Dispersal of Golgi matrix proteins during mitotic Golgi disassembly
|
|
|
- Judith Tucker
- 5 years ago
- Views:
Transcription
1 Research Article 451 Dispersal of Golgi matrix proteins during mitotic Golgi disassembly Sapna Puri, Helena Telfer, Meel Velliste, Robert F. Murphy and Adam D. Linstedt* Department of Biological Sciences, Carnegie Mellon University, th Avenue, Pittsburgh, PA 15213, USA *Author for correspondence ( Accepted 10 September 2003 Journal of Cell Science 117, Published by The Company of Biologists 2004 doi: /jcs Summary During mitosis, the mammalian Golgi disassembles into numerous vesicles and larger membrane structures referred to as clusters or remnants. Following mitosis, the vesicles and clusters reassemble to form an intact Golgi in each daughter cell. One model of Golgi biogenesis states that Golgi matrix proteins remain assembled in mitotic clusters and then serve as a template for Golgi reassembly. To test this idea, we performed a 3D-computational analysis of mitotic cells to determine the extent to which these proteins remain in mitotic clusters. As a control we used brefeldin A-induced Golgi disassembly which causes dispersal of Golgi enzymes, but leaves matrix proteins in remnant structures. Unlike brefeldin A-treated cells, in which matrix proteins were clearly sorted from non-matrix proteins, we observed extensive dispersal of matrix proteins in metaphase cells with no evidence of differential sorting of these proteins from other Golgi proteins. The extensive disassembly of matrix proteins argues against their participation in a stable template and supports a selfassembly mode of Golgi biogenesis. Key words: Golgi apparatus, Biogenesis, Golgin, GRASP, Template, De novo Introduction The mammalian Golgi apparatus mediates processing and sorting reactions in the secretory pathway. Its behavior is particularly intriguing in that although it has a complex structure, it undergoes substantial structural transformations in every cell cycle. During interphase, Golgi membranes consist of stacked cisternae. Each stack contains approximately four cisternae and there are several hundred of these stacks per cell. Each stack is also linked laterally to other stacks to form a ribbon-like membrane system positioned adjacent to the microtubule organizing center. At mitosis, the entire structure disassembles and then reassembles in each daughter cell. However, the actual extent of mitotic Golgi disassembly is controversial, and this has led to opposing models regarding the mechanism by which Golgi membranes reassemble in each daughter cell post-cytokinesis. One model suggests the use of a pre-existing template (Seemann et al., 2000; Seemann et al., 2002), whereas the other predicts reformation by self-assembly of the breakdown products (Miles et al., 2001; Ward et al., 2001; Bevis et al., 2002). The template model derives from work on two sets of Golgilocalized proteins, the golgins and GRASPs (Golgi Re- Assembly Stacking Proteins). Golgins have extensive heptad repeats predicted to form elongated coiled-coil domains on the cytoplasmic side of the Golgi membrane. GRASPs share significant sequence identity and are anchored to the cytoplasmic side of the Golgi membrane by myristoylation. One of the golgins, GM130, was discovered in a detergent insoluble extract of purified Golgi membranes (Nakamura et al., 1995). Importantly, this insoluble material yielded a pattern in electron microscope images reminiscent of stacked cisternae (Slusarewicz et al., 1994). On this basis the material was called the Golgi matrix and GM130 was termed a matrix protein. Other golgins and the GRASPs are termed matrix proteins for two reasons. The first is based on interactions among these proteins. GM130 interacts directly with GRASP65 and indirectly with giantin (Barr et al., 1997; Dirac-Svejstrup et al., 2000). GRASP55 may function in a parallel complex as it binds golgin-45 (Short et al., 2001). The second is based on the behavior of these proteins versus other Golgi proteins during treatment of cells with brefeldin A (BFA). BFA treatment induces redistribution of most Golgi-localized proteins to the ER, but giantin, GM130, GRASP65 and GRASP55 end up in membranes called BFA remnants that are distinct from the ER (Seemann et al., 2000). As BFA-induced Golgi disassembly is reversed upon drug washout, this finding is consistent with the view that these proteins remain in intact assemblies that then mediate reassembly upon drug washout. In fact, giantin, GM130, GRASP65 and GRASP55 are each required for Golgi stacking as measured using an in vitro assay (Barr et al., 1997; Shorter et al., 1999; Shorter and Warren, 1999). The mitotic Golgi disassembly/reassembly reaction offers a critical and physiologically relevant test case for this model. Most work suggests that mitotic Golgi disassembly proceeds by vesiculation and yields Golgi proteins in an abundant population of small vesicles and in some larger membranes variously termed vesicle clusters or cisternal remnants (Lucocq et al., 1989; Misteli and Warren, 1995; Shima et al., 1997; Jesch and Linstedt, 1998; Jesch et al., 2001; Jokitalo et al., 2001). The cisternal remnants are hypothesized to constitute the Golgi template. Opposed to this is evidence suggesting that during mitosis the Golgi collapses into, and partitions with, the
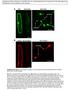
2 452 Journal of Cell Science 117 (3) ER (Zaal et al., 1999). If the entire Golgi is in the ER, then post-mitotic Golgi biogenesis must occur de novo. Interestingly, this latter work used Golgi enzymes rather than any golgin or GRASP as marker proteins. It was subsequently observed that during BFA-induced Golgi collapse, Golgi enzymes move to the ER whereas golgins and GRASPs accumulate in BFA remnants. Therefore, as suggested in several reviews (Nelson, 2000; Short and Barr, 2000; Rossanese and Glick, 2001; Jesch, 2002; Shorter and Warren, 2002), it might be possible that during mitosis redistribution to the ER is the mode for enzymes whereas golgins and GRASPs remain in mitotic clusters. This is important because the matrix template model makes a strong prediction with respect to the distribution of golgins and GRASPs. By this view mitotic Golgi clusters are the partitioning units of the Golgi (Seemann et al., 2002). They correspond roughly to the number of stacks per cell and contain golgins and GRASPs in an assembled state that will serve as template for post-mitotic Golgi reassembly. Hence, golgins and GRASPs should remain behind in mitotic Golgi clusters, whereas other Golgi markers may move into the vesicle fraction, or even into the ER. We tested this prediction by quantitative measurement of sorting of golgins and GRASPs from other Golgi proteins during mitotic Golgi breakdown. objective yielding a pixel dimension of µm was used. Sections, 15 for interphase and 40 for metaphase cells, were taken at a step size of 0.3 µm. Images were deconvolved using SoftWorx (Applied Precision). Deconvolved manually cropped sections were assembled into a 3D image using Matlab (Mathworks, Natick, MA, USA). The background value subtracted from each voxel was the most common voxel value outside the cell multiplied by a factor of 1.54 and 1.22 for the fluorescein and rhodamine channel, respectively. These factors were the average of the ratio of the most common voxel value inside the cell to the most common voxel value outside the cell determined from the 3D data set of 10 interphase and metaphase cells that were stained with secondary antibody only. Threshold values were manually chosen for sections 4, 7 and 11 for interphase cells and sections 10, 20 and 30 for metaphase cells. Using the original gray scale image as the standard, the sections were thresholded to maximize inclusion of discernable objects and minimize inclusion of non-discernable objects. The average of the three values was then used as the threshold for that cell. Threshold values varied from to 0.003% of the maximum intensity. Object finding was done on the thresholded image using 26-neighbor connectivity and the fraction of total fluorescence in objects was calculated. Given that mitotic clusters may be compartmentalized (Shima et al., 1997), we determined the percentage of mitotic clusters that were positive for a given pair of markers by considering objects identified in one channel to be positive for a marker in another channel only if object edges overlapped. Materials and Methods Cell culture and staining Growth medium for NRK cells (Dulbecco s modified Eagles medium, Life Technologies, Grand Island, NY, USA) and HeLa cells (Minimal essential medium, Sigma-Aldrich, St. Louis, MO, USA) contained 10% fetal bovine serum and 100 IU/ml penicillin-streptomycin. Where indicated, the growth medium was supplemented with 10 µg/ml BFA (Sigma). For immunofluorescence, cells were grown on 12-mm glass coverslips, treated where indicated, and either fixed in methanol ( 20 C for 10 minutes) or 3% paraformaldehyde (20 minutes at room temperature). Paraformaldehyde-fixed cells were stained as previously described (Jesch and Linstedt, 1998). After three washes with PBS, the cells were incubated for 30 minutes in blocking buffer (PBS containing 2.5% calf serum and 0.1% Tween-20), followed by consecutive 30-minute incubations in primary and secondary antibodies diluted in blocking buffer. Each antibody incubation was followed by five washes with PBS. The antibodies and their dilutions were: mouse anti-gpp130 at 1:200 (Linstedt et al., 1997); mouse anti-giantin at 1:100 (Linstedt and Hauri, 1993); rabbit anti-gpp130 at 1:500 (Puri et al., 2002); rabbit anti-giantin at 1:500 (Puthenveedu and Linstedt, 2001); rabbit anti-gm130 at 1:500 (Puthenveedu and Linstedt, 2001); rabbit anti-p115 at 1:500 (Puthenveedu and Linstedt, 2001); rabbit anti-grasp65 at 1:2000 (Sutterlin et al., 2002); rabbit anti-grasp55 at 1:2000 (a gift from Vivek Malhotra, UCSD, La Jolla, CA); mouse anti-tgn38 at 1:100 (Affinity BioReagents, Golden, CO, USA); mouse anti-mannosidase II at 1:10,000 (Sigma); fluorescein-labeled goat anti-mouse at 1:200 (Zymed, San Francisco, CA, USA); and rhodamine-labeled goat antirabbit at 1:200 (Zymed). Some experiments used a HeLa cell line stably expressing N-acetylgalactosaminyltransferase 2 tagged with GFP (Storrie et al., 1998). Image analysis Microscopy was performed using either a conventional fluorescence microscope (Linstedt et al., 1997) or a DeltaVision system (Applied Precision, Issaquah, WA, USA) essentially as previously described (Jesch et al., 2001). In the latter case a 100, 1.35 NA oil immersion Results To determine the extent of matrix marker recovery in mitotic Golgi clusters we used a 3D analysis, because 2D projections of 3D data, particularly when using the maximum value of all aligned Z-axis pixel values instead of their sum, severely under-represent dispersed fluorescence (Jesch et al., 2001). The analysis was performed by collecting a stack of images by deconvolution microscopy. Total fluorescence above background, as well as fluorescence attributed to objects, was then analyzed in three dimensions. To reduce experimental bias and to account for possible variation between cells, a statistical analysis was performed on results from multiple cells for each condition and each cell was chosen solely on the basis of its DNA staining. Also, endogenous markers for matrix and nonmatrix proteins were used to avoid the various problems associated with transgene expression. Finally, NRK cells were chosen because the presence of mitotic Golgi clusters was most apparent in these cells. If it occurs, this would increase our ability to demonstrate selective retention of matrix markers. The deconvolved images demonstrated presence of matrix and non-matrix markers in both punctate and hazy staining, previously shown to correspond to mitotic Golgi clusters and mitotic Golgi vesicles, respectively (Jesch et al., 2001). Note that the non-resolvable, or hazy, fluorescence was significantly above background and was equally present for fluorescence derived from GFP coupled to a Golgi marker (see below). For the purpose of illustration, Fig. 1 presents representative total fluorescence patterns of deconvolved optical sections as 2D projections summed along the Z-axis. In this case, interphase and metaphase NRK cells were costained for non-matrix marker mannosidase II and matrix marker GM130. For untreated interphase cells, each antibody yielded specific staining of the Golgi ribbon and the staining patterns were markedly coincident (Fig. 1A-C). In contrast, in untreated metaphase cells, both mannosidase II and GM130 yielded a
3 Mitotic dispersal of Golgi matrix proteins 453 Fig. 1. Comparison of a matrix protein and a non-matrix protein in interphase and metaphase cells. Interphase (A-C) and metaphase (D-F) NRK cells were costained for mannosidase II (A,D) and GM130 (B,E). Single-channel images are also overlayed (C,F) with mannosidase II in green and GM130 in red. The images shown are 2D projections from the deconvolved 3D data set using summation of pixel values in each plane. Bar: 10 µm. mixture of hazy and punctate staining, indicating that each marker was present in vesicular haze as well as mitotic Golgi clusters (Fig. 1D-F). Note that the marker staining of punctate structures was mostly adjacent rather than coincident (see below). Similar results were obtained for matrix markers giantin (Fig. 2A), GRASP65 (Fig. 2B) and GRASP55 (Fig. 2C), as well as a non-matrix marker, GPP130 (Fig. 2D). Identification of the NRK mitotic Golgi clusters in three dimensions indicated that there were, on a per cell basis, 203±54 (s.d., n=60) clusters and these were an average of 1 µm in diameter. Importantly, the fraction of total fluorescence comprised by mitotic Golgi clusters was at most 10% for any marker tested (Fig. 3). Thus, although mitotic Golgi clusters in NRK were larger than those previously reported in other cell types (Shima et al., 1997; Jesch et al., 2001), our analysis indicated that these clusters only represented a small fraction of the mitotic Golgi. Furthermore, the fact that the extent of marker recovery was similar for matrix and non-matrix proteins indicates a striking absence of sorting based on category of marker protein. The only exceptions were for GRASP55 and p115, which were actually recovered to an even lower extent in mitotic Golgi clusters. For p115, this can be explained as it is a peripheral membrane protein that partially dissociates from membranes during mitosis (Levine et al., 1996). Whether GRASP55 actually dissociates remains to be tested. Despite its lipid anchor, this may be possible, as there is evidence that lipid-anchored GRASP65 rapidly cycles on and off the Golgi membrane during interphase (Ward et al., 2001). Even though the recovery of Golgi markers in clusters was only 10%, this was higher than the 2% previously observed for GPP130 in metaphase HeLa cells (Jesch et al., 2001). In fact, we simultaneously analyzed this marker and several others in both cell types and found that, independent of marker tested, and independent of fixation conditions, mitotic Golgi clusters account for 2% and 10% of HeLa and NRK Golgi, respectively. Identical results in HeLa were also obtained for analysis of a Fig. 2. Presence of Golgi markers in mitotic Golgi clusters and vesicular haze. Metaphase NRK cells were stained using antibodies against giantin (A), GRASP65 (B), GRASP55 (C) and GPP130 (D). Images shown are 2D projections using summation of pixel values. Bar: 10 µm. Fig. 3. Matrix proteins are extensively dispersed in vesicular haze rather than selectively retained in mitotic Golgi clusters. Untreated interphase (open bars) and metaphase NRK cells were stained for different proteins as indicated, and the percentage fluorescence in objects of each was calculated as in Materials and Methods (±s.e.m.). The results are averages from 10 cells per condition.
4 454 Journal of Cell Science 117 (3) Fig. 4. Coincidence of Golgi proteins in mitotic Golgi clusters. Metaphase NRK cells were costained with antibodies against mannosidase II and GRASP65 and a merged image is shown with mannosidase II in green and GRASP65 in red. Metaphase cells were also costained with antibodies against TGN38 and giantin, and the merge shows TGN38 in green and giantin in red. All images shown are summed projections that were thresholded to isolate mitotic Golgi cluster staining from vesicular haze. Note that markers were frequently adjacent in clusters rather than truly coincident. Bar: 10 µm. stably expressed Golgi marker visualized on the basis of a GFP tag, arguing against inaccuracies because of staining procedures or background determination. Thus, Golgi breakdown varies depending on cell type, but at least for these two cell types, it is extensive for all categories of marker. This includes the trans-golgi network as TGN38 was also mostly present in hazy staining with only 10% recovered in NRK mitotic Golgi clusters. Interestingly, most mitotic Golgi clusters appeared to contain each Golgi marker. Mannosidase II was present in 92% of the GM130 positive clusters (n=411), 90% of the giantin positive clusters (n=146), and 88% of the GRASP65 clusters (n=147). Tests of other pairs of markers in clusters also yielded remarkably coincident patterns. However, within each cluster, marker staining was often adjacent rather than precisely coincident as is evident in metaphase cells costained for mannosidase II and GRASP65 (Fig. 4A-C). Adjacent staining was most noticeable in clusters costained for an early and late Golgi marker such as giantin and TGN38, respectively (Fig. 4D-F) consistent with previous work suggesting marker compartmentalization within individual clusters (Shima et al., 1997). Non-matrix Golgi markers redistribute to the ER upon BFA treatment, whereas matrix markers become localized in BFA remnants (Seemann et al., 2000). Consistent with this, after BFA treatment of interphase cells, mannosidase II staining collapsed from a ribbon pattern (Fig. 5A) to an ER pattern (Fig. 5C), and GM130 staining went from Golgi ribbon (Fig. 5B) to the punctate pattern of BFA remnants (Fig. 5D). Quantitative analysis showed that GM130, GRASP55 and GRASP65 were clearly enriched in BFA remnants relative to other markers (Fig. 5E). Nevertheless, matrix marker recovery in BFA remnants was less than 50% and, in the case of giantin, only 5% of the signal was in BFA remnants. These experiments with BFA make two points. First, a quantitative analysis of Golgi marker behavior in response to BFA, which had not been previously performed, revealed greater complexity than previously appreciated. Not only is recovery in BFA remnants incomplete, but also, not all golgins/grasps respond in a similar manner. As a further example, golgin 84, a protein which we could not quantitatively assess because of lack of antibodies, redistributes to the ER and not to BFA remnants (Diao et al., 2003). These complexities cast doubt regarding the stability of a Golgi matrix comprised of golgins and GRASPs. Second, the assay we used to test for selective retention of matrix markers in mitotic Golgi clusters is clearly capable of detecting sorting of proteins between larger intact golgin/grasp structures and dispersed structures. Therefore, the low and uniform recovery of all Golgi proteins, including matrix markers, in mitotic Golgi clusters is strong evidence for a self-assembly rather than a stable templatemediated mode of post-mitotic Golgi reassembly. Discussion Our test of the matrix protein model for Golgi biogenesis focused on a key prediction. Sorting during disassembly may allow dispersal and/or ER redistribution of other Golgi markers, but matrix proteins should be left in a primarily assembled state in remnant Golgi structures (Seemann et al., 2000). The presence of a stable template contrasts with de novo Golgi biogenesis in which structural elements can undergo complete disassembly followed by reassembly. It also fits with previous non-quantitative observations showing accumulation of golgins and GRASPs in BFA remnants (Seemann et al., 2000), and their presence in mitotic Golgi clusters (Shima et al., 1997). However, our quantitative assessment indicated that mitotic Golgi disassembly was accompanied by substantial dispersal of golgins and GRASPs rather than concentration in mitotic Golgi clusters. In NRK and HeLa, mitotic Golgi clusters accounted for at most 10% and 2% of any marker tested, respectively. There was also no indication that golgins
5 Mitotic dispersal of Golgi matrix proteins 455 Fig. 5. Matrix proteins are selectively, but incompletely, retained in BFA remnants. NRK cells, either untreated (A,B) or treated with BFA for 30 minutes (C,D), were costained for mannosidase II (A,C) and GM130 (B,D). Summed projections are shown. Note that for an unexplained reason, nucleolar-like staining was observed when the anti-mannosidase II antibody was used to stain interphase cells, particularly at the exposure setting necessary to visualize BFAtreated cells. This non-specific staining was excluded in quantitative analyses. Bar: 10 µm. (E) Untreated interphase (open bars) and BFAtreated NRK cells were stained for different proteins as indicated, and the percentage fluorescence in objects of each was calculated as in Materials and Methods (±s.e.m.). The results are averages from 10 cells per condition. and GRASPs are sorted differently from other categories of Golgi proteins. Even BFA remnants, which did show accumulation of golgin and GRASPs relative to other proteins, at most accounted for 50% of these markers. This finding is more consistent with BFA-induced redistribution involving cycling of golgins and GRASPs through the ER followed by their subsequent accumulation in post-er structures such as the ERGIC (Miles et al., 2001; Ward et al., 2001). Based on these observations, it can be concluded that if matrix proteins act as a template for post-mitotic Golgi assembly, it must be a dynamic template capable of considerable self-assembly. However, the evidence with respect to the actual role of matrix proteins is mixed. Two golgins, golgin-45 and golgin-84, have a profound role in Golgi biogenesis as their knockdown causes Golgi breakdown (Short et al., 2001; Diao et al., 2003). Inhibition of transport causes Golgi collapse; so it is not yet clear whether the effects observed after golgin-45 and golgin-84 knockdown are because of transport or structural defects. Note that golgin-84 is not present in BFA remnants and therefore it is arguably not a matrix protein (Diao et al., 2003). Required roles for other golgins and GRASPs are either unknown or unlikely. The principle matrix protein, GM130, as well as giantin, appears to be dispensable for Golgi biogenesis (Puthenveedu and Linstedt, 2001; Kondylis and Rabouille, 2003; Vasile et al., 2003). It may be that, at least in the case of giantin and GM130, instead of playing a role as a template, these proteins facilitate trafficking in cells in which they are expressed. If this is the case, the in vitro Golgi stacking reaction may have created dependencies for these proteins, and maybe other components, which are not required in vivo. It is noteworthy that a major factor leading to the designation of matrix proteins as such, is their presence in BFA remnants. That is, matrix proteins may not represent a functional grouping, but rather a grouping based on targeting characteristics. If matrix proteins localize to BFA remnants by cycling through the ER rather than by staying intact in a matrix (Miles et al., 2001; Ward et al., 2001), then it is certainly possible that they may mediate diverse functions, only some of which are required for Golgi biogenesis. Mitotic Golgi clusters represent a small fraction of the Golgi (Zaal et al., 1999; Jesch et al., 2001) (this study), are not selectively enriched in any particular known Golgi component (this study), and exhibit dynamic behavior suggesting that they are not stable but rather have rapid interchange with dispersed Golgi vesicles (Jesch et al., 2001). At the EM level, mitotic Golgi clusters do not exhibit any distinguishable features of Golgi structure. Nevertheless, mitotic Golgi clusters exhibit evidence of Golgi subcompartmentalization (Shima et al., 1997) (this study). Inhibition of vesicle docking and fusion at mitosis may underlie a Golgi breakdown reaction that preserves the sorting reactions that take place to maintain the Golgi apparatus during interphase. Mitotic Golgi clusters may represent incomplete breakdown and/or incomplete reassembly. Although it seems reasonable that these structures participate in post-mitotic Golgi reassembly, there is no compelling evidence that they mediate the process. Rather, it is probable that, in accordance with recent work (Bevis et al., 2002; Puri and Linstedt, 2003) the Golgi apparatus is capable of self-assembly. We thank Tina Lee and lab members for helpful comments; Vivek Malhotra for anti-grasp antibodies; J. White for the GalNAcT2 cell line; Amy Csink for the use of the DeltaVision system; and Xiang Chen for help with the Matlab scripts. This work was supported by a National Institutes of Health grant GM to A.D.L. References Barr, F. A., Puype, M., Vandekerckhove, J. and Warren, G. (1997). GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91,
6 456 Journal of Cell Science 117 (3) Bevis, B. J., Hammond, A. T., Reinke, C. A. and Glick, B. S. (2002). De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat. Cell Biol. 4, Diao, A., Rahman, D., Pappin, D. J., Lucocq, J. and Lowe, M. (2003). The coiled-coil membrane protein golgin-84 is a novel rab effector required for Golgi ribbon formation. J. Cell Biol. 160, Dirac-Svejstrup, A. B., Shorter, J., Waters, M. G. and Warren, G. (2000). Phosphorylation of the vesicle-tethering protein p115 by a casein kinase IIlike enzyme is required for Golgi reassembly from isolated mitotic fragments. J. Cell Biol. 150, Jesch, S. A. (2002). Inheriting a structural scaffold for Golgi biosynthesis. BioEssays 24, Jesch, S. A. and Linstedt, A. D. (1998). The Golgi and endoplasmic reticulum remain independent during mitosis in HeLa cells. Mol. Biol. Cell 9, Jesch, S. A., Mehta, A. J., Velliste, M., Murphy, R. F. and Linstedt, A. D. (2001). Mitotic Golgi is in a dynamic equilibrium between clustered and free vesicles independent of the ER. Traffic 2, Jokitalo, E., Cabrera-Poch, N., Warren, G. and Shima, D. T. (2001). Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J. Cell Biol. 154, Kondylis, V. and Rabouille, C. (2003). A novel role for dp115 in the organization of ter sites in Drosophila. J. Cell Biol. 162, Levine, T. P., Rabouille, C., Kieckbusch, R. H. and Warren, G. (1996). Binding of the vesicle docking protein p115 to Golgi membranes is inhibited under mitotic conditions. J. Biol. Chem. 271, Linstedt, A. D. and Hauri, H. P. (1993). Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kda. Mol. Biol. Cell 4, Linstedt, A. D., Mehta, A., Suhan, J., Reggio, H. and Hauri, H. P. (1997). Sequence and overexpression of GPP130/GIMPc: evidence for saturable ph-sensitive targeting of a type II early Golgi membrane protein. Mol. Biol. Cell 8, Lucocq, J. M., Berger, E. G. and Warren, G. (1989). Mitotic Golgi fragments in HeLa cells and their role in the reassembly pathway. J. Cell Biol. 109, Miles, S., McManus, H., Forsten, K. E. and Storrie, B. (2001). Evidence that the entire Golgi apparatus cycles in interphase HeLa cells: sensitivity of Golgi matrix proteins to an ER exit block. J. Cell Biol. 155, Misteli, T. and Warren, G. (1995). Mitotic disassembly of the Golgi apparatus in vivo. J. Cell Sci. 108, Nakamura, N., Rabouille, C., Watson, R., Nilsson, T., Hui, N., Slusarewicz, P., Kreis, T. E. and Warren, G. (1995). Characterization of a cis-golgi matrix protein, GM130. J. Cell Biol. 131, Nelson, W. J. (2000). W(h)ither the Golgi during mitosis? J. Cell Biol. 149, Puri, S. and Linstedt, A. D. (2003). Capacity of the Golgi apparatus for biogenesis from the endoplasmic reticulum. Mol. Biol. Cell 14, Puri, S., Bachert, C., Fimmel, C. J. and Linstedt, A. D. (2002). Cycling of early Golgi proteins via the cell surface and endosomes upon lumenal ph disruption. Traffic 3, Puthenveedu, M. A. and Linstedt, A. D. (2001). Evidence that Golgi structure depends on a p115 activity that is independent of the vesicle tether components giantin and GM130. J. Cell Biol. 155, Rossanese, O. W. and Glick, B. S. (2001). Deconstructing Golgi inheritance. Traffic 2, Seemann, J., Jokitalo, E., Pypaert, M. and Warren, G. (2000). Matrix proteins can generate the higher order architecture of the Golgi apparatus. Nature 407, Seemann, J., Pypaert, M., Taguchi, T., Malsam, J. and Warren, G. (2002). Partitioning of the matrix fraction of the Golgi apparatus during mitosis in animal cells. Science 295, Shima, D. T., Haldar, K., Pepperkok, R., Watson, R. and Warren, G. (1997). Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J. Cell Biol. 137, Short, B. and Barr, F. A. (2000). The Golgi apparatus. Curr. Biol. 10, R583- R585. Short, B., Preisinger, C., Korner, R., Kopajtich, R., Byron, O. and Barr, F. A. (2001). A GRASP55-rab2 effector complex linking Golgi structure to membrane traffic. J. Cell Biol. 155, Shorter, J. and Warren, G. (1999). A role for the vesicle tethering protein, p115, in the post-mitotic stacking of reassembling Golgi cisternae in a cellfree system. J. Cell Biol. 146, Shorter, J. and Warren, G. (2002). Golgi architecture and inheritance. Annu. Rev. Cell Dev. Biol. 18, Shorter, J., Watson, R., Giannakou, M. E., Clarke, M., Warren, G. and Barr, F. A. (1999). GRASP55, a second mammalian GRASP protein involved in the stacking of Golgi cisternae in a cell-free system. EMBO J. 18, Slusarewicz, P., Nilsson, T., Hui, N., Watson, R. and Warren, G. (1994). Isolation of a matrix that binds medial Golgi enzymes. J. Cell Biol. 124, Storrie, B., White, J., Rottger, S., Stelzer, E. H., Suganuma, T. and Nilsson, T. (1998). Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J. Cell Biol. 143, Sutterlin, C., Hsu, P., Mallabiabarrena, A. and Malhotra, V. (2002). Fragmentation and dispersal of the pericentriolar Golgi complex is required for entry into mitosis in mammalian cells. Cell 109, Vasile, E., Perez, T., Nakamura, N. and Krieger, M. (2003). Structural integrity of the Golgi is temperature sensitive in conditional-lethal mutants with no detectable GM130. Traffic 4, Ward, T. H., Polishchuk, R. S., Caplan, S., Hirschberg, K. and Lippincott- Schwartz, J. (2001). Maintenance of Golgi structure and function depends on the integrity of ER export. J. Cell Biol. 155, Zaal, K. J., Smith, C. L., Polishchuk, R. S., Altan, N., Cole, N. B., Ellenberg, J., Hirschberg, K., Presley, J. F., Roberts, T. H., Siggia, E. et al. (1999). Golgi membranes are absorbed into and reemerge from the ER during mitosis. Cell 99,
Running title: Golgi enzymes in mitosis Key words: Golgi apparatus, endoplasmic reticulum, mitosis, Arf1, Sar1
 Golgi inheritance in mammalian cells is mediated through ER export activities Nihal Altan-Bonnet, Rachid Sougrat, Wei Liu, Erik L. Snapp, Theresa Ward* and Jennifer Lippincott-Schwartz # Cell biology and
Golgi inheritance in mammalian cells is mediated through ER export activities Nihal Altan-Bonnet, Rachid Sougrat, Wei Liu, Erik L. Snapp, Theresa Ward* and Jennifer Lippincott-Schwartz # Cell biology and
The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression
 Traffic 2010; 11: 827 842 2010 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2010.01055.x The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression Danming Tang, Hebao Yuan and Yanzhuang
Traffic 2010; 11: 827 842 2010 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2010.01055.x The Role of GRASP65 in Golgi Cisternal Stacking and Cell Cycle Progression Danming Tang, Hebao Yuan and Yanzhuang
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins
 13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
13-3. Synthesis-Secretory pathway: Sort lumenal proteins, Secrete proteins, Sort membrane proteins Molecular sorting: specific budding, vesicular transport, fusion 1. Why is this important? A. Form and
Protein Sorting, Intracellular Trafficking, and Vesicular Transport
 Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
Protein Sorting, Intracellular Trafficking, and Vesicular Transport Noemi Polgar, Ph.D. Department of Anatomy, Biochemistry and Physiology Email: polgar@hawaii.edu Phone: 692-1422 Outline Part 1- Trafficking
The mitotic cell must divide its nucleus and cytoplasm. Nuclear
 A role for Arf1 in mitotic Golgi disassembly, chromosome segregation, and cytokinesis Nihal Altan-Bonnet*, Robert D. Phair, Roman S. Polishchuk, Roberto Weigert, and Jennifer Lippincott-Schwartz* *Cell
A role for Arf1 in mitotic Golgi disassembly, chromosome segregation, and cytokinesis Nihal Altan-Bonnet*, Robert D. Phair, Roman S. Polishchuk, Roberto Weigert, and Jennifer Lippincott-Schwartz* *Cell
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
Mapping the functional domains of the Golgi stacking factor GRASP65
 JBC Papers in Press. Published on December 2, 2004 as Manuscript M412407200 Mapping the functional domains of the Golgi stacking factor GRASP65 Yanzhuang Wang*, Ayano Satoh and Graham Warren Department
JBC Papers in Press. Published on December 2, 2004 as Manuscript M412407200 Mapping the functional domains of the Golgi stacking factor GRASP65 Yanzhuang Wang*, Ayano Satoh and Graham Warren Department
Signaling pathways controlling mitotic Golgi breakdown in mammalian cells
 Golgi Apparatus: Structure, Functions and Mechanisms. Ed. C. J. Hawkins, NOVA Science Publishers, Hauppage, N.Y., USA. Signaling pathways controlling mitotic Golgi breakdown in mammalian cells Inmaculada
Golgi Apparatus: Structure, Functions and Mechanisms. Ed. C. J. Hawkins, NOVA Science Publishers, Hauppage, N.Y., USA. Signaling pathways controlling mitotic Golgi breakdown in mammalian cells Inmaculada
A Modeling Approach to the Self-Assembly of the Golgi Apparatus
 Biophysical Journal Volume 98 June 2010 2839 2847 2839 A Modeling Approach to the Self-Assembly of the Golgi Apparatus Jens Kühnle, Julian Shillcock, Ole G. Mouritsen, and Matthias Weiss * Cellular Biophysics
Biophysical Journal Volume 98 June 2010 2839 2847 2839 A Modeling Approach to the Self-Assembly of the Golgi Apparatus Jens Kühnle, Julian Shillcock, Ole G. Mouritsen, and Matthias Weiss * Cellular Biophysics
JCB. A novel role for dp115 in the organization of ter sites in Drosophila. The Journal of Cell Biology. Article
 JCB Published Online: 21 July, 2003 Supp Info: http://doi.org/10.1083/jcb.200301136 Downloaded from jcb.rupress.org on September 30, 2018 Article A novel role for dp115 in the organization of ter sites
JCB Published Online: 21 July, 2003 Supp Info: http://doi.org/10.1083/jcb.200301136 Downloaded from jcb.rupress.org on September 30, 2018 Article A novel role for dp115 in the organization of ter sites
Heather Currinn, Benjamin Guscott, Zita Balklava, Alice Rothnie and Thomas Wassmer*
 Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
CELB40060 Membrane Trafficking in Animal Cells. Prof. Jeremy C. Simpson. Lecture 2 COPII and export from the ER
 CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
CELB40060 Membrane Trafficking in Animal Cells Prof. Jeremy C. Simpson Lecture 2 COPII and export from the ER Today s lecture... The COPII coat - localisation and subunits Formation of the COPII coat at
We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists. International authors and editors
 We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists 3,900 116,000 120M Open access books available International authors and editors Downloads Our
We are IntechOpen, the world s leading publisher of Open Access books Built by scientists, for scientists 3,900 116,000 120M Open access books available International authors and editors Downloads Our
The Golgi posttranslationally modifies proteins and sorts them
 Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression Gianni Guizzunti a and Joachim Seemann a,1 a Department of Cell Biology, University of Texas Southwestern Medical
Mitotic Golgi disassembly is required for bipolar spindle formation and mitotic progression Gianni Guizzunti a and Joachim Seemann a,1 a Department of Cell Biology, University of Texas Southwestern Medical
The Golgi is a vital membrane-bound organelle
 Golgi Biogenesis Yanzhuang Wang 1 and Joachim Seemann 2 1 Department of Molecular, Cellular and Developmental Biology, University of Michigan, Ann Arbor, Michigan 48109 2 Department of Cell Biology, University
Golgi Biogenesis Yanzhuang Wang 1 and Joachim Seemann 2 1 Department of Molecular, Cellular and Developmental Biology, University of Michigan, Ann Arbor, Michigan 48109 2 Department of Cell Biology, University
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%%
 !"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
!"#$%&'%()*%+*,,%-&,./*%01%02%/*/3452*%3&.26%&4752*,,*1%% !"#$%&'(")*++*%,*'-&'./%/,*#01#%-2)#3&)/% 4'(")*++*% % %5"0)%-2)#3&) %%% %67'2#72'*%%%%%%%%%%%%%%%%%%%%%%%4'(")0/./% % 8$+&'&,+"/7 % %,$&7&/9)7$*/0/%%%%%%%%%%
Inheritance of the mammalian Golgi apparatus during the cell cycle
 Biochimica et Biophysica Acta 1404 (1998) 139^151 Inheritance of the mammalian Golgi apparatus during the cell cycle Noemi Cabrera-Poch a, Rainer Pepperkok b, David T. Shima a; * b a Cell Biology Laboratory,
Biochimica et Biophysica Acta 1404 (1998) 139^151 Inheritance of the mammalian Golgi apparatus during the cell cycle Noemi Cabrera-Poch a, Rainer Pepperkok b, David T. Shima a; * b a Cell Biology Laboratory,
Cdc2 Kinase-dependent Disassembly of Endoplasmic Reticulum (ER) Exit Sites Inhibits ER-to-Golgi D V
 Molecular Biology of the Cell Vol. 15, 4289 4298, September 2004 Cdc2 Kinase-dependent Disassembly of Endoplasmic Reticulum (ER) Exit Sites Inhibits ER-to-Golgi D V Vesicular Transport during Mitosis Fumi
Molecular Biology of the Cell Vol. 15, 4289 4298, September 2004 Cdc2 Kinase-dependent Disassembly of Endoplasmic Reticulum (ER) Exit Sites Inhibits ER-to-Golgi D V Vesicular Transport during Mitosis Fumi
An Approach To Modeling Tracer Experiments In Metabolic Non-Steady States Robert Phair Integrative Bioinformatics Inc.
 An Approach To Modeling Tracer Experiments In Metabolic Non-Steady States Robert Phair Integrative Bioinformatics Inc. Los Altos, CA integrativebioinformatics.com Starting point: A simple mechanistic model
An Approach To Modeling Tracer Experiments In Metabolic Non-Steady States Robert Phair Integrative Bioinformatics Inc. Los Altos, CA integrativebioinformatics.com Starting point: A simple mechanistic model
Discrete, continuous, and stochastic models of protein sorting in the Golgi apparatus.
 Carnegie Mellon University Research Showcase @ CMU Department of Biological Sciences Mellon College of Science -- Discrete, continuous, and stochastic models of protein sorting in the Golgi apparatus.
Carnegie Mellon University Research Showcase @ CMU Department of Biological Sciences Mellon College of Science -- Discrete, continuous, and stochastic models of protein sorting in the Golgi apparatus.
Chapter 2 Cells and Cell Division
 Chapter 2 Cells and Cell Division MULTIPLE CHOICE 1. The process of meiosis results in: A. the production of four identical cells B. no change in chromosome number from parental cells C. a doubling of
Chapter 2 Cells and Cell Division MULTIPLE CHOICE 1. The process of meiosis results in: A. the production of four identical cells B. no change in chromosome number from parental cells C. a doubling of
arxiv: v2 [physics.bio-ph] 2 Jan 2018
![arxiv: v2 [physics.bio-ph] 2 Jan 2018 arxiv: v2 [physics.bio-ph] 2 Jan 2018](/thumbs/82/86584475.jpg) Stochastic Model of Vesicular Sorting in Cellular Organelles Quentin Vagne, Pierre Sens Institut Curie, PSL Research University, CNRS, UMR 168, 26 rue d Ulm, F-75005, Paris, France. (Dated: January 3,
Stochastic Model of Vesicular Sorting in Cellular Organelles Quentin Vagne, Pierre Sens Institut Curie, PSL Research University, CNRS, UMR 168, 26 rue d Ulm, F-75005, Paris, France. (Dated: January 3,
Multi-step down-regulation of the secretory pathway in mitosis: A fresh perspective on protein trafficking. Introduction
 Prospects & Overviews Multi-step down-regulation of the secretory pathway in mitosis: A fresh perspective on protein trafficking Foong May Yeong The secretory pathway delivers proteins synthesized at the
Prospects & Overviews Multi-step down-regulation of the secretory pathway in mitosis: A fresh perspective on protein trafficking Foong May Yeong The secretory pathway delivers proteins synthesized at the
Cell Biology Review. The key components of cells that concern us are as follows: 1. Nucleus
 Cell Biology Review Development involves the collective behavior and activities of cells, working together in a coordinated manner to construct an organism. As such, the regulation of development is intimately
Cell Biology Review Development involves the collective behavior and activities of cells, working together in a coordinated manner to construct an organism. As such, the regulation of development is intimately
16 The Cell Cycle. Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization
 The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
Selective Targeting of ER Exit Sites Supports Axon Development
 Traffic 2009; 10: 1669 1684 2009 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2009.00974.x Selective Targeting of ER Exit Sites Supports Axon Development Meir Aridor 1 and Kenneth N. Fish 2, 1 Department
Traffic 2009; 10: 1669 1684 2009 John Wiley & Sons A/S doi:10.1111/j.1600-0854.2009.00974.x Selective Targeting of ER Exit Sites Supports Axon Development Meir Aridor 1 and Kenneth N. Fish 2, 1 Department
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
SUPPLEMENTARY INFORMATION
 Supplementary Notes Downregulation of atlastin does not affect secretory traffic We investigated whether Atlastin might play a role in secretory traffic. Traffic impairment results in disruption of Golgi
Supplementary Notes Downregulation of atlastin does not affect secretory traffic We investigated whether Atlastin might play a role in secretory traffic. Traffic impairment results in disruption of Golgi
A model for the self-organization of exit sites in the endoplasmic reticulum
 Research Article 55 A model for the self-organization of exit sites in the endoplasmic reticulum Stephan Heinzer 1, Stefan Wörz 2,3, Claudia Kalla 1, Karl Rohr 2,3 and Matthias Weiss 1, * 1 Cellular Biophysics
Research Article 55 A model for the self-organization of exit sites in the endoplasmic reticulum Stephan Heinzer 1, Stefan Wörz 2,3, Claudia Kalla 1, Karl Rohr 2,3 and Matthias Weiss 1, * 1 Cellular Biophysics
The multiple facets of the Golgi reassembly stacking proteins
 Biochem. J. (2011) 433, 423 433 (Printed in Great Britain) doi:10.1042/bj20101540 423 REVIEW ARTICLE The multiple facets of the Golgi reassembly stacking proteins Fabian P. VINKE*, Adam G. GRIEVE* and
Biochem. J. (2011) 433, 423 433 (Printed in Great Britain) doi:10.1042/bj20101540 423 REVIEW ARTICLE The multiple facets of the Golgi reassembly stacking proteins Fabian P. VINKE*, Adam G. GRIEVE* and
Supplementary Information
 Supplementary Information MAP2/Hoechst Hyp.-AP ph 6.5 Hyp.-SD ph 7.2 Norm.-SD ph 7.2 Supplementary Figure 1. Mitochondrial elongation in cortical neurons by acidosis. Representative images of neuronal
Supplementary Information MAP2/Hoechst Hyp.-AP ph 6.5 Hyp.-SD ph 7.2 Norm.-SD ph 7.2 Supplementary Figure 1. Mitochondrial elongation in cortical neurons by acidosis. Representative images of neuronal
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09414 Supplementary Figure 1 FACS-isolated 8c hepatocytes are highly pure. a, Gating strategy for identifying hepatocyte populations based on DNA content. b, Detection of mchry and mchr9
doi:10.1038/nature09414 Supplementary Figure 1 FACS-isolated 8c hepatocytes are highly pure. a, Gating strategy for identifying hepatocyte populations based on DNA content. b, Detection of mchry and mchr9
Division Ave. High School AP Biology
 Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
Tour of the Cell 1 Types of cells Prokaryote bacteria cells - no organelles - organelles Eukaryote animal cells Eukaryote plant cells Why organelles? Specialized structures u specialized functions cilia
REVIEW 2: CELLS & CELL DIVISION UNIT. A. Top 10 If you learned anything from this unit, you should have learned:
 Period Date REVIEW 2: CELLS & CELL DIVISION UNIT A. Top 10 If you learned anything from this unit, you should have learned: 1. Prokaryotes vs. eukaryotes No internal membranes vs. membrane-bound organelles
Period Date REVIEW 2: CELLS & CELL DIVISION UNIT A. Top 10 If you learned anything from this unit, you should have learned: 1. Prokaryotes vs. eukaryotes No internal membranes vs. membrane-bound organelles
Introduction to Cells
 Life Science Introduction to Cells All life forms on our planet are made up of cells. In ALL organisms, cells have the same basic structure. The scientist Robert Hooke was the first to see cells under
Life Science Introduction to Cells All life forms on our planet are made up of cells. In ALL organisms, cells have the same basic structure. The scientist Robert Hooke was the first to see cells under
BIOLOGY. Chapter 10 CELL REPRODUCTION PowerPoint Image Slideshow
 BIOLOGY Chapter 10 CELL REPRODUCTION PowerPoint Image Slideshow FIGURE 10.1 A sea urchin begins life as a single cell that (a) divides to form two cells, visible by scanning electron microscopy. After
BIOLOGY Chapter 10 CELL REPRODUCTION PowerPoint Image Slideshow FIGURE 10.1 A sea urchin begins life as a single cell that (a) divides to form two cells, visible by scanning electron microscopy. After
The neuron as a secretory cell
 The neuron as a secretory cell EXOCYTOSIS ENDOCYTOSIS The secretory pathway. Transport and sorting of proteins in the secretory pathway occur as they pass through the Golgi complex before reaching the
The neuron as a secretory cell EXOCYTOSIS ENDOCYTOSIS The secretory pathway. Transport and sorting of proteins in the secretory pathway occur as they pass through the Golgi complex before reaching the
Table S1. Aspergillus nidulans strains used in this study Strain Genotype Derivation
 Supplemental Material De Souza et al., 211 Table S1. Aspergillus nidulans strains used in this study Strain Genotype Derivation CDS295 pyrg89; pyroa4; pyrg Af ::son promotor::gfp-son nup98/nup96 ; chaa1
Supplemental Material De Souza et al., 211 Table S1. Aspergillus nidulans strains used in this study Strain Genotype Derivation CDS295 pyrg89; pyroa4; pyrg Af ::son promotor::gfp-son nup98/nup96 ; chaa1
Mammalian GRIP domain proteins differ in their membrane binding properties and are recruited to distinct domains of the TGN
 Research Article 5865 Mammalian GRIP domain proteins differ in their membrane binding properties and are recruited to distinct domains of the TGN Merran C. Derby 1, Catherine van Vliet 1, Darren Brown
Research Article 5865 Mammalian GRIP domain proteins differ in their membrane binding properties and are recruited to distinct domains of the TGN Merran C. Derby 1, Catherine van Vliet 1, Darren Brown
Hybrid Gold Superstructures: Synthesis and. Specific Cell Surface Protein Imaging Applications
 Supporting Information Hybrid Gold Nanocube@Silica@Graphene-Quantum-Dot Superstructures: Synthesis and Specific Cell Surface Protein Imaging Applications Liu Deng, Ling Liu, Chengzhou Zhu, Dan Li and Shaojun
Supporting Information Hybrid Gold Nanocube@Silica@Graphene-Quantum-Dot Superstructures: Synthesis and Specific Cell Surface Protein Imaging Applications Liu Deng, Ling Liu, Chengzhou Zhu, Dan Li and Shaojun
A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces cerevisiae.
 Genetics: Published Articles Ahead of Print, published on July 30, 2012 as 10.1534/genetics.112.143818 A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces
Genetics: Published Articles Ahead of Print, published on July 30, 2012 as 10.1534/genetics.112.143818 A redundant function for the N-terminal tail of Ndc80 in kinetochore-microtubule interaction in Saccharomyces
Introduction to Cells
 Life Science Introduction to Cells All life forms on our planet are made up of cells. In ALL organisms, cells have the same basic structure. The scientist Robert Hooke was the first to see cells under
Life Science Introduction to Cells All life forms on our planet are made up of cells. In ALL organisms, cells have the same basic structure. The scientist Robert Hooke was the first to see cells under
A Few Terms: When and where do you want your cells to divide?
 Today: - Lab 4 Debrief - Mitosis - Lunch -Meiosis Other: Blood Drive Today! TIME: 11:00am 1:00pm + 2:00pm 5:00pm PLACE: Baxter Events Center Thinking About Mitosis When and where do you want your cells
Today: - Lab 4 Debrief - Mitosis - Lunch -Meiosis Other: Blood Drive Today! TIME: 11:00am 1:00pm + 2:00pm 5:00pm PLACE: Baxter Events Center Thinking About Mitosis When and where do you want your cells
The Cell. C h a p t e r. PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas
 C h a p t e r 2 The Cell PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas Copyright 2009 Pearson Education, Inc., publishing as Pearson Benjamin Cummings Introduction
C h a p t e r 2 The Cell PowerPoint Lecture Slides prepared by Jason LaPres North Harris College Houston, Texas Copyright 2009 Pearson Education, Inc., publishing as Pearson Benjamin Cummings Introduction
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and
 Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
NucView TM 488 Caspase-3 Assay Kit for Live Cells
 NucView TM 488 Caspase-3 Assay Kit for Live Cells Catalog Number: 30029 (100-500 assays) Contact Information Address: Biotium, Inc. 3423 Investment Blvd. Suite 8 Hayward, CA 94545 USA Telephone: (510)
NucView TM 488 Caspase-3 Assay Kit for Live Cells Catalog Number: 30029 (100-500 assays) Contact Information Address: Biotium, Inc. 3423 Investment Blvd. Suite 8 Hayward, CA 94545 USA Telephone: (510)
Golgin160 Recruits the Dynein Motor to Position the Golgi Apparatus
 Article Golgin160 Recruits the Dynein Motor to Position the Golgi Apparatus Smita Yadav, 1 Manojkumar A. Puthenveedu, 1 and Adam D. Linstedt 1, * 1 Department of Biological Sciences, Carnegie Mellon University,
Article Golgin160 Recruits the Dynein Motor to Position the Golgi Apparatus Smita Yadav, 1 Manojkumar A. Puthenveedu, 1 and Adam D. Linstedt 1, * 1 Department of Biological Sciences, Carnegie Mellon University,
SUPPLEMENTARY INFORMATION
 GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
Richik N. Ghosh, Linnette Grove, and Oleg Lapets ASSAY and Drug Development Technologies 2004, 2:
 1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
3.a.2- Cell Cycle and Meiosis
 Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. 3.a.2- Cell Cycle and Meiosis EU 3.A: Heritable information provides for continuity of life.
Big Idea 3: Living systems store, retrieve, transmit and respond to information essential to life processes. 3.a.2- Cell Cycle and Meiosis EU 3.A: Heritable information provides for continuity of life.
Biology. 7-2 Eukaryotic Cell Structure 10/29/2013. Eukaryotic Cell Structures
 Biology Biology 1of 49 2of 49 Eukaryotic Cell Structures Eukaryotic Cell Structures Structures within a eukaryotic cell that perform important cellular functions are known as organelles. Cell biologists
Biology Biology 1of 49 2of 49 Eukaryotic Cell Structures Eukaryotic Cell Structures Structures within a eukaryotic cell that perform important cellular functions are known as organelles. Cell biologists
7.06 Cell Biology EXAM #3 April 21, 2005
 7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
7.06 Cell Biology EXAM #3 April 21, 2005 This is an open book exam, and you are allowed access to books, a calculator, and notes but not computers or any other types of electronic devices. Please write
The Microscopic Observation of Mitosis in Plant and Animal Cells
 The Microscopic Observation of Mitosis in Plant and Animal Cells Prelab Assignment Before coming to lab, read carefully the introduction and the procedures for each part of the experiment, and then answer
The Microscopic Observation of Mitosis in Plant and Animal Cells Prelab Assignment Before coming to lab, read carefully the introduction and the procedures for each part of the experiment, and then answer
Cells and Their Organelles
 Mr. Ulrich Regents Biology Name:.. Cells and Their Organelles The cell is the basic unit of life. The following is a glossary of animal cell terms. All cells are surrounded by a cell membrane. The cell
Mr. Ulrich Regents Biology Name:.. Cells and Their Organelles The cell is the basic unit of life. The following is a glossary of animal cell terms. All cells are surrounded by a cell membrane. The cell
Cdc2 Kinase Directly Phosphorylates the cis-golgi Matrix Protein GM130 and Is Required for Golgi Fragmentation in Mitosis
 Cell, Vol. 94, 783 793, September 18, 1998, Copyright 1998 by Cell Press Cdc2 Kinase Directly Phosphorylates the cis-golgi Matrix Protein GM130 and Is Required for Golgi Fragmentation in Mitosis Martin
Cell, Vol. 94, 783 793, September 18, 1998, Copyright 1998 by Cell Press Cdc2 Kinase Directly Phosphorylates the cis-golgi Matrix Protein GM130 and Is Required for Golgi Fragmentation in Mitosis Martin
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis.
 Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Big Idea 2: Biological systems utilize free energy and molecular building blocks to grow, to reproduce and to maintain dynamic homeostasis. Enduring understanding 2.B: Growth, reproduction and dynamic
Guided Reading Activities
 Name Period Chapter 4: A Tour of the Cell Guided Reading Activities Big Idea: Introduction to the Cell Answer the following questions as you read Modules 4.1 4.4: 1. A(n) uses a beam of light to illuminate
Name Period Chapter 4: A Tour of the Cell Guided Reading Activities Big Idea: Introduction to the Cell Answer the following questions as you read Modules 4.1 4.4: 1. A(n) uses a beam of light to illuminate
Name: TF: Section Time: LS1a ICE 5. Practice ICE Version B
 Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Name: TF: Section Time: LS1a ICE 5 Practice ICE Version B 1. (8 points) In addition to ion channels, certain small molecules can modulate membrane potential. a. (4 points) DNP ( 2,4-dinitrophenol ), as
Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
 Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry 1. The study of cells has been limited by their small size, and so they were
Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry 1. The study of cells has been limited by their small size, and so they were
Dynamics of the Endoplasmic Reticulum and Golgi Apparatus during Early Sea Urchin Development
 Molecular Biology of the Cell Vol. 11, 897 914, March 2000 Dynamics of the Endoplasmic Reticulum and Golgi Apparatus during Early Sea Urchin Development Mark Terasaki* Department of Physiology, University
Molecular Biology of the Cell Vol. 11, 897 914, March 2000 Dynamics of the Endoplasmic Reticulum and Golgi Apparatus during Early Sea Urchin Development Mark Terasaki* Department of Physiology, University
ACCELERATE ITS BIOCHEMICAL PROCESSES WHICH WERE SLOWED DOWN BY MITOSIS. THE LENGTH OF THE G1 PHASE CREATES THE DIFFERENCE BETWEEN FAST DIVIDING
 CHAPTER 1: OVERVIEW OF THE CELL CYCLE THE THREE STAGES OF INTERPHASE: INTERPHASE BEFORE A CELL CAN ENTER CELL DIVISION, IT NEEDS TO PREPARE ITSELF BY REPLICATING ITS GENETIC INFORMATION AND ALL OF THE
CHAPTER 1: OVERVIEW OF THE CELL CYCLE THE THREE STAGES OF INTERPHASE: INTERPHASE BEFORE A CELL CAN ENTER CELL DIVISION, IT NEEDS TO PREPARE ITSELF BY REPLICATING ITS GENETIC INFORMATION AND ALL OF THE
In order to confirm the contribution of diffusion to the FRAP recovery curves of
 Fonseca et al., Supplementary Information FRAP data analysis ) Contribution of Diffusion to the recovery curves In order to confirm the contribution of diffusion to the FRAP recovery curves of PH::GFP
Fonseca et al., Supplementary Information FRAP data analysis ) Contribution of Diffusion to the recovery curves In order to confirm the contribution of diffusion to the FRAP recovery curves of PH::GFP
7.06 Spring 2004 PS 6 KEY 1 of 14
 7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
7.06 Spring 2004 PS 6 KEY 1 of 14 Problem Set 6. Question 1. You are working in a lab that studies hormones and hormone receptors. You are tasked with the job of characterizing a potentially new hormone
Chapter 6: A Tour of the Cell
 Chapter 6: A Tour of the Cell 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when Robert Hooke first looked at dead cells from an oak tree.
Chapter 6: A Tour of the Cell 1. The study of cells has been limited by their small size, and so they were not seen and described until 1665, when Robert Hooke first looked at dead cells from an oak tree.
Tiffany Samaroo MB&B 452a December 8, Take Home Final. Topic 1
 Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Tiffany Samaroo MB&B 452a December 8, 2003 Take Home Final Topic 1 Prior to 1970, protein and DNA sequence alignment was limited to visual comparison. This was a very tedious process; even proteins with
Name: Date: Hour:
 Name: Date: Hour: 1 2 3 4 5 6 Comprehension Questions 1. At what level of organization does life begin? 2. What surrounds all cells? 3. What is meant by semipermeable? 4. What 2 things make up the cell
Name: Date: Hour: 1 2 3 4 5 6 Comprehension Questions 1. At what level of organization does life begin? 2. What surrounds all cells? 3. What is meant by semipermeable? 4. What 2 things make up the cell
2:1 Chromosomes DNA Genes Chromatin Chromosomes CHROMATIN: nuclear material in non-dividing cell, composed of DNA/protein in thin uncoiled strands
 Human Heredity Chapter 2 Chromosomes, Mitosis, and Meiosis 2:1 Chromosomes DNA Genes Chromatin Chromosomes CHROMATIN: nuclear material in non-dividing cell, composed of DNA/protein in thin uncoiled strands
Human Heredity Chapter 2 Chromosomes, Mitosis, and Meiosis 2:1 Chromosomes DNA Genes Chromatin Chromosomes CHROMATIN: nuclear material in non-dividing cell, composed of DNA/protein in thin uncoiled strands
Supplemental Information: Govindaraghavan, Lad and Osmani
 Supplemental Information: Govindaraghavan, Lad and Osmani Table S1: List of strains used in this study Strain Name Genotype (all strains also carry vea1) Source MG190 nima7; pyrg89; argb2; ndc80-cr::pyroa
Supplemental Information: Govindaraghavan, Lad and Osmani Table S1: List of strains used in this study Strain Name Genotype (all strains also carry vea1) Source MG190 nima7; pyrg89; argb2; ndc80-cr::pyroa
7.06 Problem Set #4, Spring 2005
 7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
7.06 Problem Set #4, Spring 2005 1. You re doing a mutant hunt in S. cerevisiae (budding yeast), looking for temperaturesensitive mutants that are defective in the cell cycle. You discover a mutant strain
O.k., Now Starts the Good Stuff (Part II) Eukaryotic Cell Structure and Function
 O.k., Now Starts the Good Stuff (Part II) Eukaryotic Cell Structure and Function Eukaryotic Cells These cells have membrane-bound structures called organelles. Cell processes occur in these organelles.
O.k., Now Starts the Good Stuff (Part II) Eukaryotic Cell Structure and Function Eukaryotic Cells These cells have membrane-bound structures called organelles. Cell processes occur in these organelles.
Biology of Fungi. Fungal Structure and Function. Lecture: Structure/Function, Part A BIOL 4848/ Fall Overview of the Hypha
 Biology of Fungi Fungal Structure and Function Overview of the Hypha The hypha is a rigid tube containing cytoplasm Growth occurs at the tips of hyphae Behind the tip, the cell is aging Diagram of hyphal
Biology of Fungi Fungal Structure and Function Overview of the Hypha The hypha is a rigid tube containing cytoplasm Growth occurs at the tips of hyphae Behind the tip, the cell is aging Diagram of hyphal
Chapter 6: A Tour of the Cell
 AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
AP Biology Reading Guide Fred and Theresa Holtzclaw Chapter 6: A Tour of the Cell Name Period Chapter 6: A Tour of the Cell Concept 6.1 To study cells, biologists use microscopes and the tools of biochemistry
Organelles in Eukaryotic Cells
 Why? Organelles in Eukaryotic Cells What are the functions of different organelles in a cell? The cell is the basic unit and building block of all living things. Organisms rely on their cells to perform
Why? Organelles in Eukaryotic Cells What are the functions of different organelles in a cell? The cell is the basic unit and building block of all living things. Organisms rely on their cells to perform
AS Biology Summer Work 2015
 AS Biology Summer Work 2015 You will be following the OCR Biology A course and in preparation for this you are required to do the following for September 2015: Activity to complete Date done Purchased
AS Biology Summer Work 2015 You will be following the OCR Biology A course and in preparation for this you are required to do the following for September 2015: Activity to complete Date done Purchased
Transport between cytosol and nucleus
 of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
of 60 3 Gated trans Lectures 9-15 MBLG 2071 The n GATED TRANSPORT transport between cytoplasm and nucleus (bidirectional) controlled by the nuclear pore complex active transport for macro molecules e.g.
THE CELL 3/15/15 HUMAN ANATOMY AND PHYSIOLOGY I THE CELLULAR BASIS OF LIFE
 HUMAN ANATOMY AND PHYSIOLOGY I Lecture: M 6-9:30 Randall Visitor Center Lab: W 6-9:30 Swatek Anatomy Center, Centennial Complex Required Text: Marieb 9 th edition Dr. Trevor Lohman DPT (949) 246-5357 tlohman@llu.edu
HUMAN ANATOMY AND PHYSIOLOGY I Lecture: M 6-9:30 Randall Visitor Center Lab: W 6-9:30 Swatek Anatomy Center, Centennial Complex Required Text: Marieb 9 th edition Dr. Trevor Lohman DPT (949) 246-5357 tlohman@llu.edu
Regulation and signaling. Overview. Control of gene expression. Cells need to regulate the amounts of different proteins they express, depending on
 Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
Regulation and signaling Overview Cells need to regulate the amounts of different proteins they express, depending on cell development (skin vs liver cell) cell stage environmental conditions (food, temperature,
REVIEW 2: CELLS & CELL COMMUNICATION. A. Top 10 If you learned anything from this unit, you should have learned:
 Name AP Biology REVIEW 2: CELLS & CELL COMMUNICATION A. Top 10 If you learned anything from this unit, you should have learned: 1. Prokaryotes vs. eukaryotes No internal membranes vs. membrane-bound organelles
Name AP Biology REVIEW 2: CELLS & CELL COMMUNICATION A. Top 10 If you learned anything from this unit, you should have learned: 1. Prokaryotes vs. eukaryotes No internal membranes vs. membrane-bound organelles
Cell Structure and Function. The Development of Cell Theory
 Cell Structure and Function The Development of Cell Theory Hooke's original microscope, that he used at Oxford University, in 1665. Robert Hooke first examined a thin piece of cork (see below), and coined
Cell Structure and Function The Development of Cell Theory Hooke's original microscope, that he used at Oxford University, in 1665. Robert Hooke first examined a thin piece of cork (see below), and coined
Measuring Mitochondrial Membrane Potential with JC-1 Using the Cellometer Vision Image Cytometer
 Measuring Mitochondrial Membrane Potential with JC-1 Using the Cellometer Vision Image Cytometer Nexcelom Bioscience LLC. 360 Merrimack Street, Building 9 Lawrence, MA 01843 T: 978.327.5340 F: 978.327.5341
Measuring Mitochondrial Membrane Potential with JC-1 Using the Cellometer Vision Image Cytometer Nexcelom Bioscience LLC. 360 Merrimack Street, Building 9 Lawrence, MA 01843 T: 978.327.5340 F: 978.327.5341
Department of Cell Biology, Heidelberg Institute for Plant Sciences, University of Heidelberg, Heidelberg, Germany b
 The Plant Cell, Vol. 17, 1513 1531, May 2005, www.plantcell.org ª 2005 American Society of Plant Biologists Dynamics of COPII Vesicles and the Golgi Apparatus in Cultured Nicotiana tabacum BY-2 Cells Provides
The Plant Cell, Vol. 17, 1513 1531, May 2005, www.plantcell.org ª 2005 American Society of Plant Biologists Dynamics of COPII Vesicles and the Golgi Apparatus in Cultured Nicotiana tabacum BY-2 Cells Provides
Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER
 2924 Research Article Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER Helen Hughes 1, *, Annika Budnik 1, *, Katy Schmidt 1, *, Krysten J. Palmer 1, Judith
2924 Research Article Organisation of human ER-exit sites: requirements for the localisation of Sec16 to transitional ER Helen Hughes 1, *, Annika Budnik 1, *, Katy Schmidt 1, *, Krysten J. Palmer 1, Judith
N-acetylglucosaminyltransferase I in retention, kin recognition and structural
 Journal of Cell Science 109, 1975-1989 (1996) Printed in Great Britain The Company of Biologists Limited 1996 JCS4194 1975 The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase
Journal of Cell Science 109, 1975-1989 (1996) Printed in Great Britain The Company of Biologists Limited 1996 JCS4194 1975 The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase
GFP WxTP-GFP WxTP-GFP. stromules. DsRed WxTP-DsRed WxTP-DsRed. stromules
 Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Supplemental Data. Kitajima et al. (2009). The rice -amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. A GFP WxTP-GFP WxTP-GFP stromules stromules
Chapter 4. Table of Contents. Section 1 The History of Cell Biology. Section 2 Introduction to Cells. Section 3 Cell Organelles and Features
 Cell Structure and Function Table of Contents Section 1 The History of Cell Biology Section 2 Introduction to Cells Section 3 Cell Organelles and Features Section 4 Unique Features of Plant Cells Section
Cell Structure and Function Table of Contents Section 1 The History of Cell Biology Section 2 Introduction to Cells Section 3 Cell Organelles and Features Section 4 Unique Features of Plant Cells Section
The Discovery of Cells
 The Discovery of Cells Microscope observations! General Cell & Organelle Discovery 1600s Observations made by scientists using more powerful microscopes in the 1800s led to the formation of the cell theory.
The Discovery of Cells Microscope observations! General Cell & Organelle Discovery 1600s Observations made by scientists using more powerful microscopes in the 1800s led to the formation of the cell theory.
Stochastic Model of Maturation and Vesicular Exchange in Cellular Organelles
 Stochastic Model of Maturation and Vesicular Exchange in Cellular Organelles arxiv:1606.01075v2 [physics.bio-ph] 19 Dec 2017 Running title: Maturation and Exchange in Organelles Quentin Vagne and Pierre
Stochastic Model of Maturation and Vesicular Exchange in Cellular Organelles arxiv:1606.01075v2 [physics.bio-ph] 19 Dec 2017 Running title: Maturation and Exchange in Organelles Quentin Vagne and Pierre
Organelles in Eukaryotic Cells
 Why? Organelles in Eukaryotic Cells What are the functions of different organelles in a cell? The cell is the basic unit and building block of all living things. Organisms rely on their cells to perform
Why? Organelles in Eukaryotic Cells What are the functions of different organelles in a cell? The cell is the basic unit and building block of all living things. Organisms rely on their cells to perform
DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS. Scientific Background on the Nobel Prize in Medicine 2013
 DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS Scientific Background on the Nobel Prize in Medicine 2013 Daniela Scalet 6/12/2013 The Nobel Prize in Medicine
DISCOVERIES OF MACHINERY REGULATING VESICLE TRAFFIC, A MAJOR TRANSPORT SYSTEM IN OUR CELLS Scientific Background on the Nobel Prize in Medicine 2013 Daniela Scalet 6/12/2013 The Nobel Prize in Medicine
Supplementary information
 Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity to ER exit sites
 Journal of Cell Science 113, 2177-2185 (2000) Printed in Great Britain The Company of Biologists Limited 2000 JCS1368 2177 COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity
Journal of Cell Science 113, 2177-2185 (2000) Printed in Great Britain The Company of Biologists Limited 2000 JCS1368 2177 COPI-coated ER-to-Golgi transport complexes segregate from COPII in close proximity
Imaging the secretory pathway: The past and future impact of live cell optical techniques
 Biochimica et Biophysica Acta 1744 (2005) 259 272 Review Imaging the secretory pathway: The past and future impact of live cell optical techniques John F. Presley* McGill University, Department of Anatomy
Biochimica et Biophysica Acta 1744 (2005) 259 272 Review Imaging the secretory pathway: The past and future impact of live cell optical techniques John F. Presley* McGill University, Department of Anatomy
Lecture Series 5 Cell Cycle & Cell Division
 Lecture Series 5 Cell Cycle & Cell Division Reading Assignments Read Chapter 18 Cell Cycle & Cell Division Read Chapter 19 pages 651-663 663 only (Benefits of Sex & Meiosis sections these are in Chapter
Lecture Series 5 Cell Cycle & Cell Division Reading Assignments Read Chapter 18 Cell Cycle & Cell Division Read Chapter 19 pages 651-663 663 only (Benefits of Sex & Meiosis sections these are in Chapter
Cells and Their Organelles
 Cells and Their Organelles The cell is the basic unit of life. The following is a glossary of animal cell terms. All cells are surrounded by a cell membrane. The cell membrane is semipermeable, allowing
Cells and Their Organelles The cell is the basic unit of life. The following is a glossary of animal cell terms. All cells are surrounded by a cell membrane. The cell membrane is semipermeable, allowing
7-1 Life Is Cellular. Copyright Pearson Prentice Hall
 7-1 Life Is Cellular The Discovery of the Cell What is the cell theory? The Discovery of the Cell The cell theory states: All living things are composed of cells. Cells are the basic units of structure
7-1 Life Is Cellular The Discovery of the Cell What is the cell theory? The Discovery of the Cell The cell theory states: All living things are composed of cells. Cells are the basic units of structure
Study Guide 11 & 12 MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question.
 Study Guide 11 & 12 MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) The receptors for a group of signaling molecules known as growth factors are
Study Guide 11 & 12 MULTIPLE CHOICE. Choose the one alternative that best completes the statement or answers the question. 1) The receptors for a group of signaling molecules known as growth factors are
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Science 9 Biology. Cell Division and Reproduction Booklet 1 M. Roberts RC Palmer
 Science 9 Biology Cell Division and Reproduction Booklet 1 M. Roberts RC Palmer How do all living organisms reproduce and grow? Goal 1: Cell Review Recall and become reacquainted with the structures found
Science 9 Biology Cell Division and Reproduction Booklet 1 M. Roberts RC Palmer How do all living organisms reproduce and grow? Goal 1: Cell Review Recall and become reacquainted with the structures found
Under the Radar Screen: How Bugs Trick Our Immune Defenses
 Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Under the Radar Screen: How Bugs Trick Our Immune Defenses Session 2: Phagocytosis Marie-Eve Paquet and Gijsbert Grotenbreg Whitehead Institute for Biomedical Research Salmonella Gram negative bacteria
Inhibition of Cell-Plate Formation by Brefeldin A Inhibited the Depolymerization of Microtubules in the Central Region of the Phragmoplast
 Plant CellPhysiol. 41(3): 300-310 (2000) JSPP 2000 Inhibition of Cell-Plate Formation by Brefeldin A Inhibited the Depolymerization of Microtubules in the Central Region of the Phragmoplast Hiroki Yasuhara
Plant CellPhysiol. 41(3): 300-310 (2000) JSPP 2000 Inhibition of Cell-Plate Formation by Brefeldin A Inhibited the Depolymerization of Microtubules in the Central Region of the Phragmoplast Hiroki Yasuhara
