Molecular analysis of the taxonomic. and distributional status for the hermit crab genera Loxopagurus Forest, 1964 and Isocheles
|
|
|
- Rudolph Arnold Lawrence
- 5 years ago
- Views:
Transcription
1 Molecular analysis of the taxonomic and distributional status for the hermit crab genera Loxopagurus Forest, 1964 and Isocheles Stimpson, 1858 (Decapoda, Anomura, Diogenidae) Fernando L. M. MANTELATTO Laboratory of Bioecology and Crustacean Systematics, Department of Biology, FFCLRP, Universidade de São Paulo (USP), Av. Bandeirantes, 3900, CEP , Ribeirão Preto, SP (Brazil) Rafael ROBLES Department of Biology, Laboratory for Crustacean Research, University of Louisiana at Lafayette, Lafayette, LA (USA) Renata BIAGI Laboratory of Bioecology and Crustacean Systematics, Department of Biology, FFCLRP, Universidade de São Paulo (USP), Av. Bandeirantes, 3900, CEP , Ribeirão Preto, SP (Brazil) Darryl L. FELDER Department of Biology, Laboratory for Crustacean Research, University of Louisiana at Lafayette, Lafayette, LA (USA) Mantelatto F. L. M., Robles R., Biagi R. & Felder D. L Molecular analysis of the taxonomic and distributional status for the hermit crab genera Loxopagurus Forest, 1964 and Isocheles Stimpson, 1858 (Decapoda, Anomura, Diogenidae). Zoosystema 28 (2) : KEY WORDS Crustacea, Decapoda, Anomura, Diogenidae, Isocheles, Loxopagurus, molecular systematics, 16S rrna, South Atlantic. ABSTRACT Recent studies of adult and larval morphology have revealed characters that could justify reassignment of Loxopagurus loxochelis to the genus Isocheles. Current taxonomy of these two similar genera is based only on adult morphology and disregards morphological characters of the first zoeal stage. There has been no previous attempt to resolve evolutionary relationships among the species of these two genera using molecular tools. Herein, we include these species in a phylogenetic analysis of selected anomuran decapods based on sequences of the 16S ribosomal gene. Our present molecular analysis does not support reassignment of Loxopagurus loxochelis to the genus Isocheles. In contradiction to previous suggestions, we show that L. loxochelis and I. sawayai are indeed different species. We also corroborate the previous report of I. sawayai in Venezuelan waters. Publications Scientifiques du Muséum national d Histoire naturelle, Paris
2 Mantelatto F. L. M. et al. MOTS CLÉS Crustacea, Decapoda, Anomura, Diogenidae, Isocheles, Loxopagurus, systématique moléculaire, 16S rrna, Atlantique Sud. RÉSUMÉ Statut taxonomique et distribution des genres Loxopagurus Forest, 1964 et Isocheles Stimpson, 1858 (Decapoda, Anomura, Diogenidae) sur la base de données moléculaires. Des études récentes concernant la morphologie de l adulte et des larves ont révélé des caractères qui pourraient justifier un transfert de Loxopagurus loxochelis au genre Isocheles. La taxonomie classique de ces deux genres similaires est basée uniquement sur l adulte et ne tient aucun compte des caractères morphologiques du premier stade de la zoé. Il n y a eu jusqu à présent aucun essai de résoudre les affinités entre les espèces de ces deux genres au moyen de l analyse moléculaire. Dans ce travail, nous incluons ces espèces dans une analyse phylogénique de décapodes anomoures choisis, basée sur les séquences du gène ribosomique 16S. Notre analyse moléculaire ne justifie pas le transfert de Loxopagurus loxochelis dans le genre Isocheles. En contradiction avec les suggestions précédentes, nous montrons que L. loxochelis et I. sawayai sont bien des espèces différentes. Nous confirmons également la présence d I. sawayai dans les eaux vénézuéliennes. INTRODUCTION Loxopagurus loxochelis (Moreira, 1901) is a monotypic hermit crab genus endemic to the southwestern Atlantic coast, occurring in Brazil (from Bahia to Rio Grande do Sul states), Uruguay, and Argentina (Melo 1999). Isocheles Stimpson, 1858 is poorly known, with only a few of the constituent species treated in studies concerning biology or ecology (unpublished data). The biogeographical distribution of this genus is restricted to shallow waters of the tropical and subtropical American coasts, where five species occur: I. pilosus (Holmes, 1900), I. pacificus (Bouvier, 1907) and I. aequimanus (Dana, 1852), in the eastern Pacific Ocean; and, I. sawayai Forest & de Saint Laurent, 1968, and I. wurdemanni Stimpson, 1859, in the western Atlantic Ocean (Forest & de Saint Laurent 1968). Species of Loxopagurus and Isocheles are similar in general somatic morphology, and their current taxonomy is based solely on adult morphology. A critical study of the relationship between these genera and between constituent species of Isocheles has been long called for (Provenzano 1959; Forest & de Saint Laurent 1968). A recent, unpublished systematic review on the basis of adult and larval morphology (Nucci 2002) reported characters that potentially justify reassignment of L. loxochelis to Isocheles. Nucci examined L. loxochelis and I. sawayai and, on the basis of morphological characters of adults, and published information on larvae ( Forest & de Saint Laurent 1968; Negreiros-Fransozo & Hebling 1983; Bernardi 1986; Hebling et al. 2000), proposed that L. loxochelis be placed into synonymy with I. sawayai. To date, most systematic studies have been based solely on morphology and molecular tools have been rarely applied to solve questions of species status or to determine lower level phylogenetic relationships (Morrison et al. 2002). On the basis of partial sequences of the large ribosomal subunit 16S, we test morphologically based reassignment of Loxopagurus to Isocheles, and consequently whether L. loxochelis is a synonym of I. sawayai. MATERIAL AND METHODS Our phylogenetic analysis was based exclusively on a partial fragment of the 16S rdna gene. Mitochondrial DNA (mtdna) is maternally inherited, easy to isolate, and abundant; even so, absence of 496
3 Sequence analysis of Loxopagurus and Isocheles TABLE 1. Hermit crab species used for the phylogenetic reconstructions with respective date and site of collection, museum catalog number, and genetic database accession numbers (GenBank). Notes: a, Morrison et al. (2002); b, Zaklan & Cunningham (unpublished); c, Pérez-Losada et al. (2002); d, Schubart et al. (2000a). *, This species was referred to as Pachycheles haigae in Pérez-Losada et al. (2002); according to Harvey & De Santo (1996), P. haigae is a junior synonym of P. laevidactylus. Species Collection site, date Catalog No. GenBank accession No. Calcinus obscurus Stimpson, 1859 Panama City (Panama), 2001 AF a Calcinus tibicen (Herbst, 1791) Ubatuba (Brazil), X.2001 CCDB 769 DQ Clibanarius albidigitus Nobili, 1901 Panama City (Panama), 2001 AF b Clibanarius antillensis Stimpson, 1859 Florida (USA), VII.1998 ULLZ 4683 DQ Coenobita compressus (H. Milne Edwards, 1837) Amador Causeway (Panama), 2001 AF a Dardanus insignis (de Saussure, 1858) Caraguatatuba (Brazil), IV.2001 CCDB 774 DQ Dardanus venosus (H. Milne Edwards, 1848) Ubatuba (Brazil), XI.2001 CCDB 766 DQ Isocheles pilosus (Holmes, 1900) California (USA), 2001 AF a Isocheles sawayai Forest & de Saint Laurent, 1968 Ubatuba (Brazil), XII.2000 CCDB 302 DQ Isocheles sawayai Forest & de Saint Laurent, 1968 Margarita Is. (Venezuela), I.2002 CCDB 308 DQ Isocheles wurdemanni Stimpson, 1859 Texas (USA), X.1997 ULLZ 3890 DQ Lithodes aequispinus Benedict, 1895 Abashiri (Japan), 2001 AF b Lithodes maja (Linnaeus, 1758) Nova Scotia (Canada), 2001 AF b Lithodes santolla (Molina, 1782) Beagle Channel (Argentina), 2001 AF b Loxopagurus loxochelis (Moreira, 1901) Ubatuba (Brazil), XI.2000 CCDB 765 DQ Pachycheles laevidactylus Ortmann, 1892* Tramandaí (Brazil), 2001 AY c Paguristes calliopsis Forest & de Saint Laurent, 1968 Ubatuba (Brazil), IX.1999 CCDB 768 DQ Paguristes erythrops Holthuis, 1959 Paraty (Brazil), X.1996 CCDB 773 DQ Paguristes robustus Forest & de Saint Laurent, 1968 Ubatuba (Brazil), III.2001 CCDB 1252 DQ Paguristes tortugae complex Schmitt, 1933 Ubatuba (Brazil), XI.1999 CCDB 1251 DQ Pagurus brevidactylus (Stimpson, 1859) Ubatuba (Brazil), XI.2001 CCDB 771 DQ Pagurus criniticornis (Dana, 1852) Ubatuba (Brazil), XI CCDB 779 DQ Pagurus leptonyx Forest & de Saint Laurent, 1968 Ubatuba (Brazil), VIII.2001 CCDB 1257 DQ Petrolisthes armatus (Gibbes, 1850) Louisiana (USA), 1998 ULLZ 3779 AJ d Petrochirus diogenes (Linnaeus, 1758) Ubatuba (Brazil), III.2001 CCDB 776 DQ recombination is a limitation because phylogeny will refl ect only the maternal lineage (Hartl & Clark 1997). This gene has nonetheless shown its utility in both phylogenetic and population studies for over a decade (Bucklin et al. 1995; Schubart et al. 2000a, 2002; Stillman & Reeb 2001; Tudge & Cunningham 2002; Young et al. 2002; Harrison 2004; Morrison et al. 2004; Machordom & Macpherson 2004; Mantelatto et al. in press; Robles et al. in press), and it is a common choice for use in phylogenetic studies on decapods (Schubart et al. 2000a; Mathews et al. 2002; Harrison 2004; Morrison et al. 2004; Machordom & Macpherson 2004). Hermit crabs used in our analyses were collected between 1996 and 2002 (Table 1). Specimens selected for DNA analysis were preserved directly in 75 to 90% ethanol. Species identifications were confi rmed on the basis of morphological characters from available references (Moreira 1901; Costa 1962; Forest & de Saint Laurent 1968; Melo 1999; McLaughlin 2003). Genetic vouchers from which tissue subsamples were obtained have been deposited at the University of Louisiana at Lafayette, Zoological Collection (ULLZ) or at the University of São Paulo, Crustacean Collection of Department of Biology (CCDB) of Faculdade de Filosofia, Ciências e Letras de Ribeirão Preto (FF- CLRP) (Table 1). We included three of the five known species of Isocheles in our analysis. We were unable to obtain two species, I. aequimanus and I. pacificus. Besides the three species of Isocheles, and L. loxochelis, we included other hermit crabs from the family Diogenidae for comparison, along with representatives of other selected anomuran groups to more broadly root the analysis. Some of the comparative sequences included in the analysis were retrieved 497
4 Mantelatto F. L. M. et al. from GenBank (Table 1). In addition to Isocheles and Loxo pagurus, we used from the Diogenidae two species of Calcinus, two of Clibanarius, two of Dardanus, four species of Paguristes, and one of Petrochirus; from the Coenobitidae we used a species of Coenobita; from the Lithodidae, we used three species of Lithodes; and from the Paguridae we used three species of Pagurus. A species of Pachycheles, and one of Petrolisthes were used as representatives of the Porcellanidae (Table 1). DNA extraction, amplification and sequencing protocols follow Schubart et al. (2000a) with modifications as in Mantelatto et al. (in press) and Robles et al. (in press). Total genomic DNA was extracted from muscle tissue of walking legs or the chelipeds. Muscle was ground and incubated for 1-12h in 600 µl of lysis buffer at 65 C; protein was separated by addition of 200 µl 7.5M of ammonium acetate prior to centrifugation. DNA was precipitated by addition of 600 µl of cold isopropanol followed by centrifugation; the resultant pellet was washed with 70% ethanol, dried and resuspended in µl of TE buffer. An approximately 550-basepair region of the 16S rrna gene was amplified from diluted DNA by means of polymerase-chain-reaction (PCR) (thermal cycles: initial denaturizing for 10 min at 94 C; annealing for cycles: 1 min at 94 C, 1 min at C, 2 min at 72 C; final extension of 10 min at 72 C) with the primers designated as follows: 16SH2 (5 -AGATAGAAACCAACCTGG-3 ), 16SL2 (5 -TGCCTGTTTATCAAAAACAT-3 ), and 16Sar (5 -CGCCTGTTTATCAAAAACAT-3 ) (for references on the primers see Schubart et al. 2000a, b). PCR products were purifi ed using Microcon filters (Millipore Corp.) and sequenced with the ABI Big Dye Terminator Mix (PE Biosystems) in an ABI Prism 3 Genetic Analyzer (Applied Biosystems automated sequencer). All sequences were confirmed by sequencing both strands. Sequences were edited with the Sequencher software program (version 4.1, Genecodes, Ann Arbor, MI) and aligned on Bioedit (Hall 1999) with the following settings: pairwise parameters = gap opening 6.0, gap extension 0.2, multialignment parameters = gap opening 6.0, gap extension 0.2. Ambiguous regions of the alignment were removed. Base composition, pattern of substitution for pairwise comparison, and analysis of variability along the 16Smt DNA was performed as implemented in PAUP 4.0 beta 10 (Swofford 2003). Homogeneity of nucleotide frequency among taxa was also assessed with a χ 2 test as implemented in PAUP. Phylogenetic analyses were conducted using MR- BAYES for Bayesian analysis (BAY), and PAUP 4.0 beta 10 (Swofford 2003) for both Maximum Parsimony (MP) and Neighbor Joining (NJ) analyses. These three methods, plus Maximum Likelihood, are widely used in reconstructing phylogenies. Though they have different approaches, usually they result in the same phylogenetic tree (Hedges 2002). By using different methods we are able to evaluate the robustness of our results (Hedges 2002). Prior to conducting the BAY or NJ analyses, the model of evolution that best fits the data was determined with the software MODELTEST (Posada & Crandall 1998). The Bayesian analysis was performed by sampling one tree every generations for generations, starting with a random tree. A preliminary analysis showed that stasis was reached at approximately 7000 generations. Thus, we discarded 201 trees corresponding to generations and obtained a 50% majority rule consensus trees from the remaining saved trees. Neighbor-joining analysis was carried out with a maximum likelihood distance correction set with the parameters obtained from MOD- ELTEST (Posada & Crandall 1998). Maximum parsimony analysis was performed as a heuristic search with gaps treated as a fifth character state, multistate characters interpreted as uncertain, and all characters considered as unordered. The search was conducted with a random sequence addition of 0 random trees, including tree bisection and reconnection (TBR) as a branch swapping option. One tree was stored at every repetition, and branch swapping was performed on the best trees only. To determine confidence values for the resulting trees, for both NJ and MP, we ran 0 bootstrap replicates based on the same parameters as above. On the molecular trees, confidence values > 50% were reported for both NJ (0 bootstraps) and MP (0 bootstraps) analyses while for the BAY 498
5 Sequence analysis of Loxopagurus and Isocheles Petrolisthes armatus Pachycheles laevidactylus Coenobita compressus Petrochirus diogenes Dardanus insignis 54 Dardanus venosus Paguristes calliopsis Paguristes tortugae complex 91 Paguristes robustus Paguristes erythrops 76 Isocheles wurdemanni TEX Isocheles sawayai VEN Isocheles sawayai BRA Isocheles pilosus Loxopagurus loxochelis Calcinus tibicen Calcinus obscurus 94 Clibanarius antillensis Clibanarius albidigitus 93 Lithodes aequispinus Lithodes maja Lithodes santolla 0.05 substitutions/site Pagurus brevidactylus Pagurus leptonyx Pagurus criniticornis FIG. 1. Single phylogenetic tree obtained from NJ analysis of 16S sequences for Loxopagurus, Isocheles, and other selected anomurans. Numbers are significance values for 0 bootstraps; values 50% are not shown. Abbreviations: BRA, Brazil; TEX, Texas; VEN, Venezuela. 499
6 Mantelatto F. L. M. et al. analysis values were reported for posterior probabilities of the respective nodes among all the saved trees. Sequences have been deposited in GenBank (Table 1). RESULTS Our analysis included 25 individuals representing 24 species of anomurans (Table 1). The 16S rdna alignment contained a total of 538 basepairs excluding primer regions. From these, 84 characters were excluded (38-47; ; ) from the analysis because of uncertain alignment. The complete alignment has been submitted to Gen- Bank as a popset. The fragment used for the analysis contained 233 non-variable and 221 variable characters, of which 175 were parsimony informative. The nucleotide composition of the 16S mtdna of this dataset can be considered homogeneous (χ 2 = 26.38, df = 78, p = 0.99), with a larger percentage of A-T (0.340%; 0.337%). The best-fitting model of substitution, selected with the Akaike information criterion (AIC; Akaike 1974) as implemented in MODELTEST (Posada & Crandall 1998), was the transversional model with invariable sites and a gamma distribution (TVM+I+G) (Rodríguez et al. 1990) and with the following parameters: assumed nucleotide frequencies: A = , C = , G = , T = ; substitution rates A-C = , A-G = , A-T = , C-G = , C-T = , G-T = ; proportion of invariable sites I = ; variable sites followed a gamma distribution with shape parameter = These values were used to obtain both BAY and NJ trees. MP analysis yielded a single tree of length 680, with consistency index (CI) = and retention index (RI) = Overall, the distance, Bayesian and parsimony methods resulted in similar tree topologies, though differences were observed (Figs 1-3). In the three analyses, all representatives of the Diogenidae, Calcinus, Clibanarius, Dardanus, Paguristes, Petrochirus, Loxopagurus, and Isocheles, were clustered in a single clade along with Coenobita compressus, the only member of the Coenobitidae included in our analysis. Selected species representing each of the diogenid genera were clustered together as expected. The three species of Pagurus, representing the Paguridae, were also grouped in a single clade with another clade representating the Lithodidae as a sister group (Figs 1-3). Within the Diogenidae, all three analyses place Loxopagurus as a sister group to a clade with the species of Isocheles, but on an independent and basal branch. Differences between L. loxochelis and I. sawayai were substantial, accounting for 9.8% of the gene fragment analyzed (27 transitions, 18 transversions, and 19 indels). Among species of Isocheles in the analysis, samples from Brazilian and Venezuelan populations of I. sawayai proved to be identical. Isocheles sawayai was positioned closest to I. pilosus among its congeners in the analyses. DISCUSSION Our partial fragment analysis of 16S DNA shows that L. loxochelis is diverged at a level of genetic difference that clearly justifies its continued recognition as a separate species from I. sawayai. As result, we find no genetic justification for synonymy of Loxopagurus with Isocheles. Species of these two taxa are highly divergent in mtdna-gene sequences (9.8% of difference, with 27 transitions, 18 transversions and 19 indels on basepairs sequences), at levels comparable to or exceeding divergence typically measured between other decapod genera (Schubart et al. 2000b). This is reflected in the three molecularly based phylogenetic trees, where members of Loxopagurus and Isocheles occur on a common branch within the Diogenidae. In all three analyses, species of Isocheles are grouped into a well supported clade with Loxopagurus as the sister group. Therefore, there is sound genetic evidence to support the morphological bases for the separation of these two genera. Forest (1964) described the new genus, Loxopagurus, on the basis of specimens of Pagurus loxochelis from Bahia (Brazil) originally described by Moreira (1901). Forest reported several similarities in general body morphology and color between the new genus Loxopagurus and the genus Isocheles, particularly in the shape of the shield and peduncles, 500
7 Sequence analysis of Loxopagurus and Isocheles Petrolisthes armatus Pachycheles laevidactylus Coenobita compressus Petrochirus diogenes Dardanus insignis 86 Dardanus venosus Isocheles wurdemanni TEX Isocheles sawayai VEN Isocheles sawayai BRA Isocheles pilosus 74 Loxopagurus loxochelis 98 Clibanarius antillensis 90 Clibanarius albidigitus Calcinus tibicen Calcinus obscurus 65 Paguristes calliopsis 96 Paguristes robustus 84 Paguristes tortugae complex Paguristes erythrops 84 Lithodes aequispinus Lithodes maja Lithodes santolla Pagurus brevidactylus Pagurus leptonyx 10 changes Pagurus criniticornis FIG. 2. Single phylogenetic tree obtained from MP analysis of 16S sequences for Loxopagurus, Isocheles, and other selected anomurans. Numbers are significance values for 0 bootstraps; values 50% are not shown. Abbreviations: BRA, Brazil; TEX, Texas; VEN, Venezuela. 501
8 Mantelatto F. L. M. et al. antennal characters (pilosity of the flagellae and aciculae), and features of the mouthparts. According to Forest, the only difference was found in the shape and sculpture of the chelipeds (heterochely in Loxopagurus and isochely in Isocheles), as also mentioned by Forest & de Saint Laurent (1968). We observed such differences in chela morphology in L. loxochelis and I. sawayai. Furthermore, from previous larval descriptions by Negreiros-Fransozo & Hebling (1983) and Bernardi (1986) for both species, we recognized additional important differences that were not reported previously, especially in maxilla and maxilliped setation and structure of the antenna in zoea I, II and III. In addition, there are clear ecological differences, at least as observable in southern Atlantic shallow waters: L. loxochelis inhabits cold waters at depths as great as 35m and salinities exceeding 30 (Mantelatto et al. 2004), while I. sawayai is found in warmer shallow water generally less than 10 m deep and of salinity less than 20, often close to some freshwater influence (Martinelli et al. 2002). However, there are likewise shared attributes that suggest these genera are of close relationship; Scelzo et al. (2004) and Mantelatto et al. (unpublished data) found that members of L. loxochelis and I. sawayai are very closely allied species on the basis of spermatophore morphology. Among the species and populations of Isocheles included in our analyses, the present molecular phylogeny showed a % similarity between a sample of I. sawayai from Brazil and a sample from Venezuela, the latter of which may represent populations that were assigned to I. wurdemanni by Rodríguez (1980). At the molecular level, we are not able to separate these two populations on the basis of 16S rdna sequences, since we did not find a single basepair difference between the specimens analyzed. Ranges of the two western Atlantic species (I. wurdemanni and I. sawayai) were recently reported by Nucci & Melo (2000) to overlap in Venezuela, under the assumption that I. sawayai, previously considered endemic to Brazil, had extended its distribution to Venezuela. Without question, we can corroborate that I. sawayai occurs in Venezuela where Nucci & Melo (2000) reported it too, as our specimens came from the same site (Isla Margarita). They are clearly not identifiable with I. wurdemanni, as we sequenced specimens of the latter species from the type locality in the Gulf of Mexico (Texas) for comparison to the Venezuelan materials and found basepair divergence to exceed 4.6%. However, we also now question the supposition that ranges of these two species overlap in Venezuela. Since the materials furnished to us from there were those typically identified as I. wurdemanni, but were found in molecular analyses to be I. sawayai, we feel it is possible that only the latter species occurs there and that records to date may be in error. Our present analysis infers that the closest relative to I. sawayai is I. pilosus from the eastern Pacific, rather than any other Atlantic species of Isocheles or Loxopagurus. Differences between these two species were limited (3.1% of difference: 17 transitions, 5 transversions, 5 indels) compared to those observed among most other species of Diogenidae that we have sequenced. Provenzano (1959) previously commented on Holopagurus pilosus Holmes, 1900 bearing close resemblance to an overgrown specimen of his therein described I. wurdemanni (from Texas), noting the tendency of larger specimens in the latter species to have slightly unequal chelipeds and questioning the interpretation of this variable character. The present analysis shows that Holopagurus is correctly synonymized with Isocheles and that eastern Pacific and western Atlantic forms in this genus are closely related, rather than being separated into geographically defined branches. Our findings also argue that, in addition to reexamination of adult and larval morphological characters, phylogenetic studies can benefit from less commonly applied characters such as spermatophore morphology. The phylogenetic proximity between Loxopagurus and Clibanarius or Calcinus, demonstrated in our analysis as well as in previous works (McLaughlin 1983; Morrison et al. 2002), is corroborated by the recent observations of spermatophore (Scelzo et al. 2004; unpublished data) and sperm (Scelzo et al. in press) morphology obtained for species of the Diogenidae from the South Atlantic. Previous phylogenetic hypotheses based on limited data have suggested that Paguristes has close affinities to Pagurus (Morrison et al. 2002). However, our work suggests instead that Paguristes is more 502
9 Sequence analysis of Loxopagurus and Isocheles Petrolisthes armatus Pachycheles laevidactylus Coenobita compressus Dardanus insignis Dardanus venosus Petrochirus diogenes Isocheles wurdemanni TEX 94 Isocheles sawayai VEN Isocheles sawayai BRA Isocheles pilosus Loxopagurus loxochelis 94 Calcinus tibicen 95 Calcinus obscurus Clibanarius antillensis Clibanarius albidigitus 60 Paguristes calliopsis Paguristes robustus 87 Paguristes tortugae complex 58 Paguristes erythrops Lithodes aequispinus Lithodes maja 93 Lithodes santolla 56 Pagurus brevidactylus Pagurus criniticornis Pagurus leptonyx FIG. 3. Consensus phylogenetic tree obtained from BAY analysis (50% majority consensus of trees) of 16S sequences for Loxopagurus, Isocheles, and other selected anomurans. Numbers are significance values of posterior probabilities; values 50% are not shown. Abbreviations: BRA, Brazil; TEX, Texas; VEN, Venezuela. 503
10 Mantelatto F. L. M. et al. closely allied to the remaining diogenid species used in the analysis. Our findings are preliminary and underscore the need for comprehensive studies of Paguridae and Diogenidae throughout American waters. Dedication The authors are honored to recognize the many achievements of our friend and colleague, Patsy A. McLaughlin, by this contribution to this commemorative issue. Acknowledgements FLMM is grateful to FAPESP (Grant No. 00/ ) for financial support during a UL-Lafayette senior post-doctoral appointment. RB and FLMM thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for post-doctoral and research fellowships (Proc / and Proc /04-0, respectively). We thank Juan Bolaños for making available essential specimens of I. sawayai from Venezuela and Nilton José Hebling (Zoology Department, UNESP, Rio Claro, Brazil) whose work with hermits of Brazilian coast laid much of the groundwork for this project. Major support for analytical work on this project was provided under US National Science Foundation grants DEB and EF to D. L. Felder, along with FAPESP grants 98/ and support under the CNPq Prosul Program (Proc /2004-0) to FLMM. This is contribution No. 111 from the UL-Lafayette Laboratory for Crustacean Research. We thank Rafael Lemaitre, Chris Tudge and an anonymous reviewer for suggesting several revisions on the manuscript. All experiments conducted in this study comply with current applicable state and federal laws (IBAMA/ MMA sampling permission in Brazil Proc /98). REFERENCES AKAIKE H A new look at the statistical model identification. IEEE Transactions on Automatic Controls 19: BERNARDI J. V. E Desenvolvimento larval de Loxopagurus loxochelis (Moreira, 1901) (Crustacea, Decapoda, Diogenidae) em laboratório. Bachelor unpublished Thesis, UNESP, Rio Claro, Brazil, 33 p. BUCKLIN A., FROST B. W. & KOCHER T. D Molecular systematics of six Calanus and three Metridia species (Copepoda: Calanoida). Marine Biology 121: COSTA H. R Ocorrência do gênero Isocheles Stimpson na Costa Brasileira. Centro de Estudos Zoológicos 17/18: 1-3. FOREST J Sur un nouveau genre de Diogenidae (Crustacea Paguridea) de l Atlantique sud-américain, Loxopagurus gen. nov., établi pour Pagurus loxochelis Moreira. Zoologische Mededelingen 39: FOREST J. & SAINT LAURENT M. DE Campagne de la Calypso au large des côtes atlantiques de l Amérique du Sud ( ). 6. Crustacés décapodes : pagurides. Annales de l Institut océanographique, Monaco 45: HALL T. A BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: HARRISON J. S Evolution, biogeography, and the utility of mitochondrial 16s and COI genes in phylogenetic analysis of the crab genus Austinixa. Molecular Phylogenetic and Evolution 30: HARTL D. L. & CLARK A. G Principles of Population Genetics. 2nd ed. Sinauer Associates, Sunderland, 682 p. HARVEY A. W. & SANTO E. M. DE On the status of Pachycheles laevidactylus Ortmann, 1892 (Crustacea: Decapoda: Porcellanidae). Proceedings of Biological Society of Washington 109 (4): HEBLING N. J., NEGREIROS-FRANSOZO M. L. & BERNARDI J. V. E Larval development of Loxopagurus loxochelis (Moreira, 1901) (Crustacea, Decapoda, Diogenidae), obtained in laboratory, in I Congresso Brasileiro sobre Crustáceos 18: 51. HEDGES S. B The origin and evolution of model organisms. Nature Reviews Genetics 3: MACHORDOM A. & MACPHERSON E Rapid radiation and cryptic speciation in squat lobsters of the genus Munida (Crustacea, Decapoda) and related genera in the South West Pacific: molecular and morphological evidence. Molecular Phylogenetics and Evolution 33: MANTELATTO F. L. M., MARTINELLI J. M. & FRANSOZO A Temporal-spatial distribution of the hermit crab Loxopagurus loxochelis (Decapoda: Diogenidae) from Ubatuba Bay, São Paulo State, Brazil. Revista de Biologia Tropical 52 (1): MANTELATTO F. L. M., ROBLES R. & FELDER D. L. in press. Molecular phylogeny and taxonomic ap- 504
11 Sequence analysis of Loxopagurus and Isocheles proach of the crab genus Portunus (Crustacea, Portunidae) from Western Atlantic. Zoological Journal of the Linnean Society. MARTINELLI J. M., MANTELATTO F. L. M. & FRANSOZO A Population structure and breeding season of the South Atlantic hermit crab, Loxopagurus loxochelis (Anomura, Diogenidae) from Ubatuba region, Brazil. Crustaceana 75 (6): MATHEWS L. M., SCHUBART C. D., NEIGEL J. E. & FELDER D. L Genetic, ecological, and behavioural divergence between two sibling snapping shrimp species (Crustacea: Decapoda: Alpheus). Molecular Ecology 11: MELO G. A. S Manual de identifi cação dos Crustacea Decapoda do litoral brasileiro: Anomura, Thalassinidea, Palinuridea, Astacidea. Plêiade, São Paulo, 551 p. MCLAUGHLIN P. A Hermit crabs are they really polyphyletic? Journal of Crustacean Biology 3: MCLAUGHLIN P. A Illustrated keys to families and genera of the superfamily Paguroidea (Crustacea: Decapoda: Anomura), with diagnoses of genera of Paguridae, in LEMAITRE R. & TUDGE C. C. (eds), Biology of the Anomura. Proceedings of a symposium at the Fifth International Crustacean Congress, Melbourne, Australia, 9-13 July Memoirs of Museum Victoria 60 (1): MOREIRA C Contribuições para o conhecimento da fauna brasileira. Crustáceos do Brasil. Archivos do Museo Nacional do Rio de Janeiro 11: 1-151, pls 1-5. MORRISON C. L., HARVEY A. W., LAVERY S., TIEU K., HUANG Y. & CUNNINGHAM C. W Mitochondrial gene rearrangements confirm the parallel evolution of the crab-like form. Proceedings of the Royal Society of London, Part B, Biological Sciences 269 (1489): MORRISON C. L., RIOS R. & DUFFY J. E Phylogenetic evidence for an ancient rapid radiation of Caribbean sponge-dwelling snapping shrimps (Synalpheus). Molecular Phylogenetics and Evolution 30: NEGREIROS-FRANSOZO M. L. & HEBLING N. J Desenvolvimento pós-embrionário de Isocheles sawayai Forest & Saint Laurent, 1968 (Decapoda, Diogenidae) em laboratório. Papéis Avulsos de Zoologia 35 (4): NUCCI P. R Taxonomia e biogeografia da Superfamília Paguroidea Latreille (Crustacea, Decapoda, Anomura) no litoral brasileiro. Ph.D. unpublished Thesis, UNESP, Rio Claro, Brazil, 218 p. NUCCI P. R. & MELO G. A. S Range extensions for eight species of western atlantic hermit crabs (Crustacea, Paguroidea). Nauplius 8 (1): PÉREZ-LOSADA M., JARA C. G., BOND-BUCKUP G. & CRANDALL K. A Phylogenetic relationships among the species of Aegla (Anomura: Aeglidae) freshwater crabs from Chile. Journal of Crustacean Biology 22 (2): POSADA D. & CRANDALL K. A MODELTEST: testing the model of DNA substitution. Bioinformatics 14 (9): PROVENZANO A. J. JR The shallow-water hermit crabs of Florida. Bulletin of Marine Science of the Gulf of Caribbean 9 (4): ROBLES R., SCHUBART C. D., CONDE J. E., CARMONA- SUÁREZ C., ALVAREZ F., VILLALOBOS J. L. & FELDER D. L. in press. Molecular phylogeny of the American Callinectes Stimpson, 1860 (Brachyura: Portunidae), based on two partial mitochondrial genes. Marine Biology. RODRÍGUEZ G Los Crustaceos Decápodos de Venezuela. IVIC, Caracas, 494 p. RODRÍGUEZ R., OLIVER J. L., MARTIN A. & MEDINA J. R The general stochastic model of nucleotide substitution. Journal of Theoretical Biology 142: SCELZO M. A., MANTELATTO F. L. M. & TUDGE C. C Spermatophore morphology of the endemic hermit crab Loxopagurus loxochelis (Anomura, Diogenidae) from Southwestern Atlantic-Brazil and Argentina. Invertebrate Reproduction and Development 46 (1): 1-9. SCELZO M. A., MEDINA A. & TUDGE C. C. in press. Spermatozoal ultrastructure of the hermit crab Loxopagurus loxochelis (Moreira, 1901) (Anomura, Diogenidae) from the Southwestern Atlantic. Crustacean Research. SCHUBART C. D., NEIGEL J. E. & FELDER D. L. 2000a. Use of the mitochondrial 16S rrna gene for phylogenetic and population studies of Crustacea. Crustacean Issues 12: SCHUBART C. D., CUESTA J. A., DIESEL R. & FELDER D. L. 2000b. Molecular phylogeny, taxonomy, and evolution of nonmarine lineages within the American grapsoid crabs (Crustacea: Brachyura). Molecular Phylogenetics and Evolution 15 (2): SCHUBART C. D., CUESTA J. A. & FELDER D. L Glyptograpsidae, a new brachyuran family from Central America: larval and adult morphology, and a molecular phylogeny of the Grapsoidea. Journal of Crustacean Biology 22: STILLMAN J. H. & REEB C. A Molecular phylogeny of eastern Pacific porcelain crabs, genera Petrolisthes and Pachycheles, based on the mtdna 16S rdna sequence: phylogeographic and systematic implications. Molecular Phylogenetics and Evolution 19: SWOFFORD D. L PAUP. Phylogenetic Analysis Using Parsimony (and Other Methods). Version 4. Sinauer Associates, Sunderland, Masschusetts. 505
12 Mantelatto F. L. M. et al. TUDGE C. C. & CUNNINGHAM C Molecular phylogeny of the mud lobsters and mud shrimps (Crustacea: Decapoda: Thalassinidea) using nuclear 18S rdna and mitochondrial 16S rdna. Invertebrate Systematics 16: YOUNG A. M., TORRES C., MACK J. E. & CUNNINGHAM C Morphological and genetic evidence for vicariance and refugium in Atlantic and Gulf of Mexico populations of the hermit crab Pagurus longicarpus. Marine Biology 140: Submitted on 2 November 2005; accepted on 6 January
Supplementary Figures
 Supplementary Figures Supplementary Figure 1. Time-scaled phylogeny of Anomura, coloured to show superfamilies. This tree is the Maximum Agreement Subtree (MAST) computed from 432 MPTs of length 2548 in
Supplementary Figures Supplementary Figure 1. Time-scaled phylogeny of Anomura, coloured to show superfamilies. This tree is the Maximum Agreement Subtree (MAST) computed from 432 MPTs of length 2548 in
Molecular phylogeny of the western Atlantic species of the genus Portunus (Crustacea, Brachyura, Portunidae)
 Blackwell Publishing LtdOxford, UKZOJZoological Journal of the Linnean Society0024-4082The Linnean Society of London, 2007? 2007 1501 211220 Original Article PHYLOGENY OF PORTUNUS FROM ATLANTICF. L. MANTELATTO
Blackwell Publishing LtdOxford, UKZOJZoological Journal of the Linnean Society0024-4082The Linnean Society of London, 2007? 2007 1501 211220 Original Article PHYLOGENY OF PORTUNUS FROM ATLANTICF. L. MANTELATTO
Spatial and seasonal distribution of the hermit crab Pagurus exilis (Benedict, 1892) (Decapoda: Paguridae) in the southwestern coast of Brazil
 Revista de Biología Marina y Oceanografía 41(1): 87 95, julio de 2006 Spatial and seasonal distribution of the hermit crab Pagurus exilis (Benedict, 1892) (Decapoda: Paguridae) in the southwestern coast
Revista de Biología Marina y Oceanografía 41(1): 87 95, julio de 2006 Spatial and seasonal distribution of the hermit crab Pagurus exilis (Benedict, 1892) (Decapoda: Paguridae) in the southwestern coast
Nauplius. Gastropod shell availability as a potential resource for the hermit crab infralittoral fauna of Anchieta Island (SP), Brazil.
 11(2), 99-105, 2003 99 Gastropod shell availability as a potential resource for the hermit crab infralittoral fauna of Anchieta Island (SP), Brazil. Meireles, A. L.; Biagi, R. and Mantelatto, F. L. Departamento
11(2), 99-105, 2003 99 Gastropod shell availability as a potential resource for the hermit crab infralittoral fauna of Anchieta Island (SP), Brazil. Meireles, A. L.; Biagi, R. and Mantelatto, F. L. Departamento
Gilson Stanski 1, Fernando L. Mantelatto 2 and Antonio Leão Castilho 1
 Nauplius the Journal of the BraZIlIan crustacean society e-issn 2358-2936 www.scielo.br/nau www.crustacea.org.br original article Habitat heterogeneity in the assemblages and shell use by the most abundant
Nauplius the Journal of the BraZIlIan crustacean society e-issn 2358-2936 www.scielo.br/nau www.crustacea.org.br original article Habitat heterogeneity in the assemblages and shell use by the most abundant
Revista Chilena de Historia Natural ISSN: X Sociedad de Biología de Chile Chile
 Revista Chilena de Historia Natural ISSN: 0716-078X editorial@revchilhistnat.com Sociedad de Biología de Chile Chile BIAGI, RENATA; MEIRELES, ANDREA L.; SCELZO, MARCELO A.; MANTELATTO, FERNANDO L. Comparative
Revista Chilena de Historia Natural ISSN: 0716-078X editorial@revchilhistnat.com Sociedad de Biología de Chile Chile BIAGI, RENATA; MEIRELES, ANDREA L.; SCELZO, MARCELO A.; MANTELATTO, FERNANDO L. Comparative
Mitochondrial gene rearrangements confirm the parallel evolution of the crab-like form
 FirstCite e-publishing Received 6 July 2001 Accepted 4 October 2001 Published online Mitochondrial gene rearrangements confirm the parallel evolution of the crab-like form C. L. Morrison 1, A. W. Harvey
FirstCite e-publishing Received 6 July 2001 Accepted 4 October 2001 Published online Mitochondrial gene rearrangements confirm the parallel evolution of the crab-like form C. L. Morrison 1, A. W. Harvey
Egg production and shell relationship of the land hermit crab Coenobita scaevola (Anomura: Coenobitidae) from Wadi El-Gemal, Red Sea, Egypt
 The Journal of Basic & Applied Zoology (212) 65, 133 138 The Egyptian German Society for Zoology The Journal of Basic & Applied Zoology www.egsz.org www.sciencedirect.com Egg production and shell relationship
The Journal of Basic & Applied Zoology (212) 65, 133 138 The Egyptian German Society for Zoology The Journal of Basic & Applied Zoology www.egsz.org www.sciencedirect.com Egg production and shell relationship
Revista de Biología Tropical ISSN: Universidad de Costa Rica Costa Rica
 Revista de Biología Tropical ISSN: 0034-7744 rbt@cariari.ucr.ac.cr Universidad de Costa Rica Costa Rica Mantelatto, Fernando L.; Biagi, Renata; Meireles, Andrea L.; Scelzo, Marcelo A. Shell preference
Revista de Biología Tropical ISSN: 0034-7744 rbt@cariari.ucr.ac.cr Universidad de Costa Rica Costa Rica Mantelatto, Fernando L.; Biagi, Renata; Meireles, Andrea L.; Scelzo, Marcelo A. Shell preference
Post-doc fellowships to non-eu researchers FINAL REPORT. Home Institute: Centro de Investigaciones Marinas, Universidad de La Habana, CUBA
 Recipient: Maickel Armenteros Almanza. Post-doc fellowships to non-eu researchers FINAL REPORT Home Institute: Centro de Investigaciones Marinas, Universidad de La Habana, CUBA Promoter: Prof. Dr. Wilfrida
Recipient: Maickel Armenteros Almanza. Post-doc fellowships to non-eu researchers FINAL REPORT Home Institute: Centro de Investigaciones Marinas, Universidad de La Habana, CUBA Promoter: Prof. Dr. Wilfrida
POPULATION BIOLOGY OF THE HERMIT CRAB PETROCHIRUS DIOGENES (LINNAEUS, 1758) IN SOUTHERN BRAZIL
 POPULATION BIOLOGY OF THE HERMIT CRAB PETROCHIRUS DIOGENES (LINNAEUS, 1758) IN SOUTHERN BRAZIL Alexander Turra 1 ; Joaquim Olinto Branco 2 ; Flávio Xavier Souto 2 ABSTRACT. The aim of this study was to
POPULATION BIOLOGY OF THE HERMIT CRAB PETROCHIRUS DIOGENES (LINNAEUS, 1758) IN SOUTHERN BRAZIL Alexander Turra 1 ; Joaquim Olinto Branco 2 ; Flávio Xavier Souto 2 ABSTRACT. The aim of this study was to
Calcinus tibicen (Herbst, 1791) fail to. Silvia Sayuri Mandai 1 Raquel Corrêa Buranelli 1 Fernando Luis Mantelatto 1
 Nauplius the JoURnAl of the BRAZIlIAn crustacean society e-issn 2358-2936 www.scielo.br/nau www.crustacea.org.br This article is part of the special series offered by the Brazilian Crustacean Society in
Nauplius the JoURnAl of the BRAZIlIAn crustacean society e-issn 2358-2936 www.scielo.br/nau www.crustacea.org.br This article is part of the special series offered by the Brazilian Crustacean Society in
Dr. Amira A. AL-Hosary
 Phylogenetic analysis Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic Basics: Biological
Phylogenetic analysis Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic Basics: Biological
Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut
 Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic analysis Phylogenetic Basics: Biological
Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic analysis Phylogenetic Basics: Biological
Algorithms in Bioinformatics
 Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri Distance Methods Character Methods
Algorithms in Bioinformatics Sami Khuri Department of Computer Science San José State University San José, California, USA khuri@cs.sjsu.edu www.cs.sjsu.edu/faculty/khuri Distance Methods Character Methods
ZOOGEOGRAPHY OF HERMIT CRABS (DECAPODA: DIOGENIDAE, PAGURIDAE) FROM FOUR COASTAL LAGOONS IN THE GULF OF MEXICO
 JOURNAL OF CRUSTACEAN BIOLOGY, 24(4): 625 636, 2004 ZOOGEOGRAPHY OF HERMIT CRABS (DECAPODA: DIOGENIDAE, PAGURIDAE) FROM FOUR COASTAL LAGOONS IN THE GULF OF MEXICO A. Raz-Guzman, A. J. Sánchez, P. Peralta,
JOURNAL OF CRUSTACEAN BIOLOGY, 24(4): 625 636, 2004 ZOOGEOGRAPHY OF HERMIT CRABS (DECAPODA: DIOGENIDAE, PAGURIDAE) FROM FOUR COASTAL LAGOONS IN THE GULF OF MEXICO A. Raz-Guzman, A. J. Sánchez, P. Peralta,
Consensus Methods. * You are only responsible for the first two
 Consensus Trees * consensus trees reconcile clades from different trees * consensus is a conservative estimate of phylogeny that emphasizes points of agreement * philosophy: agreement among data sets is
Consensus Trees * consensus trees reconcile clades from different trees * consensus is a conservative estimate of phylogeny that emphasizes points of agreement * philosophy: agreement among data sets is
Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2018 University of California, Berkeley
 Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2018 University of California, Berkeley B.D. Mishler Feb. 14, 2018. Phylogenetic trees VI: Dating in the 21st century: clocks, & calibrations;
Integrative Biology 200 "PRINCIPLES OF PHYLOGENETICS" Spring 2018 University of California, Berkeley B.D. Mishler Feb. 14, 2018. Phylogenetic trees VI: Dating in the 21st century: clocks, & calibrations;
(Stevens 1991) 1. morphological characters should be assumed to be quantitative unless demonstrated otherwise
 Bot 421/521 PHYLOGENETIC ANALYSIS I. Origins A. Hennig 1950 (German edition) Phylogenetic Systematics 1966 B. Zimmerman (Germany, 1930 s) C. Wagner (Michigan, 1920-2000) II. Characters and character states
Bot 421/521 PHYLOGENETIC ANALYSIS I. Origins A. Hennig 1950 (German edition) Phylogenetic Systematics 1966 B. Zimmerman (Germany, 1930 s) C. Wagner (Michigan, 1920-2000) II. Characters and character states
Eriocheir sinensis H. Milne Edwards: 1853 or 1854 Grapsidae or Varunidae?
 Aquatic Invasions (2006) Volume 1, Issue 1: 17-27 DOI 10.3391/ai.2006.1.1.5 2006 The Author(s) Journal compilation 2006 REABIC (http://www.reabic.net) This is an Open Access article Research article Eriocheir
Aquatic Invasions (2006) Volume 1, Issue 1: 17-27 DOI 10.3391/ai.2006.1.1.5 2006 The Author(s) Journal compilation 2006 REABIC (http://www.reabic.net) This is an Open Access article Research article Eriocheir
Gulf and Caribbean Research
 Gulf and Caribbean Research Volume 19 Issue 1 January 27 Abundance and Ecological Distribution of the "Sete-Barbas" Shrimp Xiphopenaeus kroyeri (Heller, 1862) (Decapoda: Penaeoidea) in Three Bays of the
Gulf and Caribbean Research Volume 19 Issue 1 January 27 Abundance and Ecological Distribution of the "Sete-Barbas" Shrimp Xiphopenaeus kroyeri (Heller, 1862) (Decapoda: Penaeoidea) in Three Bays of the
Phylogenies Scores for Exhaustive Maximum Likelihood and Parsimony Scores Searches
 Int. J. Bioinformatics Research and Applications, Vol. x, No. x, xxxx Phylogenies Scores for Exhaustive Maximum Likelihood and s Searches Hyrum D. Carroll, Perry G. Ridge, Mark J. Clement, Quinn O. Snell
Int. J. Bioinformatics Research and Applications, Vol. x, No. x, xxxx Phylogenies Scores for Exhaustive Maximum Likelihood and s Searches Hyrum D. Carroll, Perry G. Ridge, Mark J. Clement, Quinn O. Snell
Bioinformatics tools for phylogeny and visualization. Yanbin Yin
 Bioinformatics tools for phylogeny and visualization Yanbin Yin 1 Homework assignment 5 1. Take the MAFFT alignment http://cys.bios.niu.edu/yyin/teach/pbb/purdue.cellwall.list.lignin.f a.aln as input and
Bioinformatics tools for phylogeny and visualization Yanbin Yin 1 Homework assignment 5 1. Take the MAFFT alignment http://cys.bios.niu.edu/yyin/teach/pbb/purdue.cellwall.list.lignin.f a.aln as input and
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics
 POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
POPULATION GENETICS Winter 2005 Lecture 17 Molecular phylogenetics - in deriving a phylogeny our goal is simply to reconstruct the historical relationships between a group of taxa. - before we review the
Phylogenetic diversity and conservation
 Phylogenetic diversity and conservation Dan Faith The Australian Museum Applied ecology and human dimensions in biological conservation Biota Program/ FAPESP Nov. 9-10, 2009 BioGENESIS Providing an evolutionary
Phylogenetic diversity and conservation Dan Faith The Australian Museum Applied ecology and human dimensions in biological conservation Biota Program/ FAPESP Nov. 9-10, 2009 BioGENESIS Providing an evolutionary
Applications of Genetics to Conservation Biology
 Applications of Genetics to Conservation Biology Molecular Taxonomy Populations, Gene Flow, Phylogeography Relatedness - Kinship, Paternity, Individual ID Conservation Biology Population biology Physiology
Applications of Genetics to Conservation Biology Molecular Taxonomy Populations, Gene Flow, Phylogeography Relatedness - Kinship, Paternity, Individual ID Conservation Biology Population biology Physiology
Phylogenetic analyses. Kirsi Kostamo
 Phylogenetic analyses Kirsi Kostamo The aim: To construct a visual representation (a tree) to describe the assumed evolution occurring between and among different groups (individuals, populations, species,
Phylogenetic analyses Kirsi Kostamo The aim: To construct a visual representation (a tree) to describe the assumed evolution occurring between and among different groups (individuals, populations, species,
ANALYSIS OF POPULATIONAL STRUCTURE OF THE HERMIT CRAB DARDANUS INSIGNIS
 ANALYSIS OF POPULATIONAL STRUCTURE OF THE HERMIT CRAB DARDANUS INSIGNIS (ANOMURA: DIOGENIDAE) NEAR COASTAL ISLANDS IN SOUTHEASTERN BRAZIL: A STUDY OF 14 YEARS AGO FRAMESCHI, I.F. 1 *; ANDRADE, L.S. 1 ;
ANALYSIS OF POPULATIONAL STRUCTURE OF THE HERMIT CRAB DARDANUS INSIGNIS (ANOMURA: DIOGENIDAE) NEAR COASTAL ISLANDS IN SOUTHEASTERN BRAZIL: A STUDY OF 14 YEARS AGO FRAMESCHI, I.F. 1 *; ANDRADE, L.S. 1 ;
CHACEON RAMOSAE, A NEW DEEP-WATER CRAB FROM BRAZIL (CRUSTACEA: DECAPODA: GERYONIDAE)
 CHACEON RAMOSAE, A NEW DEEP-WATER CRAB FROM BRAZIL (CRUSTACEA: DECAPODA: GERYONIDAE) Raymond B. Manning, Marcos Siqueira Tavares, and Elaine Figueiredo Albuquerque 18 October 1989 PROC. BIOL. SOC. WASH.
CHACEON RAMOSAE, A NEW DEEP-WATER CRAB FROM BRAZIL (CRUSTACEA: DECAPODA: GERYONIDAE) Raymond B. Manning, Marcos Siqueira Tavares, and Elaine Figueiredo Albuquerque 18 October 1989 PROC. BIOL. SOC. WASH.
The Phylogenetic Reconstruction of the Grass Family (Poaceae) Using matk Gene Sequences
 The Phylogenetic Reconstruction of the Grass Family (Poaceae) Using matk Gene Sequences by Hongping Liang Dissertation submitted to the Faculty of the Virginia Polytechnic Institute and State University
The Phylogenetic Reconstruction of the Grass Family (Poaceae) Using matk Gene Sequences by Hongping Liang Dissertation submitted to the Faculty of the Virginia Polytechnic Institute and State University
Anais da Academia Brasileira de Ciências ISSN: Academia Brasileira de Ciências Brasil
 Anais da Academia Brasileira de Ciências ISSN: 0001-3765 aabc@abc.org.br Academia Brasileira de Ciências Brasil Biagi, Renata; L. Meireles, Andrea; L. Mantelatto, Fernando Bio-ecological aspects of the
Anais da Academia Brasileira de Ciências ISSN: 0001-3765 aabc@abc.org.br Academia Brasileira de Ciências Brasil Biagi, Renata; L. Meireles, Andrea; L. Mantelatto, Fernando Bio-ecological aspects of the
Effects of Gap Open and Gap Extension Penalties
 Brigham Young University BYU ScholarsArchive All Faculty Publications 200-10-01 Effects of Gap Open and Gap Extension Penalties Hyrum Carroll hyrumcarroll@gmail.com Mark J. Clement clement@cs.byu.edu See
Brigham Young University BYU ScholarsArchive All Faculty Publications 200-10-01 Effects of Gap Open and Gap Extension Penalties Hyrum Carroll hyrumcarroll@gmail.com Mark J. Clement clement@cs.byu.edu See
PHYLOGENY AND SYSTEMATICS
 AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 11 Chapter 26 Activity #15 NAME DATE PERIOD PHYLOGENY AND SYSTEMATICS PHYLOGENY Evolutionary history of species or group of related species SYSTEMATICS Study
AP BIOLOGY EVOLUTION/HEREDITY UNIT Unit 1 Part 11 Chapter 26 Activity #15 NAME DATE PERIOD PHYLOGENY AND SYSTEMATICS PHYLOGENY Evolutionary history of species or group of related species SYSTEMATICS Study
Lecture 11 Friday, October 21, 2011
 Lecture 11 Friday, October 21, 2011 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean system
Lecture 11 Friday, October 21, 2011 Phylogenetic tree (phylogeny) Darwin and classification: In the Origin, Darwin said that descent from a common ancestral species could explain why the Linnaean system
Departamento de Zoologia, Universidade Federal Do Rio Grande Do Sul, Porto Alegre, Brazil 3
 Syst. Biol. 53(5):767 780, 2004 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150490522331 Molecular Systematics and Biogeography of the Southern
Syst. Biol. 53(5):767 780, 2004 Copyright c Society of Systematic Biologists ISSN: 1063-5157 print / 1076-836X online DOI: 10.1080/10635150490522331 Molecular Systematics and Biogeography of the Southern
Shell utilization by the land hermit crab Coenobita scaevola (Anomura, Coenobitidae) from Wadi El-Gemal, Red Sea
 Belg. J. Zool., 138 (1) : 13-19 January 2008 Shell utilization by the land hermit crab Coenobita scaevola (Anomura, Coenobitidae) from Wadi El-Gemal, Red Sea Sallam WS 1, Mantelatto FL 2* & Hanafy MH 1
Belg. J. Zool., 138 (1) : 13-19 January 2008 Shell utilization by the land hermit crab Coenobita scaevola (Anomura, Coenobitidae) from Wadi El-Gemal, Red Sea Sallam WS 1, Mantelatto FL 2* & Hanafy MH 1
Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition
 Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition David D. Pollock* and William J. Bruno* *Theoretical Biology and Biophysics, Los Alamos National
Assessing an Unknown Evolutionary Process: Effect of Increasing Site- Specific Knowledge Through Taxon Addition David D. Pollock* and William J. Bruno* *Theoretical Biology and Biophysics, Los Alamos National
Molecular Markers, Natural History, and Evolution
 Molecular Markers, Natural History, and Evolution Second Edition JOHN C. AVISE University of Georgia Sinauer Associates, Inc. Publishers Sunderland, Massachusetts Contents PART I Background CHAPTER 1:
Molecular Markers, Natural History, and Evolution Second Edition JOHN C. AVISE University of Georgia Sinauer Associates, Inc. Publishers Sunderland, Massachusetts Contents PART I Background CHAPTER 1:
BINF6201/8201. Molecular phylogenetic methods
 BINF60/80 Molecular phylogenetic methods 0-7-06 Phylogenetics Ø According to the evolutionary theory, all life forms on this planet are related to one another by descent. Ø Traditionally, phylogenetics
BINF60/80 Molecular phylogenetic methods 0-7-06 Phylogenetics Ø According to the evolutionary theory, all life forms on this planet are related to one another by descent. Ø Traditionally, phylogenetics
SPECIATION. REPRODUCTIVE BARRIERS PREZYGOTIC: Barriers that prevent fertilization. Habitat isolation Populations can t get together
 SPECIATION Origin of new species=speciation -Process by which one species splits into two or more species, accounts for both the unity and diversity of life SPECIES BIOLOGICAL CONCEPT Population or groups
SPECIATION Origin of new species=speciation -Process by which one species splits into two or more species, accounts for both the unity and diversity of life SPECIES BIOLOGICAL CONCEPT Population or groups
Does Petrolisthes armatus (Anomura, Porcellanidae) form a Species Complex or Are We Dealing with Just One Widely Distributed Species?
 Zoological Studies 50(3): 372-384 (2011) Does Petrolisthes armatus (Anomura, Porcellanidae) form a Species Complex or Are We Dealing with Just One Widely Distributed Species? Fernando L. Mantelatto 1,
Zoological Studies 50(3): 372-384 (2011) Does Petrolisthes armatus (Anomura, Porcellanidae) form a Species Complex or Are We Dealing with Just One Widely Distributed Species? Fernando L. Mantelatto 1,
CHUCOA ILICIFOLIA, A SPINY ONOSERIS (ASTERACEAE, MUTISIOIDEAE: ONOSERIDEAE)
 Phytologia (December 2009) 91(3) 537 CHUCOA ILICIFOLIA, A SPINY ONOSERIS (ASTERACEAE, MUTISIOIDEAE: ONOSERIDEAE) Jose L. Panero Section of Integrative Biology, 1 University Station, C0930, The University
Phytologia (December 2009) 91(3) 537 CHUCOA ILICIFOLIA, A SPINY ONOSERIS (ASTERACEAE, MUTISIOIDEAE: ONOSERIDEAE) Jose L. Panero Section of Integrative Biology, 1 University Station, C0930, The University
Letter to the Editor. Department of Biology, Arizona State University
 Letter to the Editor Traditional Phylogenetic Reconstruction Methods Reconstruct Shallow and Deep Evolutionary Relationships Equally Well Michael S. Rosenberg and Sudhir Kumar Department of Biology, Arizona
Letter to the Editor Traditional Phylogenetic Reconstruction Methods Reconstruct Shallow and Deep Evolutionary Relationships Equally Well Michael S. Rosenberg and Sudhir Kumar Department of Biology, Arizona
Constructing Evolutionary/Phylogenetic Trees
 Constructing Evolutionary/Phylogenetic Trees 2 broad categories: Distance-based methods Ultrametric Additive: UPGMA Transformed Distance Neighbor-Joining Character-based Maximum Parsimony Maximum Likelihood
Constructing Evolutionary/Phylogenetic Trees 2 broad categories: Distance-based methods Ultrametric Additive: UPGMA Transformed Distance Neighbor-Joining Character-based Maximum Parsimony Maximum Likelihood
C3020 Molecular Evolution. Exercises #3: Phylogenetics
 C3020 Molecular Evolution Exercises #3: Phylogenetics Consider the following sequences for five taxa 1-5 and the known outgroup O, which has the ancestral states (note that sequence 3 has changed from
C3020 Molecular Evolution Exercises #3: Phylogenetics Consider the following sequences for five taxa 1-5 and the known outgroup O, which has the ancestral states (note that sequence 3 has changed from
PGA: A Program for Genome Annotation by Comparative Analysis of. Maximum Likelihood Phylogenies of Genes and Species
 PGA: A Program for Genome Annotation by Comparative Analysis of Maximum Likelihood Phylogenies of Genes and Species Paulo Bandiera-Paiva 1 and Marcelo R.S. Briones 2 1 Departmento de Informática em Saúde
PGA: A Program for Genome Annotation by Comparative Analysis of Maximum Likelihood Phylogenies of Genes and Species Paulo Bandiera-Paiva 1 and Marcelo R.S. Briones 2 1 Departmento de Informática em Saúde
INCREASED RATES OF MOLECULAR EVOLUTION IN AN EQUATORIAL PLANT CLADE: AN EFFECT OF ENVIRONMENT OR PHYLOGENETIC NONINDEPENDENCE?
 Evolution, 59(1), 2005, pp. 238 242 INCREASED RATES OF MOLECULAR EVOLUTION IN AN EQUATORIAL PLANT CLADE: AN EFFECT OF ENVIRONMENT OR PHYLOGENETIC NONINDEPENDENCE? JEREMY M. BROWN 1,2 AND GREGORY B. PAULY
Evolution, 59(1), 2005, pp. 238 242 INCREASED RATES OF MOLECULAR EVOLUTION IN AN EQUATORIAL PLANT CLADE: AN EFFECT OF ENVIRONMENT OR PHYLOGENETIC NONINDEPENDENCE? JEREMY M. BROWN 1,2 AND GREGORY B. PAULY
Inferring phylogeny. Today s topics. Milestones of molecular evolution studies Contributions to molecular evolution
 Today s topics Inferring phylogeny Introduction! Distance methods! Parsimony method!"#$%&'(!)* +,-.'/01!23454(6!7!2845*0&4'9#6!:&454(6 ;?@AB=C?DEF Overview of phylogenetic inferences Methodology Methods
Today s topics Inferring phylogeny Introduction! Distance methods! Parsimony method!"#$%&'(!)* +,-.'/01!23454(6!7!2845*0&4'9#6!:&454(6 ;?@AB=C?DEF Overview of phylogenetic inferences Methodology Methods
Social shrimp (Crustacea: Decapoda: Alpheidae: Synalpheus) Resources for teaching
 Social shrimp (Crustacea: Decapoda: Alpheidae: Synalpheus) Resources for teaching All images copyright J. Emmett Duffy (jeduffy@vims.edu) These images and materials may be freely used for educational,
Social shrimp (Crustacea: Decapoda: Alpheidae: Synalpheus) Resources for teaching All images copyright J. Emmett Duffy (jeduffy@vims.edu) These images and materials may be freely used for educational,
Phylogenetics: Building Phylogenetic Trees
 1 Phylogenetics: Building Phylogenetic Trees COMP 571 Luay Nakhleh, Rice University 2 Four Questions Need to be Answered What data should we use? Which method should we use? Which evolutionary model should
1 Phylogenetics: Building Phylogenetic Trees COMP 571 Luay Nakhleh, Rice University 2 Four Questions Need to be Answered What data should we use? Which method should we use? Which evolutionary model should
DNA Barcoding and taxonomy of Glossina
 DNA Barcoding and taxonomy of Glossina Dan Masiga Molecular Biology and Biotechnology Department, icipe & Johnson Ouma Trypanosomiasis Research Centre, KARI The taxonomic problem Following ~250 years of
DNA Barcoding and taxonomy of Glossina Dan Masiga Molecular Biology and Biotechnology Department, icipe & Johnson Ouma Trypanosomiasis Research Centre, KARI The taxonomic problem Following ~250 years of
Constructing Evolutionary/Phylogenetic Trees
 Constructing Evolutionary/Phylogenetic Trees 2 broad categories: istance-based methods Ultrametric Additive: UPGMA Transformed istance Neighbor-Joining Character-based Maximum Parsimony Maximum Likelihood
Constructing Evolutionary/Phylogenetic Trees 2 broad categories: istance-based methods Ultrametric Additive: UPGMA Transformed istance Neighbor-Joining Character-based Maximum Parsimony Maximum Likelihood
Phylogenetic Tree Reconstruction
 I519 Introduction to Bioinformatics, 2011 Phylogenetic Tree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Evolution theory Speciation Evolution of new organisms is driven
I519 Introduction to Bioinformatics, 2011 Phylogenetic Tree Reconstruction Yuzhen Ye (yye@indiana.edu) School of Informatics & Computing, IUB Evolution theory Speciation Evolution of new organisms is driven
Phylogenetic inference
 Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
Phylogenetic inference Bas E. Dutilh Systems Biology: Bioinformatic Data Analysis Utrecht University, March 7 th 016 After this lecture, you can discuss (dis-) advantages of different information types
A Study of the Moss Parasite Eocronartium muscicola By: Alicia Knudson Advisor: Dr. Elizabeth Frieders
 A Study of the Moss Parasite Eocronartium muscicola By: Alicia Knudson Advisor: Dr. Elizabeth Frieders Abstract The genus Eocronartium contains a single described species of parasitic fungus on moss plants
A Study of the Moss Parasite Eocronartium muscicola By: Alicia Knudson Advisor: Dr. Elizabeth Frieders Abstract The genus Eocronartium contains a single described species of parasitic fungus on moss plants
A (short) introduction to phylogenetics
 A (short) introduction to phylogenetics Thibaut Jombart, Marie-Pauline Beugin MRC Centre for Outbreak Analysis and Modelling Imperial College London Genetic data analysis with PR Statistics, Millport Field
A (short) introduction to phylogenetics Thibaut Jombart, Marie-Pauline Beugin MRC Centre for Outbreak Analysis and Modelling Imperial College London Genetic data analysis with PR Statistics, Millport Field
Chapter 16: Reconstructing and Using Phylogenies
 Chapter Review 1. Use the phylogenetic tree shown at the right to complete the following. a. Explain how many clades are indicated: Three: (1) chimpanzee/human, (2) chimpanzee/ human/gorilla, and (3)chimpanzee/human/
Chapter Review 1. Use the phylogenetic tree shown at the right to complete the following. a. Explain how many clades are indicated: Three: (1) chimpanzee/human, (2) chimpanzee/ human/gorilla, and (3)chimpanzee/human/
Resultats DES Campagnes Musorstom: Vol 17 (Memoires Du Museum National D'Histoire Naturelle) (French Edition) READ ONLINE
 Resultats DES Campagnes Musorstom: Vol 17 (Memoires Du Museum National D'Histoire Naturelle) (French Edition) READ ONLINE If you are looking for a ebook Resultats DES Campagnes Musorstom: Vol 17 (Memoires
Resultats DES Campagnes Musorstom: Vol 17 (Memoires Du Museum National D'Histoire Naturelle) (French Edition) READ ONLINE If you are looking for a ebook Resultats DES Campagnes Musorstom: Vol 17 (Memoires
What is Phylogenetics
 What is Phylogenetics Phylogenetics is the area of research concerned with finding the genetic connections and relationships between species. The basic idea is to compare specific characters (features)
What is Phylogenetics Phylogenetics is the area of research concerned with finding the genetic connections and relationships between species. The basic idea is to compare specific characters (features)
Article. Taxonomic re-examination of the hermit crab species Pagurus forceps and Pagurus comptus (Decapoda: Paguridae) by molecular analysis
 Zootaxa 2133: 20 32 (2009) www.mapress.com/zootaxa/ opyright 2009 Magnolia Press rticle ISSN 11755326 (print edition) ZOOX ISSN 11755334 (online edition) axonomic reexamination of the hermit crab species
Zootaxa 2133: 20 32 (2009) www.mapress.com/zootaxa/ opyright 2009 Magnolia Press rticle ISSN 11755326 (print edition) ZOOX ISSN 11755334 (online edition) axonomic reexamination of the hermit crab species
Intraspecific gene genealogies: trees grafting into networks
 Intraspecific gene genealogies: trees grafting into networks by David Posada & Keith A. Crandall Kessy Abarenkov Tartu, 2004 Article describes: Population genetics principles Intraspecific genetic variation
Intraspecific gene genealogies: trees grafting into networks by David Posada & Keith A. Crandall Kessy Abarenkov Tartu, 2004 Article describes: Population genetics principles Intraspecific genetic variation
DISTRIBUTION AND ASSEMBLAGES OF ANOMURAN CRUSTACEANS IN UBATUBA BAY, NORTH COAST OF SAO PAULO STATE, BRAZIL
 ta Biol. Venez., Vol. 8 (): 5 Diciembre,8 DISTRIBUTION AND ASSEMBLAGES OF ANOMURAN CRUSTACEANS IN UBATUBA BAY, NORTH COAST OF SAO PAULO STATE, BRAZIL DISTRIBUCION Y ENSAMBLAJE DE CRUSTACEOS ANOMUROS EN
ta Biol. Venez., Vol. 8 (): 5 Diciembre,8 DISTRIBUTION AND ASSEMBLAGES OF ANOMURAN CRUSTACEANS IN UBATUBA BAY, NORTH COAST OF SAO PAULO STATE, BRAZIL DISTRIBUCION Y ENSAMBLAJE DE CRUSTACEOS ANOMUROS EN
Phylogenetics: Building Phylogenetic Trees. COMP Fall 2010 Luay Nakhleh, Rice University
 Phylogenetics: Building Phylogenetic Trees COMP 571 - Fall 2010 Luay Nakhleh, Rice University Four Questions Need to be Answered What data should we use? Which method should we use? Which evolutionary
Phylogenetics: Building Phylogenetic Trees COMP 571 - Fall 2010 Luay Nakhleh, Rice University Four Questions Need to be Answered What data should we use? Which method should we use? Which evolutionary
Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week (Jan 27 & 29):
 Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week (Jan 27 & 29): Statistical estimation of models of sequence evolution Phylogenetic inference using maximum likelihood:
Biology 559R: Introduction to Phylogenetic Comparative Methods Topics for this week (Jan 27 & 29): Statistical estimation of models of sequence evolution Phylogenetic inference using maximum likelihood:
Composition, density, and shell use of hermit crabs (Crustacea: Paguroidea) from subtidal boulder fields in southeastern Brazil
 Lat. Am. J. Aquat. Res., 46(1): 72-82, 2018 DOI: 10.3856/vol46-issue1-fulltext-9 Composition, density, and shell use of hermit crabs 721 Research Article Composition, density, and shell use of hermit crabs
Lat. Am. J. Aquat. Res., 46(1): 72-82, 2018 DOI: 10.3856/vol46-issue1-fulltext-9 Composition, density, and shell use of hermit crabs 721 Research Article Composition, density, and shell use of hermit crabs
EVOLUTIONARY DISTANCES
 EVOLUTIONARY DISTANCES FROM STRINGS TO TREES Luca Bortolussi 1 1 Dipartimento di Matematica ed Informatica Università degli studi di Trieste luca@dmi.units.it Trieste, 14 th November 2007 OUTLINE 1 STRINGS:
EVOLUTIONARY DISTANCES FROM STRINGS TO TREES Luca Bortolussi 1 1 Dipartimento di Matematica ed Informatica Università degli studi di Trieste luca@dmi.units.it Trieste, 14 th November 2007 OUTLINE 1 STRINGS:
Molecular phylogeny How to infer phylogenetic trees using molecular sequences
 Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 2009 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 2009 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Molecular phylogeny How to infer phylogenetic trees using molecular sequences
 Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 200 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Molecular phylogeny How to infer phylogenetic trees using molecular sequences ore Samuelsson Nov 200 Applications of phylogenetic methods Reconstruction of evolutionary history / Resolving taxonomy issues
Taxonomy. Content. How to determine & classify a species. Phylogeny and evolution
 Taxonomy Content Why Taxonomy? How to determine & classify a species Domains versus Kingdoms Phylogeny and evolution Why Taxonomy? Classification Arrangement in groups or taxa (taxon = group) Nomenclature
Taxonomy Content Why Taxonomy? How to determine & classify a species Domains versus Kingdoms Phylogeny and evolution Why Taxonomy? Classification Arrangement in groups or taxa (taxon = group) Nomenclature
Bayesian Inference using Markov Chain Monte Carlo in Phylogenetic Studies
 Bayesian Inference using Markov Chain Monte Carlo in Phylogenetic Studies 1 What is phylogeny? Essay written for the course in Markov Chains 2004 Torbjörn Karfunkel Phylogeny is the evolutionary development
Bayesian Inference using Markov Chain Monte Carlo in Phylogenetic Studies 1 What is phylogeny? Essay written for the course in Markov Chains 2004 Torbjörn Karfunkel Phylogeny is the evolutionary development
ESS 345 Ichthyology. Systematic Ichthyology Part II Not in Book
 ESS 345 Ichthyology Systematic Ichthyology Part II Not in Book Thought for today: Now, here, you see, it takes all the running you can do, to keep in the same place. If you want to get somewhere else,
ESS 345 Ichthyology Systematic Ichthyology Part II Not in Book Thought for today: Now, here, you see, it takes all the running you can do, to keep in the same place. If you want to get somewhere else,
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2009 University of California, Berkeley
 "PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2009 University of California, Berkeley B.D. Mishler Jan. 22, 2009. Trees I. Summary of previous lecture: Hennigian
"PRINCIPLES OF PHYLOGENETICS: ECOLOGY AND EVOLUTION" Integrative Biology 200B Spring 2009 University of California, Berkeley B.D. Mishler Jan. 22, 2009. Trees I. Summary of previous lecture: Hennigian
Supplementary Information
 Supplementary Information Supplementary Figure 1. Schematic pipeline for single-cell genome assembly, cleaning and annotation. a. The assembly process was optimized to account for multiple cells putatively
Supplementary Information Supplementary Figure 1. Schematic pipeline for single-cell genome assembly, cleaning and annotation. a. The assembly process was optimized to account for multiple cells putatively
Chapter 26: Phylogeny and the Tree of Life Phylogenies Show Evolutionary Relationships
 Chapter 26: Phylogeny and the Tree of Life You Must Know The taxonomic categories and how they indicate relatedness. How systematics is used to develop phylogenetic trees. How to construct a phylogenetic
Chapter 26: Phylogeny and the Tree of Life You Must Know The taxonomic categories and how they indicate relatedness. How systematics is used to develop phylogenetic trees. How to construct a phylogenetic
Phylogenetic Analysis. Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center
 Phylogenetic Analysis Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center Outline Basic Concepts Tree Construction Methods Distance-based methods
Phylogenetic Analysis Han Liang, Ph.D. Assistant Professor of Bioinformatics and Computational Biology UT MD Anderson Cancer Center Outline Basic Concepts Tree Construction Methods Distance-based methods
Phylogenetic Diversity and distribution patterns of the Compositae family in the high Andes of South America
 Phylogenetic Diversity and distribution patterns of the Compositae family in the high Andes of South America Scherson, R.A., Naulin,P.I., Albornoz, A., Hagemann, T., Vidal, P.M., Riveros, N., and Arroyo,
Phylogenetic Diversity and distribution patterns of the Compositae family in the high Andes of South America Scherson, R.A., Naulin,P.I., Albornoz, A., Hagemann, T., Vidal, P.M., Riveros, N., and Arroyo,
A B S T R A C T R E S U M O
 BRAZILIAN JOURNAL OF OCEANOGRAPHY, 60(3):299-310, 2012 COMPARATIVE ANALYSIS OF SHELL OCCUPATION BY TWO SOUTHERN POPULATIONS OF THE HERMIT CRAB Loxopagurus loxochelis (DECAPODA, DIOGENIDAE) Luciane Ayres-Peres
BRAZILIAN JOURNAL OF OCEANOGRAPHY, 60(3):299-310, 2012 COMPARATIVE ANALYSIS OF SHELL OCCUPATION BY TWO SOUTHERN POPULATIONS OF THE HERMIT CRAB Loxopagurus loxochelis (DECAPODA, DIOGENIDAE) Luciane Ayres-Peres
OMICS Journals are welcoming Submissions
 OMICS Journals are welcoming Submissions OMICS International welcomes submissions that are original and technically so as to serve both the developing world and developed countries in the best possible
OMICS Journals are welcoming Submissions OMICS International welcomes submissions that are original and technically so as to serve both the developing world and developed countries in the best possible
Phylogenetic Trees. Phylogenetic Trees Five. Phylogeny: Inference Tool. Phylogeny Terminology. Picture of Last Quagga. Importance of Phylogeny 5.
 Five Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu v Distance Methods v Character Methods v Molecular Clock v UPGMA v Maximum Parsimony
Five Sami Khuri Department of Computer Science San José State University San José, California, USA sami.khuri@sjsu.edu v Distance Methods v Character Methods v Molecular Clock v UPGMA v Maximum Parsimony
STEM-hy: Species Tree Estimation using Maximum likelihood (with hybridization)
 STEM-hy: Species Tree Estimation using Maximum likelihood (with hybridization) Laura Salter Kubatko Departments of Statistics and Evolution, Ecology, and Organismal Biology The Ohio State University kubatko.2@osu.edu
STEM-hy: Species Tree Estimation using Maximum likelihood (with hybridization) Laura Salter Kubatko Departments of Statistics and Evolution, Ecology, and Organismal Biology The Ohio State University kubatko.2@osu.edu
DNA Sequencing as a Method for Larval Identification in Odonates
 DNA Sequencing as a Method for Larval Identification in Odonates Adeline Harris 121 North St Apt 3 Farmington, ME 04938 Adeline.harris@maine.edu Christopher Stevens 147 Main St Apt 1 Farmington, ME 04938
DNA Sequencing as a Method for Larval Identification in Odonates Adeline Harris 121 North St Apt 3 Farmington, ME 04938 Adeline.harris@maine.edu Christopher Stevens 147 Main St Apt 1 Farmington, ME 04938
Molecular Phylogenetics of Mole Crabs (Hippidae : Emerita)
 The University of Maine DigitalCommons@UMaine Marine Sciences Faculty Scholarship School of Marine Sciences 11-1-2002 Molecular Phylogenetics of Mole Crabs (Hippidae : Emerita) P. A. Haye Y. K. Tam Irv
The University of Maine DigitalCommons@UMaine Marine Sciences Faculty Scholarship School of Marine Sciences 11-1-2002 Molecular Phylogenetics of Mole Crabs (Hippidae : Emerita) P. A. Haye Y. K. Tam Irv
"Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky
 MOLECULAR PHYLOGENY "Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky EVOLUTION - theory that groups of organisms change over time so that descendeants differ structurally
MOLECULAR PHYLOGENY "Nothing in biology makes sense except in the light of evolution Theodosius Dobzhansky EVOLUTION - theory that groups of organisms change over time so that descendeants differ structurally
Ph ylogeography. A guide to the study of the spatial distribution of Seahorses. By Leila Mougoui Bakhtiari
 Ph ylogeography A guide to the study of the spatial distribution of Seahorses By Leila Mougoui Bakhtiari Contents An Introduction to Phylogeography JT Bohem s Resarch Map of erectu s migration Conservation
Ph ylogeography A guide to the study of the spatial distribution of Seahorses By Leila Mougoui Bakhtiari Contents An Introduction to Phylogeography JT Bohem s Resarch Map of erectu s migration Conservation
8/23/2014. Phylogeny and the Tree of Life
 Phylogeny and the Tree of Life Chapter 26 Objectives Explain the following characteristics of the Linnaean system of classification: a. binomial nomenclature b. hierarchical classification List the major
Phylogeny and the Tree of Life Chapter 26 Objectives Explain the following characteristics of the Linnaean system of classification: a. binomial nomenclature b. hierarchical classification List the major
Estimating Phylogenies (Evolutionary Trees) II. Biol4230 Thurs, March 2, 2017 Bill Pearson Jordan 6-057
 Estimating Phylogenies (Evolutionary Trees) II Biol4230 Thurs, March 2, 2017 Bill Pearson wrp@virginia.edu 4-2818 Jordan 6-057 Tree estimation strategies: Parsimony?no model, simply count minimum number
Estimating Phylogenies (Evolutionary Trees) II Biol4230 Thurs, March 2, 2017 Bill Pearson wrp@virginia.edu 4-2818 Jordan 6-057 Tree estimation strategies: Parsimony?no model, simply count minimum number
Molecular phylogeny - Using molecular sequences to infer evolutionary relationships. Tore Samuelsson Feb 2016
 Molecular phylogeny - Using molecular sequences to infer evolutionary relationships Tore Samuelsson Feb 2016 Molecular phylogeny is being used in the identification and characterization of new pathogens,
Molecular phylogeny - Using molecular sequences to infer evolutionary relationships Tore Samuelsson Feb 2016 Molecular phylogeny is being used in the identification and characterization of new pathogens,
Decapod Crustacean Phylogenetics. Joel W. Martin, Keith A. Crandall, and Darryl L. Felder CRUSTACEAN ISSUES ] 3. %. m. edited by
![Decapod Crustacean Phylogenetics. Joel W. Martin, Keith A. Crandall, and Darryl L. Felder CRUSTACEAN ISSUES ] 3. %. m. edited by Decapod Crustacean Phylogenetics. Joel W. Martin, Keith A. Crandall, and Darryl L. Felder CRUSTACEAN ISSUES ] 3. %. m. edited by](/thumbs/86/94754723.jpg) CRUSTACEAN ISSUES ] 3 II %. m Decapod Crustacean Phylogenetics edited by Joel W. Martin, Keith A. Crandall, and Darryl L. Felder \ CRC Press J Taylor & Francis Group Decapod Crustacean Phylogenetics Edited
CRUSTACEAN ISSUES ] 3 II %. m Decapod Crustacean Phylogenetics edited by Joel W. Martin, Keith A. Crandall, and Darryl L. Felder \ CRC Press J Taylor & Francis Group Decapod Crustacean Phylogenetics Edited
Phylogenetic Trees. What They Are Why We Do It & How To Do It. Presented by Amy Harris Dr Brad Morantz
 Phylogenetic Trees What They Are Why We Do It & How To Do It Presented by Amy Harris Dr Brad Morantz Overview What is a phylogenetic tree Why do we do it How do we do it Methods and programs Parallels
Phylogenetic Trees What They Are Why We Do It & How To Do It Presented by Amy Harris Dr Brad Morantz Overview What is a phylogenetic tree Why do we do it How do we do it Methods and programs Parallels
Systematics - Bio 615
 Bayesian Phylogenetic Inference 1. Introduction, history 2. Advantages over ML 3. Bayes Rule 4. The Priors 5. Marginal vs Joint estimation 6. MCMC Derek S. Sikes University of Alaska 7. Posteriors vs Bootstrap
Bayesian Phylogenetic Inference 1. Introduction, history 2. Advantages over ML 3. Bayes Rule 4. The Priors 5. Marginal vs Joint estimation 6. MCMC Derek S. Sikes University of Alaska 7. Posteriors vs Bootstrap
Chapter 26 Phylogeny and the Tree of Life
 Chapter 26 Phylogeny and the Tree of Life Chapter focus Shifting from the process of how evolution works to the pattern evolution produces over time. Phylogeny Phylon = tribe, geny = genesis or origin
Chapter 26 Phylogeny and the Tree of Life Chapter focus Shifting from the process of how evolution works to the pattern evolution produces over time. Phylogeny Phylon = tribe, geny = genesis or origin
Bioinformatics 1. Sepp Hochreiter. Biology, Sequences, Phylogenetics Part 4. Bioinformatics 1: Biology, Sequences, Phylogenetics
 Bioinformatics 1 Biology, Sequences, Phylogenetics Part 4 Sepp Hochreiter Klausur Mo. 30.01.2011 Zeit: 15:30 17:00 Raum: HS14 Anmeldung Kusss Contents Methods and Bootstrapping of Maximum Methods Methods
Bioinformatics 1 Biology, Sequences, Phylogenetics Part 4 Sepp Hochreiter Klausur Mo. 30.01.2011 Zeit: 15:30 17:00 Raum: HS14 Anmeldung Kusss Contents Methods and Bootstrapping of Maximum Methods Methods
Inferring phylogeny. Constructing phylogenetic trees. Tõnu Margus. Bioinformatics MTAT
 Inferring phylogeny Constructing phylogenetic trees Tõnu Margus Contents What is phylogeny? How/why it is possible to infer it? Representing evolutionary relationships on trees What type questions questions
Inferring phylogeny Constructing phylogenetic trees Tõnu Margus Contents What is phylogeny? How/why it is possible to infer it? Representing evolutionary relationships on trees What type questions questions
Reconstructing the history of lineages
 Reconstructing the history of lineages Class outline Systematics Phylogenetic systematics Phylogenetic trees and maps Class outline Definitions Systematics Phylogenetic systematics/cladistics Systematics
Reconstructing the history of lineages Class outline Systematics Phylogenetic systematics Phylogenetic trees and maps Class outline Definitions Systematics Phylogenetic systematics/cladistics Systematics
InDel 3-5. InDel 8-9. InDel 3-5. InDel 8-9. InDel InDel 8-9
 Lecture 5 Alignment I. Introduction. For sequence data, the process of generating an alignment establishes positional homologies; that is, alignment provides the identification of homologous phylogenetic
Lecture 5 Alignment I. Introduction. For sequence data, the process of generating an alignment establishes positional homologies; that is, alignment provides the identification of homologous phylogenetic
Invertebrate Reproduction & Development. ISSN: (Print) (Online) Journal homepage:
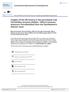 Invertebrate Reproduction & Development ISSN: 0792-4259 (Print) 2157-0272 (Online) Journal homepage: http://www.tandfonline.com/loi/tinv20 Insights of the life history in the porcellanid crab Petrolisthes
Invertebrate Reproduction & Development ISSN: 0792-4259 (Print) 2157-0272 (Online) Journal homepage: http://www.tandfonline.com/loi/tinv20 Insights of the life history in the porcellanid crab Petrolisthes
Estimating Evolutionary Trees. Phylogenetic Methods
 Estimating Evolutionary Trees v if the data are consistent with infinite sites then all methods should yield the same tree v it gets more complicated when there is homoplasy, i.e., parallel or convergent
Estimating Evolutionary Trees v if the data are consistent with infinite sites then all methods should yield the same tree v it gets more complicated when there is homoplasy, i.e., parallel or convergent
METHODS FOR DETERMINING PHYLOGENY. In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task.
 Chapter 12 (Strikberger) Molecular Phylogenies and Evolution METHODS FOR DETERMINING PHYLOGENY In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task. Modern
Chapter 12 (Strikberger) Molecular Phylogenies and Evolution METHODS FOR DETERMINING PHYLOGENY In Chapter 11, we discovered that classifying organisms into groups was, and still is, a difficult task. Modern
Amy Driskell. Laboratories of Analytical Biology National Museum of Natural History Smithsonian Institution, Wash. DC
 DNA Barcoding Amy Driskell Laboratories of Analytical Biology National Museum of Natural History Smithsonian Institution, Wash. DC 1 Outline 1. Barcoding in general 2. Uses & Examples 3. Barcoding Bocas
DNA Barcoding Amy Driskell Laboratories of Analytical Biology National Museum of Natural History Smithsonian Institution, Wash. DC 1 Outline 1. Barcoding in general 2. Uses & Examples 3. Barcoding Bocas
Project Budget: State Wildlife Grant Requested: $16,412 Project Match (UARK in kind services): $5,744 Total Project Cost: $22,156
 Project Title: Genetic examination of the Ringed Crayfish species group, with special emphasis on the endemic Gapped Ringed Crayfish (Orconectes neglectus chaenodactylus) Project Summary: Morphological
Project Title: Genetic examination of the Ringed Crayfish species group, with special emphasis on the endemic Gapped Ringed Crayfish (Orconectes neglectus chaenodactylus) Project Summary: Morphological
