NMR ANALYSIS OF PROTEIN-SMALL MOLECULE INTERACTIONS
|
|
|
- Iris Arnold
- 6 years ago
- Views:
Transcription
1 NMR ANALYSIS OF PROTEIN-SMALL MOLECULE INTERACTIONS Reported by Thomas M. Anderson April 16, 2007 INTRODUCTION Advances in NMR technology over the last 20 years 1 have made information on solution structures of proteins increasingly available. High-resolution solution structures of proteins as large as kda are tractable in most cases, 2 and recent advances allowed Kay to solve the structure of an 82 kda protein. 3 These advances in NMR analysis of protein structure have made possible the study of protein-ligand interactions by NMR. 4 From NMR data, detailed solution structures of protein-ligand complexes are accessible, and multiple methods for NMR-based analysis of protein-ligand interactions have been reported. 4 Typically, proteins are labeled with NMR-active 15 N and/or 13 C nuclei to enhance backbone NMR signals. In addition, 2 H-labels are often used to attenuate proton signals of the protein. Protein NMR spectra are analyzed in the presence and absence of ligand; based on ligand-induced changes in the NMR data, information about the location of ligand binding is obtained. The dissociation constant (K d ) for the ligand of interest is calculated directly from NMR data when the rate of ligand exchange is fast with respect to the NMR timescale. 4 For ligands with slow exchange kinetics, conventional binding assays for K d determination are more time-efficient. 1 CHEMICAL SHIFT MAPPING (CSM) CSM compares protein spectra in the presence and absence of ligand. 1,4 Ligand binding alters the local chemical environment in the binding site, resulting in chemical shift perturbations; these perturbations are correlated, or mapped, to the associated protein residues to determine the binding site. The orientation of the ligand in the binding site is determined from protein-ligand NOE correlations. A typical NMR experiment for CSM of protein binding sites is 15 N- 1 H heteronuclear single-quantum correlation ( 15 N-HSQC), 1,4-6 a two-dimensional experiment that correlates 15 N nuclei with attached protons (one-bond 15 N- 1 H coupling). Thus, amide 15 N- 1 H correlations of 15 N-labeled proteins are readily observed, and 15 N-HSQC CSM is accomplished by correlating observed chemical shift perturbations in 15 N- 1 H cross-peaks to protein residues. STRUCTURE-ACTIVITY RELATIONSHIPS (SAR) BY NMR SAR by NMR is a CSM approach to ligand optimization developed by Fesik and coworkers at Abbott Laboratories. 6 In this technique, two ligands occupying distinct, proximal sites are identified by 15 N-HSQC CSM. Optimization of the two ligands and subsequent covalent tethering of the two fragments leads to a ligand which typically exhibits improved binding over either of the first two. While this approach requires the researcher to have some structural information on the target protein, it 41
2 offers several potential advantages. First, ligands that bind to two distinct binding sites should have higher specificity than those which only target one site. Second, because the protein is 15 N-labeled, large excess of the ligand may be used, and K d values may be calculated directly from NMR data with no background signal from the ligand. 4 Thus, NMR screening can identify weak-binding ligands (K d > 1 mm), obviating lengthy synthesis of the initial library. NMR structures provide a basis for rational linker design, further reducing the amount of synthesis required for ligand optimization. Finally, targeting two separate binding sites decreases the probability of drug resistance because two sites must undergo mutation for resistance to develop. In a proof-of-concept experiment, 6 Fesik and coworkers used SAR by NMR to identify ligands optimized for binding to the FK506-binding protein (FKBP). Initially, unoptimized ligands were identified by recording 15 N-HSQC spectra of 15 N-labeled FKBP in the presence and absence of ligands. The first library screen identified pipecolic ester 1 (Figure 1, K d = 2 μm) from chemical shift perturbations in the 15 N-HSQC spectrum upon addition of 1. To identify a second, proximal binding site, 15 N-HSQC spectra of FKBP and 2.0 mm 1 were recorded in the presence of compounds from a second library. This second screen identified 2 (Figure 1), which bound to FKBP with K d = 800 μm. Proteinligand NOESY correlations established proximity of the two binding sites. SARs for 2 were investigated by variations in hydroxyl substitution patterns of the phenyl moieties, and 3 (Figure 1) showed improved binding to FKBP with K d = 100 μm. To further enhance binding of the two ligands, Fesik envisioned that covalently tethering the two ligands would create a cooperative binding effect. A series of tethered ligands of variable linker length were synthesized (Scheme 1). The optimal tether length was n = 3, resulting in a ligand with K d = 19 nm. This new ligand bound to the same site as compounds 1 and 2, as determined from protein-ligand NOESY correlations. 42
3 TARGETING ANTIAPOPTOTIC B CELL LYMPHOMA-2 (BCL-2) PROTEINS Having demonstrated the feasibility of SAR by NMR, Fesik applied the technique toward identifying inhibitors of antiapoptotic Bcl-2 proteins These proteins, which are overexpressed in several human cancer cell lines, include Bcl-2, Bcl-X L, Bcl-w, Mcl-1, and A In healthy cells, proapoptotic Bcl proteins such as Bad, Bak, Bim, and others sequester the antiapoptotic Bcl counterparts, allowing for normal cell death. Using SAR by NMR, Fesik and coworkers sought to optimize ligands for Bcl-X L. A screen of commercially available and proprietary compounds identified 4 (Figure 2), 8 which binds to Bcl-X L with K d = 300 μm. This screen revealed that the carboxyl group of 4 was crucial for effective binding, as compounds lacking the carboxyl were less active. In the presence of 4, compound 5 was found to bind to a proximal binding site (K d = 4.3 mm). NMR structures of the Bcl-X L /4/5 complex revealed two hydrophobic pockets separated by the Phe97 side chain. The binding sites for 4 and 5 correspond to the region of the protein for binding Bad and Bak. 7 For ease of reference, the binding sites for 4 and 5 will be referred to as Sites 1 and 2, respectively. To preserve the acidic Site 1 functionality and allow for fragment linking, an acylsulfonamide tether was employed (acylsulfonamides typically have pk a 3-5). To test the utility of the acysulfonamide as a carboxyl isostere, the methyl acylsulfonamide analog of 4 was synthesized via the general route shown in Scheme 2. Its activity (K d = 320 μm) is similar to that of 4. An acylsulfonamide library was 43
4 synthesized (Scheme 2), and the nitrophenyl acysulfonamide 6 was identified as the most promising hit. Because the exchange rate of 6 was slow with respect to the NMR timescale, the K d was not calculated from NMR data, and a fluorescence polarization assay was used to determine an inhibition constant (K i ) of 0.25 μm. Analysis of the NMR-derived structure of the Bcl-X L /6 complex reveals that the acylsulfonamide bridges over Phe97 as desired. Furthermore, the orientation of the nitrophenyl of 6 allows for potential π-stacking with Tyr194 and Phe97, possibly contributing to the enhanced binding. To further optimize Site 2 binding, a library of nitrophenyl acylsulfonamides was synthesized by the general route shown in Scheme 3. From 4, the corresponding 4-chloro-3-nitrophenyl acylsulfonamide was prepared via coupling with 4-chloro-3-nitrophenylsulfonamide. Nucleophilic aromatic substitution of primary and secondary amines yielded 125 compounds, of which 7 was identified with improved affinity for Bcl-X L (K i = 36 nm). From the NMR-derived structure of 7 bound to Bcl-X L, the position of the thiophenyl moiety allows for enhancement of the putative π-stacking with Phe97 and Tyr194. Thus, starting from two weak-binding ligands (4, K d = 300 μm and 3, K d = 4.3 mm), SAR by NMR was successful in identifying a nanomolar inhibitor of Bcl-X L. However, 7 had reduced affinity for Bcl-X L in the presence of 1% human serum (K i = 2.5 µm). Human serum albumin (HSA) was identified as a competitive binder for 7. REDUCING AFFINITY FOR HUMAN SERUM ALBUMIN To maintain the potential clinical relevance of 7, the researchers sought to modify the scaffold to decrease affinity for HSA while maintaining affinity for Bcl-X L. 9 The NMR-derived structure of the 44
5 HSA/7 complex showed that he biphenyl moiety interacted with HSA in a deep hydrophobic pocket that was inaccessible to solvent. 7,9 However, the analogous Site 1 of Bcl-X L was observed to be solventaccessible. Further, while the HSA binding site did not appear capable of accommodating any further steric bulk at the biphenyl, the hydrophobic Site 1 of Bcl-X L appeared to have space for extra substituents on the biphenyl. The binding mode of the 2-(phenylthio)-aminoethyl moiety was also examined. The HSA binding site was hydrophobic and solvent-inaccessible at this position, whereas Site 2 of Bcl-X L was solvent-accessible and thus would allow for functionalization at the ethyl carbons. Based on these observations, a library of functionalized biphenyl compounds was synthesized (Scheme 4) to assess the effectiveness of altering the steric environment and polarity of the biphenyl. Consistent with the observation of the deep hydrophobic pocket of HSA, charged R groups were most effective for reducing HSA affinity. Compound 10 was found to bind to Bcl-X L with K i = 0.01 μm, with K i = 0.6 μm in the presence of Bcl-X L and 1% HSA. The morpholine moiety was then incorporated in a synthesis (Scheme 5) to derivatize the α-methylene of the 2-(phenylthio)-aminoethyl group. While this introduced a chiral ligand, the R enantiomers were found to have higher Bcl-X L affinity, consistent with the position of the diastereotopic methylene protons in the Bcl-X L /7 complex. Compound 12 showed lower affinity for HSA, with K i = 0.36 μm in the presence of Bcl-X L and 10% HSA. Surprisingly, 12 inhibited Bcl-X L with K i = 8 nm, an improvement of two orders of magnitude over 10. OPTIMIZING BINDING FOR BOTH BCL-X L AND BCL-2 Given the therapeutic potential of targeting both Bcl-X L and Bcl-2, Fesik next sought to design a compound for binding to Bcl-2 without a decrease in Bcl-X L affinity. 10 Bcl-2 and Bcl-XL differ by only three residues in the binding site under consideration. The key difference, which Fesik exploited, was the presence of Met108 in Bcl-2, which had greater conformational flexibility in the Site 1 groove than the corresponding Leu108 in Bcl-X L. The conformational flexibility of Met108 allowed access to a deep 45
6 hydrophobic groove of Bcl-2 that was not observed in the structure of Bcl-X L. NMR-based structural studies of a ligand containing a Site 1 phenethyl-substituted benzothiazole docked with Bcl-2 and Bcl- X L revealed that the phenethyl moiety occupied the hydrophobic groove of Bcl-2 while maintaining a surface orientation on the face of the Bcl-X L binding site. Thus, Fesik envisioned that the Site 1 fragment could be modified to enhance Bcl-2 binding without reduction of affinity for Bcl-X L. Fesik had previously identified 13, 9 containing a Site 1 N-phenyl piperidine in place of the biphenyl, which inhibited Bcl-2 with K i = 67 nm. However, a screen of several analogous N-phenyl compounds revealed that a piperazine ring allowed for better Bcl-2 affinity while maintaining Bcl-X L affinity. Figure 3 shows four representative examples from this library. The N -benzyl piperazine 14 was chosen for further elaboration, and a library was synthesized which contained a variety of substituted benzyl groups. Compound 17, whose synthesis is shown in Scheme 6, was the most active library member with K i < 1 nm for Bcl-2 and K i < 0.5 nm for Bcl-X L. 46
7 OTHER NMR METHODS FOR ANALYZING INTERMOLECULAR INTERACTIONS While SAR by NMR represents the use of NMR as a drug discovery tool, NMR techniques have been developed and applied for studying protein-ligand interactions in other contexts. 4 Nishida has applied an NOE-based CSM technique to identify the binding site for fibrillar collagen (FC) binding to a collagen-binding protein (CBP). 11 The collagen protein contained no isotope labels, while the CBP contained 15 N- and 2 H isotope labels. Amide deuterons were exchanged in H 2 O to allow for observation of 15 N- 1 H correlations. In an experiment analogous to the standard NOESY technique, saturation of the aliphatic 1 H resonances allowed for NOE correlations to develop at the FC-CBP binding interface. After the saturation pulse, a 15 N-HSQC spectrum was obtained. 15 N-HSQC cross-peak intensities decreased for some residues (relative to the complex with no saturation pulse). Subsequent mapping of the affected residues revealed a contiguous binding domain on the CBP. Carlomagno and coworkers have developed an NMR-based method 12 for analysis of competitive binding of two weak-binding ligands. Epothilones and taxanes competitively bind to microtubules, but binding of these compounds with free tubulin has not been characterized due to the weak nature of these interactions. A NOESY spectrum of a mixture of the taxane baccatin III (18, Figure 4), epothilone A (19), and tubulin gives rise to cross-peaks between epothilone A and baccatin III which are not observed in the absence of tubulin. 47
8 CONCLUSION Advances in NMR spectroscopy have provided powerful tools for both structural analysis and structure-activity studies. While SAR by NMR is a relatively new technique, it can be utilized to discover potent protein ligands and potential drug leads. Through the use of SAR by NMR, a new drug lead targeting antiapoptotic Bcl proteins was developed. At the time of publication, 7 this ligand was 2-3 orders of magnitude more active than any previously reported Bcl-X L or Bcl-2 inhibitor. While this discovery illustrates the potential of CSM for identifying and optimizing protein-ligand binding, the NMR techniques discussed herein are not limited to study of protein-ligand interactions and could be extended to study other types of intermolecular interactions. REFERENCES 1) Cavanagh, J.; Fairbrother, W. J.; Palmer, W. G. III; Rance, M.; Skelton, N. J. Protein NMR Spectroscopy: Principles and Practices, 2 nd Ed. Academic Press, San Diego, ) Kay, L. E.; Gardner, K. H. Curr. Opin. Struct. Biol. 1997, 7, ) Tugarinov, V. ; Choy, W.-Y.; Orekhov, V. Y.; Kay, L. E. Proc. Natl. Acad. Sci. USA 2005, 102, ) Pellechia, M. Chem. Biol. 2005, 12, ) Görlach, M.; Wittekind, M.; Beckman, R. A.; Mueller, L.; Dreyfuss, G. EMBO J. 1992, 11, ) Shuker, S. B.; Hajduk, P. J.; Meadows, R. P.; Fesik, S. W. Science 1996, 274, ) Oltersdorf, T.; Elmore, S. W.; Shoemaker, A. R.; Armstrong, R. C.; Augeri, D. J.; Belli, B. A.; Bruncko, M.; Deckwerth, T. L.; Dinges, J.; Hajduk, P. J.; Joseph, M. K.; Kitada, S.; Korsmeyer, S. J.; Kunzer, A. R.; Letai, A.; Li, C.; Mitten, M. J.; Nettesheim, D. G.; Ng, S.-C.; Nimmer, P. M.; O Connor, J. M.; Oleksijew, A.; Petros, A. M.; Reed, J. C.; Shen, W.; Tahir, S. K.; Thompson, C. B.; Tomaselli, K. J.; Wang, B.; Wendt, M. D.; Zhang, H.; Fesik, S. W.; Rosenberg, S. H. Nature 2005, 435, ) Petros, A. M.; Dinges, J.; Augeri, D. J.; Baumeister, S. A.; Betebenner, D. A.; Bures, M. G.; Elmore, S. W.; Hajduk, P. J.; Joseph, M. K.; Landis, S. K.; Nettesheim, D. G.; Rosenberg, S. H.; Shen, W.; Thomas, S.; Wang, X.; Zanze, I.; Zhang, H.; Fesik, S. W. J. Med. Chem. 2006, 49, ) Wendt, M. D.; Shen, W.; Kunzer, A.; McClellan, W. J.; Bruncko, M.; Oost, T. K.; Ding, H.; Joseph, M. K.; Zhang, H.; Nimmer, P. M.; Ng, S.-C.; Shoemaker, A. R.; Petros, A. M.; Oleksijew, A.; Marsh, K.; Bauch, J.; Oltersdorf, T.; Belli, B. A.; Martineau, D.; Fesik, S. W.; Rosenberg, S. H.; Elmore, S. W. J. Med. Chem. 2006, 49, ) Bruncko, M.; Oost, T. K.; Belli, B. A.; Ding, H.; Joseph, M. K.; Kunzer, A.; Martineau, D.; McClellan, W. J.; Mitten, M.; Ng, S.-C.; Nimmer, P. M.; Oltersdorf, T.; Park, C.-M.; Petros, A. M.; Shoemaker, A. R.; Song, X.; Wang, X.; Wendt, M. D.; Zhang, H.; Fesik, S. W.; Rosenberg, S. H. ; Elmore, S. W. J. Med. Chem. 2007, 50, ) Nishida, N.; Sumikawa, H.; Sakakura, M.; Shimba, N.; Takahashi, H.; Terasawa, H.; Suzuki, E. I.; Shimada, I. Nat. Struct. Biol. 2003, 10, ) Sánchez-Pedregal, V. M. Reese, M. Meiler, J. Blommers, M. J. J. Griesinger, C. Carlomagno, T. Angew. Chem. Int. Ed. 2005, 44,
Protein Structure Determination using NMR Spectroscopy. Cesar Trinidad
 Protein Structure Determination using NMR Spectroscopy Cesar Trinidad Introduction Protein NMR Involves the analysis and calculation of data collected from multiple NMR techniques Utilizes Nuclear Magnetic
Protein Structure Determination using NMR Spectroscopy Cesar Trinidad Introduction Protein NMR Involves the analysis and calculation of data collected from multiple NMR techniques Utilizes Nuclear Magnetic
NMR study of complexes between low molecular mass inhibitors and the West Nile virus NS2B-NS3 protease
 University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2009 NMR study of complexes between low molecular mass inhibitors and the West Nile
University of Wollongong Research Online Faculty of Science - Papers (Archive) Faculty of Science, Medicine and Health 2009 NMR study of complexes between low molecular mass inhibitors and the West Nile
Supporting Information
 S-1 Supporting Information Flaviviral protease inhibitors identied by fragment-based library docking into a structure generated by molecular dynamics Dariusz Ekonomiuk a, Xun-Cheng Su b, Kiyoshi Ozawa
S-1 Supporting Information Flaviviral protease inhibitors identied by fragment-based library docking into a structure generated by molecular dynamics Dariusz Ekonomiuk a, Xun-Cheng Su b, Kiyoshi Ozawa
Supplementary Information. Overlap between folding and functional energy landscapes for. adenylate kinase conformational change
 Supplementary Information Overlap between folding and functional energy landscapes for adenylate kinase conformational change by Ulrika Olsson & Magnus Wolf-Watz Contents: 1. Supplementary Note 2. Supplementary
Supplementary Information Overlap between folding and functional energy landscapes for adenylate kinase conformational change by Ulrika Olsson & Magnus Wolf-Watz Contents: 1. Supplementary Note 2. Supplementary
Chemistry 843 "Advanced NMR Spectroscopy" Gerd Gemmecker, 1999
 Dr. Gerd Gemmecker Room 5333a Phone 2-8619 e-mail gemmecker@chem.wisc.edu Gerd.Gemmecker@ch.tum.de Chemistry 843 "Advanced NMR Spectroscopy" Gerd Gemmecker, 1999 This course will cover the theory required
Dr. Gerd Gemmecker Room 5333a Phone 2-8619 e-mail gemmecker@chem.wisc.edu Gerd.Gemmecker@ch.tum.de Chemistry 843 "Advanced NMR Spectroscopy" Gerd Gemmecker, 1999 This course will cover the theory required
Introduction to FBDD Fragment screening methods and library design
 Introduction to FBDD Fragment screening methods and library design Samantha Hughes, PhD Fragments 2013 RSC BMCS Workshop 3 rd March 2013 Copyright 2013 Galapagos NV Why fragment screening methods? Guess
Introduction to FBDD Fragment screening methods and library design Samantha Hughes, PhD Fragments 2013 RSC BMCS Workshop 3 rd March 2013 Copyright 2013 Galapagos NV Why fragment screening methods? Guess
COMBINATORIAL CHEMISTRY: CURRENT APPROACH
 COMBINATORIAL CHEMISTRY: CURRENT APPROACH Dwivedi A. 1, Sitoke A. 2, Joshi V. 3, Akhtar A.K. 4* and Chaturvedi M. 1, NRI Institute of Pharmaceutical Sciences, Bhopal, M.P.-India 2, SRM College of Pharmacy,
COMBINATORIAL CHEMISTRY: CURRENT APPROACH Dwivedi A. 1, Sitoke A. 2, Joshi V. 3, Akhtar A.K. 4* and Chaturvedi M. 1, NRI Institute of Pharmaceutical Sciences, Bhopal, M.P.-India 2, SRM College of Pharmacy,
NMR in Medicine and Biology
 NMR in Medicine and Biology http://en.wikipedia.org/wiki/nmr_spectroscopy MRI- Magnetic Resonance Imaging (water) In-vivo spectroscopy (metabolites) Solid-state t NMR (large structures) t Solution NMR
NMR in Medicine and Biology http://en.wikipedia.org/wiki/nmr_spectroscopy MRI- Magnetic Resonance Imaging (water) In-vivo spectroscopy (metabolites) Solid-state t NMR (large structures) t Solution NMR
Supporting Information. Labeled Ligand Displacement: Extending NMR-based Screening of Protein Targets
 Supporting Information Labeled Ligand Displacement: Extending NMR-based Screening of Protein Targets Steven L. Swann, Danying Song, Chaohong Sun, Philip J. Hajduk, and Andrew M. Petros Global Pharmaceutical
Supporting Information Labeled Ligand Displacement: Extending NMR-based Screening of Protein Targets Steven L. Swann, Danying Song, Chaohong Sun, Philip J. Hajduk, and Andrew M. Petros Global Pharmaceutical
Fragment based drug discovery in teams of medicinal and computational chemists. Carsten Detering
 Fragment based drug discovery in teams of medicinal and computational chemists Carsten Detering BioSolveIT Quick Facts Founded in 2001 by the developers of FlexX ~20 people Core expertise: docking, screening,
Fragment based drug discovery in teams of medicinal and computational chemists Carsten Detering BioSolveIT Quick Facts Founded in 2001 by the developers of FlexX ~20 people Core expertise: docking, screening,
György M. Keserű H2020 FRAGNET Network Hungarian Academy of Sciences
 Fragment based lead discovery - introduction György M. Keserű H2020 FRAGET etwork Hungarian Academy of Sciences www.fragnet.eu Hit discovery from screening Druglike library Fragment library Large molecules
Fragment based lead discovery - introduction György M. Keserű H2020 FRAGET etwork Hungarian Academy of Sciences www.fragnet.eu Hit discovery from screening Druglike library Fragment library Large molecules
Interpreting and evaluating biological NMR in the literature. Worksheet 1
 Interpreting and evaluating biological NMR in the literature Worksheet 1 1D NMR spectra Application of RF pulses of specified lengths and frequencies can make certain nuclei detectable We can selectively
Interpreting and evaluating biological NMR in the literature Worksheet 1 1D NMR spectra Application of RF pulses of specified lengths and frequencies can make certain nuclei detectable We can selectively
Using cryoprobes to decrease acquisition times of triple-resonance experiments used for protein resonance assignments
 Spectroscopy 17 (2003) 161 167 161 IOS Press Using cryoprobes to decrease acquisition times of triple-resonance experiments used for protein resonance assignments Michael J. Goger a,, James M. McDonnell
Spectroscopy 17 (2003) 161 167 161 IOS Press Using cryoprobes to decrease acquisition times of triple-resonance experiments used for protein resonance assignments Michael J. Goger a,, James M. McDonnell
UNIT 4 REVISION CHECKLIST CHEM 4 AS Chemistry
 UNIT 4 REVISION CHECKLIST CHEM 4 AS Chemistry Topic 4.1 Kinetics a) Define the terms: rate of a reaction, rate constant, order of reaction and overall order of reaction b) Deduce the orders of reaction
UNIT 4 REVISION CHECKLIST CHEM 4 AS Chemistry Topic 4.1 Kinetics a) Define the terms: rate of a reaction, rate constant, order of reaction and overall order of reaction b) Deduce the orders of reaction
BMB/Bi/Ch 173 Winter 2018
 BMB/Bi/Ch 173 Winter 2018 Homework Set 8.1 (100 Points) Assigned 2-27-18, due 3-6-18 by 10:30 a.m. TA: Rachael Kuintzle. Office hours: SFL 220, Friday 3/2 4-5pm and SFL 229, Monday 3/5 4-5:30pm. 1. NMR
BMB/Bi/Ch 173 Winter 2018 Homework Set 8.1 (100 Points) Assigned 2-27-18, due 3-6-18 by 10:30 a.m. TA: Rachael Kuintzle. Office hours: SFL 220, Friday 3/2 4-5pm and SFL 229, Monday 3/5 4-5:30pm. 1. NMR
NMR Spectroscopy. Guangjin Hou
 NMR Spectroscopy Guangjin Hou 22-04-2009 NMR History 1 H NMR spectra of water H NMR spectra of water (First NMR Spectra on Water, 1946) 1 H NMR spectra ethanol (First bservation of the Chemical Shift,
NMR Spectroscopy Guangjin Hou 22-04-2009 NMR History 1 H NMR spectra of water H NMR spectra of water (First NMR Spectra on Water, 1946) 1 H NMR spectra ethanol (First bservation of the Chemical Shift,
BMB/Bi/Ch 173 Winter 2018
 BMB/Bi/Ch 173 Winter 2018 Homework Set 8.1 (100 Points) Assigned 2-27-18, due 3-6-18 by 10:30 a.m. TA: Rachael Kuintzle. Office hours: SFL 220, Friday 3/2 4:00-5:00pm and SFL 229, Monday 3/5 4:00-5:30pm.
BMB/Bi/Ch 173 Winter 2018 Homework Set 8.1 (100 Points) Assigned 2-27-18, due 3-6-18 by 10:30 a.m. TA: Rachael Kuintzle. Office hours: SFL 220, Friday 3/2 4:00-5:00pm and SFL 229, Monday 3/5 4:00-5:30pm.
NMR Solutions for drug discovery
 NMR Solutions for drug discovery Dr. Matteo Pennestri London, UK Bruker Users Meeting Innovation with Integrity The principle of Fragment Based Screening from efficient fragments to Drug candidates Fragment
NMR Solutions for drug discovery Dr. Matteo Pennestri London, UK Bruker Users Meeting Innovation with Integrity The principle of Fragment Based Screening from efficient fragments to Drug candidates Fragment
COPYRIGHTED MATERIAL INTRODUCTION
 1 INTRODUCTION Nuclear magnetic resonance spectroscopy is one of the most common methods used to determine optical purity and assign the absolute configuration of chiral compounds. The strategy that has
1 INTRODUCTION Nuclear magnetic resonance spectroscopy is one of the most common methods used to determine optical purity and assign the absolute configuration of chiral compounds. The strategy that has
- Basic understandings: - Mapping interactions:
 NMR-lecture April 6th, 2009, FMP Berlin Outline: Christian Freund - Basic understandings: Relaxation Chemical exchange - Mapping interactions: -Chemical shift mapping (fast exchange) Linewidth analysis
NMR-lecture April 6th, 2009, FMP Berlin Outline: Christian Freund - Basic understandings: Relaxation Chemical exchange - Mapping interactions: -Chemical shift mapping (fast exchange) Linewidth analysis
Sensitive NMR Approach for Determining the Binding Mode of Tightly Binding Ligand Molecules to Protein Targets
 Supporting information Sensitive NMR Approach for Determining the Binding Mode of Tightly Binding Ligand Molecules to Protein Targets Wan-Na Chen, Christoph Nitsche, Kala Bharath Pilla, Bim Graham, Thomas
Supporting information Sensitive NMR Approach for Determining the Binding Mode of Tightly Binding Ligand Molecules to Protein Targets Wan-Na Chen, Christoph Nitsche, Kala Bharath Pilla, Bim Graham, Thomas
Basic principles of multidimensional NMR in solution
 Basic principles of multidimensional NMR in solution 19.03.2008 The program 2/93 General aspects Basic principles Parameters in NMR spectroscopy Multidimensional NMR-spectroscopy Protein structures NMR-spectra
Basic principles of multidimensional NMR in solution 19.03.2008 The program 2/93 General aspects Basic principles Parameters in NMR spectroscopy Multidimensional NMR-spectroscopy Protein structures NMR-spectra
Protein dynamics from NMR Relaxation data
 Protein dynamics from NMR Relaxation data Clubb 3/15/17 (S f2 ) ( e ) Nitrogen-15 relaxation ZZ-exchange R 1 = 1/T 1 Longitudinal relaxation (decay back to z-axis) R 2 = 1/T 2 Spin-spin relaxation (dephasing
Protein dynamics from NMR Relaxation data Clubb 3/15/17 (S f2 ) ( e ) Nitrogen-15 relaxation ZZ-exchange R 1 = 1/T 1 Longitudinal relaxation (decay back to z-axis) R 2 = 1/T 2 Spin-spin relaxation (dephasing
MM-GBSA for Calculating Binding Affinity A rank-ordering study for the lead optimization of Fxa and COX-2 inhibitors
 MM-GBSA for Calculating Binding Affinity A rank-ordering study for the lead optimization of Fxa and COX-2 inhibitors Thomas Steinbrecher Senior Application Scientist Typical Docking Workflow Databases
MM-GBSA for Calculating Binding Affinity A rank-ordering study for the lead optimization of Fxa and COX-2 inhibitors Thomas Steinbrecher Senior Application Scientist Typical Docking Workflow Databases
SUPPLEMENTARY INFORMATION
 Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
Figure S1. Secondary structure of CAP (in the camp 2 -bound state) 10. α-helices are shown as cylinders and β- strands as arrows. Labeling of secondary structure is indicated. CDB, DBD and the hinge are
NMR Characterization of Partially Folded and Unfolded Conformational Ensembles of Proteins
 Elisar Barbar Department of Chemistry and Biochemistry, Ohio University, Athens, OH 45701 NMR Characterization of Partially Folded and Unfolded Conformational Ensembles of Proteins Abstract: Studies of
Elisar Barbar Department of Chemistry and Biochemistry, Ohio University, Athens, OH 45701 NMR Characterization of Partially Folded and Unfolded Conformational Ensembles of Proteins Abstract: Studies of
Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination
 Lecture 9 M230 Feigon Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination Reading resources v Roberts NMR of Macromolecules, Chap 4 by Christina Redfield
Lecture 9 M230 Feigon Sequential resonance assignments in (small) proteins: homonuclear method 2º structure determination Reading resources v Roberts NMR of Macromolecules, Chap 4 by Christina Redfield
Receptor Based Drug Design (1)
 Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
Induced Fit Model For more than 100 years, the behaviour of enzymes had been explained by the "lock-and-key" mechanism developed by pioneering German chemist Emil Fischer. Fischer thought that the chemicals
17. Biomolecular Interaction
 17. Biomolecular Interaction Methods for characterizing biomolecular interactions Sequence-specific DNA binding ligands Molecular mechanisms of drug action and drug resistance In silico compound design
17. Biomolecular Interaction Methods for characterizing biomolecular interactions Sequence-specific DNA binding ligands Molecular mechanisms of drug action and drug resistance In silico compound design
Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine. JAK2 Selective Mechanism Combined Molecular Dynamics Simulation
 Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supporting Information Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine
Electronic Supplementary Material (ESI) for Molecular BioSystems. This journal is The Royal Society of Chemistry 2015 Supporting Information Enhancing Specificity in the Janus Kinases: A Study on the Thienopyridine
OXFORD H i g h e r E d u c a t i o n Oxford University Press, All rights reserved.
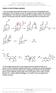 Patrick, An Introduction to dicinal hemistry 4e Answers to end-of-chapter questions 1) The mechanism below shows the release of one molecule of formaldehyde from methenamine. The mechanism can then be
Patrick, An Introduction to dicinal hemistry 4e Answers to end-of-chapter questions 1) The mechanism below shows the release of one molecule of formaldehyde from methenamine. The mechanism can then be
NMR-Based Approaches for the Identification and Optimization of Inhibitors of Protein Protein Interactions
 This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/cr Downloaded
This is an open access article published under an ACS AuthorChoice License, which permits copying and redistribution of the article or any adaptations for non-commercial purposes. pubs.acs.org/cr Downloaded
Use of deuterium labeling in NMR: overcoming a sizeable problem Michael Sattler and Stephen W Fesik*
 Ways & Means 1245 Use of deuterium labeling in NMR: overcoming a sizeable problem Michael Sattler and Stephen W Fesik* Address: Abbott Laboratories, 47G AP10,100, Abbott Park Road, Abbott Park, IL 60064-3500,
Ways & Means 1245 Use of deuterium labeling in NMR: overcoming a sizeable problem Michael Sattler and Stephen W Fesik* Address: Abbott Laboratories, 47G AP10,100, Abbott Park Road, Abbott Park, IL 60064-3500,
COMPUTER AIDED DRUG DESIGN (CADD) AND DEVELOPMENT METHODS
 COMPUTER AIDED DRUG DESIGN (CADD) AND DEVELOPMENT METHODS DRUG DEVELOPMENT Drug development is a challenging path Today, the causes of many diseases (rheumatoid arthritis, cancer, mental diseases, etc.)
COMPUTER AIDED DRUG DESIGN (CADD) AND DEVELOPMENT METHODS DRUG DEVELOPMENT Drug development is a challenging path Today, the causes of many diseases (rheumatoid arthritis, cancer, mental diseases, etc.)
Enamine Golden Fragment Library
 Enamine Golden Fragment Library 14 March 216 1794 compounds deliverable as entire set or as selected items. Fragment Based Drug Discovery (FBDD) [1,2] demonstrates remarkable results: more than 3 compounds
Enamine Golden Fragment Library 14 March 216 1794 compounds deliverable as entire set or as selected items. Fragment Based Drug Discovery (FBDD) [1,2] demonstrates remarkable results: more than 3 compounds
NMR-spectroscopy in solution - an introduction. Peter Schmieder
 NMR-spectroscopy in solution - an introduction 2/92 Advanced Bioanalytics NMR-Spectroscopy Introductory session (11:00 12:30) Basic aspects of NMR-spectroscopy NMR parameter Multidimensional NMR-spectroscopy
NMR-spectroscopy in solution - an introduction 2/92 Advanced Bioanalytics NMR-Spectroscopy Introductory session (11:00 12:30) Basic aspects of NMR-spectroscopy NMR parameter Multidimensional NMR-spectroscopy
NMR in Structural Biology
 NMR in Structural Biology Exercise session 2 1. a. List 3 NMR observables that report on structure. b. Also indicate whether the information they give is short/medium or long-range, or perhaps all three?
NMR in Structural Biology Exercise session 2 1. a. List 3 NMR observables that report on structure. b. Also indicate whether the information they give is short/medium or long-range, or perhaps all three?
Chemical Exchange and Ligand Binding
 Chemical Exchange and Ligand Binding NMR time scale Fast exchange for binding constants Slow exchange for tight binding Single vs. multiple binding mode Calcium binding process of calcium binding proteins
Chemical Exchange and Ligand Binding NMR time scale Fast exchange for binding constants Slow exchange for tight binding Single vs. multiple binding mode Calcium binding process of calcium binding proteins
Supporting Information. Synthesis and NMR Elucidation of Novel Octa-Amino Acid Resorcin[4]arenes derivatives. Glenn E. M.
![Supporting Information. Synthesis and NMR Elucidation of Novel Octa-Amino Acid Resorcin[4]arenes derivatives. Glenn E. M. Supporting Information. Synthesis and NMR Elucidation of Novel Octa-Amino Acid Resorcin[4]arenes derivatives. Glenn E. M.](/thumbs/89/97638653.jpg) Supporting Information Synthesis and NMR Elucidation of Novel cta-amino Acid Resorcin[4]arenes derivatives. Iman Elidrisi, [a] Pralav V. Bhatt, [b] Thavendran Govender, [b] Hendrik G. Kruger, [b] and Glenn
Supporting Information Synthesis and NMR Elucidation of Novel cta-amino Acid Resorcin[4]arenes derivatives. Iman Elidrisi, [a] Pralav V. Bhatt, [b] Thavendran Govender, [b] Hendrik G. Kruger, [b] and Glenn
Technical Note. Introduction
 Technical Note Characterization of Eleven 2,5-Dimethoxy-N-(2-methoxybenzyl)- phenethylamine (NBOMe) Derivatives and Differentiation from their 3- and 4- Methoxybenzyl Analogues - Part II Patrick A. Hays*,
Technical Note Characterization of Eleven 2,5-Dimethoxy-N-(2-methoxybenzyl)- phenethylamine (NBOMe) Derivatives and Differentiation from their 3- and 4- Methoxybenzyl Analogues - Part II Patrick A. Hays*,
Solving the three-dimensional solution structures of larger
 Accurate and rapid docking of protein protein complexes on the basis of intermolecular nuclear Overhauser enhancement data and dipolar couplings by rigid body minimization G. Marius Clore* Laboratory of
Accurate and rapid docking of protein protein complexes on the basis of intermolecular nuclear Overhauser enhancement data and dipolar couplings by rigid body minimization G. Marius Clore* Laboratory of
Inverse Detection in Multinuclear NMR
 Inverse Detection in Multinuclear NMR The HETCOR experiment is an example of a directly-detected heteronuclear experiment. The timing diagram for the most basic form of the HETCOR pulse sequence is shown
Inverse Detection in Multinuclear NMR The HETCOR experiment is an example of a directly-detected heteronuclear experiment. The timing diagram for the most basic form of the HETCOR pulse sequence is shown
Supporting Information
 Supporting Information Boehr et al. 10.1073/pnas.0914163107 SI Text Materials and Methods. R 2 relaxation dispersion experiments. 15 NR 2 relaxation dispersion data measured at 1 H Larmor frequencies of
Supporting Information Boehr et al. 10.1073/pnas.0914163107 SI Text Materials and Methods. R 2 relaxation dispersion experiments. 15 NR 2 relaxation dispersion data measured at 1 H Larmor frequencies of
Protein-protein interactions by NMR
 Protein-protein interactions by NMR Fast k on,off >> (ν free - ν bound ) A + B k on k off AB k on,off ~ (ν free - ν bound ) Slow k on,off
Protein-protein interactions by NMR Fast k on,off >> (ν free - ν bound ) A + B k on k off AB k on,off ~ (ν free - ν bound ) Slow k on,off
Structural basis for catalytically restrictive dynamics of a high-energy enzyme state
 Supplementary Material Structural basis for catalytically restrictive dynamics of a high-energy enzyme state Michael Kovermann, Jörgen Ådén, Christin Grundström, A. Elisabeth Sauer-Eriksson, Uwe H. Sauer
Supplementary Material Structural basis for catalytically restrictive dynamics of a high-energy enzyme state Michael Kovermann, Jörgen Ådén, Christin Grundström, A. Elisabeth Sauer-Eriksson, Uwe H. Sauer
Electronic Structure. Hammett Correlations. Linear Free Energy Relationships. Quantitative Structure Activity Relationships (QSAR)
 Electronic Structure ammett Correlations Quantitative Structure Activity Relationships (QSAR) Quantitative Structure Activity Relationships (QSAR) We have already seen that by changing groups in a drug
Electronic Structure ammett Correlations Quantitative Structure Activity Relationships (QSAR) Quantitative Structure Activity Relationships (QSAR) We have already seen that by changing groups in a drug
Targeting protein-protein interactions: A hot topic in drug discovery
 Michal Kamenicky; Maria Bräuer; Katrin Volk; Kamil Ödner; Christian Klein; Norbert Müller Targeting protein-protein interactions: A hot topic in drug discovery 104 Biomedizin Innovativ patientinnenfokussierte,
Michal Kamenicky; Maria Bräuer; Katrin Volk; Kamil Ödner; Christian Klein; Norbert Müller Targeting protein-protein interactions: A hot topic in drug discovery 104 Biomedizin Innovativ patientinnenfokussierte,
HTS by NMR of Combinatorial Libraries: A Fragment-Based Approach to Ligand Discovery
 Article HTS by NMR of Combinatorial Libraries: A Fragment-Based Approach to Ligand Discovery Bainan Wu, 1 Ziming Zhang, 1 Roberta Noberini, 1 Elisa Barile, 1 Marc Giulianotti, 2 Clemencia Pinilla, 3 Richard
Article HTS by NMR of Combinatorial Libraries: A Fragment-Based Approach to Ligand Discovery Bainan Wu, 1 Ziming Zhang, 1 Roberta Noberini, 1 Elisa Barile, 1 Marc Giulianotti, 2 Clemencia Pinilla, 3 Richard
Medicinal Chemistry/ CHEM 458/658 Chapter 3- SAR and QSAR
 Medicinal Chemistry/ CHEM 458/658 Chapter 3- SAR and QSAR Bela Torok Department of Chemistry University of Massachusetts Boston Boston, MA 1 Introduction Structure-Activity Relationship (SAR) - similar
Medicinal Chemistry/ CHEM 458/658 Chapter 3- SAR and QSAR Bela Torok Department of Chemistry University of Massachusetts Boston Boston, MA 1 Introduction Structure-Activity Relationship (SAR) - similar
FACULTY OF PHARMACY. M. Pharmacy I Semester (Suppl.) Examination, November 2015 (Common To All) Subject: Pharmaceutical Analytical Techniques
 M. Pharmacy I Semester (Suppl.) Examination, November 2015 (Common To All) Subject: Pharmaceutical Analytical Techniques Code No. 6001 / S 1 a) Describe the instrumentation and applications of UV-visible
M. Pharmacy I Semester (Suppl.) Examination, November 2015 (Common To All) Subject: Pharmaceutical Analytical Techniques Code No. 6001 / S 1 a) Describe the instrumentation and applications of UV-visible
Relaxation, Multi pulse Experiments and 2D NMR
 Relaxation, Multi pulse Experiments and 2D NMR To Do s Read Chapter 6 Complete the end of chapter problems; 6 1, 6 2, 6 3, 6 5, 6 9 and 6 10. Read Chapter 15 and do as many problems as you can. Relaxation
Relaxation, Multi pulse Experiments and 2D NMR To Do s Read Chapter 6 Complete the end of chapter problems; 6 1, 6 2, 6 3, 6 5, 6 9 and 6 10. Read Chapter 15 and do as many problems as you can. Relaxation
Solutions In each case, the chirality center has the R configuration
 CAPTER 25 669 Solutions 25.1. In each case, the chirality center has the R configuration. C C 2 2 C 3 C(C 3 ) 2 D-Alanine D-Valine 25.2. 2 2 S 2 d) 2 25.3. Pro,, Trp, Tyr, and is, Trp, Tyr, and is Arg,
CAPTER 25 669 Solutions 25.1. In each case, the chirality center has the R configuration. C C 2 2 C 3 C(C 3 ) 2 D-Alanine D-Valine 25.2. 2 2 S 2 d) 2 25.3. Pro,, Trp, Tyr, and is, Trp, Tyr, and is Arg,
FRAGMENT SCREENING IN LEAD DISCOVERY BY WEAK AFFINITY CHROMATOGRAPHY (WAC )
 FRAGMENT SCREENING IN LEAD DISCOVERY BY WEAK AFFINITY CHROMATOGRAPHY (WAC ) SARomics Biostructures AB & Red Glead Discovery AB Medicon Village, Lund, Sweden Fragment-based lead discovery The basic idea:
FRAGMENT SCREENING IN LEAD DISCOVERY BY WEAK AFFINITY CHROMATOGRAPHY (WAC ) SARomics Biostructures AB & Red Glead Discovery AB Medicon Village, Lund, Sweden Fragment-based lead discovery The basic idea:
CHEM 261 HOME WORK Lecture Topics: MODULE 1: The Basics: Bonding and Molecular Structure Text Sections (N0 1.9, 9-11) Homework: Chapter 1:
 CHEM 261 HOME WORK Lecture Topics: MODULE 1: The Basics: Bonding and Molecular Structure Atomic Structure - Valence Electrons Chemical Bonds: The Octet Rule - Ionic bond - Covalent bond How to write Lewis
CHEM 261 HOME WORK Lecture Topics: MODULE 1: The Basics: Bonding and Molecular Structure Atomic Structure - Valence Electrons Chemical Bonds: The Octet Rule - Ionic bond - Covalent bond How to write Lewis
Protein-protein interactions (PPIs) via NMR. Paola Turano
 Protein-protein interactions (PPIs) via NMR Paola Turano turano@cerm.unifi.it The magnetic field at the The chemical shift nucleus (the effective field) is generally less than the applied field by a fraction
Protein-protein interactions (PPIs) via NMR Paola Turano turano@cerm.unifi.it The magnetic field at the The chemical shift nucleus (the effective field) is generally less than the applied field by a fraction
Identifying Interaction Hot Spots with SuperStar
 Identifying Interaction Hot Spots with SuperStar Version 1.0 November 2017 Table of Contents Identifying Interaction Hot Spots with SuperStar... 2 Case Study... 3 Introduction... 3 Generate SuperStar Maps
Identifying Interaction Hot Spots with SuperStar Version 1.0 November 2017 Table of Contents Identifying Interaction Hot Spots with SuperStar... 2 Case Study... 3 Introduction... 3 Generate SuperStar Maps
Module 20: Applications of PMR in Structural Elucidation of Simple and Complex Compounds and 2-D NMR spectroscopy
 Subject Chemistry Paper No and Title Module No and Title Module Tag Paper 12: Organic Spectroscopy Module 20: Applications of PMR in Structural Elucidation of Simple and Complex Compounds and 2-D NMR spectroscopy
Subject Chemistry Paper No and Title Module No and Title Module Tag Paper 12: Organic Spectroscopy Module 20: Applications of PMR in Structural Elucidation of Simple and Complex Compounds and 2-D NMR spectroscopy
Fluorine in Peptide and Protein Engineering
 Fluorine in Peptide and Protein Engineering Rita Fernandes Porto, February 11 th 2016 Supervisor: Prof. Dr. Beate Koksch 1 Fluorine a unique element for molecule design The most abundant halogen in earth
Fluorine in Peptide and Protein Engineering Rita Fernandes Porto, February 11 th 2016 Supervisor: Prof. Dr. Beate Koksch 1 Fluorine a unique element for molecule design The most abundant halogen in earth
Protein-protein interactions and NMR: G protein/effector complexes
 Protein-protein interactions and NMR: G protein/effector complexes Helen Mott 5th CCPN Annual Conference, August 2005 Department of Biochemistry University of Cambridge Fast k on,off >> (ν free - ν bound
Protein-protein interactions and NMR: G protein/effector complexes Helen Mott 5th CCPN Annual Conference, August 2005 Department of Biochemistry University of Cambridge Fast k on,off >> (ν free - ν bound
Protein-protein interactions (PPIs) via NMR. Paola Turano
 Protein-protein interactions (PPIs) via NMR Paola Turano turano@cerm.unifi.it The magnetic field at the The chemical shift nucleus (the effective field) is generally less than the applied field by a fraction
Protein-protein interactions (PPIs) via NMR Paola Turano turano@cerm.unifi.it The magnetic field at the The chemical shift nucleus (the effective field) is generally less than the applied field by a fraction
Structural Bioinformatics (C3210) Molecular Docking
 Structural Bioinformatics (C3210) Molecular Docking Molecular Recognition, Molecular Docking Molecular recognition is the ability of biomolecules to recognize other biomolecules and selectively interact
Structural Bioinformatics (C3210) Molecular Docking Molecular Recognition, Molecular Docking Molecular recognition is the ability of biomolecules to recognize other biomolecules and selectively interact
Ping-Chiang Lyu. Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University.
 Pharmacophore-based Drug design Ping-Chiang Lyu Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University 96/08/07 Outline Part I: Analysis The analytical
Pharmacophore-based Drug design Ping-Chiang Lyu Institute of Bioinformatics and Structural Biology, Department of Life Science, National Tsing Hua University 96/08/07 Outline Part I: Analysis The analytical
1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI )
 Uses of NMR: 1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI ) 3) NMR is used as a method for determining of protein, DNA,
Uses of NMR: 1) NMR is a method of chemical analysis. (Who uses NMR in this way?) 2) NMR is used as a method for medical imaging. (called MRI ) 3) NMR is used as a method for determining of protein, DNA,
Chemical Lysine Modification at a single site. 2018/12/20 M1 Murata
 Chemical Lysine Modification at a single site 2018/12/20 M1 Murata 1 Contents Amino acid target for bioconjugation Chemical Lysine modification N-hydroxysuccinimide (NHS)-ester α, β-unsaturated sulfonamide
Chemical Lysine Modification at a single site 2018/12/20 M1 Murata 1 Contents Amino acid target for bioconjugation Chemical Lysine modification N-hydroxysuccinimide (NHS)-ester α, β-unsaturated sulfonamide
Targeted Covalent Inhibitors: A Risk-Benefit Perspective
 Targeted Covalent Inhibitors: A Risk-Benefit Perspective 2014 AAPS Annual Meeting and Exposition San Diego, CA, November 4, 2014 Thomas A. Baillie School of Pharmacy University of Washington Seattle, WA
Targeted Covalent Inhibitors: A Risk-Benefit Perspective 2014 AAPS Annual Meeting and Exposition San Diego, CA, November 4, 2014 Thomas A. Baillie School of Pharmacy University of Washington Seattle, WA
Detailed description of overall and active site architecture of PPDC- 3dThDP, PPDC-2HE3dThDP, PPDC-3dThDP-PPA and PPDC- 3dThDP-POVA
 Online Supplemental Results Detailed description of overall and active site architecture of PPDC- 3dThDP, PPDC-2HE3dThDP, PPDC-3dThDP-PPA and PPDC- 3dThDP-POVA Structure solution and overall architecture
Online Supplemental Results Detailed description of overall and active site architecture of PPDC- 3dThDP, PPDC-2HE3dThDP, PPDC-3dThDP-PPA and PPDC- 3dThDP-POVA Structure solution and overall architecture
Examples of Protein Modeling. Protein Modeling. Primary Structure. Protein Structure Description. Protein Sequence Sources. Importing Sequences to MOE
 Examples of Protein Modeling Protein Modeling Visualization Examination of an experimental structure to gain insight about a research question Dynamics To examine the dynamics of protein structures To
Examples of Protein Modeling Protein Modeling Visualization Examination of an experimental structure to gain insight about a research question Dynamics To examine the dynamics of protein structures To
C a h p a t p e t r e r 6 E z n y z m y e m s
 Chapter 6 Enzymes 4. Examples of enzymatic reactions acid-base catalysis: give and take protons covalent catalysis: a transient covalent bond is formed between the enzyme and the substrate metal ion catalysis:
Chapter 6 Enzymes 4. Examples of enzymatic reactions acid-base catalysis: give and take protons covalent catalysis: a transient covalent bond is formed between the enzyme and the substrate metal ion catalysis:
Statistical concepts in QSAR.
 Statistical concepts in QSAR. Computational chemistry represents molecular structures as a numerical models and simulates their behavior with the equations of quantum and classical physics. Available programs
Statistical concepts in QSAR. Computational chemistry represents molecular structures as a numerical models and simulates their behavior with the equations of quantum and classical physics. Available programs
Principles of Drug Design
 Advanced Medicinal Chemistry II Principles of Drug Design Tentative Course Outline Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu I. Introduction
Advanced Medicinal Chemistry II Principles of Drug Design Tentative Course Outline Instructors: Longqin Hu and John Kerrigan Direct questions and enquiries to the Course Coordinator: Longqin Hu I. Introduction
Chapter 4: Amino Acids
 Chapter 4: Amino Acids All peptides and polypeptides are polymers of alpha-amino acids. lipid polysaccharide enzyme 1940s 1980s. Lipids membrane 1960s. Polysaccharide Are energy metabolites and many of
Chapter 4: Amino Acids All peptides and polypeptides are polymers of alpha-amino acids. lipid polysaccharide enzyme 1940s 1980s. Lipids membrane 1960s. Polysaccharide Are energy metabolites and many of
10. Amines (text )
 2009, Department of Chemistry, The University of Western Ontario 10.1 10. Amines (text 10.1 10.6) A. Structure and omenclature Amines are derivatives of ammonia (H 3 ), where one or more H atoms has been
2009, Department of Chemistry, The University of Western Ontario 10.1 10. Amines (text 10.1 10.6) A. Structure and omenclature Amines are derivatives of ammonia (H 3 ), where one or more H atoms has been
CHEM 4170 Problem Set #1
 CHEM 4170 Problem Set #1 0. Work problems 1-7 at the end of Chapter ne and problems 1, 3, 4, 5, 8, 10, 12, 17, 18, 19, 22, 24, and 25 at the end of Chapter Two and problem 1 at the end of Chapter Three
CHEM 4170 Problem Set #1 0. Work problems 1-7 at the end of Chapter ne and problems 1, 3, 4, 5, 8, 10, 12, 17, 18, 19, 22, 24, and 25 at the end of Chapter Two and problem 1 at the end of Chapter Three
Name: BCMB/CHEM 8190, BIOMOLECULAR NMR FINAL EXAM-5/5/10
 Name: BCMB/CHEM 8190, BIOMOLECULAR NMR FINAL EXAM-5/5/10 Instructions: This is an open book, limited time, exam. You may use notes you have from class and any text book you find useful. You may also use
Name: BCMB/CHEM 8190, BIOMOLECULAR NMR FINAL EXAM-5/5/10 Instructions: This is an open book, limited time, exam. You may use notes you have from class and any text book you find useful. You may also use
A) at equilibrium B) endergonic C) endothermic D) exergonic E) exothermic.
 CHEM 2770: Elements of Biochemistry Mid Term EXAMINATION VERSION A Date: October 29, 2014 Instructor: H. Perreault Location: 172 Schultz Time: 4 or 6 pm. Duration: 1 hour Instructions Please mark the Answer
CHEM 2770: Elements of Biochemistry Mid Term EXAMINATION VERSION A Date: October 29, 2014 Instructor: H. Perreault Location: 172 Schultz Time: 4 or 6 pm. Duration: 1 hour Instructions Please mark the Answer
Organic Chemistry II KEY March 25, a) I only b) II only c) II & III d) III & IV e) I, II, III & IV
 rganic Chemistry II KEY March 25, 2015 Exam 2: VERSIN A 1. Which of the following compounds will give rise to an aromatic conjugate base? E a) I only b) II only c) II & III d) III & IV e) I, II, III &
rganic Chemistry II KEY March 25, 2015 Exam 2: VERSIN A 1. Which of the following compounds will give rise to an aromatic conjugate base? E a) I only b) II only c) II & III d) III & IV e) I, II, III &
The Role of Metal Ions in Processes of Conformational Selection during Ligand- Macromolecule Interactions
 102 Bulletin of Magnetic Resonance The Role of Metal Ions in Processes of Conformational Selection during Ligand- Macromolecule Interactions E.Gaggelli, N.Gaggelli, G.Valensin Department of Chemistry,
102 Bulletin of Magnetic Resonance The Role of Metal Ions in Processes of Conformational Selection during Ligand- Macromolecule Interactions E.Gaggelli, N.Gaggelli, G.Valensin Department of Chemistry,
Filtered/edited NOESY spectra
 Filtered/edited NOESY spectra NMR Seminar HS 207 Nina Ripin 22..7 Overview NMR of biomolecular complexes Problems and Solutions Filtered/edited nomenclature Experimental elements NOESY vs filtered pulse
Filtered/edited NOESY spectra NMR Seminar HS 207 Nina Ripin 22..7 Overview NMR of biomolecular complexes Problems and Solutions Filtered/edited nomenclature Experimental elements NOESY vs filtered pulse
I690/B680 Structural Bioinformatics Spring Protein Structure Determination by NMR Spectroscopy
 I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
I690/B680 Structural Bioinformatics Spring 2006 Protein Structure Determination by NMR Spectroscopy Suggested Reading (1) Van Holde, Johnson, Ho. Principles of Physical Biochemistry, 2 nd Ed., Prentice
T 1, T 2, NOE (reminder)
 T 1, T 2, NOE (reminder) T 1 is the time constant for longitudinal relaxation - the process of re-establishing the Boltzmann distribution of the energy level populations of the system following perturbation
T 1, T 2, NOE (reminder) T 1 is the time constant for longitudinal relaxation - the process of re-establishing the Boltzmann distribution of the energy level populations of the system following perturbation
DOCKING TUTORIAL. A. The docking Workflow
 2 nd Strasbourg Summer School on Chemoinformatics VVF Obernai, France, 20-24 June 2010 E. Kellenberger DOCKING TUTORIAL A. The docking Workflow 1. Ligand preparation It consists in the standardization
2 nd Strasbourg Summer School on Chemoinformatics VVF Obernai, France, 20-24 June 2010 E. Kellenberger DOCKING TUTORIAL A. The docking Workflow 1. Ligand preparation It consists in the standardization
Docking. GBCB 5874: Problem Solving in GBCB
 Docking Benzamidine Docking to Trypsin Relationship to Drug Design Ligand-based design QSAR Pharmacophore modeling Can be done without 3-D structure of protein Receptor/Structure-based design Molecular
Docking Benzamidine Docking to Trypsin Relationship to Drug Design Ligand-based design QSAR Pharmacophore modeling Can be done without 3-D structure of protein Receptor/Structure-based design Molecular
Dr. Sander B. Nabuurs. Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre
 Dr. Sander B. Nabuurs Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre The road to new drugs. How to find new hits? High Throughput
Dr. Sander B. Nabuurs Computational Drug Discovery group Center for Molecular and Biomolecular Informatics Radboud University Medical Centre The road to new drugs. How to find new hits? High Throughput
Fragment Hotspot Maps: A CSD-derived Method for Hotspot identification
 Fragment Hotspot Maps: A CSD-derived Method for Hotspot identification Chris Radoux www.ccdc.cam.ac.uk radoux@ccdc.cam.ac.uk 1 Introduction Hotspots Strongly attractive to organic molecules Organic molecules
Fragment Hotspot Maps: A CSD-derived Method for Hotspot identification Chris Radoux www.ccdc.cam.ac.uk radoux@ccdc.cam.ac.uk 1 Introduction Hotspots Strongly attractive to organic molecules Organic molecules
Effects of Chemical Exchange on NMR Spectra
 Effects of Chemical Exchange on NMR Spectra Chemical exchange refers to any process in which a nucleus exchanges between two or more environments in which its NMR parameters (e.g. chemical shift, scalar
Effects of Chemical Exchange on NMR Spectra Chemical exchange refers to any process in which a nucleus exchanges between two or more environments in which its NMR parameters (e.g. chemical shift, scalar
Implementation of novel tools to facilitate fragment-based drug discovery by NMR:
 Implementation of novel tools to facilitate fragment-based drug discovery by NMR: Automated analysis of large sets of ligand-observed NMR binding data and 19 F methods Andreas Lingel Global Discovery Chemistry
Implementation of novel tools to facilitate fragment-based drug discovery by NMR: Automated analysis of large sets of ligand-observed NMR binding data and 19 F methods Andreas Lingel Global Discovery Chemistry
pharmaceutical industry- drug discovery
 ombinatorial hemistry: molecular diversity "Synthesis and pplications of Small Molecule Libraries." Thompson, L..; llman, J.. hem. ev.,, -00. "esign, Synthesis, and valuation of Small-Molecule Libraries.
ombinatorial hemistry: molecular diversity "Synthesis and pplications of Small Molecule Libraries." Thompson, L..; llman, J.. hem. ev.,, -00. "esign, Synthesis, and valuation of Small-Molecule Libraries.
Paper 12: Organic Spectroscopy
 Subject hemistry Paper No and Title Module No and Title Module Tag Paper 12: Organic Spectroscopy 34: ombined problem on UV, IR, 1 H NMR, 13 NMR and Mass- Part 6 HE_P12_M34 TABLE OF ONTENTS 1. Learning
Subject hemistry Paper No and Title Module No and Title Module Tag Paper 12: Organic Spectroscopy 34: ombined problem on UV, IR, 1 H NMR, 13 NMR and Mass- Part 6 HE_P12_M34 TABLE OF ONTENTS 1. Learning
File: {ELS_REV}Cavanagh X/Revises/Prelims.3d Creator: / Date/Time: /9:29pm Page: 1/26 PREFACE
 PREFACE The second edition of Protein NMR Spectroscopy: Principles and Practice reflects the continued rapid pace of development of biomolecular NMR spectroscopy since the original publication in 1996.
PREFACE The second edition of Protein NMR Spectroscopy: Principles and Practice reflects the continued rapid pace of development of biomolecular NMR spectroscopy since the original publication in 1996.
Chiral Supramolecular Catalyst for Asymmetric Reaction
 Chiral Supramolecular Catalyst for Asymmetric Reaction 2017/1/21 (Sat.) Literature Seminar Taiki Fujita (B4) 1 Introduction Rational design of chiral ligands remains very difficult. Conventional chiral
Chiral Supramolecular Catalyst for Asymmetric Reaction 2017/1/21 (Sat.) Literature Seminar Taiki Fujita (B4) 1 Introduction Rational design of chiral ligands remains very difficult. Conventional chiral
C H activation of aliphatic amines without unnecessary mask M2 Takaya Togo
 C H activation of aliphatic amines without unnecessary mask 2017.11.25 M2 Takaya Togo 1 Outline 1.Introduction 2.Free amines as DG Discovery of new activation mode Mechanistic studies Application of the
C H activation of aliphatic amines without unnecessary mask 2017.11.25 M2 Takaya Togo 1 Outline 1.Introduction 2.Free amines as DG Discovery of new activation mode Mechanistic studies Application of the
Structure Investigation of Fam20C, a Golgi Casein Kinase
 Structure Investigation of Fam20C, a Golgi Casein Kinase Sharon Grubner National Taiwan University, Dr. Jung-Hsin Lin University of California San Diego, Dr. Rommie Amaro Abstract This research project
Structure Investigation of Fam20C, a Golgi Casein Kinase Sharon Grubner National Taiwan University, Dr. Jung-Hsin Lin University of California San Diego, Dr. Rommie Amaro Abstract This research project
Biochemistry 3100 Sample Problems Binding proteins, Kinetics & Catalysis
 (1) Draw an approximate denaturation curve for a typical blood protein (eg myoglobin) as a function of ph. (2) Myoglobin is a simple, single subunit binding protein that has an oxygen storage function
(1) Draw an approximate denaturation curve for a typical blood protein (eg myoglobin) as a function of ph. (2) Myoglobin is a simple, single subunit binding protein that has an oxygen storage function
Notes of Dr. Anil Mishra at 1
 Introduction Quantitative Structure-Activity Relationships QSPR Quantitative Structure-Property Relationships What is? is a mathematical relationship between a biological activity of a molecular system
Introduction Quantitative Structure-Activity Relationships QSPR Quantitative Structure-Property Relationships What is? is a mathematical relationship between a biological activity of a molecular system
Principles of NMR Protein Spectroscopy. 2) Assignment of chemical shifts in a protein ( 1 H, 13 C, 15 N) 3) Three dimensional structure determination
 1) Protein preparation (>50 aa) 2) Assignment of chemical shifts in a protein ( 1 H, 13 C, 15 N) 3) Three dimensional structure determination Protein Expression overexpression in E. coli - BL21(DE3) 1
1) Protein preparation (>50 aa) 2) Assignment of chemical shifts in a protein ( 1 H, 13 C, 15 N) 3) Three dimensional structure determination Protein Expression overexpression in E. coli - BL21(DE3) 1
Deuteration: Structural Studies of Larger Proteins
 Deuteration: Structural Studies of Larger Proteins Problems with larger proteins Impact of deuteration on relaxation rates Approaches to structure determination Practical aspects of producing deuterated
Deuteration: Structural Studies of Larger Proteins Problems with larger proteins Impact of deuteration on relaxation rates Approaches to structure determination Practical aspects of producing deuterated
Timescales of Protein Dynamics
 Timescales of Protein Dynamics From Henzler-Wildman and Kern, Nature 2007 Summary of 1D Experiment time domain data Fourier Transform (FT) frequency domain data or Transverse Relaxation Ensemble of Nuclear
Timescales of Protein Dynamics From Henzler-Wildman and Kern, Nature 2007 Summary of 1D Experiment time domain data Fourier Transform (FT) frequency domain data or Transverse Relaxation Ensemble of Nuclear
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/3/4/e1600663/dc1 Supplementary Materials for A dynamic hydrophobic core orchestrates allostery in protein kinases Jonggul Kim, Lalima G. Ahuja, Fa-An Chao, Youlin
advances.sciencemag.org/cgi/content/full/3/4/e1600663/dc1 Supplementary Materials for A dynamic hydrophobic core orchestrates allostery in protein kinases Jonggul Kim, Lalima G. Ahuja, Fa-An Chao, Youlin
Introduction to biomolecular NMR spectroscopy
 Oct 2002 Introduction to biomolecular NMR spectroscopy Michael Sattler, Structural & Computational Biology EMBL Heidelberg Contents Introduction...2 History... 3 Methodological developments for structure
Oct 2002 Introduction to biomolecular NMR spectroscopy Michael Sattler, Structural & Computational Biology EMBL Heidelberg Contents Introduction...2 History... 3 Methodological developments for structure
Chapter 6. The interaction of Src SH2 with the focal adhesion kinase catalytic domain studied by NMR
 The interaction of Src SH2 with the focal adhesion kinase catalytic domain studied by NMR 103 Abstract The interaction of the Src SH2 domain with the catalytic domain of FAK, including the Y397 SH2 domain
The interaction of Src SH2 with the focal adhesion kinase catalytic domain studied by NMR 103 Abstract The interaction of the Src SH2 domain with the catalytic domain of FAK, including the Y397 SH2 domain
