Global Quantification of Tissue Dynamics in the Developing Mouse Kidney
|
|
|
- Erin Parks
- 5 years ago
- Views:
Transcription
1 Developmental Cell, Volume 29 Supplemental Information Global Quantification of Tissue Dynamics in the Developing Mouse Kidney Kieran M. Short, Alexander N. Combes, James Lefevre, Adler L. Ju, Kylie M. Georgas, Timothy Lamberton, Oliver Cairncross, Bree A. Rumballe, Andrew P. McMahon, Nicholas A. Hamilton, Ian M. Smyth, and Melissa H. Little
2 Supplementary Figures Supplementary Figure S1: The clade structure of the ureteric tree, related to Figure 3.
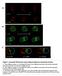
3 From ls7 to 12 ( dpc), a clearly defined ureteric tree structure was observed in all normal ureteric trees in our data. A. A schematic of the common structure is shown. From the root the tree is divided into the anterior (A) and posterior (P) branches. The anterior branch then divides into the anterior minor (Am) and anterior major (AM) branches. The Am is the smaller branch placed spatially closer to the root while the AM branch is significantly larger and further towards the anterior pole. The AM subsequently divides into the anterior major lateral (AML) and anterior major anterior (AMA) that are placed laterally and further anteriorly, respectively. The posterior minor (Pm), posterior major (PM), posterior major lateral (PML), posterior major posterior (PMP) are defined similarly. The progeny from these six key branches represent clades within the final organ. B. 3D visualisation of an ls11 (14.5dpc) and ls12 (15.5dpc) kidney with these common clades highlighted. Branches are colored according to the branch depth from the root (number of branch points between the relevant branch and the root), with red being a small branch depth and blue being large. The ls11 and ls 12 trees shown are the same as those shown in Figure 3.
4 Supplementary Figure S2: Changes in ureteric tree branch diameter, generation number and tip geometry prior to and in response to pelvis formation, related to Figure 4. A. Ureteric branch diameter. At limb stage 7, branch diameter is relatively large given the size of the organ, and branch diameter is uniform across the tree until limb stage 11 when generations closer to the pelvis begin to enlarge. The increase in diameter is most apparent between limb stages 11 and 12 for generation 1 to 4 when the renal pelvis begins to form. B. Tip geometry throughout development. Analysis of tip globallocal bifurcation angle deltas indicates that tip pairs are more outwardly curved between limb stages 7-10 than tips from limb stages 11-13, where the delta decreases (indicating a move towards a more closed geometry). C. Projection
5 of tip number versus branch generation number for kidneys of different developmental stages shows an exponential correlation (log10 tip number axis) between tip number and the number of generations that group with developmental age. This correlation is not matched by the ls13/e16.5 organs that have decreased by 3-4 branch generations (the correlation trajectory is represented by the dashed line). D. The pelvis rapidly changes in morphology between E15.5 and E16.5 kidneys, and this results in the decrease in observed branch generations through subsumption of branches nearest to the pelvis. *** p< two level nested ANOVA. Error bars represent SEM. Scale bar represents 100 µm.
6 Supplementary Figure S3. Quantitation of niches in the developing kidney, related to Figure 5. A. Six2/CytoK (red/white) OPT developmental timeseries from 11.5dpc to P3/22.5dpc. Scale bars are provided for the top (400 m) and bottom (1000 m) rows. B. Representations of distinct niche types illustrating the relationship between a CM field and the underlying ureteric epithelium. Top: at early stages the CM is broad and can cover large areas of underlying UT, often defining a niche with two terminal regions that would be identified by Tree Surveyor as two tips. Bottom: at later stages the CM is smaller and more likely to be separated between two adjacent tips, thereby defining two separate niches, and
7 aligning with Tree Surveyor tip counts. Note- Tips were manually counted in two kidneys from different litters at P3 to check the assumption that niche number still aligned with tip number at this stage. The counts were 3264 and 3065, which are in a similar area to our final niche counts at P2. C. Quantitation of niche number and tip number across development. Niche numbers were annotated as described in Supplementary methods and tip number was derived from Tree Surveyor. D. Niche and tip number plotted on a linear scale over time. Error bars represent SEM.
8 Supplementary Figure S4: Quantitation of cell numbers and rates of mitosis during development, related to Figure 5 and 6. A. Observed total cap mesenchyme cell number / organ across developmental time. Population counts based on average cap cell number per niche x average niche number. Error bars represent SEM. B. Quantitation of tip cell number
9 and cap cell number across time. n represents the number of samples used to derive the average value. C. Observed rates of mitosis (% phh3 + cells +/- SEM; n = as per relevant compartments in B) quantitated in cap mesenchyme (red) versus ureteric tip (grey) across development. D. Quantitation of cells undergoing mitosis (as labelled by phh3) across time. E. Representative optical slices from the two EdU time series experiments at 0.5, 4.5, 8.5, 12.5 and 16.5 hours post initial injection. Samples were labelled with Six2 (red), Cytokeratin (green), and EdU (white) and analysed in 3D as above. Scale bars 20 m.
10 Supplementary Figure S5: Spatial analysis of cellular behaviour across the niche, related to Figure 6. A,B. Localisation of apoptosis using immunofluorescence for Caspase 3. A. View of a 13.5dpc Six2TGC kidney analysed with antibodies to Caspase 3 (red) and anti-gfp (green) and counterstained with DAPI. Evidence for apoptosis in the CM was negligible. B. The interdigital webbing in the 13.5dpc forelimb undergoes Caspasemediated apoptosis. Robust detection of Caspase 3 protein in this sample serves as a positive control (red signal, white arrows). C-I. Spatial analysis of EdU incorporation in the CM across time. C. Analysis of relative Six2 intensity versus depth in the cap mesenchyme.
11 D-F. Overview cartoon and analysis of the incorporation of EdU relative to normalised Six2 intensity. Individual measurements are grouped by Six2 intensity bins. G-I. Overview cartoon and analysis of EdU incorporation versus depth ( ) from the periphery of the kidney/top of the cap mesenchyme.
12 Supplementary Figure S6. Imaging of cap mesenchyme during period of cessation of nephrogenesis between birth and postnatal day 3, related to Figure 7. A-D. Confocal images of the nephrogenic zone from birth (P0) to P4 illustrating the Six2 + (red) and cytokeratin + (white) compartments. Note the dramatic morphological shift in the CM compartment with the formation of large renal vesicles composed of cells still expressing Six2 protein evident from P2 (examples outlined). By P4, little remaining mesenchymal Six2 staining is evident. Time series Compartment c (hours) s (hours) c 1 (hours) c 2 (hours) a (%) 13.25dpc 17.25dpc UT 11.7 ± ± 0.6 CM 14.2 ± ± ± ± ± 5 UT 23.5 ± ± 1.3 CM 32.8 ± ± ± ± ± 6 Supplementary Table S1. Cell cycle lengths and error estimates for the CM and UT, related to Figure 6.
13
14 Supplementary Movies Supplementary Movie S1. Movies of the branching ureteric tree, related to Figure 1. Rendered optical projection tomography data collected from ls7 (12.0dpc) to ls13 (16.5dpc) embryonic kidneys with immunofluorescence staining for cytokeratin and TROP-2 illustrating the morphology of the branching ureteric epithelium. Supplementary Movie S2. Animation demonstrating methods used for both cellular quantitation within compartments of the niche and quantitation of the proportion of EdU positive cells across the EdU injection timecourses. Relates to Figures 5. A. Cellular quantitation within compartments of the niche is illustrated first using confocal data from a 15.5dpc kidney in Imaris (Bitplane). The sample was prepared using coimmunoflorescence for Six2 (red; cap mesenchyme), Calb1 (white: ureteric epithelium), nuclei (blue; DAPI) and mitosing cells (green; phh3). The sequence commences with a maximum intensity projection of the data, then displays a surface render of the ureteric epithelium, which restricts to the tip epithelium, before renderings of the cap mesenchyme appear. The renderings of these compartments are then used to isolate the fluorescent signal contained within. Six2 and Phh3 signal within cap renderings is displayed, along with DAPI signal from the rendered tips. These nuclear signals are then overlaid with the output of a spot finding algorithm (red for CM, white for UT), which enables quantitation and spatial analysis of cells within each tip and cap. B. The second portion of the movie illustrates the quantitation of the proportion of EdU positive cells in different cellular compartments. Animation sequence demonstrating the approach used for The data
15 presented represents an EdU timecourse commenced at 13.25dpc and collected after 0.5hrs. The sample was prepared using co-immunoflorescence for Six2 (red; cap mesenchyme), Cytokeratin (green: ureteric epithelium), EdU (white) and nuclei (blue; DAPI). This highlights each of the two progenitor compartments of interest (cap mesenchyme and ureteric tip epithelium) in sequence showing the fraction of cells stained for EdU in each instance. The sequence commences with a maximum intensity projection of all channels, then DAPI signal is removed, renders of Six2 positive cap cells and the underlying tip domain are displayed before removing the tip to show only the cap mesenchyme. The EdU positive cells within this region are then highlighted. Subsequently, the sequence returns to the rendered tips alone and shows EdU signal within this region.
16 Supplementary Experimental Procedures Estimating number of nephrons, CM cells and UT cells in whole kidney The mean per-niche counts of CM cells, UT cells and nephrons were multiplied by the mean niche counts at corresponding ages to obtain whole organ estimates. No niche counts were available for 22.5dpc, so the measured increase in niche number between 19.5 and 21.5dpc was extrapolated geometrically to 22.5dpc, as follows. Let and be the log niche count estimates at 19.5 and 21.5dpc, where the standard errors are obtained from the log transform of the raw counts. Then the log niche count estimate at 22.5dpc is ( ). The standard errors for the whole population estimates were calculated using the standard formula for the variance of the product of independent random variables. Defining ampullae, T-tips and tri-tips within niches. The definition of a tip within the context of OPT analyses of the ureteric epithelium was the terminal segment of the branching structure down to the closest identifiable branchpoint. While this was convenient when considering the ureteric compartment alone, when characterising the UT/CM relationships using single cell resolution confocal analyses, the ureteric tip within niches were classified into three recurring morphologies; ampulla, T-tip, and tri-tip (Fig. 2). A single terminal swelling of roughly spherical shape surrounded by a separable cap mesenchymal field was classified as an ampulla. A T-tip was defined as a terminal segment of the ureteric tree with paired swellings on either side of a
17 connecting trunk segment. This was distinguished from two ampullae when covered by a single region of Six2 + cap mesenchyme. A terminal segment with three prominences also associated with a single inseparable cap mesenchymal field was classified a tri-tip (Fig. 2). Niche rendering and cell counting within CM and UT populations Using Six2 fluorescent signal, the boundaries of cap mesenchyme were semi automatically drawn (using Isolines function) slice by slice in Surface mode. On occasion, if the boundary of one niche was immediately adjacent to another, the manual drawing function was employed. Renal vesicles and later stage nephron structures still expressing Six2 protein were excluded from the boundary. Exclusion was based on cell morphology, when polarisation and a lumen were visible. Upon completion, a surface was rendered for each cap niche and the volume recorded. Six2 and phh3 channels were masked and a spot count performed to determine the total number of Six2 + cells per niche and the percentage of cells in mitosis (phh3 + ). A co-localisation function was performed to ensure phh3 + Six2 + cells only were counted. Similar methods were employed in the tip, using Calbindin or Cytokeratin signal to demark the boundary of the tip and the isoline or manual function to segment a surface (Movie S2). Tip surfaces within a niche were cut off perpendicular to the trunk at the intersection of trunk and tip, with one tip niche corresponding to one adjacent CM niche, and a volume was recorded. DAPI and phh3 channels were masked and a spot count performed to determine the total number of nuclei per niche and the percentage of cells in mitosis. Each of these niche based metrics, and also the ratio of CM cell count to UT cell count, was averaged by sample (see Supplementary Figures S3 and S4 for sample
18 number details). These averages were then used to calculate mean and SEM by limb stage/dpc. Estimation of percentage EdU positive cells within cap mesenchyme and ureteric tip Image data from EdU timeseries labelled for Six2, cytokeratin, DAPI and EdU was loaded into Imaris (Bitplane AG) where the EdU channel was binarised (any signal above background for that channel, determined to be 35, was processed to have the maximum intensity of 255), and the binary channel inspected to ensure an accurate representation of true EdU signal. DAPI signal was then rendered to generate a population of surfaces that encompass all individual nuclei that remained after quality control (smoothing 0.62 M, background subtraction with an assumed object diameter of 2.33 M, split touching object seed detection 3.1, automatic quality threshold, filter objects by number of voxels , or approximately cubic, to exclude clusters of cells or other spurious results). These were then filtered by Six2 signal intensity (above 65 on a scale, a threshold chosen as the minimum value that reliably excluded RV and other non-cap cells). Each surface (Six2 + nucleus) within this population has a unique identifier and was interrogated for the mean intensity of binarised EdU within that surface, as well as 3 dimensional position, distance from the rendered UT, and other marker intensities. The number of surfaces with more than 128 intensity units in the binarised EdU channel (greater than one half of nucleus occupied by EdU signal above background levels) were counted and expressed as a % of the population (% incorporation). A similar method was used to determine the % incorporation of EdU in the ureteric tree, but as we did not have a nucleus
19 specific marker for that compartment we used the Calbindin or Cytokeratin signal to create a mask and excise the nuclear DAPI signal within the tree. We then created surfaces for the masked DAPI channel (nuclei within the ureteric tree) and measured the % incorporation of EdU within that population. % incorporation was assessed for >8 samples from >=2 litters for each time point. Data capture for assessment of cellular dynamics Spatial variation in EdU incorporation and Six2 intensity was examined within the CM compartment, as outlined above and in Figure S5. Within each image, an analysis was performed in R (R Development Core Team, 2013) to estimate the distance of each cap cell from the surface of the CM (depth). All images were oriented so that the confocal slices spanned this surface, with the top of the stack in the distal position. For each sample, the positions of the identified cap cells were used to generate a convex hull (Barber et al, 2012). Hull faces located on the bottom or sides of the sample were algorithmically identified and removed, and then the distance to each cell was calculated as the minimum perpendicular distance from the remaining faces. To test associations between Six2 intensity, EdU incorporation, and depth within the cap mesenchyme, data from individual cells was aggregated for each timecourse and compartment. The relationship between EdU incorporation and depth was plotted by ordering the cells at each time (hours post initial injection; hpi) by depth, grouping them into equal sized bins, and plotting mean depth against the EdU + proportion for each bin. Other comparisons were performed similarly, but with adjustments to the Six2 intensity as
20 described below. Analysis of the spatial variation of the DAPI, EdU, and Six2 signals showed a weakening of all channels closer to the cap surface and further from the tip surface. Under the assumption that this represents an image analysis artefact linked to cell density, and that there should be no true systematic variation in the DAPI intensity, we divided the mean Six2 intensity by the mean DAPI intensity in each cell to get a relative Six2 intensity metric. We also controlled for the variability in Six2 intensity between samples by ranking all cap cells within each sample by relative Six2 intensity, and assigning them a quantile score between 0 and 1. In all cases, plots were repeated with cells aggregated at a finer level to ensure that observed trends were generally consistent and not driven by outlier samples. Mathematical modelling and data analysis of cellular dynamics In this section we briefly describe the methodology for model fitting as previously given (Lefevre et al, 2013). For each timecourse and compartment measured, we fit EdU incorporation models to the timeseries of proportional EdU incorporation (litter averages), with time measured from the initial EdU injection (hpi). The models fitted were one or two population asynchronous exponential models with the second gap phase and mitosis assumed to be of negligible length (Meraldi et al, 2004). These models predict non-linear time versus EdU incorporation curves with two and four free parameters respectively, which were fitted to the timeseries using the numerical non-linear least squares fitting algorithm nls in the R statistical programming language (version ). The one population model is fully determined provided that the timeseries includes at least two time points prior to EdU saturation, while the two population model is fully determined provided
21 that there are at least two time points prior to the saturation of the faster cycling population, and two time points between the saturation of the faster cycling and slower cycling populations. Note that these conditions do not guarantee a robust fit, due to the possibility of noise in the data. Let c and s be the duration of the cell cycle and of S-phase respectively, which in the single population model are assumed to be approximately the same for all cells. Let t be the time since the initiation of the experiment (first EdU injection), N(t) be the total number of cells in the population at time t, E(t) be the number of EdU+ cells in the population at time t, and ( ) ( ) ( ) (the measured quantity). For a given cell at a given time, we refer to the time since the start of the cell cycle (initiation of G 1 ) as the age of the cell. The single population asynchronous exponential model is based on the assumption of a cell population that has been proliferating without exit or entry sufficiently long to reach an equilibrium age profile (the proportion of cells at a given age is constant). With the additional assumption that G 2 and mitosis are of negligible duration, this model predicts the near linear EdU incorporation curve ( ) The assumption that no cells enter the compartment is supported by lineage analyses (Kobayashi et al, 2008). The assumption that daughter cells remain in the compartment after mitosis as cells actively within the cell cycle is not as clear cut. The presence of a significant proportion of quiescent cells can be excluded by showing that EdU
22 incorporation eventually reaches saturation, but it is known that cells exit the CM compartment via differentiation into RV cells. If this exit occurs throughout the cell cycle without bias, then the model remains correct. However if exit occurs immediately after mitosis, the age distribution and hence the correct expression for R(V) will be altered slightly, depending on the amount of exit. The timing of exit is uncertain, however the potential impact of this issue is small (Lefevre et al, 2013). There is biological evidence of heterogeneity with respect to cellular phenotype within the cap mesenchyme (Mugford et al, 2009). Hence, we also developed a two population asynchronous exponential model with S-phase at end of cell cycle to compare for data fit. In this model, we assume that the cells within the compartment measured comprise two subpopulations, each of which follows the assumptions of the single population asynchronous exponential model with the S-phase at the end of the cell cycle. These subpopulations are assumed to have the same S-phase length s but different cell cycle lengths c 1 and c 2, where. We further assume that daughter cells remain in the same subpopulation as their parent, but that the ratio between the number of cells in each subpopulation remains constant over the time course; this implies that cells must exit from the faster cycling population (recall that this is consistent with the model provided that exit is equally common from any stage of the cell cycle). Let a be the proportion of slower cycling cells in the population. Under these assumptions the ratio of EdU + cells within each subpopulation is given by the formula for the single population asynchronous exponential model with appropriate c. Since the ratio between the subpopulations remains fixed, the
23 EdU + proportion for the combined population is a simple weighted average of these functions, ( ) { ( ) ( ) ( ) ( ) The effective average cell cycle length for the two population model is taken to be the cell cycle that would be required for a single population to have the same overall proliferation rate. The proliferation rate is inversely proportional to cell cycle length, so this is obtained by taking the harmonic mean of the two cell cycle lengths, weighted by the relative sizes of the populations. That is, ( ) EdU model fitting One and two population asynchronous models were fitted to all EdU incorporation time series (litter averages), as detailed in the supplementary methods section, with the most convincing fit selected in each case. The fitted parameters are given in Figure 6, and repeated below with standard errors where available (Supplementary Table S1). Note that standard errors are based on a linear approximation of the parameter space in the region of the fitted solution.
24 The single population model provided a good fit to the UT data at both and 17.25dpc, with no indication of a slower subpopulation, suggesting uniform proliferation within this compartment. The evidence for a substantial slowdown in proliferation between these times is very clear, with an approximate doubling of cell cycle length. The 13.25dpc CM data is clearly inconsistent with the one population model, while the two population model provides a good and robust fit. The 17.25dpc timecourse was terminated at 24.5hpi to avoid the possibility of birth. By this time, the average EdU incorporation in the CM had only reached 86%. Although a small amount of quiescence cannot be conclusively ruled out, the data is again consistent with the two population model. Modelling population exit from the ureteric tip and cap mesenchyme The rate of exit over time from each compartment was estimated using the cell population and EdU results. The niche and compartment cell counts give a timeseries of observed cell number estimates for the cap and tip compartments. Let t 1 and t 2 be two adjacent time points in this series, with cell population estimates N 1 and N 2 for a given compartment. We assume a proportional cell proliferation rate k and proportional exit rate e that are constant over the time period between t 1 and t 2 ; that is, we assume that if N(t) is the cell population at time t,, then cells are being added via mitosis at a rate of ( ) cells per unit time, and exiting at a rate of ( ) cells per unit time, so that the population follows the exponential model ( ) ( ) ( ). We use the EdU results to estimate the cell cycle length c for the period t 1 to t 2, interpolating or extrapolating as necessary. This represents
25 the time required for the population to double in the absence of exit, hence ( ). Equating to the known population values gives ( ) ( ) ( ( ) ) ( ) ( ) ( ( ) ) and hence ( ( ) )( ) Solving for e gives ( ) ( ) This implies that the total number of cells exiting the compartment over the period t 1 to t 2 is equal to ( ) ( ) ( ( ( ) ) ( ( ) ) ) ( ) Intermediate cell population estimates and the exit over each day of the period t 1 to t 2, if it spans multiple days, can be obtained from the fitted model. The standard error of the proliferation rate ( ) was obtained directly from the fitted EdU models, re-parameterised with the proliferation rate instead of the cell cycle length. Let M 1 and M 2 be the number of niches at t 1 and t 2. Then the net growth rate is ( ) ( ) ( ) ( ) ( ) Sample averaged niche counts and cell counts per niche were log-transformed, and SEMs were calculated in log space. The SEM for the net growth rate over each age interval was then calculated using the standard formula for a weighted sum of independent random
26 variables. This formula was used again to obtain the standard error for the exit rate, based on the standard errors obtained for the proliferation rate and net growth rate. The absolute cell exit estimate between t 1 and t 2 is ( ), where the exit rate is assumed to be constant over the time period. To obtain the standard error for this estimate we make the further simplifying assumption that the exit rate is independent of the integral over the population (although both terms depend on N 1 and N 2, the exit rate depends on the change in population and will have little correlation to the mean value). For the SE calculation only, we make the further conservative approximation ( ) ( )( ) ( ( ) ( )). The standard error for the absolute cell exit estimate is then obtained from the standard errors for the cell population and exit rate estimates, using the usual formula for the product of independent random variables. Application of the modelling of proliferation and exit over time Cell proliferation information is available in the form of mitosis data and the results of the EdU analysis. Mitosis results were variable, but suggested a higher rate of proliferation in the period of rapid branching elaboration prior to 15.5dpc, then transitioning to a lower rate for the remainder of the timecourse, broadly consistent with the EdU data (Figs. S4; Movie S2). On this basis we extrapolated and interpolated the estimated mean cell cycles from the EdU experiment as follows: the cell cycles obtained from the 13.25dpc EdU experiment was assumed to hold over the period dpc, the cell cycles obtained from the 17.25
27 dpc EdU experiment was assumed to hold over the period dpc, and for the period dpc we used the average of the and 17.25dpc results. The same system was used for CM and UT. Note that exit estimates are subject to this assumption. In particular, if the mitosis results indicating an increase in CM proliferation after birth reflect reality, total exit from this compartment could be significantly higher than estimated below. No later EdU experiment was possible due to the possibility of experimental stress inducing early birth. Using these cell cycle estimates and the cell population counts, exit from the CM and UT compartments was modelled as described in the supplementary methods (results in Figure 6E,F). The model gives total exit from the CM of cells by 22.5dpc. Assuming the remaining cells differentiate rapidly without significant proliferation or death gives a total exit from the CM of cells. Spatial analysis of EdU incorporation and Six2 intensity To further investigate the heterogeneity in the CM indicated by the EdU modelling, we plotted EdU incorporation against depth (distance from the outer surface of the CM), and normalised Six2 intensity, as described in the supplementary methods. In both time courses, EdU incorporation was highest in cells with low Six2 expression. Conversely, cells with high Six2 expression had the lowest average EdU incorporation (Fig. S5). The relationship between Six2 intensity and EdU incorporation is reinforced by a spatial analysis of EdU incorporation in Six2 + cells relative to depth from the top of the CM (Fig. S5). Across the and 17.25dpc experiments, cells closer to the top of the CM
28 incorporate less EdU and cells further from the top incorporate more. These results suggest a continuous variation of proliferation with Six2 intensity and depth, but they are also consistent with the two population model, with a faster cycling induced population exhibiting lower average Six2 expression relative to the uninduced population. While distinct differences can be detected at different depths in the CM, this data supports a view where the two populations are substantially intermingled or at least overlap in depth and Six2 intensity. This will be investigated in future work.
29 Supplementary References (not included in main manuscript) Meraldi, P., Draviam, V.M., and Sorger, P.K. (2004) Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7, Srinivas, S., Goldberg M.R., Watanabe T., D'Agati V., al-awqati Q., and Costantini F. (1999) Expression of green fluorescent protein in the ureteric bud of transgenic mice: a new tool for the analysis of ureteric bud morphogenesis. Dev. Genet. 24,
Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct
 SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo
 Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Fig. S1. Expression pattern of moody-gal4 in third instar. Maximum projection illustrating a dissected moody-gal4>ngfp L3 larva stained for Repo (magenta), Fas2 (blue) and GFP (green) in overview (A) and
Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration
 Cell Stem Cell Supplemental Information Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration Micah T. Webster, Uri Manor, Jennifer Lippincott-Schwartz,
Cell Stem Cell Supplemental Information Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration Micah T. Webster, Uri Manor, Jennifer Lippincott-Schwartz,
Supplemental Information. Inferring Cell-State Transition. Dynamics from Lineage Trees. and Endpoint Single-Cell Measurements
 Cell Systems, Volume 3 Supplemental Information Inferring Cell-State Transition Dynamics from Lineage Trees and Endpoint Single-Cell Measurements Sahand Hormoz, Zakary S. Singer, James M. Linton, Yaron
Cell Systems, Volume 3 Supplemental Information Inferring Cell-State Transition Dynamics from Lineage Trees and Endpoint Single-Cell Measurements Sahand Hormoz, Zakary S. Singer, James M. Linton, Yaron
Dr. Amira A. AL-Hosary
 Phylogenetic analysis Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic Basics: Biological
Phylogenetic analysis Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic Basics: Biological
In order to confirm the contribution of diffusion to the FRAP recovery curves of
 Fonseca et al., Supplementary Information FRAP data analysis ) Contribution of Diffusion to the recovery curves In order to confirm the contribution of diffusion to the FRAP recovery curves of PH::GFP
Fonseca et al., Supplementary Information FRAP data analysis ) Contribution of Diffusion to the recovery curves In order to confirm the contribution of diffusion to the FRAP recovery curves of PH::GFP
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and
 Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Supplementary Figure 1. Real time in vivo imaging of SG secretion. (a) SGs from Drosophila third instar larvae that express Sgs3-GFP (green) and Lifeact-Ruby (red) were imaged in vivo to visualize secretion
Life in an unusual intracellular niche a bacterial symbiont infecting the nucleus of amoebae
 Life in an unusual intracellular niche a bacterial symbiont infecting the nucleus of amoebae Frederik Schulz, Ilias Lagkouvardos, Florian Wascher, Karin Aistleitner, Rok Kostanjšek, Matthias Horn Supplementary
Life in an unusual intracellular niche a bacterial symbiont infecting the nucleus of amoebae Frederik Schulz, Ilias Lagkouvardos, Florian Wascher, Karin Aistleitner, Rok Kostanjšek, Matthias Horn Supplementary
protein biology cell imaging Automated imaging and high-content analysis
 protein biology cell imaging Automated imaging and high-content analysis Automated imaging and high-content analysis Thermo Fisher Scientific offers a complete portfolio of imaging platforms, reagents
protein biology cell imaging Automated imaging and high-content analysis Automated imaging and high-content analysis Thermo Fisher Scientific offers a complete portfolio of imaging platforms, reagents
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
Algorithm User Guide:
 Algorithm User Guide: Nuclear Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to use
Algorithm User Guide: Nuclear Quantification Use the Aperio algorithms to adjust (tune) the parameters until the quantitative results are sufficiently accurate for the purpose for which you intend to use
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
doi:10.1038/nature11419 Supplementary Figure 1 Schematic representation of innate immune signaling pathways induced by intracellular Salmonella in cultured macrophages. a, During the infection Salmonella
Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut
 Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic analysis Phylogenetic Basics: Biological
Amira A. AL-Hosary PhD of infectious diseases Department of Animal Medicine (Infectious Diseases) Faculty of Veterinary Medicine Assiut University-Egypt Phylogenetic analysis Phylogenetic Basics: Biological
SIGNIFICANCE OF EMBRYOLOGY
 This lecture will discuss the following topics : Definition of Embryology Significance of Embryology Old and New Frontiers Introduction to Molecular Regulation and Signaling Descriptive terms in Embryology
This lecture will discuss the following topics : Definition of Embryology Significance of Embryology Old and New Frontiers Introduction to Molecular Regulation and Signaling Descriptive terms in Embryology
5.1 Cell Division and the Cell Cycle
 5.1 Cell Division and the Cell Cycle Lesson Objectives Contrast cell division in prokaryotes and eukaryotes. Identify the phases of the eukaryotic cell cycle. Explain how the cell cycle is controlled.
5.1 Cell Division and the Cell Cycle Lesson Objectives Contrast cell division in prokaryotes and eukaryotes. Identify the phases of the eukaryotic cell cycle. Explain how the cell cycle is controlled.
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09414 Supplementary Figure 1 FACS-isolated 8c hepatocytes are highly pure. a, Gating strategy for identifying hepatocyte populations based on DNA content. b, Detection of mchry and mchr9
doi:10.1038/nature09414 Supplementary Figure 1 FACS-isolated 8c hepatocytes are highly pure. a, Gating strategy for identifying hepatocyte populations based on DNA content. b, Detection of mchry and mchr9
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Mourier et al., http://www.jcb.org/cgi/content/full/jcb.201411100/dc1 Figure S1. Size and mitochondrial content in Mfn1 and Mfn2 knockout hearts. (A) Body
Supplemental material THE JOURNAL OF CELL BIOLOGY Mourier et al., http://www.jcb.org/cgi/content/full/jcb.201411100/dc1 Figure S1. Size and mitochondrial content in Mfn1 and Mfn2 knockout hearts. (A) Body
Supporting Information Axial and transverse intensity profiles Figure S1. Relative axial coordinate of permeabilization events
 Supporting Information Localized Permeabilization of E. coli Membranes by the Antimicrobial Peptide Cecropin A Nambirajan Rangarajan, Somenath Bakshi, and James C. Weisshaar Axial and transverse intensity
Supporting Information Localized Permeabilization of E. coli Membranes by the Antimicrobial Peptide Cecropin A Nambirajan Rangarajan, Somenath Bakshi, and James C. Weisshaar Axial and transverse intensity
Analysis and Simulation of Biological Systems
 Analysis and Simulation of Biological Systems Dr. Carlo Cosentino School of Computer and Biomedical Engineering Department of Experimental and Clinical Medicine Università degli Studi Magna Graecia Catanzaro,
Analysis and Simulation of Biological Systems Dr. Carlo Cosentino School of Computer and Biomedical Engineering Department of Experimental and Clinical Medicine Università degli Studi Magna Graecia Catanzaro,
Heather Currinn, Benjamin Guscott, Zita Balklava, Alice Rothnie and Thomas Wassmer*
 Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Online Resources APP controls the formation of PI(3,5)P 2 vesicles through its binding of the PIKfyve complex. Cellular and Molecular Life Sciences Heather Currinn, Benjamin Guscott, Zita Balklava, Alice
Nature Protocols: doi: /nprot Supplementary Figure 1
 Supplementary Figure 1 Photographs of the 3D-MTC device and the confocal fluorescence microscopy. I: The system consists of a Leica SP8-Confocal microscope (with an option of STED), a confocal PC, a 3D-MTC
Supplementary Figure 1 Photographs of the 3D-MTC device and the confocal fluorescence microscopy. I: The system consists of a Leica SP8-Confocal microscope (with an option of STED), a confocal PC, a 3D-MTC
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
DOI: 10.1038/ncb2647 Figure S1 Other Rab GTPases do not co-localize with the ER. a, Cos-7 cells cotransfected with an ER luminal marker (either KDEL-venus or mch-kdel) and mch-tagged human Rab5 (mch-rab5,
7.013 Problem Set
 7.013 Problem Set 5-2013 Question 1 During a summer hike you suddenly spot a huge grizzly bear. This emergency situation triggers a fight or flight response through a signaling pathway as shown below.
7.013 Problem Set 5-2013 Question 1 During a summer hike you suddenly spot a huge grizzly bear. This emergency situation triggers a fight or flight response through a signaling pathway as shown below.
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
DOI: 10.1038/ncb2215 Figure S1 Number of egfp-vps4a bursts versus cellular expression levels. The total number of egfp-vps4a bursts, counted at the end of each movie (frame 2000, after 1h 28 min) are plotted
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Role of Mitochondrial Remodeling in Programmed Cell Death in
 Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Supplementary information
 Supplementary information doi: 10.1038/nchem.247 Amyloid!-Protein Oligomerization and the Importance of Tetramers and Dodecamers in the Aetiology of Alzheimer s Disease Summer L. Bernstein, Nicholas F.
Supplementary information doi: 10.1038/nchem.247 Amyloid!-Protein Oligomerization and the Importance of Tetramers and Dodecamers in the Aetiology of Alzheimer s Disease Summer L. Bernstein, Nicholas F.
Supplementary material: Methodological annex
 1 Supplementary material: Methodological annex Correcting the spatial representation bias: the grid sample approach Our land-use time series used non-ideal data sources, which differed in spatial and thematic
1 Supplementary material: Methodological annex Correcting the spatial representation bias: the grid sample approach Our land-use time series used non-ideal data sources, which differed in spatial and thematic
IncuCyte ZOOM NeuroTrack Fluorescent Processing
 IncuCyte ZOOM NeuroTrack Fluorescent Processing The NeuroTrack TM Software Module (Cat No 9600-0011) is used to measure the processes of neurons in monoculture or with fluorescent labeling in co-culture.
IncuCyte ZOOM NeuroTrack Fluorescent Processing The NeuroTrack TM Software Module (Cat No 9600-0011) is used to measure the processes of neurons in monoculture or with fluorescent labeling in co-culture.
Figure 1. Automated 4D detection and tracking of endosomes and polarity markers.
 A t = 0 z t = 90 B t = 0 z t = 90 C t = 0-90 t = 0-180 t = 0-300 Figure 1. Automated 4D detection and tracking of endosomes and polarity markers. A: Three different z planes of a dividing SOP shown at
A t = 0 z t = 90 B t = 0 z t = 90 C t = 0-90 t = 0-180 t = 0-300 Figure 1. Automated 4D detection and tracking of endosomes and polarity markers. A: Three different z planes of a dividing SOP shown at
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Cell Cycle and Mitosis
 Cell Cycle and Mitosis THE CELL CYCLE The cell cycle, or cell-division cycle, is the series of events that take place in a eukaryotic cell between its formation and the moment it replicates itself. These
Cell Cycle and Mitosis THE CELL CYCLE The cell cycle, or cell-division cycle, is the series of events that take place in a eukaryotic cell between its formation and the moment it replicates itself. These
Chapter 18 Lecture. Concepts of Genetics. Tenth Edition. Developmental Genetics
 Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 18 Lecture Concepts of Genetics Tenth Edition Developmental Genetics Chapter Contents 18.1 Differentiated States Develop from Coordinated Programs of Gene Expression 18.2 Evolutionary Conservation
Chapter 11. Development: Differentiation and Determination
 KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
Introduction to Embryology. He who sees things grow from the beginning will have the finest view of them.
 He who sees things grow from the beginning will have the finest view of them. Aristotle 384 322 B.C. Introduction to Embryology This lecture will introduce you to the science of developmental biology or
He who sees things grow from the beginning will have the finest view of them. Aristotle 384 322 B.C. Introduction to Embryology This lecture will introduce you to the science of developmental biology or
Lecture 10: Cyclins, cyclin kinases and cell division
 Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Chem*3560 Lecture 10: Cyclins, cyclin kinases and cell division The eukaryotic cell cycle Actively growing mammalian cells divide roughly every 24 hours, and follow a precise sequence of events know as
Nonlinear Optics. Single-Molecule Microscopy Group. Physical Optics Maria Dienerowitz.
 Single-Molecule Microscopy Group Nonlinear Optics Physical Optics 21-06-2017 Maria Dienerowitz maria.dienerowitz@med.uni-jena.de www.single-molecule-microscopy.uniklinikum-jena.de Contents Introduction
Single-Molecule Microscopy Group Nonlinear Optics Physical Optics 21-06-2017 Maria Dienerowitz maria.dienerowitz@med.uni-jena.de www.single-molecule-microscopy.uniklinikum-jena.de Contents Introduction
Introduction. Key Concepts I: Mitosis. AP Biology Laboratory 3 Mitosis & Meiosis
 Virtual Student Guide http://www.phschool.com/science/biology_place/labbench/index.html AP Biology Laboratory 3 Mitosis & Meiosis Introduction For organisms to grow and reproduce, cells must divide. Mitosis
Virtual Student Guide http://www.phschool.com/science/biology_place/labbench/index.html AP Biology Laboratory 3 Mitosis & Meiosis Introduction For organisms to grow and reproduce, cells must divide. Mitosis
Nature Methods: doi: /nmeth Supplementary Figure 1. In vitro screening of recombinant R-CaMP2 variants.
 Supplementary Figure 1 In vitro screening of recombinant R-CaMP2 variants. Baseline fluorescence compared to R-CaMP1.07 at nominally zero calcium plotted versus dynamic range ( F/F) for 150 recombinant
Supplementary Figure 1 In vitro screening of recombinant R-CaMP2 variants. Baseline fluorescence compared to R-CaMP1.07 at nominally zero calcium plotted versus dynamic range ( F/F) for 150 recombinant
Fertilization of sperm and egg produces offspring
 In sexual reproduction Fertilization of sperm and egg produces offspring In asexual reproduction Offspring are produced by a single parent, without the participation of sperm and egg CONNECTIONS BETWEEN
In sexual reproduction Fertilization of sperm and egg produces offspring In asexual reproduction Offspring are produced by a single parent, without the participation of sperm and egg CONNECTIONS BETWEEN
The Theory of HPLC. Quantitative and Qualitative HPLC
 The Theory of HPLC Quantitative and Qualitative HPLC i Wherever you see this symbol, it is important to access the on-line course as there is interactive material that cannot be fully shown in this reference
The Theory of HPLC Quantitative and Qualitative HPLC i Wherever you see this symbol, it is important to access the on-line course as there is interactive material that cannot be fully shown in this reference
Supplementary Methods
 Supplementary Methods Modeling of magnetic field In this study, the magnetic field was generated with N52 grade nickel-plated neodymium block magnets (K&J Magnetics). The residual flux density of the magnets
Supplementary Methods Modeling of magnetic field In this study, the magnetic field was generated with N52 grade nickel-plated neodymium block magnets (K&J Magnetics). The residual flux density of the magnets
The Microscopic Observation of Mitosis in Plant and Animal Cells
 The Microscopic Observation of Mitosis in Plant and Animal Cells Prelab Assignment Before coming to lab, read carefully the introduction and the procedures for each part of the experiment, and then answer
The Microscopic Observation of Mitosis in Plant and Animal Cells Prelab Assignment Before coming to lab, read carefully the introduction and the procedures for each part of the experiment, and then answer
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Cells to Tissues. Peter Takizawa Department of Cell Biology
 Cells to Tissues Peter Takizawa Department of Cell Biology From one cell to ensembles of cells. Multicellular organisms require individual cells to work together in functional groups. This means cells
Cells to Tissues Peter Takizawa Department of Cell Biology From one cell to ensembles of cells. Multicellular organisms require individual cells to work together in functional groups. This means cells
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/8/e1600060/dc1 Supplementary Materials for Analysis of neural crest derived clones reveals novel aspects of facial development Marketa Kaucka, Evgeny Ivashkin,
advances.sciencemag.org/cgi/content/full/2/8/e1600060/dc1 Supplementary Materials for Analysis of neural crest derived clones reveals novel aspects of facial development Marketa Kaucka, Evgeny Ivashkin,
Prediction of Growth Factor Dependent Cleft Formation During Branching Morphogenesis Using A Dynamic Graph-Based Growth Model
 JOURNAL OF L A T E X CLASS FILES, VOL. 13, NO. 9, SEPTEMBER 2014 1 Prediction of Growth Factor Dependent Cleft Formation During Branching Morphogenesis Using A Dynamic Graph-Based Growth Model Nimit Dhulekar
JOURNAL OF L A T E X CLASS FILES, VOL. 13, NO. 9, SEPTEMBER 2014 1 Prediction of Growth Factor Dependent Cleft Formation During Branching Morphogenesis Using A Dynamic Graph-Based Growth Model Nimit Dhulekar
Supplementary Materials for
 www.sciencemag.org/cgi/content/full/science.1244624/dc1 Supplementary Materials for Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein Sougata Roy, Hai Huang,
www.sciencemag.org/cgi/content/full/science.1244624/dc1 Supplementary Materials for Cytoneme-Mediated Contact-Dependent Transport of the Drosophila Decapentaplegic Signaling Protein Sougata Roy, Hai Huang,
Artificial Neural Networks Examination, June 2004
 Artificial Neural Networks Examination, June 2004 Instructions There are SIXTY questions (worth up to 60 marks). The exam mark (maximum 60) will be added to the mark obtained in the laborations (maximum
Artificial Neural Networks Examination, June 2004 Instructions There are SIXTY questions (worth up to 60 marks). The exam mark (maximum 60) will be added to the mark obtained in the laborations (maximum
LIST of SUPPLEMENTARY MATERIALS
 LIST of SUPPLEMENTARY MATERIALS Mir et al., Dense Bicoid Hubs Accentuate Binding along the Morphogen Gradient Supplemental Movie S1 (Related to Figure 1). Movies corresponding to the still frames shown
LIST of SUPPLEMENTARY MATERIALS Mir et al., Dense Bicoid Hubs Accentuate Binding along the Morphogen Gradient Supplemental Movie S1 (Related to Figure 1). Movies corresponding to the still frames shown
To help you complete this review activity and to help you study for your test, you should read SC State Standards B
 Name: Test Date: PAGE: Biology I: Unit 3 Cell Structure Review for Unit Test Directions: You should use this as a guide to help you study for your test. You should also read through your notes, worksheets,
Name: Test Date: PAGE: Biology I: Unit 3 Cell Structure Review for Unit Test Directions: You should use this as a guide to help you study for your test. You should also read through your notes, worksheets,
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 6, 2007 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 6, 2007 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Chapter 6: Cell Growth and Reproduction Lesson 6.1: The Cell Cycle and Mitosis
 Chapter 6: Cell Growth and Reproduction Lesson 6.1: The Cell Cycle and Mitosis No matter the type of cell, all cells come from preexisting cells through the process of cell division. The cell may be the
Chapter 6: Cell Growth and Reproduction Lesson 6.1: The Cell Cycle and Mitosis No matter the type of cell, all cells come from preexisting cells through the process of cell division. The cell may be the
Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization
 Cell Metabolism Supplemental Information Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization Prashant Mishra, Grigor Varuzhanyan,
Cell Metabolism Supplemental Information Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization Prashant Mishra, Grigor Varuzhanyan,
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing
 Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
Supplementary Figure 1. SDS-PAGE analysis of GFP oligomer variants with different linkers. Oligomer mixtures were applied to a PAGE gel containing 0.1% SDS without boiling. The gel was analyzed by a fluorescent
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/1/e1500989/dc1 Supplementary Materials for An epidermis-driven mechanism positions and scales stem cell niches in plants Jérémy Gruel, Benoit Landrein, Paul Tarr,
advances.sciencemag.org/cgi/content/full/2/1/e1500989/dc1 Supplementary Materials for An epidermis-driven mechanism positions and scales stem cell niches in plants Jérémy Gruel, Benoit Landrein, Paul Tarr,
CS 4495 Computer Vision Binary images and Morphology
 CS 4495 Computer Vision Binary images and Aaron Bobick School of Interactive Computing Administrivia PS6 should be working on it! Due Sunday Nov 24 th. Some issues with reading frames. Resolved? Exam:
CS 4495 Computer Vision Binary images and Aaron Bobick School of Interactive Computing Administrivia PS6 should be working on it! Due Sunday Nov 24 th. Some issues with reading frames. Resolved? Exam:
Supplemental Information: Govindaraghavan, Lad and Osmani
 Supplemental Information: Govindaraghavan, Lad and Osmani Table S1: List of strains used in this study Strain Name Genotype (all strains also carry vea1) Source MG190 nima7; pyrg89; argb2; ndc80-cr::pyroa
Supplemental Information: Govindaraghavan, Lad and Osmani Table S1: List of strains used in this study Strain Name Genotype (all strains also carry vea1) Source MG190 nima7; pyrg89; argb2; ndc80-cr::pyroa
Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network
 Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network Marko Tscherepanow and Franz Kummert Applied Computer Science, Faculty of Technology, Bielefeld
Subcellular Localisation of Proteins in Living Cells Using a Genetic Algorithm and an Incremental Neural Network Marko Tscherepanow and Franz Kummert Applied Computer Science, Faculty of Technology, Bielefeld
Magnetics: Fundamentals and Parameter Extraction
 : Fundamentals and Parameter Extraction Stephen Billings Magnetic module outline fundamentals Sensor systems Data examples and demo Parameter extraction Concepts Real-world examples Classification Using
: Fundamentals and Parameter Extraction Stephen Billings Magnetic module outline fundamentals Sensor systems Data examples and demo Parameter extraction Concepts Real-world examples Classification Using
Actinobacteria Relative abundance (%) Co-housed CD300f WT. CD300f KO. Colon length (cm) Day 9. Microscopic inflammation score
 y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
Name 8 Cell Cycle and Meiosis Test Date Study Guide You must know: The structure of the replicated chromosome. The stages of mitosis.
 Name 8 Cell Cycle and Meiosis Test Date Study Guide You must know: The structure of the replicated chromosome. The stages of mitosis. The role of kinases and cyclin in the regulation of the cell cycle.
Name 8 Cell Cycle and Meiosis Test Date Study Guide You must know: The structure of the replicated chromosome. The stages of mitosis. The role of kinases and cyclin in the regulation of the cell cycle.
Study Guide A. Answer Key. Cell Growth and Division. SECTION 1. THE CELL CYCLE 1. a; d; b; c 2. gaps 3. c and d 4. c 5. b and d 6.
 Cell Growth and Division Answer Key SECTION 1. THE CELL CYCLE 1. a; d; b; c 2. gaps 3. c and d 4. c 5. b and d 6. G 1 7. G 0 8. c 9. faster; too large 10. volume 11. a and b 12. repeating pattern or repetition
Cell Growth and Division Answer Key SECTION 1. THE CELL CYCLE 1. a; d; b; c 2. gaps 3. c and d 4. c 5. b and d 6. G 1 7. G 0 8. c 9. faster; too large 10. volume 11. a and b 12. repeating pattern or repetition
Unit 2: Characteristics of Living Things Lesson 25: Mitosis
 Name Unit 2: Characteristics of Living Things Lesson 25: Mitosis Objective: Students will be able to explain the phases of Mitosis. Date Essential Questions: 1. What are the phases of the eukaryotic cell
Name Unit 2: Characteristics of Living Things Lesson 25: Mitosis Objective: Students will be able to explain the phases of Mitosis. Date Essential Questions: 1. What are the phases of the eukaryotic cell
16 The Cell Cycle. Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization
 The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
The Cell Cycle 16 The Cell Cycle Chapter Outline The Eukaryotic Cell Cycle Regulators of Cell Cycle Progression The Events of M Phase Meiosis and Fertilization Introduction Self-reproduction is perhaps
Table S1. Aspergillus nidulans strains used in this study Strain Genotype Derivation
 Supplemental Material De Souza et al., 211 Table S1. Aspergillus nidulans strains used in this study Strain Genotype Derivation CDS295 pyrg89; pyroa4; pyrg Af ::son promotor::gfp-son nup98/nup96 ; chaa1
Supplemental Material De Souza et al., 211 Table S1. Aspergillus nidulans strains used in this study Strain Genotype Derivation CDS295 pyrg89; pyroa4; pyrg Af ::son promotor::gfp-son nup98/nup96 ; chaa1
Cell Division (Outline)
 Cell Division (Outline) 1. Overview of purpose and roles. Comparison of prokaryotic and eukaryotic chromosomes and relation between organelles and cell division. 2. Eukaryotic cell reproduction: asexual
Cell Division (Outline) 1. Overview of purpose and roles. Comparison of prokaryotic and eukaryotic chromosomes and relation between organelles and cell division. 2. Eukaryotic cell reproduction: asexual
Glossary. The ISI glossary of statistical terms provides definitions in a number of different languages:
 Glossary The ISI glossary of statistical terms provides definitions in a number of different languages: http://isi.cbs.nl/glossary/index.htm Adjusted r 2 Adjusted R squared measures the proportion of the
Glossary The ISI glossary of statistical terms provides definitions in a number of different languages: http://isi.cbs.nl/glossary/index.htm Adjusted r 2 Adjusted R squared measures the proportion of the
Micro Computed Tomography Based Quantification of Pore Size in Electron Beam Melted Titanium Biomaterials
 Micro Computed Tomography Based Quantification of Pore Size in Electron Beam Melted Titanium Biomaterials SJ Tredinnick* JG Chase ** *NZi3 National ICT Innovation Institute, University of Canterbury, Christchurch,
Micro Computed Tomography Based Quantification of Pore Size in Electron Beam Melted Titanium Biomaterials SJ Tredinnick* JG Chase ** *NZi3 National ICT Innovation Institute, University of Canterbury, Christchurch,
Name: Date: Period: Must-Know: Unit 6 (Cell Division) AP Biology, Mrs. Krouse. Topic #1: The Cell Cycle and Mitosis
 Name: Date: Period: Must-Know: Unit 6 (Cell Division) AP Biology, Mrs. Krouse Topic #1: The Cell Cycle and Mitosis 1. What events take place in the cell during interphase? 2. How does the amount of DNA
Name: Date: Period: Must-Know: Unit 6 (Cell Division) AP Biology, Mrs. Krouse Topic #1: The Cell Cycle and Mitosis 1. What events take place in the cell during interphase? 2. How does the amount of DNA
178 Part 3.2 SUMMARY INTRODUCTION
 178 Part 3.2 Chapter # DYNAMIC FILTRATION OF VARIABILITY WITHIN EXPRESSION PATTERNS OF ZYGOTIC SEGMENTATION GENES IN DROSOPHILA Surkova S.Yu. *, Samsonova M.G. St. Petersburg State Polytechnical University,
178 Part 3.2 Chapter # DYNAMIC FILTRATION OF VARIABILITY WITHIN EXPRESSION PATTERNS OF ZYGOTIC SEGMENTATION GENES IN DROSOPHILA Surkova S.Yu. *, Samsonova M.G. St. Petersburg State Polytechnical University,
The Kawasaki Identity and the Fluctuation Theorem
 Chapter 6 The Kawasaki Identity and the Fluctuation Theorem This chapter describes the Kawasaki function, exp( Ω t ), and shows that the Kawasaki function follows an identity when the Fluctuation Theorem
Chapter 6 The Kawasaki Identity and the Fluctuation Theorem This chapter describes the Kawasaki function, exp( Ω t ), and shows that the Kawasaki function follows an identity when the Fluctuation Theorem
Application of Micro-Flow Imaging (MFI TM ) to The Analysis of Particles in Parenteral Fluids. October 2006 Ottawa, Canada
 Application of Micro-Flow Imaging (MFI TM ) to The Analysis of Particles in Parenteral Fluids October 26 Ottawa, Canada Summary The introduction of a growing number of targeted protein-based drug formulations
Application of Micro-Flow Imaging (MFI TM ) to The Analysis of Particles in Parenteral Fluids October 26 Ottawa, Canada Summary The introduction of a growing number of targeted protein-based drug formulations
Nonlinear Optics. Single-Molecule Microscopy Group. Physical Optics Maria Dienerowitz.
 Single-Molecule Microscopy Group Nonlinear Optics Physical Optics 21-06-2017 Maria Dienerowitz maria.dienerowitz@med.uni-jena.de www.single-molecule-microscopy.uniklinikum-jena.de Contents Introduction
Single-Molecule Microscopy Group Nonlinear Optics Physical Optics 21-06-2017 Maria Dienerowitz maria.dienerowitz@med.uni-jena.de www.single-molecule-microscopy.uniklinikum-jena.de Contents Introduction
Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its
 Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its transcriptional activity in wild-type embryo. A gradient of canonical
Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its transcriptional activity in wild-type embryo. A gradient of canonical
Answer Key. Cell Growth and Division
 Cell Growth and Division Answer Key SECTION 1. THE CELL CYCLE Cell Cycle: (1) Gap1 (G 1): cells grow, carry out normal functions, and copy their organelles. (2) Synthesis (S): cells replicate DNA. (3)
Cell Growth and Division Answer Key SECTION 1. THE CELL CYCLE Cell Cycle: (1) Gap1 (G 1): cells grow, carry out normal functions, and copy their organelles. (2) Synthesis (S): cells replicate DNA. (3)
Biology 1B Evolution Lecture 2 (February 26, 2010) Natural Selection, Phylogenies
 1 Natural Selection (Darwin-Wallace): There are three conditions for natural selection: 1. Variation: Individuals within a population have different characteristics/traits (or phenotypes). 2. Inheritance:
1 Natural Selection (Darwin-Wallace): There are three conditions for natural selection: 1. Variation: Individuals within a population have different characteristics/traits (or phenotypes). 2. Inheritance:
Mitosis, development, regeneration and cell differentiation
 Mitosis, development, regeneration and cell differentiation Mitosis is a type of cell division by binary fission (splitting in two) which occurs in certain eukaryotic cells. Mitosis generates new body
Mitosis, development, regeneration and cell differentiation Mitosis is a type of cell division by binary fission (splitting in two) which occurs in certain eukaryotic cells. Mitosis generates new body
Description: Supplementary Figures, Supplementary Methods, and Supplementary References
 File Name: Supplementary Information Description: Supplementary Figures, Supplementary Methods, and Supplementary References File Name: Supplementary Movie 1 Description: Footage of time trace of seeds
File Name: Supplementary Information Description: Supplementary Figures, Supplementary Methods, and Supplementary References File Name: Supplementary Movie 1 Description: Footage of time trace of seeds
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS. Transient infection of the zebrafish notochord triggers chronic inflammation
 SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS Transient infection of the zebrafish notochord triggers chronic inflammation Mai Nguyen-Chi 1,2,5, Quang Tien Phan 1,2,5, Catherine Gonzalez 1,2, Jean-François
SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS Transient infection of the zebrafish notochord triggers chronic inflammation Mai Nguyen-Chi 1,2,5, Quang Tien Phan 1,2,5, Catherine Gonzalez 1,2, Jean-François
Monitoring neurite morphology and synapse formation in primary neurons for neurotoxicity assessments and drug screening
 APPLICATION NOTE ArrayScan High Content Platform Monitoring neurite morphology and synapse formation in primary neurons for neurotoxicity assessments and drug screening Suk J. Hong and Richik N. Ghosh
APPLICATION NOTE ArrayScan High Content Platform Monitoring neurite morphology and synapse formation in primary neurons for neurotoxicity assessments and drug screening Suk J. Hong and Richik N. Ghosh
Supplementary Information 16
 Supplementary Information 16 Cellular Component % of Genes 50 45 40 35 30 25 20 15 10 5 0 human mouse extracellular other membranes plasma membrane cytosol cytoskeleton mitochondrion ER/Golgi translational
Supplementary Information 16 Cellular Component % of Genes 50 45 40 35 30 25 20 15 10 5 0 human mouse extracellular other membranes plasma membrane cytosol cytoskeleton mitochondrion ER/Golgi translational
SUPPLEMENTARY INFORMATION
 PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis Momoko Ikeuchi 1 *, Akira Iwase 1 *, Bart Rymen 1, Hirofumi Harashima 1, Michitaro Shibata 1, Mariko Ohnuma 1, Christian Breuer 1,
PRC2 represses dedifferentiation of mature somatic cells in Arabidopsis Momoko Ikeuchi 1 *, Akira Iwase 1 *, Bart Rymen 1, Hirofumi Harashima 1, Michitaro Shibata 1, Mariko Ohnuma 1, Christian Breuer 1,
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11226 Supplementary Discussion D1 Endemics-area relationship (EAR) and its relation to the SAR The EAR comprises the relationship between study area and the number of species that are
doi:10.1038/nature11226 Supplementary Discussion D1 Endemics-area relationship (EAR) and its relation to the SAR The EAR comprises the relationship between study area and the number of species that are
Assignment 7 Due February 26
 Assignment 7 Due February 26 Cells of Multicellular organisms 1. File upload (3 points) View this electron micrograph of spinach leaf cells. The central cell has a thin cell wall; it is difficult to distinguish
Assignment 7 Due February 26 Cells of Multicellular organisms 1. File upload (3 points) View this electron micrograph of spinach leaf cells. The central cell has a thin cell wall; it is difficult to distinguish
Introduction: The Cell Cycle and Mitosis
 Contents 1 Introduction: The Cell Cycle and Mitosis 2 Mitosis Review Introduction: The Cell Cycle and Mitosis The cell cycle refers to the a series of events that describe the metabolic processes of growth
Contents 1 Introduction: The Cell Cycle and Mitosis 2 Mitosis Review Introduction: The Cell Cycle and Mitosis The cell cycle refers to the a series of events that describe the metabolic processes of growth
Developmental Biology Lecture Outlines
 Developmental Biology Lecture Outlines Lecture 01: Introduction Course content Developmental Biology Obsolete hypotheses Current theory Lecture 02: Gametogenesis Spermatozoa Spermatozoon function Spermatozoon
Developmental Biology Lecture Outlines Lecture 01: Introduction Course content Developmental Biology Obsolete hypotheses Current theory Lecture 02: Gametogenesis Spermatozoa Spermatozoon function Spermatozoon
Topic 6 Cell Cycle and Mitosis. Day 1
 Topic 6 Cell Cycle and Mitosis Day 1 Bell Ringer (5 minutes) *pick up worksheet by the door* Get out your homework and answer these questions on the back page: What do I need to do to pass my real EOC?
Topic 6 Cell Cycle and Mitosis Day 1 Bell Ringer (5 minutes) *pick up worksheet by the door* Get out your homework and answer these questions on the back page: What do I need to do to pass my real EOC?
Automated Segmentation of Low Light Level Imagery using Poisson MAP- MRF Labelling
 Automated Segmentation of Low Light Level Imagery using Poisson MAP- MRF Labelling Abstract An automated unsupervised technique, based upon a Bayesian framework, for the segmentation of low light level
Automated Segmentation of Low Light Level Imagery using Poisson MAP- MRF Labelling Abstract An automated unsupervised technique, based upon a Bayesian framework, for the segmentation of low light level
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature14242 Supplementary Methods Mathematical modelling Hematopoietic fluxes. Consider two successive cell compartments in hematopoiesis, an upstream and a reference
SUPPLEMENTARY INFORMATION doi:10.1038/nature14242 Supplementary Methods Mathematical modelling Hematopoietic fluxes. Consider two successive cell compartments in hematopoiesis, an upstream and a reference
Supporting Information
 Supporting Information Fleissner et al. 10.1073/pnas.0907039106 Fig. S1. (A) MAK-2-GFP localized to CATs tips is not bound by membrane. his-3::pccg1 mak-2-gfp; mak-2 strain labeled with membrane dye FM4
Supporting Information Fleissner et al. 10.1073/pnas.0907039106 Fig. S1. (A) MAK-2-GFP localized to CATs tips is not bound by membrane. his-3::pccg1 mak-2-gfp; mak-2 strain labeled with membrane dye FM4
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
Drosophila melanogaster- Morphogen Gradient
 NPTEL Biotechnology - Systems Biology Drosophila melanogaster- Morphogen Gradient Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by
NPTEL Biotechnology - Systems Biology Drosophila melanogaster- Morphogen Gradient Dr. M. Vijayalakshmi School of Chemical and Biotechnology SASTRA University Joint Initiative of IITs and IISc Funded by
MBios 401/501: Lecture 14.2 Cell Differentiation I. Slide #1. Cell Differentiation
 MBios 401/501: Lecture 14.2 Cell Differentiation I Slide #1 Cell Differentiation Cell Differentiation I -Basic principles of differentiation (p1305-1320) -C-elegans (p1321-1327) Cell Differentiation II
MBios 401/501: Lecture 14.2 Cell Differentiation I Slide #1 Cell Differentiation Cell Differentiation I -Basic principles of differentiation (p1305-1320) -C-elegans (p1321-1327) Cell Differentiation II
SUPPLEMENTARY INFORMATION
 Supplementary information S3 (box) Methods Methods Genome weighting The currently available collection of archaeal and bacterial genomes has a highly biased distribution of isolates across taxa. For example,
Supplementary information S3 (box) Methods Methods Genome weighting The currently available collection of archaeal and bacterial genomes has a highly biased distribution of isolates across taxa. For example,
BIOLOGY 111. CHAPTER 5: Chromosomes and Inheritance
 BIOLOGY 111 CHAPTER 5: Chromosomes and Inheritance Chromosomes and Inheritance Learning Outcomes 5.1 Differentiate between sexual and asexual reproduction in terms of the genetic variation of the offspring.
BIOLOGY 111 CHAPTER 5: Chromosomes and Inheritance Chromosomes and Inheritance Learning Outcomes 5.1 Differentiate between sexual and asexual reproduction in terms of the genetic variation of the offspring.
SUPPLEMENTARY INFORMATION
 GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
GP2 Type I-piliated bacteria FAE M cell M cell pocket idc T cell mdc Generation of antigenspecific T cells Induction of antigen-specific mucosal immune response Supplementary Figure 1 Schematic diagram
