The SHELX approach to the experimental phasing of macromolecules. George M. Sheldrick, Göttingen University
|
|
|
- Evangeline Hodges
- 5 years ago
- Views:
Transcription
1 The SHELX approach to the experimental phasing of macromolecules IUCr 2011 Madrid George M. Sheldrick, Göttingen University
2 Experimental phasing of macromolecules Except in relatively rare cases where atomic resolution data permit the phase problem to be solved by ab initio direct methods, experimental phasing usually implies the presence of heavy atoms. In order to locate the heavy atoms, we need their structure factors F A. The phases φ A calculated for the heavy atom substructure enable us to estimate starting phases φ T for the full macromolecular structure by: φ T = φ A + α After refining these phases we can use them and the native structure factors F T to calculate an electron density map of the macromolecule. As we will see, α, F A and F T can all be estimated from the experimental data.
3 Data files used for shelxc/d/e shelxc reads the experimental data and writes three files in addition to some useful statistics that are output to the console (and can be displayed graphically by Thomas Schneider s hkl2map). These files are: name.hkl reflection indices, merged native intensity for density modification with shelxe and possibly later refinement with shelxl. name_fa.ins running shelxd. instruction file for name_fa.hkl reflection indices, F A and α.
4 The analysis of MAD data Karle (1980) and Hendrickson, Smith & Sheriff (1985) showed by algebra that the measured intensities in a MAD experiment, assuming only one type of heavy atom, should be given by: F ± 2 = F T 2 + a F A 2 + b F T F A cosα ± c F T F A sinα where a = (f 2 +f 2 )/f 02, b = 2f /f 0, c = 2f /f 0 know f and f for each wavelength. and α = φ T φ A. We need to In a 2-wavelength MAD experiment, we have 4 equations for the 3 unknowns F A, F T and α, so with error-free data we should get a perfect map! For SIRAS, we have 2 equations for the derivative plus a native dataset. Given perfect isomorphism, the phase problem is solved. Unfortunately for SAD we only have 2 equations for the 3 unknowns.
5 SAD phasing For a single wavelength SAD experiment, we only have equations for F + 2 and F 2. Subtracting the second from the first, we obtain: F + 2 F 2 = 2c F A F T sinα For a small anomalous difference, we can assume: F T ~ ½ ( F + + F ) So using F + 2 F 2 = ( F + + F ) ( F + F ) we obtain: F + F = c F A sinα where c is 2f / f 0. c could be calculated but we would still require F A, not F A sinα, to find the heavy atoms. Lacking a better estimate, we have to input F + F (generated by SHELXC) into SHELXD. For MAD and SIRAS we can use F A, so there is no problem.
6 Starting atoms consistent with Patterson Dual space recycling for SHELXD substructure solution Based on the method introduced in the SnB program by Weeks, Hauptman et al. (1993) reciprocal space: refine phases SF calculation FFT Many cycles E > E min real space: select atoms First the F A values are normalized to E-values. The correlation coefficient CC between E obs and E calc is used to select the best solution. CC(weak) is also calculated for the reflections with E < E min ; these were not used for the substructure solution refine occupancies, save best solution so far
7 SAD substructure determination To find the heavy atoms using SHELXD, originally written for ab initio phasing using atomic resolution data, we would like to input F A, but we actually have to input F + F = c F A sinα. Why does this work? 1. SHELXD first normalizes F + F to get E-values. This gets rid of c and its resolution dependence! 2. Although all the reflections are used at the end for the occupancy refinement and the calculation of CC values, the direct methods location of the heavy atoms only uses the strongest reflections. These are likely to be the ones with sinα close to ±1. So to an adequate approximation, F + F is equal to F A!
8 SHELXD histograms and occupancies for Elastase 1 solution in 100 trials Diagrams from the hkl2map GUI S-SAD I-SIRAS
9 Critical parameters for SHELXD The Patterson-seeded dual-space recycling in SHELXD is effective at finding the heavy atoms, however attention needs to be paid to: 1. The resolution at which the data are truncated, e.g. where the internal CC between the signed anomalous differences of two randomly chosen reflection subsets falls below 30%. 2. The number of sites requested should be within about 20% of the true value so that the occupancy refinement works well (and reveals the true number). 3. In the case of a soak, the rejection of sites on special positions should be switched off. 4. For S-SAD, DSUL (search for disulfides in addition to atoms) can be very useful. 5. In difficult cases it may be necessary to run more trials (say 50000). The multiple-cpu version of SHELXD is recommended!
10 hits per tries CC CCweak Tendamistat CC, CCweak and hits per tries Occupancy (from SHELXD) of supersulfur peak 8 Occupancy of peak 9
11 SHELXD_MP multi-cpu heavy atom location The heavy atom search may require a large number of trials for weak SAD signals or when there are a large number of heavy atoms. It is however a good candidate for multi-tasking because only very limited communication is needed between different trials. shelxd has been parallelized using openmp. The scaling up is quite respectable (about 29 times for a computer with 32 real CPUs). In addition to reorganizing the output, an improved criterion is used for selecting the best solution: CFOM = CC + CC(weak) A beta-test is available on request.
12 The heavy atom enantiomorph problem The location of the heavy atoms from the F A -values does not define the enantiomorph of the heavy-atom substructure; there is exactly a 50% chance of getting the enantiomorph right. When the protein phases are calculated from the heavy atom reference phases, only one of the two possible maps should look like a protein, as this enables the correct heavy atom enantiomorph to be chosen. If the space group is one of an enantiomorphic pair (e.g. P and P ) the space group must be inverted as well as the atom coordinates. For three of the 65 Sohnke space groups possible for chiral molecules, the coordinates have to be inverted in a point other than the origin! These space groups and inversion operations are: I4 1 (1 x,½ y,1 z); I (1 x,½ y,¼ z); F (¼ x,¼ y,¼ z).
13 Simulations with one heavy atom in P1 MAD or SIRAS SIR SAD A perfect MAD or SIRAS experiment should give perfect phases! A centrosymmetric array of heavy atoms is fatal for SIR, but one heavy atom is enough for SAD even in space group P1, because it is easy to remove the negative image by setting negative density to zero!
14 Density modification The heavy atoms can be used to calculate reference phases; initial estimates of the protein phases can then be obtained by adding the phase shifts α to the heavy atom phases. These phases are then improved by density modification. Clearly, if we simply do an inverse Fourier transform of the unmodified density we get back the phases we put in. So we try to make a chemically sensible modification to the density before doing the inverse FFT in the hope that this will lead to improved estimates for the phases. Many such density modifications have been tried, some of them very sophisticated. Major contributions have been made by Peter Main, Kevin Cowtan and Tom Terwilliger. One of the simplest ideas, truncating negative density to zero, is actually not too bad (it is the basic idea behind the program ACORN).
15 The sphere of influence algorithm The variance V of the density on a spherical surface of radius 2.42 Å is calculated for each pixel in the map. The use of a spherical surface rather than a spherical volume was intended to save time and add a little chemical information (2.42 Å is a typical 1,3-distance in proteins and DNA). V gives an indication of the probability that a pixel corresponds to a true atomic position. Pixels with low V are flipped (ρ S = γρ where γ is usually set to 1.0). For pixels with high V, ρ is replaced by [ρ 4 /(ν 2 σ 2 (ρ)+ρ 2 )] ½ (with ν usually 0.5) if positive and by zero if negative. This has a similar effect to the procedure used in the CCP4 program ACORN. For intermediate values of V, a suitably weighted mixture of the two treatments is used. An empirical weighting scheme for phase recombination is used to combat model bias.
16 Density histograms Bernhard Rupp & Peter Zwart Although SHELXE makes no use of histogram matching, the sphere of influence algorithm is able to bring the histogram much closer to the one for the correct structure!
17 The free lunch algorithm The free lunch algorithm (FLA) is an attempt to extend the resolution of the data by including, in the density modification, reflections at higher resolution than have been measured. Although discovered independently and first published by the Bari group (Acta Cryst. D61 (2005) and ), the first successful implementation of the FLA was probably in 2001 in the program ACORN, but was not published until 2005 (Acta Cryst. D61 (2005) ). The unexpected conclusion was that if these phases are now used to recalculate the density, using very rough guesses for the (unmeasured) amplitudes, the density actually improves! The FLA is incorporated in SHELXE and tests confirm that the phases of the observed reflections improve, at least when the native data have been measured to a resolution of 2 Å or better.
18 Maps before and after a free lunch Best experimental phases after density modification (MapCC 0.57) After expansion to 1.0 Å with virtual data (MapCC 0.94) Isabel Usón et al, Acta Cryst. D63 (2007)
19 Why do we get a free lunch? It is not immediately obvious why inventing extra data improves the maps. Possible explanations are: 1. The algorithm corrects Fourier truncation errors that may have had a more serious effect on the maps than we had realised. 2. Phases are more important than amplitudes (see Kevin Cowtan s ducks and cats!), so as long as the extrapolated phases are OK any amplitudes will do. 3. Zero is a very poor estimate of the amplitude of a reflection that we did not measure.
20 The SHELXE autotracing algorithm A fast but very crude autotracing algorithm has been incorporated into the density modification in SHELXE. It is primarily designed for iterative phase improvement starting from very poor phases. The tracing proceeds as follows: 1. Find potential α-helices in the density and try to extend them at both ends. Then find other potential tripeptides and try to extend them at both ends in the same way. 2. Tidy up and splice the traces as required, applying any necessary symmetry operations. 3. Use the traced residues to estimate phases and combine these with the initial phase information using sigma-a weights, then restart density modification. The refinement of one B-value per residue provides a further opportunity to suppress wrongly traced residues.
21 Extending chains at both ends The chain extension algorithm looks two residues ahead of the residue currently being added, and employs a simplex algorithm to find a best fit to the density at the atom centers as well at holes in the chain. The quality of each completed trace is then assessed independently before accepting it. Important features of the algorithm are the generation of a no-go map that defines regions that should not be traced into, e.g. because of symmetry elements or existing atoms, and the efficient use of crystallographic symmetry. The trace is not restricted to a predefined volume, and the splicing algorithm takes symmetry equivalents into account.
22 Criteria for accepting chains The following criteria are combined into a single figure of merit for accepting traced chains: 1. The overall fit to the density should be good. 2. The chains must be long enough (in general at least 7 aminoacids); longer chains are given a higher weight. 3. There should not be too many Ramachandran outliers. 4. There should be a well defined secondary structure (φ /ϕ pairs should tend to be similar for consecutive residues). 5. On average, there should be significant positive density 2.9 Å from N in the N H direction (to a hydrogen bond acceptor): C C α N H O
23 Is the structure solved? If the CC for the structure factors calculated for the trace against the native data is better than 25%, it is extremely likely that the structure is solved. Another good indication is whether one can see side-chains.
24 Fibronectin autotracing This structure illustrates the ability of the autotracing to start from a noisy S-SAD map. Recycling the partial (but rather accurate) traces leads to better phases and an almost complete structure. N-terminus C-terminus Cycle 1: Cycle 2: Incorrectly traced C α Cycle 3: C α deviation: < 0.3Å < < 0.6Å < < 1.0Å < < 2.0Å < In the first cycle, 41% was traced with C α within 1.0Å, 33% within 0.5Å and 4% false. After 3 cycles the figures were 94%, 87% and 0%.
25 Fibronectin map quality MPE[ ] mapcc Standard S-SAD [ h s0.35]: S-SAD with FLA [ m200 h s0.5 e1]: S-SAD with autotracing [ h s0.35 a3]: S-SAD, autotracing and FLA [ h s0.35 a3 e1]: However, combining the FLA (free lunch algorithm) with autotracing did not produce much further improvement. Although the FLA had proved very useful in solving several borderline cases, with the phase improvement that arises from autotracing the FLA has almost been relegated to the role of cosmetic map improvements!
26 ACA2011 SHELXE Poly-Ala trace for 1y13-test In this case, assuming threefold NCS ( n3) found 50 residues more than without NCS. However the treatment of NCS in shelxe is quick and dirty and needs rewriting.
27 Extension of small MR fragments using SHELXE It often happens that a search model for structure solution by molecular replacement (MR) corresponds to only a small fraction of the total scattering power, and in such case expansion to the full structure can be tedious. For input to the beta-test shelxe, the PDB file of the MR solution is renamed to name.pda where the merged intensity data (e.g. from shelxc) are in the file name.hkl. Then e.g. shelxe name.pda -a30 -s0.5 -y2.0 -q -e1 can be used to run shelxe. A large number of tracing cycles may be needed (-a30)! A critical parameter is -y, the resolution at which to truncate the phases calculated from the MR solution. Experience suggests that -y2.0 is often best, with the implication that this approach works best with native data to a resolution of 2.0 Å or better.
28 Progress of fragment extension For this test, the variation of the CC value with the iteration number was unexpected. Instead of gradually improving, it meanders randomly at a about 10% and then suddenly, in the course of four or five iterations, jumps to a value well above 25%, indicating a solved structure. This strongly resembles the behavior of small-molecule direct methods, with the important differences that they involve data to about 1 Å or better, and start from random phases.
29 ARCIMBOLDO ab initio protein structure solution? Isabel Usón s arcimboldo uses a supercomputer to produce a large number of phaser MR solutions using very small but precise search fragments such as a 14-residue α-helix. Many potential solutions with (say) 1, 2 or 3 placed fragments are fed into shelxe, using the phaser TFZ as a broad-pass filter. If one or more shelxe attempts exceed the magic CC value of 25%, the structure has been solved! The only requirements are native data to about 2.1 Å or better, massive computing power and the presence of at least one α-helix (the more the better) in the structure. In principle any common fragment would do, but α-helices tend to have the most tightly conserved geometry. Since the median resolution of protein structures in the PDB is about 2.1 Å, the method should be able to solve at least 25% of the structures in the current PDB without using any experimental phase or other information! Isabel <iufcri@ibmb.csic.es> is currently setting up an arcimboldo supercomputer server.
30 ANODE The new program ANODE reads a PDB format file to calculate φ T and a file from shelxc containing F A and α. The heavy atom substructure phases are then calculated using φ A = φ T α. A Fourier map calculated with phases φ A and amplitudes F A then reveals the heavy atom substructure from a SAD, MAD or SIRAS etc. experiment. anode lyso would read the PDB format file lyso.ent and the lyso_fa.hkl file from shelxc. Lysozyme SAD-phased by four I3C 'sticky triangles' gave the following averaged anomalous densities: Averaged anomalous densities (sigma) I2_I3C I1_I3C I3_I3C SD_MET S_EPE (EPE is HEPES buffer) SG_CYS C9_EPE C4_I3C
31 ANODE (continued) This table is followed by a list of the highest unique anomalous peaks and the nearest atoms to each. In addition, a.pha file is written for displaying the anomalous density in coot. This approach always produces density with the same unit-cell origin as the original PDB file. Where alternative reflection indexing is possible it may be necessary to take it into account (with the switch -i). This figure clearly shows disulfides and other sulfurs, even though the anomalous data were too weak to find them using shelxd.
32 Acknowledgements I am particularly grateful to Isabel Usón, Thomas Schneider, Tim Gruene, Tobias Beck, Christian Grosse, Andrea Thorn and Navdeep Sidhu for discussions and testing various half-baked ideas. Andrea also prepared many of the pictures. Posters (Saturday 27th / Sunday 28th) MS58.P01/C591 Isabel Usón, Arcimboldo MS58.P05/C592 Christian Hübschle, shelxle MS58.P11/C595 Andrea Thorn, MR + shelxe MS58.P14/C596 Dayte Rodriguez Arcimboldo. References: SHELXC/D/E: Sheldrick (2010), Acta Cryst. D66, ARCIMBOLDO: Rodríguez, Grosse, Himmel, González, de Ilarduya, Becker, Sheldrick & Usón (2009), Nature Methods 6, The beta-tests of anode, shelxc, shelxd_mp and the autotracing shelxe are available on request to gsheldr@shelx.uni-ac.gwdg.de
SHELXC/D/E. Andrea Thorn
 SHELXC/D/E Andrea Thorn What is experimental phasing? Experimental phasing is what you do if MR doesn t work. What is experimental phasing? Experimental phasing methods depend on intensity differences.
SHELXC/D/E Andrea Thorn What is experimental phasing? Experimental phasing is what you do if MR doesn t work. What is experimental phasing? Experimental phasing methods depend on intensity differences.
Experimental Phasing with SHELX C/D/E
 WIR SCHAFFEN WISSEN HEUTE FÜR MORGEN Dr. Tim Grüne :: Paul Scherrer Institut :: tim.gruene@psi.ch Experimental Phasing with SHELX C/D/E CCP4 / APS School Chicago 2017 22 nd June 2017 1 - The Phase Problem
WIR SCHAFFEN WISSEN HEUTE FÜR MORGEN Dr. Tim Grüne :: Paul Scherrer Institut :: tim.gruene@psi.ch Experimental Phasing with SHELX C/D/E CCP4 / APS School Chicago 2017 22 nd June 2017 1 - The Phase Problem
CCP4 Diamond 2014 SHELXC/D/E. Andrea Thorn
 CCP4 Diamond 2014 SHELXC/D/E Andrea Thorn SHELXC/D/E workflow SHELXC: α calculation, file preparation SHELXD: Marker atom search = substructure search SHELXE: density modification Maps and coordinate files
CCP4 Diamond 2014 SHELXC/D/E Andrea Thorn SHELXC/D/E workflow SHELXC: α calculation, file preparation SHELXD: Marker atom search = substructure search SHELXE: density modification Maps and coordinate files
ANODE: ANOmalous and heavy atom DEnsity
 Bruker User s Meeting 2012 ANODE: ANOmalous and heavy atom DEnsity Andrea Thorn September 26 th, 2012 Typical SHELXC/D/E workflow The anomalous or heavy atom signal is used to find the substructure of
Bruker User s Meeting 2012 ANODE: ANOmalous and heavy atom DEnsity Andrea Thorn September 26 th, 2012 Typical SHELXC/D/E workflow The anomalous or heavy atom signal is used to find the substructure of
Macromolecular Phasing with shelxc/d/e
 Sunday, June 13 th, 2010 CCP4 Workshop APS Chicago, June 2010 http://shelx.uni-ac.gwdg.de Overview Substructure Definition and Motivation Extracting Substructure Data from measured Data Substructure Solution
Sunday, June 13 th, 2010 CCP4 Workshop APS Chicago, June 2010 http://shelx.uni-ac.gwdg.de Overview Substructure Definition and Motivation Extracting Substructure Data from measured Data Substructure Solution
Determination of the Substructure
 Monday, June 15 th, 2009 Determination of the Substructure EMBO / MAX-INF2 Practical Course http://shelx.uni-ac.gwdg.de Overview Substructure Definition and Motivation Extracting Substructure Data from
Monday, June 15 th, 2009 Determination of the Substructure EMBO / MAX-INF2 Practical Course http://shelx.uni-ac.gwdg.de Overview Substructure Definition and Motivation Extracting Substructure Data from
ANODE: ANOmalous and heavy atom DEnsity
 Bruker Webinar ANODE: ANOmalous and heavy atom DEnsity Andrea Thorn 03 December, 2013 Michael Ruf Welcome Dr. Michael Ruf Product Manager, SC-XRD Bruker AXS Inc. Madison, WI, USA Andrea Thorn Crystallographic
Bruker Webinar ANODE: ANOmalous and heavy atom DEnsity Andrea Thorn 03 December, 2013 Michael Ruf Welcome Dr. Michael Ruf Product Manager, SC-XRD Bruker AXS Inc. Madison, WI, USA Andrea Thorn Crystallographic
Experimental phasing, Pattersons and SHELX Andrea Thorn
 Experimental phasing, Pattersons and SHELX Andrea Thorn What is experimental phasing? Experimental phasing is what you do if MR doesn t work. What is experimental phasing? Experimental phasing methods
Experimental phasing, Pattersons and SHELX Andrea Thorn What is experimental phasing? Experimental phasing is what you do if MR doesn t work. What is experimental phasing? Experimental phasing methods
Practical aspects of SAD/MAD. Judit É Debreczeni
 Practical aspects of SAD/MAD Judit É Debreczeni anomalous scattering Hg sinθ/λ CuKα 0 Å - 0.4 Å - 0.6 Å - Å.5Å 0.83Å f 80 53 4 f total (θ, λ) f(θ) + f (λ) + if (λ) f f -5 8-5 8-5 8 f total increasing with
Practical aspects of SAD/MAD Judit É Debreczeni anomalous scattering Hg sinθ/λ CuKα 0 Å - 0.4 Å - 0.6 Å - Å.5Å 0.83Å f 80 53 4 f total (θ, λ) f(θ) + f (λ) + if (λ) f f -5 8-5 8-5 8 f total increasing with
PAN-modular Structure of Parasite Sarcocystis muris Microneme Protein SML-2 at 1.95 Å Resolution and the Complex with 1-Thio-β-D-Galactose
 Supplementary Material to the paper: PAN-modular Structure of Parasite Sarcocystis muris Microneme Protein SML-2 at 1.95 Å Resolution and the Complex with 1-Thio-β-D-Galactose Jürgen J. Müller, a Manfred
Supplementary Material to the paper: PAN-modular Structure of Parasite Sarcocystis muris Microneme Protein SML-2 at 1.95 Å Resolution and the Complex with 1-Thio-β-D-Galactose Jürgen J. Müller, a Manfred
Experimental phasing in Crank2
 Experimental phasing in Crank2 Pavol Skubak and Navraj Pannu Biophysical Structural Chemistry, Leiden University, The Netherlands http://www.bfsc.leidenuniv.nl/software/crank/ X-ray structure solution
Experimental phasing in Crank2 Pavol Skubak and Navraj Pannu Biophysical Structural Chemistry, Leiden University, The Netherlands http://www.bfsc.leidenuniv.nl/software/crank/ X-ray structure solution
Structure solution from weak anomalous data
 Structure solution from weak anomalous data Phenix Workshop SBGrid-NE-CAT Computing School Harvard Medical School, Boston June 7, 2014 Gábor Bunkóczi, Airlie McCoy, Randy Read (Cambridge University) Nat
Structure solution from weak anomalous data Phenix Workshop SBGrid-NE-CAT Computing School Harvard Medical School, Boston June 7, 2014 Gábor Bunkóczi, Airlie McCoy, Randy Read (Cambridge University) Nat
Protein Crystallography
 Protein Crystallography Part II Tim Grüne Dept. of Structural Chemistry Prof. G. Sheldrick University of Göttingen http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview The Reciprocal Lattice The
Protein Crystallography Part II Tim Grüne Dept. of Structural Chemistry Prof. G. Sheldrick University of Göttingen http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview The Reciprocal Lattice The
Likelihood and SAD phasing in Phaser. R J Read, Department of Haematology Cambridge Institute for Medical Research
 Likelihood and SAD phasing in Phaser R J Read, Department of Haematology Cambridge Institute for Medical Research Concept of likelihood Likelihood with dice 4 6 8 10 Roll a seven. Which die?? p(4)=p(6)=0
Likelihood and SAD phasing in Phaser R J Read, Department of Haematology Cambridge Institute for Medical Research Concept of likelihood Likelihood with dice 4 6 8 10 Roll a seven. Which die?? p(4)=p(6)=0
Experimental phasing in Crank2
 Experimental phasing in Crank2 Pavol Skubak and Navraj Pannu Biophysical Structural Chemistry, Leiden University, The Netherlands http://www.bfsc.leidenuniv.nl/software/crank/ Crank2 for experimental phasing
Experimental phasing in Crank2 Pavol Skubak and Navraj Pannu Biophysical Structural Chemistry, Leiden University, The Netherlands http://www.bfsc.leidenuniv.nl/software/crank/ Crank2 for experimental phasing
Anomalous dispersion
 Selenomethionine MAD Selenomethionine is the amino acid methionine with the Sulfur replaced by a Selenium. Selenium is a heavy atom that also has the propery of "anomalous scatter" at some wavelengths,
Selenomethionine MAD Selenomethionine is the amino acid methionine with the Sulfur replaced by a Selenium. Selenium is a heavy atom that also has the propery of "anomalous scatter" at some wavelengths,
Biology III: Crystallographic phases
 Haupt/Masterstudiengang Physik Methoden moderner Röntgenphysik II: Streuung und Abbildung SS 2013 Biology III: Crystallographic phases Thomas R. Schneider, EMBL Hamburg 25/6/2013 thomas.schneider@embl-hamburg.de
Haupt/Masterstudiengang Physik Methoden moderner Röntgenphysik II: Streuung und Abbildung SS 2013 Biology III: Crystallographic phases Thomas R. Schneider, EMBL Hamburg 25/6/2013 thomas.schneider@embl-hamburg.de
Molecular replacement. New structures from old
 Molecular replacement New structures from old The Phase Problem phase amplitude Phasing by molecular replacement Phases can be calculated from atomic model Rotate and translate related structure Models
Molecular replacement New structures from old The Phase Problem phase amplitude Phasing by molecular replacement Phases can be calculated from atomic model Rotate and translate related structure Models
Patterson Methods
 59-553 Patterson Methods 113 In 1935, Patterson showed that the unknown phase information in the equation for electron density: ρ(xyz) = 1/V h k l F(hkl) exp[iα(hkl)] exp[-2πi(h x + k y + l z)] can be
59-553 Patterson Methods 113 In 1935, Patterson showed that the unknown phase information in the equation for electron density: ρ(xyz) = 1/V h k l F(hkl) exp[iα(hkl)] exp[-2πi(h x + k y + l z)] can be
Direct Method. Very few protein diffraction data meet the 2nd condition
 Direct Method Two conditions: -atoms in the structure are equal-weighted -resolution of data are higher than the distance between the atoms in the structure Very few protein diffraction data meet the 2nd
Direct Method Two conditions: -atoms in the structure are equal-weighted -resolution of data are higher than the distance between the atoms in the structure Very few protein diffraction data meet the 2nd
Phaser: Experimental phasing
 Phaser: Experimental phasing Using SAD data in Phaser R J Read, Department of Haematology Cambridge Institute for Medical Research Diffraction with anomalous scatterers SAD: single-wavelength anomalous
Phaser: Experimental phasing Using SAD data in Phaser R J Read, Department of Haematology Cambridge Institute for Medical Research Diffraction with anomalous scatterers SAD: single-wavelength anomalous
MR model selection, preparation and assessing the solution
 Ronan Keegan CCP4 Group MR model selection, preparation and assessing the solution DLS-CCP4 Data Collection and Structure Solution Workshop 2018 Overview Introduction Step-by-step guide to performing Molecular
Ronan Keegan CCP4 Group MR model selection, preparation and assessing the solution DLS-CCP4 Data Collection and Structure Solution Workshop 2018 Overview Introduction Step-by-step guide to performing Molecular
Web-based Auto-Rickshaw for validation of the X-ray experiment at the synchrotron beamline
 Web-based Auto-Rickshaw for validation of the X-ray experiment at the synchrotron beamline Auto-Rickshaw http://www.embl-hamburg.de/auto-rickshaw A platform for automated crystal structure determination
Web-based Auto-Rickshaw for validation of the X-ray experiment at the synchrotron beamline Auto-Rickshaw http://www.embl-hamburg.de/auto-rickshaw A platform for automated crystal structure determination
Direct Methods and Many Site Se-Met MAD Problems using BnP. W. Furey
 Direct Methods and Many Site Se-Met MAD Problems using BnP W. Furey Classical Direct Methods Main method for small molecule structure determination Highly automated (almost totally black box ) Solves structures
Direct Methods and Many Site Se-Met MAD Problems using BnP W. Furey Classical Direct Methods Main method for small molecule structure determination Highly automated (almost totally black box ) Solves structures
ACORN - a flexible and efficient ab initio procedure to solve a protein structure when atomic resolution data is available
 ACORN - a flexible and efficient ab initio procedure to solve a protein structure when atomic resolution data is available Yao Jia-xing Department of Chemistry, University of York, Heslington, York, YO10
ACORN - a flexible and efficient ab initio procedure to solve a protein structure when atomic resolution data is available Yao Jia-xing Department of Chemistry, University of York, Heslington, York, YO10
Scattering by two Electrons
 Scattering by two Electrons p = -r k in k in p r e 2 q k in /λ θ θ k out /λ S q = r k out p + q = r (k out - k in ) e 1 Phase difference of wave 2 with respect to wave 1: 2π λ (k out - k in ) r= 2π S r
Scattering by two Electrons p = -r k in k in p r e 2 q k in /λ θ θ k out /λ S q = r k out p + q = r (k out - k in ) e 1 Phase difference of wave 2 with respect to wave 1: 2π λ (k out - k in ) r= 2π S r
Fast, Intuitive Structure Determination IV: Space Group Determination and Structure Solution
 Fast, Intuitive Structure Determination IV: Space Group Determination and Structure Solution November 25, 2013 Welcome I I Dr. Michael Ruf Product Manager Crystallography Bruker AXS Inc. Madison, WI, USA
Fast, Intuitive Structure Determination IV: Space Group Determination and Structure Solution November 25, 2013 Welcome I I Dr. Michael Ruf Product Manager Crystallography Bruker AXS Inc. Madison, WI, USA
SOLVE and RESOLVE: automated structure solution, density modification and model building
 Journal of Synchrotron Radiation ISSN 0909-0495 SOLVE and RESOLVE: automated structure solution, density modification and model building Thomas Terwilliger Copyright International Union of Crystallography
Journal of Synchrotron Radiation ISSN 0909-0495 SOLVE and RESOLVE: automated structure solution, density modification and model building Thomas Terwilliger Copyright International Union of Crystallography
Crystal lattice Real Space. Reflections Reciprocal Space. I. Solving Phases II. Model Building for CHEM 645. Purified Protein. Build model.
 I. Solving Phases II. Model Building for CHEM 645 Purified Protein Solve Phase Build model and refine Crystal lattice Real Space Reflections Reciprocal Space ρ (x, y, z) pronounced rho F hkl 2 I F (h,
I. Solving Phases II. Model Building for CHEM 645 Purified Protein Solve Phase Build model and refine Crystal lattice Real Space Reflections Reciprocal Space ρ (x, y, z) pronounced rho F hkl 2 I F (h,
ACORN in CCP4 and its applications
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 ACORN in CCP4 and its applications Jia-xing Yao York Structural Biology Laboratory, Department of Chemistry, University of York,
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 ACORN in CCP4 and its applications Jia-xing Yao York Structural Biology Laboratory, Department of Chemistry, University of York,
Non-merohedral Twinning in Protein Crystallography
 Bruker Users Meeting 2010 Karlsruhe, 22 nd September 2010 Non-merohedral Twinning in Protein Crystallography Regine Herbst-Irmer rherbst@shelx.uni-ac.gwdg.de http://shelx.uni-ac.gwdg.de/~rherbst/twin.html
Bruker Users Meeting 2010 Karlsruhe, 22 nd September 2010 Non-merohedral Twinning in Protein Crystallography Regine Herbst-Irmer rherbst@shelx.uni-ac.gwdg.de http://shelx.uni-ac.gwdg.de/~rherbst/twin.html
PSD '17 -- Xray Lecture 5, 6. Patterson Space, Molecular Replacement and Heavy Atom Isomorphous Replacement
 PSD '17 -- Xray Lecture 5, 6 Patterson Space, Molecular Replacement and Heavy Atom Isomorphous Replacement The Phase Problem We can t measure the phases! X-ray detectors (film, photomultiplier tubes, CCDs,
PSD '17 -- Xray Lecture 5, 6 Patterson Space, Molecular Replacement and Heavy Atom Isomorphous Replacement The Phase Problem We can t measure the phases! X-ray detectors (film, photomultiplier tubes, CCDs,
Charles Ballard (original GáborBunkóczi) CCP4 Workshop 7 December 2011
 Experimental phasing Charles Ballard (original GáborBunkóczi) CCP4 Workshop 7 December 2011 Anomalous diffraction F P protein F A anomalous substructure ano F A " -FA" A F F -* F A F P Phasing Substructure
Experimental phasing Charles Ballard (original GáborBunkóczi) CCP4 Workshop 7 December 2011 Anomalous diffraction F P protein F A anomalous substructure ano F A " -FA" A F F -* F A F P Phasing Substructure
Protein Crystallography Part II
 Molecular Biology Course 2007 Protein Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2007 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview
Molecular Biology Course 2007 Protein Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2007 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview
Molecular Biology Course 2006 Protein Crystallography Part II
 Molecular Biology Course 2006 Protein Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry December 2006 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview
Molecular Biology Course 2006 Protein Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry December 2006 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview
Electronic Supplementary Information (ESI) for Chem. Commun. Unveiling the three- dimensional structure of the green pigment of nitrite- cured meat
 Electronic Supplementary Information (ESI) for Chem. Commun. Unveiling the three- dimensional structure of the green pigment of nitrite- cured meat Jun Yi* and George B. Richter- Addo* Department of Chemistry
Electronic Supplementary Information (ESI) for Chem. Commun. Unveiling the three- dimensional structure of the green pigment of nitrite- cured meat Jun Yi* and George B. Richter- Addo* Department of Chemistry
Tutorial on how to solve a Se-substructure using
 1 Introduction Tutorial on how to solve a Se-substructure using SHELXD Thomas R. Schneider Dept. of Structural Chemistry University of Göttingen trs@shelx.uni-ac.gwdg.de July 4, 2002 The Solution of the
1 Introduction Tutorial on how to solve a Se-substructure using SHELXD Thomas R. Schneider Dept. of Structural Chemistry University of Göttingen trs@shelx.uni-ac.gwdg.de July 4, 2002 The Solution of the
Protein crystallography. Garry Taylor
 Protein crystallography Garry Taylor X-ray Crystallography - the Basics Grow crystals Collect X-ray data Determine phases Calculate ρ-map Interpret map Refine coordinates Do the biology. Nitrogen at -180
Protein crystallography Garry Taylor X-ray Crystallography - the Basics Grow crystals Collect X-ray data Determine phases Calculate ρ-map Interpret map Refine coordinates Do the biology. Nitrogen at -180
Macromolecular Crystallography Part II
 Molecular Biology Course 2009 Macromolecular Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2009 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de From
Molecular Biology Course 2009 Macromolecular Crystallography Part II Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2009 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de From
Electron Density at various resolutions, and fitting a model as accurately as possible.
 Section 9, Electron Density Maps 900 Electron Density at various resolutions, and fitting a model as accurately as possible. ρ xyz = (Vol) -1 h k l m hkl F hkl e iφ hkl e-i2π( hx + ky + lz ) Amplitude
Section 9, Electron Density Maps 900 Electron Density at various resolutions, and fitting a model as accurately as possible. ρ xyz = (Vol) -1 h k l m hkl F hkl e iφ hkl e-i2π( hx + ky + lz ) Amplitude
Twinning. Andrea Thorn
 Twinning Andrea Thorn OVERVIEW Introduction: Definitions, origins of twinning Merohedral twins: Recognition, statistical analysis: H plot, Yeates Padilla plot Example Refinement and R values Reticular
Twinning Andrea Thorn OVERVIEW Introduction: Definitions, origins of twinning Merohedral twins: Recognition, statistical analysis: H plot, Yeates Padilla plot Example Refinement and R values Reticular
Introduction to SHELXD
 SHELXD and SHELXE The structure solution program SHELXD (called XM in the Bruker SHELXTL system, but identical to SHELXD except in the logo) is able to solve larger ab initio problems than SHELXS-97, and
SHELXD and SHELXE The structure solution program SHELXD (called XM in the Bruker SHELXTL system, but identical to SHELXD except in the logo) is able to solve larger ab initio problems than SHELXS-97, and
Structure factors again
 Structure factors again Remember 1D, structure factor for order h F h = F h exp[iα h ] = I 01 ρ(x)exp[2πihx]dx Where x is fractional position along unit cell distance (repeating distance, origin arbitrary)
Structure factors again Remember 1D, structure factor for order h F h = F h exp[iα h ] = I 01 ρ(x)exp[2πihx]dx Where x is fractional position along unit cell distance (repeating distance, origin arbitrary)
Anisotropy in macromolecular crystal structures. Andrea Thorn July 19 th, 2012
 Anisotropy in macromolecular crystal structures Andrea Thorn July 19 th, 2012 Motivation Courtesy of M. Sawaya Motivation Crystal structures are inherently anisotropic. X-ray diffraction reflects this
Anisotropy in macromolecular crystal structures Andrea Thorn July 19 th, 2012 Motivation Courtesy of M. Sawaya Motivation Crystal structures are inherently anisotropic. X-ray diffraction reflects this
PHENIX Wizards and Tools
 PHENIX Wizards and Tools Tom Terwilliger Los Alamos National Laboratory terwilliger@lanl.gov l The PHENIX project Computational Crystallography Initiative (LBNL) Paul Adams, Ralf Grosse-Kunstleve, Peter
PHENIX Wizards and Tools Tom Terwilliger Los Alamos National Laboratory terwilliger@lanl.gov l The PHENIX project Computational Crystallography Initiative (LBNL) Paul Adams, Ralf Grosse-Kunstleve, Peter
What is the Phase Problem? Overview of the Phase Problem. Phases. 201 Phases. Diffraction vector for a Bragg spot. In General for Any Atom (x, y, z)
 Protein Overview of the Phase Problem Crystal Data Phases Structure John Rose ACA Summer School 2006 Reorganized by Andy Howard,, Spring 2008 Remember We can measure reflection intensities We can calculate
Protein Overview of the Phase Problem Crystal Data Phases Structure John Rose ACA Summer School 2006 Reorganized by Andy Howard,, Spring 2008 Remember We can measure reflection intensities We can calculate
Fan, Hai-fu Institute of Physics, Chinese Academy of Sciences, Beijing , China
 Direct Methods in Crystallography Fan, Hai-fu Institute of Physics, Chinese Academy of Sciences, Beijing 100080, China An important branch of crystallography is the X-ray diffraction analysis of crystal
Direct Methods in Crystallography Fan, Hai-fu Institute of Physics, Chinese Academy of Sciences, Beijing 100080, China An important branch of crystallography is the X-ray diffraction analysis of crystal
research papers Reduction of density-modification bias by b correction 1. Introduction Pavol Skubák* and Navraj S. Pannu
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Reduction of density-modification bias by b correction Pavol Skubák* and Navraj S. Pannu Biophysical Structural Chemistry, Leiden
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Reduction of density-modification bias by b correction Pavol Skubák* and Navraj S. Pannu Biophysical Structural Chemistry, Leiden
Molecular Biology Course 2006 Protein Crystallography Part I
 Molecular Biology Course 2006 Protein Crystallography Part I Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2006 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview Overview
Molecular Biology Course 2006 Protein Crystallography Part I Tim Grüne University of Göttingen Dept. of Structural Chemistry November 2006 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Overview Overview
shelxl: Refinement of Macromolecular Structures from Neutron Data
 ESS Neutron Protein Crystallography 2013 Aarhus, Denmark shelxl: Refinement of Macromolecular Structures from Neutron Data Tim Grüne University of Göttingen Dept. of Structural Chemistry http://shelx.uni-ac.gwdg.de
ESS Neutron Protein Crystallography 2013 Aarhus, Denmark shelxl: Refinement of Macromolecular Structures from Neutron Data Tim Grüne University of Göttingen Dept. of Structural Chemistry http://shelx.uni-ac.gwdg.de
research papers 1. Introduction Thomas C. Terwilliger a * and Joel Berendzen b
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy-atom solutions in the MIR and
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Discrimination of solvent from protein regions in native Fouriers as a means of evaluating heavy-atom solutions in the MIR and
Principles of Protein X-Ray Crystallography
 Principles of Protein X-Ray Crystallography Jan Drenth Principles of Protein X-Ray Crystallography Third Edition With Major Contribution from Jeroen Mesters University of Lübeck, Germany Jan Drenth Laboratory
Principles of Protein X-Ray Crystallography Jan Drenth Principles of Protein X-Ray Crystallography Third Edition With Major Contribution from Jeroen Mesters University of Lübeck, Germany Jan Drenth Laboratory
Image definition evaluation functions for X-ray crystallography: A new perspective on the phase. problem. Hui LI*, Meng HE* and Ze ZHANG
 Image definition evaluation functions for X-ray crystallography: A new perspective on the phase problem Hui LI*, Meng HE* and Ze ZHANG Beijing University of Technology, Beijing 100124, People s Republic
Image definition evaluation functions for X-ray crystallography: A new perspective on the phase problem Hui LI*, Meng HE* and Ze ZHANG Beijing University of Technology, Beijing 100124, People s Republic
Direct-method SAD phasing with partial-structure iteration: towards automation
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Editors: E. N. Baker and Z. Dauter Direct-method SAD phasing with partial-structure iteration: towards automation J. W. Wang,
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 Editors: E. N. Baker and Z. Dauter Direct-method SAD phasing with partial-structure iteration: towards automation J. W. Wang,
Space Group & Structure Solution
 Space Group & Structure Solution Determine the Space Group Space group determination can always be performed by hand by examining the intensity data. A program that can facilitate this step is the command-prompt
Space Group & Structure Solution Determine the Space Group Space group determination can always be performed by hand by examining the intensity data. A program that can facilitate this step is the command-prompt
A short history of SHELX
 Acta Crystallographica Section A Foundations of Crystallography ISSN 0108-7673 A short history of SHELX George M. Sheldrick Received 5 July 2007 Accepted 7 September 2007 Department of Structural Chemistry,
Acta Crystallographica Section A Foundations of Crystallography ISSN 0108-7673 A short history of SHELX George M. Sheldrick Received 5 July 2007 Accepted 7 September 2007 Department of Structural Chemistry,
Synthetic, Structural, and Mechanistic Aspects of an Amine Activation Process Mediated at a Zwitterionic Pd(II) Center
 Synthetic, Structural, and Mechanistic Aspects of an Amine Activation Process Mediated at a Zwitterionic Pd(II) Center Supporting Information Connie C. Lu and Jonas C. Peters* Division of Chemistry and
Synthetic, Structural, and Mechanistic Aspects of an Amine Activation Process Mediated at a Zwitterionic Pd(II) Center Supporting Information Connie C. Lu and Jonas C. Peters* Division of Chemistry and
Macromolecular Crystallography Part II
 Molecular Biology Course 2010 Macromolecular Crystallography Part II University of Göttingen Dept. of Structural Chemistry November 2010 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Crystallography
Molecular Biology Course 2010 Macromolecular Crystallography Part II University of Göttingen Dept. of Structural Chemistry November 2010 http://shelx.uni-ac.gwdg.de tg@shelx.uni-ac.gwdg.de Crystallography
The Crystallographic Process
 Experimental Phasing Macromolecular Crystallography School Madrid, May 2017 Paul Adams Lawrence Berkeley Laboratory and Department of Bioengineering UC Berkeley The Crystallographic Process Crystallization
Experimental Phasing Macromolecular Crystallography School Madrid, May 2017 Paul Adams Lawrence Berkeley Laboratory and Department of Bioengineering UC Berkeley The Crystallographic Process Crystallization
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 8, 2018 10:24 pm GMT PDB ID : 1A30 Title : HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR Authors : Louis, J.M.; Dyda, F.; Nashed, N.T.; Kimmel,
Full wwpdb X-ray Structure Validation Report i Mar 8, 2018 10:24 pm GMT PDB ID : 1A30 Title : HIV-1 PROTEASE COMPLEXED WITH A TRIPEPTIDE INHIBITOR Authors : Louis, J.M.; Dyda, F.; Nashed, N.T.; Kimmel,
BC530 Class notes on X-ray Crystallography
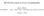 BC530 Class notes on X-ray Crystallography web material: Ethan A Merritt http://skuld.bmsc.washington.edu/~merritt/bc530/ October 11, 2016 Growing Crystals It should be self-evident that in order to do
BC530 Class notes on X-ray Crystallography web material: Ethan A Merritt http://skuld.bmsc.washington.edu/~merritt/bc530/ October 11, 2016 Growing Crystals It should be self-evident that in order to do
X-ray Crystallography. Kalyan Das
 X-ray Crystallography Kalyan Das Electromagnetic Spectrum NMR 10 um - 10 mm 700 to 10 4 nm 400 to 700 nm 10 to 400 nm 10-1 to 10 nm 10-4 to 10-1 nm X-ray radiation was discovered by Roentgen in 1895. X-rays
X-ray Crystallography Kalyan Das Electromagnetic Spectrum NMR 10 um - 10 mm 700 to 10 4 nm 400 to 700 nm 10 to 400 nm 10-1 to 10 nm 10-4 to 10-1 nm X-ray radiation was discovered by Roentgen in 1895. X-rays
Model and data. An X-ray structure solution requires a model.
 Model and data An X-ray structure solution requires a model. This model has to be consistent with: The findings of Chemistry Reflection positions and intensities Structure refinement = Model fitting by
Model and data An X-ray structure solution requires a model. This model has to be consistent with: The findings of Chemistry Reflection positions and intensities Structure refinement = Model fitting by
Recent developments in Crank. Leiden University, The Netherlands
 Recent developments in Crank Navraj js. Pannu Leiden University, The Netherlands Current developers Pavol Skubak Ruben Zubac Irakli Sikharulidze Jan Pieter Abrahams RAG de Graaff Willem-Jan Waterreus Substructure
Recent developments in Crank Navraj js. Pannu Leiden University, The Netherlands Current developers Pavol Skubak Ruben Zubac Irakli Sikharulidze Jan Pieter Abrahams RAG de Graaff Willem-Jan Waterreus Substructure
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Jan 17, 2019 09:42 AM EST PDB ID : 6D3Z Title : Protease SFTI complex Authors : Law, R.H.P.; Wu, G. Deposited on : 2018-04-17 Resolution : 2.00 Å(reported)
Full wwpdb X-ray Structure Validation Report i Jan 17, 2019 09:42 AM EST PDB ID : 6D3Z Title : Protease SFTI complex Authors : Law, R.H.P.; Wu, G. Deposited on : 2018-04-17 Resolution : 2.00 Å(reported)
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Jan 28, 2019 11:10 AM EST PDB ID : 6A5H Title : The structure of [4+2] and [6+4] cyclase in the biosynthetic pathway of unidentified natural product Authors
Full wwpdb X-ray Structure Validation Report i Jan 28, 2019 11:10 AM EST PDB ID : 6A5H Title : The structure of [4+2] and [6+4] cyclase in the biosynthetic pathway of unidentified natural product Authors
research papers HKL-3000: the integration of data reduction and structure solution from diffraction images to an initial model in minutes
 Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 HKL-3000: the integration of data reduction and structure solution from diffraction images to an initial model in minutes Wladek
Acta Crystallographica Section D Biological Crystallography ISSN 0907-4449 HKL-3000: the integration of data reduction and structure solution from diffraction images to an initial model in minutes Wladek
Phase problem: Determining an initial phase angle α hkl for each recorded reflection. 1 ρ(x,y,z) = F hkl cos 2π (hx+ky+ lz - α hkl ) V h k l
 Phase problem: Determining an initial phase angle α hkl for each recorded reflection 1 ρ(x,y,z) = F hkl cos 2π (hx+ky+ lz - α hkl ) V h k l Methods: Heavy atom methods (isomorphous replacement Hg, Pt)
Phase problem: Determining an initial phase angle α hkl for each recorded reflection 1 ρ(x,y,z) = F hkl cos 2π (hx+ky+ lz - α hkl ) V h k l Methods: Heavy atom methods (isomorphous replacement Hg, Pt)
Determining Protein Structure BIBC 100
 Determining Protein Structure BIBC 100 Determining Protein Structure X-Ray Diffraction Interactions of x-rays with electrons in molecules in a crystal NMR- Nuclear Magnetic Resonance Interactions of magnetic
Determining Protein Structure BIBC 100 Determining Protein Structure X-Ray Diffraction Interactions of x-rays with electrons in molecules in a crystal NMR- Nuclear Magnetic Resonance Interactions of magnetic
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Jan 14, 2019 11:10 AM EST PDB ID : 6GYW Title : Crystal structure of DacA from Staphylococcus aureus Authors : Tosi, T.; Freemont, P.S.; Grundling, A. Deposited
Full wwpdb X-ray Structure Validation Report i Jan 14, 2019 11:10 AM EST PDB ID : 6GYW Title : Crystal structure of DacA from Staphylococcus aureus Authors : Tosi, T.; Freemont, P.S.; Grundling, A. Deposited
Table 1. Crystallographic data collection, phasing and refinement statistics. Native Hg soaked Mn soaked 1 Mn soaked 2
 Table 1. Crystallographic data collection, phasing and refinement statistics Native Hg soaked Mn soaked 1 Mn soaked 2 Data collection Space group P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 Cell
Table 1. Crystallographic data collection, phasing and refinement statistics Native Hg soaked Mn soaked 1 Mn soaked 2 Data collection Space group P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 P2 1 2 1 2 1 Cell
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 14, 2018 02:00 pm GMT PDB ID : 3RRQ Title : Crystal structure of the extracellular domain of human PD-1 Authors : Lazar-Molnar, E.; Ramagopal, U.A.; Nathenson,
Full wwpdb X-ray Structure Validation Report i Mar 14, 2018 02:00 pm GMT PDB ID : 3RRQ Title : Crystal structure of the extracellular domain of human PD-1 Authors : Lazar-Molnar, E.; Ramagopal, U.A.; Nathenson,
From x-ray crystallography to electron microscopy and back -- how best to exploit the continuum of structure-determination methods now available
 From x-ray crystallography to electron microscopy and back -- how best to exploit the continuum of structure-determination methods now available Scripps EM course, November 14, 2007 What aspects of contemporary
From x-ray crystallography to electron microscopy and back -- how best to exploit the continuum of structure-determination methods now available Scripps EM course, November 14, 2007 What aspects of contemporary
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 8, 2018 08:34 pm GMT PDB ID : 1RUT Title : Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain Authors : Deane, J.E.; Ryan, D.P.; Maher, M.J.;
Full wwpdb X-ray Structure Validation Report i Mar 8, 2018 08:34 pm GMT PDB ID : 1RUT Title : Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain Authors : Deane, J.E.; Ryan, D.P.; Maher, M.J.;
QUADRATIC EQUATIONS M.K. HOME TUITION. Mathematics Revision Guides Level: GCSE Higher Tier
 Mathematics Revision Guides Quadratic Equations Page 1 of 8 M.K. HOME TUITION Mathematics Revision Guides Level: GCSE Higher Tier QUADRATIC EQUATIONS Version: 3.1 Date: 6-10-014 Mathematics Revision Guides
Mathematics Revision Guides Quadratic Equations Page 1 of 8 M.K. HOME TUITION Mathematics Revision Guides Level: GCSE Higher Tier QUADRATIC EQUATIONS Version: 3.1 Date: 6-10-014 Mathematics Revision Guides
X-ray Crystallography
 2009/11/25 [ 1 ] X-ray Crystallography Andrew Torda, wintersemester 2009 / 2010 X-ray numerically most important more than 4/5 structures Goal a set of x, y, z coordinates different properties to NMR History
2009/11/25 [ 1 ] X-ray Crystallography Andrew Torda, wintersemester 2009 / 2010 X-ray numerically most important more than 4/5 structures Goal a set of x, y, z coordinates different properties to NMR History
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 13, 2018 04:03 pm GMT PDB ID : 5NMJ Title : Chicken GRIFIN (crystallisation ph: 6.5) Authors : Ruiz, F.M.; Romero, A. Deposited on : 2017-04-06 Resolution
Full wwpdb X-ray Structure Validation Report i Mar 13, 2018 04:03 pm GMT PDB ID : 5NMJ Title : Chicken GRIFIN (crystallisation ph: 6.5) Authors : Ruiz, F.M.; Romero, A. Deposited on : 2017-04-06 Resolution
SUPPLEMENTARY INFORMATION
 Table of Contents Page Supplementary Table 1. Diffraction data collection statistics 2 Supplementary Table 2. Crystallographic refinement statistics 3 Supplementary Fig. 1. casic1mfc packing in the R3
Table of Contents Page Supplementary Table 1. Diffraction data collection statistics 2 Supplementary Table 2. Crystallographic refinement statistics 3 Supplementary Fig. 1. casic1mfc packing in the R3
Copyright WILEY-VCH Verlag GmbH, D Weinheim, 2000 Angew. Chem Supporting Information For Binding Cesium Ion with Nucleoside Pentamers.
 Copyright WILEY-VCH Verlag GmbH, D-69451 Weinheim, 2000 Angew. Chem. 2000 Supporting Information For Binding Cesium Ion with Nucleoside Pentamers. Templated Self-Assembly of an Isoguanosine Decamer.**
Copyright WILEY-VCH Verlag GmbH, D-69451 Weinheim, 2000 Angew. Chem. 2000 Supporting Information For Binding Cesium Ion with Nucleoside Pentamers. Templated Self-Assembly of an Isoguanosine Decamer.**
1 Introduction. Abstract
 High-resolution Structure Refinement George M. Sheldrick Institut für Anorganische Chemie der Universität Göttingen Tammannstraße 4, D-37077 Göttingen, Germany gsheldr@shelx.uni-ac.gwdg.de Abstract The
High-resolution Structure Refinement George M. Sheldrick Institut für Anorganische Chemie der Universität Göttingen Tammannstraße 4, D-37077 Göttingen, Germany gsheldr@shelx.uni-ac.gwdg.de Abstract The
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 8, 2018 06:13 pm GMT PDB ID : 5G5C Title : Structure of the Pyrococcus furiosus Esterase Pf2001 with space group C2221 Authors : Varejao, N.; Reverter,
Full wwpdb X-ray Structure Validation Report i Mar 8, 2018 06:13 pm GMT PDB ID : 5G5C Title : Structure of the Pyrococcus furiosus Esterase Pf2001 with space group C2221 Authors : Varejao, N.; Reverter,
Supplementary Material (ESI) for CrystEngComm This journal is The Royal Society of Chemistry 2010
 Electronic Supplementary Information (ESI) for: A bifunctionalized porous material containing discrete assemblies of copper-porphyrins and calixarenes metallated by ion diffusion Rita De Zorzi, Nicol Guidolin,
Electronic Supplementary Information (ESI) for: A bifunctionalized porous material containing discrete assemblies of copper-porphyrins and calixarenes metallated by ion diffusion Rita De Zorzi, Nicol Guidolin,
Pathogenic C9ORF72 Antisense Repeat RNA Forms a Double Helix with Tandem C:C Mismatches
 Supporting Information Pathogenic C9ORF72 Antisense Repeat RNA Forms a Double Helix with Tandem C:C Mismatches David W. Dodd, Diana R. Tomchick, David R. Corey, and Keith T. Gagnon METHODS S1 RNA synthesis.
Supporting Information Pathogenic C9ORF72 Antisense Repeat RNA Forms a Double Helix with Tandem C:C Mismatches David W. Dodd, Diana R. Tomchick, David R. Corey, and Keith T. Gagnon METHODS S1 RNA synthesis.
Resolution and data formats. Andrea Thorn
 Resolution and data formats Andrea Thorn RESOLUTION Motivation Courtesy of M. Sawaya Map resolution http://www.bmsc.washington.edu/people/verlinde/experiment.html Data quality indicators Resolution accounts
Resolution and data formats Andrea Thorn RESOLUTION Motivation Courtesy of M. Sawaya Map resolution http://www.bmsc.washington.edu/people/verlinde/experiment.html Data quality indicators Resolution accounts
Acta Crystallographica Section F
 Supporting information Acta Crystallographica Section F Volume 70 (2014) Supporting information for article: Chemical conversion of cisplatin and carboplatin with histidine in a model protein crystallised
Supporting information Acta Crystallographica Section F Volume 70 (2014) Supporting information for article: Chemical conversion of cisplatin and carboplatin with histidine in a model protein crystallised
The Crystallographic Process
 Phase Improvement Macromolecular Crystallography School Madrid, May 2017 Paul Adams Lawrence Berkeley Laboratory and Department of Bioengineering UC Berkeley The Crystallographic Process Crystallization
Phase Improvement Macromolecular Crystallography School Madrid, May 2017 Paul Adams Lawrence Berkeley Laboratory and Department of Bioengineering UC Berkeley The Crystallographic Process Crystallization
Full wwpdb X-ray Structure Validation Report i
 Full wwpdb X-ray Structure Validation Report i Mar 10, 2018 01:44 am GMT PDB ID : 1MWP Title : N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN Authors : Rossjohn, J.; Cappai, R.; Feil, S.C.; Henry,
Full wwpdb X-ray Structure Validation Report i Mar 10, 2018 01:44 am GMT PDB ID : 1MWP Title : N-TERMINAL DOMAIN OF THE AMYLOID PRECURSOR PROTEIN Authors : Rossjohn, J.; Cappai, R.; Feil, S.C.; Henry,
Biological Small Angle X-ray Scattering (SAXS) Dec 2, 2013
 Biological Small Angle X-ray Scattering (SAXS) Dec 2, 2013 Structural Biology Shape Dynamic Light Scattering Electron Microscopy Small Angle X-ray Scattering Cryo-Electron Microscopy Wide Angle X- ray
Biological Small Angle X-ray Scattering (SAXS) Dec 2, 2013 Structural Biology Shape Dynamic Light Scattering Electron Microscopy Small Angle X-ray Scattering Cryo-Electron Microscopy Wide Angle X- ray
Shake-and-Bake: Applications and Advances. Russ Miller & Charles M. Weeks
 Shake-and-Bake: Applications and Advances Russ Miller & Charles M. Weeks Hauptman-Woodward Med. Res. Inst. Principal Contributors: C.-S. Chang G.T. DeTitta S.M. Gallo H.A. Hauptman H.G. Khalak D.A. Langs
Shake-and-Bake: Applications and Advances Russ Miller & Charles M. Weeks Hauptman-Woodward Med. Res. Inst. Principal Contributors: C.-S. Chang G.T. DeTitta S.M. Gallo H.A. Hauptman H.G. Khalak D.A. Langs
SI Text S1 Solution Scattering Data Collection and Analysis. SI references
 SI Text S1 Solution Scattering Data Collection and Analysis. The X-ray photon energy was set to 8 kev. The PILATUS hybrid pixel array detector (RIGAKU) was positioned at a distance of 606 mm from the sample.
SI Text S1 Solution Scattering Data Collection and Analysis. The X-ray photon energy was set to 8 kev. The PILATUS hybrid pixel array detector (RIGAKU) was positioned at a distance of 606 mm from the sample.
XD2006 Primer. Document Version September 2006 Program Version 5.00 September Program authors
 XD Primer draft version dated September 2006 XD2006 Primer A Computer Program Package for Multipole Refinement, Topological Analysis of Charge Densities and Evaluation of Intermolecular Energies from Experimental
XD Primer draft version dated September 2006 XD2006 Primer A Computer Program Package for Multipole Refinement, Topological Analysis of Charge Densities and Evaluation of Intermolecular Energies from Experimental
Supporting Information
 Wiley-VCH 2007 69451 Weinheim, Germany On the polymorphism of aspirin Andrew D. Bond, Roland Boese and Gautam R. Desiraju S1. Comparison of the form I and PZ structures S2. Transforming the unit cells
Wiley-VCH 2007 69451 Weinheim, Germany On the polymorphism of aspirin Andrew D. Bond, Roland Boese and Gautam R. Desiraju S1. Comparison of the form I and PZ structures S2. Transforming the unit cells
Overview - Macromolecular Crystallography
 Overview - Macromolecular Crystallography 1. Overexpression and crystallization 2. Crystal characterization and data collection 3. The diffraction experiment 4. Phase problem 1. MIR (Multiple Isomorphous
Overview - Macromolecular Crystallography 1. Overexpression and crystallization 2. Crystal characterization and data collection 3. The diffraction experiment 4. Phase problem 1. MIR (Multiple Isomorphous
The Phase Problem of X-ray Crystallography
 163 The Phase Problem of X-ray Crystallography H.A. Hauptman Hauptman-Woodward Medical Research Institute, Inc. 73 High Street Buffalo, NY, USA hauptman@hwi.buffalo.edu ABSTRACT. The intensities of a sufficient
163 The Phase Problem of X-ray Crystallography H.A. Hauptman Hauptman-Woodward Medical Research Institute, Inc. 73 High Street Buffalo, NY, USA hauptman@hwi.buffalo.edu ABSTRACT. The intensities of a sufficient
Twinning in PDB and REFMAC. Garib N Murshudov Chemistry Department, University of York, UK
 Twinning in PDB and REFMAC Garib N Murshudov Chemistry Department, University of York, UK Contents Crystal peculiarities Problem of twinning Occurrences of twinning in PDB Recognition of potential operators
Twinning in PDB and REFMAC Garib N Murshudov Chemistry Department, University of York, UK Contents Crystal peculiarities Problem of twinning Occurrences of twinning in PDB Recognition of potential operators
X-ray Crystallography I. James Fraser Macromolecluar Interactions BP204
 X-ray Crystallography I James Fraser Macromolecluar Interactions BP204 Key take-aways 1. X-ray crystallography results from an ensemble of Billions and Billions of molecules in the crystal 2. Models in
X-ray Crystallography I James Fraser Macromolecluar Interactions BP204 Key take-aways 1. X-ray crystallography results from an ensemble of Billions and Billions of molecules in the crystal 2. Models in
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11054 Supplementary Fig. 1 Sequence alignment of Na v Rh with NaChBac, Na v Ab, and eukaryotic Na v and Ca v homologs. Secondary structural elements of Na v Rh are indicated above the
doi:10.1038/nature11054 Supplementary Fig. 1 Sequence alignment of Na v Rh with NaChBac, Na v Ab, and eukaryotic Na v and Ca v homologs. Secondary structural elements of Na v Rh are indicated above the
Detecting Dark Matter Halos Sam Beder, Brie Bunge, Adriana Diakite December 14, 2012
 Detecting Dark Matter Halos Sam Beder, Brie Bunge, Adriana Diakite December 14, 2012 Introduction Dark matter s gravitational pull affects the positions of galaxies in the universe. If galaxies were circular,
Detecting Dark Matter Halos Sam Beder, Brie Bunge, Adriana Diakite December 14, 2012 Introduction Dark matter s gravitational pull affects the positions of galaxies in the universe. If galaxies were circular,
Crystals, X-rays and Proteins
 Crystals, X-rays and Proteins Comprehensive Protein Crystallography Dennis Sherwood MA (Hons), MPhil, PhD Jon Cooper BA (Hons), PhD OXFORD UNIVERSITY PRESS Contents List of symbols xiv PART I FUNDAMENTALS
Crystals, X-rays and Proteins Comprehensive Protein Crystallography Dennis Sherwood MA (Hons), MPhil, PhD Jon Cooper BA (Hons), PhD OXFORD UNIVERSITY PRESS Contents List of symbols xiv PART I FUNDAMENTALS
