent Networks of Threshold Neurons: Basis for Associative Memory
 Physics 178/278 - David Kleinfeld - Fall 2005; Revised for Winter 2017 7 Rate-Based Recurrent etworks of Threshold eurons: Basis for Associative Memory 7.1 A recurrent network with threshold elements The
Physics 178/278 - David Kleinfeld - Fall 2005; Revised for Winter 2017 7 Rate-Based Recurrent etworks of Threshold eurons: Basis for Associative Memory 7.1 A recurrent network with threshold elements The
Food delivered. Food obtained S 3
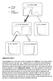 Press lever Enter magazine * S 0 Initial state S 1 Food delivered * S 2 No reward S 2 No reward S 3 Food obtained Supplementary Figure 1 Value propagation in tree search, after 50 steps of learning the
Press lever Enter magazine * S 0 Initial state S 1 Food delivered * S 2 No reward S 2 No reward S 3 Food obtained Supplementary Figure 1 Value propagation in tree search, after 50 steps of learning the
Probabilistic Models in Theoretical Neuroscience
 Probabilistic Models in Theoretical Neuroscience visible unit Boltzmann machine semi-restricted Boltzmann machine restricted Boltzmann machine hidden unit Neural models of probabilistic sampling: introduction
Probabilistic Models in Theoretical Neuroscience visible unit Boltzmann machine semi-restricted Boltzmann machine restricted Boltzmann machine hidden unit Neural models of probabilistic sampling: introduction
Linear Models for Regression. Sargur Srihari
 Linear Models for Regression Sargur srihari@cedar.buffalo.edu 1 Topics in Linear Regression What is regression? Polynomial Curve Fitting with Scalar input Linear Basis Function Models Maximum Likelihood
Linear Models for Regression Sargur srihari@cedar.buffalo.edu 1 Topics in Linear Regression What is regression? Polynomial Curve Fitting with Scalar input Linear Basis Function Models Maximum Likelihood
A graph contains a set of nodes (vertices) connected by links (edges or arcs)
 BOLTZMANN MACHINES Generative Models Graphical Models A graph contains a set of nodes (vertices) connected by links (edges or arcs) In a probabilistic graphical model, each node represents a random variable,
BOLTZMANN MACHINES Generative Models Graphical Models A graph contains a set of nodes (vertices) connected by links (edges or arcs) In a probabilistic graphical model, each node represents a random variable,
Learning Energy-Based Models of High-Dimensional Data
 Learning Energy-Based Models of High-Dimensional Data Geoffrey Hinton Max Welling Yee-Whye Teh Simon Osindero www.cs.toronto.edu/~hinton/energybasedmodelsweb.htm Discovering causal structure as a goal
Learning Energy-Based Models of High-Dimensional Data Geoffrey Hinton Max Welling Yee-Whye Teh Simon Osindero www.cs.toronto.edu/~hinton/energybasedmodelsweb.htm Discovering causal structure as a goal
Consider the following spike trains from two different neurons N1 and N2:
 About synchrony and oscillations So far, our discussions have assumed that we are either observing a single neuron at a, or that neurons fire independent of each other. This assumption may be correct in
About synchrony and oscillations So far, our discussions have assumed that we are either observing a single neuron at a, or that neurons fire independent of each other. This assumption may be correct in
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2014
 UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2014 Exam policy: This exam allows two one-page, two-sided cheat sheets (i.e. 4 sides); No other materials. Time: 2 hours. Be sure to write
UNIVERSITY of PENNSYLVANIA CIS 520: Machine Learning Final, Fall 2014 Exam policy: This exam allows two one-page, two-sided cheat sheets (i.e. 4 sides); No other materials. Time: 2 hours. Be sure to write
HST 583 FUNCTIONAL MAGNETIC RESONANCE IMAGING DATA ANALYSIS AND ACQUISITION A REVIEW OF STATISTICS FOR FMRI DATA ANALYSIS
 HST 583 FUNCTIONAL MAGNETIC RESONANCE IMAGING DATA ANALYSIS AND ACQUISITION A REVIEW OF STATISTICS FOR FMRI DATA ANALYSIS EMERY N. BROWN AND CHRIS LONG NEUROSCIENCE STATISTICS RESEARCH LABORATORY DEPARTMENT
HST 583 FUNCTIONAL MAGNETIC RESONANCE IMAGING DATA ANALYSIS AND ACQUISITION A REVIEW OF STATISTICS FOR FMRI DATA ANALYSIS EMERY N. BROWN AND CHRIS LONG NEUROSCIENCE STATISTICS RESEARCH LABORATORY DEPARTMENT
Chris Bishop s PRML Ch. 8: Graphical Models
 Chris Bishop s PRML Ch. 8: Graphical Models January 24, 2008 Introduction Visualize the structure of a probabilistic model Design and motivate new models Insights into the model s properties, in particular
Chris Bishop s PRML Ch. 8: Graphical Models January 24, 2008 Introduction Visualize the structure of a probabilistic model Design and motivate new models Insights into the model s properties, in particular
An Introductory Course in Computational Neuroscience
 An Introductory Course in Computational Neuroscience Contents Series Foreword Acknowledgments Preface 1 Preliminary Material 1.1. Introduction 1.1.1 The Cell, the Circuit, and the Brain 1.1.2 Physics of
An Introductory Course in Computational Neuroscience Contents Series Foreword Acknowledgments Preface 1 Preliminary Material 1.1. Introduction 1.1.1 The Cell, the Circuit, and the Brain 1.1.2 Physics of
The connection of dropout and Bayesian statistics
 The connection of dropout and Bayesian statistics Interpretation of dropout as approximate Bayesian modelling of NN http://mlg.eng.cam.ac.uk/yarin/thesis/thesis.pdf Dropout Geoffrey Hinton Google, University
The connection of dropout and Bayesian statistics Interpretation of dropout as approximate Bayesian modelling of NN http://mlg.eng.cam.ac.uk/yarin/thesis/thesis.pdf Dropout Geoffrey Hinton Google, University
σ(a) = a N (x; 0, 1 2 ) dx. σ(a) = Φ(a) =
 Until now we have always worked with likelihoods and prior distributions that were conjugate to each other, allowing the computation of the posterior distribution to be done in closed form. Unfortunately,
Until now we have always worked with likelihoods and prior distributions that were conjugate to each other, allowing the computation of the posterior distribution to be done in closed form. Unfortunately,
3.3 Population Decoding
 3.3 Population Decoding 97 We have thus far considered discriminating between two quite distinct stimulus values, plus and minus. Often we are interested in discriminating between two stimulus values s
3.3 Population Decoding 97 We have thus far considered discriminating between two quite distinct stimulus values, plus and minus. Often we are interested in discriminating between two stimulus values s
A Three-dimensional Physiologically Realistic Model of the Retina
 A Three-dimensional Physiologically Realistic Model of the Retina Michael Tadross, Cameron Whitehouse, Melissa Hornstein, Vicky Eng and Evangelia Micheli-Tzanakou Department of Biomedical Engineering 617
A Three-dimensional Physiologically Realistic Model of the Retina Michael Tadross, Cameron Whitehouse, Melissa Hornstein, Vicky Eng and Evangelia Micheli-Tzanakou Department of Biomedical Engineering 617
STA 4273H: Statistical Machine Learning
 STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
STA 4273H: Statistical Machine Learning Russ Salakhutdinov Department of Statistics! rsalakhu@utstat.toronto.edu! http://www.utstat.utoronto.ca/~rsalakhu/ Sidney Smith Hall, Room 6002 Lecture 3 Linear
Sean Escola. Center for Theoretical Neuroscience
 Employing hidden Markov models of neural spike-trains toward the improved estimation of linear receptive fields and the decoding of multiple firing regimes Sean Escola Center for Theoretical Neuroscience
Employing hidden Markov models of neural spike-trains toward the improved estimation of linear receptive fields and the decoding of multiple firing regimes Sean Escola Center for Theoretical Neuroscience
Neural Network Training
 Neural Network Training Sargur Srihari Topics in Network Training 0. Neural network parameters Probabilistic problem formulation Specifying the activation and error functions for Regression Binary classification
Neural Network Training Sargur Srihari Topics in Network Training 0. Neural network parameters Probabilistic problem formulation Specifying the activation and error functions for Regression Binary classification
On Bayesian Computation
 On Bayesian Computation Michael I. Jordan with Elaine Angelino, Maxim Rabinovich, Martin Wainwright and Yun Yang Previous Work: Information Constraints on Inference Minimize the minimax risk under constraints
On Bayesian Computation Michael I. Jordan with Elaine Angelino, Maxim Rabinovich, Martin Wainwright and Yun Yang Previous Work: Information Constraints on Inference Minimize the minimax risk under constraints
Basic Principles of Unsupervised and Unsupervised
 Basic Principles of Unsupervised and Unsupervised Learning Toward Deep Learning Shun ichi Amari (RIKEN Brain Science Institute) collaborators: R. Karakida, M. Okada (U. Tokyo) Deep Learning Self Organization
Basic Principles of Unsupervised and Unsupervised Learning Toward Deep Learning Shun ichi Amari (RIKEN Brain Science Institute) collaborators: R. Karakida, M. Okada (U. Tokyo) Deep Learning Self Organization
Information Theory. Mark van Rossum. January 24, School of Informatics, University of Edinburgh 1 / 35
 1 / 35 Information Theory Mark van Rossum School of Informatics, University of Edinburgh January 24, 2018 0 Version: January 24, 2018 Why information theory 2 / 35 Understanding the neural code. Encoding
1 / 35 Information Theory Mark van Rossum School of Informatics, University of Edinburgh January 24, 2018 0 Version: January 24, 2018 Why information theory 2 / 35 Understanding the neural code. Encoding
The Origin of Deep Learning. Lili Mou Jan, 2015
 The Origin of Deep Learning Lili Mou Jan, 2015 Acknowledgment Most of the materials come from G. E. Hinton s online course. Outline Introduction Preliminary Boltzmann Machines and RBMs Deep Belief Nets
The Origin of Deep Learning Lili Mou Jan, 2015 Acknowledgment Most of the materials come from G. E. Hinton s online course. Outline Introduction Preliminary Boltzmann Machines and RBMs Deep Belief Nets
Transformation of stimulus correlations by the retina
 Transformation of stimulus correlations by the retina Kristina Simmons (University of Pennsylvania) and Jason Prentice, (now Princeton University) with Gasper Tkacik (IST Austria) Jan Homann (now Princeton
Transformation of stimulus correlations by the retina Kristina Simmons (University of Pennsylvania) and Jason Prentice, (now Princeton University) with Gasper Tkacik (IST Austria) Jan Homann (now Princeton
Exam, Solutions
 Exam, - Solutions Q Constructing a balanced sequence containing three kinds of stimuli Here we design a balanced cyclic sequence for three kinds of stimuli (labeled {,, }, in which every three-element
Exam, - Solutions Q Constructing a balanced sequence containing three kinds of stimuli Here we design a balanced cyclic sequence for three kinds of stimuli (labeled {,, }, in which every three-element
DS-GA 1002 Lecture notes 12 Fall Linear regression
 DS-GA Lecture notes 1 Fall 16 1 Linear models Linear regression In statistics, regression consists of learning a function relating a certain quantity of interest y, the response or dependent variable,
DS-GA Lecture notes 1 Fall 16 1 Linear models Linear regression In statistics, regression consists of learning a function relating a certain quantity of interest y, the response or dependent variable,
Introduction to Machine Learning Midterm Exam
 10-701 Introduction to Machine Learning Midterm Exam Instructors: Eric Xing, Ziv Bar-Joseph 17 November, 2015 There are 11 questions, for a total of 100 points. This exam is open book, open notes, but
10-701 Introduction to Machine Learning Midterm Exam Instructors: Eric Xing, Ziv Bar-Joseph 17 November, 2015 There are 11 questions, for a total of 100 points. This exam is open book, open notes, but
7 Recurrent Networks of Threshold (Binary) Neurons: Basis for Associative Memory
 Physics 178/278 - David Kleinfeld - Winter 2019 7 Recurrent etworks of Threshold (Binary) eurons: Basis for Associative Memory 7.1 The network The basic challenge in associative networks, also referred
Physics 178/278 - David Kleinfeld - Winter 2019 7 Recurrent etworks of Threshold (Binary) eurons: Basis for Associative Memory 7.1 The network The basic challenge in associative networks, also referred
Neuroscience Introduction
 Neuroscience Introduction The brain As humans, we can identify galaxies light years away, we can study particles smaller than an atom. But we still haven t unlocked the mystery of the three pounds of matter
Neuroscience Introduction The brain As humans, we can identify galaxies light years away, we can study particles smaller than an atom. But we still haven t unlocked the mystery of the three pounds of matter
This cannot be estimated directly... s 1. s 2. P(spike, stim) P(stim) P(spike stim) =
 LNP cascade model Simplest successful descriptive spiking model Easily fit to (extracellular) data Descriptive, and interpretable (although not mechanistic) For a Poisson model, response is captured by
LNP cascade model Simplest successful descriptive spiking model Easily fit to (extracellular) data Descriptive, and interpretable (although not mechanistic) For a Poisson model, response is captured by
Pattern Recognition and Machine Learning
 Christopher M. Bishop Pattern Recognition and Machine Learning ÖSpri inger Contents Preface Mathematical notation Contents vii xi xiii 1 Introduction 1 1.1 Example: Polynomial Curve Fitting 4 1.2 Probability
Christopher M. Bishop Pattern Recognition and Machine Learning ÖSpri inger Contents Preface Mathematical notation Contents vii xi xiii 1 Introduction 1 1.1 Example: Polynomial Curve Fitting 4 1.2 Probability
Sparse regression. Optimization-Based Data Analysis. Carlos Fernandez-Granda
 Sparse regression Optimization-Based Data Analysis http://www.cims.nyu.edu/~cfgranda/pages/obda_spring16 Carlos Fernandez-Granda 3/28/2016 Regression Least-squares regression Example: Global warming Logistic
Sparse regression Optimization-Based Data Analysis http://www.cims.nyu.edu/~cfgranda/pages/obda_spring16 Carlos Fernandez-Granda 3/28/2016 Regression Least-squares regression Example: Global warming Logistic
Bayesian Methods for Machine Learning
 Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Bayesian Methods for Machine Learning CS 584: Big Data Analytics Material adapted from Radford Neal s tutorial (http://ftp.cs.utoronto.ca/pub/radford/bayes-tut.pdf), Zoubin Ghahramni (http://hunch.net/~coms-4771/zoubin_ghahramani_bayesian_learning.pdf),
Probabilistic & Unsupervised Learning
 Probabilistic & Unsupervised Learning Week 2: Latent Variable Models Maneesh Sahani maneesh@gatsby.ucl.ac.uk Gatsby Computational Neuroscience Unit, and MSc ML/CSML, Dept Computer Science University College
Probabilistic & Unsupervised Learning Week 2: Latent Variable Models Maneesh Sahani maneesh@gatsby.ucl.ac.uk Gatsby Computational Neuroscience Unit, and MSc ML/CSML, Dept Computer Science University College
Lecture 16 Deep Neural Generative Models
 Lecture 16 Deep Neural Generative Models CMSC 35246: Deep Learning Shubhendu Trivedi & Risi Kondor University of Chicago May 22, 2017 Approach so far: We have considered simple models and then constructed
Lecture 16 Deep Neural Generative Models CMSC 35246: Deep Learning Shubhendu Trivedi & Risi Kondor University of Chicago May 22, 2017 Approach so far: We have considered simple models and then constructed
Lecture 6. Regression
 Lecture 6. Regression Prof. Alan Yuille Summer 2014 Outline 1. Introduction to Regression 2. Binary Regression 3. Linear Regression; Polynomial Regression 4. Non-linear Regression; Multilayer Perceptron
Lecture 6. Regression Prof. Alan Yuille Summer 2014 Outline 1. Introduction to Regression 2. Binary Regression 3. Linear Regression; Polynomial Regression 4. Non-linear Regression; Multilayer Perceptron
Belief Propagation for Traffic forecasting
 Belief Propagation for Traffic forecasting Cyril Furtlehner (INRIA Saclay - Tao team) context : Travesti project http ://travesti.gforge.inria.fr/) Anne Auger (INRIA Saclay) Dimo Brockhoff (INRIA Lille)
Belief Propagation for Traffic forecasting Cyril Furtlehner (INRIA Saclay - Tao team) context : Travesti project http ://travesti.gforge.inria.fr/) Anne Auger (INRIA Saclay) Dimo Brockhoff (INRIA Lille)
External field h ext. Supplementary Figure 1. Performing the second-order maximum entropy fitting procedure on an
 Fisher information I = Susceptibility d s /dh ext 1.2 s 0.8 0.6 0.4 0.2 40 30 20 10 0 4 2 0 2 4 h ext Fit to sampled independent data Indep. model 4 2 0 2 4 External field h ext Supplementary Figure 1.
Fisher information I = Susceptibility d s /dh ext 1.2 s 0.8 0.6 0.4 0.2 40 30 20 10 0 4 2 0 2 4 h ext Fit to sampled independent data Indep. model 4 2 0 2 4 External field h ext Supplementary Figure 1.
Lecture Notes 5: Multiresolution Analysis
 Optimization-based data analysis Fall 2017 Lecture Notes 5: Multiresolution Analysis 1 Frames A frame is a generalization of an orthonormal basis. The inner products between the vectors in a frame and
Optimization-based data analysis Fall 2017 Lecture Notes 5: Multiresolution Analysis 1 Frames A frame is a generalization of an orthonormal basis. The inner products between the vectors in a frame and
Energy Landscapes of Deep Networks
 Energy Landscapes of Deep Networks Pratik Chaudhari February 29, 2016 References Auffinger, A., Arous, G. B., & Černý, J. (2013). Random matrices and complexity of spin glasses. arxiv:1003.1129.# Choromanska,
Energy Landscapes of Deep Networks Pratik Chaudhari February 29, 2016 References Auffinger, A., Arous, G. B., & Černý, J. (2013). Random matrices and complexity of spin glasses. arxiv:1003.1129.# Choromanska,
Information maximization in a network of linear neurons
 Information maximization in a network of linear neurons Holger Arnold May 30, 005 1 Introduction It is known since the work of Hubel and Wiesel [3], that many cells in the early visual areas of mammals
Information maximization in a network of linear neurons Holger Arnold May 30, 005 1 Introduction It is known since the work of Hubel and Wiesel [3], that many cells in the early visual areas of mammals
Computer Vision Group Prof. Daniel Cremers. 10a. Markov Chain Monte Carlo
 Group Prof. Daniel Cremers 10a. Markov Chain Monte Carlo Markov Chain Monte Carlo In high-dimensional spaces, rejection sampling and importance sampling are very inefficient An alternative is Markov Chain
Group Prof. Daniel Cremers 10a. Markov Chain Monte Carlo Markov Chain Monte Carlo In high-dimensional spaces, rejection sampling and importance sampling are very inefficient An alternative is Markov Chain
Gatsby Theoretical Neuroscience Lectures: Non-Gaussian statistics and natural images Parts I-II
 Gatsby Theoretical Neuroscience Lectures: Non-Gaussian statistics and natural images Parts I-II Gatsby Unit University College London 27 Feb 2017 Outline Part I: Theory of ICA Definition and difference
Gatsby Theoretical Neuroscience Lectures: Non-Gaussian statistics and natural images Parts I-II Gatsby Unit University College London 27 Feb 2017 Outline Part I: Theory of ICA Definition and difference
Recoverabilty Conditions for Rankings Under Partial Information
 Recoverabilty Conditions for Rankings Under Partial Information Srikanth Jagabathula Devavrat Shah Abstract We consider the problem of exact recovery of a function, defined on the space of permutations
Recoverabilty Conditions for Rankings Under Partial Information Srikanth Jagabathula Devavrat Shah Abstract We consider the problem of exact recovery of a function, defined on the space of permutations
CPSC 340: Machine Learning and Data Mining. Sparse Matrix Factorization Fall 2017
 CPSC 340: Machine Learning and Data Mining Sparse Matrix Factorization Fall 2017 Admin Assignment 4: Due Friday. Assignment 5: Posted, due Monday of last week of classes Last Time: PCA with Orthogonal/Sequential
CPSC 340: Machine Learning and Data Mining Sparse Matrix Factorization Fall 2017 Admin Assignment 4: Due Friday. Assignment 5: Posted, due Monday of last week of classes Last Time: PCA with Orthogonal/Sequential
