Table 1. Kinetic data obtained from SPR analysis of domain 11 mutants interacting with IGF-II. Kinetic parameters K D 1.
|
|
|
- Merry Francis
- 5 years ago
- Views:
Transcription
1 Kinetics and Thermodynamics of the Insulin-like Growth Factor II (IGF-II) Interaction with IGF-II/Mannose 6-phosphate Receptor and the function of CD and AB Loop Solvent-exposed Residues. Research Team: Oliver Zaccheo, Stuart Prince, David Miller, Christopher Williams, Lucy Catto, Matthew Crump Bassim Hassan (University of Bristol) James Brown, Yvonne Jones (University of Oxford), Fred Kemp (Biocentre Facility) Project Background: The mammalian cation-independent mannose 6-phosphate/insulin-like growth factor II receptor (IGF2R) is a type I integral membrane protein and P-type lectin, with multiple functions attributable to its wide variety of known ligands. The 270 kda glycosylated protein consists of an N-terminal signal sequence, 15 homologous extracytoplasmic repeating domains, a transmembrane region and a C-terminal cytoplasmic domain. Ligands of the IGF-II/mannose 6-phosphate receptor (IGF2R) include IGF-II and mannose 6-phosphate modified proteins, with the binding site for IGF-II being localised to domain 11. Disruption of the negative regulatory effects of IGF2R on IGF-II-induced growth can lead to embryonic lethality and cancer promotion. In order to define the functional basis of this critical biological interaction, we performed alanine mutagenesis of structurally determined solvent-exposed loop residues of the IGF-II-binding site of human domain 11, expressed these mutant forms in Pichia pastoris, and determined binding kinetics with human IGF-II using isothermal calorimetry and surface plasmon resonance with transition state thermodynamics. Two hydrophobic residues in the CD loop (F1567 and I1572) were essential for binding, with a further non-hydrophobic residue (T1570) that slows the dissociation rate. Aside from alanine mutations of AB loop residues that decrease affinity by modifying dissociation rates (e.g. Y1542), a novel mutation (E1544A) of the AB loop enhanced affinity by threefold compared to wild-type. Conversion from an acidic to a basic residue at this site (E1544K) results in a sixfold enhancement of affinity via modification principally of the association rate, with enhanced salt-dependence, decreased entropic barrier and retained specificity. These data suggest that a functional hydrophobic binding site core is formed by I1572 and F1567 located in the CD loop, which initially anchors IGF-II. Within the AB loop, residues normally act to either stabilise or function as negative regulators of the interaction. These findings have implications for the molecular architecture and evolution of the domain 11 IGF-II-binding site, and the potential interactions with other domains of IGF2R. Biocentre Contribution (Fred Kemp): Wildtype Domain 11 vs IGF-II: Following purification, the interaction of domain 11 with IGF-II was investigated by SPR. To reduce the heterogeneity of the active surface, we immobilised biotinylated IGF-II to the sensor chip surface by affinity capture to streptavidin. In order to minimise mass transport limitation, approximately 50 response units (RU) of biotinylated IGF-II was immobilised, giving a theoretical Rmax for the domain 11 products of 100 RU.
2 (1) Duplicate injections of domain 11 analyte were performed at varying concentrations and the resulting sensorgrams (Figure 1(a)) analysed. As equilibrium was achieved over the course of each injection, KD from steady-state binding was determined by plotting the response at equilibrium against analyte concentration and fitting to a 1:1 binding isotherm (Figure 1(b)). Figure 1: Kinetics of Wild type Domain 11 binding to immobilized IGF-II. The mean value of the dissociation affinity constant (KD± SEM) was 103.3(±4.4) nm with an observed Rmax (97.9(±0.7) RU) very close to the expected value of 100 RU, indicating that all immobilised IGF-II was active and accessible for binding domain 11. Kinetic parameters for the interaction were also determined, based on global fitting of the sensorgrams, providing values for the association (ka) and dissociation (kd) rate constants and the dissociation affinity constant (KD), calculated from kd/ka. Initial attempts to fit the sensorgrams to standard 1:1 Langmuir binding model indicated a slight but significant difference between the fitted model and the observed curves. Global fitting of sensorgrams to a two-state (conformational change) model produced an excellent fit to the observed data (χ2=0.6) (Figure 1(a)). The second, minor component calculated using this model had an extremely low apparent KD (5 M). Our preliminary structural NMR studies of domain 11 interacting with IGF-II have indicated binding-site loop movements upon ligand binding, supporting a conformational change that might account for a two-state model. As the second component made an insignificant contribution to the overall affinity, only the kinetic parameters of the major component were used for comparison between mutants. Using this approach, we determined mean kinetic values (±SEM) for wild-type domain 11 of ka =6.62(±0.13) 105 M 1 s 1 and kd=7.87(±0.29) 10 2 s 1. Mean values of KD calculated from the reported kinetic parameters (118.8(±3.5) nm) were in close agreement with those obtained from steady-state binding (103.3(±4.4) nm) (Table 1).
3 Table 1. Kinetic data obtained from SPR analysis of domain 11 mutants interacting with IGF-II Kinetic parameters k a 1 k d 1 K D 1 Steady state, K D Relative Loop Mutation ( 10 5 M 1 s 1 ) ( 10 2 s 1 ) ( 10 9 M) ( 10 9 M) affinity WT 6.62± ± ± ± AB Y1542A 3.74± ± ± ± S1543A 4.80± ± ± ± E1544A 12.30± ± ± ± K1545A 3.73± ± ± ± CD F1567A Q1569A 7.16± ± ± ± T1570A 4.27± ± ± ± I1572A I1572T FG S1596A 6.21± ± ± ± Figure 2. Representative sensorgrams depicting the range of observed affinities obtained from SPR analysis of mutated IGF2R domain 11 constructs binding to IGF-II. Using the same approach, binding of each mutant construct to IGF-II was assessed by SPR and compared to the wild-type domain 11. Again, sensorgrams were generated using biotinylated IGF-II immobilised to the cell surface and domain 11 mutant proteins as the free analyte (Figure 2). As before, kinetic parameters and steady-state affinity constants were determined by fitting sensorgrams to a two-state (conformational change) model (Table 1).
4 Domain 11 E1544 mutants vs IGF-II: Having established that replacement of glutamate at position 1544 with alanine caused an almost threefold increase in IGF-II affinity, via both on and off rate modification, further mutations were generated at this position in order to further evaluate this observation. Sitedirected mutagenesis was used to generate constructs in which E1544 was replaced with all possible amino acid residues. SPR analysis was conducted, using the purified domain 11 constructs, mutated at position 1544, as analyte. Where possible, kinetic parameters and steady-state affinity data were obtained from the resultant sensorgrams, as before (Figure 3a). Figure 3: Histogram showing steady-state affinities (KD) obtained from domain 11 constructs specifically mutated to replace the glutamate residue at position 1544 with the indicated amino acid. Characterisation and Thermodynamics of the E1544K enhanced affinity mutant: To investigate the relative contribution of electrostatic interactions to the binding of wild-type domain 11 and the high-affinity mutant E1544K to IGF- II, binding of each was assessed in conditions of various ionic concentrations. Kinetic parameters and steady-state affinity data were obtained from the resulting sensorgrams. For each construct, the log of 1/K D, from steady-state analysis, was plotted against the log of the concentrations of NaCl (Figure 3b). The gradient of the straight line obtained from the wild-type and the E1544K mutant was 1.0 and 1.5, respectively, suggesting that electrostatic forces make only a minor contribution to this interaction. The increased influence of ionic strength on IGF-II binding of the E1544K mutant indicates that replacement of the acidic glutamate residue at position 1544 with a basic lysine residue does indeed increase the electrostatic component of the interaction.
5 Figure 4. (a) A van't Hoff plot, (b) Eyring association and (c) dissociation plots showing the linear transformation of affinity (K D ) and kinetic (k a and k d ) parameters varying with reaction temperature for wild-type domain 11 (continuous line) and E1544K (broken line). In order to further understand the nature of the affinityenhancing E1544K mutation, transition-state thermodynamic data were obtained for both domain 11 and the E1544K mutant by analysis of SPR data obtained at a range of reaction temperatures (20-42C). For each construct, the relationship between K D and temperature was represented on a van't Hoff plot (ln(k D) versus 1/T, were T is the temperature in Kelvin, (Figure 4a) and the relationship between k a and k d and temperature on Eyring plots (ln(k a/t) versus 1/T and ln(kd/t) versus 1/T, (Figure 4b&c). In all cases, the transformed data fit well to a straight line. and TΔS ), as summarised in Figure 5. The resulting fitted straight lines were used to obtain values for changes in Gibbs free energy, enthalpy and entropy at equilibrium (ΔG, ΔH, and TΔS, respectively) and for the transition from unassociated to associated reactants (ΔG, ΔH
6 Figure 5. Summary of change in (a) Gibbs free energy (ΔG ), (b) enthalpy (ΔH ) and (c) entropy ( TΔS ) for wild-type domain 11 (continuous line) and E1544K (broken line) interacting with IGF-II, for unassociated reactants (A+B), transition from unassociated to associated binding partners (A B) and at equilibrium (AB). The activation energy that must be overcome during complex formation (Figure 5a A-B) showed only a minor difference between wild-type domain 11 and E1544K, as might be expected for interactions with only a sixfold difference in affinity. However, when the activation free energy of association (ΔG ) was resolved into enthalpy and entropy, a significant difference in the activation pattern of domain 11 versus E1544K was revealed. The activation enthalpy (Figure 5b A- B) of the E1544K mutant was elevated above that of the wildtype. This increased energy requirement might represent that needed to form the additional electrostatic interactions, evident from the enhanced effect of ionic concentration. However, the increased activation enthalpy is compensated for by the E1544K mutant having a considerably reduced activation entropy compared to the wild-type, thereby reducing the entropic barrier that must be overcome in order for the interaction to occur. The interpretation of this would be that either E1544K is less structurally constrained in the transition state, able to move rotate or vibrate more freely, or that E1544K is more able to displace solvent from the binding interface. Output: Oliver J. Zaccheo, Stuart N. Prince, David M. Miller, Christopher Williams, C. Fred Kemp, James Brown, E. Yvonne Jones, Lucy E. Catto, Matthew P. Crump and A. Bassim Hassan. Kinetics of Insulin-like Growth Factor II (IGF-II) Interaction with Domain 11 of the Human IGF-II/Mannose 6-phosphate Receptor: Function of CD and AB Loop Solvent-exposed Residues. Journal of Molecular Biology, Volume 359, Issue 2, 2 June 2006, Pages
Supplementary Figures
 1 Supplementary Figures Supplementary Figure 1 Type I FGFR1 inhibitors (a) Chemical structures of a pyrazolylaminopyrimidine inhibitor (henceforth referred to as PAPI; PDB-code of the FGFR1-PAPI complex:
1 Supplementary Figures Supplementary Figure 1 Type I FGFR1 inhibitors (a) Chemical structures of a pyrazolylaminopyrimidine inhibitor (henceforth referred to as PAPI; PDB-code of the FGFR1-PAPI complex:
Biological Thermodynamics
 Biological Thermodynamics Classical thermodynamics is the only physical theory of universal content concerning which I am convinced that, within the framework of applicability of its basic contents, will
Biological Thermodynamics Classical thermodynamics is the only physical theory of universal content concerning which I am convinced that, within the framework of applicability of its basic contents, will
A Single Outer Sphere Mutation Stabilizes apo- Mn Superoxide Dismutase by 35 C and. Disfavors Mn Binding.
 Supporting information for A Single Outer Sphere Mutation Stabilizes apo- Mn Superoxide Dismutase by 35 C and Disfavors Mn Binding. Anne-Frances Miller* and Ting Wang Department of Chemistry, University
Supporting information for A Single Outer Sphere Mutation Stabilizes apo- Mn Superoxide Dismutase by 35 C and Disfavors Mn Binding. Anne-Frances Miller* and Ting Wang Department of Chemistry, University
Isothermal Titration Calorimetry in Drug Discovery. Geoff Holdgate Structure & Biophysics, Discovery Sciences, AstraZeneca October 2017
 Isothermal Titration Calorimetry in Drug Discovery Geoff Holdgate Structure & Biophysics, Discovery Sciences, AstraZeneca October 217 Introduction Introduction to ITC Strengths / weaknesses & what is required
Isothermal Titration Calorimetry in Drug Discovery Geoff Holdgate Structure & Biophysics, Discovery Sciences, AstraZeneca October 217 Introduction Introduction to ITC Strengths / weaknesses & what is required
Kinetic & Affinity Analysis
 Kinetic & Affinity Analysis An introduction What are kinetics and affinity? Kinetics How fast do things happen? Time-dependent Association how fast molecules bind Dissociation how fast complexes fall apart
Kinetic & Affinity Analysis An introduction What are kinetics and affinity? Kinetics How fast do things happen? Time-dependent Association how fast molecules bind Dissociation how fast complexes fall apart
Energy, Enzymes, and Metabolism. Energy, Enzymes, and Metabolism. A. Energy and Energy Conversions. A. Energy and Energy Conversions
 Energy, Enzymes, and Metabolism Lecture Series 6 Energy, Enzymes, and Metabolism B. ATP: Transferring Energy in Cells D. Molecular Structure Determines Enzyme Fxn Energy is the capacity to do work (cause
Energy, Enzymes, and Metabolism Lecture Series 6 Energy, Enzymes, and Metabolism B. ATP: Transferring Energy in Cells D. Molecular Structure Determines Enzyme Fxn Energy is the capacity to do work (cause
Lecture 7. Surface Reaction Kinetics on a
 Lecture 7 Data processing in SPR Surface Reaction Kinetics on a Biochip Surface plasmon sensor Principle of affinity SP biosensor Data processing Processing steps: zero response to the base line before
Lecture 7 Data processing in SPR Surface Reaction Kinetics on a Biochip Surface plasmon sensor Principle of affinity SP biosensor Data processing Processing steps: zero response to the base line before
Problem solving steps
 Problem solving steps Determine the reaction Write the (balanced) equation ΔG K v Write the equilibrium constant v Find the equilibrium constant using v If necessary, solve for components K K = [ p ] ν
Problem solving steps Determine the reaction Write the (balanced) equation ΔG K v Write the equilibrium constant v Find the equilibrium constant using v If necessary, solve for components K K = [ p ] ν
Other Cells. Hormones. Viruses. Toxins. Cell. Bacteria
 Other Cells Hormones Viruses Toxins Cell Bacteria ΔH < 0 reaction is exothermic, tells us nothing about the spontaneity of the reaction Δ H > 0 reaction is endothermic, tells us nothing about the spontaneity
Other Cells Hormones Viruses Toxins Cell Bacteria ΔH < 0 reaction is exothermic, tells us nothing about the spontaneity of the reaction Δ H > 0 reaction is endothermic, tells us nothing about the spontaneity
An Introduction to Metabolism
 An Introduction to Metabolism I. All of an organism=s chemical reactions taken together is called metabolism. A. Metabolic pathways begin with a specific molecule, which is then altered in a series of
An Introduction to Metabolism I. All of an organism=s chemical reactions taken together is called metabolism. A. Metabolic pathways begin with a specific molecule, which is then altered in a series of
Microcalorimetry for the Life Sciences
 Microcalorimetry for the Life Sciences Why Microcalorimetry? Microcalorimetry is universal detector Heat is generated or absorbed in every chemical process In-solution No molecular weight limitations Label-free
Microcalorimetry for the Life Sciences Why Microcalorimetry? Microcalorimetry is universal detector Heat is generated or absorbed in every chemical process In-solution No molecular weight limitations Label-free
a) Write the reaction that occurs (pay attention to and label ends correctly) 5 AGCTG CAGCT > 5 AGCTG 3 3 TCGAC 5
 Chem 315 Applications Practice Problem Set 1.As you learned in Chem 315, DNA higher order structure formation is a two step process. The first step, nucleation, is entropically the least favorable. The
Chem 315 Applications Practice Problem Set 1.As you learned in Chem 315, DNA higher order structure formation is a two step process. The first step, nucleation, is entropically the least favorable. The
Protein-Ligand Interactions: hydrodynamics and calorimetry
 Protein-Ligand Interactions: hydrodynamics and calorimetry Approach Stephen E. Harding Babur Z. Chowdhry OXFORD UNIVERSITY PRESS , New York Oxford University Press, 2001 978-0-19-963746-1 List of protocols
Protein-Ligand Interactions: hydrodynamics and calorimetry Approach Stephen E. Harding Babur Z. Chowdhry OXFORD UNIVERSITY PRESS , New York Oxford University Press, 2001 978-0-19-963746-1 List of protocols
Free Energy. because H is negative doesn't mean that G will be negative and just because S is positive doesn't mean that G will be negative.
 Biochemistry 462a Bioenergetics Reading - Lehninger Principles, Chapter 14, pp. 485-512 Practice problems - Chapter 14: 2-8, 10, 12, 13; Physical Chemistry extra problems, free energy problems Free Energy
Biochemistry 462a Bioenergetics Reading - Lehninger Principles, Chapter 14, pp. 485-512 Practice problems - Chapter 14: 2-8, 10, 12, 13; Physical Chemistry extra problems, free energy problems Free Energy
schematic diagram; EGF binding, dimerization, phosphorylation, Grb2 binding, etc.
 Lecture 1: Noncovalent Biomolecular Interactions Bioengineering and Modeling of biological processes -e.g. tissue engineering, cancer, autoimmune disease Example: RTK signaling, e.g. EGFR Growth responses
Lecture 1: Noncovalent Biomolecular Interactions Bioengineering and Modeling of biological processes -e.g. tissue engineering, cancer, autoimmune disease Example: RTK signaling, e.g. EGFR Growth responses
BIOCHEMISTRY. František Vácha. JKU, Linz.
 BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
BIOCHEMISTRY František Vácha http://www.prf.jcu.cz/~vacha/ JKU, Linz Recommended reading: D.L. Nelson, M.M. Cox Lehninger Principles of Biochemistry D.J. Voet, J.G. Voet, C.W. Pratt Principles of Biochemistry
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability
 Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Proteins are not rigid structures: Protein dynamics, conformational variability, and thermodynamic stability Dr. Andrew Lee UNC School of Pharmacy (Div. Chemical Biology and Medicinal Chemistry) UNC Med
Gibbs Free Energy. Evaluating spontaneity
 Gibbs Free Energy Evaluating spontaneity Predicting Spontaneity An increase in entropy; Changing from a more structured to less structured physical state: Solid to liquid Liquid to gas Increase in temperature
Gibbs Free Energy Evaluating spontaneity Predicting Spontaneity An increase in entropy; Changing from a more structured to less structured physical state: Solid to liquid Liquid to gas Increase in temperature
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy
 Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
Lecture Series 9 Cellular Pathways That Harvest Chemical Energy Reading Assignments Review Chapter 3 Energy, Catalysis, & Biosynthesis Read Chapter 13 How Cells obtain Energy from Food Read Chapter 14
Supplementary Figure 1 Schematic overview of ASTNs in neuronal migration. (a) Schematic of roles played by ASTNs 1 and 2. ASTN-1-mediated adhesions
 Supplementary Figure 1 Schematic overview of ASTNs in neuronal migration. (a) Schematic of roles played by ASTNs 1 and 2. ASTN-1-mediated adhesions undergo endocytosis into clathrin-coated vesicles dependent
Supplementary Figure 1 Schematic overview of ASTNs in neuronal migration. (a) Schematic of roles played by ASTNs 1 and 2. ASTN-1-mediated adhesions undergo endocytosis into clathrin-coated vesicles dependent
Chapter 1. Topic: Overview of basic principles
 Chapter 1 Topic: Overview of basic principles Four major themes of biochemistry I. What are living organism made from? II. How do organism acquire and use energy? III. How does an organism maintain its
Chapter 1 Topic: Overview of basic principles Four major themes of biochemistry I. What are living organism made from? II. How do organism acquire and use energy? III. How does an organism maintain its
= (-22) = +2kJ /mol
 Lecture 8: Thermodynamics & Protein Stability Assigned reading in Campbell: Chapter 4.4-4.6 Key Terms: DG = -RT lnk eq = DH - TDS Transition Curve, Melting Curve, Tm DH calculation DS calculation van der
Lecture 8: Thermodynamics & Protein Stability Assigned reading in Campbell: Chapter 4.4-4.6 Key Terms: DG = -RT lnk eq = DH - TDS Transition Curve, Melting Curve, Tm DH calculation DS calculation van der
S2004 Methods for characterization of biomolecular interactions - classical versus modern
 S2004 Methods for characterization of biomolecular interactions - classical versus modern Isothermal Titration Calorimetry (ITC) Eva Dubská email: eva.dubska@ceitec.cz Outline Calorimetry - history + a
S2004 Methods for characterization of biomolecular interactions - classical versus modern Isothermal Titration Calorimetry (ITC) Eva Dubská email: eva.dubska@ceitec.cz Outline Calorimetry - history + a
Biology Chemistry & Physics of Biomolecules. Examination #1. Proteins Module. September 29, Answer Key
 Biology 5357 Chemistry & Physics of Biomolecules Examination #1 Proteins Module September 29, 2017 Answer Key Question 1 (A) (5 points) Structure (b) is more common, as it contains the shorter connection
Biology 5357 Chemistry & Physics of Biomolecules Examination #1 Proteins Module September 29, 2017 Answer Key Question 1 (A) (5 points) Structure (b) is more common, as it contains the shorter connection
High Specificity and Reversibility
 Lecture #8 The Cell as a Machine High Specificity and Reversibility In considering the problem of transcription factor binding in the nucleus and the great specificity that is called for to transcribe
Lecture #8 The Cell as a Machine High Specificity and Reversibility In considering the problem of transcription factor binding in the nucleus and the great specificity that is called for to transcribe
ANSWER KEY. Chemistry 25 (Spring term 2015) Midterm Examination
 Name ANSWER KEY Chemistry 25 (Spring term 2015) Midterm Examination 1 (15 pts) Short answers 1A (3 pts) Consider the following system at two time points A and B. The system is divided by a moveable partition.
Name ANSWER KEY Chemistry 25 (Spring term 2015) Midterm Examination 1 (15 pts) Short answers 1A (3 pts) Consider the following system at two time points A and B. The system is divided by a moveable partition.
Solutions and Non-Covalent Binding Forces
 Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
Chapter 6- An Introduction to Metabolism*
 Chapter 6- An Introduction to Metabolism* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. The Energy of Life
Chapter 6- An Introduction to Metabolism* *Lecture notes are to be used as a study guide only and do not represent the comprehensive information you will need to know for the exams. The Energy of Life
Problem Set 5 Question 1
 2.32 Problem Set 5 Question As discussed in class, drug discovery often involves screening large libraries of small molecules to identify those that have favorable interactions with a certain druggable
2.32 Problem Set 5 Question As discussed in class, drug discovery often involves screening large libraries of small molecules to identify those that have favorable interactions with a certain druggable
DSC Characterization of the Structure/Function Relationship for Proteins
 DSC Characterization of the Structure/Function Relationship for Proteins Differential Scanning Calorimetry (DSC) DSC is recognized as Gold Std technique for measuring molecular thermal stability and structure
DSC Characterization of the Structure/Function Relationship for Proteins Differential Scanning Calorimetry (DSC) DSC is recognized as Gold Std technique for measuring molecular thermal stability and structure
Principles of Bioenergetics. Lehninger 3 rd ed. Chapter 14
 1 Principles of Bioenergetics Lehninger 3 rd ed. Chapter 14 2 Metabolism A highly coordinated cellular activity aimed at achieving the following goals: Obtain chemical energy. Convert nutrient molecules
1 Principles of Bioenergetics Lehninger 3 rd ed. Chapter 14 2 Metabolism A highly coordinated cellular activity aimed at achieving the following goals: Obtain chemical energy. Convert nutrient molecules
FW 1 CDR 1 FW 2 CDR 2
 Supplementary Figure 1 Supplementary Figure 1: Interface of the E9:Fas structure. The two interfaces formed by V H and V L of E9 with Fas are shown in stereo. The Fas receptor is represented as a surface
Supplementary Figure 1 Supplementary Figure 1: Interface of the E9:Fas structure. The two interfaces formed by V H and V L of E9 with Fas are shown in stereo. The Fas receptor is represented as a surface
Patrick: An Introduction to Medicinal Chemistry 5e Chapter 04
 01) Which of the following statements is not true about receptors? a. Most receptors are proteins situated inside the cell. b. Receptors contain a hollow or cleft on their surface which is known as a binding
01) Which of the following statements is not true about receptors? a. Most receptors are proteins situated inside the cell. b. Receptors contain a hollow or cleft on their surface which is known as a binding
Kinetic Analysis of Interaction of BRCA1 Tandem Breast Cancer C-Terminal Domains with Phosphorylated Peptides Reveals Two Binding Conformations
 9866 Biochemistry 2008, 47, 9866 9879 Kinetic Analysis of Interaction of BRCA1 Tandem Breast Cancer C-Terminal Domains with Phosphorylated Peptides Reveals Two Binding Conformations Yves Nominé, Maria
9866 Biochemistry 2008, 47, 9866 9879 Kinetic Analysis of Interaction of BRCA1 Tandem Breast Cancer C-Terminal Domains with Phosphorylated Peptides Reveals Two Binding Conformations Yves Nominé, Maria
Membrane Protein Channels
 Membrane Protein Channels Potassium ions queuing up in the potassium channel Pumps: 1000 s -1 Channels: 1000000 s -1 Pumps & Channels The lipid bilayer of biological membranes is intrinsically impermeable
Membrane Protein Channels Potassium ions queuing up in the potassium channel Pumps: 1000 s -1 Channels: 1000000 s -1 Pumps & Channels The lipid bilayer of biological membranes is intrinsically impermeable
Full file at https://fratstock.eu
 Chapter 03 1. a. DG=DH-TDS Δ G = 80 kj ( 98 K) 0.790 kj = 44.6 kj K b. ΔG = 0 @ T m. Unfolding will be favorable at temperatures above the T m. Δ G =Δ H TΔ S 0 kj kj 80 ( xk) 0.790 K 0 Δ G = = 354.4 K
Chapter 03 1. a. DG=DH-TDS Δ G = 80 kj ( 98 K) 0.790 kj = 44.6 kj K b. ΔG = 0 @ T m. Unfolding will be favorable at temperatures above the T m. Δ G =Δ H TΔ S 0 kj kj 80 ( xk) 0.790 K 0 Δ G = = 354.4 K
Kinetic Models of Protein Binding
 Kinetic Models of Protein Binding 1:1 Langmuir Binding The ideal model of binding in which one ligand molecule interacts with one analyte molecule. assumed all binding sites are equivalent and independent
Kinetic Models of Protein Binding 1:1 Langmuir Binding The ideal model of binding in which one ligand molecule interacts with one analyte molecule. assumed all binding sites are equivalent and independent
Substrate-dependent switching of the allosteric binding mechanism of a dimeric enzyme
 Supplementary Information: Substrate-dependent switching of the allosteric binding mechanism of a dimeric enzyme Lee Freiburger, 1 Teresa Miletti, 1 Siqi Zhu, 1 Oliver Baettig, Albert Berghuis, Karine
Supplementary Information: Substrate-dependent switching of the allosteric binding mechanism of a dimeric enzyme Lee Freiburger, 1 Teresa Miletti, 1 Siqi Zhu, 1 Oliver Baettig, Albert Berghuis, Karine
What binds to Hb in addition to O 2?
 Reading: Ch5; 158-169, 162-166, 169-174 Problems: Ch5 (text); 3,7,8,10 Ch5 (study guide-facts); 1,2,3,4,5,8 Ch5 (study guide-apply); 2,3 Remember Today at 5:30 in CAS-522 is the second chance for the MB
Reading: Ch5; 158-169, 162-166, 169-174 Problems: Ch5 (text); 3,7,8,10 Ch5 (study guide-facts); 1,2,3,4,5,8 Ch5 (study guide-apply); 2,3 Remember Today at 5:30 in CAS-522 is the second chance for the MB
Supporting Information
 Supporting Information Arai et al. 10.1073/pnas.15179911 SI Text Protein Expression and Purification. Myb3 (mouse, residues 84 315) was expressed in Escherichia coli as a fusion with the B1 domain of protein
Supporting Information Arai et al. 10.1073/pnas.15179911 SI Text Protein Expression and Purification. Myb3 (mouse, residues 84 315) was expressed in Escherichia coli as a fusion with the B1 domain of protein
Chem 728 Spring 2012 April 14, 2012
 Chem 728 Spring 2012 April 14, 2012 Origin Assignment 6 Answers DUE: April 9, noon Cooperative Ligand Binding in a Dimeric Protein Origin_Assign_6_Binding_Data.xlsx contains the results of ligand binding
Chem 728 Spring 2012 April 14, 2012 Origin Assignment 6 Answers DUE: April 9, noon Cooperative Ligand Binding in a Dimeric Protein Origin_Assign_6_Binding_Data.xlsx contains the results of ligand binding
Supplementary Information
 Supplementary Information Adenosyltransferase Tailors and Delivers Coenzyme B 12 Dominique Padovani 1,2, Tetyana Labunska 2, Bruce A. Palfey 1, David P. Ballou 1 and Ruma Banerjee 1,2 * 1 Biological Chemistry
Supplementary Information Adenosyltransferase Tailors and Delivers Coenzyme B 12 Dominique Padovani 1,2, Tetyana Labunska 2, Bruce A. Palfey 1, David P. Ballou 1 and Ruma Banerjee 1,2 * 1 Biological Chemistry
Cholera Toxin Invasion
 Protein-carbohydrate interactions: Isothermal Titration Calorimetry Dr Bruce Turnbull School of Chemistry and Astbury Centre for Structural Molecular Biology University of Leeds Cholera Toxin Invasion
Protein-carbohydrate interactions: Isothermal Titration Calorimetry Dr Bruce Turnbull School of Chemistry and Astbury Centre for Structural Molecular Biology University of Leeds Cholera Toxin Invasion
BIOC : Homework 1 Due 10/10
 Contact information: Name: Student # BIOC530 2012: Homework 1 Due 10/10 Department Email address The following problems are based on David Baker s lectures of forces and protein folding. When numerical
Contact information: Name: Student # BIOC530 2012: Homework 1 Due 10/10 Department Email address The following problems are based on David Baker s lectures of forces and protein folding. When numerical
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/4/e1600663/dc1 Supplementary Materials for A dynamic hydrophobic core orchestrates allostery in protein kinases Jonggul Kim, Lalima G. Ahuja, Fa-An Chao, Youlin
advances.sciencemag.org/cgi/content/full/3/4/e1600663/dc1 Supplementary Materials for A dynamic hydrophobic core orchestrates allostery in protein kinases Jonggul Kim, Lalima G. Ahuja, Fa-An Chao, Youlin
CHAPTER 5. EQUILIBRIUM AND THERMODYNAMIC INVESTIGATION OF As(III) AND As(V) REMOVAL BY MAGNETITE NANOPARTICLES COATED SAND
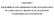 CHAPTER 5 EQUILIBRIUM AND THERMODYNAMIC INVESTIGATION OF As(III) AND As(V) REMOVAL BY MAGNETITE NANOPARTICLES COATED SAND 85 86 5.1. INTRODUCTION Since temperature plays an important role in the adsorption
CHAPTER 5 EQUILIBRIUM AND THERMODYNAMIC INVESTIGATION OF As(III) AND As(V) REMOVAL BY MAGNETITE NANOPARTICLES COATED SAND 85 86 5.1. INTRODUCTION Since temperature plays an important role in the adsorption
Effect of the Single and Double Chain Surfactant Cobalt(III) Complexes on Their Hydrophobicity, Micelle Formation,
 Electronic Supplementary Material (ESI) for Inorganic Chemistry Frontiers. This journal is the Partner Organisations 2014 Supplementary Information Effect of the Single and Double Chain Surfactant Cobalt(III)
Electronic Supplementary Material (ESI) for Inorganic Chemistry Frontiers. This journal is the Partner Organisations 2014 Supplementary Information Effect of the Single and Double Chain Surfactant Cobalt(III)
Introduction to electrophysiology. Dr. Tóth András
 Introduction to electrophysiology Dr. Tóth András Topics Transmembran transport Donnan equilibrium Resting potential Ion channels Local and action potentials Intra- and extracellular propagation of the
Introduction to electrophysiology Dr. Tóth András Topics Transmembran transport Donnan equilibrium Resting potential Ion channels Local and action potentials Intra- and extracellular propagation of the
Water. 2.1 Weak Interactions in Aqueous Sy stems Ionization of Water, Weak Acids, and Weak Bases 58
 Home http://www.macmillanhighered.com/launchpad/lehninger6e... 1 of 1 1/6/2016 3:07 PM 2 Printed Page 47 Water 2.1 Weak Interactions in Aqueous Sy stems 47 2.2 Ionization of Water, Weak Acids, and Weak
Home http://www.macmillanhighered.com/launchpad/lehninger6e... 1 of 1 1/6/2016 3:07 PM 2 Printed Page 47 Water 2.1 Weak Interactions in Aqueous Sy stems 47 2.2 Ionization of Water, Weak Acids, and Weak
3. Based on how energy is stored in the molecules, explain why ΔG is independent of the path of the reaction.
 B. Thermodynamics 1. What is "free energy"? 2. Where is this energy stored? We say that ΔG is a thermodynamic property, meaning that it is independent of the way that the conversion of reactants to products
B. Thermodynamics 1. What is "free energy"? 2. Where is this energy stored? We say that ΔG is a thermodynamic property, meaning that it is independent of the way that the conversion of reactants to products
Enzyme Enzymes are proteins that act as biological catalysts. Enzymes accelerate, or catalyze, chemical reactions. The molecules at the beginning of
 Enzyme Enzyme Enzymes are proteins that act as biological catalysts. Enzymes accelerate, or catalyze, chemical reactions. The molecules at the beginning of the process are called substrates and the enzyme
Enzyme Enzyme Enzymes are proteins that act as biological catalysts. Enzymes accelerate, or catalyze, chemical reactions. The molecules at the beginning of the process are called substrates and the enzyme
7/19/2011. Models of Solution. State of Equilibrium. State of Equilibrium Chemical Reaction
 Models of Solution Chemistry- I State of Equilibrium A covered cup of coffee will not be colder than or warmer than the room temperature Heat is defined as a form of energy that flows from a high temperature
Models of Solution Chemistry- I State of Equilibrium A covered cup of coffee will not be colder than or warmer than the room temperature Heat is defined as a form of energy that flows from a high temperature
3.1 Metabolism and Energy
 3.1 Metabolism and Energy Metabolism All of the chemical reactions in a cell To transform matter and energy Step-by-step sequences metabolic pathways Metabolic Pathways Anabolic reactions Build large molecules
3.1 Metabolism and Energy Metabolism All of the chemical reactions in a cell To transform matter and energy Step-by-step sequences metabolic pathways Metabolic Pathways Anabolic reactions Build large molecules
f) Adding an enzyme does not change the Gibbs free energy. It only increases the rate of the reaction by lowering the activation energy.
 Problem Set 2-Answer Key BILD1 SP16 1) How does an enzyme catalyze a chemical reaction? Define the terms and substrate and active site. An enzyme lowers the energy of activation so the reaction proceeds
Problem Set 2-Answer Key BILD1 SP16 1) How does an enzyme catalyze a chemical reaction? Define the terms and substrate and active site. An enzyme lowers the energy of activation so the reaction proceeds
Interpreting and evaluating biological NMR in the literature. Worksheet 1
 Interpreting and evaluating biological NMR in the literature Worksheet 1 1D NMR spectra Application of RF pulses of specified lengths and frequencies can make certain nuclei detectable We can selectively
Interpreting and evaluating biological NMR in the literature Worksheet 1 1D NMR spectra Application of RF pulses of specified lengths and frequencies can make certain nuclei detectable We can selectively
Problem Set # 1
 20.320 Problem Set # 1 September 17 th, 2010 Due on September 24 th, 2010 at 11:59am. No extensions will be granted. General Instructions: 1. You are expected to state all your assumptions and provide
20.320 Problem Set # 1 September 17 th, 2010 Due on September 24 th, 2010 at 11:59am. No extensions will be granted. General Instructions: 1. You are expected to state all your assumptions and provide
Dental Biochemistry EXAM I
 Dental Biochemistry EXAM I August 29, 2005 In the reaction below: CH 3 -CH 2 OH -~ ethanol CH 3 -CHO acetaldehyde A. acetoacetate is being produced B. ethanol is being oxidized to acetaldehyde C. acetaldehyde
Dental Biochemistry EXAM I August 29, 2005 In the reaction below: CH 3 -CH 2 OH -~ ethanol CH 3 -CHO acetaldehyde A. acetoacetate is being produced B. ethanol is being oxidized to acetaldehyde C. acetaldehyde
Ben McFarland Department of Chemistry Seattle Pacific University. Designing Proteins. The Creative Potential of Enthalpy and Entropy
 Ben McFarland Department of Chemistry Seattle Pacific University Designing Proteins The Creative Potential of Enthalpy and Entropy Proteins are chemical: they have defined atomic structures accessible
Ben McFarland Department of Chemistry Seattle Pacific University Designing Proteins The Creative Potential of Enthalpy and Entropy Proteins are chemical: they have defined atomic structures accessible
4 Examples of enzymes
 Catalysis 1 4 Examples of enzymes Adding water to a substrate: Serine proteases. Carbonic anhydrase. Restrictions Endonuclease. Transfer of a Phosphoryl group from ATP to a nucleotide. Nucleoside monophosphate
Catalysis 1 4 Examples of enzymes Adding water to a substrate: Serine proteases. Carbonic anhydrase. Restrictions Endonuclease. Transfer of a Phosphoryl group from ATP to a nucleotide. Nucleoside monophosphate
Presentation Microcalorimetry for Life Science Research
 Presentation Microcalorimetry for Life Science Research MicroCalorimetry The Universal Detector Heat is either generated or absorbed in every chemical process Capable of thermal measurements over a wide
Presentation Microcalorimetry for Life Science Research MicroCalorimetry The Universal Detector Heat is either generated or absorbed in every chemical process Capable of thermal measurements over a wide
ISoTherMal TITraTIon Calorimetry
 ISoTherMal TITraTIon Calorimetry With the Nano ITC, heat effects as small as 1 nanojoules are detectable using one nanomole or less of biopolymer. The Nano ITC uses a solid-state thermoelectric heating
ISoTherMal TITraTIon Calorimetry With the Nano ITC, heat effects as small as 1 nanojoules are detectable using one nanomole or less of biopolymer. The Nano ITC uses a solid-state thermoelectric heating
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a
 Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Supplementary figure 1 Application of tmfret in LeuT. (a) To assess the feasibility of using tmfret for distance-dependent measurements in LeuT, a series of tmfret-pairs comprised of single cysteine mutants
Potassium channel gating and structure!
 Reading: Potassium channel gating and structure Hille (3rd ed.) chapts 10, 13, 17 Doyle et al. The Structure of the Potassium Channel: Molecular Basis of K1 Conduction and Selectivity. Science 280:70-77
Reading: Potassium channel gating and structure Hille (3rd ed.) chapts 10, 13, 17 Doyle et al. The Structure of the Potassium Channel: Molecular Basis of K1 Conduction and Selectivity. Science 280:70-77
Advanced Higher Biology. Unit 1- Cells and Proteins 2c) Membrane Proteins
 Advanced Higher Biology Unit 1- Cells and Proteins 2c) Membrane Proteins Membrane Structure Phospholipid bilayer Transmembrane protein Integral protein Movement of Molecules Across Membranes Phospholipid
Advanced Higher Biology Unit 1- Cells and Proteins 2c) Membrane Proteins Membrane Structure Phospholipid bilayer Transmembrane protein Integral protein Movement of Molecules Across Membranes Phospholipid
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10458 Active Site Remodeling in the Bifunctional Fructose-1,6- bisphosphate aldolase/phosphatase Juan Du, Rafael F. Say, Wei Lü, Georg Fuchs & Oliver Einsle SUPPLEMENTARY FIGURES Figure
doi:10.1038/nature10458 Active Site Remodeling in the Bifunctional Fructose-1,6- bisphosphate aldolase/phosphatase Juan Du, Rafael F. Say, Wei Lü, Georg Fuchs & Oliver Einsle SUPPLEMENTARY FIGURES Figure
General Biology. The Energy of Life The living cell is a miniature factory where thousands of reactions occur; it converts energy in many ways
 Course No: BNG2003 Credits: 3.00 General Biology 5. An Introduction into Cell Metabolism The Energy of Life The living cell is a miniature factory where thousands of reactions occur; it converts energy
Course No: BNG2003 Credits: 3.00 General Biology 5. An Introduction into Cell Metabolism The Energy of Life The living cell is a miniature factory where thousands of reactions occur; it converts energy
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
= 16! = 16! W A = 3 = 3 N = = W B 3!3!10! = ΔS = nrln V. = ln ( 3 ) V 1 = 27.4 J.
 Answer key: Q1A Both configurations are equally likely because the particles are non-interacting (i.e., the energy does not favor one configuration over another). For A M = 16 N = 6 W A = 16! 0.9 101 =
Answer key: Q1A Both configurations are equally likely because the particles are non-interacting (i.e., the energy does not favor one configuration over another). For A M = 16 N = 6 W A = 16! 0.9 101 =
CHAPTER 2. Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules
 CHAPTER 2 Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules 2-1 Kinetics and Thermodynamics of Simple Chemical Processes Chemical thermodynamics: Is concerned with the extent that
CHAPTER 2 Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules 2-1 Kinetics and Thermodynamics of Simple Chemical Processes Chemical thermodynamics: Is concerned with the extent that
12A Entropy. Entropy change ( S) N Goalby chemrevise.org 1. System and Surroundings
 12A Entropy Entropy change ( S) A SPONTANEOUS PROCESS (e.g. diffusion) will proceed on its own without any external influence. A problem with H A reaction that is exothermic will result in products that
12A Entropy Entropy change ( S) A SPONTANEOUS PROCESS (e.g. diffusion) will proceed on its own without any external influence. A problem with H A reaction that is exothermic will result in products that
BUDE. A General Purpose Molecular Docking Program Using OpenCL. Richard B Sessions
 BUDE A General Purpose Molecular Docking Program Using OpenCL Richard B Sessions 1 The molecular docking problem receptor ligand Proteins typically O(1000) atoms Ligands typically O(100) atoms predicted
BUDE A General Purpose Molecular Docking Program Using OpenCL Richard B Sessions 1 The molecular docking problem receptor ligand Proteins typically O(1000) atoms Ligands typically O(100) atoms predicted
2. In regards to the fluid mosaic model, which of the following is TRUE?
 General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
General Biology: Exam I Sample Questions 1. How many electrons are required to fill the valence shell of a neutral atom with an atomic number of 24? a. 0 the atom is inert b. 1 c. 2 d. 4 e. 6 2. In regards
Biotechnology of Proteins. The Source of Stability in Proteins (III) Fall 2015
 Biotechnology of Proteins The Source of Stability in Proteins (III) Fall 2015 Conformational Entropy of Unfolding It is The factor that makes the greatest contribution to stabilization of the unfolded
Biotechnology of Proteins The Source of Stability in Proteins (III) Fall 2015 Conformational Entropy of Unfolding It is The factor that makes the greatest contribution to stabilization of the unfolded
Applications of Free Energy. NC State University
 Chemistry 433 Lecture 15 Applications of Free Energy NC State University Thermodynamics of glycolysis Reaction kj/mol D-glucose + ATP D-glucose-6-phosphate + ADP ΔG o = -16.7 D-glucose-6-phosphate p D-fructose-6-phosphate
Chemistry 433 Lecture 15 Applications of Free Energy NC State University Thermodynamics of glycolysis Reaction kj/mol D-glucose + ATP D-glucose-6-phosphate + ADP ΔG o = -16.7 D-glucose-6-phosphate p D-fructose-6-phosphate
Concept review: Binding equilibria
 Concept review: Binding equilibria 1 Binding equilibria and association/dissociation constants 2 The binding of a protein to a ligand at equilibrium can be written as: P + L PL And so the equilibrium constant
Concept review: Binding equilibria 1 Binding equilibria and association/dissociation constants 2 The binding of a protein to a ligand at equilibrium can be written as: P + L PL And so the equilibrium constant
A rule of seven in Watson-Crick base-pairing of mismatched sequences
 A rule of seven in Watson-Crick base-pairing of mismatched sequences Ibrahim I. Cisse 1,3, Hajin Kim 1,2, Taekjip Ha 1,2 1 Department of Physics and Center for the Physics of Living Cells, University of
A rule of seven in Watson-Crick base-pairing of mismatched sequences Ibrahim I. Cisse 1,3, Hajin Kim 1,2, Taekjip Ha 1,2 1 Department of Physics and Center for the Physics of Living Cells, University of
Gibb s Free Energy. This value represents the maximum amount of useful work (non PV-work) that can be obtained by a system.
 Gibb s Free Energy 1. What is Gibb s free energy? What is its symbol? This value represents the maximum amount of useful work (non PV-work) that can be obtained by a system. It is symbolized by G. We only
Gibb s Free Energy 1. What is Gibb s free energy? What is its symbol? This value represents the maximum amount of useful work (non PV-work) that can be obtained by a system. It is symbolized by G. We only
Biophysics 490M Project
 Biophysics 490M Project Dan Han Department of Biochemistry Structure Exploration of aa 3 -type Cytochrome c Oxidase from Rhodobacter sphaeroides I. Introduction: All organisms need energy to live. They
Biophysics 490M Project Dan Han Department of Biochemistry Structure Exploration of aa 3 -type Cytochrome c Oxidase from Rhodobacter sphaeroides I. Introduction: All organisms need energy to live. They
BIOLOGICAL SCIENCE. Lecture Presentation by Cindy S. Malone, PhD, California State University Northridge. FIFTH EDITION Freeman Quillin Allison
 BIOLOGICAL SCIENCE FIFTH EDITION Freeman Quillin Allison 8 Lecture Presentation by Cindy S. Malone, PhD, California State University Northridge Roadmap 8 In this chapter you will learn how Enzymes use
BIOLOGICAL SCIENCE FIFTH EDITION Freeman Quillin Allison 8 Lecture Presentation by Cindy S. Malone, PhD, California State University Northridge Roadmap 8 In this chapter you will learn how Enzymes use
Thermodynamics and Kinetics
 Thermodynamics and Kinetics Lecture 12 Free Energy Applications NC State University Isolated system requires DS > 0 DS sys > 0 Isolated system: Entropy increases for any spontaneous process System and
Thermodynamics and Kinetics Lecture 12 Free Energy Applications NC State University Isolated system requires DS > 0 DS sys > 0 Isolated system: Entropy increases for any spontaneous process System and
concentrations (molarity) rate constant, (k), depends on size, speed, kind of molecule, temperature, etc.
 #80 Notes Ch. 12, 13, 16, 17 Rates, Equilibriums, Energies Ch. 12 I. Reaction Rates NO 2(g) + CO (g) NO (g) + CO 2(g) Rate is defined in terms of the rate of disappearance of one of the reactants, but
#80 Notes Ch. 12, 13, 16, 17 Rates, Equilibriums, Energies Ch. 12 I. Reaction Rates NO 2(g) + CO (g) NO (g) + CO 2(g) Rate is defined in terms of the rate of disappearance of one of the reactants, but
Isothermal experiments characterize time-dependent aggregation and unfolding
 1 Energy Isothermal experiments characterize time-dependent aggregation and unfolding Technical ote Introduction Kinetic measurements have, for decades, given protein scientists insight into the mechanisms
1 Energy Isothermal experiments characterize time-dependent aggregation and unfolding Technical ote Introduction Kinetic measurements have, for decades, given protein scientists insight into the mechanisms
Intermolecular forces and enthalpies in bacterial adhesion
 Intermolecular forces and enthalpies in bacterial adhesion Henk J. Busscher, Henny C. van der Mei and Willem Norde University Medical Center Groningen and University of Groningen Department of BioMedical
Intermolecular forces and enthalpies in bacterial adhesion Henk J. Busscher, Henny C. van der Mei and Willem Norde University Medical Center Groningen and University of Groningen Department of BioMedical
Name: TF: Section Time: LS1a ICE 5. Practice ICE Version B
 Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Department of Chemistry and Biochemistry University of Lethbridge. Biochemistry II. Bioenergetics
 Department of Chemistry and Biochemistry University of Lethbridge II. Bioenergetics Slide 1 Bioenergetics Bioenergetics is the quantitative study of energy relationships and energy conversion in biological
Department of Chemistry and Biochemistry University of Lethbridge II. Bioenergetics Slide 1 Bioenergetics Bioenergetics is the quantitative study of energy relationships and energy conversion in biological
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
MCB100A/Chem130 MidTerm Exam 2 April 4, 2013
 MCBA/Chem Miderm Exam 2 April 4, 2 Name Student ID rue/false (2 points each).. he Boltzmann constant, k b sets the energy scale for observing energy microstates 2. Atoms with favorable electronic configurations
MCBA/Chem Miderm Exam 2 April 4, 2 Name Student ID rue/false (2 points each).. he Boltzmann constant, k b sets the energy scale for observing energy microstates 2. Atoms with favorable electronic configurations
I: Life and Energy. Lecture 2: Solutions and chemical potential; Osmotic pressure (B Lentz).
 I: Life and Energy Lecture 1: What is life? An attempt at definition. Energy, heat, and work: Temperature and thermal equilibrium. The First Law. Thermodynamic states and state functions. Reversible and
I: Life and Energy Lecture 1: What is life? An attempt at definition. Energy, heat, and work: Temperature and thermal equilibrium. The First Law. Thermodynamic states and state functions. Reversible and
Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition
 Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Supplementary Information to Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition Nadine Czudnochowski 1,2, *, Christian A. Bösken 1, * & Matthias Geyer 1 1 Max-Planck-Institut
Tracking Protein Allostery in Evolution
 Tracking Protein Allostery in Evolution Glycogen phosphorylase frees sugars to provide energy GP orthologs diverged 600,000,000 years can respond to transcription controls, metabolite concentrations and
Tracking Protein Allostery in Evolution Glycogen phosphorylase frees sugars to provide energy GP orthologs diverged 600,000,000 years can respond to transcription controls, metabolite concentrations and
Microteaching topics solutions Department of Biological Engineering TA Training
 Microteaching topics solutions Department of Biological Engineering TA Training Here we include brief solution outlines, so you can focus your efforts on how best to teach the topic at hand, and not on
Microteaching topics solutions Department of Biological Engineering TA Training Here we include brief solution outlines, so you can focus your efforts on how best to teach the topic at hand, and not on
Neurobiology Biomed 509 Ion Channels
 eurobiology Biomed 509 Ion hannels References: Luo 2.4, 2.4 2., (3.20 3.2), M 2.8 I. ores vs. carriers In order for hydrophilic ions to transit the hydrophobic membrane, they need to have a specific pathway
eurobiology Biomed 509 Ion hannels References: Luo 2.4, 2.4 2., (3.20 3.2), M 2.8 I. ores vs. carriers In order for hydrophilic ions to transit the hydrophobic membrane, they need to have a specific pathway
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
2013 W. H. Freeman and Company. 6 Enzymes
 2013 W. H. Freeman and Company 6 Enzymes CHAPTER 6 Enzymes Key topics about enzyme function: Physiological significance of enzymes Origin of catalytic power of enzymes Chemical mechanisms of catalysis
2013 W. H. Freeman and Company 6 Enzymes CHAPTER 6 Enzymes Key topics about enzyme function: Physiological significance of enzymes Origin of catalytic power of enzymes Chemical mechanisms of catalysis
1.8 Thermodynamics. N Goalby chemrevise.org. Definitions of enthalpy changes
 1.8 Thermodynamics Definitions of enthalpy changes Enthalpy change of formation The standard enthalpy change of formation of a compound is the energy transferred when 1 mole of the compound is formed from
1.8 Thermodynamics Definitions of enthalpy changes Enthalpy change of formation The standard enthalpy change of formation of a compound is the energy transferred when 1 mole of the compound is formed from
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
Monolayers. Factors affecting the adsorption from solution. Adsorption of amphiphilic molecules on solid support
 Monolayers Adsorption as process Adsorption of gases on solids Adsorption of solutions on solids Factors affecting the adsorption from solution Adsorption of amphiphilic molecules on solid support Adsorption
Monolayers Adsorption as process Adsorption of gases on solids Adsorption of solutions on solids Factors affecting the adsorption from solution Adsorption of amphiphilic molecules on solid support Adsorption
Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur. Lecture - 06 Protein Structure IV
 Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur Lecture - 06 Protein Structure IV We complete our discussion on Protein Structures today. And just to recap
Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur Lecture - 06 Protein Structure IV We complete our discussion on Protein Structures today. And just to recap
Protein Folding experiments and theory
 Protein Folding experiments and theory 1, 2,and 3 Protein Structure Fig. 3-16 from Lehninger Biochemistry, 4 th ed. The 3D structure is not encoded at the single aa level Hydrogen Bonding Shared H atom
Protein Folding experiments and theory 1, 2,and 3 Protein Structure Fig. 3-16 from Lehninger Biochemistry, 4 th ed. The 3D structure is not encoded at the single aa level Hydrogen Bonding Shared H atom
8 A Microscopic Approach to Entropy
 8 A Microscopic Approach to Entropy The thermodynamic approach www.xtremepapers.com Internal energy and enthalpy When energy is added to a body, its internal energy U increases by an amount ΔU. The energy
8 A Microscopic Approach to Entropy The thermodynamic approach www.xtremepapers.com Internal energy and enthalpy When energy is added to a body, its internal energy U increases by an amount ΔU. The energy
