Supplementary Materials for
|
|
|
- Jerome McKenzie
- 5 years ago
- Views:
Transcription
1 advances.sciencemag.org/cgi/content/full/2/8/e /dc1 Supplementary Materials for Analysis of neural crest derived clones reveals novel aspects of facial development Marketa Kaucka, Evgeny Ivashkin, Daniel Gyllborg, Tomas Zikmund, Marketa Tesarova, Jozef Kaiser, Meng Xie, Julian Petersen, Vassilis Pachnis, Silvia K. Nicolis, Tian Yu, Paul Sharpe, Ernest Arenas, Hjalmar Brismar, Hans Blom, Hans Clevers, Ueli Suter, Andrei S. Chagin, Kaj Fried, Andreas Hellander, Igor Adameyko Published 3 August 2016, Sci. Adv. 2, e (2016) DOI: /sciadv The PDF file includes: fig. S1. Identification of rare double-color and GFP + clones in neural crest ectomesenchyme in E9.5 to E10.5 embryonic faces. fig. S2. Clonal mixing and distribution of NCC-derived clones in the embryonic trunk and head through the development. fig. S3. Defined borders between mesoderm- and neural crest derived progenies at postnatal and embryonic stages. fig. S4. Genetic tracing of mesoderm-derived mesenchymal progenies reveals similarities with the neural crest derived ectomesenchyme. fig. S5. Live imaging of ectomesenchymal clones and progenitors in the eye shows difference between organized crowd movements and individual migrations. Legends for movies S1 to S5 Supplementary Materials and Methods Other Supplementary Material for this manuscript includes the following: (available at advances.sciencemag.org/cgi/content/full/2/8/e /dc1) movie S1 (.avi format). Live imaging of genetically traced neural crest derived progenies in Sox10-CreERT2/Ubi:zebrabow-S zebrafish larvae between 30 and 56 hpf, ventral view. movie S2 (.avi format). Live imaging of translocating ectomesenchymal clusters in Sox10-CreERT2/Ubi:zebrabow-S zebrafish larvae between 39 and 52 hpf, ventral view.
2 movie S3 (.avi format). Live imaging of genetically traced neural crest derived progenies in Sox10-CreERT2/Ubi:zebrabow-S zebrafish larvae between 30 and 88hpf, ventral view. movie S4 (.avi format). Live imaging of translocating ectomesenchymal clusters in Sox10-CreERT2/Ubi:zebrabow-S zebrafish larvae between 39 and 52 hpf, ventral view. movie S5 (.avi format). 3D EdU analysis of Col2a1aBAC:mCherry zebrafish larva s entire head at 4 dpf corresponding to Fig. 5 (O to Q).
3 Supplementary Figures and Legends fig. S1. Identification of rare double-color and GFP + clones in neural crest ectomesenchyme in E9.5 to E10.5 embryonic faces. (A-B) E9.5 mouse embryo head of PLP-CreERT2/R26Confetti embryo with rare CFP + /RFP + clone. (C-D) E10.5 mouse embryo head of PLP-CreERT2/R26Confetti embryo with rare CFP + /RFP +, RFP + /YFP + and GFP + clones. Scale bars are 200 µm.
4 fig. S2. Clonal mixing and distribution of NCC-derived clones in the embryonic trunk and head through the development. (A-C) Examples of tracing migratory neural crest in the trunks of PLP-CreERT2/R26Confetti embryos injected with tamoxifen at E8.5 and analyzed at E9.5 (A-B) and E10.5 (C). Note the absence of large clonal clusters among migrating neural crest. (D-F) Same type of genetic tracing showing branchial arches and cardiac neural crest analysed at E9.5 and E10.5. Note efficient clonal mixing and formation of clonal clusters already at E9.5 (D). Note the presence of two cardiac neural crest clones (red and blue, pointed out by arrowheads) in the area of developing heart. (G) Individual clones occupy defined areas in E16.5 genetically traced mandible (BA1). BA branchial arch. Scale bars are 200 µm.
5 fig. S3. Defined borders between mesoderm- and neural crest derived progenies at postnatal and embryonic stages. (A-Q) Genetic deletion of Sox2 from the neural crest cells using Wnt1-Cre mouse line affects the pigmentation of the skin with neural crest-derived dermis and depicts the sharpness of the borders that are, due to the stable tissue dynamics,
6 maintained till P5 of postnatal life. (A) Sox2 conditional KO mutant P5 pup with defects in pigmentation on the left next to the littermate control on the right. (B-I) Difference in pigmentation of whisker follicle and guard hair follicle between Sox2 conditional KO mutant and control conditions. (J-M) Overview of the facial skin from Sox2 conditional KO mutant and control pups. Note almost complete lack of pigmentation in Sox2 KO condition. (N-Q) Overview of the back skin from Sox2 conditional KO mutant and control pups. Note the absence of difference in pigmentation of back skin from mutant and control embryos since the dermis of the back skin is not neural-crest derived. (R-T) Neural crest genetic tracing induced in E8.5 Sox10-CreERT2/R26confetti embryo analyzed at E15.5 shows clear borders between neural crest-derived areas and those of mesodermal origin (non-traced). Scale bars are 100 µm.
7
8 fig. S4. Genetic tracing of mesoderm-derived mesenchymal progenies reveals similarities with the neural crest derived ectomesenchyme. (A-K) Genetic tracing in MesP1- Cre/R26Confetti embryos analyzed at E10.5. MIP images of confocal stacks. (A-D) Branchial arches with shown angiogenic and non-angiogenic mesodermal derivatives. Non-angiogenic derivatives of paraxial mesoderm are outlined with a dashed line in (B, D). Note that nonangiogenic derivatives localizing in the center of the arch may contain only one clonal color, for example, RFP (pointed out by arrow in B). BA1/2 branchial arches 1 and 2. (E-I) Multiple spatially overlapping clonal envelopes of mixing occipital mesoderm-derived mesenchymal clones. Arrowheads in (F-I) point out at the cells of the rare GFP + mesenchymal clone. (J-K) In the anterior face mesodermal derivatives are represented mostly by developing vessels formed via fusion of clonally heterogeneous angiogenic progenitors. (L-P) 3D analysis of mesodermal derivatives in the head represented by angiogenic and muscle progenitors. Note the examples of locations where fluorescent protein-labelled mesodermal cells were reconstructed as isosurfaces within standard region of interest (ROI) volume. Panel (P) shows the graph describing % volume of mesodermal contribution inside ROI standard volume. NP - nasal prominence. Scale bars are 150 µm.
9 fig. S5. Live imaging of ectomesenchymal clones and progenitors in the eye shows difference between organized crowd movements and individual migrations. (A-D) Clonal density tracing of neural crest progeny (4-hydroxytamoxifen administered at 16 hpf) in zebrafish ectomesenchyme of Sox10-CreERT2/Ubi:zebrabow-S zebrafish larva. Arrows point
10 out at separate compact groups of YFP + cells in the developing head. Note that during two days of development the groups still retain compact structure and defined borders. (E-P) Ventral view at YFP + ectomesenchymal clones and progenitors in the eye moving and proliferating at three different time-points. YFP-expressing recombined cells are shown as cyan for better visual compatibility with displacement arrows. (E-G) Note coordinated displacement/crowd movement of numerous cells shown by yellow displacement arrows (also see also Supplemental Video Figures). (H-M) Magnified regions from panels (E-G) showing oriented crowd movements/translocations in developing mandible/branchial arch region (H,J,L) as compared to individual cell migrations in the developing eye (I,K,M). The yellow arrows represent displacement lengths from the start to the end point without showing the exact cell track. (N-P) Magnified group of cells in front of stomodeum from outlined area in (E-G) with displacement arrows at three different timepoints. Orientations of cell divisions are shown with bars that are color-coded according to the developmental timing. Note that these ectomesenchymal cells that are relocated as a perfect group without significantly changing the distances between individual cells. Scale bars are 50 µm in A,C,E,G and 50 µm in B,D,N,P.
11 movie S1. Live imaging of genetically traced neural crest derived progenies in Sox10- CreERT2/Ubi:zebrabow-S zebrafish larvae between 30 and 56 hpf, ventral view. Note the differences in migratory behavior of labelled cell in the eyes and in ectomesenchyme. movie S2. Live imaging of translocatingectomesenchymal clusters in Sox10- CreERT2/Ubi:zebrabow-S zebrafish larvae between 39 and 52 hpf, ventral view. movie S3. Live imaging of genetically traced neural crest derived progenies in Sox10- CreERT2/Ubi:zebrabow-S zebrafish larvae between 30 and 88 hpf, ventral view. Note the translocation of labelled ectomesenchymal groups together with stomodeum migrating forward and formation of cartilaginous jaw elements seen as cell stacking. movie S4. Live imaging of translocating ectomesenchymal clusters in Sox10- CreERT2/Ubi:zebrabow-S zebrafish larvae between 39 and 52 hpf, ventral view. movie S5. 3D EdU analysis of Col2a1aBAC:mCherry zebrafish larva s entire head at 4 dpf corresponding to Fig. 5 (O to Q). Label-retaining analysis is done at 4 dpf with EdU administered at 48 hpf during 5 min.
12 Supplementary Materials and Methods Computational Model In order to model the process of clonal mixing, we developed a lattice-based stochastic model that accounts for cell division and cell migration. The computational model is an individual based model (IBM), drawing on ideas from probabilistic cellular automata. An overview of IBM modeling for tissue patterning with a review of IBMs as well as the biological conclusions they have led to in areas such as immunology, development, and cancer, can be found in (Thorne et al., 2007). Our model is stochastic because we are interested in modeling the intrinsic, naturally occurring variability that can be expected in the multicellular system arising from for example variation in cell division times, or stochasticity in direction of allocation of daughter cells after cell division. This document describes the details of the model and its implementation. The simulation code is written in Python, relies on the PyURDME package for spatial stochastic simulations ( and it is freely available for downloads from under the GPLv3 license. The individual agents - colored coded cells Each individual cell is modeled as an agent with the following properties: 1. Color (a label used to track the progeny) 2. Mean cell division time, μ p. 3. Variance in cell division time, σ 2 p. 4. Mean migration time, μ m. 5. Variance in migration time, σ 2 m.
13 In what follows, we assume that each cell have the same value of the above parameters, i.e. we account only for the intrinsic randomness caused by stochasticity in division and migration times, not the extrinsic randomness that would be caused by different cell lineages or different individuals having different characteristic values of the cell properties, something that would add another source of variability to the overall system. Also, we assume that the distributions of the cell properties do not change in time, i.e. the cell division time and cell migration distributions are constant over the course of a simulation. Each cell is assumed to occupy one voxel of a tessellation of 3D space, and the mesh resolution is chosen such that the average voxel size matches the desired cell size. In practice, the cell size has to be chosen large enough to obtain a manageable mesh size. The model does not explicitly make use the exact shape and volume of the voxels. Rather, the underlying computational grid is used to define the neighborhood of each cell, via the adjacency information of the mesh. This is illustrated in Computational Model Schematic Fig. 1A. Individual cells are thought to occupy the dual elements (dashed lines) of a unstructured triangular (2D) or tetrahedral primal mesh (3D) (solid lines). When visualized, we draw them as circles or spheres centered on the vertices of the primal mesh, with radius such that the volume corresponds to the dual element. On the unstructured mesh, there will be a size distribution for the mesh elements, i.e. there will be a small variation in the size associated with each lattice site (B). The mesh resolution is chose such that the mean cell diameter is close to the experimentally observed values, but still such that the mesh does not become too fine to permit simulations.
14 A B C j C i Computational Model Schematic 1. Individual cells are modeled by a number of properties such as their color and distributions for cell division and migration waiting times. The positions of cells in space are tracked on an underlying unstructured lattice, or grid (A). The edges in the primary mesh (solid lines) connect vertices (black dots). A biological cell is modeled by the volume made up of the dual elements (dashed lines), connecting triangle (2D) or tetrahedral (3D) centers and edge or face centers. For visualization purposes, in 3D space, we plot cell individuals as colored spheres with radius equal to the sphere with equal volume as the dual element. Size distribution for the mesh elements for the geometry and mesh used in the simulations in the main paper (B).
15 Cell division In our model, the time until a cell, or individual, divides is a random variable. The dividing cell (referred to as the mother cell) create a daughter cell after a normally distributed waiting time τ D τ p N(μ p, σ p 2 ) where the mean division time μ p and the variance σ p 2 are parameters of the model to be supplied as input to the simulation. As a measure of the degree of variability in cell division times, we use the standard deviation over the mean f = σ m After division, the daughter cell needs to be deposited on the grid. The division direction, or receiving voxel, is sampled according to a discrete distribution. In the simplest case, all directions of division are equally probable and the direction distribution is uniform. In the general case, weights are assigned according to an external, deterministic gradient. Cell division direction The division direction is a random variable. Without a polarizing field, each possible division direction is equally probable. With a polarizing field, the division probability is biased by the gradient. The weights for sampling the division direction when cell C i divides are taken to be d ij w ij = ( max(d ij ) ) j b d ij = g(x j) g(x i ) h ij
16 The parameter b 0 dictates how perfectly the cells become polarized by the concentration profile g(x). A value b = 0 leads to equal probabilities for all directions, and very large value of b means that the division direction will always be in the direction of the maximal value of the gradient (the division direction becomes deterministic in the direction of the maximal gradient in the limit b. Values in between the extremes describe an increasing precision in polarization axis alignment with the gradient field. Cell pushing Unlike related individual based models of e.g. solid tumor growth (Poleszczuk and Underling, 2014), we do not assume that cells can only divide into empty lattice sites or that cells become quiescent if they are completely surrounded by cells. Instead, a dividing cell's progeny will push neighboring cells to make room for it. If the receiving lattice site is empty, the daughter cell is simply deposited there. If it is occupied, that cell is displaced to one of its neighbors as illustrated in Computational Model Schematic 2. The probability for the displaced cell to move to a given neighboring grid point depends on the direction of pushing. Let e md be the vector along the edge connecting the mother cell C m and the daughter cell C d, pointing towards the daughter cell. Let e dk be the unit vector along the edge connecting the daughter cell C d and one of its neighbors, C k. The weight for moving the displaced cell to the neighbor with index $n$ is given by w k = max (s k (1 ci n ), 0) where s k = e md e dk That is, to account for the directionality implied by the originally sampled division direction e md, the probability of the pushed cell to move to an adjacent lattice site C k is proportional to
17 the scalar projection of the vector connecting the pushed cell and the vertex at index k onto the direction vector of the push. cis a penalty parameter in the interval [0,1] that dictates the degree of resistance in pushing the cell into an already occupied site. If c = 1, it is not possible to push a cell into an occupied site, and if c = 0 there is no resistance what so ever and only the direction of the push affects the displacement direction. Any value in between is a tradeoff between the two extremes. In Computational Model Schematic 2, for example, there is a high probability to push C j to C k, due to e dk being almost parallel to e md (highs k ), but depending on the value of c, the site C k may be sampled instead since that site is empty, even if the directionality contributions is smaller (s l < s k ). Once a neighbor has been displaced, the pushed cell moves into that lattice site and the daughter cell gets deposited on the now free lattice site of the displaced cell. The displaced cell, may in turn then go on to displace additional cells and this procedure is repeated until a cell gets pushed into an unoccupied site.
18 Computational Model Schematic 2. When a cell divides (yellow cell), the daughter cell will push surrounding cells (blue) to make room for the progeny (A). The direction of pushing (and what cell to push) is determined by a combination of the directivity of the original division or pushing direction and of a penalty for pushing an occupied site. The penalty is governed by a parameter c that dictates how much to favor pushing into a free lattice site. In the figure, pushing the blue cell at location C d to C l will be favored over C k based on the penalty if c > 0, but the site C k if favored based on the directionality of the push. If C k is selected by the probabilistic algorithm, it will in turn complete a pushing event, leading to a pushing chain that continues until a cell is pushed into an empty site. After such a pushing chain has been completed, the site sampled for the daughter cell will be free, and the newly created daughter cell can be inserted (B). This simple model for pushing gives us a practical way to model the insertion of cells without resorting to quiescent cells in the lattice model, or going to a detailed continuum description with plastic cells.
19 Cell migration Cells can migrate into a neighboring, receiving, lattice site if it is occupied by another cell or if it is empty. In the case of an empty receiving lattice site, the cell simply changes location, leaving its previous lattice site unoccupied. In the case of an occupied receiving site, the migrating cell and the cell in the receiving voxel swap places. The time to migration is modeled as a stochastic variable with a normal distribution τ m = N(μ m, σ m 2 ) The direction of a migration event is chosen uniformly amongst the neighboring lattice sites. Simulation procedure A kinetic Monte Carlo algorithm implemented in Python using a priority queue following standard methodology for event driven simulation simulates the system. To initialize the simulation, the time until the next occurrence for each possible event is sampled for all the active individuals. Then, the event with the shortest waiting time is selected, and the corresponding cell updated. In the case of a division event, a new division time is sampled for the mother cell and in the case of migration; a new migration time is sampled for the migrating cell. This procedure is repeated until the simulation has reached its final time point, or until a daughter cell cannot be inserted in the lattice after attempted pushing events. This number is selected to be high enough for this never to occur for the simulations times and parameters used in this study. Computing cell-cell distances with the chain-reaction approach To mimic the manual analysis conducted on the experimental data, we computed cell-cell distances for each colored cell (clone) using a chain reaction approach. As a starting cell, we picked the one furthest from the mass center of the clone, i.e. a cell on the periphery of the
20 clonal envelope. Then, we computed the distance to that cell's nearest neighbor. Then that cell was removed from the stack, and the same procedure was repeated for the selected nearest neighbor. This was repeated for 30 cells in the clone (as for the experimental data).
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Overexpression of YFP::GPR-1 in the germline.
 Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
Supplementary Figure 1 Overexpression of YFP::GPR-1 in the germline. The pie-1 promoter and 3 utr were used to express yfp::gpr-1 in the germline. Expression levels from the yfp::gpr-1(cai 1.0)-expressing
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
doi:10.1038/nature11589 Supplementary Figure 1 Ciona intestinalis and Petromyzon marinus neural crest expression domain comparison. Cartoon shows dorsal views of Ciona mid gastrula (left) and Petromyzon
Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration
 Cell Stem Cell Supplemental Information Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration Micah T. Webster, Uri Manor, Jennifer Lippincott-Schwartz,
Cell Stem Cell Supplemental Information Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration Micah T. Webster, Uri Manor, Jennifer Lippincott-Schwartz,
Questions in developmental biology. Differentiation Morphogenesis Growth/apoptosis Reproduction Evolution Environmental integration
 Questions in developmental biology Differentiation Morphogenesis Growth/apoptosis Reproduction Evolution Environmental integration Representative cell types of a vertebrate zygote => embryo => adult differentiation
Questions in developmental biology Differentiation Morphogenesis Growth/apoptosis Reproduction Evolution Environmental integration Representative cell types of a vertebrate zygote => embryo => adult differentiation
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/5/e1600519/dc1 Supplementary Materials for Encapsulation-free controlled release: Electrostatic adsorption eliminates the need for protein encapsulation in PLGA
advances.sciencemag.org/cgi/content/full/2/5/e1600519/dc1 Supplementary Materials for Encapsulation-free controlled release: Electrostatic adsorption eliminates the need for protein encapsulation in PLGA
In ovo time-lapse analysis after dorsal neural tube ablation shows rerouting of chick hindbrain neural crest
 In ovo time-lapse analysis after dorsal neural tube ablation shows rerouting of chick hindbrain neural crest Paul Kulesa, Marianne Bronner-Fraser and Scott Fraser (2000) Presented by Diandra Lucia Background
In ovo time-lapse analysis after dorsal neural tube ablation shows rerouting of chick hindbrain neural crest Paul Kulesa, Marianne Bronner-Fraser and Scott Fraser (2000) Presented by Diandra Lucia Background
Diffusion. CS/CME/BioE/Biophys/BMI 279 Nov. 15 and 20, 2016 Ron Dror
 Diffusion CS/CME/BioE/Biophys/BMI 279 Nov. 15 and 20, 2016 Ron Dror 1 Outline How do molecules move around in a cell? Diffusion as a random walk (particle-based perspective) Continuum view of diffusion
Diffusion CS/CME/BioE/Biophys/BMI 279 Nov. 15 and 20, 2016 Ron Dror 1 Outline How do molecules move around in a cell? Diffusion as a random walk (particle-based perspective) Continuum view of diffusion
Chapter 11. Development: Differentiation and Determination
 KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
KAP Biology Dept Kenyon College Differential gene expression and development Mechanisms of cellular determination Induction Pattern formation Chapter 11. Development: Differentiation and Determination
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/1/e1500989/dc1 Supplementary Materials for An epidermis-driven mechanism positions and scales stem cell niches in plants Jérémy Gruel, Benoit Landrein, Paul Tarr,
advances.sciencemag.org/cgi/content/full/2/1/e1500989/dc1 Supplementary Materials for An epidermis-driven mechanism positions and scales stem cell niches in plants Jérémy Gruel, Benoit Landrein, Paul Tarr,
1. Introductory Examples
 1. Introductory Examples We introduce the concept of the deterministic and stochastic simulation methods. Two problems are provided to explain the methods: the percolation problem, providing an example
1. Introductory Examples We introduce the concept of the deterministic and stochastic simulation methods. Two problems are provided to explain the methods: the percolation problem, providing an example
Supplementary Figure 1. Phenotype of the HI strain.
 Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Figure 1. Phenotype of the HI strain. (A) Phenotype of the HI and wild type plant after flowering (~1month). Wild type plant is tall with well elongated inflorescence. All four HI plants
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/6/e1700071/dc1 Supplementary Materials for Self-organization of maze-like structures via guided wrinkling The PDF file includes: Hyung Jong Bae, Sangwook Bae,
advances.sciencemag.org/cgi/content/full/3/6/e1700071/dc1 Supplementary Materials for Self-organization of maze-like structures via guided wrinkling The PDF file includes: Hyung Jong Bae, Sangwook Bae,
Developmental Zoology. Ectodermal derivatives (ZOO ) Developmental Stages. Developmental Stages
 Developmental Zoology (ZOO 228.1.0) Ectodermal derivatives 1 Developmental Stages Ø Early Development Fertilization Cleavage Gastrulation Neurulation Ø Later Development Organogenesis Larval molts Metamorphosis
Developmental Zoology (ZOO 228.1.0) Ectodermal derivatives 1 Developmental Stages Ø Early Development Fertilization Cleavage Gastrulation Neurulation Ø Later Development Organogenesis Larval molts Metamorphosis
Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its
 Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its transcriptional activity in wild-type embryo. A gradient of canonical
Supplementary Figure 1: Mechanism of Lbx2 action on the Wnt/ -catenin signalling pathway. (a) The Wnt/ -catenin signalling pathway and its transcriptional activity in wild-type embryo. A gradient of canonical
Developmental Biology 3230 Midterm Exam 1 March 2006
 Name Developmental Biology 3230 Midterm Exam 1 March 2006 1. (20pts) Regeneration occurs to some degree to most metazoans. When you remove the head of a hydra a new one regenerates. Graph the inhibitor
Name Developmental Biology 3230 Midterm Exam 1 March 2006 1. (20pts) Regeneration occurs to some degree to most metazoans. When you remove the head of a hydra a new one regenerates. Graph the inhibitor
Name. Biology Developmental Biology Winter Quarter 2013 KEY. Midterm 3
 Name 100 Total Points Open Book Biology 411 - Developmental Biology Winter Quarter 2013 KEY Midterm 3 Read the Following Instructions: * Answer 20 questions (5 points each) out of the available 25 questions
Name 100 Total Points Open Book Biology 411 - Developmental Biology Winter Quarter 2013 KEY Midterm 3 Read the Following Instructions: * Answer 20 questions (5 points each) out of the available 25 questions
7.013 Spring 2005 Problem Set 4
 MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel NAME TA 7.013 Spring 2005 Problem Set 4 FRIDAY April 8th,
MIT Department of Biology 7.013: Introductory Biology - Spring 2005 Instructors: Professor Hazel Sive, Professor Tyler Jacks, Dr. Claudette Gardel NAME TA 7.013 Spring 2005 Problem Set 4 FRIDAY April 8th,
Neural development its all connected
 Neural development its all connected How do you build a complex nervous system? How do you build a complex nervous system? 1. Learn how tissue is instructed to become nervous system. Neural induction 2.
Neural development its all connected How do you build a complex nervous system? How do you build a complex nervous system? 1. Learn how tissue is instructed to become nervous system. Neural induction 2.
Statistical Pattern Recognition
 Statistical Pattern Recognition Feature Extraction Hamid R. Rabiee Jafar Muhammadi, Alireza Ghasemi, Payam Siyari Spring 2014 http://ce.sharif.edu/courses/92-93/2/ce725-2/ Agenda Dimensionality Reduction
Statistical Pattern Recognition Feature Extraction Hamid R. Rabiee Jafar Muhammadi, Alireza Ghasemi, Payam Siyari Spring 2014 http://ce.sharif.edu/courses/92-93/2/ce725-2/ Agenda Dimensionality Reduction
Origin of mesenchymal stem cell
 Origin of mesenchymal stem cell Takumi Era Department of Cell Modulation, Institute of Molecular embryology and Genetics (IMEG) Kumamoto University Differentiation pathways of in vitro ES cell culture
Origin of mesenchymal stem cell Takumi Era Department of Cell Modulation, Institute of Molecular embryology and Genetics (IMEG) Kumamoto University Differentiation pathways of in vitro ES cell culture
Algorithms for Calculating Statistical Properties on Moving Points
 Algorithms for Calculating Statistical Properties on Moving Points Dissertation Proposal Sorelle Friedler Committee: David Mount (Chair), William Gasarch Samir Khuller, Amitabh Varshney January 14, 2009
Algorithms for Calculating Statistical Properties on Moving Points Dissertation Proposal Sorelle Friedler Committee: David Mount (Chair), William Gasarch Samir Khuller, Amitabh Varshney January 14, 2009
LIST of SUPPLEMENTARY MATERIALS
 LIST of SUPPLEMENTARY MATERIALS Mir et al., Dense Bicoid Hubs Accentuate Binding along the Morphogen Gradient Supplemental Movie S1 (Related to Figure 1). Movies corresponding to the still frames shown
LIST of SUPPLEMENTARY MATERIALS Mir et al., Dense Bicoid Hubs Accentuate Binding along the Morphogen Gradient Supplemental Movie S1 (Related to Figure 1). Movies corresponding to the still frames shown
Diffusion and cellular-level simulation. CS/CME/BioE/Biophys/BMI 279 Nov. 7 and 9, 2017 Ron Dror
 Diffusion and cellular-level simulation CS/CME/BioE/Biophys/BMI 279 Nov. 7 and 9, 2017 Ron Dror 1 Outline How do molecules move around in a cell? Diffusion as a random walk (particle-based perspective)
Diffusion and cellular-level simulation CS/CME/BioE/Biophys/BMI 279 Nov. 7 and 9, 2017 Ron Dror 1 Outline How do molecules move around in a cell? Diffusion as a random walk (particle-based perspective)
Folding of the embryo.. the embryo is becoming a tube like structure
 The embryo is a Folding of the embryo.. the embryo is becoming a tube like structure WEEK 4 EMBRYO General features Primordia of the brain Somites Primordia of the heart Branchial arches Primordia
The embryo is a Folding of the embryo.. the embryo is becoming a tube like structure WEEK 4 EMBRYO General features Primordia of the brain Somites Primordia of the heart Branchial arches Primordia
Supporting Information
 Supporting Information Visualizing the Effect of Partial Oxide Formation on Single Silver Nanoparticle Electrodissolution Vignesh Sundaresan, Joseph W. Monaghan, and Katherine A. Willets* Department of
Supporting Information Visualizing the Effect of Partial Oxide Formation on Single Silver Nanoparticle Electrodissolution Vignesh Sundaresan, Joseph W. Monaghan, and Katherine A. Willets* Department of
Mesoderm Development
 Quiz rules: Spread out across available tables No phones, text books, or (lecture) notes on your desks No consultation with your colleagues No websites open other than the Quiz page No screen snap shots
Quiz rules: Spread out across available tables No phones, text books, or (lecture) notes on your desks No consultation with your colleagues No websites open other than the Quiz page No screen snap shots
Introduction and Background
 Chapter 1 1 Introduction and Background The heart is a robust pump capable of beating rhythmically for over 2 ½ billion times in a lifetime. At very early stages of development, the embryonic heart is
Chapter 1 1 Introduction and Background The heart is a robust pump capable of beating rhythmically for over 2 ½ billion times in a lifetime. At very early stages of development, the embryonic heart is
Supplementary Figure 1
 Supplementary Figure 1 Single-cell RNA sequencing reveals unique sets of neuropeptides, transcription factors and receptors in specific types of sympathetic neurons (a) Dissection of paravertebral SGs
Supplementary Figure 1 Single-cell RNA sequencing reveals unique sets of neuropeptides, transcription factors and receptors in specific types of sympathetic neurons (a) Dissection of paravertebral SGs
Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA
 Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Himber et al. Transitivity-dependent and transitivity-independent cell-to-cell movement of RNA silencing SUPPLEMENTAL MATERIAL Supplemental material 1. Short-range movement of GFP silencing affects a nearly
Supplementary Figures
 Supplementary Figures Supplementary Fig. S1: Normal development and organization of the embryonic ventral nerve cord in Platynereis. (A) Life cycle of Platynereis dumerilii. (B-F) Axonal scaffolds and
Supplementary Figures Supplementary Fig. S1: Normal development and organization of the embryonic ventral nerve cord in Platynereis. (A) Life cycle of Platynereis dumerilii. (B-F) Axonal scaffolds and
Supplementary information
 Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
Supplementary information Statistical organelle dissection of Arabidopsis guard cells using image database LIPS Takumi Higaki *, Natsumaro Kutsuna, Yoichiroh Hosokawa, Kae Akita, Kazuo Ebine, Takashi Ueda,
SUPPLEMENTARY INFORMATION
 DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
DOI: 10.1038/ncb3267 Supplementary Figure 1 A group of genes required for formation or orientation of annular F-actin bundles and aecm ridges: RNAi phenotypes and their validation by standard mutations.
1. What are the three general areas of the developing vertebrate limb? 2. What embryonic regions contribute to the developing limb bud?
 Study Questions - Lecture 17 & 18 1. What are the three general areas of the developing vertebrate limb? The three general areas of the developing vertebrate limb are the proximal stylopod, zeugopod, and
Study Questions - Lecture 17 & 18 1. What are the three general areas of the developing vertebrate limb? The three general areas of the developing vertebrate limb are the proximal stylopod, zeugopod, and
 DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
DOI: 10.1038/ncb2819 Gαi3 / Actin / Acetylated Tubulin Gαi3 / Actin / Acetylated Tubulin a a Gαi3 a Actin Gαi3 WT Gαi3 WT Gαi3 WT b b Gαi3 b Actin Gαi3 KO Gαi3 KO Gαi3 KO # # Figure S1 Loss of protein
adhesion parameters in a cell migratory process
 1 2 Using approximate Bayesian computation to quantify cell-cell adhesion parameters in a cell migratory process 3 4 Robert J. H. Ross 1,R.E.Baker 1, Andrew Parker 1, M. J. Ford 2,R.L.Mort 3, and C. A.
1 2 Using approximate Bayesian computation to quantify cell-cell adhesion parameters in a cell migratory process 3 4 Robert J. H. Ross 1,R.E.Baker 1, Andrew Parker 1, M. J. Ford 2,R.L.Mort 3, and C. A.
Nature Neuroscience: doi: /nn.2662
 Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Supplementary Figure 1 Atlastin phylogeny and homology. (a) Maximum likelihood phylogenetic tree based on 18 Atlastin-1 sequences using the program Quicktree. Numbers at internal nodes correspond to bootstrap
Cellular Automata and Tilings
 Cellular Automata and Tilings Jarkko Kari Department of Mathematics, University of Turku, Finland TUCS(Turku Centre for Computer Science), Turku, Finland Outline of the talk (1) Cellular automata (CA)
Cellular Automata and Tilings Jarkko Kari Department of Mathematics, University of Turku, Finland TUCS(Turku Centre for Computer Science), Turku, Finland Outline of the talk (1) Cellular automata (CA)
SUPPLEMENTARY INFORMATION
 med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
med!1,2 Wild-type (N2) end!3 elt!2 5 1 15 Time (minutes) 5 1 15 Time (minutes) med!1,2 end!3 5 1 15 Time (minutes) elt!2 5 1 15 Time (minutes) Supplementary Figure 1: Number of med-1,2, end-3, end-1 and
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:1.138/nature1237 a b retinol retinal RA OH RDH (retinol dehydrogenase) O H Raldh2 O R/R.6.4.2 (retinaldehyde dehydrogenase 2) RA retinal retinol..1.1 1 Concentration (nm)
SUPPLEMENTARY INFORMATION doi:1.138/nature1237 a b retinol retinal RA OH RDH (retinol dehydrogenase) O H Raldh2 O R/R.6.4.2 (retinaldehyde dehydrogenase 2) RA retinal retinol..1.1 1 Concentration (nm)
Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization
 Cell Metabolism Supplemental Information Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization Prashant Mishra, Grigor Varuzhanyan,
Cell Metabolism Supplemental Information Mitochondrial Dynamics Is a Distinguishing Feature of Skeletal Muscle Fiber Types and Regulates Organellar Compartmentalization Prashant Mishra, Grigor Varuzhanyan,
Supplementary Materials for
 www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
www.sciencesignaling.org/cgi/content/full/6/301/ra98/dc1 Supplementary Materials for Regulation of Epithelial Morphogenesis by the G Protein Coupled Receptor Mist and Its Ligand Fog Alyssa J. Manning,
When do diffusion-limited trajectories become memoryless?
 When do diffusion-limited trajectories become memoryless? Maciej Dobrzyński CWI (Center for Mathematics and Computer Science) Kruislaan 413, 1098 SJ Amsterdam, The Netherlands Abstract Stochastic description
When do diffusion-limited trajectories become memoryless? Maciej Dobrzyński CWI (Center for Mathematics and Computer Science) Kruislaan 413, 1098 SJ Amsterdam, The Netherlands Abstract Stochastic description
Question Set # 4 Answer Key 7.22 Nov. 2002
 Question Set # 4 Answer Key 7.22 Nov. 2002 1) A variety of reagents and approaches are frequently used by developmental biologists to understand the tissue interactions and molecular signaling pathways
Question Set # 4 Answer Key 7.22 Nov. 2002 1) A variety of reagents and approaches are frequently used by developmental biologists to understand the tissue interactions and molecular signaling pathways
SUPPLEMENTARY INFORMATION
 SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION doi:10.1038/nature12791 Supplementary Figure 1 (1/3) WWW.NATURE.COM/NATURE 1 RESEARCH SUPPLEMENTARY INFORMATION Supplementary Figure 1 (2/3) 2 WWW.NATURE.COM/NATURE SUPPLEMENTARY
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
doi:10.1038/nature09450 Supplementary Table 1 Summary of kinetic parameters. Kinetic parameters were V = V / 1 K / ATP and obtained using the relationships max ( + m [ ]) V d s /( 1/ k [ ATP] + 1 k ) =,
Homotypic cell competition regulates proliferation and tiling of zebrafish pigment cells during colour pattern formation
 Received 9 Sep 205 Accepted 30 Mar 206 Published 27 Apr 206 DOI: 0.038/ncomms462 Homotypic cell competition regulates proliferation and tiling of zebrafish pigment cells during colour pattern formation
Received 9 Sep 205 Accepted 30 Mar 206 Published 27 Apr 206 DOI: 0.038/ncomms462 Homotypic cell competition regulates proliferation and tiling of zebrafish pigment cells during colour pattern formation
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
advances.sciencemag.org/cgi/content/full/1/9/e1500511/dc1 Supplementary Materials for Contractility parameters of human -cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of
Supplementary Tables and Figures
 Supplementary Tables Supplementary Tables and Figures Supplementary Table 1: Tumor types and samples analyzed. Supplementary Table 2: Genes analyzed here. Supplementary Table 3: Statistically significant
Supplementary Tables Supplementary Tables and Figures Supplementary Table 1: Tumor types and samples analyzed. Supplementary Table 2: Genes analyzed here. Supplementary Table 3: Statistically significant
MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION
 MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION Drosophila is the best understood of all developmental systems, especially at the genetic level, and although it is an invertebrate it has had an enormous
MOLECULAR CONTROL OF EMBRYONIC PATTERN FORMATION Drosophila is the best understood of all developmental systems, especially at the genetic level, and although it is an invertebrate it has had an enormous
4/19/10 More complications to Mendel
 4/19/10 More complications to Mendel Complications to the relationship between genotype to phenotype Commentary written in response to the release of the first draft of the human genome sequence From Science
4/19/10 More complications to Mendel Complications to the relationship between genotype to phenotype Commentary written in response to the release of the first draft of the human genome sequence From Science
Any live cell with less than 2 live neighbours dies. Any live cell with 2 or 3 live neighbours lives on to the next step.
 2. Cellular automata, and the SIRS model In this Section we consider an important set of models used in computer simulations, which are called cellular automata (these are very similar to the so-called
2. Cellular automata, and the SIRS model In this Section we consider an important set of models used in computer simulations, which are called cellular automata (these are very similar to the so-called
Head and Face Development
 Head and Face Development Resources: http://php.med.unsw.edu.au/embryology/ Larsen s Human Embryology The Developing Human: Clinically Oriented Embryology Dr Annemiek Beverdam School of Medical Sciences,
Head and Face Development Resources: http://php.med.unsw.edu.au/embryology/ Larsen s Human Embryology The Developing Human: Clinically Oriented Embryology Dr Annemiek Beverdam School of Medical Sciences,
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 6, 2007 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 6, 2007 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/1/8/e1500527/dc1 Supplementary Materials for A phylogenomic data-driven exploration of viral origins and evolution The PDF file includes: Arshan Nasir and Gustavo
advances.sciencemag.org/cgi/content/full/1/8/e1500527/dc1 Supplementary Materials for A phylogenomic data-driven exploration of viral origins and evolution The PDF file includes: Arshan Nasir and Gustavo
Part III. Cellular Automata Simulation of. Monolayer Surfaces
 Part III Cellular Automata Simulation of Monolayer Surfaces The concept of progress acts as a protective mechanism to shield us from the terrors of the future. the words of Paul Muad Dib 193 Chapter 6
Part III Cellular Automata Simulation of Monolayer Surfaces The concept of progress acts as a protective mechanism to shield us from the terrors of the future. the words of Paul Muad Dib 193 Chapter 6
SUPPLEMENTARY INFORMATION
 Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
Supplementary Discussion Rationale for using maternal ythdf2 -/- mutants as study subject To study the genetic basis of the embryonic developmental delay that we observed, we crossed fish with different
protein biology cell imaging Automated imaging and high-content analysis
 protein biology cell imaging Automated imaging and high-content analysis Automated imaging and high-content analysis Thermo Fisher Scientific offers a complete portfolio of imaging platforms, reagents
protein biology cell imaging Automated imaging and high-content analysis Automated imaging and high-content analysis Thermo Fisher Scientific offers a complete portfolio of imaging platforms, reagents
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/1/e1500997/dc1 Supplementary Materials for Social behavior shapes the chimpanzee pan-microbiome Andrew H. Moeller, Steffen Foerster, Michael L. Wilson, Anne E.
advances.sciencemag.org/cgi/content/full/2/1/e1500997/dc1 Supplementary Materials for Social behavior shapes the chimpanzee pan-microbiome Andrew H. Moeller, Steffen Foerster, Michael L. Wilson, Anne E.
Lecture 1: Random walk
 Lecture : Random walk Paul C Bressloff (Spring 209). D random walk q p r- r r+ Figure 2: A random walk on a D lattice. Consider a particle that hops at discrete times between neighboring sites on a one
Lecture : Random walk Paul C Bressloff (Spring 209). D random walk q p r- r r+ Figure 2: A random walk on a D lattice. Consider a particle that hops at discrete times between neighboring sites on a one
Unicellular: Cells change function in response to a temporal plan, such as the cell cycle.
 Spatial organization is a key difference between unicellular organisms and metazoans Unicellular: Cells change function in response to a temporal plan, such as the cell cycle. Cells differentiate as a
Spatial organization is a key difference between unicellular organisms and metazoans Unicellular: Cells change function in response to a temporal plan, such as the cell cycle. Cells differentiate as a
Overview of Spatial analysis in ecology
 Spatial Point Patterns & Complete Spatial Randomness - II Geog 0C Introduction to Spatial Data Analysis Chris Funk Lecture 8 Overview of Spatial analysis in ecology st step in understanding ecological
Spatial Point Patterns & Complete Spatial Randomness - II Geog 0C Introduction to Spatial Data Analysis Chris Funk Lecture 8 Overview of Spatial analysis in ecology st step in understanding ecological
Simultaneous intracellular chloride and ph measurements using a GFPbased
 nature methods Simultaneous intracellular chloride and ph measurements using a GFPbased sensor Daniele Arosio, Fernanda Ricci, Laura Marchetti, Roberta Gualdani, Lorenzo Albertazzi & Fabio Beltram Supplementary
nature methods Simultaneous intracellular chloride and ph measurements using a GFPbased sensor Daniele Arosio, Fernanda Ricci, Laura Marchetti, Roberta Gualdani, Lorenzo Albertazzi & Fabio Beltram Supplementary
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/10/eaao3530/dc1 Supplementary Materials for Topological liquid diode Jiaqian Li, Xiaofeng Zhou, Jing Li, Lufeng Che, Jun Yao, Glen McHale, Manoj K. Chaudhury,
advances.sciencemag.org/cgi/content/full/3/10/eaao3530/dc1 Supplementary Materials for Topological liquid diode Jiaqian Li, Xiaofeng Zhou, Jing Li, Lufeng Che, Jun Yao, Glen McHale, Manoj K. Chaudhury,
3D HP Protein Folding Problem using Ant Algorithm
 3D HP Protein Folding Problem using Ant Algorithm Fidanova S. Institute of Parallel Processing BAS 25A Acad. G. Bonchev Str., 1113 Sofia, Bulgaria Phone: +359 2 979 66 42 E-mail: stefka@parallel.bas.bg
3D HP Protein Folding Problem using Ant Algorithm Fidanova S. Institute of Parallel Processing BAS 25A Acad. G. Bonchev Str., 1113 Sofia, Bulgaria Phone: +359 2 979 66 42 E-mail: stefka@parallel.bas.bg
Role of Mitochondrial Remodeling in Programmed Cell Death in
 Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Developmental Cell, Vol. 12 Supplementary Data Role of Mitochondrial Remodeling in Programmed Cell Death in Drosophila melanogaster Gaurav Goyal, Brennan Fell, Apurva Sarin, Richard J. Youle, V. Sriram.
Probabilistic Cardinal Direction Queries On Spatio-Temporal Data
 Probabilistic Cardinal Direction Queries On Spatio-Temporal Data Ganesh Viswanathan Midterm Project Report CIS 6930 Data Science: Large-Scale Advanced Data Analytics University of Florida September 3 rd,
Probabilistic Cardinal Direction Queries On Spatio-Temporal Data Ganesh Viswanathan Midterm Project Report CIS 6930 Data Science: Large-Scale Advanced Data Analytics University of Florida September 3 rd,
MBios 401/501: Lecture 14.2 Cell Differentiation I. Slide #1. Cell Differentiation
 MBios 401/501: Lecture 14.2 Cell Differentiation I Slide #1 Cell Differentiation Cell Differentiation I -Basic principles of differentiation (p1305-1320) -C-elegans (p1321-1327) Cell Differentiation II
MBios 401/501: Lecture 14.2 Cell Differentiation I Slide #1 Cell Differentiation Cell Differentiation I -Basic principles of differentiation (p1305-1320) -C-elegans (p1321-1327) Cell Differentiation II
Supplemental material
 Supplemental material THE JOURNAL OF CELL BIOLOGY Civelekoglu-Scholey et al., http://www.jcb.org/cgi/content/full/jcb.200908150/dc1 Figure S1. Spindle dynamics in Drosophila embryos and in vitro gliding
Supplemental material THE JOURNAL OF CELL BIOLOGY Civelekoglu-Scholey et al., http://www.jcb.org/cgi/content/full/jcb.200908150/dc1 Figure S1. Spindle dynamics in Drosophila embryos and in vitro gliding
Description: Supplementary Figures, Supplementary Methods, and Supplementary References
 File Name: Supplementary Information Description: Supplementary Figures, Supplementary Methods, and Supplementary References File Name: Supplementary Movie 1 Description: Footage of time trace of seeds
File Name: Supplementary Information Description: Supplementary Figures, Supplementary Methods, and Supplementary References File Name: Supplementary Movie 1 Description: Footage of time trace of seeds
Chapter 4 Dynamic Bayesian Networks Fall Jin Gu, Michael Zhang
 Chapter 4 Dynamic Bayesian Networks 2016 Fall Jin Gu, Michael Zhang Reviews: BN Representation Basic steps for BN representations Define variables Define the preliminary relations between variables Check
Chapter 4 Dynamic Bayesian Networks 2016 Fall Jin Gu, Michael Zhang Reviews: BN Representation Basic steps for BN representations Define variables Define the preliminary relations between variables Check
Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct
 SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
SUPPLEMENTAL FIGURE LEGENDS Figure S1. Programmed cell death in the AB lineage occurs in temporally distinct waves. (A) A representative sub-lineage (ABala) of the C. elegans lineage tree (adapted from
How many lessons is it?
 Science Unit Learning Summary Content Eukaryotes and Prokaryotes Cells are the basic unit of all life forms. A eukaryotic cell contains genetic material enclosed within a nucleus. Plant and animal cells
Science Unit Learning Summary Content Eukaryotes and Prokaryotes Cells are the basic unit of all life forms. A eukaryotic cell contains genetic material enclosed within a nucleus. Plant and animal cells
Axis Specification in Drosophila
 Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Developmental Biology Biology 4361 Axis Specification in Drosophila November 2, 2006 Axis Specification in Drosophila Fertilization Superficial cleavage Gastrulation Drosophila body plan Oocyte formation
Comparing whole genomes
 BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
BioNumerics Tutorial: Comparing whole genomes 1 Aim The Chromosome Comparison window in BioNumerics has been designed for large-scale comparison of sequences of unlimited length. In this tutorial you will
Homework 2: MDPs and Search
 Graduate Artificial Intelligence 15-780 Homework 2: MDPs and Search Out on February 15 Due on February 29 Problem 1: MDPs [Felipe, 20pts] Figure 1: MDP for Problem 1. States are represented by circles
Graduate Artificial Intelligence 15-780 Homework 2: MDPs and Search Out on February 15 Due on February 29 Problem 1: MDPs [Felipe, 20pts] Figure 1: MDP for Problem 1. States are represented by circles
Lab 1: Numerical Solution of Laplace s Equation
 Lab 1: Numerical Solution of Laplace s Equation ELEC 3105 last modified August 27, 2012 1 Before You Start This lab and all relevant files can be found at the course website. You will need to obtain an
Lab 1: Numerical Solution of Laplace s Equation ELEC 3105 last modified August 27, 2012 1 Before You Start This lab and all relevant files can be found at the course website. You will need to obtain an
Mesoderm Induction CBT, 2018 Hand-out CBT March 2018
 Mesoderm Induction CBT, 2018 Hand-out CBT March 2018 Introduction 3. Books This module is based on the following books: - 'Principles of Developement', Lewis Wolpert, et al., fifth edition, 2015 - 'Developmental
Mesoderm Induction CBT, 2018 Hand-out CBT March 2018 Introduction 3. Books This module is based on the following books: - 'Principles of Developement', Lewis Wolpert, et al., fifth edition, 2015 - 'Developmental
Physics of Cellular materials: Filaments
 Physics of Cellular materials: Filaments Tom Chou Dept. of Biomathematics, UCLA, Los Angeles, CA 995-766 (Dated: December 6, ) The basic filamentary structures in a cell are reviewed. Their basic structures
Physics of Cellular materials: Filaments Tom Chou Dept. of Biomathematics, UCLA, Los Angeles, CA 995-766 (Dated: December 6, ) The basic filamentary structures in a cell are reviewed. Their basic structures
Predator escape: an ecologically realistic scenario for the evolutionary origins of multicellularity. Student handout
 Predator escape: an ecologically realistic scenario for the evolutionary origins of multicellularity Student handout William C. Ratcliff, Nicholas Beerman and Tami Limberg Introduction. The evolution of
Predator escape: an ecologically realistic scenario for the evolutionary origins of multicellularity Student handout William C. Ratcliff, Nicholas Beerman and Tami Limberg Introduction. The evolution of
Quantum Dots and Colors Worksheet Answers
 Quantum Dots and Colors Worksheet Answers Background Quantum dots are semiconducting nanoparticles that are able to confine electrons in small, discrete spaces. Also known as zero-dimensional electronic
Quantum Dots and Colors Worksheet Answers Background Quantum dots are semiconducting nanoparticles that are able to confine electrons in small, discrete spaces. Also known as zero-dimensional electronic
SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS. Transient infection of the zebrafish notochord triggers chronic inflammation
 SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS Transient infection of the zebrafish notochord triggers chronic inflammation Mai Nguyen-Chi 1,2,5, Quang Tien Phan 1,2,5, Catherine Gonzalez 1,2, Jean-François
SUPPLEMENTARY FIGURES AND TABLES AND THEIR LEGENDS Transient infection of the zebrafish notochord triggers chronic inflammation Mai Nguyen-Chi 1,2,5, Quang Tien Phan 1,2,5, Catherine Gonzalez 1,2, Jean-François
BIOLOGY 111. CHAPTER 5: Chromosomes and Inheritance
 BIOLOGY 111 CHAPTER 5: Chromosomes and Inheritance Chromosomes and Inheritance Learning Outcomes 5.1 Differentiate between sexual and asexual reproduction in terms of the genetic variation of the offspring.
BIOLOGY 111 CHAPTER 5: Chromosomes and Inheritance Chromosomes and Inheritance Learning Outcomes 5.1 Differentiate between sexual and asexual reproduction in terms of the genetic variation of the offspring.
OBJECTIVE: To understand the relation between electric fields and electric potential, and how conducting objects can influence electric fields.
 Name Section Question Sheet for Laboratory 4: EC-2: Electric Fields and Potentials OBJECTIVE: To understand the relation between electric fields and electric potential, and how conducting objects can influence
Name Section Question Sheet for Laboratory 4: EC-2: Electric Fields and Potentials OBJECTIVE: To understand the relation between electric fields and electric potential, and how conducting objects can influence
LINEAR ALGEBRA - CHAPTER 1: VECTORS
 LINEAR ALGEBRA - CHAPTER 1: VECTORS A game to introduce Linear Algebra In measurement, there are many quantities whose description entirely rely on magnitude, i.e., length, area, volume, mass and temperature.
LINEAR ALGEBRA - CHAPTER 1: VECTORS A game to introduce Linear Algebra In measurement, there are many quantities whose description entirely rely on magnitude, i.e., length, area, volume, mass and temperature.
LECTURE 11: Monte Carlo Methods III
 1 LECTURE 11: Monte Carlo Methods III December 3, 2012 In this last chapter, we discuss non-equilibrium Monte Carlo methods. We concentrate on lattice systems and discuss ways of simulating phenomena such
1 LECTURE 11: Monte Carlo Methods III December 3, 2012 In this last chapter, we discuss non-equilibrium Monte Carlo methods. We concentrate on lattice systems and discuss ways of simulating phenomena such
New Directions in Computer Science
 New Directions in Computer Science John Hopcroft Cornell University Time of change The information age is a revolution that is changing all aspects of our lives. Those individuals, institutions, and nations
New Directions in Computer Science John Hopcroft Cornell University Time of change The information age is a revolution that is changing all aspects of our lives. Those individuals, institutions, and nations
The Radiata-Bilateria split. Second branching in the evolutionary tree
 The Radiata-Bilateria split Second branching in the evolutionary tree Two very important characteristics are used to distinguish between the second bifurcation of metazoans Body symmetry Germinal layers
The Radiata-Bilateria split Second branching in the evolutionary tree Two very important characteristics are used to distinguish between the second bifurcation of metazoans Body symmetry Germinal layers
T H E J O U R N A L O F C E L L B I O L O G Y
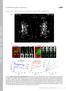 T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Eisner et al., http://www.jcb.org/cgi/content/full/jcb.201312066/dc1 Figure S1. Mitochondrial continuity in adult skeletal muscle. (A)
T H E J O U R N A L O F C E L L B I O L O G Y Supplemental material Eisner et al., http://www.jcb.org/cgi/content/full/jcb.201312066/dc1 Figure S1. Mitochondrial continuity in adult skeletal muscle. (A)
Auto-correlation of retinal ganglion cell mosaics shows hexagonal structure
 Supplementary Discussion Auto-correlation of retinal ganglion cell mosaics shows hexagonal structure Wässle and colleagues first observed that the local structure of cell mosaics was approximately hexagonal
Supplementary Discussion Auto-correlation of retinal ganglion cell mosaics shows hexagonal structure Wässle and colleagues first observed that the local structure of cell mosaics was approximately hexagonal
Lecture 7: Simple genetic circuits I
 Lecture 7: Simple genetic circuits I Paul C Bressloff (Fall 2018) 7.1 Transcription and translation In Fig. 20 we show the two main stages in the expression of a single gene according to the central dogma.
Lecture 7: Simple genetic circuits I Paul C Bressloff (Fall 2018) 7.1 Transcription and translation In Fig. 20 we show the two main stages in the expression of a single gene according to the central dogma.
I. Specialization. II. Autonomous signals
 Multicellularity Up to this point in the class we have been discussing individual cells, or, at most, populations of individual cells. But some interesting life forms (for example, humans) consist not
Multicellularity Up to this point in the class we have been discussing individual cells, or, at most, populations of individual cells. But some interesting life forms (for example, humans) consist not
Motivation. Evolution has rediscovered several times multicellularity as a way to build complex living systems
 Cellular Systems 1 Motivation Evolution has rediscovered several times multicellularity as a way to build complex living systems Multicellular systems are composed by many copies of a unique fundamental
Cellular Systems 1 Motivation Evolution has rediscovered several times multicellularity as a way to build complex living systems Multicellular systems are composed by many copies of a unique fundamental
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/10/e1600964/dc1 Supplementary Materials for Constructing 3D heterogeneous hydrogels from electrically manipulated prepolymer droplets and crosslinked microgels
advances.sciencemag.org/cgi/content/full/2/10/e1600964/dc1 Supplementary Materials for Constructing 3D heterogeneous hydrogels from electrically manipulated prepolymer droplets and crosslinked microgels
Supplemental Activity: Vectors and Forces
 Supplemental Activity: Vectors and Forces Objective: To use a force table to test equilibrium conditions. Required Equipment: Force Table, Pasco Mass and Hanger Set, String, Ruler, Polar Graph Paper, Protractor,
Supplemental Activity: Vectors and Forces Objective: To use a force table to test equilibrium conditions. Required Equipment: Force Table, Pasco Mass and Hanger Set, String, Ruler, Polar Graph Paper, Protractor,
Modelling with cellular automata
 Modelling with cellular automata Shan He School for Computational Science University of Birmingham Module 06-23836: Computational Modelling with MATLAB Outline Outline of Topics Concepts about cellular
Modelling with cellular automata Shan He School for Computational Science University of Birmingham Module 06-23836: Computational Modelling with MATLAB Outline Outline of Topics Concepts about cellular
In order to confirm the contribution of diffusion to the FRAP recovery curves of
 Fonseca et al., Supplementary Information FRAP data analysis ) Contribution of Diffusion to the recovery curves In order to confirm the contribution of diffusion to the FRAP recovery curves of PH::GFP
Fonseca et al., Supplementary Information FRAP data analysis ) Contribution of Diffusion to the recovery curves In order to confirm the contribution of diffusion to the FRAP recovery curves of PH::GFP
Alleles Notes. 3. In the above table, circle each symbol that represents part of a DNA molecule. Underline each word that is the name of a protein.
 Alleles Notes Different versions of the same gene are called alleles. Different alleles give the instructions for making different versions of a protein. This table shows examples for two human genes.
Alleles Notes Different versions of the same gene are called alleles. Different alleles give the instructions for making different versions of a protein. This table shows examples for two human genes.
Green Fluorescent Protein (GFP) Today s Nobel Prize in Chemistry
 In the news: High-throughput sequencing using Solexa/Illumina technology The copy number of each fetal chromosome can be determined by direct sequencing of DNA in cell-free plasma from pregnant women Confession:
In the news: High-throughput sequencing using Solexa/Illumina technology The copy number of each fetal chromosome can be determined by direct sequencing of DNA in cell-free plasma from pregnant women Confession:
Conclusions. The experimental studies presented in this thesis provide the first molecular insights
 C h a p t e r 5 Conclusions 5.1 Summary The experimental studies presented in this thesis provide the first molecular insights into the cellular processes of assembly, and aggregation of neural crest and
C h a p t e r 5 Conclusions 5.1 Summary The experimental studies presented in this thesis provide the first molecular insights into the cellular processes of assembly, and aggregation of neural crest and
Global Quantification of Tissue Dynamics in the Developing Mouse Kidney
 Developmental Cell, Volume 29 Supplemental Information Global Quantification of Tissue Dynamics in the Developing Mouse Kidney Kieran M. Short, Alexander N. Combes, James Lefevre, Adler L. Ju, Kylie M.
Developmental Cell, Volume 29 Supplemental Information Global Quantification of Tissue Dynamics in the Developing Mouse Kidney Kieran M. Short, Alexander N. Combes, James Lefevre, Adler L. Ju, Kylie M.
