Experimental Phasing with SHELX C/D/E
|
|
|
- Spencer Hutchinson
- 5 years ago
- Views:
Transcription
1 WIR SCHAFFEN WISSEN HEUTE FÜR MORGEN Dr. Tim Grüne :: Paul Scherrer Institut :: Experimental Phasing with SHELX C/D/E CCP4 / APS School Chicago nd June 2017
2 1 - The Phase Problem 22 nd June 2017 SHELX C/D/E 1/26
3 The Crystallographic Phase Problem 1. Crystal diffraction yields intensities I(hkl), and thus structure factor amplitude F(hkl) I(hkl) 2. Model building requires a map, i.e. ρ(x,y,z) = h,k,l F(hkl) e iφ(hkl) e 2πi(hx+ky+lz) 3. Solving a structure = determination of the phase angles φ(hkl) good enough to create an interpretable map 4. Small molecule crystallography: Problem mostly solved with direct method 5. SHELXD solves small molecule data at d < 1.2Å 6. SHELXD solves the substructure even at d = 5Å (and worse) because substructure atoms are still resolved 22 nd June 2017 SHELX C/D/E 2/26
4 The Substructure Coordinates of anomalous scatterers Anomalous difference F + (hkl) F (hkl) F sub (hkl) corresponds to small molecule data set Shelxd: solve substructure with direct methods Harker Construction: Expand phases to full data set 22 nd June 2017 SHELX C/D/E 3/26
5 SHELX C/D/E [1] in Context with Structure Solution DIALS, XDS, MOSFLM... shelxc shelxd shelxe REFMAC5 / COOT... Data Processing Anomalous Differences Substructure Solution Phasing & Density Modification Building & Refinement Molecular Replacement 22 nd June 2017 SHELX C/D/E 4/26
6 2 - Using SHELX C/D/E 22 nd June 2017 SHELX C/D/E 5/26
7 Graphical User Interface HKL2MAP 22 nd June 2017 SHELX C/D/E 6/26
8 Documentation Most shelx programs issue short usage instruction when called without an argument. shelxc SHELXC - Create input files for SHELXD and SHELXE - Version 2013/2 + + Copyright (c) George M. Sheldrick Started at 13:59:57 on 10 Jun SHELXC reads a filename stem (denoted here by xx ) on the command line plus some instructions from standard input. It writes some statistics to standard output and prepares the three files needed to run SHELXD and SHELXE. SHELXC can be called from a GUI by a command line such as: shelxc xx <t which would read the instructions from the file t, or (under most UNIX systems) by a simple shell script that includes the instructions, e.g. shelxc xx <<EOF CELL SPAG P SAD elastase.sca FIND 12 <<EOF shelxd xx_fa shelxe xx xx_fa -s0.37 -m20 -h -b shelxe xx xx_fa -s0.37 -m20 -h -b -i More information including tutorials available at 22 nd June 2017 SHELX C/D/E 7/26
9 Shelxc Data Preparation: Keywords shelxc can be used for five different phasing scenarios: SAD SIR SAD (NAT) SIR NAT SIRAS SIRA NAT MAD RIP (NAT) PEAK INFL LREM HREM BEFORE AFTER (NAT) Each keyword takes the filename of the corresponding integrated dataset. 22 nd June 2017 SHELX C/D/E 8/26
10 Running shelxc 1. Create input command file shelxc.inp with text editor CELL SPAG P FIND 12 NTRY 100 SFAC S SAD elastase.sca 2. shelxc mysad < shelxc.inp 22 nd June 2017 SHELX C/D/E 9/26
11 Shelxc Output Files The command shelxc mysad < shelxc.inp creates three files: mysad fa.ins Text file with instructions for shelxd mysad fa.hkl Artificial substructure data set from which shelxd determines substructure coordinates. contains Each line h,k,l, F + (hkl) F (hkl),α α is not used by shelxd, but by shelxe to calculate an initial phase estimate for the protein structure as φ T (hkl) = φ A (hkl) + α(hkl) MAD/SIRAS: exact α; SIR or SAD: rough estimate of α φ A is the phase angle calculated from the substructure coordinates determined by shelxd. mysad.hkl native data used by shelxe for phasing and density modification 22 nd June 2017 SHELX C/D/E 10/26
12 Shelxc: Resolution Cut-off for Anomalous Signal Reflections read from SAD file XDS_pk1pk2.HKL Unique reflections, highest resolution Angstroms Friedel pairs used on average for local scaling Resl. Inf N(data) Chi-sq <I/sig> %Complete <d"/sig> CC(1/2) anomalous signal where CC(1/2) > 30% anomalous signal where <d"/sig> > 1.3 CC > 30% usually more reliable than <d"/sig> > nd June 2017 SHELX C/D/E 11/26
13 3 - Shelxd 22 nd June 2017 SHELX C/D/E 12/26
14 Shelxd Finding the Substructure shelxd mysad_fa reads the substructure data mysad fa.hkl and its instructions from mysad fa.ins. The most important entries in mysad fa.ins: SFAC SE atom type to look for FIND 12 expected number of substructure atoms, should be within 20 % of the actual number (try several for e.g. a soak where the number is not known) SHEL resolution limits of the anomalous signal. High resolution limit can be critical, but the default of d min Å works well in many cases. NTRY number of trials. 22 nd June 2017 SHELX C/D/E 13/26
15 Shelxd Output While shelxd runs, the best solution is written to mysad fa.res which contains the substructure coordinates in fractional coordinates and which is later read by shelxe. REM Best SHELXD solution: CC CC(weak) CFOM TITL mysad_fa.ins MAD in C2 CELL LATT -7 SYMM -X, Y, -Z SFAC SE UNIT 192 SE SE [...] SE SE SE <--- SE <--- SE HKLF 3 END The sixth column contains the occupancy of the corresponding atom. A sharp drop (here between SE12 and SE13) is a promising sign of a correct solution. The correlation coefficient (CC and CCweak) in the first line measures the reliability of the solution For SAD, a CC of more than 30 % is a safe sign of a correct solution, for MAD the limit is about 40 %. 22 nd June 2017 SHELX C/D/E 14/26
16 Shelxd Output SHELXD is very robust. Attention should be paid to 1. The resolution at which the data are truncated, e.g. where the internal CC (CC1/2) between the signed anomalous differences of two randomly chosen reflection subsets falls below 30%. 2. The number of sites requested should be within about 20% of the true value. 3. In the case of a soak, the rejection of sites on special positions should be switched off. 4. For S-SAD, DSUL (search for disulfides) can be very useful. 5. In difficult cases it may be necessary to run more trials (say 50000). 22 nd June 2017 SHELX C/D/E 15/26
17 Fine tuning SHELXD substructure solution SHELXD is very fast and robust, but achieves this with the help of drastic assumptions. In borderline cases it may be worth using the LLG (log likelihood gain) to distinguish substructure solutions, e.g. using the programs SHARP, CRANK2 or PHASER. For details see: SHARP Methods Enzymol. 276 (1997) ; Acta Cryst. D59 (2003) CRANK2 Acta Cryst. D67 (2011) ; Nat. Commun. 4:2777 (2013). PHASER (for experimental phasing) Acta Cryst. D60 (2004) ; Acta Cryst. D67 (2011) These programs could also be used to refine and extend the heavy atom substructure before density modification and poly-ala tracing with SHELXE. In general LLG-based methods require more detailed information (e.g. which elements are present) than SHELXC/D/E, and they tend to be slower. 22 nd June 2017 SHELX C/D/E 16/26
18 4 - Shelxe 22 nd June 2017 SHELX C/D/E 17/26
19 Shelxe: Phasing, Density Modification, Model Building No.ins-tructions file. All parameters provided as command line options after data file names. A typical and one of the most simple command line could be shelxe mysad mysad_fa -s0.65 -h -a mysad read native data mysad.hkl mysad fa read angle estimate for α from mysad fa.hkl, substructure coordinates from mysad fa.res (the shelxd output) -s0.65 Assume a solvent content of 65%. It should be reasonably well estimated. -h substructure atoms present in native data mysad.hkl -a run 5 (default) cycles of poly-ala autotracing. 22 nd June 2017 SHELX C/D/E 18/26
20 Shelxe -i: Inverted Substructure It is impossible to distinguish the substructure from its enantiomorph with the anomalous data and there is a 50 % chance that the coordinates in mysad fa.res are inverted w.r.t. the correct substructure. Therefore shelxe must always be run twice with the direct substructure with the inverted substructure, i.e. with the same options as the direct hand plus the switch -i. This inverts the hand and takes care of everything necessary inversion of screw axes, P4 1 to P4 3 off-axis inversion for I4 1 (1-x, 1/2-y, 1-z); I (1-x, 1/2-y, 1/4-z); F (1/4-x, 1/4-y, 1/4-z) output files are automatically amended by i to distinguish the two runs. N.B. if the inverted hand turns out to be the correct hand, your space group may change - e.g. in the presence of screw axes. Keep this in mind when you convert your native data to e.g. mtz-format! 22 nd June 2017 SHELX C/D/E 19/26
21 Caveat: Substructure Resolution Normal macromolecular structure: Determine atom positions even at e.g. 5 Å resolution because of restraints. Substructure unrestrained coordinates only know within resolution of anomalous signal, often much worse than 3 Å Way out: 1. e.g. Sharp improves substructure coordinates before density modification 2. substructure recycling with shelxe 22 nd June 2017 SHELX C/D/E 20/26
22 Shelxe: Substructure Recycling shelxd mymad_fa.res input finds shelxe improves rename Better substructure = better starting phases = better map Caveat: If the inverted structure turns out to be the correct hand (i.e. mysad i.hat from the -i-run of shelxe), the second run of shelxe must be run without the -i switch: mymad.hat 22 nd June 2017 SHELX C/D/E 21/26
23 Shelxe: Did it work? Criteria to tell if phasing worked: 1. Correct hand shows better Contrast, especially at early cycles of density modification. 2. Correct hand has higher map correlation coefficient throughout resolution range: d inf <mapcc> direct <mapcc> inverse 3. A reasonable poly-ala trace (average 10 residues per chain) and a CC > 25% When using the auto-tracing option (-a) in shelxe, the first two figures (contrast/ mapcc) become meaningless, but in this case the poly-ala trace is much more conclusive. 22 nd June 2017 SHELX C/D/E 22/26
24 Shelxe: Structure Solved? Indicators from shelxe: TITLE elastase.pdb Cycle 3 CC = 41.91% 226 residues in 4 chains TITLE elastase_i.pdb Cycle 3 CC = 7.26% 61 residues in 7 chains 1. CC>25% 2. average chain length > 10 (here: 56.5 vs. 8.7) 3. jump in CC over many cycles (e.g. with -a50) 22 nd June 2017 SHELX C/D/E 23/26
25 Shelxe: Structure Solved? Coot reads mysad.pdb (poly-ala trace) and mysad.phs (map). Elastase SAD tutorial Direct hand Inverse hand 22 nd June 2017 SHELX C/D/E 24/26
26 SHELXE: Current and Future Developments SHELXE started with -x and a reference PDB file name.ent is present: the mean phase error is output at various stages. The necessary origin shift is determined on the fly. If -h is also set, the program finds the atom in the reference file nearest to each heavy atom site. particularly useful for checking the substructure. This is The density modification has been improved for SAD phasing. For 20 test structures the mean phase improvement after the first round of density modification was 4.6. These improvements are already in the current distributed version. In addition, SHELXC is being adapted to handle multiple SAD datasets and a major rewrite of SHELXE is in progress. 22 nd June 2017 SHELX C/D/E 25/26
27 References and Further Reading 1. SHELX C/D/E: G. M. Sheldrick, Acta Cryst. (2010), D66, Visit the shelx web page for documentation, tutorials, etc.: Availability SHELX is available free for academic use via the SHELX homepage Extensive documentation and many links to useful programs may also be found there. SHELX C/D/E are also distributed along with CCP4. 22 nd June 2017 SHELX C/D/E 26/26
SHELXC/D/E. Andrea Thorn
 SHELXC/D/E Andrea Thorn What is experimental phasing? Experimental phasing is what you do if MR doesn t work. What is experimental phasing? Experimental phasing methods depend on intensity differences.
SHELXC/D/E Andrea Thorn What is experimental phasing? Experimental phasing is what you do if MR doesn t work. What is experimental phasing? Experimental phasing methods depend on intensity differences.
Macromolecular Phasing with shelxc/d/e
 Sunday, June 13 th, 2010 CCP4 Workshop APS Chicago, June 2010 http://shelx.uni-ac.gwdg.de Overview Substructure Definition and Motivation Extracting Substructure Data from measured Data Substructure Solution
Sunday, June 13 th, 2010 CCP4 Workshop APS Chicago, June 2010 http://shelx.uni-ac.gwdg.de Overview Substructure Definition and Motivation Extracting Substructure Data from measured Data Substructure Solution
CCP4 Diamond 2014 SHELXC/D/E. Andrea Thorn
 CCP4 Diamond 2014 SHELXC/D/E Andrea Thorn SHELXC/D/E workflow SHELXC: α calculation, file preparation SHELXD: Marker atom search = substructure search SHELXE: density modification Maps and coordinate files
CCP4 Diamond 2014 SHELXC/D/E Andrea Thorn SHELXC/D/E workflow SHELXC: α calculation, file preparation SHELXD: Marker atom search = substructure search SHELXE: density modification Maps and coordinate files
The SHELX approach to the experimental phasing of macromolecules. George M. Sheldrick, Göttingen University
 The SHELX approach to the experimental phasing of macromolecules IUCr 2011 Madrid George M. Sheldrick, Göttingen University http://shelx.uni-ac.gwdg.de/shelx/ Experimental phasing of macromolecules Except
The SHELX approach to the experimental phasing of macromolecules IUCr 2011 Madrid George M. Sheldrick, Göttingen University http://shelx.uni-ac.gwdg.de/shelx/ Experimental phasing of macromolecules Except
Experimental phasing, Pattersons and SHELX Andrea Thorn
 Experimental phasing, Pattersons and SHELX Andrea Thorn What is experimental phasing? Experimental phasing is what you do if MR doesn t work. What is experimental phasing? Experimental phasing methods
Experimental phasing, Pattersons and SHELX Andrea Thorn What is experimental phasing? Experimental phasing is what you do if MR doesn t work. What is experimental phasing? Experimental phasing methods
Determination of the Substructure
 Monday, June 15 th, 2009 Determination of the Substructure EMBO / MAX-INF2 Practical Course http://shelx.uni-ac.gwdg.de Overview Substructure Definition and Motivation Extracting Substructure Data from
Monday, June 15 th, 2009 Determination of the Substructure EMBO / MAX-INF2 Practical Course http://shelx.uni-ac.gwdg.de Overview Substructure Definition and Motivation Extracting Substructure Data from
PAN-modular Structure of Parasite Sarcocystis muris Microneme Protein SML-2 at 1.95 Å Resolution and the Complex with 1-Thio-β-D-Galactose
 Supplementary Material to the paper: PAN-modular Structure of Parasite Sarcocystis muris Microneme Protein SML-2 at 1.95 Å Resolution and the Complex with 1-Thio-β-D-Galactose Jürgen J. Müller, a Manfred
Supplementary Material to the paper: PAN-modular Structure of Parasite Sarcocystis muris Microneme Protein SML-2 at 1.95 Å Resolution and the Complex with 1-Thio-β-D-Galactose Jürgen J. Müller, a Manfred
Phaser: Experimental phasing
 Phaser: Experimental phasing Using SAD data in Phaser R J Read, Department of Haematology Cambridge Institute for Medical Research Diffraction with anomalous scatterers SAD: single-wavelength anomalous
Phaser: Experimental phasing Using SAD data in Phaser R J Read, Department of Haematology Cambridge Institute for Medical Research Diffraction with anomalous scatterers SAD: single-wavelength anomalous
Web-based Auto-Rickshaw for validation of the X-ray experiment at the synchrotron beamline
 Web-based Auto-Rickshaw for validation of the X-ray experiment at the synchrotron beamline Auto-Rickshaw http://www.embl-hamburg.de/auto-rickshaw A platform for automated crystal structure determination
Web-based Auto-Rickshaw for validation of the X-ray experiment at the synchrotron beamline Auto-Rickshaw http://www.embl-hamburg.de/auto-rickshaw A platform for automated crystal structure determination
ANODE: ANOmalous and heavy atom DEnsity
 Bruker User s Meeting 2012 ANODE: ANOmalous and heavy atom DEnsity Andrea Thorn September 26 th, 2012 Typical SHELXC/D/E workflow The anomalous or heavy atom signal is used to find the substructure of
Bruker User s Meeting 2012 ANODE: ANOmalous and heavy atom DEnsity Andrea Thorn September 26 th, 2012 Typical SHELXC/D/E workflow The anomalous or heavy atom signal is used to find the substructure of
Likelihood and SAD phasing in Phaser. R J Read, Department of Haematology Cambridge Institute for Medical Research
 Likelihood and SAD phasing in Phaser R J Read, Department of Haematology Cambridge Institute for Medical Research Concept of likelihood Likelihood with dice 4 6 8 10 Roll a seven. Which die?? p(4)=p(6)=0
Likelihood and SAD phasing in Phaser R J Read, Department of Haematology Cambridge Institute for Medical Research Concept of likelihood Likelihood with dice 4 6 8 10 Roll a seven. Which die?? p(4)=p(6)=0
ANODE: ANOmalous and heavy atom DEnsity
 Bruker Webinar ANODE: ANOmalous and heavy atom DEnsity Andrea Thorn 03 December, 2013 Michael Ruf Welcome Dr. Michael Ruf Product Manager, SC-XRD Bruker AXS Inc. Madison, WI, USA Andrea Thorn Crystallographic
Bruker Webinar ANODE: ANOmalous and heavy atom DEnsity Andrea Thorn 03 December, 2013 Michael Ruf Welcome Dr. Michael Ruf Product Manager, SC-XRD Bruker AXS Inc. Madison, WI, USA Andrea Thorn Crystallographic
Practical aspects of SAD/MAD. Judit É Debreczeni
 Practical aspects of SAD/MAD Judit É Debreczeni anomalous scattering Hg sinθ/λ CuKα 0 Å - 0.4 Å - 0.6 Å - Å.5Å 0.83Å f 80 53 4 f total (θ, λ) f(θ) + f (λ) + if (λ) f f -5 8-5 8-5 8 f total increasing with
Practical aspects of SAD/MAD Judit É Debreczeni anomalous scattering Hg sinθ/λ CuKα 0 Å - 0.4 Å - 0.6 Å - Å.5Å 0.83Å f 80 53 4 f total (θ, λ) f(θ) + f (λ) + if (λ) f f -5 8-5 8-5 8 f total increasing with
Protein Crystallography
 Protein Crystallography Part II Tim Grüne Dept. of Structural Chemistry Prof. G. Sheldrick University of Göttingen http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview The Reciprocal Lattice The
Protein Crystallography Part II Tim Grüne Dept. of Structural Chemistry Prof. G. Sheldrick University of Göttingen http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview The Reciprocal Lattice The
Tutorial on how to solve a Se-substructure using
 1 Introduction Tutorial on how to solve a Se-substructure using SHELXD Thomas R. Schneider Dept. of Structural Chemistry University of Göttingen trs@shelx.uni-ac.gwdg.de July 4, 2002 The Solution of the
1 Introduction Tutorial on how to solve a Se-substructure using SHELXD Thomas R. Schneider Dept. of Structural Chemistry University of Göttingen trs@shelx.uni-ac.gwdg.de July 4, 2002 The Solution of the
Experimental phasing in Crank2
 Experimental phasing in Crank2 Pavol Skubak and Navraj Pannu Biophysical Structural Chemistry, Leiden University, The Netherlands http://www.bfsc.leidenuniv.nl/software/crank/ X-ray structure solution
Experimental phasing in Crank2 Pavol Skubak and Navraj Pannu Biophysical Structural Chemistry, Leiden University, The Netherlands http://www.bfsc.leidenuniv.nl/software/crank/ X-ray structure solution
Molecular Biology Course 2006 Protein Crystallography Part II
 Molecular Biology Course 2006 Protein Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry December 2006 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview
Molecular Biology Course 2006 Protein Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry December 2006 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview
Direct Methods and Many Site Se-Met MAD Problems using BnP. W. Furey
 Direct Methods and Many Site Se-Met MAD Problems using BnP W. Furey Classical Direct Methods Main method for small molecule structure determination Highly automated (almost totally black box ) Solves structures
Direct Methods and Many Site Se-Met MAD Problems using BnP W. Furey Classical Direct Methods Main method for small molecule structure determination Highly automated (almost totally black box ) Solves structures
Macromolecular Crystallography Part II
 Molecular Biology Course 2009 Macromolecular Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2009 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de From
Molecular Biology Course 2009 Macromolecular Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2009 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de From
Introduction to SHELXD
 SHELXD and SHELXE The structure solution program SHELXD (called XM in the Bruker SHELXTL system, but identical to SHELXD except in the logo) is able to solve larger ab initio problems than SHELXS-97, and
SHELXD and SHELXE The structure solution program SHELXD (called XM in the Bruker SHELXTL system, but identical to SHELXD except in the logo) is able to solve larger ab initio problems than SHELXS-97, and
Protein Crystallography Part II
 Molecular Biology Course 2007 Protein Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2007 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview
Molecular Biology Course 2007 Protein Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2007 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview
Experimental phasing in Crank2
 Experimental phasing in Crank2 Pavol Skubak and Navraj Pannu Biophysical Structural Chemistry, Leiden University, The Netherlands http://www.bfsc.leidenuniv.nl/software/crank/ Crank2 for experimental phasing
Experimental phasing in Crank2 Pavol Skubak and Navraj Pannu Biophysical Structural Chemistry, Leiden University, The Netherlands http://www.bfsc.leidenuniv.nl/software/crank/ Crank2 for experimental phasing
Molecular Biology Course 2006 Protein Crystallography Part I
 Molecular Biology Course 2006 Protein Crystallography Part I Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2006 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview Overview
Molecular Biology Course 2006 Protein Crystallography Part I Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2006 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview Overview
Direct Method. Very few protein diffraction data meet the 2nd condition
 Direct Method Two conditions: -atoms in the structure are equal-weighted -resolution of data are higher than the distance between the atoms in the structure Very few protein diffraction data meet the 2nd
Direct Method Two conditions: -atoms in the structure are equal-weighted -resolution of data are higher than the distance between the atoms in the structure Very few protein diffraction data meet the 2nd
Structure solution from weak anomalous data
 Structure solution from weak anomalous data Phenix Workshop SBGrid-NE-CAT Computing School Harvard Medical School, Boston June 7, 2014 Gábor Bunkóczi, Airlie McCoy, Randy Read (Cambridge University) Nat
Structure solution from weak anomalous data Phenix Workshop SBGrid-NE-CAT Computing School Harvard Medical School, Boston June 7, 2014 Gábor Bunkóczi, Airlie McCoy, Randy Read (Cambridge University) Nat
Fast, Intuitive Structure Determination IV: Space Group Determination and Structure Solution
 Fast, Intuitive Structure Determination IV: Space Group Determination and Structure Solution November 25, 2013 Welcome I I Dr. Michael Ruf Product Manager Crystallography Bruker AXS Inc. Madison, WI, USA
Fast, Intuitive Structure Determination IV: Space Group Determination and Structure Solution November 25, 2013 Welcome I I Dr. Michael Ruf Product Manager Crystallography Bruker AXS Inc. Madison, WI, USA
Biology III: Crystallographic phases
 Haupt/Masterstudiengang Physik Methoden moderner Röntgenphysik II: Streuung und Abbildung SS 2013 Biology III: Crystallographic phases Thomas R. Schneider, EMBL Hamburg 25/6/2013 thomas.schneider@embl-hamburg.de
Haupt/Masterstudiengang Physik Methoden moderner Röntgenphysik II: Streuung und Abbildung SS 2013 Biology III: Crystallographic phases Thomas R. Schneider, EMBL Hamburg 25/6/2013 thomas.schneider@embl-hamburg.de
Phase problem: Determining an initial phase angle α hkl for each recorded reflection. 1 ρ(x,y,z) = F hkl cos 2π (hx+ky+ lz - α hkl ) V h k l
 Phase problem: Determining an initial phase angle α hkl for each recorded reflection 1 ρ(x,y,z) = F hkl cos 2π (hx+ky+ lz - α hkl ) V h k l Methods: Heavy atom methods (isomorphous replacement Hg, Pt)
Phase problem: Determining an initial phase angle α hkl for each recorded reflection 1 ρ(x,y,z) = F hkl cos 2π (hx+ky+ lz - α hkl ) V h k l Methods: Heavy atom methods (isomorphous replacement Hg, Pt)
Molecular replacement. New structures from old
 Molecular replacement New structures from old The Phase Problem phase amplitude Phasing by molecular replacement Phases can be calculated from atomic model Rotate and translate related structure Models
Molecular replacement New structures from old The Phase Problem phase amplitude Phasing by molecular replacement Phases can be calculated from atomic model Rotate and translate related structure Models
Electronic Supplementary Information (ESI) for Chem. Commun. Unveiling the three- dimensional structure of the green pigment of nitrite- cured meat
 Electronic Supplementary Information (ESI) for Chem. Commun. Unveiling the three- dimensional structure of the green pigment of nitrite- cured meat Jun Yi* and George B. Richter- Addo* Department of Chemistry
Electronic Supplementary Information (ESI) for Chem. Commun. Unveiling the three- dimensional structure of the green pigment of nitrite- cured meat Jun Yi* and George B. Richter- Addo* Department of Chemistry
Twinning. Andrea Thorn
 Twinning Andrea Thorn OVERVIEW Introduction: Definitions, origins of twinning Merohedral twins: Recognition, statistical analysis: H plot, Yeates Padilla plot Example Refinement and R values Reticular
Twinning Andrea Thorn OVERVIEW Introduction: Definitions, origins of twinning Merohedral twins: Recognition, statistical analysis: H plot, Yeates Padilla plot Example Refinement and R values Reticular
Space Group & Structure Solution
 Space Group & Structure Solution Determine the Space Group Space group determination can always be performed by hand by examining the intensity data. A program that can facilitate this step is the command-prompt
Space Group & Structure Solution Determine the Space Group Space group determination can always be performed by hand by examining the intensity data. A program that can facilitate this step is the command-prompt
ACORN - a flexible and efficient ab initio procedure to solve a protein structure when atomic resolution data is available
 ACORN - a flexible and efficient ab initio procedure to solve a protein structure when atomic resolution data is available Yao Jia-xing Department of Chemistry, University of York, Heslington, York, YO10
ACORN - a flexible and efficient ab initio procedure to solve a protein structure when atomic resolution data is available Yao Jia-xing Department of Chemistry, University of York, Heslington, York, YO10
Homework 1 (not graded) X-ray Diffractometry CHE Multiple Choice. 1. One of the methods of reducing exposure to radiation is to minimize.
 Homework 1 (not graded) X-ray Diffractometry CHE 380.45 Multiple Choice 1. One of the methods of reducing exposure to radiation is to minimize. a) distance b) humidity c) time d) speed e) shielding 2.
Homework 1 (not graded) X-ray Diffractometry CHE 380.45 Multiple Choice 1. One of the methods of reducing exposure to radiation is to minimize. a) distance b) humidity c) time d) speed e) shielding 2.
Charles Ballard (original GáborBunkóczi) CCP4 Workshop 7 December 2011
 Experimental phasing Charles Ballard (original GáborBunkóczi) CCP4 Workshop 7 December 2011 Anomalous diffraction F P protein F A anomalous substructure ano F A " -FA" A F F -* F A F P Phasing Substructure
Experimental phasing Charles Ballard (original GáborBunkóczi) CCP4 Workshop 7 December 2011 Anomalous diffraction F P protein F A anomalous substructure ano F A " -FA" A F F -* F A F P Phasing Substructure
Electron Crystallography for Structure Solution Latest Results from the PSI Electron Diffraction Group
 WIR SCHAFFEN WISSEN HEUTE FÜR MORGEN Dr. Tim Grüne :: Paul Scherrer Institut :: tim.gruene@psi.ch Electron Crystallography for Structure Solution Latest Results from the PSI Electron Diffraction Group
WIR SCHAFFEN WISSEN HEUTE FÜR MORGEN Dr. Tim Grüne :: Paul Scherrer Institut :: tim.gruene@psi.ch Electron Crystallography for Structure Solution Latest Results from the PSI Electron Diffraction Group
What's new in SHELXL-2013?
 Bruker Webinar What's new in SHELXL-2013? Andrea Thorn 12 th September, 2013 Michael Ruf & Bruce C. Noll Welcome Dr. Michael Ruf Product Manager, SC-XRD Bruker AXS Inc. Madison, WI, USA Andrea Thorn Crystallographic
Bruker Webinar What's new in SHELXL-2013? Andrea Thorn 12 th September, 2013 Michael Ruf & Bruce C. Noll Welcome Dr. Michael Ruf Product Manager, SC-XRD Bruker AXS Inc. Madison, WI, USA Andrea Thorn Crystallographic
X-ray Crystallography. Kalyan Das
 X-ray Crystallography Kalyan Das Electromagnetic Spectrum NMR 10 um - 10 mm 700 to 10 4 nm 400 to 700 nm 10 to 400 nm 10-1 to 10 nm 10-4 to 10-1 nm X-ray radiation was discovered by Roentgen in 1895. X-rays
X-ray Crystallography Kalyan Das Electromagnetic Spectrum NMR 10 um - 10 mm 700 to 10 4 nm 400 to 700 nm 10 to 400 nm 10-1 to 10 nm 10-4 to 10-1 nm X-ray radiation was discovered by Roentgen in 1895. X-rays
Recent developments in Crank. Leiden University, The Netherlands
 Recent developments in Crank Navraj js. Pannu Leiden University, The Netherlands Current developers Pavol Skubak Ruben Zubac Irakli Sikharulidze Jan Pieter Abrahams RAG de Graaff Willem-Jan Waterreus Substructure
Recent developments in Crank Navraj js. Pannu Leiden University, The Netherlands Current developers Pavol Skubak Ruben Zubac Irakli Sikharulidze Jan Pieter Abrahams RAG de Graaff Willem-Jan Waterreus Substructure
Non-merohedral Twinning in Protein Crystallography
 Bruker Users Meeting 2010 Karlsruhe, 22 nd September 2010 Non-merohedral Twinning in Protein Crystallography Regine Herbst-Irmer rherbst@shelx.uni-ac.gwdg.de http://shelx.uni-ac.gwdg.de/~rherbst/twin.html
Bruker Users Meeting 2010 Karlsruhe, 22 nd September 2010 Non-merohedral Twinning in Protein Crystallography Regine Herbst-Irmer rherbst@shelx.uni-ac.gwdg.de http://shelx.uni-ac.gwdg.de/~rherbst/twin.html
Anisotropy in macromolecular crystal structures. Andrea Thorn July 19 th, 2012
 Anisotropy in macromolecular crystal structures Andrea Thorn July 19 th, 2012 Motivation Courtesy of M. Sawaya Motivation Crystal structures are inherently anisotropic. X-ray diffraction reflects this
Anisotropy in macromolecular crystal structures Andrea Thorn July 19 th, 2012 Motivation Courtesy of M. Sawaya Motivation Crystal structures are inherently anisotropic. X-ray diffraction reflects this
Macromolecular Crystallography Part II
 Molecular Biology Course 2010 Macromolecular Crystallography Part II University of Göttingen Dept. of Structural Chemistry November 2010 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Crystallography
Molecular Biology Course 2010 Macromolecular Crystallography Part II University of Göttingen Dept. of Structural Chemistry November 2010 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Crystallography
MR model selection, preparation and assessing the solution
 Ronan Keegan CCP4 Group MR model selection, preparation and assessing the solution DLS-CCP4 Data Collection and Structure Solution Workshop 2018 Overview Introduction Step-by-step guide to performing Molecular
Ronan Keegan CCP4 Group MR model selection, preparation and assessing the solution DLS-CCP4 Data Collection and Structure Solution Workshop 2018 Overview Introduction Step-by-step guide to performing Molecular
shelxl: Refinement of Macromolecular Structures from Neutron Data
 ESS Neutron Protein Crystallography 2013 Aarhus, Denmark shelxl: Refinement of Macromolecular Structures from Neutron Data Tim Grüne University of Göttingen Dept. of Structural Chemistry http://shelx.uni-ac.gwdg.de
ESS Neutron Protein Crystallography 2013 Aarhus, Denmark shelxl: Refinement of Macromolecular Structures from Neutron Data Tim Grüne University of Göttingen Dept. of Structural Chemistry http://shelx.uni-ac.gwdg.de
S-SAD and Fe-SAD Phasing using X8 PROTEUM
 S-SAD and Fe-SAD Phasing using X8 PROTEUM Kristina Djinovic Carugo Dept. for Structural and Computational Biology Max F. Perutz Labs Univ. Vienna, Austria Outline Fe-SAD on chlorite dismutase from Candidatus
S-SAD and Fe-SAD Phasing using X8 PROTEUM Kristina Djinovic Carugo Dept. for Structural and Computational Biology Max F. Perutz Labs Univ. Vienna, Austria Outline Fe-SAD on chlorite dismutase from Candidatus
Patterson Methods
 59-553 Patterson Methods 113 In 1935, Patterson showed that the unknown phase information in the equation for electron density: ρ(xyz) = 1/V h k l F(hkl) exp[iα(hkl)] exp[-2πi(h x + k y + l z)] can be
59-553 Patterson Methods 113 In 1935, Patterson showed that the unknown phase information in the equation for electron density: ρ(xyz) = 1/V h k l F(hkl) exp[iα(hkl)] exp[-2πi(h x + k y + l z)] can be
Garib N Murshudov MRC-LMB, Cambridge
 Garib N Murshudov MRC-LMB, Cambridge Contents Introduction AceDRG: two functions Validation of entries in the DB and derived data Generation of new ligand description Jligand for link description Conclusions
Garib N Murshudov MRC-LMB, Cambridge Contents Introduction AceDRG: two functions Validation of entries in the DB and derived data Generation of new ligand description Jligand for link description Conclusions
Anomalous dispersion
 Selenomethionine MAD Selenomethionine is the amino acid methionine with the Sulfur replaced by a Selenium. Selenium is a heavy atom that also has the propery of "anomalous scatter" at some wavelengths,
Selenomethionine MAD Selenomethionine is the amino acid methionine with the Sulfur replaced by a Selenium. Selenium is a heavy atom that also has the propery of "anomalous scatter" at some wavelengths,
Protein crystallography. Garry Taylor
 Protein crystallography Garry Taylor X-ray Crystallography - the Basics Grow crystals Collect X-ray data Determine phases Calculate ρ-map Interpret map Refine coordinates Do the biology. Nitrogen at -180
Protein crystallography Garry Taylor X-ray Crystallography - the Basics Grow crystals Collect X-ray data Determine phases Calculate ρ-map Interpret map Refine coordinates Do the biology. Nitrogen at -180
Direct-method SAD phasing with partial-structure iteration: towards automation
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Editors: E. N. Baker and Z. Dauter Direct-method SAD phasing with partial-structure iteration: towards automation J. W. Wang,
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Editors: E. N. Baker and Z. Dauter Direct-method SAD phasing with partial-structure iteration: towards automation J. W. Wang,
Scattering by two Electrons
 Scattering by two Electrons p = -r k in k in p r e 2 q k in /λ θ θ k out /λ S q = r k out p + q = r (k out - k in ) e 1 Phase difference of wave 2 with respect to wave 1: 2π λ (k out - k in ) r= 2π S r
Scattering by two Electrons p = -r k in k in p r e 2 q k in /λ θ θ k out /λ S q = r k out p + q = r (k out - k in ) e 1 Phase difference of wave 2 with respect to wave 1: 2π λ (k out - k in ) r= 2π S r
11/18/2013 SHELX. History SHELXS. .ins File for SHELXS TITL. .ins File Organization
 SHELX George Sheldrick British 1976 It is surprising that SHELX has been able to maintain a dominant position in small-molecule structure determination for the last 30 years, even though computers have
SHELX George Sheldrick British 1976 It is surprising that SHELX has been able to maintain a dominant position in small-molecule structure determination for the last 30 years, even though computers have
research papers HKL-3000: the integration of data reduction and structure solution from diffraction images to an initial model in minutes
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 HKL-3000: the integration of data reduction and structure solution from diffraction images to an initial model in minutes Wladek
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 HKL-3000: the integration of data reduction and structure solution from diffraction images to an initial model in minutes Wladek
research papers Reduction of density-modification bias by b correction 1. Introduction Pavol Skubák* and Navraj S. Pannu
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Reduction of density-modification bias by b correction Pavol Skubák* and Navraj S. Pannu Biophysical Structural Chemistry, Leiden
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Reduction of density-modification bias by b correction Pavol Skubák* and Navraj S. Pannu Biophysical Structural Chemistry, Leiden
Pipelining Ligands in PHENIX: elbow and REEL
 Pipelining Ligands in PHENIX: elbow and REEL Nigel W. Moriarty Lawrence Berkeley National Laboratory Physical Biosciences Division Ligands in Crystallography Drug design Biological function studies Generate
Pipelining Ligands in PHENIX: elbow and REEL Nigel W. Moriarty Lawrence Berkeley National Laboratory Physical Biosciences Division Ligands in Crystallography Drug design Biological function studies Generate
Rietveld Structure Refinement of Protein Powder Diffraction Data using GSAS
 Rietveld Structure Refinement of Protein Powder Diffraction Data using GSAS Jon Wright ESRF, Grenoble, France Plan This is a users perspective Cover the protein specific aspects (assuming knowledge of
Rietveld Structure Refinement of Protein Powder Diffraction Data using GSAS Jon Wright ESRF, Grenoble, France Plan This is a users perspective Cover the protein specific aspects (assuming knowledge of
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/NCHEM.1397 Crystal structures of Λ-[Ru(phen) 2 dppz] 2+ with oligonucleotides containing TA/TA and AT/AT steps show two intercalation modes Hakan Niyazi a, 1 James P. Hall a, Kyra O Sullivan
DOI: 10.1038/NCHEM.1397 Crystal structures of Λ-[Ru(phen) 2 dppz] 2+ with oligonucleotides containing TA/TA and AT/AT steps show two intercalation modes Hakan Niyazi a, 1 James P. Hall a, Kyra O Sullivan
Supporting Information
 Wiley-VCH 2007 69451 Weinheim, Germany On the polymorphism of aspirin Andrew D. Bond, Roland Boese and Gautam R. Desiraju S1. Comparison of the form I and PZ structures S2. Transforming the unit cells
Wiley-VCH 2007 69451 Weinheim, Germany On the polymorphism of aspirin Andrew D. Bond, Roland Boese and Gautam R. Desiraju S1. Comparison of the form I and PZ structures S2. Transforming the unit cells
Image definition evaluation functions for X-ray crystallography: A new perspective on the phase. problem. Hui LI*, Meng HE* and Ze ZHANG
 Image definition evaluation functions for X-ray crystallography: A new perspective on the phase problem Hui LI*, Meng HE* and Ze ZHANG Beijing University of Technology, Beijing 100124, People s Republic
Image definition evaluation functions for X-ray crystallography: A new perspective on the phase problem Hui LI*, Meng HE* and Ze ZHANG Beijing University of Technology, Beijing 100124, People s Republic
BC530 Class notes on X-ray Crystallography
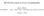 BC530 Class notes on X-ray Crystallography web material: Ethan A Merritt http://skuld.bmsc.washington.edu/~merritt/bc530/ October 11, 2016 Growing Crystals It should be self-evident that in order to do
BC530 Class notes on X-ray Crystallography web material: Ethan A Merritt http://skuld.bmsc.washington.edu/~merritt/bc530/ October 11, 2016 Growing Crystals It should be self-evident that in order to do
Crystal lattice Real Space. Reflections Reciprocal Space. I. Solving Phases II. Model Building for CHEM 645. Purified Protein. Build model.
 I. Solving Phases II. Model Building for CHEM 645 Purified Protein Solve Phase Build model and refine Crystal lattice Real Space Reflections Reciprocal Space ρ (x, y, z) pronounced rho F hkl 2 I F (h,
I. Solving Phases II. Model Building for CHEM 645 Purified Protein Solve Phase Build model and refine Crystal lattice Real Space Reflections Reciprocal Space ρ (x, y, z) pronounced rho F hkl 2 I F (h,
Materials 286C/UCSB: Class VI Structure factors (continued), the phase problem, Patterson techniques and direct methods
 Materials 286C/UCSB: Class VI Structure factors (continued), the phase problem, Patterson techniques and direct methods Ram Seshadri (seshadri@mrl.ucsb.edu) Structure factors The structure factor for a
Materials 286C/UCSB: Class VI Structure factors (continued), the phase problem, Patterson techniques and direct methods Ram Seshadri (seshadri@mrl.ucsb.edu) Structure factors The structure factor for a
The Crystallographic Process
 Experimental Phasing Macromolecular Crystallography School Madrid, May 2017 Paul Adams Lawrence Berkeley Laboratory and Department of Bioengineering UC Berkeley The Crystallographic Process Crystallization
Experimental Phasing Macromolecular Crystallography School Madrid, May 2017 Paul Adams Lawrence Berkeley Laboratory and Department of Bioengineering UC Berkeley The Crystallographic Process Crystallization
X-ray Crystallography
 2009/11/25 [ 1 ] X-ray Crystallography Andrew Torda, wintersemester 2009 / 2010 X-ray numerically most important more than 4/5 structures Goal a set of x, y, z coordinates different properties to NMR History
2009/11/25 [ 1 ] X-ray Crystallography Andrew Torda, wintersemester 2009 / 2010 X-ray numerically most important more than 4/5 structures Goal a set of x, y, z coordinates different properties to NMR History
research papers 1. Introduction Thomas C. Terwilliger a * and Joel Berendzen b
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy-atom solutions in the MIR and
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy-atom solutions in the MIR and
 Helpful resources for all X ray lectures Crystallization http://www.hamptonresearch.com under tech support: crystal growth 101 literature Spacegroup tables http://img.chem.ucl.ac.uk/sgp/mainmenu.htm Crystallography
Helpful resources for all X ray lectures Crystallization http://www.hamptonresearch.com under tech support: crystal growth 101 literature Spacegroup tables http://img.chem.ucl.ac.uk/sgp/mainmenu.htm Crystallography
X-ray Diffraction. Diffraction. X-ray Generation. X-ray Generation. X-ray Generation. X-ray Spectrum from Tube
 X-ray Diffraction Mineral identification Mode analysis Structure Studies X-ray Generation X-ray tube (sealed) Pure metal target (Cu) Electrons remover inner-shell electrons from target. Other electrons
X-ray Diffraction Mineral identification Mode analysis Structure Studies X-ray Generation X-ray tube (sealed) Pure metal target (Cu) Electrons remover inner-shell electrons from target. Other electrons
Acta Cryst. (2017). D73, doi: /s
 Acta Cryst. (2017). D73, doi:10.1107/s2059798317010932 Supporting information Volume 73 (2017) Supporting information for article: Designing better diffracting crystals of biotin carboxyl carrier protein
Acta Cryst. (2017). D73, doi:10.1107/s2059798317010932 Supporting information Volume 73 (2017) Supporting information for article: Designing better diffracting crystals of biotin carboxyl carrier protein
PSD '17 -- Xray Lecture 5, 6. Patterson Space, Molecular Replacement and Heavy Atom Isomorphous Replacement
 PSD '17 -- Xray Lecture 5, 6 Patterson Space, Molecular Replacement and Heavy Atom Isomorphous Replacement The Phase Problem We can t measure the phases! X-ray detectors (film, photomultiplier tubes, CCDs,
PSD '17 -- Xray Lecture 5, 6 Patterson Space, Molecular Replacement and Heavy Atom Isomorphous Replacement The Phase Problem We can t measure the phases! X-ray detectors (film, photomultiplier tubes, CCDs,
Resolution and data formats. Andrea Thorn
 Resolution and data formats Andrea Thorn RESOLUTION Motivation Courtesy of M. Sawaya Map resolution http://www.bmsc.washington.edu/people/verlinde/experiment.html Data quality indicators Resolution accounts
Resolution and data formats Andrea Thorn RESOLUTION Motivation Courtesy of M. Sawaya Map resolution http://www.bmsc.washington.edu/people/verlinde/experiment.html Data quality indicators Resolution accounts
SOLVE and RESOLVE: automated structure solution, density modification and model building
 Journal of Synchrotron Radiation ISSN 0909-0495 SOLVE and RESOLVE: automated structure solution, density modification and model building Thomas Terwilliger Copyright International Union of Crystallography
Journal of Synchrotron Radiation ISSN 0909-0495 SOLVE and RESOLVE: automated structure solution, density modification and model building Thomas Terwilliger Copyright International Union of Crystallography
X-ray Crystallography I. James Fraser Macromolecluar Interactions BP204
 X-ray Crystallography I James Fraser Macromolecluar Interactions BP204 Key take-aways 1. X-ray crystallography results from an ensemble of Billions and Billions of molecules in the crystal 2. Models in
X-ray Crystallography I James Fraser Macromolecluar Interactions BP204 Key take-aways 1. X-ray crystallography results from an ensemble of Billions and Billions of molecules in the crystal 2. Models in
Acta Crystallographica Section F
 Supporting information Acta Crystallographica Section F Volume 70 (2014) Supporting information for article: Chemical conversion of cisplatin and carboplatin with histidine in a model protein crystallised
Supporting information Acta Crystallographica Section F Volume 70 (2014) Supporting information for article: Chemical conversion of cisplatin and carboplatin with histidine in a model protein crystallised
Summary of Experimental Protein Structure Determination. Key Elements
 Programme 8.00-8.20 Summary of last week s lecture and quiz 8.20-9.00 Structure validation 9.00-9.15 Break 9.15-11.00 Exercise: Structure validation tutorial 11.00-11.10 Break 11.10-11.40 Summary & discussion
Programme 8.00-8.20 Summary of last week s lecture and quiz 8.20-9.00 Structure validation 9.00-9.15 Break 9.15-11.00 Exercise: Structure validation tutorial 11.00-11.10 Break 11.10-11.40 Summary & discussion
Fan, Hai-fu Institute of Physics, Chinese Academy of Sciences, Beijing , China
 Direct Methods in Crystallography Fan, Hai-fu Institute of Physics, Chinese Academy of Sciences, Beijing 100080, China An important branch of crystallography is the X-ray diffraction analysis of crystal
Direct Methods in Crystallography Fan, Hai-fu Institute of Physics, Chinese Academy of Sciences, Beijing 100080, China An important branch of crystallography is the X-ray diffraction analysis of crystal
4. Constraints and Hydrogen Atoms
 4. Constraints and ydrogen Atoms 4.1 Constraints versus restraints In crystal structure refinement, there is an important distinction between a constraint and a restraint. A constraint is an exact mathematical
4. Constraints and ydrogen Atoms 4.1 Constraints versus restraints In crystal structure refinement, there is an important distinction between a constraint and a restraint. A constraint is an exact mathematical
Linking data and model quality in macromolecular crystallography. Kay Diederichs
 Linking data and model quality in macromolecular crystallography Kay Diederichs Crystallography is highly successful Can we do better? Error in experimental data Error = random + systematic Multiplicity
Linking data and model quality in macromolecular crystallography Kay Diederichs Crystallography is highly successful Can we do better? Error in experimental data Error = random + systematic Multiplicity
Handout 13 Interpreting your results. What to make of your atomic coordinates, bond distances and angles
 Handout 13 Interpreting your results What to make of your atomic coordinates, bond distances and angles 1 What to make of the outcome of your refinement There are several ways of judging whether the outcome
Handout 13 Interpreting your results What to make of your atomic coordinates, bond distances and angles 1 What to make of the outcome of your refinement There are several ways of judging whether the outcome
TLS and all that. Ethan A Merritt. CCP4 Summer School 2011 (Argonne, IL) Abstract
 TLS and all that Ethan A Merritt CCP4 Summer School 2011 (Argonne, IL) Abstract We can never know the position of every atom in a crystal structure perfectly. Each atom has an associated positional uncertainty.
TLS and all that Ethan A Merritt CCP4 Summer School 2011 (Argonne, IL) Abstract We can never know the position of every atom in a crystal structure perfectly. Each atom has an associated positional uncertainty.
ACORN in CCP4 and its applications
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 ACORN in CCP4 and its applications Jia-xing Yao York Structural Biology Laboratory, Department of Chemistry, University of York,
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 ACORN in CCP4 and its applications Jia-xing Yao York Structural Biology Laboratory, Department of Chemistry, University of York,
Dictionary of ligands
 Dictionary of ligands Some of the web and other resources Small molecules DrugBank: http://www.drugbank.ca/ ZINC: http://zinc.docking.org/index.shtml PRODRUG: http://www.compbio.dundee.ac.uk/web_servers/prodrg_down.html
Dictionary of ligands Some of the web and other resources Small molecules DrugBank: http://www.drugbank.ca/ ZINC: http://zinc.docking.org/index.shtml PRODRUG: http://www.compbio.dundee.ac.uk/web_servers/prodrg_down.html
Refine & Validate. In the *.res file, be sure to add the following four commands after the UNIT instruction and before any atoms: ACTA CONF WPDB -2
 Refine & Validate Refinement is simply a way to improve the fit between the measured intensities and the intensities calculated from the model. The peaks in the difference map and the list of worst fitting
Refine & Validate Refinement is simply a way to improve the fit between the measured intensities and the intensities calculated from the model. The peaks in the difference map and the list of worst fitting
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 14, 2018 02:00 pm GMT PDB ID : 3RRQ Title : Crystal structure of the extracellular domain of human PD-1 Authors : Lazar-Molnar, E.; Ramagopal, U.A.; Nathenson,
Full wwpdb X-ray Structure Validation Report i Mar 14, 2018 02:00 pm GMT PDB ID : 3RRQ Title : Crystal structure of the extracellular domain of human PD-1 Authors : Lazar-Molnar, E.; Ramagopal, U.A.; Nathenson,
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Jan 14, 2019 11:10 AM EST PDB ID : 6GYW Title : Crystal structure of DacA from Staphylococcus aureus Authors : Tosi, T.; Freemont, P.S.; Grundling, A. Deposited
Full wwpdb X-ray Structure Validation Report i Jan 14, 2019 11:10 AM EST PDB ID : 6GYW Title : Crystal structure of DacA from Staphylococcus aureus Authors : Tosi, T.; Freemont, P.S.; Grundling, A. Deposited
Basic Crystallography Part 1. Theory and Practice of X-ray Crystal Structure Determination
 Basic Crystallography Part 1 Theory and Practice of X-ray Crystal Structure Determination We have a crystal How do we get there? we want a structure! The Unit Cell Concept Ralph Krätzner Unit Cell Description
Basic Crystallography Part 1 Theory and Practice of X-ray Crystal Structure Determination We have a crystal How do we get there? we want a structure! The Unit Cell Concept Ralph Krätzner Unit Cell Description
Protein Crystallography. Mitchell Guss University of Sydney Australia
 Protein Crystallography Mitchell Guss University of Sydney Australia Outline of the talk Recap some basic crystallography and history Highlight the special requirements for protein (macromolecular) structure
Protein Crystallography Mitchell Guss University of Sydney Australia Outline of the talk Recap some basic crystallography and history Highlight the special requirements for protein (macromolecular) structure
Case Studies in Molecular Replacement
 Case Studies in Molecular Replacement Melanie Vollmar Structural Genomics Consortium University of Oxford SGC Toronto SGC Oxford SGC Stockholm Overview Information about target protein Tools for model
Case Studies in Molecular Replacement Melanie Vollmar Structural Genomics Consortium University of Oxford SGC Toronto SGC Oxford SGC Stockholm Overview Information about target protein Tools for model
BCM Protein crystallography - II Isomorphous Replacement Anomalous Scattering and Molecular Replacement Model Building and Refinement
 BCM 6200 - Protein crystallography - II Isomorphous Replacement Anomalous Scattering and Molecular Replacement Model Building and Refinement Changing practice in de novo structure determination Hendrickson
BCM 6200 - Protein crystallography - II Isomorphous Replacement Anomalous Scattering and Molecular Replacement Model Building and Refinement Changing practice in de novo structure determination Hendrickson
fconv Tutorial Part 2
 fconv Tutorial Part 2 Gerd Neudert Introduction to some basic fconv features based on version 1.08 Introduction Definitions Welcome to the second part of fconv tutorials! If you have not already done part
fconv Tutorial Part 2 Gerd Neudert Introduction to some basic fconv features based on version 1.08 Introduction Definitions Welcome to the second part of fconv tutorials! If you have not already done part
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Jan 17, 2019 09:42 AM EST PDB ID : 6D3Z Title : Protease SFTI complex Authors : Law, R.H.P.; Wu, G. Deposited on : 2018-04-17 Resolution : 2.00 Å(reported)
Full wwpdb X-ray Structure Validation Report i Jan 17, 2019 09:42 AM EST PDB ID : 6D3Z Title : Protease SFTI complex Authors : Law, R.H.P.; Wu, G. Deposited on : 2018-04-17 Resolution : 2.00 Å(reported)
X-Ray structure analysis
 X-Ray structure analysis Kay Diederichs kay.diederichs@uni-konstanz.de Analysis of what? Proteins ( /ˈproʊˌtiːnz/ or /ˈproʊti.ɨnz/) are biochemical compounds consisting of one or more polypeptides typically
X-Ray structure analysis Kay Diederichs kay.diederichs@uni-konstanz.de Analysis of what? Proteins ( /ˈproʊˌtiːnz/ or /ˈproʊti.ɨnz/) are biochemical compounds consisting of one or more polypeptides typically
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 10, 2018 01:44 am GMT PDB ID : 1MWP Title : N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN Authors : Rossjohn, J.; Cappai, R.; Feil, S.C.; Henry,
Full wwpdb X-ray Structure Validation Report i Mar 10, 2018 01:44 am GMT PDB ID : 1MWP Title : N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN Authors : Rossjohn, J.; Cappai, R.; Feil, S.C.; Henry,
Protein Structure and Visualisation. Introduction to PDB and PyMOL
 Protein Structure and Visualisation Introduction to PDB and PyMOL 1 Feedback Persons http://www.bio-evaluering.dk/ 2 Program 8.00-8.15 Quiz results 8.15-8.50 Introduction to PDB & PyMOL 8.50-9.00 Break
Protein Structure and Visualisation Introduction to PDB and PyMOL 1 Feedback Persons http://www.bio-evaluering.dk/ 2 Program 8.00-8.15 Quiz results 8.15-8.50 Introduction to PDB & PyMOL 8.50-9.00 Break
Crystals, X-rays and Proteins
 Crystals, X-rays and Proteins Comprehensive Protein Crystallography Dennis Sherwood MA (Hons), MPhil, PhD Jon Cooper BA (Hons), PhD OXFORD UNIVERSITY PRESS Contents List of symbols xiv PART I FUNDAMENTALS
Crystals, X-rays and Proteins Comprehensive Protein Crystallography Dennis Sherwood MA (Hons), MPhil, PhD Jon Cooper BA (Hons), PhD OXFORD UNIVERSITY PRESS Contents List of symbols xiv PART I FUNDAMENTALS
Lattices and Symmetry Scattering and Diffraction (Physics)
 Lattices and Symmetry Scattering and Diffraction (Physics) James A. Kaduk INEOS Technologies Analytical Science Research Services Naperville IL 60566 James.Kaduk@innovene.com 1 Harry Potter and the Sorcerer
Lattices and Symmetry Scattering and Diffraction (Physics) James A. Kaduk INEOS Technologies Analytical Science Research Services Naperville IL 60566 James.Kaduk@innovene.com 1 Harry Potter and the Sorcerer
The SHELX Package. Data Flow
 The SHELX Package All Programs in the package written by George M. Sheldrick. SHELXS (XS) Structure solution by Patterson, direct methods. SHELXD (XM) Structure solution by advanced direct methods. SHELXL
The SHELX Package All Programs in the package written by George M. Sheldrick. SHELXS (XS) Structure solution by Patterson, direct methods. SHELXD (XM) Structure solution by advanced direct methods. SHELXL
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 8, 2018 08:34 pm GMT PDB ID : 1RUT Title : Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain Authors : Deane, J.E.; Ryan, D.P.; Maher, M.J.;
Full wwpdb X-ray Structure Validation Report i Mar 8, 2018 08:34 pm GMT PDB ID : 1RUT Title : Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain Authors : Deane, J.E.; Ryan, D.P.; Maher, M.J.;
Two Lectures in X-ray Crystallography
 Biochemistry 503 Michael Wiener (mwiener@virginia.edu, 3-2731, Snyder 360) Two Lectures in X-ray Crystallography Outline 1. Justification & introductory remarks 2. Experimental setup 3. Protein crystals
Biochemistry 503 Michael Wiener (mwiener@virginia.edu, 3-2731, Snyder 360) Two Lectures in X-ray Crystallography Outline 1. Justification & introductory remarks 2. Experimental setup 3. Protein crystals
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Jan 28, 2019 11:10 AM EST PDB ID : 6A5H Title : The structure of [4+2] and [6+4] cyclase in the biosynthetic pathway of unidentified natural product Authors
Full wwpdb X-ray Structure Validation Report i Jan 28, 2019 11:10 AM EST PDB ID : 6A5H Title : The structure of [4+2] and [6+4] cyclase in the biosynthetic pathway of unidentified natural product Authors
