Basics of molecular dynamics
|
|
|
- Ashlynn Hancock
- 5 years ago
- Views:
Transcription
1 Basics of molecular dynamics The basic idea of molecular dynamics (MD) simulations is to calculate how a system of particles evolves in time. The method was first used by Alder and Wainwright in 1957 to calculate properties of many-body systems. They called the particles molecules. There is an interesting parallel to classical mechanics here. The two-body motion problem was solved by Newton way back then. The three-body problem was solved by a Finnish guy, Sundman, in the early part of the last century - but the solution is utterly impractical ( terms needed in a series expansion). The N-body problem, N 3, can not be solved analytically. MD can also described to be a numerical way of solving the N-body problem. The solution is of course never exact, but if done properly it can be done arbitrarily accurately. Consider a set of atoms at positions r i and some interaction model which gives us the potential energy of the system Vr i In Newtonian mechanics we then get: dr i = v dt i, ---- d m dt i v i = F i = i V = i V 2 r i r j j + V 3 r i r j r k jk By solving the above set of equations numerically we can derive dr over some short time interval dt. + Introduction to atomistic simulations Basics of molecular dynamics 1 Basic MD algorithm (slightly simplified) Set the initial conditions r i t 0, v i t 0 Get new forces F i r i Solve the equations of motion numerically over time step t : r i t n r i t n + 1 v i t n v i t n + 1 tt + t Get desired physical quantities tt max? Calculate results and finish Introduction to atomistic simulations Basics of molecular dynamics 2
2 An alternative view MD-simulation of thermal motion over 100 time steps Zoom in on 2 time steps (5 atoms): At time t the distances r ij and hence forces F ij between nearby atoms are calculated From these forces we can solve the equations of motion, and hence get new positions and velocities. 3 2 The displacement over a time step t is denoted r. r has to be much smaller than the distance between nearby atoms. = position at t = t i = position at t = t i r vt + --at 2 F a = m 5 r 13 F 13 1 r 15 F 15 r 12 F 12 r 14 F 14 4 Introduction to atomistic simulations Basics of molecular dynamics 3 General considerations The above was the simplest possible example, the so called microcanonical or NVE ensemble. This means that the approach preserves the number of atoms N, the volume of the cell V and the energy E. Other ensembles will be dealt with later on in the course. But the NVE ensemble is the most natural one in that it is the true solution of the N-body problem, and corresponds to the real atom motion. First MD simulations: Hard spheres: B. J. Alder, T. E. Wainwright: Phase transition for a Hard Sphere-System, J. Chem. Phys. 27 (1957) 1208 Continuous potentials: J. B. Gibson, A. N. Goland, M. Milgram, G. H. Vineyard: Dynamics of Radiation Damage, Phys. Rev. 120 (1960) State-of-the-art (2008): Of the order of atoms In Finland: CSC Cray XT4 (louhi.csc.fi): some atoms with a realistic potential easily possible for thousands of time steps. If all N atoms interact with all atoms, one has to in principle calculate N 2 interactions. This is prohibitively expensive for millions of atoms. Introduction to atomistic simulations Basics of molecular dynamics 4
3 General considerations Fortunately, in practice most atomic interactions decrease rapidly in strength as r. In that case it is enough to calculate only interactions to nearby atoms. E.g. in diamond-structured semiconductors (Si, Ge, GaAs...) atoms have 4 covalent bonds, so the calculation can be reduced to 4 neighbours => 4 N interactions. In metals atoms more than ~ 5 Å far can usually be neglected => about 80 N interactions In ionic systems the interaction V 1r, i.e. decreases very slowly. It can not be cut off, but there are smart workarounds. Introduction to atomistic simulations Basics of molecular dynamics 5 Early MD simulations Introduction to atomistic simulations Basics of molecular dynamics 6
4 Early MD simulations Vr ( ) = V 0 e r cohesion: inward force on border atoms 500 atoms on IBM 704: 1 minute/time step Introduction to atomistic simulations Basics of molecular dynamics 7 Simulation cell In practice in most cases the atoms are arranged in a orthogonal simulation cell which has a size S x S y S z. It is also perfectly possible to use a simulation cell with axes than are not orthogonal. Problem: what should we do with the atoms at the borders. 1. Nothing: free boundaries This works fine if we want to deal with e.g. a molecule, nanocluster or nanotube in vacuum. If we want to describe a continuous medium, this does not work: the atoms are left hanging on the surface as if they would be on the surface. Introduction to atomistic simulations Basics of molecular dynamics 8
5 Simulation cell 2. Fix the boundary atoms: Completely unphysical, this should be avoided if possible. Sometimes it is needed and with a fairly large sacrificial region next to the fixed one can be perfectly OK. 3. Periodic boundary conditions To implement this very important boundary condition two things have to be done 1. An atom which passes over the cell boundary comes back on the other side: A A Introduction to atomistic simulations Basics of molecular dynamics 9 Simulation cell In practice this can be implemented as follows (Fortran 90) (note that atomic coordinates are between S x 2 and S x 2 ) :!x : particle coordinate!periodicx : = true periodic, false free!xsize : MD cell size (S x ) if (periodicx) then if (x < -xsize/2.0) x = x + xsize if (x >= xsize/2.0) x = x - xsize endif Similarly for y and z 2. When distances between atoms are calculated, the periodic boundaries have to be taken into account: k j r cutoff r ij i l k rij The solid box is the simulation cell, with atoms i, j, k and l. Because of the periodic boundaries, all atoms have image atoms in the repeated cells, for instance j', k', l'. S l j When we want to get the distance between atom i and j, which distance should we use? Because here r ij r ij ', we use for the distance between atoms i and j the vector r ij '. Introduction to atomistic simulations Basics of molecular dynamics 10
6 Simulation cell As a pseudo-algorithm (Fortran 90) in the x dimension: if (periodicx) then dx = x(j) - x(i) if (dx > xsize/2.0) dx = dx - xsize if (dx <= -xsize/2.0) dx = dx + xsize endif and similarly for y and z Example in 1D x j x i x j' x i S -- x S = 2 j x 2 i = x j x i S j' i j S S -- 2 Note that if the maximum distance by which atoms can interact r cutoff > xsize/2 the atom i should actually interact both with atom j and j. To prevent unphysical double interactions we need to have xsize > 2 r cutoff Introduction to atomistic simulations Basics of molecular dynamics 11 Simulation cell Thus we get a system where the simulation cell has an infinite number of image cells in all directions, and a model of an infinite system. However, be careful! Periodicity brings an artificial interaction over the simulation cell borders. For instance, a strain field arising from a point source, which is infinite, will obviously be distorted at the periodic borders. Examples: A single vacancy (one missing atom) in Si: in quantum mechanical calculations at least some 200 atoms are required to get the energy reliably [Puska 1998 Phys. Rev. B] And for instance a 5 nm Co cluster in Cu: about 10 6 atoms needed to get the strain energy reliably. Upper limit for the phonon wavelength. To test this: simulate with different N and monitor the convergence. Introduction to atomistic simulations Basics of molecular dynamics 12
7 Simulation cell Simulating surfaces: periodic boundaries only in x- and y-directions free surface: the bottom either: a) free: simulation of a free-standing thin foil with two surfaces or b) fixed to model a bulk below: very bottommost atoms fixed a few atom layers above fixed layers damped with e.g. a temperature control algorithm z x Introduction to atomistic simulations Basics of molecular dynamics 13 Simulation cell Simulation of energetic processes: In a simulation where a lot of energy is brought into the MD cell in a local region, the energy has to be scaled out from the system to model a much cooler heat bath in a realistic system. The energetic processes may also introduce a lot of momentum into the cell, which could cause the entire cell to move. Solution: fix all boundary atoms except at the surface, and do T scaling in a few atom layers above these, as above. Here also: watch out the finite-size effects! Introduction to atomistic simulations Basics of molecular dynamics 14
8 Initial conditions: creating atoms For cubic lattices (FCC, BCC, SC, DIA) it is easy to create the lattice. For instance FCC: basis(1,1)=0.0; basis(1,2)=0.0; basis(1,3)=0.0; basis(2,1)=0.5; basis(2,2)=0.5; basis(2,3)=0.0; basis(3,1)=0.5; basis(3,2)=0.0; basis(3,3)=0.5; basis(4,1)=0.0; basis(4,2)=0.5; basis(4,3)=0.5; offset(1)=0.25; offset(2)=0.25; offset(3)=0.25; nbasis=4; n=0; do i=0,nx-1 do j=0,ny-1 do k=0,nz-1 do m=1,nbasis n=n+1 x(n)=-xsize/2+(i+offset(1)+basis(m,1))*a y(n)=-ysize/2+(j+offset(2)+basis(m,2))*a z(n)=-zsize/2+(k+offset(3)+basis(m,3))*a enddo enddo enddo enddo The HCP lattice is also very common, but not orthogonal in the conventional representation. Because in the HCP structure a = b, and because cos60 = 1 2, the HCP lattice can be transformed into an equivalent orthogonal representation. Now the new unit cell (shaded area) corresponds to two of the conventional HCP unit cells. Coordinates between To refresh your memory: FCC : face centered cubic BCC : body centred cubic SC: simple cubic DIA: diamond HCP: hexagonal close- packed Introduction to atomistic simulations Basics of molecular dynamics S 2 b and S -- 2 b' 60 o a a' Initial atom velocities How do we set the cell temperature to a desired value? We have to generate initial atom velocities which correspond to the Maxwell-Boltzmann distribution (which is surprisingly well valid even in crystals): v i m i 1/ 2 1 = exp --m 2k B T 2 i v2 i k B T ; = xyz. This is just a Gaussian function with suitable scaling We usually also want to set the total momentum of the cell to zero to prevent the entire cell from starting to move: N P = m i v i i = 1 So in practice all this can be achieved with the code fragment on the right: 2 v i Note: exp , 2 k B T = m i sigma2v=sqrt(kb*2*t/(m*u))/vunit do i=1,n vx(i)=sigma2v*gaussrandom(iseed) vy(i)=sigma2v*gaussrandom(iseed) vz(i)=sigma2v*gaussrandom(iseed) vxtot=vxtot+vx(i)! If all atoms have the same mass, vytot=vytot+vy(i)! it is enough to scale the total v vztot=vztot+vz(i)! to zero enddo vxtot=vxtot/n vytot=vytot/n vztot=vztot/n do i=1,n vx(i)=vx(i)-vxtot vy(i)=vy(i)-vytot vz(i)=vz(i)-vztot enddo Introduction to atomistic simulations Basics of molecular dynamics 16
9 Initial atom velocities Note the factor of 2: if the simulation is started from perfect lattice sites, or bound equilibrium positions in a molecule, half of the initial kinetic energy will be changed to potential energy after a while. E V(r) r 0 r E kin and E pot It is also possible to get realistic initial random displacements. E kin only This can be derived from the Debye model: the thermal displacement in the direction of the axis i is a Gaussian distribution of the form w i T 2 1/ 2 e i 2 2 = 2 where T = --Å 9h 2, where = Å K 3 A k D B u D = Debye temperature of the material, A = Atomic mass The initial position can now be obtained with Gaussian-distributed random numbers as above. Note, however, that this does not account for quantum mechanical zero-point vibrations which give additional displacements near 0 K. Introduction to atomistic simulations Basics of molecular dynamics 17 Generating random numbers (This topic is dealt with in much more detail on the MC simulation course) Almost all kinds of simuations in physics use random numbers somewhere. As we saw above, MD simulations need them at least for initial velocity generation. Computer-generated random numbers are of course not truly random, but if they have been generated with a good algorithm, they start to repeat each other only after a very large (e.g ) number of iterations. If the number of random numbers used in the entire simulation is much less than the repeat number, the algorithm probably is good enough for the application. Random numbers can be generated for different distributions. This means that if we generate a large number of numbers and make statistics out of them, they will eventually approach some distribution. The most common is of course an even distribution in an interval, another very common is Gaussian-distributed numbers: P(x) P(x) Introduction to atomistic simulations Basics of molecular dynamics 18
10 Generating random numbers Evenly distributed random numbers: Many programming languages offer their own random number generator (e.g. in ANSI-C rand()). A good rule-of-thumb regarding these is: Never use them for anything serious! The reason is simply that the language standard only specifies that the generator has to be there, not that it works sensibly. Since there are no guarantees it does (there are famous examples of the opposite) it should not be used Most random number generators are based on modulo-arithmetics and iteration. In the simplest possible form: I j + 1 = ai j mod m Park and Miller minimal standard -generator: a = 16807, m = In the beginning the number I 0 i.e. the seed number is chosen randomly. This can be done e.g. by using the current system time. Introduction to atomistic simulations Basics of molecular dynamics 19 Generating random numbers One practical implementation (Fortran90): The repeat interval for this routine ~ This routine is easily good enough if for instance it is only needed for the choice of random numbers in the beginning of an MD simulation. In a long Monte Carlo integration where random numbers are used all the time, the repeat interval may be reached after which continued running will not improve on the results (and for instance the error estimate of the result will be calculated outright wrong). real function uniformrand(seed) implicit none integer :: seed,ia,im,iq,ir,mask real :: ran0,am integer :: k parameter (IA=16807,IM= ,AM=1.0/IM) parameter (IQ=127773,IR=2836,MASK= ) seed=ieor(seed,mask) k=seed/iq seed=ia*(seed-k*iq)-ir*k if (seed < 0) seed=seed+im uniformrand=am*seed seed=ieor(seed,mask) return end function uniformrand More on this topic for instance from the book: Press, Teukolsky, Vetterling, Flannery: Numerical Recipes in C/Fortran, 2nd. ed., chapter 7. The book is on-line in its entirety (see But see also: Why not use Numerical Recipes?, and the reply to this: Introduction to atomistic simulations Basics of molecular dynamics 20
11 Generating random numbers To generate Gaussian random velocities we need to be able to generate Gaussian-distributed random numbers. How to do this is dealt with in great detail in Numerical Recipes ch Here we only present the most efficient accurate algorithm for this: 1 o 2 2 Obtain two evenly distributed random numbers v 1 and v 2 between -1 and 1, then calculate w = v 1 +v 2 2 o If w 1 return to step 1 o 3 o Calculate r = 2logw 4 o Calculate x = rv 1 w and y = rv 2 w 5 o Return x and on next step y Introduction to atomistic simulations Basics of molecular dynamics 21 Choosing the MD time step Depends on the integration algorithm used, but not too strongly. The change in the atom position in the potential used should not be too strong. A practical, rough rule-of-thumb: the atoms should not move more than 1/20 of the nearest-neighbour distance. E V(r) r 0 r Thermal velocity of atoms (Maxwell-Boltzmann distribution): 3 E rms = --k 2 B T = 1 --mv 2 3kT v 2 rms = m But the distribution continues much beyond this. Rough estimate of the time step needed: 300 K Cu (m = 63.55u): 5v rms = Å/fs Nearest-neighbour distance 2.55 Å=> t = fs = 7.5 fs In practice for stability t 4 fs. Introduction to atomistic simulations Basics of molecular dynamics 22
12 Choosing the MD time step In pure MD there is no way to increase the time step above ~ 10 fs in atom systems at ordinary temperatures (77 K and up). If we would want to simulate a process which, say, takes 1 s, we would need at least time steps! This gives an easy way to estimate the order-of-magnitude of the upper limit for the time scale MD can handle in a given time: Most realistic classical MD interatomic potentials require at least of the order of 100 flops/atom/time step. Say our time step is 1 fs, and we want to simulate a atom system. Hence we need 10 6 flops/time step. To get to 1 ns = 10 9 fs we would need flops. Assuming 1 Gflop/ s processor, the simulation would thus require /10 9 seconds = 10 6 s i.e. about 11 days. To get to 1 s would require some 30 years on this processor. Hence we see that ordinary MD is restricted to 100 ns processes in practical use. Introduction to atomistic simulations Basics of molecular dynamics 23 Choosing the MD step In ordinary equilibrium MD t is usually constant throughout simulation But if the maximum velocity of atoms changes a lot during the simulation, it is best to use a variable time step, which increases as the maximum velocity decreases. Simulations of energetic processes: t n + 1 min k t E t = ( c v max F max v t t n, t max ) max k t maximum movement distance/time step (e.g. 0.1Å) E t maximum allowed energy change/time step (e.g. 300 ev) c t prevents too large sudden changes (e.g. c t = 1.1) v max F max t max maximum atom speed in system maximum force on any atom in system time step once heat bath T has been reached The example values above have been found to work well for binary collisions up to 1 GeV in many materials. Introduction to atomistic simulations Basics of molecular dynamics 24
13 Choosing the MD step What happens if t is too long? The energy is not conserved. For instance solid copper (FCC lattice, a = 3.615Å, EAM potential, code parcas) NVE simulation at 300 K: Temperature Hence the real criterion for selecting the time step becomes energy conservation: for every: new kind of system new kind of process simulated new material new interaction potential One needs to check that energy is conserved well enough by some test simulations, before starting the real production runs. Change in total energy Introduction to atomistic simulations Basics of molecular dynamics 25 Acceleration methods Speeding up MD This can be achieved at least in some cases where we are interested in transitions induced by thermally activated processes, i.e. processes which follow a behaviour of the type 0 e E A k B = T where is the rate of the process occurring. System spends most of its time in local potential energy minima B i (basins). Every once in a while it gets enough kinetic energy to go over the barrier E A : rare events. Acceleration: increase by increasing the probability for barrier crossing. Modify E A or T (??) Introduction to atomistic simulations Basics of molecular dynamics 26
14 Acceleration methods Art Voter has presented so called Hyperdynamics [A. F. Voter, J. Chem. Phys. 106 (1997) 4665; Phys. Rev. Lett. 78 (1997) 3908]. It can in some cases speed up MD by a factor of the order of , in others not at all. In this method, t does not increase, but the potential well is made shallower so that the probability of processes with a large activation energy increases. The error which is thus formed is compensated by transition state theory (which is beyond the scope of this course). The method is well suited for cases where we have to overcome a high potential energy barrier in an ordered system, e.g. vacancy and adatom diffusion. But if the energy barrier is low (e.g. interstitial migration in metals) or if we have numerous local energy minima close to each other, like in most amorphous and liquid systems, the method is useless. E pot E boost E boost Introduction to atomistic simulations Basics of molecular dynamics 27 Acceleration methods Temperature accelerated dynamics (TAD) There is of course always is the Arrhenius extrapolation method: if we know that in our system there is only one single activated process occurring, and nothing else, we can simulate at higher T and then extrapolate the Arrhenius-like exponential exp E A k B T to lower T to know the rate or time scale at lower T. A smart extension to Arrhenius extrapolation is Art Voter s TAD method [e.g. Sorensen, Phys. Rev. B 62 (2000) 3658; a review of Voters methods is given in Ann. Rev. Mater. Res. 32 (2002) 321] To understand the idea in this, let us consider a system with exactly 2 activation energies (this is just a tutorial example, the method works in principle for any number of activation energies). We want to simulate what the system does at 300 K, but the processes are so slow nothing happens there. So we will use a higher T, say 800 K. Introduction to atomistic simulations Basics of molecular dynamics 28
15 Acceleration methods Let us then assume that the Arrhenius plot of the system looks as follows: log, rate of event occurance E A,1 E A,2 EA,1 > E A,2 0 e E A k B = T E log = log A k B T 0 1/800 1/300 1 T 1/K Now when we simulate at 800 K, event type 1 will occur much more frequently than type 2. But we want to know the behaviour at 300 K, so this is wrong. The idea in TAD is to recognize every transition that occurs, determine its activation energy, and then leave out the events that would not occur at the lower T. In our example, this means that (almost) only events of type 2 would be accepted. Introduction to atomistic simulations Basics of molecular dynamics 29 Acceleration methods In principle this is an excellent idea, but in practice one needs thousands of force evaluations to recognize a transition barrier. Hence the difference between the rates of occurrance needs to be very large for a significant gain to be achieved. But the gain can be huge (Example: simulating growth of Cu (001) surface at 77 K the speedup factor is 10 7!) Like hyperdynamics, if there are lots of shallow minima TAD tends to get stuck and never really gets anywhere. TAD is developed rapidly towards wider applicability, so it will be interesting to follow the progress. Introduction to atomistic simulations Basics of molecular dynamics 30
Molecular Dynamics Simulations of Fusion Materials: Challenges and Opportunities (Recent Developments)
 Molecular Dynamics Simulations of Fusion Materials: Challenges and Opportunities (Recent Developments) Fei Gao gaofeium@umich.edu Limitations of MD Time scales Length scales (PBC help a lot) Accuracy of
Molecular Dynamics Simulations of Fusion Materials: Challenges and Opportunities (Recent Developments) Fei Gao gaofeium@umich.edu Limitations of MD Time scales Length scales (PBC help a lot) Accuracy of
3.320 Lecture 23 (5/3/05)
 3.320 Lecture 23 (5/3/05) Faster, faster,faster Bigger, Bigger, Bigger Accelerated Molecular Dynamics Kinetic Monte Carlo Inhomogeneous Spatial Coarse Graining 5/3/05 3.320 Atomistic Modeling of Materials
3.320 Lecture 23 (5/3/05) Faster, faster,faster Bigger, Bigger, Bigger Accelerated Molecular Dynamics Kinetic Monte Carlo Inhomogeneous Spatial Coarse Graining 5/3/05 3.320 Atomistic Modeling of Materials
Introduction to Molecular dynamics
 Introduction to Molecular dynamics Complete lecture notes for self-studies Professor Kai Nordlund and University lecturer Antti Kuronen 1999-2015 The idea of the course is to teach the students the basics
Introduction to Molecular dynamics Complete lecture notes for self-studies Professor Kai Nordlund and University lecturer Antti Kuronen 1999-2015 The idea of the course is to teach the students the basics
MatSci 331 Homework 4 Molecular Dynamics and Monte Carlo: Stress, heat capacity, quantum nuclear effects, and simulated annealing
 MatSci 331 Homework 4 Molecular Dynamics and Monte Carlo: Stress, heat capacity, quantum nuclear effects, and simulated annealing Due Thursday Feb. 21 at 5pm in Durand 110. Evan Reed In this homework,
MatSci 331 Homework 4 Molecular Dynamics and Monte Carlo: Stress, heat capacity, quantum nuclear effects, and simulated annealing Due Thursday Feb. 21 at 5pm in Durand 110. Evan Reed In this homework,
Potentials, periodicity
 Potentials, periodicity Lecture 2 1/23/18 1 Survey responses 2 Topic requests DFT (10), Molecular dynamics (7), Monte Carlo (5) Machine Learning (4), High-throughput, Databases (4) NEB, phonons, Non-equilibrium
Potentials, periodicity Lecture 2 1/23/18 1 Survey responses 2 Topic requests DFT (10), Molecular dynamics (7), Monte Carlo (5) Machine Learning (4), High-throughput, Databases (4) NEB, phonons, Non-equilibrium
Uniform-acceptance force-biased Monte Carlo: A cheap way to boost MD
 Dresden Talk, March 2012 Uniform-acceptance force-biased Monte Carlo: A cheap way to boost MD Barend Thijsse Department of Materials Science and Engineering Delft University of Technology, The Netherlands
Dresden Talk, March 2012 Uniform-acceptance force-biased Monte Carlo: A cheap way to boost MD Barend Thijsse Department of Materials Science and Engineering Delft University of Technology, The Netherlands
MD Thermodynamics. Lecture 12 3/26/18. Harvard SEAS AP 275 Atomistic Modeling of Materials Boris Kozinsky
 MD Thermodynamics Lecture 1 3/6/18 1 Molecular dynamics The force depends on positions only (not velocities) Total energy is conserved (micro canonical evolution) Newton s equations of motion (second order
MD Thermodynamics Lecture 1 3/6/18 1 Molecular dynamics The force depends on positions only (not velocities) Total energy is conserved (micro canonical evolution) Newton s equations of motion (second order
Semiconductor physics I. The Crystal Structure of Solids
 Lecture 3 Semiconductor physics I The Crystal Structure of Solids 1 Semiconductor materials Types of solids Space lattices Atomic Bonding Imperfection and doping in SOLIDS 2 Semiconductor Semiconductors
Lecture 3 Semiconductor physics I The Crystal Structure of Solids 1 Semiconductor materials Types of solids Space lattices Atomic Bonding Imperfection and doping in SOLIDS 2 Semiconductor Semiconductors
Electronic Structure Theory for Periodic Systems: The Concepts. Christian Ratsch
 Electronic Structure Theory for Periodic Systems: The Concepts Christian Ratsch Institute for Pure and Applied Mathematics and Department of Mathematics, UCLA Motivation There are 10 20 atoms in 1 mm 3
Electronic Structure Theory for Periodic Systems: The Concepts Christian Ratsch Institute for Pure and Applied Mathematics and Department of Mathematics, UCLA Motivation There are 10 20 atoms in 1 mm 3
Atoms, electrons and Solids
 Atoms, electrons and Solids Shell model of an atom negative electron orbiting a positive nucleus QM tells that to minimize total energy the electrons fill up shells. Each orbit in a shell has a specific
Atoms, electrons and Solids Shell model of an atom negative electron orbiting a positive nucleus QM tells that to minimize total energy the electrons fill up shells. Each orbit in a shell has a specific
Lecture 11 - Phonons II - Thermal Prop. Continued
 Phonons II - hermal Properties - Continued (Kittel Ch. 5) Low High Outline Anharmonicity Crucial for hermal expansion other changes with pressure temperature Gruneisen Constant hermal Heat ransport Phonon
Phonons II - hermal Properties - Continued (Kittel Ch. 5) Low High Outline Anharmonicity Crucial for hermal expansion other changes with pressure temperature Gruneisen Constant hermal Heat ransport Phonon
Constructing a neighbour list
 Constructing a neighbour list In MD simulations (and actually many other applications) one of the central operations is the calculation of distances between atoms. In MD this is needed in the energy and
Constructing a neighbour list In MD simulations (and actually many other applications) one of the central operations is the calculation of distances between atoms. In MD this is needed in the energy and
Module 16. Diffusion in solids II. Lecture 16. Diffusion in solids II
 Module 16 Diffusion in solids II Lecture 16 Diffusion in solids II 1 NPTEL Phase II : IIT Kharagpur : Prof. R. N. Ghosh, Dept of Metallurgical and Materials Engineering Keywords: Micro mechanisms of diffusion,
Module 16 Diffusion in solids II Lecture 16 Diffusion in solids II 1 NPTEL Phase II : IIT Kharagpur : Prof. R. N. Ghosh, Dept of Metallurgical and Materials Engineering Keywords: Micro mechanisms of diffusion,
Molecular dynamics simulation. CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror
 Molecular dynamics simulation CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror 1 Outline Molecular dynamics (MD): The basic idea Equations of motion Key properties of MD simulations Sample applications
Molecular dynamics simulation CS/CME/BioE/Biophys/BMI 279 Oct. 5 and 10, 2017 Ron Dror 1 Outline Molecular dynamics (MD): The basic idea Equations of motion Key properties of MD simulations Sample applications
Kinetic Monte Carlo (KMC)
 Kinetic Monte Carlo (KMC) Molecular Dynamics (MD): high-frequency motion dictate the time-step (e.g., vibrations). Time step is short: pico-seconds. Direct Monte Carlo (MC): stochastic (non-deterministic)
Kinetic Monte Carlo (KMC) Molecular Dynamics (MD): high-frequency motion dictate the time-step (e.g., vibrations). Time step is short: pico-seconds. Direct Monte Carlo (MC): stochastic (non-deterministic)
Molecular Dynamics and Accelerated Molecular Dynamics
 Molecular Dynamics and Accelerated Molecular Dynamics Arthur F. Voter Theoretical Division National Laboratory Lecture 3 Tutorial Lecture Series Institute for Pure and Applied Mathematics (IPAM) UCLA September
Molecular Dynamics and Accelerated Molecular Dynamics Arthur F. Voter Theoretical Division National Laboratory Lecture 3 Tutorial Lecture Series Institute for Pure and Applied Mathematics (IPAM) UCLA September
Phonons I - Crystal Vibrations (Kittel Ch. 4)
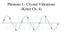 Phonons I - Crystal Vibrations (Kittel Ch. 4) Displacements of Atoms Positions of atoms in their perfect lattice positions are given by: R 0 (n 1, n 2, n 3 ) = n 10 x + n 20 y + n 30 z For simplicity here
Phonons I - Crystal Vibrations (Kittel Ch. 4) Displacements of Atoms Positions of atoms in their perfect lattice positions are given by: R 0 (n 1, n 2, n 3 ) = n 10 x + n 20 y + n 30 z For simplicity here
Condensed Matter Physics Prof. G. Rangarajan Department of Physics Indian Institute of Technology, Madras
 Condensed Matter Physics Prof. G. Rangarajan Department of Physics Indian Institute of Technology, Madras Lecture - 10 The Free Electron Theory of Metals - Electrical Conductivity (Refer Slide Time: 00:20)
Condensed Matter Physics Prof. G. Rangarajan Department of Physics Indian Institute of Technology, Madras Lecture - 10 The Free Electron Theory of Metals - Electrical Conductivity (Refer Slide Time: 00:20)
Monte Carlo. Lecture 15 4/9/18. Harvard SEAS AP 275 Atomistic Modeling of Materials Boris Kozinsky
 Monte Carlo Lecture 15 4/9/18 1 Sampling with dynamics In Molecular Dynamics we simulate evolution of a system over time according to Newton s equations, conserving energy Averages (thermodynamic properties)
Monte Carlo Lecture 15 4/9/18 1 Sampling with dynamics In Molecular Dynamics we simulate evolution of a system over time according to Newton s equations, conserving energy Averages (thermodynamic properties)
Set the initial conditions r i. Update neighborlist. r i. Get new forces F i
 Set the initial conditions r i t 0, v i t 0 Update neighborlist Get new forces F i r i Solve the equations of motion numerically over time step t : r i t n r i t n + 1 v i t n v i t n + 1 Perform T, P
Set the initial conditions r i t 0, v i t 0 Update neighborlist Get new forces F i r i Solve the equations of motion numerically over time step t : r i t n r i t n + 1 v i t n v i t n + 1 Perform T, P
Properties of Individual Nanoparticles
 TIGP Introduction technology (I) October 15, 2007 Properties of Individual Nanoparticles Clusters 1. Very small -- difficult to image individual nanoparticles. 2. New physical and/or chemical properties
TIGP Introduction technology (I) October 15, 2007 Properties of Individual Nanoparticles Clusters 1. Very small -- difficult to image individual nanoparticles. 2. New physical and/or chemical properties
Molecular Dynamics. What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: F&S Chapter 4
 Molecular Dynamics What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: F&S Chapter 4 MSE485/PHY466/CSE485 1 The Molecular Dynamics (MD) method
Molecular Dynamics What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: F&S Chapter 4 MSE485/PHY466/CSE485 1 The Molecular Dynamics (MD) method
Bulk Structures of Crystals
 Bulk Structures of Crystals 7 crystal systems can be further subdivided into 32 crystal classes... see Simon Garrett, "Introduction to Surface Analysis CEM924": http://www.cem.msu.edu/~cem924sg/lecturenotes.html
Bulk Structures of Crystals 7 crystal systems can be further subdivided into 32 crystal classes... see Simon Garrett, "Introduction to Surface Analysis CEM924": http://www.cem.msu.edu/~cem924sg/lecturenotes.html
Joint ICTP-IAEA Workshop on Physics of Radiation Effect and its Simulation for Non-Metallic Condensed Matter.
 2359-16 Joint ICTP-IAEA Workshop on Physics of Radiation Effect and its Simulation for Non-Metallic Condensed Matter 13-24 August 2012 Introduction to atomistic long time scale methods Roger Smith Loughborough
2359-16 Joint ICTP-IAEA Workshop on Physics of Radiation Effect and its Simulation for Non-Metallic Condensed Matter 13-24 August 2012 Introduction to atomistic long time scale methods Roger Smith Loughborough
Part III. Cellular Automata Simulation of. Monolayer Surfaces
 Part III Cellular Automata Simulation of Monolayer Surfaces The concept of progress acts as a protective mechanism to shield us from the terrors of the future. the words of Paul Muad Dib 193 Chapter 6
Part III Cellular Automata Simulation of Monolayer Surfaces The concept of progress acts as a protective mechanism to shield us from the terrors of the future. the words of Paul Muad Dib 193 Chapter 6
Constructing a neighbour list
 In MD simulations (and actually many other applications) one of the central operations is the calculation of distances between atoms. In MD this is needed in the energy and force calculation. Trivial calculation
In MD simulations (and actually many other applications) one of the central operations is the calculation of distances between atoms. In MD this is needed in the energy and force calculation. Trivial calculation
Partitioning 3D space for parallel many-particle simulations
 Partitioning 3D space for parallel many-particle simulations M. A. Stijnman and R. H. Bisseling Mathematical Institute, Utrecht University, PO Box 80010, 3508 TA Utrecht, The Netherlands G. T. Barkema
Partitioning 3D space for parallel many-particle simulations M. A. Stijnman and R. H. Bisseling Mathematical Institute, Utrecht University, PO Box 80010, 3508 TA Utrecht, The Netherlands G. T. Barkema
Basics of Statistical Mechanics
 Basics of Statistical Mechanics Review of ensembles Microcanonical, canonical, Maxwell-Boltzmann Constant pressure, temperature, volume, Thermodynamic limit Ergodicity (see online notes also) Reading assignment:
Basics of Statistical Mechanics Review of ensembles Microcanonical, canonical, Maxwell-Boltzmann Constant pressure, temperature, volume, Thermodynamic limit Ergodicity (see online notes also) Reading assignment:
What is Classical Molecular Dynamics?
 What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
What is Classical Molecular Dynamics? Simulation of explicit particles (atoms, ions,... ) Particles interact via relatively simple analytical potential functions Newton s equations of motion are integrated
CHAPTER 2: ENERGY BANDS & CARRIER CONCENTRATION IN THERMAL EQUILIBRIUM. M.N.A. Halif & S.N. Sabki
 CHAPTER 2: ENERGY BANDS & CARRIER CONCENTRATION IN THERMAL EQUILIBRIUM OUTLINE 2.1 INTRODUCTION: 2.1.1 Semiconductor Materials 2.1.2 Basic Crystal Structure 2.1.3 Basic Crystal Growth technique 2.1.4 Valence
CHAPTER 2: ENERGY BANDS & CARRIER CONCENTRATION IN THERMAL EQUILIBRIUM OUTLINE 2.1 INTRODUCTION: 2.1.1 Semiconductor Materials 2.1.2 Basic Crystal Structure 2.1.3 Basic Crystal Growth technique 2.1.4 Valence
Molecular Dynamics 9/6/16
 Molecular Dynamics What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: Lesar Chpt 6, F&S Chpt 4 1 The Molecular Dynamics (MD) method for classical
Molecular Dynamics What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: Lesar Chpt 6, F&S Chpt 4 1 The Molecular Dynamics (MD) method for classical
Module 5: Rise and Fall of the Clockwork Universe. You should be able to demonstrate and show your understanding of:
 OCR B Physics H557 Module 5: Rise and Fall of the Clockwork Universe You should be able to demonstrate and show your understanding of: 5.2: Matter Particle model: A gas consists of many very small, rapidly
OCR B Physics H557 Module 5: Rise and Fall of the Clockwork Universe You should be able to demonstrate and show your understanding of: 5.2: Matter Particle model: A gas consists of many very small, rapidly
BASICS OF HMT-1. This subject is always fun to teach/study in winters
 BASICS OF HMT-1 This subject is always fun to teach/study in winters #1 What is heat? Heat is a form of thermal energy which is defined ONLY when it moves from one place to other. Otherwise, the thermal
BASICS OF HMT-1 This subject is always fun to teach/study in winters #1 What is heat? Heat is a form of thermal energy which is defined ONLY when it moves from one place to other. Otherwise, the thermal
Practical calculations using first-principles QM Convergence, convergence, convergence
 Practical calculations using first-principles QM Convergence, convergence, convergence Keith Refson STFC Rutherford Appleton Laboratory September 18, 2007 Results of First-Principles Simulations..........................................................
Practical calculations using first-principles QM Convergence, convergence, convergence Keith Refson STFC Rutherford Appleton Laboratory September 18, 2007 Results of First-Principles Simulations..........................................................
2.57/2.570 Midterm Exam No. 1 April 4, :00 am -12:30 pm
 Name:.57/.570 Midterm Exam No. April 4, 0 :00 am -:30 pm Instructions: ().57 students: try all problems ().570 students: Problem plus one of two long problems. You can also do both long problems, and one
Name:.57/.570 Midterm Exam No. April 4, 0 :00 am -:30 pm Instructions: ().57 students: try all problems ().570 students: Problem plus one of two long problems. You can also do both long problems, and one
Geometry Optimisation
 Geometry Optimisation Matt Probert Condensed Matter Dynamics Group Department of Physics, University of York, UK http://www.cmt.york.ac.uk/cmd http://www.castep.org Motivation Overview of Talk Background
Geometry Optimisation Matt Probert Condensed Matter Dynamics Group Department of Physics, University of York, UK http://www.cmt.york.ac.uk/cmd http://www.castep.org Motivation Overview of Talk Background
Introduction. Chapter The Purpose of Statistical Mechanics
 Chapter 1 Introduction 1.1 The Purpose of Statistical Mechanics Statistical Mechanics is the mechanics developed to treat a collection of a large number of atoms or particles. Such a collection is, for
Chapter 1 Introduction 1.1 The Purpose of Statistical Mechanics Statistical Mechanics is the mechanics developed to treat a collection of a large number of atoms or particles. Such a collection is, for
Introductory Nanotechnology ~ Basic Condensed Matter Physics ~
 Introductory Nanotechnology ~ Basic Condensed Matter Physics ~ Atsufumi Hirohata Department of Electronics Go into Nano-Scale Lateral Size [m] 10-3 10-6 Micron-scale Sub-Micron-scale Nano-scale Human hair
Introductory Nanotechnology ~ Basic Condensed Matter Physics ~ Atsufumi Hirohata Department of Electronics Go into Nano-Scale Lateral Size [m] 10-3 10-6 Micron-scale Sub-Micron-scale Nano-scale Human hair
1 Bulk Simulations in a HCP lattice
 1 Bulk Simulations in a HCP lattice 1.1 Introduction This project is a continuation of two previous projects that studied the mechanism of melting in a FCC and BCC lattice. The current project studies
1 Bulk Simulations in a HCP lattice 1.1 Introduction This project is a continuation of two previous projects that studied the mechanism of melting in a FCC and BCC lattice. The current project studies
Local Hyperdynamics. Arthur F. Voter Theoretical Division Los Alamos National Laboratory Los Alamos, NM, USA
 Local Hyperdynamics Arthur F. Voter Theoretical Division National Laboratory, NM, USA Summer School in Monte Carlo Methods for Rare Events Division of Applied Mathematics Brown University Providence, RI
Local Hyperdynamics Arthur F. Voter Theoretical Division National Laboratory, NM, USA Summer School in Monte Carlo Methods for Rare Events Division of Applied Mathematics Brown University Providence, RI
Structure of Surfaces
 Structure of Surfaces C Stepped surface Interference of two waves Bragg s law Path difference = AB+BC =2dsin ( =glancing angle) If, n =2dsin, constructive interference Ex) in a cubic lattice of unit cell
Structure of Surfaces C Stepped surface Interference of two waves Bragg s law Path difference = AB+BC =2dsin ( =glancing angle) If, n =2dsin, constructive interference Ex) in a cubic lattice of unit cell
Optimization Methods via Simulation
 Optimization Methods via Simulation Optimization problems are very important in science, engineering, industry,. Examples: Traveling salesman problem Circuit-board design Car-Parrinello ab initio MD Protein
Optimization Methods via Simulation Optimization problems are very important in science, engineering, industry,. Examples: Traveling salesman problem Circuit-board design Car-Parrinello ab initio MD Protein
Molecular Dynamics. The Molecular Dynamics (MD) method for classical systems (not H or He)
 Molecular Dynamics What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: F&S Chapter 4 1 The Molecular Dynamics (MD) method for classical systems
Molecular Dynamics What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: F&S Chapter 4 1 The Molecular Dynamics (MD) method for classical systems
22 Path Optimisation Methods
 22 Path Optimisation Methods 204 22 Path Optimisation Methods Many interesting chemical and physical processes involve transitions from one state to another. Typical examples are migration paths for defects
22 Path Optimisation Methods 204 22 Path Optimisation Methods Many interesting chemical and physical processes involve transitions from one state to another. Typical examples are migration paths for defects
Lecture 16, February 25, 2015 Metallic bonding
 Lecture 16, February 25, 2015 Metallic bonding Elements of Quantum Chemistry with Applications to Chemical Bonding and Properties of Molecules and Solids Course number: Ch125a; Room 115 BI Hours: 11-11:50am
Lecture 16, February 25, 2015 Metallic bonding Elements of Quantum Chemistry with Applications to Chemical Bonding and Properties of Molecules and Solids Course number: Ch125a; Room 115 BI Hours: 11-11:50am
Phys 412 Solid State Physics. Lecturer: Réka Albert
 Phys 412 Solid State Physics Lecturer: Réka Albert What is a solid? A material that keeps its shape Can be deformed by stress Returns to original shape if it is not strained too much Solid structure
Phys 412 Solid State Physics Lecturer: Réka Albert What is a solid? A material that keeps its shape Can be deformed by stress Returns to original shape if it is not strained too much Solid structure
Heterogenous Nucleation in Hard Spheres Systems
 University of Luxembourg, Softmatter Theory Group May, 2012 Table of contents 1 2 3 Motivation Event Driven Molecular Dynamics Time Driven MD simulation vs. Event Driven MD simulation V(r) = { if r < σ
University of Luxembourg, Softmatter Theory Group May, 2012 Table of contents 1 2 3 Motivation Event Driven Molecular Dynamics Time Driven MD simulation vs. Event Driven MD simulation V(r) = { if r < σ
Molecular Dynamics Simulation of Nanometric Machining Under Realistic Cutting Conditions Using LAMMPS
 Molecular Dynamics Simulation of Nanometric Machining Under Realistic Cutting Conditions Using LAMMPS Rapeepan Promyoo Thesis Presentation Advisor: Dr. Hazim El-Mounayri Department of Mechanical Engineering
Molecular Dynamics Simulation of Nanometric Machining Under Realistic Cutting Conditions Using LAMMPS Rapeepan Promyoo Thesis Presentation Advisor: Dr. Hazim El-Mounayri Department of Mechanical Engineering
Investigations of the effects of 7 TeV proton beams on LHC collimator materials and other materials to be used in the LHC
 Russian Research Center Kurchatov Institute Investigations of the effects of 7 ev proton beams on LHC collimator materials and other materials to be used in the LHC A.I.Ryazanov Aims of Investigations:
Russian Research Center Kurchatov Institute Investigations of the effects of 7 ev proton beams on LHC collimator materials and other materials to be used in the LHC A.I.Ryazanov Aims of Investigations:
A Study of the Thermal Properties of a One. Dimensional Lennard-Jones System
 A Study of the Thermal Properties of a One Dimensional Lennard-Jones System Abstract In this study, the behavior of a one dimensional (1D) Lennard-Jones (LJ) system is simulated. As part of this research,
A Study of the Thermal Properties of a One Dimensional Lennard-Jones System Abstract In this study, the behavior of a one dimensional (1D) Lennard-Jones (LJ) system is simulated. As part of this research,
Ideal Gas Behavior. NC State University
 Chemistry 331 Lecture 6 Ideal Gas Behavior NC State University Macroscopic variables P, T Pressure is a force per unit area (P= F/A) The force arises from the change in momentum as particles hit an object
Chemistry 331 Lecture 6 Ideal Gas Behavior NC State University Macroscopic variables P, T Pressure is a force per unit area (P= F/A) The force arises from the change in momentum as particles hit an object
Defects in Crystals. Image borrowed from who got it from Davis & Reynolds 1996.
 Defects in Crystals Image borrowed from http://comp.uark.edu/~pjansma/geol3513_25_defmechs1_04.ppt who got it from Davis & Reynolds 1996. It's easy to think of real crystals as having these ideal structures.
Defects in Crystals Image borrowed from http://comp.uark.edu/~pjansma/geol3513_25_defmechs1_04.ppt who got it from Davis & Reynolds 1996. It's easy to think of real crystals as having these ideal structures.
The electronic structure of materials 1
 Quantum mechanics 2 - Lecture 9 December 18, 2013 1 An overview 2 Literature Contents 1 An overview 2 Literature Electronic ground state Ground state cohesive energy equilibrium crystal structure phase
Quantum mechanics 2 - Lecture 9 December 18, 2013 1 An overview 2 Literature Contents 1 An overview 2 Literature Electronic ground state Ground state cohesive energy equilibrium crystal structure phase
Introduction to Solid State Physics or the study of physical properties of matter in a solid phase
 Introduction to Solid State Physics or the study of physical properties of matter in a solid phase Prof. Germar Hoffmann 1. Crystal Structures 2. Reciprocal Lattice 3. Crystal Binding and Elastic Constants
Introduction to Solid State Physics or the study of physical properties of matter in a solid phase Prof. Germar Hoffmann 1. Crystal Structures 2. Reciprocal Lattice 3. Crystal Binding and Elastic Constants
Statistical Thermodynamics and Monte-Carlo Evgenii B. Rudnyi and Jan G. Korvink IMTEK Albert Ludwig University Freiburg, Germany
 Statistical Thermodynamics and Monte-Carlo Evgenii B. Rudnyi and Jan G. Korvink IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals From Micro to Macro Statistical Mechanics (Statistical
Statistical Thermodynamics and Monte-Carlo Evgenii B. Rudnyi and Jan G. Korvink IMTEK Albert Ludwig University Freiburg, Germany Preliminaries Learning Goals From Micro to Macro Statistical Mechanics (Statistical
Lecture 1 - Electrons, Photons and Phonons. September 4, 2002
 6.720J/3.43J - Integrated Microelectronic Devices - Fall 2002 Lecture 1-1 Lecture 1 - Electrons, Photons and Phonons Contents: September 4, 2002 1. Electronic structure of semiconductors 2. Electron statistics
6.720J/3.43J - Integrated Microelectronic Devices - Fall 2002 Lecture 1-1 Lecture 1 - Electrons, Photons and Phonons Contents: September 4, 2002 1. Electronic structure of semiconductors 2. Electron statistics
PHYS485 Materials Physics
 5/11/017 PHYS485 Materials Physics Dr. Gregory W. Clar Manchester University LET S GO ON A (TEK)ADVENTURE! WHAT? TRIP TO A MAKER S SPACE IN FORT WAYNE WHEN? THURSDAY, MAY 11 TH @ 5PM WHERE? TEKVENTURE
5/11/017 PHYS485 Materials Physics Dr. Gregory W. Clar Manchester University LET S GO ON A (TEK)ADVENTURE! WHAT? TRIP TO A MAKER S SPACE IN FORT WAYNE WHEN? THURSDAY, MAY 11 TH @ 5PM WHERE? TEKVENTURE
Atomic Arrangement. Primer in Materials Spring
 Atomic Arrangement Primer in Materials Spring 2017 30.4.2017 1 Levels of atomic arrangements No order In gases, for example the atoms have no order, they are randomly distributed filling the volume to
Atomic Arrangement Primer in Materials Spring 2017 30.4.2017 1 Levels of atomic arrangements No order In gases, for example the atoms have no order, they are randomly distributed filling the volume to
Energy Barriers and Rates - Transition State Theory for Physicists
 Energy Barriers and Rates - Transition State Theory for Physicists Daniel C. Elton October 12, 2013 Useful relations 1 cal = 4.184 J 1 kcal mole 1 = 0.0434 ev per particle 1 kj mole 1 = 0.0104 ev per particle
Energy Barriers and Rates - Transition State Theory for Physicists Daniel C. Elton October 12, 2013 Useful relations 1 cal = 4.184 J 1 kcal mole 1 = 0.0434 ev per particle 1 kj mole 1 = 0.0104 ev per particle
Set the initial conditions r i. Update neighborlist. Get new forces F i
 Set the initial conditions r i ( t 0 ), v i ( t 0 ) Update neighborlist Get new forces F i ( r i ) Solve the equations of motion numerically over time step Δt : r i ( t n ) r i ( t n + 1 ) v i ( t n )
Set the initial conditions r i ( t 0 ), v i ( t 0 ) Update neighborlist Get new forces F i ( r i ) Solve the equations of motion numerically over time step Δt : r i ( t n ) r i ( t n + 1 ) v i ( t n )
Introduction To Materials Science FOR ENGINEERS, Ch. 5. Diffusion. MSE 201 Callister Chapter 5
 Diffusion MSE 201 Callister Chapter 5 1 Goals: Diffusion - how do atoms move through solids? Fundamental concepts and language Diffusion mechanisms Vacancy diffusion Interstitial diffusion Impurities Diffusion
Diffusion MSE 201 Callister Chapter 5 1 Goals: Diffusion - how do atoms move through solids? Fundamental concepts and language Diffusion mechanisms Vacancy diffusion Interstitial diffusion Impurities Diffusion
Statistical Mechanics
 Statistical Mechanics Newton's laws in principle tell us how anything works But in a system with many particles, the actual computations can become complicated. We will therefore be happy to get some 'average'
Statistical Mechanics Newton's laws in principle tell us how anything works But in a system with many particles, the actual computations can become complicated. We will therefore be happy to get some 'average'
Plot the interatomic distances as a function of time and characterize the reactants and products through the plot. w
 Module 7 : Theories of Reaction Rates Lecture 35 : Potential Energy Surfaces (PES) II Objectives After studying this Lecture you will learn to do the following Relate a trajectory on a PES to a collision
Module 7 : Theories of Reaction Rates Lecture 35 : Potential Energy Surfaces (PES) II Objectives After studying this Lecture you will learn to do the following Relate a trajectory on a PES to a collision
arxiv:nucl-th/ v1 12 Jun 2000
 Microcanonical Lattice Gas Model for Nuclear Disassembly C. B. Das 1, S. Das Gupta 1 and S. K. Samaddar 2 1 Physics Department, McGill University, 3600 University St., Montréal, Québec Canada H3A 2T8 2
Microcanonical Lattice Gas Model for Nuclear Disassembly C. B. Das 1, S. Das Gupta 1 and S. K. Samaddar 2 1 Physics Department, McGill University, 3600 University St., Montréal, Québec Canada H3A 2T8 2
Modeling of high-dose radiation damage in semiconductors
 UNIVERSITY OF HELSINKI REPORT SERIES IN PHYSICS HU-P-107 Modeling of high-dose radiation damage in semiconductors Janne Nord Accelerator Laboratory Department of Physical Sciences Faculty of Science University
UNIVERSITY OF HELSINKI REPORT SERIES IN PHYSICS HU-P-107 Modeling of high-dose radiation damage in semiconductors Janne Nord Accelerator Laboratory Department of Physical Sciences Faculty of Science University
ATOMISTIC MODELING OF DIFFUSION IN ALUMINUM
 ATOMISTIC MODELING OF DIFFUSION IN ALUMINUM S. GRABOWSKI, K. KADAU and P. ENTEL Theoretische Physik, Gerhard-Mercator-Universität Duisburg, 47048 Duisburg, Germany (Received...) Abstract We present molecular-dynamics
ATOMISTIC MODELING OF DIFFUSION IN ALUMINUM S. GRABOWSKI, K. KADAU and P. ENTEL Theoretische Physik, Gerhard-Mercator-Universität Duisburg, 47048 Duisburg, Germany (Received...) Abstract We present molecular-dynamics
Chapter 2 Experimental sources of intermolecular potentials
 Chapter 2 Experimental sources of intermolecular potentials 2.1 Overview thermodynamical properties: heat of vaporization (Trouton s rule) crystal structures ionic crystals rare gas solids physico-chemical
Chapter 2 Experimental sources of intermolecular potentials 2.1 Overview thermodynamical properties: heat of vaporization (Trouton s rule) crystal structures ionic crystals rare gas solids physico-chemical
For this activity, all of the file labels will begin with a Roman numeral IV.
 I V. S O L I D S Name Section For this activity, all of the file labels will begin with a Roman numeral IV. A. In Jmol, open the SCS file in IV.A.1. Click the Bounding Box and Axes function keys. Use the
I V. S O L I D S Name Section For this activity, all of the file labels will begin with a Roman numeral IV. A. In Jmol, open the SCS file in IV.A.1. Click the Bounding Box and Axes function keys. Use the
Simulations with MM Force Fields. Monte Carlo (MC) and Molecular Dynamics (MD) Video II.vi
 Simulations with MM Force Fields Monte Carlo (MC) and Molecular Dynamics (MD) Video II.vi Some slides taken with permission from Howard R. Mayne Department of Chemistry University of New Hampshire Walking
Simulations with MM Force Fields Monte Carlo (MC) and Molecular Dynamics (MD) Video II.vi Some slides taken with permission from Howard R. Mayne Department of Chemistry University of New Hampshire Walking
Two simple lattice models of the equilibrium shape and the surface morphology of supported 3D crystallites
 Bull. Nov. Comp. Center, Comp. Science, 27 (2008), 63 69 c 2008 NCC Publisher Two simple lattice models of the equilibrium shape and the surface morphology of supported 3D crystallites Michael P. Krasilnikov
Bull. Nov. Comp. Center, Comp. Science, 27 (2008), 63 69 c 2008 NCC Publisher Two simple lattice models of the equilibrium shape and the surface morphology of supported 3D crystallites Michael P. Krasilnikov
Temperature and Pressure Controls
 Ensembles Temperature and Pressure Controls 1. (E, V, N) microcanonical (constant energy) 2. (T, V, N) canonical, constant volume 3. (T, P N) constant pressure 4. (T, V, µ) grand canonical #2, 3 or 4 are
Ensembles Temperature and Pressure Controls 1. (E, V, N) microcanonical (constant energy) 2. (T, V, N) canonical, constant volume 3. (T, P N) constant pressure 4. (T, V, µ) grand canonical #2, 3 or 4 are
From Random Numbers to Monte Carlo. Random Numbers, Random Walks, Diffusion, Monte Carlo integration, and all that
 From Random Numbers to Monte Carlo Random Numbers, Random Walks, Diffusion, Monte Carlo integration, and all that Random Walk Through Life Random Walk Through Life If you flip the coin 5 times you will
From Random Numbers to Monte Carlo Random Numbers, Random Walks, Diffusion, Monte Carlo integration, and all that Random Walk Through Life Random Walk Through Life If you flip the coin 5 times you will
Random Walks A&T and F&S 3.1.2
 Random Walks A&T 110-123 and F&S 3.1.2 As we explained last time, it is very difficult to sample directly a general probability distribution. - If we sample from another distribution, the overlap will
Random Walks A&T 110-123 and F&S 3.1.2 As we explained last time, it is very difficult to sample directly a general probability distribution. - If we sample from another distribution, the overlap will
Accelerated Quantum Molecular Dynamics
 Accelerated Quantum Molecular Dynamics Enrique Martinez, Christian Negre, Marc J. Cawkwell, Danny Perez, Arthur F. Voter and Anders M. N. Niklasson Outline Quantum MD Current approaches Challenges Extended
Accelerated Quantum Molecular Dynamics Enrique Martinez, Christian Negre, Marc J. Cawkwell, Danny Perez, Arthur F. Voter and Anders M. N. Niklasson Outline Quantum MD Current approaches Challenges Extended
Direct and Indirect Semiconductor
 Direct and Indirect Semiconductor Allowed values of energy can be plotted vs. the propagation constant, k. Since the periodicity of most lattices is different in various direction, the E-k diagram must
Direct and Indirect Semiconductor Allowed values of energy can be plotted vs. the propagation constant, k. Since the periodicity of most lattices is different in various direction, the E-k diagram must
Copyright 2001 University of Cambridge. Not to be quoted or copied without permission.
 Course MP3 Lecture 4 13/11/2006 Monte Carlo method I An introduction to the use of the Monte Carlo method in materials modelling Dr James Elliott 4.1 Why Monte Carlo? The name derives from the association
Course MP3 Lecture 4 13/11/2006 Monte Carlo method I An introduction to the use of the Monte Carlo method in materials modelling Dr James Elliott 4.1 Why Monte Carlo? The name derives from the association
Physics 141 Dynamics 1 Page 1. Dynamics 1
 Physics 141 Dynamics 1 Page 1 Dynamics 1... from the same principles, I now demonstrate the frame of the System of the World.! Isaac Newton, Principia Reference frames When we say that a particle moves
Physics 141 Dynamics 1 Page 1 Dynamics 1... from the same principles, I now demonstrate the frame of the System of the World.! Isaac Newton, Principia Reference frames When we say that a particle moves
Molecular Dynamics Simulation of Argon
 Molecular Dynamics Simulation of Argon The fundamental work on this problem was done by A. Rahman, Phys. Rev. 136, A405 (1964). It was extended in many important ways by L. Verlet, Phys. Rev. 159, 98 (1967),
Molecular Dynamics Simulation of Argon The fundamental work on this problem was done by A. Rahman, Phys. Rev. 136, A405 (1964). It was extended in many important ways by L. Verlet, Phys. Rev. 159, 98 (1967),
Mat E 272 Lecture 25: Electrical properties of materials
 Mat E 272 Lecture 25: Electrical properties of materials December 6, 2001 Introduction: Calcium and copper are both metals; Ca has a valence of +2 (2 electrons per atom) while Cu has a valence of +1 (1
Mat E 272 Lecture 25: Electrical properties of materials December 6, 2001 Introduction: Calcium and copper are both metals; Ca has a valence of +2 (2 electrons per atom) while Cu has a valence of +1 (1
Properties of Liquids and Solids. Vaporization of Liquids. Vaporization of Liquids. Aims:
 Properties of Liquids and Solids Petrucci, Harwood and Herring: Chapter 13 Aims: To use the ideas of intermolecular forces to: Explain the properties of liquids using intermolecular forces Understand the
Properties of Liquids and Solids Petrucci, Harwood and Herring: Chapter 13 Aims: To use the ideas of intermolecular forces to: Explain the properties of liquids using intermolecular forces Understand the
Properties of Liquids and Solids. Vaporization of Liquids
 Properties of Liquids and Solids Petrucci, Harwood and Herring: Chapter 13 Aims: To use the ideas of intermolecular forces to: Explain the properties of liquids using intermolecular forces Understand the
Properties of Liquids and Solids Petrucci, Harwood and Herring: Chapter 13 Aims: To use the ideas of intermolecular forces to: Explain the properties of liquids using intermolecular forces Understand the
Communications with Optical Fibers
 Communications with Optical Fibers In digital communications, signals are generally sent as light pulses along an optical fiber. Information is first converted to an electrical signal in the form of pulses
Communications with Optical Fibers In digital communications, signals are generally sent as light pulses along an optical fiber. Information is first converted to an electrical signal in the form of pulses
Energy bands in solids. Some pictures are taken from Ashcroft and Mermin from Kittel from Mizutani and from several sources on the web.
 Energy bands in solids Some pictures are taken from Ashcroft and Mermin from Kittel from Mizutani and from several sources on the web. we are starting to remind p E = = mv 1 2 = k mv = 2 2 k 2m 2 Some
Energy bands in solids Some pictures are taken from Ashcroft and Mermin from Kittel from Mizutani and from several sources on the web. we are starting to remind p E = = mv 1 2 = k mv = 2 2 k 2m 2 Some
Set the initial conditions r i. Update neighborlist. r i. Get new forces F i
 Set the initial conditions r i t 0, v i t 0 Update neighborlist Get new forces F i r i Solve the equations of motion numerically over time step t : r i t n r i t n + v i t n v i t n + Perform T, P scaling
Set the initial conditions r i t 0, v i t 0 Update neighborlist Get new forces F i r i Solve the equations of motion numerically over time step t : r i t n r i t n + v i t n v i t n + Perform T, P scaling
Comparisons of DFT-MD, TB- MD and classical MD calculations of radiation damage and plasmawallinteractions
 CMS Comparisons of DFT-MD, TB- MD and classical MD calculations of radiation damage and plasmawallinteractions Kai Nordlund Department of Physics and Helsinki Institute of Physics University of Helsinki,
CMS Comparisons of DFT-MD, TB- MD and classical MD calculations of radiation damage and plasmawallinteractions Kai Nordlund Department of Physics and Helsinki Institute of Physics University of Helsinki,
Kinetic Monte Carlo: from transition probabilities to transition rates
 Kinetic Monte Carlo: from transition probabilities to transition rates With MD we can only reproduce the dynamics of the system for 100 ns. Slow thermallyactivated processes, such as diffusion, cannot
Kinetic Monte Carlo: from transition probabilities to transition rates With MD we can only reproduce the dynamics of the system for 100 ns. Slow thermallyactivated processes, such as diffusion, cannot
Monte Carlo Simulations in Statistical Physics
 Part II Monte Carlo Simulations in Statistical Physics By D.Stauffer Introduction In Statistical Physics one mostly deals with thermal motion of a system of particles at nonzero temperatures. For example,
Part II Monte Carlo Simulations in Statistical Physics By D.Stauffer Introduction In Statistical Physics one mostly deals with thermal motion of a system of particles at nonzero temperatures. For example,
Lecture 2. Unit Cells and Miller Indexes. Reading: (Cont d) Anderson 2 1.8,
 Lecture 2 Unit Cells and Miller Indexes Reading: (Cont d) Anderson 2 1.8, 2.1-2.7 Unit Cell Concept The crystal lattice consists of a periodic array of atoms. Unit Cell Concept A building block that can
Lecture 2 Unit Cells and Miller Indexes Reading: (Cont d) Anderson 2 1.8, 2.1-2.7 Unit Cell Concept The crystal lattice consists of a periodic array of atoms. Unit Cell Concept A building block that can
Lecture 2: Fundamentals. Sourav Saha
 ME 267: Mechanical Engineering Fundamentals Credit hours: 3.00 Lecture 2: Fundamentals Sourav Saha Lecturer Department of Mechanical Engineering, BUET Email address: ssaha09@me.buet.ac.bd, souravsahame17@gmail.com
ME 267: Mechanical Engineering Fundamentals Credit hours: 3.00 Lecture 2: Fundamentals Sourav Saha Lecturer Department of Mechanical Engineering, BUET Email address: ssaha09@me.buet.ac.bd, souravsahame17@gmail.com
Temperature and Pressure Controls
 Ensembles Temperature and Pressure Controls 1. (E, V, N) microcanonical (constant energy) 2. (T, V, N) canonical, constant volume 3. (T, P N) constant pressure 4. (T, V, µ) grand canonical #2, 3 or 4 are
Ensembles Temperature and Pressure Controls 1. (E, V, N) microcanonical (constant energy) 2. (T, V, N) canonical, constant volume 3. (T, P N) constant pressure 4. (T, V, µ) grand canonical #2, 3 or 4 are
dynamics computer code, whiccan simulate the motion of atoms at a surface and We coembininewton's equation for the nuclei,
 DUI FILE COpy T DTIC ANNUAL LETTER REPORT BY 0. F. ELEC33 20KE AD-A224 069 P.1. FOR N00014-85-K-0442 U Theory of electronic states and formations energies of.defect comp exes, interstitial defects, and
DUI FILE COpy T DTIC ANNUAL LETTER REPORT BY 0. F. ELEC33 20KE AD-A224 069 P.1. FOR N00014-85-K-0442 U Theory of electronic states and formations energies of.defect comp exes, interstitial defects, and
Rate of Heating and Cooling
 Rate of Heating and Cooling 35 T [ o C] Example: Heating and cooling of Water E 30 Cooling S 25 Heating exponential decay 20 0 100 200 300 400 t [sec] Newton s Law of Cooling T S > T E : System S cools
Rate of Heating and Cooling 35 T [ o C] Example: Heating and cooling of Water E 30 Cooling S 25 Heating exponential decay 20 0 100 200 300 400 t [sec] Newton s Law of Cooling T S > T E : System S cools
From Atoms to Materials: Predictive Theory and Simulations
 From Atoms to Materials: Predictive Theory and Simulations Week 3 Lecture 4 Potentials for metals and semiconductors Ale Strachan strachan@purdue.edu School of Materials Engineering & Birck anotechnology
From Atoms to Materials: Predictive Theory and Simulations Week 3 Lecture 4 Potentials for metals and semiconductors Ale Strachan strachan@purdue.edu School of Materials Engineering & Birck anotechnology
The Morse Potential. ChE Reactive Process Engineering. Let s have a look at a very simple model reaction :
 The Morse Potential L7-1 Let s have a look at a very simple model reaction : We can describe the F + H-H energy of this system based on the relative position of the atoms. This energy is (in the most
The Morse Potential L7-1 Let s have a look at a very simple model reaction : We can describe the F + H-H energy of this system based on the relative position of the atoms. This energy is (in the most
Chem 728 Introduction to Solid Surfaces
 Chem 728 Introduction to Solid Surfaces Solids: hard; fracture; not compressible; molecules close to each other Liquids: molecules mobile, but quite close to each other Gases: molecules very mobile; compressible
Chem 728 Introduction to Solid Surfaces Solids: hard; fracture; not compressible; molecules close to each other Liquids: molecules mobile, but quite close to each other Gases: molecules very mobile; compressible
Interatomic Potentials. The electronic-structure problem
 Interatomic Potentials Before we can start a simulation, we need the model! Interaction between atoms and molecules is determined by quantum mechanics: Schrödinger Equation + Born-Oppenheimer approximation
Interatomic Potentials Before we can start a simulation, we need the model! Interaction between atoms and molecules is determined by quantum mechanics: Schrödinger Equation + Born-Oppenheimer approximation
Ab Ini'o Molecular Dynamics (MD) Simula?ons
 Ab Ini'o Molecular Dynamics (MD) Simula?ons Rick Remsing ICMS, CCDM, Temple University, Philadelphia, PA What are Molecular Dynamics (MD) Simulations? Technique to compute statistical and transport properties
Ab Ini'o Molecular Dynamics (MD) Simula?ons Rick Remsing ICMS, CCDM, Temple University, Philadelphia, PA What are Molecular Dynamics (MD) Simulations? Technique to compute statistical and transport properties
Road map (Where are we headed?)
 Road map (Where are we headed?) oal: Fairly high level understanding of carrier transport and optical transitions in semiconductors Necessary Ingredients Crystal Structure Lattice Vibrations Free Electron
Road map (Where are we headed?) oal: Fairly high level understanding of carrier transport and optical transitions in semiconductors Necessary Ingredients Crystal Structure Lattice Vibrations Free Electron
Hill climbing: Simulated annealing and Tabu search
 Hill climbing: Simulated annealing and Tabu search Heuristic algorithms Giovanni Righini University of Milan Department of Computer Science (Crema) Hill climbing Instead of repeating local search, it is
Hill climbing: Simulated annealing and Tabu search Heuristic algorithms Giovanni Righini University of Milan Department of Computer Science (Crema) Hill climbing Instead of repeating local search, it is
André Schleife Department of Materials Science and Engineering
 André Schleife Department of Materials Science and Engineering Length Scales (c) ICAMS: http://www.icams.de/cms/upload/01_home/01_research_at_icams/length_scales_1024x780.png Goals for today: Background
André Schleife Department of Materials Science and Engineering Length Scales (c) ICAMS: http://www.icams.de/cms/upload/01_home/01_research_at_icams/length_scales_1024x780.png Goals for today: Background
