BIOC : Homework 1 Due 10/10
|
|
|
- Jordan Morgan
- 5 years ago
- Views:
Transcription
1 Contact information: Name: Student # BIOC : Homework 1 Due 10/10 Department address The following problems are based on David Baker s lectures of forces and protein folding. When numerical values are not specified, you should be able to find appropriate values in lecture notes. 1. Using the Lennard-Jones parameters from the lecture notes sketch out or plot the attractive and repulsive contributions (and their sum) to the interaction energy of a hydrogen and a oxygen atom as a function distance. 2. What is the Lennard-Jones interaction energy between a nitrogen and an oxygen
2 when these are 6 Å apart? Use the parameter table from the lecture notes. The equation for Van der Waal s interaction energy is: EVdW = εij [( σij/rij ) 12 2( σij/rij ) 6 ] εij = sqrt(e n x e o ) = sqrt( 0.24 x 0.16 ) = σij = r n + r o = = 3.2 rij = distance between the atoms = 8 EVdW = 0.196[( 3.2/6 ) 12 2( 3.2/6 ) 6 ] = kcal/mol 3. What is the electrostatic interaction energy of a lysine nitrogen and a glutamate oxygen when they are separated by 5 Å (Assume the charge on the nitrogen is +1 and the charge on the oxygen is -1.) (a) in water? (b) in the protein interior (assumed to be a homogeneous medium with dielectric constant 4.) Where r=5, q1=1, q2 = -1 k = 332 (kcal/mol) Å/e (this is just a conversion factor, given in the notes. It is not the same as the k in the entropy equations, which is Boltzmann s constant) in water. ε 80 E = 332(-1/80*5) = kcal/mol in protein interior, ε = 4 E = 332(-1/4*5) = kcal/mol 4. Calculate the ΔG for moving two charges (+1 and -1) from an organic solvent with dielectric constant 1 into water, keeping their distance fixed at: a) r ij = 3.5 Å b) r ij = 7.5 Å The equation for electrostatic interaction energy between two charges is:
3 Where r=3.5, q1=1, q2 = -1, and remember that k = 332 (kcal/mol) Å/e Remember from the notes that the energy associated with transferring a charge q from a vacuum (dielectric constant of 1) to a dielectric medium is thus: (1/ε - 1) k q2/2r (this is the solvation energy) Moving the charges from organic solvent (ε=1) to water (ε=80): But remember, we must add the solvation energy (for both charges): Remember that R is the distance from the center of the ion to the solvent. Solvation energy will be the same for both ions, so I double the term in the calculations below: ΔG=332(-1/(80*3.5))-332(-1/(1*3.5))+2*(((1/80)-1)*332*((1)/(2*1.6)) ΔG= kcal/mol b) Same equations, using r=7.5: ΔG=332(-1/(80*7.5))-332(-1/(1*7.5))+2*(((1/80)-1)*332*((1)/(2*1.6)) ΔG= kcal/mol 5. In mass spectrometers, proteins are dispersed into the gas phase. Would you expect proteins to be more or less stable in the gas phase? (Answer this by listing how the contributions to protein stability of each of the major forces would change for a protein in gas phase compared to a protein in water.) Solution Remember that folding intrinsically involves a decrease in entropy. When a disordered peptide
4 folds into an ordered structure, there is an entropic cost associated. The more stable a protein, the better this cost is offset by the other energies involved. There are four forces to consider: a. Hydrogen bonds (dipole-dipole interactions): In water, an unfolded protein has ideal hydrogen bonds, since the water is polar and it will interact favorably with any dipole in the protein. The folded protein will very rarely (if ever) have ideal hydrogen bonds because the process of folding will inevitably cause some of the polar groups within the protein to be in less-than-ideal environments where they don t interact with surrounding atoms as favorably as they would if they were surrounded with water. However, typical proteins fold such that most of the h-bonds between the water and the protein that are broken by folding are replaced by h-bonds within the protein. If there were too many h-bond donors & acceptors that were left unaccommodated within the protein interior, the protein wouldn t fold into a stable structure. So in water, h-bond interactions favor the unfolded state slightly, but are reasonably favorable in the folded state as well. In gas, the unfolded protein would have no polar solvent to interact with, so hydrogen bonds are exclusive to the folded state. In gas, h-bond interactions favor the folded state much more than the unfolded state, so, considering only h-bonds, the fold is more stable in gas than water. b. Electrostatics (interactions between fixed charges): The same reasoning as with hydrogen bonds applies here. In water (or, more realistically, in a buffer that has salt ions dissolved in it), an unfolded protein has ideal electrostatic interactions with dissolved salts. The folded protein will very rarely (if ever) have perfectly ideal electrostatic interactions because the process of folding will inevitably cause a lessthanperfect environment for some charges. However, typical proteins fold such that the charges that are internalized by folding are interacting nearly as well with their environments as they would in water so that the increase in energy of these charges as the protein folds is minimized; otherwise, the protein wouldn t fold! So in water, electrostatic interactions favor the unfolded state slightly, but are reasonably favorable in the folded state as well. In gas, there is no solvent to interact with the charges of the unfolded protein, so favorable interactions that may have been going on in the interior of the folded protein will be lost (and replaced with nothing) when the protein unfolds. In gas, electrostatic interactions favor the folded state much more than the unfolded state. c. Van der Waals interactions: Again, we follow the same reasoning: VdW s are ideal in the unfolded state, less than ideal but almost as favorable in the folded state. In water, VdW s interactions are slightly more favorable in the unfolded state than in the folded state. In gas, VdW s favor the folded state much more than the unfolded state, since when the protein unfolds, there s no solvent for the protein to interact with. In gas, there is no solvent to interact with the charges of the unfold d. Hydrophobic effect The hydrophobic effect is the energy associated with opening a cavity in water and placing the protein into it. Cavities in water disrupt the H-bonding of the water molecules that were next to each other are now separated and can t interact with each other. Furthermore, whereas the water molecules, if left to themselves, could adopt many favorable conformations, when they interact with protein, they have to align with the polar groups on
5 the surface of the protein (which exacts a significant entropy cost). By folding up, the protein sequesters its hydrophobic groups away from the water into the center of the protein, which decreases the disruption of H-bonding in the water. The hydrophobic effect therefore greatly favors the folded state to the unfolded state. In fact, this is the primary driving force for the folding of the protein. The decrease in free energy associated with decreasing the interface between hydrophobic groups and water is the primary offset to the entropic cost of folding. In gas, of course, there is no hydrophobic effect. There is no water around to prefer the sequestration of hydrophobic groups into the center of the protein. The hydrophobic effect doesn t exist, and hence the primary driving force of folding doesn t exist. The entropic cost of folding in gas will be slightly offset by the other three forces listed above, but without the hydrophobic effect, the protein fold will be very unstable: the protein will denature in gas. 6. A lysine side-chain on the protein surface can adopt one of 81 possible rotamer conformations with equal probabilities. When the protein forms a complex with anther protein, only one rotamer is possible (the others would bump into the other protein). What is the free energy loss associated with this entropy loss at room temperature? The free energy change associated with entropy changes is: -TΔS Now all you need to know is the temperature (given as room temperature which is about 300K), and the change in entropy. The change in entropy is given in the lecture notes: ΔS = k ln(ω2) k ln(ω1) The Ω refers to the number of possible conformations; this goes from an initial value of 81 to a final value of 1. The constant k is Boltzmann s constant, given in the lecture notes as This is different from the conversion factor k used earlier! = ln (1) ln(81) = kcal/molk Since this is a negative number, the entropy decreased (i.e. the disorder decreased).when you apply the free energy equation, you see that the free energy change is positive, as it should be for a decrease in disorder. ΔG=-TΔs = -300 Å~ ( ) = kcal/mol
6 7. If the free energy difference between unfolded state and folded state of protein X is 4.0 kcal/mol, what is the ratio of the two populations (the equilibrium constant) at 300K? According to the Boltzmann equation, the ratio of folded to unfolded molecules is: ΔG = -k T lnkeq Rearranging this to isolate Keq (which is the equilibrium ratio of one state over the other) Keq = e^(-δg/kt) 8. A small protein has a ΔH of folding=3 kcal/mol and ΔS of folding=50 cal/k mol. a) Calculate the temperature at which the ratio of folded to unfolded protein is 10:1 b) Calculate the melting temperature (the temperature at which half the protein will be folded and half will be unfolded). Keq = e^- (ΔG/kT) Substituting in the Gibbs free energy equation: Note: Remember that ΔG in this case will be for the protein going from the unfolded state to the folded state, so you must reverse its sign. Keq = e^- (- ΔG/kT) Keq = e^- ((- (ΔH- TΔS))/kt)=e^- ((TΔS- ΔH))/kt) Solve for T: T=ΔH/(ln(Keq)K+ΔS) Remember to change ΔS from cal to kcal: ΔH=3 kcal/mol, ΔS=50 cal/k mol = 0.05 kcal/k mol, K= kcal / K mol a) Keq=folded/unfolded=10/1=10 T=ΔH/(ln(Keq)K+ΔS) T=3/(ln(10)* ) T=54.98 K b) Keq=folded/unfolded=1/1=1 T=ΔH/(ln(Keq)K+ΔS) T=3/(ln(1)* ) T=60 K
7 9. A protein has three states: Unfolded state--200 configurations, all with a +1 charge and a -1 charge 12Å apart, and two carbon atoms 10Å apart. Intermediate state--50 configurations, all with a +1 charge and a -1 charge 8Å apart, and two carbon atoms 7Å apart. Native state--1 configuration, with a +1 charge and a -1 charge 4Å apart, and two carbon atoms 4Å apart, and 3 hydrogen bonds worth 1kcal/M. The charges are exposed to solvent in all three states. Is the protein in the native state a) at 300K? b) at 400K? Hint: in the native state means has a probability of >50% For each state Van Der Waals and Coulombic interaction energy needs to calculated using: The equation for Van der Waal s interaction energy in the native is: EVdW = εij [( σij/rij ) 12 2( σij/rij ) 6 ] εij = sqrt(e n x e o ) = sqrt( 0.12 x 0.12 ) = 0.12 σij = r n + r o = = 4.2 rij = distance between the atoms = 4 EVdW = 0.12[( 4.2/6 ) 12 2( 4.2/6 ) 6 ] = kcal/mol Where r=5, q1=1, q2 = -1 k = 332 (kcal/mol) Å/ in water. ε 80 E = 332(-1/80*4) = kcal/mol
8 Interaction energy(native) = (hydrogen bonds = kcal/mol Before partition function = configurations * exp(-interaction energy/kt) = 1 * exp(4.112/(.6)) = 9.90E+02 But remember that this number is meaningless unless you use a partition function that includes all of the available states. Actual_Prob native = Prob native / (Prob native + Prob intermeidate + Prob unfolded ) So you have to calculate all of the probabilities for all the states: interaction hydrogen before partition 300K energy bonds configurations function probability unfolded E intermediate E native E At 300K the probability of being in the native state is 68%.68 = 9.90E+02/(9.90E E E+02) And recalculated at 400K: interaction hydrogen before partition 400K energy bonds configurations funtion probability unfolded E intermediate E native E At 300K the probability of being in the native state is 31%
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Lecture 2-3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2-3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions Van der Waals Interactions
Lecture 2-3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions Van der Waals Interactions
Why Proteins Fold. How Proteins Fold? e - ΔG/kT. Protein Folding, Nonbonding Forces, and Free Energy
 Why Proteins Fold Proteins are the action superheroes of the body. As enzymes, they make reactions go a million times faster. As versatile transport vehicles, they carry oxygen and antibodies to fight
Why Proteins Fold Proteins are the action superheroes of the body. As enzymes, they make reactions go a million times faster. As versatile transport vehicles, they carry oxygen and antibodies to fight
= (-22) = +2kJ /mol
 Lecture 8: Thermodynamics & Protein Stability Assigned reading in Campbell: Chapter 4.4-4.6 Key Terms: DG = -RT lnk eq = DH - TDS Transition Curve, Melting Curve, Tm DH calculation DS calculation van der
Lecture 8: Thermodynamics & Protein Stability Assigned reading in Campbell: Chapter 4.4-4.6 Key Terms: DG = -RT lnk eq = DH - TDS Transition Curve, Melting Curve, Tm DH calculation DS calculation van der
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Free energy, electrostatics, and the hydrophobic effect
 Protein Physics 2016 Lecture 3, January 26 Free energy, electrostatics, and the hydrophobic effect Magnus Andersson magnus.andersson@scilifelab.se Theoretical & Computational Biophysics Recap Protein structure
Protein Physics 2016 Lecture 3, January 26 Free energy, electrostatics, and the hydrophobic effect Magnus Andersson magnus.andersson@scilifelab.se Theoretical & Computational Biophysics Recap Protein structure
Other Cells. Hormones. Viruses. Toxins. Cell. Bacteria
 Other Cells Hormones Viruses Toxins Cell Bacteria ΔH < 0 reaction is exothermic, tells us nothing about the spontaneity of the reaction Δ H > 0 reaction is endothermic, tells us nothing about the spontaneity
Other Cells Hormones Viruses Toxins Cell Bacteria ΔH < 0 reaction is exothermic, tells us nothing about the spontaneity of the reaction Δ H > 0 reaction is endothermic, tells us nothing about the spontaneity
Biophysics II. Hydrophobic Bio-molecules. Key points to be covered. Molecular Interactions in Bio-molecular Structures - van der Waals Interaction
 Biophysics II Key points to be covered By A/Prof. Xiang Yang Liu Biophysics & Micro/nanostructures Lab Department of Physics, NUS 1. van der Waals Interaction 2. Hydrogen bond 3. Hydrophilic vs hydrophobic
Biophysics II Key points to be covered By A/Prof. Xiang Yang Liu Biophysics & Micro/nanostructures Lab Department of Physics, NUS 1. van der Waals Interaction 2. Hydrogen bond 3. Hydrophilic vs hydrophobic
Solutions and Non-Covalent Binding Forces
 Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
Chapter 3 Solutions and Non-Covalent Binding Forces 3.1 Solvent and solution properties Molecules stick together using the following forces: dipole-dipole, dipole-induced dipole, hydrogen bond, van der
Protein Folding & Stability. Lecture 11: Margaret A. Daugherty. Fall How do we go from an unfolded polypeptide chain to a
 Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2004 How do we go from an unfolded polypeptide chain to a compact folded protein? (Folding of thioredoxin, F. Richards) Structure - Function
Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2004 How do we go from an unfolded polypeptide chain to a compact folded protein? (Folding of thioredoxin, F. Richards) Structure - Function
Lecture 1. Conformational Analysis in Acyclic Systems
 Lecture 1 Conformational Analysis in Acyclic Systems Learning Outcomes: by the end of this lecture and after answering the associated problems, you will be able to: 1. use Newman and saw-horse projections
Lecture 1 Conformational Analysis in Acyclic Systems Learning Outcomes: by the end of this lecture and after answering the associated problems, you will be able to: 1. use Newman and saw-horse projections
Lecture 11: Protein Folding & Stability
 Structure - Function Protein Folding: What we know Lecture 11: Protein Folding & Stability 1). Amino acid sequence dictates structure. 2). The native structure represents the lowest energy state for a
Structure - Function Protein Folding: What we know Lecture 11: Protein Folding & Stability 1). Amino acid sequence dictates structure. 2). The native structure represents the lowest energy state for a
Protein Folding & Stability. Lecture 11: Margaret A. Daugherty. Fall Protein Folding: What we know. Protein Folding
 Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2003 Structure - Function Protein Folding: What we know 1). Amino acid sequence dictates structure. 2). The native structure represents
Lecture 11: Protein Folding & Stability Margaret A. Daugherty Fall 2003 Structure - Function Protein Folding: What we know 1). Amino acid sequence dictates structure. 2). The native structure represents
16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit. Lecture 2 (9/11/17)
 16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit By Anthony Quintano - https://www.flickr.com/photos/quintanomedia/15071865580, CC BY 2.0, https://commons.wikimedia.org/w/index.php?curid=38538291
16 years ago TODAY (9/11) at 8:46, the first tower was hit at 9:03, the second tower was hit By Anthony Quintano - https://www.flickr.com/photos/quintanomedia/15071865580, CC BY 2.0, https://commons.wikimedia.org/w/index.php?curid=38538291
Biochemistry 530: Introduction to Structural Biology. Autumn Quarter 2014 BIOC 530
 Biochemistry 530: Introduction to Structural Biology Autumn Quarter 2014 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic
Biochemistry 530: Introduction to Structural Biology Autumn Quarter 2014 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic
MCB100A/Chem130 MidTerm Exam 2 April 4, 2013
 MCBA/Chem Miderm Exam 2 April 4, 2 Name Student ID rue/false (2 points each).. he Boltzmann constant, k b sets the energy scale for observing energy microstates 2. Atoms with favorable electronic configurations
MCBA/Chem Miderm Exam 2 April 4, 2 Name Student ID rue/false (2 points each).. he Boltzmann constant, k b sets the energy scale for observing energy microstates 2. Atoms with favorable electronic configurations
Protein Folding experiments and theory
 Protein Folding experiments and theory 1, 2,and 3 Protein Structure Fig. 3-16 from Lehninger Biochemistry, 4 th ed. The 3D structure is not encoded at the single aa level Hydrogen Bonding Shared H atom
Protein Folding experiments and theory 1, 2,and 3 Protein Structure Fig. 3-16 from Lehninger Biochemistry, 4 th ed. The 3D structure is not encoded at the single aa level Hydrogen Bonding Shared H atom
CHAPTER 2. Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules
 CHAPTER 2 Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules 2-1 Kinetics and Thermodynamics of Simple Chemical Processes Chemical thermodynamics: Is concerned with the extent that
CHAPTER 2 Structure and Reactivity: Acids and Bases, Polar and Nonpolar Molecules 2-1 Kinetics and Thermodynamics of Simple Chemical Processes Chemical thermodynamics: Is concerned with the extent that
Aqueous solutions. Solubility of different compounds in water
 Aqueous solutions Solubility of different compounds in water The dissolution of molecules into water (in any solvent actually) causes a volume change of the solution; the size of this volume change is
Aqueous solutions Solubility of different compounds in water The dissolution of molecules into water (in any solvent actually) causes a volume change of the solution; the size of this volume change is
Chapter 1. Topic: Overview of basic principles
 Chapter 1 Topic: Overview of basic principles Four major themes of biochemistry I. What are living organism made from? II. How do organism acquire and use energy? III. How does an organism maintain its
Chapter 1 Topic: Overview of basic principles Four major themes of biochemistry I. What are living organism made from? II. How do organism acquire and use energy? III. How does an organism maintain its
BIOC 530 Fall, 2011 BIOC 530
 Fall, 2011 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic acids, control of enzymatic reactions. Please see the course
Fall, 2011 Course Information Course Description Graduate-level discussion of the structure, function, and chemistry of proteins and nucleic acids, control of enzymatic reactions. Please see the course
Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur. Lecture - 06 Protein Structure IV
 Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur Lecture - 06 Protein Structure IV We complete our discussion on Protein Structures today. And just to recap
Biochemistry Prof. S. DasGupta Department of Chemistry Indian Institute of Technology Kharagpur Lecture - 06 Protein Structure IV We complete our discussion on Protein Structures today. And just to recap
Protein Structure. W. M. Grogan, Ph.D. OBJECTIVES
 Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Biological Thermodynamics
 Biological Thermodynamics Classical thermodynamics is the only physical theory of universal content concerning which I am convinced that, within the framework of applicability of its basic contents, will
Biological Thermodynamics Classical thermodynamics is the only physical theory of universal content concerning which I am convinced that, within the framework of applicability of its basic contents, will
MCB100A/Chem130 MidTerm Exam 2 April 4, 2013
 MCB1A/Chem13 MidTerm Exam 2 April 4, 213 Name Student ID True/False (2 points each). 1. The Boltzmann constant, k b T sets the energy scale for observing energy microstates 2. Atoms with favorable electronic
MCB1A/Chem13 MidTerm Exam 2 April 4, 213 Name Student ID True/False (2 points each). 1. The Boltzmann constant, k b T sets the energy scale for observing energy microstates 2. Atoms with favorable electronic
Structural Bioinformatics (C3210) Molecular Mechanics
 Structural Bioinformatics (C3210) Molecular Mechanics How to Calculate Energies Calculation of molecular energies is of key importance in protein folding, molecular modelling etc. There are two main computational
Structural Bioinformatics (C3210) Molecular Mechanics How to Calculate Energies Calculation of molecular energies is of key importance in protein folding, molecular modelling etc. There are two main computational
schematic diagram; EGF binding, dimerization, phosphorylation, Grb2 binding, etc.
 Lecture 1: Noncovalent Biomolecular Interactions Bioengineering and Modeling of biological processes -e.g. tissue engineering, cancer, autoimmune disease Example: RTK signaling, e.g. EGFR Growth responses
Lecture 1: Noncovalent Biomolecular Interactions Bioengineering and Modeling of biological processes -e.g. tissue engineering, cancer, autoimmune disease Example: RTK signaling, e.g. EGFR Growth responses
BCMP 201 Protein biochemistry
 BCMP 201 Protein biochemistry BCMP 201 Protein biochemistry with emphasis on the interrelated roles of protein structure, catalytic activity, and macromolecular interactions in biological processes. The
BCMP 201 Protein biochemistry BCMP 201 Protein biochemistry with emphasis on the interrelated roles of protein structure, catalytic activity, and macromolecular interactions in biological processes. The
3. Solutions W = N!/(N A!N B!) (3.1) Using Stirling s approximation ln(n!) = NlnN N: ΔS mix = k (N A lnn + N B lnn N A lnn A N B lnn B ) (3.
 3. Solutions Many biological processes occur between molecules in aqueous solution. In addition, many protein and nucleic acid molecules adopt three-dimensional structure ( fold ) in aqueous solution.
3. Solutions Many biological processes occur between molecules in aqueous solution. In addition, many protein and nucleic acid molecules adopt three-dimensional structure ( fold ) in aqueous solution.
Macromolecule Stability Curves
 Chem728 page 1 Spring 2012 Macromolecule Stability Curves Macromolecule Transitions - We have discussed in class the factors that determine the spontaneity of processes using conformational transitions
Chem728 page 1 Spring 2012 Macromolecule Stability Curves Macromolecule Transitions - We have discussed in class the factors that determine the spontaneity of processes using conformational transitions
q 2 Da This looks very similar to Coulomb doesn t it? The only difference to Coulomb is the factor of 1/2.
 Born Lets now think about a somewhat different, but related problem that is associated with the name Born. Using a name (usually the name of the guy who first discovered or solved the problem) to describe
Born Lets now think about a somewhat different, but related problem that is associated with the name Born. Using a name (usually the name of the guy who first discovered or solved the problem) to describe
An introduction to Molecular Dynamics. EMBO, June 2016
 An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
a) Write the reaction that occurs (pay attention to and label ends correctly) 5 AGCTG CAGCT > 5 AGCTG 3 3 TCGAC 5
 Chem 315 Applications Practice Problem Set 1.As you learned in Chem 315, DNA higher order structure formation is a two step process. The first step, nucleation, is entropically the least favorable. The
Chem 315 Applications Practice Problem Set 1.As you learned in Chem 315, DNA higher order structure formation is a two step process. The first step, nucleation, is entropically the least favorable. The
Molecular Modeling -- Lecture 15 Surfaces and electrostatics
 Molecular Modeling -- Lecture 15 Surfaces and electrostatics Molecular surfaces The Hydrophobic Effect Electrostatics Poisson-Boltzmann Equation Electrostatic maps Electrostatic surfaces in MOE 15.1 The
Molecular Modeling -- Lecture 15 Surfaces and electrostatics Molecular surfaces The Hydrophobic Effect Electrostatics Poisson-Boltzmann Equation Electrostatic maps Electrostatic surfaces in MOE 15.1 The
Supplementary Information
 1 Supplementary Information Figure S1 The V=0.5 Harker section of an anomalous difference Patterson map calculated using diffraction data from the NNQQNY crystal at 1.3 Å resolution. The position of the
1 Supplementary Information Figure S1 The V=0.5 Harker section of an anomalous difference Patterson map calculated using diffraction data from the NNQQNY crystal at 1.3 Å resolution. The position of the
Some properties of water
 Some properties of water Hydrogen bond network Solvation under the microscope 1 Water solutions Oil and water does not mix at equilibrium essentially due to entropy Substances that does not mix with water
Some properties of water Hydrogen bond network Solvation under the microscope 1 Water solutions Oil and water does not mix at equilibrium essentially due to entropy Substances that does not mix with water
Lecture 34 Protein Unfolding Thermodynamics
 Physical Principles in Biology Biology 3550 Fall 2018 Lecture 34 Protein Unfolding Thermodynamics Wednesday, 21 November c David P. Goldenberg University of Utah goldenberg@biology.utah.edu Clicker Question
Physical Principles in Biology Biology 3550 Fall 2018 Lecture 34 Protein Unfolding Thermodynamics Wednesday, 21 November c David P. Goldenberg University of Utah goldenberg@biology.utah.edu Clicker Question
Lecture Notes 1: Physical Equilibria Vapor Pressure
 Lecture Notes 1: Physical Equilibria Vapor Pressure Our first exploration of equilibria will examine physical equilibria (no chemical changes) in which the only changes occurring are matter changes phases.
Lecture Notes 1: Physical Equilibria Vapor Pressure Our first exploration of equilibria will examine physical equilibria (no chemical changes) in which the only changes occurring are matter changes phases.
From Amino Acids to Proteins - in 4 Easy Steps
 From Amino Acids to Proteins - in 4 Easy Steps Although protein structure appears to be overwhelmingly complex, you can provide your students with a basic understanding of how proteins fold by focusing
From Amino Acids to Proteins - in 4 Easy Steps Although protein structure appears to be overwhelmingly complex, you can provide your students with a basic understanding of how proteins fold by focusing
Dihedral Angles. Homayoun Valafar. Department of Computer Science and Engineering, USC 02/03/10 CSCE 769
 Dihedral Angles Homayoun Valafar Department of Computer Science and Engineering, USC The precise definition of a dihedral or torsion angle can be found in spatial geometry Angle between to planes Dihedral
Dihedral Angles Homayoun Valafar Department of Computer Science and Engineering, USC The precise definition of a dihedral or torsion angle can be found in spatial geometry Angle between to planes Dihedral
BIBC 100. Structural Biochemistry
 BIBC 100 Structural Biochemistry http://classes.biology.ucsd.edu/bibc100.wi14 Papers- Dialogue with Scientists Questions: Why? How? What? So What? Dialogue Structure to explain function Knowledge Food
BIBC 100 Structural Biochemistry http://classes.biology.ucsd.edu/bibc100.wi14 Papers- Dialogue with Scientists Questions: Why? How? What? So What? Dialogue Structure to explain function Knowledge Food
= 16! = 16! W A = 3 = 3 N = = W B 3!3!10! = ΔS = nrln V. = ln ( 3 ) V 1 = 27.4 J.
 Answer key: Q1A Both configurations are equally likely because the particles are non-interacting (i.e., the energy does not favor one configuration over another). For A M = 16 N = 6 W A = 16! 0.9 101 =
Answer key: Q1A Both configurations are equally likely because the particles are non-interacting (i.e., the energy does not favor one configuration over another). For A M = 16 N = 6 W A = 16! 0.9 101 =
Short Announcements. 1 st Quiz today: 15 minutes. Homework 3: Due next Wednesday.
 Short Announcements 1 st Quiz today: 15 minutes Homework 3: Due next Wednesday. Next Lecture, on Visualizing Molecular Dynamics (VMD) by Klaus Schulten Today s Lecture: Protein Folding, Misfolding, Aggregation
Short Announcements 1 st Quiz today: 15 minutes Homework 3: Due next Wednesday. Next Lecture, on Visualizing Molecular Dynamics (VMD) by Klaus Schulten Today s Lecture: Protein Folding, Misfolding, Aggregation
Applications of Free Energy. NC State University
 Chemistry 433 Lecture 15 Applications of Free Energy NC State University Thermodynamics of glycolysis Reaction kj/mol D-glucose + ATP D-glucose-6-phosphate + ADP ΔG o = -16.7 D-glucose-6-phosphate p D-fructose-6-phosphate
Chemistry 433 Lecture 15 Applications of Free Energy NC State University Thermodynamics of glycolysis Reaction kj/mol D-glucose + ATP D-glucose-6-phosphate + ADP ΔG o = -16.7 D-glucose-6-phosphate p D-fructose-6-phosphate
Lec.1 Chemistry Of Water
 Lec.1 Chemistry Of Water Biochemistry & Medicine Biochemistry can be defined as the science concerned with the chemical basis of life. Biochemistry can be described as the science concerned with the chemical
Lec.1 Chemistry Of Water Biochemistry & Medicine Biochemistry can be defined as the science concerned with the chemical basis of life. Biochemistry can be described as the science concerned with the chemical
Biology Chemistry & Physics of Biomolecules. Examination #1. Proteins Module. September 29, Answer Key
 Biology 5357 Chemistry & Physics of Biomolecules Examination #1 Proteins Module September 29, 2017 Answer Key Question 1 (A) (5 points) Structure (b) is more common, as it contains the shorter connection
Biology 5357 Chemistry & Physics of Biomolecules Examination #1 Proteins Module September 29, 2017 Answer Key Question 1 (A) (5 points) Structure (b) is more common, as it contains the shorter connection
Chapter 7 Chemical Reactions: Energy, Rates, and Equilibrium
 Chapter 7 Chemical Reactions: Energy, Rates, and Equilibrium Introduction This chapter considers three factors: a) Thermodynamics (Energies of Reactions) a reaction will occur b) Kinetics (Rates of Reactions)
Chapter 7 Chemical Reactions: Energy, Rates, and Equilibrium Introduction This chapter considers three factors: a) Thermodynamics (Energies of Reactions) a reaction will occur b) Kinetics (Rates of Reactions)
Carbohydrate- Protein interac;ons are Cri;cal in Life and Death. Other Cells. Hormones. Viruses. Toxins. Cell. Bacteria
 ther Cells Carbohydrate- Protein interac;ons are Cri;cal in Life and Death ormones Viruses Toxins Cell Bacteria ow to Model Protein- ligand interac;ons? Protein Protein Protein DNA/RNA Protein Carbohydrate
ther Cells Carbohydrate- Protein interac;ons are Cri;cal in Life and Death ormones Viruses Toxins Cell Bacteria ow to Model Protein- ligand interac;ons? Protein Protein Protein DNA/RNA Protein Carbohydrate
Chapter 2 - Water 9/8/2014. Water exists as a H-bonded network with an average of 4 H-bonds per molecule in ice and 3.4 in liquid. 104.
 Chapter 2 - Water Water exists as a -bonded network with an average of 4 -bonds per molecule in ice and 3.4 in liquid. 104.5 o -bond: An electrostatic attraction between polarized molecules containing
Chapter 2 - Water Water exists as a -bonded network with an average of 4 -bonds per molecule in ice and 3.4 in liquid. 104.5 o -bond: An electrostatic attraction between polarized molecules containing
Energy functions and their relationship to molecular conformation. CS/CME/BioE/Biophys/BMI 279 Oct. 3 and 5, 2017 Ron Dror
 Energy functions and their relationship to molecular conformation CS/CME/BioE/Biophys/BMI 279 Oct. 3 and 5, 2017 Ron Dror Outline Energy functions for proteins (or biomolecular systems more generally)
Energy functions and their relationship to molecular conformation CS/CME/BioE/Biophys/BMI 279 Oct. 3 and 5, 2017 Ron Dror Outline Energy functions for proteins (or biomolecular systems more generally)
Full file at https://fratstock.eu
 Chapter 03 1. a. DG=DH-TDS Δ G = 80 kj ( 98 K) 0.790 kj = 44.6 kj K b. ΔG = 0 @ T m. Unfolding will be favorable at temperatures above the T m. Δ G =Δ H TΔ S 0 kj kj 80 ( xk) 0.790 K 0 Δ G = = 354.4 K
Chapter 03 1. a. DG=DH-TDS Δ G = 80 kj ( 98 K) 0.790 kj = 44.6 kj K b. ΔG = 0 @ T m. Unfolding will be favorable at temperatures above the T m. Δ G =Δ H TΔ S 0 kj kj 80 ( xk) 0.790 K 0 Δ G = = 354.4 K
Why study protein dynamics?
 Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Thermodynamics. Entropy and its Applications. Lecture 11. NC State University
 Thermodynamics Entropy and its Applications Lecture 11 NC State University System and surroundings Up to this point we have considered the system, but we have not concerned ourselves with the relationship
Thermodynamics Entropy and its Applications Lecture 11 NC State University System and surroundings Up to this point we have considered the system, but we have not concerned ourselves with the relationship
Proteins polymer molecules, folded in complex structures. Konstantin Popov Department of Biochemistry and Biophysics
 Proteins polymer molecules, folded in complex structures Konstantin Popov Department of Biochemistry and Biophysics Outline General aspects of polymer theory Size and persistent length of ideal linear
Proteins polymer molecules, folded in complex structures Konstantin Popov Department of Biochemistry and Biophysics Outline General aspects of polymer theory Size and persistent length of ideal linear
WHAT IS A SOLUTION? PROPERTIES OF SOLUTIONS SOLUTION TYPES. Possible answers BRAINSTORM: CH. 13
 WHAT IS A SOLUTION? PROPERTIES OF SOLUTIONS BRAINSTORM: What do you already know about solutions? CH. 13 Possible answers SOLUTION TYPES Homogeneous Composed of solute and solvent Solvent is commonly the
WHAT IS A SOLUTION? PROPERTIES OF SOLUTIONS BRAINSTORM: What do you already know about solutions? CH. 13 Possible answers SOLUTION TYPES Homogeneous Composed of solute and solvent Solvent is commonly the
Life Sciences 1a Lecture Slides Set 10 Fall Prof. David R. Liu. Lecture Readings. Required: Lecture Notes McMurray p , O NH
 Life ciences 1a Lecture lides et 10 Fall 2006-2007 Prof. David R. Liu Lectures 17-18: The molecular basis of drug-protein binding: IV protease inhibitors 1. Drug development and its impact on IV-infected
Life ciences 1a Lecture lides et 10 Fall 2006-2007 Prof. David R. Liu Lectures 17-18: The molecular basis of drug-protein binding: IV protease inhibitors 1. Drug development and its impact on IV-infected
The change in free energy on transferring an ion from a medium of low dielectric constantε1 to one of high dielectric constant ε2:
 The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
Atomic and molecular interaction forces in biology
 Atomic and molecular interaction forces in biology 1 Outline Types of interactions relevant to biology Van der Waals interactions H-bond interactions Some properties of water Hydrophobic effect 2 Types
Atomic and molecular interaction forces in biology 1 Outline Types of interactions relevant to biology Van der Waals interactions H-bond interactions Some properties of water Hydrophobic effect 2 Types
reduction kj/mol
 1. Glucose is oxidized to water and CO 2 as a result of glycolysis and the TCA cycle. The net heat of reaction for the oxidation is -2870 kj/mol. a) How much energy is required to produce glucose from
1. Glucose is oxidized to water and CO 2 as a result of glycolysis and the TCA cycle. The net heat of reaction for the oxidation is -2870 kj/mol. a) How much energy is required to produce glucose from
Energy functions and their relationship to molecular conformation. CS/CME/BioE/Biophys/BMI 279 Oct. 3 and 5, 2017 Ron Dror
 Energy functions and their relationship to molecular conformation CS/CME/BioE/Biophys/BMI 279 Oct. 3 and 5, 2017 Ron Dror Yesterday s Nobel Prize: single-particle cryoelectron microscopy 2 Outline Energy
Energy functions and their relationship to molecular conformation CS/CME/BioE/Biophys/BMI 279 Oct. 3 and 5, 2017 Ron Dror Yesterday s Nobel Prize: single-particle cryoelectron microscopy 2 Outline Energy
Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions. Let's get together.
 Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions Let's get together. Most gases are NOT ideal except at very low pressures: Z=1 for ideal gases Intermolecular interactions come
Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions Let's get together. Most gases are NOT ideal except at very low pressures: Z=1 for ideal gases Intermolecular interactions come
Biochemistry 675, Lecture 8 Electrostatic Interactions
 Biochemistry 675, Lecture 8 Electrostatic Interactions Previous Classes Hydrophobic Effect Hydrogen Bonds Today: Electrostatic Interactions Reading: Handout:P.R. Bergethon & E.R. Simons, Biophysical Chemistry,
Biochemistry 675, Lecture 8 Electrostatic Interactions Previous Classes Hydrophobic Effect Hydrogen Bonds Today: Electrostatic Interactions Reading: Handout:P.R. Bergethon & E.R. Simons, Biophysical Chemistry,
Energy is the capacity to do work
 1 of 10 After completing this chapter, you should, at a minimum, be able to do the following. This information can be found in my lecture notes for this and other chapters and also in your text. Correctly
1 of 10 After completing this chapter, you should, at a minimum, be able to do the following. This information can be found in my lecture notes for this and other chapters and also in your text. Correctly
Chemical thermodynamics and bioenergetics
 Chemical thermodynamics and bioenergetics Thermodynamics is a branch of physics that studies energy, the forms of its transformation, and the laws controlling its properties. Basic Concepts and Definitions.
Chemical thermodynamics and bioenergetics Thermodynamics is a branch of physics that studies energy, the forms of its transformation, and the laws controlling its properties. Basic Concepts and Definitions.
Chemical thermodynamics the area of chemistry that deals with energy relationships
 Chemistry: The Central Science Chapter 19: Chemical Thermodynamics Chemical thermodynamics the area of chemistry that deals with energy relationships 19.1: Spontaneous Processes First law of thermodynamics
Chemistry: The Central Science Chapter 19: Chemical Thermodynamics Chemical thermodynamics the area of chemistry that deals with energy relationships 19.1: Spontaneous Processes First law of thermodynamics
Due in class on Thursday Sept. 8 th
 Problem Set #1 Chem 391 Due in class on Thursday Sept. 8 th ame Solutions A note: To help me read quickly, I structure the problem set like an exam. I have made small spaces for answers where I d like
Problem Set #1 Chem 391 Due in class on Thursday Sept. 8 th ame Solutions A note: To help me read quickly, I structure the problem set like an exam. I have made small spaces for answers where I d like
LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor
 LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
The energy of oxidation of 11 g glucose = kj = kg/m 2 s 2
 Chem 350 thermo problems Key 1. How many meters of stairway could a 70kg man climb if all the energy available in metabolizing an 11 g spoonful of sugar to carbon dioxide and water could be converted to
Chem 350 thermo problems Key 1. How many meters of stairway could a 70kg man climb if all the energy available in metabolizing an 11 g spoonful of sugar to carbon dioxide and water could be converted to
THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION
 THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION AND CALIBRATION Calculation of turn and beta intrinsic propensities. A statistical analysis of a protein structure
THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION AND CALIBRATION Calculation of turn and beta intrinsic propensities. A statistical analysis of a protein structure
Problems from Previous Class
 1 Problems from Previous lass 1. What is K m? What are the units of K m? 2. What is V max? What are the units of V max? 3. Write down the Michaelis-Menten equation. 4. What order of reaction is the reaction
1 Problems from Previous lass 1. What is K m? What are the units of K m? 2. What is V max? What are the units of V max? 3. Write down the Michaelis-Menten equation. 4. What order of reaction is the reaction
Water, water everywhere,; not a drop to drink. Consumption resulting from how environment inhabited Deforestation disrupts water cycle
 Chapter 3 Water: The Matrix of Life Overview n n n Water, water everywhere,; not a drop to drink Only 3% of world s water is fresh How has this happened Consumption resulting from how environment inhabited
Chapter 3 Water: The Matrix of Life Overview n n n Water, water everywhere,; not a drop to drink Only 3% of world s water is fresh How has this happened Consumption resulting from how environment inhabited
Water. 2.1 Weak Interactions in Aqueous Sy stems Ionization of Water, Weak Acids, and Weak Bases 58
 Home http://www.macmillanhighered.com/launchpad/lehninger6e... 1 of 1 1/6/2016 3:07 PM 2 Printed Page 47 Water 2.1 Weak Interactions in Aqueous Sy stems 47 2.2 Ionization of Water, Weak Acids, and Weak
Home http://www.macmillanhighered.com/launchpad/lehninger6e... 1 of 1 1/6/2016 3:07 PM 2 Printed Page 47 Water 2.1 Weak Interactions in Aqueous Sy stems 47 2.2 Ionization of Water, Weak Acids, and Weak
Lattice protein models
 Lattice protein models Marc R. Roussel epartment of Chemistry and Biochemistry University of Lethbridge March 5, 2009 1 Model and assumptions The ideas developed in the last few lectures can be applied
Lattice protein models Marc R. Roussel epartment of Chemistry and Biochemistry University of Lethbridge March 5, 2009 1 Model and assumptions The ideas developed in the last few lectures can be applied
Dana Alsulaibi. Jaleel G.Sweis. Mamoon Ahram
 15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
LS1a Midterm Exam 1 Review Session Problems
 LS1a Midterm Exam 1 Review Session Problems 1. n aqueous mixture of a weak acid and its conjugate base is often used in the laboratory to prepare solutions referred to as buffers. ne commonly used acid
LS1a Midterm Exam 1 Review Session Problems 1. n aqueous mixture of a weak acid and its conjugate base is often used in the laboratory to prepare solutions referred to as buffers. ne commonly used acid
Scoring functions. Talk Overview. Eran Eyal. Scoring functions what and why
 Scoring unctions Talk Overview Scoring unctions what and why Force ields based on approximation o molecular orces as we understand them Knowledge-based potentials let the data speak May 2011 Eran Eyal
Scoring unctions Talk Overview Scoring unctions what and why Force ields based on approximation o molecular orces as we understand them Knowledge-based potentials let the data speak May 2011 Eran Eyal
Protein folding. Today s Outline
 Protein folding Today s Outline Review of previous sessions Thermodynamics of folding and unfolding Determinants of folding Techniques for measuring folding The folding process The folding problem: Prediction
Protein folding Today s Outline Review of previous sessions Thermodynamics of folding and unfolding Determinants of folding Techniques for measuring folding The folding process The folding problem: Prediction
High Specificity and Reversibility
 Lecture #8 The Cell as a Machine High Specificity and Reversibility In considering the problem of transcription factor binding in the nucleus and the great specificity that is called for to transcribe
Lecture #8 The Cell as a Machine High Specificity and Reversibility In considering the problem of transcription factor binding in the nucleus and the great specificity that is called for to transcribe
= (+)206 (kj mol 1) 206 scores 1 only Units not essential if ans in kj mol 1 but penalise incorrect units
 M.(a) (i) ΔH = Σ(enthalpies formation products) Σ(enthalpies formation reactants) Or correct cycle with enthalpy changes labelled = ( 75 242) = (+)206 (kj mol ) 206 scores only Units not essential if ans
M.(a) (i) ΔH = Σ(enthalpies formation products) Σ(enthalpies formation reactants) Or correct cycle with enthalpy changes labelled = ( 75 242) = (+)206 (kj mol ) 206 scores only Units not essential if ans
Chem. 27 Section 1 Conformational Analysis Week of Feb. 6, TF: Walter E. Kowtoniuk Mallinckrodt 303 Liu Laboratory
 Chem. 27 Section 1 Conformational Analysis TF: Walter E. Kowtoniuk wekowton@fas.harvard.edu Mallinckrodt 303 Liu Laboratory ffice hours are: Monday and Wednesday 3:00-4:00pm in Mallinckrodt 303 Course
Chem. 27 Section 1 Conformational Analysis TF: Walter E. Kowtoniuk wekowton@fas.harvard.edu Mallinckrodt 303 Liu Laboratory ffice hours are: Monday and Wednesday 3:00-4:00pm in Mallinckrodt 303 Course
The protein folding problem consists of two parts:
 Energetics and kinetics of protein folding The protein folding problem consists of two parts: 1)Creating a stable, well-defined structure that is significantly more stable than all other possible structures.
Energetics and kinetics of protein folding The protein folding problem consists of two parts: 1)Creating a stable, well-defined structure that is significantly more stable than all other possible structures.
Flexibility of Protein Structure
 Flexibility of Protein Structure Proteins show varying degree of conformational flexibility Due to movements of atoms in molecules vibration in bond length and angles Reflects the existence of populations
Flexibility of Protein Structure Proteins show varying degree of conformational flexibility Due to movements of atoms in molecules vibration in bond length and angles Reflects the existence of populations
S and G Entropy and Gibbs Free Energy
 Week 3 problem solving + equa'ons, + applica'ons S and G Entropy and Gibbs Free Energy Classical defini,on of ΔS = q/t Problem: A calorimeter measured heat of 120J absorbed by a drug specimen while T increased
Week 3 problem solving + equa'ons, + applica'ons S and G Entropy and Gibbs Free Energy Classical defini,on of ΔS = q/t Problem: A calorimeter measured heat of 120J absorbed by a drug specimen while T increased
Fundamentals of Distribution Separations (III)
 Fundamentals of Distribution Separations (III) (01/16/15) K = exp -Δμ 0 ext i - Δμ i RT distribution coefficient C i = exp -Δμ 0 RT - - Δμ i = ΔH i TΔS i 0 0 0 solubility q A---B A + B 0 0 0 ΔH i = ΔH
Fundamentals of Distribution Separations (III) (01/16/15) K = exp -Δμ 0 ext i - Δμ i RT distribution coefficient C i = exp -Δμ 0 RT - - Δμ i = ΔH i TΔS i 0 0 0 solubility q A---B A + B 0 0 0 ΔH i = ΔH
Back to molecular interac/ons Quantum theory and molecular structure
 Back to molecular interac/ons Quantum theory and molecular structure Atoms are arranged in 3D to cons1tute a molecule. Atoms in one molecule are connected by strong covalent bonds that are not easily broken
Back to molecular interac/ons Quantum theory and molecular structure Atoms are arranged in 3D to cons1tute a molecule. Atoms in one molecule are connected by strong covalent bonds that are not easily broken
Thermodynamics: Free Energy and Entropy. Suggested Reading: Chapter 19
 Thermodynamics: Free Energy and Entropy Suggested Reading: Chapter 19 System and Surroundings System: An object or collection of objects being studied. Surroundings: Everything outside of the system. the
Thermodynamics: Free Energy and Entropy Suggested Reading: Chapter 19 System and Surroundings System: An object or collection of objects being studied. Surroundings: Everything outside of the system. the
CHEM 203. Topics Discussed on Sept. 16
 EM 203 Topics Discussed on Sept. 16 Special case of Lewis acid-base reactions: proton transfer ( protonation) reactions, i.e. Bronsted acid-base reactions. Example: N large lobe of σ* small lobe of σ*
EM 203 Topics Discussed on Sept. 16 Special case of Lewis acid-base reactions: proton transfer ( protonation) reactions, i.e. Bronsted acid-base reactions. Example: N large lobe of σ* small lobe of σ*
Section Week 3. Junaid Malek, M.D.
 Section Week 3 Junaid Malek, M.D. Biological Polymers DA 4 monomers (building blocks), limited structure (double-helix) RA 4 monomers, greater flexibility, multiple structures Proteins 20 Amino Acids,
Section Week 3 Junaid Malek, M.D. Biological Polymers DA 4 monomers (building blocks), limited structure (double-helix) RA 4 monomers, greater flexibility, multiple structures Proteins 20 Amino Acids,
Molecular Modelling. part of Bioinformatik von RNA- und Proteinstrukturen. Sonja Prohaska. Leipzig, SS Computational EvoDevo University Leipzig
 part of Bioinformatik von RNA- und Proteinstrukturen Computational EvoDevo University Leipzig Leipzig, SS 2011 Protein Structure levels or organization Primary structure: sequence of amino acids (from
part of Bioinformatik von RNA- und Proteinstrukturen Computational EvoDevo University Leipzig Leipzig, SS 2011 Protein Structure levels or organization Primary structure: sequence of amino acids (from
First Law of Thermodynamics
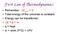 First Law of Thermodynamics Remember: ΔE univ = 0 Total energy of the universe is constant. Energy can be transferred: ΔE = q + w q = heat w = work (F*D) = ΔPV 1 st Law, review For constant volume process:
First Law of Thermodynamics Remember: ΔE univ = 0 Total energy of the universe is constant. Energy can be transferred: ΔE = q + w q = heat w = work (F*D) = ΔPV 1 st Law, review For constant volume process:
Concept review: Binding equilibria
 Concept review: Binding equilibria 1 Binding equilibria and association/dissociation constants 2 The binding of a protein to a ligand at equilibrium can be written as: P + L PL And so the equilibrium constant
Concept review: Binding equilibria 1 Binding equilibria and association/dissociation constants 2 The binding of a protein to a ligand at equilibrium can be written as: P + L PL And so the equilibrium constant
Chapter 1 1) Biological Molecules a) Only a small subset of the known elements are found in living systems i) Most abundant- C, N, O, and H ii) Less
 Chapter 1 1) Biological Molecules a) Only a small subset of the known elements are found in living systems i) Most abundant- C, N, O, and H ii) Less abundant- Ca, P, K, S, Cl, Na, and Mg b) Cells contain
Chapter 1 1) Biological Molecules a) Only a small subset of the known elements are found in living systems i) Most abundant- C, N, O, and H ii) Less abundant- Ca, P, K, S, Cl, Na, and Mg b) Cells contain
Chemical Thermodynamics
 Page III-16-1 / Chapter Sixteen Lecture Notes Chemical Thermodynamics Thermodynamics and Kinetics Chapter 16 Chemistry 223 Professor Michael Russell How to predict if a reaction can occur, given enough
Page III-16-1 / Chapter Sixteen Lecture Notes Chemical Thermodynamics Thermodynamics and Kinetics Chapter 16 Chemistry 223 Professor Michael Russell How to predict if a reaction can occur, given enough
Scuola di Chimica Computazionale
 Societa Chimica Italiana Gruppo Interdivisionale di Chimica Computazionale Scuola di Chimica Computazionale Introduzione, per Esercizi, all Uso del Calcolatore in Chimica Organica e Biologica Modellistica
Societa Chimica Italiana Gruppo Interdivisionale di Chimica Computazionale Scuola di Chimica Computazionale Introduzione, per Esercizi, all Uso del Calcolatore in Chimica Organica e Biologica Modellistica
Gibb s Free Energy. This value represents the maximum amount of useful work (non PV-work) that can be obtained by a system.
 Gibb s Free Energy 1. What is Gibb s free energy? What is its symbol? This value represents the maximum amount of useful work (non PV-work) that can be obtained by a system. It is symbolized by G. We only
Gibb s Free Energy 1. What is Gibb s free energy? What is its symbol? This value represents the maximum amount of useful work (non PV-work) that can be obtained by a system. It is symbolized by G. We only
ANSWER KEY. Chemistry 25 (Spring term 2017) Midterm Examination
 Name ANSWER KEY Chemistry 25 (Spring term 2017) Midterm Examination Distributed Thursday, May 4, 2017 Due Thursday, May 11, 2017 by 1 pm in class or by 12:45 pm in 362 Broad a drop box will be left outside
Name ANSWER KEY Chemistry 25 (Spring term 2017) Midterm Examination Distributed Thursday, May 4, 2017 Due Thursday, May 11, 2017 by 1 pm in class or by 12:45 pm in 362 Broad a drop box will be left outside
Essential Forces in Protein Folding
 Essential Forces in Protein Folding Dr. Mohammad Alsenaidy Department of Pharmaceutics College of Pharmacy King Saud University Office: AA 101 msenaidy@ksu.edu.sa Previously on PHT 426!! Amino Acid sequence
Essential Forces in Protein Folding Dr. Mohammad Alsenaidy Department of Pharmaceutics College of Pharmacy King Saud University Office: AA 101 msenaidy@ksu.edu.sa Previously on PHT 426!! Amino Acid sequence
Chapter 19 Chemical Thermodynamics Entropy and free energy
 Chapter 19 Chemical Thermodynamics Entropy and free energy Learning goals and key skills: Explain and apply the terms spontaneous process, reversible process, irreversible process, and isothermal process.
Chapter 19 Chemical Thermodynamics Entropy and free energy Learning goals and key skills: Explain and apply the terms spontaneous process, reversible process, irreversible process, and isothermal process.
Some properties of water
 Some properties of water Hydrogen bond network Solvation under the microscope 1 NB Queste diapositive sono state preparate per il corso di Biofisica tenuto dal Dr. Attilio V. Vargiu presso il Dipartimento
Some properties of water Hydrogen bond network Solvation under the microscope 1 NB Queste diapositive sono state preparate per il corso di Biofisica tenuto dal Dr. Attilio V. Vargiu presso il Dipartimento
3rd Advanced in silico Drug Design KFC/ADD Molecular mechanics intro Karel Berka, Ph.D. Martin Lepšík, Ph.D. Pavel Polishchuk, Ph.D.
 3rd Advanced in silico Drug Design KFC/ADD Molecular mechanics intro Karel Berka, Ph.D. Martin Lepšík, Ph.D. Pavel Polishchuk, Ph.D. Thierry Langer, Ph.D. Jana Vrbková, Ph.D. UP Olomouc, 23.1.-26.1. 2018
3rd Advanced in silico Drug Design KFC/ADD Molecular mechanics intro Karel Berka, Ph.D. Martin Lepšík, Ph.D. Pavel Polishchuk, Ph.D. Thierry Langer, Ph.D. Jana Vrbková, Ph.D. UP Olomouc, 23.1.-26.1. 2018
