PM atopy 72 (24%) Latent class
|
|
|
- Gerard Stevens
- 6 years ago
- Views:
Transcription
1 Supplementary figures 100 Low no sensitization 226 (76%) PM atopy 72 (24%) Sensitized (%) Allergen Alternaria Bla g 2 Can f 1 Der f 1 Egg Fel d 1 Milk Peanut Common ragweed Timothy grass Latent class 1. Low no sensitization 2. Highly sensitized 3. Milk and egg 4. Peanut and inhalant(s) Latent class Fig. 1. Atopy status at age two years is defined by latent class analysis, which clusters participants into one of four groups based on their pattern of specific IgE response to 10 common allergens. For the purpose of this study, latent classes 2,3 and 4 were grouped as the predominately multi sensitized (PM atopy) group. Number of participants in each group and percent of population is provided.
2 Fig. 2. Bacterial and fungal α diversity correlate with age of participant at the time of stool sample collection. (a) Bacterial diversity positively correlated with increasing age at the time of stool sample collection (n = 298; Pearson s correlation; r = 0.47; P < 0.001). (b) Fungal diversity negatively correlated with increasing age of stool sample collection (n = 188; Pearson s correlation; r = 0.23; P = ).
3 Model fit Number of dirichlet components Fig. 3 Dirichlet multinomial mixture model identifies three compositionally distinct bacterial NGMs as the best model fit. Model fit was based on the Laplace approximation to the negative log model where a lower value indicates a better model fit.
4 Fig. 4. NGM2 and NGM3 gut microbiota exhibit significant differences in bacterial taxonomic content. Zero inflated negative binomial regression model corrected using Benjamini Hochberg method for false discovery, identified taxa present in significantly different relative abundance between NGM2 and NGM3, q < 0.05 (n = 130). Between group differences in relative abundance were natural log transformed prior to plotting on phylogenetic tree (height of bars indicating magnitude of between group difference).
5 Fig. 5. Bacterial pathways predicted to be encoded by taxa that significantly differ in relative abundance between NGM3 compared with NGM1 or NGM2. In silico metagenomic predictions were based specifically on those taxa that significantly differentiated the three microbiota states based on zero inflated negative binomial regression (n = 130). Bacterial amino acid (n = 23), xenobiotic (n = 20) and lipid (n = 17) metabolism pathways represented a large proportion of bacterial pathways relatively depleted in NGM3 microbiota.
6 Fig. 6. Inter NGMS comparisons reveal evidence of distinct programs of metabolism associated with each NGM. Comparative UPLC MS/MS based metabolic profiling of representative neonatal feces from each of the three NGMs indicates that compared to the high-risk NGM3 (n=8), lower risk NGM1 (n = 10) and NGM2 (n = 10) subjects consistently exhibit significant relative increases in docosapentaenoate, dihomo-γ-linoleate, 3fucosyllactose, lacto-n-fucopentaose II and succinate concentrations. In comparison, NGM3 subjects are consistently relatively enriched for stigmasterol, β-sitosterol, α-cehc sulfate, γ-tocopherol, and 12, 13 DiHOME compared to the lower risk NGMs (Welch s t test; P < 0.05).
7 20 P = CD4 + IFN + :CD4 + IL PBS NGM1 NGM3 Fig. 7. Sterile fecal water from NGM3 participants induces a CD4 + IL 4 + cell skew. Dendritic cells from serum of two healthy adult donors (biological replicates), were incubated with sterile fecal water from NGM1 (n = 7; three biological replicates per sample) or NGM3 (n = 5; three biological replicates per sample) participants, prior to co-incubation with autologously purified naïve CD4 + cells. NGM3 fecal water induces a trend toward a CD4 + IL 4 + cell skew compared with NGM1 (LME; P = 0.095).
8 Fig. 8. Weighted correlation network analysis identifies 32 modules of co-associated fecal metabolites. Pearson s correlation was used to determine inter metabolite relationships. Each module, represented by a distinct color, corresponds to a group of positively co-associated metabolites (minimum five metabolites per module). Number of metabolites per each module is provided below each bar; grey bars represent unassigned biochemicals.
9 8 P = Scaled intensity 4 0 NGM1 Microbial state Fig. 9. Confirmation that the concentration of the dihydroxy fatty acid 12, 13 DiHOME, is significantly increased in the NGM3 sample subset used for ex vivo assays. Using the subset of samples employed in the ex vivo DC T cell assay and based on metabolite scaled intensity data, 12, 13 DiHOME is significantly increased in relative concentration in NGM3 (n = 7) compared to NGM1 (n = 5) samples (Welch s t test; P = 0.033). NGM3
10 Supplementary tables Table 1. Allergens used to determine PM atopy status of participants in this study. Mean and median of allergen specific IgE (IU ml 1 ) is provided for each. Allergen n Mean (SD) Median [Min, Max] Alternaria (0.86) 0.05 [0.05, 12.8] German cockroach (Bla g 2) (1.09) 0.05 [0.05, 14.7] Dog (Can f 1) (0.65) 0.05 [0.05, 6.22] House dust mite (Der f 1) (0.72) 0.05 [0.05, 7.55] Egg (10.64) 0.05 [0.05, 170] Cat (Fel d 1) (1.27) 0.05 [0.05, 14.8] Milk (3.65) 0.05 [0.05, 59.0] Peanut (34.07) 0.05 [0.05, 572] Common ragweed (0.12) 0.05 [0.05, 1.08] Timothy grass (0.23) 0.05 [0.05, 2.22] Table 2. Risk ratio of IGMs (infants > 6 months old) for developing atopy or having parental report of doctor s diagnosis of asthma. Risk ratios were calculated based on log binomial regression. DMM community state IGM1 (n = 89) IGM2 (n = 79) RR (95% CI) IGM2 versus IGM1 P value Atopy (PM) 21 (23.6%) 19 (24.1%) 1.02 (0.59,1.75) 0.94 Parental report of doctor diagnosed asthma Atopy (IgE > 0.35 IU ml 1 ) 15 (19.2%) 7 (9.7%) 0.51 (0.22, 1.17) (55.1%) 38 (48.1%) 0.87 (0.65, 1.17) 0.37 Table 3: Association between early life factors and IGMs. Table 4: Association between early life factors and NGMs. Table 5. Factors tested for possible confounding effect on the risk of developing PM atopy for NGM. Table 6. Bacterial taxa exhibiting significantly increased relative abundance in lower risk NGM1 versus the higher risk NGM3 neonatal gut microbiota. Table 7. Bacterial taxa exhibiting significantly increased relative abundance in lower risk NGM2 versus the higher risk NGM3 neonatal gut microbiota.
11 Table 8. Fungal taxa exhibiting significantly increased relative abundance in lower risk NGM1 versus higher risk NGM3 neonatal gut microbiota. Significant difference in relative abundance was determined using a zero inflated negative binomial regression model (q < 0.20). White background indicates taxa enriched in NGM1 (compared with NGM3), gray background indicates taxa enriched in NGM3 (compared with NGM1). Findings are ranked by difference in relative abundance (NGM1 NGM3). OTU NGM1 NGM3 q value Phylum Order Family Genus E 11 Ascomycota Saccharomycetales Unclassified Unclassified E 03 Basidiomycota Malasseziales Incertae sedis Malassezia E 05 Basidiomycota Malasseziales Incertae sedis Malassezia E 127 Unclassified Unclassified Unclassified Unclassified E 02 Ascomycota Capnodiales Davidiellaceae Unclassified E 03 Ascomycota Eurotiales Trichocomaceae Unclassified E 49 Unclassified Unclassified Unclassified Unclassified E 11 Basidiomycota Trichosporonales Trichosporonaceae Unclassified E 05 Ascomycota Chaetothyriales Unclassified Unclassified E 01 Unclassified Unclassified Unclassified Unclassified E 01 Basidiomycota Malasseziales Incertae sedis Malassezia E 02 Ascomycota Saccharomycetales Incertae sedis Candida E 04 Basidiomycota Sporidiobolales Incertae sedis Rhodotorula E 29 Ascomycota Hypocreales Nectriaceae Unclassified E 14 Basidiomycota Polyporales Phanerochaetaceae Phanerochaete E 17 Ascomycota Pleosporales Pleosporaceae Unclassified E 07 Ascomycota Unclassified Unclassified Unclassified E 06 Ascomycota Saccharomycetales Incertae sedis Unclassified E 06 Ascomycota Saccharomycetales Incertae sedis Candida E 12 Ascomycota Saccharomycetales Incertae sedis Cyberlindnera E 06 Ascomycota Saccharomycetales Debaryomycetaceae Meyerozyma E 03 Ascomycota Unclassified Unclassified Unclassified E 43 Ascomycota Pleosporales Incertae sedis Unclassified E 19 Ascomycota Saccharomycetales Saccharomycetaceae Saccharomyces E 03 Basidiomycota Sporidiobolales Incertae sedis Rhodotorula E 03 Basidiomycota Unclassified Unclassified Unclassified E 03 Ascomycota Pleosporales Cucurbitariaceae Pyrenochaetopsis E 03 Ascomycota Saccharomycetales Incertae sedis Candida E 02 Ascomycota Saccharomycetales Saccharomycetaceae Saccharomyces
12 Table 9. Fungal taxa exhibiting significantly increased relative abundance in lower risk NGM2 versus higher risk NGM3 neonatal gut microbiota. Significant difference in relative abundance was determined using zero inflated negative binomial regression model (q < 0.20). White background indicates taxa enriched in NGM2 (compared with NGM3), gray background indicates taxa enriched in NGM3 (compared with NGM2). Findings are ranked by difference in relative abundance (NGM2 NGM3). OTU NGM2 NGM3 q value Phylum Order Family Genus E 05 Basidiomycota Malasseziales Incertae sedis Malassezia E 05 Basidiomycota Malasseziales Incertae sedis Malassezia E 01 Basidiomycota Malasseziales Incertae sedis Malassezia E 02 Basidiomycota Malasseziales Incertae sedis Malassezia E 01 Ascomycota Capnodiales Davidiellaceae Unclassified E 02 Ascomycota Eurotiales Trichocomaceae Unclassified E 140 Unclassified Unclassified Unclassified Unclassified E 56 Basidiomycota Trichosporonales Trichosporonaceae Trichosporon E 01 Basidiomycota Malasseziales Incertae sedis Malassezia E 01 Ascomycota Eurotiales Trichocomaceae Unclassified E 02 Unclassified Unclassified Unclassified Unclassified E 05 Unclassified Unclassified Unclassified Unclassified E 03 Unclassified Unclassified Unclassified Unclassified E 02 Unclassified Unclassified Unclassified Unclassified E 02 Unclassified Unclassified Unclassified Unclassified E 04 Basidiomycota Sporidiobolales Incertae sedis Rhodotorula E 70 Ascomycota Hypocreales Nectriaceae Unclassified E 01 Ascomycota Saccharomycetales Incertae sedis Candida E 15 Ascomycota Saccharomycetales Incertae sedis Debaryomyces E 04 Ascomycota Unclassified Unclassified Unclassified E 22 Ascomycota Saccharomycetales Incertae sedis Unclassified E 03 Ascomycota Saccharomycetales Debaryomycetaceae Meyerozyma E 12 Ascomycota Trichosphaeriales Incertae sedis Nigrospora E 20 Ascomycota Pleosporales Incertae sedis Unclassified E 13 Ascomycota Saccharomycetales Saccharomycetaceae Saccharomyces E 03 Ascomycota Pleosporales Cucurbitariaceae Pyrenochaetopsis E 13 Unclassified Unclassified Unclassified Unclassified E 02 Unclassified Unclassified Unclassified Unclassified E 02 Ascomycota Saccharomycetales Incertae sedis Candida E 02 Unclassified Unclassified Unclassified Unclassified
13 Table 10: Procrustes analyses of 16S rrna based β diversity, PICRUSt and metabolomic datasets. Results from Procrustes analyses indicate that bacterial β diversity, PICRUSt and metabolomic data is highly and significantly correlated. Comparison r* (M 2 ) P value 16S rrna versus PICRUSt 0.72 (0.48) < S rrna versus Metabolomics PICRUSt versus Metabolomics 0.87 (0.24) 0.66 (0.56) < *r = correlation between data sources. Unweighted UniFrac distance used for 16S rrna; Canberra distance used for PICRUSt and Metabolomic datasets. Table 11. Metabolites significantly enriched in lower risk NGM1 versus higher risk NGM3 neonatal gut microbiota. Table 12. Metabolites significantly enriched in lower risk NGM2 versus higher risk NGM3 neonatal gut microbiota.
14 Supplementary note Gut microbiota state validation in an independent cohort To assess the validity of our DMM modeling, the published 16S rrna data of Arrieta et al. 1 was used (n = 319 independent fecal samples collected at approximately 3 12 months of age in the Canadian Healthy Infant Longitudinal Development (CHILD) Study). The specific age of each participant was unavailable and the youngest participants in this cohort were 3 months of age, substantially older than neonates in the WHEALS cohort. Hence the dataset could not be segregated into samples that were > or < 6 months of age, as had been performed for our Wayne County Health, Environment, Allergy and Asthma Longitudinal Study (WHEALS) cohort. This limited our capacity to identify neonatal microbiota states associated with subsequent childhood atopy and asthma outcomes. Nonetheless, we used the cohort to determine whether any of the microbiota states identified in our study were replicated in the CHILD cohort. Because of the age range of the CHILD cohort, we applied both our NGM and IGM model parameters to the entire data set. A better model fit (i.e., smaller laplace approximation to the negative log model evidence) was obtained when the CHILD data was fit to the NGM model compared with the IGM model (model fit: 32,502 versus 174,610, respectively) and a two group solution represented the best fit for the CHILD data. Group 1 (G1) included 221 (69%) participants and group 2 (G2) 98 (31%). The posterior probabilities were on average higher for G1 compared to G2 (0.98 vs. 0.95, respectively). Consistent with our findings, CHILD participants assigned to G1 were typically defined by high Bifidobacteriaceae relative abundance (average relative abundance (ara): 75%). G2 participants were characterized by Lachnospiraceae (ara: 39%), Clostridiaceae (ara: 29%), and Ruminococcaceae (ara: 12%), more reflective of the IGM2 cluster identified in our cohort. Code availability The following script may be used to calculate a representative multiply rarefied OTU table from an unrarefied OTU table, an alterative to singly rarefied tables. This approach stabilizes the effect of random sampling and results in an OTU table that is more representative of community composition. Multiple single rarefied OTU tables are calculated for each sample, and the distance between the subject specific rarefied vectors calculated. The rarefied vector that is the minimum average (or median) distance from itself to all other rarefied vectors is considered the most representative for
15 that subject and used to represent community composition for that sample in the resulting multiply rarified OTU table. library(vegan) library(guniffrac) ##Parameters # specify the raw OTU count table, with samples = rows, taxa = columns # rawtab = otu_tab_t # specify the depth you would like to rarefy your tables to the default is to just use the minimum sequencing #depth raredepth = min(rowsums(rawtab)) # specify the number of rarefied tables you would like to generate to calculate your representatiave rarefied #table from ntables = 100 # specify the distance measure to use to calculate distance between rarefied data sets, for each subject #can be any of the methods available in the vegdist function of vegan distmethod = "euclidean" # specify the method to summarize across distances if mean distance, then summarymeasure = mean #if median distance, then summarymeasure = median # summarymeasure = mean # specify the seed start for the rarefied tables # for each subsequent table, 1 will be added that the previous seed # for reproducibility, always save your seedstart value (or just use the default for simplicity). # seedstart = 500 # specify if you want progress updates to be printed # verbose = TRUE
16 ### returns a representative rarefied OTU table of class matrix. ##functions reprare < function(rawtab = otu_tab_t, raredepth = min(rowsums(otu_tab_t)), ntables = 100, distmethod = euclidean", summarymeasure=mean, seedstart = 500, verbose = TRUE) { raretabs = list() for (z in 1:ntables) { if (verbose == TRUE) { print(paste("calculating rarefied table number", z, sep = " ")) } set.seed(seedstart + z) raretabs[[z]] = Rarefy(rawtab, depth = raredepth)[[1]] } raretabsa = array(unlist(raretabs), dim = c(nrow(raretabs[[z]]), ncol(rawtab), ntables)) final_tab = c() for (y in 1:nrow(raretabs[[z]])) { if (verbose == TRUE) { print(paste("determining rep rarefied vector for subject number", y, sep = " ")) } distmat = as.matrix(vegdist(t(raretabsa[y,,]), method = distmethod)) # distance across reps for subject y distsummary = apply(distmat, 2, summarymeasure) whichbestrep = which(distsummary == min(distsummary))[1] # the best rep is the one with the minimum average/median distance to all other reps. (in case of ties, just select the first) bestrep = raretabsa[y,,whichbestrep] # select that rep only for subject y final_tab = rbind(final_tab, bestrep) # build that rep for subject y into final table } rownames(final_tab) = rownames(raretabs[[z]]) colnames(final_tab) = colnames(rawtab) return(final_tab) }
17 ###### example runs of the function: ###### ### dummy data set for example ### ntaxa = 200 nsubj = 50 set.seed(444) dummyotu < matrix(sample(0:500, ntaxa*nsubj, prob = c(0.7,0.1,0.1,rep(0.1/498, 498)), replace = TRUE), ncol = ntaxa) colnames(dummyotu) = paste("otu", 1:ntaxa, sep = "") rownames(dummyotu) = paste("subj", 1:nsubj, sep = "") sort(rowsums(dummyotu)) # sequencing depth is uneven # specify the minimum depth repraretable = reprare(rawtab = dummyotu, raredepth = min(rowsums(dummyotu)), ntables = 100, distmethod = "euclidean", summarymeasure = mean, seedstart = 500, verbose = TRUE) dim(repraretable) sort(rowsums(repraretable)) # sequencing depth is now even # specify a depth other than the minimum repraretable = reprare(rawtab = dummyotu, raredepth = 3380, ntables = 100, distmethod = "euclidean", summarymeasure = mean, seedstart = 500, verbose = TRUE) dim(repraretable) # subjects with less than the minimum are no longer in the table sort(rowsums(repraretable)) # sequencing depth is now even
Microbiota: Its Evolution and Essence. Hsin-Jung Joyce Wu "Microbiota and man: the story about us
 Microbiota: Its Evolution and Essence Overview q Define microbiota q Learn the tool q Ecological and evolutionary forces in shaping gut microbiota q Gut microbiota versus free-living microbe communities
Microbiota: Its Evolution and Essence Overview q Define microbiota q Learn the tool q Ecological and evolutionary forces in shaping gut microbiota q Gut microbiota versus free-living microbe communities
Supplementary Information
 Supplementary Information Altitudinal patterns of diversity and functional traits of metabolically active microorganisms in stream biofilms Linda Wilhelm 1, Katharina Besemer 2, Lena Fragner 3, Hannes
Supplementary Information Altitudinal patterns of diversity and functional traits of metabolically active microorganisms in stream biofilms Linda Wilhelm 1, Katharina Besemer 2, Lena Fragner 3, Hannes
Taxonomy and Clustering of SSU rrna Tags. Susan Huse Josephine Bay Paul Center August 5, 2013
 Taxonomy and Clustering of SSU rrna Tags Susan Huse Josephine Bay Paul Center August 5, 2013 Primary Methods of Taxonomic Assignment Bayesian Kmer Matching RDP http://rdp.cme.msu.edu Wang, et al (2007)
Taxonomy and Clustering of SSU rrna Tags Susan Huse Josephine Bay Paul Center August 5, 2013 Primary Methods of Taxonomic Assignment Bayesian Kmer Matching RDP http://rdp.cme.msu.edu Wang, et al (2007)
Outline Classes of diversity measures. Species Divergence and the Measurement of Microbial Diversity. How do we describe and compare diversity?
 Species Divergence and the Measurement of Microbial Diversity Cathy Lozupone University of Colorado, Boulder. Washington University, St Louis. Outline Classes of diversity measures α vs β diversity Quantitative
Species Divergence and the Measurement of Microbial Diversity Cathy Lozupone University of Colorado, Boulder. Washington University, St Louis. Outline Classes of diversity measures α vs β diversity Quantitative
Other resources. Greengenes (bacterial) Silva (bacteria, archaeal and eukarya)
 General QIIME resources http://qiime.org/ Blog (news, updates): http://qiime.wordpress.com/ Support/forum: https://groups.google.com/forum/#!forum/qiimeforum Citing QIIME: Caporaso, J.G. et al., QIIME
General QIIME resources http://qiime.org/ Blog (news, updates): http://qiime.wordpress.com/ Support/forum: https://groups.google.com/forum/#!forum/qiimeforum Citing QIIME: Caporaso, J.G. et al., QIIME
Lecture 2: Descriptive statistics, normalizations & testing
 Lecture 2: Descriptive statistics, normalizations & testing From sequences to OTU table Sequencing Sample 1 Sample 2... Sample N Abundances of each microbial taxon in each of the N samples 2 1 Normalizing
Lecture 2: Descriptive statistics, normalizations & testing From sequences to OTU table Sequencing Sample 1 Sample 2... Sample N Abundances of each microbial taxon in each of the N samples 2 1 Normalizing
Statistical methods for the analysis of microbiome compositional data in HIV studies
 1/ 56 Statistical methods for the analysis of microbiome compositional data in HIV studies Javier Rivera Pinto November 30, 2018 Outline 1 Introduction 2 Compositional data and microbiome analysis 3 Kernel
1/ 56 Statistical methods for the analysis of microbiome compositional data in HIV studies Javier Rivera Pinto November 30, 2018 Outline 1 Introduction 2 Compositional data and microbiome analysis 3 Kernel
SUPPLEMENTARY INFORMATION
 City of origin as a confounding variable. The original study was designed such that the city where sampling was performed was perfectly confounded with where the DNA extractions and sequencing was performed.
City of origin as a confounding variable. The original study was designed such that the city where sampling was performed was perfectly confounded with where the DNA extractions and sequencing was performed.
Supplementary Information
 Supplementary Information Table S1. Per-sample sequences, observed OTUs, richness estimates, diversity indices and coverage. Samples codes as follows: YED (Young leaves Endophytes), MED (Mature leaves
Supplementary Information Table S1. Per-sample sequences, observed OTUs, richness estimates, diversity indices and coverage. Samples codes as follows: YED (Young leaves Endophytes), MED (Mature leaves
Lecture: Mixture Models for Microbiome data
 Lecture: Mixture Models for Microbiome data Lecture 3: Mixture Models for Microbiome data Outline: - - Sequencing thought experiment Mixture Models (tangent) - (esp. Negative Binomial) - Differential abundance
Lecture: Mixture Models for Microbiome data Lecture 3: Mixture Models for Microbiome data Outline: - - Sequencing thought experiment Mixture Models (tangent) - (esp. Negative Binomial) - Differential abundance
DETAILED RESULTS ON FUNGAL AND BACTERIAL COMMUNITIES COLONIZING THE COMPOST-INCUBATED POLYMER CARD SURFACE
 DETAILED RESULTS ON FUNGAL AND BACTERIAL COMMUNITIES COLONIZING THE COMPOST-INCUBATED POLYMER CARD SURFACE PART 1: FUNGAL COMMUNITIES COLONIZING THE COMPOST-INCUBATED POLYMER CARD SURFACE Fungal OTUs richness,
DETAILED RESULTS ON FUNGAL AND BACTERIAL COMMUNITIES COLONIZING THE COMPOST-INCUBATED POLYMER CARD SURFACE PART 1: FUNGAL COMMUNITIES COLONIZING THE COMPOST-INCUBATED POLYMER CARD SURFACE Fungal OTUs richness,
Lecture 3: Mixture Models for Microbiome data. Lecture 3: Mixture Models for Microbiome data
 Lecture 3: Mixture Models for Microbiome data 1 Lecture 3: Mixture Models for Microbiome data Outline: - Mixture Models (Negative Binomial) - DESeq2 / Don t Rarefy. Ever. 2 Hypothesis Tests - reminder
Lecture 3: Mixture Models for Microbiome data 1 Lecture 3: Mixture Models for Microbiome data Outline: - Mixture Models (Negative Binomial) - DESeq2 / Don t Rarefy. Ever. 2 Hypothesis Tests - reminder
Supplemental Online Results:
 Supplemental Online Results: Functional, phylogenetic, and computational determinants of prediction accuracy using reference genomes A series of tests determined the relationship between PICRUSt s prediction
Supplemental Online Results: Functional, phylogenetic, and computational determinants of prediction accuracy using reference genomes A series of tests determined the relationship between PICRUSt s prediction
Compositional data methods for microbiome studies
 Compositional data methods for microbiome studies M.Luz Calle Dept. of Biosciences, UVic-UCC http://mon.uvic.cat/bms/ http://mon.uvic.cat/master-omics/ 1 Important role of the microbiome in human health
Compositional data methods for microbiome studies M.Luz Calle Dept. of Biosciences, UVic-UCC http://mon.uvic.cat/bms/ http://mon.uvic.cat/master-omics/ 1 Important role of the microbiome in human health
Supplementary Materials for
 advances.sciencemag.org/cgi/content/full/2/1/e1500997/dc1 Supplementary Materials for Social behavior shapes the chimpanzee pan-microbiome Andrew H. Moeller, Steffen Foerster, Michael L. Wilson, Anne E.
advances.sciencemag.org/cgi/content/full/2/1/e1500997/dc1 Supplementary Materials for Social behavior shapes the chimpanzee pan-microbiome Andrew H. Moeller, Steffen Foerster, Michael L. Wilson, Anne E.
An Adaptive Association Test for Microbiome Data
 An Adaptive Association Test for Microbiome Data Chong Wu 1, Jun Chen 2, Junghi 1 Kim and Wei Pan 1 1 Division of Biostatistics, School of Public Health, University of Minnesota; 2 Division of Biomedical
An Adaptive Association Test for Microbiome Data Chong Wu 1, Jun Chen 2, Junghi 1 Kim and Wei Pan 1 1 Division of Biostatistics, School of Public Health, University of Minnesota; 2 Division of Biomedical
Package milineage. October 20, 2017
 Type Package Package milineage October 20, 2017 Title Association Tests for Microbial Lineages on a Taxonomic Tree Version 2.0 Date 2017-10-18 Author Zheng-Zheng Tang Maintainer Zheng-Zheng Tang
Type Package Package milineage October 20, 2017 Title Association Tests for Microbial Lineages on a Taxonomic Tree Version 2.0 Date 2017-10-18 Author Zheng-Zheng Tang Maintainer Zheng-Zheng Tang
Package LDM. R topics documented: March 19, Type Package
 Type Package Package LDM March 19, 2018 Title Testing Hypotheses about the Microbiome using an Ordination-based Linear Decomposition Model Version 1.0 Date 2018-3-19 Depends GUniFrac, vegan Suggests R.rsp
Type Package Package LDM March 19, 2018 Title Testing Hypotheses about the Microbiome using an Ordination-based Linear Decomposition Model Version 1.0 Date 2018-3-19 Depends GUniFrac, vegan Suggests R.rsp
concentration ( mol l -1 )
 concentration ( mol l -1 ) 8 10 0 20 40 60 80 100 120 140 160 180 methane sulfide ammonium oxygen sulfate (/10) b depth (m) 12 14 Supplementary Figure 1. Water column parameters from August 2011. Chemical
concentration ( mol l -1 ) 8 10 0 20 40 60 80 100 120 140 160 180 methane sulfide ammonium oxygen sulfate (/10) b depth (m) 12 14 Supplementary Figure 1. Water column parameters from August 2011. Chemical
Microbial analysis with STAMP
 Microbial analysis with STAMP Conor Meehan cmeehan@itg.be A quick aside on who I am Tangents already! Who I am A postdoc at the Institute of Tropical Medicine in Antwerp, Belgium Mycobacteria evolution
Microbial analysis with STAMP Conor Meehan cmeehan@itg.be A quick aside on who I am Tangents already! Who I am A postdoc at the Institute of Tropical Medicine in Antwerp, Belgium Mycobacteria evolution
FIG S1: Rarefaction analysis of observed richness within Drosophila. All calculations were
 Page 1 of 14 FIG S1: Rarefaction analysis of observed richness within Drosophila. All calculations were performed using mothur (2). OTUs were defined at the 3% divergence threshold using the average neighbor
Page 1 of 14 FIG S1: Rarefaction analysis of observed richness within Drosophila. All calculations were performed using mothur (2). OTUs were defined at the 3% divergence threshold using the average neighbor
Fundamentals to Biostatistics. Prof. Chandan Chakraborty Associate Professor School of Medical Science & Technology IIT Kharagpur
 Fundamentals to Biostatistics Prof. Chandan Chakraborty Associate Professor School of Medical Science & Technology IIT Kharagpur Statistics collection, analysis, interpretation of data development of new
Fundamentals to Biostatistics Prof. Chandan Chakraborty Associate Professor School of Medical Science & Technology IIT Kharagpur Statistics collection, analysis, interpretation of data development of new
Systems biology. Abstract
 Bioinformatics, 31(10), 2015, 1607 1613 doi: 10.1093/bioinformatics/btu855 Advance Access Publication Date: 6 January 2015 Original Paper Systems biology Selection of models for the analysis of risk-factor
Bioinformatics, 31(10), 2015, 1607 1613 doi: 10.1093/bioinformatics/btu855 Advance Access Publication Date: 6 January 2015 Original Paper Systems biology Selection of models for the analysis of risk-factor
Microbiome: 16S rrna Sequencing 3/30/2018
 Microbiome: 16S rrna Sequencing 3/30/2018 Skills from Previous Lectures Central Dogma of Biology Lecture 3: Genetics and Genomics Lecture 4: Microarrays Lecture 12: ChIP-Seq Phylogenetics Lecture 13: Phylogenetics
Microbiome: 16S rrna Sequencing 3/30/2018 Skills from Previous Lectures Central Dogma of Biology Lecture 3: Genetics and Genomics Lecture 4: Microarrays Lecture 12: ChIP-Seq Phylogenetics Lecture 13: Phylogenetics
Supplementary Figure 1. Chao 1 richness estimator of microbial OTUs (16S rrna
 Supplementary Information Supplementary Figure 1. Chao 1 richness estimator of microbial OTUs (16S rrna gene survey) from skin and hindgut of wild vultures and from feces of zoo birds. Supplementary Note
Supplementary Information Supplementary Figure 1. Chao 1 richness estimator of microbial OTUs (16S rrna gene survey) from skin and hindgut of wild vultures and from feces of zoo birds. Supplementary Note
Assigning Taxonomy to Marker Genes. Susan Huse Brown University August 7, 2014
 Assigning Taxonomy to Marker Genes Susan Huse Brown University August 7, 2014 In a nutshell Taxonomy is assigned by comparing your DNA sequences against a database of DNA sequences from known taxa Marker
Assigning Taxonomy to Marker Genes Susan Huse Brown University August 7, 2014 In a nutshell Taxonomy is assigned by comparing your DNA sequences against a database of DNA sequences from known taxa Marker
SPECIATION. REPRODUCTIVE BARRIERS PREZYGOTIC: Barriers that prevent fertilization. Habitat isolation Populations can t get together
 SPECIATION Origin of new species=speciation -Process by which one species splits into two or more species, accounts for both the unity and diversity of life SPECIES BIOLOGICAL CONCEPT Population or groups
SPECIATION Origin of new species=speciation -Process by which one species splits into two or more species, accounts for both the unity and diversity of life SPECIES BIOLOGICAL CONCEPT Population or groups
Istituto di Microbiologia. Università Cattolica del Sacro Cuore, Roma. Gut Microbiota assessment and the Meta-HIT program.
 Istituto di Microbiologia Università Cattolica del Sacro Cuore, Roma Gut Microbiota assessment and the Meta-HIT program Giovanni Delogu 1 Most of the bacteria species living in the gut cannot be cultivated
Istituto di Microbiologia Università Cattolica del Sacro Cuore, Roma Gut Microbiota assessment and the Meta-HIT program Giovanni Delogu 1 Most of the bacteria species living in the gut cannot be cultivated
1. Poisson distribution is widely used in statistics for modeling rare events.
 Discrete probability distributions - Class 5 January 20, 2014 Debdeep Pati Poisson distribution 1. Poisson distribution is widely used in statistics for modeling rare events. 2. Ex. Infectious Disease
Discrete probability distributions - Class 5 January 20, 2014 Debdeep Pati Poisson distribution 1. Poisson distribution is widely used in statistics for modeling rare events. 2. Ex. Infectious Disease
Microbes and you ON THE LATEST HUMAN MICROBIOME DISCOVERIES, COMPUTATIONAL QUESTIONS AND SOME SOLUTIONS. Elizabeth Tseng
 Microbes and you ON THE LATEST HUMAN MICROBIOME DISCOVERIES, COMPUTATIONAL QUESTIONS AND SOME SOLUTIONS Elizabeth Tseng Dept. of CSE, University of Washington Johanna Lampe Lab, Fred Hutchinson Cancer
Microbes and you ON THE LATEST HUMAN MICROBIOME DISCOVERIES, COMPUTATIONAL QUESTIONS AND SOME SOLUTIONS Elizabeth Tseng Dept. of CSE, University of Washington Johanna Lampe Lab, Fred Hutchinson Cancer
Using Topological Data Analysis to find discrimination between microbial states in human microbiome data
 Using Topological Data Analysis to find discrimination between microbial states in human microbiome data Mehrdad Yazdani 1,2, Larry Smarr 1,3 and Rob Knight 4 1 California Institute for Telecommunications
Using Topological Data Analysis to find discrimination between microbial states in human microbiome data Mehrdad Yazdani 1,2, Larry Smarr 1,3 and Rob Knight 4 1 California Institute for Telecommunications
Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 Title ghost-tree: creating hybrid-gene phylogenetic trees for diversity analyses
Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes. - Supplementary Information -
 Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes - Supplementary Information - Martin Bartl a, Martin Kötzing a,b, Stefan Schuster c, Pu Li a, Christoph Kaleta b a
Dynamic optimisation identifies optimal programs for pathway regulation in prokaryotes - Supplementary Information - Martin Bartl a, Martin Kötzing a,b, Stefan Schuster c, Pu Li a, Christoph Kaleta b a
Package DirichletMultinomial
 Type Package Package DirichletMultinomial April 11, 2018 Title Dirichlet-Multinomial Mixture Model Machine Learning for Microbiome Data Version 1.20.0 Author Martin Morgan
Type Package Package DirichletMultinomial April 11, 2018 Title Dirichlet-Multinomial Mixture Model Machine Learning for Microbiome Data Version 1.20.0 Author Martin Morgan
* Author to whom correspondence should be addressed; Tel.: ; Fax:
 Agronomy 2013, 3, 704-731; doi:10.3390/agronomy3040704 Article OPEN ACCESS agronomy ISSN 2073-4395 www.mdpi.com/journal/agronomy Introduction of Aureobasidium pullulans to the Phyllosphere of Organically
Agronomy 2013, 3, 704-731; doi:10.3390/agronomy3040704 Article OPEN ACCESS agronomy ISSN 2073-4395 www.mdpi.com/journal/agronomy Introduction of Aureobasidium pullulans to the Phyllosphere of Organically
LDM Package. 1 Overview. Yi-Juan Hu and Glen A. Satten March 19, 2018
 LDM Package Yi-Juan Hu and Glen A. Satten March 19, 2018 1 Overview The LDM package implements the Linear Decomposition Model (Hu and Satten 2018), which provides a single analysis path that includes distance-based
LDM Package Yi-Juan Hu and Glen A. Satten March 19, 2018 1 Overview The LDM package implements the Linear Decomposition Model (Hu and Satten 2018), which provides a single analysis path that includes distance-based
*Equal contribution Contact: (TT) 1 Department of Biomedical Engineering, the Engineering Faculty, Tel Aviv
 Supplementary of Complementary Post Transcriptional Regulatory Information is Detected by PUNCH-P and Ribosome Profiling Hadas Zur*,1, Ranen Aviner*,2, Tamir Tuller 1,3 1 Department of Biomedical Engineering,
Supplementary of Complementary Post Transcriptional Regulatory Information is Detected by PUNCH-P and Ribosome Profiling Hadas Zur*,1, Ranen Aviner*,2, Tamir Tuller 1,3 1 Department of Biomedical Engineering,
Amplicon Sequencing. Dr. Orla O Sullivan SIRG Research Fellow Teagasc
 Amplicon Sequencing Dr. Orla O Sullivan SIRG Research Fellow Teagasc What is Amplicon Sequencing? Sequencing of target genes (are regions of ) obtained by PCR using gene specific primers. Why do we do
Amplicon Sequencing Dr. Orla O Sullivan SIRG Research Fellow Teagasc What is Amplicon Sequencing? Sequencing of target genes (are regions of ) obtained by PCR using gene specific primers. Why do we do
Diversity, Productivity and Stability of an Industrial Microbial Ecosystem
 Diversity, Productivity and Stability of an Industrial Microbial Ecosystem Doruk Beyter 1, Pei-Zhong Tang 2, Scott Becker 2, Tony Hoang 3, Damla Bilgin 3, Yan Wei Lim 4, Todd C. Peterson 2, Stephen Mayfield
Diversity, Productivity and Stability of an Industrial Microbial Ecosystem Doruk Beyter 1, Pei-Zhong Tang 2, Scott Becker 2, Tony Hoang 3, Damla Bilgin 3, Yan Wei Lim 4, Todd C. Peterson 2, Stephen Mayfield
Zhiguang Huo 1, Chi Song 2, George Tseng 3. July 30, 2018
 Bayesian latent hierarchical model for transcriptomic meta-analysis to detect biomarkers with clustered meta-patterns of differential expression signals BayesMP Zhiguang Huo 1, Chi Song 2, George Tseng
Bayesian latent hierarchical model for transcriptomic meta-analysis to detect biomarkers with clustered meta-patterns of differential expression signals BayesMP Zhiguang Huo 1, Chi Song 2, George Tseng
An introduction to biostatistics: part 1
 An introduction to biostatistics: part 1 Cavan Reilly September 6, 2017 Table of contents Introduction to data analysis Uncertainty Probability Conditional probability Random variables Discrete random
An introduction to biostatistics: part 1 Cavan Reilly September 6, 2017 Table of contents Introduction to data analysis Uncertainty Probability Conditional probability Random variables Discrete random
Supervised Learning to Predict Geographic Origin of Human Metagenomic Samples
 Supervised Learning to Predict Geographic Origin of Human Metagenomic Samples Christopher Malow cmalow@stanford.edu Abstract Metagenomic studies of human fecal samples have used whole genome shotgun sequencing
Supervised Learning to Predict Geographic Origin of Human Metagenomic Samples Christopher Malow cmalow@stanford.edu Abstract Metagenomic studies of human fecal samples have used whole genome shotgun sequencing
Taxonomical Classification using:
 Taxonomical Classification using: Extracting ecological signal from noise: introduction to tools for the analysis of NGS data from microbial communities Bergen, April 19-20 2012 INTRODUCTION Taxonomical
Taxonomical Classification using: Extracting ecological signal from noise: introduction to tools for the analysis of NGS data from microbial communities Bergen, April 19-20 2012 INTRODUCTION Taxonomical
In this investigation you will use the statistics skills that you learned the to display and analyze a cup of peanut M&Ms.
 M&M Madness In this investigation you will use the statistics skills that you learned the to display and analyze a cup of peanut M&Ms. Part I: Categorical Analysis: M&M Color Distribution 1. Record the
M&M Madness In this investigation you will use the statistics skills that you learned the to display and analyze a cup of peanut M&Ms. Part I: Categorical Analysis: M&M Color Distribution 1. Record the
Niche Modeling. STAMPS - MBL Course Woods Hole, MA - August 9, 2016
 Niche Modeling Katie Pollard & Josh Ladau Gladstone Institutes UCSF Division of Biostatistics, Institute for Human Genetics and Institute for Computational Health Science STAMPS - MBL Course Woods Hole,
Niche Modeling Katie Pollard & Josh Ladau Gladstone Institutes UCSF Division of Biostatistics, Institute for Human Genetics and Institute for Computational Health Science STAMPS - MBL Course Woods Hole,
Lab 3: Two levels Poisson models (taken from Multilevel and Longitudinal Modeling Using Stata, p )
 Lab 3: Two levels Poisson models (taken from Multilevel and Longitudinal Modeling Using Stata, p. 376-390) BIO656 2009 Goal: To see if a major health-care reform which took place in 1997 in Germany was
Lab 3: Two levels Poisson models (taken from Multilevel and Longitudinal Modeling Using Stata, p. 376-390) BIO656 2009 Goal: To see if a major health-care reform which took place in 1997 in Germany was
BIOL 51A - Biostatistics 1 1. Lecture 1: Intro to Biostatistics. Smoking: hazardous? FEV (l) Smoke
 BIOL 51A - Biostatistics 1 1 Lecture 1: Intro to Biostatistics Smoking: hazardous? FEV (l) 1 2 3 4 5 No Yes Smoke BIOL 51A - Biostatistics 1 2 Box Plot a.k.a box-and-whisker diagram or candlestick chart
BIOL 51A - Biostatistics 1 1 Lecture 1: Intro to Biostatistics Smoking: hazardous? FEV (l) 1 2 3 4 5 No Yes Smoke BIOL 51A - Biostatistics 1 2 Box Plot a.k.a box-and-whisker diagram or candlestick chart
MiGA: The Microbial Genome Atlas
 December 12 th 2017 MiGA: The Microbial Genome Atlas Jim Cole Center for Microbial Ecology Dept. of Plant, Soil & Microbial Sciences Michigan State University East Lansing, Michigan U.S.A. Where I m From
December 12 th 2017 MiGA: The Microbial Genome Atlas Jim Cole Center for Microbial Ecology Dept. of Plant, Soil & Microbial Sciences Michigan State University East Lansing, Michigan U.S.A. Where I m From
Carlo Vittorio Cannistraci. Minimum Curvilinear Embedding unveils nonlinear patterns in 16S metagenomic data
 Carlo Vittorio Cannistraci Minimum Curvilinear Embedding unveils nonlinear patterns in 16S metagenomic data Biomedical Cybernetics Group Biotechnology Center (BIOTEC) Technische Universität Dresden (TUD)
Carlo Vittorio Cannistraci Minimum Curvilinear Embedding unveils nonlinear patterns in 16S metagenomic data Biomedical Cybernetics Group Biotechnology Center (BIOTEC) Technische Universität Dresden (TUD)
Microbial Taxonomy and the Evolution of Diversity
 19 Microbial Taxonomy and the Evolution of Diversity Copyright McGraw-Hill Global Education Holdings, LLC. Permission required for reproduction or display. 1 Taxonomy Introduction to Microbial Taxonomy
19 Microbial Taxonomy and the Evolution of Diversity Copyright McGraw-Hill Global Education Holdings, LLC. Permission required for reproduction or display. 1 Taxonomy Introduction to Microbial Taxonomy
SUPPLEMENTARY INFORMATION
 doi:10.1038/nature25973 Power Simulations We performed extensive power simulations to demonstrate that the analyses carried out in our study are well powered. Our simulations indicate very high power for
doi:10.1038/nature25973 Power Simulations We performed extensive power simulations to demonstrate that the analyses carried out in our study are well powered. Our simulations indicate very high power for
Supplementary Information
 Supplementary Information Supplementary Figure 1. Schematic pipeline for single-cell genome assembly, cleaning and annotation. a. The assembly process was optimized to account for multiple cells putatively
Supplementary Information Supplementary Figure 1. Schematic pipeline for single-cell genome assembly, cleaning and annotation. a. The assembly process was optimized to account for multiple cells putatively
Nature Biotechnology: doi: /nbt Supplementary Figure 1. Detailed overview of the primer-free full-length SSU rrna library preparation.
 Supplementary Figure 1 Detailed overview of the primer-free full-length SSU rrna library preparation. Detailed overview of the primer-free full-length SSU rrna library preparation. Supplementary Figure
Supplementary Figure 1 Detailed overview of the primer-free full-length SSU rrna library preparation. Detailed overview of the primer-free full-length SSU rrna library preparation. Supplementary Figure
Math 102 Test Chapters 5, 6,7, 8 Name: Problems Chosen: Extra Credit Chosen:
 Math 102 Test Chapters 5, 6,7, 8 Name: Problems Chosen: Extra Credit Chosen: Directions: Choose 8 problems. Each will be worth 25 points for a total of 200 points. You must include two from {1,2,3,4},
Math 102 Test Chapters 5, 6,7, 8 Name: Problems Chosen: Extra Credit Chosen: Directions: Choose 8 problems. Each will be worth 25 points for a total of 200 points. You must include two from {1,2,3,4},
arxiv: v2 [stat.ap] 17 Mar 2017
![arxiv: v2 [stat.ap] 17 Mar 2017 arxiv: v2 [stat.ap] 17 Mar 2017](/thumbs/83/87096152.jpg) A phylogenetic scan test on Dirichlet-tree multinomial model for microbiome data Yunfan Tang, Li Ma, and Dan L. Nicolae University of Chicago and Duke University arxiv:1610.08974v2 [stat.ap] 17 Mar 2017
A phylogenetic scan test on Dirichlet-tree multinomial model for microbiome data Yunfan Tang, Li Ma, and Dan L. Nicolae University of Chicago and Duke University arxiv:1610.08974v2 [stat.ap] 17 Mar 2017
BIOSTATISTICS METHODS FOR TRANSLATIONAL & CLINICAL RESEARCH DAY #4: REGRESSION APPLICATIONS, PART C MULTILE REGRESSION APPLICATIONS
 BIOSTATISTICS METHODS FOR TRANSLATIONAL & CLINICAL RESEARCH DAY #4: REGRESSION APPLICATIONS, PART C MULTILE REGRESSION APPLICATIONS This last part is devoted to Multiple Regression applications; it covers
BIOSTATISTICS METHODS FOR TRANSLATIONAL & CLINICAL RESEARCH DAY #4: REGRESSION APPLICATIONS, PART C MULTILE REGRESSION APPLICATIONS This last part is devoted to Multiple Regression applications; it covers
PubH 7405: REGRESSION ANALYSIS MLR: BIOMEDICAL APPLICATIONS
 PubH 7405: REGRESSION ANALYSIS MLR: BIOMEDICAL APPLICATIONS Multiple Regression allows us to get into two new areas that were not possible with Simple Linear Regression: (i) Interaction or Effect Modification,
PubH 7405: REGRESSION ANALYSIS MLR: BIOMEDICAL APPLICATIONS Multiple Regression allows us to get into two new areas that were not possible with Simple Linear Regression: (i) Interaction or Effect Modification,
Supplementary Information
 The ISME Journal Supplementary Information Ciobanu et al. SI Tables Table S1 List of studied samples and depths Sample name Core depth below the seafloor (m) 1H3 4 2H1* 6 3H1* 15 3H5 21 4H1 25 4H5* 31
The ISME Journal Supplementary Information Ciobanu et al. SI Tables Table S1 List of studied samples and depths Sample name Core depth below the seafloor (m) 1H3 4 2H1* 6 3H1* 15 3H5 21 4H1 25 4H5* 31
Advanced Statistical Methods: Beyond Linear Regression
 Advanced Statistical Methods: Beyond Linear Regression John R. Stevens Utah State University Notes 3. Statistical Methods II Mathematics Educators Worshop 28 March 2009 1 http://www.stat.usu.edu/~jrstevens/pcmi
Advanced Statistical Methods: Beyond Linear Regression John R. Stevens Utah State University Notes 3. Statistical Methods II Mathematics Educators Worshop 28 March 2009 1 http://www.stat.usu.edu/~jrstevens/pcmi
On Procedures Controlling the FDR for Testing Hierarchically Ordered Hypotheses
 On Procedures Controlling the FDR for Testing Hierarchically Ordered Hypotheses Gavin Lynch Catchpoint Systems, Inc., 228 Park Ave S 28080 New York, NY 10003, U.S.A. Wenge Guo Department of Mathematical
On Procedures Controlling the FDR for Testing Hierarchically Ordered Hypotheses Gavin Lynch Catchpoint Systems, Inc., 228 Park Ave S 28080 New York, NY 10003, U.S.A. Wenge Guo Department of Mathematical
Normalization of metagenomic data A comprehensive evaluation of existing methods
 MASTER S THESIS Normalization of metagenomic data A comprehensive evaluation of existing methods MIKAEL WALLROTH Department of Mathematical Sciences CHALMERS UNIVERSITY OF TECHNOLOGY UNIVERSITY OF GOTHENBURG
MASTER S THESIS Normalization of metagenomic data A comprehensive evaluation of existing methods MIKAEL WALLROTH Department of Mathematical Sciences CHALMERS UNIVERSITY OF TECHNOLOGY UNIVERSITY OF GOTHENBURG
Chapter 2 Solutions Page 15 of 28
 Chapter Solutions Page 15 of 8.50 a. The median is 55. The mean is about 105. b. The median is a more representative average" than the median here. Notice in the stem-and-leaf plot on p.3 of the text that
Chapter Solutions Page 15 of 8.50 a. The median is 55. The mean is about 105. b. The median is a more representative average" than the median here. Notice in the stem-and-leaf plot on p.3 of the text that
IS FUNGAL COMMUNITY OF CHESTNUT ORCHARD SOILS
 9 SEPTEMBER, 2015 IS FUNGAL COMMUNITY OF CHESTNUT ORCHARD SOILS RESILIENT TO FUNGAL INOCULATION? ÂNGELA SOARES, FRANCISCA REIS, ERIC PEREIRA, PAULA BAPTISTA, TERESA LINO-NETO The relationships between
9 SEPTEMBER, 2015 IS FUNGAL COMMUNITY OF CHESTNUT ORCHARD SOILS RESILIENT TO FUNGAL INOCULATION? ÂNGELA SOARES, FRANCISCA REIS, ERIC PEREIRA, PAULA BAPTISTA, TERESA LINO-NETO The relationships between
Taxonomy. Content. How to determine & classify a species. Phylogeny and evolution
 Taxonomy Content Why Taxonomy? How to determine & classify a species Domains versus Kingdoms Phylogeny and evolution Why Taxonomy? Classification Arrangement in groups or taxa (taxon = group) Nomenclature
Taxonomy Content Why Taxonomy? How to determine & classify a species Domains versus Kingdoms Phylogeny and evolution Why Taxonomy? Classification Arrangement in groups or taxa (taxon = group) Nomenclature
Bacterial Communities in Women with Bacterial Vaginosis: High Resolution Phylogenetic Analyses Reveal Relationships of Microbiota to Clinical Criteria
 Bacterial Communities in Women with Bacterial Vaginosis: High Resolution Phylogenetic Analyses Reveal Relationships of Microbiota to Clinical Criteria Seminar presentation Pierre Barbera Supervised by:
Bacterial Communities in Women with Bacterial Vaginosis: High Resolution Phylogenetic Analyses Reveal Relationships of Microbiota to Clinical Criteria Seminar presentation Pierre Barbera Supervised by:
STATISTICAL LEARNING OF INTEGRATIVE ANALYSIS. Meilei Jiang
 STATISTICAL LEARNING OF INTEGRATIVE ANALYSIS Meilei Jiang A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree
STATISTICAL LEARNING OF INTEGRATIVE ANALYSIS Meilei Jiang A dissertation submitted to the faculty of the University of North Carolina at Chapel Hill in partial fulfillment of the requirements for the degree
Network hubs in root-associated fungal metacommunities
 Toju et al. Microbiome (2018) 6:116 https://doi.org/10.1186/s40168-018-0497-1 RESEARCH Network hubs in root-associated fungal metacommunities Hirokazu Toju 1,2*, Akifumi S. Tanabe 3 and Hirotoshi Sato
Toju et al. Microbiome (2018) 6:116 https://doi.org/10.1186/s40168-018-0497-1 RESEARCH Network hubs in root-associated fungal metacommunities Hirokazu Toju 1,2*, Akifumi S. Tanabe 3 and Hirotoshi Sato
Package CausalFX. May 20, 2015
 Type Package Package CausalFX May 20, 2015 Title Methods for Estimating Causal Effects from Observational Data Version 1.0.1 Date 2015-05-20 Imports igraph, rcdd, rje Maintainer Ricardo Silva
Type Package Package CausalFX May 20, 2015 Title Methods for Estimating Causal Effects from Observational Data Version 1.0.1 Date 2015-05-20 Imports igraph, rcdd, rje Maintainer Ricardo Silva
Human Microbiome Project
 Human Microbiome Project Definitions Microbiome: the collective genomes of the community of organisms that share our space Metagenome: culture independent study of genomes of many organisms in order to
Human Microbiome Project Definitions Microbiome: the collective genomes of the community of organisms that share our space Metagenome: culture independent study of genomes of many organisms in order to
Non-specific filtering and control of false positives
 Non-specific filtering and control of false positives Richard Bourgon 16 June 2009 bourgon@ebi.ac.uk EBI is an outstation of the European Molecular Biology Laboratory Outline Multiple testing I: overview
Non-specific filtering and control of false positives Richard Bourgon 16 June 2009 bourgon@ebi.ac.uk EBI is an outstation of the European Molecular Biology Laboratory Outline Multiple testing I: overview
Charles J. Fisk * NAVAIR-Point Mugu, CA 1. INTRODUCTION
 76. CLUSTER ANALYSIS OF PREFERRED MONTH-TO-MONTH PRECIPITATION ANOMALY PATTERNS FOR LOS ANGLES/SAN DIEGO AND SAN FRANCISCO WITH BAYESIAN ANALYSES OF THEIR OCCURRENCE PROBABILITIES REALTIVE TO EL NINO,
76. CLUSTER ANALYSIS OF PREFERRED MONTH-TO-MONTH PRECIPITATION ANOMALY PATTERNS FOR LOS ANGLES/SAN DIEGO AND SAN FRANCISCO WITH BAYESIAN ANALYSES OF THEIR OCCURRENCE PROBABILITIES REALTIVE TO EL NINO,
Generalized Linear Models for Non-Normal Data
 Generalized Linear Models for Non-Normal Data Today s Class: 3 parts of a generalized model Models for binary outcomes Complications for generalized multivariate or multilevel models SPLH 861: Lecture
Generalized Linear Models for Non-Normal Data Today s Class: 3 parts of a generalized model Models for binary outcomes Complications for generalized multivariate or multilevel models SPLH 861: Lecture
Exploring Microbes in the Sea. Alma Parada Postdoctoral Scholar Stanford University
 Exploring Microbes in the Sea Alma Parada Postdoctoral Scholar Stanford University Cruising the ocean to get us some microbes It s all about the Microbe! Microbes = microorganisms an organism that requires
Exploring Microbes in the Sea Alma Parada Postdoctoral Scholar Stanford University Cruising the ocean to get us some microbes It s all about the Microbe! Microbes = microorganisms an organism that requires
Community Health Needs Assessment through Spatial Regression Modeling
 Community Health Needs Assessment through Spatial Regression Modeling Glen D. Johnson, PhD CUNY School of Public Health glen.johnson@lehman.cuny.edu Objectives: Assess community needs with respect to particular
Community Health Needs Assessment through Spatial Regression Modeling Glen D. Johnson, PhD CUNY School of Public Health glen.johnson@lehman.cuny.edu Objectives: Assess community needs with respect to particular
Practical Statistics for the Analytical Scientist Table of Contents
 Practical Statistics for the Analytical Scientist Table of Contents Chapter 1 Introduction - Choosing the Correct Statistics 1.1 Introduction 1.2 Choosing the Right Statistical Procedures 1.2.1 Planning
Practical Statistics for the Analytical Scientist Table of Contents Chapter 1 Introduction - Choosing the Correct Statistics 1.1 Introduction 1.2 Choosing the Right Statistical Procedures 1.2.1 Planning
More Statistics tutorial at Logistic Regression and the new:
 Logistic Regression and the new: Residual Logistic Regression 1 Outline 1. Logistic Regression 2. Confounding Variables 3. Controlling for Confounding Variables 4. Residual Linear Regression 5. Residual
Logistic Regression and the new: Residual Logistic Regression 1 Outline 1. Logistic Regression 2. Confounding Variables 3. Controlling for Confounding Variables 4. Residual Linear Regression 5. Residual
Characterizing and predicting cyanobacterial blooms in an 8-year
 1 2 3 4 5 Characterizing and predicting cyanobacterial blooms in an 8-year amplicon sequencing time-course Authors Nicolas Tromas 1*, Nathalie Fortin 2, Larbi Bedrani 1, Yves Terrat 1, Pedro Cardoso 4,
1 2 3 4 5 Characterizing and predicting cyanobacterial blooms in an 8-year amplicon sequencing time-course Authors Nicolas Tromas 1*, Nathalie Fortin 2, Larbi Bedrani 1, Yves Terrat 1, Pedro Cardoso 4,
You may use your calculator and a single page of notes. The room is crowded. Please be careful to look only at your own exam.
 LAST NAME (Please Print): KEY FIRST NAME (Please Print): HONOR PLEDGE (Please Sign): Statistics 111 Midterm 1 This is a closed book exam. You may use your calculator and a single page of notes. The room
LAST NAME (Please Print): KEY FIRST NAME (Please Print): HONOR PLEDGE (Please Sign): Statistics 111 Midterm 1 This is a closed book exam. You may use your calculator and a single page of notes. The room
Supplementary Appendix
 Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Downs SH, Schindler C, Liu L-JS, et al. Reduced exposure to
Supplementary Appendix This appendix has been provided by the authors to give readers additional information about their work. Supplement to: Downs SH, Schindler C, Liu L-JS, et al. Reduced exposure to
Name Block Date Final Exam Study Guide
 Name Block Date Final Exam Study Guide Unit 7: DNA & Protein Synthesis List the 3 building blocks of DNA (sugar, phosphate, base) Use base-pairing rules to replicate a strand of DNA (A-T, C-G). Transcribe
Name Block Date Final Exam Study Guide Unit 7: DNA & Protein Synthesis List the 3 building blocks of DNA (sugar, phosphate, base) Use base-pairing rules to replicate a strand of DNA (A-T, C-G). Transcribe
Phylogenetic diversity and conservation
 Phylogenetic diversity and conservation Dan Faith The Australian Museum Applied ecology and human dimensions in biological conservation Biota Program/ FAPESP Nov. 9-10, 2009 BioGENESIS Providing an evolutionary
Phylogenetic diversity and conservation Dan Faith The Australian Museum Applied ecology and human dimensions in biological conservation Biota Program/ FAPESP Nov. 9-10, 2009 BioGENESIS Providing an evolutionary
Sections 3.4, 3.5. Timothy Hanson. Department of Statistics, University of South Carolina. Stat 770: Categorical Data Analysis
 Sections 3.4, 3.5 Timothy Hanson Department of Statistics, University of South Carolina Stat 770: Categorical Data Analysis 1 / 22 3.4 I J tables with ordinal outcomes Tests that take advantage of ordinal
Sections 3.4, 3.5 Timothy Hanson Department of Statistics, University of South Carolina Stat 770: Categorical Data Analysis 1 / 22 3.4 I J tables with ordinal outcomes Tests that take advantage of ordinal
Hierarchical Factor Analysis.
 Hierarchical Factor Analysis. As published in Benchmarks RSS Matters, April 2013 http://web3.unt.edu/benchmarks/issues/2013/04/rss-matters Jon Starkweather, PhD 1 Jon Starkweather, PhD jonathan.starkweather@unt.edu
Hierarchical Factor Analysis. As published in Benchmarks RSS Matters, April 2013 http://web3.unt.edu/benchmarks/issues/2013/04/rss-matters Jon Starkweather, PhD 1 Jon Starkweather, PhD jonathan.starkweather@unt.edu
Modeling conditional dependence among multiple diagnostic tests
 Received: 11 June 2017 Revised: 1 August 2017 Accepted: 6 August 2017 DOI: 10.1002/sim.7449 RESEARCH ARTICLE Modeling conditional dependence among multiple diagnostic tests Zhuoyu Wang 1 Nandini Dendukuri
Received: 11 June 2017 Revised: 1 August 2017 Accepted: 6 August 2017 DOI: 10.1002/sim.7449 RESEARCH ARTICLE Modeling conditional dependence among multiple diagnostic tests Zhuoyu Wang 1 Nandini Dendukuri
Actinobacteria Relative abundance (%) Co-housed CD300f WT. CD300f KO. Colon length (cm) Day 9. Microscopic inflammation score
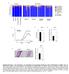 y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
y groups y individuals 9 Actinobacteria Relative abundance (%) acteroidetes Cyanobacteria Deferribacteres Firmicutes Proteobacteria TM Tenericutes Unclassified CDf CDf Co-housed CDf Co-housed CDf CDf CDf
High-Throughput Protein Quantitation Using Multiple Reaction Monitoring
 High-Throughput Protein Quantitation Using Multiple Reaction Monitoring Application Note Authors Ning Tang, Christine Miller, Joe Roark, Norton Kitagawa and Keith Waddell Agilent Technologies, Inc. Santa
High-Throughput Protein Quantitation Using Multiple Reaction Monitoring Application Note Authors Ning Tang, Christine Miller, Joe Roark, Norton Kitagawa and Keith Waddell Agilent Technologies, Inc. Santa
Fixed Effects, Invariance, and Spatial Variation in Intergenerational Mobility
 American Economic Review: Papers & Proceedings 2016, 106(5): 400 404 http://dx.doi.org/10.1257/aer.p20161082 Fixed Effects, Invariance, and Spatial Variation in Intergenerational Mobility By Gary Chamberlain*
American Economic Review: Papers & Proceedings 2016, 106(5): 400 404 http://dx.doi.org/10.1257/aer.p20161082 Fixed Effects, Invariance, and Spatial Variation in Intergenerational Mobility By Gary Chamberlain*
Competition-induced starvation drives large-scale population cycles in Antarctic krill
 In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 1 ARTICLE NUMBER: 0177 Competition-induced starvation drives large-scale population cycles in Antarctic krill Alexey
In the format provided by the authors and unedited. SUPPLEMENTARY INFORMATION VOLUME: 1 ARTICLE NUMBER: 0177 Competition-induced starvation drives large-scale population cycles in Antarctic krill Alexey
Statistical Methods III Statistics 212. Problem Set 2 - Answer Key
 Statistical Methods III Statistics 212 Problem Set 2 - Answer Key 1. (Analysis to be turned in and discussed on Tuesday, April 24th) The data for this problem are taken from long-term followup of 1423
Statistical Methods III Statistics 212 Problem Set 2 - Answer Key 1. (Analysis to be turned in and discussed on Tuesday, April 24th) The data for this problem are taken from long-term followup of 1423
Overview of Dispersion. Standard. Deviation
 15.30 STATISTICS UNIT II: DISPERSION After reading this chapter, students will be able to understand: LEARNING OBJECTIVES To understand different measures of Dispersion i.e Range, Quartile Deviation, Mean
15.30 STATISTICS UNIT II: DISPERSION After reading this chapter, students will be able to understand: LEARNING OBJECTIVES To understand different measures of Dispersion i.e Range, Quartile Deviation, Mean
Chapter Six: Two Independent Samples Methods 1/51
 Chapter Six: Two Independent Samples Methods 1/51 6.3 Methods Related To Differences Between Proportions 2/51 Test For A Difference Between Proportions:Introduction Suppose a sampling distribution were
Chapter Six: Two Independent Samples Methods 1/51 6.3 Methods Related To Differences Between Proportions 2/51 Test For A Difference Between Proportions:Introduction Suppose a sampling distribution were
Clustering using Mixture Models
 Clustering using Mixture Models The full posterior of the Gaussian Mixture Model is p(x, Z, µ,, ) =p(x Z, µ, )p(z )p( )p(µ, ) data likelihood (Gaussian) correspondence prob. (Multinomial) mixture prior
Clustering using Mixture Models The full posterior of the Gaussian Mixture Model is p(x, Z, µ,, ) =p(x Z, µ, )p(z )p( )p(µ, ) data likelihood (Gaussian) correspondence prob. (Multinomial) mixture prior
Bayesian Inference on Joint Mixture Models for Survival-Longitudinal Data with Multiple Features. Yangxin Huang
 Bayesian Inference on Joint Mixture Models for Survival-Longitudinal Data with Multiple Features Yangxin Huang Department of Epidemiology and Biostatistics, COPH, USF, Tampa, FL yhuang@health.usf.edu January
Bayesian Inference on Joint Mixture Models for Survival-Longitudinal Data with Multiple Features Yangxin Huang Department of Epidemiology and Biostatistics, COPH, USF, Tampa, FL yhuang@health.usf.edu January
Chad Burrus April 6, 2010
 Chad Burrus April 6, 2010 1 Background What is UniFrac? Materials and Methods Results Discussion Questions 2 The vast majority of microbes cannot be cultured with current methods Only half (26) out of
Chad Burrus April 6, 2010 1 Background What is UniFrac? Materials and Methods Results Discussion Questions 2 The vast majority of microbes cannot be cultured with current methods Only half (26) out of
Frailty Modeling for Spatially Correlated Survival Data, with Application to Infant Mortality in Minnesota By: Sudipto Banerjee, Mela. P.
 Frailty Modeling for Spatially Correlated Survival Data, with Application to Infant Mortality in Minnesota By: Sudipto Banerjee, Melanie M. Wall, Bradley P. Carlin November 24, 2014 Outlines of the talk
Frailty Modeling for Spatially Correlated Survival Data, with Application to Infant Mortality in Minnesota By: Sudipto Banerjee, Melanie M. Wall, Bradley P. Carlin November 24, 2014 Outlines of the talk
Biostatistics for biomedical profession. BIMM34 Karin Källen & Linda Hartman November-December 2015
 Biostatistics for biomedical profession BIMM34 Karin Källen & Linda Hartman November-December 2015 12015-11-02 Who needs a course in biostatistics? - Anyone who uses quntitative methods to interpret biological
Biostatistics for biomedical profession BIMM34 Karin Källen & Linda Hartman November-December 2015 12015-11-02 Who needs a course in biostatistics? - Anyone who uses quntitative methods to interpret biological
Supplementary Figure 1 Annual number of F0-F5 (grey) and F2-F5 (black) tornado observations over 30 years ( ) for Canada and United States.
 SUPPLEMENTARY FIGURES Supplementary Figure 1 Annual number of F0-F5 (grey) and F2-F5 (black) tornado observations over 30 years (1980-2009) for Canada and United States. Supplementary Figure 2 Differences
SUPPLEMENTARY FIGURES Supplementary Figure 1 Annual number of F0-F5 (grey) and F2-F5 (black) tornado observations over 30 years (1980-2009) for Canada and United States. Supplementary Figure 2 Differences
Supplementary Figure 3
 Supplementary Figure 3 a 1 (i) (ii) (iii) (iv) (v) log P gene Q group, % ~ ε nominal 2 1 1 8 6 5 A B C D D' G J L M P R U + + ε~ A C B D D G JL M P R U -1 1 ε~ (vi) Z group 2 1 1 (vii) (viii) Z module
Supplementary Figure 3 a 1 (i) (ii) (iii) (iv) (v) log P gene Q group, % ~ ε nominal 2 1 1 8 6 5 A B C D D' G J L M P R U + + ε~ A C B D D G JL M P R U -1 1 ε~ (vi) Z group 2 1 1 (vii) (viii) Z module
Chapter 2. Mean and Standard Deviation
 Chapter 2. Mean and Standard Deviation The median is known as a measure of location; that is, it tells us where the data are. As stated in, we do not need to know all the exact values to calculate the
Chapter 2. Mean and Standard Deviation The median is known as a measure of location; that is, it tells us where the data are. As stated in, we do not need to know all the exact values to calculate the
Unit Two: Biodiversity. Chapter 4
 Unit Two: Biodiversity Chapter 4 A. Classifying Living Things (Ch.4 - page 100) Scientific knowledge is constantly evolving ( changing ): new evidence is discovered laws and theories are tested and possibly
Unit Two: Biodiversity Chapter 4 A. Classifying Living Things (Ch.4 - page 100) Scientific knowledge is constantly evolving ( changing ): new evidence is discovered laws and theories are tested and possibly
