Nature Methods: doi: /nmeth Supplementary Figure 1. Principle of force-distance (FD) curve based AFM.
|
|
|
- Garey Kennedy
- 6 years ago
- Views:
Transcription
1 Supplementary Figure 1 Principle of force-distance (FD) curve based AFM. (a) FD-based AFM contours the sample surface while oscillating the AFM tip with a sine wave at a frequency of 0.25 khz. Pixel-by-pixel the AFM tip is approached (blue curve) and retracted (red curve) from the sample. The AFM cantilever deflection measures the force interacting between AFM tip and sample. During these approach and retraction cycles the force vs time (b) and force vs distance (c) is recorded. Thereby, the maximal force (imaging force) used to touch the sample F i is kept constant using a feedback loop. (c) The mechanical deformation or distance of deformation D Def of a soft biological sample is described by the indentation of a much stiffer AFM tip. This indentation is detected at a certain repulsive force. (d) During retraction, adhesive force F Adh is recorded between the tip and the sample. Using a functionalized cantilever F Adh can detect the rupture of specific interactions between for example a functionalized tip and sample. (e) The parameters extracted from individual force curves can be displayed as maps such as the sample topography (height image) contoured at a given imaging force, the adhesion force or sample deformation Medalsy, I., Hensen, U. & Muller, D.J. Imaging and quantifying chemical and physical properties of native proteins at molecular resolution by force-volume AFM. Angew Chem Int Ed Engl 50, (2011).
2 Supplementary Figure 2 SDS-PAGE of PAR1 liposomes. SDS-PAGE was performed on a 12% acrylamide gel with 150 V and 60 min in a loading buffer (50 mm Tris, ph 7, 6% SDS, 50 mm DTT, 8% glycerol, 0.1% bromphenol blue) and stained with Coomassie blue. Lane 1, molecular weight marker; Lane 2, 10 µl of empty lipid vesicles (liposomes made of 0.5 mg ml 1 DOPC and 0.05 mg ml 1 CHS); Lane 3, 10 µl of PAR1 proteoliposomes (10 µm PAR1 reconstituted in liposomes made of 0.5 mg ml 1 DOPC and 0.05 mg ml 1 CHS). The band at 40 kda is in good agreement with the expected size of 43.9 kda calculated from the PAR1 sequence.
3 Supplementary Figure 3 SMFS of human PAR1 and human β 2 AR reconstituted in liposomes. Selection of FD curves of recorded upon unfolding of single PAR1 (a) and β 2 AR (b) embedded in lipid membranes composed of DOPC and CHS. Superimpositions of FD curves recorded of PAR1 (c) and β 2 AR (d) 2,3. For each GPCR the FD curves recorded were superimposed following the procedure previously described for β 2 AR 3,4. n gives the number of FD curves superimposed. For SMFS, PAR1 proteoliposomes were adsorbed for 1 h at room temperature to freshly cleaved mica in SMFS buffer solution (300 mm NaCl, 25 mm MgCl 2, 25 mm Tris, ph 7.0) and β 2 AR proteoliposomes were adsorbed over night at 4 C to freshly cleaved mica in SMFS buffer solution. It has been shown that the stability of human and animal GPCRs embedded in lipid membranes and adsorbed onto mica in buffer solution does not alter within 24 h 3-5. Thus, samples were newly prepared before becoming 24 h old. Within this 24 h time frame we could not observe any alteration of the FD curves, which would indicate denaturation of the GPCR. For each GPCR characterized by SMFS we had to prepare at least 20 different samples. Each time a new sample has been prepared a new AFM cantilever was taken. SMFS of both GPCRs was conducted as described 3 and carried out using automated AFM-based SMFS (ForceRobot 300; JPK Instruments) 2. SMFS data of both β 2 AR and PAR1 were recorded in SMFS buffer solution, at room temperature and at pulling velocities of nm s Struckmeier, J. et al. Fully automated single-molecule force spectroscopy for screening applications. Nanotechnology 19, (2008). 3. Zocher, M., Fung, J.J., Kobilka, B.K. & Muller, D.J. Ligand-specific interactions modulate kinetic, energetic, and mechanical properties of the human beta2 adrenergic receptor. Structure 20, (2012). 4. Zocher, M., Zhang, C., Rasmussen, S.G., Kobilka, B.K. & Muller, D.J. Cholesterol increases kinetic, energetic, and mechanical stability of the human beta2-adrenergic receptor. Proc Natl Acad Sci U S A 109, E (2012). 5. Sapra, K.T. et al. Detecting molecular interactions that stabilize bovine rhodopsin. J Mol Biol 358, (2006).
4 Supplementary Figure 4 Overview AFM topograph of PAR1 proteoliposome. (a) Topography (height image, 2.5 x 2.5 µm 2 ) showing membrane patches on mica. Proteoliposomes were adsorbed to freshly cleaved mica in buffer solution. To remove weakly attached membrane patches, the sample was rinsed several times with the buffer (see Online Methods). After adsorption to mica the proteoliposomes break open so that they showed single-layered membrane patches. (b) Cross-section (white dashed line in (a)) showing a lipid membrane protruding 4.5 ± 0.7 nm (average ± S.D., n=10) from the supporting mica. The sparsely distributed single protrusions originating from single or clustered PAR1. The FD-based AFM topograph was recorded in imaging buffer (300 mm NaCl, 20 mm Hepes, 25 mm MgCl 2, ph 7.0) at room temperature.
5 Supplementary Figure 5 Structural analysis of PAR1 reconstituted in proteoliposomes. (a) Topography of PAR1s sparsely distributed in lipid membranes made of 0.5 mg ml 1 DOPC and 0.05 mg ml 1 CHS. Histogram of diameter (b) and height (c) of PAR1 particles imaged in (a). (b) The diameter distribution showed two peaks centered at 8.1 ± 1.3 nm (average ± S.D.) and 14.7 ± 0.6 nm. Diameters were measured at full-width half maximum of particle heights. (c) The height distribution showed two peaks centered at 1.2 ± 0.2 nm (average ± S.D.) and 2.0 ± 0.3 nm, which could correspond to the height of the extracellular or intracellular surface emerging from the DOPS/CHS membrane, respectively (d). The FD-based AFM topograph was recorded in imaging buffer (300 mm NaCl, 20 mm Hepes, 25 mm MgCl2, ph 7.0) at room temperature. n gives the number of PAR1 particles analyzed.
6 Supplementary Figure 6 AFM topograph and multiparametric maps of PAR1 reconstituted in liposomes. (a) Topograph showing single and clustered PAR1 molecules protruding from the lipid bilayer. (b) Applied force error map showing low errors of <20 pn. (c) Adhesion map showing the SFLLRN functionalized AFM tip interacting sparsely with the lipid bilayer and mainly with PAR1 (see (a)). (d) Deformation map showing enhanced deformation values of PAR1 molecules. The FD-based AFM data was recorded as described (Online Methods).
7 Supplementary Figure 7 Mapping the binding of the SFLLRN ligand to human PAR1 proteoliposomes using FD-based AFM. Topographs (left column) of human PAR1 reconstituted in proteoliposomes taken with the SFLLRN-ligand functionalized AFM tip oscillated at 0.25 khz and amplitude of 50 nm. As described, the SFLLRN has been attached via the PEG-polypeptide linker to the AFM tip (Fig. 1c). Corresponding adhesion maps (right column). To increase their visibility adhesive pixel were enlarged by a factor 2. The FD-based AFM data was recorded as described (Online Methods).
8 Supplementary Figure 8 Validating that SFLLRN ligand functionalized AFM tips detect specific interactions with PAR1. Height images (a,c,e,g) and corresponding adhesion images (b,d,e,h) recorded with either (a,b) a bare AFM tip, (c,d) a hexa-glycine, (e,f) a scrambled peptide (FLLNSR) or (g,h) a SFLLRN functionalized tip in the presence of peptide mimetic antagonist (1 µm BMS). Every functional group (hexa-glycine, FLLNSR, or SFLLRN) was attached via the same PEG-polypeptide linker to the AFM tip (Fig. 1c). This supplementary figure correlates to Fig. 2, which maps receptor-ligand interactions on PAR1 using SFLLRN functionalized AFM tips. The FD-based AFM data was recorded as described (Online Methods).
9 Supplementary Figure 9 Determining the effective spring constant k eff of the cantilever-peg-polypeptide linker system. (a) The effective spring constant k eff is the combination of the cantilever stiffness k c and the stiffness k L of the PEG-polypeptide linker. As described (Supplementary Note), k L was estimated using the combination of the elasticity of the PEG linker (k PEG ) and the elasticity of the polypeptide linker (k polypeptide ). The linker consists of the 27-unit long PEG spacer and the 28 aa long polypeptide, which mimics the thrombin cleaved N-terminal end of PAR1 (comp. Fig. 1). (b) The extension of the PEG-polypeptide linker applied to a stretching force F in water is described by the combination of both the PEG elasticity model 6 and the WLC model 7. (c) Calculated stiffness k L of the PEG-polypeptide linker (blue curve) vs extension and effective stiffness k eff of cantilever and linker (red curve). (d) The force vs k eff relationship was used to determine the effective spring constant of cantilever-peg-polypeptide linker system exposed to a given force F eq and to subsequently calculate ΔG bu. 6. Sulchek, T., Friddle, R.W. & Noy, A. Strength of multiple parallel biological bonds. Biophys. J. 90, (2006). 7. Bustamante, C., Marko, J.F., Siggia, E.D. & Smith, S. Entropic elasticity of lambda-phage DNA. Science 265, (1994).
10 Supplementary Figure 10 Validating the elastic potential of the PEG-polypeptide linker attaching the ligand to the AFM tip. Shown are individual FD curves, each one detecting the rupture of a specific SFLLRN-PAR1 bond. Each of these representative FD curves was fitted using the effective spring constant k eff of the cantilever-peg-polypeptide linker system (Supplementary Fig. 9). The individual fits were shifted along the distance axis to minimize the sum of squared residuals between the fit and the adhesion force peak. The d values given correspond to the shifted distance. FD curves were recorded as described (Fig. 2) using a SFLLRN functionalized AFM cantilever and imaging a PAR1 proteoliposome by FD-based AFM.
11 Supplementary Figure 11 The position of the PEG-polypeptide linker bound to the tip affects the distance of the force peak stretching the linker. The stretching of the PEG-polypeptide linker under force is described combining the model describing the PEG elasticity and the WLC model describing the polypeptide elasticity (see Supplementary Note). Independent of the position at which the PEG-polypeptide linker is chemically anchored to the AFM tip the force-distance characteristics describing the stretching of the linker is always similar since in all cases the linker is bound via one terminal end to the AFM tip and with the other terminal end adheres to the receptor. However, due to the vertical position at which the linker is anchored to the tip, the first part of the force-distance characteristics describing the stretching of the PEG-polypeptide linker can be hidden by relative distance " d" value (dashed line).
12 Supplementary Figure 12 Influence of the AFM tip drive frequency on ligand-receptor unbinding force. DFS plot showing that the tip drive frequency (0.25 khz, red or 0.5 khz, blue) has no influence on the force required to separate SFLLRN-ligand and PAR1. Although the higher oscillation frequency reduced the contact time between tip and sample and, thus, lowered the frequency of bonds formed between ligand and PAR1, we could record a sufficient amount of specific unbinding events. Data were recorded in imaging buffer (300 mm NaCl, 20 mm Hepes, 25 mm MgCl 2, ph 7.0) at room temperature and fitted using the Friddle-Noy-de Yoreo model (thin red line) 8. Each circle represents one measurement. Darker red shaded areas represent 99% confidence intervals and lighter red shaded areas represent 99% of prediction intervals. 8. Friddle, R.W., Noy, A. & De Yoreo, J.J. Interpreting the widespread nonlinear force spectra of intermolecular bonds. Proc Natl Acad Sci U S A 109, (2012).
13 Supplementary Information Supplementary note Establishing a realistic measure of the elastic potential and experimental fit To establish a direct connection between F eq and G bu, we need to establish a realistic measure of the elastic potential E s (F eq ) of the linker tethering the hexa- amino acid sequence (e.g., SFLLRN or other) to the AFM tip. This linker consists of a 27- unit PEG spacer and a 28 aa long polypeptide, which mimics the thrombin cleaved native N- terminal end of PAR1 (Fig. 1c and Supplementary Fig. 9a). To properly describe the entire PEG- polypeptide linker, we depicted the extension of polypeptide using the worm- like- chain (WLC) model 1 (with a persistence length of l p = 0.4 nm and a contour length of L c = 28 aa) and the extension of the PEG spacer containing 27 monomers (n m ) in water 2 using the model for PEG elasticity 2 (with a Kuhn length of L k = 0.7 nm, a segment elasticity of K s = 150 N/m, a monomer length in planar configuration of L planar = nm, a monomer length in helical configuration of L helical = 0.28 nm and a free energy difference between planar and helical configuration of G = 3 k b T). Worm- like- chain model: " % $ ' F = k T $ b 1 l P " 4$ 1 x 2 % + x ' $ 0.25 L ' c ' $ # # L ' c & & Model for PEG elasticity: L(F) = L C (F) coth Fl k k b T k bt Fl k + n mf K s L C (F) = n m L planar ΔG(F ) k e b T +1 + L helical ΔG(F ) k e b T ( ) ΔG(F) = ΔG F L planar L helical +1 Combining both models, we obtained the force- extension relationship of the PEG- polypeptide linker (Supplementary Fig. 9b). Derivative of this force- extension relationship results in the stiffness- extension k L of the entire linker (Supplementary Fig. 9c, blue curve). Finally, using the relationship for two springs in series (cantilever spring k c and linker spring k L ) (Supplementary Fig. 9a), we obtained the stiffness k eff of the cantilever- PEG- polypeptide system (Supplementary Fig. 9c, red curve). After having established a more realistic measure of the elastic potential stressing the ligand- receptor bond we applied this relationship to fit unbinding (or
14 adhesion) events recorded in single force- distance (FD) curves. As shown in Supplementary Fig. 10 this relationship describes very well the specific adhesion events. Thereby, every unbinding event is fitted using the same fitting parameters and the fitted curves were simply shifted along the distance axis by Δd. As explained schematically in Supplementary Fig. 11, this Δd- value refers to the vertical distance between the tip apex and the position at which the linker chemically anchored to the tip. As shown in the schematics, a linker anchored by a distance Δd above the tip apex will lead to the hiding of the first Δd of the model curve fit. Analyzing the specific unbinding events detected in FD curves, which were recorded with the same functionalized AFM tip, in average showed 1 3 different Δd values. This suggests that an average AFM tip only bears a few functional groups at the tip apex. These Δd values were in the range of 0 12 nm, which is compatible with the expected length of the PEG- polypeptide linker. Therefore, we validate the above- mentioned model to realistically describe the elastic potential of the PEG- polypeptide linker.
15 Supplementary Table 1 Overview of peptides probed for their specific binding to PAR1. Peptide Inhibition Parameters extracted from DFS plots F eq (pn) x u (Å) k 0 off (s - 1 ) ΔG bu (kcal/mol) K d Affinity Data shown in ± ± ± ± nm High Fig. 4 SFLLRN BMS None Supp. Fig. 8 Vorapaxar 26.4 ± ± ± ± µm Low Fig. 4 SFLLAN ± ± ± ± µm Low Fig. 4 Vorapaxar 27.1 ± ± ± ± µm Low Fig. 4 SALLRN ± ± ± ± µm None Fig. 4 A(pF- F)RChahRY FLLNSR (scrambled) ± ± ± ± nm High Fig None Supp. Fig. 8 GGGGGG None Supp. Fig. 8
16 Supplementary References 1. Bustamante, C., Marko, J.F., Siggia, E.D. & Smith, S. Entropic elasticity of lambda- phage DNA. Science 265, (1994). 2. Sulchek, T., Friddle, R.W. & Noy, A. Strength of multiple parallel biological bonds. Biophys. J. 90, (2006).
FD-based AFM: The tool to image and simultaneously map multiple properties of biological systems
 force-distance curve-based atomic force microscopy FD-based AFM: The tool to image and simultaneously map multiple properties of biological systems Technical Journal Club 1. Sept 215 Valeria Eckhardt Overview
force-distance curve-based atomic force microscopy FD-based AFM: The tool to image and simultaneously map multiple properties of biological systems Technical Journal Club 1. Sept 215 Valeria Eckhardt Overview
Structural investigation of single biomolecules
 Structural investigation of single biomolecules NMR spectroscopy and X-ray crystallography are currently the most common techniques capable of determining the structures of biological macromolecules like
Structural investigation of single biomolecules NMR spectroscopy and X-ray crystallography are currently the most common techniques capable of determining the structures of biological macromolecules like
Single molecule force spectroscopy reveals a highly compliant helical
 Supplementary Information Single molecule force spectroscopy reveals a highly compliant helical folding for the 30 nm chromatin fiber Maarten Kruithof, Fan-Tso Chien, Andrew Routh, Colin Logie, Daniela
Supplementary Information Single molecule force spectroscopy reveals a highly compliant helical folding for the 30 nm chromatin fiber Maarten Kruithof, Fan-Tso Chien, Andrew Routh, Colin Logie, Daniela
Lecture 12: Biomaterials Characterization in Aqueous Environments
 3.051J/20.340J 1 Lecture 12: Biomaterials Characterization in Aqueous Environments High vacuum techniques are important tools for characterizing surface composition, but do not yield information on surface
3.051J/20.340J 1 Lecture 12: Biomaterials Characterization in Aqueous Environments High vacuum techniques are important tools for characterizing surface composition, but do not yield information on surface
Supporting Information for: Mechanism of Reversible Peptide-Bilayer. Attachment: Combined Simulation and Experimental Single-Molecule Study
 Supporting Information for: Mechanism of Reversible Peptide-Bilayer Attachment: Combined Simulation and Experimental Single-Molecule Study Nadine Schwierz a,, Stefanie Krysiak c, Thorsten Hugel c,d, Martin
Supporting Information for: Mechanism of Reversible Peptide-Bilayer Attachment: Combined Simulation and Experimental Single-Molecule Study Nadine Schwierz a,, Stefanie Krysiak c, Thorsten Hugel c,d, Martin
3.052 Nanomechanics of Materials and Biomaterials Tuesday 04/03/07 Prof. C. Ortiz, MIT-DMSE I LECTURE 13: MIDTERM #1 SOLUTIONS REVIEW
 I LECTURE 13: MIDTERM #1 SOLUTIONS REVIEW Outline : HIGH RESOLUTION FORCE SPECTROSCOPY...2-10 General Experiment Description... 2 Verification of Surface Functionalization:Imaging of Planar Substrates...
I LECTURE 13: MIDTERM #1 SOLUTIONS REVIEW Outline : HIGH RESOLUTION FORCE SPECTROSCOPY...2-10 General Experiment Description... 2 Verification of Surface Functionalization:Imaging of Planar Substrates...
Measurements of interaction forces in (biological) model systems
 Measurements of interaction forces in (biological) model systems Marina Ruths Department of Chemistry, UMass Lowell What can force measurements tell us about a system? Depending on the technique, we might
Measurements of interaction forces in (biological) model systems Marina Ruths Department of Chemistry, UMass Lowell What can force measurements tell us about a system? Depending on the technique, we might
Mapping Elastic Properties of Heterogeneous Materials in Liquid with Angstrom-Scale Resolution
 Supporting information Mapping Elastic Properties of Heterogeneous Materials in Liquid with Angstrom-Scale Resolution Carlos A. Amo, Alma. P. Perrino, Amir F. Payam, Ricardo Garcia * Materials Science
Supporting information Mapping Elastic Properties of Heterogeneous Materials in Liquid with Angstrom-Scale Resolution Carlos A. Amo, Alma. P. Perrino, Amir F. Payam, Ricardo Garcia * Materials Science
Detecting Molecular Interactions that Stabilize Native Bovine Rhodopsin
 doi:10.1016/j.jmb.2006.02.008 J. Mol. Biol. (2006) 358, 255 269 Detecting Molecular Interactions that Stabilize Native Bovine Rhodopsin K. Tanuj Sapra 1, Paul S.-H. Park 2 *, Slawomir Filipek 3, Andreas
doi:10.1016/j.jmb.2006.02.008 J. Mol. Biol. (2006) 358, 255 269 Detecting Molecular Interactions that Stabilize Native Bovine Rhodopsin K. Tanuj Sapra 1, Paul S.-H. Park 2 *, Slawomir Filipek 3, Andreas
Supporting Information
 Supporting Information The force of transporting amino-acid into the living cell measured by atomic force microscopy Xin Shang a,, Yuping Shan b,, Yangang Pan a,, Mingjun Cai a, Junguang Jiang a, Hongda
Supporting Information The force of transporting amino-acid into the living cell measured by atomic force microscopy Xin Shang a,, Yuping Shan b,, Yangang Pan a,, Mingjun Cai a, Junguang Jiang a, Hongda
Biological Sciences 11 Spring Experiment 4. Protein crosslinking
 Biological Sciences 11 Spring 2000 Experiment 4. Protein crosslinking = C - CH 2 - CH 2 - CH 2 - C = H H GA Cl - H 2 N N H 2 Cl - C - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - C DMS CH 3 CH 3 N - - C -
Biological Sciences 11 Spring 2000 Experiment 4. Protein crosslinking = C - CH 2 - CH 2 - CH 2 - C = H H GA Cl - H 2 N N H 2 Cl - C - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - CH 2 - C DMS CH 3 CH 3 N - - C -
Multimedia : Fibronectin and Titin unfolding simulation movies.
 I LECTURE 21: SINGLE CHAIN ELASTICITY OF BIOMACROMOLECULES: THE GIANT PROTEIN TITIN AND DNA Outline : REVIEW LECTURE #2 : EXTENSIBLE FJC AND WLC... 2 STRUCTURE OF MUSCLE AND TITIN... 3 SINGLE MOLECULE
I LECTURE 21: SINGLE CHAIN ELASTICITY OF BIOMACROMOLECULES: THE GIANT PROTEIN TITIN AND DNA Outline : REVIEW LECTURE #2 : EXTENSIBLE FJC AND WLC... 2 STRUCTURE OF MUSCLE AND TITIN... 3 SINGLE MOLECULE
Supporting information
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 Supporting information Self-assembled nanopatch with peptide-organic multilayers and mechanical
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2014 Supporting information Self-assembled nanopatch with peptide-organic multilayers and mechanical
Mechanical Proteins. Stretching imunoglobulin and fibronectin. domains of the muscle protein titin. Adhesion Proteins of the Immune System
 Mechanical Proteins F C D B A domains of the muscle protein titin E Stretching imunoglobulin and fibronectin G NIH Resource for Macromolecular Modeling and Bioinformatics Theoretical Biophysics Group,
Mechanical Proteins F C D B A domains of the muscle protein titin E Stretching imunoglobulin and fibronectin G NIH Resource for Macromolecular Modeling and Bioinformatics Theoretical Biophysics Group,
SUPPLEMENTARY INFORMATION
 Ultrafast proton transport in sub-1-nm diameter carbon nanotube porins Ramya H. Tunuguntla, 1 Frances I. Allen, 2 Kyunghoon Kim, 1, Allison Belliveau, 1 Aleksandr Noy 1,3, * * Correspondence to AN: noy1@llnl.gov
Ultrafast proton transport in sub-1-nm diameter carbon nanotube porins Ramya H. Tunuguntla, 1 Frances I. Allen, 2 Kyunghoon Kim, 1, Allison Belliveau, 1 Aleksandr Noy 1,3, * * Correspondence to AN: noy1@llnl.gov
Magnetic tweezers and its application to DNA mechanics
 Biotechnological Center Research group DNA motors (Seidel group) Handout for Practical Course Magnetic tweezers and its application to DNA mechanics When: 9.00 am Where: Biotec, 3 rd Level, Room 317 Tutors:
Biotechnological Center Research group DNA motors (Seidel group) Handout for Practical Course Magnetic tweezers and its application to DNA mechanics When: 9.00 am Where: Biotec, 3 rd Level, Room 317 Tutors:
Models for Single Molecule Experiments
 Models for Single Molecule Experiments Binding Unbinding Folding - Unfolding -> Kramers Bell theory 1 Polymer elasticity: - Hooke s law (?) - FJC (Freely joined chain) - WLC (Worm-like chain) 3 Molecular
Models for Single Molecule Experiments Binding Unbinding Folding - Unfolding -> Kramers Bell theory 1 Polymer elasticity: - Hooke s law (?) - FJC (Freely joined chain) - WLC (Worm-like chain) 3 Molecular
Lecture 34 Protein Unfolding Thermodynamics
 Physical Principles in Biology Biology 3550 Fall 2018 Lecture 34 Protein Unfolding Thermodynamics Wednesday, 21 November c David P. Goldenberg University of Utah goldenberg@biology.utah.edu Clicker Question
Physical Principles in Biology Biology 3550 Fall 2018 Lecture 34 Protein Unfolding Thermodynamics Wednesday, 21 November c David P. Goldenberg University of Utah goldenberg@biology.utah.edu Clicker Question
Substrate-induced changes in the structural properties of LacY. Significance
 Substrate-induced changes in the structural properties of LacY Tetiana Serdiuk a, M. Gregor Madej b, Junichi Sugihara b, Shiho Kawamura a, Stefania A. Mari a, H. Ronald Kaback b,c,d,1, and Daniel J. Müller
Substrate-induced changes in the structural properties of LacY Tetiana Serdiuk a, M. Gregor Madej b, Junichi Sugihara b, Shiho Kawamura a, Stefania A. Mari a, H. Ronald Kaback b,c,d,1, and Daniel J. Müller
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis):
 SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis): Aim: SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is one of the common methods used in the molecular biology
SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis): Aim: SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis) is one of the common methods used in the molecular biology
Supplementary Figures
 Supplementary Figures Supplementary Figure 1: Stacking fault density is direction dependent: Illustration of the stacking fault multiplicity: lattice disorder is clearly direction specific, gradually zooming
Supplementary Figures Supplementary Figure 1: Stacking fault density is direction dependent: Illustration of the stacking fault multiplicity: lattice disorder is clearly direction specific, gradually zooming
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1.
 Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Table S1. Overview of used PDZK1 constructs and their binding affinities to peptides. Related to figure 1. PDZK1 constru cts Amino acids MW [kda] KD [μm] PEPT2-CT- FITC KD [μm] NHE3-CT- FITC KD [μm] PDZK1-CT-
Basic Laboratory. Materials Science and Engineering. Atomic Force Microscopy (AFM)
 Basic Laboratory Materials Science and Engineering Atomic Force Microscopy (AFM) M108 Stand: 20.10.2015 Aim: Presentation of an application of the AFM for studying surface morphology. Inhalt 1.Introduction...
Basic Laboratory Materials Science and Engineering Atomic Force Microscopy (AFM) M108 Stand: 20.10.2015 Aim: Presentation of an application of the AFM for studying surface morphology. Inhalt 1.Introduction...
Mechanical Proteins. Stretching imunoglobulin and fibronectin. domains of the muscle protein titin. Adhesion Proteins of the Immune System
 Mechanical Proteins F C D B A domains of the muscle protein titin E Stretching imunoglobulin and fibronectin G NIH Resource for Macromolecular Modeling and Bioinformatics Theoretical Biophysics Group,
Mechanical Proteins F C D B A domains of the muscle protein titin E Stretching imunoglobulin and fibronectin G NIH Resource for Macromolecular Modeling and Bioinformatics Theoretical Biophysics Group,
Supporting Information. for. Angew. Chem. Int. Ed. Z WILEY-VCH Verlag GmbH & Co. KGaA
 Supporting Information for Angew. Chem. Int. Ed. Z19447 2002 WILEY-VCH Verlag GmbH & Co. KGaA 69451 Weinheim, Germany Spatially Directed Protein Adsorption By Using a Novel, Nanoscale Surface Template
Supporting Information for Angew. Chem. Int. Ed. Z19447 2002 WILEY-VCH Verlag GmbH & Co. KGaA 69451 Weinheim, Germany Spatially Directed Protein Adsorption By Using a Novel, Nanoscale Surface Template
Announcements. Homework 3 (Klaus Schulten s Lecture): Due Wednesday at noon. Next homework assigned. Due Wednesday March 1.
 Announcements Homework 3 (Klaus Schulten s Lecture): Due Wednesday at noon. Next homework assigned. Due Wednesday March 1. No lecture next Monday, Feb. 27 th! (Homework is a bit longer.) Marco will have
Announcements Homework 3 (Klaus Schulten s Lecture): Due Wednesday at noon. Next homework assigned. Due Wednesday March 1. No lecture next Monday, Feb. 27 th! (Homework is a bit longer.) Marco will have
BMB November 17, Single Molecule Biophysics (I)
 BMB 178 2017 November 17, 2017 14. Single Molecule Biophysics (I) Goals 1. Understand the information SM experiments can provide 2. Be acquainted with different SM approaches 3. Learn to interpret SM results
BMB 178 2017 November 17, 2017 14. Single Molecule Biophysics (I) Goals 1. Understand the information SM experiments can provide 2. Be acquainted with different SM approaches 3. Learn to interpret SM results
NIS: what can it be used for?
 AFM @ NIS: what can it be used for? Chiara Manfredotti 011 670 8382/8388/7879 chiara.manfredotti@to.infn.it Skype: khiaram 1 AFM: block scheme In an Atomic Force Microscope (AFM) a micrometric tip attached
AFM @ NIS: what can it be used for? Chiara Manfredotti 011 670 8382/8388/7879 chiara.manfredotti@to.infn.it Skype: khiaram 1 AFM: block scheme In an Atomic Force Microscope (AFM) a micrometric tip attached
Single-Molecule Force Spectroscopy on Poly(acrylic acid) by AFM
 2120 Langmuir 1999, 15, 2120-2124 Single-Molecule Force Spectroscopy on Poly(acrylic acid) by AFM Hongbin Li, Bingbing Liu, Xi Zhang,*, Chunxiao Gao, Jiacong Shen, and Guangtian Zou Key Lab for Supramolecular
2120 Langmuir 1999, 15, 2120-2124 Single-Molecule Force Spectroscopy on Poly(acrylic acid) by AFM Hongbin Li, Bingbing Liu, Xi Zhang,*, Chunxiao Gao, Jiacong Shen, and Guangtian Zou Key Lab for Supramolecular
Recently developed single-molecule atomic force microscope
 Stepwise unfolding of titin under force-clamp atomic force microscopy Andres F. Oberhauser*, Paul K. Hansma, Mariano Carrion-Vazquez*, and Julio M. Fernandez* *Department of Physiology and Biophysics,
Stepwise unfolding of titin under force-clamp atomic force microscopy Andres F. Oberhauser*, Paul K. Hansma, Mariano Carrion-Vazquez*, and Julio M. Fernandez* *Department of Physiology and Biophysics,
SUPPLEMENTARY INFORMATION. microscopy of membrane proteins
 SUPPLEMENTARY INFORMATION Glass is a viable substrate for precision force microscopy of membrane proteins Nagaraju Chada 1, Krishna P. Sigdel 1, Raghavendar Reddy Sanganna Gari 1, Tina Rezaie Matin 1,
SUPPLEMENTARY INFORMATION Glass is a viable substrate for precision force microscopy of membrane proteins Nagaraju Chada 1, Krishna P. Sigdel 1, Raghavendar Reddy Sanganna Gari 1, Tina Rezaie Matin 1,
Scanning Force Microscopy. i.e. Atomic Force Microscopy. Josef A. Käs Institute for Soft Matter Physics Physics Department. Atomic Force Microscopy
 Scanning Force Microscopy i.e. Atomic Force Microscopy Josef A. Käs Institute for Soft Matter Physics Physics Department Atomic Force Microscopy Single Molecule Force Spectroscopy Zanjan 4 Hermann E. Gaub
Scanning Force Microscopy i.e. Atomic Force Microscopy Josef A. Käs Institute for Soft Matter Physics Physics Department Atomic Force Microscopy Single Molecule Force Spectroscopy Zanjan 4 Hermann E. Gaub
SECOND PUBLIC EXAMINATION. Honour School of Physics Part C: 4 Year Course. Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS
 2757 SECOND PUBLIC EXAMINATION Honour School of Physics Part C: 4 Year Course Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS TRINITY TERM 2013 Monday, 17 June, 2.30 pm 5.45 pm 15
2757 SECOND PUBLIC EXAMINATION Honour School of Physics Part C: 4 Year Course Honour School of Physics and Philosophy Part C C7: BIOLOGICAL PHYSICS TRINITY TERM 2013 Monday, 17 June, 2.30 pm 5.45 pm 15
Scanning Force Microscopy
 Scanning Force Microscopy Roland Bennewitz Rutherford Physics Building 405 Phone 398-3058 roland.bennewitz@mcgill.ca Scanning Probe is moved along scan lines over a sample surface 1 Force Microscopy Data
Scanning Force Microscopy Roland Bennewitz Rutherford Physics Building 405 Phone 398-3058 roland.bennewitz@mcgill.ca Scanning Probe is moved along scan lines over a sample surface 1 Force Microscopy Data
Biophysics of Macromolecules
 Biophysics of Macromolecules Lecture 11: Dynamic Force Spectroscopy Rädler/Lipfert SS 2014 - Forced Ligand-Receptor Unbinding - Bell-Evans Theory 22. Mai. 2014 AFM experiments with single molecules custom-built
Biophysics of Macromolecules Lecture 11: Dynamic Force Spectroscopy Rädler/Lipfert SS 2014 - Forced Ligand-Receptor Unbinding - Bell-Evans Theory 22. Mai. 2014 AFM experiments with single molecules custom-built
Identification of core amino acids stabilizing rhodopsin
 Identification of core amino acids stabilizing rhodopsin A. J. Rader, Gulsum Anderson, Basak Isin, H. Gobind Khorana, Ivet Bahar, and Judith Klein- Seetharaman Anes Aref BBSI 2004 Outline Introduction
Identification of core amino acids stabilizing rhodopsin A. J. Rader, Gulsum Anderson, Basak Isin, H. Gobind Khorana, Ivet Bahar, and Judith Klein- Seetharaman Anes Aref BBSI 2004 Outline Introduction
Supplementary Figure 1 a) Scheme of microfluidic device fabrication by photo and soft lithography,
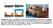 a b 1 mm Supplementary Figure 1 a) Scheme of microfluidic device fabrication by photo and soft lithography, (a1, a2) 50nm Pd evaporated on Si wafer with 100 nm Si 2 insulating layer and 5nm Cr as an adhesion
a b 1 mm Supplementary Figure 1 a) Scheme of microfluidic device fabrication by photo and soft lithography, (a1, a2) 50nm Pd evaporated on Si wafer with 100 nm Si 2 insulating layer and 5nm Cr as an adhesion
Atomic and molecular interactions. Scanning probe microscopy.
 Atomic and molecular interactions. Scanning probe microscopy. Balázs Kiss Nanobiotechnology and Single Molecule Research Group, Department of Biophysics and Radiation Biology 27. November 2013. 2 Atomic
Atomic and molecular interactions. Scanning probe microscopy. Balázs Kiss Nanobiotechnology and Single Molecule Research Group, Department of Biophysics and Radiation Biology 27. November 2013. 2 Atomic
Supplementary Information
 Supplementary Information Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism Duyoung Min, Kipom Kim, Changbong Hyeon, Yong Hoon Cho, Yeon-Kyun
Supplementary Information Mechanical unzipping and rezipping of a single SNARE complex reveals hysteresis as a force-generating mechanism Duyoung Min, Kipom Kim, Changbong Hyeon, Yong Hoon Cho, Yeon-Kyun
Electrically controlled DNA adhesion
 SUPPLEMENTARY INFORMATION Electrically controlled DNA adhesion Matthias Erdmann, Ralf David +, Ann Fornof and Hermann E. Gaub* Chair for Applied Physics and Center for NanoScience, Ludwigs-Maximilians-Universität
SUPPLEMENTARY INFORMATION Electrically controlled DNA adhesion Matthias Erdmann, Ralf David +, Ann Fornof and Hermann E. Gaub* Chair for Applied Physics and Center for NanoScience, Ludwigs-Maximilians-Universität
SUPPLEMENTARY INFORMATION
 doi:10.108/nature11899 Supplementar Table 1. Data collection and refinement statistics (+TPMP, native) (-TPMP, native) (+TPMP, recombinant) (MgCl ) (MgSO ) Data collection Space group C P 1 C P 1 1 P 1
doi:10.108/nature11899 Supplementar Table 1. Data collection and refinement statistics (+TPMP, native) (-TPMP, native) (+TPMP, recombinant) (MgCl ) (MgSO ) Data collection Space group C P 1 C P 1 1 P 1
The four forces of nature. Intermolecular forces, surface forces & the Atomic Force Microscope (AFM) Force- and potential curves
 Intermolecular forces, surface forces & the Atomic Force Microscope (AFM) The four forces of nature Strong interaction Holds neutrons and protons together in atomic nuclei. Weak interaction β and elementary
Intermolecular forces, surface forces & the Atomic Force Microscope (AFM) The four forces of nature Strong interaction Holds neutrons and protons together in atomic nuclei. Weak interaction β and elementary
Scale in the biological world
 Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Scale in the biological world 2 A cell seen by TEM 3 4 From living cells to atoms 5 Compartmentalisation in the cell: internal membranes and the cytosol 6 The Origin of mitochondria: The endosymbion hypothesis
Molecular dynamics simulations of anti-aggregation effect of ibuprofen. Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov
 Biophysical Journal, Volume 98 Supporting Material Molecular dynamics simulations of anti-aggregation effect of ibuprofen Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov Supplemental
Biophysical Journal, Volume 98 Supporting Material Molecular dynamics simulations of anti-aggregation effect of ibuprofen Wenling E. Chang, Takako Takeda, E. Prabhu Raman, and Dmitri Klimov Supplemental
Stiffness versus architecture of single helical polyisocyanopeptides
 This journal is The Royal Society of Chemistry 213 Supplementary information Stiffness versus architecture of single helical polyisocyanopeptides Arend M. van Buul, a Erik Schwartz, b Patrick Brocorens,
This journal is The Royal Society of Chemistry 213 Supplementary information Stiffness versus architecture of single helical polyisocyanopeptides Arend M. van Buul, a Erik Schwartz, b Patrick Brocorens,
AFM Imaging In Liquids. W. Travis Johnson PhD Agilent Technologies Nanomeasurements Division
 AFM Imaging In Liquids W. Travis Johnson PhD Agilent Technologies Nanomeasurements Division Imaging Techniques: Scales Proteins 10 nm Bacteria 1μm Red Blood Cell 5μm Human Hair 75μm Si Atom Spacing 0.4nm
AFM Imaging In Liquids W. Travis Johnson PhD Agilent Technologies Nanomeasurements Division Imaging Techniques: Scales Proteins 10 nm Bacteria 1μm Red Blood Cell 5μm Human Hair 75μm Si Atom Spacing 0.4nm
Digital processing of multi-mode nano-mechanical cantilever data
 IOP Publishing Journal of Physics: Conference Series 61 (2007) 341 345 doi:10.1088/1742-6596/61/1/069 International Conference on Nanoscience and Technology (ICN&T 2006) Digital processing of multi-mode
IOP Publishing Journal of Physics: Conference Series 61 (2007) 341 345 doi:10.1088/1742-6596/61/1/069 International Conference on Nanoscience and Technology (ICN&T 2006) Digital processing of multi-mode
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10458 Active Site Remodeling in the Bifunctional Fructose-1,6- bisphosphate aldolase/phosphatase Juan Du, Rafael F. Say, Wei Lü, Georg Fuchs & Oliver Einsle SUPPLEMENTARY FIGURES Figure
doi:10.1038/nature10458 Active Site Remodeling in the Bifunctional Fructose-1,6- bisphosphate aldolase/phosphatase Juan Du, Rafael F. Say, Wei Lü, Georg Fuchs & Oliver Einsle SUPPLEMENTARY FIGURES Figure
Atomic Force Microscopy imaging and beyond
 Atomic Force Microscopy imaging and beyond Arif Mumtaz Magnetism and Magnetic Materials Group Department of Physics, QAU Coworkers: Prof. Dr. S.K.Hasanain M. Tariq Khan Alam Imaging and beyond Scanning
Atomic Force Microscopy imaging and beyond Arif Mumtaz Magnetism and Magnetic Materials Group Department of Physics, QAU Coworkers: Prof. Dr. S.K.Hasanain M. Tariq Khan Alam Imaging and beyond Scanning
Supplementary Information
 Supplementary Information Switching of myosin-v motion between the lever-arm swing and Brownian search-and-catch Keisuke Fujita 1*, Mitsuhiro Iwaki 2,3*, Atsuko H. Iwane 1, Lorenzo Marcucci 1 & Toshio
Supplementary Information Switching of myosin-v motion between the lever-arm swing and Brownian search-and-catch Keisuke Fujita 1*, Mitsuhiro Iwaki 2,3*, Atsuko H. Iwane 1, Lorenzo Marcucci 1 & Toshio
The Powerful Diversity of the AFM Probe
 The Powerful Diversity of the AFM Probe Stefan B. Kaemmer, Bruker Nano Surfaces Division, Santa Barbara, CA 93117 stefan.kaemmer@bruker-nano.com March 21, 2011 Introduction The tip allows us to measure
The Powerful Diversity of the AFM Probe Stefan B. Kaemmer, Bruker Nano Surfaces Division, Santa Barbara, CA 93117 stefan.kaemmer@bruker-nano.com March 21, 2011 Introduction The tip allows us to measure
In Situ Gelation-Induced Death of Cancer Cells Based on Proteinosomes
 Supporting information for In Situ Gelation-Induced Death of Cancer Cells Based on Proteinosomes Yuting Zhou, Jianmin Song, Lei Wang*, Xuting Xue, Xiaoman Liu, Hui Xie*, and Xin Huang* MIIT Key Laboratory
Supporting information for In Situ Gelation-Induced Death of Cancer Cells Based on Proteinosomes Yuting Zhou, Jianmin Song, Lei Wang*, Xuting Xue, Xiaoman Liu, Hui Xie*, and Xin Huang* MIIT Key Laboratory
Intermittent-Contact Mode Force Microscopy & Electrostatic Force Microscopy (EFM)
 WORKSHOP Nanoscience on the Tip Intermittent-Contact Mode Force Microscopy & Electrostatic Force Microscopy (EFM) Table of Contents: 1. Motivation... 1. Simple Harmonic Motion... 1 3. AC-Mode Imaging...
WORKSHOP Nanoscience on the Tip Intermittent-Contact Mode Force Microscopy & Electrostatic Force Microscopy (EFM) Table of Contents: 1. Motivation... 1. Simple Harmonic Motion... 1 3. AC-Mode Imaging...
Detailed AFM Force Spectroscopy of the Interaction between. CD44-IgG-Fusion Protein and Hyaluronan
 Electronic Supplementary Material to: Detailed AFM Force Spectroscopy of the Interaction between CD44-IgG-Fusion Protein and Hyaluronan Aernout A. Martens a, Marcel Bus a, Peter C. Thüne b,, Tjerk H. Oosterkamp,
Electronic Supplementary Material to: Detailed AFM Force Spectroscopy of the Interaction between CD44-IgG-Fusion Protein and Hyaluronan Aernout A. Martens a, Marcel Bus a, Peter C. Thüne b,, Tjerk H. Oosterkamp,
BA, BSc, and MSc Degree Examinations
 Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes
Examination Candidate Number: Desk Number: BA, BSc, and MSc Degree Examinations 2017-8 Department : BIOLOGY Title of Exam: Molecular Biology and Biochemistry Part I Time Allowed: 1 hour and 30 minutes
Amphiphilic diselenide-containing supramolecular polymers
 Electronic Supplementary Material (ESI) for Polymer Chemistry. This journal is The Royal Society of Chemistry 2014 Amphiphilic diselenide-containing supramolecular polymers Xinxin Tan, Liulin Yang, Zehuan
Electronic Supplementary Material (ESI) for Polymer Chemistry. This journal is The Royal Society of Chemistry 2014 Amphiphilic diselenide-containing supramolecular polymers Xinxin Tan, Liulin Yang, Zehuan
Dana Alsulaibi. Jaleel G.Sweis. Mamoon Ahram
 15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
15 Dana Alsulaibi Jaleel G.Sweis Mamoon Ahram Revision of last lectures: Proteins have four levels of structures. Primary,secondary, tertiary and quaternary. Primary structure is the order of amino acids
Elasticity of the human red blood cell skeleton
 Biorheology 40 (2003) 247 251 247 IOS Press Elasticity of the human red blood cell skeleton G. Lenormand, S. Hénon, A. Richert, J. Siméon and F. Gallet Laboratoire de Biorhéologie et d Hydrodynamique Physico-Chimique,
Biorheology 40 (2003) 247 251 247 IOS Press Elasticity of the human red blood cell skeleton G. Lenormand, S. Hénon, A. Richert, J. Siméon and F. Gallet Laboratoire de Biorhéologie et d Hydrodynamique Physico-Chimique,
Measure mass, thickness and structural properties of molecular layers Automated and fully integrated turn-key system
 Product Information Q-Sense Omega Auto Real-time interface characterization Measure mass, thickness and structural properties of molecular layers Automated and fully integrated turn-key system 30 µl sample
Product Information Q-Sense Omega Auto Real-time interface characterization Measure mass, thickness and structural properties of molecular layers Automated and fully integrated turn-key system 30 µl sample
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
It s really this simple.
 Background Light harvesting complexes exist to facilitate and maximize the absorption capacity of the reaction centers (RC) as well as PSI and PSII Purple bacteria utilize these functions by having an
Background Light harvesting complexes exist to facilitate and maximize the absorption capacity of the reaction centers (RC) as well as PSI and PSII Purple bacteria utilize these functions by having an
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor
 LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
LS1a Fall 2014 Problem Set #2 Due Monday 10/6 at 6 pm in the drop boxes on the Science Center 2 nd Floor Note: Adequate space is given for each answer. Questions that require a brief explanation should
MAE 545: Lecture 4 (9/29) Statistical mechanics of proteins
 MAE 545: Lecture 4 (9/29) Statistical mechanics of proteins Ideal freely jointed chain N identical unstretchable links (Kuhn segments) of length a with freely rotating joints ~r 2 a ~r ~R ~r N In first
MAE 545: Lecture 4 (9/29) Statistical mechanics of proteins Ideal freely jointed chain N identical unstretchable links (Kuhn segments) of length a with freely rotating joints ~r 2 a ~r ~R ~r N In first
BioAFM spectroscopy for mapping of Young s modulus of living cells
 BioAFM spectroscopy for mapping of Young s modulus of living cells Jan Přibyl pribyl@nanobio.cz Optical microscopy AFM Confocal microscopy Young modulus Content Introduction (theory) Hook s law, Young
BioAFM spectroscopy for mapping of Young s modulus of living cells Jan Přibyl pribyl@nanobio.cz Optical microscopy AFM Confocal microscopy Young modulus Content Introduction (theory) Hook s law, Young
Viscoelasticity of a Single Polymer Chain
 Netsu Sokutei 33 4 183-190 2006 7 10 2006 7 22 Viscoelasticity of a Single Polymer Chain Ken Nakajima and Toshio Nishi (Received July 10, 2006; Accepted July 22, 2006) Atomic force microscopy (AFM) enabled
Netsu Sokutei 33 4 183-190 2006 7 10 2006 7 22 Viscoelasticity of a Single Polymer Chain Ken Nakajima and Toshio Nishi (Received July 10, 2006; Accepted July 22, 2006) Atomic force microscopy (AFM) enabled
thiol monolayers by means of high-rate dynamic force spectroscopy
 1) Max Planck Institute for Polymer Research 2) Poznan University of Technology Adhesion on self-assembled thiol monolayers by means of high-rate dynamic force spectroscopy Hubert Gojżewski 1,2, Arkadiusz
1) Max Planck Institute for Polymer Research 2) Poznan University of Technology Adhesion on self-assembled thiol monolayers by means of high-rate dynamic force spectroscopy Hubert Gojżewski 1,2, Arkadiusz
Intensity (a.u.) Intensity (a.u.) Raman Shift (cm -1 ) Oxygen plasma. 6 cm. 9 cm. 1mm. Single-layer graphene sheet. 10mm. 14 cm
 Intensity (a.u.) Intensity (a.u.) a Oxygen plasma b 6 cm 1mm 10mm Single-layer graphene sheet 14 cm 9 cm Flipped Si/SiO 2 Patterned chip Plasma-cleaned glass slides c d After 1 sec normal Oxygen plasma
Intensity (a.u.) Intensity (a.u.) a Oxygen plasma b 6 cm 1mm 10mm Single-layer graphene sheet 14 cm 9 cm Flipped Si/SiO 2 Patterned chip Plasma-cleaned glass slides c d After 1 sec normal Oxygen plasma
Chain-configuration and rate dependent rheological properties in transient networks
 Electronic Supplementary Material (ESI) for Soft Matter. This journal is The Royal Society of Chemistry 204 Supplementary Information Chain-configuration and rate dependent rheological properties in transient
Electronic Supplementary Material (ESI) for Soft Matter. This journal is The Royal Society of Chemistry 204 Supplementary Information Chain-configuration and rate dependent rheological properties in transient
Bringing optics into the nanoscale a double-scanner AFM brings advanced optical experiments within reach
 Bringing optics into the nanoscale a double-scanner AFM brings advanced optical experiments within reach Beyond the diffraction limit The resolution of optical microscopy is generally limited by the diffraction
Bringing optics into the nanoscale a double-scanner AFM brings advanced optical experiments within reach Beyond the diffraction limit The resolution of optical microscopy is generally limited by the diffraction
Supplementary Information. The Solution Structural Ensembles of RNA Kink-turn Motifs and Their Protein Complexes
 Supplementary Information The Solution Structural Ensembles of RNA Kink-turn Motifs and Their Protein Complexes Xuesong Shi, a Lin Huang, b David M. J. Lilley, b Pehr B. Harbury a,c and Daniel Herschlag
Supplementary Information The Solution Structural Ensembles of RNA Kink-turn Motifs and Their Protein Complexes Xuesong Shi, a Lin Huang, b David M. J. Lilley, b Pehr B. Harbury a,c and Daniel Herschlag
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability
 Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Lecture 2 and 3: Review of forces (ctd.) and elementary statistical mechanics. Contributions to protein stability Part I. Review of forces Covalent bonds Non-covalent Interactions: Van der Waals Interactions
Outline Scanning Probe Microscope (SPM)
 AFM Outline Scanning Probe Microscope (SPM) A family of microscopy forms where a sharp probe is scanned across a surface and some tip/sample interactions are monitored Scanning Tunneling Microscopy (STM)
AFM Outline Scanning Probe Microscope (SPM) A family of microscopy forms where a sharp probe is scanned across a surface and some tip/sample interactions are monitored Scanning Tunneling Microscopy (STM)
Dynamic Force Spectroscopy of the Silicon Carbon Bond
 Hochschule für angewandte Wissenschaften FH München Fakultät 06 Feinwerk- und Mikrotechnik, Physikalische Technik Mikro- und Nanotechnik Master Thesis Dynamic Force Spectroscopy of the Silicon Carbon Bond
Hochschule für angewandte Wissenschaften FH München Fakultät 06 Feinwerk- und Mikrotechnik, Physikalische Technik Mikro- und Nanotechnik Master Thesis Dynamic Force Spectroscopy of the Silicon Carbon Bond
Multimedia : Podcast : Sacrificial Bonds in Biological Materials; Fantner, et al. Biophys. J , 1411
 3.52 Nanomechanics o Materials and Biomaterials Tuesday 5/1/7 Pro. C. Ortiz, MIT-DMSE I LECTURE 2: THEORETICAL ASPECTS O SINGLE MOLECULE ORCE SPECTROSCOPY 2 : EXTENSIBILITY AND THE WORM LIKE CHAIN (WLC)
3.52 Nanomechanics o Materials and Biomaterials Tuesday 5/1/7 Pro. C. Ortiz, MIT-DMSE I LECTURE 2: THEORETICAL ASPECTS O SINGLE MOLECULE ORCE SPECTROSCOPY 2 : EXTENSIBILITY AND THE WORM LIKE CHAIN (WLC)
SUPPLEMENTARY MATERIAL FOR
 SUPPLEMENTARY MATERIAL FOR THE LIPID-BINDING DOMAIN OF WILD TYPE AND MUTANT ALPHA- SYNUCLEIN: COMPACTNESS AND INTERCONVERSION BETWEEN THE BROKEN- AND EXTENDED-HELIX FORMS. Elka R. Georgieva 1, Trudy F.
SUPPLEMENTARY MATERIAL FOR THE LIPID-BINDING DOMAIN OF WILD TYPE AND MUTANT ALPHA- SYNUCLEIN: COMPACTNESS AND INTERCONVERSION BETWEEN THE BROKEN- AND EXTENDED-HELIX FORMS. Elka R. Georgieva 1, Trudy F.
Lecture 5: Macromolecules, polymers and DNA
 1, polymers and DNA Introduction In this lecture, we focus on a subfield of soft matter: macromolecules and more particularly on polymers. As for the previous chapter about surfactants and electro kinetics,
1, polymers and DNA Introduction In this lecture, we focus on a subfield of soft matter: macromolecules and more particularly on polymers. As for the previous chapter about surfactants and electro kinetics,
Lab Week 2 Module α 2. AFM/DSC Study of Protein Denaturation. Instructor: Gretchen DeVries
 3.014 Materials Laboratory October 24-28, 2005 Lab Week 2 Module α 2 AFM/DSC Study of Protein Denaturation Instructor: Gretchen DeVries Objectives Understand the theory and instrumentation in Differential
3.014 Materials Laboratory October 24-28, 2005 Lab Week 2 Module α 2 AFM/DSC Study of Protein Denaturation Instructor: Gretchen DeVries Objectives Understand the theory and instrumentation in Differential
Microcalorimetry for the Life Sciences
 Microcalorimetry for the Life Sciences Why Microcalorimetry? Microcalorimetry is universal detector Heat is generated or absorbed in every chemical process In-solution No molecular weight limitations Label-free
Microcalorimetry for the Life Sciences Why Microcalorimetry? Microcalorimetry is universal detector Heat is generated or absorbed in every chemical process In-solution No molecular weight limitations Label-free
THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION
 THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION AND CALIBRATION Calculation of turn and beta intrinsic propensities. A statistical analysis of a protein structure
THE TANGO ALGORITHM: SECONDARY STRUCTURE PROPENSITIES, STATISTICAL MECHANICS APPROXIMATION AND CALIBRATION Calculation of turn and beta intrinsic propensities. A statistical analysis of a protein structure
SUPPLEMENTARY INFORMATION
 1. Supplementary Methods Characterization of AFM resolution We employed amplitude-modulation AFM in non-contact mode to characterize the topography of the graphene samples. The measurements were performed
1. Supplementary Methods Characterization of AFM resolution We employed amplitude-modulation AFM in non-contact mode to characterize the topography of the graphene samples. The measurements were performed
A General Equation for Fitting Contact Area and Friction vs Load Measurements
 Journal of Colloid and Interface Science 211, 395 400 (1999) Article ID jcis.1998.6027, available online at http://www.idealibrary.com on A General Equation for Fitting Contact Area and Friction vs Load
Journal of Colloid and Interface Science 211, 395 400 (1999) Article ID jcis.1998.6027, available online at http://www.idealibrary.com on A General Equation for Fitting Contact Area and Friction vs Load
Lecture Note October 1, 2009 Nanostructure characterization techniques
 Lecture Note October 1, 29 Nanostructure characterization techniques UT-Austin PHYS 392 T, unique # 5977 ME 397 unique # 1979 CHE 384, unique # 151 Instructor: Professor C.K. Shih Subjects: Applications
Lecture Note October 1, 29 Nanostructure characterization techniques UT-Austin PHYS 392 T, unique # 5977 ME 397 unique # 1979 CHE 384, unique # 151 Instructor: Professor C.K. Shih Subjects: Applications
Atomic Force Microscopy (AFM) Part I
 Atomic Force Microscopy (AFM) Part I CHEM-L2000 Eero Kontturi 6 th March 2018 Lectures on AFM Part I Principles and practice Imaging of native materials, including nanocellulose Part II Surface force measurements
Atomic Force Microscopy (AFM) Part I CHEM-L2000 Eero Kontturi 6 th March 2018 Lectures on AFM Part I Principles and practice Imaging of native materials, including nanocellulose Part II Surface force measurements
Supporting information
 Supporting information Fluorescent derivatives of AC-42 to probe bitopic orthosteric/allosteric binding mechanisms on muscarinic M1 receptors Sandrine B. Daval, Céline Valant, Dominique Bonnet, Esther
Supporting information Fluorescent derivatives of AC-42 to probe bitopic orthosteric/allosteric binding mechanisms on muscarinic M1 receptors Sandrine B. Daval, Céline Valant, Dominique Bonnet, Esther
Instrumentation and Operation
 Instrumentation and Operation 1 STM Instrumentation COMPONENTS sharp metal tip scanning system and control electronics feedback electronics (keeps tunneling current constant) image processing system data
Instrumentation and Operation 1 STM Instrumentation COMPONENTS sharp metal tip scanning system and control electronics feedback electronics (keeps tunneling current constant) image processing system data
Potassium channel gating and structure!
 Reading: Potassium channel gating and structure Hille (3rd ed.) chapts 10, 13, 17 Doyle et al. The Structure of the Potassium Channel: Molecular Basis of K1 Conduction and Selectivity. Science 280:70-77
Reading: Potassium channel gating and structure Hille (3rd ed.) chapts 10, 13, 17 Doyle et al. The Structure of the Potassium Channel: Molecular Basis of K1 Conduction and Selectivity. Science 280:70-77
SDS-polyacrylamide gel electrophoresis
 SDS-polyacrylamide gel electrophoresis Protein Isolation and Purification Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells,
SDS-polyacrylamide gel electrophoresis Protein Isolation and Purification Protein purification is a series of processes intended to isolate one or a few proteins from a complex mixture, usually cells,
Structure of the α-helix
 Structure of the α-helix Structure of the β Sheet Protein Dynamics Basics of Quenching HDX Hydrogen exchange of amide protons is catalyzed by H 2 O, OH -, and H 3 O +, but it s most dominated by base
Structure of the α-helix Structure of the β Sheet Protein Dynamics Basics of Quenching HDX Hydrogen exchange of amide protons is catalyzed by H 2 O, OH -, and H 3 O +, but it s most dominated by base
Isothermal experiments characterize time-dependent aggregation and unfolding
 1 Energy Isothermal experiments characterize time-dependent aggregation and unfolding Technical ote Introduction Kinetic measurements have, for decades, given protein scientists insight into the mechanisms
1 Energy Isothermal experiments characterize time-dependent aggregation and unfolding Technical ote Introduction Kinetic measurements have, for decades, given protein scientists insight into the mechanisms
Higher harmonic atomic force microscopy: Imaging of biological membranes in liquid
 Institute for Biophysics Johannes Kepler University of Linz Altenbergerstr. 69 A-4040 Linz, Austria Higher harmonic atomic force microscopy: Imaging of biological membranes in liquid Peter Hinterdorfer
Institute for Biophysics Johannes Kepler University of Linz Altenbergerstr. 69 A-4040 Linz, Austria Higher harmonic atomic force microscopy: Imaging of biological membranes in liquid Peter Hinterdorfer
Quiz 2 Morphology of Complex Materials
 071003 Quiz 2 Morphology of Complex Materials 1) Explain the following terms: (for states comment on biological activity and relative size of the structure) a) Native State b) Unfolded State c) Denatured
071003 Quiz 2 Morphology of Complex Materials 1) Explain the following terms: (for states comment on biological activity and relative size of the structure) a) Native State b) Unfolded State c) Denatured
Mechanical Properties of Biological Materials M.Richetta and R.Montanari
 Mechanical Properties of Biological Materials M.Richetta and R.Montanari Department of Industrial Engineering University of Rome-Tor Vergata- Italy, Mechanical Properties of Biological Materials 1. Growing
Mechanical Properties of Biological Materials M.Richetta and R.Montanari Department of Industrial Engineering University of Rome-Tor Vergata- Italy, Mechanical Properties of Biological Materials 1. Growing
Interpreting and evaluating biological NMR in the literature. Worksheet 1
 Interpreting and evaluating biological NMR in the literature Worksheet 1 1D NMR spectra Application of RF pulses of specified lengths and frequencies can make certain nuclei detectable We can selectively
Interpreting and evaluating biological NMR in the literature Worksheet 1 1D NMR spectra Application of RF pulses of specified lengths and frequencies can make certain nuclei detectable We can selectively
Biomolecular Interactions Measured by Atomic Force Microscopy
 Biophysical Journal Volume 79 December 2000 3267 3281 3267 Biomolecular Interactions Measured by Atomic Force Microscopy Oscar H. Willemsen,* Margot M. E. Snel,* Alessandra Cambi, Jan Greve,* Bart G. De
Biophysical Journal Volume 79 December 2000 3267 3281 3267 Biomolecular Interactions Measured by Atomic Force Microscopy Oscar H. Willemsen,* Margot M. E. Snel,* Alessandra Cambi, Jan Greve,* Bart G. De
SUPPLEMENTARY INFORMATION
 Supplementary Information for Manuscript: Nanoscale wear as a stress-assisted chemical reaction Supplementary Methods For each wear increment, the diamond indenter was slid laterally relative to the silicon
Supplementary Information for Manuscript: Nanoscale wear as a stress-assisted chemical reaction Supplementary Methods For each wear increment, the diamond indenter was slid laterally relative to the silicon
Supporting Information
 Supporting Information Beyond the Helix Pitch: Direct Visualization of Native DNA in Aqueous Solution Shinichiro Ido, Kenjiro Kimura, Noriaki Oyabu, Kei Kobayashi, Masaru Tsukada, Kazumi Matsushige and
Supporting Information Beyond the Helix Pitch: Direct Visualization of Native DNA in Aqueous Solution Shinichiro Ido, Kenjiro Kimura, Noriaki Oyabu, Kei Kobayashi, Masaru Tsukada, Kazumi Matsushige and
Supplementary Materials
 DNA-Linker Induced Surface Assembly of Ultra Dense Parallel Single Walled Carbon Nanotube Arrays Si-ping Han,, Hareem T. Maune, #, Robert D. Barish,, Marc Bockrath,@ and William A. Goddard III, 3 * Materials
DNA-Linker Induced Surface Assembly of Ultra Dense Parallel Single Walled Carbon Nanotube Arrays Si-ping Han,, Hareem T. Maune, #, Robert D. Barish,, Marc Bockrath,@ and William A. Goddard III, 3 * Materials
Supporting Information
 Supporting Information Mullins et al. 10.1073/pnas.0906781106 SI Text Detection of Calcium Binding by 45 Ca 2 Overlay. The 45 CaCl 2 (1 mci, 37 MBq) was obtained from NEN. The general method of 45 Ca 2
Supporting Information Mullins et al. 10.1073/pnas.0906781106 SI Text Detection of Calcium Binding by 45 Ca 2 Overlay. The 45 CaCl 2 (1 mci, 37 MBq) was obtained from NEN. The general method of 45 Ca 2
Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides
 Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides Zaixing
Electronic Supplementary Material (ESI) for Nanoscale. This journal is The Royal Society of Chemistry 2015 Destruction of Amyloid Fibrils by Graphene through Penetration and Extraction of Peptides Zaixing
