Bcl::ChemInfo - Qualitative Analysis Of Machine Learning Models For Activation Of HSD Involved In Alzheimer s Disease
|
|
|
- Arlene Long
- 6 years ago
- Views:
Transcription
1 Bcl::ChemInfo - Qualitative Analysis Of Machine Learning Models For Activation Of HSD Involved In Alzheimer s Disease Mariusz Butkiewicz, Edward W. Lowe, Jr., Jens Meiler Abstract In this case study, a ligand-based virtual high throughput screening suite, bcl::cheminfo, was applied to screen for activation of the protein target 17-beta hydroxysteroid dehydrogenase type 10 (HSD) involved in Alzheimer s Disease. bcl::cheminfo implements a diverse set of machine learning techniques such as artificial neural networks (ANN), support vector machines (SVM) with the extension for regression, kappa nearest neighbor (KNN), and decision trees (DT). Molecular structures were converted into a distinct collection of descriptor groups involving 2D- and 3Dautocorrelation, and radial distribution functions. A confirmatory high-throughput screening data set contained over 72,000 experimentally validated compounds, available through PubChem. Here, the systematical model development was achieved through optimization of feature sets and algorithmic parameters resulting in a theoretical enrichment of 11 (44% of maximal enrichment), and an area under the ROC curve (AUC) of 0.75 for the best performing machine learning technique on an independent data set. In addition, consensus combinations of all involved predictors were evaluated and achieved the best enrichment of 13 (50%), and AUC of All models were computed in silico and represent a viable option in guiding the drug discovery process through virtual library screening and compound prioritization a priori to synthesis and biological testing. The best consensus predictor will be made accessible for the academic community at Keywords Machine Learning, Quantitative Structure Activity Relation (QSAR), Artificial Neural Network (ANN), Support Vector Machine (SVM), Decision Trees (DT), Kohonen Network, kappa - Nearest Neighbor (KNN), Area under the curve (AUC), Receiver Operator Characteristics (ROC), Enrichment, highthroughput screening (HTS) Alzheimer s Disease puts a financial burden on society with over $150 billion annually, making it the 3rd most costly disease after heart disease and cancer [1]. In modern drug design, compounds with undesirable biological activity can be eliminated from the available chemical space while optimizing efficacy. The ability to predict active compounds related to cognitive disorders such as Alzheimer s Disease has the potential to reduce the medical cost involved. The protein target 17-beta hydroxysteroid dehydrogenase This work is supported by 1R21MH and 1R01MH to Jens Meiler. Edward W. Lowe, Jr. acknowledges NSF support through the CI-TraCS Fellowship (OCI ). Mariusz Butkiewicz is a graduate student in Chemistry at Vanderbilt University, Nashville TN, 37232; Edward W. Lowe, Jr., PhD, is a research assistant professor at the Center for Structural Biology at Vanderbilt University, Nashville TN, 37232; Jens Meiler, PhD, is Associate Professor of Chemistry, Pharmacology, and Biomedical Informatics, Institute for Chemical Biology Corresponding author: Jens Meiler, PhD, Center for Structural Biology, Vanderbilt University, Nashville, TN type 10 (HSD) has been found in elevated concentrations in the hippocampi of Alzheimer s disease patients. HSD may play a role in the degradation of neuroprotective agents. The inhibition of HSD has been indicated as a possible mean s of treating Alzheimer's disease. Dysfunctions in human 17 betahydroxysteroid dehydrogenases result in disorders of biology of reproduction and neuronal diseases, the enzymes are also involved in the pathogenesis of various cancers. HSD has a high affinity for amyloid proteins. Thus, it has been proposed that HSD may contribute to the amyloid plaques found in Alzheimer's patients [2]. Furthermore, HSD degrades neuroprotective agents like allopregnanolone which may lead to memory loss. Therefore, it has been postulated that inhibition of HSD may help lessen the symptoms associated with Alzheimer's. High-throughput screening (HTS) has become a key technology of pharmaceutical research [3], often more than one million compounds per biological target are screened [2]. At the same time, the number of compounds testable in a HTS experiment remains limited and costs increase linearly with size of the screen [4]. This challenge motivates the development of virtual screening methods which search large compound libraries in silico and identify novel chemical entities with a desired biological activity [2]. Machine learning techniques play a crucial role in modeling quantitative structure activity relationships (QSAR) by correlating chemical structure with its biological activity for a specific biological target [3, 5-7]. In recent years the potential of approaches such as Support Vector Machines (SVM) and Artificial Neural Networks (ANN) for establishing highly non-linear relations has become apparent. [18-22]. The algorithms learn to recognize complex patterns and make intelligent decisions based on an established compound library. Imposing such acquired sets of patterns obtained by a learning process, the algorithms are able to recognize not yet tested molecules and categorize them towards a given outcome. In this study, a cheminformatics software suite named bcl::cheminfo, incorporates several predictive models using supervised machine learning techniques including artificial neural networks [8], support vector machines with the extension for regression estimation (SVR) [9], decision trees [10], and unsupervised techniques such as kappa nearest neighbors (KNN) [11], and kohonen networks (Kohonen) [12].
2 I. MACHINE LEARNING TECHNIQUES A. Unsupervised Learning The kohonen network represents an unsupervised learning algorithm [12-14]. It is conceptually derived from artificial neural networks consisting of an input layer and a two-dimensional grid of neurons, the kohonen network. The second unsupervised learning method is the Kappa Nearest Neighbors [15-17]. This method uses a distance function to calculate pair-wise distances between query points and reference points, where query points are those to be classified. Figure 1: The schematic view of three unsupervised machine learning techniques is presented. A) A Kohonen network is represented by a input layer connected to a grid of nodes, each fully connected with its neighbors. B) The kappa nearestneighbors represents the predicted value of a query point as the weighted average of its kappa nearest reference points. B. Supervised Learning Artificial Neural Networks are successful attempting classification and regression problems in chemistry and biology. The structure of ANNs resembles the neuron structure of a human neural net. Layers of neurons are connected by weighted edges w ji. The input data x i are summed according to their weights, activation function applied, and output used as the input to neurons of the next layer (Figure 2). Support Vector Machine learning with extension for regression estimation [18, 19] represents a supervised machine learning approach successfully applied in the past [3, 7]. The core principles lay in linear functions defined in high-dimensional hyperspace [20], risk minimization according to Vapnik s - intensive loss function, and structural risk minimization [21] of a risk function consisting of the empirical error and the regularized term. The Decision Tree learning algorithm [10, 22] determines sets of rules to partition a given training data set. The outcome is a tree diagram or dendrogram (Figure 2) that describes how a given dataset can be classified by assessing a number of predictor variables and a dependent variable. Figure 2: Depictions of Decision Trees (A), Artificial Neural Networks (B), and Support Vector Machines (C) are presented. In A), the partitioning algorithm determines each predictor forecast, the value of the dependent variable. The dataset is then successively split into subsets (nodes) by the descriptor that produces the greater purity in the resulting subsets. In B), a three-layer feed forward network is shown using a sigmoid activation function in each neuron. In C), the prediction process of a support vector machine is shown for an unknown vector. II. TRAINING DATA The protein target 17-beta hydroxysteroid dehydrogenase type 10 (HSD) is part of the family of eleven 17-β hydroxysteroid dehydrogenases that oxidize or reduce steroids at the 17 position [23]. Thus, the biological activity of these steroids is modulated. HSD catalyzes the oxidation of the positive allosteric modulators of GABA, allopregnanolone and allotetrahydrodeoxycorticosterone (3, 5 -THDOC), to 5α-DHP, 5α-DHDOC, respectively [21]. It also inactivates 17β-estradiol [24]. When first identified, HSD was known as endoplasmic reticulum-associated amyloid binding protein (ERAB) [2]. HSD has since been identified as being the only member of the 17β-HSD family to be found in mitochondria [24]. HSD has been found in increased concentrations in the mitochondria of the hippocampi of Alzheimer's disease mice [24] and humans [25]. Several possible relationships between HSD and Alzheimer s disease have been proposed in the literature [23]. HSD has a high affinity for amyloid proteins. Therefore, it has been suggested that HSD may contribute to the amyloid plaques found in Alzheimer's patients [2]. 17βestradiol is a neuroprotective agent which prevents the degradation of existing neurons via its regulation of the β- amyloid precursor protein metabolism [24]. HSD has been
3 shown to degrade 17β-estradiol which may lead to neuronal degradation and the accumulation of β-amyloid, forming characteristic plaques [21]. It was also suggested that allopregnanolone reverses memory loss and dementia in the mouse model of Alzheimer's disease [26]. HSD is involved in the degradation of allopregnanolone which may lead to memory loss. Therefore, it has been postulated that inhibition of HSD may help lessen the symptoms associated with Alzheimer's. The data set for the protein target HSD used in this study was obtained through PubChem [27] (AID 886) and resulted in a final data set of 72,066 molecules. A confirmatory high throughput screen revealed 2,463 molecules which activate the enzyme, as experimentally determined by dose-response curves. Among the actives, 495 compounds with a concentration <= 1 µm were identified. All molecules in the data set were numerically encoded using a series of transformation-invariant descriptors which serve as unique fingerprints. The descriptors (Table I) were calculated using in-house code. III. IMPLEMENTATION / METHOD Our in-house developed C++ class library, the BioChemistryLibrary (BCL), was employed to implement all machine learning algorithms, and descriptor calculations used for this study. A third-party 3D conformation generator, CORINA [28], was used to calculate 3D coordinates for molecules prior to descriptor calculation. The here applied ligand-based virtual high throughput screening suite, bcl::cheminfo, is part of the BCL library. A. Dataset Generation During the training of the models, 10% of the data set was used for monitoring and 10% were used for independent testing of the trained models, leaving 80% for the training data set. The independent data set is put aside and not used during the training process. Each trained model is evaluated by a given quality measure based on this independent data set. B. Quality Measures The machine learning approaches are evaluated by means of a receiver operating characteristic (ROC) curve using cross-validated models. ROC curves plot the rate of true positives ( ), or sensitivity versus the rate of false positives ( ), or (1 - specificity). The diagonal represents performance of a random predictor and has an integral or area under the curve ( ) of 0.5. The QSAR model progressively improves as the -value increases. Often is normalized with the background fraction of active compounds through computation of the enrichment measure: ( ) ( ) ( ) (9) The value represents the factor by which the fraction of active compounds can be theoretically increased above the fraction observed in the original screen through in silico ranking. Another introduced measure is the root mean square deviation ( ). ( ) with and the predicted value. C. Feature Selection A total of 60 descriptor groups resulting in 1,284 descriptor values were generated using the BCL. These 60 categories consisted of scalar descriptors, as well as 2D- and 3D autocorrelation functions, radial distribution functions, and van der Waals surface area weighted variations of each of the non-scalar descriptors (see Table I). Sequential forward feature selection [29] was used for feature optimization for each machine learning technique individually. It describes a deterministic greedy search algorithm among all features. First, every single feature is selected and five-fold cross-validated models are trained followed by the evaluation of respective objective functions. The top performing feature is elected as the starting subset for the next round. Next, each remaining feature is added to the current subset in an iterative fashion resulting in N-1 feature sets. The best performing feature set is chosen again as the starting set for the next round. This process is repeated until all features are selected and the best descriptor combination is determined. Additionally, each feature set was trained with 5-fold cross-validation evaluated on an independent data set. The number of models generated during this process for each training method was ( ). Upon identification of the optimized feature set for each algorithm, any algorithmspecific parameters were optimized using the entire training data set and using 5-fold cross-validation. Every cross-validation model was evaluated by its quality measure on the independent data set. IV. RESULTS Various machine learning methods were evaluated as single predictors, highlighted in Table II. Given the independent data set, a perfect predictor would achieve a theoretical enrichment of 27.
4 TABLE I THE MOLECULAR DESCRIPTORS BY NAME AND DESCRIPTION Descriptor Description Name Scalar descriptors Weight Molecular weight of compound H-Bond donors Number of hydrogen bonding acceptors derived from the sum of nitrogen and oxygen atoms in the H-Bond acceptors molecule Number of hydrogen bonding donors derived from the sum of N- H and O-H groups in the molecule TPSA Topological polar surface area in [Å 2 ] of the molecule derived from polar 2D fragments Vector descriptors Identity weighted by atom identities 2D Autocorrelation (11 descriptors) 3D Autocorrelation (12 descriptors) Sigma Charge Pi Charge Total Charge Sigma Electronegativity Radial Distribution Pi Electronegativity Function (48 descriptors) Lone Pair Electronegativity Polarizibility weighted by σ atom charges weighted by π atom charges weighted by sum of σ and π charges weighted by σ atom electronegativities weighted by π atom electronegativities weighted by lone pair electronegativities weighted by effective atom polarizabilities ANNs were trained applying simple propagation as a weight update algorithm each iteration. The was evaluated as the objective function every step during the feature optimization process. ANNs as a single predictor achieved a theoretical enrichment of 10 (37% of possible maximal enrichment) on an independent dataset. An integral under the ROC curve of 0.83 was obtained. SVMs were trained using an initial cost parameter C of 1.0 and the kernel parameter γ of 0.5 during the feature optimization process. Upon identification of the optimal feature set, the cost and γ parameters were optimized to 2 and 32, respectively. As a single predictor, SVMs achieved a theoretical enrichment of ~12 (44%) and an AUC of 0.75 on an independent dataset. The KNN algorithm was used to predict the biological activity values of the training, monitoring, and independent data sets. The value of kappa, the number of neighbors to consider, was optimized with the full data set using the optimized feature set determined during the feature selection process. KNNs, as a single predictor, achieved a theoretical enrichment of ~11 (41%), AUC of 0.77 using an optimal kappa of 4. The Kohonen networks were trained with a network grid dimension of 10 x 10 nodes and a neighbor radius of 4 using the Gaussian neighbor kernel. The best result achieved by Kohonen networks was a theoretical enrichment of ~7 (28%) and an area under the ROC curve of TABLE II SINGLE AND CONSENSUS PREDICTOR RESULTS Method Enrichment (% max) AUC ANN/ Kohonen / KNN / SVM (50) 0.86 ANN/DT/Kohonen/KNN/SVM (50) 0.86 ANN / KNN / SVM (49) 0.86 ANN / DT / KNN / SVM (49) 0.86 ANN / DT / SVM (48) 0.85 ANN / SVM (48) 0.85 Kohonen / KNN / SVM (47) 0.81 DT / Kohonen / KNN / SVM (47) 0.81 ANN / Kohonen / KNN (47) 0.85 ANN / DT / Kohonen / KNN (47) 0.85 ANN / KNN (47) 0.85 ANN / DT / KNN (47) 0.85 DT / KNN / SVM (46) 0.77 KNN / SVM (46) 0.77 ANN / Kohonen / SVM (46) 0.85 ANN / DT / Kohonen / SVM (46) 0.85 DT / SVM (44) 0.75 SVM (44) 0.75 Kohonen / KNN (44) 0.78 DT / Kohonen / KNN (44) 0.78 Kohonen / SVM (43) 0.80 DT / Kohonen / SVM (43) 0.80 DT / KNN (41) 0.77 KNN (41) 0.77 ANN / DT (38) 0.83 ANN (37) 0.83 ANN / Kohonen (37) 0.83 ANN / DT / Kohonen (37) 0.83 Kohonen (28) 0.74 DT / Kohonen (27) 0.74 DT (18) 0.70 The assessed Decision Trees were cross-validated and evaluated resulting in a theoretical enrichment of ~5 (18%) and an AUC of To further evaluate the predictive models, ensemble predictors were also created by averaging the predictions using all possible combinations of models. The best consensus resulted in a of 0.86 and a theoretical enrichment of 13 (50%) achieved by the ensemble predictor model ANN/Kohonen/KNN/SVM (Table II) (Figure 3). Additionally, Table II lists all possible combinations introducing consensus predictors. All entries in Table II are sorted by Enrichment in descending order. V. DISCUSSION Among single predictors, SVM, KNN, and ANN achieved a comparable enrichment performance (37% to 44%). Kohonen networks (28%) and DTs (18%) underperformed
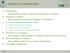
5 True Positive Rate ROC Curve Figure 3: A receiver operating characteristic (ROC) curve for the best consensus model ANN/Kohonen/KNN/SVM is shown with a theoretical enrichment of 13 (50% max enrichment) and an area under the curve (AUC) of The sub graph plots the same ROC curve on a logarithmic scale. in comparison. In contrast, consensus models clearly outperform single technique predictive models. The majority of ensemble models rank above the average (Table II). Among the best ensemble predictors, ANNs, SVMs, and KNN in combination yield the best enrichment result (50%). Adding each one of the mentioned techniques to an ensemble increased the predictive accuracy, respectively. This implies that each single technique supplies a distinct contribution to the predictive accuracy of the final model (ANN/Kohonen/KNN/SVM). The inclusion of decision trees into the final ensemble did not change the overall performance (see ANN/DT/Kohonen/KNN/SVM). Both predictors achieved the same enrichment (50%)., as a quality measure, discriminates distinguished predictive models by precision rather than accuracy of the predictions. Sorting Table II by indicates that KNNs contribute among all top ranking predictors. VI False Positive Rate CONCLUSIONS In this research, we present a case study application of the ligand-based virtual high throughput screening suite, bcl::cheminfo, on the target protein 17-beta hydroxysteroid dehydrogenase type 10 (HSD). HSD is involved in Alzheimer s Disease which puts a tremendous financial burden on today s society. QSARs models were developed to identify biologically active compounds for the activation of HSD. We have shown that the best consensus predictor achieved 50% of the maximal enrichment on an independent dataset. The best consensus predictor will be made accessible for the academic community at ACKNOWLEDGEMENTS The authors thank the Advanced Computing Center for Research & Education (ACCRE) at Vanderbilt University for hardware support. REFERENCES [1] D. L. Leslie and S. K. Inouye, "The Importance of Delirium: Economic and Societal Costs," Journal of the American Geriatrics Society, vol. 59, pp. S241-S243, [2] S. D. Yan, J. Fu, C. Soto, X. Chen, H. Zhu, F. Al-Mohanna, K. Collison, A. Zhu, E. Stern, T. Saido, M. Tohyama, S. Ogawa, A. Roher, and D. Stern, "An intracellular protein that binds amyloid-beta peptide and mediates neurotoxicity in Alzheimer's disease," Nature, vol. 389, pp , Oct [3] R. Mueller, A. L. Rodriguez, E. S. Dawson, M. Butkiewicz, T. T. Nguyen, S. Oleszkiewicz, A. Bleckmann, C. D. Weaver, C. W. Lindsley, P. J. Conn, and J. Meiler, "Identification of Metabotropic Glutamate Receptor Subtype 5 Potentiators Using Virtual High-Throughput Screening," ACS Chemical Neuroscience. [4] R. D. Cramer, 3rd, D. E. Patterson, and J. D. Bunce, "Recent advances in comparative molecular field analysis (CoMFA)," Prog Clin Biol Res, vol. 291, pp , [5] V. V. Zernov, K. V. Balakin, A. A. Ivaschenko, N. P. Savchuk, and I. V. Pletnev, "Drug discovery using support vector machines. The case studies of drug-likeness, agrochemicallikeness, and enzyme inhibition predictions," J Chem Inf Comput Sci, vol. 43, pp , Nov-Dec [6] A. Bleckmann and J. Meiler, "Epothilones: Quantitative Structure Activity Relations Studied by Support Vector Machines and Artificial Neural Networks," QSAR Comb. Sci., vol. 22, pp , [7] M. Butkiewicz, R. Mueller, D. Selic, E. Dawson, and J. Meiler, "Application of Machine Learning Approaches on Quantitative Structure Activity Relationships," presented at the IEEE Symposium on Computational Intelligence in Bioinformatics and Computational Biology, Nashville, [8] D. Winkler, "Neural networks as robust tools in drug lead discovery and development," Molecular Biotechnology, vol. 27, pp , [9] B. Schoelkopf, "SVM and Kernel Methods," www, [10] J. R. Quinlan, "Induction of Decision Trees," Machine Learning, vol. 1, pp , [11] M. Shen, Y. Xiao, A. Golbraikh, V. K. Gombar, and A. Tropsha, "Development and Validation of k-nearest-neighbor QSPR Models of Metabolic Stability of Drug Candidates," Journal of Medicinal Chemistry, vol. 46, pp , [12] T. Kohonen, "Self-organized formation of topologically correct feature maps," Biological cybernetics, vol. 43, pp , [13] T. Kohonen, "The self-organizing map," Proceedings of the IEEE, vol. 78, pp , [14] T. Kohonen, "Self-organization and associative memory," Self- Organization and Associative Memory, 100 figs. XV, 312 pages.. Springer-Verlag Berlin Heidelberg New York. Also Springer Series in Information Sciences, volume 8, vol. 1, [15] M. Shen, Y. Xiao, A. Golbraikh, V. K. Gombar, and A. Tropsha, "Development and validation of k-nearest-neighbor QSPR models of metabolic stability of drug candidates," J. Med. Chem, vol. 46, pp , [16] F. Nigsch, A. Bender, B. van Buuren, J. Tissen, E. Nigsch, and J. B. O. Mitchell, "Melting point prediction employing k-nearest neighbor algorithms and genetic parameter optimization," J. Chem. Inf. Model, vol. 46, pp , [17] S. Ajmani, K. Jadhav, and S. A. Kulkarni, "Three-dimensional QSAR using the k-nearest neighbor method and its interpretation," J. Chem. Inf. Model, vol. 46, pp , 2006.
6 [18] L. M. Berezhkovskiy, "Determination of volume of distribution at steady state with complete consideration of the kinetics of protein and tissue binding in linear pharmacokinetics," Journal of pharmaceutical sciences, vol. 93, pp , Feb [19] J. C. Kalvass, D. A. Tess, C. Giragossian, M. C. Linhares, and T. S. Maurer, "Influence of microsomal concentration on apparent intrinsic clearance: implications for scaling in vitro data," Drug metabolism and disposition: the biological fate of chemicals, vol. 29, pp , Oct [20] B. Schoelkopf and A. J. Smola, Learning with Kernels. Cambridge, Massachusetts: The MIT Press, [21] X. Y. He, J. Wegiel, Y. Z. Yang, R. Pullarkat, H. Schulz, and S. Y. Yang, "Type 10 17beta-hydroxysteroid dehydrogenase catalyzing the oxidation of steroid modulators of gammaaminobutyric acid type A receptors," Mol Cell Endocrinol, vol. 229, pp , Jan [22] M. R. H. M. Maruf Hossain, James Bailey, "ROC-tree: A Novel Decision Tree Induction Algorithm Based on Receiver Operating Characteristics to Classify Gene Expression Data," [23] J. Adamski and F. J. Jakob, "A guide to 17beta-hydroxysteroid dehydrogenases," Mol Cell Endocrinol, vol. 171, pp. 1-4, Jan [24] X. Y. He, G. Y. Wen, G. Merz, D. Lin, Y. Z. Yang, P. Mehta, H. Schulz, and S. Y. Yang, "Abundant type betahydroxysteroid dehydrogenase in the hippocampus of mouse Alzheimer's disease model," Brain Res Mol Brain Res, vol. 99, pp , Feb [25] P. Hovorkova, Z. Kristofikova, A. Horinek, D. Ripova, E. Majer, P. Zach, P. Sellinger, and J. Ricny, "Lateralization of 17beta-hydroxysteroid dehydrogenase type 10 in hippocampi of demented and psychotic people," Dement Geriatr Cogn Disord, vol. 26, pp , [26] J. M. Wang, C. Singh, L. Liu, R. W. Irwin, S. Chen, E. J. Chung, R. F. Thompson, and R. D. Brinton, "Allopregnanolone reverses neurogenic and cognitive deficits in mouse model of Alzheimer's disease," Proc Natl Acad Sci U S A, vol. 107, pp , Apr 6. [27] "PubChem ed. [28] J. Gasteiger, C. Rudolph, and J. Sadowski, "Automatic Generation of 3D-Atomic Coordinates for Organic Molecules," Tetrahedron Comput. Method., vol. 3, pp , [29] K. Z. Mao, "Orthogonal forward selection and backward elimination algorithms for feature subset selection," IEEE Trans Syst Man Cybern B Cybern, vol. 34, pp , Feb 2004.
Comparative Analysis of Machine Learning Techniques for the Prediction of LogP
 Comparative Analysis of Machine Learning Techniques for the Prediction of LogP Edward W. Lowe, Jr., Mariusz Butkiewicz, Matthew Spellings, Albert Omlor, Jens Meiler Abstract Several machine learning techniques
Comparative Analysis of Machine Learning Techniques for the Prediction of LogP Edward W. Lowe, Jr., Mariusz Butkiewicz, Matthew Spellings, Albert Omlor, Jens Meiler Abstract Several machine learning techniques
bcl::cheminfo Suite Enables Machine Learning-Based Drug Discovery Using GPUs Edward W. Lowe, Jr. Nils Woetzel May 17, 2012
 bcl::cheminfo Suite Enables Machine Learning-Based Drug Discovery Using GPUs Edward W. Lowe, Jr. Nils Woetzel May 17, 2012 Outline Machine Learning Cheminformatics Framework QSPR logp QSAR mglur 5 CYP
bcl::cheminfo Suite Enables Machine Learning-Based Drug Discovery Using GPUs Edward W. Lowe, Jr. Nils Woetzel May 17, 2012 Outline Machine Learning Cheminformatics Framework QSPR logp QSAR mglur 5 CYP
Comparative Analysis of Machine Learning Techniques for the Prediction of the DMPK Parameters Intrinsic Clearance and Plasma Protein Binding
 Comparative Analysis of Machine Learning Techniques for the Prediction of the DMPK Parameters Intrinsic Clearance and Plasma Protein Binding Edward W. Lowe, Jr., Mariusz Butkiewicz, Zollie White III, Matthew
Comparative Analysis of Machine Learning Techniques for the Prediction of the DMPK Parameters Intrinsic Clearance and Plasma Protein Binding Edward W. Lowe, Jr., Mariusz Butkiewicz, Zollie White III, Matthew
GPU-Accelerated Machine Learning Techniques Enable QSAR Modeling Of Large HTS Data
 GPU-Accelerated Machine Learning Techniques Enable QSAR Modeling Of Large HTS Data Edward W. Lowe, Jr., Mariusz Butkiewicz, Nils Woetzel, Jens Meiler Abstract Quantitative structure activity relationship
GPU-Accelerated Machine Learning Techniques Enable QSAR Modeling Of Large HTS Data Edward W. Lowe, Jr., Mariusz Butkiewicz, Nils Woetzel, Jens Meiler Abstract Quantitative structure activity relationship
In silico pharmacology for drug discovery
 In silico pharmacology for drug discovery In silico drug design In silico methods can contribute to drug targets identification through application of bionformatics tools. Currently, the application of
In silico pharmacology for drug discovery In silico drug design In silico methods can contribute to drug targets identification through application of bionformatics tools. Currently, the application of
Structure-Activity Modeling - QSAR. Uwe Koch
 Structure-Activity Modeling - QSAR Uwe Koch QSAR Assumption: QSAR attempts to quantify the relationship between activity and molecular strcucture by correlating descriptors with properties Biological activity
Structure-Activity Modeling - QSAR Uwe Koch QSAR Assumption: QSAR attempts to quantify the relationship between activity and molecular strcucture by correlating descriptors with properties Biological activity
Introduction to Chemoinformatics and Drug Discovery
 Introduction to Chemoinformatics and Drug Discovery Irene Kouskoumvekaki Associate Professor February 15 th, 2013 The Chemical Space There are atoms and space. Everything else is opinion. Democritus (ca.
Introduction to Chemoinformatics and Drug Discovery Irene Kouskoumvekaki Associate Professor February 15 th, 2013 The Chemical Space There are atoms and space. Everything else is opinion. Democritus (ca.
Machine Learning Concepts in Chemoinformatics
 Machine Learning Concepts in Chemoinformatics Martin Vogt B-IT Life Science Informatics Rheinische Friedrich-Wilhelms-Universität Bonn BigChem Winter School 2017 25. October Data Mining in Chemoinformatics
Machine Learning Concepts in Chemoinformatics Martin Vogt B-IT Life Science Informatics Rheinische Friedrich-Wilhelms-Universität Bonn BigChem Winter School 2017 25. October Data Mining in Chemoinformatics
Plan. Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics.
 Plan Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics. Exercise: Example and exercise with herg potassium channel: Use of
Plan Lecture: What is Chemoinformatics and Drug Design? Description of Support Vector Machine (SVM) and its used in Chemoinformatics. Exercise: Example and exercise with herg potassium channel: Use of
QSAR Modeling of Human Liver Microsomal Stability Alexey Zakharov
 QSAR Modeling of Human Liver Microsomal Stability Alexey Zakharov CADD Group Chemical Biology Laboratory Frederick National Laboratory for Cancer Research National Cancer Institute, National Institutes
QSAR Modeling of Human Liver Microsomal Stability Alexey Zakharov CADD Group Chemical Biology Laboratory Frederick National Laboratory for Cancer Research National Cancer Institute, National Institutes
Virtual affinity fingerprints in drug discovery: The Drug Profile Matching method
 Ágnes Peragovics Virtual affinity fingerprints in drug discovery: The Drug Profile Matching method PhD Theses Supervisor: András Málnási-Csizmadia DSc. Associate Professor Structural Biochemistry Doctoral
Ágnes Peragovics Virtual affinity fingerprints in drug discovery: The Drug Profile Matching method PhD Theses Supervisor: András Málnási-Csizmadia DSc. Associate Professor Structural Biochemistry Doctoral
HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH
 HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH Hoang Trang 1, Tran Hoang Loc 1 1 Ho Chi Minh City University of Technology-VNU HCM, Ho Chi
HYPERGRAPH BASED SEMI-SUPERVISED LEARNING ALGORITHMS APPLIED TO SPEECH RECOGNITION PROBLEM: A NOVEL APPROACH Hoang Trang 1, Tran Hoang Loc 1 1 Ho Chi Minh City University of Technology-VNU HCM, Ho Chi
Structural biology and drug design: An overview
 Structural biology and drug design: An overview livier Taboureau Assitant professor Chemoinformatics group-cbs-dtu otab@cbs.dtu.dk Drug discovery Drug and drug design A drug is a key molecule involved
Structural biology and drug design: An overview livier Taboureau Assitant professor Chemoinformatics group-cbs-dtu otab@cbs.dtu.dk Drug discovery Drug and drug design A drug is a key molecule involved
Bioengineering & Bioinformatics Summer Institute, Dept. Computational Biology, University of Pittsburgh, PGH, PA
 Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Pharmacophore Model Development for the Identification of Novel Acetylcholinesterase Inhibitors Edwin Kamau Dept Chem & Biochem Kennesa State Uni ersit Kennesa GA 30144 Dept. Chem. & Biochem. Kennesaw
Early Stages of Drug Discovery in the Pharmaceutical Industry
 Early Stages of Drug Discovery in the Pharmaceutical Industry Daniel Seeliger / Jan Kriegl, Discovery Research, Boehringer Ingelheim September 29, 2016 Historical Drug Discovery From Accidential Discovery
Early Stages of Drug Discovery in the Pharmaceutical Industry Daniel Seeliger / Jan Kriegl, Discovery Research, Boehringer Ingelheim September 29, 2016 Historical Drug Discovery From Accidential Discovery
QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression
 APPLICATION NOTE QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression GAINING EFFICIENCY IN QUANTITATIVE STRUCTURE ACTIVITY RELATIONSHIPS ErbB1 kinase is the cell-surface receptor
APPLICATION NOTE QSAR Modeling of ErbB1 Inhibitors Using Genetic Algorithm-Based Regression GAINING EFFICIENCY IN QUANTITATIVE STRUCTURE ACTIVITY RELATIONSHIPS ErbB1 kinase is the cell-surface receptor
Next Generation Computational Chemistry Tools to Predict Toxicity of CWAs
 Next Generation Computational Chemistry Tools to Predict Toxicity of CWAs William (Bill) Welsh welshwj@umdnj.edu Prospective Funding by DTRA/JSTO-CBD CBIS Conference 1 A State-wide, Regional and National
Next Generation Computational Chemistry Tools to Predict Toxicity of CWAs William (Bill) Welsh welshwj@umdnj.edu Prospective Funding by DTRA/JSTO-CBD CBIS Conference 1 A State-wide, Regional and National
Dr. Sander B. Nabuurs. Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre
 Dr. Sander B. Nabuurs Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre The road to new drugs. How to find new hits? High Throughput
Dr. Sander B. Nabuurs Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre The road to new drugs. How to find new hits? High Throughput
Machine learning for ligand-based virtual screening and chemogenomics!
 Machine learning for ligand-based virtual screening and chemogenomics! Jean-Philippe Vert Institut Curie - INSERM U900 - Mines ParisTech In silico discovery of molecular probes and drug-like compounds:
Machine learning for ligand-based virtual screening and chemogenomics! Jean-Philippe Vert Institut Curie - INSERM U900 - Mines ParisTech In silico discovery of molecular probes and drug-like compounds:
Xia Ning,*, Huzefa Rangwala, and George Karypis
 J. Chem. Inf. Model. XXXX, xxx, 000 A Multi-Assay-Based Structure-Activity Relationship Models: Improving Structure-Activity Relationship Models by Incorporating Activity Information from Related Targets
J. Chem. Inf. Model. XXXX, xxx, 000 A Multi-Assay-Based Structure-Activity Relationship Models: Improving Structure-Activity Relationship Models by Incorporating Activity Information from Related Targets
Quantitative structure activity relationship and drug design: A Review
 International Journal of Research in Biosciences Vol. 5 Issue 4, pp. (1-5), October 2016 Available online at http://www.ijrbs.in ISSN 2319-2844 Research Paper Quantitative structure activity relationship
International Journal of Research in Biosciences Vol. 5 Issue 4, pp. (1-5), October 2016 Available online at http://www.ijrbs.in ISSN 2319-2844 Research Paper Quantitative structure activity relationship
QSAR/QSPR modeling. Quantitative Structure-Activity Relationships Quantitative Structure-Property-Relationships
 Quantitative Structure-Activity Relationships Quantitative Structure-Property-Relationships QSAR/QSPR modeling Alexandre Varnek Faculté de Chimie, ULP, Strasbourg, FRANCE QSAR/QSPR models Development Validation
Quantitative Structure-Activity Relationships Quantitative Structure-Property-Relationships QSAR/QSPR modeling Alexandre Varnek Faculté de Chimie, ULP, Strasbourg, FRANCE QSAR/QSPR models Development Validation
Retrieving hits through in silico screening and expert assessment M. N. Drwal a,b and R. Griffith a
 Retrieving hits through in silico screening and expert assessment M.. Drwal a,b and R. Griffith a a: School of Medical Sciences/Pharmacology, USW, Sydney, Australia b: Charité Berlin, Germany Abstract:
Retrieving hits through in silico screening and expert assessment M.. Drwal a,b and R. Griffith a a: School of Medical Sciences/Pharmacology, USW, Sydney, Australia b: Charité Berlin, Germany Abstract:
ARTIFICIAL NEURAL NETWORKS گروه مطالعاتي 17 بهار 92
 ARTIFICIAL NEURAL NETWORKS گروه مطالعاتي 17 بهار 92 BIOLOGICAL INSPIRATIONS Some numbers The human brain contains about 10 billion nerve cells (neurons) Each neuron is connected to the others through 10000
ARTIFICIAL NEURAL NETWORKS گروه مطالعاتي 17 بهار 92 BIOLOGICAL INSPIRATIONS Some numbers The human brain contains about 10 billion nerve cells (neurons) Each neuron is connected to the others through 10000
Statistical concepts in QSAR.
 Statistical concepts in QSAR. Computational chemistry represents molecular structures as a numerical models and simulates their behavior with the equations of quantum and classical physics. Available programs
Statistical concepts in QSAR. Computational chemistry represents molecular structures as a numerical models and simulates their behavior with the equations of quantum and classical physics. Available programs
Predicting Protein Functions and Domain Interactions from Protein Interactions
 Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Chemoinformatics and information management. Peter Willett, University of Sheffield, UK
 Chemoinformatics and information management Peter Willett, University of Sheffield, UK verview What is chemoinformatics and why is it necessary Managing structural information Typical facilities in chemoinformatics
Chemoinformatics and information management Peter Willett, University of Sheffield, UK verview What is chemoinformatics and why is it necessary Managing structural information Typical facilities in chemoinformatics
Ignasi Belda, PhD CEO. HPC Advisory Council Spain Conference 2015
 Ignasi Belda, PhD CEO HPC Advisory Council Spain Conference 2015 Business lines Molecular Modeling Services We carry out computational chemistry projects using our selfdeveloped and third party technologies
Ignasi Belda, PhD CEO HPC Advisory Council Spain Conference 2015 Business lines Molecular Modeling Services We carry out computational chemistry projects using our selfdeveloped and third party technologies
Virtual Libraries and Virtual Screening in Drug Discovery Processes using KNIME
 Virtual Libraries and Virtual Screening in Drug Discovery Processes using KNIME Iván Solt Solutions for Cheminformatics Drug Discovery Strategies for known targets High-Throughput Screening (HTS) Cells
Virtual Libraries and Virtual Screening in Drug Discovery Processes using KNIME Iván Solt Solutions for Cheminformatics Drug Discovery Strategies for known targets High-Throughput Screening (HTS) Cells
Receptor Based Drug Design (1)
 Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Data Mining. Preamble: Control Application. Industrial Researcher s Approach. Practitioner s Approach. Example. Example. Goal: Maintain T ~Td
 Data Mining Andrew Kusiak 2139 Seamans Center Iowa City, Iowa 52242-1527 Preamble: Control Application Goal: Maintain T ~Td Tel: 319-335 5934 Fax: 319-335 5669 andrew-kusiak@uiowa.edu http://www.icaen.uiowa.edu/~ankusiak
Data Mining Andrew Kusiak 2139 Seamans Center Iowa City, Iowa 52242-1527 Preamble: Control Application Goal: Maintain T ~Td Tel: 319-335 5934 Fax: 319-335 5669 andrew-kusiak@uiowa.edu http://www.icaen.uiowa.edu/~ankusiak
Discriminative Direction for Kernel Classifiers
 Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Discriminative Direction for Kernel Classifiers Polina Golland Artificial Intelligence Lab Massachusetts Institute of Technology Cambridge, MA 02139 polina@ai.mit.edu Abstract In many scientific and engineering
Principles of Drug Design
 Advanced Medicinal Chemistry II Principles of Drug Design Tentative Course Outline Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu I. Introduction
Advanced Medicinal Chemistry II Principles of Drug Design Tentative Course Outline Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu I. Introduction
Generalization to a zero-data task: an empirical study
 Generalization to a zero-data task: an empirical study Université de Montréal 20/03/2007 Introduction Introduction and motivation What is a zero-data task? task for which no training data are available
Generalization to a zero-data task: an empirical study Université de Montréal 20/03/2007 Introduction Introduction and motivation What is a zero-data task? task for which no training data are available
hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference
 CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
CS 229 Project Report (TR# MSB2010) Submitted 12/10/2010 hsnim: Hyper Scalable Network Inference Machine for Scale-Free Protein-Protein Interaction Networks Inference Muhammad Shoaib Sehgal Computer Science
De Novo molecular design with Deep Reinforcement Learning
 De Novo molecular design with Deep Reinforcement Learning @olexandr Olexandr Isayev, Ph.D. University of North Carolina at Chapel Hill olexandr@unc.edu http://olexandrisayev.com About me Ph.D. in Chemistry
De Novo molecular design with Deep Reinforcement Learning @olexandr Olexandr Isayev, Ph.D. University of North Carolina at Chapel Hill olexandr@unc.edu http://olexandrisayev.com About me Ph.D. in Chemistry
Applications of multi-class machine
 Applications of multi-class machine learning models to drug design Marvin Waldman, Michael Lawless, Pankaj R. Daga, Robert D. Clark Simulations Plus, Inc. Lancaster CA, USA Overview Applications of multi-class
Applications of multi-class machine learning models to drug design Marvin Waldman, Michael Lawless, Pankaj R. Daga, Robert D. Clark Simulations Plus, Inc. Lancaster CA, USA Overview Applications of multi-class
Advanced Medicinal Chemistry SLIDES B
 Advanced Medicinal Chemistry Filippo Minutolo CFU 3 (21 hours) SLIDES B Drug likeness - ADME two contradictory physico-chemical parameters to balance: 1) aqueous solubility 2) lipid membrane permeability
Advanced Medicinal Chemistry Filippo Minutolo CFU 3 (21 hours) SLIDES B Drug likeness - ADME two contradictory physico-chemical parameters to balance: 1) aqueous solubility 2) lipid membrane permeability
Holdout and Cross-Validation Methods Overfitting Avoidance
 Holdout and Cross-Validation Methods Overfitting Avoidance Decision Trees Reduce error pruning Cost-complexity pruning Neural Networks Early stopping Adjusting Regularizers via Cross-Validation Nearest
Holdout and Cross-Validation Methods Overfitting Avoidance Decision Trees Reduce error pruning Cost-complexity pruning Neural Networks Early stopping Adjusting Regularizers via Cross-Validation Nearest
Applied Machine Learning Annalisa Marsico
 Applied Machine Learning Annalisa Marsico OWL RNA Bionformatics group Max Planck Institute for Molecular Genetics Free University of Berlin 22 April, SoSe 2015 Goals Feature Selection rather than Feature
Applied Machine Learning Annalisa Marsico OWL RNA Bionformatics group Max Planck Institute for Molecular Genetics Free University of Berlin 22 April, SoSe 2015 Goals Feature Selection rather than Feature
Drug Design 2. Oliver Kohlbacher. Winter 2009/ QSAR Part 4: Selected Chapters
 Drug Design 2 Oliver Kohlbacher Winter 2009/2010 11. QSAR Part 4: Selected Chapters Abt. Simulation biologischer Systeme WSI/ZBIT, Eberhard-Karls-Universität Tübingen Overview GRIND GRid-INDependent Descriptors
Drug Design 2 Oliver Kohlbacher Winter 2009/2010 11. QSAR Part 4: Selected Chapters Abt. Simulation biologischer Systeme WSI/ZBIT, Eberhard-Karls-Universität Tübingen Overview GRIND GRid-INDependent Descriptors
Introduction to Support Vector Machines
 Introduction to Support Vector Machines Andreas Maletti Technische Universität Dresden Fakultät Informatik June 15, 2006 1 The Problem 2 The Basics 3 The Proposed Solution Learning by Machines Learning
Introduction to Support Vector Machines Andreas Maletti Technische Universität Dresden Fakultät Informatik June 15, 2006 1 The Problem 2 The Basics 3 The Proposed Solution Learning by Machines Learning
Classification Ensemble That Maximizes the Area Under Receiver Operating Characteristic Curve (AUC)
 Classification Ensemble That Maximizes the Area Under Receiver Operating Characteristic Curve (AUC) Eunsik Park 1 and Y-c Ivan Chang 2 1 Chonnam National University, Gwangju, Korea 2 Academia Sinica, Taipei,
Classification Ensemble That Maximizes the Area Under Receiver Operating Characteristic Curve (AUC) Eunsik Park 1 and Y-c Ivan Chang 2 1 Chonnam National University, Gwangju, Korea 2 Academia Sinica, Taipei,
Prediction and Classif ication of Human G-protein Coupled Receptors Based on Support Vector Machines
 Article Prediction and Classif ication of Human G-protein Coupled Receptors Based on Support Vector Machines Yun-Fei Wang, Huan Chen, and Yan-Hong Zhou* Hubei Bioinformatics and Molecular Imaging Key Laboratory,
Article Prediction and Classif ication of Human G-protein Coupled Receptors Based on Support Vector Machines Yun-Fei Wang, Huan Chen, and Yan-Hong Zhou* Hubei Bioinformatics and Molecular Imaging Key Laboratory,
Quantitative Structure-Activity Relationship (QSAR) computational-drug-design.html
 Quantitative Structure-Activity Relationship (QSAR) http://www.biophys.mpg.de/en/theoretical-biophysics/ computational-drug-design.html 07.11.2017 Ahmad Reza Mehdipour 07.11.2017 Course Outline 1. 1.Ligand-
Quantitative Structure-Activity Relationship (QSAR) http://www.biophys.mpg.de/en/theoretical-biophysics/ computational-drug-design.html 07.11.2017 Ahmad Reza Mehdipour 07.11.2017 Course Outline 1. 1.Ligand-
Similarity Search. Uwe Koch
 Similarity Search Uwe Koch Similarity Search The similar property principle: strurally similar molecules tend to have similar properties. However, structure property discontinuities occur frequently. Relevance
Similarity Search Uwe Koch Similarity Search The similar property principle: strurally similar molecules tend to have similar properties. However, structure property discontinuities occur frequently. Relevance
Data Mining in the Chemical Industry. Overview of presentation
 Data Mining in the Chemical Industry Glenn J. Myatt, Ph.D. Partner, Myatt & Johnson, Inc. glenn.myatt@gmail.com verview of presentation verview of the chemical industry Example of the pharmaceutical industry
Data Mining in the Chemical Industry Glenn J. Myatt, Ph.D. Partner, Myatt & Johnson, Inc. glenn.myatt@gmail.com verview of presentation verview of the chemical industry Example of the pharmaceutical industry
#33 - Genomics 11/09/07
 BCB 444/544 Required Reading (before lecture) Lecture 33 Mon Nov 5 - Lecture 31 Phylogenetics Parsimony and ML Chp 11 - pp 142 169 Genomics Wed Nov 7 - Lecture 32 Machine Learning Fri Nov 9 - Lecture 33
BCB 444/544 Required Reading (before lecture) Lecture 33 Mon Nov 5 - Lecture 31 Phylogenetics Parsimony and ML Chp 11 - pp 142 169 Genomics Wed Nov 7 - Lecture 32 Machine Learning Fri Nov 9 - Lecture 33
Introduction. OntoChem
 Introduction ntochem Providing drug discovery knowledge & small molecules... Supporting the task of medicinal chemistry Allows selecting best possible small molecule starting point From target to leads
Introduction ntochem Providing drug discovery knowledge & small molecules... Supporting the task of medicinal chemistry Allows selecting best possible small molecule starting point From target to leads
CH MEDICINAL CHEMISTRY
 CH 458 - MEDICINAL CHEMISTRY SPRING 2011 M: 5:15pm-8 pm Sci-1-089 Prerequisite: Organic Chemistry II (Chem 254 or Chem 252, or equivalent transfer course) Instructor: Dr. Bela Torok Room S-1-132, Science
CH 458 - MEDICINAL CHEMISTRY SPRING 2011 M: 5:15pm-8 pm Sci-1-089 Prerequisite: Organic Chemistry II (Chem 254 or Chem 252, or equivalent transfer course) Instructor: Dr. Bela Torok Room S-1-132, Science
The Role of Network Science in Biology and Medicine. Tiffany J. Callahan Computational Bioscience Program Hunter/Kahn Labs
 The Role of Network Science in Biology and Medicine Tiffany J. Callahan Computational Bioscience Program Hunter/Kahn Labs Network Analysis Working Group 09.28.2017 Network-Enabled Wisdom (NEW) empirically
The Role of Network Science in Biology and Medicine Tiffany J. Callahan Computational Bioscience Program Hunter/Kahn Labs Network Analysis Working Group 09.28.2017 Network-Enabled Wisdom (NEW) empirically
October 6 University Faculty of pharmacy Computer Aided Drug Design Unit
 October 6 University Faculty of pharmacy Computer Aided Drug Design Unit CADD@O6U.edu.eg CADD Computer-Aided Drug Design Unit The development of new drugs is no longer a process of trial and error or strokes
October 6 University Faculty of pharmacy Computer Aided Drug Design Unit CADD@O6U.edu.eg CADD Computer-Aided Drug Design Unit The development of new drugs is no longer a process of trial and error or strokes
COMPARISON OF SIMILARITY METHOD TO IMPROVE RETRIEVAL PERFORMANCE FOR CHEMICAL DATA
 http://www.ftsm.ukm.my/apjitm Asia-Pacific Journal of Information Technology and Multimedia Jurnal Teknologi Maklumat dan Multimedia Asia-Pasifik Vol. 7 No. 1, June 2018: 91-98 e-issn: 2289-2192 COMPARISON
http://www.ftsm.ukm.my/apjitm Asia-Pacific Journal of Information Technology and Multimedia Jurnal Teknologi Maklumat dan Multimedia Asia-Pasifik Vol. 7 No. 1, June 2018: 91-98 e-issn: 2289-2192 COMPARISON
Introduction to Chemoinformatics
 Introduction to Chemoinformatics Dr. Igor V. Tetko Helmholtz Zentrum München - German Research Center for Environmental Health (GmbH) Institute of Bioinformatics & Systems Biology (HMGU) Kyiv, 10 August
Introduction to Chemoinformatics Dr. Igor V. Tetko Helmholtz Zentrum München - German Research Center for Environmental Health (GmbH) Institute of Bioinformatics & Systems Biology (HMGU) Kyiv, 10 August
Topology based deep learning for biomolecular data
 Topology based deep learning for biomolecular data Guowei Wei Departments of Mathematics Michigan State University http://www.math.msu.edu/~wei American Institute of Mathematics July 23-28, 2017 Grant
Topology based deep learning for biomolecular data Guowei Wei Departments of Mathematics Michigan State University http://www.math.msu.edu/~wei American Institute of Mathematics July 23-28, 2017 Grant
Microarray Data Analysis: Discovery
 Microarray Data Analysis: Discovery Lecture 5 Classification Classification vs. Clustering Classification: Goal: Placing objects (e.g. genes) into meaningful classes Supervised Clustering: Goal: Discover
Microarray Data Analysis: Discovery Lecture 5 Classification Classification vs. Clustering Classification: Goal: Placing objects (e.g. genes) into meaningful classes Supervised Clustering: Goal: Discover
Drug Informatics for Chemical Genomics...
 Drug Informatics for Chemical Genomics... An Overview First Annual ChemGen IGERT Retreat Sept 2005 Drug Informatics for Chemical Genomics... p. Topics ChemGen Informatics The ChemMine Project Library Comparison
Drug Informatics for Chemical Genomics... An Overview First Annual ChemGen IGERT Retreat Sept 2005 Drug Informatics for Chemical Genomics... p. Topics ChemGen Informatics The ChemMine Project Library Comparison
Classification Based on Logical Concept Analysis
 Classification Based on Logical Concept Analysis Yan Zhao and Yiyu Yao Department of Computer Science, University of Regina, Regina, Saskatchewan, Canada S4S 0A2 E-mail: {yanzhao, yyao}@cs.uregina.ca Abstract.
Classification Based on Logical Concept Analysis Yan Zhao and Yiyu Yao Department of Computer Science, University of Regina, Regina, Saskatchewan, Canada S4S 0A2 E-mail: {yanzhao, yyao}@cs.uregina.ca Abstract.
Plan. Day 2: Exercise on MHC molecules.
 Plan Day 1: What is Chemoinformatics and Drug Design? Methods and Algorithms used in Chemoinformatics including SVM. Cross validation and sequence encoding Example and exercise with herg potassium channel:
Plan Day 1: What is Chemoinformatics and Drug Design? Methods and Algorithms used in Chemoinformatics including SVM. Cross validation and sequence encoding Example and exercise with herg potassium channel:
MSc Drug Design. Module Structure: (15 credits each) Lectures and Tutorials Assessment: 50% coursework, 50% unseen examination.
 Module Structure: (15 credits each) Lectures and Assessment: 50% coursework, 50% unseen examination. Module Title Module 1: Bioinformatics and structural biology as applied to drug design MEDC0075 In the
Module Structure: (15 credits each) Lectures and Assessment: 50% coursework, 50% unseen examination. Module Title Module 1: Bioinformatics and structural biology as applied to drug design MEDC0075 In the
Comparison of Shannon, Renyi and Tsallis Entropy used in Decision Trees
 Comparison of Shannon, Renyi and Tsallis Entropy used in Decision Trees Tomasz Maszczyk and W lodzis law Duch Department of Informatics, Nicolaus Copernicus University Grudzi adzka 5, 87-100 Toruń, Poland
Comparison of Shannon, Renyi and Tsallis Entropy used in Decision Trees Tomasz Maszczyk and W lodzis law Duch Department of Informatics, Nicolaus Copernicus University Grudzi adzka 5, 87-100 Toruń, Poland
Notes of Dr. Anil Mishra at 1
 Introduction Quantitative Structure-Activity Relationships QSPR Quantitative Structure-Property Relationships What is? is a mathematical relationship between a biological activity of a molecular system
Introduction Quantitative Structure-Activity Relationships QSPR Quantitative Structure-Property Relationships What is? is a mathematical relationship between a biological activity of a molecular system
Chemogenomic: Approaches to Rational Drug Design. Jonas Skjødt Møller
 Chemogenomic: Approaches to Rational Drug Design Jonas Skjødt Møller Chemogenomic Chemistry Biology Chemical biology Medical chemistry Chemical genetics Chemoinformatics Bioinformatics Chemoproteomics
Chemogenomic: Approaches to Rational Drug Design Jonas Skjødt Møller Chemogenomic Chemistry Biology Chemical biology Medical chemistry Chemical genetics Chemoinformatics Bioinformatics Chemoproteomics
Kernel-based Machine Learning for Virtual Screening
 Kernel-based Machine Learning for Virtual Screening Dipl.-Inf. Matthias Rupp Beilstein Endowed Chair for Chemoinformatics Johann Wolfgang Goethe-University Frankfurt am Main, Germany 2008-04-11, Helmholtz
Kernel-based Machine Learning for Virtual Screening Dipl.-Inf. Matthias Rupp Beilstein Endowed Chair for Chemoinformatics Johann Wolfgang Goethe-University Frankfurt am Main, Germany 2008-04-11, Helmholtz
In Silico Investigation of Off-Target Effects
 PHARMA & LIFE SCIENCES WHITEPAPER In Silico Investigation of Off-Target Effects STREAMLINING IN SILICO PROFILING In silico techniques require exhaustive data and sophisticated, well-structured informatics
PHARMA & LIFE SCIENCES WHITEPAPER In Silico Investigation of Off-Target Effects STREAMLINING IN SILICO PROFILING In silico techniques require exhaustive data and sophisticated, well-structured informatics
Principles of Drug Design
 (16:663:502) Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu For more current information, please check WebCT at https://webct.rutgers.edu
(16:663:502) Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu For more current information, please check WebCT at https://webct.rutgers.edu
A Support Vector Regression Model for Forecasting Rainfall
 A Support Vector Regression for Forecasting Nasimul Hasan 1, Nayan Chandra Nath 1, Risul Islam Rasel 2 Department of Computer Science and Engineering, International Islamic University Chittagong, Bangladesh
A Support Vector Regression for Forecasting Nasimul Hasan 1, Nayan Chandra Nath 1, Risul Islam Rasel 2 Department of Computer Science and Engineering, International Islamic University Chittagong, Bangladesh
Data Quality Issues That Can Impact Drug Discovery
 Data Quality Issues That Can Impact Drug Discovery Sean Ekins 1, Joe Olechno 2 Antony J. Williams 3 1 Collaborations in Chemistry, Fuquay Varina, NC. 2 Labcyte Inc, Sunnyvale, CA. 3 Royal Society of Chemistry,
Data Quality Issues That Can Impact Drug Discovery Sean Ekins 1, Joe Olechno 2 Antony J. Williams 3 1 Collaborations in Chemistry, Fuquay Varina, NC. 2 Labcyte Inc, Sunnyvale, CA. 3 Royal Society of Chemistry,
Iterative Laplacian Score for Feature Selection
 Iterative Laplacian Score for Feature Selection Linling Zhu, Linsong Miao, and Daoqiang Zhang College of Computer Science and echnology, Nanjing University of Aeronautics and Astronautics, Nanjing 2006,
Iterative Laplacian Score for Feature Selection Linling Zhu, Linsong Miao, and Daoqiang Zhang College of Computer Science and echnology, Nanjing University of Aeronautics and Astronautics, Nanjing 2006,
COMBINATORIAL CHEMISTRY IN A HISTORICAL PERSPECTIVE
 NUE FEATURE T R A N S F O R M I N G C H A L L E N G E S I N T O M E D I C I N E Nuevolution Feature no. 1 October 2015 Technical Information COMBINATORIAL CHEMISTRY IN A HISTORICAL PERSPECTIVE A PROMISING
NUE FEATURE T R A N S F O R M I N G C H A L L E N G E S I N T O M E D I C I N E Nuevolution Feature no. 1 October 2015 Technical Information COMBINATORIAL CHEMISTRY IN A HISTORICAL PERSPECTIVE A PROMISING
3D QSAR studies of Substituted Benzamides as Nonacidic Antiinflammatory Agents by knn MFA Approach
 INTERNATIONAL JOURNAL OF ADVANCES IN PARMACY, BIOLOGY AND CEMISTRY Research Article 3D QSAR studies of Substituted Benzamides as Nonacidic Antiinflammatory Agents by knn MFA Approach Anupama A. Parate*
INTERNATIONAL JOURNAL OF ADVANCES IN PARMACY, BIOLOGY AND CEMISTRY Research Article 3D QSAR studies of Substituted Benzamides as Nonacidic Antiinflammatory Agents by knn MFA Approach Anupama A. Parate*
Kinome-wide Activity Models from Diverse High-Quality Datasets
 Kinome-wide Activity Models from Diverse High-Quality Datasets Stephan C. Schürer*,1 and Steven M. Muskal 2 1 Department of Molecular and Cellular Pharmacology, Miller School of Medicine and Center for
Kinome-wide Activity Models from Diverse High-Quality Datasets Stephan C. Schürer*,1 and Steven M. Muskal 2 1 Department of Molecular and Cellular Pharmacology, Miller School of Medicine and Center for
Protein Structure Prediction Using Multiple Artificial Neural Network Classifier *
 Protein Structure Prediction Using Multiple Artificial Neural Network Classifier * Hemashree Bordoloi and Kandarpa Kumar Sarma Abstract. Protein secondary structure prediction is the method of extracting
Protein Structure Prediction Using Multiple Artificial Neural Network Classifier * Hemashree Bordoloi and Kandarpa Kumar Sarma Abstract. Protein secondary structure prediction is the method of extracting
Feature gene selection method based on logistic and correlation information entropy
 Bio-Medical Materials and Engineering 26 (2015) S1953 S1959 DOI 10.3233/BME-151498 IOS Press S1953 Feature gene selection method based on logistic and correlation information entropy Jiucheng Xu a,b,,
Bio-Medical Materials and Engineering 26 (2015) S1953 S1959 DOI 10.3233/BME-151498 IOS Press S1953 Feature gene selection method based on logistic and correlation information entropy Jiucheng Xu a,b,,
An Integrated Approach to in-silico
 An Integrated Approach to in-silico Screening Joseph L. Durant Jr., Douglas. R. Henry, Maurizio Bronzetti, and David. A. Evans MDL Information Systems, Inc. 14600 Catalina St., San Leandro, CA 94577 Goals
An Integrated Approach to in-silico Screening Joseph L. Durant Jr., Douglas. R. Henry, Maurizio Bronzetti, and David. A. Evans MDL Information Systems, Inc. 14600 Catalina St., San Leandro, CA 94577 Goals
Analysis of a Large Structure/Biological Activity. Data Set Using Recursive Partitioning and. Simulated Annealing
 Analysis of a Large Structure/Biological Activity Data Set Using Recursive Partitioning and Simulated Annealing Student: Ke Zhang MBMA Committee: Dr. Charles E. Smith (Chair) Dr. Jacqueline M. Hughes-Oliver
Analysis of a Large Structure/Biological Activity Data Set Using Recursive Partitioning and Simulated Annealing Student: Ke Zhang MBMA Committee: Dr. Charles E. Smith (Chair) Dr. Jacqueline M. Hughes-Oliver
Computational chemical biology to address non-traditional drug targets. John Karanicolas
 Computational chemical biology to address non-traditional drug targets John Karanicolas Our computational toolbox Structure-based approaches Ligand-based approaches Detailed MD simulations 2D fingerprints
Computational chemical biology to address non-traditional drug targets John Karanicolas Our computational toolbox Structure-based approaches Ligand-based approaches Detailed MD simulations 2D fingerprints
Artificial Intelligence
 Artificial Intelligence Jeff Clune Assistant Professor Evolving Artificial Intelligence Laboratory Announcements Be making progress on your projects! Three Types of Learning Unsupervised Supervised Reinforcement
Artificial Intelligence Jeff Clune Assistant Professor Evolving Artificial Intelligence Laboratory Announcements Be making progress on your projects! Three Types of Learning Unsupervised Supervised Reinforcement
Structure-based maximal affinity model predicts small-molecule druggability
 Structure-based maximal affinity model predicts small-molecule druggability Alan Cheng alan.cheng@amgen.com IMA Workshop (Jan 17, 2008) Druggability prediction Introduction Affinity model Some results
Structure-based maximal affinity model predicts small-molecule druggability Alan Cheng alan.cheng@amgen.com IMA Workshop (Jan 17, 2008) Druggability prediction Introduction Affinity model Some results
Using AutoDock for Virtual Screening
 Using AutoDock for Virtual Screening CUHK Croucher ASI Workshop 2011 Stefano Forli, PhD Prof. Arthur J. Olson, Ph.D Molecular Graphics Lab Screening and Virtual Screening The ultimate tool for identifying
Using AutoDock for Virtual Screening CUHK Croucher ASI Workshop 2011 Stefano Forli, PhD Prof. Arthur J. Olson, Ph.D Molecular Graphics Lab Screening and Virtual Screening The ultimate tool for identifying
Lecture 3: Decision Trees
 Lecture 3: Decision Trees Cognitive Systems - Machine Learning Part I: Basic Approaches of Concept Learning ID3, Information Gain, Overfitting, Pruning last change November 26, 2014 Ute Schmid (CogSys,
Lecture 3: Decision Trees Cognitive Systems - Machine Learning Part I: Basic Approaches of Concept Learning ID3, Information Gain, Overfitting, Pruning last change November 26, 2014 Ute Schmid (CogSys,
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie
 A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
A short introduction to supervised learning, with applications to cancer pathway analysis Dr. Christina Leslie Computational Biology Program Memorial Sloan-Kettering Cancer Center http://cbio.mskcc.org/leslielab
Exploring the black box: structural and functional interpretation of QSAR models.
 EMBL-EBI Industry workshop: In Silico ADMET prediction 4-5 December 2014, Hinxton, UK Exploring the black box: structural and functional interpretation of QSAR models. (Automatic exploration of datasets
EMBL-EBI Industry workshop: In Silico ADMET prediction 4-5 December 2014, Hinxton, UK Exploring the black box: structural and functional interpretation of QSAR models. (Automatic exploration of datasets
Chapter 6: Classification
 Chapter 6: Classification 1) Introduction Classification problem, evaluation of classifiers, prediction 2) Bayesian Classifiers Bayes classifier, naive Bayes classifier, applications 3) Linear discriminant
Chapter 6: Classification 1) Introduction Classification problem, evaluation of classifiers, prediction 2) Bayesian Classifiers Bayes classifier, naive Bayes classifier, applications 3) Linear discriminant
Translating Methods from Pharma to Flavours & Fragrances
 Translating Methods from Pharma to Flavours & Fragrances CINF 27: ACS National Meeting, New Orleans, LA - 18 th March 2018 Peter Hunt, Edmund Champness, Nicholas Foster, Tamsin Mansley & Matthew Segall
Translating Methods from Pharma to Flavours & Fragrances CINF 27: ACS National Meeting, New Orleans, LA - 18 th March 2018 Peter Hunt, Edmund Champness, Nicholas Foster, Tamsin Mansley & Matthew Segall
FRAUNHOFER IME SCREENINGPORT
 FRAUNHOFER IME SCREENINGPORT Design of screening projects General remarks Introduction Screening is done to identify new chemical substances against molecular mechanisms of a disease It is a question of
FRAUNHOFER IME SCREENINGPORT Design of screening projects General remarks Introduction Screening is done to identify new chemical substances against molecular mechanisms of a disease It is a question of
Evidence for the Existence of Non-monotonic Dose-response: Does it or Doesn t it?
 Evidence for the Existence of Non-monotonic Dose-response: Does it or Doesn t it? Scott M. Belcher, PhD University of Cincinnati Department of Pharmacology and Cell Biophysics Evidence for the Existence
Evidence for the Existence of Non-monotonic Dose-response: Does it or Doesn t it? Scott M. Belcher, PhD University of Cincinnati Department of Pharmacology and Cell Biophysics Evidence for the Existence
Reading, UK 1 2 Abstract
 , pp.45-54 http://dx.doi.org/10.14257/ijseia.2013.7.5.05 A Case Study on the Application of Computational Intelligence to Identifying Relationships between Land use Characteristics and Damages caused by
, pp.45-54 http://dx.doi.org/10.14257/ijseia.2013.7.5.05 A Case Study on the Application of Computational Intelligence to Identifying Relationships between Land use Characteristics and Damages caused by
Chemical Space. Space, Diversity, and Synthesis. Jeremy Henle, 4/23/2013
 Chemical Space Space, Diversity, and Synthesis Jeremy Henle, 4/23/2013 Computational Modeling Chemical Space As a diversity construct Outline Quantifying Diversity Diversity Oriented Synthesis Wolf and
Chemical Space Space, Diversity, and Synthesis Jeremy Henle, 4/23/2013 Computational Modeling Chemical Space As a diversity construct Outline Quantifying Diversity Diversity Oriented Synthesis Wolf and
Support Vector Machines (SVM) in bioinformatics. Day 1: Introduction to SVM
 1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
1 Support Vector Machines (SVM) in bioinformatics Day 1: Introduction to SVM Jean-Philippe Vert Bioinformatics Center, Kyoto University, Japan Jean-Philippe.Vert@mines.org Human Genome Center, University
Artificial Neural Network
 Artificial Neural Network Contents 2 What is ANN? Biological Neuron Structure of Neuron Types of Neuron Models of Neuron Analogy with human NN Perceptron OCR Multilayer Neural Network Back propagation
Artificial Neural Network Contents 2 What is ANN? Biological Neuron Structure of Neuron Types of Neuron Models of Neuron Analogy with human NN Perceptron OCR Multilayer Neural Network Back propagation
Data Mining Part 5. Prediction
 Data Mining Part 5. Prediction 5.5. Spring 2010 Instructor: Dr. Masoud Yaghini Outline How the Brain Works Artificial Neural Networks Simple Computing Elements Feed-Forward Networks Perceptrons (Single-layer,
Data Mining Part 5. Prediction 5.5. Spring 2010 Instructor: Dr. Masoud Yaghini Outline How the Brain Works Artificial Neural Networks Simple Computing Elements Feed-Forward Networks Perceptrons (Single-layer,
Active Sonar Target Classification Using Classifier Ensembles
 International Journal of Engineering Research and Technology. ISSN 0974-3154 Volume 11, Number 12 (2018), pp. 2125-2133 International Research Publication House http://www.irphouse.com Active Sonar Target
International Journal of Engineering Research and Technology. ISSN 0974-3154 Volume 11, Number 12 (2018), pp. 2125-2133 International Research Publication House http://www.irphouse.com Active Sonar Target
Logistic Regression and Boosting for Labeled Bags of Instances
 Logistic Regression and Boosting for Labeled Bags of Instances Xin Xu and Eibe Frank Department of Computer Science University of Waikato Hamilton, New Zealand {xx5, eibe}@cs.waikato.ac.nz Abstract. In
Logistic Regression and Boosting for Labeled Bags of Instances Xin Xu and Eibe Frank Department of Computer Science University of Waikato Hamilton, New Zealand {xx5, eibe}@cs.waikato.ac.nz Abstract. In
CSE 352 (AI) LECTURE NOTES Professor Anita Wasilewska. NEURAL NETWORKS Learning
 CSE 352 (AI) LECTURE NOTES Professor Anita Wasilewska NEURAL NETWORKS Learning Neural Networks Classifier Short Presentation INPUT: classification data, i.e. it contains an classification (class) attribute.
CSE 352 (AI) LECTURE NOTES Professor Anita Wasilewska NEURAL NETWORKS Learning Neural Networks Classifier Short Presentation INPUT: classification data, i.e. it contains an classification (class) attribute.
Machine Learning Linear Classification. Prof. Matteo Matteucci
 Machine Learning Linear Classification Prof. Matteo Matteucci Recall from the first lecture 2 X R p Regression Y R Continuous Output X R p Y {Ω 0, Ω 1,, Ω K } Classification Discrete Output X R p Y (X)
Machine Learning Linear Classification Prof. Matteo Matteucci Recall from the first lecture 2 X R p Regression Y R Continuous Output X R p Y {Ω 0, Ω 1,, Ω K } Classification Discrete Output X R p Y (X)
Quantitative Structure Activity Relationships: An overview
 Quantitative Structure Activity Relationships: An overview Prachi Pradeep Oak Ridge Institute for Science and Education Research Participant National Center for Computational Toxicology U.S. Environmental
Quantitative Structure Activity Relationships: An overview Prachi Pradeep Oak Ridge Institute for Science and Education Research Participant National Center for Computational Toxicology U.S. Environmental
CSE 417T: Introduction to Machine Learning. Final Review. Henry Chai 12/4/18
 CSE 417T: Introduction to Machine Learning Final Review Henry Chai 12/4/18 Overfitting Overfitting is fitting the training data more than is warranted Fitting noise rather than signal 2 Estimating! "#$
CSE 417T: Introduction to Machine Learning Final Review Henry Chai 12/4/18 Overfitting Overfitting is fitting the training data more than is warranted Fitting noise rather than signal 2 Estimating! "#$
Contents 1 Open-Source Tools, Techniques, and Data in Chemoinformatics
 Contents 1 Open-Source Tools, Techniques, and Data in Chemoinformatics... 1 1.1 Chemoinformatics... 2 1.1.1 Open-Source Tools... 2 1.1.2 Introduction to Programming Languages... 3 1.2 Chemical Structure
Contents 1 Open-Source Tools, Techniques, and Data in Chemoinformatics... 1 1.1 Chemoinformatics... 2 1.1.1 Open-Source Tools... 2 1.1.2 Introduction to Programming Languages... 3 1.2 Chemical Structure
Improving EASI Model via Machine Learning and Regression Techniques
 Improving EASI Model via Machine Learning and Regression Techniques P. Kaewfoongrungsi, D.Hormdee Embedded System R&D Group, Computer Engineering, Faculty of Engineering, Khon Kaen University, 42, Thailand.
Improving EASI Model via Machine Learning and Regression Techniques P. Kaewfoongrungsi, D.Hormdee Embedded System R&D Group, Computer Engineering, Faculty of Engineering, Khon Kaen University, 42, Thailand.
