Binary Regulation of Hippo Pathway by Merlin/NF2, Kibra, Lgl, and Melted Specifies and Maintains Postmitotic Neuronal Fate
|
|
|
- Franklin Fields
- 5 years ago
- Views:
Transcription
1 Article Binary Regulation of Hippo Pathway by Merlin/NF2, Kibra, Lgl, and Melted Specifies and Maintains Postmitotic Neuronal Fate David Jukam 1 and Claude Desplan 1, * 1 Center for Developmental Genetics, Department of Biology, New York University, 100 Washington Square East, New York, NY , USA *Correspondence: cd38@nyu.edu DOI /j.devcel SUMMARY Patterning the Drosophila retina for color vision relies on postmitotic specification of photoreceptor subtypes. R8 photoreceptors express one of two lightsensing Rhodopsins, or. This fate decision involves a bistable feedback loop between Melted, a PH-domain protein, and Warts, a kinase in the Hippo growth pathway. Here, we show that a subset of the Hippo pathway Merlin, Kibra, and Lethal(2) giant larvae (Lgl), but not Expanded or Fat is required for Warts expression and activity in R8 to specify fate. Melted represses warts transcription to disrupt Hippo pathway activity and specify fate. Therefore, R8 Hippo signaling exhibits ON-or-OFF regulation, promoting mutually exclusive fates. Furthermore, Merlin and Lgl are continuously required to maintain R8 neuronal subtypes. These results reveal roles for Merlin, Kibra, and Lgl in neuronal specification and maintenance and show that the Hippo pathway is reimplemented for sensory neuron fate by combining canonical and noncanonical regulatory steps. INTRODUCTION In neural development, neurons progressively differentiate toward terminal fates, culminating in the postmitotic specialization of subtypes with subtle, yet critical differences. For example, sensory neurons of the same class often express one sensory receptor from among several options to endow each neuron with a precise functional identity. Although neural progenitor specification (Jessell, 2000) and the role of transcription factors in generating neural diversity (Hobert, 2008) are well studied, the mechanisms by which signaling pathways control terminal neuronal subtype specification events, like mutually exclusive sensory receptor expression, are less well understood. Here, we use terminal differentiation of R8 photoreceptor subtypes in the Drosophila eye as a model to study how an otherwise equipotent precursor cell uses a signaling pathway to instruct one of two alternate fates. The Drosophila compound eye comprises 800 ommatidia, or unit eyes, that each contains 8 photoreceptor neurons (Hardie, 1985) that express light-sensitive Rh proteins and transmit visual information to the brain. Six outer photoreceptors (R1 R6) express Rh1 and, like vertebrate rods, mediate motion detection and dim light vision (Yamaguchi et al., 2008). Two inner photoreceptors, R7 and R8, are involved in color vision, analogous to vertebrate cones, and detect light by expressing one of four Rhs of different wavelength sensitivity that define ommatidial subtypes (reviewed in Rister and Desplan, 2011). In yellow (y) ommatidia, yr7 expresses long UV-sensitive Rh4, and yr8 expresses green-sensitive, whereas in pale (p) ommatidia, pr7 expresses short UV-sensitive Rh3, and pr8 expresses blue-sensitive. y and p subtypes are distributed stochastically in a roughly 70% y to 30% p ratio (Figures 1A and 1D). R7 and R8 coordinate their subtype identities in two sequential postmitotic fate decisions. First, stochastic expression of the transcription factor gene spineless (ss) in 70% of R7 cells induces yr7 fate and rh4 (Wernet et al., 2006). pr7s without ss express rh3 and instruct the underlying R8 to become pr8 and express rh5. The remaining yr8s express rh6 by default (Chou et al., 1999). Two proteins previously known as growth regulators the NDR Ser/Thr kinase Warts (Wts), and a PH domain-containing protein, Melted (Melt) control postmitotic specification of R8 subtypes (Figure 1B) (Mikeladze-Dvali et al., 2005). melt is necessary and sufficient for pr8 fate and rh5 expression (Figure 1E), whereas wts is necessary and sufficient for yr8 and rh6 (Figure 1F). wts and melt repress each other s transcription in a double-negative, bistable feedback loop that directs robust expression of either rh5 or rh6 in R8. The bistable loop ensures an unambiguous R8 fate decision in response to an instructive signal from the R7 cell. Wts is a tumor suppressor and nexus of the Hippo pathway, which coordinates the balance between proliferation and apoptosis (Figure 1C) (reviewed in Pan, 2010; Halder and Johnson, 2011). Wts inhibits growth by phosphorylating the transcriptional coactivator Yorkie (Yki) in the cytoplasm, preventing it from entering the nucleus (Huang et al., 2005; Dong et al., 2007; Oh and Irvine, 2008) where Yki would otherwise interact with transcription factors such as Scalloped (Sd) or Homothorax (Hth) (Goulev et al., 2008; Wu et al., 2008; Zhang et al., 2008; Peng et al., 2009) to upregulate pro-proliferation and antiapoptosis target genes. Wts is activated upstream by another Ser/Thr kinase, Hippo (Hpo), bound by the scaffolding protein Salvador (Sav) and cofactor Mob as tumor suppressor (Mats) (Harvey and Tapon, 21, 1 14, November 15, 2011 ª2011 Elsevier Inc. 1
2 A B C R7 R8 Rh3 Rh1 Rh1 R1 R6 ~30% p Rh4 Rh1 Rh1 R1 R6 ~70% y R7 R8 Rh3 melt wts Rh4 melt wts dco fat dachs crb ex kib mer lgl hpo sav mats wts apkc pro-proliferation & anti-apoptosis genes drassf D E F G H Wild-type I melt 1 J wts P1 K L mats RNAi mer 3 M kib RNAi ex AP50 fat 8 dachs GC13 dco 3 N 100 Hippo core complex Merlin, Kibra Expanded Fat, Dachs, Dco drassf % % Wild-Type range { 0 y 1 w 67 (control) melt 1 warts P1 hpo sav shrp1 ey,lgmr>matsrnai KK, Dcr2 ey,lgmr>matsrnai TRiP, Dcr2 ey,lgmr>dcr2 (control) ey,lgmr>kibrnai KK, Dcr2 mer 3 mer 4 mer 4 /+ (control) ex e1 ex e1 /+ ex AP49 fat 8 fat 8 /+ Dachs GC13 Dachs GC13 /+ dco 3 dco 3 /+ fat 8, Dachs GC13 fat 8, Dachs GC13 /+ ex e1 Dachs GC13 ex e1 Dachs GC13 /+ drassf44.2 Figure 1. A Mer/Kib Branch of the Hippo Pathway Regulates R8 Subtype Specification (A) R7/R8 Rhodopsin pairing defines ommatidial subtypes. Outer photoreceptors (R1 R6) express rh1, whereas R7/R8 primarily express either rh3/rh5 or rh4/rh6. (B) A bistable feedback loop regulates mutually exclusive and expression in R8: warts and melted are expressed in y and p R8, respectively, and repress each other s transcription. (C) The Hippo-signaling network regulates proliferation and cell death. Upstream inputs converge onto the Hpo/Sav/Mats/Wts core complex to regulate Wts activity. (D M) Confocal images of adult retinas stained with (blue) and (red) antibodies that label R8 subtypes. (E), (F), (H), and (J) (M) are whole-mutant retinas. Genotypes are listed in the Supplemental Experimental Procedures. (D) Wild-type. Note the roughly 70:30 ratio of :. (E) melt 1. (F) wts P1. (G) Retina expressing mats-rnai. (H) mer 3. (I) Retina expressing kib-rnai. All R8s in kib del null mutants also express, but the kib del chromosome contains a mutation in rh6 preventing analysis of expression (data not shown). (J) ex AP50. (K) fat 8. (L) dachs GC13. (M) dco 3. (N) Quantification of R8 subtypes in Hippo pathway mutants. Graph presents proportion of R8s (y axis) that contains (blue), (red), or coexpression (pink). Dashed lines indicate the wild-type range of 20% 45% based on counts in wild-type or balancer strains of various genetic backgrounds (data not shown). Heterozygote controls contained either a wild-type chromosome (chr), or, for mutants on chr X, FM7c; chr II, the CyO balancer; chr III, TM2 or TM6B. Mean % was compared with a two-tailed, unpaired t test; p < Wild-type: n = 9 retinas, n = 3055 ommatidia; mats-rnai: n = 8 retinas, n = 1561 ommatidia; kib-rnai: n = 11 retinas, n = 3214 ommatidia; ex e1 : n = 8 retinas, n = 1869 ommatidia; all others genotypes: n R 4 retinas, n R 800 ommatidia. Error bars are ±1 standard deviation. 2 21, 1 14, November 15, 2011 ª2011 Elsevier Inc.
3 2007). Further upstream are several independent or semiredundant input branches (Figure 1C): FERM domain proteins Expanded (Ex) and Merlin (Mer) (Hamaratoglu et al., 2006; Pellock et al., 2007; Tyler and Baker, 2007) can act with the WW-domain protein Kibra (Kib) to activate Hpo (Baumgartner et al., 2010; Genevet et al., 2010; Yu et al., 2010). The membrane protein Crumbs (Crb) mediates upstream activation of Ex (Chen et al., 2010; Ling et al., 2010; Robinson et al., 2010), and Ex can also directly inhibit Yki (Badouel et al., 2009; Oh and Irvine, 2009). Another input consists of the atypical cadherin, Fat, and the kinase Discs overgrown (Dco), which act through the unconventional myosin Dachs to control Wts protein levels (Bennett and Harvey, 2006; Cho et al., 2006; Silva et al., 2006; Willecke et al., 2006; Tyler and Baker, 2007). Finally, apicalbasal polarity regulators Lethal (2) giant larvae (Lgl) and atypical protein kinase C (apkc) act antagonistically to influence the Hippo pathway by unknown mechanisms, possibly by affecting the inhibition of Hpo by drassf (Polesello et al., 2006; Grzeschik et al., 2010). Mutations in fat, lgl, crb, ex, mer, hpo, sav, mats, and wts, or overexpression of yki, cause overgrowth in developing tissues, whereas pathway activation or yki depletion blocks proliferation and induces cell death (Pan, 2010). Many Hippo pathway members have orthologs implicated in human cancers, in particular, the mer ortholog, NF2, whose mutations cause Neurofibromatosis type 2 (NF-2) (MacCollin et al., 1993). The Hippo pathway also regulates nongrowth processes in Drosophila, implying that pathway regulation can be heterogeneous and context dependent. The Hippo pathway functions in autophagic cell death in salivary glands (Dutta and Baehrecke, 2008), apicobasal polarity (Hamaratoglu et al., 2009; Genevet et al., 2009), establishment and maintenance of dendritic arborizations (Emoto et al., 2006), and differentiation of the optic lobe neuroepithelia (Reddy et al., 2010). In these processes, wts is believed to be ubiquitously expressed and controlled posttranscriptionally to mediate its activation. R8 subtype specification is the only known cellular process in which wts is controlled transcriptionally through a genetic interaction with Melt. Melt modulates the TOR/Insulin-signaling pathway in fat metabolism (Teleman et al., 2005), yet mutants in other TOR/Insulin pathway genes do not exhibit R8 phenotypes (Mikeladze-Dvali et al., 2005), suggesting that Melt acts independently to regulate R8 subtypes. Whether Wts acts with its canonical signaling partners besides Hpo and Sav for cell fate determination, and how diverse biological processes such as tissue growth and sensory receptor expression can reuse the same signaling modules, is not known. Here, we report that Wts is regulated by the integration of canonical (tumor suppressor pathway) and noncanonical mechanisms for R8 subtype specification. mer, kib, lgl, and the hpo/ sav/mats/wts core complex specify yr8 subtypes, while Ex and Fat upstream inputs are not involved. mer and kib are required for wts expression and to repress melt such that the state of the wts-melt transcriptional feedback loop is coupled to upstream Hippo pathway signaling. Conditional inactivation of mer or lgl in adult flies reveals that continuous Hippo pathway activity also maintains mutually exclusive and expression, and melt can disrupt Hippo signaling long after R8 subtype specification has occurred. Therefore, constitutive and continuous activation by upstream Hippo inputs keeps Wts ON to specify yr8, while repression of wts transcription by Melt acts as a circuit breaker to switch pathway activity OFF to specify pr8. Such regulation allows Hippo pathway activity to depend on the presence or absence of Wts, which ensures the binary output of and. RESULTS To test whether the upstream Hippo growth network regulates Wts in R8 photoreceptor subtype specification, we genetically manipulated Mats, Hpo, Sav, Ex, Kib, Mer, Fat, Fj, Dco, Dachs, drassf, apkc, and Lgl and assayed for changes in the ratio of R8 subtypes as defined by the expression of and. The Hippo Core Complex Module Is Functionally Conserved in R8 Subtype Specification Wts, Hpo, Sav, and Mats physically interact to form the core complex of Hippo growth signaling (reviewed in Zhao et al., 2010). Yet core complex members can behave differently in distinct contexts. For instance, Hpo activates another NDR kinase, Tricornered (Trc), instead of Wts, to establish dendritic fields of larval body wall neurons (Emoto et al., 2006). In R8, mutations in hpo and sav phenocopy wts mutants (Mikeladze- Dvali et al., 2005). To confirm that the core complex functions canonically during R8 photoreceptor subtype specification, we analyzed mats and performed epistasis among mats, wts, and hpo. We generated wts, hpo, or sav whole-mutant eyes with the FLP/FRT system using eyeless-flp (ey-flp) and a chromosome containing a cell lethal mutation and GMR-hid that eliminates all nonhomozygous mutant cells in the eye (Stowers and Schwarz, 1999; Newsome et al., 2000). In these mutants, all R8s adopted the pr8 subtype fate and expressed (Figures 1F and 1N; see Figures S1D and S1E available online) (Mikeladze- Dvali et al., 2005). Because homozygous mats mutant eyes do not fully differentiate photoreceptors (Lai et al., 2005), we examined late mats function in the retina by expressing mats- RNAi with either the lgmr-gal4 driver, in cells posterior to the morphogenetic furrow (Wernet et al., 2003), or a combination of lgmr-gal4 and ey-gal4 in the entire eye. Three different mats-rnai transgenes resulted in loss of and expansion of (Figures 1G and 1N; Figures S1A, S1B, and S1F). Thus, all four members of the Hippo core complex are required to specify yr8. Expression of hpo in all photoreceptors (GMR-hpo) induced expression in all R8s (Figure S1G) (Mikeladze-Dvali et al., 2005). wts P1 hypomorphic mutant eyes suppressed the hpo misexpression phenotype, and about half of R8s expressed (Figure S1H). mats-rnai also suppressed the all hpo gain-of-function phenotype; ey,lgmr>matsrnai; GMRhpo retinas contained both and (80% of R8s expressed, 60% coexpressed and, and 20% only ) (Figure S1I). Similarly, removing hpo from lgmr>wts retinas reduced the proportion of R8s that expressed exclusively (data not shown). Thus, wts, hpo, and mats are required for each other s ability to promote yr8 fate and, consistent with functional conservation of the Hippo core complex in R8 subtype specification. 21, 1 14, November 15, 2011 ª2011 Elsevier Inc. 3
4 mer and kib Are Required to Specify R8 Photoreceptor Subtypes Independently of Upstream Hippo Pathway Regulators Ex and Fat To test whether upstream regulators of wts act in R8, we first generated whole-mer mutant eyes. Hypomorphic mer 3 led to expression in almost all (>99%) R8 cells, with concomitant loss of (Figure 1H). Similar results were obtained with null alleles (mer 1, mer 2, mer 4 ), in mer 4 mutant clones, or after eyespecific expression of mer-rnai (Figure 1N). Wild-type UASmer expressed in all photoreceptors using lgmr-gal4 rescued the mer 4 mutant phenotype. We next analyzed mutants for Kib, which acts in a complex with Mer and Ex to promote Hippo pathway activity. kib del null mutant retinas contained mostly -expressing R8s, indicating that kib is also required for yr8 fate. Eye-specific kib-rnai expression (ey+lgmr-gal4; UASkib-RNAi, UAS-Dcr2) led to similar results (Figure 1I). Thus, mer and kib are required to specify yr8 fate and induce expression. We also generated eyes mutant for ex, which acts upstream of wts for growth regulation, in part redundantly with mer (McCartney et al., 2000; Hamaratoglu et al., 2006). Three different alleles of ex (ex e1, ex AP49, ex AP50 ) did not significantly affect the R8 subtype ratio (Figures 1J and 1N; Figures S2A and S2B). To test for redundancy with mer, we removed one copy of mer in homozygous ex mutant clones and again observed no detectable difference in the : ratio (Figures S2C S2C 00 ), consistent with mer functioning nonredundantly in the Hippo pathway to specify R8 subtypes. Several other studies have suggested that mer has functions separate from ex in the Hippo pathway (Pellock et al., 2007; Bennett and Harvey, 2006; Cho et al., 2006; Silva et al., 2006; Willecke et al., 2006), but to our knowledge, this is the first example where mer mutant phenotypes perfectly match hpo, sav, and wts mutant phenotypes with ex completely dispensable (Figure 1N). Null or strong alleles of fat (fat 8, fat Grv, fat fd ), dco (dco 3 ), dachs (d GC13 ), and drassf (drassf X16, drassf 44.2 ), or ex e1, fat Gr-v double mutants, also all had normal pr8:yr8 ratios (Figures 1K 1N; Figures S2D S2D 0 ). Therefore, the Mer/Kib upstream branch of the Hippo pathway is required for R8 subtype specification, whereas the Fat/Fj/Dco/Dachs pathway, Ex, and drassf are not involved. Mer Requires Its BB Domain and C Terminus for Activity in R8 Subtype Specification For viability, Mer requires the N-terminal FERM domain, as well as portions of the protein C-terminal half (LaJeunesse et al., 1998). We tested whether these domains are also required for Mer in R8. Mer constructs missing either the C-terminal half (UAS-mer DC ) or the 7 aa Blue Box sequence ( 170 YQMTPEM 177 ) in the FERM domain (UAS-mer DBB ) were ectopically expressed in photoreceptors with lgmr-gal4. Almost all R8s expressed and not (Figures 2C and 2G), suggesting that these proteins act as dominant negatives. Furthermore, mer DBB and mer DC constructs expressed in mer whole-mutant eyes failed to rescue the loss of yr8s (>99% of R8s expressed ) (Figures 2D 2G). Thus, the 7 aa Blue Box domain, which is perfectly conserved in human NF2, and the C-terminal half of the protein are essential for Mer function in R8 subtype specification. Mer Acts Autonomously in R8 for Subtype Specification The R8 mer mutant phenotype is not due to general photoreceptor fate defects because mer retinas did not aberrantly express the cell fate markers Spalt (R7 and R8) and Rh1 (R1 R6) (Figures S3D S3D 0 and S3G S3G 0 ). Because pr8 fate depends on signaling from pr7, mer could affect R7 subtype specification, and only indirectly affect R8 rh expression. However, Rh3 and Rh4 protein expression was normal in mer or kib mutant retinas (Figures 2J 2J 000 ; Figures S3A S3B 0 ), leading to extensive Rh4/ (yr7/pr8) miscoupling (Figure 2I) never observed in wild-type. To assess whether mer acts cell autonomously in R8, we misexpressed the dominant-negative UAS-mer DBB using PanR8- Gal4, which is expressed exclusively in all R8s starting at 70% pupation, after subtypes are specified (Mikeladze-Dvali et al., 2005). PanR8>Mer DBB retinas exhibited a dramatic gain of (Figures 2K 2K 0 ), and partial loss of (Figure 2K 00 ), leading many R8s to coexpress and (Figures 2H and 2K). This is likely because the PanR8 driver is expressed late, after has been expressed. protein levels appeared lower in R8 cells that coexpressed (Figure 2K 00 ), confirming that yr8 subtype fate had been respecified to pr8. Thus, mer is specifically required in R8 to regulate R8 subtype fate. Kib and the Core Complex Are Necessary and Sufficient to Specify yr8 Fate We next tested if Mer was sufficient to induce yr8 fate in wildtype animals by using lgmr-gal4 to misexpress either a fulllength (UAS-mer + ) or a 35-aa-truncated protein (UAS -mer ) proposed to be an activated form (LaJeunesse et al., 1998). Neither Mer construct affected R8 subtypes (Figures 2A, 2B, and 2L). In contrast, lgmr>kib retinas exhibited a complete fate transformation, and all R8s expressed (Figure 2M), similar to misexpression of wts, sav, or hpo (Mikeladze-Dvali et al., 2005). Thus, kib is necessary and sufficient to induce yr8 fate. An activated, membrane-tethered Mats containing a myristoylation sequence (UAS-Myr-matsGFP)(Ho et al., 2010) driven with lgmr-gal4 also induced, with a corresponding loss of (Figure 2N). Late expression of Myr-matsGFP with PanR8-Gal4 was also sufficient to reprogram pr8 subtypes to express (Figure 2H). In contrast, expression of a dominant-negative construct with a mutated myristoylation sequence (lgmr-gal4, UAS-Myr G2A -matsgfp) induced (Figure S1C). Therefore, kib and all four core complex members are sufficient to induce yr8 subtypes. These results also support the idea that membrane localization of core complex proteins is important for their activation (Ho et al., 2010). mer might be permissively required for Hippo pathway activation in R8 because mer is necessary, but not sufficient, to induce the yr8 fate when ectopically expressed at high levels. Wts and Sav Require Mer to Induce the yr8 Subtype In the tumor suppressor pathway, Mer acts upstream of Wts to promote Wts activity (Hamaratoglu et al., 2006). However, removing mer in clonal patches of GMR-wts retinas completely suppressed the ability of wts to activate and led to a mer mutant phenotype (>99% of R8s expressed ) (Figures 3A 3A 00 ). All R8s in neighboring GMR-wts only tissue expressed 4 21, 1 14, November 15, 2011 ª2011 Elsevier Inc.
5 A B C lgmr-gal4 mer 4 ;lgmr-gal4 lgmr>mer + lgmr>mer ΔC D E F G mer + mer ΔC mer BB mer Merlin WT (635 aa) FERM (1-330) mer 4 ;lgmr>mer + % (wt) mer 4, lgmr> lgmr> 38% 43% 96% >99% 99% >99% 42% n.d. Rh4 Rh3 Rh4 GFP(wt) I J L R7 R8 mer 4 PanR8>mer BB lgmr>mer M H mer 4 ;lgmr>mer ΔC % 60 % 40 mer 4 clones lgmr>kibra Rh3 N 20 0 Rh4 PanR8-Gal4 (control) K K K PanR8>mer BB (DomNeg) J J J PanR8>>Myr-mats (activated Mats) merge lgmr>mats myr Figure 2. Mer Acts Specifically in R8 and Requires the FERM Domain and C Terminus to Regulate yr8 Photoreceptor Subtypes (A F and J N) Adult retinas stained for (blue) and (red). Full genotypes are listed in Supplemental Experimental Procedures. (A) lgmr-gal4 control. exclusively (Figure 3A 00 ). Similar results were obtained in mer mutant clones when sav (mer; GMR-sav) or sav and wts (mer; GMR-sav,wts) were misexpressed in all photoreceptors (Figures 3B 3B 00 ; data not shown). Therefore, mer might act downstream of the pathway, or it might act upstream but be required for the ability of sav and wts to regulate R8 subtype fate. To address these possibilities, we used Hpo to activate the pathway independently of mer. If mer acts upstream of hpo and wts, as in growth signaling, activated Hpo should suppress the mer phenotype. Hpo can auto-activate when expressed at high levels (Praskova et al., 2004). Indeed, lgmr-hpo led all R8s to express (data not shown; see Figure S1G) and suppressed the mer phenotype, inducing in most R8s (Figure 3C). Activated Mats (lgmr>myr-mats) also suppressed the mer mutant phenotype (Figures 3D and 3E). These results suggest that mer acts upstream of the hpo/sav/mats/wts core complex in R8, as in growth control. Moreover, because mer is required for wts and sav activity in R8, even when wts and sav are expressed at high levels, the Mer upstream branch is a critical activator of Wts for R8 subtype specification. ex and drassf Are Not Required for R8 Subtype Specification Intriguingly, two upstream genes whose functions are not required for R8 subtype determination, ex and drassf, were sufficient to alter R8 subtypes when misexpressed. lgmr>drassf induced, which led to partial coexpression of and (Figure S4B), whereas lgmr>ex mimicked Hippo pathway activation and induced gain of and loss of (Figure S4C). ex and drassf are likely sufficient to artificially perturb the Hippo pathway by performing their canonical activities Ex to activate Hpo or sequester Yki, drassf to inhibit Hpo when ectopically expressed at high levels. This (B) lgmr>mer+. (C) lgmr>mer DC. (D) mer 4 ; lgmr-gal4 (negative control for rescue experiments). (E) mer 4 ; lgmr>mer + (rescue of R8 subtype specification). (F) mer 4 ; lgmr>mer DC. (G) Mer deletion constructs (LaJeunesse et al., 1998) misexpressed in wildtype and mer 4 mutant retinas. Columns indicate percent of R8s that express. n.d., not determined. (H) Proportion of R8s that expresses (blue), (red), or both (purple) in PanR8>mer DBB (comparing %; n = 8 retinas, n = 2329 ommatidia), Pan- R8>Myr-mats (comparing %; n = 7 retinas, n = 2355 ommatidia), and PanR8-Gal4 controls (n = 7 retinas, n = 2131 ommatidia). Two-tailed, unpaired t test; p < Error bars are ±1 standard deviation. (I) Side view of mer 4 mutant ommatidia showing Rh4 (red) and (blue) miscoupling in the same ommatidium. Alexa Fluor 488-conjugated Phalloidin (green) stains actin and labels rhabdomeres (outer photoreceptors in image). (J J 00 ) mer 4 mutant clones (absence of GFP) stained with antibodies for R7 Rhs, Rh3 (blue), and Rh4 (red). (J 0 J 000 ) Dorsal third yr7 ommatidia, where Rh3 and Rh4 are coexpressed (white arrowheads) (Mazzoni et al., 2008). White box in (J) indicates area shown in adjacent panels. Dashed line indicates clone boundary. (K K 00 ) PanR8>mer DBB.(K 0 ). (K 00 ). Arrowheads show coexpression of and. Note the decrease in intensity in coexpressing cells (K 00 ). (L) lgmr>mer (M) lgmr>kib. (N) lgmr>myr-mats-gfp (activated mats containing a myristoylation sequence) The disorganized ommatidia and altered rhabdomere structure in some R8s are likely due to earlier growth functions of mats (Ho et al., 2010). 21, 1 14, November 15, 2011 ª2011 Elsevier Inc. 5
6 GFP GFP A A A mer 4 ; GMR-wts B mer 4 ; GMR-wts,sav C B D E mer 4 ; lgmr>hpo lgmr>mats myr mer 4 ; lgmr>mats myr Figure 3. Wts Requires Mer to Induce yr8 Subtype (A B 00 ) Adult retina stained with antibodies for (blue), (red), and GFP (green). Dashed lines indicate clonal boundaries. (A) mer 4 ; GMR-wts (mer 4 mutant clones [marked by absence of GFP] in retina that misexpresses wts in all photoreceptors). Note the presence of only in GMR-wts tissue when mer is wild-type (GFP + ), and absence of when mer is removed (non-gfp). (A 0 ) GFP only. (A 00 ) and only. (B) mer 4 ; GMR-sav,wts (mer 4 mutant clones marked by absence of GFP in retinas that misexpresses sav and wts in all photoreceptors). (B 0 ) GFP only. (B 00 ) and only. (C E) Adult retinas stained with antibodies for (blue) and (red). (C) mer 4 ; lgmr>hpo. Compare to Figure 2D (mer 4 mutants) and Figure S1H (GMR-hpo). Experiment was performed at 18 C because lgmr>hpo flies do not eclose at 25 C. (D) lgmr>myr-mats. (E) mer 4 ;lgmr>myr-mats. Compare to mer 4 mutants in Figure 2D. suggests that normal Ex and drassf protein function is not prohibited in R8 subtypes; rather, they might be sequestered from the Wts/Hpo complex, or not highly expressed in postmitotic R8s. Indeed, Ex levels are lower posterior to the morphogenetic furrow and in pupation relative to the larval eye disc (Milton et al., 2010). Supporting this hypothesis, we were unable to detect a difference in Ex immunoreactivity between wild-type and ex mutant clones in adult photoreceptors (data not shown), and temporal RNA-seq data from Drosophila modencode indicate that drassf reaches its lowest expression level during late pupation (Graveley et al., 2011). A Role for Lgl and apkc in R8 Subtype Specification We next tested lgl, a neoplastic tumor suppressor and apicobasal cell polarity regulator that genetically interacts with the Hippo B pathway in the developing eye disc (Grzeschik et al., 2010). lgl 4 mutant clones contained R8s that expressed almost exclusively, with little or no (Figures 4A 4A 00 ). lgl-rnai also led to induction of and loss of (ey,lgmr>lgl-rnai: 80.6%, 11.7%, 7.7% coexpression), as did a conditional allele (lgl ts3 ) at the nonpermissive temperature (Figures 4B 4D and 4G). The lgl mutant defect is specific to R8, because R7 and outer photoreceptor Rh expression was unaffected (Figures S3C S3C 0 and S3F S3F 0 ). Loss of lgl, or RNAi targeting another apicobasal polarity regulator, bazooka, severely disrupted photoreceptor polarity and rhabdomere morphology, as measured by F-actin localization (Figures S5A S5A 00 ). But only lgl affected the : ratio, suggesting that the lgl mutant yr8 specification defect is not due to general cell polarity aberrations. We investigated another polarity regulator, apkc, which acts antagonistically to Lgl (Lee et al., 2006), and is genetically linked to the Hippo growth pathway (Grzeschik et al., 2010). Overexpression of an activated apkc targeted to the plasma membrane with a CAAX prenylation motif (lgmr>apkc CAAX-WT ) (Sotillos et al., 2004) disrupted photoreceptor polarity, and converted all R8s to pr8 (Figure 4F) without affecting R7 Rhs (Figure S5B). We conclude that apkc kinase function is required for this phenotype because overexpression of a membrane-tethered kinase-dead version (lgmr>apkc CAAX- KD ) did not strongly increase (Figure 4E). Wts Requires Lgl to Specify yr8 in Parallel to Mer Because lgl, mer, and wts have identical Rh phenotypes, we examined genetic interactions between lgl and core complex members, wts and sav. lgl mutant clones suppressed the ability of ectopic Wts to induce yr8, and most R8s expressed in lgl;gmr-wts tissue (Figures 4H 4H 00 ), indicating that lgl, likemer, isrequired for Wts activity. But when sav was co-overexpressed with wts in lgl mutants, all R8s expressed (Figures 4I 4I 00 ). Thus, the ability of Wts to become activated in lgl mutant R8s is sensitive to levels of sav.furthermore,because mer can fully suppress the GMR-sav,wts overexpression phenotype while lgl cannot (compare Figures 3B 3B 00 to Figures 4I 4I 00 ), Lgl might promote Wts activity in parallel to the Mer/Kib branch. lgl mutants and apkc gain of function have identical R8 phenotypes, suggesting that apkc also influences the Hippo pathway to induce pr8 fate. Wts misexpression significantly suppressed induction, but not the polarity phenotype of activated apkc (Figure S5C), consistent with apkc acting upstream or in parallel to Wts. These data are consistent with Lgl and apkc acting upstream of Wts, likely in parallel to Mer/ Kib, to regulate the core complex, although other possibilities cannot be excluded. Upstream Hippo Pathway Regulators Control wts and melt Expression In R8, melt represses wts while wts represses melt transcription, and wts is the output of the bistable feedback loop for Rh expression (Mikeladze-Dvali et al., 2005). To test the epistatic relationship between mer and melt, we generated retinas double mutant for mer and melt (Figures 5A 5C), which phenocopied mer single mutants: most R8 cells expressed, and less than 1% expressed (Figure 5C). Similarly, loss of lgl suppressed 6 21, 1 14, November 15, 2011 ª2011 Elsevier Inc.
7 A lgl 4 clones B GFP A 18ºC C 29ºC lgl ts lgl ts lgl RNAi E F A D G % 60 % Figure 4. Lgl Is Required, and apkc Is Sufficient, to Specify yr8 Subtype (A F and H I 00 ) Adult retinas stained for (blue), (red), and GFP (green). Dashed lines indicate clonal boundaries. (A A 00 ) lgl 4 clones (non-gfp). (A 0 ) GFP only. (A 00 ) and only. Dashed line indicates clone boundary. (B) lgl ts3 raised at 18 C. (C) lgl ts3 shifted to 29 C during early pupation. (D) Retina expressing lgl-rnai. (E) lgmr>apkc CAAX-KD. (F) lgmr>apkc CAAX-WT (active). (G) Proportion of R8 that express (blue), (red), or both (purple) in listed genotypes. For all genotypes: n R 5 retinas, n R 1100 ommatidia; % means compared with two-tailed, unpaired t test; p < Error bars are ±1 standard deviation. (H) lgl 4 clones in GMR-wts retina background. (H 0 ) GFP only. (H 00 ) and only. (I) lgl 4 clones in GMR-wts,sav background. (I 0 ) GFP only. (I 00 ) and only. lgmr>apkc CAAX-KD GFP GFP H H H lgl 4 ; GMR-wts I I I lgl 4 ; GMR-wts,sav lgmr>apkc CAAX-WT the melt phenotype, and lgl mutant clones in melt mutant retinas expressed only (Figures 5D 5D 00 ). Thus, like wts, mer and lgl are required for the melt phenotype, consistent with a role in R8 to activate Wts genetically downstream of melt. To determine if upstream Hippo signaling influences the transcriptional feedback loop between wts and melt, we asked 0 ey,lgmr>dcr2 (control) lgmr-gal4 (control) lgmr>apkc CAAX-WT ey,lgmr>lgl RNAi,Dcr2 whether loss of mer affected wts or melt transcription. wts-lacz expression was lost in mer mutant clones, and instead, all R8s expressed melt-lacz (Figures 5E 5F 00 ). Similar results were obtained for kib mutants, with slightly less expressivity (Figures 5G 5G 00 ; data not shown). These data are consistent with a model wherein the upstream Mer/Kib branch represses melt and promotes wts transcription by keeping Wts protein active. In the absence of mer or kib, Wts protein kinase is inactive and unable to promote its own expression through the bistable feedback loop. Therefore, transcriptional feedback between wts and melt is coupled to the ON/OFF state of the entire upstream Mer/Kib activation branch of the Hippo pathway. Mer and Hippo Pathway Are First Required in R8 at 40% Pupation The genetic program that defines p and y ommatidial subtypes begins in early pupation with stochastic expression of ss in R7 cells (Wernet et al., 2006) followed by an R7-to-R8 signal that induces R8 subtypes, and then and expression around 70% 80% pupation (Earl and Britt, 2006). To examine temporal dynamics of the Hippo pathway in R8 subtype specification, we took advantage of a temperature-sensitive mer ts allele, which at the nonpermissive temperature behaves genetically as a null allele (MacDougall et al., 2001). To define the earliest requirement for mer, we moved flies to the nonpermissive temperature at 10% pupation to remove mer function, and 21, 1 14, November 15, 2011 ª2011 Elsevier Inc. 7
8 wts-lacz melt-lacz melt-lacz A mer B melt C mer; melt GFP GFP (melt-/-) D D D lgl clones; melt merge GFP LacZ E E E mer clones F F F mer clones G G G kib clones Figure 5. Mer and Kib Regulate the Expression of Wts and Melt in R8 (A C) Adult retinas stained for (blue) and (red). (A) Hemizygous mer ts /Y mutants kept at the nonpermissive temperature from early pupation. (B) melt 1. (C) mer ts /Y; melt 1 double mutants. (D D 00 ) lgl 4 clones in melt 1 mutant background. (blue), (red), and GFP (green). melt 1 tissue is GFP-positive; lgl 4 ; melt 1 double mutant tissue is GFPnegative. Dashed lines indicate clonal boundaries. (E G 00 ) Adult retinas showing either mer or kib mutant clones (GFP-negative) stained with b-gal antibody that labels nuclearly localized warts- or melt-lacz. Confocal image taken in the R8 nucleus layer focal plane. In (E) (G), laczpositive R8 cells that overlap GFP represent wild-type R8 nuclei. Dashed lines indicate clonal boundaries. (E) mer 4 clones (GFP-negative) and warts-lacz. Note the presence of warts- LacZ expression in wild-type tissue (GFP+). (E 0 ) Grayscale of GFP in (E). (E 00 ) wts-lacz of (E). (F) mer 4 mutant clones (GFP-negative) and melt-lacz. Note that melt-lacz expression is present in roughly one-third of wild-type R8 cells, but in almost all mer 4 mutant R8s. (F 0 ) Grayscale of GFP in (F). (F 00 ) melt-lacz of (F). (G) kib del mutant clones (GFP-negative) and melt-lacz. (G 0 ) Grayscale of GFP in (G). (G 00 ) melt-lacz of (G). then shifted them permanently to the permissive temperature (18 C) to restore mer function at successively later time points. When mer was inactivated until up to 38% pupation, we observed a wild-type : ratio in R8 (Figure 6A). Yet when inactivation persisted until 44% pupation, the proportion of R8 cells expressing increased, and continued to increase as mer function was removed for longer periods (Figure 6A). Complete transformation into pr8 was achieved when mer function was restored later than 70% pupation, when R8 subtypes are fully established. Therefore, between 38% and 44% pupation (or even later because it might take time for wild-type mer function to recover), mer and upstream Hippo signaling become required in presumptive yr8 photoreceptors. Subtypes appear fully specified after 70% 80% pupation, when melt/ and wts/ are expressed in p or y R8 subtypes, respectively (Figure 6B). Mer, Lgl, and Hippo Pathway Signaling Are Continuously Required to Maintain yr8 Photoreceptor Neuron Subtypes We next attempted to identify the latest point at which mer is required by using mer ts to remove mer function at successively later time points. Even when we inactivated mer as late as 90% pupation after and are expressed retinas always exhibited some derepression of into yr8s. Retinas from mer ts adult flies reared at 18 C and shifted to 29 C for 7 days (starting when flies were 7 ± 2 days old) also showed an increase in expressing R8s, which often coexpressed, revealing that is derepressed into yr8 cells as R8s shift to pr8 fate upon loss of mer (Figure 6H). Strikingly, when mer ts flies were kept at 29 C for 19 days, almost all R8s expressed, and often coexpressed (Figures 6E 6E 000 and 6H; compare to Figures 6C 6D 000 ). Thus, in addition to its role in yr8 establishment, the mer upstream branch of the Hippo pathway is also required to maintain yr8 fate in adult flies. To determine whether yr8 fate maintenance was specific to the Mer branch, or if it involved the entire Hippo pathway, we performed a similar experiment using lgl ts. Homozygous lgl ts flies reared at 18 C until several days posteclosure and shifted to 29 C for 16 days derepressed into the yr8 subtype, which coexpressed and in 14.4% of R8s (Figures 6G 6G 000 ). Control lgl ts flies reared continuously at 18 Corlgl ts /+ flies shifted to 29 C as adults exhibited a wild-type : ratio (Figures 6F 6F 000 and 6I). Thus, two different upstream regulators, mer and lgl, are required to specify as well as maintain yr8 subtypes, suggesting that the entire R8 Hippo pathway is required to keep Wts protein active for the life of the neuron. Melt Is Sufficient to Respecify Adult yr8 into pr8 Because wts expression and (i.e., yr8) are the default state for R8 in the absence of an R7 signal, the role of melt in specifying pr8 appears to be as an OFF switch that can disrupt Hippo pathway activity (Figures 7A 7A 0 ). This model predicts that ectopic Melt should still be able to turn off Hippo pathway activity in adult photoreceptors. To test this hypothesis, we misexpressed Melt long after R8 subtype specification, starting in 7-day-old flies using an inducible flp-out cassette (hs-flp; GMR-FRT-w+, STOP-FRT-Gal4). This led almost all R8s in the 8 21, 1 14, November 15, 2011 ª2011 Elsevier Inc.
9 m er ts me r m er ts / Y m er ts / e r er m er ts / FM 7 contro l m er ts me r m er ts / Y ts l gl lg l ts l gl / Cy O s l gl s l g t Please cite this article in press as: Jukam and Desplan, Binary Regulation of Hippo Pathway by Merlin/NF2, Kibra, Lgl, and Melted Specifies and A % R8 cells that express % Pupal development 18ºC control 29ºC to 18ºC eclosure B & warts and melt in subtypes mer first required in R eclosure % Pupal development adult 19 days at 29 C 18 C (control) +/- (control) -/- C D E 16 days at 29 C +/- (control) -/- F G mer ts /mer ts mer ts /+ mer ts /mer ts lgl ts /+ lgl ts /lgl ts C D E F G C D E F G C D E F G H % % C / ts 7 days 29 C m ts t /Y m s 19 days 29 C / ts I % % C / ts 16 days 29 C * t / l Figure 6. Continuous Mer and Lgl Signaling Is Required to Maintain the yr8 Subtype Fate (A) mer is first required for yr8 specification at 38% 44% pupation (shaded gray bar). Graph shows proportion of R8s expressing in adult retinas of mer ts flies where mer function was restored after shift from the restrictive to permissive temperature at various time points during pupation (black squares). (B) R8 subtype specification timeline during development. mer is required as early as 40% pupation, followed by R8 subtype-specific expression of wts and melt, which define y and p R8, respectively, and induce and expression around 70% 80% pupation. R8 subtypes and mutually exclusive Rh expression are maintained in adults. (C G 000 ) All images are adult retinas stained for (blue) and (red). (C C 000 ) Homozygous mer ts flies reared at the permissive temperature. (C 0 ) of (C). (C 00 ) Close-up of (C). (C 000 ) of (C 00 ). (D) Heterozygous mer ts /+ control flies shifted to the restrictive temperature (29 C) for 19 days during adulthood. (D 0 ) of (D). (D 00 ) Close-up of (D). and are coexpressed in fewer than 2% of R8 cells (dashed circle). (D 000 ) of (D 00 ). Dashed circle indicates the rare yr8 that has 21, 1 14, November 15, 2011 ª2011 Elsevier Inc. 9
10 dorsal half of the retina to express, while 70% still contained (Figures 7B 7C 0 ). This phenotype is similar to late removal of mer or lgl function and demonstrates that melt is sufficient to abrogate Hippo signaling in yr8, to allow reactivation of even in adult flies. DISCUSSION This work describes a mechanism for a postmitotic neuronal subtype decision wherein a complex signaling pathway is regulated both canonically and noncanonically to instruct either of two cell states. The control point for Hippo pathway signaling in R8 is the expression or repression of Wts, whose activity is promoted by constitutive upstream Hippo pathway activity (Figure 7D). Several lines of evidence support this model. First, two upstream inputs, mer/kib and lgl, and the core complex of hpo/sav/mats/wts are required for R8 subtype specification, and all have identical Rh phenotypes. Second, mer is required for Wts activity and expression, indicating that the entire Hippo pathway is coupled to the wts-melt transcriptional feedback loop and binary fate mechanism. Third, mer and lgl temperature-sensitive experiments showed that upstream Hippo pathway signaling is continuously required to maintain yr8 fate. Finally, Hippo signaling can be abrogated long after the fate decision by late induction of melt, which respecifies pr8 fate, demonstrating that melt can disrupt active Hippo signaling. Given that wts acts genetically downstream of melt in R8 subtype specification, why engage transcriptional regulation of wts instead of simply inactivating Wts kinase? Regulating signal transduction members by transcription might be an efficient way to arrest signaling permanently, especially if the regulation is mediated by a bistable loop, a common genetic motif used by cells to transition from one cell state to another, often irreversibly (reviewed in Ferrell, 2002). Thus, it is well suited to promote mutually exclusive gene expression and switch-like fate decisions during neural specification (reviewed in Jukam and Desplan, 2010). Regulation of wts by melt acts as an OFF switch for Hippo pathway activity, and the bistable feedback loop reinforces this cell state transition, despite the activity of multiple upstream Hippo regulators. Hippo Pathway and R8 Subtype Specification Dynamics These results deepen our understanding of key temporal steps in R8 subtype specification. Ectopic kib, mats, and hpo can induce the yr8 fate in all R8s but require Wts for their function, indicating that some basal level of Wts must be present initially in all R8s. Such Wts is likely dispensable for yr8 specification until at least 40% pupation, when mer is first required. wts is derepressed in melt mutants, and melt is derepressed in wts mutants. Thus, a general factor or factors must be competent to activate transcription of wts and melt in all R8s, independent from the R7 signal that coordinates R7/R8 subtypes. This activation would be restricted to mutually exclusive R8s by the bistable feedback loop. In presumptive yr8s, this general activator might boost Wts levels to allow sustained activation by constitutive upstream Hippo signaling. In presumptive pr8, the R7 signal could induce a further increase in melt expression or protein activity that allows Melt to inactivate the Hippo pathway and repress wts. Alternatively, the R7 signal might inhibit upstream activators of the Hippo pathway like Mer or Lgl, which likely act near the membrane and are candidates for substrates that mediate reception of the R7 signal. Melt could then be activated by loss of Wts, and reinforce the loss in Wts activity by transcriptionally repressing wts. Mer, Lgl, and the Hippo Pathway in Neuronal Maintenance Neural subtypes, once specified, appear to require active genetic programs to maintain their fate (Lesch and Bargmann, 2010). A second role for the Hippo pathway in R8, in addition to specifying yr8, is to maintain exclusion of the opposite fate in old R8 neurons, by continuously repressing rh5 in adult yr8 subtypes. When mer or lgl was removed late, the yr8 character was partially lost, because many R8 cells contained both and. Photoreceptors must coordinate sensory receptor expression with hardwired axon projections to the visual processing centers (reviewed in Sanes and Zipursky, 2010), so the functional identity of the R8 neuron should be stable, or the fly risks sensory confusion. Context Specificity and Mer, Kib, and Lgl Protein Function in the Hippo-Signaling Network Upstream inputs and downstream transcriptional targets of Wts signaling in development are context dependent (Pellock et al., 2007; Cho et al., 2006; Peng et al., 2009; Milton et al., 2010; Grusche et al., 2010). Evidence that Kib can bind separately to either Ex or Mer, as well as synergistically in a complex to both, implies that Ex, Kib, and Mer act semi-redundantly in growth control (Genevet et al., 2010; Grusche et al., 2010). Our results demonstrate that Mer/Kib can operate as a branch completely independent from Ex upstream of Wts for R8 specification. mer and lgl mutants phenocopy wts mutants with respect to R8 subtype specification. If Mer and Lgl do not act in a linear pathway or complex, why would Hippo signaling require two nonredundant upstream inputs to keep Wts protein active? Mer/Kib and Lgl might simply provide complementary means weakly derepressed. (E) Homozygous mer ts flies shifted to the restrictive temperature (29 C) for 19 days during adulthood. (E 0 ) of (E). (E 00 ) Close-up of (E 0 ). Arrowheads indicate examples of derepressed in the former yr8 subtype. (E 000 ) of (E 00 ). (F) Heterozygous lgl ts /+ flies shifted to the restrictive temperature for 16 days during adulthood. (F 0 ) of (F). (F 00 ) Close-up of (F 0 ). (F 000 ) of (F 00 ). (G) Homozygous lgl ts flies shifted to the restricted temperature for 16 days during adulthood. (G 0 ) of (G). (G 00 ) Close-up of (G 0 ). Many -expressing cells coexpress (pink); examples are indicated by arrowheads. (G 000 ) of (G 00 ). (H) Proportion of R8s that expresses when mer function is removed in adult flies, as compared to controls. y axis shows percent exclusively (blue), exclusively (red), and / coexpression (purple). Two-tailed, unpaired t test; p < (I) Proportion of R8s that expresses when lgl function is removed in adult flies, as compared to controls. y axis shows percent exclusively (blue), exclusively (red), and / coexpression (purple). Two-tailed, unpaired t test; p < 0.001; *p < Error bars are ±1 standard deviation , 1 14, November 15, 2011 ª2011 Elsevier Inc.
11 D A PanR8>melt PanR8>melt A active Hippo pathway as default state PanR8>melt Specification Maintenance continuous pathway activity maintains yr8 subtype. Ss Ss yellow B Late expression of melt hs-flp; GMR>>melt B Rh3 Rh4 R7 signal apkc lgl kib mer hpo sav mats wts Rh3 Rh4 apkc lgl kib mer hpo sav mats wts hs-flp; GMR>>melt melt melt R7 R8 pale hs-flp; GMR>>melt hs-flp; GMR>>melt to promote Wts function in R8. The protein-protein interactions between Mer, Kib and Sav (Yu et al., 2010) and Kib and Wts (Genevet et al., 2010) support a localization or scaffolding role for Mer and Kib in activation of Hpo and Wts, perhaps by C C Rh3 Rh4 R7 signal apkc lgl Ss kib mer hpo sav mats wts apkc lgl Rh3 Rh4 kib mer hpo sav mats wts melt? melt mid-late pupation Melt induction and disruption of Hippo pathway adult Figure 7. Model for Regulation of Hippo Pathway Signaling in a Postmitotic Fate Decision (A C 0 ) All panels show adult retinas stained for (blue) and (red). (A) PanR8>melt. melt expressed immediately after subtype-specific Rh expression can reset the bistable feedback loop and induce the pr8 fate. (A 0 ) Close-up of (A). Note the increase in -only expressing pr8s and decrease in -expressing yr8 cells. (B) Ectopic melt expressed in adult flies (see Experimental Procedures). (B 0 ) Close-up of (B). Note that staining appears weaker in cells that coexpress and. (C) of (B). (C 0 ) of (B 0 ). (D) Model of Hippo pathway regulation for binary fate specification of R8. Constitutive and continuous upstream pathway signaling by Mer, Kib, and Lgl promotes yr8 fate, while repression of warts by Melt disrupts Hippo signaling to specify pr8 fate. Genes in black or colored font are expressed and active in that particular cell; genes in gray font are not expressed or not active. recruiting Hpo, Sav, and Mats near the membrane where Hpo can phosphorylate Wts. Membrane-tethered Mats promotes Wts activation in our system and in growth (Ho et al., 2010). Kib, like Mats, Hpo, Sav, or Wts, is sufficient to induce the yr8 subtype. Dramatically increasing the expression level of kib, hpo, mats, sav, or wts might promote synergistic localization and binding of all six proteins near the membrane, resulting in phosphorylated and active Wts. Lgl could function to inhibit a negative pathway regulator, such as the dstripak PP2A phosphatase complex that acts on Hpo (Ribeiro et al., 2010). Lgl would then promote activation of Wts by preventing inactivation of Wts or Hpo. Such regulation would be useful where a continuously active Hippo pathway is necessary, as in the maintenance of yr8 subtype fate. Alternatively, the intracellular polarity roles of Lgl and apkc might generically affect localization of Hippo pathway components. Perhaps the molecular signal from R7 that promotes Melt and represses Wts activity shares features with nonautonomous signals that affect polarity regulators of Hippo signaling in dividing cells. R8 Subtypes as a Model for Studying Mer/NF2 Function Several reports suggest that Hippo pathway signaling might underlie some pathology of NF-2. Mer/NF2 can regulate contact-dependent growth inhibition and nuclear localization of the Yki ortholog and oncogene, YAP, in human meningioma cell lines (Striedinger et al., 2008), as well as genetically interact with YAP during tumorigenesis in the mouse liver (Zhang et al., 2010). Other studies have implicated misregulated EGFR signaling in NF-2 mutant tumors (Benhamouche et al., 2010) or suggested that Hippo pathway-like overgrowth phenotypes in mer;ex double mutants are a secondary consequence of EGFR misregulation in Drosophila (Maitra et al., 2006). Our results support a role for Mer upstream of the Hippo pathway in the context of R8 subtype specification. The binary assay of or in R8 photoreceptors can be used to dissect Mer protein function in Hippo signaling without complications from redundant upstream regulators of Wts. Reuse of Signaling Pathways in Development The modular reuse of signaling pathway components for different purposes in development is widespread, and is likely a key molecular conduit for the evolution of cellular and morphological diversity (Wilkins, 2002). The uncoupling of mer, kib, lgl, and the core Hippo-signaling cassette from growth control allows the genes to contribute to a broader array of developmental decisions. It will be interesting to see if the Lgl/Mer/Kib/ Hpo/Sav/Mats/Wts module is regulated by Melt in other postmitotic fate-specification events during neural development. EXPERIMENTAL PROCEDURES Drosophila Stocks For Drosophila genotypes and fly strains, see Supplemental Experimental Procedures. Drosophila Genetics and Induction of Mutant Tissue Flies were raised on corn meal-molasses-agar medium under standard laboratory conditions. y 1, w 67 ;+;+ flies were considered wild-type and used as a control for rh expression. For details on Gal4 drivers, generating 21, 1 14, November 15, 2011 ª2011 Elsevier Inc. 11
Chapter 4 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays.
 Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
Evaluating a potential interaction between deltex and git in Drosophila: genetic interaction, gene overexpression and cell biology assays. The data described in chapter 3 presented evidence that endogenous
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
Supplementary Information
 Supplementary Information Supplementary Figure 1. JAK/STAT in early wing development (a-f) Wing primordia of second instar larvae of the indicated genotypes labeled to visualize expression of upd mrna
Supplementary Information Supplementary Figure 1. JAK/STAT in early wing development (a-f) Wing primordia of second instar larvae of the indicated genotypes labeled to visualize expression of upd mrna
L œil de la Drosophile pour l étude du développement et de l apoptose
 L œil de la Drosophile pour l étude du développement et de l apoptose Pr Bertrand Mollereau Apoptosis and Neurogenetic LBMC UMR5239 ENS Lyon http://www.ens-lyon.fr/lbmc/apodroso/ Drosophila is a powerful
L œil de la Drosophile pour l étude du développement et de l apoptose Pr Bertrand Mollereau Apoptosis and Neurogenetic LBMC UMR5239 ENS Lyon http://www.ens-lyon.fr/lbmc/apodroso/ Drosophila is a powerful
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
Spatial Organization of Hippo Signaling at the Plasma Membrane Mediated by the Tumor Suppressor Merlin/NF2
 Spatial Organization of Hippo Signaling at the Plasma Membrane Mediated by the Tumor Suppressor Merlin/NF2 Feng Yin, Jianzhong Yu, Yonggang Zheng, Qian Chen, Nailing Zhang, and Duojia Pan, * Department
Spatial Organization of Hippo Signaling at the Plasma Membrane Mediated by the Tumor Suppressor Merlin/NF2 Feng Yin, Jianzhong Yu, Yonggang Zheng, Qian Chen, Nailing Zhang, and Duojia Pan, * Department
Developmental Biology
 Developmental Biology 350 (2011) 255 266 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology The Salvador/Warts/Hippo pathway controls
Developmental Biology 350 (2011) 255 266 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology The Salvador/Warts/Hippo pathway controls
Three Distinct Roles for Notch in Drosophila R7 Photoreceptor Specification
 Three Distinct Roles for Notch in Drosophila R7 Photoreceptor Specification Andrew Tomlinson 1. *, Yannis Emmanuel Mavromatakis 1., Gary Struhl 1,2 1 Department of Genetics and Development, College of
Three Distinct Roles for Notch in Drosophila R7 Photoreceptor Specification Andrew Tomlinson 1. *, Yannis Emmanuel Mavromatakis 1., Gary Struhl 1,2 1 Department of Genetics and Development, College of
MOB AS TUMOR SUPPRESSOR IS ACTIVATED AT THE PLASMA MEMBRANE TO CONTROL TISSUE GROWTH AND ORGAN SIZE IN DROSOPHILA
 The Pennsylvania State University The Graduate School Graduate Program in Integrative Biosciences MOB AS TUMOR SUPPRESSOR IS ACTIVATED AT THE PLASMA MEMBRANE TO CONTROL TISSUE GROWTH AND ORGAN SIZE IN
The Pennsylvania State University The Graduate School Graduate Program in Integrative Biosciences MOB AS TUMOR SUPPRESSOR IS ACTIVATED AT THE PLASMA MEMBRANE TO CONTROL TISSUE GROWTH AND ORGAN SIZE IN
purpose of this Chapter is to highlight some problems that will likely provide new
 119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
119 Chapter 6 Future Directions Besides our contributions discussed in previous chapters to the problem of developmental pattern formation, this work has also brought new questions that remain unanswered.
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila
 Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
Baz, Par-6 and apkc are not required for axon or dendrite specification in Drosophila Melissa M. Rolls and Chris Q. Doe, Inst. Neurosci and Inst. Mol. Biol., HHMI, Univ. Oregon, Eugene, Oregon 97403 Correspondence
Midterm 1. Average score: 74.4 Median score: 77
 Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Midterm 1 Average score: 74.4 Median score: 77 NAME: TA (circle one) Jody Westbrook or Jessica Piel Section (circle one) Tue Wed Thur MCB 141 First Midterm Feb. 21, 2008 Only answer 4 of these 5 problems.
Bypass and interaction suppressors; pathway analysis
 Bypass and interaction suppressors; pathway analysis The isolation of extragenic suppressors is a powerful tool for identifying genes that encode proteins that function in the same process as a gene of
Bypass and interaction suppressors; pathway analysis The isolation of extragenic suppressors is a powerful tool for identifying genes that encode proteins that function in the same process as a gene of
A complementation test would be done by crossing the haploid strains and scoring the phenotype in the diploids.
 Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
Problem set H answers 1. To study DNA repair mechanisms, geneticists isolated yeast mutants that were sensitive to various types of radiation; for example, mutants that were more sensitive to UV light.
Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain
 Development 127, 367-379 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV5342 367 Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain Wei Du
Development 127, 367-379 (2000) Printed in Great Britain The Company of Biologists Limited 2000 DEV5342 367 Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain Wei Du
Reading: Chapter 5, pp ; Reference chapter D, pp Problem set F
 Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Mosaic Analysis Reading: Chapter 5, pp140-141; Reference chapter D, pp820-823 Problem set F Twin spots in Drosophila Although segregation and recombination in mitosis do not occur at the same frequency
Eukaryotic Gene Expression
 Eukaryotic Gene Expression Lectures 22-23 Several Features Distinguish Eukaryotic Processes From Mechanisms in Bacteria 123 Eukaryotic Gene Expression Several Features Distinguish Eukaryotic Processes
Eukaryotic Gene Expression Lectures 22-23 Several Features Distinguish Eukaryotic Processes From Mechanisms in Bacteria 123 Eukaryotic Gene Expression Several Features Distinguish Eukaryotic Processes
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Regulation of Organ Size: Insights From the Drosophila Hippo Signaling Pathway
 DEVELOPMENTAL DYNAMICS 238:1627 1637, 2009 REVIEWS A PEER REVIEWED FORUM Regulation of Organ Size: Insights From the Drosophila Hippo Signaling Pathway Madhuri Kango-Singh 1 * and Amit Singh 2,3 * Organ
DEVELOPMENTAL DYNAMICS 238:1627 1637, 2009 REVIEWS A PEER REVIEWED FORUM Regulation of Organ Size: Insights From the Drosophila Hippo Signaling Pathway Madhuri Kango-Singh 1 * and Amit Singh 2,3 * Organ
Linking Cell Fate, Trajectory Choice, and Target Selection: Genetic Analysis of Sema-2b in Olfactory Axon Targeting
 Article Linking Cell Fate, Trajectory Choice, and Target Selection: Genetic Analysis of Sema-2b in Olfactory Axon Targeting William J. Joo, 1,2 Lora B. Sweeney, 1,2,4 Liang Liang, 1,3 and Liqun Luo 1,2,
Article Linking Cell Fate, Trajectory Choice, and Target Selection: Genetic Analysis of Sema-2b in Olfactory Axon Targeting William J. Joo, 1,2 Lora B. Sweeney, 1,2,4 Liang Liang, 1,3 and Liqun Luo 1,2,
Evolution of the Complex Eye and Pax6 Gene
 Evolution of the Complex Eye and Pax6 Gene Rachel Thomsen Sarah Kim Jenia Ostrovskaya Key Points Definition of an Eye o Types of Eyes Origin of Species: Difficulties on Theory Pax family Pax6 gene why
Evolution of the Complex Eye and Pax6 Gene Rachel Thomsen Sarah Kim Jenia Ostrovskaya Key Points Definition of an Eye o Types of Eyes Origin of Species: Difficulties on Theory Pax family Pax6 gene why
(Write your name on every page. One point will be deducted for every page without your name!)
 POPULATION GENETICS AND MICROEVOLUTIONARY THEORY FINAL EXAMINATION (Write your name on every page. One point will be deducted for every page without your name!) 1. Briefly define (5 points each): a) Average
POPULATION GENETICS AND MICROEVOLUTIONARY THEORY FINAL EXAMINATION (Write your name on every page. One point will be deducted for every page without your name!) 1. Briefly define (5 points each): a) Average
Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Tuesday, December 27, 16
 Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Enduring understanding 3.B: Expression of genetic information involves cellular and molecular
Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. Enduring understanding 3.B: Expression of genetic information involves cellular and molecular
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1244624/dc1 Supplementary Materials for Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein Sougata Roy, Hai Huang,
www.sciencemag.org/cgi/content/full/science.1244624/dc1 Supplementary Materials for Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein Sougata Roy, Hai Huang,
Solutions to Problem Set 4
 Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
Question 1 Solutions to 7.014 Problem Set 4 Because you have not read much scientific literature, you decide to study the genetics of garden peas. You have two pure breeding pea strains. One that is tall
Types of biological networks. I. Intra-cellurar networks
 Types of biological networks I. Intra-cellurar networks 1 Some intra-cellular networks: 1. Metabolic networks 2. Transcriptional regulation networks 3. Cell signalling networks 4. Protein-protein interaction
Types of biological networks I. Intra-cellurar networks 1 Some intra-cellular networks: 1. Metabolic networks 2. Transcriptional regulation networks 3. Cell signalling networks 4. Protein-protein interaction
Axis determination in flies. Sem 9.3.B.5 Animal Science
 Axis determination in flies Sem 9.3.B.5 Animal Science All embryos are in lateral view (anterior to the left). Endoderm, midgut; mesoderm; central nervous system; foregut, hindgut and pole cells in yellow.
Axis determination in flies Sem 9.3.B.5 Animal Science All embryos are in lateral view (anterior to the left). Endoderm, midgut; mesoderm; central nervous system; foregut, hindgut and pole cells in yellow.
S Phase Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues
 S Phase Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues Jean M. Davidson 1, Robert J. Duronio 1,2,3 * 1 Department of Biology, The University of North Carolina at Chapel Hill, Chapel
S Phase Coupled E2f1 Destruction Ensures Homeostasis in Proliferating Tissues Jean M. Davidson 1, Robert J. Duronio 1,2,3 * 1 Department of Biology, The University of North Carolina at Chapel Hill, Chapel
10/2/2015. Chapter 4. Determination and Differentiation. Neuroanatomical Diversity
 Chapter 4 Determination and Differentiation Neuroanatomical Diversity 1 Neurochemical diversity: another important aspect of neuronal fate Neurotransmitters and their receptors Excitatory Glutamate Acetylcholine
Chapter 4 Determination and Differentiation Neuroanatomical Diversity 1 Neurochemical diversity: another important aspect of neuronal fate Neurotransmitters and their receptors Excitatory Glutamate Acetylcholine
Introduction. Gene expression is the combined process of :
 1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
1 To know and explain: Regulation of Bacterial Gene Expression Constitutive ( house keeping) vs. Controllable genes OPERON structure and its role in gene regulation Regulation of Eukaryotic Gene Expression
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
Report. Ajuba LIM Proteins Are Negative Regulators of the Hippo Signaling Pathway
 Current Biology 20, 657 662, April 13, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2010.02.035 Ajuba LIM Proteins Are Negative Regulators of the Hippo Signaling Pathway Report Meghna
Current Biology 20, 657 662, April 13, 2010 ª2010 Elsevier Ltd All rights reserved DOI 10.1016/j.cub.2010.02.035 Ajuba LIM Proteins Are Negative Regulators of the Hippo Signaling Pathway Report Meghna
Sang-Chul Nam a,1, Bibhash Mukhopadhyay b, Kwang-Wook Choi a,b,c,
 Developmental Biology 306 (2007) 624 635 www.elsevier.com/locate/ydbio Antagonistic functions of Par-1 kinase and protein phosphatase 2A are required for localization of Bazooka and photoreceptor morphogenesis
Developmental Biology 306 (2007) 624 635 www.elsevier.com/locate/ydbio Antagonistic functions of Par-1 kinase and protein phosphatase 2A are required for localization of Bazooka and photoreceptor morphogenesis
Reception The target cell s detection of a signal coming from outside the cell May Occur by: Direct connect Through signal molecules
 Why Do Cells Communicate? Regulation Cells need to control cellular processes In multicellular organism, cells signaling pathways coordinate the activities within individual cells that support the function
Why Do Cells Communicate? Regulation Cells need to control cellular processes In multicellular organism, cells signaling pathways coordinate the activities within individual cells that support the function
Developmental Biology
 Developmental Biology 350 (2011) 139 151 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Regulation of Hippo signaling by Jun kinase
Developmental Biology 350 (2011) 139 151 Contents lists available at ScienceDirect Developmental Biology journal homepage: www.elsevier.com/developmentalbiology Regulation of Hippo signaling by Jun kinase
Lecture 10: Cyclins, cyclin kinases and cell division
 Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature10923 Supplementary Figure 1 Ten-a and Ten-m antibody and cell type specificities. a c, Representative single confocal sections of a Drosophila NMJ stained with antibodies to Ten-a (red),
doi:10.1038/nature10923 Supplementary Figure 1 Ten-a and Ten-m antibody and cell type specificities. a c, Representative single confocal sections of a Drosophila NMJ stained with antibodies to Ten-a (red),
Chapter 18 Lecture. Concepts of Genetics. Tenth Edition. Developmental Genetics
 Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Nature Neuroscience: doi: /nn.2717
 Supplementary Fig. 1. Dendrite length is not secondary to body length. Dendrite growth proceeds independently of the rate of body growth and decreases in rate in adults. n 20 on dendrite measurement, n
Supplementary Fig. 1. Dendrite length is not secondary to body length. Dendrite growth proceeds independently of the rate of body growth and decreases in rate in adults. n 20 on dendrite measurement, n
Iroquois Complex Genes Induce Co-Expression of rhodopsins in Drosophila
 Iroquois Complex Genes Induce Co-Expression of rhodopsins in Drosophila Esteban O. Mazzoni a, Arzu Celik b, Mathias F. Wernet c, Daniel Vasiliauskas, Robert J. Johnston, Tiffany A. Cook d, Franck Pichaud
Iroquois Complex Genes Induce Co-Expression of rhodopsins in Drosophila Esteban O. Mazzoni a, Arzu Celik b, Mathias F. Wernet c, Daniel Vasiliauskas, Robert J. Johnston, Tiffany A. Cook d, Franck Pichaud
Review Article Hippo and rassf1a Pathways: A Growing Affair
 Molecular Biology International Volume 2012, Article ID 307628, 12 pages doi:10.1155/2012/307628 Review Article Hippo and rassf1a Pathways: A Growing Affair Francesca Fausti, 1 Silvia Di Agostino, 2 Andrea
Molecular Biology International Volume 2012, Article ID 307628, 12 pages doi:10.1155/2012/307628 Review Article Hippo and rassf1a Pathways: A Growing Affair Francesca Fausti, 1 Silvia Di Agostino, 2 Andrea
Lecture 7. Development of the Fruit Fly Drosophila
 BIOLOGY 205/SECTION 7 DEVELOPMENT- LILJEGREN Lecture 7 Development of the Fruit Fly Drosophila 1. The fruit fly- a highly successful, specialized organism a. Quick life cycle includes three larval stages
BIOLOGY 205/SECTION 7 DEVELOPMENT- LILJEGREN Lecture 7 Development of the Fruit Fly Drosophila 1. The fruit fly- a highly successful, specialized organism a. Quick life cycle includes three larval stages
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and
 Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Conclusions. The experimental studies presented in this thesis provide the first molecular insights
 C h a p t e r 5 Conclusions 5.1 Summary The experimental studies presented in this thesis provide the first molecular insights into the cellular processes of assembly, and aggregation of neural crest and
C h a p t e r 5 Conclusions 5.1 Summary The experimental studies presented in this thesis provide the first molecular insights into the cellular processes of assembly, and aggregation of neural crest and
Supporting Information
 Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
Supporting Information Cao et al. 10.1073/pnas.1306220110 Gram - bacteria Hemolymph Cytoplasm PGRP-LC TAK1 signalosome Imd dfadd Dredd Dnr1 Ikk signalosome P Relish Nucleus AMP and effector genes Fig.
Chapter 11. Development: Differentiation and Determination
 KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
AP Biology Unit 6 Practice Test 1. A group of cells is assayed for DNA content immediately following mitosis and is found to have an average of 8
 AP Biology Unit 6 Practice Test Name: 1. A group of cells is assayed for DNA content immediately following mitosis and is found to have an average of 8 picograms of DNA per nucleus. How many picograms
AP Biology Unit 6 Practice Test Name: 1. A group of cells is assayed for DNA content immediately following mitosis and is found to have an average of 8 picograms of DNA per nucleus. How many picograms
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A)
 Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Fig. S1. Proliferation and cell cycle exit are affected by the med mutation. (A,B) M-phase nuclei are visualized by a-ph3 labeling in wild-type (A) and mutant (B) 4 dpf retinae. The central retina of the
Analysis and Simulation of Biological Systems
 Analysis and Simulation of Biological Systems Dr. Carlo Cosentino School of Computer and Biomedical Engineering Department of Experimental and Clinical Medicine Università degli Studi Magna Graecia Catanzaro,
Analysis and Simulation of Biological Systems Dr. Carlo Cosentino School of Computer and Biomedical Engineering Department of Experimental and Clinical Medicine Università degli Studi Magna Graecia Catanzaro,
Enduring understanding 1.A: Change in the genetic makeup of a population over time is evolution.
 The AP Biology course is designed to enable you to develop advanced inquiry and reasoning skills, such as designing a plan for collecting data, analyzing data, applying mathematical routines, and connecting
The AP Biology course is designed to enable you to develop advanced inquiry and reasoning skills, such as designing a plan for collecting data, analyzing data, applying mathematical routines, and connecting
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
Regulation and signaling. Overview. Control of gene expression. Cells need to regulate the amounts of different proteins they express, depending on
 Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
AP Curriculum Framework with Learning Objectives
 Big Ideas Big Idea 1: The process of evolution drives the diversity and unity of life. AP Curriculum Framework with Learning Objectives Understanding 1.A: Change in the genetic makeup of a population over
Big Ideas Big Idea 1: The process of evolution drives the diversity and unity of life. AP Curriculum Framework with Learning Objectives Understanding 1.A: Change in the genetic makeup of a population over
Stochastic simulations
 Stochastic simulations Application to molecular networks Literature overview Noise in genetic networks Origins How to measure and distinguish between the two types of noise (intrinsic vs extrinsic)? What
Stochastic simulations Application to molecular networks Literature overview Noise in genetic networks Origins How to measure and distinguish between the two types of noise (intrinsic vs extrinsic)? What
A Screen for Conditional Growth Suppressor Genes Identifies the Drosophila Homolog of HD-PTP as a Regulator of the Oncoprotein Yorkie
 Article A Screen for Conditional Growth Suppressor Genes Identifies the Drosophila Homolog of HD-PTP as a Regulator of the Oncoprotein Yorkie M. Melissa Gilbert, 1, * Marla Tipping, 2 Alexey Veraksa, 2
Article A Screen for Conditional Growth Suppressor Genes Identifies the Drosophila Homolog of HD-PTP as a Regulator of the Oncoprotein Yorkie M. Melissa Gilbert, 1, * Marla Tipping, 2 Alexey Veraksa, 2
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
- interactions between alleles. - multiple phenotypic effects of one gene. - phenotypic variability in single genes. - interactions between genes
 Genotype Phenotype - interactions between alleles - dominance - multiple phenotypic effects of one gene - phenotypic variability in single genes - interactions between genes amount of product threshold
Genotype Phenotype - interactions between alleles - dominance - multiple phenotypic effects of one gene - phenotypic variability in single genes - interactions between genes amount of product threshold
targets. clustering show that different complex pathway
 Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
Supplementary Figure 1. CLICR allows clustering and activation of cytoplasmic protein targets. (a, b) Upon light activation, the Cry2 (red) and LRP6c (green) components co-cluster due to the heterodimeric
16 CONTROL OF GENE EXPRESSION
 16 CONTROL OF GENE EXPRESSION Chapter Outline 16.1 REGULATION OF GENE EXPRESSION IN PROKARYOTES The operon is the unit of transcription in prokaryotes The lac operon for lactose metabolism is transcribed
16 CONTROL OF GENE EXPRESSION Chapter Outline 16.1 REGULATION OF GENE EXPRESSION IN PROKARYOTES The operon is the unit of transcription in prokaryotes The lac operon for lactose metabolism is transcribed
4) Please cite Dagda et al J Biol Chem 284: , for any publications or presentations resulting from use or modification of the macro.
 Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Supplement Figure S1. Algorithmic quantification of mitochondrial morphology in SH- SY5Y cells treated with known fission/fusion mediators. Parental SH-SY5Y cells were transiently transfected with an empty
Two Frizzled Planar Cell Polarity Signals in the Drosophila Wing Are Differentially Organized by the Fat/Dachsous Pathway
 Two Frizzled Planar Cell Polarity Signals in the Drosophila Wing Are Differentially Organized by the Fat/Dachsous Pathway Justin Hogan, Meagan Valentine, Chris Cox, Kristy Doyle, Simon Collier* Department
Two Frizzled Planar Cell Polarity Signals in the Drosophila Wing Are Differentially Organized by the Fat/Dachsous Pathway Justin Hogan, Meagan Valentine, Chris Cox, Kristy Doyle, Simon Collier* Department
Drosophila IAP1-mediated ubiquitylation controls activation of the initiator caspase DRONC independent of protein degradation
 University of Massachusetts Medical School escholarship@umms Molecular, Cell and Cancer Biology Publications Molecular, Cell and Cancer Biology 9-1-2011 Drosophila IAP1-mediated ubiquitylation controls
University of Massachusetts Medical School escholarship@umms Molecular, Cell and Cancer Biology Publications Molecular, Cell and Cancer Biology 9-1-2011 Drosophila IAP1-mediated ubiquitylation controls
Big Idea 1: The process of evolution drives the diversity and unity of life.
 Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
AP Biology Gene Regulation and Development Review
 AP Biology Gene Regulation and Development Review 1. What does the regulatory gene code for? 2. Is the repressor by default active/inactive? 3. What changes the repressor activity? 4. What does repressor
AP Biology Gene Regulation and Development Review 1. What does the regulatory gene code for? 2. Is the repressor by default active/inactive? 3. What changes the repressor activity? 4. What does repressor
Role of Mitochondrial Remodeling in Programmed Cell Death in
 Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Autophagy-independent function of Atg1 for apoptosis-induced compensatory proliferation
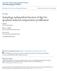 University of Massachusetts Medical School escholarship@umms Molecular, Cell and Cancer Biology Publications Molecular, Cell and Cancer Biology 8-19-2016 Autophagy-independent function of Atg1 for apoptosis-induced
University of Massachusetts Medical School escholarship@umms Molecular, Cell and Cancer Biology Publications Molecular, Cell and Cancer Biology 8-19-2016 Autophagy-independent function of Atg1 for apoptosis-induced
A A A A B B1
 LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
Epigenetics and Flowering Any potentially stable and heritable change in gene expression that occurs without a change in DNA sequence
 Epigenetics and Flowering Any potentially stable and heritable change in gene expression that occurs without a change in DNA sequence www.plantcell.org/cgi/doi/10.1105/tpc.110.tt0110 Epigenetics Usually
Epigenetics and Flowering Any potentially stable and heritable change in gene expression that occurs without a change in DNA sequence www.plantcell.org/cgi/doi/10.1105/tpc.110.tt0110 Epigenetics Usually
 DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
TRANSCRIPTIONAL CONTROL OF PHOTORECEPTOR AXON GROWTH AND TARGETING IN DROSOPHILA MELANOGASTER
 TRANSCRIPTIONAL CONTROL OF PHOTORECEPTOR AXON GROWTH AND TARGETING IN DROSOPHILA MELANOGASTER by JONATHAN S. KNISS A DISSERTATION Presented to the Department of Biology and the Graduate School of the University
TRANSCRIPTIONAL CONTROL OF PHOTORECEPTOR AXON GROWTH AND TARGETING IN DROSOPHILA MELANOGASTER by JONATHAN S. KNISS A DISSERTATION Presented to the Department of Biology and the Graduate School of the University
Segment boundary formation in Drosophila embryos
 Segment boundary formation in Drosophila embryos Development 130, August 2003 Camilla W. Larsen, Elizabeth Hirst, Cyrille Alexandre and Jean Paul Vincent 1. Introduction: - Segment boundary formation:
Segment boundary formation in Drosophila embryos Development 130, August 2003 Camilla W. Larsen, Elizabeth Hirst, Cyrille Alexandre and Jean Paul Vincent 1. Introduction: - Segment boundary formation:
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 6, 2007 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 6, 2007 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Why Flies? stages of embryogenesis. The Fly in History
 The Fly in History 1859 Darwin 1866 Mendel c. 1890 Driesch, Roux (experimental embryology) 1900 rediscovery of Mendel (birth of genetics) 1910 first mutant (white) (Morgan) 1913 first genetic map (Sturtevant
The Fly in History 1859 Darwin 1866 Mendel c. 1890 Driesch, Roux (experimental embryology) 1900 rediscovery of Mendel (birth of genetics) 1910 first mutant (white) (Morgan) 1913 first genetic map (Sturtevant
Signal Transduction. Dr. Chaidir, Apt
 Signal Transduction Dr. Chaidir, Apt Background Complex unicellular organisms existed on Earth for approximately 2.5 billion years before the first multicellular organisms appeared.this long period for
Signal Transduction Dr. Chaidir, Apt Background Complex unicellular organisms existed on Earth for approximately 2.5 billion years before the first multicellular organisms appeared.this long period for
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo
 Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
The Pax and large Maf families of genes in mammalian eye development
 The Pax and large Maf families of genes in mammalian eye development Vertebrate eye development is dependent on the coordinated action of thousands of genes. A specific group of over one hundred of regulatory
The Pax and large Maf families of genes in mammalian eye development Vertebrate eye development is dependent on the coordinated action of thousands of genes. A specific group of over one hundred of regulatory
5- Semaphorin-Plexin-Neuropilin
 5- Semaphorin-Plexin-Neuropilin 1 SEMAPHORINS-PLEXINS-NEUROPILINS ligands receptors co-receptors semaphorins and their receptors are known signals for: -axon guidance -cell migration -morphogenesis -immune
5- Semaphorin-Plexin-Neuropilin 1 SEMAPHORINS-PLEXINS-NEUROPILINS ligands receptors co-receptors semaphorins and their receptors are known signals for: -axon guidance -cell migration -morphogenesis -immune
Cell Death & Trophic Factors II. Steven McLoon Department of Neuroscience University of Minnesota
 Cell Death & Trophic Factors II Steven McLoon Department of Neuroscience University of Minnesota 1 Remember? Neurotrophins are cell survival factors that neurons get from their target cells! There is a
Cell Death & Trophic Factors II Steven McLoon Department of Neuroscience University of Minnesota 1 Remember? Neurotrophins are cell survival factors that neurons get from their target cells! There is a
Visual pigments. Neuroscience, Biochemistry Dr. Mamoun Ahram Third year, 2019
 Visual pigments Neuroscience, Biochemistry Dr. Mamoun Ahram Third year, 2019 References Webvision: The Organization of the Retina and Visual System (http://www.ncbi.nlm.nih.gov/books/nbk11522/#a 127) The
Visual pigments Neuroscience, Biochemistry Dr. Mamoun Ahram Third year, 2019 References Webvision: The Organization of the Retina and Visual System (http://www.ncbi.nlm.nih.gov/books/nbk11522/#a 127) The
Supplementary Figure 1. Nature Genetics: doi: /ng.3848
 Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Supplementary Figure 1 Phenotypes and epigenetic properties of Fab2L flies. A- Phenotypic classification based on eye pigment levels in Fab2L male (orange bars) and female (yellow bars) flies (n>150).
Biol403 - Receptor Serine/Threonine Kinases
 Biol403 - Receptor Serine/Threonine Kinases The TGFβ (transforming growth factorβ) family of growth factors TGFβ1 was first identified as a transforming factor; however, it is a member of a family of structurally
Biol403 - Receptor Serine/Threonine Kinases The TGFβ (transforming growth factorβ) family of growth factors TGFβ1 was first identified as a transforming factor; however, it is a member of a family of structurally
MicroRNA mir-34 provides robustness to environmental stress response via
 MicroRNA mir-34 provides robustness to environmental stress respoe via the DAF-16 network in C. elega Meltem Isik 1,2, T. Keith Blackwell 2, and Eugene Berezikov 1,3 1 Hubrecht Ititute-KNAW and University
MicroRNA mir-34 provides robustness to environmental stress respoe via the DAF-16 network in C. elega Meltem Isik 1,2, T. Keith Blackwell 2, and Eugene Berezikov 1,3 1 Hubrecht Ititute-KNAW and University
Regulation of gene expression. Premedical - Biology
 Regulation of gene expression Premedical - Biology Regulation of gene expression in prokaryotic cell Operon units system of negative feedback positive and negative regulation in eukaryotic cell - at any
Regulation of gene expression Premedical - Biology Regulation of gene expression in prokaryotic cell Operon units system of negative feedback positive and negative regulation in eukaryotic cell - at any
Nature Genetics: doi: /ng Supplementary Figure 1. The phenotypes of PI , BR121, and Harosoy under short-day conditions.
 Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Supplementary Figure 1 The phenotypes of PI 159925, BR121, and Harosoy under short-day conditions. (a) Plant height. (b) Number of branches. (c) Average internode length. (d) Number of nodes. (e) Pods
Three different fusions led to three basic ideas: 1) If one fuses a cell in mitosis with a cell in any other stage of the cell cycle, the chromosomes
 Section Notes The cell division cycle presents an interesting system to study because growth and division must be carefully coordinated. For many cells it is important that it reaches the correct size
Section Notes The cell division cycle presents an interesting system to study because growth and division must be carefully coordinated. For many cells it is important that it reaches the correct size
downstream (0.8 kb) homologous sequences to the genomic locus of DIC. A DIC mutant strain (ro- 6
 A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
A B C D ts Figure S1 Generation of DIC- mcherry expressing N.crassa strain. A. N. crassa colony morphology. When a cot1 (top, left panel) strain is grown at permissive temperature (25 C), it exhibits straight
9/4/2015 INDUCTION CHAPTER 1. Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology. Fig 1.
 INDUCTION CHAPTER 1 Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology Fig 1.1 1 EVOLUTION OF METAZOAN BRAINS GASTRULATION MAKING THE 3 RD GERM LAYER
INDUCTION CHAPTER 1 Neurons are similar across phyla Thus, many different model systems are used in developmental neurobiology Fig 1.1 1 EVOLUTION OF METAZOAN BRAINS GASTRULATION MAKING THE 3 RD GERM LAYER
Supplementary Figure 1. Biochemical and sequence alignment analyses the
 Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Supplementary Figure 1. Biochemical and sequence alignment analyses the interaction of OPTN and TBK1. (a) Analytical gel filtration chromatography analysis of the interaction between TBK1 CTD and OPTN(1-119).
Cell Cell Communication in Development
 Biology 4361 Developmental Biology Cell Cell Communication in Development June 25, 2008 Cell Cell Communication Concepts Cells in developing organisms develop in the context of their environment, including
Biology 4361 Developmental Biology Cell Cell Communication in Development June 25, 2008 Cell Cell Communication Concepts Cells in developing organisms develop in the context of their environment, including
Constitutively active Protein kinase D acts as negative regulator of the Slingshot-phosphatase in Drosophila
 Hereditas 147: 237 242 (2010) Constitutively active Protein kinase D acts as negative regulator of the Slingshot-phosphatase in Drosophila ANJA C. NAGEL, JENS SCHMID, JASMIN S. AUER, ANETTE PREISS and
Hereditas 147: 237 242 (2010) Constitutively active Protein kinase D acts as negative regulator of the Slingshot-phosphatase in Drosophila ANJA C. NAGEL, JENS SCHMID, JASMIN S. AUER, ANETTE PREISS and
Written Exam 15 December Course name: Introduction to Systems Biology Course no
 Technical University of Denmark Written Exam 15 December 2008 Course name: Introduction to Systems Biology Course no. 27041 Aids allowed: Open book exam Provide your answers and calculations on separate
Technical University of Denmark Written Exam 15 December 2008 Course name: Introduction to Systems Biology Course no. 27041 Aids allowed: Open book exam Provide your answers and calculations on separate
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family
 Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Leucine-rich repeat receptor-like kinases (LRR-RLKs), HAESA, ERECTA-family GENES & DEVELOPMENT (2000) 14: 108 117 INTRODUCTION Flower Diagram INTRODUCTION Abscission In plant, the process by which a plant
Chapter 18 Regulation of Gene Expression
 Chapter 18 Regulation of Gene Expression Differential gene expression Every somatic cell in an individual organism contains the same genetic information and replicated from the same original fertilized
Chapter 18 Regulation of Gene Expression Differential gene expression Every somatic cell in an individual organism contains the same genetic information and replicated from the same original fertilized
PRACTICE EXAM. 20 pts: 1. With the aid of a diagram, indicate how initial dorsal-ventral polarity is created in fruit fly and frog embryos.
 PRACTICE EXAM 20 pts: 1. With the aid of a diagram, indicate how initial dorsal-ventral polarity is created in fruit fly and frog embryos. No Low [] Fly Embryo Embryo Non-neural Genes Neuroectoderm Genes
PRACTICE EXAM 20 pts: 1. With the aid of a diagram, indicate how initial dorsal-ventral polarity is created in fruit fly and frog embryos. No Low [] Fly Embryo Embryo Non-neural Genes Neuroectoderm Genes
Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc
 OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
OPTIMIZATION OF IMMUNOBLOT PROTOCOL 121 Optimization of Immunoblot Protocol for Use with a Yeast Strain Containing the CDC7 Gene Tagged with myc Jacqueline Bjornton and John Wheeler Faculty Sponsor: Anne
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering
 Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
Photoreceptor Regulation of Constans Protein in Photoperiodic Flowering by Valverde et. Al Published in Science 2004 Presented by Boyana Grigorova CBMG 688R Feb. 12, 2007 Circadian Rhythms: The Clock Within
3/8/ Complex adaptations. 2. often a novel trait
 Chapter 10 Adaptation: from genes to traits p. 302 10.1 Cascades of Genes (p. 304) 1. Complex adaptations A. Coexpressed traits selected for a common function, 2. often a novel trait A. not inherited from
Chapter 10 Adaptation: from genes to traits p. 302 10.1 Cascades of Genes (p. 304) 1. Complex adaptations A. Coexpressed traits selected for a common function, 2. often a novel trait A. not inherited from
Lesson Overview. Gene Regulation and Expression. Lesson Overview Gene Regulation and Expression
 13.4 Gene Regulation and Expression THINK ABOUT IT Think of a library filled with how-to books. Would you ever need to use all of those books at the same time? Of course not. Now picture a tiny bacterium
13.4 Gene Regulation and Expression THINK ABOUT IT Think of a library filled with how-to books. Would you ever need to use all of those books at the same time? Of course not. Now picture a tiny bacterium
