Bioorganic & Medicinal Chemistry Letters
|
|
|
- Gilbert Peters
- 5 years ago
- Views:
Transcription
1 Bioorganic & Medicinal Chemistry Letters 23 (2013) Contents lists available at ciencedirect Bioorganic & Medicinal Chemistry Letters journal homepage: The discovery of inhibitors of Fas-mediated cell death pathway using the combined computational method eo ee Jung a, Ji ee uh b, Eun ee Kim c, Jun Tae Kim a, eung-eun Yoo a, am ook Kang a, a Graduate chool of ew Drug Discovery and Development, Chungnam ational University, Daehakno 99, Yuseong-gu, Daejeon , Republic of Korea b Bio-organic cience Division, Korea Research Institute of Chemical Technology, Yuseong-gu, Daejeon , Republic of Korea c College of Biological cience and Biotechnology, Chungnam ational University, Daehakno 99, Yuseong-gu, Daejeon , Republic of Korea article info abstract Article history: Received 5 March 2013 Revised 25 June 2013 Accepted 12 July 2013 Available online 19 July 2013 Keywords: Fas-mediated cell death pathway Pharmacophore 3D-QAR In this study, pharmacophore and 3D-QAR models were developed for analogues of 3-substituted-benzofuran-2-carboxylate as inhibitors of Fas-mediated cell death pathways. ur pharmacophore model has good correspondence with experimental results and can explain the variance in biological activities coherently with respect to the structure of the data set compounds. The predictive power for our synthesized compounds were 0.96 for the pharmacophore model, 0.58 for the comparative molecular field analysis (CoMFA) model, and 0.57 for the comparative molecular similarity analysis (CoMIA) model. Ó 2013 The Authors. Published by Elsevier Ltd. pen access under CC BY-C-D license. The contraction or occlusion of blood vessels results in ischemia, which is frequently associated with cardiovascular diseases, angina pectoris, coronary artery diseases, and other heart conditions. 1,2 Recent studies have shown that caspase-independent cell death (CICD) mediators including RIP1K, JK, and PARP are significant in ischemic cell death. 3 Alternatively, caspase-dependent cell death (CDCD) is also an important alternative pathway for ischemic cell death. 4,5 In particular, Fas-mediated cell death has an important function in CDCD as a component of the Fas-death inducing signaling complex (DIC) and it also sensitizes cells to chemotherapeutics in chemotherapeutics-induced cell death. 4 Following the Fas-DIC formation, caspase-8 is activated, which then triggers programmed cell death, that is, apoptosis. 5 Blocking the Fas-mediated cell death pathway has been reported to be effective in the treatment of ischemic strokes, myocardial hypertrophy, and traumatic brain injuries. In this study, we optimized the inhibitors of the Fas-mediated cell death pathway screened using pharmacophore and 3D-QAR models. We built a combined qualitative pharmacophore and 3D- QAR, comparative molecular field analysis (CoMFA)/comparative molecular similarity analysis (CoMIA) models of heterocyclic carboxylate derivatives (3-substituted-benzofuran, furo[2,3-b]pyridine, thiophene and pyrazole derivatives). 5 9 The pharmacophore Corresponding author. address: nskang@cnu.ac.kr (.. Kang). model and 3D-QAR, CoMFA/CoMIA calculative analyses were performed using Catalyst and Accerlys Discovery tudio 3.1 and Tripos ybyl 2.0, respectively. All chemical inhibitors and their biological data (e.g., cell death values) were obtained from high-throughput screening experiments including the work in our previous paper. 5 The cell death (%) values ranged from 4.00% to 30.86% for the 32 compounds selected for the theoretical model in this research. The biological activities (EC 50 ) were obtained from the known cell death (%) and EC 50 data using a simple regression analysis, and then the EC 50 was converted to pec 50 (= logec 50 )in order to obtain useful values. The biological activity values (pec 50 ) of the training set and test set compounds cover a range of more than 3log units (pec 50 = 5 7), as shown in Table 1. The training set of the pharmacophore model included 7 compounds with high-ranked activity and the remaining 25 compounds were used as a test data set. The common pharmacophore features were applied using the ipop algorithm in order to generate a qualitative common features model, which was implemented in the Catalyst molecular modeling software. 10,11 Because this algorithm is a useful computational tool for building a 3D pharmacophore, we performed the ipop pharmacophore modeling using a highly active compound. In the ipop run, the most active compound (compound 27) was considered as the reference compound, which specifies a principal value of 2 and a maximum omitted feat (MF) value of 0. The initial features were specified based on an overview of the training set including the hydrogen-bond donor (BD), hydrogen-bond acceptor (BA), ring aromatic feature (RA), and hydrophobic feature (Y) X Ó 2013 The Authors. Published by Elsevier Ltd. pen access under CC BY-C-D license.
2 Table 1 tructure and biological activity of the training and test set Compound tructure pec 50 Compound tructure pec Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013)
3 Table 1 (continued) Compound tructure pec 50 Compound tructure pec (continued on next page).. Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013)
4 Table 1 (continued) Compound tructure pec 50 Compound tructure pec Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013)
5 .. Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013) Table 2 ummary of CoMFA and CoMIA models CoMFA CoMIA tatistical results q 2a r 2b EE c F value components 6 6 Fraction (%) teric Electrostatic ydrophobic Donor Acceptor Grid spacing: 2.0 Å Figure 1. The pharmacophore mapping of the most active compound 27. The pharmacophoric points are color coded with green; hydrogen-bond acceptor (BA), magenta; hydrogen-bond donor (BD), orange; ring aromatic (RA), Cyan; hydrophobic (Y). Fit value Training set R 2 = 0.97 Test set R 2 = Experimental pec 50 Figure 2. The relationship between experimental data versus pharmacophore fit value. Training set 7 compounds and test set 25 compounds. The correlation was calculated using spearman s correlation efficient. a b c Cross validated correlation coefficient. Conventional correlation coefficient. tandard error of estimate. The predictive power of the quantitative model was verified using the 25 compounds from the test data set. The best qualitative pharmacophore model contained six features: one hydrogen-bond donor (BD), two hydrogen-bond acceptors (BA), two ring aromatic features (RA), and one hydrophobic feature (Y), with each distance represented in Figure 1. This model had a maximum correlation coefficient of 0.97 with a high goodness of fit. Also, an independent test set that contained 25 external compounds was used to validate the established model and the obtained predictive score (0.74). The experimental and predicted activities of the training and test data set compounds are shown in Figure 2. The carboxylate group nearby the 2-position of the thiophene or the benzofuran ring indicated the importance of the hydrophobic group in conferring the biological activity. In contrast, when the carboxyl group was located in this region, it decreased the biological activity (e.g., compounds 9, 18, 23, and 30). This model proposes that the hydrophobic interactions between ligands and proteins are important. In order to clarify this further, the pharmacophore parameters of the training data set compounds are reported in upplementary data Table 1. The 3D-QAR, CoMFA, and CoMIA models were constructed using derivatives based on benzofuran analogues, as shown in Figure 3. Group 1 contains a training data set of 23 compounds that produce the 3D-QAR model (compounds 1 23 in Table 1), and Group 2 is a test data set of 9 compounds (compounds in Table 1) in order to validate the model. This model can be used Figure 3. tereo-view of the CoMFA/CoMIA training and test set compounds superimposed in molecular shape analysis.
6 Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013) (A) 6.5 (B) Predicted pec Predicted pec Training set R 2 = 0.98 Test set R 2 = Training set R 2 = 0.97 Test set R 2 = Experimental pec Experimental pec 50 Figure 4. Plots of the experimental versus predicted pec 50 values of the training ( ) and test ( ) compounds based on the best (A) CoMFA model and (B) CoMIA model. Figure 5. Contour maps of (A) CoMFA and (B) CoMIA analysis based on compound 12, high scored conformation in training set. Green contours signify steric fields where bulky groups increase activity, while yellow contours indicate fields where less bulky groups decrease activity. CoMFA steric fields were displayed in favored level 80% (green) and disfavored level 20% (yellow), CoMIA fields were in 85% and 15% contributions by using TDEV CEFF field. Compound 12 is depicted in capped stick type, referentially. Figure 6. Contour maps of (A) CoMFA and (B) CoMIA analysis are based on compound 12. Blue contours signify electrostatic fields where positive charged groups increase activity, while red contours indicate fields where negative charged groups increase activity. CoMFA electrostatic fields were displayed in favored level 85% (blue) and disfavored level 15% (red), fraction of CoMIA fields was identical. to obtain numerical values for statistical results such as TDEV CEFF at individual lattice points. Partial atomic charges were calculated using the Gasteiger ückel charge. 12 The results were statistically significant and the data from the 3D-QAR analyses are summarized in Table 2 and Figure 4. The best crossvalidated L values for the CoMFA and CoMIA models were computed as q 2 = 0.59 and q 2 = 0.60, respectively, using six components. The non-cross-validated values for the CoMFA and CoMIA
7 .. Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013) Figure 7. Contour map of CoMIA analysis is based on compound 12. Yellow and white contours (A) signify favorable and unfavorable hydrophobic fields in same proportion. ydrogen bond donor contours (B) were displayed in favored level 75% (cyan) and disfavored level 25% (purple). And hydrogen bond acceptor contours (C) were indicated favorable fraction 90% (magenta) and unfavorable fraction 10% (red). Table 3 Biological activity and predicted values of synthesized compounds Compound tructure Cell death (%) pec 50 Fit value CoMFA CoMIA (continued on next page)
8 Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013) Table 3 (continued) Compound tructure Cell death (%) pec 50 Fit value CoMFA CoMIA
9 .. Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013) Table 3 (continued) Compound tructure Cell death (%) pec 50 Fit value CoMFA CoMIA X X X R 2 Y Y Y R R R R a b c d 33: R =, X =, Y = 36: R =, X =, Y = C 3 34: R = C 3, X =, Y = 35: R =, X =, Y = n 37: n = 1, X = 38: n = 1, X = 39: n = 2, X = X A n Z R 40: A = C, n = 1, R =, X =, Z = 3,5-di- 41: A =, n = 2, R = C 3, Z = 3,5-di- 42: A =, n = 2, R = C 3, Z = 3,4,5-tri-C 3 X n R A 43: A =, n = 1, R = 44: A =, n = 2, R = C 3 2 e, f 2 R' a g 45 R' R' R' = Ph(C 2 ) 2 h 46 R' cheme 1. Reagents and conditions: (a) bromoacethyl bromide, Et 3, TF, rt, 83 93%; (b) (i) benzene thiol, Et 3, TF, rt or (ii) -methyaniline, K 2 C 3, DMF, rt, 42 91%; (c) 2 a, TF, reflux, 90%; (d) (i) di(2-pyridyl)carbonate, DMAP, TF, rt (ii) 40 wt % dimethylamine, rt, 50%; (e) (2-bromoethyl)benzene, Cs 2 C 3, DMF, rt, 74%; (f) 2 (g), 10% Pd/C, Me, 50 PI, rt, 94%; (g) hydrocinnamoyl chloride, Et 3, TF, rt, 87%; (h) 4-bromophenol, K 2 C 3, DMF, rt, 69%. And refer to upplementary data for details. models were calculated to be r 2 = 0.98 and r 2 = 0.97 with an estimated standard error of 0.03, 0.04 and F values of and 94.28, respectively. The CoMFA and CoMIA models were validated by substituting the test set compounds and obtaining sufficient prediction (r 2 predictive = 0.72 and 0.73, respectively). The CoMFA and CoMIA models were developed based on a superimposition obtained using the pharmacophoric map. The contours provide an interpretation of the correlation obtained in terms of the field contributions. The CoMFA model, which includes steric and electrostatic fields, is presented as 3D contour map using the TDEV CEFF field in Figure 5a and Figure 6a. The respective contributions of the steric and electrostatic fields for the CoMFA model are 70% and 30%. The green contours signify the steric fields where the bulky groups increase activity, while the yellow contours indicate fields where the less bulky groups decrease activity. As mentioned above, the green contours shown near the
10 Jung et al. / Bioorg. Med. Chem. Lett. 23 (2013) meta-position of the benzofuran signify that any bulky group at this site could increase activity. The yellow contours near the 4 0 -position of the benzene ring suggest that any bulky group could decrease activity. Compounds 14 and 25 showed a negative relationship between the predicted pec 50 and the character of the contours. The blue contours signify electrostatic fields where positively charged groups increase activity, while the red contours indicate fields where the negatively charged groups increase activity. Compounds 1 and 10 were accurately interpreted as red contours; thus, nitro-benzene, which contains a negative group, had the highest activity. imilar to the CoMFA fields, the CoMIA model was also developed using the TDEV CEFF field, as shown in Figure 5(b), Figure 6(b), and Figure 7. The respective contributions of the steric and electrostatic fields for the CoMIA model were 10% and 17%. The CoMIA fields were aligned similarly to the CoMFA fields. In addition, the CoMIA model contained hydrophobic, hydrogenbond donor, and hydrogen-bond acceptor fields. The respective contributions of these three fields were 40%, 16%, and 17%. In the hydrophobic contour map, the yellow contours indicate the sites where the hydrophobic groups are favorable and the white contours show unfavorable positions. In Figure 7a, the two yellow contours are located in the vicinity of the benzofuran and amine groups, which signify that the hydrophobic groups facilitate higher activity. Compounds are well fitted to our model, which had highly predictive ability. The yellow contours near the 1 0 -position of the pyrazole ring suggest that any hydrophobic substitution could increase activity. In the hydrogen bond donor contour map, the cyan contours indicate the regions where the hydrogen bond donor groups are favorable and the purple contours show unfavorable positions in Figure 7(b). Figure 7(c) depicts the hydrogen bond acceptor contour map. The magenta contours indicate the sites where the hydrogen bond acceptor groups are favorable for activity, while the red contours present the regions where hydrogen bond acceptors are unfavorable. erein, all contour maps were depicted with compound 12 in Figures 5 7. Based on the well-accepted pharmacophore and 3D-QAR models, the 14 new compounds shown in Table 3 were designed and synthesized in order to predict accurate activities, as presented in upplementary data Table 2. Based on our computational modeling and investigation of the AR, two aromatic features were key points in the biological activity. According to Figure 1, the distance between two RA central points was 9.14 Å and we synthesized 14 novel compounds with a carbon linker (n = 4, 5) while adhering to the distance rule. For this reason, the thiophene and pyrazole derivatives were synthesized from the corresponding 3-amino-thiophene-2-carboxylic acid methyl ester and 4-nitro-pyrazole-3-carboxylate as readily available starter materials, as described in cheme 1 and the upplementary data. The following compounds were prepared according to the general procedures described above employing the appropriate methyl 3-aminothiophene-2- carboxylate, benzenethiol, and pyrazole. The modification of the aromatic ring or benzofuran core was achieved using a similar method as described for the synthesis of compound 33. Following these procedures, numerous analogues (e.g., benzofuran and benzene derivatives) were prepared in a short period in a parallel synthesis fashion. Consequently, the 14 synthesized compounds were well-matched with two key ring aromatic centers and they obtained reasonable fitted values compared with the experimental activity, as summarized in Table 3. In summary, this study developed pharmacophore and 3D- QAR models for analogues of 3-substituted-benzofuran-2-carboxylic ester as inhibitors of the Fas-mediated cell death pathway. The proposed pharmacophore model has good agreement with the synthesized compounds and is capable of explaining the variance in biological activities coherently with respect to the structures of the data set compounds. The predictive power for the synthesized compounds was 0.96 for the pharmacophore model, 0.58 for the CoMFA model, and 0.57 for the CoMIA model. Consequentially, the prediction of the ligand-based pharmacophore hypothesis provided significant structural insights and also illuminated the important binding features of heterocyclic carboxylic ester analogues. In future research, we will develop strategies for the optimization of the inhibitors of the Fas-mediated cell death pathway based on these results. upplementary data upplementary data associated with this article can be found, in the online version, at References and notes 1. earse, D. J. Cardiovasc. Res. 1994, 28, Margaix Muñoz, M.; Jiménez oriano, Y.; Poveda Roda, R.; arrión, G. Med. ral. Patol. ral. Cir. Bucal. 2008, 13, E Ryu,. W.; Chae,. K.; Lee, K. J.; Kim, E.. Biochem. Biophys. Res. Commun. 1999, 262, Menges, C. W.; Altomare, D. A.; Testa, J. R. Cell Cycle 2009, 8, uh, J..; Yi, K. Y.; Lee, Y..; Kim, E..; Yum, E. K.; Yoo,. E. Bioorg. Med. Chem. Lett. 2010, 20, Cramer, R. D.; Patterson, D. E.; Bunce, J. D. J. Med. Chem. oc. 1988, 110, Klebe, G.; Abraham, U.; Mietzner, T. J. Med. Chem. 1994, 37, Kim,..; Cho, I..; Gu,. K.; Lee, D..; Lim,.; Yoo,. E. Eur. J. Pharmacol. 2004, 487, uh, J..; Yoo,. E.; Yi, K. Y.; Kim,. J.; Kim, E..; Jung, Y..; Lee, Y..; Kim,. Y. Patent PCT W 20,09,048,274, Barnum, D.; Greene, J.; mellie, A.; prague, P. J. Chem. Inf. Comput. 1996, 36, ecker, E. A.; Duraiswani, C.; Andrea, T. A.; Diller, D. J. J. Chem. Inf. Comput. 2002, 42, Gasteiger, J.; Marsili, M. Tetrahedron 1980, 36, 3219.
Ligand-based QSAR Studies on the Indolinones Derivatives Bull. Korean Chem. Soc. 2004, Vol. 25, No
 Ligand-based QSAR Studies on the Indolinones Derivatives Bull. Korean Chem. Soc. 2004, Vol. 25, No. 12 1801 Ligand-based QSAR Studies on the Indolinones Derivatives as Inhibitors of the Protein Tyrosine
Ligand-based QSAR Studies on the Indolinones Derivatives Bull. Korean Chem. Soc. 2004, Vol. 25, No. 12 1801 Ligand-based QSAR Studies on the Indolinones Derivatives as Inhibitors of the Protein Tyrosine
T. J. Hou, Z. M. Li, Z. Li, J. Liu, and X. J. Xu*,
 1002 J. Chem. Inf. Comput. Sci. 2000, 40, 1002-1009 Three-Dimensional Quantitative Structure-Activity Relationship Analysis of the New Potent Sulfonylureas Using Comparative Molecular Similarity Indices
1002 J. Chem. Inf. Comput. Sci. 2000, 40, 1002-1009 Three-Dimensional Quantitative Structure-Activity Relationship Analysis of the New Potent Sulfonylureas Using Comparative Molecular Similarity Indices
3D-QSAR Studies on Angiotensin-Converting Enzyme (ACE) Inhibitors: a Molecular Design in Hypertensive Agents
 952 Bull. Korean Chem. Soc. 2005, Vol. 26, No. 6 Amor A. San Juan and Seung Joo Cho 3D-QSAR Studies on Angiotensin-Converting Enzyme (ACE) Inhibitors: a Molecular Design in Hypertensive Agents Amor A.
952 Bull. Korean Chem. Soc. 2005, Vol. 26, No. 6 Amor A. San Juan and Seung Joo Cho 3D-QSAR Studies on Angiotensin-Converting Enzyme (ACE) Inhibitors: a Molecular Design in Hypertensive Agents Amor A.
Characterization of Binding Mode for Human Coagulation Factor XI (FXI) Inhibitors
 1212 Bull. Korean Chem. Soc. 2013, Vol. 34, No. 4 Jae Eun Cho et al. http://dx.doi.org/10.5012/bkcs.2013.34.4.1212 Characterization of Binding Mode for Human Coagulation Factor XI (FXI) Inhibitors Jae
1212 Bull. Korean Chem. Soc. 2013, Vol. 34, No. 4 Jae Eun Cho et al. http://dx.doi.org/10.5012/bkcs.2013.34.4.1212 Characterization of Binding Mode for Human Coagulation Factor XI (FXI) Inhibitors Jae
Structural biology and drug design: An overview
 Structural biology and drug design: An overview livier Taboureau Assitant professor Chemoinformatics group-cbs-dtu otab@cbs.dtu.dk Drug discovery Drug and drug design A drug is a key molecule involved
Structural biology and drug design: An overview livier Taboureau Assitant professor Chemoinformatics group-cbs-dtu otab@cbs.dtu.dk Drug discovery Drug and drug design A drug is a key molecule involved
Molecular docking, 3D-QSAR studies of indole hydrazone as Staphylococcus aureus pyruvate kinase inhibitor
 World Journal of Pharmaceutical Sciences ISSN (Print): 2321-3310; ISSN (Online): 2321-3086 Published by Atom and Cell Publishers All Rights Reserved Available online at: http://www.wjpsonline.org/ Original
World Journal of Pharmaceutical Sciences ISSN (Print): 2321-3310; ISSN (Online): 2321-3086 Published by Atom and Cell Publishers All Rights Reserved Available online at: http://www.wjpsonline.org/ Original
Plan. Day 2: Exercise on MHC molecules.
 Plan Day 1: What is Chemoinformatics and Drug Design? Methods and Algorithms used in Chemoinformatics including SVM. Cross validation and sequence encoding Example and exercise with herg potassium channel:
Plan Day 1: What is Chemoinformatics and Drug Design? Methods and Algorithms used in Chemoinformatics including SVM. Cross validation and sequence encoding Example and exercise with herg potassium channel:
Hologram and Receptor-Guided 3D QSAR Analysis of Anilinobipyridine JNK3 Inhibitors
 3D QSAR Analysis of Anilinobipyridine JK3 Inhibitors Bull. Korean Chem. Soc. 2009, Vol. 30, o. 11 2739 Hologram and Receptor-Guided 3D QSAR Analysis of Anilinobipyridine JK3 Inhibitors Jae Yoon Chung,,
3D QSAR Analysis of Anilinobipyridine JK3 Inhibitors Bull. Korean Chem. Soc. 2009, Vol. 30, o. 11 2739 Hologram and Receptor-Guided 3D QSAR Analysis of Anilinobipyridine JK3 Inhibitors Jae Yoon Chung,,
3D QSAR and Pharmacophore Modelling of Selected Benzimidazole Derivatives as Factor IXa Inhibitors
 Research Paper 3D QSAR and Pharmacophore Modelling of Selected Benzimidazole Derivatives as Factor IXa Inhibitors S. S. KUMBHAR*, P. B. CHOUDHARI AD M. S. BHATIA Drug Design and Development Group, Department
Research Paper 3D QSAR and Pharmacophore Modelling of Selected Benzimidazole Derivatives as Factor IXa Inhibitors S. S. KUMBHAR*, P. B. CHOUDHARI AD M. S. BHATIA Drug Design and Development Group, Department
Three Dimensional Pharmacophore Modelling of Monoamine oxidase-a (MAO-A) inhibitors
 Int. J. Mol. ci. 2007, 8, 894-919 pecial Issue The Chemical Bond and Bonding International Journal of Molecular ciences I 1422-0067 2007 by MDPI http://www.mdpi.org/ijms ull Research Paper Three Dimensional
Int. J. Mol. ci. 2007, 8, 894-919 pecial Issue The Chemical Bond and Bonding International Journal of Molecular ciences I 1422-0067 2007 by MDPI http://www.mdpi.org/ijms ull Research Paper Three Dimensional
* Author to whom correspondence should be addressed; Tel.: ; Fax:
 Int. J. Mol. Sci. 2011, 12, 946-970; doi:10.3390/ijms12020946 OPEN ACCESS Article International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Structural Determination of Three
Int. J. Mol. Sci. 2011, 12, 946-970; doi:10.3390/ijms12020946 OPEN ACCESS Article International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Structural Determination of Three
Hydrogen Bonding & Molecular Design Peter
 Hydrogen Bonding & Molecular Design Peter Kenny(pwk.pub.2008@gmail.com) Hydrogen Bonding in Drug Discovery & Development Interactions between drug and water molecules (Solubility, distribution, permeability,
Hydrogen Bonding & Molecular Design Peter Kenny(pwk.pub.2008@gmail.com) Hydrogen Bonding in Drug Discovery & Development Interactions between drug and water molecules (Solubility, distribution, permeability,
Using Phase for Pharmacophore Modelling. 5th European Life Science Bootcamp March, 2017
 Using Phase for Pharmacophore Modelling 5th European Life Science Bootcamp March, 2017 Phase: Our Pharmacohore generation tool Significant improvements to Phase methods in 2016 New highly interactive interface
Using Phase for Pharmacophore Modelling 5th European Life Science Bootcamp March, 2017 Phase: Our Pharmacohore generation tool Significant improvements to Phase methods in 2016 New highly interactive interface
Mohmed et al., IJPSR, 2016; Vol. 7(3): E-ISSN: ; P-ISSN:
 IJPSR (2016), Vol. 7, Issue 3 (Research Article) Received on 23 September, 2015; received in revised form, 05 November, 2015; accepted, 17 December, 2015; published 01 March, 2016 3D-QSAR STUDY OF BENZOTHIAZOLE
IJPSR (2016), Vol. 7, Issue 3 (Research Article) Received on 23 September, 2015; received in revised form, 05 November, 2015; accepted, 17 December, 2015; published 01 March, 2016 3D-QSAR STUDY OF BENZOTHIAZOLE
5.1. Hardwares, Softwares and Web server used in Molecular modeling
 5. EXPERIMENTAL The tools, techniques and procedures/methods used for carrying out research work reported in this thesis have been described as follows: 5.1. Hardwares, Softwares and Web server used in
5. EXPERIMENTAL The tools, techniques and procedures/methods used for carrying out research work reported in this thesis have been described as follows: 5.1. Hardwares, Softwares and Web server used in
Analogue and Structure Based Drug Designing of Prenylated Flavonoid Derivatives as PKB/Akt1 Inhibitors
 Available online at www.ijpcr.com International Journal of Pharmaceutical and Clinical esearch 2016; 8(8): 1205-1211 esearch Article ISS- 0975 1556 Analogue and Structure Based Drug Designing of Prenylated
Available online at www.ijpcr.com International Journal of Pharmaceutical and Clinical esearch 2016; 8(8): 1205-1211 esearch Article ISS- 0975 1556 Analogue and Structure Based Drug Designing of Prenylated
Bioengineering & Bioinformatics Summer Institute, Dept. Computational Biology, University of Pittsburgh, PGH, PA
 Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
CHAPTER 3. Vibrational Characteristics of PTP-1B Inhibitors
 CHAPTER 3 Vibrational Characteristics of PTP-1B Inhibitors 3.1 Preface Theoretical Frequency calculations were performed on the molecules chosen in this study. Initially all the geometries were optimized
CHAPTER 3 Vibrational Characteristics of PTP-1B Inhibitors 3.1 Preface Theoretical Frequency calculations were performed on the molecules chosen in this study. Initially all the geometries were optimized
3D QSAR Analyses of Novel Tyrosine Kinase Inhibitors Based on Pharmacophore Alignment
 1032 J. Chem. Inf. Comput. Sci. 2001, 41, 1032-1040 3D QSAR Analyses of Novel Tyrosine Kinase Inhibitors Based on Pharmacophore Alignment L. L. Zhu, T. J. Hou, L. R. Chen, and X. J. Xu*, College of Chemistry
1032 J. Chem. Inf. Comput. Sci. 2001, 41, 1032-1040 3D QSAR Analyses of Novel Tyrosine Kinase Inhibitors Based on Pharmacophore Alignment L. L. Zhu, T. J. Hou, L. R. Chen, and X. J. Xu*, College of Chemistry
Nonlinear QSAR and 3D QSAR
 onlinear QSAR and 3D QSAR Hugo Kubinyi Germany E-Mail kubinyi@t-online.de HomePage www.kubinyi.de onlinear Lipophilicity-Activity Relationships drug receptor Possible Reasons for onlinear Lipophilicity-Activity
onlinear QSAR and 3D QSAR Hugo Kubinyi Germany E-Mail kubinyi@t-online.de HomePage www.kubinyi.de onlinear Lipophilicity-Activity Relationships drug receptor Possible Reasons for onlinear Lipophilicity-Activity
Quinazolinone Derivatives as Growth Hormone Secretagogue Receptor Inhibitors: 3D-QSAR study
 International Journal of ChemTech Research CODE (USA): IJCRGG, ISS: 0974-4290, ISS(Online):2455-9555 Vol.9, o.05 pp 896-903, 2016 Quinazolinone Derivatives as Growth Hormone Secretagogue Receptor Inhibitors:
International Journal of ChemTech Research CODE (USA): IJCRGG, ISS: 0974-4290, ISS(Online):2455-9555 Vol.9, o.05 pp 896-903, 2016 Quinazolinone Derivatives as Growth Hormone Secretagogue Receptor Inhibitors:
A 3D-QSAR Study of Celebrex-based PDK1 Inhibitors Using CoMFA Method
 Journal of the Chinese Chemical Society, 2009, 56, 59-64 59 A 3D-QSAR Study of Celebrex-based PDK1 Inhibitors Using CoMFA Method Wen-Hung Wang a ( ),N.R.Jena a ( ), Yi-Ching Wang b ( ) and Ying-Chieh Sun
Journal of the Chinese Chemical Society, 2009, 56, 59-64 59 A 3D-QSAR Study of Celebrex-based PDK1 Inhibitors Using CoMFA Method Wen-Hung Wang a ( ),N.R.Jena a ( ), Yi-Ching Wang b ( ) and Ying-Chieh Sun
ISSN: ; CODEN ECJHAO E-Journal of Chemistry 2009, 6(3),
 ISS: 0973-4945; CODE ECJHAO E- Chemistry http://www.e-journals.net 2009, 6(3), 651-658 Comparative Molecular Field Analysis (CoMFA) for Thiotetrazole Alkynylacetanilides, a on-ucleoside Inhibitor of HIV-1
ISS: 0973-4945; CODE ECJHAO E- Chemistry http://www.e-journals.net 2009, 6(3), 651-658 Comparative Molecular Field Analysis (CoMFA) for Thiotetrazole Alkynylacetanilides, a on-ucleoside Inhibitor of HIV-1
Using Bayesian Statistics to Predict Water Affinity and Behavior in Protein Binding Sites. J. Andrew Surface
 Using Bayesian Statistics to Predict Water Affinity and Behavior in Protein Binding Sites Introduction J. Andrew Surface Hampden-Sydney College / Virginia Commonwealth University In the past several decades
Using Bayesian Statistics to Predict Water Affinity and Behavior in Protein Binding Sites Introduction J. Andrew Surface Hampden-Sydney College / Virginia Commonwealth University In the past several decades
3D QSAR analysis of quinolone based s- triazines as antimicrobial agent
 International Journal of PharmTech Research CODEN (USA): IJPRIF ISSN : 0974-4304 Vol.4, No.3, pp 1096-1100, July-Sept 2012 3D QSAR analysis of quinolone based s- triazines as antimicrobial agent Ramesh
International Journal of PharmTech Research CODEN (USA): IJPRIF ISSN : 0974-4304 Vol.4, No.3, pp 1096-1100, July-Sept 2012 3D QSAR analysis of quinolone based s- triazines as antimicrobial agent Ramesh
Keywords: anti-coagulants, factor Xa, QSAR, Thrombosis. Introduction
 PostDoc Journal Vol. 2, No. 3, March 2014 Journal of Postdoctoral Research www.postdocjournal.com QSAR Study of Thiophene-Anthranilamides Based Factor Xa Direct Inhibitors Preetpal S. Sidhu Department
PostDoc Journal Vol. 2, No. 3, March 2014 Journal of Postdoctoral Research www.postdocjournal.com QSAR Study of Thiophene-Anthranilamides Based Factor Xa Direct Inhibitors Preetpal S. Sidhu Department
Drug Design 2. Oliver Kohlbacher. Winter 2009/ QSAR Part 4: Selected Chapters
 Drug Design 2 Oliver Kohlbacher Winter 2009/2010 11. QSAR Part 4: Selected Chapters Abt. Simulation biologischer Systeme WSI/ZBIT, Eberhard-Karls-Universität Tübingen Overview GRIND GRid-INDependent Descriptors
Drug Design 2 Oliver Kohlbacher Winter 2009/2010 11. QSAR Part 4: Selected Chapters Abt. Simulation biologischer Systeme WSI/ZBIT, Eberhard-Karls-Universität Tübingen Overview GRIND GRid-INDependent Descriptors
* Author to whom correspondence should be addressed; Tel.: ; Fax:
 Int. J. Mol. Sci. 2011, 12, 1807-1835; doi:10.3390/ijms12031807 OPEN ACCESS International Journal of Molecular Sciences Article ISSN 1422-0067 www.mdpi.com/journal/ijms Combined 3D-QSAR, Molecular Docking
Int. J. Mol. Sci. 2011, 12, 1807-1835; doi:10.3390/ijms12031807 OPEN ACCESS International Journal of Molecular Sciences Article ISSN 1422-0067 www.mdpi.com/journal/ijms Combined 3D-QSAR, Molecular Docking
Structure-Activity Modeling - QSAR. Uwe Koch
 Structure-Activity Modeling - QSAR Uwe Koch QSAR Assumption: QSAR attempts to quantify the relationship between activity and molecular strcucture by correlating descriptors with properties Biological activity
Structure-Activity Modeling - QSAR Uwe Koch QSAR Assumption: QSAR attempts to quantify the relationship between activity and molecular strcucture by correlating descriptors with properties Biological activity
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
doi:10.1038/nature11085 Supplementary Tables: Supplementary Table 1. Summary of crystallographic and structure refinement data Structure BRIL-NOP receptor Data collection Number of crystals 23 Space group
Prediction of the Antagonistic Activity of Aryl Benzyl Ethers against LTD4 by Using 3D-CoMFA Model Developed with Pranlukast Analogues
 3D-CoMFA Study of LTD4 Antagonist Bull. Korean Chem. Soc. 2006, Vol. 27, No. 7 1025 Prediction of the Antagonistic Activity of Aryl Benzyl Ethers against LTD4 by Using 3D-CoMFA Model Developed with Pranlukast
3D-CoMFA Study of LTD4 Antagonist Bull. Korean Chem. Soc. 2006, Vol. 27, No. 7 1025 Prediction of the Antagonistic Activity of Aryl Benzyl Ethers against LTD4 by Using 3D-CoMFA Model Developed with Pranlukast
Synthetic organic compounds
 Synthetic organic compounds for research and drug discovery Compounds for TS Fragment libraries Target-focused libraries Chemical building blocks Custom synthesis Drug discovery services Contract research
Synthetic organic compounds for research and drug discovery Compounds for TS Fragment libraries Target-focused libraries Chemical building blocks Custom synthesis Drug discovery services Contract research
Ligand-receptor interactions
 University of Silesia, Katowice, Poland 11 22 March 2013 Ligand-receptor interactions Dr. Pavel Polishchuk A.V. Bogatsky Physico-Chemical Institute of National Academy of Sciences of Ukraine Odessa, Ukraine
University of Silesia, Katowice, Poland 11 22 March 2013 Ligand-receptor interactions Dr. Pavel Polishchuk A.V. Bogatsky Physico-Chemical Institute of National Academy of Sciences of Ukraine Odessa, Ukraine
Data Quality Issues That Can Impact Drug Discovery
 Data Quality Issues That Can Impact Drug Discovery Sean Ekins 1, Joe Olechno 2 Antony J. Williams 3 1 Collaborations in Chemistry, Fuquay Varina, NC. 2 Labcyte Inc, Sunnyvale, CA. 3 Royal Society of Chemistry,
Data Quality Issues That Can Impact Drug Discovery Sean Ekins 1, Joe Olechno 2 Antony J. Williams 3 1 Collaborations in Chemistry, Fuquay Varina, NC. 2 Labcyte Inc, Sunnyvale, CA. 3 Royal Society of Chemistry,
QSAR Study of Quinazoline Derivatives as Inhibitor of Epidermal Growth Factor Receptor-Tyrosine Kinase (EGFR-TK)
 3rd International Conference on Computation for cience and Technology (ICCT-3) QAR tudy of Quinazoline Derivatives as Inhibitor of Epidermal Growth Factor Receptor-Tyrosine Kinase (EGFR-TK) La de Aman1*,
3rd International Conference on Computation for cience and Technology (ICCT-3) QAR tudy of Quinazoline Derivatives as Inhibitor of Epidermal Growth Factor Receptor-Tyrosine Kinase (EGFR-TK) La de Aman1*,
March 08 Dr. Abdullah Saleh
 March 08 Dr. Abdullah Saleh 1 Effects of Substituents on Reactivity and Orientation The nature of groups already on an aromatic ring affect both the reactivity and orientation of future substitution Activating
March 08 Dr. Abdullah Saleh 1 Effects of Substituents on Reactivity and Orientation The nature of groups already on an aromatic ring affect both the reactivity and orientation of future substitution Activating
3D QSAR studies of Substituted Benzamides as Nonacidic Antiinflammatory Agents by knn MFA Approach
 INTERNATIONAL JOURNAL OF ADVANCES IN PARMACY, BIOLOGY AND CEMISTRY Research Article 3D QSAR studies of Substituted Benzamides as Nonacidic Antiinflammatory Agents by knn MFA Approach Anupama A. Parate*
INTERNATIONAL JOURNAL OF ADVANCES IN PARMACY, BIOLOGY AND CEMISTRY Research Article 3D QSAR studies of Substituted Benzamides as Nonacidic Antiinflammatory Agents by knn MFA Approach Anupama A. Parate*
3D-QSAR study on heterocyclic topoisomerase II inhibitors using CoMSIAy
 SAR and QSAR in Environmental Research Vol. 17, No. 2, April 2006, 121 132 3D-QSAR study on heterocyclic topoisomerase II inhibitors using CoMSIAy B. TEKINER-GULBAS, O. TEMIZ-ARPACI, I. YILDIZ*, E. AKI-SENER
SAR and QSAR in Environmental Research Vol. 17, No. 2, April 2006, 121 132 3D-QSAR study on heterocyclic topoisomerase II inhibitors using CoMSIAy B. TEKINER-GULBAS, O. TEMIZ-ARPACI, I. YILDIZ*, E. AKI-SENER
Dispensing Processes Profoundly Impact Biological, Computational and Statistical Analyses
 Dispensing Processes Profoundly Impact Biological, Computational and Statistical Analyses Sean Ekins 1, Joe Olechno 2 Antony J. Williams 3 1 Collaborations in Chemistry, Fuquay Varina, NC. 2 Labcyte Inc,
Dispensing Processes Profoundly Impact Biological, Computational and Statistical Analyses Sean Ekins 1, Joe Olechno 2 Antony J. Williams 3 1 Collaborations in Chemistry, Fuquay Varina, NC. 2 Labcyte Inc,
Introduction to Chemoinformatics and Drug Discovery
 Introduction to Chemoinformatics and Drug Discovery Irene Kouskoumvekaki Associate Professor February 15 th, 2013 The Chemical Space There are atoms and space. Everything else is opinion. Democritus (ca.
Introduction to Chemoinformatics and Drug Discovery Irene Kouskoumvekaki Associate Professor February 15 th, 2013 The Chemical Space There are atoms and space. Everything else is opinion. Democritus (ca.
Molecular Modeling Study of Some Anthelmintic 2-phenyl Benzimidazole-1- Acetamides as β-tubulin Inhibitor
 Sawant et al : Molecular Modeling Study of Some Anthelmintic 2-phenyl Benzimidazole-1-Acetamides as -tubulin Inhibitor 1269 International Journal of Drug Design and Discovery Volume 5 Issue 1 January March
Sawant et al : Molecular Modeling Study of Some Anthelmintic 2-phenyl Benzimidazole-1-Acetamides as -tubulin Inhibitor 1269 International Journal of Drug Design and Discovery Volume 5 Issue 1 January March
Direct Oxidative Heck Cyclizations: Intramolecular Fujiwara-Moritani Arylations for the Synthesis of Functionalized Benzofurans and Dihydrobenzofurans
 Direct xidative eck Cyclizations: Intramolecular Fujiwara-Moritani Arylations for the Synthesis of Functionalized Benzofurans and Dihydrobenzofurans by Zhang,.; Ferreira, E. M.; Stoltz, B. M. Angewandte
Direct xidative eck Cyclizations: Intramolecular Fujiwara-Moritani Arylations for the Synthesis of Functionalized Benzofurans and Dihydrobenzofurans by Zhang,.; Ferreira, E. M.; Stoltz, B. M. Angewandte
CHAPTER-2. Drug discovery is a comprehensive approach wherein several disciplines
 36 CAPTER-2 Molecular Modeling Analog Based Studies Drug discovery is a comprehensive approach wherein several disciplines are used to design or discover the drugs. The R&D expenditure incurred to bring
36 CAPTER-2 Molecular Modeling Analog Based Studies Drug discovery is a comprehensive approach wherein several disciplines are used to design or discover the drugs. The R&D expenditure incurred to bring
Biologically Relevant Molecular Comparisons. Mark Mackey
 Biologically Relevant Molecular Comparisons Mark Mackey Agenda > Cresset Technology > Cresset Products > FieldStere > FieldScreen > FieldAlign > FieldTemplater > Cresset and Knime About Cresset > Specialist
Biologically Relevant Molecular Comparisons Mark Mackey Agenda > Cresset Technology > Cresset Products > FieldStere > FieldScreen > FieldAlign > FieldTemplater > Cresset and Knime About Cresset > Specialist
Self-Organizing Molecular Field Analysis on a New Series of COX-2 Selective Inhibitors: 1,5-Diarylimidazoles
 Int. J. Mol. Sci. 2006, 7, 220-229 International Journal of Molecular Sciences ISSN 1422-0067 2006 by MDPI www.mdpi.org/ijms/ Self-Organizing Molecular Field Analysis on a New Series of COX-2 Selective
Int. J. Mol. Sci. 2006, 7, 220-229 International Journal of Molecular Sciences ISSN 1422-0067 2006 by MDPI www.mdpi.org/ijms/ Self-Organizing Molecular Field Analysis on a New Series of COX-2 Selective
Saika Siddiqui, Bruce Gustafson, and Ramachandra, S. Hosmane*
 Third International Electronic Conference on Synthetic Organic Chemistry (ECSOC-3), www.mdpi.org/ecsoc-3.htm, September 1-30, 1999 [A0029] A New Synthetic Approach toward Ring-Expanded ("Fat") Purine Nucleobases:
Third International Electronic Conference on Synthetic Organic Chemistry (ECSOC-3), www.mdpi.org/ecsoc-3.htm, September 1-30, 1999 [A0029] A New Synthetic Approach toward Ring-Expanded ("Fat") Purine Nucleobases:
Overview. Descriptors. Definition. Descriptors. Overview 2D-QSAR. Number Vector Function. Physicochemical property (log P) Atom
 verview D-QSAR Definition Examples Features counts Topological indices D fingerprints and fragment counts R-group descriptors ow good are D descriptors in practice? Summary Peter Gedeck ovartis Institutes
verview D-QSAR Definition Examples Features counts Topological indices D fingerprints and fragment counts R-group descriptors ow good are D descriptors in practice? Summary Peter Gedeck ovartis Institutes
OXFORD H i g h e r E d u c a t i o n Oxford University Press, All rights reserved.
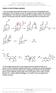 Patrick, An Introduction to dicinal hemistry 4e Answers to end-of-chapter questions 1) The mechanism below shows the release of one molecule of formaldehyde from methenamine. The mechanism can then be
Patrick, An Introduction to dicinal hemistry 4e Answers to end-of-chapter questions 1) The mechanism below shows the release of one molecule of formaldehyde from methenamine. The mechanism can then be
SAR. Structure - Activity Relationships (alkoholy, amíny, aldehydy, ketóny, estery, amidy, kyseliny, uhľovodíky) 2/28/2016
 Structure Activity elationships (SA) identifies which functional groups are important for binding and activity SA Structure - Activity elationships (alkoholy, amíny, aldehydy, ketóny, estery, amidy, kyseliny,
Structure Activity elationships (SA) identifies which functional groups are important for binding and activity SA Structure - Activity elationships (alkoholy, amíny, aldehydy, ketóny, estery, amidy, kyseliny,
QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression
 APPLICATION NOTE QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression GAINING EFFICIENCY IN QUANTITATIVE STRUCTURE ACTIVITY RELATIONSHIPS ErbB1 kinase is the cell-surface receptor
APPLICATION NOTE QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression GAINING EFFICIENCY IN QUANTITATIVE STRUCTURE ACTIVITY RELATIONSHIPS ErbB1 kinase is the cell-surface receptor
Quantum Mechanical Models of P450 Metabolism to Guide Optimization of Metabolic Stability
 Quantum Mechanical Models of P450 Metabolism to Guide Optimization of Metabolic Stability Optibrium Webinar 2015, June 17 2015 Jonathan Tyzack, Matthew Segall, Peter Hunt Optibrium, StarDrop, Auto-Modeller
Quantum Mechanical Models of P450 Metabolism to Guide Optimization of Metabolic Stability Optibrium Webinar 2015, June 17 2015 Jonathan Tyzack, Matthew Segall, Peter Hunt Optibrium, StarDrop, Auto-Modeller
Alkane/water partition coefficients and hydrogen bonding. Peter Kenny
 Alkane/water partition coefficients and hydrogen bonding Peter Kenny (pwk.pub.2008@gmail.com) Neglect of hydrogen bond strength: A recurring theme in medicinal chemistry Rule of 5 Rule of 3 Scoring functions
Alkane/water partition coefficients and hydrogen bonding Peter Kenny (pwk.pub.2008@gmail.com) Neglect of hydrogen bond strength: A recurring theme in medicinal chemistry Rule of 5 Rule of 3 Scoring functions
Design Synthesis and SAR of Non-peptidic Urotensin II Agonists
 MEDICIAL CEMISTY Design Synthesis and SA of on-peptidic Urotensin II Agonists C Val Cys Tyr Lys Trp 2 Glu Thr Pro Asp Cys Phe uman urotensin II (S) pec 50 7.49 () pec 50 5.84 FL-104 AC-7954 Fredrik Lehmann
MEDICIAL CEMISTY Design Synthesis and SA of on-peptidic Urotensin II Agonists C Val Cys Tyr Lys Trp 2 Glu Thr Pro Asp Cys Phe uman urotensin II (S) pec 50 7.49 () pec 50 5.84 FL-104 AC-7954 Fredrik Lehmann
Identifying Interaction Hot Spots with SuperStar
 Identifying Interaction Hot Spots with SuperStar Version 1.0 November 2017 Table of Contents Identifying Interaction Hot Spots with SuperStar... 2 Case Study... 3 Introduction... 3 Generate SuperStar Maps
Identifying Interaction Hot Spots with SuperStar Version 1.0 November 2017 Table of Contents Identifying Interaction Hot Spots with SuperStar... 2 Case Study... 3 Introduction... 3 Generate SuperStar Maps
Targeting protein-protein interactions: A hot topic in drug discovery
 Michal Kamenicky; Maria Bräuer; Katrin Volk; Kamil Ödner; Christian Klein; Norbert Müller Targeting protein-protein interactions: A hot topic in drug discovery 104 Biomedizin Innovativ patientinnenfokussierte,
Michal Kamenicky; Maria Bräuer; Katrin Volk; Kamil Ödner; Christian Klein; Norbert Müller Targeting protein-protein interactions: A hot topic in drug discovery 104 Biomedizin Innovativ patientinnenfokussierte,
Notes of Dr. Anil Mishra at 1
 Introduction Quantitative Structure-Activity Relationships QSPR Quantitative Structure-Property Relationships What is? is a mathematical relationship between a biological activity of a molecular system
Introduction Quantitative Structure-Activity Relationships QSPR Quantitative Structure-Property Relationships What is? is a mathematical relationship between a biological activity of a molecular system
Similarity Search. Uwe Koch
 Similarity Search Uwe Koch Similarity Search The similar property principle: strurally similar molecules tend to have similar properties. However, structure property discontinuities occur frequently. Relevance
Similarity Search Uwe Koch Similarity Search The similar property principle: strurally similar molecules tend to have similar properties. However, structure property discontinuities occur frequently. Relevance
NMR study of complexes between low molecular mass inhibitors and the West Nile virus NS2B-NS3 protease
 University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2009 NMR study of complexes between low molecular mass inhibitors and the West Nile
University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2009 NMR study of complexes between low molecular mass inhibitors and the West Nile
1. Theoretical Investigation of Mechanisms and Stereoselectivities of Synthetic Organic Reactions
 1. Theoretical Investigation of Mechanisms and Stereoselectivities of Synthetic Organic Reactions 2. Copper Catalyzed One-Pot Synthesis of Multisubstituted Quinolinones Hao Wang Denmark Group Presentation
1. Theoretical Investigation of Mechanisms and Stereoselectivities of Synthetic Organic Reactions 2. Copper Catalyzed One-Pot Synthesis of Multisubstituted Quinolinones Hao Wang Denmark Group Presentation
Retrieving hits through in silico screening and expert assessment M. N. Drwal a,b and R. Griffith a
 Retrieving hits through in silico screening and expert assessment M.. Drwal a,b and R. Griffith a a: School of Medical Sciences/Pharmacology, USW, Sydney, Australia b: Charité Berlin, Germany Abstract:
Retrieving hits through in silico screening and expert assessment M.. Drwal a,b and R. Griffith a a: School of Medical Sciences/Pharmacology, USW, Sydney, Australia b: Charité Berlin, Germany Abstract:
Heterocyclic Chemistry Midterm Examination. 3 May Professor Baran Department of Chemistry The Scripps Research Institute
 eterocyclic Chemistry Midterm Examination 3 May 2013 Professor Baran Department of Chemistry The cripps Research Institute ame: Last 4 digits of your ocial ecurity #: This is a 2-hour test that you have
eterocyclic Chemistry Midterm Examination 3 May 2013 Professor Baran Department of Chemistry The cripps Research Institute ame: Last 4 digits of your ocial ecurity #: This is a 2-hour test that you have
Bio-elements. Living organisms requires only 27 of the 90 common chemical elements found in the crust of the earth, to be as its essential components.
 Bio-elements Living organisms requires only 27 of the 90 common chemical elements found in the crust of the earth, to be as its essential components. Most of the chemical components of living organisms
Bio-elements Living organisms requires only 27 of the 90 common chemical elements found in the crust of the earth, to be as its essential components. Most of the chemical components of living organisms
Ignasi Belda, PhD CEO. HPC Advisory Council Spain Conference 2015
 Ignasi Belda, PhD CEO HPC Advisory Council Spain Conference 2015 Business lines Molecular Modeling Services We carry out computational chemistry projects using our selfdeveloped and third party technologies
Ignasi Belda, PhD CEO HPC Advisory Council Spain Conference 2015 Business lines Molecular Modeling Services We carry out computational chemistry projects using our selfdeveloped and third party technologies
Supporting Information
 Supporting Information Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies Johannes T. Cramer 1,2, Jana I. Führing 1, Petra Baruch 2, Christian Brütting 3,
Supporting Information Decoding Allosteric Networks in Biocatalysts: Rational Approach to Therapies and Biotechnologies Johannes T. Cramer 1,2, Jana I. Führing 1, Petra Baruch 2, Christian Brütting 3,
In silico pharmacology for drug discovery
 In silico pharmacology for drug discovery In silico drug design In silico methods can contribute to drug targets identification through application of bionformatics tools. Currently, the application of
In silico pharmacology for drug discovery In silico drug design In silico methods can contribute to drug targets identification through application of bionformatics tools. Currently, the application of
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
SUPPLEMENTARY INFORMATION doi:10.1038/nature11524 Supplementary discussion Functional analysis of the sugar porter family (SP) signature motifs. As seen in Fig. 5c, single point mutation of the conserved
Molecular Docking and 3D-QSAR Based Design of Novel Imidazopyridinone Derivatives as Pseudomonas Aeruginosa Thymidylate Kinase Inhibitors
 http://www.e-journals.in Chemical Science Transactions DI:10.7598/cst2014.704 2014, 3(2), 498-509 RESEARC ARTICLE Molecular Docking and 3D-QSAR Based Design of ovel Imidazopyridinone Derivatives as Pseudomonas
http://www.e-journals.in Chemical Science Transactions DI:10.7598/cst2014.704 2014, 3(2), 498-509 RESEARC ARTICLE Molecular Docking and 3D-QSAR Based Design of ovel Imidazopyridinone Derivatives as Pseudomonas
COMBINATORIAL CHEMISTRY: CURRENT APPROACH
 COMBINATORIAL CHEMISTRY: CURRENT APPROACH Dwivedi A. 1, Sitoke A. 2, Joshi V. 3, Akhtar A.K. 4* and Chaturvedi M. 1, NRI Institute of Pharmaceutical Sciences, Bhopal, M.P.-India 2, SRM College of Pharmacy,
COMBINATORIAL CHEMISTRY: CURRENT APPROACH Dwivedi A. 1, Sitoke A. 2, Joshi V. 3, Akhtar A.K. 4* and Chaturvedi M. 1, NRI Institute of Pharmaceutical Sciences, Bhopal, M.P.-India 2, SRM College of Pharmacy,
1. (18) Multiple choice questions. Please place your answer on the line preceding each question.
 CEM 5720 ame KEY Exam 2 ctober 21, 2015 Read all questions carefully and attempt those questions you are sure of first. Remember to proof your work; art work carries as much importance as written responses.
CEM 5720 ame KEY Exam 2 ctober 21, 2015 Read all questions carefully and attempt those questions you are sure of first. Remember to proof your work; art work carries as much importance as written responses.
Institute of Physical and Theoretical Chemistry Wroclaw University of Technology Wyb. Wyspiańskiego Wrocław, Poland
 COMPUTATIONAL METHODS IN SCIENCE AND TECHNOLOGY 3, 55-62 (1997) THE STRUCTURE, PROTON AFFINITY, ELECTROSTATIC PROPERTIES AND ITS RELATION TO BIOLOGICAL ACTIVITY OF COCAINE AND ITS DERIVATIVES. S. Roszak
COMPUTATIONAL METHODS IN SCIENCE AND TECHNOLOGY 3, 55-62 (1997) THE STRUCTURE, PROTON AFFINITY, ELECTROSTATIC PROPERTIES AND ITS RELATION TO BIOLOGICAL ACTIVITY OF COCAINE AND ITS DERIVATIVES. S. Roszak
Description of Molecules with Molecular Interaction Fields (MIF)
 Description of Molecules with Molecular Interaction Fields (MIF) Introduction to Ligand-Based Drug Design Chimica Farmaceutica 2 Reduction of Dimensionality into Few New Highly Informative Entities -----
Description of Molecules with Molecular Interaction Fields (MIF) Introduction to Ligand-Based Drug Design Chimica Farmaceutica 2 Reduction of Dimensionality into Few New Highly Informative Entities -----
Receptor Based Drug Design (1)
 Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Studies of New Fused Benzazepine as Selective Dopamine D3 Receptor Antagonists Using 3D-QSAR, Molecular Docking and Molecular Dynamics
 Int. J. Mol. Sci. 2011, 12, 1196-1221; doi:10.3390/ijms12021196 OPEN ACCESS Article International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Studies of New Fused Benzazepine
Int. J. Mol. Sci. 2011, 12, 1196-1221; doi:10.3390/ijms12021196 OPEN ACCESS Article International Journal of Molecular Sciences ISSN 1422-0067 www.mdpi.com/journal/ijms Studies of New Fused Benzazepine
Practical Synthesis of PC190723, an Inhibitor of the Bacterial Cell Division Protein FtsZ!
 Practical ynthesis of PC190723, an Inhibitor of the Bacterial Cell Division Protein tsz! orto,. A.; lmstead, M. M.; haw, J. T. J. rg. Chem. 2010, 75, 7946.! Ammonia ynthons for the Multicomponent Assembly
Practical ynthesis of PC190723, an Inhibitor of the Bacterial Cell Division Protein tsz! orto,. A.; lmstead, M. M.; haw, J. T. J. rg. Chem. 2010, 75, 7946.! Ammonia ynthons for the Multicomponent Assembly
SUPPLEMENTARY INFORMATION
 Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
Predicting Alkylbenzimidazole Derivatives as Angiotensin II Receptor Antagonists: 3d Qsar by knn-mfa Approach
 Advances in Biological Research 5 (3): 161-169, 011 ISSN 199-0067 IDOSI Publications, 011 Predicting Alkylbenzimidazole Derivatives as Angiotensin II Receptor Antagonists: 3d Qsar by knn-mfa Approach Mukesh
Advances in Biological Research 5 (3): 161-169, 011 ISSN 199-0067 IDOSI Publications, 011 Predicting Alkylbenzimidazole Derivatives as Angiotensin II Receptor Antagonists: 3d Qsar by knn-mfa Approach Mukesh
Medicinal Chemist s Relationship with Additivity: Are we Taking the Fundamentals for Granted?
 Medicinal Chemist s Relationship with Additivity: Are we Taking the Fundamentals for Granted? J. Guy Breitenbucher Streamlining Drug Discovery Conference San Francisco, CA ct. 25, 2018 Additivity as the
Medicinal Chemist s Relationship with Additivity: Are we Taking the Fundamentals for Granted? J. Guy Breitenbucher Streamlining Drug Discovery Conference San Francisco, CA ct. 25, 2018 Additivity as the
Hit Finding and Optimization Using BLAZE & FORGE
 Hit Finding and Optimization Using BLAZE & FORGE Kevin Cusack,* Maria Argiriadi, Eric Breinlinger, Jeremy Edmunds, Michael Hoemann, Michael Friedman, Sami Osman, Raymond Huntley, Thomas Vargo AbbVie, Immunology
Hit Finding and Optimization Using BLAZE & FORGE Kevin Cusack,* Maria Argiriadi, Eric Breinlinger, Jeremy Edmunds, Michael Hoemann, Michael Friedman, Sami Osman, Raymond Huntley, Thomas Vargo AbbVie, Immunology
Ping-Chiang Lyu. Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University.
 Pharmacophore-based Drug design Ping-Chiang Lyu Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University 96/08/07 Outline Part I: Analysis The analytical
Pharmacophore-based Drug design Ping-Chiang Lyu Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University 96/08/07 Outline Part I: Analysis The analytical
Journal of Molecular Graphics and Modelling
 Journal of Molecular Graphics and Modelling 30 (2011) 67 81 Contents lists available at ScienceDirect Journal of Molecular Graphics and Modelling journal homepage: www.elsevier.com/locate/jmgm Development
Journal of Molecular Graphics and Modelling 30 (2011) 67 81 Contents lists available at ScienceDirect Journal of Molecular Graphics and Modelling journal homepage: www.elsevier.com/locate/jmgm Development
LigandScout. Automated Structure-Based Pharmacophore Model Generation. Gerhard Wolber* and Thierry Langer
 LigandScout Automated Structure-Based Pharmacophore Model Generation Gerhard Wolber* and Thierry Langer * E-Mail: wolber@inteligand.com Pharmacophores from LigandScout Pharmacophores & the Protein Data
LigandScout Automated Structure-Based Pharmacophore Model Generation Gerhard Wolber* and Thierry Langer * E-Mail: wolber@inteligand.com Pharmacophores from LigandScout Pharmacophores & the Protein Data
Supporting Information
 Discovery of kinase inhibitors by high-throughput docking and scoring based on a transferable linear interaction energy model Supporting Information Peter Kolb, Danzhi Huang, Fabian Dey and Amedeo Caflisch
Discovery of kinase inhibitors by high-throughput docking and scoring based on a transferable linear interaction energy model Supporting Information Peter Kolb, Danzhi Huang, Fabian Dey and Amedeo Caflisch
Abstracts VII. Phthalocyanines and Related Compounds. M. S. Rodr guez-morgade and T. Torres
 VII p1 thalocyanines and Related Compounds 17.9.24 M.. Rodr guez-morgade and T. Torres This review updates the original cience of ynthesis chapter (ection 17.9) on phthalocyanines and various ring-fused,
VII p1 thalocyanines and Related Compounds 17.9.24 M.. Rodr guez-morgade and T. Torres This review updates the original cience of ynthesis chapter (ection 17.9) on phthalocyanines and various ring-fused,
Chapter 15 Reactions of Aromatic Compounds
 Chapter 15 1 Chapter 15 Reactions of Aromatic Compounds Electrophilic Aromatic Substitution Arene (Ar-H) is the generic term for an aromatic hydrocarbon The aryl group (Ar) is derived by removal of a hydrogen
Chapter 15 1 Chapter 15 Reactions of Aromatic Compounds Electrophilic Aromatic Substitution Arene (Ar-H) is the generic term for an aromatic hydrocarbon The aryl group (Ar) is derived by removal of a hydrogen
12/27/2010. Chapter 15 Reactions of Aromatic Compounds
 Chapter 15 Reactions of Aromatic Compounds Electrophilic Aromatic Substitution Arene (Ar-H) is the generic term for an aromatic hydrocarbon The aryl group (Ar) is derived by removal of a hydrogen atom
Chapter 15 Reactions of Aromatic Compounds Electrophilic Aromatic Substitution Arene (Ar-H) is the generic term for an aromatic hydrocarbon The aryl group (Ar) is derived by removal of a hydrogen atom
Chemical Space. Space, Diversity, and Synthesis. Jeremy Henle, 4/23/2013
 Chemical Space Space, Diversity, and Synthesis Jeremy Henle, 4/23/2013 Computational Modeling Chemical Space As a diversity construct Outline Quantifying Diversity Diversity Oriented Synthesis Wolf and
Chemical Space Space, Diversity, and Synthesis Jeremy Henle, 4/23/2013 Computational Modeling Chemical Space As a diversity construct Outline Quantifying Diversity Diversity Oriented Synthesis Wolf and
Computational chemical biology to address non-traditional drug targets. John Karanicolas
 Computational chemical biology to address non-traditional drug targets John Karanicolas Our computational toolbox Structure-based approaches Ligand-based approaches Detailed MD simulations 2D fingerprints
Computational chemical biology to address non-traditional drug targets John Karanicolas Our computational toolbox Structure-based approaches Ligand-based approaches Detailed MD simulations 2D fingerprints
Virtual Screening: How Are We Doing?
 Virtual Screening: How Are We Doing? Mark E. Snow, James Dunbar, Lakshmi Narasimhan, Jack A. Bikker, Dan Ortwine, Christopher Whitehead, Yiannis Kaznessis, Dave Moreland, Christine Humblet Pfizer Global
Virtual Screening: How Are We Doing? Mark E. Snow, James Dunbar, Lakshmi Narasimhan, Jack A. Bikker, Dan Ortwine, Christopher Whitehead, Yiannis Kaznessis, Dave Moreland, Christine Humblet Pfizer Global
Introduction. OntoChem
 Introduction ntochem Providing drug discovery knowledge & small molecules... Supporting the task of medicinal chemistry Allows selecting best possible small molecule starting point From target to leads
Introduction ntochem Providing drug discovery knowledge & small molecules... Supporting the task of medicinal chemistry Allows selecting best possible small molecule starting point From target to leads
11/30/ Substituent Effects in Electrophilic Substitutions. Substituent Effects in Electrophilic Substitutions
 Chapter 9 Problems: 9.1-29, 32-34, 36-37, 39-45, 48-56, 58-59, 61-69, 71-72. 9.8 Substituent effects in the electrophilic substitution of an aromatic ring Substituents affect the reactivity of the aromatic
Chapter 9 Problems: 9.1-29, 32-34, 36-37, 39-45, 48-56, 58-59, 61-69, 71-72. 9.8 Substituent effects in the electrophilic substitution of an aromatic ring Substituents affect the reactivity of the aromatic
Creating a Pharmacophore Query from a Reference Molecule & Scaffold Hopping in CSD-CrossMiner
 Table of Contents Creating a Pharmacophore Query from a Reference Molecule & Scaffold Hopping in CSD-CrossMiner Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Features
Table of Contents Creating a Pharmacophore Query from a Reference Molecule & Scaffold Hopping in CSD-CrossMiner Introduction... 2 CSD-CrossMiner Terminology... 2 Overview of CSD-CrossMiner... 3 Features
Using AutoDock for Virtual Screening
 Using AutoDock for Virtual Screening CUHK Croucher ASI Workshop 2011 Stefano Forli, PhD Prof. Arthur J. Olson, Ph.D Molecular Graphics Lab Screening and Virtual Screening The ultimate tool for identifying
Using AutoDock for Virtual Screening CUHK Croucher ASI Workshop 2011 Stefano Forli, PhD Prof. Arthur J. Olson, Ph.D Molecular Graphics Lab Screening and Virtual Screening The ultimate tool for identifying
Introduction to Spark
 1 As you become familiar or continue to explore the Cresset technology and software applications, we encourage you to look through the user manual. This is accessible from the Help menu. However, don t
1 As you become familiar or continue to explore the Cresset technology and software applications, we encourage you to look through the user manual. This is accessible from the Help menu. However, don t
Patrick: An Introduction to Medicinal Chemistry 5e Chapter 01
 Questions Patrick: An Introduction to Medicinal Chemistry 5e 01) Which of the following molecules is a phospholipid? a. i b. ii c. iii d. iv 02) Which of the following statements is false regarding the
Questions Patrick: An Introduction to Medicinal Chemistry 5e 01) Which of the following molecules is a phospholipid? a. i b. ii c. iii d. iv 02) Which of the following statements is false regarding the
Protein Structure. W. M. Grogan, Ph.D. OBJECTIVES
 Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Protein Structure W. M. Grogan, Ph.D. OBJECTIVES 1. Describe the structure and characteristic properties of typical proteins. 2. List and describe the four levels of structure found in proteins. 3. Relate
Highlights of Schmidt Reaction in the Last Ten Years
 ighlights of Schmidt eaction in the Last Ten Years Dendrobates histrionicus Jack Liu ov. 18, 2003 Introduction Classical Schmidt reaction of aldehydes and carboxylic acids Classical Schmidt reaction of
ighlights of Schmidt eaction in the Last Ten Years Dendrobates histrionicus Jack Liu ov. 18, 2003 Introduction Classical Schmidt reaction of aldehydes and carboxylic acids Classical Schmidt reaction of
Structure Activity Relationships (SAR) and Pharmacophore Discovery Using Inductive Logic Programming (ILP)
 Structure Activity Relationships (SAR) and Pharmacophore Discovery Using Inductive Logic Programming (ILP) Structure Activity Relationships (SAR) and Pharmacophore Discovery Using Inductive Logic Programming
Structure Activity Relationships (SAR) and Pharmacophore Discovery Using Inductive Logic Programming (ILP) Structure Activity Relationships (SAR) and Pharmacophore Discovery Using Inductive Logic Programming
Drug Discovery. Zainab Al Kharusi Office: 33-10
 Drug Discovery Zainab Al Kharusi Office: 33-10 Objectives: To understand the processes of Drug Development To be able to develop a plan for drug discovery Reference: Patrick G L. An Introduction to Medicinal
Drug Discovery Zainab Al Kharusi Office: 33-10 Objectives: To understand the processes of Drug Development To be able to develop a plan for drug discovery Reference: Patrick G L. An Introduction to Medicinal
Nature Structural & Molecular Biology: doi: /nsmb Supplementary Figure 1
 Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
Supplementary Figure 1 Crystallization. a, Crystallization constructs of the ET B receptor are shown, with all of the modifications to the human wild-type the ET B receptor indicated. Residues interacting
75. A This is a Markovnikov addition reaction. In these reactions, the pielectrons in the alkene act as a nucleophile. The strongest electrophile will
 71. B SN2 stands for substitution nucleophilic bimolecular. This means that there is a bimolecular rate-determining step. Therefore, the reaction will follow second-order kinetics based on the collision
71. B SN2 stands for substitution nucleophilic bimolecular. This means that there is a bimolecular rate-determining step. Therefore, the reaction will follow second-order kinetics based on the collision
