Probabilistic Gene Network
|
|
|
- Virgil Johns
- 5 years ago
- Views:
Transcription
1 Probabilistic Gene Network Kristine Joy E. Carpio,3, Gilles Bernot 2, Jean-Paul Comet 2 and Francine Diener Laboratoire J.A. Dieudonné, Université de Nice - Sophia Antipolis, France 2 CNRS UMR 727, Laboratoire I3S, Université de Nice - Sophia Antipolis, France Abstract In this article we present a modelling framework that links the well known modelling framework of gene network introduced by R. Thomas and Markov chains. In a first development we introduce a Markov chain having as state space the set of all possible states of the R. Thomas models: we generate the transition probabilities by examining all the possible parameterizations of the interaction graph. The second development focuses on a stochastic framework where several parameterizations of a same qualitative gene interaction graph are considered and transition probabilities allow one to jump from a state to another one which can potentially be in another parameterized model. The idea is to consider only parameterized qualitative models of R. Thomas which abstract biological knowledge, and to use transition probabilities to allow to jump from one to another, if information coming from biological experiments reinforces the belief in a particular model. Introduction Regulatory networks are models based on graphs which are used to obtain a simpler view of gene regulation [6, p. ]. Gene regulation is defined as the process of turning genes on and off which is made possible by a network of interactions that includes chemically modifying genes and using regulatory proteins. Gene regulation guarantees that appropriate genes are expressed at proper times specially during early development where cells begin to take on specific functions; it also helps an organism respond to its environment [8]. The different frameworks for modelling gene networks can be classified into three main groups. The systems of differential equations have been largely used in order to represent a lot of systems with a lot of details (transcription, traduction, transports...). The second group consists of stochastic frameworks like Markov chains. The Markov modelling framework supposes that, given the past and the present, the future only depends on the present [9, p. 63]. 3 On research leave from De La Salle University, Philippines. Funded by Emma in the framework of the EU Erasmus Mundus Action 2.
2 This framework is well adapted to biological systems but supposes a strong effort in the enumeration of all entities (and interactions between them) that play a role in the system. The third group of approaches consists of qualitative frameworks in which details have been abstracted and only main causalities have been taken into account. Two paradigmatic frameworks can be classified in this group: the Boolean networks first introduced by Kauffman [5] and the multi-valuated modelling framework first introduced by R. Thomas [3]. In this paper we present a modelling framework that links the well known modelling framework of gene network introduced by R. Thomas and Markov chains. In a first development we introduce a Markov chain having as state space the set of all possible states of the R. Thomas models: we generate the transition probabilities by examining all the possible parameterizations of the interaction graph. Thus the Markov chain represents the possible behaviours obtained by superposition of all parameterized models. We then extend this stochastic framework to a Markov chain in which we distinguish the states of each parameterized model and where the probabilities are computed on a smaller set of parameterizations. The idea is to consider only parameterized qualitative models of R. Thomas which represent well the biological knowledge and to use transition probabilities to allow the system to jump from a particular dynamics to another one. The earliest qualitative model for a gene regulatory network was introduced by Kauffman [5]. In Kauffman s model, a gene is modelled as a binary variable ( or ) which takes only one of the possible Boolean functions of its inputs. When the gene is on it takes the value of, otherwise it takes. The outputs of a gene at time t + depends only on the activity at time t. In this group of qualitative modelling frameworks, we can also cite the framework of R. Thomas in which each gene can have several levels of expression [3]. Thomas model allows the gene to be represented as a multilevel logical variable (,, 2,...); the number of possible values depends on the number of distinct actions it does on the network. In this case, the actions refer to a gene acting as an activator or repressor of some of the genes in the network. For each distinct action, a threshold value is assigned to specify from which expression level the influence takes place. So a variable with n distinct actions has n thresholds and this variable becomes an (n + )-level variable. Allowing multilevel logical variables guarantees that no two distinct actions can happen simultaneously. In order to illustrate our modelling approach, we focus on the gene regulatory network of the pathogen Pseudomonas aeruginosa, more specifically on the subsystem which is responsible for mucus production in the lungs of individuals with cystic fibrosis. Although the global gene regulatory network of
3 Promotor algu muca mucb mucc mucd + mucus synthesis + AlgU anti-algu Figure : Portion of the gene regulatory network of the pathogen Pseudomonas aeruginosa, responsible for the mucus production; an arrow indicates activation or stimulation while a T-symbol represents repression [, 2]. this pathogen consists of 69 genes and 2 regulatory interactions between their products [3], the subsystem controlling the mucus production consists of some genes and proteins, see Figure. Because the mucus production worsens the respiratory problem of the patients which is often the cause of death [2, p. 75], elucidating the behaviour of this subsystem may be of great help to address this outcome. The paper is organised as follows. Section 2 is devoted to sketch the qualitative modeling framework of R. Thomas. Section 3 explains how to build a Markov chain from the set of all possible parameterizations of an interaction graph. We can then push this idea forward and propose, when biological knowledge allows to reduce the set of possible parameterizations, a unique stochastic model where it becomes possible to jump from one qualitative model to another, see Section 4. Finally Section 5 is devoted to conclusion and discussion. 2 Reminding of R. Thomas Modelling Framework The biological regulatory network controlling the mucus production in Pseudomonas aeruginosa can be abstracted by the simple directed graphs of Figure 2 in which positive and negative signs indicate activation and repression, respectively, following the direction of the edge they label. These interaction graphs would suffice if we are only interested in applying Kauffman s model but if we want to apply Thomas model there must be a threshold indicator for each distinct action of the gene as seen in Figure 2. Such interaction graphs, as those in Figure 2, are called biological regulatory graphs [, Definition ] and are represented by graphs G = (V, E), where V is the set of genes in the network and E represents the set of interactions between the genes in V. Each vertex v V has a boundary b v that is less than or equal to its out-degree (unless its out-degree is zero in which case we take the boundary to be one) while each edge is labelled by an ordered pair
4 containing the threshold t and action ε (activation + or repression ). a) b) x y + x y 2+ mucus 2+ mucus Figure 2: Two simple interaction graphs representing the system controlling the production of mucus in Pseudomonas aeruginosa (see Figure ). The variable x denotes the gene algu and the protein AlgU while y denotes the gene mucb and the anti-algu. Both biological regulatory graphs differ by the labelling of edges outgoing from node x: the thresholds are not the same. In Figure 2, we have V = {x, y} and E = {(x x), (x y), (y x)}. The interaction x mucus is not taken into consideration since mucus has no action backward toward x and y: it is a by-product of x which is produced when x is at its highest level (the regulation (x mucus) is labelled by the threshold 2 since x has two distinct actions on y and on itself). The variable y has a unique action (repression of x) so the only possible threshold of the regulation (y x) is. Lastly note that Figure 2 does consider two possible biological regulatory graphs because the ordering between thresholds labelling edges outgoing from node x is not well known: we have to consider the two possible orderings. In order to build the the dynamics of a biological regulatory graph, we first introduce the states of the network. A state of a regulatory network is a tuple denoted by (n v,..., n vp ), where p denotes the number of genes and for each n vi N (natural numbers / non negative integers) n vi b v [, Definition 3]. We have now to define the resources of a vertex v i with respect to a state (n v,..., n vp ). Given a regulatory network, a state (n v,..., n vp ) and an edge (v i v j ) with label (t, ε), the vertex v i is a resource of v j if and only if n vi t and ε = + or n vi < t and ε = [, Definition 4]. The intuition is that the absence of an inhibitor plays the same role as the presence of an activator. Finally, a biological regulatory network refers to the biological regulatory graph G = (V, E) together with a set of parameters K = {k v,ω }, where v V, ω G (v) = {u (u v) is an edge in G} and k v,ω b v [, Definition 2]. The parameter k v,ω gives the value towards which v is attracted when the set of resources of v is ω. An easy way to represent the dynamics of a regulatory network is to associate with each state, the state towards which the system is attracted, when considering that each variable v changes at the same time to its current attrac-
5 tion value k v,ω (ω being the current set of ressources of v). This defines the so-called synchronous state graph S = (S, T ): The set of vertices S contains all possible states, and the edges of T are of the form (n v,..., n vp ) (k (v,ω ),..., k (vp,ω p)) such that for every i, ω i is the set of resources of v i at the state (n v,..., n vp ) [, Definition 5]. Unfortunately, the parameters k v,ω are not measurable in vivo [, p. 342] so we are left with several possibilities which results to obtaining several synchronous state graphs. The synchronous state graph is not well adapted to represent evolution of the biological system because it is improbable that two (or more) genes reach their thresholds exactly at the same time and because a gene cannot directly jump two or more consecutive thresholds. To correct these drawbacks, one has to desynchronize each transition. Each transition (n v,..., n vp ) (n v,..., n v p ) is replaced by the set of its desynchronizations which are of the form (n v,..., n vi, n vi, n vi +,..., n vp ) (n v,..., n vi, n vi + δ, n vi +,..., n vp ) for i such that n vi n v i and δ = when n vi < n v i, otherwise δ = [, Definition 6]. The desynchronization step allows some states to transition to more than one other state. Thus, the dynamics of the regulatory graph is represented by the asynchronous state graph S = (S, T ) where the set S of vertices is the set of states and the set T of transitions contains all desynchronized transitions of the synchronous state graph [, Definition 7]. Note that two different synchronous state graphs may lead to the same asynchronous state graph since the desynchronization step can reduce two distinct synchronous transitions to the same set of desynchronized transitions. y y y x x x Figure 3: A possible qualitative dynamics in the modelling framework of R. Thomas (left). This asynchronous state graph can correspond to an inward spiral (centre) or to an outward spiral (right). The global modelling approach consists of identifying all variables of the system, as well as their interactions and then the identification of parameters. Unfortunately, sometimes, it is not clear which parameters to choose. Consider the possible qualitative dynamics shown in Figure 3 (left) where we see that the state (2, ) is a stable state and that the system presents a counterclockwise oscillation between states which have a level of x less or equal to. It is clear that both the inward spiral (Figure 3 centre) and the outward spiral (right) are
6 represented by the same qualitative model. But it could be more convenient to represent the inward spiral by the model where transition (, ) (2, ) does not appear: the small part of the domain (, ) from which the stable domain (2, ) is reachable, is integrated in the domain (2, ), and when a grand tour is done in the inward spiral, it become impossible to reach the stable domain (2, ). 3 Markov Chains in Gene Regulatory Networks When Kauffman proposed the Boolean model, the output of the genes at time t + were only dependent on the activity of its inputs at time t [5, p. 44] which resembles the Markov property where given the past and the present, the future only depends on the present [9, p. 63]. In Kauffman s model a gene at time t transitions only to exactly one state at t + while in a Markov chain, the transition probabilities allows the system to go from a state at time t to more than one other state at t +. In this section, we discuss briefly several ways of setting up Markov chains for gene regulatory networks based on available literature (see the works of Skornyakov et al. [2], Kim et al. [7] and Shmulevich et al. []) and then we give a basic Markov chain that represents the asynchronous dynamics of the interaction graphs of Figure 2. In these three articles [7,, 2], a state can be thought of as a snapshot of the activity level of all the genes with respect to a given time. In [], these states were referred to as maps. The Markov chain was applied to Kauffman s Boolean model of a gene regulatory network but it requires that the cooperation between interactions are well specified. In [7], the Markov chain allowed each gene to take three states, namely - (under-expressed), (equivalentlyexpressed) and (over-expressed) and it makes use of conditional probabilities to compute the transition probabilities. The Probabilistic Boolean Network (PBN) [] addresses the deterministic nature of Kauffman s Boolean model. Both works [7, ] can be easily extended to Thomas model. However, the updates on all the genes in a PBN are done synchronously to simplify computation while preserving the generic properties of global network dynamics []. In Thomas modelling framework, there are several possible values for the parameters k vi,ω i, where v i denotes the ith gene while ω i denotes the ith gene s resources. The variability of these parameters results to potentially enormous (exponential) number of synchronous state graphs, but this number can be largely trimmed by considering the following constraints: k v, = and ω ω k v,ω k v,ω. () When a gene v has no resources, its expression level is not supposed to increase. Hence, a value of zero is assigned to k v,. When a gene loses some of
7 its resources its expression level may drop while increasing its resources may increase its expression level. Because of the constraints in (), the number of synchronous state graphs for each regulatory graph in Figure 2 is reduced to 28 which is now a reasonable number of graphs to work with. These synchronous state graphs can be obtained by playing with the different values of the parameters of Table with the previous constraints of inclusion of resources in mind. a) State Next State x y x y k x,{y} k x,{y} k y,{x} k y,{x} 2 k x,{x,y} k y,{x} 2 k x,{x} k y,{x} b) State Next State x y x y k x,{y} k x,{x,y} k x,{x} 2 k x,{x,y} k y,{x} 2 k x,{x} k y,{x} Table : Tables giving the parameters of interaction graphs according to the current state: Tables a) and b) correspond to Figures 2(a) and 2(b), respectively. We now recall the foundations of Markov chains in discrete time on a countable state space. Let p n ij denote the probability that state j can be reached from state i in n steps. If n = we have the entries p ij of the transition matrix P while the n-step transition probabilities p n ij (n > ) are contained in the matrix P n. If for any couple of states (i, j) we can find an n N + (positive integers) such that p n ij > and pn ji >, then we say that the states communicate with each other. This indicates that all the states belong to a unique class (Markov chain is irreducible). A state i has period d if p n ii = whenever n is not divisible by d and d is the greatest integer with this property [9, p. 69]. In an irreducible aperiodic Markov chain the states are either all transient or null recurrent (finite number of visits) or positive recurrent (infinite number of visits) with a unique stationary distribution {π j, j =, 2,...}, where π j = lim n p n ij > [9, Theorem 4.3.3]. Note that an irreducible Markov chain with a finite state space cannot have transient states because the chain will eventually stop once it has visited all the states in a finite number of time which should not be the case [9, p. 7]. Thus, in such a chain, all the states must be positive recurrent. Let µ jj denote the expected number of transitions needed to return to state j starting from j. When state j is positive recurrent µ jj < [9, p. 73] and when the Markov chain is aperiodic and irreducible, we have lim n p n ij = /µ jj [9, Theorem 4.3.]. It follows that in such a Markov chain, we have π j = /µ jj. An aperiodic irreducible positive recurrent Markov chain is called ergodic [9, p. 77]. In an ergodic Markov chain we have a limiting matrix
8 Π = lim n P n with all rows having the same vector π = (π, π 2,...) of positive probabilities with sum equal to ; the probability π i denotes the long-run proportion of time that the Markov chain stays in state i (A) (,) (,) (,) (,) (2,) (2,) (,) 2 6 (,) 28 (,) (,) 2 7 (2,) (2,) (B) (,) (,) (,) (,) (2,) (2,) (,) 2 6 (,) 28 (,) (,) (2,) (2,) Table 2: Sum of the probabilities assigned to each possible transition over all asynchronous state graphs. (A) and (B) are built from Tables (a) and (b) respectively. These numbers take into account the multiplicity of asynchronous state graphs. To model the asynchronous dynamics by a Markov chain, we examine all the possible asynchronous state graphs. Since different synchronous state graphs may lead to an identical asynchronous state graph, the number of distinct asynchronous state graphs can be less than the number of distinct synchronous state graphs. In that case, we also have to take into account the multiplicity (number of occurrences) of each distinct asynchronous state graph. In each asynchronous state graph, we assign appropriate probabilities to transitions (the transitions outgoing from a same state receive the same probability if no knowledge contradicts this hypothesis). Once this is done for each asynchronous state graph, we multiply the probabilities assigned to each possible transition by the multiplicity of the asynchronous state graph and take the sum of all such terms over all the possible asynchronous state graphs. We now set-up the transition probability matrices for Tables (a) and (b) which are obtained by simply dividing the entries of Table 2 by the total number of synchronous state graphs which is 28 as already mentioned. We have: P a = and P b = The order of the entries in P a and P b follow the order given in Table 2. These Markov chains are ergodic, which guarantees the existence of a unique stationary probability π i which gives the long-run proportion of time in i.
9 Evaluating lim n P n a, we have π (,) =.339, π (,) =.96, π (,) =.34, π (,) =.28, π (2,) =.9, and π (2,) =.42. In the longrun, the most visited states are (,), (,), and (,). Given the long-run probabilities, we can also compute the average number of transitions required to return to each state. Recall that µ ii denotes the expected number of transitions needed to return to state i starting at state i. For this chain, the mean return times for the states (, ), (, ), (, ), (, ), (2, ) and (2, ) are given by µ = 2.949, µ 22 =.424, µ 33 = 3.285, µ 44 = 7.88, µ 55 =.4, and µ 66 = , respectively. For P b, we obtain lim n P n b which gives the stationary distributions π (,) =.73, π (,) =.22, π (,) =.285, π (,) =., π (2,) =.347, and π (2,) =.72. The most visited states are (2, ), (, ), and (2, ). For this chain, the mean return times are given by µ = 3.592, µ 22 = 46.25, µ 33 = 3.58, µ 44 = 9.884, µ 55 = 2.883, and µ 66 = Both chains show that 7% of the time in the long-run y = which means that x is not inhibited; we then expect that in the long-run x most of the time. On one hand, this is true for P b since the most visited states in the long-run are the states (2, ) and (, ). On the other hand, in the case of P a, these both states are visited only close to 4% of the time. Moreover a drawback of setting up a Markov chain this way is the inability to show the steady-states or circuits of the asynchronous state graphs. 4 Probabilistic Gene Network (PGN) In the previous model, the Markov chain comes from a superposition of all the parameterized qualitative models. In order to distinguish the different asynchronous state graphs, we introduce a Markov chain which memorizes the asynchronous state graph a particular state is in. Because all asynchronous state graphs can differ drastically, we limit this Markov chain to a set of asynchronous state graphs that behave closely (see below). In PGNs, we take into consideration attractors. The attractors of a network are the smallest sets of states from which one cannot escape [, Section 2.5]. This can be a stable state which is a state without successors or a group of states that demonstrates sustained oscillations without exits. These latter attractors are said to be cyclic and, naturally, it is not possible to reach a stable state starting from a cyclic attractor. Note that every asynchronous state graph has at least an attractor. The stable states of an asynchronous state graph results to having absorbing states in the Markov chain built on it. The presence of absorbing states may result to obtaining an absorbing Markov chain; this
10 happens when it is possible to eventually reach an absorbing state from every state [4, p. 46]. To show a Markov chain is absorbing, we need to find an n N + such that all the entries of P n are non zero, P being the transition probability matrix. An absorbing Markov chain give the expected times of absorption and the probability of absorption from every transient state. Let N = {N,...} denote a subset of Thomas networks (asynchronous state graphs). This set can correspond to all asynchronous state graphs that are coherent with some biological knowledge, in that sense, the set is supposed to be largely smaller than the total number of asynchronous state graphs. This set can result e.g. from a filtering step which selects only asynchronous state graphs which are coherent with behavioural properties expressed in a formal language []. We introduce an ordering relation between Thomas networks: for distinct networks N and N, we have N > N if and only if N is a subgraph of N. This ordering relation leads to consider the set of models equipped with this relation as a lattice with possibly several minimal elements. Denote by S = {s, s 2,...} a set of states (this set is common to all asynchronous state graphs). The intuition is the following. A biological system can be represented by a set of different dynamics (asynchronous state graphs). In a particular environment, the biological system can behave exactly as one of these dynamics but according to some changes of the environment, the behaviour of the biological system can adopt the dynamics of another asynchronous state graph. It becomes natural to allow the Markov chain to jump from one asynchronous state graph to another. But it is unlikely that the biological system jumps from a state of a certain network toward another state in another network with a very different dynamics from the initial network. This is the reason why the jumps are possible only under some conditions on the ordering relation between Thomas networks. A probabilistic gene network (PGN for short) on N satisfies the following conditions: i. For each N N and each transition s s on the asynchronous state graph N a probability of P N (s s ) is attached in such a way that for every s, s S P N(s s ) =. ii. For each pair of networks N, N N such that N > N and there exists no other network N such that N > N > N, a probability is attached to (N N ) in such a way that the sum of all such probability for a given N is less than. Once a probabilistic gene network has been established, we define its cor-
11 responding probabilistic state graph. Let (N, s) denote the set of nodes of the probabilistic state graph, where N N and s S. The set of edges (transitions) is defined by (N, s) (N, s ) if and only if s s (with s s) is a transition of N and P (N N ). The probability P ((N, s) (N, s )) is defined by P ((N, s) (N, s )) W (N ) [(s s = ) N](N)P N (s s ) W (N ) [(s s ) N ](N )P N (s s ), (2) N N s S where A ( ) is the indicator function and W (N) denotes the weight of the network N which pertains to its multiplicity in the set of all possible asynchronous state graphs of N. When the transition does not involve a change of network, we replace N by N to compute for the corresponding probability. The probabilistic state graph is used to set up a Markov chain that would hopefully give a better representation of the dynamics of a gene regulatory network. More precisely the probabilistic state graph allows one to walk inside the set of considered asynchronous state graphs. Thus, a probabilistic gene network gives information not only on the state of each genes but also the network containing the state. We present an illustration of a Probabilistic Gene Network by considering the networks of Figure 4. For N = N (resp. N 2 ), the value of P N (s s ) is computed making the assumption that in the network N the probability of transitioning from s to any of its successor is equally likely which is divided by the number of transitions outgoing from s in N. Moreover we suppose that N contains twice N and once N 2, which can be interpreted as: staying in N is more likely than jumping from N to N 2. The transition probability matrix is: N, (, ) N, (, ) 2/3 /3 N, (, ) 2/3 /3 N, (, ) /3 /3 /6 /6 N, (2, ) N, (2, ) /3 /3 /3. N 2, (, ) N 2, (, ) N 2, (, ) N 2, (, ) /2 /2 N 2, (2, ) N 2, (2, )
12 We can note that it is not possible to escape from (N, (, )) and (N, (2, )). This is due to the fact that to jump from an asynchronous state graph to another one, it is mandatory to be in a state which is not stable in the initial asynchronous state graph. Thus stable states in non minimal asynchronous state graphs (in the lattice) become absorbing states of the chain. In this illustration, the resulting Markov chain is an absorbing Markov chain so we can generate the expected time of absorption from each transient state and its corresponding probability of absorption in the stable states as shown in Tables 3 and 4, respectively. y Network (N ) (, ) (, ) 2 2 (2, ) N y 2 Network 2 (N 2 ) x (, ) (, ) 2 (, ) (, ) 2 (2, ) (2, ) N 2 2 x (, ) (, ) (2, ) Figure 4: Two asynchronous state graphs (left) associated with interaction graph of Figure 2(a) on which we illustrate the construction of the probabilistic gene network (right). The numbers labelling the transitions (right) correspond to the numerator of Equation 2. When involved networks have only cyclic attractors, the resulting Markov chain gets divided into several classes whose number would depend on the number of networks in the subset and on the structure of the lattice. Because the chain only allows a transition from a larger network to a smaller one, all the states in a network form a class of transient states except the states in the smallest networks (according to the lattice structure of the set of networks) which constitute different classes of recurrent states. Transient Expected Time of States Absorption (N,(, )) (N,(, )) (N,(, )) 2 (N,(2, )) 5/3 =.666 Transient Expected Time of States Absorption (N 2,(, )) (N 2,(, )) (N 2,(, )) 2 (N 2,(2, )) Table 3: Expected time of absorption (number of transitions) from any of transient state to any of the absorbing states for the networks in Figure 4.
13 Transient Probability of Absorption States (N, (, )) (N, (2, )) (N 2, (, )) (N 2, (2, )) (N, (, )) 2/3 /3 (N, (, )) 2/3 /3 (N, (, )) 4/9 5/9 (N, (2, )) 4/27 /3 5/27 /3 (N 2, (, )) (N 2, (, )) (N 2, (, )) (N 2, (2, )) Table 4: The transient states in the smaller network can only be absorbed in its own steady states because it is not allowed to leave the network. The transient states in the bigger network can be absorbed by any of the absorbing states of the networks under consideration, see Figure 4. 5 Discussion and conclusion In this article we mixed two different frameworks of gene networks allowing to take advantage of the formal framework of R. Thomas modelling theory and to use transition probabilities of Markov chains to change the parameterized model. According to Figure 3, the modelling framework of R. Thomas leads to a single unique model, both the inward spiral and the outward spiral. In a natural way, it could be more efficient to represent the inward spiral by the model where transition (, ) (2, ) does not appear. In such a case, it could be interesting to consider in a unique framework both discrete state graphs and to allow the trajectory to jump from one to another, if information coming from biological experiments reinforces the belief in a particular model. For example, longer are the observed traces around the qualitative cycle, bigger the belief in the model representing the inward spiral. In such a way, it becomes natural to consider each asynchronous state graph as the dynamics of the biological system in a particular context. When the environment changes the context, the qualitative dynamics can also change. Probabilistic Gene Networks presented in this article, constitute a first framework allowing to jump from a qualitative dynamics to another one. References [] Bernot, G., J-P. Comet, A. Richard & J. Guespin, (24) Application of formal methods to biological regulatory networks: extending Thomas asynchronous logical approach with temporal logic. Journal of Theoretical Biology 229:
14 [2] Bernot, G., J-P. Comet, A. Richard, M. Chaves, J-L. Gouzé, J-L. & F. Dayan, (23) Modeling and analysis of gene regulatory networks. In Modeling in Computational Biology and Biomedicine: A Multidisciplinary Endeavor, eds F. Cazals, P. Kornprobst, pp Springer- Verlag Berlin Heidelberg. [3] Galán-Vásquez, E., B. Luna, & A. Martinez-Antonio, (2) The regulatory network of Pseudomonas aeruginosa. Microbial Informatics and Experimentation (), 3. doi:.86/ [4] Grinstead, C.M. & J.L. Snell, (997) Introduction to Probability, 2nd edn (pp ). American Mathematical Society. [5] Kauffman, S.A., (969). Metabolic stability and epigenesis in randomly constructed genetic nets. Journal of Theoretical Biology 22: [6] Klipp, E., R. Herwig, A. Kowad, C. Wierling & H. Lehrach, (25) Systems Biology in Practice: Concepts, Implementation and Application. Wiley-VCH, Weinheim. [7] Kim, S., H. Li, H., E.R. Dougherty, N. Cao, Y. Chen, M. Bittner, & E.B. Suh, (22) Can Markov chain models mimic biological regulations? Journal of Biological Systems (4): [8] National Institutes of Health. Talking Glossary of Genetic Terms. National Human Genome Research Institute. [9] Ross, S.M., (996) Stochastic Processes, 2nd edn. John Wiley & Sons, Inc., USA. [] Richard, A., J-P. Comet & G. Bernot, (28) R. Thomas logical model. Available from: bernot/teaching/28- LilleSchool-RichardCometBernot.pdf [] Shmulevich, I., E.R. Dougherty, S. Kim & W. Zhang, (22) Probabilistic Boolean networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics 8(2), [2] Skornyakov, V.P., M.V. Skoryakova, A.V. Shurygina & P.V. Skornyakov, (24) Finite-state discrete-time Markov chain models of gene regulatory networks. [Preprint] Available from: [3] Thomas, R., (99). Regulatory networks seen as asynchronous automata: a logical description. Journal of Theoretical Biology 53: -23.
Lecture 20 : Markov Chains
 CSCI 3560 Probability and Computing Instructor: Bogdan Chlebus Lecture 0 : Markov Chains We consider stochastic processes. A process represents a system that evolves through incremental changes called
CSCI 3560 Probability and Computing Instructor: Bogdan Chlebus Lecture 0 : Markov Chains We consider stochastic processes. A process represents a system that evolves through incremental changes called
Markov Processes Hamid R. Rabiee
 Markov Processes Hamid R. Rabiee Overview Markov Property Markov Chains Definition Stationary Property Paths in Markov Chains Classification of States Steady States in MCs. 2 Markov Property A discrete
Markov Processes Hamid R. Rabiee Overview Markov Property Markov Chains Definition Stationary Property Paths in Markov Chains Classification of States Steady States in MCs. 2 Markov Property A discrete
Markov Chains Handout for Stat 110
 Markov Chains Handout for Stat 0 Prof. Joe Blitzstein (Harvard Statistics Department) Introduction Markov chains were first introduced in 906 by Andrey Markov, with the goal of showing that the Law of
Markov Chains Handout for Stat 0 Prof. Joe Blitzstein (Harvard Statistics Department) Introduction Markov chains were first introduced in 906 by Andrey Markov, with the goal of showing that the Law of
Markov chains. Randomness and Computation. Markov chains. Markov processes
 Markov chains Randomness and Computation or, Randomized Algorithms Mary Cryan School of Informatics University of Edinburgh Definition (Definition 7) A discrete-time stochastic process on the state space
Markov chains Randomness and Computation or, Randomized Algorithms Mary Cryan School of Informatics University of Edinburgh Definition (Definition 7) A discrete-time stochastic process on the state space
Synchronous state transition graph
 Heike Siebert, FU Berlin, Molecular Networks WS10/11 2-1 Synchronous state transition graph (0, 2) (1, 2) vertex set X (state space) edges (x,f(x)) every state has only one successor attractors are fixed
Heike Siebert, FU Berlin, Molecular Networks WS10/11 2-1 Synchronous state transition graph (0, 2) (1, 2) vertex set X (state space) edges (x,f(x)) every state has only one successor attractors are fixed
Markov Chains CK eqns Classes Hitting times Rec./trans. Strong Markov Stat. distr. Reversibility * Markov Chains
 Markov Chains A random process X is a family {X t : t T } of random variables indexed by some set T. When T = {0, 1, 2,... } one speaks about a discrete-time process, for T = R or T = [0, ) one has a continuous-time
Markov Chains A random process X is a family {X t : t T } of random variables indexed by some set T. When T = {0, 1, 2,... } one speaks about a discrete-time process, for T = R or T = [0, ) one has a continuous-time
Outlines. Discrete Time Markov Chain (DTMC) Continuous Time Markov Chain (CTMC)
 Markov Chains (2) Outlines Discrete Time Markov Chain (DTMC) Continuous Time Markov Chain (CTMC) 2 pj ( n) denotes the pmf of the random variable p ( n) P( X j) j We will only be concerned with homogenous
Markov Chains (2) Outlines Discrete Time Markov Chain (DTMC) Continuous Time Markov Chain (CTMC) 2 pj ( n) denotes the pmf of the random variable p ( n) P( X j) j We will only be concerned with homogenous
LIMITING PROBABILITY TRANSITION MATRIX OF A CONDENSED FIBONACCI TREE
 International Journal of Applied Mathematics Volume 31 No. 18, 41-49 ISSN: 1311-178 (printed version); ISSN: 1314-86 (on-line version) doi: http://dx.doi.org/1.173/ijam.v31i.6 LIMITING PROBABILITY TRANSITION
International Journal of Applied Mathematics Volume 31 No. 18, 41-49 ISSN: 1311-178 (printed version); ISSN: 1314-86 (on-line version) doi: http://dx.doi.org/1.173/ijam.v31i.6 LIMITING PROBABILITY TRANSITION
Lecture 9 Classification of States
 Lecture 9: Classification of States of 27 Course: M32K Intro to Stochastic Processes Term: Fall 204 Instructor: Gordan Zitkovic Lecture 9 Classification of States There will be a lot of definitions and
Lecture 9: Classification of States of 27 Course: M32K Intro to Stochastic Processes Term: Fall 204 Instructor: Gordan Zitkovic Lecture 9 Classification of States There will be a lot of definitions and
Probability, Random Processes and Inference
 INSTITUTO POLITÉCNICO NACIONAL CENTRO DE INVESTIGACION EN COMPUTACION Laboratorio de Ciberseguridad Probability, Random Processes and Inference Dr. Ponciano Jorge Escamilla Ambrosio pescamilla@cic.ipn.mx
INSTITUTO POLITÉCNICO NACIONAL CENTRO DE INVESTIGACION EN COMPUTACION Laboratorio de Ciberseguridad Probability, Random Processes and Inference Dr. Ponciano Jorge Escamilla Ambrosio pescamilla@cic.ipn.mx
arxiv: v2 [cs.dm] 29 Mar 2013
![arxiv: v2 [cs.dm] 29 Mar 2013 arxiv: v2 [cs.dm] 29 Mar 2013](/thumbs/72/67770593.jpg) arxiv:1302.6346v2 [cs.dm] 29 Mar 2013 Fixed point theorems for Boolean networks expressed in terms of forbidden subnetworks Adrien Richard Laboratoire I3S, CNRS & Université de Nice-Sophia Antipolis, France.
arxiv:1302.6346v2 [cs.dm] 29 Mar 2013 Fixed point theorems for Boolean networks expressed in terms of forbidden subnetworks Adrien Richard Laboratoire I3S, CNRS & Université de Nice-Sophia Antipolis, France.
Reinforcement Learning
 Reinforcement Learning March May, 2013 Schedule Update Introduction 03/13/2015 (10:15-12:15) Sala conferenze MDPs 03/18/2015 (10:15-12:15) Sala conferenze Solving MDPs 03/20/2015 (10:15-12:15) Aula Alpha
Reinforcement Learning March May, 2013 Schedule Update Introduction 03/13/2015 (10:15-12:15) Sala conferenze MDPs 03/18/2015 (10:15-12:15) Sala conferenze Solving MDPs 03/20/2015 (10:15-12:15) Aula Alpha
IEOR 6711: Professor Whitt. Introduction to Markov Chains
 IEOR 6711: Professor Whitt Introduction to Markov Chains 1. Markov Mouse: The Closed Maze We start by considering how to model a mouse moving around in a maze. The maze is a closed space containing nine
IEOR 6711: Professor Whitt Introduction to Markov Chains 1. Markov Mouse: The Closed Maze We start by considering how to model a mouse moving around in a maze. The maze is a closed space containing nine
8. Statistical Equilibrium and Classification of States: Discrete Time Markov Chains
 8. Statistical Equilibrium and Classification of States: Discrete Time Markov Chains 8.1 Review 8.2 Statistical Equilibrium 8.3 Two-State Markov Chain 8.4 Existence of P ( ) 8.5 Classification of States
8. Statistical Equilibrium and Classification of States: Discrete Time Markov Chains 8.1 Review 8.2 Statistical Equilibrium 8.3 Two-State Markov Chain 8.4 Existence of P ( ) 8.5 Classification of States
TMA 4265 Stochastic Processes Semester project, fall 2014 Student number and
 TMA 4265 Stochastic Processes Semester project, fall 2014 Student number 730631 and 732038 Exercise 1 We shall study a discrete Markov chain (MC) {X n } n=0 with state space S = {0, 1, 2, 3, 4, 5, 6}.
TMA 4265 Stochastic Processes Semester project, fall 2014 Student number 730631 and 732038 Exercise 1 We shall study a discrete Markov chain (MC) {X n } n=0 with state space S = {0, 1, 2, 3, 4, 5, 6}.
Statistics 150: Spring 2007
 Statistics 150: Spring 2007 April 23, 2008 0-1 1 Limiting Probabilities If the discrete-time Markov chain with transition probabilities p ij is irreducible and positive recurrent; then the limiting probabilities
Statistics 150: Spring 2007 April 23, 2008 0-1 1 Limiting Probabilities If the discrete-time Markov chain with transition probabilities p ij is irreducible and positive recurrent; then the limiting probabilities
STOCHASTIC PROCESSES Basic notions
 J. Virtamo 38.3143 Queueing Theory / Stochastic processes 1 STOCHASTIC PROCESSES Basic notions Often the systems we consider evolve in time and we are interested in their dynamic behaviour, usually involving
J. Virtamo 38.3143 Queueing Theory / Stochastic processes 1 STOCHASTIC PROCESSES Basic notions Often the systems we consider evolve in time and we are interested in their dynamic behaviour, usually involving
5.3 METABOLIC NETWORKS 193. P (x i P a (x i )) (5.30) i=1
 5.3 METABOLIC NETWORKS 193 5.3 Metabolic Networks 5.4 Bayesian Networks Let G = (V, E) be a directed acyclic graph. We assume that the vertices i V (1 i n) represent for example genes and correspond to
5.3 METABOLIC NETWORKS 193 5.3 Metabolic Networks 5.4 Bayesian Networks Let G = (V, E) be a directed acyclic graph. We assume that the vertices i V (1 i n) represent for example genes and correspond to
Chapter 16 focused on decision making in the face of uncertainty about one future
 9 C H A P T E R Markov Chains Chapter 6 focused on decision making in the face of uncertainty about one future event (learning the true state of nature). However, some decisions need to take into account
9 C H A P T E R Markov Chains Chapter 6 focused on decision making in the face of uncertainty about one future event (learning the true state of nature). However, some decisions need to take into account
Discrete Markov Chain. Theory and use
 Discrete Markov Chain. Theory and use Andres Vallone PhD Student andres.vallone@predoc.uam.es 2016 Contents 1 Introduction 2 Concept and definition Examples Transitions Matrix Chains Classification 3 Empirical
Discrete Markov Chain. Theory and use Andres Vallone PhD Student andres.vallone@predoc.uam.es 2016 Contents 1 Introduction 2 Concept and definition Examples Transitions Matrix Chains Classification 3 Empirical
2 Discrete-Time Markov Chains
 2 Discrete-Time Markov Chains Angela Peace Biomathematics II MATH 5355 Spring 2017 Lecture notes follow: Allen, Linda JS. An introduction to stochastic processes with applications to biology. CRC Press,
2 Discrete-Time Markov Chains Angela Peace Biomathematics II MATH 5355 Spring 2017 Lecture notes follow: Allen, Linda JS. An introduction to stochastic processes with applications to biology. CRC Press,
Discrete time Markov chains. Discrete Time Markov Chains, Limiting. Limiting Distribution and Classification. Regular Transition Probability Matrices
 Discrete time Markov chains Discrete Time Markov Chains, Limiting Distribution and Classification DTU Informatics 02407 Stochastic Processes 3, September 9 207 Today: Discrete time Markov chains - invariant
Discrete time Markov chains Discrete Time Markov Chains, Limiting Distribution and Classification DTU Informatics 02407 Stochastic Processes 3, September 9 207 Today: Discrete time Markov chains - invariant
CS145: Probability & Computing Lecture 18: Discrete Markov Chains, Equilibrium Distributions
 CS145: Probability & Computing Lecture 18: Discrete Markov Chains, Equilibrium Distributions Instructor: Erik Sudderth Brown University Computer Science April 14, 215 Review: Discrete Markov Chains Some
CS145: Probability & Computing Lecture 18: Discrete Markov Chains, Equilibrium Distributions Instructor: Erik Sudderth Brown University Computer Science April 14, 215 Review: Discrete Markov Chains Some
Networks in systems biology
 Networks in systems biology Matthew Macauley Department of Mathematical Sciences Clemson University http://www.math.clemson.edu/~macaule/ Math 4500, Spring 2017 M. Macauley (Clemson) Networks in systems
Networks in systems biology Matthew Macauley Department of Mathematical Sciences Clemson University http://www.math.clemson.edu/~macaule/ Math 4500, Spring 2017 M. Macauley (Clemson) Networks in systems
Math Homework 5 Solutions
 Math 45 - Homework 5 Solutions. Exercise.3., textbook. The stochastic matrix for the gambler problem has the following form, where the states are ordered as (,, 4, 6, 8, ): P = The corresponding diagram
Math 45 - Homework 5 Solutions. Exercise.3., textbook. The stochastic matrix for the gambler problem has the following form, where the states are ordered as (,, 4, 6, 8, ): P = The corresponding diagram
Lecture 11: Introduction to Markov Chains. Copyright G. Caire (Sample Lectures) 321
 Lecture 11: Introduction to Markov Chains Copyright G. Caire (Sample Lectures) 321 Discrete-time random processes A sequence of RVs indexed by a variable n 2 {0, 1, 2,...} forms a discretetime random process
Lecture 11: Introduction to Markov Chains Copyright G. Caire (Sample Lectures) 321 Discrete-time random processes A sequence of RVs indexed by a variable n 2 {0, 1, 2,...} forms a discretetime random process
Countable state discrete time Markov Chains
 Countable state discrete time Markov Chains Tuesday, March 18, 2014 2:12 PM Readings: Lawler Ch. 2 Karlin & Taylor Chs. 2 & 3 Resnick Ch. 1 Countably infinite state spaces are of practical utility in situations
Countable state discrete time Markov Chains Tuesday, March 18, 2014 2:12 PM Readings: Lawler Ch. 2 Karlin & Taylor Chs. 2 & 3 Resnick Ch. 1 Countably infinite state spaces are of practical utility in situations
Logic-Based Modeling in Systems Biology
 Logic-Based Modeling in Systems Biology Alexander Bockmayr LPNMR 09, Potsdam, 16 September 2009 DFG Research Center Matheon Mathematics for key technologies Outline A.Bockmayr, FU Berlin/Matheon 2 I. Systems
Logic-Based Modeling in Systems Biology Alexander Bockmayr LPNMR 09, Potsdam, 16 September 2009 DFG Research Center Matheon Mathematics for key technologies Outline A.Bockmayr, FU Berlin/Matheon 2 I. Systems
On the convergence of Boolean automata networks without negative cycles
 On the convergence of Boolean automata networks without negative cycles Tarek Melliti, Damien Regnault, Adrien Richard, and Sylvain Sené,3 Laboratoire IBISC, EA456, Université d Évry Val-d Essonne, 9000
On the convergence of Boolean automata networks without negative cycles Tarek Melliti, Damien Regnault, Adrien Richard, and Sylvain Sené,3 Laboratoire IBISC, EA456, Université d Évry Val-d Essonne, 9000
2. Transience and Recurrence
 Virtual Laboratories > 15. Markov Chains > 1 2 3 4 5 6 7 8 9 10 11 12 2. Transience and Recurrence The study of Markov chains, particularly the limiting behavior, depends critically on the random times
Virtual Laboratories > 15. Markov Chains > 1 2 3 4 5 6 7 8 9 10 11 12 2. Transience and Recurrence The study of Markov chains, particularly the limiting behavior, depends critically on the random times
Let (Ω, F) be a measureable space. A filtration in discrete time is a sequence of. F s F t
 2.2 Filtrations Let (Ω, F) be a measureable space. A filtration in discrete time is a sequence of σ algebras {F t } such that F t F and F t F t+1 for all t = 0, 1,.... In continuous time, the second condition
2.2 Filtrations Let (Ω, F) be a measureable space. A filtration in discrete time is a sequence of σ algebras {F t } such that F t F and F t F t+1 for all t = 0, 1,.... In continuous time, the second condition
Example: physical systems. If the state space. Example: speech recognition. Context can be. Example: epidemics. Suppose each infected
 4. Markov Chains A discrete time process {X n,n = 0,1,2,...} with discrete state space X n {0,1,2,...} is a Markov chain if it has the Markov property: P[X n+1 =j X n =i,x n 1 =i n 1,...,X 0 =i 0 ] = P[X
4. Markov Chains A discrete time process {X n,n = 0,1,2,...} with discrete state space X n {0,1,2,...} is a Markov chain if it has the Markov property: P[X n+1 =j X n =i,x n 1 =i n 1,...,X 0 =i 0 ] = P[X
Stochastic modelling of epidemic spread
 Stochastic modelling of epidemic spread Julien Arino Centre for Research on Inner City Health St Michael s Hospital Toronto On leave from Department of Mathematics University of Manitoba Julien Arino@umanitoba.ca
Stochastic modelling of epidemic spread Julien Arino Centre for Research on Inner City Health St Michael s Hospital Toronto On leave from Department of Mathematics University of Manitoba Julien Arino@umanitoba.ca
Characterization of Fixed Points in Sequential Dynamical Systems
 Characterization of Fixed Points in Sequential Dynamical Systems James M. W. Duvall Virginia Polytechnic Institute and State University Department of Mathematics Abstract Graph dynamical systems are central
Characterization of Fixed Points in Sequential Dynamical Systems James M. W. Duvall Virginia Polytechnic Institute and State University Department of Mathematics Abstract Graph dynamical systems are central
Measures for information propagation in Boolean networks
 Physica D 227 (2007) 100 104 www.elsevier.com/locate/physd Measures for information propagation in Boolean networks Pauli Rämö a,, Stuart Kauffman b, Juha Kesseli a, Olli Yli-Harja a a Institute of Signal
Physica D 227 (2007) 100 104 www.elsevier.com/locate/physd Measures for information propagation in Boolean networks Pauli Rämö a,, Stuart Kauffman b, Juha Kesseli a, Olli Yli-Harja a a Institute of Signal
MATH 56A: STOCHASTIC PROCESSES CHAPTER 1
 MATH 56A: STOCHASTIC PROCESSES CHAPTER. Finite Markov chains For the sake of completeness of these notes I decided to write a summary of the basic concepts of finite Markov chains. The topics in this chapter
MATH 56A: STOCHASTIC PROCESSES CHAPTER. Finite Markov chains For the sake of completeness of these notes I decided to write a summary of the basic concepts of finite Markov chains. The topics in this chapter
Any live cell with less than 2 live neighbours dies. Any live cell with 2 or 3 live neighbours lives on to the next step.
 2. Cellular automata, and the SIRS model In this Section we consider an important set of models used in computer simulations, which are called cellular automata (these are very similar to the so-called
2. Cellular automata, and the SIRS model In this Section we consider an important set of models used in computer simulations, which are called cellular automata (these are very similar to the so-called
In biological terms, memory refers to the ability of neural systems to store activity patterns and later recall them when required.
 In biological terms, memory refers to the ability of neural systems to store activity patterns and later recall them when required. In humans, association is known to be a prominent feature of memory.
In biological terms, memory refers to the ability of neural systems to store activity patterns and later recall them when required. In humans, association is known to be a prominent feature of memory.
Stochastic processes. MAS275 Probability Modelling. Introduction and Markov chains. Continuous time. Markov property
 Chapter 1: and Markov chains Stochastic processes We study stochastic processes, which are families of random variables describing the evolution of a quantity with time. In some situations, we can treat
Chapter 1: and Markov chains Stochastic processes We study stochastic processes, which are families of random variables describing the evolution of a quantity with time. In some situations, we can treat
SMSTC (2007/08) Probability.
 SMSTC (27/8) Probability www.smstc.ac.uk Contents 12 Markov chains in continuous time 12 1 12.1 Markov property and the Kolmogorov equations.................... 12 2 12.1.1 Finite state space.................................
SMSTC (27/8) Probability www.smstc.ac.uk Contents 12 Markov chains in continuous time 12 1 12.1 Markov property and the Kolmogorov equations.................... 12 2 12.1.1 Finite state space.................................
Markov Chains (Part 3)
 Markov Chains (Part 3) State Classification Markov Chains - State Classification Accessibility State j is accessible from state i if p ij (n) > for some n>=, meaning that starting at state i, there is
Markov Chains (Part 3) State Classification Markov Chains - State Classification Accessibility State j is accessible from state i if p ij (n) > for some n>=, meaning that starting at state i, there is
= P{X 0. = i} (1) If the MC has stationary transition probabilities then, = i} = P{X n+1
 Properties of Markov Chains and Evaluation of Steady State Transition Matrix P ss V. Krishnan - 3/9/2 Property 1 Let X be a Markov Chain (MC) where X {X n : n, 1, }. The state space is E {i, j, k, }. The
Properties of Markov Chains and Evaluation of Steady State Transition Matrix P ss V. Krishnan - 3/9/2 Property 1 Let X be a Markov Chain (MC) where X {X n : n, 1, }. The state space is E {i, j, k, }. The
The Theory behind PageRank
 The Theory behind PageRank Mauro Sozio Telecom ParisTech May 21, 2014 Mauro Sozio (LTCI TPT) The Theory behind PageRank May 21, 2014 1 / 19 A Crash Course on Discrete Probability Events and Probability
The Theory behind PageRank Mauro Sozio Telecom ParisTech May 21, 2014 Mauro Sozio (LTCI TPT) The Theory behind PageRank May 21, 2014 1 / 19 A Crash Course on Discrete Probability Events and Probability
4.7.1 Computing a stationary distribution
 At a high-level our interest in the rest of this section will be to understand the limiting distribution, when it exists and how to compute it To compute it, we will try to reason about when the limiting
At a high-level our interest in the rest of this section will be to understand the limiting distribution, when it exists and how to compute it To compute it, we will try to reason about when the limiting
Optimal State Estimation for Boolean Dynamical Systems using a Boolean Kalman Smoother
 Optimal State Estimation for Boolean Dynamical Systems using a Boolean Kalman Smoother Mahdi Imani and Ulisses Braga-Neto Department of Electrical and Computer Engineering Texas A&M University College
Optimal State Estimation for Boolean Dynamical Systems using a Boolean Kalman Smoother Mahdi Imani and Ulisses Braga-Neto Department of Electrical and Computer Engineering Texas A&M University College
Property preservation along embedding of biological regulatory networks
 Property preservation along embedding of biological regulatory networks Mbarka Mabrouki 12, Marc Aiguier 12, Jean-Paul Comet 3, and Pascale Le Gall 12 1 École Centrale Paris Laboratoire de Mathématiques
Property preservation along embedding of biological regulatory networks Mbarka Mabrouki 12, Marc Aiguier 12, Jean-Paul Comet 3, and Pascale Le Gall 12 1 École Centrale Paris Laboratoire de Mathématiques
Random Boolean Networks
 Random Boolean Networks Boolean network definition The first Boolean networks were proposed by Stuart A. Kauffman in 1969, as random models of genetic regulatory networks (Kauffman 1969, 1993). A Random
Random Boolean Networks Boolean network definition The first Boolean networks were proposed by Stuart A. Kauffman in 1969, as random models of genetic regulatory networks (Kauffman 1969, 1993). A Random
1 Random Walks and Electrical Networks
 CME 305: Discrete Mathematics and Algorithms Random Walks and Electrical Networks Random walks are widely used tools in algorithm design and probabilistic analysis and they have numerous applications.
CME 305: Discrete Mathematics and Algorithms Random Walks and Electrical Networks Random walks are widely used tools in algorithm design and probabilistic analysis and they have numerous applications.
Lecture Notes: Markov chains Tuesday, September 16 Dannie Durand
 Computational Genomics and Molecular Biology, Lecture Notes: Markov chains Tuesday, September 6 Dannie Durand In the last lecture, we introduced Markov chains, a mathematical formalism for modeling how
Computational Genomics and Molecular Biology, Lecture Notes: Markov chains Tuesday, September 6 Dannie Durand In the last lecture, we introduced Markov chains, a mathematical formalism for modeling how
Lecture Notes: Markov chains
 Computational Genomics and Molecular Biology, Fall 5 Lecture Notes: Markov chains Dannie Durand At the beginning of the semester, we introduced two simple scoring functions for pairwise alignments: a similarity
Computational Genomics and Molecular Biology, Fall 5 Lecture Notes: Markov chains Dannie Durand At the beginning of the semester, we introduced two simple scoring functions for pairwise alignments: a similarity
Markov chains and the number of occurrences of a word in a sequence ( , 11.1,2,4,6)
 Markov chains and the number of occurrences of a word in a sequence (4.5 4.9,.,2,4,6) Prof. Tesler Math 283 Fall 208 Prof. Tesler Markov Chains Math 283 / Fall 208 / 44 Locating overlapping occurrences
Markov chains and the number of occurrences of a word in a sequence (4.5 4.9,.,2,4,6) Prof. Tesler Math 283 Fall 208 Prof. Tesler Markov Chains Math 283 / Fall 208 / 44 Locating overlapping occurrences
Recap. Probability, stochastic processes, Markov chains. ELEC-C7210 Modeling and analysis of communication networks
 Recap Probability, stochastic processes, Markov chains ELEC-C7210 Modeling and analysis of communication networks 1 Recap: Probability theory important distributions Discrete distributions Geometric distribution
Recap Probability, stochastic processes, Markov chains ELEC-C7210 Modeling and analysis of communication networks 1 Recap: Probability theory important distributions Discrete distributions Geometric distribution
Markov Chains, Stochastic Processes, and Matrix Decompositions
 Markov Chains, Stochastic Processes, and Matrix Decompositions 5 May 2014 Outline 1 Markov Chains Outline 1 Markov Chains 2 Introduction Perron-Frobenius Matrix Decompositions and Markov Chains Spectral
Markov Chains, Stochastic Processes, and Matrix Decompositions 5 May 2014 Outline 1 Markov Chains Outline 1 Markov Chains 2 Introduction Perron-Frobenius Matrix Decompositions and Markov Chains Spectral
ISE/OR 760 Applied Stochastic Modeling
 ISE/OR 760 Applied Stochastic Modeling Topic 2: Discrete Time Markov Chain Yunan Liu Department of Industrial and Systems Engineering NC State University Yunan Liu (NC State University) ISE/OR 760 1 /
ISE/OR 760 Applied Stochastic Modeling Topic 2: Discrete Time Markov Chain Yunan Liu Department of Industrial and Systems Engineering NC State University Yunan Liu (NC State University) ISE/OR 760 1 /
Markov Chains Absorption (cont d) Hamid R. Rabiee
 Markov Chains Absorption (cont d) Hamid R. Rabiee 1 Absorbing Markov Chain An absorbing state is one in which the probability that the process remains in that state once it enters the state is 1 (i.e.,
Markov Chains Absorption (cont d) Hamid R. Rabiee 1 Absorbing Markov Chain An absorbing state is one in which the probability that the process remains in that state once it enters the state is 1 (i.e.,
Markov Chains and MCMC
 Markov Chains and MCMC Markov chains Let S = {1, 2,..., N} be a finite set consisting of N states. A Markov chain Y 0, Y 1, Y 2,... is a sequence of random variables, with Y t S for all points in time
Markov Chains and MCMC Markov chains Let S = {1, 2,..., N} be a finite set consisting of N states. A Markov chain Y 0, Y 1, Y 2,... is a sequence of random variables, with Y t S for all points in time
Simulation of Gene Regulatory Networks
 Simulation of Gene Regulatory Networks Overview I have been assisting Professor Jacques Cohen at Brandeis University to explore and compare the the many available representations and interpretations of
Simulation of Gene Regulatory Networks Overview I have been assisting Professor Jacques Cohen at Brandeis University to explore and compare the the many available representations and interpretations of
INTRODUCTION TO MARKOV CHAINS AND MARKOV CHAIN MIXING
 INTRODUCTION TO MARKOV CHAINS AND MARKOV CHAIN MIXING ERIC SHANG Abstract. This paper provides an introduction to Markov chains and their basic classifications and interesting properties. After establishing
INTRODUCTION TO MARKOV CHAINS AND MARKOV CHAIN MIXING ERIC SHANG Abstract. This paper provides an introduction to Markov chains and their basic classifications and interesting properties. After establishing
30 Classification of States
 30 Classification of States In a Markov chain, each state can be placed in one of the three classifications. 1 Since each state falls into one and only one category, these categories partition the states.
30 Classification of States In a Markov chain, each state can be placed in one of the three classifications. 1 Since each state falls into one and only one category, these categories partition the states.
An Application of Graph Theory in Markov Chains Reliability Analysis
 An Application of Graph Theory in Markov Chains Reliability Analysis Pavel SKALNY Department of Applied Mathematics, Faculty of Electrical Engineering and Computer Science, VSB Technical University of
An Application of Graph Theory in Markov Chains Reliability Analysis Pavel SKALNY Department of Applied Mathematics, Faculty of Electrical Engineering and Computer Science, VSB Technical University of
DESIGN OF EXPERIMENTS AND BIOCHEMICAL NETWORK INFERENCE
 DESIGN OF EXPERIMENTS AND BIOCHEMICAL NETWORK INFERENCE REINHARD LAUBENBACHER AND BRANDILYN STIGLER Abstract. Design of experiments is a branch of statistics that aims to identify efficient procedures
DESIGN OF EXPERIMENTS AND BIOCHEMICAL NETWORK INFERENCE REINHARD LAUBENBACHER AND BRANDILYN STIGLER Abstract. Design of experiments is a branch of statistics that aims to identify efficient procedures
7.32/7.81J/8.591J: Systems Biology. Fall Exam #1
 7.32/7.81J/8.591J: Systems Biology Fall 2013 Exam #1 Instructions 1) Please do not open exam until instructed to do so. 2) This exam is closed- book and closed- notes. 3) Please do all problems. 4) Use
7.32/7.81J/8.591J: Systems Biology Fall 2013 Exam #1 Instructions 1) Please do not open exam until instructed to do so. 2) This exam is closed- book and closed- notes. 3) Please do all problems. 4) Use
Chapter 2: Markov Chains and Queues in Discrete Time
 Chapter 2: Markov Chains and Queues in Discrete Time L. Breuer University of Kent 1 Definition Let X n with n N 0 denote random variables on a discrete space E. The sequence X = (X n : n N 0 ) is called
Chapter 2: Markov Chains and Queues in Discrete Time L. Breuer University of Kent 1 Definition Let X n with n N 0 denote random variables on a discrete space E. The sequence X = (X n : n N 0 ) is called
Lecture #5. Dependencies along the genome
 Markov Chains Lecture #5 Background Readings: Durbin et. al. Section 3., Polanski&Kimmel Section 2.8. Prepared by Shlomo Moran, based on Danny Geiger s and Nir Friedman s. Dependencies along the genome
Markov Chains Lecture #5 Background Readings: Durbin et. al. Section 3., Polanski&Kimmel Section 2.8. Prepared by Shlomo Moran, based on Danny Geiger s and Nir Friedman s. Dependencies along the genome
Lecture notes for Analysis of Algorithms : Markov decision processes
 Lecture notes for Analysis of Algorithms : Markov decision processes Lecturer: Thomas Dueholm Hansen June 6, 013 Abstract We give an introduction to infinite-horizon Markov decision processes (MDPs) with
Lecture notes for Analysis of Algorithms : Markov decision processes Lecturer: Thomas Dueholm Hansen June 6, 013 Abstract We give an introduction to infinite-horizon Markov decision processes (MDPs) with
At the boundary states, we take the same rules except we forbid leaving the state space, so,.
 Birth-death chains Monday, October 19, 2015 2:22 PM Example: Birth-Death Chain State space From any state we allow the following transitions: with probability (birth) with probability (death) with probability
Birth-death chains Monday, October 19, 2015 2:22 PM Example: Birth-Death Chain State space From any state we allow the following transitions: with probability (birth) with probability (death) with probability
Lectures on Markov Chains
 Lectures on Markov Chains David M. McClendon Department of Mathematics Ferris State University 2016 edition 1 Contents Contents 2 1 Markov chains 4 1.1 The definition of a Markov chain.....................
Lectures on Markov Chains David M. McClendon Department of Mathematics Ferris State University 2016 edition 1 Contents Contents 2 1 Markov chains 4 1.1 The definition of a Markov chain.....................
 http://www.math.uah.edu/stat/markov/.xhtml 1 of 9 7/16/2009 7:20 AM Virtual Laboratories > 16. Markov Chains > 1 2 3 4 5 6 7 8 9 10 11 12 1. A Markov process is a random process in which the future is
http://www.math.uah.edu/stat/markov/.xhtml 1 of 9 7/16/2009 7:20 AM Virtual Laboratories > 16. Markov Chains > 1 2 3 4 5 6 7 8 9 10 11 12 1. A Markov process is a random process in which the future is
Treball final de grau GRAU DE MATEMÀTIQUES Facultat de Matemàtiques Universitat de Barcelona MARKOV CHAINS
 Treball final de grau GRAU DE MATEMÀTIQUES Facultat de Matemàtiques Universitat de Barcelona MARKOV CHAINS Autor: Anna Areny Satorra Director: Dr. David Márquez Carreras Realitzat a: Departament de probabilitat,
Treball final de grau GRAU DE MATEMÀTIQUES Facultat de Matemàtiques Universitat de Barcelona MARKOV CHAINS Autor: Anna Areny Satorra Director: Dr. David Márquez Carreras Realitzat a: Departament de probabilitat,
Lecture 5: Random Walks and Markov Chain
 Spectral Graph Theory and Applications WS 20/202 Lecture 5: Random Walks and Markov Chain Lecturer: Thomas Sauerwald & He Sun Introduction to Markov Chains Definition 5.. A sequence of random variables
Spectral Graph Theory and Applications WS 20/202 Lecture 5: Random Walks and Markov Chain Lecturer: Thomas Sauerwald & He Sun Introduction to Markov Chains Definition 5.. A sequence of random variables
Discrete quantum random walks
 Quantum Information and Computation: Report Edin Husić edin.husic@ens-lyon.fr Discrete quantum random walks Abstract In this report, we present the ideas behind the notion of quantum random walks. We further
Quantum Information and Computation: Report Edin Husić edin.husic@ens-lyon.fr Discrete quantum random walks Abstract In this report, we present the ideas behind the notion of quantum random walks. We further
Méthode logique multivaluée de René Thomas et logique temporelle
 Méthode logique multivaluée de René Thomas et logique temporelle Gilles Bernot Universit of Nice sophia antipolis, I3S laborator, France Menu 1. Models and formal logic 2. Thomas models for gene networks
Méthode logique multivaluée de René Thomas et logique temporelle Gilles Bernot Universit of Nice sophia antipolis, I3S laborator, France Menu 1. Models and formal logic 2. Thomas models for gene networks
State-Feedback Control of Partially-Observed Boolean Dynamical Systems Using RNA-Seq Time Series Data
 State-Feedback Control of Partially-Observed Boolean Dynamical Systems Using RNA-Seq Time Series Data Mahdi Imani and Ulisses Braga-Neto Department of Electrical and Computer Engineering Texas A&M University
State-Feedback Control of Partially-Observed Boolean Dynamical Systems Using RNA-Seq Time Series Data Mahdi Imani and Ulisses Braga-Neto Department of Electrical and Computer Engineering Texas A&M University
MATH 564/STAT 555 Applied Stochastic Processes Homework 2, September 18, 2015 Due September 30, 2015
 ID NAME SCORE MATH 56/STAT 555 Applied Stochastic Processes Homework 2, September 8, 205 Due September 30, 205 The generating function of a sequence a n n 0 is defined as As : a ns n for all s 0 for which
ID NAME SCORE MATH 56/STAT 555 Applied Stochastic Processes Homework 2, September 8, 205 Due September 30, 205 The generating function of a sequence a n n 0 is defined as As : a ns n for all s 0 for which
Examples of Countable State Markov Chains Thursday, October 16, :12 PM
 stochnotes101608 Page 1 Examples of Countable State Markov Chains Thursday, October 16, 2008 12:12 PM Homework 2 solutions will be posted later today. A couple of quick examples. Queueing model (without
stochnotes101608 Page 1 Examples of Countable State Markov Chains Thursday, October 16, 2008 12:12 PM Homework 2 solutions will be posted later today. A couple of quick examples. Queueing model (without
Lecture 5. If we interpret the index n 0 as time, then a Markov chain simply requires that the future depends only on the present and not on the past.
 1 Markov chain: definition Lecture 5 Definition 1.1 Markov chain] A sequence of random variables (X n ) n 0 taking values in a measurable state space (S, S) is called a (discrete time) Markov chain, if
1 Markov chain: definition Lecture 5 Definition 1.1 Markov chain] A sequence of random variables (X n ) n 0 taking values in a measurable state space (S, S) is called a (discrete time) Markov chain, if
Automata on linear orderings
 Automata on linear orderings Véronique Bruyère Institut d Informatique Université de Mons-Hainaut Olivier Carton LIAFA Université Paris 7 September 25, 2006 Abstract We consider words indexed by linear
Automata on linear orderings Véronique Bruyère Institut d Informatique Université de Mons-Hainaut Olivier Carton LIAFA Université Paris 7 September 25, 2006 Abstract We consider words indexed by linear
Attractor computation using interconnected Boolean networks: testing growth rate models in E. Coli
 Attractor computation using interconnected Boolean networks: testing growth rate models in E. Coli Madalena Chaves, Alfonso Carta To cite this version: Madalena Chaves, Alfonso Carta. Attractor computation
Attractor computation using interconnected Boolean networks: testing growth rate models in E. Coli Madalena Chaves, Alfonso Carta To cite this version: Madalena Chaves, Alfonso Carta. Attractor computation
Chapter 7. Markov chain background. 7.1 Finite state space
 Chapter 7 Markov chain background A stochastic process is a family of random variables {X t } indexed by a varaible t which we will think of as time. Time can be discrete or continuous. We will only consider
Chapter 7 Markov chain background A stochastic process is a family of random variables {X t } indexed by a varaible t which we will think of as time. Time can be discrete or continuous. We will only consider
Google PageRank. Francesco Ricci Faculty of Computer Science Free University of Bozen-Bolzano
 Google PageRank Francesco Ricci Faculty of Computer Science Free University of Bozen-Bolzano fricci@unibz.it 1 Content p Linear Algebra p Matrices p Eigenvalues and eigenvectors p Markov chains p Google
Google PageRank Francesco Ricci Faculty of Computer Science Free University of Bozen-Bolzano fricci@unibz.it 1 Content p Linear Algebra p Matrices p Eigenvalues and eigenvectors p Markov chains p Google
Stochastic Petri Net
 Stochastic Petri Net Serge Haddad LSV ENS Cachan & CNRS & INRIA haddad@lsv.ens-cachan.fr Petri Nets 2013, June 24th 2013 1 Stochastic Petri Net 2 Markov Chain 3 Markovian Stochastic Petri Net 4 Generalized
Stochastic Petri Net Serge Haddad LSV ENS Cachan & CNRS & INRIA haddad@lsv.ens-cachan.fr Petri Nets 2013, June 24th 2013 1 Stochastic Petri Net 2 Markov Chain 3 Markovian Stochastic Petri Net 4 Generalized
A REACHABLE THROUGHPUT UPPER BOUND FOR LIVE AND SAFE FREE CHOICE NETS VIA T-INVARIANTS
 A REACHABLE THROUGHPUT UPPER BOUND FOR LIVE AND SAFE FREE CHOICE NETS VIA T-INVARIANTS Francesco Basile, Ciro Carbone, Pasquale Chiacchio Dipartimento di Ingegneria Elettrica e dell Informazione, Università
A REACHABLE THROUGHPUT UPPER BOUND FOR LIVE AND SAFE FREE CHOICE NETS VIA T-INVARIANTS Francesco Basile, Ciro Carbone, Pasquale Chiacchio Dipartimento di Ingegneria Elettrica e dell Informazione, Università
arxiv:quant-ph/ v1 15 Apr 2005
 Quantum walks on directed graphs Ashley Montanaro arxiv:quant-ph/0504116v1 15 Apr 2005 February 1, 2008 Abstract We consider the definition of quantum walks on directed graphs. Call a directed graph reversible
Quantum walks on directed graphs Ashley Montanaro arxiv:quant-ph/0504116v1 15 Apr 2005 February 1, 2008 Abstract We consider the definition of quantum walks on directed graphs. Call a directed graph reversible
Optimism in the Face of Uncertainty Should be Refutable
 Optimism in the Face of Uncertainty Should be Refutable Ronald ORTNER Montanuniversität Leoben Department Mathematik und Informationstechnolgie Franz-Josef-Strasse 18, 8700 Leoben, Austria, Phone number:
Optimism in the Face of Uncertainty Should be Refutable Ronald ORTNER Montanuniversität Leoben Department Mathematik und Informationstechnolgie Franz-Josef-Strasse 18, 8700 Leoben, Austria, Phone number:
Fixed point theorems for Boolean networks expressed in terms of forbidden subnetworks
 Fixed point theorems for Boolean networks expressed in terms of forbidden subnetworks Adrien Richard To cite this version: Adrien Richard. Fixed point theorems for Boolean networks expressed in terms of
Fixed point theorems for Boolean networks expressed in terms of forbidden subnetworks Adrien Richard To cite this version: Adrien Richard. Fixed point theorems for Boolean networks expressed in terms of
The State Explosion Problem
 The State Explosion Problem Martin Kot August 16, 2003 1 Introduction One from main approaches to checking correctness of a concurrent system are state space methods. They are suitable for automatic analysis
The State Explosion Problem Martin Kot August 16, 2003 1 Introduction One from main approaches to checking correctness of a concurrent system are state space methods. They are suitable for automatic analysis
Undirected Graphical Models
 Outline Hong Chang Institute of Computing Technology, Chinese Academy of Sciences Machine Learning Methods (Fall 2012) Outline Outline I 1 Introduction 2 Properties Properties 3 Generative vs. Conditional
Outline Hong Chang Institute of Computing Technology, Chinese Academy of Sciences Machine Learning Methods (Fall 2012) Outline Outline I 1 Introduction 2 Properties Properties 3 Generative vs. Conditional
INTRODUCTION TO MARKOV CHAIN MONTE CARLO
 INTRODUCTION TO MARKOV CHAIN MONTE CARLO 1. Introduction: MCMC In its simplest incarnation, the Monte Carlo method is nothing more than a computerbased exploitation of the Law of Large Numbers to estimate
INTRODUCTION TO MARKOV CHAIN MONTE CARLO 1. Introduction: MCMC In its simplest incarnation, the Monte Carlo method is nothing more than a computerbased exploitation of the Law of Large Numbers to estimate
Noisy Attractors and Ergodic Sets in Models. of Genetic Regulatory Networks
 Noisy Attractors and Ergodic Sets in Models of Genetic Regulatory Networks Andre S. Ribeiro Institute for Biocomplexity and Informatics, Univ. of Calgary, Canada Department of Physics and Astronomy, Univ.
Noisy Attractors and Ergodic Sets in Models of Genetic Regulatory Networks Andre S. Ribeiro Institute for Biocomplexity and Informatics, Univ. of Calgary, Canada Department of Physics and Astronomy, Univ.
Notes for Math 450 Stochastic Petri nets and reactions
 Notes for Math 450 Stochastic Petri nets and reactions Renato Feres Petri nets Petri nets are a special class of networks, introduced in 96 by Carl Adam Petri, that provide a convenient language and graphical
Notes for Math 450 Stochastic Petri nets and reactions Renato Feres Petri nets Petri nets are a special class of networks, introduced in 96 by Carl Adam Petri, that provide a convenient language and graphical
Coupled Random Boolean Network Forming an Artificial Tissue
 Coupled Random Boolean Network Forming an Artificial Tissue M. Villani, R. Serra, P.Ingrami, and S.A. Kauffman 2 DSSC, University of Modena and Reggio Emilia, via Allegri 9, I-4200 Reggio Emilia villani.marco@unimore.it,
Coupled Random Boolean Network Forming an Artificial Tissue M. Villani, R. Serra, P.Ingrami, and S.A. Kauffman 2 DSSC, University of Modena and Reggio Emilia, via Allegri 9, I-4200 Reggio Emilia villani.marco@unimore.it,
arxiv: v1 [q-bio.mn] 7 Nov 2018
![arxiv: v1 [q-bio.mn] 7 Nov 2018 arxiv: v1 [q-bio.mn] 7 Nov 2018](/thumbs/90/102608433.jpg) Role of self-loop in cell-cycle network of budding yeast Shu-ichi Kinoshita a, Hiroaki S. Yamada b a Department of Mathematical Engineering, Faculty of Engeneering, Musashino University, -- Ariake Koutou-ku,
Role of self-loop in cell-cycle network of budding yeast Shu-ichi Kinoshita a, Hiroaki S. Yamada b a Department of Mathematical Engineering, Faculty of Engeneering, Musashino University, -- Ariake Koutou-ku,
6 Markov Chain Monte Carlo (MCMC)
 6 Markov Chain Monte Carlo (MCMC) The underlying idea in MCMC is to replace the iid samples of basic MC methods, with dependent samples from an ergodic Markov chain, whose limiting (stationary) distribution
6 Markov Chain Monte Carlo (MCMC) The underlying idea in MCMC is to replace the iid samples of basic MC methods, with dependent samples from an ergodic Markov chain, whose limiting (stationary) distribution
AARMS Homework Exercises
 1 For the gamma distribution, AARMS Homework Exercises (a) Show that the mgf is M(t) = (1 βt) α for t < 1/β (b) Use the mgf to find the mean and variance of the gamma distribution 2 A well-known inequality
1 For the gamma distribution, AARMS Homework Exercises (a) Show that the mgf is M(t) = (1 βt) α for t < 1/β (b) Use the mgf to find the mean and variance of the gamma distribution 2 A well-known inequality
Automata, Logic and Games: Theory and Application
 Automata, Logic and Games: Theory and Application 1. Büchi Automata and S1S Luke Ong University of Oxford TACL Summer School University of Salerno, 14-19 June 2015 Luke Ong Büchi Automata & S1S 14-19 June
Automata, Logic and Games: Theory and Application 1. Büchi Automata and S1S Luke Ong University of Oxford TACL Summer School University of Salerno, 14-19 June 2015 Luke Ong Büchi Automata & S1S 14-19 June
Markov Chains Absorption Hamid R. Rabiee
 Markov Chains Absorption Hamid R. Rabiee Absorbing Markov Chain An absorbing state is one in which the probability that the process remains in that state once it enters the state is (i.e., p ii = ). A
Markov Chains Absorption Hamid R. Rabiee Absorbing Markov Chain An absorbing state is one in which the probability that the process remains in that state once it enters the state is (i.e., p ii = ). A
Powerful tool for sampling from complicated distributions. Many use Markov chains to model events that arise in nature.
 Markov Chains Markov chains: 2SAT: Powerful tool for sampling from complicated distributions rely only on local moves to explore state space. Many use Markov chains to model events that arise in nature.
Markov Chains Markov chains: 2SAT: Powerful tool for sampling from complicated distributions rely only on local moves to explore state space. Many use Markov chains to model events that arise in nature.
Markov chains (week 6) Solutions
 Markov chains (week 6) Solutions 1 Ranking of nodes in graphs. A Markov chain model. The stochastic process of agent visits A N is a Markov chain (MC). Explain. The stochastic process of agent visits A
Markov chains (week 6) Solutions 1 Ranking of nodes in graphs. A Markov chain model. The stochastic process of agent visits A N is a Markov chain (MC). Explain. The stochastic process of agent visits A
Coalescing Cellular Automata
 Coalescing Cellular Automata Jean-Baptiste Rouquier 1 and Michel Morvan 1,2 1 ENS Lyon, LIP, 46 allée d Italie, 69364 Lyon, France 2 EHESS and Santa Fe Institute {jean-baptiste.rouquier, michel.morvan}@ens-lyon.fr
Coalescing Cellular Automata Jean-Baptiste Rouquier 1 and Michel Morvan 1,2 1 ENS Lyon, LIP, 46 allée d Italie, 69364 Lyon, France 2 EHESS and Santa Fe Institute {jean-baptiste.rouquier, michel.morvan}@ens-lyon.fr
Transience: Whereas a finite closed communication class must be recurrent, an infinite closed communication class can be transient:
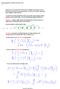 Stochastic2010 Page 1 Long-Time Properties of Countable-State Markov Chains Tuesday, March 23, 2010 2:14 PM Homework 2: if you turn it in by 5 PM on 03/25, I'll grade it by 03/26, but you can turn it in
Stochastic2010 Page 1 Long-Time Properties of Countable-State Markov Chains Tuesday, March 23, 2010 2:14 PM Homework 2: if you turn it in by 5 PM on 03/25, I'll grade it by 03/26, but you can turn it in
