Toxicogenomics. S. Amala
|
|
|
- Denis Bishop
- 6 years ago
- Views:
Transcription
1 Journal of Bioinformatics and Sequence Analysis Vol. 2(4), pp , August 2010 Available online at DOI: /JBSA ISSN Academic Journals Review Toxicogenomics S. Amala Department of Biotechnology and Bioinformatics, Dhanalakshmi Srinivasan, College of Arts and Science for Women, Perambalur , Tamil Nadu, India. Accepted 21 June, 2010 The field of toxicology is defined as the study of stressors and their adverse effects. One discipline should deals with hazard identification, mechanistic toxicology, and risk assessment. Thus emerge a new field called toxicogenomics. Toxicogenomics is a rapidly developing discipline that promises to aid scientists in understanding the molecular and cellular effects of chemicals in biological systems. This is a comparatively new field of biological inquiry now providing insights into the toxic effects of chemicals on biological systems and helping investigators to predict risks associated with exposure to these agents. This field encompasses global assessment of biological effects using technologies such as DNA micro arrays or high throughput NMR and protein expression analysis. Key words: Genomics, toxicogenomics, toxicology. INTRODUCTION The rapid evolution of genome-based technologies has greatly accelerated the application of gene expression profiling in toxicology studies. These technological advances have led to the development of the field of toxicogenomics, which proposes to apply mrna expression technologies to study effects of hazards in biological sys-tems. Application of genomics to toxicology, toxicogeno-mics, yield a number of substantial dividends, including assisting predevelopment toxicology by facilitating more rapid screens for compound toxicity, allowing compound selection decisions to be based on safety as well as efficacy, the provision of new research leads; a more detailed appreciation of molecular mechanisms of toxicity, and an enhanced ability to extrapolate accurately between experimental animals and humans in the context of risk assessment. Toxicogenomics combines traditional toxicology using appropriate pharmacological and toxicological models with global omics technologies to provide a comprehensive view of the functioning of the genetic and biochemical machinery in organisms under stress. Applications of these technologies help in predicting the potential toxicity of a drug or chemical be-fore functional damages are recognized, in classification of toxicants, and in screening human susceptibility to diseases, drugs or environmental hazards. WHAT IS TOXICOGENOMICS? Toxicogenomics is a field of science that deals with the collection, interpretation, and storage of information about gene and protein activity within particular cell or tissue of an organism in response to toxic substances. Toxicogenomics combines toxicology with genomics or other high throughput molecular profiling technologies such as transcriptomics, proteomics and metabolomics. Toxicogenomics endeavors to elucidate molecular mechanisms evolved in the expression of toxicity, and to derive molecular expression patterns that predict toxicity or the genetic susceptibility to it. Toxicogenomics is a rapidly developing discipline that promises to aid scientists in understanding the molecular and cellular effects of chemicals in biological systems. This field encompasses global assessment of biological effects using technologies such as DNA micro arrays or high throughput NMR and protein expression analysis. Toxicogenomics represents the merging of toxicology with technologies that have been developed, together with bioinformatics, to identify and quantify global gene expression changes. It represents a new paradigm in drug development and risk assessment, which promises to generate a wealth of information towards an increased understanding of the molecular mechanisms that lead to
2 Amala 43 drug toxicity and efficacy, and of DNA polymorphisms responsible for individual susceptibility to toxicity. Gene expression profiling, through the use of DNA micro array and proteomic technologies will aid in establishing links between expression profiles, mode of action and traditional toxic endpoints. Such patterns of gene expression, or 'molecular fingerprints' could be used as diagnostic or predictive markers of exposure that is characteristic of a specific mechanism of induction of that toxic or efficacious effect. It is anticipated that toxicogenomics will be increasingly integrated into all phases of the drug development process particularly in mechanistic and predictive toxicology, and biomarker discovery. DEFINITION OF TOXICOGENOMICS United States environmental protection agency stating that "the term "genomics" encompasses a broader scope of scientific inquiry and associated technologies than when genomics was initially considered. A genome is the sum total of all an individual organism's genes. Thus, genomics is the study of all the genes of a cell, or tissue, at the DNA (genotype), mrna (transcriptome), or protein (proteome) levels. Genomics methodologies are expected to provide valuable insights for evaluating how environmental stressors affect cellular/tissue function and how changes in gene expression may relate to adverse effects. However, the relationships between changes in gene expression and adverse effects are unclear at this time and may likely be difficult to elucidate. In pharmaceutical research, toxicogenomics is more narrowly defined as the study of the structure and function of the genome as it responds to adverse xenobiotic exposure. It is the toxicological sub discipline of pharmacogenomics, which is broadly defined as the study of inter-individual variations in whole-genome or candidate gene single-nucleotide polymorphism maps, haplotype markers, and alterations in gene expression that might correlate with drug responses. TECHNOLOGIES IN TOXICOGENOMICS Gene expression profiling Gene expression changes are associated with signal pathway. Activation can provide compound-specific information on the pharmacological or toxicological effects of a chemical. An advantage of this traditional molecular technique is that it definitively shows the expression level of all transcripts for a particular gene. Alternate technologies, including DNA microarrays, can measure the expression of tens of thousands of genes in an equivalent amount of time. There are two basic types of micro arrays used in gene expression analyses: Oligonucleotide-based arrays and cdna arrays For example, one can compare tissue extracted from toxicant treated organism versus that of vehicle exposed animals. In addition, other scenarios may include the analysis of healthy versus diseased tissue or susceptible versus resistant tissue. One can combine micro arrays with quantitative polymerase chain reaction (QPCR) or Taqman and other technologies in development to monitor the expression of hundreds of genes in a high throughput fashion. Protein expression Gene expression alone is not adequate to serve the understanding of toxicant action and the disease outcomes they induce. Abnormalities in protein production or function are expected in response to toxicant exposure and the onset of disease states. To understand the complete mechanism of toxicant action, it is necessary to identify the protein alterations associated with that exposure and to understand how these changes affect protein/cellular function. Unlike classical genomic approaches that discover genes related to toxicant induced disease, proteomics can aid to characterize the disease process directly by capturing proteins that participate in the disease. The lack of a direct functional correlation between gene transcripts and their corresponding proteins necessitates the use of proteomics as a tool in toxicology. Proteomics, under the umbrella of toxicogenomics, involves the comprehensive functional annotation and validation of proteins in response to toxicant exposure. Understanding the functional characteristics of proteins and their activity requires a determination of cellular localization and quantization, tissue distribution, post-translational modification state, domain modules and their effect on protein interactions, protein complexes, ligand binding sites and structural representation. Currently, the most commonly used technologies for proteomics research are 2-dimensional (2D) gel electrophoresis for protein separation followed by mass spectrometry analysis of proteins of interest. Matrix-assisted laser desorption mass spectrometry (MALDI-MS) has become a widely used method for determination of biomolecules including peptides. Metabolite analysis by NMR Genomic and proteomic methods do not offer the information needed to gain understanding of the resulting output function in a living system. Neither approach addresses the dynamic metabolic status of the whole animal. The metabonomic approach is based on the premise that toxicant-induced pathological or physiological alterations result in changes in relative concentrations of endogenous biochemical. Metabolites in body fluids such
3 44 J. Bioinform. Seq. Anal. as urine, blood, or cerebrospinal fluid (CSF), are in dynamic equilibrium with those inside cells and tissues, thus toxicant-induced cellular abnormalities in tissues should be reflected in altered biofluid compositions. TOXICOGENOMIC COMPONENTS Comparative/predictive toxicogenomics Comparative genomic, proteomic, or metabonomic studies measure the number and types of genes, protein, and metabolites, respectively, that is present in normal and toxicant-exposed cells, tissues, or biofluids. This approach is useful in defining the composition of the assayed samples in terms of genetic, proteomic or metabolic variables. Thus a biological sample derived from toxicant, or sham treated animals can be regarded as an n-dimensional vector in gene expression space with genes as variables along each dimension. The possibility that a specific group or class of compounds (grouped by toxic endpoint, mechanism, structure, target organ etc.) may induce signature patterns of gene expression changes is the basis for the application of toxicogenomics to predictive toxicology. The use of these technologies to analyze genome-wide changes in mrna expression following treatment of in vitro systems with known reference toxicants may permit the identification of diagnostic gene expression patterns. Pattern recognition allows the design and construction of mini-arrays, customized to detect specific toxicity endpoints or pathways. Functional toxicogenomics Functional toxicogenomics is the study of genes and proteins biological activities in the context of compound effects on an organism. Gene and protein expression profiles are analyzed for information that might provide insight into specific mechanistic pathways. Mechanistic inference is complex when the sequence of events following toxicant exposure is viewed in both dose and time space. Gene and protein expression patterns can indeed be highly dependent on the toxicant concentrations furnished at the assessed tissue and the time of exposure to the agent. Expression patterns are only a snapshot in time and dose space. Thus, a comprehensive understanding of potential mechanisms of action of a compound requires establishing patterns at various combinations of time and dose. Studies that target temporal expression of specific genes and protein in response to toxicant exposure will lead to a better under-standing of the sequence of events in complex regulatory networks. An area of study which is of great interest to toxicologists is the mechanistic understanding of toxicant induced pathological endpoints. The premise that perturbations in gene, protein, or metabolite levels are reflective of adverse phenotypic effects of toxicants offers an opportunity to phenotypically anchor these perturbations. This is quite challenging due to the fact that phenotypic effects often vary in the time-dose space of the studied agent and may have regional variations in the tissue. Furthermore, very few compounds exist that result in only one phenotypic alteration at a given coordinate in dose and time. Thus, objective assignment of measured variables to multiple phenotypic events is not possible under these circumstances. APPLICATIONS OF TOXICOGENOMICS Toxicogenomics in drug safety New drugs are screened for adverse reactions using a laborious, costly process and still some promising therapeutics is withdrawn from the marketplace because of unforeseen human toxicity. Novel higher throughput methods in toxicology need to be developed. These new approaches should provide more insight into potential human toxicity than current methods. Toxicogenomics, the examination of changes in gene expression following exposure to a toxicant, offers the potential to identify a human toxicant earlier in drug development and to detect human-specific toxicants that cause no adverse reaction in rats. To understand the mechanisms of drug-induced hepatotoxicity during drug discovery and development Hepatotoxicity is a common cause of failure in drug discovery and development and is also frequently the source of adverse drug reactions. Therefore, a better prediction, characterization and understanding of druginduced hepatotoxicity could result in safer drugs and a more efficient drug discovery and development process. Toxicogenomics represents an attractive approach to predict toxicity and to gain a mechanistic understanding of toxic changes. Ecotoxicogenomics Rapid progress in the field of genomics (the study of how an individual's entire genetic make-up, the genome, translates into biological functions) is beginning to provide tools that may assist our understanding of how chemicals can impact on human and ecosystem health. Given the parallel implications for ecological (environmental) risk assessment, a term ecotoxicoge-nomics is there to describe the integration of genomics (transcriptomics, proteomics and metabolomics) into ecotoxicology. Ecotoxicogenomics is defined as the study of gene and protein expression in non-target organisms that is important
4 Amala 45 in responses to environmental toxicant exposures. The potential of ecotoxicogenomic tools in ecological risk assessment seems great. Many phenomenological approaches that are useful for identifying chemicals of potential concern provide little understanding of the mechanism of chemical toxicity. Without this understanding, it will be difficult to address some of the key challenges that currently face aquatic ecotoxicology. Ecotoxicogenomic tools may provide us with a better mechanistic understanding of aquatic ecotoxico-logy (Snape et al., 2004). In endocrine disruption Toxicogenomics can be expected to be a useful method for detecting the carcinogenic potential of endocrine active substances (EASs) in the short term with the generation of understanding of mode-of-action and mechanisms when a reliable database with information about proteomics and informatics is established. At present, there are no concrete epidemiological data supporting any exogenous EAS contribution to hormone-related organ carcinogenesis in humans. However, with the establishment of appropriate animal models and analysis of genomic-scale gene expression, risk identification and evaluation should be facilitated within a relatively short period, and this approach eventually promises to contribute a great deal of risk management regarding EASs (Tomoyuki and Makoto, 2003). In assessment of immunotoxicity Microarray analysis is used for simultaneous measurement of expression of thousands of genes in a given sample and as such extends and deepens our understanding of biological processes. Application of the technique in toxicology is referred to as toxicogenomics. The assessment of immunotoxicity by gene expression profiling show that micro array analysis is able to detect known and novel effects of a wide range of immunomodulating agents. Besides the elucidation of mechanisms of action, toxicogenomics is also applied to predict consequences of exposing biological systems to toxic agents. Successful attempts to classify compounds using signature gene expression profiles have been reported. The application of toxicogenomics in evaluation of immunotoxicity contributes to the understanding of immunotoxic processes and the development of in vitro screening assays, though, and is therefore expected to be of value for mechanistic insight into immunotoxicity and hazard identification of existing and novel compounds (Baken et al., 2007). TOXICOGENOMICS DATABASE AND RESOURCES Toxicogenomics studies are generally built on standard toxicology studies generating biological end point data, and as such, one goal of toxicogenomics is to detect relationships between changes in gene expression and in those biological parameters. These challenges are best addressed through data collection into a well-designed toxicogenomics database (Mattes et al., 2004). The chemical effects in biological systems (CEBS) knowledge base The CEBS knowledge base ( is under development by the NIEHS NCT as a public toxicogenomics information resource combining data sets from tran-scri-ptomics, proteomics, metabonomics, and conventional toxicology with pathway and network information relevant to environmental toxicology and human disease. The overall goal of CEBS is to support hypothesis-driven and discovery research in environmental toxicology and the research needs of risk assessment. Specific objectives are a) to compare toxicogenomic effects of chemicals/stressors across species yielding signatures of altered molecular expression; b) to phenotypically anchor these changes with conventional toxicology data classifying biological effects as well as disease phenotypes; and c) to delineate global changes as adaptive, pharmacologic, or toxic outcomes defining early biomarkers, the sequence of key events, and mechanisms of toxicant action. CEBS is designed to meet the information needs of systems toxicology and involves study of chemical or stressor perturbations, monitoring changes in molecular expression, and iteratively integrating biological response data to describe the functioning organism. Comparative toxicogenomics database (CTD) The NIEHS DERT supports an international public database devoted primarily to comparative toxicogenomics in aquatic and mammalian species, the CTD ( The Mount Desert Island Biological Laboratory is developing CTD as a communitysupported genomic resource devoted to genes of human toxicological significance. CTD will be the first publicly available database to a) provide annotated associations between genes, references, and toxic agents; b) include nucleotide and protein sequences from diverse species with a focus on aquatic and mammalian organisms; c) offer a range of analytical tools for customized comparative studies; and d) provide information to investigators on available molecular reagents. The primary goals of CTD are to advance the understanding of the effects of environmental chemicals on human health. DbZach The Molecular and Genomic Toxicology Laboratory, Michigan State University (East Lansing, MI) has developed the dbzach System ( a multifacteted toxicogenomics
5 46 J. Bioinform. Seq. Anal. bioinformatics infrastructure. The goal of the dbzach System is to provide a) facilities for the modeling of toxicogenomics data; b) a centralized source of biological knowledge to facilitate data mining and allow full knowledge-based understanding of the toxicological mechanisms; and c) an environment for bioinformatics algorithmic and analysis tools development. DbZach, designed in a modular structure to handle multispecies array-based toxicogen-omics information, is the core database implemented in Oracle. Array track The NCTR ( toxicoinformatics/tools.htm) is developing TIS to integrate genomics, proteomics, and metabonomics data with conventional in vivo and in vitro toxicology data. TIS is designed to meet the challenge of data management, analysis, and inter-pretation through the integration of toxicogenomics data, gene function, and pathways to enable hypothesis generation. EDGE The EDGE database ( was developed at the McArdle Laboratory for Cancer Research, University of Wisconsin (Madison, WI) as a resource for toxicology-related gene expression information. It is based on experiments conducted using custom cdna micro arrays that include unique ESTs identified as regulated under conditions of toxicity. Public toxicogenomics projects - Chemical Effects in Biological Systems (CEBS) - Project hosted by the National Institute of Environmental Health Sciences (NIEHS) is building a knowledgebase of toxicology studies including study design, clinical pathology, and histopathology and toxicogenomics data. - InnoMed PredTox for assessing the value of combining results from omics technologies together with the results from more conventional toxicology methods in more informed decision making in preclinical safety evaluation. - Predictive Safety Testing Consortium aimed at identifying and clinically qualifying safety biomarkers for regulatory use as part of the FDA's Critical Path Initiative - ToxCast program for Predicting Hazard, Characterizing Toxicity Pathways, and Prioritizing the Toxicity Testing of Environmental Chemicals at the United States Environmental Protection Agency Conclusion Toxicogenomics is an information- and informaticsintensive field. An important aspect of toxicogenomics research is the development and application of bioinformatics tools and databases in order to facilitate the analysis, mining, visualizing and sharing of the vast amount of biological information being generated in this field. This rapidly growing research area will have a large impact on many other scientific and medical disciplines, including systems biology, as researchers strive to generate complete descriptions of how components of biological systems work together and across organisms to respond to specific stresses, drugs, or toxicants. By establishing associations between the unique genetic makeup of individuals and their responsiveness to specific drugs, we expect to discover better therapies and improve prospects for providing individuals with personalized medicine. And by combining this knowledge with technology for high-throughput screening of candidate drugs early on, we also expect to streamline and enhance the process of drug discovery REFERENCES Baken KA, Vandebriel RJ, Pennings JL, Kleinjans JC, van Loveren H (2007). Toxicogenomics in the assessment of immunotoxicity Methods. 41: Snape JR, Maund SJ, Pickford DB, Hutchinson TH, (2004) Ecotoxicogenomics: the challenge of integrating genomics into aquatic and terrestrial ecotoxicology. Aquat. Toxicol., 67 : Tomoyuki S, Akoto A (2003). Application of toxicogenomics to the endocrine disruption issue IUPAC, Pure and Applied Chemistry 75: Zhou T, Chou J, Watkins PB, Kaufmann WK (2009), Toxicogenomics: transcription profiling for toxicology assessment EXS. 99:
An introduction to SYSTEMS BIOLOGY
 An introduction to SYSTEMS BIOLOGY Paolo Tieri CNR Consiglio Nazionale delle Ricerche, Rome, Italy 10 February 2015 Universidade Federal de Minas Gerais, Belo Horizonte, Brasil Course outline Day 1: intro
An introduction to SYSTEMS BIOLOGY Paolo Tieri CNR Consiglio Nazionale delle Ricerche, Rome, Italy 10 February 2015 Universidade Federal de Minas Gerais, Belo Horizonte, Brasil Course outline Day 1: intro
FRAUNHOFER IME SCREENINGPORT
 FRAUNHOFER IME SCREENINGPORT Design of screening projects General remarks Introduction Screening is done to identify new chemical substances against molecular mechanisms of a disease It is a question of
FRAUNHOFER IME SCREENINGPORT Design of screening projects General remarks Introduction Screening is done to identify new chemical substances against molecular mechanisms of a disease It is a question of
Proteomics Systems Biology
 Dr. Sanjeeva Srivastava IIT Bombay Proteomics Systems Biology IIT Bombay 2 1 DNA Genomics RNA Transcriptomics Global Cellular Protein Proteomics Global Cellular Metabolite Metabolomics Global Cellular
Dr. Sanjeeva Srivastava IIT Bombay Proteomics Systems Biology IIT Bombay 2 1 DNA Genomics RNA Transcriptomics Global Cellular Protein Proteomics Global Cellular Metabolite Metabolomics Global Cellular
Application of Toxicogenomics for Toxicity Assessment and Screening of Nanomaterials
 NEN Summit 2010 Lowell MA June 22-24 Application of Toxicogenomics for Toxicity Assessment and Screening of Nanomaterials April Z. Gu, Na Gou and Annalisa Onnis-Hayden Civil and Environmental Engineering
NEN Summit 2010 Lowell MA June 22-24 Application of Toxicogenomics for Toxicity Assessment and Screening of Nanomaterials April Z. Gu, Na Gou and Annalisa Onnis-Hayden Civil and Environmental Engineering
Introduction Biology before Systems Biology: Reductionism Reduce the study from the whole organism to inner most details like protein or the DNA.
 Systems Biology-Models and Approaches Introduction Biology before Systems Biology: Reductionism Reduce the study from the whole organism to inner most details like protein or the DNA. Taxonomy Study external
Systems Biology-Models and Approaches Introduction Biology before Systems Biology: Reductionism Reduce the study from the whole organism to inner most details like protein or the DNA. Taxonomy Study external
Bioinformatics. Dept. of Computational Biology & Bioinformatics
 Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Bioinformatics Dept. of Computational Biology & Bioinformatics 3 Bioinformatics - play with sequences & structures Dept. of Computational Biology & Bioinformatics 4 ORGANIZATION OF LIFE ROLE OF BIOINFORMATICS
Introduction to Bioinformatics. Shifra Ben-Dor Irit Orr
 Introduction to Bioinformatics Shifra Ben-Dor Irit Orr Lecture Outline: Technical Course Items Introduction to Bioinformatics Introduction to Databases This week and next week What is bioinformatics? A
Introduction to Bioinformatics Shifra Ben-Dor Irit Orr Lecture Outline: Technical Course Items Introduction to Bioinformatics Introduction to Databases This week and next week What is bioinformatics? A
Adverse Outcome Pathways in Ecotoxicology Research
 Adverse Outcome Pathways in Ecotoxicology Research Michael W. Hornung US Environmental Protection Agency, Mid-Continent Ecology Division, Duluth, MN Meeting of the Northland Chapter of SOT October 7, 2010
Adverse Outcome Pathways in Ecotoxicology Research Michael W. Hornung US Environmental Protection Agency, Mid-Continent Ecology Division, Duluth, MN Meeting of the Northland Chapter of SOT October 7, 2010
Biological Concepts and Information Technology (Systems Biology)
 Biological Concepts and Information Technology (Systems Biology) Janaina de Andréa Dernowsek Postdoctoral at Center for Information Technology Renato Archer Janaina.dernowsek@cti.gov.br Division of 3D
Biological Concepts and Information Technology (Systems Biology) Janaina de Andréa Dernowsek Postdoctoral at Center for Information Technology Renato Archer Janaina.dernowsek@cti.gov.br Division of 3D
Curriculum Links. AQA GCE Biology. AS level
 Curriculum Links AQA GCE Biology Unit 2 BIOL2 The variety of living organisms 3.2.1 Living organisms vary and this variation is influenced by genetic and environmental factors Causes of variation 3.2.2
Curriculum Links AQA GCE Biology Unit 2 BIOL2 The variety of living organisms 3.2.1 Living organisms vary and this variation is influenced by genetic and environmental factors Causes of variation 3.2.2
Course Descriptions Biology
 Course Descriptions Biology BIOL 1010 (F/S) Human Anatomy and Physiology I. An introductory study of the structure and function of the human organ systems including the nervous, sensory, muscular, skeletal,
Course Descriptions Biology BIOL 1010 (F/S) Human Anatomy and Physiology I. An introductory study of the structure and function of the human organ systems including the nervous, sensory, muscular, skeletal,
BEFORE TAKING THIS MODULE YOU MUST ( TAKE BIO-4013Y OR TAKE BIO-
 2018/9 - BIO-4001A BIODIVERSITY Autumn Semester, Level 4 module (Maximum 150 Students) Organiser: Dr Harriet Jones Timetable Slot:DD This module explores life on Earth. You will be introduced to the major
2018/9 - BIO-4001A BIODIVERSITY Autumn Semester, Level 4 module (Maximum 150 Students) Organiser: Dr Harriet Jones Timetable Slot:DD This module explores life on Earth. You will be introduced to the major
MEDLINE Clinical Laboratory Sciences Journals
 Source Type Publication Name ISSN Peer-Reviewed Academic Journal Acta Biochimica et Biophysica Sinica 1672-9145 Y Academic Journal Acta Physiologica 1748-1708 Y Academic Journal Aging Cell 1474-9718 Y
Source Type Publication Name ISSN Peer-Reviewed Academic Journal Acta Biochimica et Biophysica Sinica 1672-9145 Y Academic Journal Acta Physiologica 1748-1708 Y Academic Journal Aging Cell 1474-9718 Y
Life Sciences
 3.3.3.5 Life Sciences Hosted by the Department of Biological Sciences, Faculty of Science (FoS), the NUS Life Sciences Undergraduate Programme offers the Life Sciences Major. The curriculum is taught by
3.3.3.5 Life Sciences Hosted by the Department of Biological Sciences, Faculty of Science (FoS), the NUS Life Sciences Undergraduate Programme offers the Life Sciences Major. The curriculum is taught by
VCE BIOLOGY Relationship between the key knowledge and key skills of the Study Design and the Study Design
 VCE BIOLOGY 2006 2014 Relationship between the key knowledge and key skills of the 2000 2005 Study Design and the 2006 2014 Study Design The following table provides a comparison of the key knowledge (and
VCE BIOLOGY 2006 2014 Relationship between the key knowledge and key skills of the 2000 2005 Study Design and the 2006 2014 Study Design The following table provides a comparison of the key knowledge (and
In Silico Investigation of Off-Target Effects
 PHARMA & LIFE SCIENCES WHITEPAPER In Silico Investigation of Off-Target Effects STREAMLINING IN SILICO PROFILING In silico techniques require exhaustive data and sophisticated, well-structured informatics
PHARMA & LIFE SCIENCES WHITEPAPER In Silico Investigation of Off-Target Effects STREAMLINING IN SILICO PROFILING In silico techniques require exhaustive data and sophisticated, well-structured informatics
mrna proteins DNA predicts certain possible future health issues Limited info concerning evolving health; advanced measurement technologies
 DA predicts certain possible future health issues mra Limited info concerning evolving health; advanced measurement technologies proteins Potentially comprehensive information concerning evolving health;
DA predicts certain possible future health issues mra Limited info concerning evolving health; advanced measurement technologies proteins Potentially comprehensive information concerning evolving health;
What is Systems Biology
 What is Systems Biology 2 CBS, Department of Systems Biology 3 CBS, Department of Systems Biology Data integration In the Big Data era Combine different types of data, describing different things or the
What is Systems Biology 2 CBS, Department of Systems Biology 3 CBS, Department of Systems Biology Data integration In the Big Data era Combine different types of data, describing different things or the
Development Plan 2025
 Development Plan 2025 4.15 Faculty of Life Sciences 4.15.1 Objectives The Faculty of Life Sciences encompasses the disciplines of biology, pharmacy and nutritional sciences. The Faculty aims at acquiring
Development Plan 2025 4.15 Faculty of Life Sciences 4.15.1 Objectives The Faculty of Life Sciences encompasses the disciplines of biology, pharmacy and nutritional sciences. The Faculty aims at acquiring
Next Generation Computational Chemistry Tools to Predict Toxicity of CWAs
 Next Generation Computational Chemistry Tools to Predict Toxicity of CWAs William (Bill) Welsh welshwj@umdnj.edu Prospective Funding by DTRA/JSTO-CBD CBIS Conference 1 A State-wide, Regional and National
Next Generation Computational Chemistry Tools to Predict Toxicity of CWAs William (Bill) Welsh welshwj@umdnj.edu Prospective Funding by DTRA/JSTO-CBD CBIS Conference 1 A State-wide, Regional and National
Biological Mass Spectrometry
 Biochemistry 412 Biological Mass Spectrometry February 13 th, 2007 Proteomics The study of the complete complement of proteins found in an organism Degrees of Freedom for Protein Variability Covalent Modifications
Biochemistry 412 Biological Mass Spectrometry February 13 th, 2007 Proteomics The study of the complete complement of proteins found in an organism Degrees of Freedom for Protein Variability Covalent Modifications
Enduring understanding 1.A: Change in the genetic makeup of a population over time is evolution.
 The AP Biology course is designed to enable you to develop advanced inquiry and reasoning skills, such as designing a plan for collecting data, analyzing data, applying mathematical routines, and connecting
The AP Biology course is designed to enable you to develop advanced inquiry and reasoning skills, such as designing a plan for collecting data, analyzing data, applying mathematical routines, and connecting
TEST SUMMARY AND FRAMEWORK TEST SUMMARY
 Washington Educator Skills Tests Endorsements (WEST E) TEST SUMMARY AND FRAMEWORK TEST SUMMARY BIOLOGY Copyright 2014 by the Washington Professional Educator Standards Board 1 Washington Educator Skills
Washington Educator Skills Tests Endorsements (WEST E) TEST SUMMARY AND FRAMEWORK TEST SUMMARY BIOLOGY Copyright 2014 by the Washington Professional Educator Standards Board 1 Washington Educator Skills
Integrated Approaches to Testing and Assessment in Developmental Toxicology
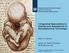 Integrated Approaches to Testing and Assessment in Developmental Toxicology Aldert H. Piersma Center for Health Protection RIVM Bilthoven-NL 1 European Teratology Society Conference, Budapest, Hungary,
Integrated Approaches to Testing and Assessment in Developmental Toxicology Aldert H. Piersma Center for Health Protection RIVM Bilthoven-NL 1 European Teratology Society Conference, Budapest, Hungary,
1.1. KEY CONCEPT Biologists study life in all its forms. 4 Reinforcement Unit 1 Resource Book. Biology in the 21st Century CHAPTER 1
 1.1 THE STUDY OF LIFE KEY CONCEPT Biologists study life in all its forms. Biology is the scientific study of all forms of life. Living things are found almost everywhere on Earth, from very hot environments
1.1 THE STUDY OF LIFE KEY CONCEPT Biologists study life in all its forms. Biology is the scientific study of all forms of life. Living things are found almost everywhere on Earth, from very hot environments
Cheminformatics Role in Pharmaceutical Industry. Randal Chen Ph.D. Abbott Laboratories Aug. 23, 2004 ACS
 Cheminformatics Role in Pharmaceutical Industry Randal Chen Ph.D. Abbott Laboratories Aug. 23, 2004 ACS Agenda The big picture for pharmaceutical industry Current technological/scientific issues Types
Cheminformatics Role in Pharmaceutical Industry Randal Chen Ph.D. Abbott Laboratories Aug. 23, 2004 ACS Agenda The big picture for pharmaceutical industry Current technological/scientific issues Types
ADVANCED PLACEMENT BIOLOGY
 ADVANCED PLACEMENT BIOLOGY Description Advanced Placement Biology is designed to be the equivalent of a two-semester college introductory course for Biology majors. The course meets seven periods per week
ADVANCED PLACEMENT BIOLOGY Description Advanced Placement Biology is designed to be the equivalent of a two-semester college introductory course for Biology majors. The course meets seven periods per week
Introduction to Bioinformatics
 CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
CSCI8980: Applied Machine Learning in Computational Biology Introduction to Bioinformatics Rui Kuang Department of Computer Science and Engineering University of Minnesota kuang@cs.umn.edu History of Bioinformatics
Map of AP-Aligned Bio-Rad Kits with Learning Objectives
 Map of AP-Aligned Bio-Rad Kits with Learning Objectives Cover more than one AP Biology Big Idea with these AP-aligned Bio-Rad kits. Big Idea 1 Big Idea 2 Big Idea 3 Big Idea 4 ThINQ! pglo Transformation
Map of AP-Aligned Bio-Rad Kits with Learning Objectives Cover more than one AP Biology Big Idea with these AP-aligned Bio-Rad kits. Big Idea 1 Big Idea 2 Big Idea 3 Big Idea 4 ThINQ! pglo Transformation
SCOTCAT Credits: 20 SCQF Level 7 Semester 1 Academic year: 2018/ am, Practical classes one per week pm Mon, Tue, or Wed
 Biology (BL) modules BL1101 Biology 1 SCOTCAT Credits: 20 SCQF Level 7 Semester 1 10.00 am; Practical classes one per week 2.00-5.00 pm Mon, Tue, or Wed This module is an introduction to molecular and
Biology (BL) modules BL1101 Biology 1 SCOTCAT Credits: 20 SCQF Level 7 Semester 1 10.00 am; Practical classes one per week 2.00-5.00 pm Mon, Tue, or Wed This module is an introduction to molecular and
Receptor Based Drug Design (1)
 Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Mole_Oce Lecture # 24: Introduction to genomics
 Mole_Oce Lecture # 24: Introduction to genomics DEFINITION: Genomics: the study of genomes or he study of genes and their function. Genomics (1980s):The systematic generation of information about genes
Mole_Oce Lecture # 24: Introduction to genomics DEFINITION: Genomics: the study of genomes or he study of genes and their function. Genomics (1980s):The systematic generation of information about genes
Proteomics. 2 nd semester, Department of Biotechnology and Bioinformatics Laboratory of Nano-Biotechnology and Artificial Bioengineering
 Proteomics 2 nd semester, 2013 1 Text book Principles of Proteomics by R. M. Twyman, BIOS Scientific Publications Other Reference books 1) Proteomics by C. David O Connor and B. David Hames, Scion Publishing
Proteomics 2 nd semester, 2013 1 Text book Principles of Proteomics by R. M. Twyman, BIOS Scientific Publications Other Reference books 1) Proteomics by C. David O Connor and B. David Hames, Scion Publishing
Proteomics. Areas of Interest
 Introduction to BioMEMS & Medical Microdevices Proteomics and Protein Microarrays Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
Introduction to BioMEMS & Medical Microdevices Proteomics and Protein Microarrays Companion lecture to the textbook: Fundamentals of BioMEMS and Medical Microdevices, by Prof., http://saliterman.umn.edu/
THE ROLE OF BBSRC IN BIODIVERSITY RESEARCH
 THE ROLE OF BBSRC IN BIODIVERSITY RESEARCH INTRODUCTION The aim of this document is to explain the role of biodiversity research in the delivery of the BBSRC mission, and thereby to provide guidance to
THE ROLE OF BBSRC IN BIODIVERSITY RESEARCH INTRODUCTION The aim of this document is to explain the role of biodiversity research in the delivery of the BBSRC mission, and thereby to provide guidance to
Aplicació de la proteòmica a la cerca de Biomarcadors proteics Barcelona, 08 de Juny 2010
 Aplicació de la proteòmica a la cerca de Biomarcadors proteics Barcelona, 8 de Juny 21 Eliandre de Oliveira Plataforma de Proteòmica Parc Científic de Barcelona Protein Chemistry Proteomics Hypothesis-free
Aplicació de la proteòmica a la cerca de Biomarcadors proteics Barcelona, 8 de Juny 21 Eliandre de Oliveira Plataforma de Proteòmica Parc Científic de Barcelona Protein Chemistry Proteomics Hypothesis-free
In silico pharmacology for drug discovery
 In silico pharmacology for drug discovery In silico drug design In silico methods can contribute to drug targets identification through application of bionformatics tools. Currently, the application of
In silico pharmacology for drug discovery In silico drug design In silico methods can contribute to drug targets identification through application of bionformatics tools. Currently, the application of
Unravelling the biochemical reaction kinetics from time-series data
 Unravelling the biochemical reaction kinetics from time-series data Santiago Schnell Indiana University School of Informatics and Biocomplexity Institute Email: schnell@indiana.edu WWW: http://www.informatics.indiana.edu/schnell
Unravelling the biochemical reaction kinetics from time-series data Santiago Schnell Indiana University School of Informatics and Biocomplexity Institute Email: schnell@indiana.edu WWW: http://www.informatics.indiana.edu/schnell
Keystone Exams: Biology Assessment Anchors and Eligible Content. Pennsylvania Department of Education
 Assessment Anchors and Pennsylvania Department of Education www.education.state.pa.us 2010 PENNSYLVANIA DEPARTMENT OF EDUCATION General Introduction to the Keystone Exam Assessment Anchors Introduction
Assessment Anchors and Pennsylvania Department of Education www.education.state.pa.us 2010 PENNSYLVANIA DEPARTMENT OF EDUCATION General Introduction to the Keystone Exam Assessment Anchors Introduction
Big Idea 1: The process of evolution drives the diversity and unity of life.
 Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
Big Idea 1: The process of evolution drives the diversity and unity of life. understanding 1.A: Change in the genetic makeup of a population over time is evolution. 1.A.1: Natural selection is a major
AP Curriculum Framework with Learning Objectives
 Big Ideas Big Idea 1: The process of evolution drives the diversity and unity of life. AP Curriculum Framework with Learning Objectives Understanding 1.A: Change in the genetic makeup of a population over
Big Ideas Big Idea 1: The process of evolution drives the diversity and unity of life. AP Curriculum Framework with Learning Objectives Understanding 1.A: Change in the genetic makeup of a population over
MSc Drug Design. Module Structure: (15 credits each) Lectures and Tutorials Assessment: 50% coursework, 50% unseen examination.
 Module Structure: (15 credits each) Lectures and Assessment: 50% coursework, 50% unseen examination. Module Title Module 1: Bioinformatics and structural biology as applied to drug design MEDC0075 In the
Module Structure: (15 credits each) Lectures and Assessment: 50% coursework, 50% unseen examination. Module Title Module 1: Bioinformatics and structural biology as applied to drug design MEDC0075 In the
Goal 1: Develop knowledge and understanding of core content in biology
 Indiana State University» College of Arts & Sciences» Biology BA/BS in Biology Standing Requirements s Library Goal 1: Develop knowledge and understanding of core content in biology 1: Illustrate and examine
Indiana State University» College of Arts & Sciences» Biology BA/BS in Biology Standing Requirements s Library Goal 1: Develop knowledge and understanding of core content in biology 1: Illustrate and examine
The Globally Harmonized System of Classification and Labelling of Chemicals (GHS) Purpose, scope and application
 The Globally Harmonized System of Classification and Labelling of Chemicals (GHS) Purpose, scope and application Purpose of the GHS (1) (a) (b) (c) (d) To enhance the protection of human health and the
The Globally Harmonized System of Classification and Labelling of Chemicals (GHS) Purpose, scope and application Purpose of the GHS (1) (a) (b) (c) (d) To enhance the protection of human health and the
Introduction to Chemoinformatics
 Introduction to Chemoinformatics Dr. Igor V. Tetko Helmholtz Zentrum München - German Research Center for Environmental Health (GmbH) Institute of Bioinformatics & Systems Biology (HMGU) Kyiv, 10 August
Introduction to Chemoinformatics Dr. Igor V. Tetko Helmholtz Zentrum München - German Research Center for Environmental Health (GmbH) Institute of Bioinformatics & Systems Biology (HMGU) Kyiv, 10 August
October 08-11, Co-Organizer Dr. S D Samantaray Professor & Head,
 A Journey towards Systems system Biology biology :: Biocomputing of of Hi-throughput Omics omics Data data October 08-11, 2018 Coordinator Dr. Anil Kumar Gaur Professor & Head Department of Molecular Biology
A Journey towards Systems system Biology biology :: Biocomputing of of Hi-throughput Omics omics Data data October 08-11, 2018 Coordinator Dr. Anil Kumar Gaur Professor & Head Department of Molecular Biology
BIOCHEMISTRY AND MOLECULAR BIOLOGY ELLIOTT
 page 1 / 5 page 2 / 5 biochemistry and molecular biology pdf Journal Impact Factor: 1.48. ISSN: 2471-8084. Biochemistry and Molecular Biology Journal is an international scholarly, peer reviewed journal
page 1 / 5 page 2 / 5 biochemistry and molecular biology pdf Journal Impact Factor: 1.48. ISSN: 2471-8084. Biochemistry and Molecular Biology Journal is an international scholarly, peer reviewed journal
Proteomics. Yeast two hybrid. Proteomics - PAGE techniques. Data obtained. What is it?
 Proteomics What is it? Reveal protein interactions Protein profiling in a sample Yeast two hybrid screening High throughput 2D PAGE Automatic analysis of 2D Page Yeast two hybrid Use two mating strains
Proteomics What is it? Reveal protein interactions Protein profiling in a sample Yeast two hybrid screening High throughput 2D PAGE Automatic analysis of 2D Page Yeast two hybrid Use two mating strains
A A A A B B1
 LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
LEARNING OBJECTIVES FOR EACH BIG IDEA WITH ASSOCIATED SCIENCE PRACTICES AND ESSENTIAL KNOWLEDGE Learning Objectives will be the target for AP Biology exam questions Learning Objectives Sci Prac Es Knowl
STAAR Biology Assessment
 STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules as building blocks of cells, and that cells are the basic unit of
STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules as building blocks of cells, and that cells are the basic unit of
Computational Systems Biology
 Computational Systems Biology Vasant Honavar Artificial Intelligence Research Laboratory Bioinformatics and Computational Biology Graduate Program Center for Computational Intelligence, Learning, & Discovery
Computational Systems Biology Vasant Honavar Artificial Intelligence Research Laboratory Bioinformatics and Computational Biology Graduate Program Center for Computational Intelligence, Learning, & Discovery
Genom, transkriptom in proteom
 Genom, transkriptom in proteom The conceptual relationship of the genome, transcriptome, proteome and metabolome Fenotipi/Phenotypes Morphological phenotypes are fixed in distinct dog breeds. Fizikalna
Genom, transkriptom in proteom The conceptual relationship of the genome, transcriptome, proteome and metabolome Fenotipi/Phenotypes Morphological phenotypes are fixed in distinct dog breeds. Fizikalna
The Role of Network Science in Biology and Medicine. Tiffany J. Callahan Computational Bioscience Program Hunter/Kahn Labs
 The Role of Network Science in Biology and Medicine Tiffany J. Callahan Computational Bioscience Program Hunter/Kahn Labs Network Analysis Working Group 09.28.2017 Network-Enabled Wisdom (NEW) empirically
The Role of Network Science in Biology and Medicine Tiffany J. Callahan Computational Bioscience Program Hunter/Kahn Labs Network Analysis Working Group 09.28.2017 Network-Enabled Wisdom (NEW) empirically
Compare and contrast the cellular structures and degrees of complexity of prokaryotic and eukaryotic organisms.
 Subject Area - 3: Science and Technology and Engineering Education Standard Area - 3.1: Biological Sciences Organizing Category - 3.1.A: Organisms and Cells Course - 3.1.B.A: BIOLOGY Standard - 3.1.B.A1:
Subject Area - 3: Science and Technology and Engineering Education Standard Area - 3.1: Biological Sciences Organizing Category - 3.1.A: Organisms and Cells Course - 3.1.B.A: BIOLOGY Standard - 3.1.B.A1:
Driving Serendipity: the Era of Active Ingredients Discovery. Valentina Sedini Scientific Marketing
 Driving Serendipity: the Era of Active Ingredients Discovery Valentina Sedini Scientific Marketing Active Ingredients Discovery in cosmetics HOW Active Ingredients Discovery: the drug ancestor Historically,
Driving Serendipity: the Era of Active Ingredients Discovery Valentina Sedini Scientific Marketing Active Ingredients Discovery in cosmetics HOW Active Ingredients Discovery: the drug ancestor Historically,
Canada s Experience with Chemicals Assessment and Management and its Application to Nanomaterials
 Canada s Experience with Chemicals Assessment and Management and its Application to Nanomaterials European Chemicals Agency (ECHA) Topical Scientific Workshop: Regulatory Challenges in Risk Assessment
Canada s Experience with Chemicals Assessment and Management and its Application to Nanomaterials European Chemicals Agency (ECHA) Topical Scientific Workshop: Regulatory Challenges in Risk Assessment
COURSE NUMBER: EH 590R SECTION: 1 SEMESTER: Fall COURSE TITLE: Computational Systems Biology: Modeling Biological Responses
 DEPARTMENT: Environmental Health COURSE NUMBER: EH 590R SECTION: 1 SEMESTER: Fall 2017 CREDIT HOURS: 2 COURSE TITLE: Computational Systems Biology: Modeling Biological Responses COURSE LOCATION: TBD PREREQUISITE:
DEPARTMENT: Environmental Health COURSE NUMBER: EH 590R SECTION: 1 SEMESTER: Fall 2017 CREDIT HOURS: 2 COURSE TITLE: Computational Systems Biology: Modeling Biological Responses COURSE LOCATION: TBD PREREQUISITE:
Bioinformatics 2. Yeast two hybrid. Proteomics. Proteomics
 GENOME Bioinformatics 2 Proteomics protein-gene PROTEOME protein-protein METABOLISM Slide from http://www.nd.edu/~networks/ Citrate Cycle Bio-chemical reactions What is it? Proteomics Reveal protein Protein
GENOME Bioinformatics 2 Proteomics protein-gene PROTEOME protein-protein METABOLISM Slide from http://www.nd.edu/~networks/ Citrate Cycle Bio-chemical reactions What is it? Proteomics Reveal protein Protein
Biology Assessment. Eligible Texas Essential Knowledge and Skills
 Biology Assessment Eligible Texas Essential Knowledge and Skills STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules
Biology Assessment Eligible Texas Essential Knowledge and Skills STAAR Biology Assessment Reporting Category 1: Cell Structure and Function The student will demonstrate an understanding of biomolecules
USING THE COMPARATIVE TOXICOGENOMICS DATABASE TO UNDERSTAND THE IMPACT OF HIGH-RISK TOXICANTS ON BIOLOGICAL PATHWAYS. Samantha E.
 USING THE COMPARATIVE TOXICOGENOMICS DATABASE TO UNDERSTAND THE IMPACT OF HIGH-RISK TOXICANTS ON BIOLOGICAL PATHWAYS Samantha E. Tulenko A technical report submitted to the faculty of the University of
USING THE COMPARATIVE TOXICOGENOMICS DATABASE TO UNDERSTAND THE IMPACT OF HIGH-RISK TOXICANTS ON BIOLOGICAL PATHWAYS Samantha E. Tulenko A technical report submitted to the faculty of the University of
Abiotic Stress in Crop Plants
 1 Abiotic Stress in Crop Plants Mirza Hasanuzzaman, PhD Professor Department of Agronomy Sher-e-Bangla Agricultural University E-mail: mhzsauag@yahoo.com Stress Stress is usually defined as an external
1 Abiotic Stress in Crop Plants Mirza Hasanuzzaman, PhD Professor Department of Agronomy Sher-e-Bangla Agricultural University E-mail: mhzsauag@yahoo.com Stress Stress is usually defined as an external
Clustering and Network
 Clustering and Network Jing-Dong Jackie Han jdhan@picb.ac.cn http://www.picb.ac.cn/~jdhan Copy Right: Jing-Dong Jackie Han What is clustering? A way of grouping together data samples that are similar in
Clustering and Network Jing-Dong Jackie Han jdhan@picb.ac.cn http://www.picb.ac.cn/~jdhan Copy Right: Jing-Dong Jackie Han What is clustering? A way of grouping together data samples that are similar in
Canadian Advanced Senior High
 Canadian Advanced Senior High Department: Science Course Development Date: November 2017 Course Title: Biology Grade: 12 Course Type: Ministry Course Code: University SBI4U Credit Value: 1 Hours: 110 Ministry
Canadian Advanced Senior High Department: Science Course Development Date: November 2017 Course Title: Biology Grade: 12 Course Type: Ministry Course Code: University SBI4U Credit Value: 1 Hours: 110 Ministry
Prereq: Concurrent 3 CH
 0201107 0201101 General Biology (1) General Biology (1) is an introductory course which covers the basics of cell biology in a traditional order, from the structure and function of molecules to the structure
0201107 0201101 General Biology (1) General Biology (1) is an introductory course which covers the basics of cell biology in a traditional order, from the structure and function of molecules to the structure
COMBINATORIAL CHEMISTRY: CURRENT APPROACH
 COMBINATORIAL CHEMISTRY: CURRENT APPROACH Dwivedi A. 1, Sitoke A. 2, Joshi V. 3, Akhtar A.K. 4* and Chaturvedi M. 1, NRI Institute of Pharmaceutical Sciences, Bhopal, M.P.-India 2, SRM College of Pharmacy,
COMBINATORIAL CHEMISTRY: CURRENT APPROACH Dwivedi A. 1, Sitoke A. 2, Joshi V. 3, Akhtar A.K. 4* and Chaturvedi M. 1, NRI Institute of Pharmaceutical Sciences, Bhopal, M.P.-India 2, SRM College of Pharmacy,
Exploration of alternative methods for toxicity assessment of pesticide metabolites
 Exploration of alternative methods for toxicity assessment of pesticide metabolites Alternative in vitro methods to characterize the role of endocrine active substances (EAS) in hormone-targeted tissues,
Exploration of alternative methods for toxicity assessment of pesticide metabolites Alternative in vitro methods to characterize the role of endocrine active substances (EAS) in hormone-targeted tissues,
Predicting Protein Functions and Domain Interactions from Protein Interactions
 Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
Predicting Protein Functions and Domain Interactions from Protein Interactions Fengzhu Sun, PhD Center for Computational and Experimental Genomics University of Southern California Outline High-throughput
BIOLOGY 111. CHAPTER 1: An Introduction to the Science of Life
 BIOLOGY 111 CHAPTER 1: An Introduction to the Science of Life An Introduction to the Science of Life: Chapter Learning Outcomes 1.1) Describe the properties of life common to all living things. (Module
BIOLOGY 111 CHAPTER 1: An Introduction to the Science of Life An Introduction to the Science of Life: Chapter Learning Outcomes 1.1) Describe the properties of life common to all living things. (Module
The Institute of Cancer Research PHD STUDENTSHIP PROJECT PROPOSAL
 The Institute of Cancer Research PHD STUDENTSHIP PROJECT PROPOSAL PROJECT DETAILS Project Title: Design and synthesis of libraries of bifunctional degraders for the discovery of new cancer targets SUPERVISORY
The Institute of Cancer Research PHD STUDENTSHIP PROJECT PROPOSAL PROJECT DETAILS Project Title: Design and synthesis of libraries of bifunctional degraders for the discovery of new cancer targets SUPERVISORY
Priority Setting of Endocrine Disruptors Using QSARs
 Priority Setting of Endocrine Disruptors Using QSARs Weida Tong Manager of Computational Science Group, Logicon ROW Sciences, FDA s National Center for Toxicological Research (NCTR), U.S.A. Thanks for
Priority Setting of Endocrine Disruptors Using QSARs Weida Tong Manager of Computational Science Group, Logicon ROW Sciences, FDA s National Center for Toxicological Research (NCTR), U.S.A. Thanks for
Host-Pathogen Interaction. PN Sharma Department of Plant Pathology CSK HPKV, Palampur
 Host-Pathogen Interaction PN Sharma Department of Plant Pathology CSK HPKV, Palampur-176062 PATHOGEN DEFENCE IN PLANTS A BIOLOGICAL AND MOLECULAR VIEW Two types of plant resistance response to potential
Host-Pathogen Interaction PN Sharma Department of Plant Pathology CSK HPKV, Palampur-176062 PATHOGEN DEFENCE IN PLANTS A BIOLOGICAL AND MOLECULAR VIEW Two types of plant resistance response to potential
Supplementary information. A proposal for a novel impact factor as an alternative to the JCR impact factor
 Supplementary information A proposal for a novel impact factor as an alternative to the JCR impact factor Zu-Guo Yang a and Chun-Ting Zhang b, * a Library, Tianjin University, Tianjin 300072, China b Department
Supplementary information A proposal for a novel impact factor as an alternative to the JCR impact factor Zu-Guo Yang a and Chun-Ting Zhang b, * a Library, Tianjin University, Tianjin 300072, China b Department
Chemists are from Mars, Biologists from Venus. Originally published 7th November 2006
 Chemists are from Mars, Biologists from Venus Originally published 7th November 2006 Chemists are from Mars, Biologists from Venus Andrew Lemon and Ted Hawkins, The Edge Software Consultancy Ltd Abstract
Chemists are from Mars, Biologists from Venus Originally published 7th November 2006 Chemists are from Mars, Biologists from Venus Andrew Lemon and Ted Hawkins, The Edge Software Consultancy Ltd Abstract
School of Biology. Biology (BL) modules. Biology & 2000 Level /8 - August BL1101 Biology 1
 School of Biology Biology (BL) modules BL1101 Biology 1 SCOTCAT Credits: 20 SCQF Level 7 Semester: 1 10.00 am; Practical classes one per week 2.00-5.00 pm Mon, Tue, or Wed This module is an introduction
School of Biology Biology (BL) modules BL1101 Biology 1 SCOTCAT Credits: 20 SCQF Level 7 Semester: 1 10.00 am; Practical classes one per week 2.00-5.00 pm Mon, Tue, or Wed This module is an introduction
Biology Scope and Sequence Student Outcomes (Objectives Skills/Verbs)
 C-4 N.12.A 1-6 N.12.B.1-4 Scientific Literacy/ Nature of (embedded throughout course) Scientific Inquiry is the process by which humans systematically examine the natural world. Scientific inquiry is a
C-4 N.12.A 1-6 N.12.B.1-4 Scientific Literacy/ Nature of (embedded throughout course) Scientific Inquiry is the process by which humans systematically examine the natural world. Scientific inquiry is a
LIFE-EDESIA: the role of functional assays to implement the substitution principle for EDCs. Stefano Lorenzetti
 2 nd LIFE-EDESIA workshop The role of in vitro functional assays for the assessment of Endocrine Disruptors / EDCs Ranco (VA), July 20 th 2014 LIFE-EDESIA: the role of functional assays to implement the
2 nd LIFE-EDESIA workshop The role of in vitro functional assays for the assessment of Endocrine Disruptors / EDCs Ranco (VA), July 20 th 2014 LIFE-EDESIA: the role of functional assays to implement the
Exhaustive search. CS 466 Saurabh Sinha
 Exhaustive search CS 466 Saurabh Sinha Agenda Two different problems Restriction mapping Motif finding Common theme: exhaustive search of solution space Reading: Chapter 4. Restriction Mapping Restriction
Exhaustive search CS 466 Saurabh Sinha Agenda Two different problems Restriction mapping Motif finding Common theme: exhaustive search of solution space Reading: Chapter 4. Restriction Mapping Restriction
Studying Life. Lesson Overview. Lesson Overview. 1.3 Studying Life
 Lesson Overview 1.3 Characteristics of Living Things What characteristics do all living things share? Living things are made up of basic units called cells, are based on a universal genetic code, obtain
Lesson Overview 1.3 Characteristics of Living Things What characteristics do all living things share? Living things are made up of basic units called cells, are based on a universal genetic code, obtain
FORENSIC TOXICOLOGY SCREENING APPLICATION SOLUTION
 FORENSIC TOXICOLOGY SCREENING APPLICATION SOLUTION A purpose-built collection of the best-inclass components for forensic toxicology Whether you re challenged to present reliable and secure forensic sample
FORENSIC TOXICOLOGY SCREENING APPLICATION SOLUTION A purpose-built collection of the best-inclass components for forensic toxicology Whether you re challenged to present reliable and secure forensic sample
Reaxys Medicinal Chemistry Fact Sheet
 R&D SOLUTIONS FOR PHARMA & LIFE SCIENCES Reaxys Medicinal Chemistry Fact Sheet Essential data for lead identification and optimization Reaxys Medicinal Chemistry empowers early discovery in drug development
R&D SOLUTIONS FOR PHARMA & LIFE SCIENCES Reaxys Medicinal Chemistry Fact Sheet Essential data for lead identification and optimization Reaxys Medicinal Chemistry empowers early discovery in drug development
I. Molecules & Cells. A. Unit One: The Nature of Science. B. Unit Two: The Chemistry of Life. C. Unit Three: The Biology of the Cell.
 I. Molecules & Cells A. Unit One: The Nature of Science a. How is the scientific method used to solve problems? b. What is the importance of controls? c. How does Darwin s theory of evolution illustrate
I. Molecules & Cells A. Unit One: The Nature of Science a. How is the scientific method used to solve problems? b. What is the importance of controls? c. How does Darwin s theory of evolution illustrate
RECENT TRENDS IN PHARMACEUTICAL CHEMISTRY FOR DRUG DISCOVERY
 INTERNATIONAL JOURNAL OF RESEARCH IN PHARMACY AND CHEMISTRY Available online at www.ijrpc.com Review Article RECENT TRENDS IN PHARMACEUTICAL CHEMISTRY FOR DRUG DISCOVERY Sathyaraj A Department of Chemistry,
INTERNATIONAL JOURNAL OF RESEARCH IN PHARMACY AND CHEMISTRY Available online at www.ijrpc.com Review Article RECENT TRENDS IN PHARMACEUTICAL CHEMISTRY FOR DRUG DISCOVERY Sathyaraj A Department of Chemistry,
Valley Central School District 944 State Route 17K Montgomery, NY Telephone Number: (845) ext Fax Number: (845)
 Valley Central School District 944 State Route 17K Montgomery, NY 12549 Telephone Number: (845)457-2400 ext. 18121 Fax Number: (845)457-4254 Advance Placement Biology Presented to the Board of Education
Valley Central School District 944 State Route 17K Montgomery, NY 12549 Telephone Number: (845)457-2400 ext. 18121 Fax Number: (845)457-4254 Advance Placement Biology Presented to the Board of Education
Extrapolating New Approaches into a Tiered Approach to Mixtures Risk Assessment
 Extrapolating New into a Tiered Approach to Mixtures Risk Assessment Michael L. Dourson, PhD, DABT, FATS, FSRA Toxicology Excellence for Risk Assessment (TERA) dourson@tera.org Conflict of Interest Statement
Extrapolating New into a Tiered Approach to Mixtures Risk Assessment Michael L. Dourson, PhD, DABT, FATS, FSRA Toxicology Excellence for Risk Assessment (TERA) dourson@tera.org Conflict of Interest Statement
Gene Network Science Diagrammatic Cell Language and Visual Cell
 Gene Network Science Diagrammatic Cell Language and Visual Cell Mr. Tan Chee Meng Scientific Programmer, System Biology Group, Bioinformatics Institute Overview Introduction Why? Challenges Diagrammatic
Gene Network Science Diagrammatic Cell Language and Visual Cell Mr. Tan Chee Meng Scientific Programmer, System Biology Group, Bioinformatics Institute Overview Introduction Why? Challenges Diagrammatic
Miller & Levine Biology 2010
 A Correlation of 2010 to the Pennsylvania Assessment Anchors Grades 9-12 INTRODUCTION This document demonstrates how 2010 meets the Pennsylvania Assessment Anchors, grades 9-12. Correlation page references
A Correlation of 2010 to the Pennsylvania Assessment Anchors Grades 9-12 INTRODUCTION This document demonstrates how 2010 meets the Pennsylvania Assessment Anchors, grades 9-12. Correlation page references
The Globally Harmonized System (GHS) for Hazard Classification and Labelling. Development of a Worldwide System for Hazard Communication
 The Globally Harmonized System (GHS) for Hazard Classification and Labelling Development of a Worldwide System for Hazard Communication What is the GHS? A common and coherent approach to defining and classifying
The Globally Harmonized System (GHS) for Hazard Classification and Labelling Development of a Worldwide System for Hazard Communication What is the GHS? A common and coherent approach to defining and classifying
Bioinformatics. Genotype -> Phenotype DNA. Jason H. Moore, Ph.D. GECCO 2007 Tutorial / Bioinformatics.
 Bioinformatics Jason H. Moore, Ph.D. Frank Lane Research Scholar in Computational Genetics Associate Professor of Genetics Adjunct Associate Professor of Biological Sciences Adjunct Associate Professor
Bioinformatics Jason H. Moore, Ph.D. Frank Lane Research Scholar in Computational Genetics Associate Professor of Genetics Adjunct Associate Professor of Biological Sciences Adjunct Associate Professor
KLARA Risk assessment
 KLARA Risk assessment - When working with laboratory chemicals Ulrika Olsson, Chemical Safety Coordinator ulrika.olsson@ki.se Introduction SWEA develop regulations and the aim is to prevent us from illness
KLARA Risk assessment - When working with laboratory chemicals Ulrika Olsson, Chemical Safety Coordinator ulrika.olsson@ki.se Introduction SWEA develop regulations and the aim is to prevent us from illness
HAWAII CONTENT AND PERFORMANCE STANDARDS BIOLOGICAL SCIENCE
 HAWAII CONTENT AND PERFORMANCE STANDARDS BIOLOGICAL SCIENCE Correlated to BIOLOGY: CYCLES OF LIFE 2006 5910 Rice Creek Parkway, Suite 1000 Shoreview, Minnesota 55126 Telephone (800) 328-2560 www.agsglobe.com
HAWAII CONTENT AND PERFORMANCE STANDARDS BIOLOGICAL SCIENCE Correlated to BIOLOGY: CYCLES OF LIFE 2006 5910 Rice Creek Parkway, Suite 1000 Shoreview, Minnesota 55126 Telephone (800) 328-2560 www.agsglobe.com
Informatics Framework to Support Integrative
 Informatics Framework to Support Integrative Science and Engineering in Nanotechnology Stacey Lynn Harper, Ph.D. Environmental and Molecular Toxicology Chemical, Biological and Environmental Engineering
Informatics Framework to Support Integrative Science and Engineering in Nanotechnology Stacey Lynn Harper, Ph.D. Environmental and Molecular Toxicology Chemical, Biological and Environmental Engineering
Draft Guideline on Bioanalytical Method (Ligand Binding Assay) Validation in Pharmaceutical Development. (24 January, 2014, MHLW, Japan)
 Draft Guideline on Bioanalytical Method (Ligand Binding Assay) Validation in Pharmaceutical Development (24 January, 2014, MHLW, Japan) Table of Contents 1. Introduction 2. Scope 3. Reference Standard
Draft Guideline on Bioanalytical Method (Ligand Binding Assay) Validation in Pharmaceutical Development (24 January, 2014, MHLW, Japan) Table of Contents 1. Introduction 2. Scope 3. Reference Standard
Gene Control Mechanisms at Transcription and Translation Levels
 Gene Control Mechanisms at Transcription and Translation Levels Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by MHRD Page 1 of 9
Gene Control Mechanisms at Transcription and Translation Levels Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by MHRD Page 1 of 9
Evidence for the Existence of Non-monotonic Dose-response: Does it or Doesn t it?
 Evidence for the Existence of Non-monotonic Dose-response: Does it or Doesn t it? Scott M. Belcher, PhD University of Cincinnati Department of Pharmacology and Cell Biophysics Evidence for the Existence
Evidence for the Existence of Non-monotonic Dose-response: Does it or Doesn t it? Scott M. Belcher, PhD University of Cincinnati Department of Pharmacology and Cell Biophysics Evidence for the Existence
Bioimage Informatics. Lecture 23, Spring Emerging Applications: Molecular Imaging
 Bioimage Informatics Lecture 23, Spring 2012 Emerging Applications: Molecular Imaging Lecture 23 April 25, 2012 1 Outline Overview of molecular imaging Molecular imaging modalities Molecular imaging applications
Bioimage Informatics Lecture 23, Spring 2012 Emerging Applications: Molecular Imaging Lecture 23 April 25, 2012 1 Outline Overview of molecular imaging Molecular imaging modalities Molecular imaging applications
Bioinformatics and Computerscience
 Bioinformatics and Computerscience Systems Biology Data collection Network Inference Network-based dataintegration 1. ARRAY BASED 2. NEXT-GEN SEQUENCING RNA-Seq analysis ChIP-seq Bulked segregant analysis
Bioinformatics and Computerscience Systems Biology Data collection Network Inference Network-based dataintegration 1. ARRAY BASED 2. NEXT-GEN SEQUENCING RNA-Seq analysis ChIP-seq Bulked segregant analysis
BMD645. Integration of Omics
 BMD645 Integration of Omics Shu-Jen Chen, Chang Gung University Dec. 11, 2009 1 Traditional Biology vs. Systems Biology Traditional biology : Single genes or proteins Systems biology: Simultaneously study
BMD645 Integration of Omics Shu-Jen Chen, Chang Gung University Dec. 11, 2009 1 Traditional Biology vs. Systems Biology Traditional biology : Single genes or proteins Systems biology: Simultaneously study
Controlled Substances: TSCA the Toxic Substances Control Act
 The select agent list and relevant regulatory program information are maintained by the CDC and can be found on the select agent web site http://www.selectagents.gov/. The regulations covering select agents
The select agent list and relevant regulatory program information are maintained by the CDC and can be found on the select agent web site http://www.selectagents.gov/. The regulations covering select agents
Richik N. Ghosh, Linnette Grove, and Oleg Lapets ASSAY and Drug Development Technologies 2004, 2:
 1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
1 3/1/2005 A Quantitative Cell-Based High-Content Screening Assay for the Epidermal Growth Factor Receptor-Specific Activation of Mitogen-Activated Protein Kinase Richik N. Ghosh, Linnette Grove, and Oleg
Fundamentals Of Analytical Toxicology PDF
 Fundamentals Of Analytical Toxicology PDF The analytical toxicologist may be required to detect, identify, and in many cases measure a wide variety of compounds in samples from almost any part of the body
Fundamentals Of Analytical Toxicology PDF The analytical toxicologist may be required to detect, identify, and in many cases measure a wide variety of compounds in samples from almost any part of the body
