Set the initial conditions r i. Update neighborlist. Get new forces F i
|
|
|
- Candice Henry
- 5 years ago
- Views:
Transcription
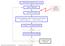
1 Set the initial conditions r i ( t 0 ), v i ( t 0 ) Update neighborlist Get new forces F i ( r i ) Solve the equations of motion numerically over time step Δt : r i ( t n ) r i ( t n + 1 ) v i ( t n ) v i ( t n + 1 ) Perform T, P scaling (ensembles) t t+ Δt Get desired physical quantities t > t max? Calculate results and finish Introduction to molecular dynamics Calculating the forces 1
2 Calculating the forces between atoms The forces between atoms can be calculated in many different ways This lecture: classical potentials. pair potentials, many-body potentials Quantum mechanics A classical potential can be written in the form: V = V 1 ( r i ) + i, j V 2 ( r i, r j ) + V 3 ( r i, r j, r k ) + i, j, k i V is the total potential energy of an N atom system. In principle all sums loop from 1 to N V 1 : single particle potential: external forces V 2 : pair potential which only depends on the distance between atoms r ij direct dependence on the vectors r i, r j => dependence on the choice of the origin V 3 : three-body potential which may have an angular dependence depends only on three variables, i.e. V 3 = V 3 ( r ij, r ik, θ ijk ) Four-body potentials, even five-body terms: chemical and biological applications Introduction to molecular dynamics Calculating the forces 2
3 Calculating the forces between atoms V 2 and V 3 enough to describe the basic mechanical and structural properties of most elements and simple compounds In order that things would not be too straightforward, in many cases a environment-dependence (i.e. implicit three-body term) is embedded into the two-body term V 2. We will give examples on these later. All terms which are not pure single particle or pair potentials are called many-body terms. Introduction to molecular dynamics Calculating the forces 3
4 Calculating the forces between atoms Classification of empirical interatomic potentials [A. E. Carlsson, Solid State Physics: Advances in Research and Applications, 43 (1990) 1] Pair Potential V = i, j V P ( r ij ) Pair Functional Potential V = V PF ( ρ i ), ρ = fr ( ) i ij i j i If fr ( ) = ar back to pair potential Cluster Potential V = V CP ( r ij, r ik, r jk ) Only clusters of three atoms here i j k Cluster Functional Potential V = V CF ( ρ i ), ρ i = gr ( ij, r ik, r jk ) i j, k i j k Real potentials often combinations of these: e.g. EAM for metals V = i V PF ( ρ i ) + i, j V P ( r ij ) Introduction to molecular dynamics Calculating the forces 4
5 Force calculation for pair potentials Pure pair potential V( r ij ). The force acting on atom i from atom j f ij f ij V V V = ri Vr ( ij ) = rij Vr ( ij ) = xˆ + ŷ + ẑ, x ij y ij z ij r ij i (xˆ, ŷ, ẑ unit vectors) -f ij j r ij = r i r j, x ij x i x j V = etc., xij dv r ij =, dr x ij r ij = x ij x ij r ij f ij = dv dr r = r ij r ij r ij To be precise operates on the position r i of atom i. (Makes a difference for many-body potentials.) Cut-off radius r c : atom pairs with r ij > r c do not interact, r c a few Å. Introduction to molecular dynamics Calculating the forces 5
6 Force calculation for pair potentials In case the potential extends to infinity, an analytical correction can be made to the energy, and other quantities of interest: V tot = V 2 + V corr = E c + 2πNρ r 2 Vr ( ) dr where ρ is the atom density of the system. This obviously assumes that when r work when for example a surface is present. r c > r c the atom density is constant everywhere, and thus does not Introduction to molecular dynamics Calculating the forces 6
7 Force calculation for pair potentials Discontinuity at r c jumps in energy Solution: take the potential to zero in [ r c, r c + Δr] potential and the force are continuous (3 rd order polynomial) or displace the potential, as the zero point of V is arbitrary but this changes the value of V tot Many modern potentials are in fact defined so that they have a well-defined cutoff r c where V and at least the first derivative are 0. V(r) r c r Introduction to molecular dynamics Calculating the forces 7
8 Force calculation for pair potentials Example: cut-off of Lennard-Jones potential r c = 2.3 Å Δr c = 0.2 Å V LJ ( r) = 4ε σ -- r 12 σ -- r 6 shift and tilt Shift and tilt the potential: V( r) and V' ( r) continuous at r c : polynomial Vr ( ) = V LJ ( r) ( r r c )V' LJ ( r c ) V LJ ( r c ) Problem: may change the potential at smaller r values Fit a polynomial P( r) = ar 3 + br 2 + cr + d from [ r c, r c + Δr c ]: V LJ ( r) Pr ( ) Pr ( c ) = V LJ ( r c ) Pr ( c + Δr c ) = 0 P' ( r c + Δr c ) = 0 P' ( r c ) = V' LJ ( r c ) Introduction to molecular dynamics Calculating the forces 8
9 Force calculation for pair potentials Problem: high forces may result (see below) Brenner potential for carbon (Well, this is not a pair potential): Potential quickly to zero; doesn t look too bad However: huge forces; effect seen in fracture simulations (see also M. Sammalkorpi et al., Phys. Rev. B 70 (2004) ) potential force T. Belytschko et al., Phys. Rev. B 65 (2002) Introduction to molecular dynamics Calculating the forces 9
10 Force calculation for pair potentials Force calculation without periodic boundaries or neighbour list: do i=1,n do j=1,n if (i==j) cycle rijx = rx(j)-rx(i) rijy = ry(j)-ry(i) rijz = rz(j)-rz(i) rijsq = rijx**2+rijy**2+rijz**2 rij = sqrt(rijsq) if (rij < rcut) then V = (Potential energy per atom)/2 dvdr =...derivative of potential energy with respect to its only argument r... a = -dvdr/m/2.0! Unit transformations may be needed. Note the factor 1/2!! ax(i) = ax(i)-rijx/rij*a! The application on both ax(j) = ax(j)+rijx/rij*a! i and j ensures that ay(i) = ay(i)-rijy/rij*a! Newton s third law is ay(j) = ay(j)+rijy/rij*a! fulfilled az(i) = az(i)-rijz/rij*a az(j) = az(j)+rijz/rij*a endif enddo enddo Introduction to molecular dynamics Calculating the forces 10
11 Force calculation for pair potentials Use of Verlet neighbour list (cf. lecture 3): startofineighbourlist=1 do i=1,n nneighboursi=neighbourlist(startofineighbourlist) do jj=1,nneighboursi j=neighbourlist(startofineighbourlist+jj) rijx = rx(j)-rx(i) rijy = ry(j)-ry(i) rijz = rz(j)-rz(i) rijsq = rijx**2+rijy**2+rijz**2 rij = sqrt(rijsq) if (rij < rcut) then V = (Potential energy per atom)/2 dvdr =...derivative of potential energy with respect to its only argument r... a = -dvdr/m/2.0! Plus unit transformations! Note the factor 1/2!! ax(i) = ax(i)-rijx/rij*a ax(j) = ax(j)+rijx/rij*a ay(i) = ay(i)-rijy/rij*a ay(j) = ay(j)+rijy/rij*a az(i) = az(i)-rijz/rij*a az(j) = az(j)+rijz/rij*a endif enddo startofineighbourlist=startofineighbourlist+nneighboursi+1 enddo Introduction to molecular dynamics Calculating the forces 11
12 Force calculation for pair potentials Note that in the sum above every interaction is counted twice: do i=1,n do j=1,n if (i==j) cycle... That is, e.g. interaction 1-3 is counted both as 1-3 and 3-1. Hence the factor 1/2 in front of the potential energy summation and forces (this actually depends on the exact definition of the potentials, some already have a factor of 1/2 in front). A straightforward solution: do i=1,n-1 do j=i+1,n... (either in constructing the neighbour list or forces) reduces the calculation time to one half. For some many-body potentials this does not work. V( r) often is defined to give the total energy for a pair of atoms. When one wants the potential energy per atom one thus may have to include one more factor of 1/2. But this additional factor is not needed in the force calculation since the force always affects both atoms (Newton s III law). Note that the sign conventions in defining r ij in the literature may vary. Introduction to molecular dynamics Calculating the forces 12
13 Force calculation for pair potentials One practical way of checking that you have correctly derived the forces from the potential energy and that all signs and factors of ½ are OK in you potential implementation: 1. Calculate E pot at 0 K and compare with an analytical prediction for some simple system, e.g. a dimer or perfect lattice. 2. Simulate a two-atom system starting from a very small distance, so that E pot is very large, much larger than the equilibrium energy per atom (say ev). When you run the simulation with a very small time step the atoms should explode outwards from each other so that the final E kin /atom is the same as the original E pot /atom. If you are uncertain what a very small time step is, keep decreasing it until the answer doesn t change. 3. Another good test: numerical derivation of potential energy: Move one atom in direction ŝ amount Δs. Directional derivative of the potential (assume ŝ = 1 ): V( r) ŝ V( r + hŝ) V( r) = lim = V( r) ŝ = h 0 h Computed from potential energy as ΔV Δs Fr ( ) ŝ Computed from forces as F x s x + F y s y + F z s z Fr ( ) s r s F Introduction to molecular dynamics Calculating the forces 13
14 Force calculation for a three-body potential For a pure pair potential for an interaction between atoms i and j V ij = V ji because Vr ( ij ) = Vr ( ji ) and hence also i V ij = i V ji as described above. This symmetry simplifies the force calculation. For a three-body potential things get trickier because V ij may not = V ji. To get the force F i acting on an atom i one instead has to calculate F i = i ( V ij + V ji ) + V jki = ( i V + V ij i ji ) + j j k j j Many practical three-body potentials have been written such that V 3 ( r ij, r ik, θ ijk ) = V 3 ( r ij, r ik, cosθ ijk ) i V jki k i.e. all angular information is in a cosine term. Introduction to molecular dynamics Calculating the forces 14
15 Force calculation for a three-body potential In this case one can utilize the following equalities: r cosθ ij r ik ijk = r ij r ik i r ij θ ijk r ik j k r ij r ik cosθ i cosθ ijk = ijk i = = r ij r ik 2 r ij that is, no need to evaluate cos function. 1 cosθ r ijk r ij r ij ik r ik r r ij r ik ik In many-body potentials there are often symmetries which can be used to reduce the number of operations needed in the force calculation even more. Introduction to molecular dynamics Calculating the forces 15
16 The physical/chemical origin of interactions Qualitatively a two-atom interaction looks like the following: The minimum, i.e. equilibrium distance, is r 0. At small separations there is a strong repulsion. Just below r 0 this derives primarily from the Pauli rule preventing electrons being in states with the same quantum numbers, and from the electronelectron repulsion, whereas when the nuclei are very close to each other, the Coulombic repulsion between the nuclei dominates completely. V(r) r 0 r At larger distances there may be an attraction, which can have different reasons: van der Waals attraction, Coulomb attraction, a covalent bond, (due to pairing of valence electrons) or metallic bonding Potential may also be purely repulsive Introduction to molecular dynamics Calculating the forces 16
17 A few examples (1 bohr = 0.53 Å) Introduction to molecular dynamics Calculating the forces 17
18 Overview of bonding expected in different cases, and illustration of electron distributions [Kittel, Introduction to Solid State physics] Introduction to molecular dynamics Calculating the forces 18
19 So for the pure elements we get the familiar division: Introduction to molecular dynamics Calculating the forces 19
20 Idealized potentials for theoretical and qualitative studies Hard sphere: V HS ( r) =, r < σ 0, r σ First MD simulations were carried out with this potential. The equations of motion reduce to calculating where the next collision occurs: true billiard ball physics Applications in packing problems Square well:, r < σ 1 V SW ( r) = ε, σ 1 r < σ 2 0, r σ 2 soft sphere: V SS ( r) = ε σ -- ν r Source: Allen-Tildesley Introduction to molecular dynamics Calculating the forces 20
21 Realistic pair potentials Lennard-Jones (LJ) Vr ( ) = 4ε σ -- r 12 σ -- r 6 The attractive 1 r 6 - term can be derived from the dipole-dipole interaction, or as the interactions between two oscillators (QM) [Kittel, Introduction to Solid State Physics, 7th edition, p. 62]. It is also known as the Van der Waals or London interaction. The repulsive term 1 r 12 chosen for convenience. Also other exponents used; notation for any two exponents A and B is LJ (A-B) potential. ε and σ are usually chosen by fitting into experimental data. σ gives the equilibrium distance ε the cohesive energy. A few Lennard-Jones-parameters for gases [Ashcroft-Mermin s. 398]: Ne Ar Kr Xe ε (ev) σ (Å) Very weak interaction: e.g. V min = 3.1 mev for Ne. LJ (12-6) potentials have proven to be good for noble gases (filled electron shells almost always neutral) close to equilibrium. But they are obviously terrible for very small r (r 1 Å) since the true interaction is about e r r and not 1 r 12. Introduction to molecular dynamics Calculating the forces 21
22 Realistic pair potentials LJ potentials have been, and are used a lot, for instance in molecular modelling, in many cases even in systems where there is no physical motivation to using the LJ functional form. But if the fit is good for some purpose, using it may still be justified as long as the limitations are kept in mind. Reduced units If a potential only has a couple of parameters, evaluating it can be really efficient in reduced units Also, in reduced units the results are always the same, so the results can be transferred to different systems with straightforward scaling. For instance for the Lennard-Jones-potential: σ Vr ( ) 4ε -- r 12 σ = -- r 6 [or any V( r) = εfr ( σ) )] Natural length unit = σ natural energy unit = ε V * ( x) = 4[ x 12 x 6 ] Introduction to molecular dynamics Calculating the forces 22
23 Realistic pair potentials other units: t * = t [( mσ 2 ) ε] 1 2 ρ * = ρσ 3 T * = k B T ε P * = Pσ 3 ε f * = fσ ε v * = v [ ε m] 1 2 Reduced units were very popular when one had to save CPU time in every single multiplication, and when potentials were still as simple as LJ. Introduction to molecular dynamics Calculating the forces 23
24 Realistic pair potentials Morse potential Simple metals (sp-metals, e.g. Na, Mg, Al; and metals with the fcc- or hcp-structure), are at least to some extent describable with a pair potential A popular choice: the Morse potential [P. M. Morse, Phys. Rev. 34 (1930) 57.]: Vr ( ) De 2α ( r r 0) = 2De α ( r r 0) Designed originally to describe vibrations in molecules. The Schrödinger equation happens to have an analytical solution for this functional form. Efficient to evaluate, in the form above only one exponential function needs to be evaluated. Decays faster at large r than Lennard-Jones: less problems with cut-off. A fit for many metals [Girifalco and Weizer, Phys. Rev. 114 (1959) 687.] Works decently for being a pair potential. Girifalco and Weizer, Phys. Rev. 114 (1959) 687. Introduction to molecular dynamics Calculating the forces 24
25 Realistic pair potentials An ordinary pair potential has a close-packed structure as the ground state. (usually either face-centered cubic, FCC or hexagonal close packed, HCP). HCP FCC Introduction to molecular dynamics Calculating the forces 25
26 Realistic pair potentials A pair potential can thus not describe well elements with other structures than FCC or HCP. But this doesn t mean people haven t tried: Diamond lattice: open structure, four nearest neighbours, very far from close packed. Still, it is actually possible to make diamond stable locally with a pair potential, but this will become rather pathological (Mazzone potential for Si, [Phys. Stat. Sol (b) 165 (1991) 395.]): Does actually work close to perfect lattice. But what happens when atoms leave the harmonic well due to e.g. a high temperature? System will collapse to close-packed structure => applicability of potential extremely limited V(r) Morse harmonic well Unfortunately this is not uncommon regarding interatomic potentials: one has to be very critical of any new potential! Even well-respected physicists have presented potentials which have some very pathological features... r 0 r Introduction to molecular dynamics Calculating the forces 26
27 Realistic pair potentials Ionic compounds Different ions, between which the electron density is very small. The ions have filled electron shells, and are thus unlikely to change their electron configuration An extreme examples: NaCl: A pair potential approximation works quite well, and potentials abound in the literature, as there is much experimental data available for the alkali halides which can be used in potential fitting. Potentials typically contain a short-range (SR) term and the Coulomb interaction: z Vr ( ij ) V SR ( r ij ) 1 z 2 e 2 = ; z 4πε 0 r i = ion charges ij V SR : repulsive force between electrons packed closely together and an attractive van der Waals (vdw) interaction Introduction to molecular dynamics Calculating the forces 27
28 Realistic pair potentials Most common forms for the short range potential: Buckingham: V SR r ( ) = Ae r/ ρ ---- C r 6 Born-Huggins-Mayer: V SR r ( ) Ae Br ( σ) C = ---- r 6 D ---- r 8 Morse: V SR ( r) De 2α ( r r 0) = 2De α ( r r 0) 1 r 6 -term comes from the dipole-dipole interaction (again) The repulsion is usually significant only for nearest neighbours, and the vdw interaction for next-nearest neighbours. Frequently for instance in oxides the only interaction assumed between cations is their Coulombic repulsion. Introduction to molecular dynamics Calculating the forces 28
29 Fitting of potential parameters In almost all classical potentials there is a number of free parameters, e.g. in Lennard-Jones 2 (ε and σ), Morse 3 (D, α, r 0 ) etc. An extreme example: the ReaxFF model for hydrocarbons: A.C.T. van Duin et al., J. Chem. Phys. A 105 (2001) Introduction to molecular dynamics Calculating the forces 29
30 Fitting of potential parameters Two main approaches to develop a potential exist: 1. Derivation from so called ab initio (quantum mechanical) calculations 2. Fit to empirical and/or ab initio data Although the previous approach is better motivated physically, in practice the latter approach, or a combination of the two, often works better. A good classical potential is one which with a small number of free parameters can describe a wide range of properties well (usually 5-20 % accuracy in condensed matter physics is considered to be well, since experiments seldom are much more accurate than this). A related concept is that a good potential should be transferable, which means that it should be able to describe properties of other states of the material than those it was originally fitted to. Introduction to molecular dynamics Calculating the forces 30
31 Fitting of potential parameters Regarding fitting the parameters in a potential of type 2, there are two opposite extreme approaches: 1. Blind fitting : choose a functional form and a set of data to which the parameters are fit. Then use some fitting routine to obtain a best fit to all the data. 2. Parameter choice by hand : use reliable experimental or ab initio data of crucial data to set as many potential parameters as possible exactly, then fit only the remaining (if any) parameters. For instance, the equilibrium separation, binding energy and vibration frequency for a dimer can be used to fix all the 3 Morse potential parameters. A pure approach 1 is dangerous in that quantities which are outside the original parameter set may obtain completely pathological values. Example: some Si bulk potentials predict that the Si dimer is non-bonding. Also, if some potential parameter happens to be insensitive to all quantities in the data set, the fit may give ridiculously small or large values for it, which may cause trouble elsewhere. To obtain transferable potentials, approach 2 is thus usually to be preferred. On the other hand, if optimal precision in a limited set of systems (say, elastic properties of a perfect bulk crystal) is desirable, approach 1 may still be the better way to go. Most authors use approaches somewhere between 1 and 2. Introduction to molecular dynamics Calculating the forces 31
32 Fitting of potential parameters A functional form can sometimes be derived from experimental equations of state P V ( ). Example: solid Ne and Ar: Introduction to molecular dynamics Calculating the forces 32
33 Fitting of potential parameters Here is a short list of macroscopic, physical, properties which can and often are used to derive or fit interatomic potentials: mechanical Physical property Crystal structure Cohesive energy Elastic constants c ρσ Equation of state Neutron scattering PV ( ) Atom-level property Balance of atomic forces. Potential energy at the equilibrium atom positions Long-wavelength acoustic vibrations Elastic distortions of unit cell. Compression or expansion of material Phonon ω( k) in the Brillouin zone. Dielectric constant ε Electronic polarizability electric ε Dielectric constant 0 Infrared absorption Raman scattering Polarizarization of electrons and lattice; long-wavelength optical vibration modes; Long-wavelength vibrations with a dipole moment. Long-wavelength vibrations which change the polarizability. Out of these, the first five depend purely on the mechanical properties of the material, and are relevant to almost all solids. The latter four involve electric properties and may or may not be relevant depending on what kind of materials and properties are studied. Introduction to molecular dynamics Calculating the forces 33
34 Fitting of potential parameters Crystal structure: The equilibrium crystal structure should be stable if one wants to describe any process where large atom displacements may occur (melting, surfaces, deposition, etc. etc.). In equilibrium the force acting on every atom in the unit cell i should vanish: f ij = 0 j Here the potential is only tested at a few r ij values. (The smaller the crystal symmetry, the more values.) Any potential has a minimum potential energy configuration, or many configurations with the same energy. Example: Tersoff potential for Si [J. Tersoff, Phys. Rev. B 38 (1988) 9902.] Introduction to molecular dynamics Calculating the forces 34
35 Fitting of potential parameters Local stability is easy to achieve in a classical potential. But global stability (that is, that the real crystal structure is indeed the global minimum of the potential) may be surprisingly difficult. Even well-known authors make mistakes. For instance, the first Si potential of Tersoff [Tersoff, Phys. Rev. Lett. 56 (1986) 632.] was well motivated, well derived, and published in the best journal in physics. But the formation energy of the vacancy turned out to be negative, which means it did not have the right ground state structure... A good way to test the minimum energy: start from random atom positions, and quench the cell slowly enough so that it crystallizes. If the structure is the correct one, it probably is indeed the ground state. Unfortunately doing this may take forever. Another test: simulate a liquid and solid in equilibrium at the melting point, and check that the solid remains stable and the liquid recrystallizes to the same structure on slight cooling below T melt. Introduction to molecular dynamics Calculating the forces 35
36 Fitting of potential parameters Cohesive energy (E coh = energy difference between free atoms and the solid): Directly related to the potential minimum energy level Often easy to get right exactly. Elastic constants 1 c ρσ Related to deformation in the material Rr ( ) = r' r = u 1 ( r)xˆ + u 2 ( r)ŷ + u 3 ( r)ẑ and to the external stress (pressure) σ : σ ρ = c ρσ e σ σ Voigt notation for ρ - and σ -indexing: xx 1, yy 2, zz 3, yz 4, zx 5, xy 6 Here the strain (crystal distortion) components e ij are e ii u i 1 u = ; e x ij = -- i + i 2 x j u j x i 1. See e.g. Kittel, Introduction to solid state physics, 7th edition, ch. 3. Introduction to molecular dynamics Calculating the forces 36
37 Fitting of potential parameters The stress component σ ij is the force which acts on the plane with the normal x j in the direction x i In principle there are 36 stress and strain components, but their number reduces to much smaller numbers in practice. For instance in a cubic crystal there are only three independent elastic constants c 11 = c xxxx, c 12 = c xxyy and c 44 = c xyxy. Particularly important if there are deformations (compression, shear, melting) in the simulations. Also related to defect properties and the melting point if we get the elastic constants about right we are already on a good way to a good potential. Introduction to molecular dynamics Calculating the forces 37
38 An example of an (unusually) good fit: F. Ercolessi, J. B. Adams, Europhys. Lett. 26 (1994) 583. Introduction to molecular dynamics Calculating the forces 38
39 Weaknesses of pair potentials A pair potential can never describe well the directional properties of covalent bonds. For instance in the diamond/zincblende structure (C, Si, Ge, α-sn, many compound semiconductors) the ideal angle between bonds = o. Similarly, in almost all molecules the directional properties of covalent bonds is of crucial importance. Also longer-range angular dependence is completely neglected. For instance in the structure of polymers torsional terms are important. Also, recent calculations of BCC metals have shown that 4-particle interactions are about 50 % of the bond. Pair potentials also do not account for the environmental dependence. They predict that the strength of the two-atom bond is as strong in a dimer as inside a material, which almost never is true. Introduction to molecular dynamics Calculating the forces 39
40 Weaknesses of pair potentials For instance the Ga-As interaction: Introduction to molecular dynamics Calculating the forces 40
41 Weaknesses of pair potentials Moreover, a pair potential always predicts that the elastic constants c 12 = c for 44 cubic crystals. but in reality: Also, vacancy formation energies are often completely wrong in pair potentials (see below). Pair potentials also usually give bad surface properties. Summa summarum: the pair potential approximation: may work well close to equilibrium structure in many materials is good for noble gases is rather good for ionic compounds such as alkali halides is rather bad for FCC and HCP metals is terrible for covalently bonded materials Source: Ashcroft-Mermin But for all these groups much better, and only slightly slower, models exist. These will be described later on this course. Introduction to molecular dynamics Calculating the forces 41
42 Weaknesses of pair potentials Simple estimate of vacancy formation energy using pair potentials: f E vac = E tot ( vacancy, N) E tot ( perfect, N) nearest neighbor pair potential, energy/bond=v( r nn ) φ no relaxation fcc structure 12 neighbors 1 E tot ( vacancy, N) = -- [( N 12)12φ + 12( 12 1)φ] = 6( N 1)φ 2 1 E tot ( perfect, N) = --N12φ = 6Nφ 2 f E vac = 6φ = E coh However, ab initio calculations 1 : f Element E coh (ev) E vac (ev) V ± 0.2 Nb ± 0.3 W ± 0.2 Relaxation: only minor effect (far less than 1 ev). 1. A. E. Carlsson, Solid State Physics: Advances in Research and Applications, 43 (1990) 1. Introduction to molecular dynamics Calculating the forces 42
Set the initial conditions r i. Update neighborlist. r i. Get new forces F i
 Set the initial conditions r i t 0, v i t 0 Update neighborlist Get new forces F i r i Solve the equations of motion numerically over time step t : r i t n r i t n + 1 v i t n v i t n + 1 Perform T, P
Set the initial conditions r i t 0, v i t 0 Update neighborlist Get new forces F i r i Solve the equations of motion numerically over time step t : r i t n r i t n + 1 v i t n v i t n + 1 Perform T, P
Pair Potentials and Force Calculations
 Molecular Dynamics simulations Lecture 04: Pair Potentials and Force Calculations Dr. Olli Pakarinen University of Helsinki Fall 01 Lecture notes based on notes by Dr. Jani Kotakoski, 010 CONSTRUCTING
Molecular Dynamics simulations Lecture 04: Pair Potentials and Force Calculations Dr. Olli Pakarinen University of Helsinki Fall 01 Lecture notes based on notes by Dr. Jani Kotakoski, 010 CONSTRUCTING
Potentials, periodicity
 Potentials, periodicity Lecture 2 1/23/18 1 Survey responses 2 Topic requests DFT (10), Molecular dynamics (7), Monte Carlo (5) Machine Learning (4), High-throughput, Databases (4) NEB, phonons, Non-equilibrium
Potentials, periodicity Lecture 2 1/23/18 1 Survey responses 2 Topic requests DFT (10), Molecular dynamics (7), Monte Carlo (5) Machine Learning (4), High-throughput, Databases (4) NEB, phonons, Non-equilibrium
Lecture 6 - Bonding in Crystals
 Lecture 6 onding in Crystals inding in Crystals (Kittel Ch. 3) inding of atoms to form crystals A crystal is a repeated array of atoms Why do they form? What are characteristic bonding mechanisms? How
Lecture 6 onding in Crystals inding in Crystals (Kittel Ch. 3) inding of atoms to form crystals A crystal is a repeated array of atoms Why do they form? What are characteristic bonding mechanisms? How
Interatomic Potentials. The electronic-structure problem
 Interatomic Potentials Before we can start a simulation, we need the model! Interaction between atoms and molecules is determined by quantum mechanics: Schrödinger Equation + Born-Oppenheimer approximation
Interatomic Potentials Before we can start a simulation, we need the model! Interaction between atoms and molecules is determined by quantum mechanics: Schrödinger Equation + Born-Oppenheimer approximation
From Atoms to Materials: Predictive Theory and Simulations
 From Atoms to Materials: Predictive Theory and Simulations Week 3 Lecture 4 Potentials for metals and semiconductors Ale Strachan strachan@purdue.edu School of Materials Engineering & Birck anotechnology
From Atoms to Materials: Predictive Theory and Simulations Week 3 Lecture 4 Potentials for metals and semiconductors Ale Strachan strachan@purdue.edu School of Materials Engineering & Birck anotechnology
CE 530 Molecular Simulation
 1 CE 530 Molecular Simulation Lecture 14 Molecular Models David A. Kofke Department of Chemical Engineering SUNY Buffalo kofke@eng.buffalo.edu 2 Review Monte Carlo ensemble averaging, no dynamics easy
1 CE 530 Molecular Simulation Lecture 14 Molecular Models David A. Kofke Department of Chemical Engineering SUNY Buffalo kofke@eng.buffalo.edu 2 Review Monte Carlo ensemble averaging, no dynamics easy
The broad topic of physical metallurgy provides a basis that links the structure of materials with their properties, focusing primarily on metals.
 Physical Metallurgy The broad topic of physical metallurgy provides a basis that links the structure of materials with their properties, focusing primarily on metals. Crystal Binding In our discussions
Physical Metallurgy The broad topic of physical metallurgy provides a basis that links the structure of materials with their properties, focusing primarily on metals. Crystal Binding In our discussions
Introductory Nanotechnology ~ Basic Condensed Matter Physics ~
 Introductory Nanotechnology ~ Basic Condensed Matter Physics ~ Atsufumi Hirohata Department of Electronics Go into Nano-Scale Lateral Size [m] 10-3 10-6 Micron-scale Sub-Micron-scale Nano-scale Human hair
Introductory Nanotechnology ~ Basic Condensed Matter Physics ~ Atsufumi Hirohata Department of Electronics Go into Nano-Scale Lateral Size [m] 10-3 10-6 Micron-scale Sub-Micron-scale Nano-scale Human hair
Reactive potentials and applications
 1.021, 3.021, 10.333, 22.00 Introduction to Modeling and Simulation Spring 2011 Part I Continuum and particle methods Reactive potentials and applications Lecture 8 Markus J. Buehler Laboratory for Atomistic
1.021, 3.021, 10.333, 22.00 Introduction to Modeling and Simulation Spring 2011 Part I Continuum and particle methods Reactive potentials and applications Lecture 8 Markus J. Buehler Laboratory for Atomistic
Atoms & Their Interactions
 Lecture 2 Atoms & Their Interactions Si: the heart of electronic materials Intel, 300mm Si wafer, 200 μm thick and 48-core CPU ( cloud computing on a chip ) Twin Creeks Technologies, San Jose, Si wafer,
Lecture 2 Atoms & Their Interactions Si: the heart of electronic materials Intel, 300mm Si wafer, 200 μm thick and 48-core CPU ( cloud computing on a chip ) Twin Creeks Technologies, San Jose, Si wafer,
Chapter 2 Experimental sources of intermolecular potentials
 Chapter 2 Experimental sources of intermolecular potentials 2.1 Overview thermodynamical properties: heat of vaporization (Trouton s rule) crystal structures ionic crystals rare gas solids physico-chemical
Chapter 2 Experimental sources of intermolecular potentials 2.1 Overview thermodynamical properties: heat of vaporization (Trouton s rule) crystal structures ionic crystals rare gas solids physico-chemical
Materials 218/UCSB: Class III Cohesion in solids van der Waals, ionic, covalent, metallic
 Materials 218/UCSB: Class III Cohesion in solids van der Waals, ionic, covalent, metallic Ram Seshadri (seshadri@mrl.ucsb.edu) Introduction There are four forces in nature. The strong and the weak interactions
Materials 218/UCSB: Class III Cohesion in solids van der Waals, ionic, covalent, metallic Ram Seshadri (seshadri@mrl.ucsb.edu) Introduction There are four forces in nature. The strong and the weak interactions
Lecture 11 - Phonons II - Thermal Prop. Continued
 Phonons II - hermal Properties - Continued (Kittel Ch. 5) Low High Outline Anharmonicity Crucial for hermal expansion other changes with pressure temperature Gruneisen Constant hermal Heat ransport Phonon
Phonons II - hermal Properties - Continued (Kittel Ch. 5) Low High Outline Anharmonicity Crucial for hermal expansion other changes with pressure temperature Gruneisen Constant hermal Heat ransport Phonon
Chapter 3. Crystal Binding
 Chapter 3. Crystal Binding Energy of a crystal and crystal binding Cohesive energy of Molecular crystals Ionic crystals Metallic crystals Elasticity What causes matter to exist in three different forms?
Chapter 3. Crystal Binding Energy of a crystal and crystal binding Cohesive energy of Molecular crystals Ionic crystals Metallic crystals Elasticity What causes matter to exist in three different forms?
Atoms, electrons and Solids
 Atoms, electrons and Solids Shell model of an atom negative electron orbiting a positive nucleus QM tells that to minimize total energy the electrons fill up shells. Each orbit in a shell has a specific
Atoms, electrons and Solids Shell model of an atom negative electron orbiting a positive nucleus QM tells that to minimize total energy the electrons fill up shells. Each orbit in a shell has a specific
The electronic structure of materials 1
 Quantum mechanics 2 - Lecture 9 December 18, 2013 1 An overview 2 Literature Contents 1 An overview 2 Literature Electronic ground state Ground state cohesive energy equilibrium crystal structure phase
Quantum mechanics 2 - Lecture 9 December 18, 2013 1 An overview 2 Literature Contents 1 An overview 2 Literature Electronic ground state Ground state cohesive energy equilibrium crystal structure phase
 lectures accompanying the book: Solid State Physics: An Introduction, by Philip ofmann (2nd edition 2015, ISBN-10: 3527412824, ISBN-13: 978-3527412822, Wiley-VC Berlin. www.philiphofmann.net 1 Bonds between
lectures accompanying the book: Solid State Physics: An Introduction, by Philip ofmann (2nd edition 2015, ISBN-10: 3527412824, ISBN-13: 978-3527412822, Wiley-VC Berlin. www.philiphofmann.net 1 Bonds between
Ionic Bonding. Example: Atomic Radius: Na (r = 0.192nm) Cl (r = 0.099nm) Ionic Radius : Na (r = 0.095nm) Cl (r = 0.181nm)
 Ionic Bonding Ion: an atom or molecule that gains or loses electrons (acquires an electrical charge). Atoms form cations (+charge), when they lose electrons, or anions (- charge), when they gain electrons.
Ionic Bonding Ion: an atom or molecule that gains or loses electrons (acquires an electrical charge). Atoms form cations (+charge), when they lose electrons, or anions (- charge), when they gain electrons.
CHEM 103 Quantum Mechanics and Periodic Trends
 CHEM 103 Quantum Mechanics and Periodic Trends Lecture Notes April 11, 2006 Prof. Sevian Agenda Predicting electronic configurations using the QM model Group similarities Interpreting measured properties
CHEM 103 Quantum Mechanics and Periodic Trends Lecture Notes April 11, 2006 Prof. Sevian Agenda Predicting electronic configurations using the QM model Group similarities Interpreting measured properties
Structure and Dynamics : An Atomic View of Materials
 Structure and Dynamics : An Atomic View of Materials MARTIN T. DOVE Department ofearth Sciences University of Cambridge OXFORD UNIVERSITY PRESS Contents 1 Introduction 1 1.1 Observations 1 1.1.1 Microscopic
Structure and Dynamics : An Atomic View of Materials MARTIN T. DOVE Department ofearth Sciences University of Cambridge OXFORD UNIVERSITY PRESS Contents 1 Introduction 1 1.1 Observations 1 1.1.1 Microscopic
Classical potentials for metals
 Classical potentials for metals About 80 % of all elements are metals. The crystal structures of the elements are distributed as follows: FCC 15 HCP 26 BCC 16 All other metals 13 So if we can describe
Classical potentials for metals About 80 % of all elements are metals. The crystal structures of the elements are distributed as follows: FCC 15 HCP 26 BCC 16 All other metals 13 So if we can describe
Physics of Condensed Matter I
 Physics of Condensed Matter I 1100-4INZ`PC Solid State 1 Faculty of Physics UW Jacek.Szczytko@fuw.edu.pl Chemical bonding and molecules Born Oppenheimer approximation Max Born (1882-1970) Jacob R. Oppenheimer
Physics of Condensed Matter I 1100-4INZ`PC Solid State 1 Faculty of Physics UW Jacek.Szczytko@fuw.edu.pl Chemical bonding and molecules Born Oppenheimer approximation Max Born (1882-1970) Jacob R. Oppenheimer
Materials for Civil and Construction Engineers CHAPTER 2. Nature of Materials
 Materials for Civil and Construction Engineers CHAPTER 2 Nature of Materials Bonds 1. Primary Bond: forms when atoms interchange or share electrons in order to fill the outer (valence) shells like noble
Materials for Civil and Construction Engineers CHAPTER 2 Nature of Materials Bonds 1. Primary Bond: forms when atoms interchange or share electrons in order to fill the outer (valence) shells like noble
E12 UNDERSTANDING CRYSTAL STRUCTURES
 E1 UNDERSTANDING CRYSTAL STRUCTURES 1 Introduction In this experiment, the structures of many elements and compounds are rationalized using simple packing models. The pre-work revises and extends the material
E1 UNDERSTANDING CRYSTAL STRUCTURES 1 Introduction In this experiment, the structures of many elements and compounds are rationalized using simple packing models. The pre-work revises and extends the material
Atoms, Molecules and Solids (selected topics)
 Atoms, Molecules and Solids (selected topics) Part I: Electronic configurations and transitions Transitions between atomic states (Hydrogen atom) Transition probabilities are different depending on the
Atoms, Molecules and Solids (selected topics) Part I: Electronic configurations and transitions Transitions between atomic states (Hydrogen atom) Transition probabilities are different depending on the
2. As gas P increases and/or T is lowered, intermolecular forces become significant, and deviations from ideal gas laws occur (van der Waal equation).
 A. Introduction. (Section 11.1) CHAPTER 11: STATES OF MATTER, LIQUIDS AND SOLIDS 1. Gases are easily treated mathematically because molecules behave independently. 2. As gas P increases and/or T is lowered,
A. Introduction. (Section 11.1) CHAPTER 11: STATES OF MATTER, LIQUIDS AND SOLIDS 1. Gases are easily treated mathematically because molecules behave independently. 2. As gas P increases and/or T is lowered,
Introduction to Condensed Matter Physics
 Introduction to Condensed Matter Physics Crystalline Solids - Introduction M.P. Vaughan Overview Overview of course Crystal solids Crystal structure Crystal symmetry The reciprocal lattice Band theory
Introduction to Condensed Matter Physics Crystalline Solids - Introduction M.P. Vaughan Overview Overview of course Crystal solids Crystal structure Crystal symmetry The reciprocal lattice Band theory
Atomic Structure & Interatomic Bonding
 Atomic Structure & Interatomic Bonding Chapter Outline Review of Atomic Structure Atomic Bonding Atomic Structure Atoms are the smallest structural units of all solids, liquids & gases. Atom: The smallest
Atomic Structure & Interatomic Bonding Chapter Outline Review of Atomic Structure Atomic Bonding Atomic Structure Atoms are the smallest structural units of all solids, liquids & gases. Atom: The smallest
Physics 211B : Problem Set #0
 Physics 211B : Problem Set #0 These problems provide a cross section of the sort of exercises I would have assigned had I taught 211A. Please take a look at all the problems, and turn in problems 1, 4,
Physics 211B : Problem Set #0 These problems provide a cross section of the sort of exercises I would have assigned had I taught 211A. Please take a look at all the problems, and turn in problems 1, 4,
"My name is Bond." N2 (Double 07)
 "My name is Bond." N2 (Double 07) "Metallic Bond." Element 3: Lithium [He]2s 1 "My name is Bond." In the last lecture we identified three types of molecular bonding: van der Waals Interactions (Ar) Covalent
"My name is Bond." N2 (Double 07) "Metallic Bond." Element 3: Lithium [He]2s 1 "My name is Bond." In the last lecture we identified three types of molecular bonding: van der Waals Interactions (Ar) Covalent
Introduction to molecular dynamics
 1 Introduction to molecular dynamics Yves Lansac Université François Rabelais, Tours, France Visiting MSE, GIST for the summer Molecular Simulation 2 Molecular simulation is a computational experiment.
1 Introduction to molecular dynamics Yves Lansac Université François Rabelais, Tours, France Visiting MSE, GIST for the summer Molecular Simulation 2 Molecular simulation is a computational experiment.
The Periodic Table and Chemical Reactivity
 The and Chemical Reactivity Noble gases Less electronegative elements More electronegative elements Then what is electronegativity? The tendency of an atom to attract an electron (or electron density)
The and Chemical Reactivity Noble gases Less electronegative elements More electronegative elements Then what is electronegativity? The tendency of an atom to attract an electron (or electron density)
Scientific Computing II
 Scientific Computing II Molecular Dynamics Simulation Michael Bader SCCS Summer Term 2015 Molecular Dynamics Simulation, Summer Term 2015 1 Continuum Mechanics for Fluid Mechanics? Molecular Dynamics the
Scientific Computing II Molecular Dynamics Simulation Michael Bader SCCS Summer Term 2015 Molecular Dynamics Simulation, Summer Term 2015 1 Continuum Mechanics for Fluid Mechanics? Molecular Dynamics the
CHAPTER 1 Atoms and bonding. Ionic bonding Covalent bonding Metallic bonding van der Waals bonding
 CHAPTER 1 Atoms and bonding The periodic table Ionic bonding Covalent bonding Metallic bonding van der Waals bonding Atoms and bonding In order to understand the physics of semiconductor (s/c) devices,
CHAPTER 1 Atoms and bonding The periodic table Ionic bonding Covalent bonding Metallic bonding van der Waals bonding Atoms and bonding In order to understand the physics of semiconductor (s/c) devices,
Defects in Crystals. Image borrowed from who got it from Davis & Reynolds 1996.
 Defects in Crystals Image borrowed from http://comp.uark.edu/~pjansma/geol3513_25_defmechs1_04.ppt who got it from Davis & Reynolds 1996. It's easy to think of real crystals as having these ideal structures.
Defects in Crystals Image borrowed from http://comp.uark.edu/~pjansma/geol3513_25_defmechs1_04.ppt who got it from Davis & Reynolds 1996. It's easy to think of real crystals as having these ideal structures.
Quantum Condensed Matter Physics Lecture 5
 Quantum Condensed Matter Physics Lecture 5 detector sample X-ray source monochromator David Ritchie http://www.sp.phy.cam.ac.uk/drp2/home QCMP Lent/Easter 2019 5.1 Quantum Condensed Matter Physics 1. Classical
Quantum Condensed Matter Physics Lecture 5 detector sample X-ray source monochromator David Ritchie http://www.sp.phy.cam.ac.uk/drp2/home QCMP Lent/Easter 2019 5.1 Quantum Condensed Matter Physics 1. Classical
Intermolecular Forces and Monte-Carlo Integration 열역학특수연구
 Intermolecular Forces and Monte-Carlo Integration 열역학특수연구 2003.3.28 Source of the lecture note. J.M.Prausnitz and others, Molecular Thermodynamics of Fluid Phase Equiliria Atkins, Physical Chemistry Lecture
Intermolecular Forces and Monte-Carlo Integration 열역학특수연구 2003.3.28 Source of the lecture note. J.M.Prausnitz and others, Molecular Thermodynamics of Fluid Phase Equiliria Atkins, Physical Chemistry Lecture
CHAPTER 2 INTERATOMIC FORCES. atoms together in a solid?
 CHAPTER 2 INTERATOMIC FORCES What kind of force holds the atoms together in a solid? Interatomic Binding All of the mechanisms which cause bonding between the atoms derive from electrostatic interaction
CHAPTER 2 INTERATOMIC FORCES What kind of force holds the atoms together in a solid? Interatomic Binding All of the mechanisms which cause bonding between the atoms derive from electrostatic interaction
Set the initial conditions r i. Update neighborlist. Get new forces F i
 Set the initial conditions r i ( t 0 ), v i ( t 0 ) Update neighborlist Potential models ionic compounds Get new forces F i ( r i ) Solve the equations of motion numerically over time step Δt : r i ( t
Set the initial conditions r i ( t 0 ), v i ( t 0 ) Update neighborlist Potential models ionic compounds Get new forces F i ( r i ) Solve the equations of motion numerically over time step Δt : r i ( t
Ionic Bonding - Electrostatic Interactions and Polarization
 Ionic Bonding - Electrostatic Interactions and Polarization Chemistry 754 Solid State Chemistry Dr. Patrick Woodward Lecture #13 Born-Haber Cycle for NaCl It is energetically unfavorable for Na metal and
Ionic Bonding - Electrostatic Interactions and Polarization Chemistry 754 Solid State Chemistry Dr. Patrick Woodward Lecture #13 Born-Haber Cycle for NaCl It is energetically unfavorable for Na metal and
T. Egami. Model System of Dense Random Packing (DRP)
 Introduction to Metallic Glasses: How they are different/similar to other glasses T. Egami Model System of Dense Random Packing (DRP) Hard Sphere vs. Soft Sphere Glass transition Universal behavior History:
Introduction to Metallic Glasses: How they are different/similar to other glasses T. Egami Model System of Dense Random Packing (DRP) Hard Sphere vs. Soft Sphere Glass transition Universal behavior History:
Quantum Condensed Matter Physics Lecture 4
 Quantum Condensed Matter Physics Lecture 4 David Ritchie QCMP Lent/Easter 2019 http://www.sp.phy.cam.ac.uk/drp2/home 4.1 Quantum Condensed Matter Physics 1. Classical and Semi-classical models for electrons
Quantum Condensed Matter Physics Lecture 4 David Ritchie QCMP Lent/Easter 2019 http://www.sp.phy.cam.ac.uk/drp2/home 4.1 Quantum Condensed Matter Physics 1. Classical and Semi-classical models for electrons
Lecture Outline: Atomic Structure
 Lecture Outline: Atomic Structure Electronic Structure of the Atom Periodic Table Types of Atomic Bonding, primary/secondary bonds Coordination and next neighbors Binding Energy, Interatomic Spacing, &
Lecture Outline: Atomic Structure Electronic Structure of the Atom Periodic Table Types of Atomic Bonding, primary/secondary bonds Coordination and next neighbors Binding Energy, Interatomic Spacing, &
1.4 Crystal structure
 1.4 Crystal structure (a) crystalline vs. (b) amorphous configurations short and long range order only short range order Abbildungen: S. Hunklinger, Festkörperphysik, Oldenbourg Verlag represenatives of
1.4 Crystal structure (a) crystalline vs. (b) amorphous configurations short and long range order only short range order Abbildungen: S. Hunklinger, Festkörperphysik, Oldenbourg Verlag represenatives of
PH 548 Atomistic Simulation Techniques
 PH 548 Atomistic Simulation Techniques Lectures: Lab: 2-0-2-6 Tuesday 12-1 PM Thursday 12-1 PM Monday 2-5 PM P. K. Padmanabhan PH548: Two Excellent Books How to do it? 1 2 F 12 F 23 3 F i = F ij j i F
PH 548 Atomistic Simulation Techniques Lectures: Lab: 2-0-2-6 Tuesday 12-1 PM Thursday 12-1 PM Monday 2-5 PM P. K. Padmanabhan PH548: Two Excellent Books How to do it? 1 2 F 12 F 23 3 F i = F ij j i F
1 Bulk Simulations in a HCP lattice
 1 Bulk Simulations in a HCP lattice 1.1 Introduction This project is a continuation of two previous projects that studied the mechanism of melting in a FCC and BCC lattice. The current project studies
1 Bulk Simulations in a HCP lattice 1.1 Introduction This project is a continuation of two previous projects that studied the mechanism of melting in a FCC and BCC lattice. The current project studies
2. As gas P increases and/or T is lowered, intermolecular forces become significant, and deviations from ideal gas laws occur (van der Waal equation).
 A. Introduction. (Section 11.1) CHAPTER 11: STATES OF MATTER, LIQUIDS AND SOLIDS 1. Gases are easily treated mathematically because molecules behave independently. 2. As gas P increases and/or T is lowered,
A. Introduction. (Section 11.1) CHAPTER 11: STATES OF MATTER, LIQUIDS AND SOLIDS 1. Gases are easily treated mathematically because molecules behave independently. 2. As gas P increases and/or T is lowered,
PART CHAPTER2. Atomic Bonding
 PART O N E APTER2 Atomic Bonding The scanning tunneling microscope (Section 4.7) allows the imaging of individual atoms bonded to a material surface. In this case, the microscope was also used to manipulate
PART O N E APTER2 Atomic Bonding The scanning tunneling microscope (Section 4.7) allows the imaging of individual atoms bonded to a material surface. In this case, the microscope was also used to manipulate
VIRTUAL LATTICE TECHNIQUE AND THE INTERATOMIC POTENTIALS OF ZINC-BLEND-TYPE BINARY COMPOUNDS
 Modern Physics Letters B, Vol. 16, Nos. 5 & 6 (2002) 187 194 c World Scientific Publishing Company VIRTUAL LATTICE TECHNIQUE AND THE INTERATOMIC POTENTIALS OF ZINC-BLEND-TYPE BINARY COMPOUNDS LIU YING,
Modern Physics Letters B, Vol. 16, Nos. 5 & 6 (2002) 187 194 c World Scientific Publishing Company VIRTUAL LATTICE TECHNIQUE AND THE INTERATOMIC POTENTIALS OF ZINC-BLEND-TYPE BINARY COMPOUNDS LIU YING,
Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland
 Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland 1) Question. Two methods which are widely used for the optimization of molecular geometies are the Steepest descents and Newton-Raphson
Example questions for Molecular modelling (Level 4) Dr. Adrian Mulholland 1) Question. Two methods which are widely used for the optimization of molecular geometies are the Steepest descents and Newton-Raphson
Electronic Structure Theory for Periodic Systems: The Concepts. Christian Ratsch
 Electronic Structure Theory for Periodic Systems: The Concepts Christian Ratsch Institute for Pure and Applied Mathematics and Department of Mathematics, UCLA Motivation There are 10 20 atoms in 1 mm 3
Electronic Structure Theory for Periodic Systems: The Concepts Christian Ratsch Institute for Pure and Applied Mathematics and Department of Mathematics, UCLA Motivation There are 10 20 atoms in 1 mm 3
Phys 412 Solid State Physics. Lecturer: Réka Albert
 Phys 412 Solid State Physics Lecturer: Réka Albert What is a solid? A material that keeps its shape Can be deformed by stress Returns to original shape if it is not strained too much Solid structure
Phys 412 Solid State Physics Lecturer: Réka Albert What is a solid? A material that keeps its shape Can be deformed by stress Returns to original shape if it is not strained too much Solid structure
Material Surfaces, Grain Boundaries and Interfaces: Structure-Property Relationship Predictions
 Material Surfaces, Grain Boundaries and Interfaces: Structure-Property Relationship Predictions Susan B. Sinnott Department of Materials Science and Engineering Penn State University September 16, 2016
Material Surfaces, Grain Boundaries and Interfaces: Structure-Property Relationship Predictions Susan B. Sinnott Department of Materials Science and Engineering Penn State University September 16, 2016
Imperfect Gases. NC State University
 Chemistry 431 Lecture 3 Imperfect Gases NC State University The Compression Factor One way to represent the relationship between ideal and real gases is to plot the deviation from ideality as the gas is
Chemistry 431 Lecture 3 Imperfect Gases NC State University The Compression Factor One way to represent the relationship between ideal and real gases is to plot the deviation from ideality as the gas is
Introduction to Materials Science Prof. Michael Roth
 Introduction to Materials Science Prof. Michael Roth Chapter 3 Crystal Binding and Elasticity Introduction What holds a crystal together? Atoms in a solid are bound together by Coulomb forces; Others,
Introduction to Materials Science Prof. Michael Roth Chapter 3 Crystal Binding and Elasticity Introduction What holds a crystal together? Atoms in a solid are bound together by Coulomb forces; Others,
Machine learning the Born-Oppenheimer potential energy surface: from molecules to materials. Gábor Csányi Engineering Laboratory
 Machine learning the Born-Oppenheimer potential energy surface: from molecules to materials Gábor Csányi Engineering Laboratory Interatomic potentials for molecular dynamics Transferability biomolecular
Machine learning the Born-Oppenheimer potential energy surface: from molecules to materials Gábor Csányi Engineering Laboratory Interatomic potentials for molecular dynamics Transferability biomolecular
Structures of Solids. Unit Cells - Not(?) Chapter 4 Ionic and Other Inorganic Solids. CHEM 462 Wednesday, September 22 T.
 Chapter 4 Ionic and Other Inorganic Solids CHEM 462 Wednesday, September 22 T. Hughbanks Structures of Solids Many dense solids are described in terms of packing of atoms or ions. Although these geometric
Chapter 4 Ionic and Other Inorganic Solids CHEM 462 Wednesday, September 22 T. Hughbanks Structures of Solids Many dense solids are described in terms of packing of atoms or ions. Although these geometric
An Introduction to Lattice Vibrations
 An Introduction to Lattice Vibrations Andreas Wacker 1 Mathematical Physics, Lund University November 3, 2015 1 Introduction Ideally, the atoms in a crystal are positioned in a regular manner following
An Introduction to Lattice Vibrations Andreas Wacker 1 Mathematical Physics, Lund University November 3, 2015 1 Introduction Ideally, the atoms in a crystal are positioned in a regular manner following
Charge equilibration
 Charge equilibration Taylor expansion of energy of atom A @E E A (Q) =E A0 + Q A + 1 @Q A 0 2 Q2 A @ 2 E @Q 2 A 0 +... The corresponding energy of cation/anion and neutral atom E A (+1) = E A0 + @E @Q
Charge equilibration Taylor expansion of energy of atom A @E E A (Q) =E A0 + Q A + 1 @Q A 0 2 Q2 A @ 2 E @Q 2 A 0 +... The corresponding energy of cation/anion and neutral atom E A (+1) = E A0 + @E @Q
The change in free energy on transferring an ion from a medium of low dielectric constantε1 to one of high dielectric constant ε2:
 The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
The Born Energy of an Ion The free energy density of an electric field E arising from a charge is ½(ε 0 ε E 2 ) per unit volume Integrating the energy density of an ion over all of space = Born energy:
1.1 Atoms. 1.1 Atoms
 1. Chemical bonding and crystal structure 19 21 Hydrogen atom Scanning electron microscopy Ni surface Cleaved surface ZnO, TiO 2, NiO, NaCl, Si, Ge, GaAs, InP Crystals are build by small repeating units
1. Chemical bonding and crystal structure 19 21 Hydrogen atom Scanning electron microscopy Ni surface Cleaved surface ZnO, TiO 2, NiO, NaCl, Si, Ge, GaAs, InP Crystals are build by small repeating units
Chapter 1: Chemical Bonding
 Chapter 1: Chemical Bonding Linus Pauling (1901 1994) January 30, 2017 Contents 1 The development of Bands and their filling 3 2 Different Types of Bonds 7 2.1 Covalent Bonding.............................
Chapter 1: Chemical Bonding Linus Pauling (1901 1994) January 30, 2017 Contents 1 The development of Bands and their filling 3 2 Different Types of Bonds 7 2.1 Covalent Bonding.............................
Chem 442 Review for Exam 2. Exact separation of the Hamiltonian of a hydrogenic atom into center-of-mass (3D) and relative (3D) components.
 Chem 44 Review for Exam Hydrogenic atoms: The Coulomb energy between two point charges Ze and e: V r Ze r Exact separation of the Hamiltonian of a hydrogenic atom into center-of-mass (3D) and relative
Chem 44 Review for Exam Hydrogenic atoms: The Coulomb energy between two point charges Ze and e: V r Ze r Exact separation of the Hamiltonian of a hydrogenic atom into center-of-mass (3D) and relative
Physics of Materials: Bonding and Material Properties On The basis of Geometry and Bonding (Intermolecular forces) Dr.
 : Bonding and Material Properties On The basis of Geometry and Bonding (Intermolecular forces) Dr. Anurag Srivastava Atal Bihari Vajpayee Indian Institute of Information Technology and Manegement, Gwalior
: Bonding and Material Properties On The basis of Geometry and Bonding (Intermolecular forces) Dr. Anurag Srivastava Atal Bihari Vajpayee Indian Institute of Information Technology and Manegement, Gwalior
Accuracy and transferability of GAP models for tungsten
 Accuracy and transferability of GAP models for tungsten Wojciech J. Szlachta Albert P. Bartók Gábor Csányi Engineering Laboratory University of Cambridge 5 November 214 Motivation Number of atoms 1 1 2
Accuracy and transferability of GAP models for tungsten Wojciech J. Szlachta Albert P. Bartók Gábor Csányi Engineering Laboratory University of Cambridge 5 November 214 Motivation Number of atoms 1 1 2
Molecular Dynamics Simulations. Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia
 Molecular Dynamics Simulations Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia 1 An Introduction to Molecular Dynamics Simulations Macroscopic properties
Molecular Dynamics Simulations Dr. Noelia Faginas Lago Dipartimento di Chimica,Biologia e Biotecnologie Università di Perugia 1 An Introduction to Molecular Dynamics Simulations Macroscopic properties
Bonding in solids The interaction of electrons in neighboring atoms of a solid serves the very important function of holding the crystal together.
 Bonding in solids The interaction of electrons in neighboring atoms of a solid serves the very important function of holding the crystal together. For example Nacl In the Nacl lattice, each Na atom is
Bonding in solids The interaction of electrons in neighboring atoms of a solid serves the very important function of holding the crystal together. For example Nacl In the Nacl lattice, each Na atom is
T6.2 Molecular Mechanics
 T6.2 Molecular Mechanics We have seen that Benson group additivities are capable of giving heats of formation of molecules with accuracies comparable to those of the best ab initio procedures. However,
T6.2 Molecular Mechanics We have seen that Benson group additivities are capable of giving heats of formation of molecules with accuracies comparable to those of the best ab initio procedures. However,
Atoms, Molecules and Solids (selected topics)
 Atoms, Molecules and Solids (selected topics) Part I: Electronic configurations and transitions Transitions between atomic states (Hydrogen atom) Transition probabilities are different depending on the
Atoms, Molecules and Solids (selected topics) Part I: Electronic configurations and transitions Transitions between atomic states (Hydrogen atom) Transition probabilities are different depending on the
Molecular Dynamics of Covalent Crystals
 Molecular Dynamics of Covalent Crystals J. Hahn and H.-R. Trebin Institut für Theoretische und Angewandte Physik, Universität Stuttgart, D-70550 Stuttgart, Germany Abstract. A molecular mechanics-like
Molecular Dynamics of Covalent Crystals J. Hahn and H.-R. Trebin Institut für Theoretische und Angewandte Physik, Universität Stuttgart, D-70550 Stuttgart, Germany Abstract. A molecular mechanics-like
1. Introduction to Clusters
 1. Introduction to Clusters 1.1 The Field of Clusters Atomic clusters are aggregates of atoms containing from few to a few thousand atoms. Due to their small size, the properties of the clusters are, in
1. Introduction to Clusters 1.1 The Field of Clusters Atomic clusters are aggregates of atoms containing from few to a few thousand atoms. Due to their small size, the properties of the clusters are, in
Complicated, short range. þq 1 Q 2 /4p3 0 r (Coulomb energy) Q 2 u 2 /6(4p3 0 ) 2 ktr 4. u 2 1 u2 2 =3ð4p3 0Þ 2 ktr 6 ðkeesom energyþ
 Bonding ¼ Type of interaction Interaction energy w(r) Covalent, metallic Complicated, short range Charge charge þq 1 Q 2 /4p3 0 r (Coulomb energy) Charge dipole Qu cos q/4p3 0 r 2 Q 2 u 2 /6(4p3 0 ) 2
Bonding ¼ Type of interaction Interaction energy w(r) Covalent, metallic Complicated, short range Charge charge þq 1 Q 2 /4p3 0 r (Coulomb energy) Charge dipole Qu cos q/4p3 0 r 2 Q 2 u 2 /6(4p3 0 ) 2
Pre-yield non-affine fluctuations and a hidden critical point in strained crystals
 Supplementary Information for: Pre-yield non-affine fluctuations and a hidden critical point in strained crystals Tamoghna Das, a,b Saswati Ganguly, b Surajit Sengupta c and Madan Rao d a Collective Interactions
Supplementary Information for: Pre-yield non-affine fluctuations and a hidden critical point in strained crystals Tamoghna Das, a,b Saswati Ganguly, b Surajit Sengupta c and Madan Rao d a Collective Interactions
Set the initial conditions r i. Update neighborlist. r i. Get new forces F i. Solve the equations of motion numerically over time step t : r i t n
 Set the initial conditions r i t 0, v i t 0 Update neighborlist Get new forces F i r i Potential models for diamond and zincblende structures Solve the equations of motion numerically over time step t
Set the initial conditions r i t 0, v i t 0 Update neighborlist Get new forces F i r i Potential models for diamond and zincblende structures Solve the equations of motion numerically over time step t
Atomic Structure. Atomic weight = m protons + m neutrons Atomic number (Z) = # of protons Isotope corresponds to # of neutrons
 Atomic Structure Neutrons: neutral Protons: positive charge (1.6x10 19 C, 1.67x10 27 kg) Electrons: negative charge (1.6x10 19 C, 9.11x10 31 kg) Atomic weight = m protons + m neutrons Atomic number (Z)
Atomic Structure Neutrons: neutral Protons: positive charge (1.6x10 19 C, 1.67x10 27 kg) Electrons: negative charge (1.6x10 19 C, 9.11x10 31 kg) Atomic weight = m protons + m neutrons Atomic number (Z)
MP464: Solid State Physics Problem Sheet
 MP464: Solid State Physics Problem Sheet 1 Write down primitive lattice vectors for the -dimensional rectangular lattice, with sides a and b in the x and y-directions respectively, and a face-centred rectangular
MP464: Solid State Physics Problem Sheet 1 Write down primitive lattice vectors for the -dimensional rectangular lattice, with sides a and b in the x and y-directions respectively, and a face-centred rectangular
4. Thermal properties of solids. Time to study: 4 hours. Lecture Oscillations of the crystal lattice
 4. Thermal properties of solids Time to study: 4 hours Objective After studying this chapter you will get acquainted with a description of oscillations of atoms learn how to express heat capacity for different
4. Thermal properties of solids Time to study: 4 hours Objective After studying this chapter you will get acquainted with a description of oscillations of atoms learn how to express heat capacity for different
The Solid State. Phase diagrams Crystals and symmetry Unit cells and packing Types of solid
 The Solid State Phase diagrams Crystals and symmetry Unit cells and packing Types of solid Learning objectives Apply phase diagrams to prediction of phase behaviour Describe distinguishing features of
The Solid State Phase diagrams Crystals and symmetry Unit cells and packing Types of solid Learning objectives Apply phase diagrams to prediction of phase behaviour Describe distinguishing features of
3.091 Introduction to Solid State Chemistry. Lecture Notes No. 5a ELASTIC BEHAVIOR OF SOLIDS
 3.091 Introduction to Solid State Chemistry Lecture Notes No. 5a ELASTIC BEHAVIOR OF SOLIDS 1. INTRODUCTION Crystals are held together by interatomic or intermolecular bonds. The bonds can be covalent,
3.091 Introduction to Solid State Chemistry Lecture Notes No. 5a ELASTIC BEHAVIOR OF SOLIDS 1. INTRODUCTION Crystals are held together by interatomic or intermolecular bonds. The bonds can be covalent,
Classical Theory of Harmonic Crystals
 Classical Theory of Harmonic Crystals HARMONIC APPROXIMATION The Hamiltonian of the crystal is expressed in terms of the kinetic energies of atoms and the potential energy. In calculating the potential
Classical Theory of Harmonic Crystals HARMONIC APPROXIMATION The Hamiltonian of the crystal is expressed in terms of the kinetic energies of atoms and the potential energy. In calculating the potential
CHAPTER 2: BONDING AND PROPERTIES
 CHAPTER 2: BONDING AND PROPERTIES ISSUES TO ADDRESS... What promotes bonding? What types of bonds are there? What properties are inferred from bonding? Chapter 2 1 Fundamental concepts Proton and electron,
CHAPTER 2: BONDING AND PROPERTIES ISSUES TO ADDRESS... What promotes bonding? What types of bonds are there? What properties are inferred from bonding? Chapter 2 1 Fundamental concepts Proton and electron,
Molecular Dynamics Simulation of a Nanoconfined Water Film
 Molecular Dynamics Simulation of a Nanoconfined Water Film Kyle Lindquist, Shu-Han Chao May 7, 2013 1 Introduction The behavior of water confined in nano-scale environment is of interest in many applications.
Molecular Dynamics Simulation of a Nanoconfined Water Film Kyle Lindquist, Shu-Han Chao May 7, 2013 1 Introduction The behavior of water confined in nano-scale environment is of interest in many applications.
Structural Bioinformatics (C3210) Molecular Mechanics
 Structural Bioinformatics (C3210) Molecular Mechanics How to Calculate Energies Calculation of molecular energies is of key importance in protein folding, molecular modelling etc. There are two main computational
Structural Bioinformatics (C3210) Molecular Mechanics How to Calculate Energies Calculation of molecular energies is of key importance in protein folding, molecular modelling etc. There are two main computational
Everything starts with atomic structure and bonding
 Everything starts with atomic structure and bonding not all energy values can be possessed by electrons; e- have discrete energy values we call energy levels or states. The energy values are quantized
Everything starts with atomic structure and bonding not all energy values can be possessed by electrons; e- have discrete energy values we call energy levels or states. The energy values are quantized
Force Fields in Molecular Mechanics
 Force Fields in Molecular Mechanics Rajarshi Guha (9915607) and Rajesh Sardar (9915610) March 21, 2001 1 Introduction With the advent of computers chemists have realized the utility of carrying out simulations
Force Fields in Molecular Mechanics Rajarshi Guha (9915607) and Rajesh Sardar (9915610) March 21, 2001 1 Introduction With the advent of computers chemists have realized the utility of carrying out simulations
Introduction to model potential Molecular Dynamics A3hourcourseatICTP
 Introduction to model potential Molecular Dynamics A3hourcourseatICTP Alessandro Mattoni 1 1 Istituto Officina dei Materiali CNR-IOM Unità di Cagliari SLACS ICTP School on numerical methods for energy,
Introduction to model potential Molecular Dynamics A3hourcourseatICTP Alessandro Mattoni 1 1 Istituto Officina dei Materiali CNR-IOM Unità di Cagliari SLACS ICTP School on numerical methods for energy,
Molecular Dynamics 9/6/16
 Molecular Dynamics What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: Lesar Chpt 6, F&S Chpt 4 1 The Molecular Dynamics (MD) method for classical
Molecular Dynamics What to choose in an integrator The Verlet algorithm Boundary Conditions in Space and time Reading Assignment: Lesar Chpt 6, F&S Chpt 4 1 The Molecular Dynamics (MD) method for classical
Why study protein dynamics?
 Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Why study protein dynamics? Protein flexibility is crucial for function. One average structure is not enough. Proteins constantly sample configurational space. Transport - binding and moving molecules
Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions. Let's get together.
 Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions Let's get together. Most gases are NOT ideal except at very low pressures: Z=1 for ideal gases Intermolecular interactions come
Lecture C2 Microscopic to Macroscopic, Part 2: Intermolecular Interactions Let's get together. Most gases are NOT ideal except at very low pressures: Z=1 for ideal gases Intermolecular interactions come
Journal of Theoretical Physics
 1 Journal of Theoretical Physics Founded and Edited by M. Apostol 53 (2000) ISSN 1453-4428 Ionization potential for metallic clusters L. C. Cune and M. Apostol Department of Theoretical Physics, Institute
1 Journal of Theoretical Physics Founded and Edited by M. Apostol 53 (2000) ISSN 1453-4428 Ionization potential for metallic clusters L. C. Cune and M. Apostol Department of Theoretical Physics, Institute
Ab initio phonon calculations in mixed systems
 Ab initio phonon calculations in mixed systems Andrei Postnikov apostnik@uos.de Outline: Experiment vs. ab initio theory Ways of theory: linear response and frozen phonon approaches Applications: Be x
Ab initio phonon calculations in mixed systems Andrei Postnikov apostnik@uos.de Outline: Experiment vs. ab initio theory Ways of theory: linear response and frozen phonon approaches Applications: Be x
Atomic and molecular interaction forces in biology
 Atomic and molecular interaction forces in biology 1 Outline Types of interactions relevant to biology Van der Waals interactions H-bond interactions Some properties of water Hydrophobic effect 2 Types
Atomic and molecular interaction forces in biology 1 Outline Types of interactions relevant to biology Van der Waals interactions H-bond interactions Some properties of water Hydrophobic effect 2 Types
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015,
 Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
Biochemistry,530:,, Introduc5on,to,Structural,Biology, Autumn,Quarter,2015, Course,Informa5on, BIOC%530% GraduateAlevel,discussion,of,the,structure,,func5on,,and,chemistry,of,proteins,and, nucleic,acids,,control,of,enzyma5c,reac5ons.,please,see,the,course,syllabus,and,
INTERMEDIATE BONDING AND INTERMOLECULAR FORCES. Electronegativity
 INTERMEDIATE BNDING AND INTERMLECULAR FRCES Electronegativity is defined as follows Electronegativity Electronegativity is the ability of an atom within a covalent bond to attract the bonding pair of electrons.
INTERMEDIATE BNDING AND INTERMLECULAR FRCES Electronegativity is defined as follows Electronegativity Electronegativity is the ability of an atom within a covalent bond to attract the bonding pair of electrons.
An introduction to Molecular Dynamics. EMBO, June 2016
 An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
An introduction to Molecular Dynamics EMBO, June 2016 What is MD? everything that living things do can be understood in terms of the jiggling and wiggling of atoms. The Feynman Lectures in Physics vol.
PHYS 3313 Section 001 Lecture # 22
 PHYS 3313 Section 001 Lecture # 22 Dr. Barry Spurlock Simple Harmonic Oscillator Barriers and Tunneling Alpha Particle Decay Schrodinger Equation on Hydrogen Atom Solutions for Schrodinger Equation for
PHYS 3313 Section 001 Lecture # 22 Dr. Barry Spurlock Simple Harmonic Oscillator Barriers and Tunneling Alpha Particle Decay Schrodinger Equation on Hydrogen Atom Solutions for Schrodinger Equation for
Intermolecular Forces I
 I How does the arrangement of atoms differ in the 3 phases of matter (solid, liquid, gas)? Why doesn t ice just evaporate into a gas? Why does liquid water exist at all? There must be some force between
I How does the arrangement of atoms differ in the 3 phases of matter (solid, liquid, gas)? Why doesn t ice just evaporate into a gas? Why does liquid water exist at all? There must be some force between
CHAPTER 2. Atomic Structure And Bonding 2-1
 CHAPTER 2 Atomic Structure And Bonding 2-1 Structure of Atoms ATOM Basic Unit of an Element Diameter : 10 10 m. Neutrally Charged Nucleus Diameter : 10 14 m Accounts for almost all mass Positive Charge
CHAPTER 2 Atomic Structure And Bonding 2-1 Structure of Atoms ATOM Basic Unit of an Element Diameter : 10 10 m. Neutrally Charged Nucleus Diameter : 10 14 m Accounts for almost all mass Positive Charge
Metallic and Ionic Structures and Bonding
 Metallic and Ionic Structures and Bonding Ionic compounds are formed between elements having an electronegativity difference of about 2.0 or greater. Simple ionic compounds are characterized by high melting
Metallic and Ionic Structures and Bonding Ionic compounds are formed between elements having an electronegativity difference of about 2.0 or greater. Simple ionic compounds are characterized by high melting
